This study advances our understanding of the contribution of type III polyketide synthase (PKS) in phlorotannin biosynthesis of the marine brown algae Ectocarpus. It provides structural insights into the specificity of PKS1 for phloroglucinol formation. The evolution of the type III PKS gene family in Stramenopiles also suggests a lateral gene transfer event from an actinobacterium.
Abstract
Brown algal phlorotannins are structural analogs of condensed tannins in terrestrial plants and, like plant phenols, they have numerous biological functions. Despite their importance in brown algae, phlorotannin biosynthetic pathways have been poorly characterized at the molecular level. We found that a predicted type III polyketide synthase in the genome of the brown alga Ectocarpus siliculosus, PKS1, catalyzes a major step in the biosynthetic pathway of phlorotannins (i.e., the synthesis of phloroglucinol monomers from malonyl-CoA). The crystal structure of PKS1 at 2.85-Å resolution provided a good quality electron density map showing a modified Cys residue, likely connected to a long chain acyl group. An additional pocket not found in other known type III PKSs contains a reaction product that might correspond to a phloroglucinol precursor. In vivo, we also found a positive correlation between the phloroglucinol content and the PKS III gene expression level in cells of a strain of Ectocarpus adapted to freshwater during its reacclimation to seawater. The evolution of the type III PKS gene family in Stramenopiles suggests a lateral gene transfer event from an actinobacterium.
INTRODUCTION
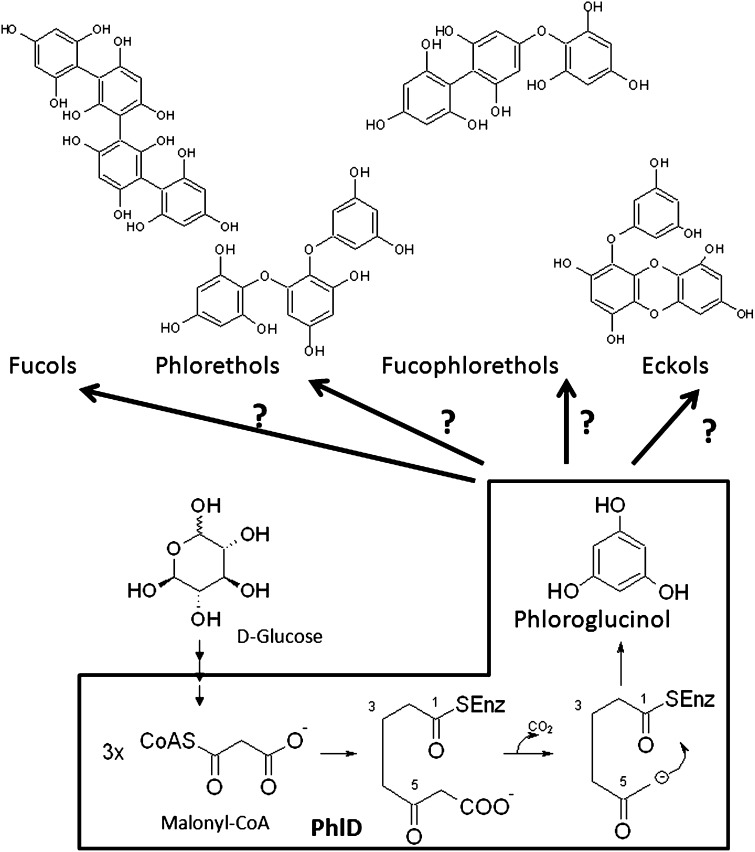
Brown algal phenols attract considerable attention due to the wide variety of biological activities and potential beneficial health effects of these so-called phlorotannins (Stengel et al., 2011; Thomas and Kim, 2011). Phlorotannins are known only from brown algae (Phaeophyceae) and are structural analogs of condensed tannins, such as anthocyanidins and other flavonoid derivatives, a diverse class of metabolites with a vast array of functions in terrestrial plants (Dakora, 1995). They are also suggested to play multiple ecological roles in brown algae, such as antifouling substances (Sieburth and Conover 1965) and chemical defenses against herbivory (Toth and Pavia, 2000), in addition to providing UV sunscreens to intertidal seaweeds (Amsler and Fairhead, 2006). Their chemical structure is based on aryl-aryl and/or diaryl ether linkages of phloroglucinol (1,3,5-trihydroxybenzene) units and is rather complex. Polymerization processes lead to a wide range of molecular sizes generally between 10 and 100 kD (Boettcher and Targett, 1993; McClintock and Baker, 2001; Le Lann et al., 2012).
Numerous dehydro-oligomers of phloroglucinol (fucols, fuhalols, phlorethols, fucophloretols, and eckols) have been characterized by chemical analyses in the three past decades (Figure 1) and are essentially from Fucales (Ascophyllum, Fucus, and Sargassum species) and Laminariales (Ecklonia and Eisenia species) (Ragan and Glombitza, 1986; Amsler and Fairhead, 2006; Le Lann et al., 2012). These compounds, which can represent up to 25% dry weight, are found as soluble forms in cellular compartments (Amsler and Fairhead, 2006), as insoluble forms cross-linked to cell walls (Koivikko et al., 2005, 2007), and in extracellular exudates (Shibata et al., 2006). Phlorotannins accumulate especially in specialized secretion vesicles named physodes and may be synthesized in chloroplasts or endoplasmic reticulum (Schoenwaelder and Clayton, 1998; Arnold and Targett, 2002; Parys et al., 2007). Despite the importance of phlorotannins in brown algal biology and ecology, their corresponding biosynthetic pathways have been little characterized at the biochemical and molecular levels.
Figure 1.
Chemical Structures of Phloroglucinol and Subunits of Four Different Structural Classes of Phlorotannins.
Boxed area shows the biosynthesis of a phloroglucinol by a bacterial type III PKS (PhlD) in P. fluorescens from malonyl-CoA as the sole substrate. The polymerization steps of phloroglucinol (1,3,5-trihydroxybenzene) condensation are not known, and only the phloroglucinol synthesis from the acetate-malonate pathway was hypothesized by a process that may involve a polyketide synthase-type enzyme complex in brown algae.
Brown algal phlorotannins were proposed to be generated via the condensation of acetate and malonate units in a manner similar to the synthesis of fatty acids (Arnold and Targett, 2002; Amsler and Fairhead, 2006; Pelletreau and Targett, 2007; Pelletreau, 2008). In addition, other routes that produce aromatic compounds and tannins in land plants, such as the shikimate pathway and the phenylpropanoid pathway, have been suggested (Chen et al., 1997) as alternative conserved pathways and experimentally investigated without success in brown algae (Pelletreau, 2008). The sequential condensation of acetate and malonate units is catalyzed by type III polyketide synthase (PKS III; Abe and Morita, 2010), and the resulting polyketide chain could undergo cyclization and tautomerization to form phloroglucinol (Waterman and Mole, 1994). Previous work in bacteria has demonstrated the biosynthesis of phloroglucinol following this mechanism via the phlorogucinol synthase D (PhlD; Figure 1), a PKS III from Pseudomonas fluorescens (Achkar et al., 2005; Zha et al., 2006). The PKS III protein family is specialized in the production of important secondary metabolites corresponding to aromatic polyketides that are of increasing economic interest (Abe and Morita, 2010). The biological roles of these molecules have been related to defense (phytoalexins and antibiotics; Abe and Morita, 2010), development (differentiation factor; Sankaranarayanan, 2006), and pigmentation (chalcones and flavones; Abe and Morita, 2010).
PKS III proteins have been reported to date in many organisms, including bacteria, land plants, fungi, and amoebas. The best described are probably the chalcone synthases (CHSs) and stilbene synthases (STSs), which produce precursors of the flavonoid and stilbenoid pathways in terrestrial plants by catalyzing the sequential decarboxylative addition of three acetate units from malonyl-CoA to a p-coumaryl-CoA starter molecule derived from the general phenylpropanoid pathway (Pfeifer and Khosla, 2001). The first crystal structure of a PKS III was resolved for the alfalfa (Medicago sativa) CHS (Ferrer et al., 1999), and a dozen new structures have been described to date for plants (Abe and Morita, 2010) and bacteria (Austin et al., 2004a). Among them, a type III PKS from Mycobacterium tuberculosis, Mtu-PKS18, displays an unusual broad specificity for aliphatic long-chain acyl-CoA starter units (C6–C20) to produce tri- and tetraketide pyrones (Sankaranarayanan et al., 2004).
The active state of PKS III enzymes is always found in dimeric form and they structurally and functionally differ from the multimodular type I PKS and the multisubunit complexes of type II PKS (Pfeifer and Khosla, 2001). These structural studies revealed that the catalytic residues of PKS III are structurally conserved (Abe and Morita, 2010) and correspond to the catalytic triad (Cys-164, His-303, and Asn-336; following the numbering in alfalfa CHS). However, the active site cavity in each protein is drastically different, thus explaining the diversity of PKS III in terms of their substrate specificity and polyketide length (Jez et al., 2001). The amino acid residues in the active site of type III PKS (i.e., Thr-197, Gly-256, and Ser-338) (following numbering in CHS of alfalfa) are involved in the selectivity of starter substrate and control of product chain length (Ferrer et al., 1999; Jez et al., 2002; Abe et al., 2005a, 2005b).
The filamentous alga Ectocarpus siliculosus is becoming a reference model for studying brown algal biology (Charrier et al., 2008; Cock and Coelho, 2011) due to the molecular and genetic tools that have recently been made available, including a complete genome sequence (Cock et al., 2010). During the evolution of Eukarya, the Stramenopiles, the lineage that encompasses brown algae (Brown and Sorhannus, 2010), have been evolving for over a billion years independently compared with the most commonly studied multicellular eukaryotes comprising opisthokonts (represented by animals and fungi) and Plantae (red algae, green algae, and plants). Interestingly, the E. siliculosus genome was predicted to encode three PKS III, which like P. fluorescens PhlD, may direct phloroglucinol biosynthesis (Cock et al., 2010). Moreover, one isoform of PKS III is highly represented in the expressed sequence tags libraries of E. siliculosus, probably reflecting a key role in a biosynthetic pathway (Cock et al., 2010). Previous attempts in the brown seaweed Sargassum binderi to characterize the biochemical function of a putative type III PKS failed to express a soluble active enzyme, and it was reported that a crude extract from a Escherichia coli recombinant clone incubated with malonyl-CoA yielded tetraketide pyrone but no phloroglucinol as products (Baharum et al., 2011). So far, the expression of recombinant soluble and active enzymes from brown algae remains highly challenging.
In this study, we provide phylogenetic, biochemical, and structural evidence that support a primary role of PKS1 in the condensation of malonyl-CoA to produce phloroglucinol, the direct precursor of phlorotannins unique to brown algae. We also link the phloroglucinol biosynthesis to physiological processes involved in the acclimation to salinity of a freshwater strain of Ectocarpus.
RESULTS
Phylogenetic Analysis of the Predicted E. siliculosus Type III PKS
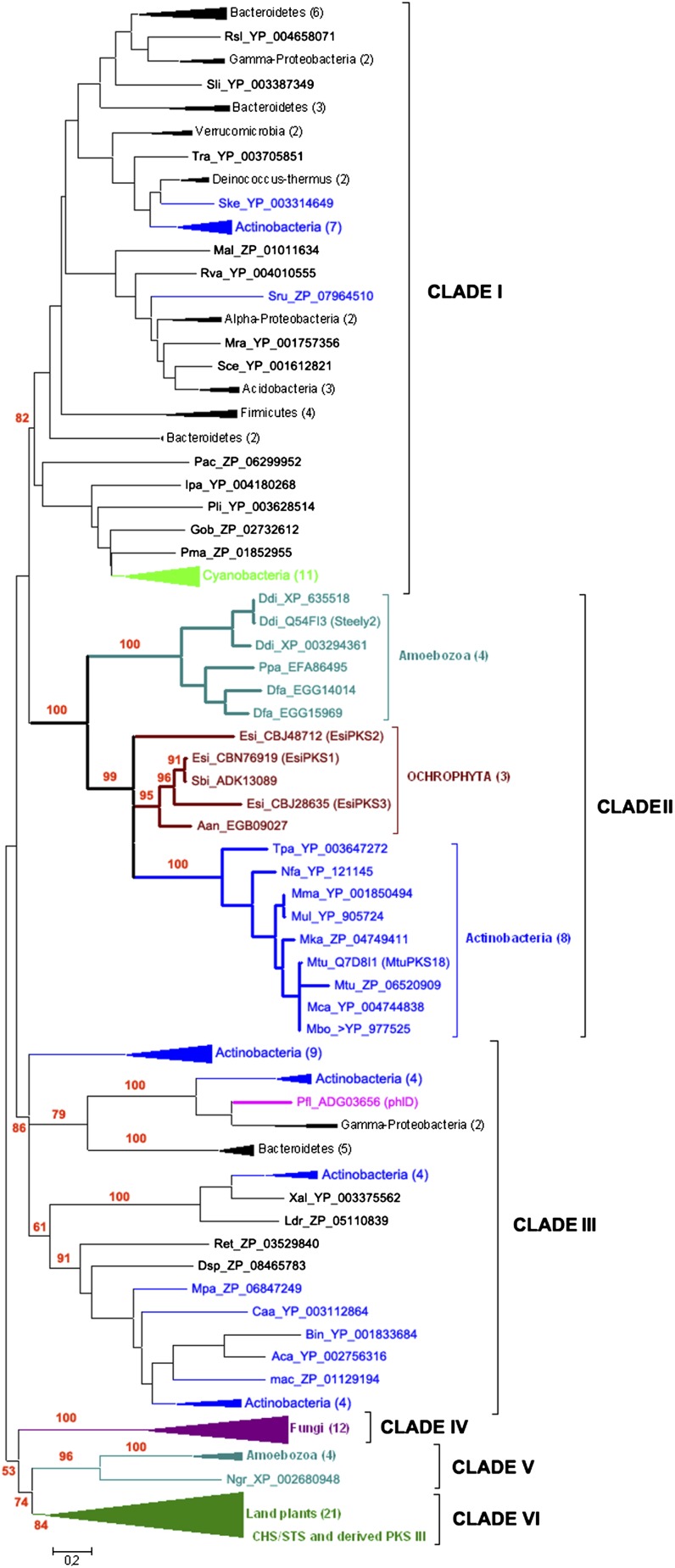
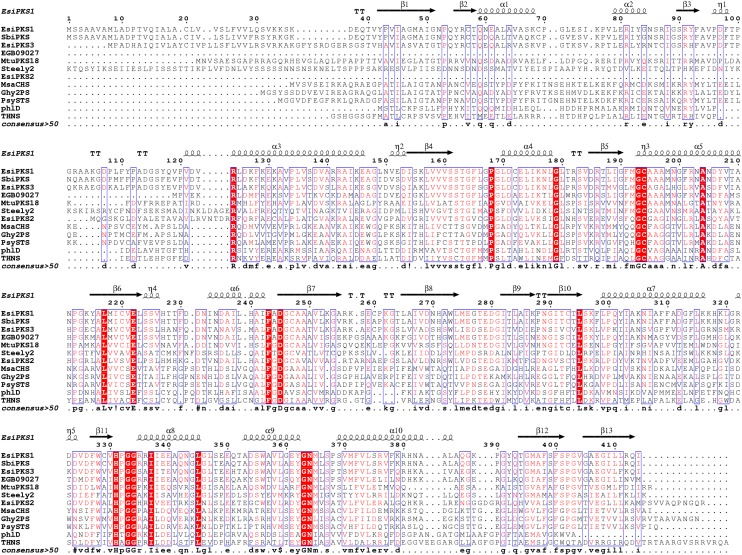
Three genes encoding proteins similar to type III PKS (loci Esi0024_0032, Esi0110_0075, and Esi0046_0100 in the ORCAE database, named E. siliculosus (Esi) PKS1 [CBN76919.1], EsiPKS2 [CBJ48712], and EsiPKS3 [CBJ28635.1], respectively; http://bioinformatics.psb.ugent.be/orcae/overview/Ectsi), were predicted during the annotation of the Ectocarpus genome (Cock et al., 2010). The amino acid sequences of Esi-PKS1 and Esi-PKS3 were highly similar to the PKS cDNA recently cloned in S. binderi that codes for a putative type III PKS. (Baharum et al., 2011). By contrast, the Esi-PKS2 cDNA was only a partial sequence of the gene. Indeed, Sb-PKS and Esi-PKS1 open reading frames were 85% identical at the nucleotide level, and the two proteins of 414 amino acids in length shared 92% identical residues and up to 97% similar residues (see Supplemental Figure 1 online). A phylogenetic analysis was performed on 161 representative type III PKS proteins from eukaryotes and bacteria, including functionally characterized enzymes accepting a diversity of CoA thioester starter units. The global topology of the maximum likelihood tree showed major clades of PKS III, based on bootstrap analyses (Figure 2 and Supplemental Figure 1 online).
Figure 2.
Unrooted Phylogenetic Tree of PKS III.
The phylogenetic tree presented here was constructed using the maximum likelihood approach. Numbers indicate the bootstrap values in the maximum likelihood analysis. The full listing of the aligned proteins is reported in Supplemental Data Set 1 and Supplemental Table 1 online. Rsl, Runella slithyformis; Sli, Spirosoma linguale; Tra, Truepera radiovictrix; Ske, Sanguibacter keddieii; Mal, Maritimibacter alkaliphilus; Rva, Rhodomicrobium vannielii; Sru, Segniliparus rugosus; Mra, Methylobacterium radiotolerans; Sce, Sorangium cellulosum; Pac, Parachlamydia acanthamoebae; Ipa, Isosphaera pallida; Pli, Planctomyces limnophilus; Gob, Gemmata obscuriglobus; Pma, Planctomyces maris; Ddi, Dictyostelium discoideum; Ppa, Polysphondylium pallidum; Dfa, Dictyostelium fasciculatum; Esi, Ectocarpus siliculosus; Sbi, Sargassum binderi; Aan, Aureococcus anophagefferens; Tpa, Tsukamurella paurometabola; Nfa, Nocardia farcinica; Mma, Mycobacterium marinum; Mul, Mycobacterium ulcerans; Mka, Mycobacterium kansasii; Mtu, Mycobacterium tuberculosis; Mca, Mycobacterium canettii; Mbo, Mycobacterium bovis; Pfl, Pseudomonas fluorescens; Xal, Xanthomonas albilineans; Ldr, Legionella drancourtii; Ret, Rhizobium etli; Dsp: Desmospora sp; Mpa, Mycobacterium parascrofulaceum; Caa, Catenulispora acidiphila; Bin, Beijerinckia indica; Aca, Acidobacterium capsulatum; mac, marine actinobacterium.
Clade I, supported by a bootstrap value of 82, contains only prokaryote enzymes from a large panel of bacterial groups (mainly belonging to cyanobacteria, proteobacteria, bacteroidetes, actinobacteria, and firmicutes). In clade I, none of the proteins have been functionally characterized, and most of them were discovered from environmental samples. Clade II corresponds to proteins from both eukaryotic and prokaryotic organisms and is clearly split into two well-supported groups. The first one contains only proteins from Amoebozoa (a sister lineage of opisthokonts), in which the PKS III steely 2 has been well characterized to produce acylphloroglucinols in vitro and in vivo (Sankaranarayanan, 2006). The second branch, strongly supported by a high bootstrap value of 99% leads to Actinobacteria proteins (in particular Corynebacteriaceae), such as Mtu-PKS18, which is closely related to PKS III of photosynthetic Stramenopiles (Ochrophyta) represented by two brown algae (E. siliculosus and S. binderi) and the pelagophyte Aureococcus anophagefferens.
Clade III contains exclusively prokaryotic proteins from actinobacteria, bacteroidetes, proteobacteria, and acidobacteria, but, in contrast with the first bacterial clade, this one contains numerous proteins that have been biochemically studied in the past 10 years. Most of the characterized enzymes are present in Actinobacteria, in particular Streptomyces species (1,3,6,8-tetrahydroxynaphthlene synthase [THNS], RppA and RppB, encoded in the same gene cluster and SrsA, and SrsB encoded in the same gene cluster in another species) and Mycobacterium species (PKS10 and PKS11). Another well-characterized PKS III activity has been described in the γ-proteobacterium P. fluorescens (PhlD). The compounds produced by enzymes from the corresponding gene clusters have antibiotic function (napyradiomycine A in Streptomyces coelicolor and 2,4-diacylphloroglucinol in P. fluorescens; Bangera and Thomashow, 1999; Funa et al., 2002) or confer resistance to antibiotics (penicillin resistance in Streptomyces lividans; Funabashi et al., 2008) or virulence (complex lipids in M. tuberculosis; Waddell et al., 2005).
Finally, Clades IV, V, and VI, supported with lower bootstrap values, grouped together eukaryotic PKS III proteins belonging to fungi, Amoebozoa, and land plants. Among them, plant PKS III enzymes have only been well studied, especially those involved in the flavonoid pathway (Abe and Morita, 2010, Ferrer et al., 1999; Jez et al., 2001, 2002; Austin et al., 2004b).
Expression and Enzymatic Activities of Recombinant PKS1 Protein
To characterize the type III PKS function of PKS1, we produced the enzyme as recombinant protein with a His-tag tail at the N terminus. In SDS-PAGE gels (see Supplemental Figure 2A online), the purified PKS1 migrated as a 40-kD protein, consistent with the predicted theoretical mass of 40.975 kD calculated without 34 amino acids of the putative N-terminal signal peptide. The identity of the purified protein was verified by matrix-assisted laser desorption/ionization–time-of-flight (MALDI-TOF) mass spectrometry analysis (see Supplemental Figure 2B online). An additional band was detected on the SDS-PAGE; however, the dynamic light scattering analysis showed that a homogenous pure protein was present in the sample. Both results suggest that PKS1 forms a physiologically active, dimeric complex, as already shown for most type III PKS, including PKS18 (Sankaranarayanan et al., 2004).
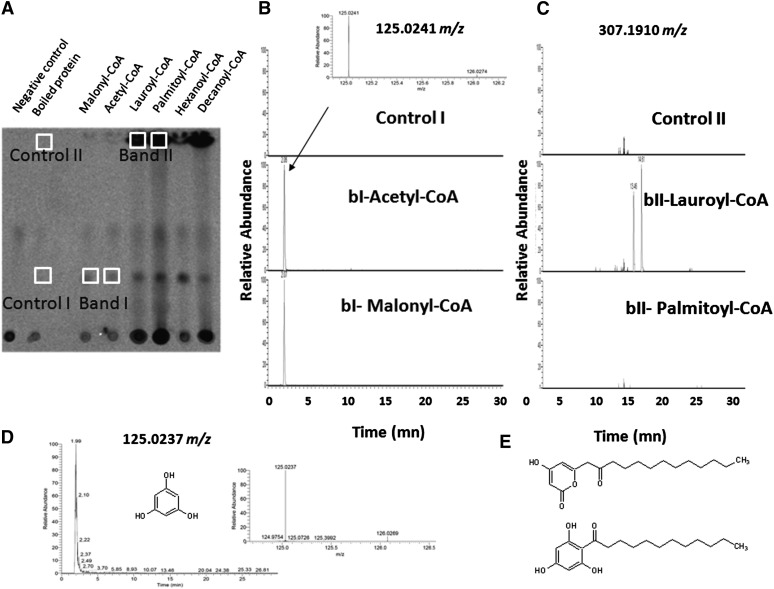
Activity of the recombinant PKS1 protein was assayed in presence of various fatty acyl-CoA esters as starter substrates and malonyl-CoA as the extender molecule. We first employed 14C malonyl-CoA as a tracer to visualize radioactive products of condensation reaction. After extraction, compounds were fractionated by thin layer chromatography (TLC) and revealed by exposure to a phosphor imager plate (Figure 3A).
Figure 3.
Identification of PKS1 Reaction Products by TLC Separation and UPLC-ESI-MS Analysis.
Recombinant protein was incubated with different CoA esters as starters and 14C-malonyl-CoA or unlabeled malonyl-CoA as extender.
(A) TLC analysis of radiolabeled reaction products of PKS1, detected using a phosphor imager. Chemically synthesized CoA esters used as starters are indicated above each lane. No product was observed without addition of PKS1 (negative control) or after addition of boiled enzyme. Controls display only one radioactive spot, which corresponds to the unreacted 14C-malonyl-CoA at the origin of the migration.
(B) Ultra-HPLC-ESI-MS chromatogram of a precursor ion at 125.0241 m/z of TLC-separated products corresponding to band I (bI), from incubation of unlabeled malonyl-CoA with acetyl-CoA or malonyl-CoA as starters, fitting with authentic phloroglucinol standard (see [D]). Inset displays the mass spectrum of the extracted ion at retention times (rt) 2.08 and 125.0241 m/z. The control I corresponds to an incubation of band I scraped from TLC in the lane corresponding to the incubation of boiled enzyme with malonyl-CoA as a starter and unlabeled malonyl-CoA as an extender.
(C) Ultra-HPLC-ESI-MS chromatogram of a precursor ion at 307.1910 m/z of TLC-separated products corresponding to band II (bII), from incubation of unlabeled malonyl-CoA as extender with lauroyl-CoA and palmitoyl-CoA as starter molecules. The control II corresponds to an incubation of band II scraped from a TLC in the lane corresponding to the incubation of boiled enzyme with lauroyl-CoA as a starter and unlabeled malonyl-CoA as an extender.
(D) Ultra-HPLC-ESI-MS chromatogram of a precursor ion at 125.0237 m/z of an authentic phloroglucinol standard. Insert displays the mass spectrum of the extracted ion at rt 2.10 min and 125.0237 m/z.
(E) Putative structures corresponding to the formula C18H28O4 and 307.1910 m/z are drawn corresponding to a tetraketide pyrone and an acylphloroglucinol, respectively.
When nonradiolabeled malonyl-CoA or acetyl-CoA was used as a starter molecule in the reaction mixture, only one radioactive product was observed (labeled “Band I” on Figure 3A) that was absent in the two negative controls consisting in incubations of 14C malonyl-CoA with boiled enzyme and without enzyme addition. When synthetic lauroyl-CoA (C16:0-CoA) and palmitoyl-CoA (C18:0-CoA) were used as starters (Figure 3A), additional reaction products were detected in incubation mixtures (labeled “Band II” on Figure 3A). The identity of some of the corresponding products was further investigated. For this purpose, similar experiments were conducted using nonradioactive malonyl-CoA as extender. With either acetyl-CoA or malonyl-CoA as starters, the major reaction TLC-separated product was scraped from the spot corresponding to “Band I” in the silica plate and extracted in methanol and was identified as phloroglucinol by liquid chromatography–electrospray ionization–mass spectrometry (LC-ESI-MS) analysis (Figures 3B and 3D) and confirmed by gas chromatography–mass spectrometry (GC-MS) using authentic standard (see Supplemental Figure 3 online).
A similar observation was previously described for the recombinant PhlD enzyme from the bacteria P. fluorescens (Zha et al., 2006). By contrast, Baharum et al. (2011) did not observe phloroglucinol formation after incubation of brown alga S. binderi PKS in presence of various acyl-CoAs and proposed the formation of triketide and tetraketide pyrone derivatives. In the presence of lauroyl-CoA, scraped from the spot corresponding to “band II” on the silica plate and extracted in methanol, only two major metabolites were detected by LC-ESI-MS analysis (Figure 3C; see Supplemental Figure 4 online). These two products feature the same [M-H]− 307.1910 mass-to-charge ratio (m/z) at retention time (rt) 13.46 and 14.61 min with a putative identification as C18H28O4 (Δppm −1.571). They are likely to be two different tetraketide derivatives from lauroyl-CoA: 1-(2,4,6-trihydroxyphenyl)-dodecan-1-one and 4-hydroxy-6-(2-oxotridecyl)-2H-pyran-2-one, that are also identified in ultra performance liquid chromatography (UPLC)-mass spectrometry (MS) analysis of whole incubation medium (Figure 3E; see Supplemental Figure 4 and Supplemental Table 2 online). Moreover, they were not detected in the presence of palmitoyl-CoA (Figure 3C). This indicates that the enzyme formed specific metabolites depending on the acyl chain of the starter units of the reaction.
X-Ray Crystallographic Structure of Ectocarpus PKS1
To explore the potential structural basis for the observed substrate specificity and catalytic mechanism of PKS1, we solved its crystal structure. The structure of Esi-PKS1 was solved at 2.85-Å resolution using molecular replacement (Figure 4). The space group of the crystals was determined to be P212121, with unit cell dimensions a = 61.3 Å, b = 83.4 Å, and c = 152.7 Å (Table 1). The asymmetric unit contains two molecules, giving a crystal volume per protein mass (VM) of 2.39 Å3 D−1 and a solvent content of 48.5% by volume (Matthews, 1968).
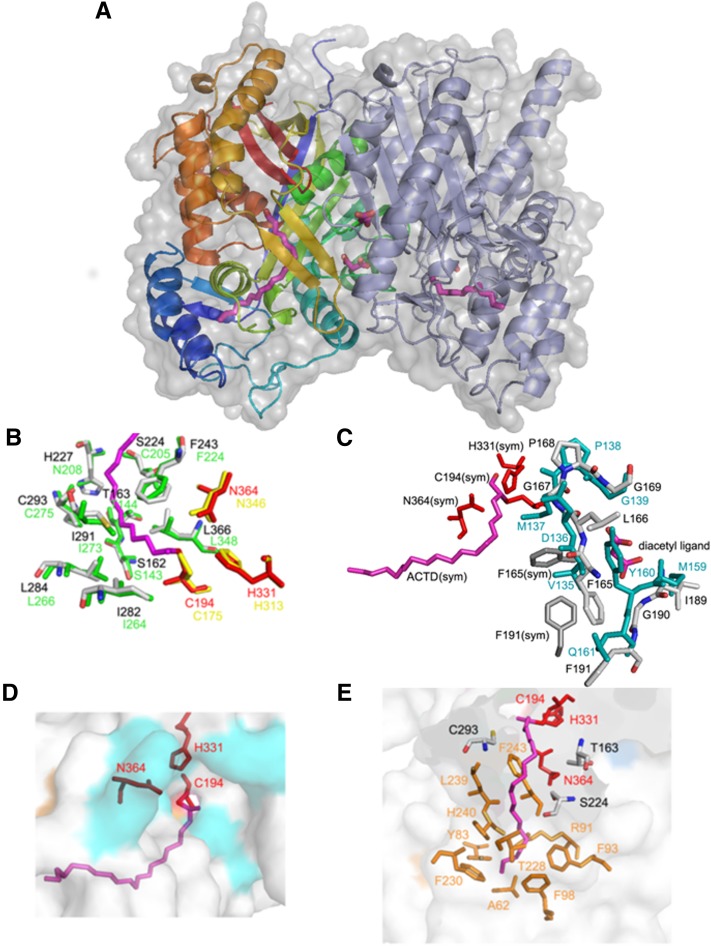
Figure 4.
Overall Structure of Esi-PKS1 and Details of the Structural Insights of the Active Site and of the Tunnel for Binding of Long-Chain Acyl-CoAs.
(A) A ribbon representation of the dimeric Esi-PKS1 molecule superposed with its surface view. The two monomers are represented as ribbons that are rainbow-colored from the N terminus (blue) to the C terminus (red) for molecule A and gray for molecule B of the dimer.
(B) Superimposition of the initiation and elongation pocket of Esi-PKS1 (gray) and Mtu-PKS18 (green). Residues surrounding the substrate binding pocket in the tunnel region are labeled pinpointing the two differences between Esi-PKS1 and Mtu-PKS18, Ser-224/Cys-205 and His-227/Asn-208, respectively. These residues are likely important in the different catalytic machinery. Residue labels are gray and red for Esi-PKS1 and green and yellow for Mtu-PKS1.
(C) Structural basis for substrate specificity at the dimer interface (near the putative phloroglucinol product): Two successive GF stretches are present in each monomer, the second Phe (Phe-191) is in an atypical position. These residues are located in two adjacent loops near the surface of the enzyme, allowing the formation of a new pocket at the dimer interface. A product has been entrapped in the crystal structure that is represented as a diketide, given the resolution of the crystal structure.
(D) Surface view of the entrance of the substrate in the CoA binding site that is highlighted by the side chains of Cys-194, His-331, and Asn-364, which form the catalytic triad and the acyl binding site of CoA. Esi-PKS1 possesses a tunnel where long chain fatty acid acyl-CoA starter molecules can be inserted before ligation to the catalytic Cys and elongation by addition of acetate units provided from malonyl-CoA.
(E) Surface view and a close-up view of the residues surrounding the binding tunnel of the acyl-CoA starter where long fatty acid chains of starter acyl-CoA molecules can be inserted before ligation to the catalytic Cys-194.
Table 1. Data Collection, Phasing, and Refinement Statistics for Esi-PKS1.
| Beamline at ESRF | ID23-I |
|---|---|
| Wavelength (Å) | 0.979 |
| Unit cell parameters (in Å) | a = 61.3, b = 83.4, c = 152.7 |
| Space group | P212121 |
| Resolution range (Å) | 49.4–2.85 (3.02–2.85)a |
| Number of observations | 70,191 (11602) |
| Number of unique reflections | 16,506 (2670) |
| Completeness (%) | 91.7 (77.4) |
| <I/σ(I)> | 8.5 (3.4) |
| Redundancy | 4.2 (4.3) |
| Rsymb;Rpimc (%) | 10.0 (33.3); 5.3 (17.7) |
| Map correlation coefficient (CC1/2) | 0.994 (0.989) |
| Refinement statistics | |
| Resolution range | 49.4–2.85 |
| R factor (Rfree on 5%) | 21.2 (23.6) |
| Overall B factor (Å2) | 39.97 |
| No. of protein atoms (mean B-factor in Å2) | 5,746 (A 42.2; B 39.4) |
| No. of ions/ligand atoms (mean B-factor in Å2) | 110 (21.6) |
| No. of solvent atoms (mean B-factor in Å2) | 345 (23.5) |
| Rms deviation in bond lengths (Å)d | 0.01 |
| Rms deviation in bond angles (°) | 1.369 |
| Ramachandran plot, most favored (%) | 96.2 |
| Ramachandran plot, additional allowed (%) | 2.5 |
Values for the highest resolution shell are given in parenthesis.
Rsym = Σ | I-Iav | / Σ| I |, where the summation is over all symmetry-equivalent reflections.
Rpim corresponds to the multiplicity weighted Rsym.
Rms, root mean square.
The quaternary structure of PKS1 is a dimer, formed by two monomers present in the asymmetric unit of PKS1 crystal structure (Figure 4A). The dimeric arrangement is perfectly superimposable with those observed in other polyketide synthase crystal structures. In Figure 5, the primary sequence of PKS1 is compared with those of various other polyketide synthases and the residue positions are marked with respect to their secondary structure positions in the overall PKS fold. Structural alignment scores of several structures of type III PKSs deposited in the Protein Data Bank (PDB), computed by the Dali server, are provided in Supplemental Table 3 online.
Figure 5.
Structure Alignment of the Full Sequences of Esi-PKS1 and 11 Other PKS III Exhibiting Various Specificities of Products.
The sequences are compared with the secondary structures of Esi-PKS1. α-Helices and β-strands are represented above the alignment as helices and arrows, respectively, and β-turns are marked with TT.
A major part of the residues corresponding to the initiation and elongation pockets of the active site are identical between Esi-PKS1 and Mtu-PKS18 (Ser-143/Ser-162, Thr-144/Thr-163, Phe-224/Phe-243, Ile-264/Ile-282, Leu-266/Leu-284, Ile-273/Ile-291, and Cys-275/Cys-293). The superimposition of the two structures shows that the positions are also well conserved (Figure 4B), and globally the volume of the cavity is equivalent. Nevertheless, two of these amino acids differ (Cys-205/Ser-224 and Asn-208/His-227), suggesting that these residues may contribute to the functional specificity of cyclization exhibited by the two enzymes. The most evident change in Esi-PKS1 is the presence of His-227, which occupies more space than the equivalent Asn-208 in Mtu-PKS18. As it was described for Mtu-PKS18, Asn-208, Leu-266, and Leu-348 significantly affect the active site volume. Therefore, the presence of His227 instead of an Asn residue might be crucial to produce acylphloroglucinols. For the synthesis of phloroglucinol, the cyclization of a shorter polyketide (triketide) should be more likely related to the proximal amino acids. This set of residues is completely identical, suggesting a fine-tuning of the production of the phloroglucinol core in Esi-PKS1, whereas lactonization is found in Mtu-PKS18. For example, a Thr is responsible for either the Claisen cyclization C6→C1 giving rise to a phloroglucinol core in CHS or the aldol switch C2→C7 mechanism in STS (Austin et al., 2004b). The difference is due to the distance of the hydroxyl group oxygen that can activate a water molecule in the case of STS and a decarboxylation of the polyketide. However, the resolution of our structure is not sufficient to conclude more precisely on the reaction mechanism.
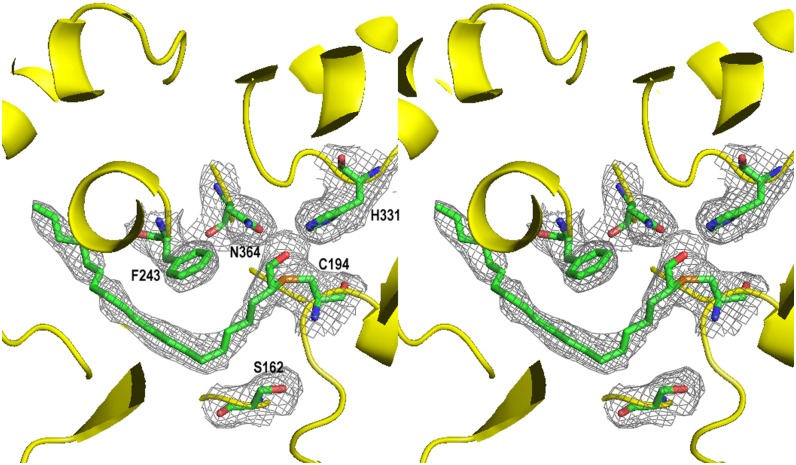
Similar to the presence of myristic acid in the crystal structure of PKS18 (Sankaranarayanan et al., 2004), the electron density in the active site of the Esi-PKS1 crystal structure revealed the presence of an aliphatic chain (Figure 6). The molecule was modeled as a C20 acyl group in order to match the electron density maxima that coincided well with chain length and distribution of double bonds of this acyl compound. The final structural model of Esi-PKS1 consists of 379 residues, representing residues 36 to 414 of each monomer, both of which bind an acetate ion and an arachidonyl group (ACTD group). The atomic distances of the acetate oxygen and the proximal Cys sulfur (Cys-194) are indicative of a covalent bond formation (Figures 4B, 4D, and 6). Moreover, the interpretation of the electron density suggests the presence of a malonic acid (MLA) that is practically superimposable with the positions of water molecules in PKS18 (pdbcode 1ted) and that comes to lie on the position of Tyr-165 in MsaPKS (pdbcode 1bq6). In addition to the amino acids and the two ligands of each monomer, the final Esi-PKS1 model contains 321 crystallographically defined water molecules.
Figure 6.
Wall-Eyed Stereo Representation of the Electron Density (2Fo-Fc Map; Cutoff Level 1σ) Showing the Covalently Attached Aliphatic Ligand That Was Identified in the Catalytic Tunnel of Each Molecule of the Dimeric PKS.
The correlation coefficient when calculating an omit map of this ligand is 0.90.
The pocket accommodating the MLA ligand has a volume of 210 Å3 and is formed by Phe-128, Val-159, Phe-165, Leu-166, Leu-170, Leu-188, and Phe-191, close to the dimer interface. This interface displays an interesting feature, consisting of a cluster of two successive Gly and Phe (164 to 165 and 190 to 191) in each monomer. The second Phe (Phe-191) is in a constraint conformer. These residues, making an interface between the two pockets, seem to represent a barrier to ligand exit and entry (Figure 4C). We can also notice that residues Phe-230, Leu-366, and Gly-403 are additionally allowed or outlier regions of the Ramachandran plot, probably due to strong interactions with the ligand.
Most importantly and similar to Mtu-PKS18, Esi-PKS1 possesses a tunnel for insertion of long-chain acyl-CoA starter molecules to guide the ligation to the catalytic Cys and further elongation by addition of acetate units provided by malonyl-CoA. This pocket accommodating the ACTD ligand has a volume of 1300 Å3 and is formed by Ala-62, Tyr-83, Arg-91, Phe-93, Phe-98, Thr-163, Cys-194, Ser-224, Thr-228, Phe-230, Leu-239, His-240, Phe-243, Cys-293, His-331, and Asn-364 (Figure 4E). Within this pocket, three residues, Cys-175, His-313, and Asn-346 in the PKS18, form the catalytic triad that is conserved at the following positions in Esi-PKS1: Cys-194, His-331, and Asn-364 (Figure 4D).
Accumulation of Phloroglucinol and Correlation with PSK1 Transcripts in the Acclimation to Seawater of a Freshwater Strain of Ectocarpus
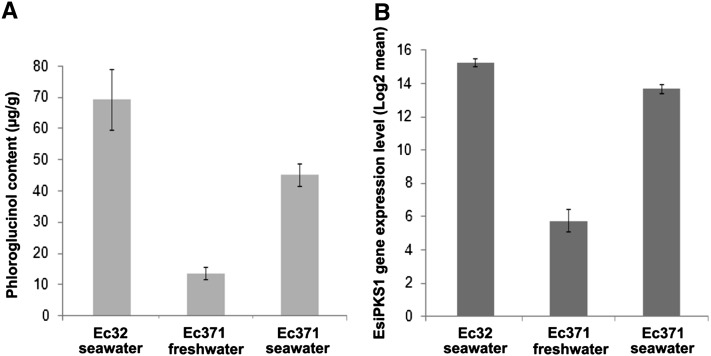
To further explore the biological relevance of phloroglucinol biosynthesis in Ectocarpus, methanol extracts prepared from two strains of Ectocarpus were analyzed by GC-MS for the presence of phloroglucinol. These strains are adapted to marine (Ectocarpus [Ec] 32 genome-sequenced strain) and freshwater (Ec 371) environments, respectively, and were recently compared at a transcriptional and metabolic level to study the mechanisms that may allow the transition between these habitats (Dittami et al., 2012a). As shown in Supplemental Figure 3 online, phloroglucinol was identified from total ion chromatograms of extracts prepared from these isolates by extracted ion monitoring at a m/z of 342 and retention time of 46.8 min, and the corresponding mass spectra for the peaks revealed characteristic fragment ions supporting this identification and similar retention time as the authentic standard (see Supplemental Figure 3C online). Interestingly, absolute quantification revealed that the extract from the Ec 371 strain cultivated in freshwater exhibited a very low level of phloroglucinol compared with marine strain Ec 32 (Figure 7A). By contrast, when transferred in seawater, phloroglucinol accumulated in Ec 371 to a concentration that was comparable to those measured in the seawater strain Ec 32. To determine if the characterized Esi-PKS1 and the two other predicted PKS III sequences in Ectocarpus exhibit expression patterns correlating with the accumulation of phloroglucinol (Figure 7), we examined microarray data recently published for these two strains of Ectocarpus (Dittami et al., 2012a). As shown in Figure 7B, the characterized Esi-PKS1 exhibited overexpression in the freshwater isolate Ec 371 reacclimated to seawater with a 244-fold increase (P value of 0.03). The two PKS III–like candidates, Esi-PKS2 and Esi-PKS3, did not show any significant changes of expression patterns (Dittami et al., 2012a).
Figure 7.
Phloroglucinol Accumulation and Expression Profiles of PKS1 in Two Isolates of Ectocarpus Adapted to Contrasting Environments.
(A) The phloroglucinol levels were determined by GC-MS analysis in methanol extracts prepared from the two isolates of Ectocarpus: marine Ec 32 cultured in undiluted seawater and freshwater Ec 371 in undiluted (saline stress) and highly diluted seawater (control).
(B) The relative expression levels of PKS1 determined by microarray analysis (Dittami et al., 2012a) were plotted for Ec 32 cultured in seawater and Ec 371, both in undiluted and highly diluted seawater. Relative changes are means of three replicates and error bars represent the corresponding standard deviations.
DISCUSSION
Until now, it has not been possible to experimentally identify genes or enzymes responsible for phlorotannin biosynthesis in any species of brown algae (Arnold and Targett, 2002; Amsler and Fairhead, 2006; Pelletreau and Targett, 2007; Parys et al., 2007). Interestingly, homologs of some of the land plant flavonoid pathway genes were found in Ectocarpus genome, but these are completely absent from diatom or green algal genomes (Cock et al., 2010). The shikimate pathway is also fully conserved in Ectocarpus genome, but some of the pathways that branch off the shikimate pathway in plants are absent, including routes for important compounds such as phenylpropanoids and salicylic acid (Cock et al., 2010). However, recent genomic approaches have predicted that several species of brown algae display gene homologs of type III polyketide synthases (Wong et al., 2007; Pearson et al., 2010; Baharum et al., 2011).
In this work, we demonstrate that similar to the bacterial enzyme PhlD (Achkar et al., 2005; Zha et al., 2006), Esi-PKS1 functions as a phloroglucinol synthase, using the malonyl-CoA substrate. The differences in products produced from malonyl-CoA in Esi-PKS1 versus Sb-PKS (Baharum et al., 2011) might be a result of an overinterpretation of the HPLC and LC-ESI-MS profiles by Baharum et al. (2011). Alternatively, it can be explained by interfering reactions due to the use of a recombinant protein that was only partially purified in the previous study. A major product of the incubation of Esi-PKS1 with malonyl-CoA as an extender and lauroyl-CoA as a starter molecule was also tentatively identified by LC-ESI-MS as an acylphloroglucinol or a tetraketide that are reminiscent of the phenolic lipids (Gerwick and Fenical, 1982) that have been proposed to be derived from various long-chain fatty acids, precursor of phloroglucinol derivatives in the brown seaweed Zonaria tournefortii (El Hattab et al., 2009).
As suggested by El Hattab et al. (2009), acylphloroglucinol derivatives could be generated by a polyketide-type biosynthesis beginning with the condensation of malonyl-CoA and acyl-CoA in order to furnish a tetraketide or a pentaketide intermediate. Therefore, as suggested by the production in vitro of long-chain acylphloroglucinol by recombinant ESi-PKS1, its homologs might be involved in the synthesis of long-chain acylphloroglucinol in orders of brown algae, such as Dictyotales and Fucales (Gerwick and Fenical, 1982; Wisespongpand and Kuniyoshi, 2003), which are phylogenetically distant from the Ectocarpales. Presently, there is no report of the occurrence of acylphloroglucinols in Ectocarpus.
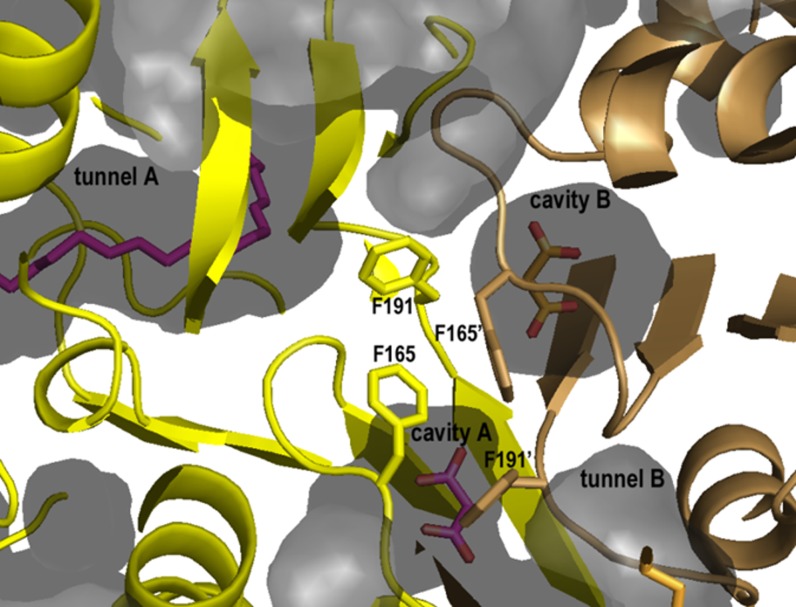
The crystal structure of Esi-PKS1 demonstrates that, similar to Mtu-PKS18 (Sankaranarayanan et al., 2004), Esi-PKS1 uses the tunnel created in the thiolase fold of the protein for substrate binding (Figure 4E). Interestingly, an additional cavity is found at the dimeric interface of Esi-PKS1 (Figures 4C and 8). In plant CHS or STS (Ferrer et al., 1999; Abe and Morita, 2010), this cavity is filled with a bulky aromatic amino acid instead of Gly-190 in Esi-PKS1. The absence of this bulky residue results in an additional cavity that allows the insertion of a small molecule. A product has been trapped in the crystal structure, but the electron density is not clear enough to unambiguously identify the nature of the molecule. We suggest that there is enough space to accommodate a phloroglucinol precursor. However, the resolution of the structure at 2.85 Å is too low to confirm this hypothesis. A diketide molecule (MLA) can be stably refined against the electron density (Figure 4C). Moreover, this additional pocket is adjacent to a cavity that, like in CHS, could accommodate a malonyl-CoA molecule. These supplemental cavities are covered by two adjacent and symmetric loops containing the Gly-Phe stretch, located near the surface of the enzyme. The movement of these loops might enhance the release of phloroglucinol units out of the molecule.
Figure 8.
Clipped Cartoon and Surface Representation at the Interface of the Dimeric Esi-PKS1 Molecule, Highlighting Additional Pockets.
The independent monomers are colored in yellow and light-brown, respectively. The ligands belonging to the same molecule have the same color (i.e., magenta for those in molecule A and orange in molecule B). The clipped surface highlights the tunnels that contain the aliphatic lipid chain (tunnels A and B) and the additional cavities formed at the dimeric interface (cavities A and B). Cavity B is closer to the tunnel of molecule A. Two of a group of four Phe residues at the interface are in sterically constrained conformations and thus could open the cavity to the exterior through a change of conformation.
Most of the plant PKS III have evolved to use the phenylpropanoid product coumaroyl-CoA in condensation reactions, but some functionally divergent PKS III are specialized to use long-chain acyl-CoA. The binding site of CoA, acetyl-CoA, or hexanoyl-CoA has been identified in CHS complexed with these substrates, and it corresponds to a 16-Å tunnel connected with the active-site cavity (Ferrer et al., 1999). MsaCHS and Ghy2PS share 74% amino acid identity, but they are drastically different in function and the study of the corresponding internal active site cavities demonstrated that those of 2PS is reduced of about two-thirds in volume compared with CHS (Jez et al., 2001). These observations underline the importance of the environment of specific amino acids in the initiation/elongation/cyclization pockets constituting the internal cavity and the pressure of the bulky residues on the corresponding mechanisms. For example, the replacement of three small amino acids by cumbersome ones in CHS (T197L/G256L/S338I) abolished the use of coumaroyl-CoA and promoted the use of acetyl-CoA and the synthesis of CTAL instead of chalcone (Jez et al., 2001). In this mutant, there is a change of regiospecificity of the cyclization reaction that corresponds to a derailment of the Claisen condensation to lactonization. Recently, Arabidopsis PKS-A and PKS-B have been described to produce alkylresorcinols important for the biosynthesis of pollen envelope (Kim et al., 2010). In these enzymes, a long tunnel similar to PKS18 is present (Mizuuchi et al., 2008). Further investigations of mutants of Esi-PKS1 might reveal interesting features of the regiospecificity of this enzyme.
Interestingly, in living algae, phloroglucinol contents differed strongly between two strains of Ectocarpus isolated from different environments (Figure 7A). Indeed, the freshwater isolate Ec 371, that belongs to a clade of Ectocarpus broadly distributed in habitats spanning a wide range of salinities, exhibited very low levels of phloroglucinol when maintained in highly diluted seawater. However, when transferred in undiluted seawater, phloroglucinol accumulated to a concentration comparable to that measured in the seawater isolate Ec 32. Noteworthy, the characterized Esi-PKS1 exhibits gene expression patterns (Dittami et al., 2012a) correlating with the accumulation of phloroglucinol in the freshwater strain Ec 371 acclimated to seawater. By contrast, it is likely that its expression is constitutive in the marine strain Ec 32, as also observed in a tiling array experiment and expressed sequence tag collections of E. siliculosus (Cock et al., 2010). The limited increase in phloroglucinol concentration in comparison with the level of transcripts is very likely representative of the activation of the whole metabolism of phlorotannins with yet unidentified compounds that are condensed from the precursor phloroglucinol (Amsler and Fairhead, 2006). However, the exact pathway of phlorotannin condensation is not known (Figure 1). The involvement of apoplastic vanadium-dependent haloperoxidases, in the presence of halide ions and hydrogen peroxide, has been demonstrated for in vitro cross-linking of phlorotannins (Berglin et al., 2004; Bitton et al., 2007), but the presence of halogenated oligomers during the oxidation process is still speculative and is awaiting confirmation by kinetic studies and mass spectral analysis.
The high levels of phenolic compounds in brown algae are thought to be involved in a number of secondary roles (Toth and Pavia, 2000), as well as being integral components of cell walls (Schoenwaelder and Clayton, 1998). This latter primary function in cell wall strengthening is particularly relevant in the context of adaptation to salinity that seems to involve a large reprogramming at the transcriptional and metabolome level in Ec 371 (Dittami et al., 2012a), involving the upregulation of sulfotransferases, which are tuning cell wall polysaccharide sulfation, of several mannuronan Carbon 5 (C5) epimerases that are involved in the control of cell wall rigidity through alginate mannuronan epimerization (Nyvall et al., 2003), and of Esi-vBPO, the vanadium bromoperoxidase that is thought to be involved in the cross-linking of secreted phenolic polymers and cell adhesion (Berglin et al., 2004; Bitton et al., 2007).
In contrast with the large multigenic families of PKS III present in land plants (Abe and Morita, 2010), among other phyla in the Plantae, no PKS III gene homolog was found in the available genomes for green and red algae. This suggests that these genes have been acquired later in evolution and probably after the land conquest, and it might implicate a lateral gene transfer from an uncharacterized bacterial class or from fungi. No PKS III genes were found either in the genomes of the diatoms Phaeodactylum tricornutum and Thalassiosira pseudonana or in nonphotosynthetic stramenopile oomycetes. However, a study has recently confirmed the presence of at least two different putative PKS III in Pseudochattonella farcimen from another class of ochrophytes, the Dichtyochophyta (Dittami et al., 2012b). These data corroborate the fact that a lateral transfer of PKS III gene has occurred after separation of diatoms from other ochrophytes, but before the divergence of brown algae with pelagophytes and dichtyochophytes. It was reported that several enzymes of major specific pathways found in brown algae and absent in diatoms (synthesis of mannitol, alginate, and hemicelluloses) are also derived from a lateral gene transfer involving an ancestral actinobacterium (Michel et al., 2010a, 2010b). It was proposed that a single massive transfer event allowed the acquisition of all these key genes and the emergence of brown algae. In addition, the phylogenetic tree suggests that the duplication of E. siliculosus PKS1 and PKS3 from an ancestral gene is very recent and posterior to the separation from Aureococcus, whereas PKS2 has been duplicated prior to this split (Figure 1). The acquisition of two genes of type III PKS (corresponding to Esi-PKS1 and Esi-PKS2) from actinobacteria that would imply a secondary loss in pelagophytes might also be possible.
In summary, this study deepens our understanding of the contribution of type III PKS in phlorotannin biosynthesis in E. siliculosus and provides structural insights into the specificity of PKS1 for phloroglucinol formation. Taken together, the above results suggest a conserved function of phlorotannin monomer biosynthesis for PSK1 and its orthologs in other brown algae and indicate that this function arose in Ochrophyta after the divergence of diatoms with the common ancestor of the other groups of photosynthetic stramenopiles, through a lateral gene transfer. This study also provides molecular tools to further investigate the regulation of phlorotannin biosynthesis, providing strong evidence that in vivo this type III PKS is also involved in the synthesis of phloroglucinol, the precursor of brown algal phlorotannins, and that the activation of this pathway is related to the acclimation and adaptation to salinity of euryhaline brown algae. Finally, it is likely that these tools will promote novel biotechnological developments to exploit the pharmaceutical potential of brown algal phenolic compounds.
METHODS
Chemicals
Malonyl-CoA, acetyl-CoA, hexanoyl-CoA, lauroyl-CoA, palmytoyl-CoA, decanoyl-CoA, phloroglucinol, vanillin, and all chemicals were purchased from Sigma-Aldrich. [2-14C] Malonyl-CoA (55 mCi/mmol) was purchased from Perkin-Elmer.
Algal Cultures and Saline Stress Experiments
The two strains used for these experiments were the Ectocarpus siliculosus marine strain Ec 32 (accession CCAP 1310/4, origin San Juan de Marcona, Peru) and the freshwater strain Ec 371 (accession CCAP 1310/196, origin Hopkins River Falls, Victoria, Australia). Cultures and experimental conditions were detailed previously (Dittami et al., 2012a). These experiments were done to study the acclimation of the freshwater stain, Ec 371, to saline stress. This strain was cultivated, during several months, in 100% seawater and in a mixture of 5% seawater and 95% distilled water with a final salinity of ∼1.6 ppt. The Ec 32 strain was only cultivated in 100% seawater and did not survive in 1.6 ppt medium. Samples for GC-MS analysis were obtained in similar conditions than the samples used for microarray experiments in previous experiments (Dittami et al., 2012a).
Bacterial Strains
Escherichia coli DH5α [fhuA2 Δ(argF-lacZ)U169 phoA glnV44 Φ80 Δ(lacZ)M15 gyrA96 recA1 relA1 endA1 thi-1 hsdR17 ] (Stratagene) was used as a host strain for maintaining plasmids. For protein expression, cis-repressed pQE-80L (Qiagen) derivatives were transformed into E. coli BL21 (DE3) codon Plus RIPL [E. coli B F– ompT hsdS(rB– mB–) dcm+ Tetr gal λ (DE3) endA Hte [argU ileY leuW Camr]] (Stratagene) containing extra copies for Arg, Ile, Pro, and Leu tRNAs.
Expression and Purification of Recombinant Proteins
The E. siliculosus PKS1 gene, locus Esi0024_0032 in the Ectocarpus ORCAE database (http://bioinformatics.psb.ugent.be/webtools/bogas/overview/Ectsi), was amplified by PCR with the corresponding cDNA n°LQ0AAA5YM06FM1 as template, without signal peptide at the beginning (Cock et al., 2010). The pQE-80L (Qiagen) expression vector containing the isopropyl-β-d-thiogalactopyranoside–inducible bacteriophage T5 promoter was used to express all proteins. E. siliculosus was cloned using SphI and HindIII restriction sites and the following primers: PQECHSFowBis, 5′-GGCGGATCCGCATGCATGTCCAAGGACGAGCAGACGGTATACCCGGTCATCGCC-3′, and pQECHSRev, 5′-GGCTAAGCTTTTACTAGATCTGCCTGAGAAGGATGCCCTCTGCCCC-3′. Expression constructs encode full-length proteins with a six-His tag on their N termini. The obtained clones were transformed into recombinant E. coli BL21-CodonPlus-RIPL cells (Stratagene) and then selected using Luria-Bertani solid medium, supplemented with 100 µg/mL of ampicillin. Protein expression was induced in ZIP medium (Studier, 2005) at 20°C. After 48 h of culture, induction was maintained by adding 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG) at a temperature of 20°C for 2 h. Cells were then collected by centrifugation and resuspended in buffer A (20 mM Tris-HCl, pH 7.5, 300 mM NaCl, and 50 mM imidazole) supplemented with protease inhibitor mixture, lyzozyme, and DNase. Cell lysis was performed by two passes on a French press to decrease viscosity of supernatants. Cellular debris was eliminated by centrifugation at 45,000g at 4°C during 90 min. All chromatographic procedures were performed on an Äkta Avant system at 20°C (GE Healthcare). Tagged proteins were purified on immobilized Ni-nickel tetradentate absorbent (NTA) medium, according to the manufacturer’s instructions (GE Healthcare), followed by purification on a Sephacryl-200 gel filtration column (GE Healthcare). All protein samples were analyzed for purity and integrity using 12% SDS-PAGE and by MALDI-TOF analyses (MetaboMER platform; SB Roscoff).
Dynamic light scattering analysis was performed using a DynaPro-801 molecular-sizing instrument (Structural Biology platform; SB Roscoff) equipped with a microsampler (Protein Solutions). A 50-μL sample was passed through a filtering assembly containing a 0.02-mm filter into a 12-μL chamber quartz cuvette. The data were analyzed using the Dynamics 4.0 and DynaLS software.
Enzyme Assays
TLC
Experiments were performed by individually testing five different starter acyl-CoAs at 200 µM (acetyl-CoA, hexanoyl-CoA, lauroyl-CoA, palmytoyl-CoA, and decanoyl-CoA) in an assay mixture of 500 μL containing also 20 µM [2-14C] malonyl-CoA (55 mCi/mmol), 50 µg of purified enzyme, 50 mM Tris HCl, pH 7.5, and 1 mM EDTA (final concentrations). Incubations were performed at room temperature for 1 or 3 h and stopped by adding 10 μL of 37% HCl. The products were then extracted with 1 mL of ethyl acetate and separated by TLC (Merck Art. 1.11798 silica gel 60 F254; ethyl acetate/hexane/AcOH 65:25:5, v/v/v). Radioactive signals were detected and quantified with a Typhoon imaging system (Molecular Dynamics-GE Healthcare).
GC-MS
Assays were performed as described for TLC. An internal standard of 2.50 µg of vanillin was added. Samples were vortexed for 5 min and centrifuged at 1000g for 5 min. The organic phase was transferred to a glass vial and evaporated under a stream of nitrogen. Trimethylsilyl-ethers were formed by addition of 100 μL acetonitrile and 100 μL of bis(trimethylsilyl)trifluoroacetamide (Sylon-BFTR) for 60 min at 60°C and evaporated under a stream of nitrogen. Metabolites were resuspended in 100 μL hexane and analyzed by GC-MS in the EI mode on an Agilent GC 6890 coupled to a 5973 MS detector (Agilent) and equipped with a DB-5MS column (30 m × 0.25 mm i.d. × 0.25-µm film thickness; J and W Scientific, Agilent). Temperatures of the injection port and interface were 250 and 280°C, respectively; those of the ion source and MS analyzer were set at 230 and 150°C, respectively. The samples were injected in splitless mode. The oven temperature was first set at 60°C for 5 min and then raised at 10°C/min to 100°C, elevated at 1°C/min to 150°C, and finally elevated at 290°C at the rate of 8°C/min and held for 5 min. The compounds were ionized by electron impact at 70 eV energy. Analytes were detected by total ion current from m/z 50 to 850.
LC-MS
The products were separated by reverse-phase U-HPLC (Dionex Ultimate 3000) on a Acclaim RSLC 120, C18, 2.2 µm (2.1 × 100 mm) column at a flow rate of 250 µL/min. Gradient elution was performed with water and acetonitrile, both containing 1% acetic acid, from 20 to 100% acetonitrile in 25 min. Elution was monitored by a multichannel UV detector at 280 or 360 nm. Online LC-ESI-MS spectral analyses were performed using a Thermo LTQ Orbitrap Discovery. Identification of the enzyme reaction products was performed by direct comparison with the authentic compounds or proposed from accurate m/z determination and fragmentation patterns.
Crystallization, Data Collection, Structure Determination, and Refinement
PKS1 crystals were grown by the hanging drop vapor diffusion method at room temperature (19°C) in 4.5-μL drops containing a 2:1 mixture of 7 mg/mL protein and crystallization buffer (0.1 M Bicine, pH 8, and 18% polyethylene glycol 6000).
Prior to data collection, the crystals were rapidly soaked in a cryo-buffer that was identical to the reservoir solution supplemented with 10% glycerol and subsequently frozen in a nitrogen gas stream at 100K. X-ray diffraction data were then collected from PKS1 crystals at 100K on beamline ID23-I at the European Synchrotron Radiation Facilities (ESRF; Grenoble, France) using an ADSC Quantum Q315r charge-coupled device detector. The crystals were rotated through 110° with a 1° oscillation range per frame at a wavelength of 0.979 Å. All raw data were processed using the program Mosflm (Leslie, 2006), and the resultant data were merged and scaled using the program Scala (Evans, 2006). All further data collection statistics are given in Table 1. Potential models for structure solution by molecular replacement were selected by a sequence search using BLAST (Altschul et al., 1990) against the PDB sequence database, indicating that the crystal structure of 1ted was the best model (see Supplemental Table 3 online). Consequently, we used the protein coordinates of the dimeric unit of 1ted as a search model to solve the structure of PKS1 by molecular replacement, which was performed with the program Molrep (Vagin and Teplyakov, 2010). A single solution with a weighted R-factor of 0.53 and a correlation coefficient of 0.34 was obtained using the complete data set at 2.85-Å resolution.
The starting phases obtained with Molrep were subsequently used to automatically build the initial model using ARP/wARP and REFMAC (Murshudov et al., 1997; Perrakis et al., 1999) as part of the CCP4 suite (Potterton et al., 2003, 2004). The initial electron density map was displayed with Coot (Emsley et al., 2010). Roughly 70% of the helices were constructed by the automatic procedure. The subsequent manual adjustment and model building was performed with Coot and alternated with refinement cycles using REFMAC. Water molecules were added automatically with the REFMAC-ARP/wARP option and visually verified, one by one, using Coot. The phasing and final refinement statistics are given in Table 1.
Molecular Phylogenetic Analysis of PKS III Sequences
The unrooted maximum likelihood phylogenetic tree and evolutionary analyses were conducted using MEGA5 (Tamura et al., 2007). A preliminary step was done to find the best-fit model among 48 different amino acid substitution models where the WAG+G+I was selected. The tree with the highest log likelihood (−38977,7031) is shown in Figure 2. Initial trees for the heuristic search were obtained automatically as follows. When the number of common sites was <100 or less than one-fourth of the total number of sites, the maximum parsimony method was used; otherwise, the BIONJ method with MCL distance matrix was used. A discrete gamma distribution was used to model evolutionary rate differences among sites (five categories; +G, parameter = 2.3239). The rate variation model allowed for some sites to be evolutionarily invariable ([+I], 3.9969% sites). The analysis involved 160 amino acid sequences. All positions containing gaps and missing data were eliminated. There were a total of 219 positions in the final data set.
Accession Numbers
Sequence data from this article can be found in the ORCAE Ectocarpus database (Sterck et al., 2012) (http://bioinformatics.psb.ugent.be/orcae/overview/Ectsi) or GenBank/EMBL data libraries under the following accession numbers: EsiPKS1 (CBN76919.1), EsiPKS2 (CBJ48712), and EsiPKS3 (CBJ28635.1), respectively. Accession numbers for the sequences used in the phylogenetic analysis of Figure 2 are provided in Supplemental Figure 1 online. Accession numbers are also labeled on the alignment in Figure 5. The coordinates and structure factors of Esi-PKS1 structure have been deposited in the Protein Data Bank (www.pdb.org; PDB ID 4b0n). Raw data and publicly available Gene Ontology annotations for the Ectocarpus microarrays are available at http://www.sb-roscoff.fr/UMR7139/ectocarpus/transcriptomics/.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Full Alignment Used to Draw the Phylogenetic Tree of Figure 1.
Supplemental Figure 2. SDS-PAGE of Recombinant PKS1 from E. siliculosus and MALDI-TOF Mass Spectrometry Spectrum of the Purified Recombinant PKS1.
Supplemental Figure 3. GC-MS Chromatograms of Phloroglucinol-TMS Products.
Supplemental Figure 4. U-HPLC-ESI-MS Chromatograms.
Supplemental Table 1. Accession Numbers, Organismal Origin, and Short Name of the Organism of the Proteins That Were Aligned to Construct the Phylogeny of Figure 2.
Supplemental Table 2. Formula and Chemical Structures of the Products Identified by UPLC-MS Analysis in the Incubation of Malonyl-CoA with Different Starter Molecules.
Supplemental Table 3. Structural Alignment Scores of Several Structures of Type III PKS Deposited in the PDB Database, Computed by the Dali-Server.
Supplemental Data Set 1. Text File of the Sequences and Alignment Used for the Phylogenetic Analysis Shown in Figure 2 and Supplemental Figure 1 Online.
Acknowledgments
Crystal structure determination was performed at the crystallography platform of the Station Biologique de Roscoff, supported by the Centre National de la Recherche Scientifique and Université Pierre et Marie Curie, Paris 06. We are indebted to the staff of the ESRF (Grenoble, France), beamlines ID23-I and BM30A, for technical support during data collection and treatment. We thank Thierry Tonon for providing biological material and access to microarrays data before publication and for helpful discussions and advice, Fanny Gaillard for MALDI-TOF mass spectrometry analysis, and Laurence Dartevelle for technical assistance. This project was supported by a Grant-in-Aid for Scientific Research from Groupement d'Interet Scientifique Europole MER to V.S.-P. and P.P. (Phlorotann’ING project). L.M.-C. and S.G. were also partly supported by the project IDEALG (ANR-10-BTBR-04-02) “Investissements d’avenir, Biotechnologies-Bioresources.” We also thank Groupement d'Interet Scientifique BiogenOuest for supporting MetaboMER facilities through the CORSAIRE metabolomics network.
AUTHOR CONTRIBUTIONS
P.P., V.S.-P., E.A.G., C.L., and M.C. designed research. L.M.-C., L.D., S.G., M.C., and C.J.-J.L. performed research. S.G., C.J.-J.L, E.C., and M.C. contributed new reagents/analytic tools. L.M.-C., L.D., S.G., M.C., C.J.-J.L., C.L., and P.P. analyzed data. L.M.-C, L.D., M.C., and P.P. wrote the article.
Glossary
- PKS III
type III polyketide synthase
- CHS
chalcone synthase
- STS
stilbene synthase
- MALDI-TOF
matrix-assisted laser desorption/ionization–time-of-flight
- TLC
thin layer chromatography
- LC-ESI-MS
liquid chromatography–electrospray ionization–mass spectrometry
- GC-MS
gas chromatography–mass spectrometry
- m/z
mass-to-charge ratio
- MLA
malonic acid
- ESRF
European Synchrotron Radiation Facilities
References
- Abe I., Morita H. (2010). Structure and function of the chalcone synthase superfamily of plant type III polyketide synthases. Nat. Prod. Rep. 27: 809–838 [DOI] [PubMed] [Google Scholar]
- Abe I., Oguro S., Utsumi Y., Sano Y., Noguchi H. (2005a). Engineered biosynthesis of plant polyketides: Chain length control in an octaketide-producing plant type III polyketide synthase. J. Am. Chem. Soc. 127: 12709–12716 [DOI] [PubMed] [Google Scholar]
- Abe I., Utsumi Y., Oguro S., Morita H., Sano Y., Noguchi H. (2005b). A plant type III polyketide synthase that produces pentaketide chromone. J. Am. Chem. Soc. 127: 1362–1363 [DOI] [PubMed] [Google Scholar]
- Achkar J., Xian M., Zhao H., Frost J.W. (2005). Biosynthesis of phloroglucinol. J. Am. Chem. Soc. 127: 5332–5333 [DOI] [PubMed] [Google Scholar]
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. (1990). Basic local alignment search tool. J. Mol. Biol. 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Amsler C.D., Fairhead V.A. (2006). Defensive and sensory chemical ecology of brown algae. Adv. Bot. Res. 43: 1–91 [Google Scholar]
- Arnold T.M., Targett N.M. (2002). Marine tannins: The importance of a mechanistic framework for predicting ecological roles. J. Chem. Ecol. 28: 1919–1934 [DOI] [PubMed] [Google Scholar]
- Austin M.B., Bowman M.E., Ferrer J.L., Schröder J., Noel J.P. (2004b). An aldol switch discovered in stilbene synthases mediates cyclization specificity of type III polyketide synthases. Chem. Biol. 11: 1179–1194 [DOI] [PubMed] [Google Scholar]
- Austin M.B., Izumikawa M., Bowman M.E., Udwary D.W., Ferrer J.L., Moore B.S., Noel J.P. (2004a). Crystal structure of a bacterial type III polyketide synthase and enzymatic control of reactive polyketide intermediates. J. Biol. Chem. 279: 45162–45174 [DOI] [PubMed] [Google Scholar]
- Baharum H., Morita H., Tomitsuka A., Lee F.C., Ng K.Y., Rahim R.A., Abe I., Ho C.L. (2011). Molecular cloning, modeling, and site-directed mutagenesis of type III polyketide synthase from Sargassum binderi (Phaeophyta). Mar. Biotechnol. (NY) 13: 845–856 [DOI] [PubMed] [Google Scholar]
- Bangera M.G., Thomashow L.S. (1999). Identification and characterization of a gene cluster for synthesis of the polyketide antibiotic 2,4-diacetylphloroglucinol from Pseudomonas fluorescens Q2-87. J. Bacteriol. 181: 3155–3163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berglin M., Delage L., Potin P., Vilter H., Elwing H. (2004). Enzymatic cross-linking of a phenolic polymer extracted from the marine alga Fucus serratus. Biomacromolecules 5: 2376–2383 [DOI] [PubMed] [Google Scholar]
- Bitton R., Berglin M., Elwing H., Colin C., Delage L., Potin P., Bianco-Peled H. (2007). The influence of halide-mediated oxidation on algae-born adhesives. Macromol. Biosci. 7: 1280–1289 [DOI] [PubMed] [Google Scholar]
- Boettcher A.A., Targett N.M. (1993). Role of polyphenolic molecular size in reduction of assimilation efficiency in Xiphister mucosus. Ecology 74: 891–903 [Google Scholar]
- Brown J.W., Sorhannus U. (2010). A molecular genetic timescale for the diversification of autotrophic stramenopiles (Ochrophyta): Substantive underestimation of putative fossil ages. PLoS ONE 5: e12759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charrier B., Coelho S.M., Le Bail A., Tonon T., Michel G., Potin P., Kloareg B., Boyen C., Peters A.F., Cock J.M. (2008). Development and physiology of the brown alga Ectocarpus siliculosus: Two centuries of research. New Phytol. 177: 319–332 [DOI] [PubMed] [Google Scholar]
- Chen Y., Yan X., Fan X. (1997). A hypothesis on Phaeophyceae polyphenols: Their structural unit and mechanism of the formation (in Chinese with English summary). Oceanol. Limnol. Sin. 28: 225–232 [Google Scholar]
- Cock J.M., Coelho S.M. (2011). Algal models in plant biology. J. Exp. Bot. 62: 2425–2430 [DOI] [PubMed] [Google Scholar]
- Cock J.M., et al. (2010). The Ectocarpus genome and the independent evolution of multicellularity in brown algae. Nature 465: 617–621 [DOI] [PubMed] [Google Scholar]
- Dakora F.D. (1995). Plant flavonoid: Biological molecules for useful exploitation. Aust. J. Phys. 22: 87–99 [Google Scholar]
- Dittami S.M., Gravot A., Goulitquer S., Rousvoal S., Peters A.F., Bouchereau A., Boyen C., Tonon T. (2012a). Towards deciphering dynamic changes and evolutionary mechanisms involved in the adaptation to low salinities in Ectocarpus (brown algae). Plant J. 71: 366–377 [DOI] [PubMed] [Google Scholar]
- Dittami S.M., Riisberg I., John U., Orr R.J., Jakobsen K.S., Edvardsen B. (2012b). Analysis of expressed sequence tags from the marine microalga Pseudochattonella farcimen (Dictyochophyceae). Protist 163: 143–161 [DOI] [PubMed] [Google Scholar]
- El Hattab M., Bouzidi N., Ortalo-Magne A., Daghbouche Y., Richou M., Chitour S.E., de Reviers B., Piovetti L. (2009). Eicosapentaenoic acid: Possible precursor of the phloroglucinol derivatives isolated from the brown alga Zonaria tournefortii (J.V. Lamouroux) Montagne. Biochem. Syst. Ecol. 37: 55–58 [Google Scholar]
- Emsley P., Lohkamp B., Scott W.G., Cowtan K. (2010). Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66: 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans P. (2006). Scaling and assessment of data quality. Acta Crystallogr. D Biol. Crystallogr. 62: 72–82 [DOI] [PubMed] [Google Scholar]
- Ferrer J.L., Jez J.M., Bowman M.E., Dixon R.A., Noel J.P. (1999). Structure of chalcone synthase and the molecular basis of plant polyketide biosynthesis. Nat. Struct. Biol. 6: 775–784 [DOI] [PubMed] [Google Scholar]
- Funa N., Ohnishi Y., Ebizuka Y., Horinouchi S. (2002). Alteration of reaction and substrate specificity of a bacterial type III polyketide synthase by site-directed mutagenesis. Biochem. J. 367: 781–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funabashi M., Funa N., Horinouchi S. (2008). Phenolic lipids synthesized by type III polyketide synthase confer penicillin resistance on Streptomyces griseus. J. Biol. Chem. 283: 13983–13991 [DOI] [PubMed] [Google Scholar]
- Gerwick W., Fenical W. (1982). Phenolic lipids from related marine algae of the order Dictyotales. Phytochemistry 21: 633–637 [Google Scholar]
- Jez J.M., Bowman M.E., Noel J.P. (2001). Structure-guided programming of polyketide chain-length determination in chalcone synthase. Biochemistry 40: 14829–14838 [DOI] [PubMed] [Google Scholar]
- Jez J.M., Bowman M.E., Noel J.P. (2002). Expanding the biosynthetic repertoire of plant type III polyketide synthases by altering starter molecule specificity. Proc. Natl. Acad. Sci. USA 99: 5319–5324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.S., et al. (2010). LAP6/POLYKETIDE SYNTHASE A and LAP5/POLYKETIDE SYNTHASE B encode hydroxyalkyl α-pyrone synthases required for pollen development and sporopollenin biosynthesis in Arabidopsis thaliana. Plant Cell 22: 4045–4066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koivikko R., Loponen J., Honkanen T., Jormalainen V. (2005). Contents of soluble, cell-wall-bound and exuded phlorotannins in the brown alga Fucus vesiculosus, with implications on their ecological functions. J. Chem. Ecol. 31: 195–212 [DOI] [PubMed] [Google Scholar]
- Koivikko R., Loponen J., Pihlaja K., Jormalainen V. (2007). High-performance liquid chromatographic analysis of phlorotannins from the brown alga Fucus vesiculosus. Phytochem. Anal. 18: 326–332 [DOI] [PubMed] [Google Scholar]
- Le Lann K., Connan S., Stiger-Pouvreau V. (2012). Phenology, TPC and size-fractioning phenolics variability in temperate Sargassaceae (Phaeophyceae, Fucales) from Western Brittany: Native versus introduced species. Mar. Environ. Res. 80: 1–11 [DOI] [PubMed] [Google Scholar]
- Leslie A.G. (2006). The integration of macromolecular diffraction data. Acta Crystallogr. D Biol. Crystallogr. 62: 48–57 [DOI] [PubMed] [Google Scholar]
- Matthews B.W. (1968). Solvent content of protein crystals. J. Mol. Biol. 33: 491–497 [DOI] [PubMed] [Google Scholar]
- McClintock, J.B., and Baker, B.J. (2001). Marine Chemical Ecology. (Boca Rotan, FL: CRC Press). [Google Scholar]
- Michel G., Tonon T., Scornet D., Cock J.M., Kloareg B. (2010a). Central and storage carbon metabolism of the brown alga Ectocarpus siliculosus: Insights into the origin and evolution of storage carbohydrates in Eukaryotes. New Phytol. 188: 67–81 [DOI] [PubMed] [Google Scholar]
- Michel G., Tonon T., Scornet D., Cock J.M., Kloareg B. (2010b). The cell wall polysaccharide metabolism of the brown alga Ectocarpus siliculosus. Insights into the evolution of extracellular matrix polysaccharides in Eukaryotes. New Phytol. 188: 82–97 [DOI] [PubMed] [Google Scholar]
- Mizuuchi Y., Shimokawa Y., Wanibuchi K., Noguchi H., Abe I. (2008). Structure function analysis of novel type III polyketide synthases from Arabidopsis thaliana. Biol. Pharm. Bull. 31: 2205–2210 [DOI] [PubMed] [Google Scholar]
- Murshudov G.N., Vagin A.A., Dodson E.J. (1997). Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53: 240–255 [DOI] [PubMed] [Google Scholar]
- Nyvall P., Corre E., Boisset C., Barbeyron T., Rousvoal S., Scornet D., Kloareg B., Boyen C. (2003). Characterization of mannuronan C-5-epimerase genes from the brown alga Laminaria digitata. Plant Physiol. 133: 726–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parys S., Rosenbaum A., Kehraus S., Reher G., Glombitza K.-W., König G.M. (2007). Evaluation of quantitative methods for the determination of polyphenols in algal extracts. J. Nat. Prod. 70: 1865–1870 [DOI] [PubMed] [Google Scholar]
- Pearson G.A., Hoarau G., Lago-Leston A., Coyer J.A., Kube M., Reinhardt R., Henckel K., Serrão E.T., Corre E., Olsen J.L. (2010). An expressed sequence tag analysis of the intertidal brown seaweeds Fucus serratus (L.) and F. vesiculosus (L.) (Heterokontophyta, Phaeophyceae) in response to abiotic stressors. Mar. Biotechnol. (NY) 12: 195–213 [DOI] [PubMed] [Google Scholar]
- Pelletreau, K.N. (2008). The Application of Molecular Tools towards the Study of Brown Algal Chemical Ecology and the Production of Phlorotannins. PhD dissertation (University of Delaware, Newark). [Google Scholar]
- Pelletreau, K.N., and Targett, N.M. (2007). New perspectives for addressing patterns of secondary metabolites in marine macroalgae. In Algal Chemical Ecology, C.D. Amsler, ed (Berlin: Springer-Verlag), pp. 121–146. [Google Scholar]
- Perrakis A., Morris R., Lamzin V.S. (1999). Automated protein model building combined with iterative structure refinement. Nat. Struct. Biol. 6: 458–463 [DOI] [PubMed] [Google Scholar]
- Pfeifer B.A., Khosla C. (2001). Biosynthesis of polyketides in heterologous hosts. Microbiol. Mol. Biol. Rev. 65: 106–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potterton E., Briggs P., Turkenburg M., Dodson E. (2003). A graphical user interface to the CCP4 program suite. Acta Crystallogr. D Biol. Crystallogr. 59: 1131–1137 [DOI] [PubMed] [Google Scholar]
- Potterton L., McNicholas S., Krissinel E., Gruber J., Cowtan K., Emsley P., Murshudov G.N., Cohen S., Perrakis A., Noble M. (2004). Developments in the CCP4 molecular-graphics project. Acta Crystallogr. D Biol. Crystallogr. 60: 2288–2294 [DOI] [PubMed] [Google Scholar]
- Ragan, M.A., and Glombitza, K.W. 1986. Phlorotannins, brown algal polyphenols. In Progress in Phycological Research, F.E. Round and D.J. Chapman, eds (Bristol, UK: Biopress), pp. 129–241. [Google Scholar]
- Sankaranarayanan R. (2006). A type III PKS makes the DIFference. Nat. Chem. Biol. 2: 451–452 [DOI] [PubMed] [Google Scholar]
- Sankaranarayanan R., Saxena P., Marathe U.B., Gokhale R.S., Shanmugam V.M., Rukmini R. (2004). A novel tunnel in mycobacterial type III polyketide synthase reveals the structural basis for generating diverse metabolites. Nat. Struct. Mol. Biol. 11: 894–900 [DOI] [PubMed] [Google Scholar]
- Schoenwaelder M.E.A., Clayton M.N. (1998). Secretion of phenolic substances into the zygote wall and cell plate in embryos of Hormosira and Acrocarpia (Fucales, Phaeophyceae). J. Phycol. 34: 969–980 [Google Scholar]
- Shibata T., Hama Y., Miyasaki T., Ito M., Nakamura T. (2006). Extracellular secretion of phenolic substances from living brown algae. J. Appl. Phycol. 18: 787–794 [Google Scholar]
- Sieburth J.McN., Conover J.T. (1965). Sargassum tannin, an antibiotic which retards fouling. Nature 208: 52–53 [Google Scholar]
- Stengel D.B., Connan S., Popper Z.A. (2011). Algal chemodiversity and bioactivity: Sources of natural variability and implications for commercial application. Biotechnol. Adv. 29: 483–501 [DOI] [PubMed] [Google Scholar]
- Sterck L., Billiau K., Abeel T., Rouzé P., Van de Peer Y. (2012). ORCAE: Online resource for community annotation of eukaryotes. Nat. Methods 9: 1041. [DOI] [PubMed] [Google Scholar]
- Studier F.W. (2005). Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 41: 207–234 [DOI] [PubMed] [Google Scholar]
- Tamura K., Dudley J., Nei M., Kumar S. (2007). MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24: 1596–1599 [DOI] [PubMed] [Google Scholar]
- Thomas N.V., Kim S.K. (2011). Potential pharmacological applications of polyphenolic derivatives from marine brown algae. Environ. Toxicol. Pharmacol. 32: 325–335 [DOI] [PubMed] [Google Scholar]
- Toth G.B., Pavia H. (2000). Water-borne cues induce chemical defense in a marine alga (Ascophyllum nodosum). Proc. Natl. Acad. Sci. USA 97: 14418–14420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin A., Teplyakov A. (2010). Molecular replacement with MOLREP. Acta Crystallogr. D Biol. Crystallogr. 66: 22–25 [DOI] [PubMed] [Google Scholar]
- Waddell S.J., Chung G.A., Gibson K.J., Everett M.J., Minnikin D.E., Besra G.S., Butcher P.D. (2005). Inactivation of polyketide synthase and related genes results in the loss of complex lipids in Mycobacterium tuberculosis H37Rv. Lett. Appl. Microbiol. 40: 201–206 [DOI] [PubMed] [Google Scholar]
- Waterman, P.G., and Mole, S. (1994). Analysis of Plant Metabolites, Methods in Ecology Series. (Oxford, UK: Blackwell). [Google Scholar]
- Wisespongpand P., Kuniyoshi M. (2003). Bioactive phloroglucinols from the brown alga Zonaria diesingiana. J. Appl. Phycol. 15: 225–228 [Google Scholar]
- Wong T.K.-M., Ho C.-L., Lee W.-W., Rahim R.A., Phang S.-M. (2007). Analyses of expressed sequence tags (ESTs) from Sargassum binderi (Phaeophyta). J. Phycol. 43: 528–534 [Google Scholar]
- Zha W., Rubin-Pitel S.B., Zhao H. (2006). Characterization of the substrate specificity of PhlD, a type III polyketide synthase from Pseudomonas fluorescens. J. Biol. Chem. 281: 32036–32047 [DOI] [PubMed] [Google Scholar]