This work explores the regulation of polyunsaturated fatty acid content in Arabidopsis seed oil. It shows that a transcriptional complex containing the basic Leu zipper protein bZIP67 and LEAFY COTYLEDON1-LIKE is responsible for activation of FATTY ACID DESATURASE3, which encodes an enzyme whose activity determines the level of omega-3 polyunsaturated fatty acids.
Abstract
Arabidopsis thaliana seed maturation is accompanied by the deposition of storage oil, rich in the essential ω-3 polyunsaturated fatty acid α-linolenic acid (ALA). The synthesis of ALA is highly responsive to the level of FATTY ACID DESATURASE3 (FAD3) expression, which is strongly upregulated during embryogenesis. By screening mutants in LEAFY COTYLEDON1 (LEC1)–inducible transcription factors using fatty acid profiling, we identified two mutants (lec1-like and bzip67) with a seed lipid phenotype. Both mutants share a substantial reduction in seed ALA content. Using a combination of in vivo and in vitro assays, we show that bZIP67 binds G-boxes in the FAD3 promoter and enhances FAD3 expression but that activation is conditional on bZIP67 association with LEC1-LIKE (L1L) and NUCLEAR FACTOR-YC2 (NF-YC2). Although FUSCA3 and ABSCISIC ACID INSENSITIVE3 are required for L1L and bZIP67 expression, neither protein is necessary for [bZIP67:L1L:NF-YC2] to activate FAD3. We conclude that a transcriptional complex containing L1L, NF-YC2, and bZIP67 is induced by LEC1 during embryogenesis and specifies high levels of ALA production for storage oil by activating FAD3 expression.
INTRODUCTION
Seed maturation in higher plants is associated with the deposition of storage reserves, such as oil (triacylglycerol), carbohydrates, and proteins (Baud and Lepiniec, 2010). The physiological role of these reserves is to serve as a source of carbon (and nitrogen) to fuel postgerminative growth, thereby enabling seedling establishment and completion of the plant’s life cycle (Graham, 2008). However, seed storage reserves also provide a major source of nutrition for humans and livestock, and serve as feedstock for a broad variety of industrial applications (Lu et al., 2011). As such, seeds have a significant social and economic importance. Understanding how seed storage reserve content and composition are controlled is of considerable basic and strategic interest.
Genetic studies have revealed that a complex network of transcriptional master regulators orchestrates the seed maturation program, of which storage reserve deposition is an integral part (Vicente-Carbajosa and Carbonero, 2005; Santos-Mendoza et al., 2008). In Arabidopsis thaliana, forward genetic screens have identified four loci, in particular, that act as positive regulators: namely LEAFY COTYLEDON1 (LEC1), LEC2, FUSCA3 (FUS3), and ABSCISIC ACID INSENSITIVE3 (ABI3) (Giraudat et al., 1992; Meinke, 1992; Keith et al., 1994; West et al., 1994; Lotan et al., 1998; Luerssen et al., 1998; Stone et al., 2001). LEC1 encodes a protein that is homologous to the Saccharomyces cerevisiae HEME ACTIVATOR PROTEIN3 or mammalian NUCLEAR FACTOR YB subunit of the heterotrimeric CCAAT box binding factor (Lotan et al., 1998; Lee et al., 2003). LEC2, FUS3, and ABI3 encode plant-specific transcription factors (TFs) that are closely related and contain a conserved B3 DNA binding domain (Giraudat et al., 1992; Luerssen et al., 1998; Stone et al., 2001).
Ectopic embryogenesis can be induced in vegetative tissues of Arabidopsis by expression of LEC1 or LEC2 (Lotan et al., 1998; Santos Mendoza et al., 2005; Mu et al., 2008). This developmental shift is accompanied by the differential expression of several hundred genes, including many that encode the metabolic apparatus for reserve synthesis and storage (Santos Mendoza et al., 2005; Mu et al., 2008). The regulation of seed storage protein (SSP) synthesis has been studied in some detail, and there is evidence to support a role for B3 domain proteins (such as ABI3) in the transactivation of SSP gene expression, either directly by binding RY/Sph cis-elements (Kroj et al., 2003; Stone et al., 2008) or through association with TFs from other families, such as basic Leu zippers (bZIP) domain proteins (Alonso et al., 2009). LEC1 has also been implicated in transactivation of SSP genes through an association with bZIPs (Yamamoto et al., 2009).
In contrast with SSPs, the transcriptional regulation of many key genes involved in storage lipid biosynthesis is less well understood. These genes may be direct targets of LEC1, LEC2, ABI3, and FUS3, or they may be controlled by other TFs that interact directly or lie downstream in the regulatory network (Baud and Lepiniec, 2010). An important example is WRINKLED1 (WRI1) (Focks and Benning, 1998; Cernac and Benning, 2004). WRI1 is a TF from the APETALA2/ethylene-responsive element binding family, which has been shown to be regulated by LEC2 and LEC1 in developing seeds (Baud et al., 2007; Mu et al., 2008). WRI1 governs the flux of carbon through glycolysis and fatty acid synthesis by regulating the expression of a suite of genes encoding enzymes in these pathways (Cernac and Benning, 2004; Baud et al., 2007). However, WRI1 is not required for the expression of several key enzymes in the pathways of fatty acid modification and triacylglycerol (TAG) assembly in the endoplasmic reticulum (Baud et al., 2007; To et al., 2012). These include FATTY ACID DESATURASE2 (FAD2), FAD3, FATTY ACID ELONGASE1 (FAE1), and DIACYLGLYCEROL ACYLTRANSFERASE1 (DGAT1) (To et al., 2012), which are critical for determining the composition and/or quantity of seed storage oil in Arabidopsis (Li-Beisson et al., 2013).
The aim of this study was to identify and characterize TFs that lie downstream of or function together with LEC1 and are responsible for regulating key enzymes of TAG synthesis during Arabidopsis seed maturation. To do this, TF genes that are substantially upregulated both during wild-type embryo maturation and following ectopic expression of LEC1 were selected using public microarray data (Winter et al., 2007; Mu et al., 2008). T-DNA insertion mutants were then obtained (Alonso et al., 2003) and mature seeds were screened for informative changes in fatty acid composition. Arabidopsis seed oil contains six major fatty acid species, namely, palmitic acid (16:0), stearic acid (18:0), oleic acid (18:1n9), α-linoleic acid (18:2n6), α-linolenic acid (18:3n3), and eicosenoic acid (20:1n11), and a number of minor species (<3 mol % each). The composition is highly heritable and genetic variation (both induced and natural) has been used extensively as a tool to elucidate gene function in seed oil metabolism (Lemieux et al., 1990; O’Neill et al., 2003). Two TF mutants were identified that showed a seed lipid phenotype. Further experimentation revealed the molecular mechanism by which both TFs cooperate to regulate the expression of FAD3 and, therefore, the level of the ω-3 polyunsaturated fatty acid 18:3n3, which is an essential fatty acid for human and livestock nutrition.
RESULTS
L1L and bZIP67 Are Regulators of Seed Storage Oil Composition
Several studies have established that ectopic expression of LEC1 leads to the induction of genes associated with the embryo maturation program, and results in accumulation of storage oil (Lotan et al., 1998; Mu et al., 2008). To identify TFs that might lie downstream of LEC1 in the regulatory network that controls seed oil content and composition, published Affymetrix Ath1 chip microarray data were used to select those genes listed on the Arabidopsis Transcription Factor Database (http://Arabidopsis.med.ohio-state.edu/AtTFDB/) that are more than fourfold upregulated in both LEC1-overexpressing seedlings (Mu et al., 2008) and wild-type developing seeds (stages 7 to 9; Winter et al., 2007). In total, 30 genes were shortlisted (see Supplemental Table 1 online), including ABI3, FUS3, and WRI1, which have been well characterized and are known to play a role in storage oil synthesis (Baud and Lepiniec, 2010). LEC2 is absent from this list because it is not upregulated in LEC1-overexpressing seedlings (Mu et al., 2008).
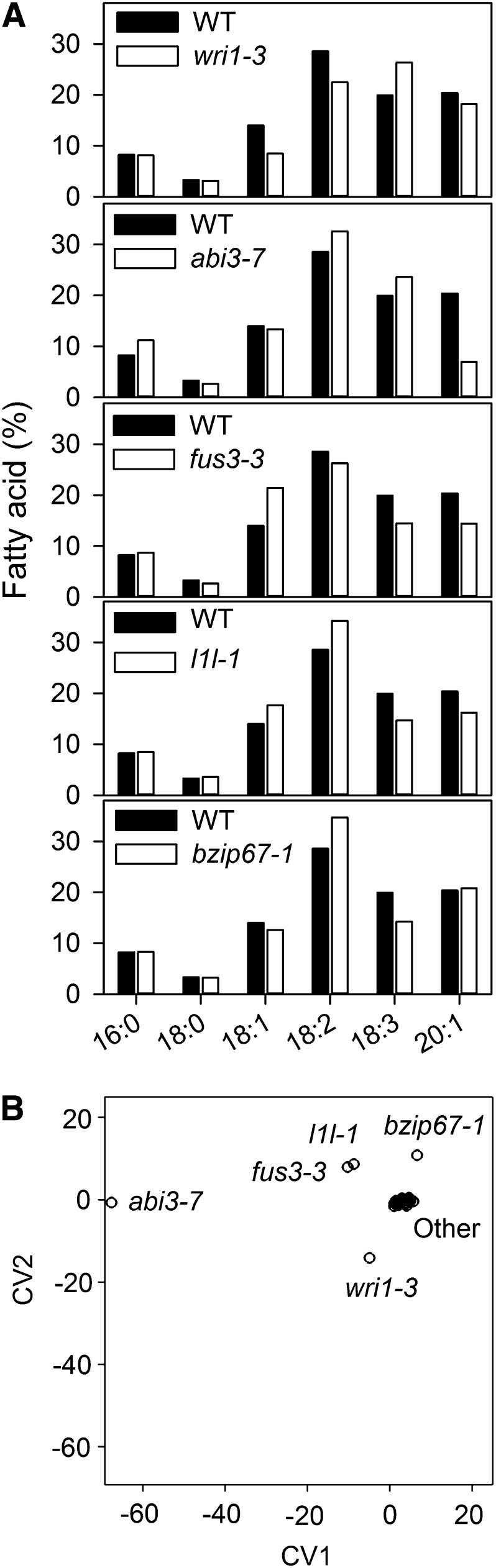
To screen the shortlisted TFs for a role in storage oil synthesis, publically available T-DNA mutants (see Supplemental Table 1 online) were identified on the SIGnAL T-DNA Express website (Alonso et al., 2003), four homozygous plants of each genotype were grown in a controlled environment, and their seeds were profiled for alterations in fatty acid composition (Lemieux et al., 1990). Five mutants (fus3-3, abi3-7, wri1-3, l1l-1, and bzip67-1) were found to exhibit significant differences in their seed fatty acid composition (P < 0.05) in comparison to their wild-type genetic background Columbia-0 (Col-0; Figure 1A). The alterations in fatty acid profiles are relatively complex; therefore, canonical variate analysis (CVA) was used to assist in the assessment of overall differences (and similarities) between the genotypes (Figure 1B). CVA showed that the first two CVs were sufficient to account for the majority of the variance (93.21%) and possible discrimination. The loadings indicated that 20:1n11, followed by 18:3n3 and 18:3n6, were most important in the discrimination observed in the CV1 direction. 18:3n6 and 18:3n3 also had influence in the CV2 direction, along with 18:0. It is clear that abi3-7 is very different from all other genotypes in the CV1 direction, with a large negative CV1 score indicating a strong influence of 18:3n3 and 18:3n6. The four other genotypes appear to be different in the CV2 direction. The most different of these is wri1-3, with a negative CV2 score. Interestingly, the other three genotypes (bzip67-1, fus3-3, and l1l-1) have similar positive CV2 scores, and fus3-3 and l1l-1 are not significantly different given their 95% confidence circles, while bzip67-1 is separated from these two on CV1 with a positive score (Figure 1B), suggesting a stronger influence of 20:1n11 for this genotype.
Figure 1.
Analysis of Fatty Acid Composition of Seeds from LEC1-Inducible TF Mutants.
(A) Fatty acid composition of five mutant genotypes that had significant (P < 0.05) differences in profile compared with wild-type (WT) ecotype Col-0. Values are the mean of measurements on seed from four plants of each genotype. All se are <2% of the mean.
(B) CVA plot assessing overall differences between all genotypes tested. The plot shows the 95% confidence circles around the CV mean for each genotype. Most genotypes, including wild-type ecotypes Col-0 and Col-3, are marked collectively as “Other” and are clustered at the intersection of the axis. The first two CVs were sufficient to account for 93.12% of the variance and possible discrimination between the genotypes.
L1L (LEC1-LIKE), ABI3, FUS3, and WRI1 have all been implicated in storage oil synthesis previously (Baud and Lepiniec, 2010), and abi3, fus3, and wri1 seeds have already been reported to exhibit substantial changes in fatty acid content/composition (Finkelstein and Somerville, 1990; Keith et al., 1994; Focks and Benning, 1998). However, a lipid phenotype previously has not been reported for l1l (Yamamoto et al., 2009), and bzip67 has not been reported to have any anatomical or biochemical phenotype (Bensmihen et al., 2005; Le et al., 2010). L1L is closely related to LEC1 and is capable of functionally complementing the lec1 mutant, when expressed under the LEC1 promoter (Kwong et al., 2003). However, the spatial and temporal pattern of L1L expression during seed development is substantially different from that of LEC1 (Winter et al., 2007), and L1L induction is also dependent on both LEC1 and FUS3 (Mu et al., 2008). The dependence of L1L expression on FUS3 may explain why the fatty acid profiles of l1l-1 and fus3-3 cannot be discriminated by CVA (Figure 1B).
bZIP67 is a seed-specific basic Leu zipper protein from group A that is closely related to ABI5 (Jakoby et al., 2002). Despite the lack of a reported phenotype of bzip67 (Bensmihen et al., 2005; Le et al., 2010), modeling studies have placed bZIP67 as a central hub in the gene regulatory networks that govern seed maturation (Belmonte et al., 2013). CVA suggests that the fatty acid profile of bzip67-1 is similar (but not identical) to that of l1l-1 and fus3-3 (Figure 1B). Because little is known about the physiological role of bZIP67, we decided to characterize mutants in this gene in greater detail and investigate its relationship with L1L and FUS3, within the context of seed lipid metabolism.
Molecular Characterization of Two T-DNA Mutants in bZIP67
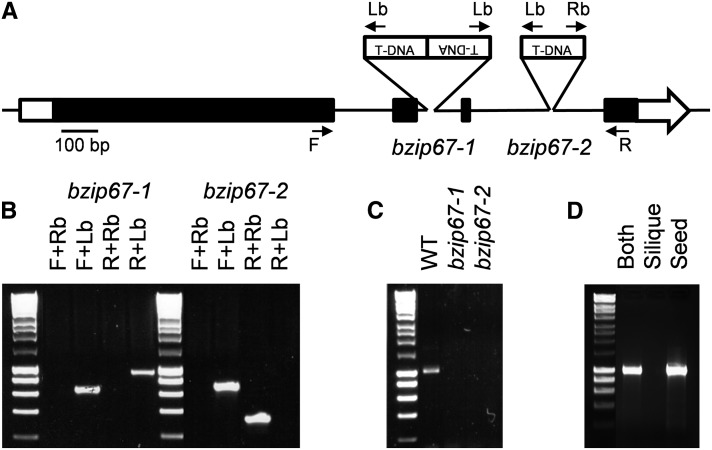
To confirm that disruption of bZIP67 does alter seed metabolism, two independent T-DNA mutant alleles were characterized for this locus (Figure 2A). PCR was performed on genomic DNA using gene-specific primers straddling the insertion site alone and in combination with T-DNA boarder primers (Figure 2B) and the flanking sequences were sequenced. In bzip67-1 (SALK_085497), the insertion was found to consist of an inverted T-DNA repeat situated in the second intron at +1033 bp 3′ of the start codon and results in a further 10-bp deletion. In bzip67-2 (GABI314D04), the T-DNA insertion site is in the third intron at +1356 bp 3′ of the start codon and results in a further 22-bp deletion.
Figure 2.
Characterization of Two T-DNA Mutants in bZIP67.
(A) A diagram showing the position of T-DNA insertions in the bZIP67 gene. The bzip67-1 and bzip67-2 alleles are SALK_085497 and GK314D04, respectively. Exons are presented as black bars and introns as lines. The 5′ and 3′ untranslated regions are a white bar and a white arrow, respectively.
(B) PCR on bzip67-1 and bzip67-2 genomic DNA using primers spanning the T-DNA insertion sites in combination with T-DNA boarder primers (marked in [A]).
(C) RT-PCR on RNA isolated from whole developing siliques of bzip67-1, bzip67-2 and wild-type (WT) plants using bZIP67 primers that amplify the full-length cDNA (∼1 kb).
(D) RT-PCR on RNA isolated from whole developing siliques (both) and also separated silique and seed tissue of wild-type plants using bZIP67 primers that amplify the full-length cDNA. The PCR data are from single samples for each genotype but are representative of the results obtained from multiple independent experiments.
To investigate what impact the two T-DNA insertions have on bZIP67 transcript, RT-PCR was performed on RNA extracted from stage 8 (Winter et al., 2007) developing siliques of the wild type (Col-0), bzip67-1, and bzip67-2 (Figure 2C). No PCR product was amplified from bzip67-1 and bzip67-2 using primers that straddle the T-DNA insertion sites, showing that either no transcript is present or that the RNA is incorrectly spliced. Even if mis-spliced transcripts are produced by these alleles, they are likely to be null because the conserved basic Leu zipper domain of bZIP67 is partially coded for by the last exon (Bensmihen et al., 2002) that lies downstream of the insertion sites. RT-PCR performed on separated seed and silique tissue of wild-type plants showed that bZIP67 transcripts are only detected in the seed (Figure 2D). Public microarray data also suggests that bZIP67 expression is restricted to seed tissues (Winter et al., 2007).
To investigate whether bzip67-1 and bzip67-2 exhibit the same seed fatty acid composition phenotype, they were grown together with the wild type in a controlled environment and the seed was analyzed (Li et al., 2006). As observed previously (Figure 1A), the fatty acid composition of seed from bzip67-1 was clearly different from the wild type, and there was also no significant difference (P > 0.05) between the two bzip67 alleles (see Supplemental Figure 1A online). On a percentage basis, the levels of 18:1n9 and 18:3n3 are decreased in both bzip67 mutants, and the level of 18:2n6 is increased (Figure 1A; see Supplemental Figure 1A online). Finally, a ProbZIP67:GFP-bZIP67 construct from a transgenic line (Bensmihen et al., 2005) was introduced into the bzip67-1 background by crossing in order to perform a complementation test. Expression of bZIP67 recovered the seed fatty acid composition of bzip67-1 to a profile that is very similar to the wild type (see Supplemental Figure 1B online).
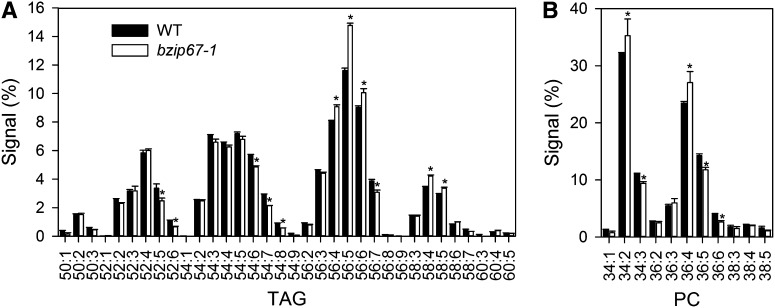
Disruption of bZIP67 Alters TAG and Phosphatidylcholine Composition, but Total Fatty Acid, Protein Content, and Seed Weight Are Unchanged
To investigate the effect of bZIP67 disruption on seed lipid composition in more detail, electrospray ionization–tandem mass spectrometry (Devaiah et al., 2006; Krank et al., 2007) was used to profile neutral and polar lipid species in mature and developing (stage 8) seeds, respectively, of bzip67-1 and the wild type (Figure 3). Analysis of TAG showed that molecular species containing 18:3n3 (e.g., 52:6, 54:7, and 56:7) are significantly (P < 0.05) less abundant in bzip67 seeds and those containing 18:2n6 (e.g., 56:4, 56:5, and 58:4) are more abundant (Figure 3A). Similarly, analysis of phosphatidylcholine (PC) molecular species also showed that 36:4 is significantly more abundant in bzip67-1 (P < 0.05), while 36:5 and 36:6 are less abundant (Figure 3B). Smaller increases in 36:4 were also observed in phosphatidylethanolamide and phosphatidic acid, but the compositions of phosphatidylglycerol and mono- and digalactosyldiacylglycerol did not appear to be changed (see Supplemental Table 2 online). In contrast with the changes in lipid composition detected in bzip67-1 seed, the total quantity of fatty acids was not significantly different (P > 0.05) from the wild type (see Supplemental Table 3 online). Measurements of total protein content and fresh weight also suggested that there are no significant differences (P > 0.05) from the wild type (see Supplemental Table 3 online).
Figure 3.
Effect of bZIP67 Disruption on Seed Lipid Composition.
(A) Mature seed TAG composition. WT, the wild type.
(B) Developing seed PC composition. Classes are defined by the total number of carbons and double bonds within the acyl groups.
Values are the mean ± se of measurements on seed batches from four plants of each genotype. Asterisk denotes a significant difference between the wild type and bzip67-1 (P < 0.05).
Disruption of bZIP67 Suppresses FAD3 and Enhances ROD1 Expression
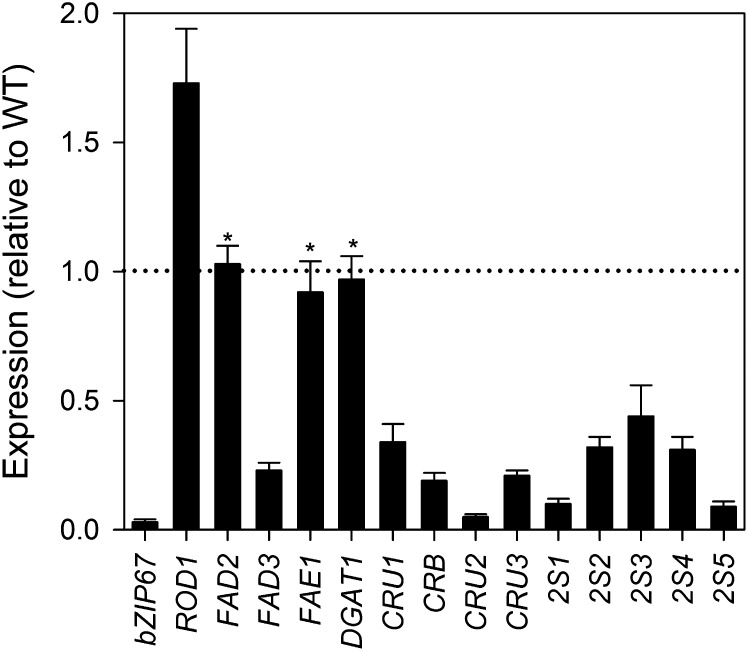
To identify genes that may be targets of bZIP67 regulation, microarray experiments were performed on RNA from whole siliques of wild-type and bzip67-1 plants using the Affymetrix Ath1 chip. Siliques were selected that contained developing seeds at stage 8 when bZIP67 (Winter et al., 2007) and many genes involved in storage lipid synthesis are strongly expressed (Baud and Lepiniec, 2010). In whole siliques, bZIP67 expression is only present in the seed (Figure 2D); therefore, any changes in transcript abundance that are detected in bzip67-1 siliques are likely to have arisen from the seed. Analysis of data from three biological replicates suggested that disruption of bZIP67 leads to a relatively small number of changes in gene expression at this stage in development (see Supplemental Data Set 1 online). Using Extraction of Differential Gene Expression software (Leek et al., 2006) only 37 genes were found with significant changes in expression (P < 0.01) that were also >1.5-fold up or downregulated. The signal for bZIP67 was reduced 12-fold in bzip67-1, which is consistent with the characterized gene defect in this mutant (Figure 2).
Cross-referencing those transcripts that are significantly (P < 0.01) up- or downregulated with a recent census of Arabidopsis genes involved in storage oil metabolism (Li-Beisson et al., 2013) flagged a single gene: FAD3. FAD3 transcript abundance was approximately twofold lower in whole bzip67-1 siliques (see Supplemental Data Set 1 online). FAD3 encodes a microsomal 18:2n6 desaturase that uses 18:2n6 esterified to PC as its substrate and is required for the majority of 18:3n3 produced in Arabidopsis seeds (Lemieux et al., 1990; Browse et al., 1993). The abundance of 18:3n3 is known to be highly responsive to the level of FAD3 expression (Shah et al., 1997; Puttick et al., 2009; O’Neill et al., 2011) and FAD3 transcript abundance increases approximately fivefold during the maturation stage of embryo development (Winter et al., 2007). Reduced expression of FAD3 could therefore explain the higher level of 18:2n6 and lower level of 18:3n3-containing species in both PC and TAG, in bzip67 seeds (Figure 3). FAD3 is also expressed in vegetative tissues; therefore, the change in gene expression detected in whole siliques might underrepresent the size of the effect in seeds. To measure FAD3 expression in stage 8 developing seeds, real-time RT-PCR experiments were performed (Figure 4). These experiments suggest that FAD3 expression is reduced fourfold in bzip67-1 seeds.
Figure 4.
Analysis of Gene Expression in Developing bzip67-1 Seeds.
Quantitative real-time-PCR analysis of expression was performed on selected genes. Values are the mean ± se of measurements performed on three separate RNA samples prepared from isolated stage 8 seeds of each genotype. 18S expression was used as a control for normalization. Asterisk denotes no significant difference from the wild type (WT) (P > 0.05).
A small but significant (P < 0.05) reduction in 18:1n9 level is also observed in bzip67 seeds (Figure 1A), and this cannot easily be explained by misregulation of FAD3 (Browse et al., 1993; O’Neill et al., 2011). Therefore, real-time RT-PCR was also used to measure the transcript abundance of several other important genes associated with 18:1n9 metabolism in stage 8 developing seeds (i.e., ROD1, FAD2, FAE1, and DGAT1). None of these genes appeared to be differentially expressed in whole siliques, based on our microarray data (see Supplemental Data Set 1 online). However, the transcript abundance of ROD1 was significantly higher (P < 0.05) in real-time RT-PCR experiments performed on seeds (Figure 4). ROD1 encodes a PC:diacylglycerol cholinephosphotransferase that provides FAD2 with 18:1n9-PC substrate for desaturation to 18:2n6-PC (Lu et al., 2009). Mutations in both ROD1 and FAD2 lead to an increase in the 18:1n9 content of seed oil (Okuley et al., 1994; Lu et al., 2009). It is therefore possible that increased expression of ROD1 might contribute to the lower level of 18:1n9 in bzip67 seeds.
Disruption of bZIP67 Also Reduces Seed Storage Protein Gene Expression
Microarray analysis on bzip67-1 siliques also revealed that, unlike storage lipid synthesis, many SSP genes and several late embryogenesis abundant (LEA) genes were significantly repressed (P < 0.01) at stage 8 (see Supplemental Data Set 1 online). In Arabidopsis seeds, four genes encode the major 12S globulins/cruciferins (CRU1/CRA1, CRB, CRU2, and CRU3/CRC) and five genes encode the 2S albumins (2S1 to 2S5) (Baud et al., 2008). The microarray data suggested that transcript abundance of all of them was reduced in bzip67-1, and in particular CRU2, 2S1, and 2S5 were downregulated more than 10-fold (see Supplemental Data Set 1 online). Ectopic expression of bZIP67, in combination with other seed-specific TFs, has been reported to activate CRU3 expression (Yamamoto et al., 2009). Real-time PCR analysis of the 12S globulins (including CRU3) and the 2S albumin genes in bzip67-1 stage 8 developing seeds confirmed that the transcript abundance of all nine genes is significantly (P < 0.05) reduced (Figure 4). Analysis of the bzip67-1 mutant therefore provides genetic evidence that bZIP67 plays a nonredundant physiological role in regulating SSP expression (Yamamoto et al., 2009).
bZIP67 Can Transactivate FAD3 in the Presence of L1L + NF-YC2
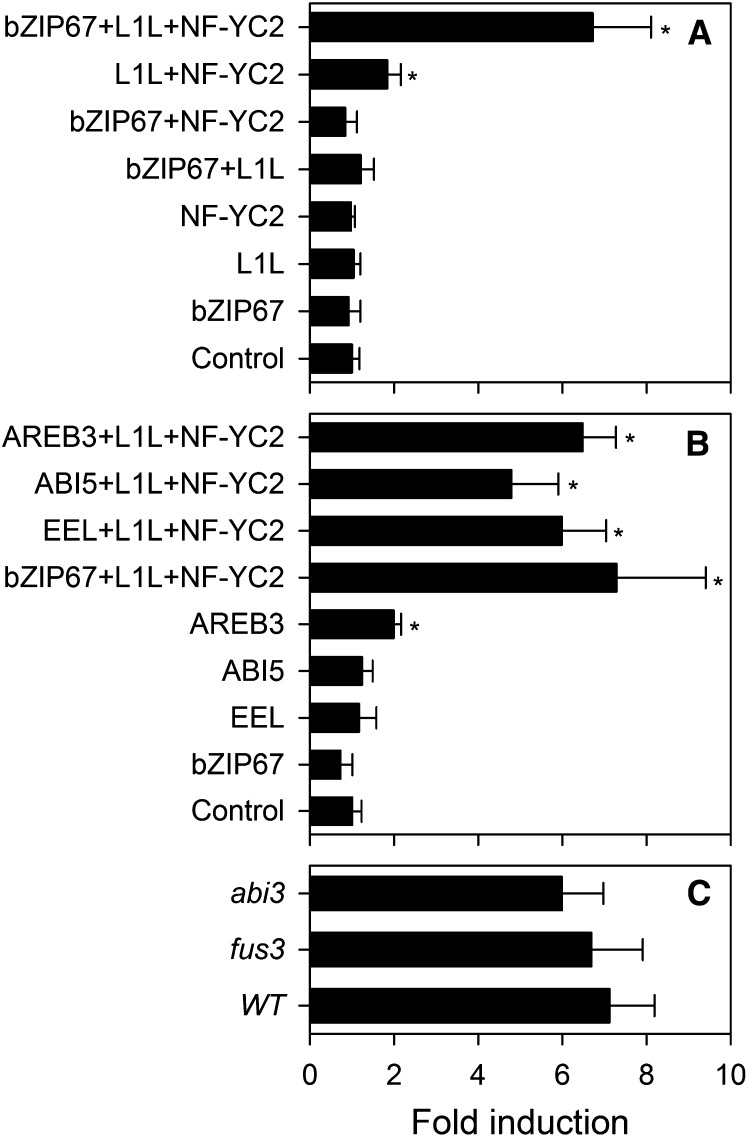
Yamamoto et al. (2009) have previously shown that several bZIP TFs from group A, including bZIP67, can transactivate the CRU3 SSP promoter in Arabidopsis protoplasts, but only when coexpressed with LEC1 (or L1L) and a NF-YC subunit, such as NF-YC2, which is seed specific. Our own analysis of the bzip67-1 mutant shows that bZIP67 is necessary for full CRU3 expression in stage 8 developing seeds (Figure 4). Furthermore, it is apparent from seed fatty acid profiling that both bzip67-1 and l1l-1 share common features; in particular, a substantial reduction in the level of 18:3n3 verses 18:2n6 in their seed oil (Figure 1A). We therefore decided to investigate whether bZIP67 might transactivate FAD3, together with L1L and NF-YC2. A 600-bp region of the FAD3 promoter was cloned upstream of β-glucuronidase (GUS), and the construct was transfected into Arabidopsis mesophyll protoplasts, alone and in combination with bZIP67, L1L, and NF-YC2 constructs driven by the 35S promoter (Yamamoto et al., 2009). Transfection with ProFAD3:GUS alone resulted in significant (P < 0.05) GUS activity, which is consistent with the fact that FAD3 is expressed and functions in vegetative tissues as well as in seeds (Browse et al., 1993). Cotransfection with bZIP67 individually did not enhance ProFAD3:GUS expression significantly (P > 0.05), while cotransfection with L1L plus NF-YC2 led to a twofold increase (Figure 5A). When bZIP67, L1L, and NF-YC2 were cotransfected, ProFAD3:GUS activity was enhanced more than sixfold (Figure 5A).
Figure 5.
Effect of bZIPs, L1L, and NF-YC2 on FAD3 Expression in the Wild Type, fus3-3, and abi3-7.
(A) Combined effect of bZIP67, L1L, and NF-YC2 transgenic expression in the wild type.
(B) Comparative effect of bZIP67, EEL, ABI5, and AREB3 transgenic expression in the wild type.
(C) Effect of bZIP67+L1L+NF-YC2 in abi3-7 and fus3-3. Mesophyll protoplasts were transfected with a FAD3 promoter-GUS reporter plasmid and effector plasmids for the indicated TFs. Transfected cells were cultured for 16 h and assayed for GUS and LUC activities. WT, the wild type.
Values are the mean ± se of measurements performed on three separate transfections and are normalized relative to LUC. Asterisk denotes a significant difference from the control (P < 0.05).
Other Group A bZIPs Can Also Transactivate FAD3 in the Presence of L1L + NF-YC2
ENHANCED EM LEVEL (EEL), ABSCISIC ACID-RESPONSIVE ELEMENT BINDING PROTEIN3 (AREB3), bZIP66, and ABI5 are group A bZIPs that are closely related to bZIP67 and are also expressed in maturing seeds (Bensmihen et al., 2005). To determine whether these TFs could also function as activators of ProFAD3:GUS, each was cotransfected together with L1L and NF-YC2. As with bZIP67, EEL, AREB3, and ABI5 could enhance FAD3 expression in the presence of L1L and NF-YC2 (Figure 5B). These data suggest that strong transactivation of FAD3 can potentially be achieved by the combined action of one of several group A bZIPs, L1L, and NF-YC2. Of the three additional group A bZIPs that are known to be expressed in developing seeds, EEL is the only one that is also more than fourfold upregulated in both LEC1-overexpressing seedlings and wild-type developing seeds (see Supplemental Table 1 online). However, analysis of a bzip67-1 eel-3 double mutant showed that disruption of EEL has no additional effect on seed fatty acid composition, suggesting that this gene is unlikely to contribute to the regulation of FAD3 in vivo (see Supplemental Figure 1C online).
FUS3 and ABI3 Are Not Required for Activation of FAD3 by bZIP67-L1L-NF-YC2
Mu et al. (2008) have shown that FUS3 (and to a lesser extent ABI3) are required for LEC1 induction of FAD3. However, FUS3 and ABI3 are also required for bZIP67 and L1L expression (Kagaya et al., 2005; Mu et al., 2008). Therefore, it is not clear whether FUS3 and ABI3 participate directly in transactivation of the FAD3 promoter or act through their control of bZIP67 and L1L expression. To investigate this question, cotransfection experiments were performed using mesophyll protoplasts from wild-type, fus3-3, and abi3-7 leaves. When bZIP67, L1L, and NF-YC2 were cotransfected together, ProFAD3:GUS activity was enhanced in all three genetic backgrounds (Figure 5C). Therefore, FUS3 and ABI3 are not directly required for induction of FAD3 by bZIP67-L1L-NF-YC2.
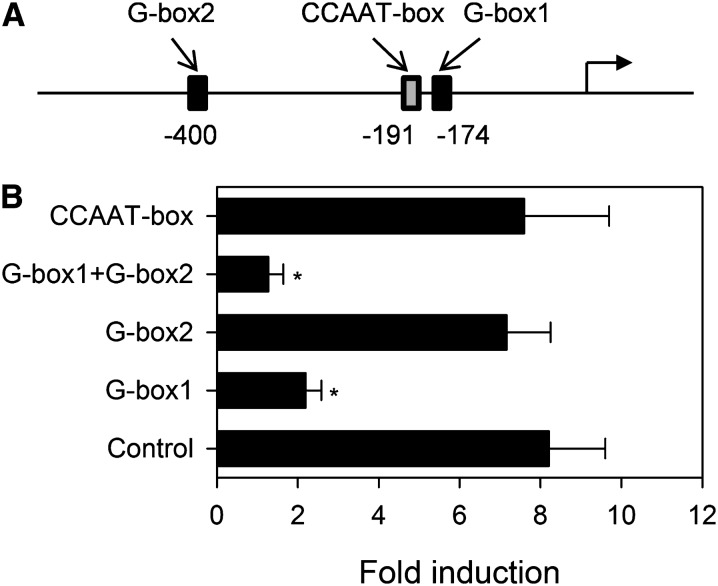
Transactivation of FAD3 by bZIP67-L1L-NF-YC2 Is Dependent on G-Box Elements
Transactivation of CRU3 by bZIP67, LEC1 (or L1L), and NF-YC2 has been shown to rely on G-box elements in the promoter (Yamamoto et al., 2009). The FAD3 promoter contains two such elements with an ACGT core within 600 bp upstream of the transcriptional start site, situated at −174 to −171 (G-box1) and −400 to −397 bp (G-box2) (Figure 6A; see Supplemental Figure 2 online). To investigate whether these G-boxes are required for transactivation of FAD3, the ACGT cores of the elements were systematically mutated in the ProFAD3:GUS construct, and its ability to drive GUS expression when cotransfected with bZIP67-L1L-NF-YC2 was investigated. Disruption of G-box1 reduced GUS expression by ∼80%, and mutations in both G-boxes effectively blocked transactivation altogether (Figure 6B). A putative CCAAT-box was also identified in the FAD3 promoter at −191 to −187 bp, in close proximity to G-box1, which could be a potential L1L binding site (Figure 6A). Disruption of this element had no significant (P > 0.05) effect on GUS expression (Figure 6B). bZIPs are known to interact with B3 domain proteins, such as ABI3, FUS3, and LEC2, which in turn can bind RY elements in SSP promoters (Nakamura et al., 2001). However, no putative RY elements (CATGCA[T/G]) were identified in the 600 bp FAD3 promoter (see Supplemental Figure 2 online).
Figure 6.
Effect of G-Box and CCAAT-Box Deletions on FAD3 Expression.
(A) Schematic of the 600-bp FAD3 promoter showing the position of the putative cis-elements. Positions correspond to the start of the elements with the distance upstream of the transcriptional start site marked in base pairs.
(B) Effect of disruption of the elements on gene expression. Mesophyll protoplasts were transfected with FAD3 promoter-GUS reporter plasmids containing mutations in the G-boxes (ACGT to AAGG) and CCAAT-box (CCAAT to CCTAT) and effector plasmids for bZIP67, L1L, and NF-YC2. Transfected cells were cultured for 16 h and were assayed for GUS and LUC activities. Values are the mean ± se of measurements performed on three separate transfections and are normalized relative to LUC. Asterisk denotes a significant difference from the control (P < 0.05).
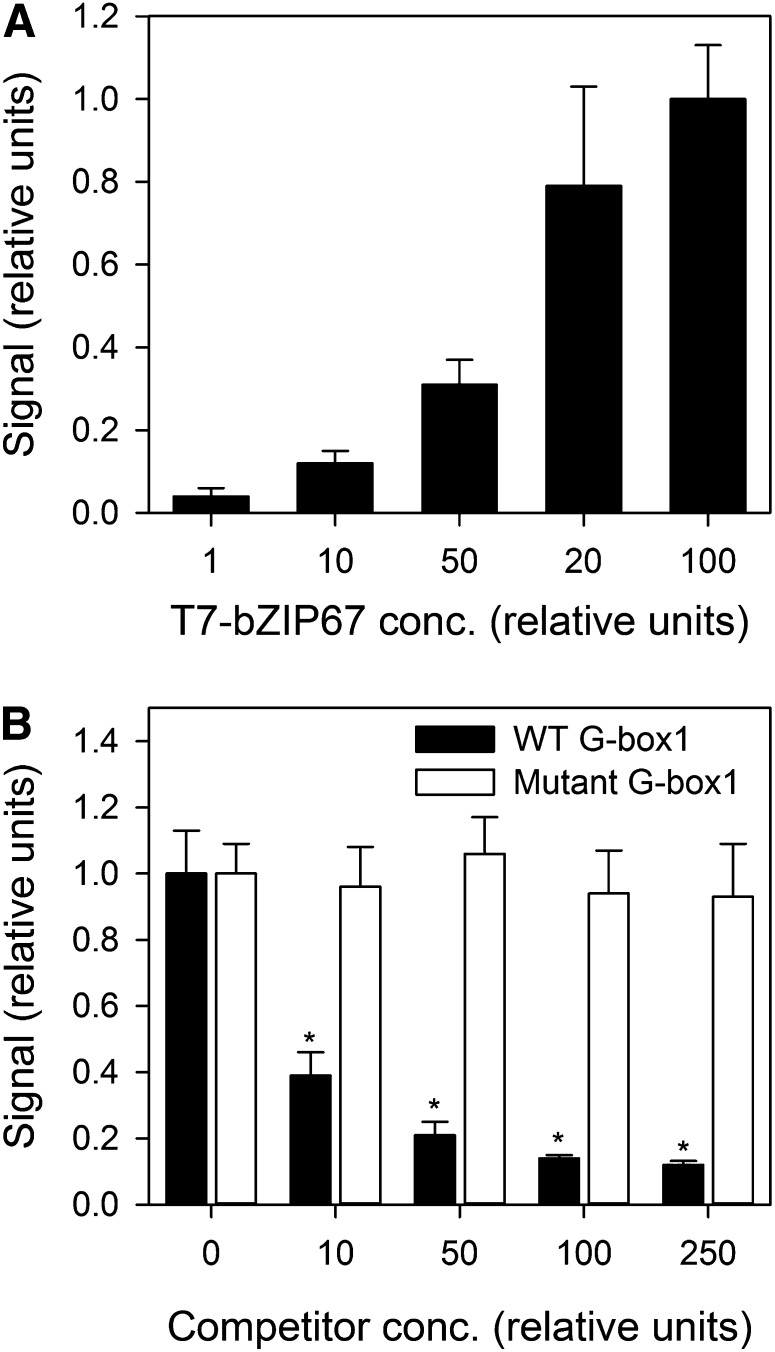
bZIP67 Can Bind FAD3 G-box1 in Vitro
Although it has been established that several members of bZIP group A can bind G-boxes in vitro, this has not been demonstrated previously for bZIP67 (Bensmihen et al., 2002; Kim et al., 2002). To confirm that bZIP67 is able to bind G-box1 from the FAD3 promoter, in vitro experiments were performed using an ELISA-based DNA binding assay (Alonso et al., 2009). Epitope-tagged recombinant bZIP67 was incubated with immobilized double-stranded DNA oligonucleotides containing the putative cis-element (G-box1), and binding was determined by immunodetection. The strength of the ELISA signal increased with the concentration of T7-tagged bZIP67 applied to the G-box1 oligonucleotide, indicating that the protein can bind to it (Figure 7A). Furthermore, in competition experiments, the addition of free G-box1 oligonucleotides could significantly suppress the ELISA signal (P < 0.05), while addition of G-box1 oligonucleotides with a mutated ACGT core could not (Figure 7B).
Figure 7.
In Vitro Determination of bZIP67 FAD3 Promoter Binding Using an ELISA-Based Assay.
(A) Biotinylated oligonucleotide containing the FAD3 G-box1 sequence was bound to streptavidin-coated wells and incubated with increasing amounts of a T7-tagged bZIP67 protein expressed in relative units. Unbound proteins were removed, and the amount of T7-bZIP67 protein was quantified by immunodetection with an anti-T7 antibody.
(B) The binding specificity of bZIP67 to G-box1 was analyzed in competition experiments where increasing amounts of unlabeled competitor containing a wild-type (WT) or a mutated G-box1 (ACGT to AAGG) were incubated with a fixed amount of the T7-bZIP67 protein and the biotinylated wild-type G-box1. Asterisk denotes a significant difference from the wild type (P < 0.05).
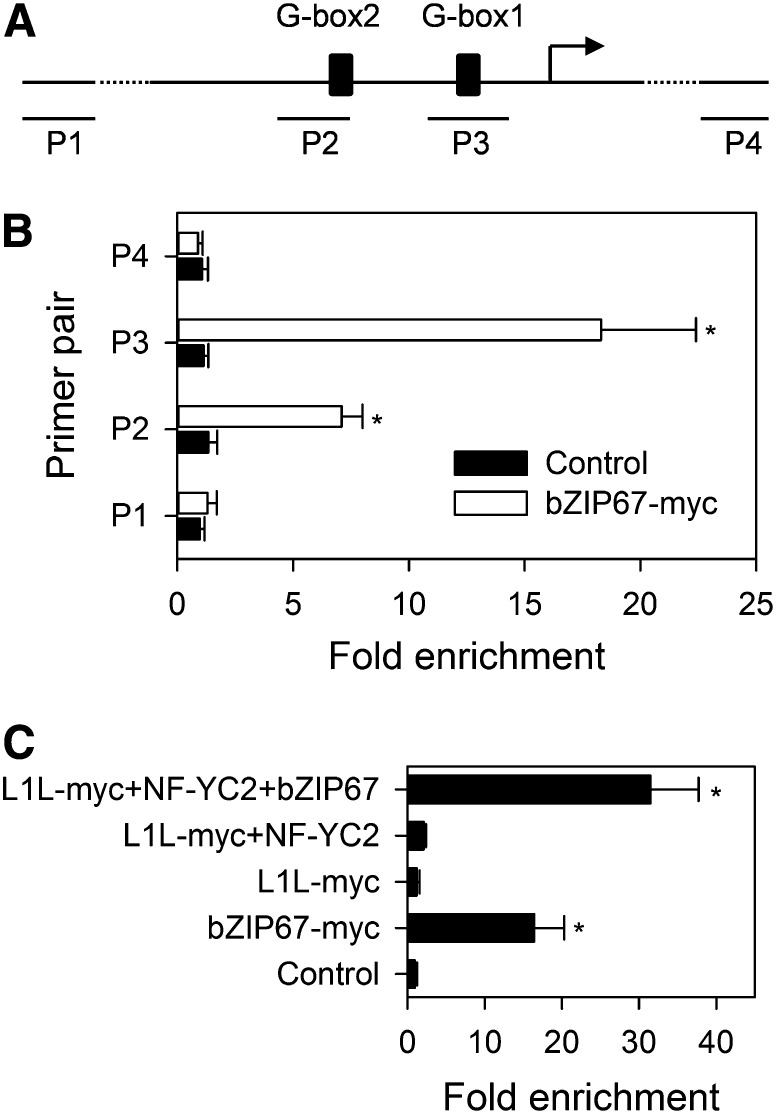
In Vivo Association of L1L-NF-YC2 with the FAD3 Promoter Requires bZIP67 Binding
Yamamoto et al. (2009) have shown via coimmunoprecipitation experiments that L1L and bZIP67 associate with one another in vivo. However, binding with the CRU3 promoter was not demonstrated directly. To investigate whether L1L, NF-YC2, and bZIP67 can bind the FAD3 promoter, in vivo chromatin immunoprecipitation (ChIP) experiments were performed on transfected Arabidopsis mesophyll protoplasts using the method of Lee et al. (2007). Constructs containing either myc-tagged or untagged bZIP67, L1L, and NF-YC2 were transfected into protoplasts, and after 24 h, chromatin complexes were cross-linked using formaldehyde. After shearing by sonication, the fragmented chromatin was incubated with monoclonal anti-myc antibodies and immunoprecipitated complexes were captured using magnetic protein G beads. DNA eluted from the beads was analyzed by real-time PCR using primers corresponding to regions of the FAD3 promoter and a negative control gene (Figure 8A; see Supplemental Table 4 online).
Figure 8.
In Vivo Determination of FAD3 Promoter Binding by bZIP67, L1L, and NF-YC2 Using Chromatin Immunoprecipitation.
(A) Schematic showing the positions of the PCR amplicons (P1, P2, P3, and P4) from the FAD3 promoter and surrounding DNA.
(B) Relative enrichment of DNA fragments detected using PCR amplicons depicted in (A).
(C) Relative enrichment of DNA fragments detected using P3. Mesophyll protoplasts were transfected with myc-tagged or untagged effector plasmids for bZIP67, L1L, and NF-YC2, as indicated. Real-time PCR was used to determine the fold enrichment of immunoprecipitated DNA fragments. Asterisk denotes a significant (P < 0.05) difference from the control (ACTIN7).
PCR performed on immunoprecipitated chromatin from bZIP67-myc transfected protoplasts indicated that promoter regions of FAD3 containing G-box1 and G-box2 are significantly enriched (P < 0.05) in comparison to the controls, with the greatest enrichment at G-box1 (Figure 8B). Very little enrichment of the G-box1–specific product was obtained when protoplasts were transfected with L1L-myc[NF-YC2]. However, when protoplasts were cotransfected with untagged bZIP67 and L1L-myc[NF-YC2], a substantial enrichment of the FAD3 promoter G-box1 region was again observed (Figure 8C). These data suggest that bZIP67 preferentially binds the FAD3 promoter in vivo at G-box1 and that binding enables L1L-NF-YC2 to associate with the DNA/protein complex, most likely via protein–protein interaction between L1L and bZIP67 (Yamamoto et al., 2009).
bZIP67 Expression Does Not Limit 18:3n3 Production in Wild-Type Seeds
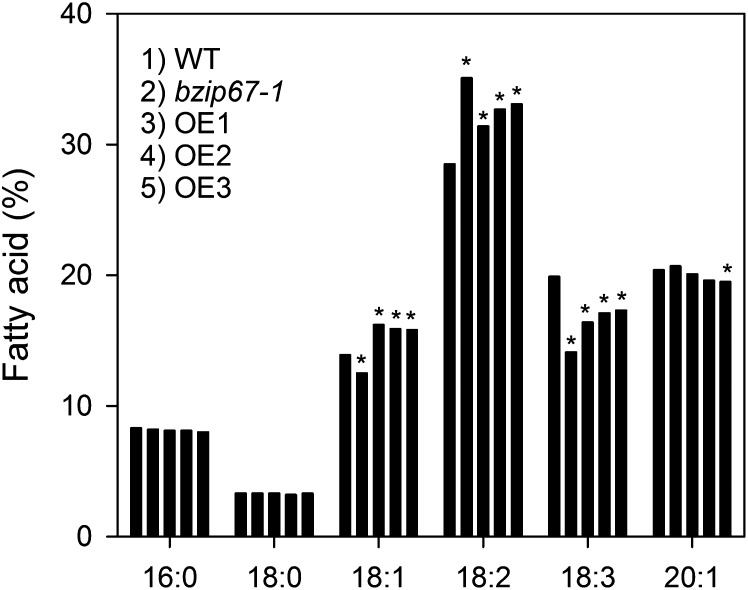
Our data suggest that bZIP67 is required for full activation of FAD3 expression and 18:3n3 production in developing seeds but that additional proteins, including L1L (or LEC1) and an NF-YC subunit, are also necessary. We expressed bZIP67 in wild-type plants under the control of a strong seed-specific promoter (glycinin) to investigate whether the availability of bZIP67 is limiting for 18:3n3 production in seeds. Multiple independent transgenic lines were isolated, and the fatty acid compositions of T3 homozygous seeds from three separate lines with the strongest effect are shown in Figure 9. The fatty acid composition of bZIP67 overexpressors was significantly different (P < 0.05) from that of the wild type. In the case of 18:1n9, levels were increased to values significantly greater than the wild type (P < 0.05), consistent with a predicted gain-of-function effect. However, 18:3n3 levels were not increased and were actually significantly decreased, relative to the wild type (P < 0.05). The total fatty acid, protein, and fresh weight of seeds from overexpressor lines was not altered (see Supplemental Table 3 online), and the plants have no obvious morphological phenotypes. These data confirm that changes in the level of bZIP67 expression do affect storage oil composition. However, the alterations caused by reduction and increase in bZIP67 expression do not simply mirror each other. The data can best be explained by a model in which bZIP67 abundance is colimiting for 18:3n3 production and where changes influence multiple protein and DNA interactions, which give rise to both positive and negative effects on gene expression.
Figure 9.
Effect of bZIP67 Overexpression on Seed Fatty Acid Composition.
Values are the mean of measurements of seeds from six plants of each genotype. All se are <1% of the mean. For each fatty acid, the columns from left to right correspond to the numbered genotypes (1 to 5). Wild-type (Col-0) plants were transformed with a construct containing bZIP67 under the control of the glycinin promoter, and three independent homozygous T3 lines with the strongest effect were selected and analyzed (OE1-3). Asterisk denotes a significant difference from the wild type (WT) (P < 0.05).
DISCUSSION
Seed storage reserves not only serve as a major source of calories in the human diet but also provide essential metabolic precursors that we lack the capacity to synthesize for ourselves. The ω-3 polyunsaturated fatty acid α-linolenic acid (18:3n3) is an important example. The enzyme primarily responsible for the production of 18:3n3 in seeds is the microsomal ω-3 fatty acid desaturase FAD3 (Browse et al., 1993). In Arabidopsis seeds, the expression level of FAD3 determines the abundance of the fatty acid in storage oil (Puttick et al., 2009; O’Neill et al., 2011), but the precise molecular mechanism for the transcriptional regulation of FAD3 is not known. In this study, we addressed this question by identifying three components of a transcriptional complex that governs FAD3 expression in developing seeds.
We used a targeted reverse genetic screen to identify five TFs that are induced during embryo maturation, act downstream of LEC1, and lead to significant changes in seed fatty acid profile when they are knocked out. The fatty acid profiles of three mutants exhibited a unifying feature. Namely, fus3, l1l, and bzip67 all share a characteristic reduction in the ratio of 18:3n3 to 18:2n6, potentially implicating them in the regulation of FAD3. Disruption of FUS3 is known to affect seed lipid metabolism and block FAD3 expression (Mu et al., 2008). However, l1l and bzip67 have not been shown to exhibit a seed lipid phenotype previously, and in the case of bzip67, no phenotype of any kind has so far been ascribed to the mutant (Bensmihen et al., 2005; Le et al., 2010). This is despite models that place bZIP67 near the center of gene regulatory networks that govern seed maturation (Belmonte et al., 2013). Given this fact, we focused primarily on characterizing bzip67.
Microarray and real-time PCR experiments performed on developing siliques and seeds of bzip67-1 confirmed that FAD3 is downregulated, while the expression of many other genes involved in lipid metabolism is unaffected. Disruption of bZIP67 also had a pronounced negative impact on the expression of all the major SSP genes. Interestingly, Yamamoto et al. (2009) have shown that bZIP TFs from group A, including bZIP67, can transactivate the CRU3 SSP promoter in Arabidopsis protoplasts, but only when coexpressed with LEC1 (or L1L) and a NF-YC subunit such as NF-YC2. Given that l1l and bzip67 share a reduction in the ratio of 18:3n3 to 18:2n6, we hypothesized that FAD3 might also be regulated by the cooperative action of these three proteins. Using a combination of in vivo and in vitro assays, we show that bZIP67 binds G-box cis-elements in the FAD3 promoter and enhances FAD3 expression but that DNA binding and enhanced transcription are conditional on bZIP67 association with L1L and NF-YC2. Furthermore, although FUS3 (and to a lesser extent ABI3) are known to be required for L1L, bZIP67, and FAD3 expression during seed maturation (Mu et al., 2008), using fus3 and abi3 null backgrounds, we show that these proteins need not participate directly in transactivation of FAD3 by [L1L:NF-YC2:bZIP67].
Based on our data, we propose a model (Figure 10) whereby LEC1 specifies high levels of 18:3n3 production during Arabidopsis seed development by inducing FUS3 which then, either independently or cooperatively with LEC1, triggers the induction of L1L, NF-YC2, and bZIP67. The latter three gene products then combine to form part of a transcriptional complex that binds the FAD3 promoter via bZIP67 interaction with G-boxes and drives FAD3 expression. It is likely that some redundancy exists among the isoforms that make up the complex, since l1l and bzip67 seeds only show a partial reduction in 18:3n3 content and nf-yc2 does not exhibit a significant change in fatty acid composition (Figure 1).
Figure 10.
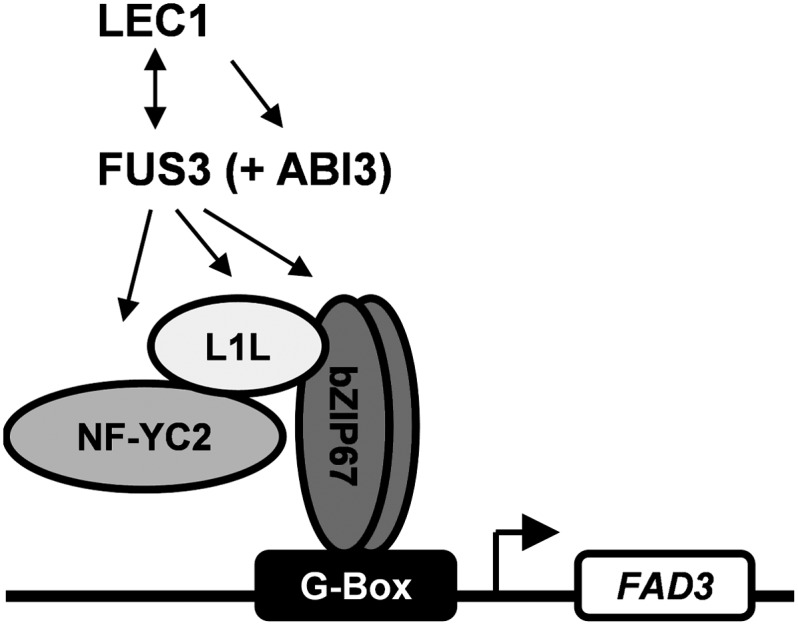
A Model Illustrating the Proposed Role of LEC1, L1L, FUS3, ABI3, NF-YC2, and bZIP67 in the Transcriptional Regulation of FAD3 during Arabidopsis Embryo Maturation.
Loss- and gain-of-function genetic studies have established that L1L, FUS3, ABI3, NF-YC2, bZIP67, and FAD3 expression is induced by LEC1 (Mu et al., 2008). FAD3 expression is also dependent on FUS3 and ABI3 (Mu et al., 2008). Here, we show that bZIP67 binds the FAD3 promoter via G-box elements and that trans-activation depends on complex formation between bZIP67, L1L, and NF-YC2 (and not ABI3 and FUS3). Analysis of l1l seeds suggests that the complex contains L1L rather than LEC1. This would be consistent with expression data that show that L1L is expressed more strongly in embryonic tissues than LEC1 during the stages when FAD3 is also expressed (Winter et al., 2007).
LEC1 and L1L are considered to be functionally equivalent (Kwong et al., 2003), and the complex may potentially contain either protein. However, the temporal and spatial patterns of expression of the two genes during seed development differs (Kwong et al., 2003), and while LEC1 expression peaks sharply in heart stage embryos (stage 4) and also endosperm, L1L is most strongly expressed in torpedo and walking stick stage embryos (stage 6-7), when both bZIP67 and FAD3 expression are also approaching their peak (see Supplemental Figure 3 online; Winter et al., 2007). Other group A bZIPs, in addition to bZIP67, may also form part of the complex that regulates FAD3 expression. We show that EEL, AREB3, and ABI5 can also transactivate FAD3 expression in combination with L1L and NF-YC2. EEL is the only one of these three genes that has a similar temporal pattern of expression during seed development to bZIP67 (Winter et al., 2007) and is also more than fourfold induced by ectopic expression of LEC1 (Mu et al., 2008). However, analysis of the bzip67-1 eel-3 double mutant seed suggested that EEL does not participate in the regulation of fatty acid composition. Bensmihen et al. (2005) reported that simultaneous RNA interference–targeted suppression of bZIP67, EEL, and AREB3 leads to no macroscopic changes in seed morphology, but no analysis of storage reserves was described.
Although we show that bZIP67 plays a specific role in regulating the level of 18:3n3 in seeds through its regulation of FAD3, it is apparent from our analysis of bzip67 mutants and bZIP67 overexpressor lines that manipulating the abundance of this TF has broader and more complex effects on lipid metabolism. Seeds of bzip67 contain slightly less 18:1n9 as well as less 18:3n3, and this cannot be explained simply by misregulation of FAD3 (Puttick et al., 2009; O’Neill et al., 2011). Real-time PCR experiments performed on developing seeds suggest that ROD1 is upregulated in bzip67, and this might explain the low 18:1n9 content, since the rod1 mutant has elevated 18:1n9 levels (Lu et al., 2009). Interestingly, ROD1 expression peaks earlier in seed development than bZIP67 and the transcript abundance of the two genes is inversely correlated (Winter et al., 2007). To et al. (2012) recently showed that ROD1 expression is reduced in the wri1 wri3 wri4 mutant, suggesting that WRI family TFs regulate this gene. How bZIP67 represses ROD1 expression is currently unclear. However, it is possible that bZIP67 might block the interaction of other TFs (such as WRI1) with the ROD1 promoter via either DNA or protein binding. Group A bZIPs are thought to be capable of forming dimers with one another (Deppmann et al., 2004), and both negative and positive regulatory roles have been shown previously. For example, the abi5 mutant has reduced expression of the LEA genes EM1 and EM6, while expression of these genes is enhanced in the eel mutant (Finkelstein and Lynch, 2000; Bensmihen et al., 2002). Overexpression of bZIP67 in wild-type plants leads to an increase in 18:1n9, but, surprisingly, 18:3n3 is not increased and is actually slightly decreased. This result suggests that bZIP67 is not the rate-limiting component for trans-activation of FAD3 expression. Indeed, an overabundance of bZIP67 might even inhibit FAD3 expression by sequestering other components of the complex. Interestingly, the fatty acid profile of l1l seeds mimics that of bZIP67 overexpression with increased 18:1n9 and decreased 18:3n3, so L1L function might be negatively affected (Figures 1A and 9).
The observation that all major SSP genes are substantially downregulated in stage 8 developing bzip67 seeds supports a physiological role of this TF in regulating SSP expression (Yamamoto et al., 2009) as well as FAD3. Although abi5 and eel are affected in the expression of certain LEA genes, such as EM1 and EM6 (Finkelstein and Lynch, 2000; Bensmihen et al., 2002), these genes do not appear to be misregulated in bzip67, while some other LEA genes are affected (see Supplemental Data Set 1 online). These data suggest some specificity of function for bZIP67, ABI5, and EEL. The impact of bzip67 on SSP expression does not translate into a reduced level of total protein in mature seeds. However, SSPs are still expressed in bzip67-1. It is probable that other TFs partially compensate for a defect in bZIP67. Other group A bZIPs might contribute in this instance (Yamamoto et al., 2009). However, bZIP53 (from group S1) has also been reported to activate CRU3 via G-box binding as a heterodimer with group B bZIPs, and a bzip53 mutant was also shown to have reduced CRU3 and 2S2 expression during seed development (Alonso et al., 2009). It appears that bZIP53 is not induced by ectopic expression of LEC1 based on microarray analysis (Mu et al., 2008) and was therefore not investigated in this study. Interestingly, severe disruption of SSPs in soybean (Glycine max) seeds does not alter the protein content of the seed because other proteins are produced to compensate (Schmidt et al., 2011). It is therefore possible that detailed proteomic analysis of seeds from mutants such as bzip53 and bzip67 might reveal quantitative differences in specific SSPs. In addition to FAD3 and SSPs, our microarray data suggest that several other genes with well-defined developmental and metabolic functions in seeds might be positively regulated by bZIP67, and these await further study (see Supplemental Data Set 1 online). Among the genes is SUCROSE SYNTHASE2, which Yamamoto et al. (2009) previously showed could be activated by [L1L:NF-YC2:bZIP67]. We investigated two previously characterized T-DNA mutants in this gene (Bieniawska et al., 2007) but did not observe any effect on seed fatty acid composition.
In conclusion, we show that the LEC1-inducible transcriptional regulators L1L and bZIP67 both play a significant and nonredundant role in storage reserve accumulation during Arabidopsis seed maturation. Focusing on the regulation of storage oil composition, we demonstrate that bZIP67 is required for FAD3 expression and, thus, the production of the essential fatty acid 18:3n3, which is a major component of many seed oils. We show that bZIP67 binds the FAD3 promoter directly via interaction with a G-box but that bZIP67 also requires both L1L and a NF-YC subunit, such as the seed-specific NF-YC2, to drive gene expression. LEC1 induction of bZIP67, L1L, and NF-YC2 is known to require FUS3 and ABI3, but we show that neither protein is directly necessary for the protein complex to trans-activate FAD3. Hence, we provide a model to explain how the production of 18:3n3 for incorporation into seed oil is ultimately coupled to embryogenesis in Arabidopsis. Further work will be required to address how the expression of other important structural genes involved in fatty acid modification and TAG assembly are integrated with the embryogenesis developmental program.
METHODS
Plant Material and Growth Conditions
All T-DNA insertion mutants were identified on the SIGnAL T-DNA Express website (http://signal.salk.edu/cgi-bin/tdnaexpress), and seeds were obtained from the European Arabidopsis Stock Centre (NASC; University of Nottingham, UK). All mutants were from the SALK, GABI-Kat, or SAIL collections, and the appropriate wild-type genetic backgrounds were used for comparison. The bZIP67p:GFP-bZIP67 reporter line used in this study is described by Bensmihen et al. (2005). For plant growth experiments, the seeds were sown on moist Levingtons F2 compost in 7-cm2 pots, and the pots were stored in the dark for 4 d before being transferred to a growth chamber set to 21°C, 70% relative humidity (16 h light [22°C]/8 h dark [18°C]; PPFD = 150 µmol m−2 s−1). The genotype of the T-DNA lines was confirmed by genomic PCR using gene-specific primers in combination with T-DNA left and right border primers (Alonso et al., 2003). Primers are listed in Supplemental Table 4 online.
Metabolite Measurements
The fatty acid content and composition of seeds was measured by gas chromatographic analysis after combined digestion and fatty acid methyl ester formation using the method of Browse et al. (1986). Pentadecanoin (Sigma-Aldrich) was added to the samples as an internal control. Seed total protein and storage protein content were determined using the methods of Baud et al. (2002). TAG and phospholipid analyses were performed on developing seeds (between stages 7 and 9; Winter et al., 2007) and mature dry seeds using a 4000 QTRAP LC-MS/MS (ABSiex). Seeds were ground in 1 mL of ethanol/chloroform (9:1 v/v), shaken for 1 h, and centrifuged for 2 min at 3500 rpm, and the supernatant was transferred to a new vial and dried down under N2. The lipids were dissolved in chloroform/methanol/0.3 M ammonium acetate (300:665:35 v/v) for phospholipid analysis following the methods described by Devaiah et al. (2006). For TAG, the lipids were dissolved in IMAD buffer (isopropyl alcohol/methanol/50 mM ammonium acetate/dichloromethane [4:3:2:1 v/v]), and analysis was performed following the method of Krank et al. (2007), except that 12 periods of 2 min were used for neutral loss scans. The data were subsequently analyzed using Lipidview (V1.1) software.
Gene Expression Analysis
DNase-treated total RNA was isolated from developing siliques and seeds using the RNeasy kit from Qiagen with modifications described by Wu et al. (2002). The synthesis of single-stranded cDNA was performed using SuperScript II RNase H- reverse transcriptase from Invitrogen. Real-time PCR was performed in a MyiQ Single-Color real-time PCR detection system (Bio-Rad) using qPCR Mastermix Plus from Eurogentec. Data were analyzed using Bio-Rad iQ5, Optical System Software, version 2.0. The primer pairs used for real-time PCR are listed in Supplemental Table 4 online. Microarray analysis was performed by NASC using the Affymetrix Ath1 chip, according to the manufacturer’s protocols. Raw data were normalized by MAS5 (www.Affymetrix.com) to a target signal of 500. P and Q values for the comparison of genotypes were calculated using Extraction of Differential Gene Expression software (http://www.genomine.org/edge/), and a P = 0.1 cutoff was used to select the genes that are differentially regulated (Liu and Howell, 2010). All key findings were replicated in independent experiments using real-time PCR. The microarray data (NASCARRAYS-606) are publically available at the NASC website (http://Arabidopsis.info/).
In Vitro Protein-DNA Binding Assays
Protein-DNA binding assays were performed using the method of Alonso et al. (2009), with the following modifications. A cDNA encoding bZIP67 was cloned into pET23a (Novagen), and the T7 epitope–tagged protein was expressed in Escherichia coli. Biotinylated complementary oligonucleotides derived from the FAD3 promoter G-box1 sequence (5′-GTTTTCACTTAACAACGTAACCAAAAGTATTAAAC-3′ and 5′-GTTTAATACTTTTGGTTACGTTGTTAAGTGAAAAC-3′) or a mutated version (5′-GTTTTCACTTAACAAaGgAACCAAAAGTATTAAAC-3′ and 5′- GTTTAATACTTTTGGTTcCtTTGTTAAGTGAAAAC-3′) were annealed and used for binding assays.
Transient Expression and Chromatin Immunoprecipitation
Arabidopsis thaliana mesophyll protoplasts were prepared, transfected, and cultured according to the methods of Yoo et al. (2007). After polyethylene glycol–calcium transfection with plasmid DNA carrying reporter gene constructs (ProFAD3:GUS and Pro35S:LUC) and combinations of effector constructs (Pro35S:bZIP67, Pro35S:EEL, Pro35S:ABI5, Pro35S:AREB3, Pro35S:L1L, and Pro35S:NF-YC2), cells were cultured for 16 h before luciferase (LUC) and GUS activities were determined using methods described by Yamamoto et al. (2009).
For ChIP assays, Arabidopsis mesophyll protoplasts were transfected with various combinations of Pro35S:bZIP67-myc, Pro35S:L1L-myc, and Pro35S:NF-YC2. After 24 h, protoplasts were harvested and ChIP assays were conducted following the procedure of Lee et al. (2007), with minor modifications. After formaldehyde fixation, the chromatin of the protoplasts was isolated and extensively sheared by sonication to obtain fragment sizes between 300 and 400 bp. Rat anti-myc monoclonal antibodies (Roche) and Dynabeads Protein G magnetic beads (Invitrogen) were used to immunoprecipitate the genomic fragments. Real-time PCR was performed on the immunoprecipitated DNA as described above using primer sets corresponding to four regions of FAD3 and a control gene (ACTIN7) and were corrected for their individual PCR amplification efficiencies. The primer pairs are listed in Supplemental Table 4 online.
Creation of DNA Constructs and Arabidopsis Transformation
bZIP67 was amplified by PCR from cDNA using primer pairs 5′-CGTCTAGAATGTCGGTTTTCGAATCGGAGAC-3′ and 5′-CGCCCGGGTTACCACCCGGCACTGGCCATCCTC-3′. The product was cloned into the TA vector pCR2.1 (Invitrogen) following the manufacturer’s instructions. The gene was then excised using XbaI and SmaI (restriction sites underlined) and inserted into the destination vector pBinGlyRed3 using T4 DNA ligase. The construct was transformed into Agrobacterium tumefaciens strain GV3101 by heat shock and subsequently into bzip67-1 using the floral dip method (Clough and Bent, 1998). Transformed seeds were identified by detection of the DsRed marker using fluorescence microscopy.
Statistical Analysis
One-way analysis of variance was used to assess the differences between genotypes or treatments. Following significant (P < 0.05) F-test results, means of interest were compared using the appropriate LSD value at the 5% (P = 0.05) level of significance, on the corresponding df. CVA was also used to assess overall differences in seed fatty acid composition between genotypes. As the data were compositional, account of this dependence was taken using a centering transformation. The GenStat (2011, 14th edition; VSN International) statistical system was used for these analyses.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: LEC1, At1g21970; L1L, At5g47670; ABI3, At3g24650; FUS3, At3g26790; bZIP67, At3g44460; WRI1, At3g54320; FAD3, At2g29980; ROD1, At3g15820; CRU1, At5g44120; CRB, At1g03880; CRU2, At1g03890; CRU3, At4g28520; 2S1, At4g27140; 2S2, At4g27150; 2S3, At4g27160; 2S4, At4g27170; 2S5, At5g54740; NF-YC2, At1g56170; ABI5, At2g36270; EEL, At2g41070; and AREB3, At3g56850. The microarray data (NASCARRAYS-606) are publically available at the NASC website (http://Arabidopsis.info/).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Seed Fatty Acid Composition of Various Genotypes.
Supplemental Figure 2. The FAD3 Promoter Region Used for Promoter-Reporter Studies.
Supplemental Figure 3. Temporal and Spatial Expression Patterns of LEC1, L1L, bZIP67, and FAD3 in Seeds.
Supplemental Table 1. A List of Putative Transcription Factors That Are >4-Fold Upregulated in LEC1-Overexpressing Seedlings and Maturing Wild-Type Embryos.
Supplemental Table 2. Composition of Polar Lipid Classes in Developing bzip67-1 Seeds.
Supplemental Table 3. Effect bZIP67 Disruption and Overexpression on Seed Weight and Storage Reserve Content.
Supplemental Table 4. List of Primers Used in This Study.
Supplemental Data Set 1. ATH1 Microarray Data from Developing Siliques of bzip67-1.
Acknowledgments
We thank Tsukaho Hattori for providing bZIP, LEC1, L1L, and NF-YC2 constructs for transient assays. François Parcy and Sandra Bensmihen kindly supplied the ProbZIP67:GFP-bZIP67 reporter line used for complementation of bzip67-1. We also thank Ed Cahoon for providing the pBinGlyRed3 vector for seed-specific expression. This work was funded by the UK Biotechnology and Biological Sciences Research Council through a studentship at the University of Warwick to A.M. and through an Institute Strategic Program grant at Rothamsted Research.
AUTHOR CONTRIBUTIONS
P.J.E. conceived and designed the experiments. A.M., A.A.K., H.v.E., E.S., S.K., and P.J.E. performed the experiments. S.J.P. carried out the statistical analysis. P.J.E. wrote the article.
Glossary
- TF
transcription factor
- SSP
seed storage protein
- TAG
triacylglycerol
- CVA
canonical variate analysis
- Col-0
Columbia-0
- PC
phosphatidylcholine
- ChIP
chromatin immunoprecipitation
- NASC
European Arabidopsis Stock Centre
References
- Alonso J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Alonso R., Oñate-Sánchez L., Weltmeier F., Ehlert A., Diaz I., Dietrich K., Vicente-Carbajosa J., Dröge-Laser W. (2009). A pivotal role of the basic leucine zipper transcription factor bZIP53 in the regulation of Arabidopsis seed maturation gene expression based on heterodimerization and protein complex formation. Plant Cell 21: 1747–1761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baud S., Boutin J.-P., Miquel M., Lepiniec L., Rochat C. (2002). An integrated overview of seed development in Arabidopsis thaliana ecotype WS. Plant Physiol. Biochem. 40: 151–160 [Google Scholar]
- Baud, S., Dubreucq, B., Miquel, M., Rochat, C., and Lepiniec, L. (2008). Storage reserve accumulation in Arabidopsis: Metabolic and developmental control of seed filling. In The Arabidopsis Book 6:e0113, /10.1199/tab.0113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baud S., Lepiniec L. (2010). Physiological and developmental regulation of seed oil production. Prog. Lipid Res. 49: 235–249 [DOI] [PubMed] [Google Scholar]
- Baud S., Mendoza M.S., To A., Harscoët E., Lepiniec L., Dubreucq B. (2007). WRINKLED1 specifies the regulatory action of LEAFY COTYLEDON2 towards fatty acid metabolism during seed maturation in Arabidopsis. Plant J. 50: 825–838 [DOI] [PubMed] [Google Scholar]
- Belmonte M.F., et al. (2013). Comprehensive developmental profiles of gene activity in regions and subregions of the Arabidopsis seed. Proc. Natl. Acad. Sci. USA 110: E435–E444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensmihen S., Giraudat J., Parcy F. (2005). Characterization of three homologous basic leucine zipper transcription factors (bZIP) of the ABI5 family during Arabidopsis thaliana embryo maturation. J. Exp. Bot. 56: 597–603 [DOI] [PubMed] [Google Scholar]
- Bensmihen S., Rippa S., Lambert G., Jublot D., Pautot V., Granier F., Giraudat J., Parcy F. (2002). The homologous ABI5 and EEL transcription factors function antagonistically to fine-tune gene expression during late embryogenesis. Plant Cell 14: 1391–1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieniawska, Z., Paul Barratt, D.H., Garlick, A.P., Thole, V., Kruger, N.J., Martin, C., Zrenner, R. and Smith, A.M. (2007). Analysis of the sucrose synthase gene family in Arabidopsis Plant J. 49: 810–828. [DOI] [PubMed]
- Browse J., McConn M., James D., Jr, Miquel M. (1993). Mutants of Arabidopsis deficient in the synthesis of alpha-linolenate. Biochemical and genetic characterization of the endoplasmic reticulum linoleoyl desaturase. J. Biol. Chem. 268: 16345–16351 [PubMed] [Google Scholar]
- Browse J., McCourt P.J., Somerville C.R. (1986). Fatty acid composition of leaf lipids determined after combined digestion and fatty acid methyl ester formation from fresh tissue. Anal. Biochem. 152: 141–145 [DOI] [PubMed] [Google Scholar]
- Cernac A., Benning C. (2004). WRINKLED1 encodes an AP2/EREB domain protein involved in the control of storage compound biosynthesis in Arabidopsis. Plant J. 40: 575–585 [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Deppmann C.D., Acharya A., Rishi V., Wobbes B., Smeekens S., Taparowsky E.J., Vinson C. (2004). Dimerization specificity of all 67 B-ZIP motifs in Arabidopsis thaliana: A comparison to Homo sapiens B-ZIP motifs. Nucleic Acids Res. 32: 3435–3445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaiah S.P., Roth M.R., Baughman E., Li M., Tamura P., Jeannotte R., Welti R., Wang X. (2006). Quantitative profiling of polar glycerolipid species from organs of wild-type Arabidopsis and a phospholipase Dalpha1 knockout mutant. Phytochemistry 67: 1907–1924 [DOI] [PubMed] [Google Scholar]
- Finkelstein R.R., Lynch T.J. (2000). The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell 12: 599–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein R.R., Somerville C.R. (1990). Three classes of abscisic acid (ABA)-insensitive mutations of Arabidopsis define genes that control overlapping subsets of ABA responses. Plant Physiol. 94: 1172–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Focks N., Benning C. (1998). wrinkled1: A novel, low-seed-oil mutant of Arabidopsis with a deficiency in the seed-specific regulation of carbohydrate metabolism. Plant Physiol. 118: 91–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraudat J., Hauge B.M., Valon C., Smalle J., Parcy F., Goodman H.M. (1992). Isolation of the Arabidopsis ABI3 gene by positional cloning. Plant Cell 4: 1251–1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham I.A. (2008). Seed storage oil mobilization. Annu. Rev. Plant Biol. 59: 115–142 [DOI] [PubMed] [Google Scholar]
- Jakoby, M., Weisshaar, B., Dröge-Laser, W., Vicente-Carbajosa, J., Tiedemann, J., Kroj, T., and Parcy, F.; bZIP Research Group (2002). bZIP transcription factors in Arabidopsis Trends Plant Sci. 7: 106–111. [DOI] [PubMed]
- Kagaya Y., Okuda R., Ban A., Toyoshima R., Tsutsumida K., Usui H., Yamamoto A., Hattori T. (2005). Indirect ABA-dependent regulation of seed storage protein genes by FUSCA3 transcription factor in Arabidopsis. Plant Cell Physiol. 46: 300–311 [DOI] [PubMed] [Google Scholar]
- Keith K., Kraml M., Dengler N.G., McCourt P. (1994). fusca3: A heterochronic mutation affecting late embryo development in Arabidopsis. Plant Cell 6: 589–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.Y., Ma J., Perret P., Li Z., Thomas T.L. (2002). Arabidopsis ABI5 subfamily members have distinct DNA-binding and transcriptional activities. Plant Physiol. 130: 688–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krank J., Murphy R.C., Barkley R.M., Duchoslav E., McAnoy A. (2007). Qualitative analysis and quantitative assessment of changes in neutral glycerol lipid molecular species within cells. Methods Enzymol. 432: 1–20 [DOI] [PubMed] [Google Scholar]
- Kroj T., Savino G., Valon C., Giraudat J., Parcy F. (2003). Regulation of storage protein gene expression in Arabidopsis. Development 130: 6065–6073 [DOI] [PubMed] [Google Scholar]
- Kwong R.W., Bui A.Q., Lee H., Kwong L.W., Fischer R.L., Goldberg R.B., Harada J.J. (2003). LEAFY COTYLEDON1-LIKE defines a class of regulators essential for embryo development. Plant Cell 15: 5–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le B.H., et al. (2010). Global analysis of gene activity during Arabidopsis seed development and identification of seed-specific transcription factors. Proc. Natl. Acad. Sci. USA 107: 8063–8070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H., Fischer R.L., Goldberg R.B., Harada J.J. (2003). Arabidopsis LEAFY COTYLEDON1 represents a functionally specialized subunit of the CCAAT binding transcription factor. Proc. Natl. Acad. Sci. USA 100: 2152–2156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.H., Yoo S.J., Park S.H., Hwang I., Lee J.S., Ahn J.H. (2007). Role of SVP in the control of flowering time by ambient temperature in Arabidopsis. Genes Dev. 21: 397–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leek J.T., Monsen E.C., Dabney A.R., Storey J.D. (2006). EDGE: Extraction and analysis of differential gene expression. Bioinformatics 22: 507–508 [DOI] [PubMed] [Google Scholar]
- Lemieux B., Miquel M., Somerville C., Browse J. (1990). Mutants of Arabidopsis with alterations in seed lipid fatty acid composition. Theor. Appl. Genet. 80: 234–240 [DOI] [PubMed] [Google Scholar]
- Li Y., Beisson F., Pollard M., Ohlrogge J. (2006). Oil content of Arabidopsis seeds: The influence of seed anatomy, light and plant-to-plant variation. Phytochemistry 67: 904–915 [DOI] [PubMed] [Google Scholar]
- Li-Beisson, Y., et al. (2013). Acyl-lipid metabolism. In The Arabidopsis Book 11:e0161, /10.1199/tab.0161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J.X., Howell S.H. (2010). bZIP28 and NF-Y transcription factors are activated by ER stress and assemble into a transcriptional complex to regulate stress response genes in Arabidopsis. Plant Cell 22: 782–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotan T., Ohto M., Yee K.M., West M.A., Lo R., Kwong R.W., Yamagishi K., Fischer R.L., Goldberg R.B., Harada J.J. (1998). Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell 93: 1195–1205 [DOI] [PubMed] [Google Scholar]
- Lu C., Napier J.A., Clemente T.E., Cahoon E.B. (2011). New frontiers in oilseed biotechnology: Meeting the global demand for vegetable oils for food, feed, biofuel, and industrial applications. Curr. Opin. Biotechnol. 22: 252–259 [DOI] [PubMed] [Google Scholar]
- Lu C., Xin Z., Ren Z., Miquel M., Browse J. (2009). An enzyme regulating triacylglycerol composition is encoded by the ROD1 gene of Arabidopsis. Proc. Natl. Acad. Sci. USA 106: 18837–18842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luerssen H., Kirik V., Herrmann P., Miséra S. (1998). FUSCA3 encodes a protein with a conserved VP1/AB13-like B3 domain which is of functional importance for the regulation of seed maturation in Arabidopsis thaliana. Plant J. 15: 755–764 [DOI] [PubMed] [Google Scholar]
- Meinke D.W. (1992). A homoeotic mutant of Arabidopsis thaliana with leafy cotyledons. Science 258: 1647–1650 [DOI] [PubMed] [Google Scholar]
- Mu J., Tan H., Zheng Q., Fu F., Liang Y., Zhang J., Yang X., Wang T., Chong K., Wang X.J., Zuo J. (2008). LEAFY COTYLEDON1 is a key regulator of fatty acid biosynthesis in Arabidopsis. Plant Physiol. 148: 1042–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura S., Lynch T.J., Finkelstein R.R. (2001). Physical interactions between ABA response loci of Arabidopsis. Plant J. 26: 627–635 [DOI] [PubMed] [Google Scholar]
- Okuley J., Lightner J., Feldmann K., Yadav N., Lark E., Browse J. (1994). Arabidopsis FAD2 gene encodes the enzyme that is essential for polyunsaturated lipid synthesis. Plant Cell 6: 147–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill C.M., Baker D., Bennett G., Clarke J., Bancroft I. (2011). Two high linolenic mutants of Arabidopsis thaliana contain megabase-scale genome duplications encompassing the FAD3 locus. Plant J. 68: 912–918 [DOI] [PubMed] [Google Scholar]
- O’Neill C.M., Gill S., Hobbs D., Morgan C., Bancroft I. (2003). Natural variation for seed oil composition in Arabidopsis thaliana. Phytochemistry 64: 1077–1090 [DOI] [PubMed] [Google Scholar]
- Puttick D., Dauk M., Lozinsky S., Smith M.A. (2009). Overexpression of a FAD3 desaturase increases synthesis of a polymethylene-interrupted dienoic fatty acid in seeds of Arabidopsis thaliana L. Lipids 44: 753–757 [DOI] [PubMed] [Google Scholar]
- Santos-Mendoza M., Dubreucq B., Baud S., Parcy F., Caboche M., Lepiniec L. (2008). Deciphering gene regulatory networks that control seed development and maturation in Arabidopsis. Plant J. 54: 608–620 [DOI] [PubMed] [Google Scholar]
- Santos Mendoza M., Dubreucq B., Miquel M., Caboche M., Lepiniec L. (2005). LEAFY COTYLEDON 2 activation is sufficient to trigger the accumulation of oil and seed specific mRNAs in Arabidopsis leaves. FEBS Lett. 579: 4666–4670 [DOI] [PubMed] [Google Scholar]
- Schmidt M.A., Barbazuk W.B., Sandford M., May G., Song Z., Zhou W., Nikolau B.J., Herman E.M. (2011). Silencing of soybean seed storage proteins results in a rebalanced protein composition preserving seed protein content without major collateral changes in the metabolome and transcriptome. Plant Physiol. 156: 330–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah S., Xin Z., Browse J. (1997). Overexpression of the FAD3 desaturase gene in a mutant of Arabidopsis. Plant Physiol. 114: 1533–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone S.L., Braybrook S.A., Paula S.L., Kwong L.W., Meuser J., Pelletier J., Hsieh T.F., Fischer R.L., Goldberg R.B., Harada J.J. (2008). Arabidopsis LEAFY COTYLEDON2 induces maturation traits and auxin activity: Implications for somatic embryogenesis. Proc. Natl. Acad. Sci. USA 105: 3151–3156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone S.L., Kwong L.W., Yee K.M., Pelletier J., Lepiniec L., Fischer R.L., Goldberg R.B., Harada J.J. (2001). LEAFY COTYLEDON2 encodes a B3 domain transcription factor that induces embryo development. Proc. Natl. Acad. Sci. USA 98: 11806–11811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- To A., Joubès J., Barthole G., Lécureuil A., Scagnelli A., Jasinski S., Lepiniec L., Baud S. (2012). WRINKLED transcription factors orchestrate tissue-specific regulation of fatty acid biosynthesis in Arabidopsis. Plant Cell 24: 5007–5023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente-Carbajosa J., Carbonero P. (2005). Seed maturation: Developing an intrusive phase to accomplish a quiescent state. Int. J. Dev. Biol. 49: 645–651 [DOI] [PubMed] [Google Scholar]
- West M., Yee K.M., Danao J., Zimmerman J.L., Fischer R.L., Goldberg R.B., Harada J.J. (1994). LEAFY COTYLEDON1 is an essential regulator of late embryogenesis and cotyledon identity in Arabidopsis. Plant Cell 6: 1731–1745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter D., Vinegar B., Nahal H., Ammar R., Wilson G.V., Provart N.J. (2007). An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Llewellyn D.J., Dennis E.S. (2002). A quick and easy method for isolating good quality RNA from cotton (Gossypium hirsutum L.) tissues. Plant Mol. Biol. Rep. 20: 213–218 [Google Scholar]
- Yamamoto A., Kagaya Y., Toyoshima R., Kagaya M., Takeda S., Hattori T. (2009). Arabidopsis NF-YB subunits LEC1 and LEC1-LIKE activate transcription by interacting with seed-specific ABRE-binding factors. Plant J. 58: 843–856 [DOI] [PubMed] [Google Scholar]
- Yoo S.D., Cho Y.H., Sheen J. (2007). Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2: 1565–1572 [DOI] [PubMed] [Google Scholar]