Fertilization in flowering plants requires the coordination of many developmental processes. We propose that adaptor protein complex 2–dependent endocytosis plays a crucial role in this coordination by modulating cellular auxin levels through the regulation of the amount and polarity of auxin efflux carriers, such as PIN FORMED2.
Abstract
Fertilization in flowering plants requires the temporal and spatial coordination of many developmental processes, including pollen production, anther dehiscence, ovule production, and pollen tube elongation. However, it remains elusive as to how this coordination occurs during reproduction. Here, we present evidence that endocytosis, involving heterotetrameric adaptor protein complex 2 (AP-2), plays a crucial role in fertilization. An Arabidopsis thaliana mutant ap2m displays multiple defects in pollen production and viability, as well as elongation of staminal filaments and pollen tubes, all of which are pivotal processes needed for fertilization. Of these abnormalities, the defects in elongation of staminal filaments and pollen tubes were partially rescued by exogenous auxin. Moreover, DR5rev:GFP (for green fluorescent protein) expression was greatly reduced in filaments and anthers in ap2m mutant plants. At the cellular level, ap2m mutants displayed defects in both endocytosis of N-(3-triethylammonium-propyl)-4-(4-diethylaminophenylhexatrienyl) pyridinium dibromide, a lypophilic dye used as an endocytosis marker, and polar localization of auxin-efflux carrier PIN FORMED2 (PIN2) in the stamen filaments. Moreover, these defects were phenocopied by treatment with Tyrphostin A23, an inhibitor of endocytosis. Based on these results, we propose that AP-2–dependent endocytosis plays a crucial role in coordinating the multiple developmental aspects of male reproductive organs by modulating cellular auxin level through the regulation of the amount and polarity of PINs.
INTRODUCTION
Endocytosis was originally regarded as a simple process by which extracellular nutrients and plasma membrane (PM) proteins were internalized in a clathrin-dependent manner (Sigismund et al., 2012). However, current knowledge indicates that endocytosis occurs via multiple routes with or without clathrin and plays a pivotal role in various essential cellular processes, such as cell signaling, cell fate determination, cell division, and cell mobility, thus being considered a major player in the regulation of cellular signaling circuits (Fischer et al., 2006; Chen et al., 2011; Andersson, 2012). The underlying principle by which endocytosis plays a role in these processes is to orchestrate spatially and temporally the level and type of proteins localized at the PM and endosomes according to the cellular environment (Polo and Di Fiore, 2006; Sigismund et al., 2012).
Endocytosis is a complex process consisting of multiple distinct steps: budding of a vesicle, packaging cargoes into the vesicle, and release of the vesicle from the PM followed by its fusion to endosomes (Sigismund et al., 2012). All of these steps have to be precisely and closely coordinated for successful endocytosis to occur. A large number of molecules are involved in each of these steps. Notably, the adaptor proteins play a pivotal role in the coordination between the formation of a vesicle at the PM by recruiting cytosolic coat proteins and the loading of the cargo proteins into the budding vesicle by interacting with both clathrin and cargo receptors (Nakatsu and Ohno, 2003). Multiple types of adaptor proteins exist, including the heterotetrameric adaptor protein complexes (AP) and monomeric adaptors such as epsin and related proteins (Robinson, 2004). Epsin and epsin-related proteins contain within a single polypeptide multiple domains that interact with various proteins, such as clathrin, cargo receptors, and other adaptor proteins as well as being able to bind to phospholipids. By contrast, in the endocytosis-specific AP complex AP-2 (Traub, 2003), the tasks of the interaction with other molecules are distributed among the four subunits, called adaptins. The two large subunits, α- and β-adaptins, interact with clathrin and the other adaptins to form the clathrin cage around the vesicle membrane, whereas the medium subunit μ-adaptin recognizes a specific sorting motif, YXXΦ motif (Y, Tyr; X, any amino acid; phi, bulky hydrophobic amino acid), of cargo receptors for the interaction (Robinson and Bonifacino, 2001). Thus, AP-2 adaptors play a crucial role in endocytosis. Consistent with this observation, knockouts of μ-adaptin produce an embryonic lethal phenotype in mice (Mitsunari et al., 2005; Ohno, 2006), confirming their essential role in animal development. On the other hand, certain adaptor proteins exhibit specificity toward cargo receptors/cargo proteins.
In contrast with the vast knowledge on the role of endocytosis in various cellular processes in animal cells, only recently has it become clear that endocytosis plays a similar crucial role in plants (Friml, 2010; Chen et al., 2011). The physiological importance of endocytosis in plants has been demonstrated by the polar distribution of PINs and boron transporters in root cells, which are mediated by endocytosis (Takano et al., 2005; Dhonukshe et al., 2007; Friml, 2010; Grunewald and Friml, 2010). The polar distribution of PINs is pivotal for polar root growth and its gravitropism. The colocalization of PIN1/2 and clathrin to the PM has been demonstrated in Arabidopsis thaliana root cells (Dhonukshe et al., 2007). In addition, homeostasis of K+ is also modulated by an endocytosis-mediated regulation of potassium channel levels at the PM (Sutter et al., 2007). However, it remains elusive as to how broadly endocytosis is involved in regulating cellular processes in plants. In addition, the molecular machineries of endocytosis in plants are not perfectly characterized. It is likely that the basic mechanisms of endocytosis in plant cells are the same as those in other eukaryotic cells, since plants contain homologs of almost all of the molecular components in endocytosis in animal cells (Happel et al., 2004). Moreover, the endocytic inhibitor Tyrphostin A23 has the same effect on endocytosis of cargo proteins in plant cells (Ortiz-Zapater et al., 2006).
In this study, we demonstrate that AP2A1 and AP2M are the components of the AP-2 complex and localize to the PM and the cytosol in Arabidopsis. Furthermore, we provide evidence that AP-2–mediated endocytosis plays an essential role in fertilization by regulating pollen biogenesis and the growth of both pollen tubes and stamen filaments.
RESULTS
AP2A1 and AP2M Are Components of the AP-2 Complex and Localize to the PM and Cytosol in Arabidopsis
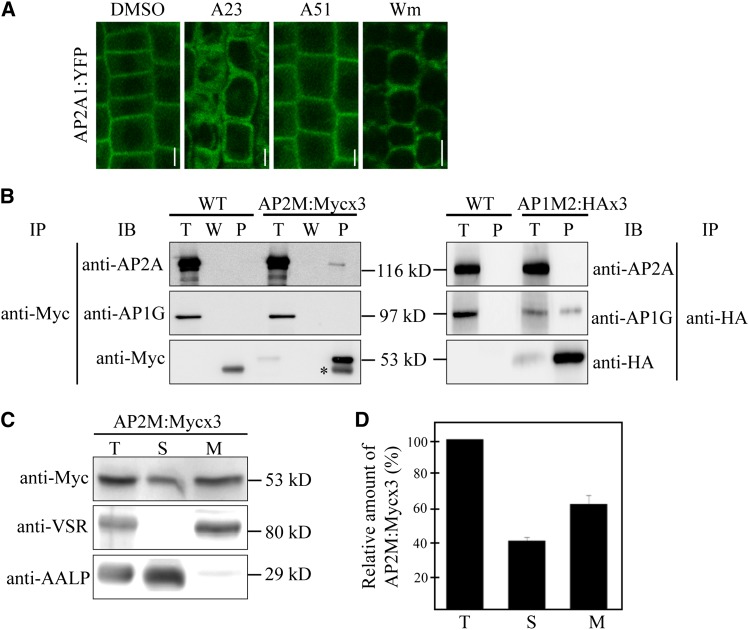
To gain insight into the physiological role of AP-2–mediated endocytosis in plants, we initially identified the components of AP-2 and their localization. In animals, the endocytic adaptor AP-2 complex is formed by assembly of four distinct types of subunits, α-adaptin, β-adaptin, μ-adaptin, and σ-adaptin (Boehm and Bonifacino, 2010). The Arabidopsis genome contains two genes, named AP2A1 and AP2A2, which show high sequence homology and structural similarity to animal α-adaptins (Barth and Holstein, 2004). To test whether these two proteins are α-adaptin of AP-2 in plants, we tagged AP2A1 with yellow fluorescent protein (YFP) at the C terminus and examined its localization. Previously, the C-terminally YFP-tagged α-adaptin, α-adaptin:YFP, localized to the PM and cytosol in animals (Rappoport et al., 2005). We generated the transgenic plants expressing AP2A1:YFP from the native promoter or cauliflower mosaic virus 35S promoter and confirmed that AP2A1:YFP was detected with both anti–green fluorescent protein (GFP) and anti-AP2A antibodies at 143 kD, at the expected size (see Supplemental Figure 1 online). Moreover, the immunocomplex obtained with anti-GFP antibody was recognized by anti-AP2A antibody (see Supplemental Figure 1 online). Next, we examined the localization of AP2A1:YFP expressed from the native promoter in root tissues. Similar to the localization pattern of AP-2 in animals, AP2A1:YFP was localized to the PM in addition to the cytosol (Figure 1A). These results indicate that AP2A1 is the α-adaptin subunit of AP-2 in Arabidopsis. Moreover, the PM-localized AP2A1:YFP signals disappeared when root tissues were treated with Tyrphostin A23, an endocytosis inhibitor that prevents interaction between the YXXΦ motif of a cargo protein and μ-adaptin (Ortiz-Zapater et al., 2006) but not with Tyrphostin A51 used as a control (Figure 1A). In addition, the PM localization of AP2A1:YFP was also affected by Wortmannin, an inhibitor of phosphatidyl inositol 3-kinase known to inhibit endocytosis (Corvera et al.,1999). These results indicate that the interaction of AP-2 with its cargo is critical for localization of AP-2 to the PM.
Figure 1.
AP-2 Localizes to the Plasma Membrane and Cytosol.
(A) Localization of AP2A1:YFP. The localization of AP2A1:YFP expressed in transgenic plants was examined in root cells after treating with DMSO, 30 μM Tyrphostin A23 (A23), or 30 μM Tyrphostin A51 (A51) for 2 h or 33 μM Wortmannin (Wm) for 1 h. Images were taken using a confocal laser scanning microscope. Bar = 10 μm.
(B) Specific interaction of AP2M with AP2A but not AP1G. Protein extracts from transgenic plants expressing AP2M:Mycx3 or AP1M:HAx3 were subjected to immunoprecipitation with anti-Myc or anti-HA antibodies, respectively, and the precipitates were analyzed by protein gel blotting using anti-AP2A, anti-AP1G, anti-Myc, or anti-HA antibodies. T, total protein extracts; W, washing-off solution; P; immunoprecipitates. IB, immunoblot; IP, immunoprecipitation; asterisk, Ig-G heavy chain; WT, the wild type.
(C) Subcellular fractionation of AP2M. Protein extracts from AP2M:Mycx3 transgenic plants were separated into soluble and membrane fractions by ultracentrifugation at 100,000g for 1 h. Soluble and pellet fractions were detected by anti-Myc, anti-AALP, or anti-VSR antibodies. T, total plant extracts; S, soluble fraction; M, membrane fraction.
(D) Quantification of soluble and membrane fractions of AP2M. To quantify the expression level of soluble and membrane fraction of AP2M:Mycx3, the signal intensity of protein on immunoblots was measured using software associated with the Luminescent Images Analyzer LAS3000 (Fujifilm). Error bar indicates sd (n = 3).
Next, we identified μ-adaptin of AP-2. The Arabidopsis genome contains five μ-adaptin homologs. Recently, it has been shown that muB1 (AP1M1) and muB2 (AP1M2) are μ-adaptin subunits of AP-1 and μD is the μ-adaptin subunit of AP-3 (Happel et al., 2004; Niihama et al., 2009; Zwiewka et al., 2011; Park et al., 2013). Sequence homology analysis revealed that AP2M (aka muA) displayed the highest sequence homology to the animal μ2 of AP-2 (see Supplemental Figures 2A and 2B online), raising the possibility that AP2M is the μ-adaptin subunit of AP-2 in Arabidopsis. To test this idea, we generated transgenic plants expressing AP2M:Mycx3 or AP1M2:HAx3, and protein extracts from these plants were used for immunoprecipitation with anti-Myc or anti-hemagglutinin (HA) antibodies. The precipitates were analyzed by protein gel blotting using anti-AP2A (α-adaptin) or anti-AP1G (γ-adaptin) antibodies. AP2A but not AP1G was coprecipitated with anti-Myc antibody, whereas AP1G but not AP2A was coprecipitated with anti-HA antibody (Figure 1B), indicating that AP2M is μ-adaptin of plant AP-2. To examine the subcellular distribution of AP2M, protein extracts from transgenic plants expressing AP2M:Mycx3 were separated into soluble and membrane fractions by ultracentrifugation, and these fractions were analyzed by protein gel blotting using anti-Myc antibody. AP2M was detected in both fractions, with 40% in the soluble fraction and 60% in the membrane fraction (Figures 1C and 1D), indicating that AP2M exists as soluble and membrane-associated forms.
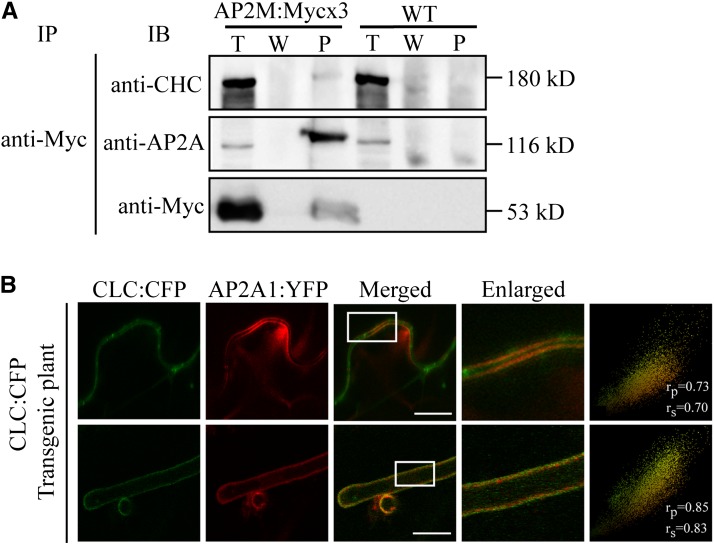
AP-2 is the adaptor of clathrin for clathrin-coated vesicles (CCVs) (Kirchhausen, 2002; Traub, 2003; Robinson, 2004). To test this in plants, we examined the interaction of AP-2 with clathrin by coimmunoprecipitation. Protein extracts from transgenic plants expressing AP2M:Mycx3 were used for coimmunoprecipitation with anti-Myc antibody, and the precipitates were probed with anti–clathrin heavy chain (CHC) antibody. CHC was detected in the precipitates of anti-Myc antibody (Figure 2A), indicating that AP-2 interacts with clathrin. Next, we examined the place where this interaction occurs by in vivo imaging using AP2A1:YFP and cyan fluorescent protein (CFP)–tagged clathrin light chain (CLC:CFP) after Agrobacterium tumefaciens infiltration-mediated transient expression of AP2A1:YFP in CLC:CFP transgenic plants. Indeed, both proteins localized to the PM in leaf and root hair cells (Figure 2B). To quantify their colocalization, we used the linear Pearson moment-product correlation coefficient (rp) and the nonlinear Spearman rank correlation coefficient (rs) (French et al., 2008). AP2A1:YFP closely colocalized with CLC:YFP in both cells (Figure 2B; rp/rs = 0.73/0.70 for leaf cells and rp/rs = 0.85/0.83 for root hair cells). Taken together, these data suggest that AP-2 is involved in CCV formation at the PM.
Figure 2.
AP2M Interacts with CHC and Colocalizes with CLC Tagged with CFP at the PM.
(A) Interaction of AP2M with clathrin. Protein extracts from transgenic plants expressing AP2M:Mycx3 or wild-type plants were subjected to immunoprecipitation with anti-Myc antibody, and the precipitates were analyzed by protein gel blotting using anti-AP2A, anti-Myc, and anti-CHC antibodies. Note that the migration of CHC, AP2A, and AP2M:Mycx3 in the lane of the immunoprecipitates (P) was slightly retarded compared with total protein extracts lane (T). W, washing-off solution; WT, the wild type; IB, immunoblot; IP, immunoprecipitation.
(B) Colocalization of AP2A1:YFP with CLC:CFP at the PM. AP2A1:YFP was transiently expressed in transgenic plants expressing CLC:CFP by Agrobacterium-mediated infiltration and localization of CLC:CFP and AP2A1:YFP in leaf (top panels) and root hair (bottom panels) cells was observed with a confocal laser scanning microscope. The boxes indicate the areas shown by the enlarged images. To quantify their colocalization, 30 independent cells were analyzed using the Pearson and Spearman correlation coefficient colocalization plug-in of ImageJ software. The results are shown as scatterplots (rightmost panels). Bar = 20 μm.
ap2m-1 Mutant Plants Exhibit a Severe Defect in Seed Production and Many Developmental Defects
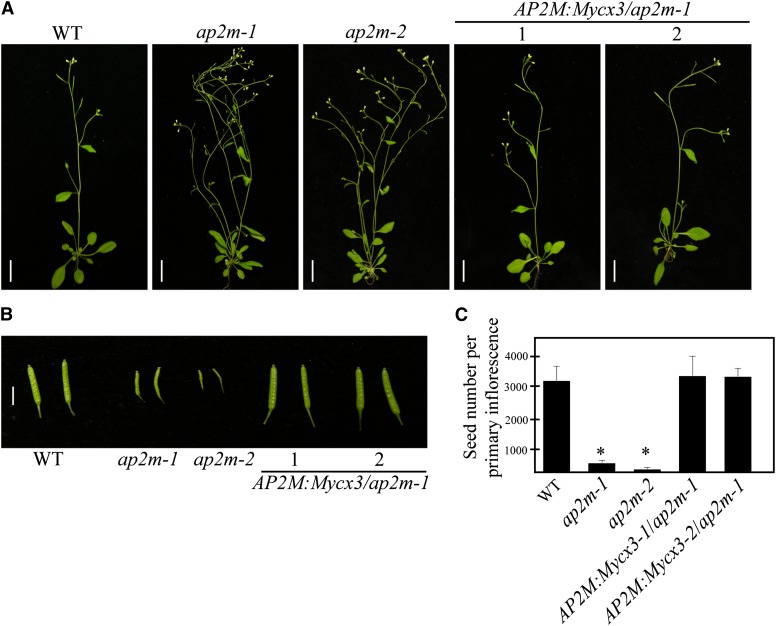
The physiological role of AP-2 in plants was examined genetically using knockout mutants or RNA interference (RNAi) plants of AP-2 components. First, we identified two independent mutant alleles, ap2m-1 and ap2m-2, which had a T-DNA insertion in exons 4 and 7 of AP2M, respectively (see Supplemental Figures 3A and 3B online). The absence of AP2M transcripts was confirmed by RT-PCR (see Supplemental Figure 3C online). Next, we examined the morphology of plants bearing the two alleles, ap2m-1 and ap2m-2. Both of them had more branches and produced small siliques (Figures 3A and 3B). When quantified, these small siliques in ap2m plants contained only 100 to 200 seeds compared with 3100 seeds per primary inflorescence in the wild type (Figure 3C). These results suggest that AP2M has a crucial role in fertilization.
Figure 3.
The ap2m Mutant Plants Show Multiple Morphological Abnormalities.
(A) Images of whole plants. Wild-type (WT), ap2m mutants (two alleles, ap2m-1 and ap2m-2), and rescued ap2m-1 plants with AP2M:Mycx3 (two independent lines, AP2Mpro:AP2M:Mycx3-1 and AP2Mpro:AP2M:Mycx3-2) were grown on soil and images were taken 6 weeks after planting. Bars = 10 cm.
(B) Siliques of wild-type, two alleles of ap2m, and rescued ap2m-1 plants. Images were taken from the indicated types of plants. Bar = 2 cm.
(C) Reduction of seed production in ap2m mutants. Seeds were counted from 25 siliques/primary inflorescence from the indicated plants. Error bar indicates se (n = 20 plants). Statistical analysis was performed between wild-type and ap2m-1 mutants using Student’s t test (*P < 0.01).
In addition to this, ap2m-1 mutants showed many developmental defects, including abnormal phyllotaxis, an increase in the number of shoots and branches, smaller leaves, an increase in the leaf number, and shorter root hairs (see Supplemental Figure 4 online). To confirm that these phenotypes were caused by the mutation in AP2M, we examined whether AP2M:Mycx3 complements these phenotypes of the ap2m-1 plants. Expression of AP2M:Mycx3 under the native promoter in the background of ap2m-1 rescued all of these phenotypes, confirming that these developmental defects are caused by the mutation in AP2M (Figure 3). The genotype and expression of AP2M:Mycx3 in the complemented plants were confirmed by PCR (see Supplemental Figure 3D online).
Since AP-2 is a heterotetrameric complex, we tested whether mutation in AP2A encoding α-adaptin, the largest subunit of AP-2, exhibits the same phenotype. We employed the RNAi approach to knockdown expression of AP2A using an AP2A1 RNAi construct that targeted both AP2A genes, AP2A1 and AP2A2, because AP2A1 and AP2A2 are tandemly repeated next to each other on the same chromosome, which makes it very difficult to obtain a double knockout mutant of AP2A1 and AP2A2 (see Supplemental Figure 5A online). The AP2A levels in AP2A1 RNAi plants were examined by protein gel blot analysis using anti-AP2A antibody. Multiple AP2A1 RNAi transgenic lines showed significantly reduced AP2A protein levels (see Supplemental Figure 5B online). As a negative control, we examined the AP1G level using anti-AP1G antibody and found that the AP1G levels in the RNAi plants were the same as those in the wild-type plants, confirming the specificity of the AP2A1 RNAi construct. These AP2A1 RNAi plants exhibited the same phenotype as ap2m mutants, namely, small siliques with only few seeds and abnormal phyllotaxis (see Supplemental Figure 5C online). These results confirm that AP-2 plays an essential role in fertilization.
ap2m-1 Mutants Have Multiple Defects in Processes Involved in Fertilization
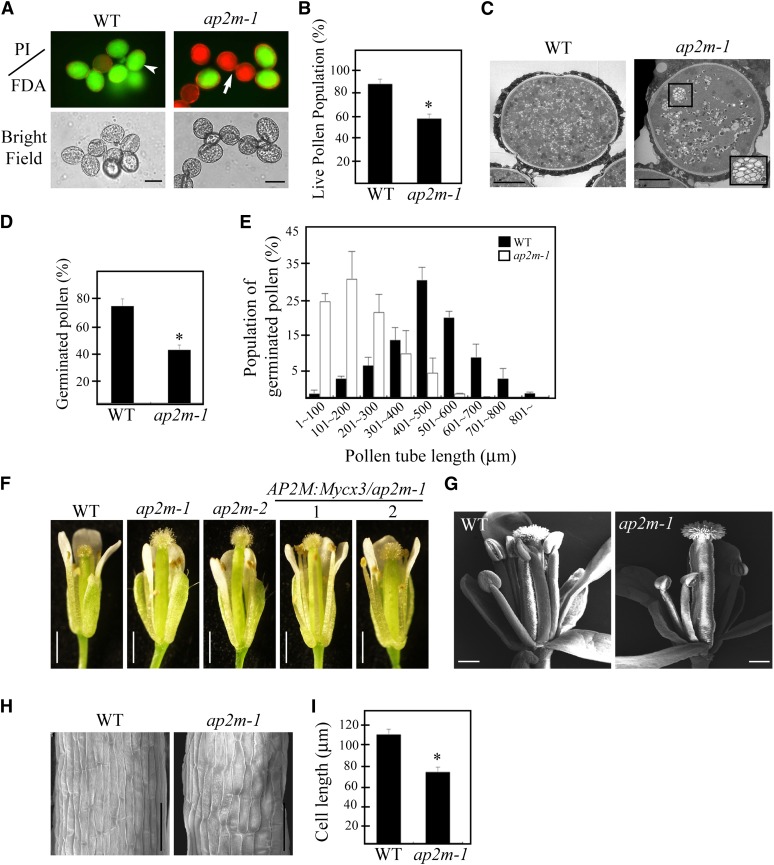
Of multiple abnormalities in ap2m mutants, we focused on defects in fertilization. Fertilization involves many complex developmental processes, such as pollen biogenesis in anthers, the proper elongation of staminal filaments, pollen tube elongation, and the fertilization of the egg cells by sperm cells (Hiscock and Allen, 2008; Suzuki, 2009). To gain insights into the role(s) of AP2M in fertilization, we examined which of these processes is affected in ap2m plants. The amount of pollen produced in the anthers appeared to be greatly reduced in ap2m plants. To look for defects in pollen quality, the viability of pollen grains was tested using propidium iodide and fluorescein diacetate (FDA), which stain dead and live pollen, respectively (Pinillos and Cuevas, 2008) (Figure 4A). Dead pollen grains amounted to 42% in ap2m-1 plants, compared with only 15% in the wild type (Figure 4B). Consistent with this finding, pollination of wild-type plants with ap2m-1 pollen produced only a few seeds, as observed in ap2m-1 plants (see Supplemental Figure 6 online). Interestingly, the plant growth conditions affected the degree of defects in pollen viability; the defect was much more severe in plants grown in a rooftop greenhouse than in those grown in an indoor greenhouse (see Supplemental Figure 7 online). Previous studies have shown that growth conditions such as relative humidity affect pollen viability (Ma et al., 2012). In plants, pollen biogenesis occurs through a lengthy pollen developmental process in the anther and can be divided into 13 different stages (Borg and Twell, 2010). To define the exact step at which AP2M is involved during pollen biogenesis, ultrathin sections of anthers prepared at various developmental stages were examined by electron microscopy. Until stage 10, the ap2m-1 mutant plants did not display any noticeable difference to the wild type (see Supplemental Figure 8 online). However, the most prominent difference was detected at stage 13 when the central vacuole (CV) undergoes degeneration (Yamamoto et al., 2003; Pacini et al., 2011). In wild-type pollen grains, the CV was fragmented into numerous small vacuoles <0.3 μm in diameter and uniformly distributed in the entire cellular space. In ap2m-1 pollen grains, the CV was also fragmented into small vacuoles. However, some of them were still as large as 1 μm in diameter, and often the smaller vacuoles were attached to each other (Figure 4C), indicating that AP2M plays a role in pollen biogenesis via degeneration of the CV. Thus, it is possible that incomplete degeneration of the CV may lead to dead pollen. However, this defect in pollen biogenesis may not account fully for the severe reduction in seed production, since 58% of ap2m-1 pollen was still alive according to FDA staining. To identify additional defects, we examined pollen tube growth in vitro. Indeed, ap2m-1 pollen exhibited a severe defect in tube growth ex planta: The average length of a pollen tube was 200 μm with ap2m-1 pollen, compared with 500 μm with wild-type pollen, indicating that AP2M also plays a crucial role in pollen tube elongation (Figures 4D and 4E).
Figure 4.
ap2m-1 Plants Have Defects in Pollen Biogenesis, Pollen Tube Elongation, and Growth of Stamen Filaments.
(A) and (B) Defect in pollen viability.
(A) Viability of pollen grains from wild-type (WT) or ap2m-1 plants was tested by staining with propidium iodide (PI; 1 µg/mL) (red fluorescence, arrow) or FDA (0.5 µg/mL) (green fluorescence, arrowhead). Bar = 10 µm.
(B) To quantify the viability of pollen grains, pollen grains that stained with FDA were counted. Error bar indicates se (n = 3500 for wild-type and 4130 for ap2m-1 plants). Statistical analysis was performed between wild-type and ap2m-1 mutants using Student’s t test (*P < 0.01).
(C) A defect in vacuole degeneration in ap2m-1 mutant plants. Ultrathin sections from anthers at the flower stage 13 were prepared and the cross sections of pollen mother cells were examined by electron microscopy. Inset: the enlarged image of the boxed area. Bar = 5 μm.
(D) and (E) Defects of pollen germination and pollen tube growth.
(D) To quantify germination rate, pollen grains were germinated on agarose medium and incubated for 4 h at room temperature for the elongation of pollen tubes. The number of pollen grains with pollen tube regardless of its length was counted. Error bar indicates se (n = 30). Statistical analysis was performed between wild-type and ap2m-1 mutants using Student’s t test (*P < 0.01).
(E) The length of individual pollen tubes was measured and the distribution pattern of the average pollen tube length in a group of 100 pollen grains was presented. Three independent measurements were performed. Error bar indicates se (n = 1500 for the wild type, and n = 2110 for ap2m-1).
(F) and (G) A defect in growth of stamen filaments.
(F) Image of a whole flower with filaments; the wild type, two independent alleles (ap2m-1 and ap2m-2), and two lines of complemented ap2m-1 plants with AP2M:Mycx3-1 or AP2M:Mycx3-2. Bar = 1 cm.
(G) Scanning electron microscopy image of flowers. Bar = 200 μm.
(H) Scanning electron microscopy images of filaments showing a defect in cell elongation. Bar = 50 μm.
(I) The length of single cells was quantified. Error bar indicates se (n = 30). Statistical analysis was performed between the wild type and ap2m-1 mutants using Student’s t test (*P < 0.01).
Another noticeable abnormality was the shorter length of the staminal filaments; in ap2m-1 plants, the length of stamen filaments was shorter than those of wild-type plants so that pollen grains cannot be easily delivered from the anther to the stigma (Figures 4F and 4G). To determine the underlying cause of the shorter filament phenotype, we examined the size of individual cells in the filaments. The length of individual cells was shorter in ap2m-1 plants than in wild-type plants (Figures 4H and 4I), indicating that AP2M plays a role in cell elongation in the staminal filaments. Taken together, these results suggest that ap2m-1 plants have defects in at least three different developmental processes, including pollen biogenesis, pollen tube elongation, and elongation of staminal filaments, which contribute collectively to the phenotype of severe reduction in seed production. Consistent with this notion, AP2M was expressed at high levels in anthers, filaments, and pollen tubes, as determined by β-glucuronidase (GUS) staining from transgenic plants harboring the AP2Mp:GUS construct (see Supplemental Figure 9 online).
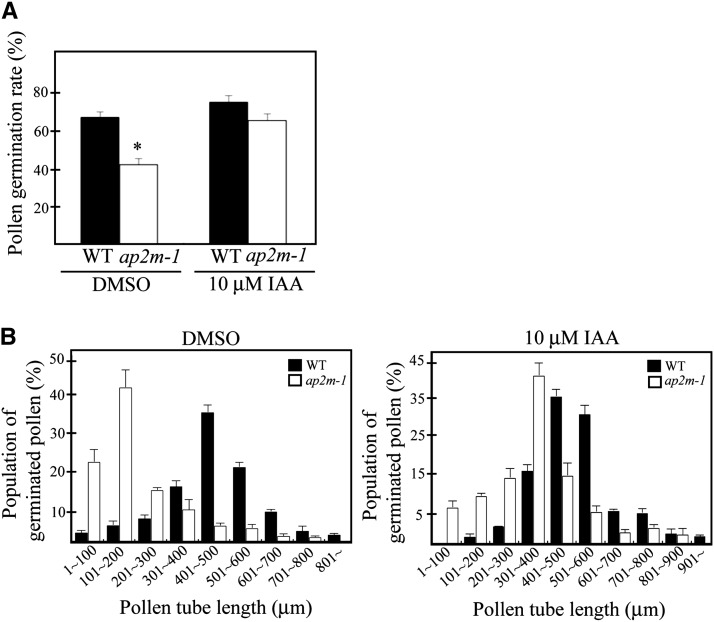
The phenotypes of ap2m-1 plants, in particular, the defects in pollen production and filament elongation, were reminiscent of the phenotypes of yuc2 yuc6, tir1 afb2 afb3, tir1 abf1 abf2 abf3, and arf6 arf8, all of which have a defect in auxin biosynthesis or signaling (Nagpal et al., 2005; Cheng et al., 2006; Cecchetti et al., 2008), raising the possibility that ap2m mutants may also have a defect in maintaining auxin levels. To test whether the failure of pollen elongation is also caused by low levels of auxin, we examined the effect of exogenous auxin application on pollen tube elongation. Indeed, pollen germination and tube growth were restored upon application of auxin (Figures 5A and 5B). These results suggest that AP2M is involved in maintaining auxin levels in anthers and during the growth of staminal filaments and pollen tube elongation.
Figure 5.
Application of Exogenous Auxin Rescues Partially the Defects of Pollen Germination and Pollen Tube Elongation in the ap2m-1 Plants.
(A) Quantification of the effect of exogenous auxin on the pollen germination rate. To quantify the pollen germination rate, pollen grains with a pollen tube were counted 4 h after treating with DMSO or 10 μM IAA. Error bar indicates se (n = 800 for the wild type [WT], and n = 1100 for ap2m-1). Statistical analysis was performed between wild-type and ap2m-1 mutant plants using Student’s t test (*P < 0.01).
(B) Quantification of pollen tube elongation. To quantify the effect of exogenously applied IAA on pollen tube growth, the length of individual pollens tubes was measured and the average length of pollen tube was presented for a group of 100 pollen grains. Error bar indicates se (n = 1200 for the wild type, and n = 1800 for ap2m-1).
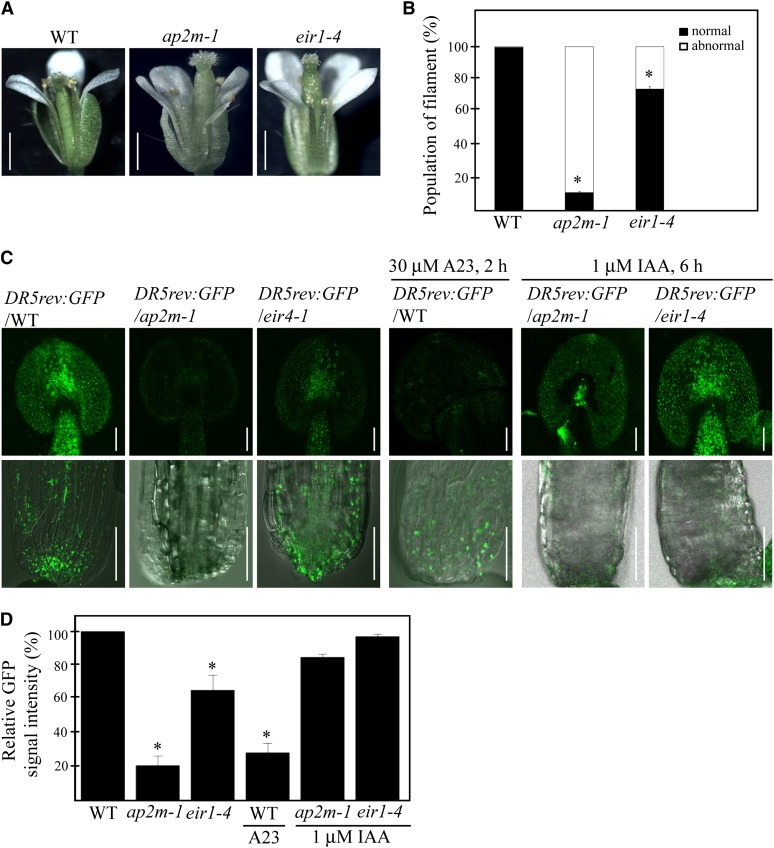
To further test this hypothesis, we examined whether the ap2m-1 mutant plants had any alteration in cellular auxin levels. As a control, we included a pin2 mutant, ethylene insensitive root1-4 (eir1-4), that had a T-DNA insertion in PIN2 (Abas et al., 2006). eir1-4 plants also showed the shorter filament phenotype in 30% of flowers, thus at a lesser degree compared with the ap2m-1 plants, which had defective flowers at 90% (Figures 6A and 6B). Next, to confirm this finding, we examined the expression of auxin-inducible genes in anthers and filaments. The expression of GFP under the control of DR5rev, an auxin-inducible promoter (Ulmasov et al., 1997), was greatly reduced in anthers and filaments of ap2m-1 plants compared with wild-type plants (Figures 6C and 6D). In both ap2m-1 and eir1-4 plants, the GFP signals expressed from DR5rev:GFP in anthers and basal sides of the filaments were dramatically reduced to 20%and 65%, respectively, compared with the wild-type plants (Figures 6C and 6D), indicating that auxin levels are altered in both ap2m-1 and eir1-4 mutants, albeit to a lesser degree in eir1-4 plants (Figure 6D). To further confirm that the reduced expression of DR5rev:GFP in ap2m-1 and eir1-4 plants is caused by a defect in auxin distribution, we tested whether application of exogenous auxin can restore the expression of DR5rev:GFP in these mutants. Indeed, the treatment of 1 μM indole-3-acetic acid (IAA) for 6 h restored the expression of DR5rev:GFP partially in ap2m-1 plants and almost completely in eir1-4 plants (Figures 6C and 6D), confirming that ap2m-1 plants have a defect in auxin distribution. The difference in severity of defects between the ap2m-1 and eir1-4 mutants raises the possibility that auxin levels are also modulated by other PIN proteins in addition to PIN2 in stamens.
Figure 6.
ap2m-1 Mutant Plants Have Low Levels of Auxin in Anthers and Filaments.
(A) Defects in elongation of stamen filaments in ap2m-1 and eir1-4 plants. Images of a whole flower were taken from wild-type (WT), ap2m-1, and eir1-4 plants grown on soil for 6 weeks. Bar = 1 cm.
(B) Quantification of defective flowers in wild-type, ap2m-1, and eir1-4 plants. Flowers that had abnormal growth of filaments were counted. Error bar indicates se (n = 300 flowers). Statistical analysis was performed between the wild type and ap2m-1 and eir1-4 mutants using Student’s t test (*P < 0.01).
(C) Expression of DR5rev:GFP in stamens in wild-type, ap2m-1, and eir1-4 plants. The DR5rev:GFP transgenic plants were crossed with ap2m-1 or eir1-4, respectively, and the expression levels of GFP were examined in wild-type background or the indicated mutant backgrounds. The stamens of wild-type, ap2m-1, and eir1-4 plants were also treated with the indicated chemicals for the indicated periods of time, and DR5rev:GFP expression was examined. Bars = 50 μm (top panels) and 100 μm (bottom panels).
(D) Quantification of GFP signals intensity in wild-type, ap2m-1, and eir1-4 plants. Fluorescence signal intensity of the whole stamens shown in (C) (top panels) was measured using LSM 510 (Zeiss) and expressed as relative values to that of the wild-type plants. Error bar indicates se (n = 50 stamens). Statistical analysis was performed between the wild type and ap2m-1 and eir1-4 mutants using Student’s t test (*P < 0.01).
In addition, transcript levels of endogenous TIR1 and ARF6 were significantly reduced (see Supplemental Figure 10 online). By contrast, two gibberellin- and two jasmonate-inducible genes used as controls showed only slight or no reduction in their expression (see Supplemental Figure 10 online). Intriguingly, the reduced length of ap2m-1 filaments was partially recovered by treatment with IAA, indicating that the ap2m-1 mutant was defective in endogenous auxin levels (see Supplemental Figure 11 online).
ap2m-1 Plants Are Defective in Endocytosis and Localization of PIN2:GFP in Filaments
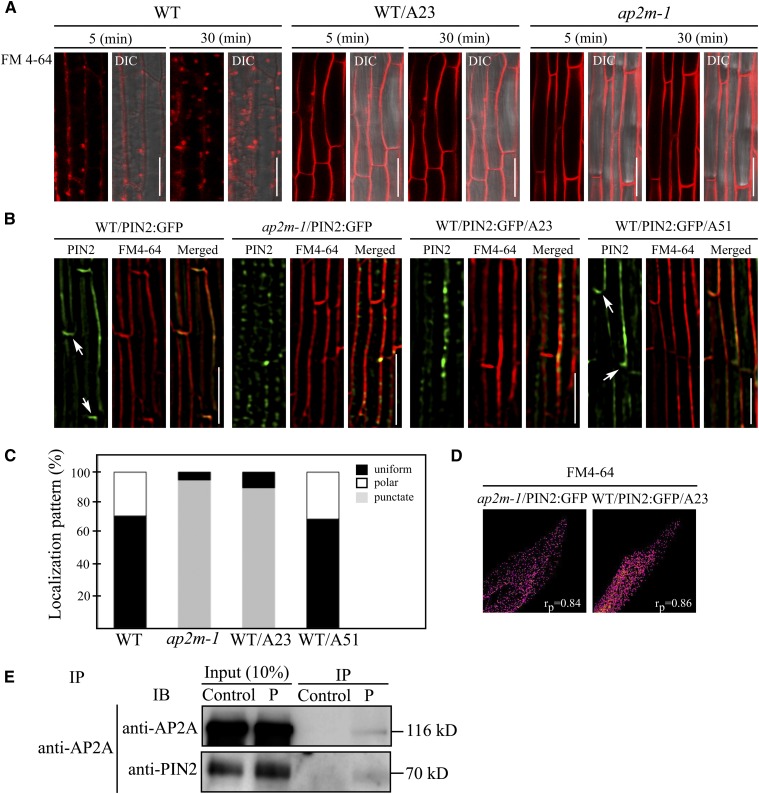
Since AP-2 is an adaptor protein complex for clathrin-mediated endocytosis (Nakatsu and Ohno, 2003; Traub, 2003), we monitored endocytosis in the filaments of ap2m-1 plants using a lipophilic dye, N-(3-triethylammonium-propyl)-4-(4-diethylaminophenylhexatrienyl) pyridinium dibromide (FM4-64) (Bolte et al., 2004). As observed in the root cells of wild-type plants (Dettmer et al., 2006; Dhonukshe et al., 2008), FM4-64 rapidly enters endosomes as early as 30 min after its application to the cells and accumulates in endosomes with almost no signal remaining at the PM (Figure 7A). By contrast, in ap2m-1 plants, FM4-64 was almost exclusively restricted to the PM even 30 min after application (Figure 7A), indicating that endocytosis is impaired in the ap2m-1 plants. Moreover, Tyrphostin A23 nearly completely inhibited endocytosis of FM4-64 in wild-type filaments as observed in ap2m-1 plants (Figure 7A), supporting the idea that AP-2 plays an important role in endocytosis in filaments.
Figure 7.
Cells in Filaments of ap2m-1 Plants Exhibit a Defect in Endocytosis and Polar Localization of PIN 2:GFP.
(A) Defect in uptake of endocytic marker FM4-64 in filaments of the ap2m-1 plants. Filaments of wild-type (WT) and ap2m-1 plants that had been treated with or without Tyrphostin A23, respectively, were stained with FM4-64, and internalization of FM4-64 was examined. Images were obtained at 5 or 30 min after staining. DIC, differential interference contrast. Bars = 20 μm.
(B) Distribution of PIN2:GFP in wild-type and ap2m-1 filaments. Wild-type and ap2m-1 plants expressing PIN2:GFP were briefly stained with FM4-64 and localization of PIN2:GFP was examined by super-resolution structured illumination microscopy. Wild-type plants expressing PIN2:GFP were also treated with Tyrphostin A23 or Tyrphostin A51 for 2 h before PIN2:GFP observation. Arrows indicate the basal localization of PIN2:GFP. Bar = 50 μm.
(C) Quantification of PIN2:GFP localization patterns. To quantify the localization pattern of PIN2:GFP in the images shown in (B), the cells were counted according to their localization patterns (polar, uniform, and punctate staining patterns). More than 30 cells in six independent plants were analyzed for each sample.
(D) Colocalization of PIN2:GFP at punctate stains with FM4-64 in the PM. To quantify colocalization between PIN2:GFP, the punctate stains, and FM4-64 in the PM, 200 PIN2:GFP foci were selected and analyzed using NIS-E software for the Pearson correlation coefficient value (rp) associated with the super-resolution imaging microscope (SIM; Nikon). The results are shown as scatterplots.
(E) Interaction of PIN2 with AP-2. Protein extracts from wild-type plants were incubated with anti-AP2A antibody, and the immunocomplexes were precipitated and analyzed by protein gel blotting using anti-PIN2 and anti-AP2A antibodies. Control, no AP2A antibody; IP, immunoprecipitation. IB, immunoblot.
Endocytosis plays an important role in polar localization of the auxin efflux transporters of the PIN family (Friml, 2010; Grunewald and Friml, 2010). Thus, it is possible that the level and/or localization of PINs are affected in staminal filaments and pollen tubes of ap2m-1 plants. According to publicly available microarray data, PIN8 showed the highest expression in pollen and filaments, whereas the expression levels of PIN2 and PIN1 are moderate or barely detectable, respectively. However, PIN8 is reported to localize to the endoplasmic reticulum (Ding et al., 2012). Accordingly, to reveal a possible relationship to AP2M function in filament growth, we initially examined localization of GFP-tagged PIN2 in the filaments. In wild-type plants, PIN2:GFP localized to the PM. Interestingly, PIN2:GFP showed polarity in its distribution with strong basal signals in 28% of the cells (Figures 7B and 7C). However, in ap2m-1 filaments, PIN2:GFP showed a striking alteration in its localization, thus revealing a punctate staining pattern along the PM (Figure 7B). These results indicate that AP2M plays a crucial role in polar localization of PIN2 at the PM of the filaments. This is similar to the case with cells in the root cortex, which also show a basal distribution in the PM (Grunewald and Friml, 2010). To confirm that the PIN2:GFP localization pattern in ap2m-1 plants is caused by a defect in endocytosis, wild-type filaments were treated with Tyrphostin A23, or Tyrphostin A51 as a control, and the localization of PIN2:GFP was examined after briefly staining with FM4-64 to visualize the PM. As observed in ap2m-1 plants, Tyrphostin A23 but not Tyrphostin A51 treatment induced a punctate staining pattern of PIN2:GFP along the PM. To test whether PIN2:GFP punctate stains localize at the PM, we compared its localization with FM4-64. PIN2:GFP-positive punctate stains closely overlapped with FM4-64. To quantify the colocalization, we used the Pearson correlation coefficient value and confirmed their close colocalization (rp = 0.84 and 0.86 for ap2m-1 and wild type/A23, respectively; Figure 7D), which in turn confirmed the localization of PIN2:GFP at the PM. These results suggest that AP-2–mediated endocytosis plays a crucial role in the proper distribution of PIN2 in the filaments.
These results suggested that internalization of PIN2:GFP from the PM is defective in ap2m-1 mutant plants. Furthermore, the punctate stains on the PM raised the possibility that PIN2:GFP accumulates at certain sites on the PM but cannot be internalized. Moreover, this localization pattern confirms the idea that PIN2:GFP accumulates at specific site on the PM in the intact filaments. To confirm that PIN2 is internalized by endocytosis involving AP-2, we examined the interaction of PIN2 with AP-2 by coimmunoprecipitation using anti-AP2A (α-adaptin) antibody. PIN2 was detected in the immunocomplexes obtained with the anti-AP2A antibody (Figure 7C), confirming that AP-2 interacts with PIN2.
DISCUSSION
AP2A1 and AP2M Are the α-Adaptin and μ-Adaptin, Respectively, of AP-2 Involved in Clathrin-Mediated Endocytosis at the PM
In this study, we demonstrated that AP2A1 and AP2M are the α- and μ-adaptin, respectively, of AP-2 in Arabidopsis. This conclusion is based on several lines of evidence. AP2A1:YFP expressed from its native promoter in transgenic plants localized to the PM and cytosol and also colocalized with clathrin at the PM in leaf and root hair cells. In addition, AP2M was coimmunoprecipitated with α-adaptin but not γ-adaptin. Consistent with the biochemical and subcellular localization data, ap2m-1 plants exhibit a defect in endocytosis of FM4-64 in the filaments, which was phenocopied by the Tyrphostin A23 treatment of wild-type filaments. This finding is consistent with recent observations that μD is a component of AP-3 and AP1M1 (muB1) and AP1M2 (muB2) are components of AP-1 complexes (Happel et al., 2004; Niihama et al., 2009; Zwiewka et al., 2011, Park et al., 2013). Another piece of supporting evidence for AP2M as a component of AP-2 is that AP2A1 RNAi transgenic plants showed phenotypes that were similar to those of ap2m mutant plants in defects in seed production and phyllotaxis. Thus, our results suggest that AP2A1 and AP2M of AP-2 in plant cells also localize to the PM for CCV formation, similar to AP-2 in animals.
AP-2–Mediated Endocytosis Plays a Crucial Role in Reproductive Organ Development
In this study, we demonstrated that endocytosis mediated by AP-2 plays an essential role in reproductive organ development. Detailed analysis of ap2m mutant plants revealed multiple abnormalities in reproductive organs, such as shorter filaments of the stamen, a significant reduction in the number of pollen grains, their reduced viability, and a defect in pollen tube growth, which led to small siliques with only few seeds. This conclusion is based on the phenotype of ap2m mutant plants. In AP-2, μ-adaptin is responsible for the recognition of endocytic cargoes or cargo receptors (Traub, 2009). Thus, the mutant phenotypes of the ap2m plants are likely to be caused by a defect in endocytosis involving AP-2. Most of the cellular processes in which endocytosis is implicated have previously been studied using cargo proteins such as PINs and ion transporters. Polar localization of PINs that is achieved via endocytosis-mediated recycling is crucial for proper root development (Grunewald and Friml, 2010). In addition, regulation of boron transporter levels by endocytosis plays an important role in the homeostasis of boron in plant cells (Takano et al., 2005). In ap2m mutant plants, the most prominent defect was small siliques with only few seeds, although the severity of the defects in pollen viability and the silique size varied depending on the growth conditions. It has been shown that relative humidity affects pollen viability (Ma et al., 2012). Thus, the difference in relative humidity between the rooftop and indoor greenhouses might be the underlying cause of the differential defect in pollen viability. Moreover, ap2m mutants displayed additional morphological alterations: an increase in the number of shoots, branches and leaves, shorter root hairs, and abnormal phyllotaxis. Thus, these results revealed that AP-2 mediated endocytosis plays an essential role in normal development of reproductive tissues in male organs. However, surprisingly few seeds developed into nearly normal looking mature plants with increased numbers of secondary shoots and branches. This is in contrast with what is observed in mice, where depletion of μ2-adaptin of AP-2 causes an embryonic lethal phenotype (Mitsunari et al., 2005). Similar to animal cells, where endocytic routes can be classified on the basis of the type of coat protein (Hsu et al., 2012), multiple routes employing different adaptor proteins or different coat proteins may also exist in plants. Multiple adaptor proteins exist for CCVs. Thus, the absence of AP-2 may be compensated for by other routes or other adaptor proteins. In fact, a large number of epsin-related proteins including ECA1 exist in Arabidopsis, and they have been proposed to play a role in endocytosis in plants (Song et al., 2012).
As in the case of filament growth, impaired endocytosis is likely the underlying cause of the multiple defects associated with pollen in the ap2m plants. Consistent with the proposed role of AP2M in pollen biogenesis, AP2M was strongly expressed in anthers at the late stages (stages 12 and 13) of flowering as well as germinating pollen. Ultrastructural analysis of anthers revealed that pollen undergoing maturation still contains some large vacuoles or large aggregations of vacuoles derived from the lytic vacuole during pollen biogenesis even after stage 12 of flowering, suggesting that AP2M plays a role in the fragmentation of the lytic vacuole during pollen biogenesis. However, the mechanism by which AP2M plays a role in the degeneration of the lytic vacuole from the maturing pollen cells remains unclear. This result suggests that impairment of endocytosis directly or indirectly affects processes related to the degeneration or maintenance of vacuoles. In addition to pollen biogenesis in the anther, pollen grains from the ap2m plants also showed a defect in pollen tube elongation. Indeed, a recent study has provided evidence that an ARF-GEF involved in recycling plays a crucial role in pollen tube elongation in Arabidopsis (Richter et al., 2012).
AP2M Plays a Crucial Role in Modulating Cellular Auxin Levels by Regulating the Localization and Levels of PINs at the PM in Filaments and Pollen Tubes
Consistent with the expected role of AP2M, the cells in the filaments of the ap2m-1 plants exhibit a severe defect in endocytosis of FM4-64, the lypophilic dye used as an endocytic tracer (Jelínková et al., 2010). In addition, the Tyrphostin A23 treatment also blocked internalization of the dye into endosomes. This raises the question as to which cargo proteins are regulated by AP-2–mediated endocytosis in the cells of reproductive tissues or pollen. In plants, one of the best understood developmental processes that are regulated by endocytosis is the polar growth of roots (Grunewald and Friml, 2010). The polar distribution of PINs, which occurs by endocytosis-mediated recycling of PINs, is essential to this process. Thus, the defect in endocytosis of PINs from the PM in root cells results in a failure to regulate cellular auxin levels, which in turn causes a failure of polar root growth (Dhonukshe et al., 2008; Friml, 2010; Grunewald and Friml, 2010). Similarly, the defects in filament growth, pollen germination, and pollen tube elongation appeared to be associated with a defect in homeostasis of the cellular auxin level. This conclusion is based on several lines of evidence; application of exogenous auxin rescued the short filament phenotype and the pollen tube elongation defect, and auxin-inducible genes were expressed at lower levels in mutant than in wild-type filaments. Consistent with this hypothesis, the PIN2:GFP localization pattern was strikingly altered in the ap2m-1 mutant filaments; PIN2:GFP localized to the PM as punctate stains, suggesting that PIN2:GFP accumulated in certain domains of the PM but failed to be internalized. The accumulation of PIN2:GFP at specific sites of the PM in ap2m plants and in the presence of Tyrphostin A23 is different from the behavior of cargo proteins in animal cells, where cargo proteins do not accumulate at specific sites of the PM under the same conditions (Banbury et al., 2003).
This finding raises an intriguing possibility that the mechanism of PIN2:GFP accumulation to the budding vesicles may be different from that of cargo accumulation in animal cells. The aggregation of PIN2:GFP at certain sites of the PM is similar to the behavior of DRP1A and 1C under the Tyrphostin A23 treatment. Dynamics of these proteins was disturbed and the size of the foci at the PM to which fluorescent protein–tagged DRP1s localize was increased (Konopka et al., 2008; Fujimoto et al., 2010). Moreover, the localization pattern of PIN2:GFP in the ap2m-1 mutant filaments was phenocopied by treatment of the wild type with Tyrphostin A23. In wild-type plants, PIN2:GFP showed stronger signals at the basal PM, indicating that the polar localization of PIN2 at the basal PM may be achieved by the recycling of PIN2 from the PM via endocytosis as observed in root cells (Blilou et al., 2005; Vieten et al., 2007). Consistent with the role of PINs in auxin homeostasis and in filament growth, previous studies have demonstrated that various hormones, in particular auxin, play a crucial role in pollen biogenesis and filament growth, thereby regulating fertilization in Arabidopsis (Cecchetti et al., 2008). Moreover, similar to the ap2m-1 plants, eir1-4 plants, which have loss-of-function mutation in PIN2, showed defects in filament growth and DR5rev:GFP expression in stamens. However, the eir1-4 mutants showed less severe defects than the ap2m plants, indicating that additional PINs are also affected in the ap2m plants. Moreover, in the presence of Tyrphostin A23, DR5rev:GFP expression was also impaired, as observed in ap2m mutants, suggesting that AP-2–mediated polar distribution of PIN2 is required for the maintenance of auxin homeostasis in reproductive organs.
However, the defect in pollen tube elongation may not be directly related to the polar localization of PINs at the PM of the pollen tube. Previously, it has been demonstrated that overexpression of various PINs in root hair cells results in a short root hair phenotype (Ganguly et al., 2010). Thus, one possible scenario is that in the pollen tubes the defect in endocytosis may result in higher levels of PINs at the PM as observed with the overexpression of PINs, thereby facilitating auxin efflux from the pollen tube. The lower cellular auxin levels may not support pollen tube elongation. Consistent with this idea, application of exogenous auxin rescued the pollen tube elongation defect. Thus, the levels of PINs at the PM cannot be maintained properly in the ap2m plants. Indeed, the levels of PINs at the PM are known to be regulated by endocytosis-mediated transport to the lytic vacuole through the trans-Golgi network/ early endosome (Kleine-Vehn et al., 2008). Currently, it is not known which PINs are involved in maintaining auxin homeostasis in the pollen tube.
For efficient fertilization in flowering plants, it is essential to achieve proper coordination of multiple developmental processes, including pollen biogenesis, pollen tube elongation, and filament growth, leading to the generation of male reproductive organs. However, these developmental processes are spatially and temporally separated, which leads to the question of how this coordination operates to achieve maximal fertilization. In this study, we provide evidence that auxin levels in these male organs are crucial in determining their development. The cellular auxin levels are regulated by PINs whose localization and levels at the PM are regulated by endocytosis-mediated recycling. Further studies are necessary to elucidate the detailed mechanism(s) of how endocytosis defines PIN localization in filaments and pollen tubes and also how PIN-regulated auxin levels contribute to the coordination of various cellular processes spatially and temporally to achieve maximal seed output.
METHODS
Plant Material and Growth Conditions
Wild-type and ap2m mutant Arabidopsis thaliana (ecotype Columbia-0) plants were grown under a 16-h/8-h photoperiod at 22°C in rooftop (40% relative humidity) or indoor (70% relative humidity) greenhouses. The AP2M knockout mutant ap2m-1 (SALK_083693) and ap2m-2 (CS807972) were obtained from Salk Institute Genomic Analysis Laboratory (http://signal.Salk.edu/).
Construction of Plasmid DNAs
To generate the AP2A1Pro:AP2A1:YFP and AP2A1:YFP construct, the promoter region and the AP2A1 cDNA were amplified by PCR (Bioneer) using specific primers AP2A1Pro(Bam HI)-5′ and AP2A1Pro(Spe I)-3′, and AP2A1(Xba I)-5′ and AP2A1(Sal I)-3′, respectively. The products were inserted into the vector PCAMBIA3301 (Invitrogen) using BamHI and SpeI, XbaI and SalI, respectively. The AP2M cDNA was isolated by PCR using the gene-specific primers AP2M-5′ and AP2M-3′ (see Supplemental Table 1 online). To generate the complementation line of ap2m-1 mutants, the Myc epitope was added right behind to the N-terminal spanning domain of AP2M by overlapping PCR using the primers AP2M-5′, AP2M-3′, AP2M:Mycx3-5′, and AP2M:Mycx3-3′ in such a way that the first-round PCR was performed using the primers AP2M-5′ and AP2M:Mycx3-3′ for the 5′ fragment, and AP2M:Mycx3-5′ and AP2M-3′ for the 3′ fragment. Subsequently, PCR products were ligated by the second-round PCR using primers AP2M-5′ and AP2M-3′. The products were inserted to PCAMBIA3301 (Invitrogen) and 326-sGFP (Jin et al., 2001) using XbaI and BamHI restriction sites, respectively. To obtain the promoter region of AP2M, PCR was performed using primers AP2Mpro(SpeI)-5′ and AP2Mpro(SpeI)-3′ with genomic DNA as template, and the PCR product was inserted into the upstream of AP2M:Mycx3 using the SpeI restriction site. To generate the AP2M promoter (AP2Mp):GUS construct, a genomic DNA fragment containing a 2.4-kb AP2M promoter region (−2400 to −1 positions) was amplified by PCR using specific primers AP2Mpro(BamHI):GUS-5′ and AP2Mpro(NcoI):GUS-3′, and the PCR product was inserted into the vector PCAMBIA3301 using BamHI and NcoI restriction sites. The AP2A1 cDNA was amplified by PCR using gene-specific primers (see Supplemental Table 1 online). The PCR product was ligated 326-YFP using XbaI and SalI restriction sites. To generate transgenic plants with suppression of both AP2A1 (At5g22770) and AP2A2 (At5g22780) that encode α-adaptin simultaneously, an RNAi construct was designed using a DNA fragment from nucleotide positions 150 to 699 of AP2A1 that shows identical sequence to the corresponding region of AP2A2. The DNA fragment was isolated by PCR using primers AP2ARNAi-5′ and AP2ARNAi-3′ and inserted into pENTER vector using XhoI and Acc65I restriction sites. Subsequently, the AP2A1 RNAi construct in the pENTER vector was transferred to pHellgate8 with LR clonase (Invitrogen) to generate the pHellsgate-AP2A1 RNAi construct. The generation of the AP1M2:HAx3 construct was performed as described (Park et al., 2013).
Sequence Alignment
The amino acid sequences were aligned using the program ClustalW2. Protein sequences were downloaded from the National Center for Biotechnology Information website. Sequence alignments data are provided in Supplemental Figure 2 online.
Generation of Transgenic Plants
The binary vectors were used to transform wild-type and ap2m heterozygote plants, according to a published protocol (Clough and Bent, 1998). Transgenic plants were screened on Murashige and Skoog plates supplemented with hygromycin (25 mg/L) or phosphinothricin (25 mg/L).
Immunoprecipitation
Immunoprecipitation was performed as described previously (Park et al., 2005) with some modification. Protein extracts were prepared in immunoprecipitation buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 5 mM EDTA, 1 mM DTT, and 1% [v/v] Triton X-100) supplemented with an EDTA-free protease inhibitor cocktail and incubated with 4 µg of Myc antibody (Cell Signaling) and AP2A (α-adaptin) antibody for 4 h at 4°C. For coimmunoprecipitation experiment, anti-AP2A antibody (4 μg) was used as bait. Immunocomplexes were precipitated by incubating them with Protein A agarose beads for 1 h at 4°C. The pellet was washed three times with immunoprecipitation buffer, suspended in homogenization buffer, and subjected to immunoblot analysis using anti-AP2A (Song et al., 2012), anti-AP1G, anti-PIN2 (Agrisera), or anti-CHC antibodies, respectively (Kim et al., 2001).
Subcellular Fractionation
Protein extracts in sonication buffer (100 mM Tris-HCl, 1 mM EDTA, 1 mM DTT, and 150 mM NaCl) were separated into soluble and membrane fractions by ultracentrifugation at 100,000g for 1 h. Soluble and pellet fractions were collected separately and placed in denaturation buffer supplemented with 2.5% SDS and 2% β-mercaptoethanol. These fractions were analyzed by protein gel blotting using Myc antibody. AALP (Xu et al., 2012) and VSR (Kang et al., 2012) antibodies were used to detect soluble or membrane fractions, respectively.
Transient Expression by Agrobacterium tumefaciens–Mediated Infiltration
Agrobacteria harboring the AP2A1:YFP construct were grown overnight at 28°C in 5 mL Luria-Bertani medium containing 50 μg/mL rifampicin and 50 μg/mL kanamycin. The overnight culture was inoculated into fresh Luria-Bertani medium containing 50 μg/mL rifampicin and 50 μg/mL kanamycin, and Agrobacteria were grown to OD 1 to 2 at 600 nm. Bacteria were harvested by centrifugation at 3000g and resuspended in new medium (10 mM MgCl2, 10 mM MES, pH 5.6, and 150 μM acetosyringone) to OD 0.5 at 600 nm and then incubated at 28°C for 4 h (Yang et al. 2000). For infiltration of Agrobacterium into CLC:CFP transgenic plant leaves, the culture was injected into leaves using a 1-mL syringe without a needle (Wroblewski et al., 2005). Agrobacterium-infiltrated plants were kept in the dark for 1 d, and infected leaves were observed 3 d after infiltration using a confocal laser scanning microscope. For agroinfiltration into root hair cells, the Agrobacterium culture solution was applied twice to the root tissues under vacuum (10 mmHg) for 1 min. Excess infiltration medium was removed, and the plates were incubated in a culture room for 3 d (Marion et al., 2008).
Chemical Treatment and In Vivo Imaging
For chemical treatments, FM4-64 (Invitrogen), Tyrphostin A23 (MP-Biomedical), Tyrphostin A51 (MP-Biomedical), and Wortmannin (Enzo Life Science) were dissolved in dimethylsulfoxide and added to half-strength Murashige and Skoog liquid medium to give the final concentration of 4 μM FM4-64, 30 μM Tyrphostin A23, 30 μM Tyrphostin A51, and 33 μM Wortmannin.
Images were captured using a Zeiss LSM 510 META confocal laser scanning microscope. The combinations of excitation wavelength/emission filter were 488 nm (argon-ion laser)/515 to 530 band-pass for GFP; 458 nm (argon-ion laser)/475 to 525 band-pass for CFP; 514 nm (argon-ion laser)/530 to 560 band-pass for YFP; 543 nm (HeNe laser)/560 to 615 band-pass for monomeric RFP and FM4-64. In addition, images were obtained using a Zeiss Axioplan fluorescence microscope equipped with a cooled charge-coupled device camera. The images were processed using the LSM 5 image browser and Adobe Photoshop 7 and presented as pseudo images. The overlap of images was analyzed using Pearson and Spearman correlation coefficients colocalization plug-in of the ImageJ analysis program (French et al., 2008). A threshold level of 10 was set, under which pixel values were considered noised and not included in the statistical analysis.
Expression of GUS under the AP2M Promoter
To detect expression of GUS from AP2Mpro:GUS in transgenic plants, T2 plants from 10 independent transgenic lines were stained with X-GLUC as described (Xu et al., 2012). Whole plants were soaked in X-GLUC solution at 37°C for 3 h.
In Vitro Pollen Germination, Pollen Tube Growth, and Length Measurement
For pollen germination and pollen tube growth, wild-type and ap2m mutant plants were grown with a 16-h/8-h light/dark cycle at 22°C in a rooftop greenhouse. Open flowers were obtained from wild-type plants 2 weeks after bolting and dehydrated for 1 h at room temperature. Pollen grains were transferred by brushing the flowers onto an germination agar plate [18% Suc, 0.01% boric acid, 1 mM MgSO4, 1 mM CaCl2, 1 mM Ca(NO3)2, and 1% low-melting agarose, pH 7.0] (Suzuki, 2009). Pollen grains were germinated at 28°C for 5 h, examined with a light microscope, and photographed (Axioskop 2) with a charge-coupled device camera (Axio Cam; Carl Zeiss). Pollen tube length was measured from photographs using the Interactive Measurement software package AxioVision 3.0.6 (Carl Zeiss).
To test viability of pollen, grains were released from anthers and soaked in a staining solution containing 1 µg/mL propidium iodide and 0.5 µg/mL FDA, and pollen grains were observed under an epifluorescence microscope (Axioskop 2; Carl Zeiss).
Image Analysis of Developing Anthers by Light, Transmission Electron, and Super-Resolution Structured Illumination Microscopy
Anthers from wild-type and ap2m-1 knockout mutant plants were fixed with solution containing 2.5% glutaraldehyde and 4% paraformaldehyde in a 0.1 M phosphate buffer, pH 7.4, at 4°C for 4 h, rinsed with the phosphate buffer, and fixed in 1% (w/v) osmium tetroxide for another 4 h at 4°C. The samples were dehydrated in an alcohol series and embedded in LR White resin (London Resin Company). For light or transmission electron microscopy, 1-μm or 40- to 50-nm sections were obtained using an ultramicrotome (Bromma 2088; LKB), respectively. The sections were collected on a nickel grid (1-GN, 150 mesh) and stained with uranyl acetate and lead citrate. For electron microscopy, a JEM-100CX-1 transmission electron microscope was used. The PIN2 polarity in filaments was observed using a super-resolution structured illumination microscope (SIM; Nikon N-SIM). The raw images were reconstructed to 3D-SIM image using NIS-E software (Nikon). Images were taken with an Eclipse Ti-E research inverted microscope with Nikon's CFI Apo TIRF ×100 oil objective lens (numerical aperture of 1.49) and 512 × 512 pixel resolution with an iXon DU-897 EMCCD camera (Andor Technology). Multicolor fluorescence was acquired using a diode laser (488 and 561 nm).
Quantitative Real-Time RT-PCR
The excised filaments or total tissues from wild-type plants and ap2m-1 mutant plants were rapidly frozen in liquid nitrogen. Total RNA was extracted using a Qiagen RNeasy plant mini kit (Ambion) and digested with TURBO DNase (Ambion). Total RNA (2 μg) was reverse transcribed into cDNA using a high-capacity cDNA reverse transcription kit (Applied Biosystems), and 50 ng of cDNA was used to detect transcript levels of marker genes by quantitative RT-PCR (Applied Biosystems) using a SYBR Green Kit (Applied Biosystems). ACT2 was used as an internal control. Primer sequences were as follows (Supplemental Table 1 online): ACT2-5′ and ACT2-3′ for ACT2, AP2MqRT-5′ and AP2MqRT-3′ for AP2M, ARF6-5′ and ARF6-3′ for ARF6, ARF8-5′ and ARF8-3′ for ARF8, AFB2-5′ and AFB2-3′ for AFB2, AFB3-5′ and AFB3-3′ for AFB3, TIR1-5′ and TIR1-3′ for TIR1, MYB21-5′ and MYB21-3′ for MYB21, MYB24-5′ and MYB24-3′ for MYB24, MYB57-5′ and MYB57-3′ for MYB57, and RGA-5′ and RGA-3′ for RGA.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: AP2M, At5g46630; AP2A1, At5g22770; AP2A2, At5g22780; PIN1, At1g73590; PIN2, At5g57090; SYP61, At1g28490; SYP41, At5g26980; YUCCA2, At4g13260; YUCCA6, At5g25620; TIR1, At3g62980; AFB1, At1g49720; AFB2, At3g26810; AFB3, At1g12820; ARF6, At1g30330; ARF8, At5g37020; VSR, At3g52850; ACT2, At3g18780; MYB21, At3g27810; MYB24, At5g40350; MYB57, At3g01530; RGA, At2g01570; CHC1, At3g11130; and γ-COP (Sec21), At4g34450.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Expression of AP2A1:YFP in Transgenic Plants.
Supplemental Figure 2. Sequence Alignment of AP2M with Other μ-Adaptin Isoforms.
Supplemental Figure 3. Screening of Two Independent Alleles of ap2m-1 Mutants.
Supplemental Figure 4. ap2m-1 Plants Exhibit Various Developmental Defects.
Supplemental Figure 5. AP2A1 RNAi Transgenic Plants Exhibit Small Siliques with Reduction in Seed Production.
Supplemental Figure 6. Cross-Pollination of ap2m-1 and Wild-Type Plants.
Supplemental Figure 7. Plant Growth Conditions Affect the Severity of the Defects in the Silique Size and Pollen Viability of ap2m-1 Plants.
Supplemental Figure 8. The ap2m-1 Mutant Plants Do Not Exhibit a Defect at the Early Stages of Pollen Biogenesis.
Supplemental Figure 9. The Spatial Expression Pattern of AP2M.
Supplemental Figure 10. qRT-PCR Analysis of Auxin-Inducible Genes in Filaments of ap2m-1 Plants.
Supplemental Figure 11. Exogenous Application of IAA Partially Rescues the Short Filament Phenotype of ap2m-1 Plants.
Supplemental Table 1. Primer Sequences Used in This Study.
Acknowledgments
We appreciate the technical assistance with the use of the SIM facility supported by the Center for Evaluation of Biomaterials at Pohang Technopark. This work was supported in part by a grant (20110031340) from the Advanced Biomass R&D Center, the Ministry of Education, Science, and Technology, and a grant (609004-05-3-HD240) from the Ministry for Agriculture, Food, and Forestry (Korea). E.J.S. and S.Y.K. were supported by NRF2011-0012880 and NRF355-2011-1-C00146 from the National Research Foundation (Korea), respectively.
AUTHOR CONTRIBUTIONS
S.Y.K., Z.-Y.X., and I.H. conceived the project and designed the research strategies. S.Y.K. and Z.-Y.X. isolated ap2m mutants, generated most of the transgenic plants, and performed the majority of the genetic assays. S.Y.K. conducted most of the imaging-related experiments, pollen germination, and pollen viability assays. Z.-Y.X. conducted most of the biochemical experiments and quantitative RT-PCR analysis. K.S., D.H.K., H.K., and E.J.S. generated the AP2A1-RNAi transgenic plants. I.R. and G.J. made the AP2M:3xMyc construct. J.F. generated the PIN2:PIN2:GFP and DR5rev:GFP/eir1-4 transgenic plants. S.Y.K., Z.-Y.X., and I.H. wrote the article.
Glossary
- PM
plasma membrane
- CCV
clathrin-coated vesicle
- CHC
clathrin heavy chain
- RNAi
RNA interference
- FDA
fluorescein diacetate
- CV
central vacuole
- IAA
indole-3-acetic acid
- FM4-64
N-(3-triethylammonium-propyl)-4-(4-diethylaminophenylhexatrienyl) pyridinium dibromide
References
- Abas L., Benjamins R., Malenica N., Paciorek T., Wiśniewska J., Moulinier-Anzola J.C., Sieberer T., Friml J., Luschnig C. (2006). Intracellular trafficking and proteolysis of the Arabidopsis auxin-efflux facilitator PIN2 are involved in root gravitropism. Nat. Cell Biol. 8: 249–256 [DOI] [PubMed] [Google Scholar]
- Andersson E.R. (2012). The role of endocytosis in activating and regulating signal transduction. Cell. Mol. Life Sci. 69: 1755–1771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banbury D.N., Oakley J.D., Sessions R.B., Banting G. (2003). Tyrphostin A23 inhibits internalization of the transferrin receptor by perturbing the interaction between tyrosine motifs and the medium chain subunit of the AP-2 adaptor complex. J. Biol. Chem. 278: 12022–12028 [DOI] [PubMed] [Google Scholar]
- Barth M., Holstein S.E. (2004). Identification and functional characterization of Arabidopsis AP180, a binding partner of plant alphaC-adaptin. J. Cell Sci. 117: 2051–2062 [DOI] [PubMed] [Google Scholar]
- Blilou I., Xu J., Wildwater M., Willemsen V., Paponov I., Friml J., Heidstra R., Aida M., Palme K., Scheres B. (2005). The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433: 39–44 [DOI] [PubMed] [Google Scholar]
- Boehm M., Bonifacino J.S. (2010). Adaptins: The final recount. Mol. Biol. Cell 12: 2907–2920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolte S., Talbot C., Boutte Y., Catrice O., Read N.D., Satiat-Jeunemaitre B. (2004). FM-dyes as experimental probes for dissecting vesicle trafficking in living plant cells. J. Microsc. 214: 159–173 [DOI] [PubMed] [Google Scholar]
- Borg M., Twell D. (2010). Life after meiosis: Patterning the angiosperm male gametophyte. Biochem. Soc. Trans. 38: 577–582 [DOI] [PubMed] [Google Scholar]
- Cecchetti V., Altamura M.M., Falasca G., Costantino P., Cardarelli M. (2008). Auxin regulates Arabidopsis anther dehiscence, pollen maturation, and filament elongation. Plant Cell 20: 1760–1774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Irani N.G., Friml J. (2011). Clathrin-mediated endocytosis: The gateway into plant cells. Curr. Opin. Plant Biol. 14: 674–682 [DOI] [PubMed] [Google Scholar]
- Cheng Y., Dai X., Zhao Y. (2006). Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev. 20: 1790–1799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Corvera S., D’Arrigo A., Stenmark H. (1999). Phosphoinositides in membrane traffic. Curr. Opin. Cell Biol. 11: 460–465 [DOI] [PubMed] [Google Scholar]
- Dettmer J., Hong-Hermesdorf A., Stierhof Y.D., Schumacher K. (2006). Vacuolar H+-ATPase activity is required for endocytic and secretory trafficking in Arabidopsis. Plant Cell 18: 715–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Z., et al. (2012). ER-localized auxin transporter PIN8 regulates auxin homeostasis and male gametophyte development in Arabidopsis. Nat. Commun. 3: 941–949 [DOI] [PubMed] [Google Scholar]
- Dhonukshe P., Aniento F., Hwang I., Robinson D.G., Mravec J., Stierhof Y.D., Friml J. (2007). Clathrin-mediated constitutive endocytosis of PIN auxin efflux carriers in Arabidopsis. Curr. Biol. 17: 520–527 [DOI] [PubMed] [Google Scholar]
- Dhonukshe P., et al. (2008). Generation of cell polarity in plants links endocytosis, auxin distribution and cell fate decisions. Nature 456: 962–966 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Fischer J.A., Eun S.H., Doolan B.T. (2006). Endocytosis, endosome trafficking, and the regulation of Drosophila development. Annu. Rev. Cell Dev. Biol. 22: 181–206 [DOI] [PubMed] [Google Scholar]
- Friml J. (2010). Subcellular trafficking of PIN auxin efflux carriers in auxin transport. Eur. J. Cell Biol. 89: 231–235 [DOI] [PubMed] [Google Scholar]
- Fujimoto M., Arimura S., Ueda T., Takanashi H., Hayashi Y., Nakano A., Tsutsumi N. (2010). Arabidopsis dynamin-related proteins DRP2B and DRP1A participate together in clathrin-coated vesicle formation during endocytosis. Proc. Natl. Acad. Sci. USA 107: 6094–6099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- French A.P., Mills S., Swarup R., Bennett M.J., Pridmore T.P. (2008). Colocalization of fluorescent markers in confocal microscope images of plant cells. Nat. Protoc. 3: 619–628 [DOI] [PubMed] [Google Scholar]
- Ganguly A., Lee S.H., Cho M., Lee O.R., Yoo H., Cho H.T. (2010). Differential auxin-transporting activities of PIN-FORMED proteins in Arabidopsis root hair cells. Plant Physiol. 153: 1046–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunewald W., Friml J. (2010). The march of the PINs: Developmental plasticity by dynamic polar targeting in plant cells. EMBO J. 29: 2700–2714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Happel N., Höning S., Neuhaus J.-M., Paris N., Robinson D.G., Holstein S.E. (2004). Arabidopsis mu A-adaptin interacts with the tyrosine motif of the vacuolar sorting receptor VSR-PS1. Plant J. 37: 678–693 [DOI] [PubMed] [Google Scholar]
- Hiscock S.J., Allen A.M. (2008). Diverse cell signalling pathways regulate pollen-stigma interactions: The search for consensus. New Phytol. 179: 286–317 [DOI] [PubMed] [Google Scholar]
- Hsu V.W., Bai M., Li J. (2012). Getting active: Protein sorting in endocytic recycling. Nat. Rev. Mol. Cell Biol. 13: 323–328 [DOI] [PubMed] [Google Scholar]
- Jelínková A., Malínská K., Simon S., Kleine-Vehn J., Parezová M., Pejchar P., Kubeš M., Martinec J., Friml J., Zazímalová E., Petrásek J. (2010). Probing plant membranes with FM dyes: Tracking, dragging or blocking? Plant J. 61: 883–892 [DOI] [PubMed] [Google Scholar]
- Jin J.B., Kim Y.A., Kim S.J., Lee S.H., Kim D.H., Cheong G.W., Hwang I. (2001). A new dynamin-like protein, ADL6, is involved in trafficking from the trans-Golgi network to the central vacuole in Arabidopsis. Plant Cell 13: 1511–1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H., Kim S.Y., Song K., Sohn E.J., Lee Y., Lee D.W., Hara-Nishimura I., Hwang I. (2012). Trafficking of vacuolar proteins: The crucial role of Arabidopsis vacuolar protein sorting 29 in recycling vacuolar sorting receptor. Plant Cell 24: 5058–5073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.W., Park D.S., Park S.C., Kim S.H., Cheong G.W., Hwang I. (2001). Arabidopsis dynamin-like 2 that binds specifically to phosphatidylinositol 4-phosphate assembles into a high-molecular weight complex in vivo and in vitro. Plant Physiol. 127: 1243–1255 [PMC free article] [PubMed] [Google Scholar]
- Kirchhausen T. (2002). Clathrin adaptors really adapt. Cell 109: 413–416 [DOI] [PubMed] [Google Scholar]
- Kleine-Vehn J., Leitner J., Zwiewka M., Sauer M., Abas L., Luschnig C., Friml J. (2008). Differential degradation of PIN2 auxin efflux carrier by retromer-dependent vacuolar targeting. Proc. Natl. Acad. Sci. USA 105: 17812–17817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konopka C.A., Backues S.K., Bednarek S.Y. (2008). Dynamics of Arabidopsis dynamin-related protein 1C and a clathrin light chain at the plasma membrane. Plant Cell 20: 1363–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J.F., Liu Z.H., Chu C.P., Hu Z.Y., Wang X.L., Zhang X.S. (2012). Different regulatory processes control pollen hydration and germination in Arabidopsis. Sex. Plant Reprod. 25: 77–82 [DOI] [PubMed] [Google Scholar]
- Marion J., Bach L., Bellec Y., Meyer C., Gissot L., Faure J.D. (2008). Systematic analysis of protein subcellular localization and interaction using high-throughput transient transformation of Arabidopsis seedlings. Plant J. 56: 169–179 [DOI] [PubMed] [Google Scholar]
- Mitsunari T., Nakatsu F., Shioda N., Love P.E., Grinberg A., Bonifacino J.S., Ohno H. (2005). Clathrin adaptor AP-2 is essential for early embryonal development. Mol. Cell. Biol. 25: 9318–9323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsu F., Ohno H. (2003). Adaptor protein complexes as the key regulators of protein sorting in the post-Golgi network. Cell Struct. Funct. 28: 419–429 [DOI] [PubMed] [Google Scholar]
- Nagpal P., Ellis C.M., Weber H., Ploense S.E., Barkawi L.S., Guilfoyle T.J., Hagen G., Alonso J.M., Cohen J.D., Farmer E.E., Ecker J.R., Reed J.W. (2005). Auxin response factors ARF6 and ARF8 promote jasmonic acid production and flower maturation. Development 132: 4107–4118 [DOI] [PubMed] [Google Scholar]
- Niihama M., Takemoto N., Hashiguchi Y., Tasaka M., Morita M.T. (2009). ZIP genes encode proteins involved in membrane trafficking of the TGN-PVC/vacuoles. Plant Cell Physiol. 50: 2057–2068 [DOI] [PubMed] [Google Scholar]
- Ohno H. (2006). Physiological roles of clathrin adaptor AP complexes: Lessons from mutant animals. J. Biochem. 139: 943–948 [DOI] [PubMed] [Google Scholar]
- Ortiz-Zapater E., Soriano-Ortega E., Marcote M.J., Ortiz-Masiá D., Aniento F. (2006). Trafficking of the human transferrin receptor in plant cells: Effects of tyrphostin A23 and brefeldin A. Plant J. 48: 757–770 [DOI] [PubMed] [Google Scholar]
- Pacini E., Jacquard C., Clément C. (2011). Pollen vacuoles and their significance. Planta 234: 217–227 [DOI] [PubMed] [Google Scholar]
- Park M., Lee D., Lee G.J., Hwang I. (2005). AtRMR1 functions as a cargo receptor for protein trafficking to the protein storage vacuole. J. Cell Biol. 170: 757–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park M., Song K., Reichardt I., Kim H., Mayer U., Stierhof Y.-D., Hwang I., Jürgens G. (2013). Arabidopsis μ-adaptin subunit AP1M of adaptor protein complex 1 mediates late secretory and vacuolar traffic and is required for growth. Proc. Natl. Acad. Sci. USA 110: 10318–10323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinillos V., Cuevas J. (2008). Standardization of the fluorochromatic reaction test to assess pollen viability. Biotech. Histochem. 83: 15–21 [DOI] [PubMed] [Google Scholar]
- Polo S., Di Fiore P.P. (2006). Endocytosis conducts the cell signaling orchestra. Cell 124: 897–900 [DOI] [PubMed] [Google Scholar]
- Rappoport J.Z., Benmerah A., Simon S.M. (2005). Analysis of the AP-2 adaptor complex and cargo during clathrin-mediated endocytosis. Traffic 6: 539–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter S., Müller L.M., Stierhof Y.D., Mayer U., Takada N., Kost B., Vieten A., Geldner N., Koncz C., Jürgens G. (2012). Polarized cell growth in Arabidopsis requires endosomal recycling mediated by GBF1-related ARF exchange factors. Nat. Cell Biol. 14: 80–86 [DOI] [PubMed] [Google Scholar]
- Robinson M.S. (2004). Adaptable adaptors for coated vesicles. Trends Cell Biol. 14: 167–174 [DOI] [PubMed] [Google Scholar]
- Robinson M.S., Bonifacino J.S. (2001). Adaptor-related proteins. Curr. Opin. Cell Biol. 13: 444–453 [DOI] [PubMed] [Google Scholar]
- Sigismund S., Confalonieri S., Ciliberto A., Polo S., Scita G., Di Fiore P.P. (2012). Endocytosis and signaling: Cell logistics shape the eukaryotic cell plan. Physiol. Rev. 92: 273–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song K., Jang M., Kim S.Y., Lee G., Lee G.-J., Kim D.H., Lee Y., Cho W., Hwang I. (2012). An A/ENTH domain-containing protein functions as an adaptor for clathrin-coated vesicles on the growing cell plate in Arabidopsis root cells. Plant Physiol. 159: 1013–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutter J.U., Sieben C., Hartel A., Eisenach C., Thiel G., Blatt M.R. (2007). Abscisic acid triggers the endocytosis of the arabidopsis KAT1 K+ channel and its recycling to the plasma membrane. Curr. Biol. 17: 1396–1402 [DOI] [PubMed] [Google Scholar]
- Suzuki G. (2009). Recent progress in plant reproduction research: The story of the male gametophyte through to successful fertilization. Plant Cell Physiol. 50: 1857–1864 [DOI] [PubMed] [Google Scholar]
- Takano J., Miwa K., Yuan L., von Wirén N., Fujiwara T. (2005). Endocytosis and degradation of BOR1, a boron transporter of Arabidopsis thaliana, regulated by boron availability. Proc. Natl. Acad. Sci. USA 102: 12276–12281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traub L.M. (2003). Sorting it out: AP-2 and alternate clathrin adaptors in endocytic cargo selection. J. Cell Biol. 163: 203–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traub L.M. (2009). Tickets to ride: Selecting cargo for clathrin-regulated internalization. Nat. Rev. Mol. Cell Biol. 10: 583–596 [DOI] [PubMed] [Google Scholar]
- Ulmasov T., Murfett J., Hagen G., Guilfoyle T.J. (1997). Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9: 1963–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieten A., Sauer M., Brewer P.B., Friml J. (2007). Molecular and cellular aspects of auxin-transport-mediated development. Trends Plant Sci. 12: 160–168 [DOI] [PubMed] [Google Scholar]
- Wroblewski T., Tomczak A., Michelmore R. (2005). Optimization of Agrobacterium-mediated transient assays of gene expression in lettuce, tomato and Arabidopsis. Plant Biotechnol. J. 3: 259–273 [DOI] [PubMed] [Google Scholar]
- Xu Z.Y., Lee K.H., Dong T., Jeong J.C., Jin J.B., Kanno Y., Kim D.H., Kim S.Y., Seo M., Bressan R.A., Yun D.J., Hwang I. (2012). A vacuolar β-glucosidase homolog that possesses glucose-conjugated abscisic acid hydrolyzing activity plays an important role in osmotic stress responses in Arabidopsis. Plant Cell 24: 2184–2199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto Y., Nishimura M., Hara-Nishimura I., Noguchi T. (2003). Behavior of vacuoles during microspore and pollen development in Arabidopsis thaliana. Plant Cell Physiol. 44: 1192–1201 [DOI] [PubMed] [Google Scholar]
- Yang Y., Li R., Qi M. (2000). In vivo analysis of plant promoters and transcription factors by agroinfiltration of tobacco leaves. Plant J. 22: 543–551 [DOI] [PubMed] [Google Scholar]
- Zwiewka M., Feraru E., Möller B., Hwang I., Feraru M.I., Kleine-Vehn J., Weijers D., Friml J. (2011). The AP-3 adaptor complex is required for vacuolar function in Arabidopsis. Cell Res. 21: 1711–1722 [DOI] [PMC free article] [PubMed] [Google Scholar]