Plants strictly regulate uptake of the essential micronutrient iron by protein turnover of the major ferrous iron transporter IRON-REGULATED TRANSPORTER1 (IRT1). This study revealed that IRT1 DEGRADATION FACTOR1 directly functions in IRT1 turnover through its RING E3 ubiquitin ligase activity.
Abstract
Fe is an essential micronutrient for plant growth and development; plants have developed sophisticated strategies to acquire ferric Fe from the soil. Nongraminaceous plants acquire Fe by a reduction-based mechanism, and graminaceous plants use a chelation-based mechanism. In Arabidopsis thaliana, which uses the reduction-based method, IRON-REGULATED TRANSPORTER1 (IRT1) functions as the most important transporter for ferrous Fe uptake. Rapid and constitutive degradation of IRT1 allows plants to quickly respond to changing conditions to maintain Fe homeostasis. IRT1 degradation involves ubiquitination. To identify the specific E3 ubiquitin ligases involved in IRT1 degradation, we screened a set of insertional mutants in RING-type E3 ligases and identified a mutant that showed delayed degradation of IRT1 and loss of IRT1-ubiquitin complexes. The corresponding gene was designated IRT1 DEGRADATION FACTOR1 (IDF1). Evidence of direct interaction between IDF1 and IRT1 in the plasma membrane supported the role of IDF1 in IRT1 degradation. IRT1 accumulation was reduced when coexpressed with IDF1 in yeast or Xenopus laevis oocytes. IDF1 function was RING domain dependent. The idf1 mutants showed increased tolerance to Fe deficiency, resulting from increased IRT1 levels. This evidence indicates that IDF1 directly regulates IRT1 degradation through its RING-type E3 ligase activity.
INTRODUCTION
Fe is an essential micronutrient for plant growth and is indispensable for large numbers of proteins involved in various metabolic pathways, including respiration and photosynthesis (Jeong and Guerinot, 2009). Fe deficiency limits growth, causes leaf chlorosis, and reduces agricultural yields (Marschner, 1995; Jeong and Guerinot, 2009; Kobayashi and Nishizawa, 2012); conversely, Fe at high levels can be toxic. Therefore, plants strictly regulate Fe homeostasis. Plants are highly sensitive to changes in Fe supply and must react to Fe deficiency as well as Fe overload in the environment (Connolly and Guerinot, 2002; Kim and Guerinot, 2007; Jeong and Guerinot, 2009; Kobayashi and Nishizawa, 2012). Fe is abundant in the earth’s crust, but most Fe in the plant environment is found in silicate minerals or as iron oxide and hydroxide, forms that are not directly available to plants. Therefore, plant Fe deficiency is a common occurrence in agriculture. Thus, elucidating the mechanism of Fe homeostasis may help improve the efficiency of Fe usage and increase crop yields.
To date, two different Fe uptake strategies have been described in plants: a reduction mechanism (strategy I) and a chelation mechanism (strategy II) (Hell and Stephan, 2003). Members of the Poaceae family use strategy II, and all other plants are strategy I species. Strategy I has three steps induced by Fe deficiency in roots (Hell and Stephan, 2003). First, Fe deficiency induces activation of a P-type H+-ATPase (AHA2 in Arabidopsis thaliana) that pumps protons out of the cell and leads to a pH reduction that solubilizes ferric Fe in the rhizosphere (Santi and Schmidt, 2009). Then, a low Fe-inducible ferric chelate reductase (FRO2 in Arabidopsis) reduces Fe on the root surface from ferric to ferrous Fe (Robinson et al., 1999; Jeong and Connolly, 2009). Finally, surface ferrous Fe is transported into root epidermal cells by high-affinity ferrous Fe uptake transporters (IRT1 in Arabidopsis).
IRT1, a member of the Zinc-Regulated Transporter (ZRT)/IRT-like protein family, plays a crucial role in Fe homeostasis in Arabidopsis (Korshunova et al., 1999; Connolly et al., 2002; Vert et al., 2002). Loss-of-function mutant irt1 exhibits a severe Fe deficiency phenotype with chlorotic symptoms and growth arrest (Vert et al., 2002). This phenotype can be specifically rescued by Fe resupply but not by other metals (Vert et al., 2002). Thus, IRT1 is an important Fe transporter required for Fe uptake from soil. IRT1 can be induced by Fe deficiency (Connolly and Guerinot, 2002). IRT1 protein accumulation is at its highest level after Fe starvation for 3 d and rapidly decreases upon Fe resupply (Connolly et al., 2002). IRT1 is a broad substrate range metal ion transporter that can transport Fe2+, Zn2+, Mn2+, Co2+, and Cd2+ either expressed in yeast or in plants (Korshunova et al., 1999). Previous studies have suggested that in Arabidopsis, Fe-induced turnover of IRT1 is regulated by ubiquitination (Kerkeb et al., 2008; Barberon et al., 2011). The second intracellular loop of IRT1 is thought to be an E3 interacting and ubiquitination domain (Kerkeb et al., 2008). Several residues in this domain (His-154, His-156, His-158, His-160, Lys-146, and Lys-171) have been examined for their function in the degradation process (Kerkeb et al., 2008). Lys-146 or Lys-171 were found to be required for the degradation of IRT1. Plants expressing IRT1 with substitutions at these two residues accumulated more Fe than the wild type (Kerkeb et al., 2008). This report was the first to support the involvement of ubiquitination in the degradation of IRT1. More recently, the existence of monoubiquitin-conjugated IRT1 was identified. Monoubiquitin-conjugated IRT1 was found to undergo endocytosis-dependent trafficking to lytic vacuoles for constitutive degradation (Barberon et al., 2011). This study suggested that IRT1 degradation is ubiquitination-dependent in Arabidopsis.
Ubiquitination is a posttranslational modification through which the highly conserved 8-kD, 76–amino acid protein, ubiquitin (Ub) is attached to specific substrates, typically to Lys residues (Mazzucotelli et al., 2006). This modification can alter the stability, localization, activity, or protein interaction of the targeted proteins (Deshaies and Joazeiro, 2009). Ubiquitination of proteins is performed in three sequential steps: Ub activation by an E1 enzyme, Ub conjugation by an E2 enzyme, and Ub ligation by an E3 enzyme. The specific recognition of E2s and substrates by E3s is the mediated, critical last step in ubiquitination. In Arabidopsis, more than 1400 genes encode proteins demonstrated or predicted to be involved in Ub-dependent degradation pathways and ∼90% of these genes encode E3 ubiquitin ligases (Moon et al., 2004). Certain E3s ubiquitinate specific substrates in response to changes in environmental stimuli and are therefore important in stress responses (Hua and Vierstra, 2011; Lee and Kim, 2011). One major type of E3 is the Really Interesting New Gene (RING)/U-box-type whose conserved Really Interesting New Gene (RING) domain serves as the Ub∼E2 interaction motif, facilitating ubiquitin transfer directly from E2 to the substrate (Deshaies and Joazeiro, 2009). RING domains are characterized by a spatially conserved octet of His or Cys residues that chelate two zinc atoms in a cross-brace structure (Stone et al., 2005; Deshaies and Joazeiro, 2009). More than 400 RING domain–containing proteins have been predicted in Arabidopsis (Stone et al., 2005), but it is not clear whether they all have ligase activities (Stone et al., 2005; Randow and Lehner, 2009).
To identify candidate E3s involved in IRT1 degradation, we screened 120 lines, each homozygous for T-DNA insertion a gene encoding a putative RING-type E3 ligase (see Supplemental Data Set 1 online) for defects in IRT1 degradation. The gene defective in one line that exhibited delayed degradation of IRT1 protein and loss of IRT1-ubiquitin complexes was designated as IRT1 DEGRADATION FACTOR1 (IDF1). To shed light on the biological role of IDF1 and its relationship to IRT1 degradation in plants, we investigated the interaction and promotion of degradation of IRT1 by IDF1. We found that IDF1 interacts with IRT1 in the plasma membrane and promotes IRT1 degradation in vivo. Our results establish a crucial role for IDF1 in IRT1 regulation as well as in plant growth and development.
RESULTS
Identification of IDF1
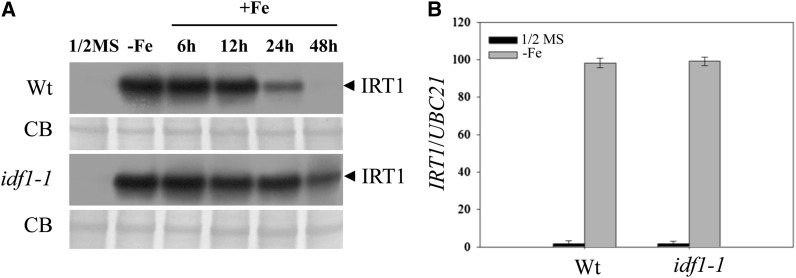
It has been proposed that IRT1 degradation involves ubiquitination (Kerkeb et al., 2008). To identify the E3 ubiquitin ligases potentially involved in IRT1 degradation, we investigated 120 E3 ligase T-DNA insertion lines. We examined IRT1 degradation in these lines during Fe resupply after Fe deficiency. The 120 E3 mutant lines were verified as homozygous, but lacked other experimental data (ABRC stock center; see Supplemental Data Set 1 online). One line with defective IRT1 degradation was identified and the gene affected was designated IDF1. Accumulation of IRT1 was higher in the idf1-1 mutant at various times after Fe resupply than in the wild type (Figure 1A). In the wild type, accumulation of IRT1 decreased significantly after resupply of Fe, whereas in the idf1-1 mutant, IRT1 accumulation was consistently higher than in the wild type at every time point. After Fe resupply for 48 h, IRT1 protein could still be observed in the idf1-1 mutant but not in the wild type (Figure 1A). IRT1 transcript accumulated at similar levels in both wild type and the idf1-1 mutant, confirming that the high accumulation of IRT1 protein in the idf1-1 mutant is not due to different IRT1 mRNA levels (Figure 1B). Accumulation of IRT1 is reduced after a long Fe deficiency treatment (Connolly et al., 2002). We observed that the level of IRT1 protein in the idf1-1 mutant was much higher than that in the wild type after 6 d of Fe deficiency (see Supplemental Figure 1 online). Together, these data suggest that IDF1 is involved in the degradation of IRT1 protein.
Figure 1.
IRT1 Accumulation in the idf1-1 Mutant.
(A) Fourteen-day-old wild-type (Wt) and idf1-1 plants were grown on half-strength MS or Fe-deficient conditions for 3 d (-Fe) and then resupplied with Fe for 6, 12, 24, and 48 h. IRT1 was detected by the anti-IRT1 antibody (IRT1). CB, Coomassie blue stain.
(B) Real-time quantitative RT-PCR monitoring of IRT1 expression in roots of the wild type and idf1-1 under half-strength MS and deficient conditions (-Fe). The expression of ubiquitin21 (UBC21) was used as an internal control. Means and error bars (se) are calculated from four biological repeats.
Ubiquitination of IRT1 in the idf1 Mutant
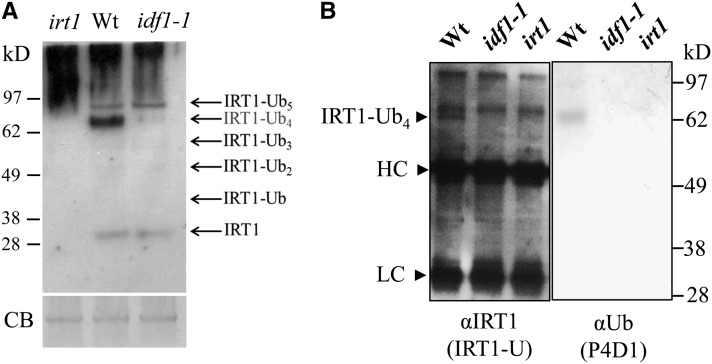
Mono- and multiubiquitinated forms of IRT1 have been identified (Ciechanover, 2005; Barberon et al., 2011). In order to investigate whether IDF1 is involved in mono- and multiubiquitination, the antibody IRT1-U against both IRT1 and ubiquitinated IRT1 (Barberon et al., 2011) was generated and used to detect ubiquitinated IRT1. Intriguingly, the putative IRT1-Ub4 complex was dramatically reduced in the idf1-1 mutant (Figure 2A). In the irt1 mutant, high molecular mass background signals could be seen, but IRT1 and its ubiquitin conjugates could not be detected (Figure 2A). The IRT1-Ub4 complex was immunoprecipitated using the IRT1-U antibody and confirmed to be a Ub conjugate by probing with the anti-Ub antibody P4D1 (Figure 2B). We thus conclude that IDF1 is essential for the formation of the IRT1-Ub4 complex.
Figure 2.
IRT1 Ubiquitination in the idf1-1 Mutant.
(A) IRT1 and its ubiquitin conjugates were detected by immunoblot with IRT1-U antibody on protein extracts from the roots of the wild type (Wt) and idf1-1 after Fe deficiency for 3 d. Predicted IRT1-ubquitin conjugates are indicated. Coomassie blue stain (CB) indicates equal loading.
(B) Immunoprecipitation was performed on 3-d Fe-deficient root protein extracts with anti-IRT1 (IRT1-U) antibody and subjected to immunoblot with anti-IRT1 (IRT1-U) antibody or the anti-ubiquitin (P4D1) antibody. Heavy chain (HC) and light chain (LC) of IgG are indicated. The signal of IRT1 with four ubiquitin molecules (IRT1-Ub4) was not observed in the idf1-1 and irt1 mutants.
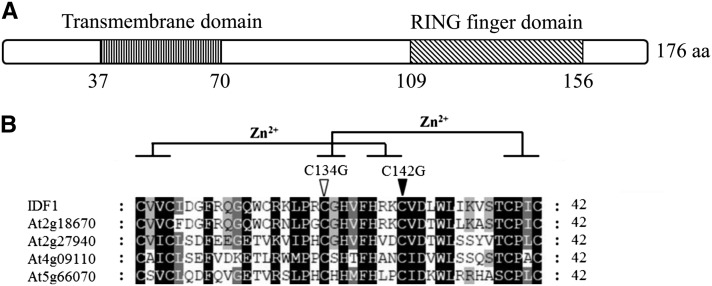
IDF1 is encoded by At4g30370 and predicted to be a 20.8-kD protein with an N-terminal transmembrane domain and a C-terminal single RING domain (Figure 3A). IDF1 shares 90, 53, 28, and 19.6% amino acid sequence identities with the Arabidopsis RING-H2 proteins At2g18670, At2g27940, At4g09110, and At5g66070, respectively (Figure 3B). Their RING domains are highly conserved (Figure 3B). A double amino acid substitution mutation of IDF1 (IDF1-CG) in which the conserved Cys-134 and Cys-142 in the RING domain were replaced by Gly residues was constructed for functional analysis (Figure 3B). IDF1 exhibited RING-dependent autocatalytic ubiquitination activity, but IDF-CG exhibited severely reduced activity, indicating that it is an active ubiquitin E3, dependent on Zn chelating residues as previously observed for many RING type E3s (see Supplemental Figure 2 online) (Kraft et al., 2005; Stone et al., 2005; Deshaies and Joazeiro, 2009).
Figure 3.
Sequence Analysis of IDF1.
(A) Schematic structure of the predicted IDF1 protein. The predicted N-terminal transmembrane domain and C-terminal RING domain are indicated. aa, amino acids.
(B) Sequence alignment of the RING domain of IDF1 and other RING proteins. Putative Zn2+-interacting amino acid residues are marked. Amino acid residues Cys-134 and Cys-142 in the RING domain of IDF1 that were used for the substitution mutations are indicated.
Expression of IDF1
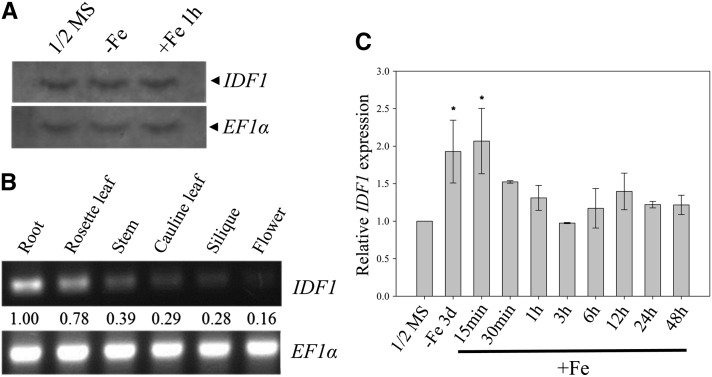
IDF1 expression patterns based on mRNA accumulation profiles were obtained by RNA gel blot analysis. Accumulation of the IDF1 transcript, which is ∼1 kb, was not markedly changed by Fe deficiency or Fe resupply treatments in comparison with half-strength Murashige and Skoog (MS) (Figure 4A). IDF1 was expressed ubiquitously in all organs, including roots, stem, leaves, flowers, and siliques that showed pronounced expression in the roots (Figure 4B). To acquire quantitative data, expression of IDF1 was measured by quantitative RT-PCR under various Fe treatments. IDF1 transcript levels were slightly but significantly induced by ∼1.5-fold under Fe deficiency for 3 d and gradually reduced by Fe resupply over a 48-h time course (Figure 4C), suggesting that IDF1 functions under Fe deficiency.
Figure 4.
Expression of IDF1.
(A) Accumulation of IDF1 transcripts in the roots of 14-d-old plants treated with half-strength MS or in Fe-deficient conditions for 3 d (-Fe), followed by resupply of Fe for 1 h (+Fe 1 h). EF1α transcript levels were used as the loading control.
(B) RT-PCR (30 cycles) of IDF1 in various tissues as indicated. The expression of EF1α was used as an internal control. Relative expression levels are indicated.
(C) Real-time quantitative RT-PCR monitoring of IDF1 transcript accumulation in roots of the wild type under various treatments as indicated. *Relative expression >1.5-fold compared with the wild type in half-strength MS.
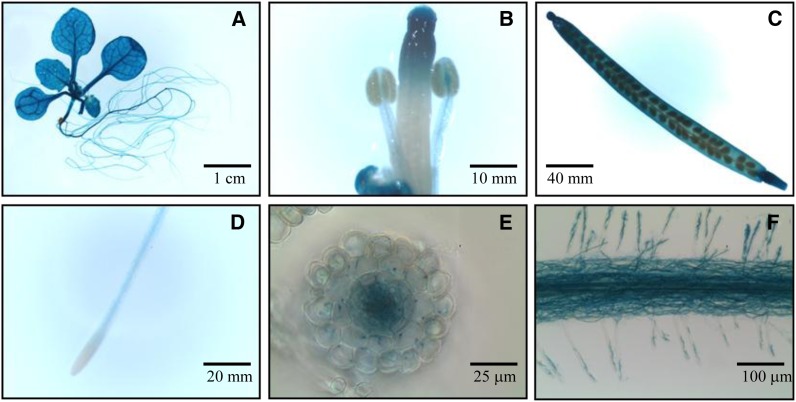
To reveal the IDF1 expression patterns at different developmental stages and tissues, transgenic plants that expressed β-glucuronidase (GUS) driven by the 1.2-kb upstream region of the IDF1 promoter were generated and analyzed. GUS expression indicated that IDF1 promoter activity occurs across all developmental stages (Figure 5). Consistent with this, the expression of GUS was slightly induced by Fe deficiency without changes in temporal and spatial expression patterns. Of note, GUS staining was weak in the meristematic zone of the root and strong in the root hairs (Figure 5). Moreover, GUS signals were detected in the epidermis and cortex in root cross sections (Figures 5E and 5F).
Figure 5.
Spatial Expression of IDF1.
GUS staining was performed in six independent transgenic lines expressing the IDF1 promoter:GUS construct. Transgenic plants were grown in half-strength MS. GUS staining of the transgenic lines in various tissues, including the whole plant (A), reproductive organs (B), silique (C), root tip (D), root cross section (E), and root hairs (F), are shown. Bars indicate scale.
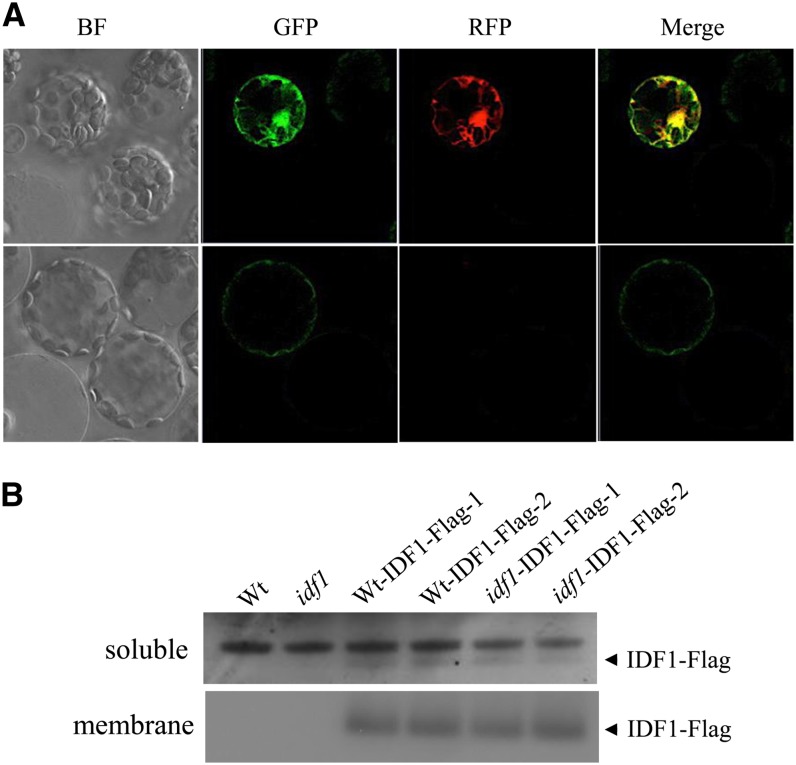
To examine the subcellular localization of IDF1, green fluorescent protein (GFP) was fused to the C terminus of IDF1 (IDF1-GFP) and overexpressed in Arabidopsis mesophyll protoplasts. Green fluorescence from IDF1-GFP colocalized with signals from the cytoplasm marker mCherry (Figure 6A). In addition, the IDF1-GFP signal was also detected at the plasma membrane (Figure 6A). By protein fractionation, IDF1 was consistently detected in both the membrane and soluble fractions of both transgenic idf1-1 mutants and wild-type plants with expression of C-terminal Flag epitope–tagged IDF1 (Figure 6B; see Supplemental Figure 3 online). These results clearly indicate that IDF1 is localized in both the cytoplasm and the plasma membrane.
Figure 6.
Subcellular Localization of IDF1.
(A) Subcellular localization of IDF1 in protoplasts. 35S:IDF1-GFP and 35S:mCherry were cotransfected into Arabidopsis protoplasts, and the transient expression of the fluorescent reporter proteins was detected by confocal microscopy. Two sets of images of different focal planes (top and bottom rows) represent the bright-field (BF) GFP-specific signal for IDF1-GFP (excitation, 488 nm; emission, 500 to 530 nm), the red fluorescent protein (RFP)–specific signal for mCherry (excitation, 561 nm; emission, 575-630 nm), and a merged image of the GFP and red fluorescent protein images.
(B) IDF1-Flag was detected with anti-Flag antibody in both the soluble and membrane fractions of root proteins from the transgenic wild-type (Wt) and idf1-1 (idf1) expressing IDF1 promoter:IDF1-Flag (Wt-IDF1-Flag-1,-2 and idf1-IDF1-Flag-1,-2). Two representatives of at least three lines of each transgenic construct characterized are shown.
Interaction between IDF1 and IRT1
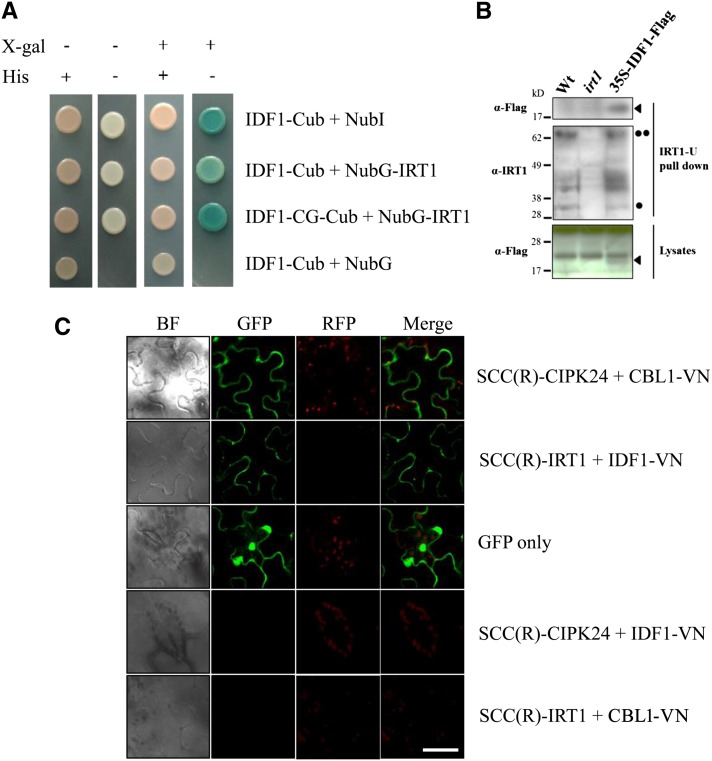
A split-ubiquitin membrane-based yeast two-hybrid system was used to examine the interaction between IDF1 and IRT1. The C terminus of ubiquitin (Cub) linked to a transcription factor Lambda excision A/herpes simplex virus viral protein16 transcriptional activator was fused to the C-terminal end of IDF1 (IDF1-Cub) or IDF1-CG (IDF1-CG-Cub) and expressed in yeast together with the mutated N-terminal fragment of ubiquitin (NubG) fused to the N-terminal end of IRT1 (NubG-IRT1) to test for interaction. If the Cub and NubG constructs are forced into close proximity, Lambda excision A/herpes simplex virus viral protein16 transcriptional activator is released which activates the reporter genes. The wild-type ubiquitin N-terminal fragment (NubI) was used as a positive control. As shown in Figure 7A, IDF1 and IDF1-CG interacted with IRT1 in yeast. The interaction resulted in survival in His dropout media and β-galactosidase activity (Figure 7A). The direct interaction between IRT1 and IDF1 was also supported by a pull-down experiment in Arabidopsis with overexpression of IDF1-Flag (Figure 7B). The IRT1 and its interaction complex in the IDF1-Flag overexpressor were pulled down by the IRT1-U antibody (Figure 7B, middle panel). The IDF1-Flag protein was clearly detected in the pull-down fraction (Figure 7B, top panel).
Figure 7.
IDF1 Directly Interacts with IRT1.
(A) The interaction between IDF1 and IRT1 was detected using in the DUALmembrane yeast two-hybrid system. IDF1 was used as the fused bait protein (IDF1-Cub and IDF1-CG-Cub). IRT1 was used as the fused prey protein (NubG-IRT1). The presence (+) or absence (−) of His or 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-gal) are indicated.
(B) The antibody IRT-U was used to pull down the protein complex with IRT1 from the wild type (Wt), irt1, and IDF1-Flag overexpressor (35S-IDF1-Flag) extracts. Anti-Flag (M2, αFlag) and anti-IRT1 (IRT1-U, αIRT1) antibodies were used to detect the target proteins in the IRT1-U pull-down sample and lysates. Arrowheads indicate IDF1-Flag, the single dot indicates IRT1, and the double dot indicates IRT1-Ub4.
(C) The interaction between IDF1 and IRT1 [SCC(R)-IRT1/IDF1-VN] was observed by BiFC conducted in the leaves of N. benthamiana. The green fluorescence of SCC(R)-CIPK24/CBL1-VN is presented as a control for interaction at the plasma membrane, and GFP shows the cytoplasm. The combination of SCC(R)-CIPK24/IDF1-VN and SCC(R)-IRT1/CBL1-VN were used for false-positive tests. BF, bright field. Bar = 50 μm.
The interaction between IDF1 and IRT1 was also observed in a bimolecular fluorescence complementation (BiFC) analysis in the mesophyll cells of Nicotiana benthamiana (Waadt et al., 2008). The C-terminal fragment of super cyan fluorescent protein 3A (SCFP3A) was linked in frame to the N terminus of IRT1 [SCC(R)-IRT1], and the N-terminal fragment of Venus was fused to the C terminus of IDF1 (IDF1-VN). These two constructs together were introduced into N. benthamiana mesophyll cells by Agrobacterium tumefaciens–mediated infiltration. Intense fluorescence emission produced by the SCFP3A and Venus heterodimer indicated direct interaction of IRT1 and IDF1 (Figure 7C). The pattern of fluorescence emission was identical to that of positive control CBL-interacting protein kinase24 (CIPK24)/calcineurin B-like protein1 (CBL1) protein complexes used as a marker for the plasma membrane (Waadt et al., 2008). This result strongly suggests that IDF1 interacts with IRT1 at the plasma membrane (Figure 7C).
IDF1 Function and the RING Domain
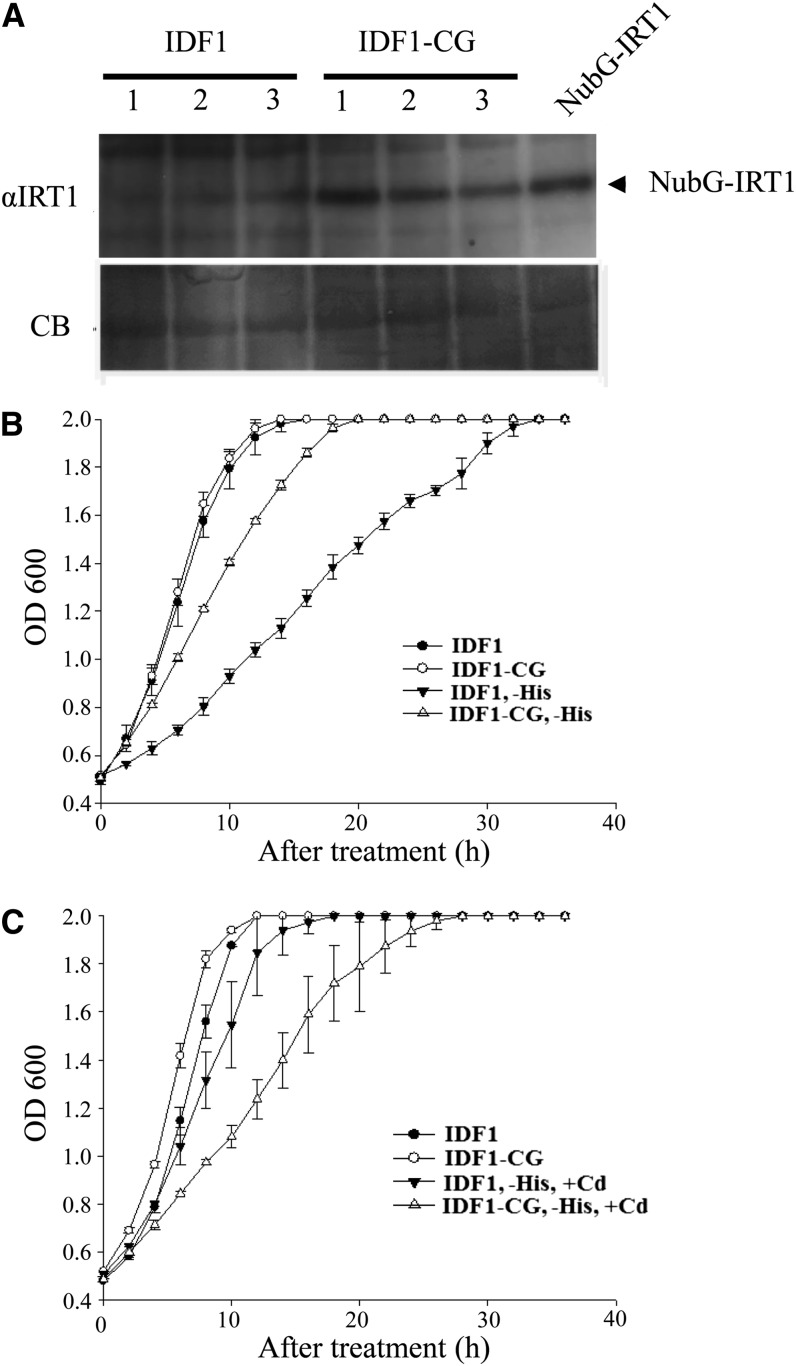
We anticipated that IDF1 would promote IRT1 degradation and the RING domain would be important for this function. In the yeast two-hybrid system, we found that more IRT1 accumulated in yeast cells containing IDF1-CG-Cub and NubG-IRT1 than in IDF1-Cub and NubG-IRT1 (Figure 8A). Reduction in IRT1 accumulation with IDF1 coexpression was also seen in a Xenopus laevis oocyte system. The reduction of IRT1 accumulation was clearly ameliorated upon coexpression with IDF1-CG, although both types of IDF1 were ubiquitinated in oocytes (see Supplemental Figure 4 online). Furthermore, yeast cells containing IDF1-CG-Cub and NubG-IRT1 grew better than those containing IDF1-Cub and NubG-IRT1 in His dropout media (Figure 8B), suggesting less IRT1 degradation following more Cub-NubG complexes activating the His3 gene against the His dropout selection. As a consequence of higher IRT1 accumulation in the selection media, yeast cells containing IDF1-CG-Cub and NubG-IRT1 were more sensitive to the presence of cadmium than those containing IDF1-Cub and NubG-IRT1 (Figure 8C) (Connolly et al., 2002; Barberon et al., 2011). These results suggest that IDF1 stimulates IRT1 degradation, and the RING domain of IDF1 is important in this IRT1 degradation. Collectively, these results indicate that IDF1 is able to interact with IRT1 at the plasma membrane and promote the degradation of IRT1 through its E3 ligase function.
Figure 8.
IRT1 Protein Accumulation and Growth Curves in Yeast Expressing IDF1 and IRT1.
(A) The protein level of IRT1 detected by IRT1 antibody in total protein (20 μg) isolated from three yeast strains grown in His dropout media and expressing NubG-IRT1 with IDF1-Cub (IDF1) or IDF1(CG)-Cub (IDF1-CG), respectively. Total protein (20 μg) extracted from yeast expressing NubG-IRT1 as a positive control. Coomassie blue staining (CB) of protein was used to verify the loadings.
(B) and (C) The growth curves of yeast expressing IDF1 or IDF1-CG with IRT1 were grown in SD media (B) or in SD with cadmium (+Cd) (C). Data are means ± sd of four replicates.
Characterization of IRT1 Degradation
Ubiquitinated membrane proteins can be degraded through the proteasome system and/or endocytosis (Paciorek et al., 2005; Lee et al., 2009). It has been suggested that IRT1 degradation is regulated by endocytosis (Barberon et al., 2011), but the possible involvement of the proteasome system cannot be ruled out. Therefore, next, we investigated IRT1 degradation in plants treated with an endocytosis inhibitor E-64d, a proteasome inhibitor carbobenzoxy-Leu-Leu-leucinal (MG132), or both. After Fe deficiency for 3 d, wild-type and idf1-1 plants were treated with different inhibitors and resupplied with Fe in the presence of the protein synthesis inhibitor cycloheximide (CHX). We found that E-64d or MG132 treatment alone could slow down but not completely block the degradation of IRT1 in the wild type (Table 1; see Supplemental Figure 5 online). The half-life of IRT1, however, was comparable in wild-type plants and idf1 mutants treated with both E-64d and MG132 (Table 1; see Supplemental Figure 5 online), suggesting that both endocytosis and proteasome activity are involved in IRT1 degradation. The longer half-life of IRT1 in the idf1-1 mutant than in wild-type plants with either MG132 or E-64d treatment suggests that IDF1 is involved in IRT1 degradation via both the proteasome and endocytosis pathways.
Table 1. Half-Life of IRT1 Protein in Fe Resupply.
| IRT1 t1/2 (h) |
||
|---|---|---|
| Treatments | Wild Type | idf1-1 |
| CHX | 15.0 ± 0.6 | 17.0 ± 0.7 |
| CHX/MG132 | 20.0 ± 0.2 | 21.6 ± 0.1 |
| CHX/E-64d | 17.8 ± 1.1 | 18.2 ± 0.7 |
| CHX/MG132/E-64d | 22.6 ± 0.5 | 22.6 ± 0.4 |
Data are means ± sd of three independent experiments in the presence of CHX, MG132, or E-64D as indicated. t1/2, half-life was quantified and analyzed by ImageJ after immunoblotting shown in Supplemental Figure 5 online.
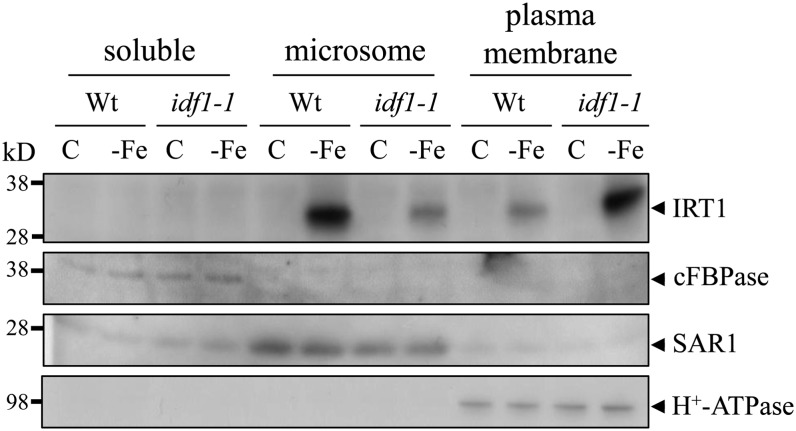
Subcellular fractionation studies showed that nondegraded IRT1 detected by the IRT1 antibody was present at a higher level in the plasma membrane–enriched fraction in the idf1-1 mutant than in the wild type. By contrast, there was a lower level of IRT1 in the microsome fraction in the idf1-1 mutant than in the wild type (Figure 9). This result implies that IDF1 is involved in endocytosis from the plasma membrane. Furthermore, the localization of IRT1 in the wild type and idf1-1 were investigated using an immunogold localization study. These results indicate that IRT1 is predominantly located in the plasma membrane in the absence of IDF1. IRT1 gold particle signals during both Fe deficiency and resupply were readily observed in the plasma membrane in the idf1-1 mutant (Table 2; see Supplemental Figure 6 online). By contrast, fewer membrane-localized IRT1 gold particle signals were observed under Fe resupply conditions in the wild type. Together, these data strongly suggest that IDF1 is important for the degradation of IRT1 from the plasma membrane.
Figure 9.
Proteins in Cytosolic, Microsome, and Plasma Membrane–Enriched Fractions.
Soluble, microsome, and plasma membrane proteins in the root were isolated from 14-d-old wild type (Wt) and idf1-1 treated for 3 d in Fe-sufficient (C) or -deficient conditions (-Fe). IRT1 was detected with the IRT1 antibody. Anti-cFBPase, anti-SAR1, and anti-H+-ATPase antibodies (Agrisera antibodies AS04-043, AS08-326, and AS07-260) were used for the detection of the cytosolic protein cFBPase, endosome protein SAR1, and the plasma membrane protein H+-ATPase, respectively. The corresponding proteins are indicated with arrowheads.
Table 2. Statistical Data of IRT1 Gold Particle Signals.
| Plant | Half-Strength MS | −Fe | +Fe (12 h) | |||
|---|---|---|---|---|---|---|
| Membrane | Cytosol | Membrane | Cytosol | Membrane | Cytosol | |
| Wild type | 2.4 ± 1.1 | 5.5 ± 2.9 | 22.1 ± 2.7 | 7.4 ± 1.4 | 4.3 ± 2.2 | 12.5 ± 3.1 |
| idf1-1 | 4.5 ± 1.2 | 4.8 ± 2.4 | 32.6 ± 4.5 | 3.2 ± 1.7 | 13.5 ± 1.0 | 6.5 ± 1.0 |
Data are means ± sd of three independent epidermis root cells.
The idf1 Mutant Is Tolerant to Fe Deficiency
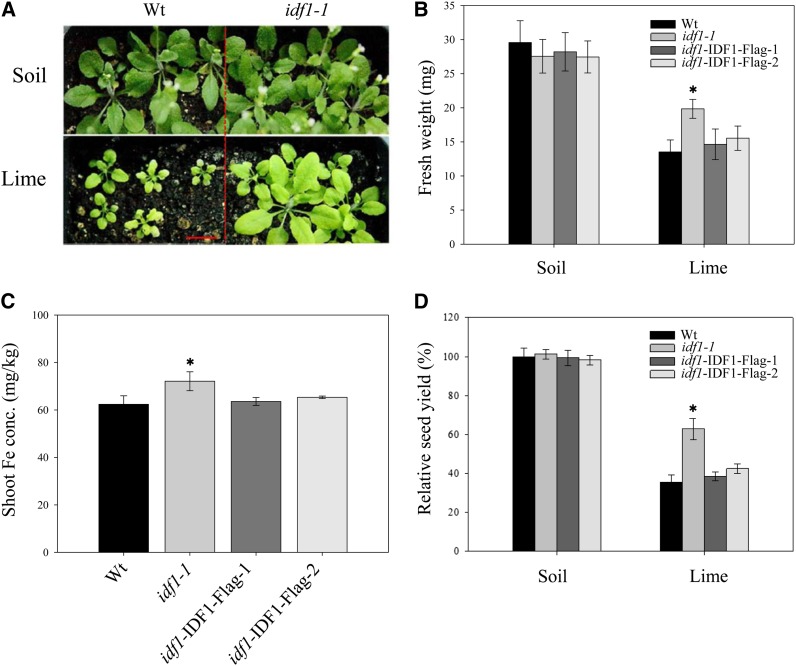
To examine the biological function of IDF1, we examined the phenotypes of Arabidopsis mutants with T-DNA insertion in the IDF1 locus, SALK_038445 (idf1-1), and SALK_129958 (idf1-2) (see Supplemental Figures 7A and 7B online). Both idf1-1 and idf1-2 are IDF1 null mutants (see Supplemental Figure 7C online). No obvious phenotype was observed when these mutants were grown in Fe-sufficient soil (Figures 10A and 10B), but tolerance to Fe-deficient conditions was seen in both idf1-1 and idf1-2 at both the vegetative and productive stages (Figures 10A and 10B; see Supplemental Figure 8A online). The idf1-1 mutant acquired slightly more Fe than the wild type under Fe-deficient conditions (Figure 10C). Seed yield and bolting were approximately twofold higher in the idf1-1 mutant than in the wild type under Fe deficiency (Figure 10D; see Supplemental Figure 8B online). The phenotype in the idf1-1 mutant was restored by IDF1 complementation (Figures 10B to 10D). The idf1 Fe deficiency tolerance phenotype may result from the high IRT1 accumulation in idf1-1 and idf1-2 mutants (see Supplemental Figure 8C online). In summary, loss of function of IDF1 led to overaccumulation of IRT1 and then tolerance to Fe deficiency. These data again support the hypothesis that IDF1 plays a role in IRT1 degradation and influences Fe accumulation in plants.
Figure 10.
Phenotype of idf1-1 and IDF1 Complemented Plants Grown in Alkaline Soil.
(A) Seeds of wild type (Wt) and idf1-1 were directly grown in soil (organic substrate, vermiculite, and mica at a ratio of 9:1:1) without (Soil) or with 3.9 g lime/kg soil (Lime) for 21 d with watering twice a week. Bar = 1 cm.
(B) Fresh weights of the wild type (idf1-1) and two complementation lines (idf1-IDF1-Flag-1 and -2) were measured. Data are means ± sd of four replicates with six seedlings each.
(C) Shoot Fe concentration was determined. Data are means ± sd of four replicates with 20 seedlings each.
(D) The seed yield per silique of soil grown plants. Data are means ± sd of four replicates with six siliques each. *P < 0.05 compared with the wild type under the same conditions.
[See online article for color version of this figure.]
DISCUSSION
In this study, we identified a RING-type E3 ubiquitin ligase, designated as IDF1, which is directly involved in IRT1 degradation. Previous reports indicated that ubiquitination could be one mechanism responsible for the loss of IRT1 protein (Kerkeb et al., 2008), and mono- and multiubiquitin-dependent endocytosis has been found to be involved in constitutive downregulation of IRT1 (Barberon et al., 2011). The putative IRT1-Ub4 complex observed previously (Barberon et al., 2011) disappeared in the idf1-1 mutant (Figure 3A). Therefore, IDF1 plays an important role in the formation of the putative IRT1-Ub4 conjugate. However, IRT1 degradation was not completely arrested, and residual IRT1-Ub complexes were observed in the idf1-1 mutant (Figure 2A). The simplest explanation of this phenomenon is that IDF1 is not the only E3 ubiquitin ligase involved in the ubiquitination process. Degradation of many other proteins is known to involve more than one E3 ubiquitin ligase (Deshaies and Joazeiro, 2009). Our results suggest the existence of additional E3 ligase(s) or degradation pathway(s) for IRT1 degradation. Possible candidates could be the IDF1-related RING proteins, which are as yet uncharacterized. It may be worth undertaking further screening to explore more E3 ubiquitin ligases involved in IRT1 degradation.
In Arabidopsis, RING MEMBRANE-ANCHOR1 (RMA1), a homolog of the human E3 ubiquitin ligase, promotes degradation of aquaporin isoform plasma membrane intrinsic protein2;1 (PIP2;1). Therefore, RMA1 is suspected to have a role in the endocytic degradation of PIP2;1 (Paciorek et al., 2005; Lee et al., 2009). However, the effect of RMA1 could be inhibited by treatment with proteasome inhibitor MG132, suggesting that multiple degradation pathways are involved in PIP2;1 degradation (Lee et al., 2009). In addition, the activated erythropoietin receptor (EpoR) was degraded by both a proteasomal and a lysosomal pathway (Walrafen et al., 2005). Upon erythropoietin (Epo) binding to the EpoR, this complex is Tyr phosphorylated and polyubiquitinated in the intracellular domain. The intracellular domain is degraded by the 26S proteasome and the cleaved EpoR is subsequently internalized and degraded by lysosomes (Walrafen et al., 2005). Based on the loss of IRT1-Ub4 and the remaining plasma membrane–localized IRT1 in the idf1-1 mutant, we suggest that IDF1 may function as one of several E3 ligases involved in IRT1 degradation (Figures 2 and 9, Table 2; see Supplemental Figure 6 online). The proteasome inhibitor MG132 can further reduce the degradation in the idf1-1 mutant (Table 1; see Supplemental Figure 5 online), suggesting that the proteasome degradation pathway is also involved in IRT1 degradation in parallel with the endocytosis pathway. Further identification of additional components involved in IRT1 degradation is needed to elucidate the detailed regulation of IRT1 stability in roots.
The overexpression of IRT1 with the K146R, K171R double substitution presumably impaired ubiquitin-dependent degradation of IRT1, resulting in accumulation of more IRT1 and Fe compared with the wild type (Kerkeb et al., 2008). In our study, tolerance to Fe deficiency and higher accumulation of Fe were observed in the idf1-1 mutant (Figure 10; see Supplemental Figure 8 online). These observations agree with the conclusion of the study conducted by Connolly and colleagues (Kerkeb et al., 2008). Our findings also suggest that IDF1 plays a regulatory role in Fe acquisition through its function in regulating IRT1 degradation. Under Fe sufficient conditions, IRT1 is expressed at extremely low levels, when IDF1 may not be essential. However, IDF1 is upregulated and IRT1 is induced by Fe deficiency (Figure 4C). The same progression of response to Fe status for IDF1 and IRT1 suggests the importance of quick downregulation of IRT1. IDF1 induction may also provide a defense against the uptake of toxic metals from the environment and function as part of a negative feedback loop.
In mammals, Akt/protein kinase B plays a key role in multiple cellular processes, such as Glc metabolism, apoptosis, cell proliferation, transcription, and cell migration (Lu and Hunter, 2009). Akt is primarily localized in the cytosol; however, the ubiquitinated form of Akt can be recruited to the membrane, ready for its role in signal transduction. Ubiquitination and translocation are important for Akt function in mammalian systems (Fan et al., 2013). IDF1 protein was detected in both the cytoplasm and plasma membrane (Figure 6), while the interaction between IDF1 and IRT1 was identified in the plasma membrane (Figure 7C). Currently, it is not clear whether IDF1 is dynamically distributed or recruits some other cooperative components to fulfill its function(s) in the cytoplasm and plasma membrane. A previous study indicated that endogenous IRT1 was colocalized with the trans-Golgi network/early endosome. These intracellular dynamics could also play a role in optimizing Fe uptake under various Fe conditions (Barberon et al., 2011). IDF1 may also affect IRT1 translocation from the plasma membrane to trans-Golgi network/early endosome (Figure 9, Table 2; see Supplemental Figure 6 online). The relationship between IDF1 and IRT1 definitely provides an opportunity to clarify the cellular mechanism of IRT1 trafficking in the future. In summary, we demonstrated a biological function for IDF1, an active RING-type E3 ligase, in Arabidopsis. IDF1 directly interacts with IRT1 in the plasma membrane and contributes to the ubiquitination and degradation of IRT1. This study therefore provides insight into the molecular mechanism of IRT1 regulation.
METHODS
Plant Materials and Growth Conditions
Plant growth procedures were modified from a previous study (Shin et al., 2012). Arabidopsis thaliana ecotype Columbia-0 and the idf1 T-DNA insertion lines (SALK_038445, idf1-1; and SALK_129958, idf1-2) seeds were obtained from the ABRC. Individuals homozygous for their respective T-DNA insertions were used in these studies, verified by PCR. Seeds were surface sterilized, grown (after stratification for 3 d) in pots containing organic substrate, vermiculite, and mica at a ratio of 9:1:1 at a light intensity of 100 μmol m−2 s−1 under a 16-h-light/8-h-dark cycle at 22°C. For expression, plants were grown in half-strength MS for the Fe-sufficient condition and in 300 μM 3-(2-pyridyl)-5,6-diphenyl-1,2,4-triazine sulfonate (ferrozine) for the Fe-deficient condition.
Protein Extraction
The protein extraction procedure was as described previously (Shin et al., 2012). Plant or Xenopus laevis oocyte samples were extracted with extraction buffer (2× SDS sample buffer containing 20 mM N-ethylmaleimide, 100 mM Na2S2O5, and one tablet of protease inhibitor cocktail [Roche Applied Science] per 50 mL). Samples were centrifuged at 12,000g for 10 min, and the protein concentration was determined using the BCA Protein Assay Kit (Thermo Scientific).
Fractionation of plasma membrane, microsomal, and cytosolic proteins was modified from a previous report (Zhang and Peck, 2011). Root samples (0.3 g) were frozen with liquid nitrogen and ground. Homogenized proteins were incubated in 300 μL ice-cold buffer H (330 mM Suc, 50 mM HEPES/KOH, pH 7.5, 50 mM Na4P2O7, 5% glycerol, 0.5% polyvinylpyrrolidone, 10 mM EDTA, 10 μM leupeptin, 3 mM DTT, and protease inhibitor cocktail [Roche]) for 30 min. After removal of cell debris by centrifugation for 10 min at 10,000g at 4°C, cytosolic proteins were isolated as the supernatant after ultracentrifugation for 30 min at 100,000g at 4°C. To isolate microsomes, pellets after 30 min at 100,000g at 4°C were washed once with DTT excluded buffer H and incubated in 150 μL buffer B (DTT excluded buffer H with 0.02% [w/v] Brij-58) on ice for 45 min. After incubation, microsomes were isolated as the supernatant after ultracentrifugation for 30 min at 100,000g at 4°C, and the resulting pellets were washed twice with 150 μL 100 mM Na2CO3, pH 11 to 12) in DTT excluded buffer H to yield the plasma membrane–enriched fraction, which was dissolved in 2× SDS sample buffer for protein analysis.
Antibody and Immunoblot Analysis
The IRT1-specific antibody, IRT1, was generated in a previous study (Shanmugam et al., 2011). The IRT1 polyclonal antibody, IRT1-U, against IRT1 and IRT1-ubiquitin conjugates was raised in rabbits immunized with two synthetic peptides, H2N-CMASNSALLMKTIFLV-CONH2 and H2N-CPANDVTLPIKEDDSS-CONH2 (Barberon et al., 2011). Ubiquitin conjugates and HA fusion protein were detected using the P4D1 mouse monoclonal antibody and HA probe antibody (Y-11; Santa Cruz Biotechnology), respectively. Flag fusion protein was detected using monoclonal ANTI-FLAG M2 antibody (Sigma-Aldrich).
Proteins (20 μg) were separated on NuPAGE 4 to 12% Bis-Tris Gel (Invitrogen) and transferred to a polyvinylidene difluoride membrane (Immobilon-P; Millipore), which was blocked with 5% fat-free milk and 0.1% Tween 20 in phosphate-buffered saline (PBS) for 1 h, incubated with 1:5000-diluted antibody against tested proteins, washed with PBS buffer containing 0.1% Tween 20, and incubated for 1 h with 1:10,000-diluted secondary antibody (POX-conjugated goat anti-rabbit IgG; Millipore). The membrane was washed five times for 10 min each with PBS buffer containing 0.1% Tween 20 solution before development. Specific protein bands were visualized by use of the Immobilon Western Chemiluminescent horseradish peroxidase substrate (Millipore).
Ultrathin Section Immunolocalization
Arabidopsis root tissues were used for the localization of IRT1 protein. Root tissues were fixed with 0.1% gluteraldehyde and 4% paraformaldehyde in 0.1 M phosphate buffer, pH 7.0, dehydrated in a graded alcohol series, and embedded in Lowicryl HM20. Ultrathin sections were taken with a diamond knife by the Ultracut-E ultramicrotome (Reichert Jung) and collected on nickel grids coated with Formvar and stained as described previously (Lending et al., 1988). Tissues were incubated with primary antiserum (IRT1) against the IRT1 protein (1:300 dilutions) at 37°C for 1 h. After washing, the sections were treated with 1:20 diluted goat anti-mouse colloid gold conjugates (12-nm colloidal gold-Affinipure goat anti-mouse IgG; Jackson Immuno Research Lab) for 30 min at room temperature. Sections were counterstained with uranyl acetate and lead citrate before examination under an electron microscope (Philips CM 100) at 80 kV.
Immunoprecipitation
Fourteen-day-old seedlings were grown under Fe deficiency for a further 3 d before protein extraction. The roots of 20 plants were extracted in 1 mL 2× RadioImmunoPrecipitation Assay (RIPA) buffer (1× RIPA buffer is 50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.5% sodium deoxycholate, 1% Nonidet P-40, 0.1% SDS, 10 mM N-ethylmaleimide, and protease inhibitor [Roche] mixture). Extract (100 μL) mixed with the same volume of 2× SDS buffer, boiled for 10 min, and 20 μL was used for the input. Protein extract (100 μL) diluted to 1 mL with RIPA buffer was used for the immunoprecipitation reaction. IRT1-U antibody (4 μL) and 40 μL protein A beads were added to the immunoprecipitation reaction. After overnight incubation, beads were collected by centrifugation and washed four times with RIPA buffer minus SDS. Washed beads were boiled with 100 μL 1× SDS buffer. Then, 20 μL was used for the immunoblot analysis.
For pull-down assay, IRT1-U antibody cross-linked protein A Mag sepharose (GE Healthcare Life Science) was used. The eluted sample was concentrated by Vivaspin 500 (Sartorius Stedim Biotech) before electrophoresis and immunoblot analysis.
Elemental Analysis
Elemental analysis was as previously described (Shin et al., 2012). Harvested plant samples were washed with CaCl2 and water and dried for 3 d before digestion. Microwave-digested samples were analyzed by inductively coupled plasma–optical emission spectrometry (OPTIMA 5300; Perkin-Elmer).
RNA Isolation and Quantification
The procedure was described previously (Shin et al., 2012). Frozen root tissues (0.5 g) were ground in liquid nitrogen using a tissue homogenizer (SH-48; J&H Technology). Total RNA was isolated by the TRIzol method. RNA was precipitated by adding 0.5 mL of isopropanol and incubating at −80°C for 30 min. After centrifugation at 15,000g at 4°C for 15 min, the resulting pellet was washed twice with 75% ethanol. RNA was redissolved in 30 mL of diethylpyrocarbonate-treated water. The RNA concentration was determined at 260 nm on a NanoDrop ND-1000 spectrophotometer (Isogen Life Science). Subsequently, 2 μg of RNA was treated with RQ1 RNase-free DNase (Promega), and the reaction buffer was replaced with 5× first-strand RT buffer (Invitrogen). The cDNA was synthesized by use of SuperScript III reverse transcriptase (Invitrogen). Quantitative RT-PCR analyses used KAPA SYBR FAST qPCR Kits (Kapa Biosystems). UBC21 expression was used as the internal control for all tested genes. The sequences of primers are given in Supplemental Table 1 online.
For RT-PCR, first-strand cDNA synthesis was performed using SuperScript III reverse transcriptase system (Invitrogen). Amplification of EF1α was used as an internal control. The primers EF1α-F, EF1α-R, IDF1-RT-F, and IDF1-RT-R (see Supplemental Table 1 online) were used.
For RNA gel blots, RNA was separated by electrophoresis and transferred to a 0.45-μm nylon membrane (Micron Separations) overnight in 10× SSC/0.1% (w/v) SDS. Membranes were hybridized with random primer-labeled cDNA probes made from the coding region of PCR products. Final washings were performed at 65°C in 0.1× SSC/0.1% (w/v) SDS. Probes were made by PCR with following primers. For EF1α, EF1α-NB-F and EF1-NB-R (see Supplemental Table 1 online) were used. For IDF1, IDF1-NB-F and IDF1-NB-R (see Supplemental Table 1 online) were used. Membranes were exposed to film (XAR-5, Eastman-Kodak) with intensifying screens at −70°C.
Functional Analysis of IDF1 in X. laevis Oocytes
In vivo IDF1-HA and IRT1-Flag expression in X. laevis oocytes was performed as previously described (Wang and Tsay, 2011). The 531-bp IDF1 or the 1044-bp IRT1 cDNA was cloned into the oocyte expression vector pGEMHA2 or pGEMHE and linearized with NheI. Capped mRNA was transcribed in vitro using mMESSAGE mMACHINE kits (Ambion). Oocytes were isolated and injected with the desired amount of IDF1-HA with or without IRT1-Flag cRNA in 50 nL of water, except that the Barth solution was replaced with ND96 solution (96 mM NaCl, 2 mM KCl, 1 mM MgCl2, 1.8 mM CaCl2, and 5 mM HEPES, pH 7.4). The oocytes were then incubated for 2 d in ND96 solution containing 10 mg/L of gentamycin before further immunoblot analysis.
Subcellular Localization of IDF1 in Arabidopsis Protoplasts
IDF1 cDNA was amplified by PCR using the primer FP-IDF1-XbaI and RP-IDF1-BamHI (see Supplemental Table 1 online). The amplified PCR fragment was then subcloned in frame in front of the GFP coding region in the vector 326-GFP (Rus et al., 2001), producing the IDF1-GFP construct driven by 35S promoter. The IDF1-GFP fusion constructs with or without mCherry constructs were then transfected into Arabidopsis protoplasts. Arabidopsis protoplast transformation was performed by following the previously described protocol (Sheen, 2001). Arabidopsis protoplasts were isolated from leaf tissues of 3- to 4-week-old plants grown on soil. IDF1-GFP plasmids were purified with a Qiagen Plasmid Maxi Kit and transformed into protoplasts by the polyethylene glycol–mediated method. After incubation in W5 solution (154 mM NaCl, 125 mM CaCl2, 5 mM KCl, 2 mM MES, pH 5.7, and 5 mM Glc) under light for 12 h, transfected protoplasts were detected for florescence signals by confocal microscopy (Zeiss LSM510, META) with excitation at 488 nm and emission at 500 to 530 nm for GFP, excitation at 561 nm and emission at 575 to 630 nm for mCherry, or at 650 to 710 nm for chloroplast autofluorescence.
Plant Transformation for IDF1 Promoter:GUS, Overexpression, and idf1-1 Complementation
To construct IDF1 promoter:GUS/pCAMBIA1305.1, a 1.2-kb IDF1 promoter was amplified by PCR using the primers FP-OP-IDF1-EcoR I and RP-OP-IDF1-Nco I (see Supplemental Table 1 online). The amplified PCR fragment including the 5′ untranslated region was subcloned to fuse the GUS coding region in the binary vector pCAMBIA1305.1. T2 transgenic lines were subjected to GUS staining as described previously (Lin et al., 2009).
To construct IDF1 promoter:IDF1⁄pCAMBIA1305.1, the PCR-amplified IDF1 coding sequence was subcloned to IDF1 promoter:GUS/pCAMBIA1305.1 with the replacement of GUS. The primers FP-IDF1-NcoI and RP-IDF1-PmlI (see Supplemental Table 1 online) were used. To construct 35S and IDF1 promoter:IDF1-Flag⁄pCAMBIA1305.1, two primers, FP-IDF1-Flag-NcoI and RP-IDF1-Flag-PmlI (see Supplemental Table 1 online), were used to amplify IDF1-Flag from IDF1-Flag/pGEMHE and subcloned to 35S promoter/pCAMBIA1305.1 and IDF1 promoter:GUS/pCAMBIA1305.1 with the replacement of GUS.
Agrobacterium tumefaciens strain GV3101 harboring the plasmids IDF1 promoter:IDF1⁄pCAMBIA1305.1 and IDF1 promoter:IDF1-Flag⁄pCAMBIA1305.1 were used to transform the wild type and the idf1-1 mutant.
Agrobacterium-Mediated Transient Expression in Nicotiana benthamiana
Transient expression in N. benthamiana was as described previously (Waadt et al., 2008). Agrobacterium strains (GV3101) carrying the BiFC constructs and the p19 strain were used for infiltration (Voinnet et al., 2003) into ∼6-week-old N. benthamiana leaves. The Agrobacterium strains were infiltrated at an OD600 of 0.5 for the BiFC constructs and an OD600 of 0.3 for the p19 strain as described by Waadt et al. (2008). All infiltrated leaves were incubated for 12 h before observation. All constructs were driven by 35S promoter. For triple infiltration experiments, SCYCE(R):CIPK constructs were driven by the MAS promoter.
Yeast Two-Hybrid System and Constructs
Cloning and tests were according to the manufacturer’s manual (Dualsystem Biotech; DUAL membrane kit 1, P01001). All PCR products were amplified with Phusion DNA-Polymerase (Finnzymes), and all constructs were verified by sequencing. The primers used for the DUAL membrane yeast two-hybrid system are listed in Supplemental Table 1 online.
Statistical Analysis
Student’s t test was used for statistical analysis. P < 0.05 was considered statistically significant.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: IDF1 (At4g30370), IRT1 (At4g19690), and UBC21 (At5g25760).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. IRT1 Accumulation in the idf1-1 Mutant.
Supplemental Figure 2. In Vitro Self-Ubiquitination Assay of IDF1.
Supplemental Figure 3. Proteins in Soluble and Membrane-Enriched Fractions.
Supplemental Figure 4. IDF1 Promotes IRT1 Degradation in Oocytes.
Supplemental Figure 5. The Kinetics of IRT1 Degradation in Plants.
Supplemental Figure 6. Localization of IRT1 Protein by Immunogold Labeling.
Supplemental Figure 7. IDF1 T-DNA Insertion Mutants idf1-1 (Salk_038445) and idf1-2 (Salk_129958).
Supplemental Figure 8. Phenotypes of the idf1 Mutants (idf1-1 and idf1-2) and the IDF1 Complementation Line idf1-IDF1-Flag-1 Grown in Soil with Lime.
Supplemental Table 1. Primers used in this work.
Supplemental Data Set 1. Putative RING-Type E3 Mutants Analyzed.
Acknowledgments
This work was supported by grants from the National Science Council (NSC 101-2311-B-001-014-MY3), Academia Sinica, and the National Science Foundation (Arabidopsis 2010 Grant MCB-0929100 to J.C.). We thank Yi-Fang Tsay and her lab members for assistance with oocyte and DUAL membrane yeast two-hybrid systems; Wann-Neng Jane in the Plant Cell Biology Core Lab, Institute of Plant and Microbial Biology, Academia Sinica, for the ultrathin-section immunolocalization; Shu-Chen Shen in the Confocal Microscope Core Facility for assistance with confocal microscopy; Pai-Tsang Chang and Ping Kao for technical support; Jill Herschleb, Andy Troy, and Michael Kerber for assistance in identification of homozygous T-DNA insertion lines; many UC Davis, undergraduates for assistance in growing Arabidopsis plants; and the UC Davis–Controlled Environment Facility for providing growth facilities. We also thank Miranda Loney for editing the article.
AUTHOR CONTRIBUTIONS
K.-C.Y. and L.-J.S. designed research, analyzed data, and wrote the article. L.-J.S., J.-C.L., and G.-H.C. performed research. J.C. and H.F. provided materials and analyzed data.
Glossary
- MS
Murashige and Skoog
- GUS
β-glucuronidase
- BiFC
bimolecular fluorescence complementation
- CHX
cycloheximide
- PBS
phosphate-buffered saline
References
- Barberon M., Zelazny E., Robert S., Conéjéro G., Curie C., Friml J., Vert G. (2011). Monoubiquitin-dependent endocytosis of the iron-regulated transporter 1 (IRT1) transporter controls iron uptake in plants. Proc. Natl. Acad. Sci. USA 108: E450–E458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciechanover A. (2005). Proteolysis: From the lysosome to ubiquitin and the proteasome. Nat. Rev. Mol. Cell Biol. 6: 79–87 [DOI] [PubMed] [Google Scholar]
- Connolly E.L., Fett J.P., Guerinot M.L. (2002). Expression of the IRT1 metal transporter is controlled by metals at the levels of transcript and protein accumulation. Plant Cell 14: 1347–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly E.L., Guerinot M. (2002). Iron stress in plants. Genome Biol. 3: REVIEWS1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshaies R.J., Joazeiro C.A. (2009). RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 78: 399–434 [DOI] [PubMed] [Google Scholar]
- Fan C.D., Lum M.A., Xu C., Black J.D., Wang X. (2013). Ubiquitin-dependent regulation of phospho-AKT dynamics by the ubiquitin E3 ligase, NEDD4-1, in the insulin-like growth factor-1 response. J. Biol. Chem. 288: 1674–1684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hell R., Stephan U.W. (2003). Iron uptake, trafficking and homeostasis in plants. Planta 216: 541–551 [DOI] [PubMed] [Google Scholar]
- Hua Z., Vierstra R.D. (2011). The cullin-RING ubiquitin-protein ligases. Annu. Rev. Plant Biol. 62: 299–334 [DOI] [PubMed] [Google Scholar]
- Jeong J., Connolly E.L. (2009). Iron uptake mechanisms in plants: Functions of the FRO family of ferric reductases. Plant Sci. 176: 709–714 [Google Scholar]
- Jeong J., Guerinot M.L. (2009). Homing in on iron homeostasis in plants. Trends Plant Sci. 14: 280–285 [DOI] [PubMed] [Google Scholar]
- Kerkeb L., Mukherjee I., Chatterjee I., Lahner B., Salt D.E., Connolly E.L. (2008). Iron-induced turnover of the Arabidopsis IRON-REGULATED TRANSPORTER1 metal transporter requires lysine residues. Plant Physiol. 146: 1964–1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.A., Guerinot M.L. (2007). Mining iron: Iron uptake and transport in plants. FEBS Lett. 581: 2273–2280 [DOI] [PubMed] [Google Scholar]
- Kobayashi T., Nishizawa N.K. (2012). Iron uptake, translocation, and regulation in higher plants. Annu. Rev. Plant Biol. 63: 131–152 [DOI] [PubMed] [Google Scholar]
- Korshunova Y.O., Eide D., Clark W.G., Guerinot M.L., Pakrasi H.B. (1999). The IRT1 protein from Arabidopsis thaliana is a metal transporter with a broad substrate range. Plant Mol. Biol. 40: 37–44 [DOI] [PubMed] [Google Scholar]
- Kraft E., Stone S.L., Ma L., Su N., Gao Y., Lau O.S., Deng X.W., Callis J. (2005). Genome analysis and functional characterization of the E2 and RING-type E3 ligase ubiquitination enzymes of Arabidopsis. Plant Physiol. 139: 1597–1611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H.K., Cho S.K., Son O., Xu Z., Hwang I., Kim W.T. (2009). Drought stress-induced Rma1H1, a RING membrane-anchor E3 ubiquitin ligase homolog, regulates aquaporin levels via ubiquitination in transgenic Arabidopsis plants. Plant Cell 21: 622–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.H., Kim W.T. (2011). Regulation of abiotic stress signal transduction by E3 ubiquitin ligases in Arabidopsis. Mol. Cells 31: 201–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lending C.R., Kriz A.L., Larkins B.A., Bracker C.E. (1988). Structure of maize protein bodies and immunocytochemical localization of zeins. Protoplasma 143: 51–62 [Google Scholar]
- Lin Y.F., Liang H.M., Yang S.Y., Boch A., Clemens S., Chen C.C., Wu J.F., Huang J.L., Yeh K.C. (2009). Arabidopsis IRT3 is a zinc-regulated and plasma membrane localized zinc/iron transporter. New Phytol. 182: 392–404 [DOI] [PubMed] [Google Scholar]
- Lu Z., Hunter T. (2009). Degradation of activated protein kinases by ubiquitination. Annu. Rev. Biochem. 78: 435–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marschner, H. (1995). Mineral Nutrition of Higher Plants. (London: Academic Press). [Google Scholar]
- Mazzucotelli E., Belloni S., Marone D., De Leonardis A., Guerra D., Di Fonzo N., Cattivelli L., Mastrangelo A. (2006). The e3 ubiquitin ligase gene family in plants: Regulation by degradation. Curr. Genomics 7: 509–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon J., Parry G., Estelle M. (2004). The ubiquitin-proteasome pathway and plant development. Plant Cell 16: 3181–3195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paciorek T., Zazímalová E., Ruthardt N., Petrásek J., Stierhof Y.D., Kleine-Vehn J., Morris D.A., Emans N., Jürgens G., Geldner N., Friml J. (2005). Auxin inhibits endocytosis and promotes its own efflux from cells. Nature 435: 1251–1256 [DOI] [PubMed] [Google Scholar]
- Randow F., Lehner P.J. (2009). Viral avoidance and exploitation of the ubiquitin system. Nat. Cell Biol. 11: 527–534 [DOI] [PubMed] [Google Scholar]
- Robinson N.J., Procter C.M., Connolly E.L., Guerinot M.L. (1999). A ferric-chelate reductase for iron uptake from soils. Nature 397: 694–697 [DOI] [PubMed] [Google Scholar]
- Rus A., Yokoi S., Sharkhuu A., Reddy M., Lee B.H., Matsumoto T.K., Koiwa H., Zhu J.K., Bressan R.A., Hasegawa P.M. (2001). AtHKT1 is a salt tolerance determinant that controls Na(+) entry into plant roots. Proc. Natl. Acad. Sci. USA 98: 14150–14155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santi S., Schmidt W. (2009). Dissecting iron deficiency-induced proton extrusion in Arabidopsis roots. New Phytol. 183: 1072–1084 [DOI] [PubMed] [Google Scholar]
- Shanmugam V., Lo J.C., Wu C.L., Wang S.L., Lai C.C., Connolly E.L., Huang J.L., Yeh K.C. (2011). Differential expression and regulation of iron-regulated metal transporters in Arabidopsis halleri and Arabidopsis thaliana - The role in zinc tolerance. New Phytol. 190: 125–137 [DOI] [PubMed] [Google Scholar]
- Sheen J. (2001). Signal transduction in maize and Arabidopsis mesophyll protoplasts. Plant Physiol. 127: 1466–1475 [PMC free article] [PubMed] [Google Scholar]
- Shin L.J., Lo J.C., Yeh K.C. (2012). Copper chaperone antioxidant protein1 is essential for copper homeostasis. Plant Physiol. 159: 1099–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone S.L., Hauksdóttir H., Troy A., Herschleb J., Kraft E., Callis J. (2005). Functional analysis of the RING-type ubiquitin ligase family of Arabidopsis. Plant Physiol. 137: 13–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vert G., Grotz N., Dédaldéchamp F., Gaymard F., Guerinot M.L., Briat J.F., Curie C. (2002). IRT1, an Arabidopsis transporter essential for iron uptake from the soil and for plant growth. Plant Cell 14: 1223–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet O., Rivas S., Mestre P., Baulcombe D. (2003). An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J. 33: 949–956 [DOI] [PubMed] [Google Scholar]
- Waadt R., Schmidt L.K., Lohse M., Hashimoto K., Bock R., Kudla J. (2008). Multicolor bimolecular fluorescence complementation reveals simultaneous formation of alternative CBL/CIPK complexes in planta. Plant J. 56: 505–516 [DOI] [PubMed] [Google Scholar]
- Walrafen P., Verdier F., Kadri Z., Chrétien S., Lacombe C., Mayeux P. (2005). Both proteasomes and lysosomes degrade the activated erythropoietin receptor. Blood 105: 600–608 [DOI] [PubMed] [Google Scholar]
- Wang Y.Y., Tsay Y.F. (2011). Arabidopsis nitrate transporter NRT1.9 is important in phloem nitrate transport. Plant Cell 23: 1945–1957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z.J., Peck S.C. (2011). Simplified enrichment of plasma membrane proteins for proteomic analyses in Arabidopsis thaliana. Proteomics 11: 1780–1788 [DOI] [PubMed] [Google Scholar]