The activity of NON-SMOKY GLYCOSYLTRANSFERASE1 (NSGT1) mediates the conversion of hydrolysis-susceptible glycosides of phenylpropanoid volatiles into hydrolysis-resistant forms and prevents damage-induced release of these volatiles in tomato fruit. This leads to a perceivable reduction in the smoky aroma intensity of these fruits compared to fruits of cultivars containing a truncated NSGT1 gene.
Abstract
Phenylpropanoid volatiles are responsible for the key tomato fruit (Solanum lycopersicum) aroma attribute termed “smoky.” Release of these volatiles from their glycosylated precursors, rather than their biosynthesis, is the major determinant of smoky aroma in cultivated tomato. Using a combinatorial omics approach, we identified the NON-SMOKY GLYCOSYLTRANSFERASE1 (NSGT1) gene. Expression of NSGT1 is induced during fruit ripening, and the encoded enzyme converts the cleavable diglycosides of the smoky-related phenylpropanoid volatiles into noncleavable triglycosides, thereby preventing their deglycosylation and release from tomato fruit upon tissue disruption. In an nsgt1/nsgt1 background, further glycosylation of phenylpropanoid volatile diglycosides does not occur, thereby enabling their cleavage and the release of corresponding volatiles. Using reverse genetics approaches, the NSGT1-mediated glycosylation was shown to be the molecular mechanism underlying the major quantitative trait locus for smoky aroma. Sensory trials with transgenic fruits, in which the inactive nsgt1 was complemented with the functional NSGT1, showed a significant and perceivable reduction in smoky aroma. NSGT1 may be used in a precision breeding strategy toward development of tomato fruits with distinct flavor phenotypes.
INTRODUCTION
Volatile organic compounds (VOC) produced and emitted by plants are extremely diverse and ubiquitous. In nature, they can play a role in the chemical language that plants use to communicate to each other and to other living organisms. VOCs also determine the aroma of fruits and vegetables. More than 400 VOCs have been identified in tomato (Solanum lycopersicum) fruit, but only a fraction of those occur in high enough concentrations to be detectable by the human nose (Buttery et al., 1987; Petro-Turza, 1987; Baldwin et al., 1998, 2000; Krumbein et al., 2004). It has been shown that changes in volatile composition can affect human perception and preference of tomato fruit (Tieman et al., 2012).
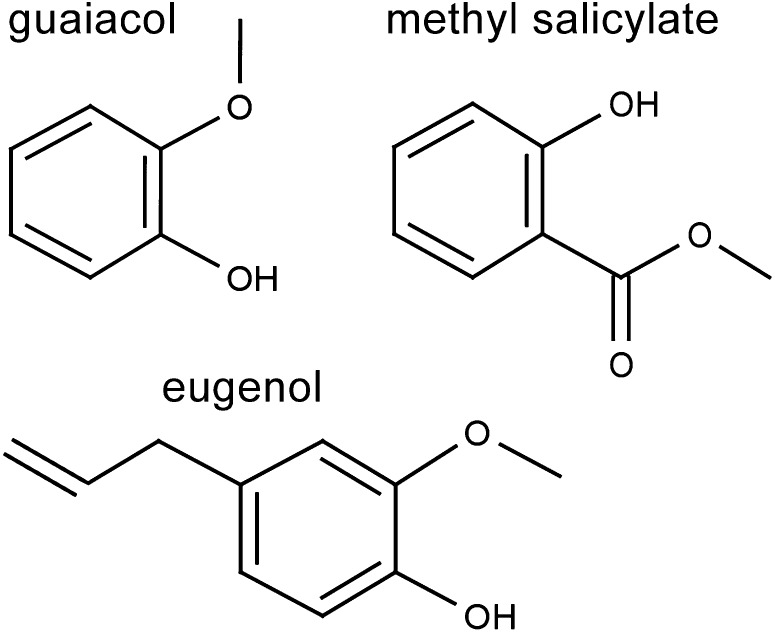
The phenylpropanoid volatiles (PhP-Vs) guaiacol, methyl salicylate (MeSA), and eugenol (Figure 1) have been previously shown to contribute to aroma of tomato fruit and tomato fruit products (Buttery et al., 1987, 1990b). The aroma of guaiacol is often described as “pharmaceutical” or “smoky” (Krumbein and Auerswald, 1998; Robin et al., 1999; Jensen and Whitfield, 2003; Lee and Noble, 2003; Morales et al., 2005; Akakabe et al., 2006; Kennison et al., 2007; Arumugam et al., 2010). A range of enzymes that catalyze different steps in the biosynthetic pathway leading from Phe or isochorismate to both PhP-V and other phenolic volatiles has been described in the literature (Dudareva and Pichersky, 2000; Wildermuth et al., 2001; Boatright et al., 2004; Dudareva et al., 2004; Gang, 2005). Some of the underlying genes have also been cloned and characterized (Koeduka et al., 2006, 2009; Tieman et al., 2010; Mageroy et al., 2012).
Figure 1.
In contrast with the biosynthesis of volatile molecules, little is known about the logistical processes, which include their conjugation, transport of these conjugated forms, compartmentalization, storage, deconjugation, and release. Glycoconjugation is a common feature of plant secondary metabolite modification. It facilitates transport, deactivation, and storage of chemical compounds within the plant cell (Bowles et al., 2006). The enzymatic machinery responsible for the synthesis of glycoconjugates and their cleavage consists of two major enzyme families: glycosyltransferases and glycosyl hydrolases (glycosidases), respectively. In plants, these are two of the largest and most diverse enzyme families. This enzymatic variation is responsible for the large structural diversity of glycoconjugates, which may lead to different physical and chemical properties and, as a result, different physiological functions. Taste and color of plant products, as well as biological activity and assimilation efficiency, are known to depend on glycoconjugation of secondary metabolites (Frydman et al., 2004; Morita et al., 2005; Veitch and Grayer, 2011; Cobucci-Ponzano and Moracci, 2012).
Several reports demonstrate that the amount of nonvolatile glycoconjugated VOC may considerably exceed the total amount of free VOC in fruit of modern tomato germplasm (Buttery et al., 1990a; Ortiz-Serrano and Gil, 2007; Tikunov et al., 2010). This considerable volatile reserve is of fundamental importance for production of aroma and represents an obvious target for its modification.
Our previous biochemical studies on fruits of a collection of 94 tomato cultivars showed that PhP-Vs are predominantly present, in intact fruit, as two major types of glycoconjugates (Tikunov et al., 2010). The first type is a PhP-V diglycoside present in mature green fruits and consisting of a volatile aglycone conjugated with a hexose-pentose sugar moiety. These diglycosides are rapidly cleaved upon fruit tissue damage (e.g., by blending), and the corresponding volatiles are released. During fruit ripening, the PhP-V diglycosides can be further converted into more complex triglycosides containing a dihexose-pentose moiety. These triglycosides are resistant to cleavage and, as a result, ripe tomato fruits do not show any damage-induced PhP-V release. In fruits of roughly a half of the tomato cultivars studied, the ripening-induced conversion of cleavable diglycosides into noncleavable triglycosides did not occur. Therefore, the ripe fruits of these cultivars retained the ability to release the corresponding volatiles upon fruit tissue damage. This release of PhP-V has been associated with the smoky aroma of tomato as determined using a trained fruit tasting panel (Menéndez et al., 2012).
Here, we present NON-SMOKY GLYCOSYLTRANSFERASE1 (NSGT1), which prevents the damage-induced release of the smoky aroma–associated PhP-V in ripening tomato fruit by means of structural modification of their glycoconjugates. This glycosyltransferase is the key factor determining the presence or absence of a smoky aroma in fruits of cultivated tomato.
RESULTS
Expression of a Candidate Glycosyltransferase Correlates with the Release of PhP-Vs from Damaged Tomato Fruit
We hypothesized that the conversion of PhP-V diglycosides into triglycosides occurring in non-smoky fruits during ripening might be a result of induction of expression of a gene encoding a glycosyltransferase (GT) enzyme. In order to find a candidate GT, an untargeted gene expression analysis using next-generation transcript sequencing called digital gene expression (DGE) was performed. Two groups, consisting of 25 ‘smoky’ and 25 ‘non-smoky’ cultivars, were selected from a set of commercial cultivars characterized previously (Tikunov et al., 2005, 2010; Menéndez et al., 2012). Fruit material of each of the groups was pooled at two ripening stages: mature green (MG) and turning (T). In total, four pooled samples were prepared: (1) mature green ‘smoky’ (MG-S), (2) turning ‘smoky’ (TN-S), (3) mature green ‘non-smoky’ (MG-NS), and (4) turning ‘non-smoky’ (TN-NS). Expression of a hypothetical GT was expected to be higher in TN-NS, since this fruit material contains noncleavable PhP-V triglycosides, whereas the other three fruit pools contain the diglycoside precursors.
The DGE method produces a 21-bp read of each transcript (a transcript tag) starting from the most distal NlaIII restriction site at the 3′ part of its corresponding cDNA sequence. The number of reads of a transcript tag is used as a measure of this transcript’s abundance in a given sample. In total, 22,994 unique transcript tags (each 21-bp long) with a relative frequency equal to or greater than six reads per million were detected in the four pooled samples analyzed. Of these, 20,383 matched to the tomato genome (cv Heinz; Tomato Genome Consortium SL2.40) sequence at unique positions with 100% identity and 100% tag coverage. To determine which genes these tags represented, genomic sequence fragments, each containing a 1-kb sequence upstream of each tag, were extracted from the tomato genome sequence and annotated using BLAST against the non-redundant protein database of the National Center for Biotechnology Information, TAIR10, and Uniprot plant protein databases and specific tomato unigene and EST databases: SGN-Unigene v.5 (Sol Genomics Network) and Tomato Gene Index v.13 (The Gene Index Project) databases. A total of 199 of these annotated gene sequence fragments were located within the genomic region corresponding to the ‘smoky’/PhP volatile quantitative trait locus region reported previously to be located on chromosome IX (Chaïb et al., 2006), between the restriction fragment length polymorphism markers TG348 and TG008, at positions 62,797 and 65,657 kb, respectively. One of these gene fragments (687 bp) corresponding to tag 403908 contained a 360-bp open reading frame (ORF). The encoded protein sequence was similar to the C-terminal domain of several plant GTs (see Supplemental Figure 1 online). The corresponding DGE tag 403908 showed an 11.6-fold higher frequency in the TN-NS (1160 reads per million) sample compared with its earlier ripening stage MG-NS (100 reads per million). The frequency of this tag in the MG-S and TN-S samples was below six reads per million.
Thus, the untargeted gene expression analysis revealed a transcript of a putative NSGT that showed considerably higher abundance in tomato fruit material containing the noncleavable PhP-V triglycosides compared with fruits containing their cleavable PhP-V diglycoside precursors.
Obtaining NSGT Sequences, Their Comparative Analysis, and Organization in the Tomato Genome
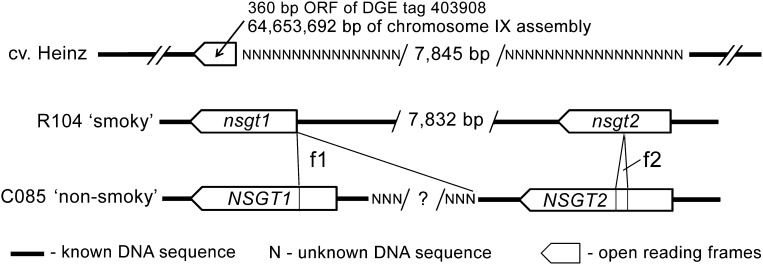
Only an incomplete DNA sequence encoding the C-terminal domain of the hypothetical NSGT protein could be extracted from the tomato genome assembly (cv Heinz) of chromosome IX, since a large fragment of unknown sequence (approximately 8 kb) was located upstream of this partial NSGT gene sequence (Figure 2). This suggested that this unknown sequence could contain the missing part of the gene encoding the N-terminal domain of the GT. To obtain the full-length coding sequence of the putative NSGT gene, genome walking was performed in two homozygous tomato lines, ‘smoky’ R104 (‘smoky’ fruit) and C085 (‘non-smoky’ fruit), using the 360-bp partial candidate NSGT ORF sequence as starting template. Sequences of two intronless genes were obtained from ‘non-smoky’ C085. The first sequence containing a 1353-bp ORF encoding a 450–amino acid GT protein was further denoted as NSGT1 (Figure 2; see Supplemental Figures 2 and 3 online). The second sequence contained a 1350-bp ORF, which showed 93% sequence similarity to that of NSGT1 and encoded a 449–amino acid GT protein. This sequence was denoted as NSGT2. Compared with NSGT1, NSGT2 had a number of single nucleotide polymorphisms, a deletion and an insertion, which all together did not result in an ORF shift (see Supplemental Figure 2 online).
Figure 2.
Organization of the NSGT Genes in Chromosome IX of Tomato.
DGE tag 403908 overexpressed in the ‘non-smoky’ tomato sample was mapped at ∼64,654 kb of chromosome IX assembly (cv Heinz, SL2.40). In cv Heinz, the genome sequence fragment corresponding to this tag revealed a 360-bp ORF terminated by a predicted 7845-bp fragment of unknown sequence located upstream. In the ‘non-smoky’ tomato line C085, two genes, NSGT1 and NSGT2, obtained by genome walking, are separated by a fragment of unknown length and composition. This fragment (f1) containing the 5′-end of the NSGT1 ORF and probably the promoter region is absent in the consensus sequence of this region obtained for the ‘smoky’ tomato line R104 by combining genome walking and tomato genome sequence information (cv Heinz). The size of the consensus sequence obtained in R104 was 7832 bp, which is close to the length of the unknown 7845-bp sequence predicted in the Heinz genome. Another ORF present in the R104 consensus sequence, nsgt2, is missing the 3′-end compared with NSGT2 of line C085 due to deletion of a 38-bp fragment f2, which causes an ORF shift and a premature stop codon (see Supplemental Figure 2 online for details). All three models of cv Heinz, R104, and C085 have the same scale.
The two NSGT-like sequences were also found in the ‘smoky’ tomato line R104, but they had severe alterations compared with those found in the ‘non-smoky’ C085. The first sequence of R104 lacked the first 306 bp of the 5′ coding region, including the start codon (see Supplemental Figure 2 online). The remaining aligned part of this sequence had only four nucleotide mismatches compared with NSGT1 and 72 mismatches to NSGT2. Hence, it was denoted further as nsgt1. The resulting protein translated from the next possible start codon of nsgt1 missed a large part of the N-terminal domain, in which enzyme active sites, including a possible substrate binding pocket, were predicted to be located (see Supplemental Figure 3 online). The second sequence obtained from R104 was more similar to NSGT2 (51 mismatches) than to NSGT1 (68 mismatches). Besides, the nucleotide substitutions in the 3′ part of this sequence were identical to those distinguishing NSGT2 from NSGT1. Hence, this sequence was further denoted as nsgt2. Compared with NSGT2 of C085, nsgt2 of R104 had a 38-bp deletion, which caused an ORF shift and a premature stop codon. This premature stop codon would lead to a truncated protein lacking Plant Secondary Product Glycosyltransferase domain, which is thought to be the UDP-sugar donor binding site (see Supplemental Figure 3 online).
A hypothetical model of the organization of the NSGT locus was produced by combining the genome walking data with the tomato chromosome assembly (Tomato Genome Consortium, SL2.40) as well as with the tomato genome sequence contigs and scaffolds, which have not yet been assembled into chromosomes. According to this model, in the ‘smoky’ tomato background, R104 and in cv Heinz nsgt1 and nsgt2 are located in tandem and ∼6 kb apart (Figure 2). The same tandem organization could be observed in ‘non-smoky’ tomato C085, but sequencing of the NSGT1-NSGT2 intragenic region of ‘non-smoky’ tomato C085 proved not to be possible due to presence of dinucleotide repeats. Therefore, the composition and the exact length of this region in the ‘non-smoky’ background remained unknown.
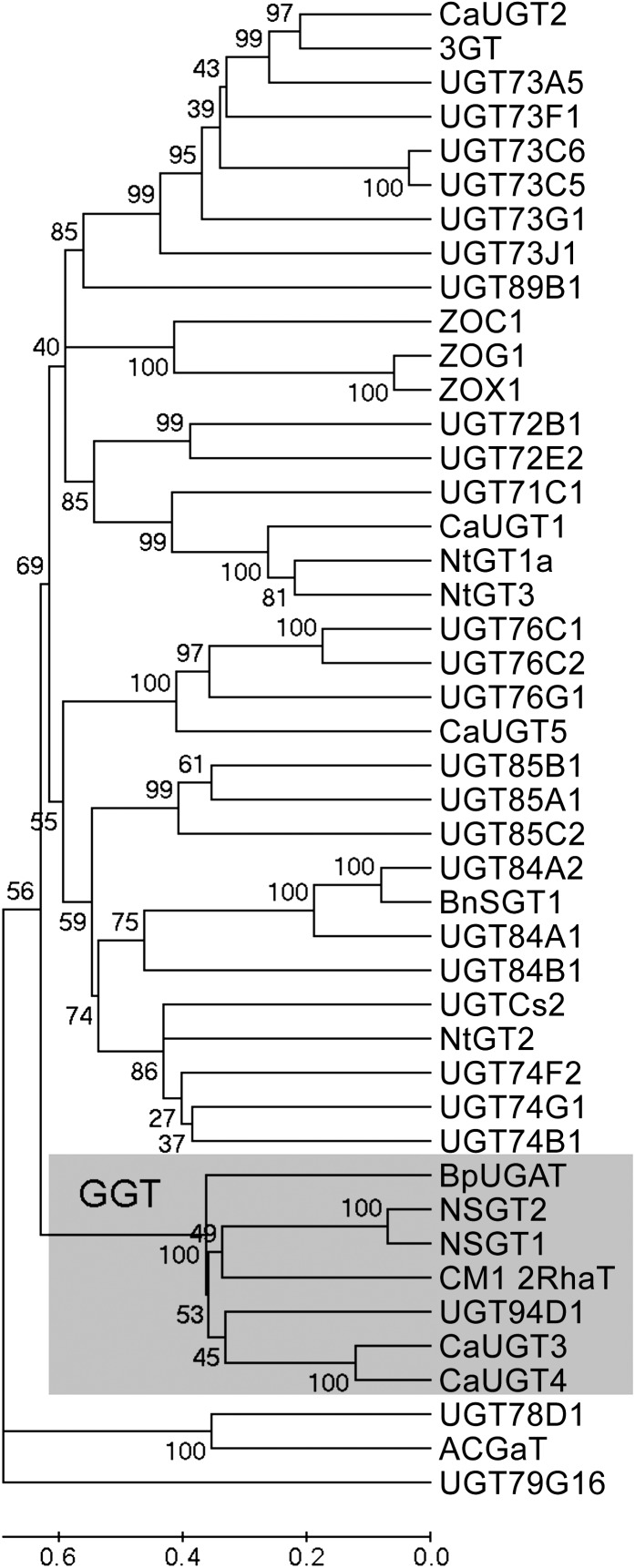
A comparative sequence analysis of the nontruncated NSGT1 and NSGT2 proteins of the ‘non-smoky’ line C085 with plant GTs for which a function has been established (see Supplemental Table 1 online) showed that they belonged to the glycoside glycosyltransferase (GGT) clade (Figure 3). GTs of this clade have revealed a branch forming activity; they recognize glycosides as acceptor molecules and add a sugar to an already existing moiety of a glycoside (Bowles et al., 2006).
Figure 3.
A Phylogenetic Tree of NSGT Proteins and 42 Plant GTs with Known Function (Published by Bowles et al. [2005] and Masada et al. [2009]; See Supplemental Table 1 Online).
The evolutionary history was inferred using the UPGMA method. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) is shown next to each branch. The evolutionary distances were computed using the Poisson correction method and are in the units of the number of amino acid substitutions per site. All positions containing gaps and missing data were eliminated. There were a total of 296 positions in the final data set. Evolutionary analyses were conducted in MEGA5 (Tamura et al., 2011). The alignment data used to build the phylogenetic tree are presented in Supplemental Data Set 1 online.
In summary, two GT genes, NSGT1 and NSGT2, were obtained from a ‘non-smoky’ tomato genome. Structural alterations in both genes in a ‘smoky’ tomato background lead to predicted truncated proteins lacking important functional domains that likely render them inactive.
Transcription Analysis of NSGT1 and NSGT2 during Fruit Ripening
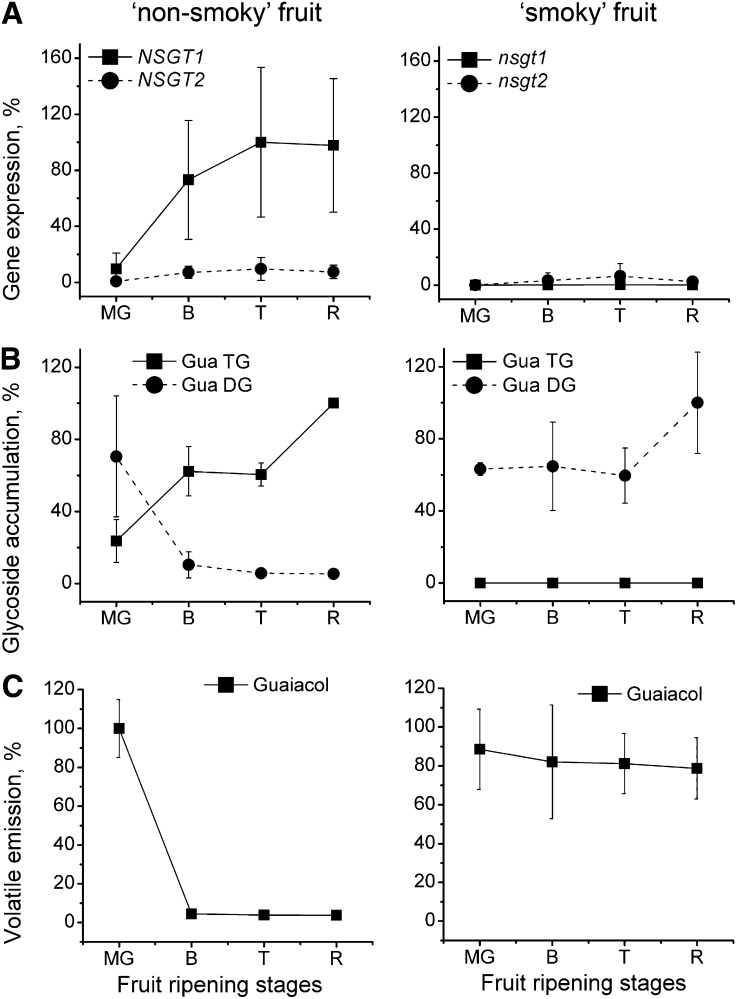
The expression of NSGT1 and NSGT2 in tomato fruit was analyzed using quantitative real-time PCR (qRT-PCR). Six ‘smoky’ and six ‘non-smoky’ cultivars were selected from each of the 25-cultivar pools previously used in the DGE experiment. Expression of NSGT1 and NSGT2 was determined in fruits of these 12 cultivars, at four different stages of ripening: mature green, breaker, turning, and ripe (see Methods for the ripening stage term definition). A strong induction of expression (up to 15-fold) was only observed for NSGT1 in ripening ‘non-smoky’ fruits (Figure 4A). This induction corresponded well to the change in glycosylation pattern of these fruits: The cleavable PhP-V diglycosides were converted into noncleavable PhP-V triglycosides during ripening (Figure 4B). This process was also accompanied by the arrest of the release of the corresponding volatiles from disrupted ‘non-smoky’ fruit material (Figure 4C). The expression of nsgt1 in ‘smoky’ fruit samples remained very low (threshold cycle [Ct] = 30.0 or higher) at all ripening stages, and no metabolic changes were observed (Figures 3B and 3C). NSGT2 also showed very low expression in all fruit samples analyzed (Ct = 30.0 or higher).
Figure 4.
Expression Analysis of NSGT Genes and Metabolite Levels in Ripening Fruits of Six ‘Non-Smoky’ and Six ‘Smoky’ Tomato Cultivars.
Relative expression of NSGT genes in ‘non-smoky’ and ‘smoky’ fruits (A). The metabolic levels exemplified by guaiacol diglycoside (Gua DG) and guaiacol triglycoside (Gua TG) (B), and guaiacol release from disrupted fruits (C). MeSA, eugenol, and their corresponding glycoconjugated moieties behave in a similar way to guaiacol/guaiacol glycosides (Tikunov et al., 2010). Fruit ripening stages are as follows: MG, mature green fruit; B, breaker fruit; T, turning fruit; and R, ripe fruit. Each data point represents the average gene expression or the average metabolite abundance in six tomato fruit samples. The maximum average expression is set at 100%. The error bars represent standard deviations of gene expression or metabolite abundance levels (%) measured in six tomato fruit samples. Graphs on the left, ‘non-smoky’ cultivars; graphs on the right, ‘smoky’ cultivars.
Thus, only NSGT1 from a ‘non-smoky’ tomato variety was expressed at a considerable level, and the increase of its expression in ripening ‘non-smoky’ fruits was correlated with the PhP-V glycoside conversion and the release of the corresponding volatiles from disrupted fruit tissue.
NSGT1 Converts PhP-V Diglycosides of ‘Smoky’ Tomato Fruit into Noncleavable Triglycosides of ‘Non-Smoky’ Fruit
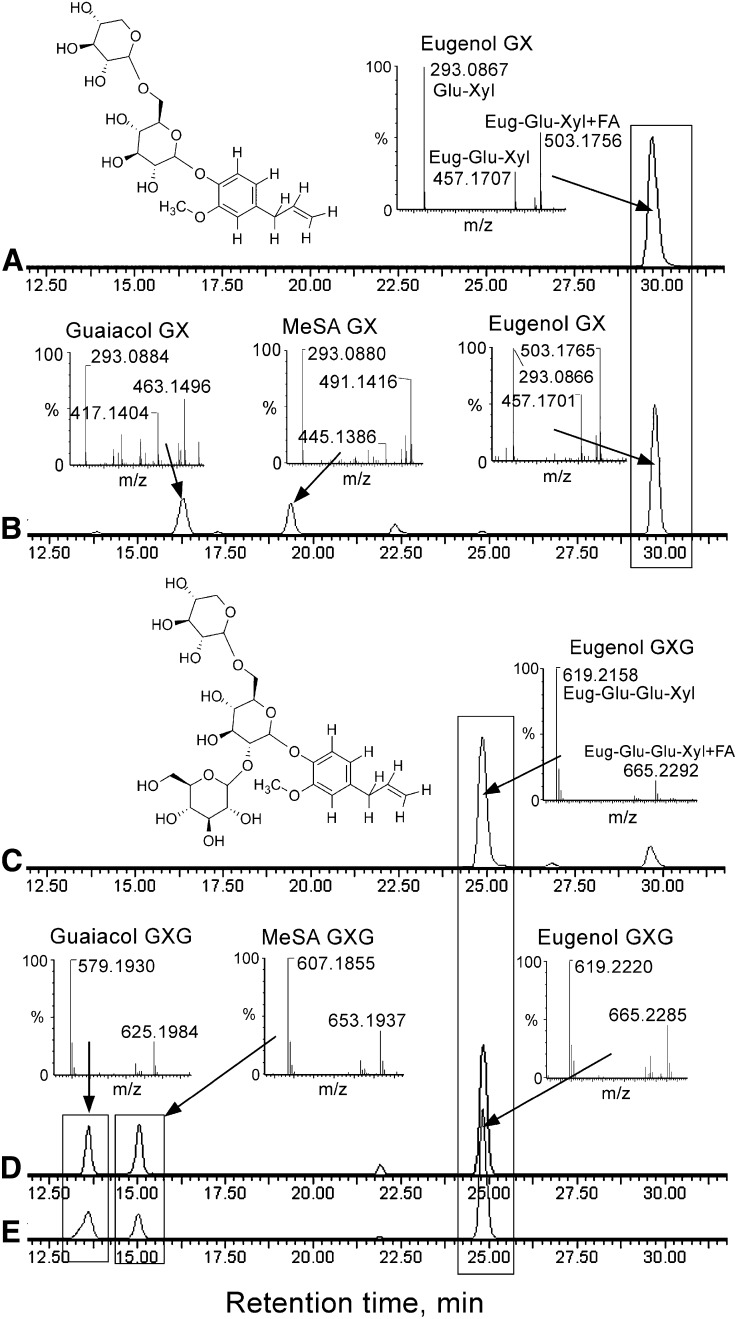
In order to determine whether the NSGT1 enzyme might indeed be responsible for the diglycoside-to-triglycoside conversion, the NSGT1 coding sequence cloned from ‘non-smoky’ tomato line C085 was expressed in Escherichia coli, and the product was tested with PhP-V diglycoside substrates. Due to the large complexity of tomato fruit metabolic extracts, we were unable to derive pure fractions of the PhP-V diglycosides from ‘smoky’ tomato fruit. Instead, we produced a pure PhP-V diglycoside substrate. Based on the study of Ono et al. (2010), who elucidated the structure of several tomato fruit glycosides, we hypothesized that PhP-V triglycosides and their precursor diglycosides could consist of 2-O-β-d-glucopyranosyl-(1→2)-[O-β-d-xylopyranosyl-(1→6)]-O-β-d-glucopyranoside (GXG) and O-β-d-xylopyranosyl-(1→6)]-O-β-d-glucopyranoside (GX) moieties, respectively. Eugenol O-β-d-xylopyranosyl-(1→6)]-O-β-d-glucopyranoside (eugenol-GX) was produced enzymatically, and its structure was confirmed using NMR spectroscopy (Figure 5A; see Supplemental Methods 1 online). A comparative liquid chromatography–mass spectrometry (LC-MS) analysis showed that the retention time and the mass spectrum of produced eugenol-GX were identical to those of eugenol diglycoside present in ‘smoky’ tomato fruit (Figure 5B): Both eluted at 29.75 min and the mass spectra consisted of a parent molecule ion [M – H]– with mass/charge (m/z) 457 [457 = 164(eugenol) + 162(Glc – water) + 132(Xyl – water) – H]–, its formic acid adduct m/z 503 [503 = 457 + 46(formic acid)] and a sugar moiety fragment with m/z 293 [162(Glc – water) + 132(Xyl – water)]–. Pure MeSA-GX and guaiacol-GX substrates could not be produced due to specificity of the enzymes used (see Supplemental Methods 1 online). However, the fragmentation patterns of diglycosides of MeSA and guaiacol present in ‘smoky’ fruits were identical to that of eugenol-GX (Figures 5A and 5B). Both contained the characteristic largest peak of the sugar moiety fragment with m/z 293, which, being conjugated with the corresponding volatile aglycone molecules of guaiacol ([M] = 124) and MeSA ([M] = 152), gave parent ions with m/z 417 and m/z 445, respectively. This suggested that similar to eugenol, MeSA and guaiacol were bound to the same GX moiety in ‘smoky’ tomato fruit.
Figure 5.
LC-MS Analysis of Glycosylation Products of NSGT1 Expressed in E. coli.
Each chromatogram in (A) to (E) is represented in selected ion mode and displays intensity signals of specific masses (m/z) of diglycosides of ‘smoky’ fruits, m/z 293.0878 ± 20 ppm (a common mass fragment of all PhP-V diglycosides representing GX sugar moiety); and triglycosides of ‘non-smoky’ fruits, 579.1930 ± 20 ppm (guaiacol-GXG), 607.1879 ± 20 ppm (MeSA-GXG), and 619.2243 ± 20 ppm (eugenol-GXG). All chromatograms were set at the same intensity scale.
(A) Enzymatically synthesized eugenol-GX, compound (1), is the structure of eugenol-GX resolved using NMR.
(B) A crude glycoside extract from ‘smoky’ tomato containing peaks of guaiacol-GX, MeSA-GX, and eugenol-GX: Based on retention time and mass spectra, eugenol-GX of ‘smoky’ fruit corresponds to enzymatically produced eugenol-GX in chromatogram (A).
(C) Chromatogram, mass spectrum, and NMR-derived structure of eugenol-GXG, compound (2), produced by NSGT1-mediated glycosylation of eugenol-GX [chromatogram in (A), compound (1)].
(D) A crude glycoside extract from ‘non-smoky’ fruits displaying peaks of PhP-V triglycosides: guaiacol-GXG, MeSA-GXG, and eugenol-GXG.
(E) A crude glycoside extract from ‘smoky’ tomato incubated with the protein extract from E. coli transformed with NSGT1 and a donor sugar UDP-Glc. Peaks of eugenol-GX, guaiacol-GX, and MeSA-GX (shown in chromatogram [B]) can be no longer observed in this chromatogram as a result of their conversion into the corresponding eugenol-GXG, guaiacol-GXG, and MeSA-GXG. All three triglycosides have identical retention time and mass spectra to PhP-V triglycosides present in ‘non-smoky’ tomato fruit (shown in chromatogram [D]), and eugenol-GXG is identical to eugenol-GXG confirmed by NMR [compound (2) in chromatogram (C)].
To test whether NSGT1 could convert PhP-V GX into the corresponding GXG, produced eugenol-GX was further used as an acceptor substrate and UDP-Glc was used as a sugar donor. Although we were unable to detect any activity of the NSGT1 enzyme purified using Ni-affinity chromatography, a crude enzyme extract of E. coli transformed with NSGT1 did convert eugenol-GX into a new compound corresponding to eugenol GXG (Figure 5C), since its parent ion mass [M – H]– m/z 619 corresponded to the eugenol-GX acceptor molecule [M – H]– m/z 457 conjugated with an additional dehydrated Glc moiety [M] m/z 162. Further structural analysis of this compound using NMR spectroscopy showed that it was indeed eugenol 2-O-β-d-glucopyranosyl-(1→2)-[O-β-d-xylopyranosyl-(1→6)]-O-β-d-glucopyranoside (eugenol-GXG) (Figure 5C; see Supplemental Methods 1 online). LC-MS characteristics of this eugenol-GXG matched those of the eugenol triglycoside present in ripe ‘non-smoky’ tomato fruits (Figure 5D). Since pure MeSA-GX and guaiacol-GX substrates could not be produced enzymatically, a crude glycoside extract from ‘smoky’ fruit containing these two compounds as well as eugenol-GX was used as an acceptor substrate. All three PhP-V GX were converted by the enzyme extract from NSGT1-transformed E. coli into their corresponding GXG forms (Figure 5E). LC-MS characteristics of these compounds were in agreement with those of PhP-V GXG present in ‘non-smoky’ tomato fruit (Figure 5D).
A protein extract from E. coli transformed with an empty vector was used as a negative control in this and in all further in vitro experiments. This extract did not show any glycosylation activity on the PhP-V acceptor glycoside substrates used.
Pure aglycone MeSA, guaiacol, and eugenol were also tested as acceptor substrates for NSGT1 to test its suggested phylogenetic classification in the clade of branch-forming GTs, which prefer glycoside sugar moieties as acceptor substrates rather than pure aglycone molecules. No activity on glycosylating PhP-V aglycones was detected.
In conclusion, these results support the hypothesis that NSGT1 is capable of converting PhP-V diglycosides typical of ‘smoky’ tomato fruit, GX, into the PhP-V triglycosides typical of ‘non-smoky’ fruit, GXG.
Metabolic and Gene Expression Analysis of NSGT1 Transgenic Fruits
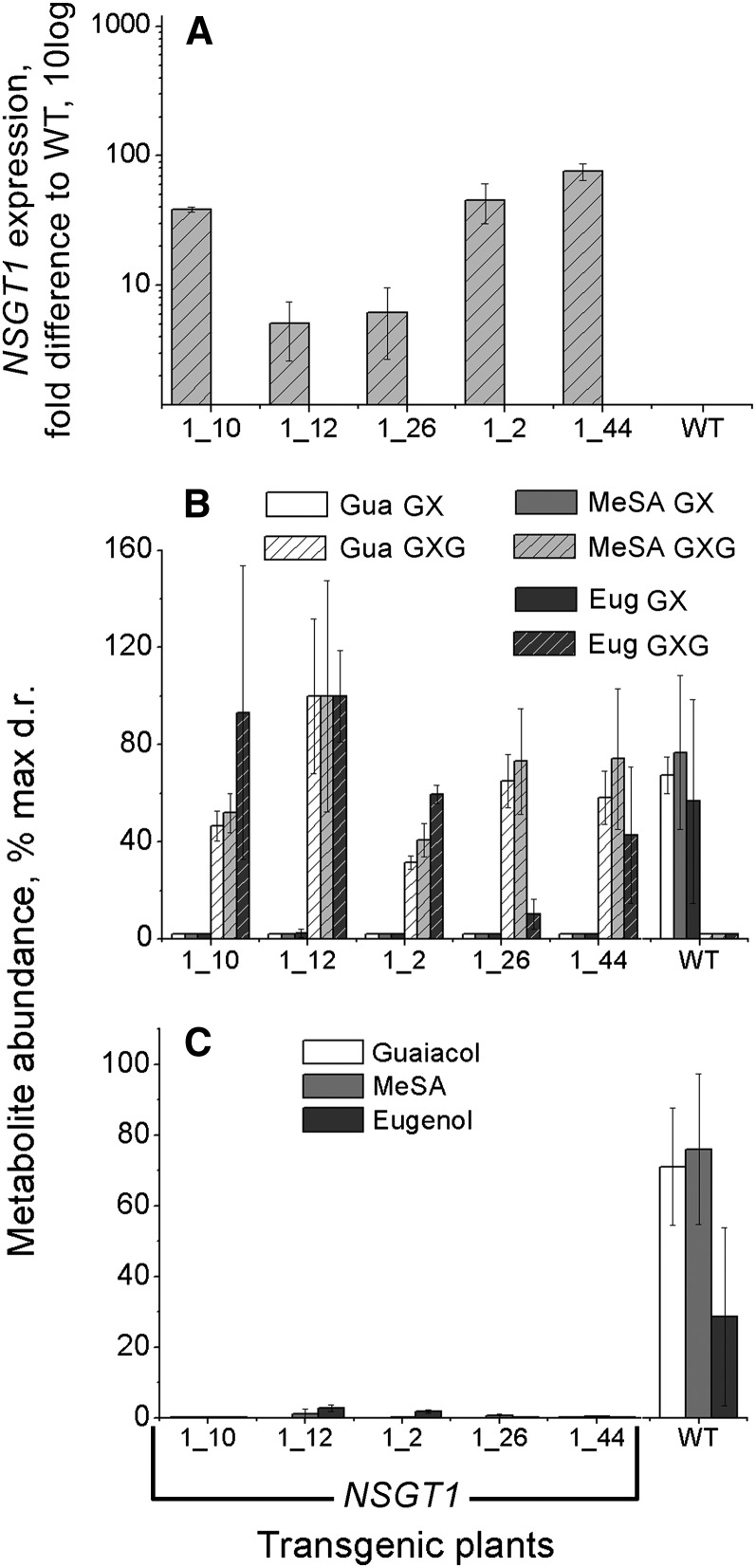
To study the function of NSGT1 in planta, the coding sequence of this gene obtained from ‘non-smoky’ line C085 was constitutively expressed in a ‘smoky’ tomato background, cv Moneymaker (Figure 6; see Supplemental Figure 4 online), under control of the cauliflower mosaic virus double 35S promoter. Ripe fruits of wild-type and transgenic plants were analyzed for NSGT1 expression using qRT-PCR. In parallel, PhP-V glycoside composition of intact fruits was determined using LC-MS, and PhP-V release from disrupted fruits was determined using headspace gas chromatography–mass spectrometry (GC-MS). NSGT1 appeared to be highly overexpressed in fruits of the transgenic lines compared with the wild-type fruits, in which the endogenous mutant gene was barely expressed (an average Ct = 29.6) (Figure 6A). In transgenic fruits overexpressing NSGT1, the levels of MeSA-GX, guaiacol-GX, and eugenol-GX were reduced to nondetectable levels (Figure 6B). Instead, GXG forms of these volatiles were detected which were not present in the wild-type Moneymaker fruits. The release of guaiacol, MeSA, and eugenol from disrupted fruits of the transgenic lines was strongly decreased compared with that detected in the wild-type fruits and in most cases dropped to nondetectable levels (Figure 6C).
Figure 6.
Accumulation of PhP-V Glycosides in, and Release of Corresponding Volatiles from, NSGT1 Transgenic Fruits (Ripe Stage).
(A) NSGT1 expression difference between wild-type Moneymaker fruits (WT) and NSGT1-overexpressing fruits.
(B) Accumulation of PhP-V diglycosides (GX) and PhP-V triglycosides (GXG) in wild-type and transgenic fruits.
(C) Release of PhP-V.
Bars in plots in (B) and (C) represent metabolite abundance expressed as percentage to the highest accumulation (glycosides) or release (volatiles) measured as LC-MS or GC-MS detector response, respectively. Nondetectable glycoside levels in graph (B) were set at 2.0 just to indicate a bar position. Each data bar and corresponding error bar represent average or standard deviation of three individual fruit samples, respectively.
Thus, the nonfunctional nsgt1 gene of a ‘smoky’ tomato was complemented by constitutive overexpression of NSGT1 cloned from a ‘non-smoky’ tomato. This resulted in conversion of the cleavable GX forms of guaiacol, MeSA, and eugenol into noncleavable GXG forms, thereby preventing their damage-induced deglycosylation and release from tomato fruit.
Sensory Analysis of NSGT1 Transgenic Fruits
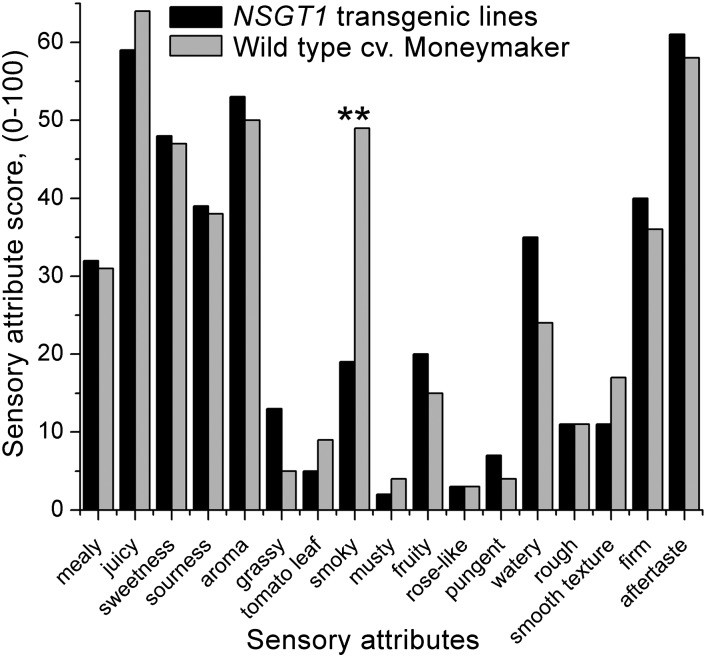
To study whether transgenic overexpression of NSGT1 also affected the aroma of tomato fruit, a sensory analysis was performed. Fully ripe fruits of wild-type cv Moneymaker and of NSGT1 overexpressing transgenic lines were harvested and subjected to a blind evaluation for taste and flavor by a sensory panel consisting of 13 trained judges. Seventeen different flavor and texture attributes were evaluated. NSGT1-overexpressing fruits showed a significant 2.8-fold (Student’s t test, P < 0.01) reduction in “smoky” aroma compared with the wild-type Moneymaker fruits (Figure 7). No other sensory attributes were significantly affected in NSGT1-overexpressing fruits compared with the wild-type fruits.
Figure 7.
Sensory Analysis of NSGT1 Transgenic Fruits.
Each bar represents the score (average of 13 judges) for a particular sensory attribute in NSGT1-overexpressing transgenic lines and in the wild-type cv Moneymaker. NSGT1-overexpressing fruits showed a significant decrease in ‘smoky’ aroma (Student’s t test, **P < 0.01) compared with the cv Moneymaker wild-type fruits. Other attributes did not reveal significant differences between the transgenic and wild-type fruits.
Segregation Analysis of the Smoky Aroma Trait Using NSGT1-Based Genetic Markers
Two PCR-based markers were developed based on polymorphism found between NSGT1 and nsgt1 sequences. These markers enable codominant scoring of tomato plants for “smoky” and “non-smoky” backgrounds. Marker performance was validated in a segregating F6 recombinant inbred line (RIL) population derived from breeding lines R104 and C085 as parents. All RILs were genotyped using the two NSGT1 markers and their ripe fruits were analyzed for volatile release. The marker scores corresponded perfectly with PhP-V release (see Supplemental Figure 5 online). In addition, we genotyped a diverse set of commercial tomato cultivars (F1 hybrids), which had previously been profiled for taste and aroma and for volatile and nonvolatile metabolites (Tikunov et al., 2005, 2010; Menéndez et al., 2012). The two markers revealed 100% cosegregation with the different types of PhP-V glycoconjugates and the resulting difference in the release of the corresponding volatiles (see Supplemental Figure 6 online). The ‘smoky’ aroma intensity, as measured in ripe fruits of the F1 cultivars by a sensory panel, also revealed a high correlation with the marker score (see Supplemental Figure 6 online). Tomato varieties Moneymaker and M82 are often used in tomato research programs to create segregating populations or as wild-type plants for reverse genetics experiments. According to the metabolic profiling data, Moneymaker produces “smoky” fruits and M82 produces “non-smoky” fruits. NSGT1 marker data perfectly correlated with metabolic data in these two reference cultivars (see Supplemental Figure 4 online).
DISCUSSION
NSGT1 Plays a Major Role in the Determination of Smoky Aroma of Tomato Fruit
Using a combinatorial approach, including (1) complementary metabolomics methods (Tikunov et al., 2010), (2) the available quantitative trait locus mapping information (Chaïb et al., 2006; Zanor et al., 2009), (3) tomato genome sequence data (Tomato Genome Consortium, 2012), and (4) untargeted next-generation sequencing–based transcriptomics, we identified the NSGT1 gene. NSGT1 is a key factor determining PhP-V release in tomato fruit and hence is a key determinant of the smoky aroma of commercial tomato varieties. It functions as an off switch for the release of PhP-V during ripening through structural modification of the corresponding PhP-V glycoconjugate precursors in ‘non-smoky’ genotypes. A loss-of-function mutation of NSGT1 in ‘smoky’ tomato fruit leads to the accumulation of PhP-V GXs (also called β-primeverosides). Subsequently, fruit damage–induced hydrolysis of these β-primeverosides releases the corresponding volatiles. β-Primeverosides have already been shown to be major precursors of aroma formed in tea leaves during processing (Mizutani et al., 2002). In ‘non-smoky’ fruits, which express a functional NSGT1, these β-primeverosides are further converted into the more complex and hydrolysis-resistant GXG. Consequently, PhP-V can no longer be released. The transgenic overexpression of the NSGT1 coding sequence in a ‘smoky’ tomato background led to a complete conversion of cleavable PhP-V β-primeverosides into noncleavable GXGs, a reduction in damage-induced PhP-V release, and a significant decrease in ‘smoky’ aroma intensity of transgenic fruits.
There are a few examples of human sensory evaluation of fresh, unprocessed transgenic plant material, including transgenic calcium biofortified lettuce (Lactuca sativa; Park et al., 2009), LoxC antisense transgenic tomato (Tieman et al., 2012), and geraniol synthase–overexpressing tomato (Davidovich-Rikanati et al., 2007; Lewinsohn et al., 2010). Similar to these studies, our sensory experiment, in which transgenic tomato fruits were evaluated by trained panelists, provided an invaluable link between the discovery of the molecular function of NSGT1 and its impact on tomato aroma, as perceived by humans. The sensory evaluation showed that NSGT1 specifically affected “smoky” aroma and did not affect other important sensory characteristics in transgenic tomato fruit. The NSGT1-driven glycosylation affected the release of all three PhP volatiles: guaiacol, MeSA, and eugenol. Guaiacol is probably the most important candidate to contribute to “smoky” aroma, since the odor of pure guaiacol is described as “smoky,” and it has been shown to contribute to smoky aroma in other food products (Robin et al., 1999; Jensen and Whitfield, 2003; Lee and Noble, 2003; Morales et al., 2005; Akakabe et al., 2006; Kennison et al., 2007; Arumugam et al., 2010).
Phylogenetic analysis of NSGT1 showed that it belongs to a specific clade of plant GGTs or branch-forming glycosyltransferases. These GTs are involved in the further elongation of the glycosidic moiety of glycosides but are not active on the metabolite aglycones (Figure 3). The lack of activity of NSGT1 on the volatile PhP aglycones supports the protein sequence based classification of NSGT1. Several other GGTs with an established function have been described: A GGT of pummelo (Citrus maxima), Cm1,2RhaT, converts flavanone-7-glucosides into flavanone-7-neohesperidoside, which directly contributes to the characteristic bitter taste of grapefruit and pummelo (Frydman et al., 2004). Other GGTs have been shown to further glycosylate various flavonoid glycosides (including anthocyanins) in flowers of Madagascar periwinkle (Catharanthus roseus) (Masada et al., 2009), daisy (Bellis perennis) (Sawada et al., 2005), and Japanese morning glory (Ipomoea nil) (Morita et al., 2005), where they affect flower color.
Organization and Evolution of NSGT1 in the Tomato Genome
The NSGT1 gene sequence was not represented in any of the publicly available tomato unigene or EST databases (Sol Genomics Network Unigene or Tomato Gene Index v.13 [The Gene Index Project]). This is likely due to the fact that most tomato varieties used for EST and genome sequencing (e.g., Heinz, Moneymaker, and Microtom) are of the ‘smoky’ type and hence do not express a functional NSGT1. Interestingly, NSGT1 coding and protein sequences had also not been predicted by the International Tomato Annotation Group (at the moment of article preparation) due to the fact that a large part of the gene sequence was missing in a large ∼8-kb fragment of unknown genomic sequence. Further attempts to isolate the complete sequence of the transcribed region of the gene revealed not just one, but two DNA sequences encoding two very similar proteins, NSGT1 and NSGT2, which were located within this fragment (Figure 2). The high similarity (90%) and tandem colocalization of NSGT1 and NSGT2 sequences suggests that these genes may have originated from a common ancestral gene by tandem duplication. The draft genome of the proposed progenitor of domesticated tomato, Solanum pimpinellifolium (accession number CGN G1.1589, Sol Genomics Network), also contains full-length NSGT1 and NSGT2 gene sequences, which were almost identical to those we cloned from the ‘non-smoky’ background. At the protein level, NSGT1 from S. pimpinellifolium had just three amino acid residue substitutions (see Supplemental Figure 7 online). It is unclear whether the further mutations resulting in truncation of the NSGT genes present in ‘smoky’ commercial tomato varieties occurred in a wild progenitor or in the domesticated tomato. We observed a clear overrepresentation of the ‘non-smoky’ fruit metabolic phenotype and of the non-smoky NSGT1 genotype in a set of 54 S. pimpinellifolium accessions (see Supplemental Figure 8 online), while in the commercial tomato germplasm described previously (Tikunov et al., 2005, 2010), ‘smoky’ and ‘non-smoky’ genotypes were equally represented (see Supplemental Figure 6 online). This might indicate that in the natural habitats of the wild accessions, there may have been a preferential selection for ‘non-smoky’ fruit phenotype. We can only speculate which natural factor could mediate the selection for ‘non-smokiness’ of tomato fruit and what physiological or ecological role this could play. For example, the volatile composition of fruits may attract or repel seed dispersing animals (Borges et al., 2008), and in this respect, one can hypothesize that production of “smoky” volatiles in immature fruits, when seeds are unripe, may repel seed dispersers and prevent the fruits from being prematurely eaten. Arresting release of these volatiles in ripe fruits may then, in turn, stimulate their consumption and hence facilitate seed dispersal.
The Use of Metabolic Logistics for Aroma Determination
The biosynthesis of phenolic volatiles has been explored thoroughly over the past decade (Dudareva and Pichersky, 2000; Boatright et al., 2004; Dudareva et al., 2004; Gang, 2005; Koeduka et al., 2006, 2009; Tieman et al., 2010; Mageroy et al., 2012). This study illustrates the importance of the metabolic logistical processes, conjugation and deconjugation, in determining how and when these volatiles can be accumulated in and released from tomato fruit. Depending on their glycoconjugate structure, PhP volatiles can either contribute to tomato fruit aroma upon specific circumstances (e.g., when they are being consumed by the human), or these volatiles and the corresponding aroma can be sequestered. Besides the PhP volatiles, the destiny of which is described in this study, many other volatiles are accumulated predominantly as glycoconjugates in tomato fruit, thus representing a considerable locked-in flavor resource (Buttery et al., 1990a; Baldwin et al., 1998; Krumbein and Auerswald, 1998; Krumbein et al., 2004; Ortiz-Serrano and Gil, 2007). Therefore, knowledge of the molecular factors regulating these logistical processes will pave the way to a more efficient use of plant metabolic resources.
METHODS
Plant Material
The set of commercial tomato (Solanum lycopersicum) cultivars (F1 hybrids) referred to in this article was provided by four different breeding companies and has been characterized biochemically in Tikunov et al. (2005, 2010). Two elite homozygous breeding lines, R104 (round type tomato, ‘smoky’ fruit) and C085 (cherry type tomato, ‘non-smoky’ fruit), were used to clone the NSGT genes. When tomato fruit ripening stages were studied, fruits were harvested according to a standardized fruit color chart consisting of 12 stages provided by The Greenery (Valstar Holland): stage 1, mature green, fruit surface is completely green, and the shade of green may vary from light to dark; stage 3, breaker, there is a definite break in color from green to tannish-yellow, pink, or red on not more than 10% of the surface; stage 5, turning, 10 to 30% of the surface is not green, in the aggregate, shows a definite change from green to tannish-yellow, pink, red, or a combination thereof; stage 9-10, ripe, more than 90% of the surface is not green, in the aggregate, shows red color. The cv Moneymaker plants were used as a wild type for transformation with the NSGT genes.
Material Availability
Seeds of NSGT1 transgenic plants and seeds of F6 RILs obtained by crossing the ‘smoky’ R104 and ‘non-smoky’ C085 parental lines (see Supplemental Figure 5 online) are available upon request. Seeds of tomato cv Moneymaker (smoky) and tomato cv M82 (non-smoky) can be obtained from genetic stock centers.
Sensory Analyses
The taste trials with genetically engineered tomatoes were performed by trained tasting panels after approval by the Dutch Ministry of Economic Affairs and the Plant Science Group of Wageningen University. Thirteen trained experts volunteered to participate in the transgenic tomato fruit taste panel. They were fully informed about the nature of the transgenic fruits they were going to taste and signed a document relating to this and to the conditions of the tasting experiment with transgenic tomato fruits. Tomato fruits were cut into segments and seeds were removed, collected, and autoclaved to prevent spreading of transgenic materials. The experts were allowed to spit out the fruit material (seedless fruit pericarp tissue) after tasting.
The sensory panel experiment was run equivalent to the Quantitative Descriptive Analysis methodology (Stone and Sidel, 2004) but following a more extensive training stage. The tomatoes were presented to the panelists according to a Williams Latin Square design (MacFie, 1990).
DGE Profiling
Total RNA was isolated from whole tomato fruit samples according to Chang et al. (1993). RNA samples were treated with DNase to eliminate the presence of genomic DNA and purified with an RNAeasy spin column (Qiagen). The DGE analysis was performed using an Illumina DGE platform by ServiceXS.
RNA Isolation and Gene Expression Analysis Using qRT-PCR
For the expression analysis of NSGT genes during fruit ripening, six ‘smoky’ and six ‘non-smoky’ tomato cultivars (all F1 hybrids) were used as biological replicates. Fruit material (whole fruit) from nine plants was pooled to make a representative sample for each cultivar at each of the four ripening stages: mature green, breaker, turning, and ripe.
For the expression analysis of NSGT1 in fruits of transgenic plants, three fruits were analyzed independently for each transgenic plant and the wild-type control (cv Moneymaker).
All fruit materials were harvested, immediately frozen in liquid nitrogen, ground in liquid nitrogen, and stored at −80°C before RNA extraction. RNA was extracted essentially following the protocol for RNA extraction from tissues recalcitrant to extraction in guanidine (Bugos et al., 1995). Digestion with Ambion DNase (Invitrogen) was performed according to the manufacturer’s instructions. RNA concentration was measured in a Nanodrop ND-1000 spectrophotometer, and RNA integrity was checked by means of a 2100 BioAnalyzer (Agilent Technologies). cDNA was synthesized from 1 µg RNA using Superscript II reverse transcriptase (Invitrogen) following the manufacturer’s instructions. Gene expression analysis was performed using qRT-PCR using SYBR Green in an ABI 7500 Fast Real-Time PCR system (Applied Biosystems). Ubiquitin was used as the reference gene. Primers used for amplification were designed using Primer Express version 2.0 software (Applied Biosystems) and are listed in Supplemental Table 2 online. Triplicate analyses were performed for each biological replicate. Standard curves were constructed both for target and reference genes. The amount of RNA in the samples was calculated using the Ct value and the corresponding standard curve and further normalized to a calibrator sample. Expression data for NSGT genes are expressed as levels relative to the wild type.
Phylogenetic Analysis of GTs
An alignment (see Supplemental Data Set 1 online) and a comparative phylogenetic analysis of plant GT protein sequences was conducted using the ClustalW program (Larkin et al., 2007) and the UPGMA method (Sneath and Sokal, 1973), respectively, implemented in MEGA5 software (Tamura et al., 2011). Statistical significance of the phylogenetic trees was calculated using the bootstrap method (1000 replicates) (Felsenstein, 1985). The evolutionary distances were computed using the Poisson correction method (Zuckerkandl and Pauling, 1965). All positions containing gaps and missing data were eliminated.
Preparation of Tomato Fruit Extracts Enriched with Crude Glycosides
Crude metabolic extracts enriched with glycosides were prepared from tomato fruit material using ion exchange preparative chromatography on Amberlite XAD-2 resin, as described by Tikunov et al. (2010). The presence of diglycosides of guaiacol, MeSA, and eugenol in the eluate was confirmed using accurate mass LC-MS analysis.
Metabolite Analyses
Solid-Phase Microextraction Gas-Chromatography Mass Spectrometry
The profiling of volatile metabolites was performed using the headspace solid-phase microextraction gas-chromatography mass spectrometry (SPME-GC-MS) method described by Tikunov et al. (2005). Frozen fruit powder (1 g fresh weight) was weighed into a 5-mL screw-cap vial, closed, and incubated for 10 min at 30°C. An aqueous EDTA-sodium hydroxide (NaOH) solution was prepared by adjusting 100 mM EDTA to pH of 7.5 with NaOH. Then, 1 mL of the EDTA-NaOH solution was added to the sample to give a final EDTA concentration of 50 mM. Solid CaCl2 was then immediately added to give a final concentration of 5 M. The closed vials were then sonicated for 5 min. A 1-mL aliquot of the pulp was transferred into a 10-mL crimp cap vial (Waters), capped, and used for SPME GC-MS analysis. Volatiles were automatically extracted from the vial headspace and injected into the GC-MS via a Combi PAL autosampler (CTC Analytics). Headspace volatiles were extracted by exposing a 65-μm polydimethylsiloxane-divenylbenzene SPME fiber (Supelco) to the vial headspace for 20 min under continuous agitation and heating at 50°C. The fiber was desorbed in the GC-MS injection port and compounds were separated on an HP-5 (50 m × 0.32 mm × 1.05 μm) column with helium as carrier gas (37 kPa). Mass spectra in the 35 to 400 m/z range were recorded by an MD800 electron impact MS (Fisons Instruments) at a scanning speed of 2.8 scans/s and an ionization energy of 70 eV. The chromatography and spectral data were evaluated using Xcalibur software (Thermo Scientific).
PhP volatiles were identified by matching compound mass spectra to the NIST mass spectral library and using pure authentic chemical standards of guaiacol, MeSA, and eugenol purchased from Sigma-Aldrich.
Liquid Chromatography-Quadrupole Time-of-Flight Mass Spectrometry
The extraction and liquid chromatography-quadrupole time- of-flight mass-spectrometry (LC-QTOF-MS) analysis of semipolar compounds was performed according to the protocol described (Moco et al., 2006); 0.5 g frozen tomato fruit powder (fresh weight) was extracted with a 1.5-mL formic acid:methanol (1:1000, v/v) solution. The extracts were sonicated for 15 min and filtered through a 0.2-μm inorganic membrane filter (Anotop 10; Whatman).The LC-QTOF-MS platform consisted of a Waters Alliance 2795 HT HPLC system equipped with a Luna C18(2) analytical column (2.0 × 150 mm, 100 Å, particle size 3 μm; Phenomenex), held at 40°C, connected to an Ultima V4.00.00 QTOF mass spectrometer (Waters). Formic acid:ultrapure water (1:1000, v/v; eluent A) and formic acid:acetonitrile (1:1000, v/v; eluent B) were pumped into the HPLC system at 190 μL min–1 and the gradient linearly increased from 5 to 35% eluent B over a 45-min period, followed by 15 min of washing and equilibration of the column. Ionization was performed using an electrospray ionization source, and masses were detected in negative mode. A collision energy of 10 eV was used for full-scan LC-MS in the range of m/z 100 to 1500. Leu enkephalin, [M – H]– = 554.2620 was used for online mass calibration (lock mass).
Isolation and Cloning of NSGT Genes from Tomato
For bacterial overexpression, the coding sequence of NSGT1 was amplified from tomato fruit cDNA (turning stage) of tomato elite line C085 (non-smoky fruits) using high fidelity Phusion polymerase (Finnzymes). The primers used for cloning and their Ta°C are listed in Supplemental Table 3 online. PCR was performed using one cycle of 98°C (45 s) followed by 30 cycles of 98°C (10 s)/Ta°C (20 s)/72°C (2 min). BamHI-PstI restriction sites were used to clone the PCR product in frame with the N-terminal His6-Tag of pACYCDuet-1 expression vector (Novagen). Four individual clones were fully sequenced and showed no sequence variation.
Glycosylation of PhP-V Glycosides in Vitro Using Protein Extract from Escherichia coli Transformed with NSGT1
The pACYCDUET-NSGT1 plasmid was transformed into E. coli BL21(DE3) cells and used for production of a recombinant enzyme. A starter culture was grown overnight at 37°C in 10 mL of Luria-Bertani medium containing 50 µg/mL chloroamphenicol and 1% Glc. The starter culture was diluted 100 times in 25 mL 2× YT medium supplemented with 50 µg/mL chloroamphenicol and incubated for 2 h at 37°C to an OD600 of 0.6. Then, protein expression was induced by adding 1 mM isopropyl-1-thio-β-d-galactopyranoside, and the culture was incubated overnight at 18°C. Cells were harvested by centrifugation for 15 min at 6500g, and the pellet was frozen at −20°C. E. coli BL21(DE3) cells harboring an empty pACYCDUET-1 plasmid were used as negative control.
Frozen cells were re-suspended in 0.1 M Tris-HCl buffer, pH 7.5, and disrupted by sonication on ice. The cell lysate was clarified twice by centrifugation for 5 min (16,000g) at 4°C. Further purification of the enzymes using nickel-nitrilotriacetic acid chromatography was initially included but was observed to destroy the activity of the proteins and therefore was omitted from the procedure. The reaction mixture (1000 µL) contained 1.4 μM eugenol-GX (produced as described in Supplemental Methods 1 online), 10 mM UDP-Glc (Sigma-Aldrich), 100 μL clarified enzyme extract in 0.1 M Tris-HCl buffer, pH 7.5, and 5 mM β-mercaptoethanol. The glycosylation reaction was performed overnight at 37°C. After centrifugation for 5 min at 16,000g, the supernatant was purified using an Oasis hydrophilic-lipophilic-balanced 3 cc extraction cartridge (Waters). A part of the methanol eluate was filtered using Minisart SPR4 filters (Sartorius Stedim Biotech) and subjected to a comparative metabolic analysis using LC-QTOF-MS. Another part was dried, and the resulting eugenol-xylosyl-diglucoside (0.2 µM) was analyzed using NMR (see Supplemental Methods 1 online). For NSGT-mediated glycosylation of PhP-diglycosides present in ‘smoky’ tomato fruit, a crude glycoside fraction extracted from 2 g of fresh weight tomato fruit was used as an acceptor substrate.
Tomato Transformation
The coding sequence of NSGT1 was amplified from fruits of tomato elite line C085 using the primers listed in Supplemental Table 3 online. The amplification product was digested with BamHI and SalI restriction enzymes and was subcloned into pFLAP50 containing a fusion of the double cauliflower mosaic virus 35S promoter and the Agrobacterium tumefaciens nos terminator (Tnos). The resulting plasmid construct was digested with PacI and AscI, and the fragment containing Pd35-NSGT1-Tnos fusion was subsequently transferred into the corresponding sites of the binary vector pBBC50. The resulting plasmid construct was verified by a restriction enzyme analysis and sequencing and then transferred to Agrobacterium LBA4404 using electroporation (Shen and Forde, 1989) before being used for plant transformation. Tomato cv Moneymaker plants were transformed using the Agrobacterium-mediated transformation protocol of tomato cotyledon explants as described (Martí et al., 2007). The ploidy level of regenerated plants was analyzed and only diploids were selected for further characterization. More than 20 independent transformation events per construct were selected. The presence of the corresponding transgene was tested using PCR amplification with NSGT1-specific primers (see Supplemental Table 3 online).
PCR-Based Markers for the Smoky Trait in Tomato
Homozygous and heterozygous NSGT1 tomato plants were distinguished using two PCR reactions. The first reaction was performed to amplify a 1350-bp fragment of the functional NSGT1 sequence using the following primers: a forward primer, 5′-GAGAGGATCCATGGAGAGAATTAAGGAAAATAGTCC-3′, and a reverse primer, 5′-GAGAGTCGACTCAATATAATAGCTTCAACAACTT-3′. The PCR program was conducted as follows: (1) 1 min at 95°C; (2) seven cycles of 30 s at 94°C and 2 min 30 s at 68°C; (3) 25 cycles of 30 s at 94°C, 30 s at 64°C, and 30 s at 68°C; and (4) 3 min at 68°C. The second reaction is performed to amplify a 1100-bp fragment of the mutant nsgt1 sequence using a forward primer, 5′-GAGAGGATCCATGGAGAGAATTAAGGAAAATAGTCC-3′, and a reverse primer, 5′-GAGAGTCGACTCAATATAATAGCTTCAACAACTT-3′. The PCR program as conducted as follows: (1) 1 min at 95°C; (2) 35 cycles of 30 s at 95°C, 30 s at 55°C, and 1 min 30 s at 68°C; and (3) 3 min at 68°C.
Accession Numbers
Sequence data from this article can be found in the EMBL/GenBank data libraries under the following accession numbers: NSGT1 (KC696865), nsgt1 (KC696866), NSGT2 (KC696867), and nsgt2 (KC696868).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. BLASTX Hits and an Alignment Map of the 120–Amino Acid Protein Sequence Corresponding to DGE Tag 403908.
Supplemental Figure 2. Alignment of Coding Sequences of NSGT Genes.
Supplemental Figure 3. Alignment of NSGT1 and NSGT2 Proteins of a ‘Smoky’ and a ‘Non-Smoky’ Tomato.
Supplemental Figure 4. Metabolic and Genetic Marker Analysis of Tomato Cultivars Commonly Used in Tomato Research: cv M82 and cv Moneymaker.
Supplemental Figure 5. Genetic and Metabolic Analysis of F6 RILs.
Supplemental Figure 6. NSGT1 SCAR Marker Performance in Commercial Tomato Cultivars.
Supplemental Figure 7. Coding Sequence and Protein Alignment of NSGT1 and NSGT2 Derived from the ‘Non-Smoky’ Variety C085 and Pimp-NSGT1 and Pimp-NSGT2 Derived from the Draft Genome Sequence of S. pimpinellifolium CGN G1.1589.
Supplemental Figure 8. Relative Abundance of Guaiacol-GX (Blue Bars) and Guaiacol-GXG (Red Bars) in S. pimpinellifolium Accessions.
Supplemental Table 1. Plant Glycosyltransferases with Known Function.
Supplemental Table 2. Primers Used for qRT-PCR Analysis of NSGT1 and NSGT2 Genes.
Supplemental Table 3. Primers Used for Cloning NSGT Genes.
Supplemental Methods 1. Enzymatic Production and Structural Elucidation of PhP-V Glycosides Using NMR.
Supplemental Data Set 1. Data Used for Sequence Alignment.
Acknowledgments
We thank Syngenta Seeds, Seminis, Enza Zaden, Rijk Zwaan, Vilmorin, and de Ruiter Seeds for providing seeds of the 94 tomato cultivars. We acknowledge financial support from the Center for BioSystems Genomics, provided under the auspices of the Netherlands Genomics Initiative. R.C.H.d.V. and R.D.H. thank the Netherlands Metabolomics Centre for additional funding. We also thank the Metabolomics lab for assistance in volatile determination and Rafael Martinez at the Instituto de Biología Molecular y Celular de Plantas for excellent plant management. We thank Gerco Angenent for discussions on the research and Ruud de Maagd for critical reading of the article. We also thank Fien Meijer-Dekens and A.W. van Heusden for excellent greenhouse management and plant cultivation. Finally, we thank Harry Jonker and Bert Schipper for preparation and analyses of the samples for LC-QTOF-MS.
AUTHOR CONTRIBUTIONS
Y.M.T. designed and performed research and wrote the article. J.M., R.C.H.d.V., J.B., A.v.H., J.J.J.v.d.H., M.N.-d.V., C.W.L., W.V. , M.V.Z., S.P., and J.L.R. performed research. H.v.d.G. provided bioinformatics support. A.G. designed and performed research. R.D.H. designed research. A.G.B. designed research and wrote the article.
Glossary
- VOC
volatile organic compound
- PhP-V
phenylpropanoid volatile
- MeSA
methyl salicylate
- GT
glycosyltransferase
- DGE
digital gene expression
- ORF
open reading frame
- GGT
glycoside glycosyltransferase
- qRT-PCR
quantitative real-time PCR
- Ct
threshold cycle
- GXG
2-O-β-d-glucopyranosyl-(1→2)-[O-β-d-xylopyranosyl-(1→6)]-O-β-d-glucopyranoside
- eugenol-GX
eugenol O-β-d-xylopyranosyl-(1→6)]-O-β-d-glucopyranoside
- GX
O-β-d-xylopyranosyl-(1→6)]-O-β-d-glucopyranoside
- LC-MS
liquid chromatography–mass spectrometry
- m/z
mass/charge
- eugenol-GXG
eugenol 2-O-β-d-glucopyranosyl-(1→2)-[O-β-d-xylopyranosyl-(1→6)]-O-β-d-glucopyranoside
- GC-MS
gas chromatography–mass spectrometry
- RIL
recombinant inbred line
- SPME
solid-phase microextraction
- QTOF
quadrupole time-of-flight
References
- Akakabe Y., Tamura Y., Iwamoto S., Takabayashi M., Nyuugaku T. (2006). Volatile organic compounds with characteristic odor in bamboo vinegar. Biosci. Biotechnol. Biochem. 70: 2797–2799 [DOI] [PubMed] [Google Scholar]
- Arumugam M., Mitra A., Jaisankar P., Dasgupta S., Sen T., Gachhui R., Kumar Mukhopadhyay U., Mukherjee J. (2010). Isolation of an unusual metabolite 2-allyloxyphenol from a marine actinobacterium, its biological activities and applications. Appl. Microbiol. Biotechnol. 86: 109–117 [DOI] [PubMed] [Google Scholar]
- Baldwin E.A., Scott J.W., Einstein M.A., Malundo T.M.M., Carr B.T., Shewfelt R.L., Tandon K.S. (1998). Relationship between sensory and instrumental analysis for tomato flavor. J. Am. Soc. Hortic. Sci. 123: 906–915 [Google Scholar]
- Baldwin E.A., Scott J.W., Shewmaker C.K., Schuch W. (2000). Flavor trivia and tomato aroma: Biochemistry and possible mechanisms for control of important aroma components. HortScience 35: 1013–1022 [Google Scholar]
- Boatright J., Negre F., Chen X., Kish C.M., Wood B., Peel G., Orlova I., Gang D., Rhodes D., Dudareva N. (2004). Understanding in vivo benzenoid metabolism in petunia petal tissue. Plant Physiol. 135: 1993–2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borges R.M., Bessière J.M., Hossaert-McKey M. (2008). The chemical ecology of seed dispersal in monoecious and dioecious figs. Funct. Ecol. 22: 484–493 [Google Scholar]
- Bowles D., Isayenkova J., Lim E.-K., Poppenberger B. (2005). Glycosyltransferases: Managers of small molecules. Curr. Opin. Plant Biol. 8: 254–263 [DOI] [PubMed] [Google Scholar]
- Bowles D., Lim E.K., Poppenberger B., Vaistij F.E. (2006). Glycosyltransferases of lipophilic small molecules. Annu. Rev. Plant Biol. 57: 567–597 [DOI] [PubMed] [Google Scholar]
- Bugos R.C., Chiang V.L., Zhang X.H., Campbell E.R., Podila G.K., Campbell W.H. (1995). RNA isolation from plant tissues recalcitrant to extraction in guanidine. Biotechniques 19: 734–737 [PubMed] [Google Scholar]
- Buttery R.G., Ling L.C., Light D.M. (1987). Tomato leaf volatile aroma components. J. Agric. Food Chem. 35: 1039–1042 [Google Scholar]
- Buttery R.G., Takeoka G., Teranishi R., Ling L.C. (1990a). Tomato aroma components: Identification of glycoside hydrolysis volatiles. J. Agric. Food Chem. 38: 2050–2053 [Google Scholar]
- Buttery R.G., Teranishi R., Ling L.C., Turnbaugh J.G. (1990b). Quantitative and sensory studies on tomato paste volatiles. J. Agric. Food Chem. 38: 336–340 [Google Scholar]
- Chaïb J., Lecomte L., Buret M., Causse M. (2006). Stability over genetic backgrounds, generations and years of quantitative trait locus (QTLs) for organoleptic quality in tomato. Theor. Appl. Genet. 112: 934–944 [DOI] [PubMed] [Google Scholar]
- Chang S., Puryear J., Cairney J. (1993). A simple and efficient method for isolating RNA from pine trees. Plant Mol. Biol. Rep. 11: 113–116 [Google Scholar]
- Cobucci-Ponzano B., Moracci M. (2012). Glycosynthases as tools for the production of glycan analogs of natural products. Nat. Prod. Rep. 29: 697–709 [DOI] [PubMed] [Google Scholar]
- Davidovich-Rikanati R., Sitrit Y., Tadmor Y., Iijima Y., Bilenko N., Bar E., Carmona B., Fallik E., Dudai N., Simon J.E., Pichersky E., Lewinsohn E. (2007). Enrichment of tomato flavor by diversion of the early plastidial terpenoid pathway. Nat. Biotechnol. 25: 899–901 [DOI] [PubMed] [Google Scholar]
- Dudareva N., Pichersky E. (2000). Biochemical and molecular genetic aspects of floral scents. Plant Physiol. 122: 627–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudareva N., Pichersky E., Gershenzon J. (2004). Biochemistry of plant volatiles. Plant Physiol. 135: 1893–1902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. (1985). Confidence limits on phylogenies: An approach using the bootstrap. Evolution 39: 783–791 [DOI] [PubMed] [Google Scholar]
- Frydman A., Weisshaus O., Bar-Peled M., Huhman D.V., Sumner L.W., Marin F.R., Lewinsohn E., Fluhr R., Gressel J., Eyal Y. (2004). Citrus fruit bitter flavors: Isolation and functional characterization of the gene Cm1,2RhaT encoding a 1,2 rhamnosyltransferase, a key enzyme in the biosynthesis of the bitter flavonoids of citrus. Plant J. 40: 88–100 [DOI] [PubMed] [Google Scholar]
- Gang D.R. (2005). Evolution of flavors and scents. Annu. Rev. Plant Biol. 56: 301–325 [DOI] [PubMed] [Google Scholar]
- Jensen N., Whitfield F.B. (2003). Role of Alicyclobacillus acidoterrestris in the development of a disinfectant taint in shelf-stable fruit juice. Lett. Appl. Microbiol. 36: 9–14 [DOI] [PubMed] [Google Scholar]
- Kennison K.R., Wilkinson K.L., Williams H.G., Smith J.H., Gibberd M.R. (2007). Smoke-derived taint in wine: Effect of postharvest smoke exposure of grapes on the chemical composition and sensory characteristics of wine. J. Agric. Food Chem. 55: 10897–10901 [DOI] [PubMed] [Google Scholar]
- Koeduka T., et al. (2006). Eugenol and isoeugenol, characteristic aromatic constituents of spices, are biosynthesized via reduction of a coniferyl alcohol ester. Proc. Natl. Acad. Sci. USA 103: 10128–10133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koeduka T., Orlova I., Baiga T.J., Noel J.P., Dudareva N., Pichersky E. (2009). The lack of floral synthesis and emission of isoeugenol in Petunia axillaris subsp. parodii is due to a mutation in the isoeugenol synthase gene. Plant J. 58: 961–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krumbein A., Auerswald H. (1998). Characterization of aroma volatiles in tomatoes by sensory analyses. Nahrung 42: 395–399 [DOI] [PubMed] [Google Scholar]
- Krumbein A., Peters P., Brückner B. (2004). Flavour compounds and a quantitative descriptive analysis of tomatoes (Lycopersicon esculentum Mill.) of different cultivars in short-term storage. Postharvest Biol. Technol. 32: 15–28 [Google Scholar]
- Larkin M.A., Blackshields G., Brown N.P., Chenna R., McGettigan P.A., McWilliam H., Valentin F., Wallace I.M., Wilm A., Lopez R., Thompson J.D., Gibson T.J., Higgins D.G. (2007). Clustal W and Clustal X version 2. Bioinformatics 23: 2947–2948 [DOI] [PubMed] [Google Scholar]
- Lee S.J., Noble A.C. (2003). Characterization of odor-active compounds in Californian chardonnay wines using GC-olfactometry and GC-mass spectrometry. J. Agric. Food Chem. 51: 8036–8044 [DOI] [PubMed] [Google Scholar]
- Lewinsohn E., Davidovich-Rikanati R., Iijima Y., Pichersky E., Sitrit Y. (2010). Functional genomics for the discovery of genes affecting lemon basil aroma and their use in flavor engineering of tomato. Acta Hortic. 860: 205–210 [Google Scholar]
- MacFie H.J.H. (1990). Assessment of the sensory properties of food. Nutr. Rev. 48: 87–93, discussion 114–131. [DOI] [PubMed] [Google Scholar]
- Mageroy M.H., Tieman D.M., Floystad A., Taylor M.G., Klee H.J. (2012). A Solanum lycopersicum catechol-O-methyltransferase involved in synthesis of the flavor molecule guaiacol. Plant J. 69: 1043–1051 [DOI] [PubMed] [Google Scholar]
- Martí C., Orzáez D., Ellul P., Moreno V., Carbonell J., Granell A. (2007). Silencing of DELLA induces facultative parthenocarpy in tomato fruits. Plant J. 52: 865–876 [DOI] [PubMed] [Google Scholar]
- Masada S., Terasaka K., Oguchi Y., Okazaki S., Mizushima T., Mizukami H. (2009). Functional and structural characterization of a flavonoid glucoside 1,6-glucosyltransferase from Catharanthus roseus. Plant Cell Physiol. 50: 1401–1415 [DOI] [PubMed] [Google Scholar]
- Menéndez P., Eilers P., Tikunov Y., Bovy A., van Eeuwijk F. (2012). Penalized regression techniques for modeling relationships between metabolites and tomato taste attributes. Euphytica 183: 379–387 [Google Scholar]
- Mizutani M., Nakanishi H., Ema J., Ma S.J., Noguchi E., Inohara-Ochiai M., Fukuchi-Mizutani M., Nakao M., Sakata K. (2002). Cloning of β-primeverosidase from tea leaves, a key enzyme in tea aroma formation. Plant Physiol. 130: 2164–2176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moco S., Bino R.J., Vorst O., Verhoeven H.A., de Groot J., van Beek T.A., Vervoort J., de Vos C.H. (2006). A liquid chromatography-mass spectrometry-based metabolome database for tomato. Plant Physiol. 141: 1205–1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales M.T., Luna G., Aparicio R. (2005). Comparative study of virgin olive oil sensory defects. Food Chem. 91: 293–301 [Google Scholar]
- Morita Y., Hoshino A., Kikuchi Y., Okuhara H., Ono E., Tanaka Y., Fukui Y., Saito N., Nitasaka E., Noguchi H., Iida S. (2005). Japanese morning glory dusky mutants displaying reddish-brown or purplish-gray flowers are deficient in a novel glycosylation enzyme for anthocyanin biosynthesis, UDP-glucose:anthocyanidin 3-O-glucoside-2″-O-glucosyltransferase, due to 4-bp insertions in the gene. Plant J. 42: 353–363 [DOI] [PubMed] [Google Scholar]
- Ono M., Shiono Y., Tanaka T., Masuoka C., Yasuda S., Ikeda T., Okawa M., Kinjo J., Yoshimitsu H., Nohara T. (2010). Three new aromatic glycosides from the ripe fruit of cherry tomato. J. Nat. Med. 64: 500–505 [DOI] [PubMed] [Google Scholar]
- Ortiz-Serrano P., Gil J.V. (2007). Quantitation of free and glycosidically bound volatiles in and effect of glycosidase addition on three tomato varieties (Solanum lycopersicum L.). J. Agric. Food Chem. 55: 9170–9176 [DOI] [PubMed] [Google Scholar]
- Park S., Elless M.P., Park J., Jenkins A., Lim W., Chambers E., IV, Hirschi K.D. (2009). Sensory analysis of calcium-biofortified lettuce. Plant Biotechnol. J. 7: 106–117 [DOI] [PubMed] [Google Scholar]
- Petro-Turza M. (1987). Flavour of tomato and tomato products. Food Rev. Int. 2: 309–351 [Google Scholar]
- Robin O., Alaoui-Ismaïli O., Dittmar A., Vernet-Maury E. (1999). Basic emotions evoked by eugenol odor differ according to the dental experience. A neurovegetative analysis. Chem. Senses 24: 327–335 [DOI] [PubMed] [Google Scholar]
- Sawada S., Suzuki H., Ichimaida F., Yamaguchi M.A., Iwashita T., Fukui Y., Hemmi H., Nishino T., Nakayama T. (2005). UDP-glucuronic acid: Anthocyanin glucuronosyltransferase from red daisy (Bellis perennis) flowers. Enzymology and phylogenetics of a novel glucuronosyltransferase involved in flower pigment biosynthesis. J. Biol. Chem. 280: 899–906 [DOI] [PubMed] [Google Scholar]
- Shen W.J., Forde B.G. (1989). Efficient transformation of Agrobacterium spp. by high voltage electroporation. Nucleic Acids Res. 17: 8385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sneath, P.H.A., and Sokal, R.R. (1973). Numerical Taxonomy. (San Francisco, CA: Freeman). [Google Scholar]
- Stone H., Sidel J. (2004). Descriptive Analysis. In Sensory Evaluation Practices. 3rd ed. (London: Academic Press), pp. 201–244 [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (2011). MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28: 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tieman D., et al. (2012). The chemical interactions underlying tomato flavor preferences. Curr. Biol. 22: 1035–1039 [DOI] [PubMed] [Google Scholar]
- Tieman D., Zeigler M., Schmelz E., Taylor M.G., Rushing S., Jones J.B., Klee H.J. (2010). Functional analysis of a tomato salicylic acid methyl transferase and its role in synthesis of the flavor volatile methyl salicylate. Plant J. 62: 113–123 [DOI] [PubMed] [Google Scholar]
- Tikunov Y., Lommen A., de Vos C.H.R., Verhoeven H.A., Bino R.J., Hall R.D., Bovy A.G. (2005). A novel approach for nontargeted data analysis for metabolomics. Large-scale profiling of tomato fruit volatiles. Plant Physiol. 139: 1125–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tikunov Y.M., de Vos R.C.H., González Paramás A.M., Hall R.D., Bovy A.G. (2010). A role for differential glycoconjugation in the emission of phenylpropanoid volatiles from tomato fruit discovered using a metabolic data fusion approach. Plant Physiol. 152: 55–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomato Genome Consortium (2012). The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485: 635–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veitch N.C., Grayer R.J. (2011). Flavonoids and their glycosides, including anthocyanins. Nat. Prod. Rep. 28: 1626–1695 [DOI] [PubMed] [Google Scholar]
- Wildermuth M.C., Dewdney J., Wu G., Ausubel F.M. (2001). Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 414: 562–565 [DOI] [PubMed] [Google Scholar]
- Zanor M.I., Rambla J.L., Chaïb J., Steppa A., Medina A., Granell A., Fernie A.R., Causse M. (2009). Metabolic characterization of loci affecting sensory attributes in tomato allows an assessment of the influence of the levels of primary metabolites and volatile organic contents. J. Exp. Bot. 60: 2139–2154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuckerkandl, E., and Pauling, L. (1965). Evolutionary divergence and convergence in proteins. In Evolving Genes and Proteins, V. Bryson and H.J. Vogel, eds (New York: Academic Press), pp. 97–166. [Google Scholar]