A triple mutant of MYC2, MYC3, and MYC4 transcription factors is devoid of glucosinolates and more sensitive to attack by a generalist insect. These MYCs physically interact with six MYB transcription factors (MYB28, MYB29, MYB34, MYB51, MYB76, and MYB122) and regulate both basal and inducible glucosinolate levels by directly activating the expression of glucosinolate biosynthesis genes.
Abstract
Arabidopsis thaliana plants fend off insect attack by constitutive and inducible production of toxic metabolites, such as glucosinolates (GSs). A triple mutant lacking MYC2, MYC3, and MYC4, three basic helix-loop-helix transcription factors that are known to additively control jasmonate-related defense responses, was shown to have a highly reduced expression of GS biosynthesis genes. The myc2 myc3 myc4 (myc234) triple mutant was almost completely devoid of GS and was extremely susceptible to the generalist herbivore Spodoptera littoralis. On the contrary, the specialist Pieris brassicae was unaffected by the presence of GS and preferred to feed on wild-type plants. In addition, lack of GS in myc234 drastically modified S. littoralis feeding behavior. Surprisingly, the expression of MYB factors known to regulate GS biosynthesis genes was not altered in myc234, suggesting that MYC2/MYC3/MYC4 are necessary for direct transcriptional activation of GS biosynthesis genes. To support this, chromatin immunoprecipitation analysis showed that MYC2 binds directly to the promoter of several GS biosynthesis genes in vivo. Furthermore, yeast two-hybrid and pull-down experiments indicated that MYC2/MYC3/MYC4 interact directly with GS-related MYBs. This specific MYC–MYB interaction plays a crucial role in the regulation of defense secondary metabolite production and underlines the importance of GS in shaping plant interactions with adapted and nonadapted herbivores.
INTRODUCTION
Plants resist attack by herbivorous insects with a combination of constitutive and inducible defenses. In addition to the presence of physical barriers like cuticle, spines, or hairs, plants produce defense proteins and secondary metabolites that are toxic to arthropods (Howe and Jander, 2008). Glucosinolates (GSs) constitute a class of nitrogen- and sulfur-containing thioglucosides characteristic of the Brassicaceae family. During insect feeding, GSs get mixed with specific β-glucosidases termed myrosinases, releasing an aglucone that spontaneously rearranges into toxic thiocyanates, isothiocyanates, or nitriles (Bones and Rossiter, 2006). GSs are classified into aliphatic, aromatic, and indole GSs depending on the origin of the amino acid–derived side chain (Wittstock and Halkier, 2002). They are present constitutively (Wittstock and Gershenzon, 2002) but also accumulate in response to stresses including wounding or herbivory (Mikkelsen et al., 2003; Mewis et al., 2006; Schlaeppi et al., 2008). In recent years, most genes involved in Arabidopsis thaliana GS biosynthesis have been identified and are involved in amino acid chain elongation, core GS structure biosynthesis, sulfate assimilation pathway, and secondary modifications (Sønderby et al., 2010b). GS biosynthesis genes are regulated by six R2R3-MYB transcription factors. MYB28, MYB29, and MYB76 act in concert in a complex interaction module to control aliphatic-GS genes (Hirai et al., 2007; Gigolashvili et al., 2008; Sønderby et al., 2010a), whereas MYB34, MYB51, and MYB122 control indole-GS genes (Gigolashvili et al., 2007a). Mutant and overexpression analyses have clearly shown that GS levels are directly and specifically correlated with the activity of these GS-related MYBs (Celenza et al., 2005; Gigolashvili et al., 2007a, 2008; Sønderby et al., 2007, 2010a). Trans-activation assays with promoters of GS-reporter genes and the presence of MYB binding sites in their promoter suggest that GS biosynthesis genes are direct targets of MYBs (Gigolashvili et al., 2007b, 2008).
Different GS classes play a defensive role against generalist herbivores. High levels of aliphatic-GS and insect performance were negatively correlated in different A. thaliana genotypes (Kliebenstein et al., 2005). Overexpression of MYB28 in A. thaliana increased aliphatic-GS contents and reduced growth of Spodoptera exigua (Gigolashvili et al., 2007b). On the contrary, a myb28 myb29 double mutant lacking aliphatic-GS was more susceptible to feeding by Mamestra brassicae (Beekwilder et al., 2008). Overexpression of MYB51 in A. thaliana increased the production of indole-GS and reduced herbivory by S. exigua (Gigolashvili et al., 2007a), whereas the cyp79B2 cyp79B3 double mutant, impaired in the first step of indole-GS biosynthesis, was more susceptible to Spodoptera littoralis (Schlaeppi et al., 2008). By contrast, specialist insects like Plutella xylostella have evolved a sulfatase activity that prevents the formation of toxic GS hydrolysis products (Ratzka et al., 2002). Pieris rapae and Pieris brassicae larvae are equipped with a nitrile-specifier protein in their midgut that allows the formation of less toxic nitriles instead of isothiocyanates (Wittstock et al., 2004). As a consequence, these species feed equally on wild-type A. thaliana plants or on mutants with altered GS contents (Schlaeppi et al., 2008; Müller et al., 2010). Specialists even use GS to identify their host plants, as both butterflies and larvae are attracted by GS-containing plants (Renwick et al., 2006; Sun et al., 2009).
The jasmonate (JA) pathway is involved in the regulation of several processes, including plant growth, fertility, and defense against insects and necrotroph fungi (Howe and Jander, 2008; Browse, 2009). In response to herbivory, JA accumulates and triggers large changes in gene expression (Reymond et al., 2000, 2004; De Vos et al., 2005; Ehlting et al., 2008). Key players in signaling events leading to this transcriptional reprogramming have recently been identified. In response to stress, JA is conjugated to Ile and binds to a receptor complex consisting of CORONATINE INSENSITIVE1 (COI1) and JASMONATE ZIM-domain (JAZ) repressors (Chini et al., 2007; Thines et al., 2007; Yan et al., 2007; Katsir et al., 2008; Fonseca et al., 2009; Sheard et al., 2010). The F-box protein COI1 is part of a Skip/Cullin/Fbox (SCF)-type E3 ubiquitin ligase complex that participates in ubiquitination of target proteins and subsequent degradation by the 26S proteasome. In the absence of JA-Ile, JAZ repressors bind to transcription factors (TFs) and block their activity by interacting with the adaptor protein NINJA and the corepressor TOPLESS (Pauwels et al., 2010). Upon JA-Ile accumulation and its binding to COI1, JAZ repressors are degraded by SCFCOI1, allowing transcription of early JA-responsive genes (Chini et al., 2007; Thines et al., 2007; Sheard et al., 2010). Until recently, the basic helix-loop-helix (bHLH) TF MYC2/JASMONATE INSENSITIVE1 (JIN1) was the only known target of JAZ repressors (Chini et al., 2007), but phenotypes of myc2/jin1 mutants were difficult to reconcile with a unique TF regulating the diversity of JA-mediated processes. However, new studies have unveiled additional factors that might control specific JA-dependent responses. Two R2R3-MYB factors, MYB21 and MYB24, were identified as direct targets of JAZs in the regulation of male fertility (Song et al., 2011), whereas the combined interaction of JAZs with bHLHs (TRANSPARENT TESTA8 [TT8], GLABRA3 [GL3], and ENHANCER OF GLABRA3 [EGL3]) and MYBs (MYB75 and GL1) was shown to control anthocyanin accumulation and trichome formation (Qi et al., 2011). MYC3 and MYC4, two close homologs of MYC2, interact with JAZs (Fernández-Calvo et al., 2011; Niu et al., 2011) and act additively with MYC2 to regulate defense against insect herbivory (Fernández-Calvo et al., 2011). This finding, besides the known compromised resistance of coi1 mutants to arthropod herbivores in different plant species (Li et al., 2004; Reymond et al., 2004; Bodenhausen and Reymond, 2007; Paschold et al., 2007), established that the JA signaling module is central for plant resistance to herbivory. Promoter analysis and protein binding microarray studies have shown that MYC2, MYC3, and MYC4 bind with similar affinities to the core CACGTG motif, named the G-box, and its variants (Dombrecht et al., 2007; Fernández-Calvo et al., 2011; Godoy et al., 2011). However, which defense genes are regulated by MYC2/MYC3/MYC4 is largely unknown.
Methyl jasmonate treatment induces the expression of GS biosynthesis genes (Mikkelsen et al., 2003; Reymond et al., 2004) and triggers GS accumulation (Kliebenstein et al., 2002). Given that resistance to S. littoralis herbivory requires the additive action of MYC2/MYC3/MYC4 (Fernández-Calvo et al., 2011), we decided to further explore the involvement of these factors in the regulation of GS biosynthesis and to test their specific contribution in resistance to herbivory. In this study, we show that the myc2 myc3 myc4 mutant has a highly reduced expression of GS biosynthesis genes and is practically devoid of GS. Lack of GS in this mutant is correlated with an increased feeding performance of the generalist S. littoralis but not of the specialist P. brassicae. Furthermore, we demonstrate that MYC2 binds in vivo to the promoter of several GS genes and that MYC2/MYC3/MYC4 interact directly with GS-related MYBs.
RESULTS
Reduced Expression of GS Pathway Genes in myc234
The bHLH TFs MYC2/MYC3/MYC4 act additively to control JA responses, including root growth inhibition and defense against herbivory (Fernández-Calvo et al., 2011). Indeed, the generalist herbivore S. littoralis gained equally more weight on myc234 and coi1-1 than on Columbia-0 (Col-0), suggesting that the expression of defense-related genes was severely compromised in myc234 (Fernández-Calvo et al., 2011). We therefore analyzed the transcriptome of myc234 plants with an A. thaliana whole-genome microarray and compared it to that of Col-0 plants. Using a threshold of twofold change in gene expression (P < 0.05, false discovery rate < 10%), we identified 84 genes whose expression was significantly reduced in myc234 (see Supplemental Data Set 1 online). Interestingly, among the 50 most differentially expressed genes between Col-0 and myc234, 27 genes are involved in the GS pathway, including most genes responsible for the synthesis of core aliphatic- and indole-GS structures, genes involved in side-chain modification, and genes involved in primary sulfate assimilation (Figure 1; see Supplemental Data Set 1 online). Moreover, the expression of a large majority of all known or predicted genes involved in GS biosynthesis was significantly reduced in myc234 (see Supplemental Table 1 online). Since GSs are known to accumulate in response to herbivory, we selected nine genes from the list of differentially expressed genes and measured their expression by quantitative real-time PCR in Col-0, myc234, and coi1-1 after challenge with S. littoralis larvae (Figure 2). Expression of these genes was significantly reduced in myc234 compared with Col-0 plants, validating our microarray data. Surprisingly, the basal expression of most of these genes was significantly higher in coi1-1 than in myc234. In addition, although constitutive expression of GS genes in myc234 and coi1-1 was strikingly different, these genes were not induced by herbivory in these mutants (Figure 2). These data indicate that the JA pathway controls the induction of GS genes in response to herbivory but that MYC2/MYC3/MYC4 have additional roles in regulating their basal expression.
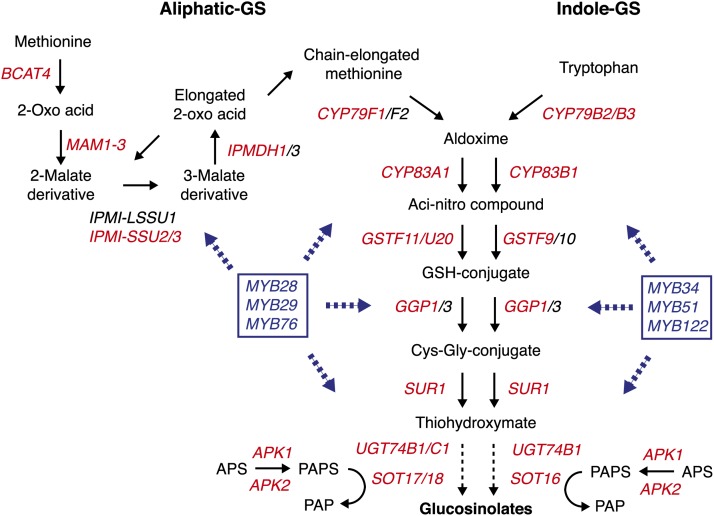
Figure 1.
Aliphatic and Indolic GS Biosynthetic Pathway in A. thaliana.
The biosynthesis of Met-derived aliphatic and Trp-derived indolic GSs is depicted. A. thaliana genes involved in distinct and common steps are indicated as well as genes involved in primary sulfate assimilation. MYB TFs known to regulate the expression of aliphatic- or indole-GS genes are indicated in blue. Genes in red were differentially expressed between Col-0 and myc234 plants (this study). The figure is adapted from Sønderby et al. (2010b) and Yatusevich et al. (2010).
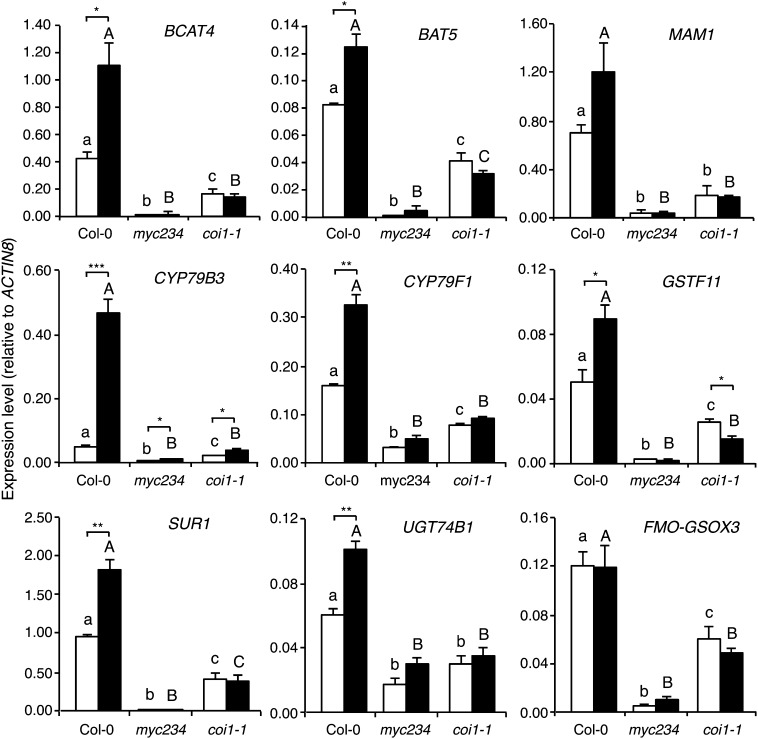
Figure 2.
Expression of GS Pathway Genes in Response to Herbivory.
Expression of nine GS pathway genes was measured in different genotypes by real-time PCR in untreated plants (white bars) and in plants challenged for 48 h with S. littoralis larvae (black bars). Values are the mean (±se) of three biological replicates. myc234 stands for the myc2 myc3 myc4 triple mutant. Different letters within each treatment indicate significant differences at P < 0.05 (Tukey’s highly significant difference test); capital letters compare with each other, and lowercase letters compare with each other. Asterisks denote statistically significant differences between indicated samples (**P < 0.01 and *P < 0.05; Student's t test).
MYC2/MYC3/MYC4 were shown to regulate JA-dependent genes like VEGETATIVE STORAGE PROTEIN2 or JAZ10, and only the myc234 triple mutant showed a complete loss of expression of these genes (Fernández-Calvo et al., 2011), suggesting that each one of the three MYCs is required to regulate the expression of GS pathway genes. To validate this hypothesis, we measured the expression of three GS biosynthesis genes (BRANCHED-CHAIN AMINOTRANSFERASE4 [BCAT4], CYP79B3, and SUPERROOT1 [SUR1]) in myc2, myc3, and myc4 single mutants. As postulated, expression levels of these GS genes were very similar in Col-0 and single myc mutants (see Supplemental Figure 1 online), contrasting with their reduced expression in myc234 (Figure 2). To further support the finding that MYC factors regulate the expression of GS biosynthesis genes, expression of BCAT4, CYP79B3, and SUR1 was significantly higher in a MYC2-overexpressing line compared with Col-0 (see Supplemental Figure 2 online).
Greatly Reduced GS Levels in myc234
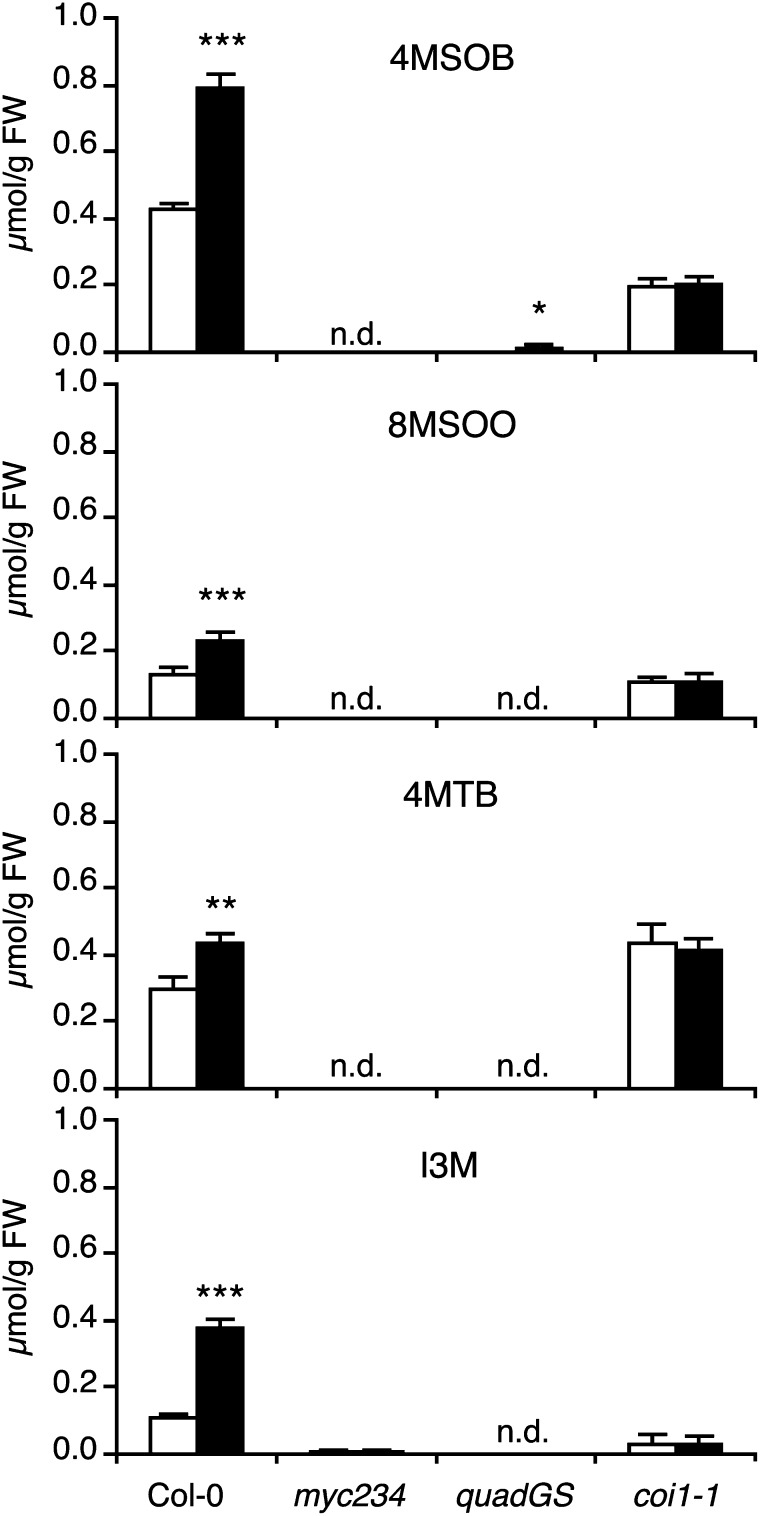
Having observed that expression of key GS biosynthesis genes was severely affected in myc234, we quantified GS levels by ultrahigh-pressure liquid chromatography coupled to quadrupole time-of-flight mass spectrometry and compared the results with the quadruple mutant cyp79B2 cyp79B3 myb28 myb29 (quadGS), which is nearly devoid of major classes of foliar GSs (Müller et al., 2010). Consistent with expression data, myc234 had greatly reduced levels of 16 GS, 14 of them being undetectable even with a highly sensitive quadrupole time-of-flight mass spectrometry detection system (Figure 3; see Supplemental Table 2 online). Total GS amounts in myc234 and quadGS were <1% of Col-0 (see Supplemental Table 2 online). In addition, S. littoralis herbivory caused a significant increase in GS levels in Col-0 plants but not in myc234 or quadGS (Figure 3; see Supplemental Table 2 online). Thus, MYC2/MYC3/MYC4 are essential factors for GS accumulation.
Figure 3.
Quantification GSs in Response to Herbivory.
Levels of four abundant GS were quantified in different plant genotypes. Plants were challenged for 2 d with S. littoralis larvae (black bars). Unchallenged plants were used as controls (white bars). Values are the mean (±se) of four biological replicates. Significant differences between control and treated plants are indicated (Student's t test; * P < 0.05, **P < 0.01, and ***P < 0.001). 4MSOB, 4-methylsulfinylbutyl-GS; 8MSOO, 8-methylsulfinyloctyl-GS; 4MTB, 4-methylthiobutyl-GS; I3M, indol-3-ylmethyl-GS. quadGS stands for the cyp79b2 cyp79b3 myb28 myb29 quadruple mutant. FW, fresh weight.
Since coi1-1 had residual expression of GS biosynthesis genes, we hypothesized that it might still contain significant amounts of GS. Indeed, all GS were clearly more abundant in coi1-1 than in myc234 or quadGS, reaching even wild-type levels for the main aliphatic-GS 8MSOO, 8MTO, and 4MTB (Figure 3; see Supplemental Table 2 online). Total GS content in control coi1-1 plants was 83% of Col-0 levels (see Supplemental Table 2 online). However, GS levels in coi1-1 did not change significantly in response to S. littoralis, in accordance with the lack of induction of GS biosynthesis genes (Figure 2). Taken together, our results indicate that the JA pathway is absolutely required for insect-induced GS accumulation. However, an important proportion of constitutive GS accumulate in a COI1-independent but MYC2/MYC3/MYC4-dependent manner.
Contrasting Performance and Behavior of Specialist and Generalist Herbivores on myc234 Plants
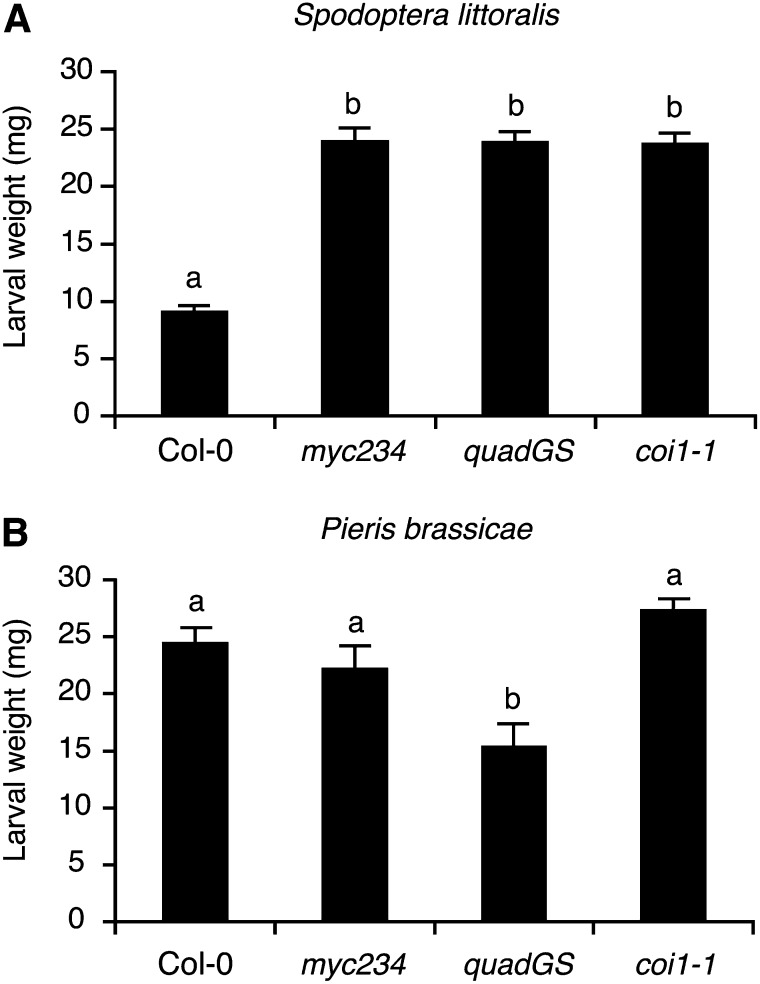
In order to evaluate the contribution of MYC2/MYC3/MYC4 to larval performance, we challenged myc234, quadGS, and coi1-1 plants with the generalist S. littoralis and the specialist P. brassicae. S. littoralis larvae grew significantly bigger on myc234 and quadGS than on Col-0, in accordance with their highly reduced GS content. But, surprisingly, larval weight was also significantly higher on coi1-1 and was not different from the two GS-deficient mutants, although coi1-1 still contains significant GS levels (Figure 4A). On the contrary, P. brassicae larvae grew equally on Col-0, myc234, and coi1-1 but were smaller on quadGS plants (Figure 4B).
Figure 4.
Performance of a Generalist and a Specialist Herbivore on GS Mutants.
Larval performance was tested in a no-choice test on wild-type plants, on JA signaling myc234 and coi1-1 mutants, and on the GS-deficient quadGS mutant. Freshly hatched S. littoralis (A) or P. brassicae (B) larvae were placed on 3-week-old plants and larval weight (mean ± se) was measured after 7 d of feeding. Different letters indicate significant differences at P < 0.05 (Tukey’s highly significant difference test).
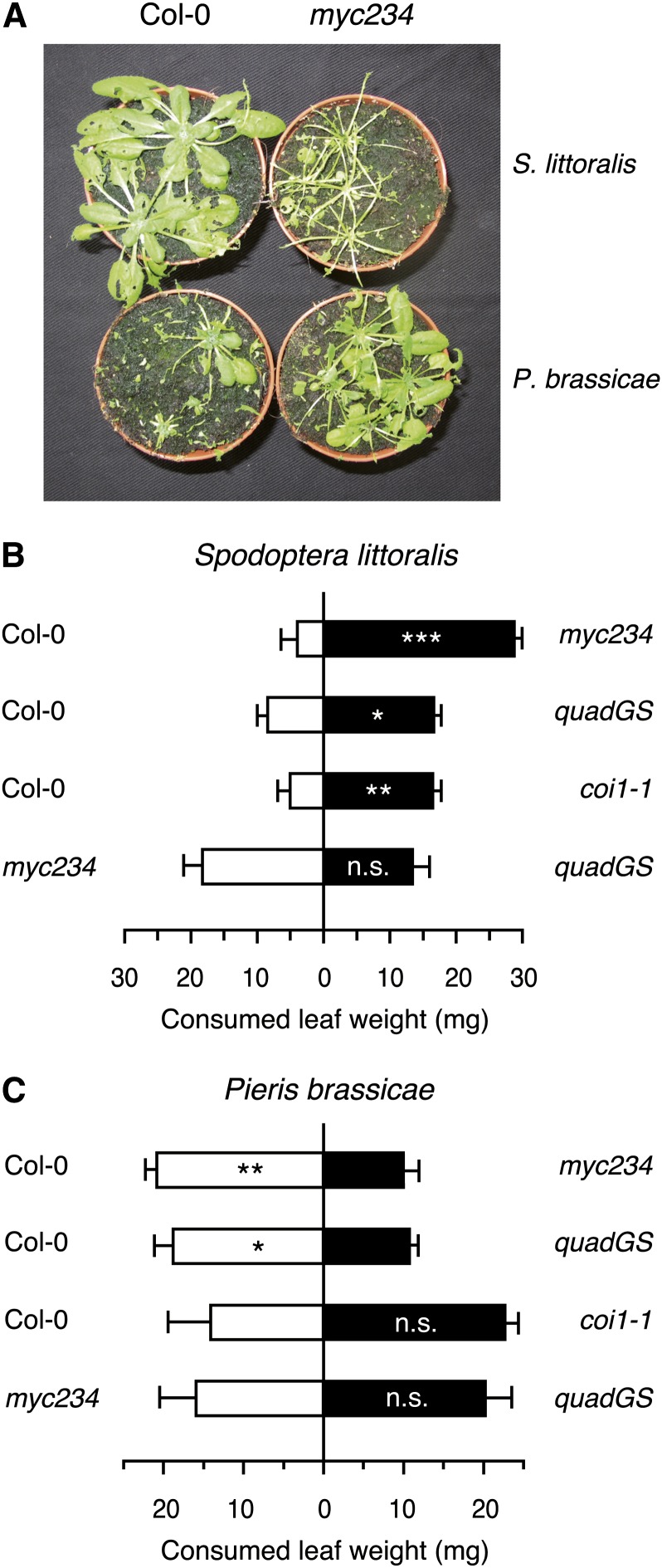
GSs are known for their repellent effect on generalist insects, but they can also act as feeding attractants for host recognition by specialized herbivores or as oviposition cues (Renwick et al., 2006; de Vos et al., 2008). We therefore tested GS mutants in dual-choice experiments with S. littoralis or P. brassicae. Consistent with the increased weight gain in the no-choice experiment, S. littoralis larvae consumed significantly more leaf material on myc234, quadGS, or coi1-1 than on wild-type plants (Figures 5A and 5B). In addition, larvae did not show any feeding preferences between myc234 and quadGS. Dual-choice assays with the specialist P. brassicae revealed an opposite pattern, as larvae consumed significantly more leaf material on wild-type plants than on the two GS-deficient mutants myc234 and quadGS. Moreover, P. brassicae did not show any feeding preference between myc234 and quadGS and did not discriminate wild-type from coi1-1 plants (Figures 5A and 5C). When offered different genotypes for oviposition, P. brassicae butterflies clearly selected Col-0 as a host and deposited almost no eggs on myc234 or quadGS (see Supplemental Figure 3 online).
Figure 5.
Contrasting Feeding Preferences of S. littoralis and P. brassicae Larvae in Dual-Choice Tests.
(A) Dual-choice tests between Col-0 and myc234 intact plants. Newly hatched larvae were allowed to feed for 7 d on both genotypes, and the picture was taken at the end of the feeding period.
(B) and (C) Dual-choice tests between Col-0 and JA signaling or GS biosynthesis mutants. Feeding preference of S. littoralis (B) and P. brassicae (C) larvae on two leaves of each genotype was measured in Petri dishes after 4 h. Values are the mean (±se) of 25 to 30 independent samples. Asterisks indicate statistically significant differences between the tested genotypes (Student’s t test; *P < 0.05, **P < 0.01, and ***P < 0.001).
Leaf GS Contents Influence the Feeding Behavior of Generalist and Specialist Herbivores
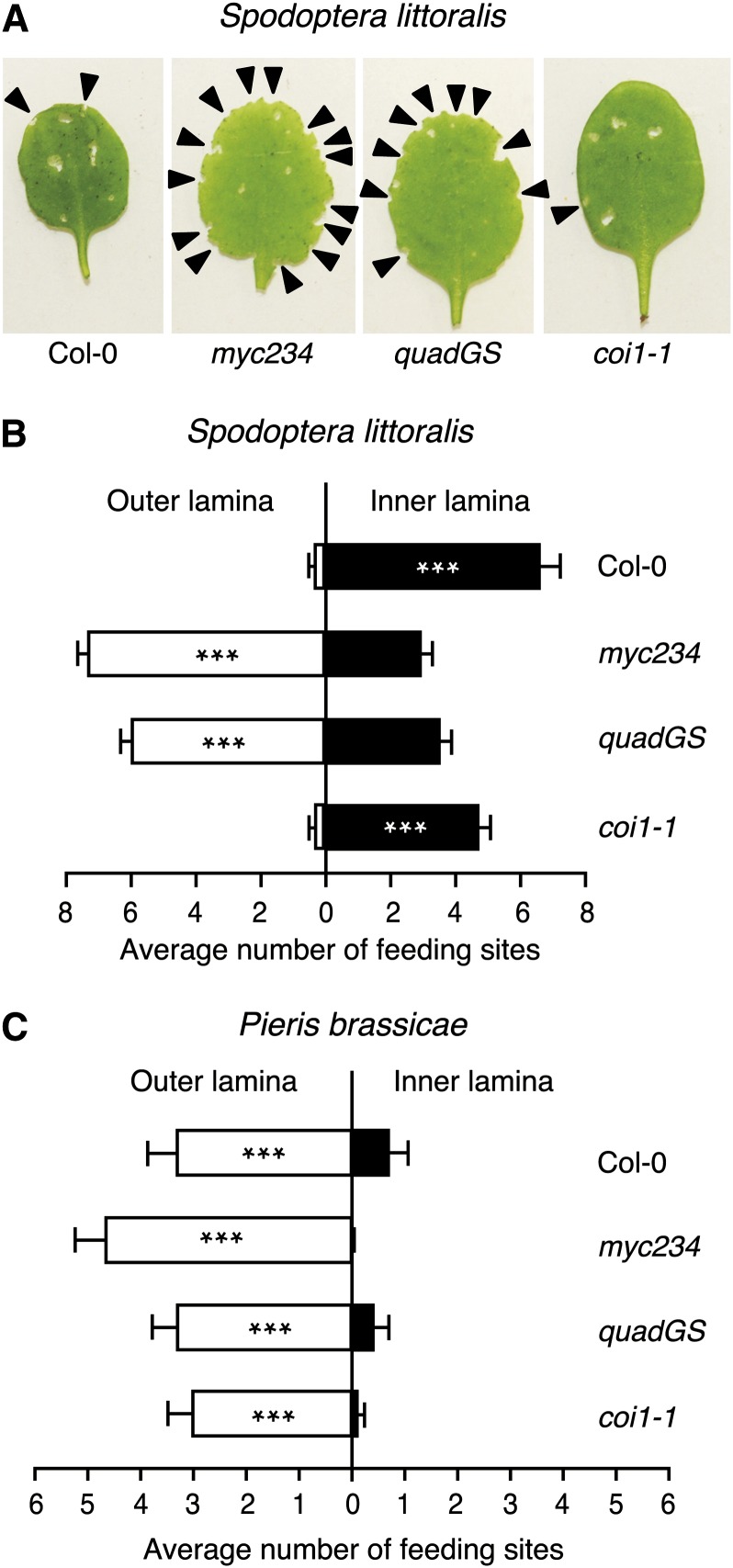
By carefully observing insects during our bioassays, we noticed that S. littoralis and P. brassicae had a different feeding behavior depending on plant genotypes. On GS-containing Col-0 and coi1-1, S. littoralis larvae fed almost exclusively on the leaf inner lamina, whereas they drastically changed behavior on myc234 and quadGS where they preferentially attacked the leaf outer lamina (Figures 6A and 6B). A similar experiment with P. brassicae showed a different trend. Specialist larvae always chose the outer leaf lamina, independently of which genotype they were allowed to feed on (Figure 6C). Given that highest GS concentrations are found in midvein and leaf periphery (Shroff et al., 2008), these results indicate that generalists tend to avoid feeding on GS-rich tissues, but when leaves lack these compounds they prefer to feed on leaf edges. Similarly, although they are adapted to GS and use them as feeding stimulants, specialists still prefer leaf edges in GS-deficient plants. This is an interesting observation, suggesting that leaf periphery has a particular property that attracts insects.
Figure 6.
S. littoralis and P. brassicae Prefer to Feed on the Leaf Margin of GS-Lacking Plants.
(A) Two neonate S. littoralis larvae were placed on a 4-week-old plant and allowed to feed for 36 h. The picture is taken at the end of the feeding period. Arrows indicate feeding sites.
(B) Number of feeding sites on inner and outer leaf blade was scored on each eaten leaf.
(C) Experiment with P. brassicae was done similarly except that one neonate larva was used per plant and feeding time was reduced to 24 h.
Values are the mean (±se) from 28 plants for each A. thaliana genotype. Asterisks indicate statistically significant differences between inner and outer lamina (Student’s t test; *P < 0.05, **P < 0.01, and ***P < 0.001). Experiments were repeated three times independently with similar results.
MYC2, MYC3, and MYC4 Interact Directly with GS-Related MYB Factors
Having found that myc234 shows a highly reduced expression of GS biosynthesis genes and is consequently devoid of GS, we hypothesized that MYC2/MYC3/MYC4 might control GS biosynthesis indirectly by regulating the expression of GS-related MYBs. Surprisingly, expression of these MYBs in control plants was not significantly different between Col-0 and myc234 plants (see Supplemental Figure 4 online) and therefore did not correlate with the reduced expression of their target genes and the absence of GS in myc234. Similarly, their expression in response to S. littoralis herbivory was not significantly lower in myc234 compared with Col-0, except for MYB29 and MYB34, and is difficult to reconcile with the large difference in GS levels between Col-0 and myc234 after insect attack. Finally, expression of these MYBs did not differ significantly between Col-0 and coi1-1, except for MYB51, which showed higher expression in the mutant, indicating that most of them are not controlled by the JA pathway (see Supplemental Figure 4 online). Taken together, our results show that MYC2/MYC3/MYC4 are unlikely to regulate the GS pathway via transcriptional activation of MYBs.
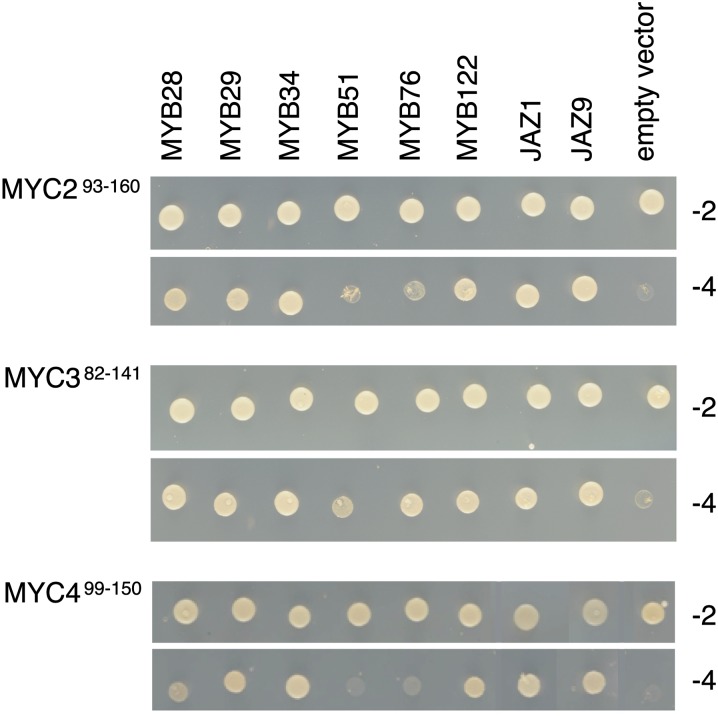
A recent study demonstrated that JAZ repressors target bHLH TFs (GL3, EGL3, and TT8) and MYB factors (MYB75 and GL1) to control JA-dependent anthocyanin accumulation and trichome initiation (Qi et al., 2011). We reasoned that a similar complex might be formed to control GS accumulation. To test this hypothesis, we first looked for physical interaction between all 12 JAZs and any of the six GS-related MYBs by yeast two-hybrid assay. However, no direct interaction between JAZs and MYBs was detected (see Supplemental Figure 5 online). An alternative scenario could be that MYCs bind to these MYBs directly. Initial experiments with the C-terminal part of MYC2 and MYC3 showed that this fragment was autoactivating the reporter gene in the yeast assay. However, when we tested a 51– to 67–amino acid N-terminal fragment containing the JAZ interaction domain (JID) of MYC2, MYC3, and MYC4, we detected an interaction with GS-related MYB proteins (Figure 7). MYC3-JID interacted strongly with all MYBs, whereas MYC2-JID and MYC4-JID interacted strongly with MYB28, MYB29, MYB34, and MYB122, but weakly (MYC2-JID) or not (MYC4-JID) with MYB51 and MYB76. As a positive control, the three MYC2, MYC3, and MYC4 JID-containing fragments interacted with JAZ1 and JAZ9 (Figure 7).
Figure 7.
The JID Domain of MYC2, MYC3, and MYC4 Is Sufficient for Interaction with MYB Proteins.
MYC293-160 (top panel), MYC382-141 (middle panel), and MYC499-150 (bottom panel) derivatives in GAL4-BD were tested for interaction with MYB28, MYB29, MYB34, MYB51, MYB76, and MYB122 in GAL4-AD. JAZ1 and JAZ9 were used as positive interaction controls. Yeast cells cotransformed with pGBKT7g- MYC293-160, pGBKT7g- MYC382-141, or pGBKT7g- MYC499-150 (bait), and pGADT7-MYBs (prey) were selected and subsequently grown on yeast synthetic dropout lacking Leu and Trp (-2) or on selective media lacking Ade, His, Leu, and Trp (-4) to test protein interactions. pGBKT7g-MYC293-160, pGBKT7g MYC382-141, and pGBKT7g MYC499-150 cotransformations with the pGADT7g vector were included as negative controls.
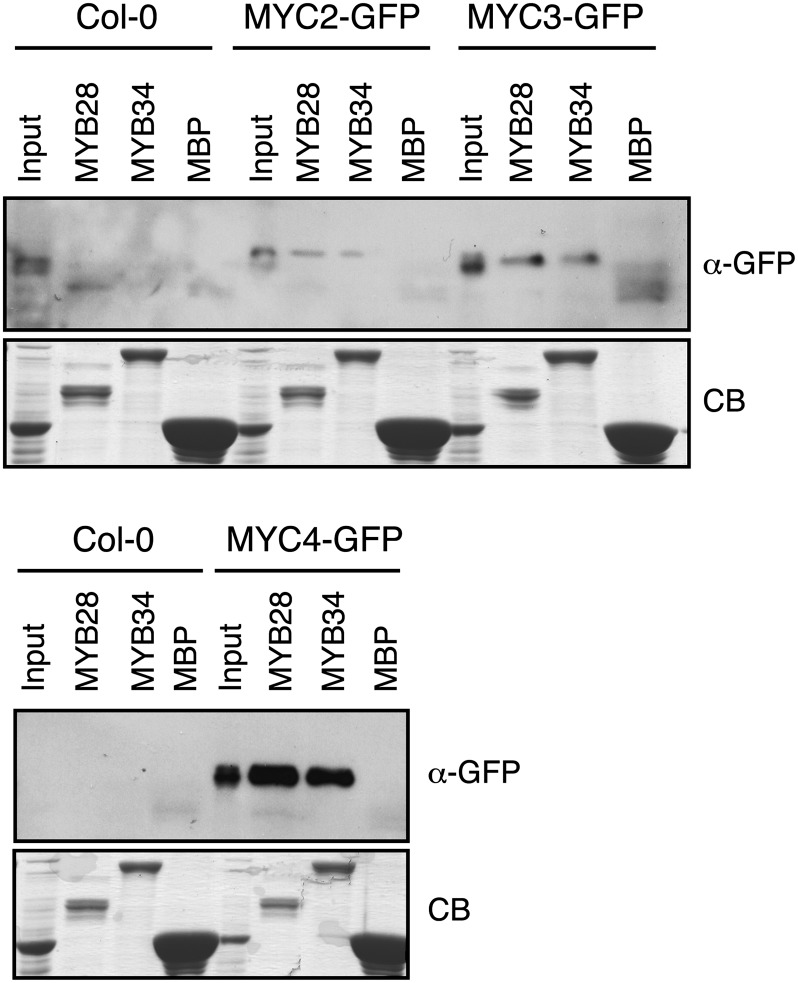
To confirm further that MYC factors associate with GS-related MYB factors, we performed pull-down experiments with each recombinant MBP-MYB fusion protein and extracts of transgenic plants expressing MYC2-, MYC3-, or MYC4-GFP (for green fluorescent protein). MYC2, MYC3, and MYC4 were pulled down by the six MBP-MYBs (Figure 8; see Supplemental Figure 6 online). Thus, although yeast two-hybrid assays did not show a clear interaction between MYC2 and MYC4 with both MYB51 and MYB76, the pull-down results were fully convincing. Collectively, these data strongly suggest that MYC2, MYC3, and MYC4 can form complexes with all known GS-related MYBs to regulate GS biosynthesis. The exact composition of complexes that can be formed in vivo will require further analyses.
Figure 8.
MYC2, MYC3, and MYC4 Interact with MYB28 and MYB34 in Pull-Down Assays.
Immunoblots with anti-GFP antibody of recovered MYC2-GFP, MYC3-GFP, and MYC4-GFP after pull-down reactions using crude protein extracts from 35S:MYC2-GFP, 35S:MYC3-GFP, 35S:MYC4-GFP, or the wild type (Col-0) A. thaliana plants and resin-bound recombinant MBP-MYB proteins. Input lanes show the level of expression of recombinant proteins in transgenic and control plants. Coomassie blue staining shows the amount of recombinant proteins used (CB).
MYC2 Binds to the Promoter of GS Biosynthesis Genes
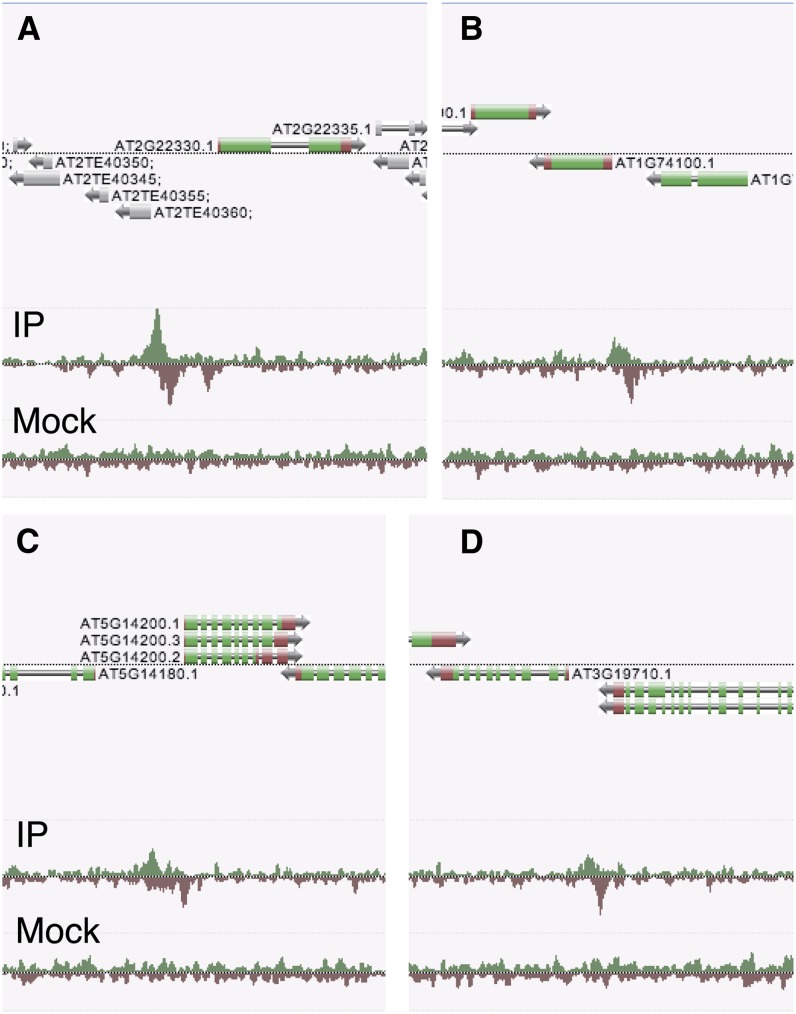
Interestingly, analysis of GS genes revealed that there is a significant overrepresentation of a canonical G-box motif in their promoter (see Supplemental Figure 7 online). We also found that this G-box is conserved in homologous GS genes of the related Arabidopsis lyrata (see Supplemental Figure 8 online). To test whether MYCs bind directly to the promoter of GS biosynthesis genes, we screened the 27 MYC-regulated genes by chromatin immunoprecipitation sequencing (ChIP-Seq). Transgenic A. thaliana plants expressing MYC2:FLAG under its native promoter (Hou et al., 2010) were treated with JA and their chromatin was immunoprecipitated with anti-FLAG antibody, after which bound DNA was analyzed by high-throughput sequencing (see Methods). Focusing on the 27 GS biosynthesis genes that are differentially expressed between Col-0 and myc234, a significant enrichment of reads (greater than or equal to threefold, P < 10−10) was found in the promoter region of 14 genes (Figures 9A to 9D; see Supplemental Table 3 online). This overrepresentation of genes containing a MYC2 binding site among GS genes was highly significant (hypergeometrical distribution, P = 2.05*10−08). In addition, the G-box bound by MYC2 (Fernández-Calvo et al., 2011) and its variants were found at least once in the MYC2 binding site of the 14 GS biosynthesis genes (see Supplemental Table 4 online). Thus, a majority of GS biosynthesis genes that are downregulated in myc234 are direct targets of MYC2 and possess a G-box motif in their promoter.
Figure 9.
MYC2 Binds to the Promoter of GS Biosynthesis Genes in Vivo.
ChIP-Seq was conducted on MYC2 using a MYC2-FLAG construct under native expression (Hou et al., 2010). Reads from high-throughput sequencing of immunoprecipitation from JA-treated plants (IP) and the mock experiment (mock) are shown. Screenshots from in-house genome browser (Anno-J, www.annoj.org) include four genes involved in the core biosynthetic pathways for indole-GSs ([A], CYP79B3; [B], SOT16) and aliphatic-GSs ([C], IPMDH1; [D], BCAT4).
DISCUSSION
The bHLH TFs MYC2, MYC3, and MYC4 control additively subsets of JA-dependent responses, including root growth inhibition, defense against bacterial pathogens, and defense against insect herbivory (Fernández-Calvo et al., 2011). These three TFs can form homo- and heterodimers and have almost identical DNA binding specificities for the canonical G-box (CACGTG) and some of its variants (Fernández-Calvo et al., 2011). This suggests that they constitute a transcriptional module that recognizes target genes involved in JA responses. However, the number and identity of MYC2/MYC3/MYC4-regulated genes is largely unknown. Our whole-genome microarray analysis revealed that more than half of the 50 most differentially expressed genes between myc234 and Col-0 belong to the GS pathway. This enrichment of GS pathway genes was remarkable and pointed to a crucial role of MYC2, MYC3, and MYC4 in the control of GS accumulation. Indeed, GS analysis showed that myc234 has undetectable levels of 14 GSs and contains only traces of two indole-GSs. Although GS levels in myc234 are <1% of wild-type levels, expression of GS biosynthesis genes was not completely abolished in myc234. It is thus plausible that another MYC factor contributes to this residual activity. There are at least 162 bHLH proteins in the A. thaliana genome, and MYC2, MYC3, and MYC4 belong to a subfamily (IIId+e) of eight related homologs (Pires and Dolan, 2010). At5g46830 (bHLH028), the closest homolog to MYC2, does not interact with JAZ proteins (Fernández-Calvo et al., 2011). Nevertheless, it would be interesting to assess the role of other closely related members of this subfamily.
Contrary to myc234, single myc mutants retained wild-type expression of GS biosynthesis genes, indicating that there is functional redundancy between MYC2, MYC3, and MYC4 in the control of GS gene expression. A genome-wide transcript analysis of myc2/jin1-9 identified a list of MYC2-regulated genes, including genes involved in wound/insect response, flavonoid biosynthesis, and oxidative stress tolerance (Dombrecht et al., 2007). Intriguingly, the authors found that genes involved in indole-GS biosynthesis were upregulated in jin1-9 and that the mutant accumulated more indole-GS in response to methyl jasmonate treatment. They concluded that MYC2 was a negative regulator of indole-GS accumulation. In light of our results, we rather interpret these data as being a consequence of compensatory effects exerted by MYC3 and MYC4 in jin1-9. In support of this hypothesis, gene expression analysis of MYC2, MYC3, and MYC4 showed that expression of the two other MYCs was affected in single myc mutants (Fernández-Calvo et al., 2011). In addition, single myc2 mutant is only partially more sensitive to herbivory than Col-0 (Fernández-Calvo et al., 2011).
S. littoralis larvae gained much more weight on myc234 and quadGS than on Col-0, and, when given the choice, clearly preferred to feed on mutants that lack GS. On the contrary, P. brassicae gained the same weight on myc234 and Col-0 and preferred to feed on GS-containing plants. These findings confirm the important deterrent function of these secondary metabolites against nonadapted herbivores and their role as attractant for adapted herbivores (Barth and Jander, 2006; Beekwilder et al., 2008; Schlaeppi et al., 2008; Müller et al., 2010). The observation that S. littoralis larvae had the same weight on myc234 and quadGS plants and that they did not discriminate between the two genotypes indicates that larval performance and host preference in this insect is mainly dictated by the production of plant GS. This significant contribution of one biosynthetic pathway is remarkable given the hundreds of genes that are differentially induced in response to herbivory (Reymond et al., 2004; De Vos et al., 2005; Ehlting et al., 2008). The transcriptome analysis identified GS pathway genes as the most differentially expressed genes between myc234 and Col-0, but the list contained other putative defense genes, including lectins and protease inhibitors. Whether these genes play a defense role in other environmental conditions or against other insects remains to be established.
Surprisingly, we found that S. littoralis larvae gained as much weight on coi1-1 as they did on myc234 and quadGS. Considering that coi1-1 still contains 83% of constitutive and 44% of insect-induced GS compared with Col-0 (see Supplemental Table 2 online), it is plausible that these levels contribute to partial defense and that other defense compounds/proteins do not accumulate in coi1-1 but still accumulate in myc234. This clearly indicates that insect performance on A. thaliana is not strictly correlated with GS levels. Indeed, we recently showed that the transcriptome of myc234 after challenge with S. littoralis is different from coi1-1 (Schweizer et al., 2013). Many COI1-dependent genes were normally expressed in myc234 or showed an intermediate induction. To further confirm this finding, we analyzed the expression of two marker genes of the ethylene/JA pathway, PLANT DEFENSIN1.2 and HEVEIN-LIKE, and one anti-insect Cys proteinase gene (At4g11320) by quantitative PCR. These genes were significantly induced by S. littoralis in Col-0 and myc234, whereas they were not induced in coi1-1 (see Supplemental Figure 9 online). Whether they contribute quantitatively to defense against herbivory in myc234 has yet to be shown, but our findings are consistent with the hypothesis that the enhanced performance of S. littoralis on myc234 and coi1-1 results from a complete lack of GS in the case of myc234 and from a combination of reduced GS levels and impaired expression of some defense genes in coi1-1. This strongly suggests that other TFs are controlled by the COI1/JA-Ile/JAZ signaling complex to modulate defense against herbivory. In support of this hypothesis, we recently identified TFs that control resistance to S. littoralis in a partially MYC2/3/4-independent manner (Schweizer et al., 2013). Moreover, a complex of bHLH factors (TT8, GL3, and EGL3) and MYB factors (MYB75 and GL1) was found to interact with JAZs and to be required for JA-dependent anthocyanin accumulation and trichome initiation (Qi et al., 2011). Thus, there is increasing evidence for multiple targets of JAZ repressors controlling subsets of JA-dependent responses.
Interestingly, we observed that S. littoralis larvae preferentially feed on the inner leaf lamina of Col-0 and coi1-1 plants, avoiding leaf edge and main vein. On the contrary, larvae clearly chose to feed on the outer leaf lamina in myc234 and quadGS. For the specialist P. brassicae, larvae always consumed the outer leaf lamina, irrespective of the genotype. GSs are not distributed uniformly in A. thaliana leaves and accumulate to higher levels in leaf margins and veins (Shroff et al., 2008; Sønderby et al., 2010a). Shroff et al. (2008) observed that larvae of the generalist Helicoverpa armigera avoided leaf margins and veins of A. thaliana rosette leaves. Herbivorous insects often consume specific parts of their host and have evolved recognition cues to avoid poison-containing areas and structural defenses or to select nutritious tissues (Kester et al., 2002; Schoonhoven et al., 2005). The nonuniform GS distribution could thus explain the feeding behavior of S. littoralis larvae, which avoided tissues with high GS contents, and that of P. brassicae larvae, which were attracted by GS-containing tissues. We found that MYC2/3/4 and GS biosynthesis genes are not differentially expressed between the leaf edge or the leaf inner lamina, indicating that this does not contribute to uneven GS accumulation (data not shown). On the contrary, GS-related MYBs are expressed predominantly in the veins (Gigolashvili et al., 2007b, 2008). These factors play a role in leaf GS distribution, and it was postulated that they regulate a specific GS transport from the vein to the leaf edge (Sønderby et al., 2010a). Recently, the first GS transporters, belonging to the family of nitrate/peptide transporters, were identified. They are expressed in veins and surrounding mesophyll cells and were shown to transport GS from the leaves to the seeds (Nour-Eldin et al., 2012). Whether these or related transporters are involved in local redistribution of GS deserves further research. Another intriguing finding is that both generalist and specialist larvae fed on the outer leaf lamina of GS-lacking plants. This behavior might be explained either by a better nutritive quality of the leaf margin or by other defense compounds located in the leaf inner lamina that repels both generalists and specialists. Clearly, more work will be needed to understand this feeding pattern, but GS-lacking mutants have unveiled an intriguing aspect of A. thaliana defense against herbivores and will provide useful tools to investigate this phenomenon in the future.
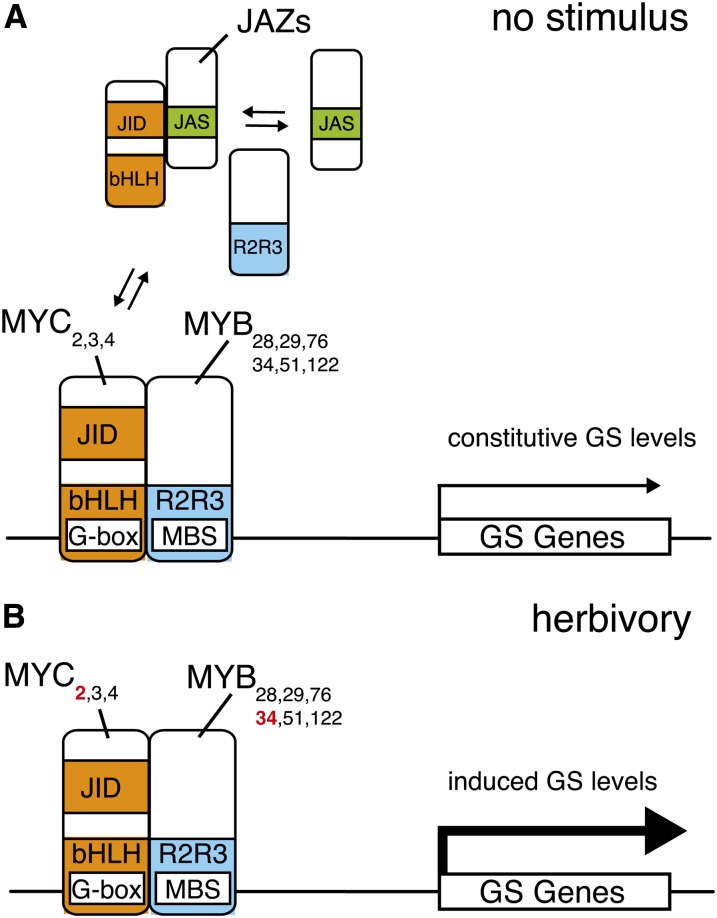
Physical interaction between bHLH and MYB factors is a well-known example of combinatorial gene regulation in plants and plays a role in different plant processes (Feller et al., 2011). Here, we show that MYC2/MYC3/MYC4 interact directly with GS-related MYBs, that MYC2 binds directly to the promoter of more than half of GS biosynthesis genes in vivo, and that these genes contain a canonical MYC2 binding motif. This suggests that the GS pathway is under tight transcriptional control and that the coordinated expression of several biosynthesis genes is required to accumulate GS. Whether all GS genes are regulated by MYCs and whether MYC3 or MYC4 bind to the promoters of GS genes that not are direct targets of MYC2 remains to be investigated. Based on the observations that GS-related MYBs positively control the expression of GS biosynthesis genes (Gigolashvili et al., 2007b, 2008), that MYC2/3/4 bind directly to these MYBs, and that MYC2 binds to the promoters of several GS biosynthesis genes (this study), we propose a model where both MYCs and MYBs are recruited to the promoter of GS genes to mediate transcription (Figure 10). In absence of stimulus, JAZ proteins bind to the JID domain of MYC2/MYC3/MYC4 (Fernández-Calvo et al., 2011) and inhibit most MYC transcriptional activity. However, since the JID domain of these MYCs also binds to GS-related MYBs, MYBs would prevent the JID domain interaction with JAZs. This might allow the formation of some MYC-MYB complexes that bind to the promoter of GS genes and activate transcription, thus explaining the presence of constitutive expression of GS biosynthesis genes and basal GS levels in Col-0 and in coi1-1. In particular, the fact that coi1-1 shows basal levels of GSs strongly supports the existence of a pool of active MYCs (the MYC-MYB complexes) that are still able to regulate GS biosynthesis genes in the absence of JA signaling. Upon herbivory and activation of the JA pathway, degradation of JAZ proteins by the SCFCOI1 complex might trigger a stronger MYC-MYB association and an increased transcriptional activation of GS biosynthesis genes, resulting in higher GS levels. This is illustrated by the findings that expression of GS biosynthesis genes and GS levels are induced in response to herbivory in Col-0 but not in coi1-1. Additionally, whereas the expression of MYC2, MYC3, and MYC4 is relatively similar in unchallenged plants, MYC2 shows a strong induction after herbivory (see Supplemental Figure 10 online). Interestingly, MYB34 is also induced by herbivory (see Supplemental Figure 4 online), thus indicating that after stimulus the increasing amounts of MYC2 and MYB34 will contribute to enhance the amount of active MYC-MYB complexes and might potentiate GS synthesis. Given that MYC2, MYC3, and MYC4 were shown to form homo- and heterodimers in vivo (Fernández-Calvo et al., 2011), it is likely that these three factors cooperate in a regulatory complex with GS-related MYBs to control the expression of GS biosynthesis genes. Whether this alters the stoichiometry of the MYC-MYB complexes and contributes to increased GS accumulation is an attractive hypothesis that will require further investigation. In addition, although GS biosynthesis genes contain MYB binding sites in their promoters (see Supplemental Figure 8 online), a direct binding has not yet been demonstrated. It will be interesting in future studies to test whether these sites are necessary to establish MYC-MYB complexes. Finally, our model does not exclude the contribution of additional factors. For instance, the DELLA proteins, which are repressors of the gibberellin-signaling pathway, have been shown to interact with JAZ1 and prevent JAZ1–MYC2 interaction (Hou et al., 2010). They might thus also interfere with the inhibition of MYCs by JAZs and lead to a release of free MYCs that could form active MYC-MYB complexes. This hypothesis will be addressed in future work.
Figure 10.
A Working Model for Regulation of GS Accumulation in A. thaliana.
(A) In unstimulated plants, JAZ repressors bind to the JID domain of bHLH MYC2/MYC3/MYC4 TFs through their Jas domain and inhibit the interaction between these MYCS and GS-related R2R3-MYB TFs. This attenuates the transcriptional activity of MYC-MYB complexes at the promoters of GS pathway genes. Competitive binding to JID domain by JAZs and MYBs might allow some basal transcriptional activity and therefore could explain the presence of constitutive GS levels in wild-type plants.
(B) Upon herbivory and activation of the JA pathway, JAZ repressors are degraded by the SCFCOI1 complex. A strong interaction between MYCs and MYBs leads to enhanced transcription of GS pathway genes and results in elevated GS levels. In addition, increased expression of MYC2 and MYB34 (red) might enhance the amount of MYC-MYB complexes and potentiate GS biosynthesis.
The exact composition of MYC-MYB complexes is not known, but homo- and heterodimerization of MYC2/MYC3/MYC4 have been reported (Fernández-Calvo et al., 2011). Direct binding of GS-related MYBs to the promoter of GS genes has not yet been demonstrated, but several GS genes contain MBS in their promoter. G-box, MYC binding site; MBS, MYB binding site.
Besides JAZs and MYBs, there is increasing evidence for the binding of MYC2 with several proteins involved in signaling, including DELLAs (Hong et al., 2012), the regulator of circadian rhythm TIME FOR COFFEE (Shin et al., 2012), the regulator of G-protein signaling ACIREDUCTONE DIOXYGENASE1 (Klopffleisch et al., 2011), and the regulator of cytokinin signaling HISTIDINE-CONTAINING PHOSPHOTRANSFER FACTOR5 (Yamashino et al., 2003). Thus, MYC2, and potentially MYC3 and MYC4, emerges as a central component that physically interacts with multiple factors to control defense, hormonal, and development programs (Kazan and Manners, 2013). In conclusion, this study reveals that MYC2/MYC3/MYC4 are essential for constitutive and insect-inducible GS biosynthesis. These factors contribute to a robust A. thaliana defense against nonadapted herbivores and have a marked effect on their feeding behavior. Their interaction with GS-related MYBs to regulate GS biosynthesis provides a novel example of how JA signaling specificity is achieved to control multiple plant processes.
METHODS
Plant and Insect Growth Conditions
Arabidopsis thaliana wild-type (Col-0) and all mutants were vernalized in water for 4 d at 4°C, except myc2 myc3 myc4 (myc234) triple mutants, which were vernalized in water containing 0.1 mM gibberellic acid (Duchefa) to synchronize germination (Fernández-Calvo et al., 2011). Light and growth conditions were reported previously (Reymond et al., 2000). Seeds of coi1-1 (nonglabrous) were obtained from Jane Glazebrook (University of Minnesota, St. Paul, MN), the myb28 myb29 double mutant from Piero Morandini (University of Milan, Milano, Italy), and the cyp79b2 cyp79b3 double mutant from Yunde Zhao (University of California, San Diego, CA). Both double mutants were crossed to obtain the quadruple mutant cyp79b2 cyp79b3 myb28 myb29 (quadGS). Homozygous seedlings of the male sterile coi1-1 were selected as described previously (Xie et al., 1998). Spodoptera littoralis (Egyptian cotton worm) eggs were obtained from Syngenta. Pieris brassicae (large white butterfly) were reared as described previously (Schlaeppi et al., 2008).
Insect Bioassays
No-choice insect bioassays with S. littoralis and P. brassicae were as described (Bodenhausen and Reymond, 2007). Experiments were performed with 3-week-old plants in transparent plastic boxes (Fernández-Calvo et al., 2011). Briefly, 40 freshly hatched S. littoralis, or 15 neonate P. brassicae, were placed on plants of each genotype for 7 d of feeding. Larvae were then weighed on a precision balance Mettler-Toledo MT5 (Mettler-Toledo). Similar results were obtained in three independent experiments.
For dual-choice experiments, an initial test was done on potted plants. Five pots per genotype were placed in a transparent plastic box and challenged with 40 freshly hatched S. littoralis or 15 neonate P. brassicae larvae for 7 d. In follow-up experiments, two leaves of 4-week-old plants of different A. thaliana genotypes were weighed and laid on a moistened filter paper in a 60 × 15-mm Petri dish. Freshly hatched S. littoralis larvae were fed for 6 d on artificial diet (Syngenta) and then starved for 12 h at 5°C to prevent cannibalism. One larva was then placed in each Petri dish for 4 h in the dark at room temperature. Leaves were weighed again, and consumed tissue was calculated by subtracting the final weight from the initial weight. Dual-choice experiments with P. brassicae were performed as described above except that two 48-h-old larvae fed on Col-0 were used per Petri dish. In both experiments, four different combinations were tested: Col-0 versus myc234, Col-0 versus quadGS, Col-0 versus coi1-1, and myc234 versus quadGS. Data represent the average values of 25 to 30 independent samples.
Feeding preference of S. littoralis between the inner and the outer leaf blade of different A. thaliana genotypes was determined as follows: two neonate larvae were placed on a 4-week-old plant and allowed to feed for 36 h. The number of feeding sites on inner and outer leaf blade was scored on each eaten leaf. Twenty-eight plants were used per A. thaliana genotype. Experiments with P. brassicae were done similarly except that one neonate larva was used per plant and feeding time was reduced to 24 h. Experiments were repeated three times independently with similar results.
Oviposition two-choice tests were performed with 4-week-old plants under constant light. Three female and two male P. brassicae butterflies were placed in an 11-liter Plexiglas box containing four plants of each genotype. Number of laid eggs was assessed after 12 h. Each comparison was repeated 10 times.
Microarray Experiments and Data Analysis
For microarray analyses, leaves of 35 4-week-old Col-0 and myc2 myc3 myc4 plants were harvested and flash-frozen in liquid nitrogen. Total RNA was extracted, reverse-transcribed, and processed according to a previously published procedure (Bodenhausen and Reymond, 2007). Each experiment was replicated three times independently. Labeled probes were hybridized onto CATMA v4 microarrays containing 32,998 A. thaliana gene-specific tags and gene family tags (Sclep et al., 2007). Hybridization and scanning have been described previously (Reymond et al., 2004). Data analysis was carried out using an interface developed at the University of Lausanne (Gene Expression Data Analysis Interface) (Liechti et al., 2010). Differentially expressed genes between Col-0 and myc2 myc3 myc4 were identified by fitting a linear model for each gene and evaluating the fold change and moderated t statistic P values (Smyth, 2004). Adjusted P values were obtained by correcting for multiple testing using the false discovery rate method of Storey and Tibshirani (2003). Microarray data have been submitted to the ArrayExpress database (www.ebi.ac.uk/arrayexpress) under accession number E-MTAB-1777.
Quantitative Real-Time PCR
Leaf samples from five 10 plants were harvested and pooled after 48 h of herbivory by S. littoralis. Tissue samples were ground in liquid nitrogen, and total RNA was extracted using the RNeasy plant mini kit and treated with DNaseI (Qiagen). Afterwards, cDNA was synthesized from RNA using M-MLV reverse transcriptase (Invitrogen) and subsequently diluted fourfold with water. Gene-specific primers were designed to produce amplicons between 80 and 120 bp (see Supplemental Table 5 online). Primer efficiencies (E) were evaluated by five-step dilution regression. Quantitative real-time PCR reactions were performed using Brilliant II Fast SYBR-Green QPCR Master Mix on an Mx3000P real-time PCR instrument (Agilent) with the following program: 95°C for 5 min, then 40 cycles of 10 s at 95°C, 20 s at 55°C, and 30 s at 60°C. Values were normalized to the housekeeping gene ACTIN8. The expression level of a target gene (TG) was normalized to the reference gene (RG) and calculated as normalized relative quantity (NRQ) as follows: NRQ = ECtRG/ECtTG. For each experiment, three biological replicates were analyzed.
GS Analysis
For GS analysis, seven 3-week-old plants were challenged for 48 h with two neonate S. littoralis larvae per plant. Unchallenged plants were used as controls. Samples from four biologically independent replicates were analyzed. Extraction method, ultrahigh-pressure liquid chromatography coupled to quadrupole time-of-flight mass spectrometry measurements, and analysis have been recently described (Glauser et al., 2012). List of GS analyzed in this study are found in Supplemental Table 6 online.
Yeast Two-Hybrid Assays
MYC293-160, MYC382-141, and MYC499-150 derivatives and JAZ1 and JAZ9 full-length proteins in pGBKT7gateway (GAL4 binding domain) were generated as previously described by Fernández-Calvo et al. (2011) and Chini et al. (2009), respectively. Full-length sequences of MYB28, MYB29, MYB34, MYB51, MYB76, and MYB122 were amplified with Expand High Fidelity polymerase (Roche) using Gateway-compatible primers. PCR products were cloned into pDONR207 with a Gateway BP II kit (Invitrogen) and sequence verified. These MYB constructs were used in Gateway LR reactions, in combination with the destination high-copy yeast expression vector pGADT7gateway (Gal4 activation domain). To assess protein interactions, the corresponding plasmids were cotransformed into Saccharomyces cerevisiae AH109 cells following standard heat shock protocols (Chini et al., 2009). Successfully transformed colonies were identified on yeast synthetic dropout lacking Leu and Trp. At 3 d after transformation, yeast colonies were grown in selective -Leu, -Trp liquid media for 6 h, and the cell density was adjusted to 3 × 107 cells mL−1 (OD600 = 1). A 3-μL sample of the cell suspensions was plated out on yeast synthetic dropout lacking Ade, His, Leu, and Trp to test protein interaction. Plates were incubated at 28°C for 2 to 4 d. The empty vectors pGADT7gateway or pGBKT7gateway were also cotransformed as negative controls.
For JAZ–MYB interactions, full-length MYB proteins were cloned in pDEST22 (Invitrogen) and full-length JAZ proteins were cloned in pDEST32 (Invitrogen). Transformation and selection was then carried out as mentioned above.
Recombinant Proteins
Coding sequences for full-length MYB28, MYB29, MYB34, MYB51, MYB76, and MYB122 proteins were PCR amplified from cDNA, maintaining both the frame and the stop codons. Using the Gateway system (Invitrogen), these amplicons were cloned into pDONR207 and recombined in pDEST-TH1 (Hammarström et al., 2002) to obtain N-terminal maltose binding protein (MBP) fusions. All constructs were verified by sequencing prior to protein expression. Recombinant MBP fusion proteins were expressed in Escherichia coli BL21 cells and were then purified in amylose resin columns (New England Biolabs), following a method previously described (Chini et al., 2007). Protein purity was assessed by Coomassie blue gel staining, and quantification was performed in gels by comparison with known concentrations of BSA.
Protein Extracts and Pull-Down Assays
Ten-day-old A. thaliana wild-type seedlings (Col-0) and lines expressing 35S:MYC2:GFP, 35S:MYC3-GFP, or 35S:MYC4-GFP were ground in liquid nitrogen and homogenized in extraction buffer containing 50 mM Tris-HCl, pH 7.4, 80 mM NaCl, 10% glycerol, 0.1% Tween 20, 1 mM DTT, 1 mM phenylmethylsulphonyl fluoride, 50 μM MG132 (Sigma-Aldrich), and complete protease inhibitor (Roche). After centrifugation (16,000g at 4°C), the supernatant was collected. For in vivo pull-down experiments, 6 μg of resin-bound MBP fusion protein (MBP-MYB28, MBP-MYB29, MBP-MYB34, MBP-MYB51, MBP-MYB76, and MBP-MYB122) was added to 1 mg of total protein extract and incubated for 1 h at 4°C with rotation. After washing, samples were denaturalized, loaded onto 8% SDS-PAGE gels, transferred to polyvinylidene fluoride membranes (Millipore), and incubated with anti-GFP–horseradish peroxidase (Milteny Biotec) antibody. A 3-μL aliquot of MBP-fused protein of each sample was run into SDS-PAGE gels and stained with Coomassie Brilliant Blue to confirm equal protein loading.
ChIP-Seq Analyses
Three-day-old etiolated jin1-8 pMYC2:MYC2-FLAG (Hou et al., 2010) seedlings were treated with JA via the gaseous phase for 8 h. The JA treatment was conducted in a closed container with a concentration of 1 μL methyl jasmonate (95% purity; Sigma-Aldrich) per 1 liter of container volume. Approximately 2 g of seedlings was cross-linked for 20 min in 1% formaldehyde. ChIP assays were performed as previously described with minor modifications (Gendrel et al., 2002). The sonicated chromatin was immunoprecipitated with an anti-FLAG antibody (Sigma-Aldrich; F1804) overnight. Incubation of chromatin with mouse IgG (Jackson Immuno Research Laboratories) served as our mock immunoprecipitation control. Protein G Dynabeads (Life Technologies) were used to capture the immunocomplexes. After reverse cross-linking and proteinase-K digestion, the DNA was extracted with phenol-chloroform and then precipitated with ethanol. The immunoprecipitated DNA was subsequently used for ChIP-Seq library construction using the TruSeq DNA Sample preparation kit (Illumina) and single-end sequenced (100 base reads) on the Illumina HiSequation 2500 according to the manufacturer's instructions.
Reads were aligned to a TAIR10 index using Bowtie 2 version 2-2.0.5 (Langmead and Salzberg, 2012) with default parameters. Enriched peaks of MYC2 reads were determined using MACS2 version 2.0.10.20130306 (Zhang et al., 2008; https://github.com/taoliu/MACS/). Statistically significant peaks were identified by applying a cutoff of P ≤ 10−10 and fold enrichment ≥3. Data were visualized using the Anno-J viewer (http://www.annoj.org). Biological replicate data showed comparable results, and the strongest replicate was selected for further analyses. Statistical significance of the overlap between MYC2-bound GS genes and significantly differentially regulated GS transcripts was calculated using a hypergeometric distribution. The number of occurrences of the core MYC2 binding motif within MYC2 peaks associated with GS genes was calculated using FIMO (Grant et al., 2011) against the central six nucleotides of a MYC2 motif previously determined in vitro (Fernández-Calvo et al., 2011).
Accession Numbers
Sequence data from this article can be found in The Arabidopsis Information Resource (www.Arabidopsis.org) under the following accession numbers: MYC2 (At1g32640), MYC3 (At5g46760), MYC4 (At4g17880), PDF1.2 (At5g44420), HEL (At3g04720), CYS PROT (At4g11320), MYB28 (At5g61420), MYB29 (At5g07690), MYB34 (At5g60890), MYB51 (At1g18570), MYB76 (At5g07700), MYB122 (At1g74080), JAZ1 (At1g19180), JAZ2 (At1g74950), JAZ3 (At3g17860), JAZ4 (At2g48500), JAZ5 (At1g17380), JAZ6 (At1g72450), JAZ7 (At2g34600), JAZ8 (At1g30135), JAZ9 (At1g70700), JAZ10 (At5g13220), JAZ11 (At3g43440), and JAZ12 (At5g20900). Accession numbers for other genes discussed in this article are listed in Table 1.
Table 1. Genes Differentially Expressed Between Col-0 and myc2myc3myc4 Triple Mutants.
| AGI Code | Gene Namea | Description | Col-0/myc234b | Adj. P Value |
|---|---|---|---|---|
| At3g19710 | BCAT4 | Branched-chain aminotransferase | 62.81 | 0.001 |
| At1g16410 | CYP79F1 | Cytochrome P450 | 47.70 | 0.000 |
| At4g39940 | APK2 | APS-kinase | 35.27 | 0.000 |
| At3g58990 | IPMI-SSU3 | Aconitase | 32.75 | 0.000 |
| At5g14200 | IPMDH1 | 3-Isopropylmalate dehydrogenase | 30.01 | 0.006 |
| At3g02020 | AK3 | Asp kinase | 25.80 | 0.000 |
| At4g13770 | CYP83A1 | Cytochrome P450 | 20.77 | 0.002 |
| At1g62560 | FMO-GSOX3 | Flavin-containing monooxygenase | 16.08 | 0.003 |
| At1g18590 | SOT17 | Sulfotransferase family protein | 15.50 | 0.001 |
| At4g12030 | BAT5 | Bile acid:sodium symporter family protein | 15.30 | 0.002 |
| At1g52400 | BG1 | β-Glucosidase | 15.17 | 0.001 |
| At5g23010 | MAM1 | 2-Isopropylmalate synthase | 14.76 | 0.006 |
| At1g14250 | GDA1/CD39 nucleoside phosphatase | 14.11 | 0.006 | |
| At1g65860 | FMO-GSOX1 | Flavin-containing monooxygenase | 13.48 | 0.002 |
| At2g39330 | Jacalin lectin | 11.85 | 0.004 | |
| At4g39950 | CYP79B2 | Cytochrome P450 | 11.42 | 0.007 |
| At1g52000 | Jacalin lectin | 11.34 | 0.010 | |
| At5g23020 | MAM3 | 2-Isopropylmalate synthase | 10.58 | 0.001 |
| At4g31500 | CYP83B1 | Cytochrome P450 | 10.33 | 0.005 |
| At2g46650 | ATCB5-C | Cytochrome b5 | 8.63 | 0.002 |
| At4g17470 | Palmitoyl protein thioesterase | 8.38 | 0.026 | |
| At4g03060 | AOP2 | 2-Oxoglutarate-dependent dioxygenase | 8.19 | 0.013 |
| At2g39030 | NATA1 | Orn acetyltransferase | 7.74 | 0.020 |
| At2g14750 | APK1 | APS-kinase | 7.66 | 0.006 |
| At4g17880 | MYC4 | bHLH TF | 6.68 | 0.003 |
| At3g53490 | Unknown protein | 6.61 | 0.004 | |
| At4g14040 | SBP2 | Selenium binding protein | 6.44 | 0.017 |
| At1g52030 | MBP2 | Myrosinase binding protein | 6.00 | 0.003 |
| At1g52040 | MBP1 | Myrosinase binding protein | 5.92 | 0.010 |
| At3g03190 | GSTF11 | Glutathione S-transferase | 5.69 | 0.004 |
| At3g25760 | AOC1 | Allene oxide cyclase | 5.31 | 0.011 |
| At1g61120 | TPS04 | Terpene synthase | 5.22 | 0.019 |
| At1g78370 | GSTU20 | Glutathione S-transferase | 4.66 | 0.039 |
| At4g04830 | MSRB5 | Met sulfoxide reductase | 4.66 | 0.039 |
| At2g22330 | CYP79B3 | Cytochrome P450 | 4.55 | 0.026 |
| At2g38240 | 2-Oxoglutarate- and Fe(II)-dependent oxygenase | 4.47 | 0.035 | |
| At3g54600 | Gln-amido transferase | 4.38 | 0.050 | |
| At1g73325 | Trypsin and protease inhibitor | 4.31 | 0.040 | |
| At4g04610 | APR1 | PAPS reductase | 4.21 | 0.050 |
| At1g74100 | SOT16 | Sulfotransferase | 4.12 | 0.006 |
| At3g22840 | ELIP1 | Early light-inducible protein | 4.10 | 0.010 |
| At5g05600 | 2-Oxoglutarate- and Fe(II)-dependent oxygenase | 3.97 | 0.013 | |
| At1g24100 | UGT74B1 | UDP-Glc:thiohydroximate S-glucosyltransferase | 3.93 | 0.006 |
| At2g43100 | IPMI-SSU2 | Aconitase | 3.90 | 0.045 |
| At1g74090 | SOT18 | Sulfotransferase | 3.76 | 0.010 |
| At4g29700 | Alkaline phosphatase | 3.59 | 0.031 | |
| At1g52410 | TSA1 | TSK-associating protein, calcium ion binding | 3.46 | 0.040 |
| At2g14247 | Unknown protein | 3.39 | 0.038 | |
| At2g06050 | OPR3 | 12-Oxophytodienoate reductase | 3.36 | 0.013 |
| At2g20610 | SUR1 | C-S lyase | 3.33 | 0.019 |
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Expression of Glucosinolate Genes in Single myc Mutants.
Supplemental Figure 2. MYC2 Overexpression Enhances the Expression of GS Biosynthesis Genes.
Supplemental Figure 3. P. brassicae Oviposition on Col-0, myc234, and quadGS.
Supplemental Figure 4. Expression of GS-Related MYBs.
Supplemental Figure 5. JAZ Repressors Do Not Interact with GS-Related MYBs.
Supplemental Figure 6. MYC2, MYC3, and MYC4 Interact with MYB29, MYB76, MYB51, and MYB122 in Pull-Down Assays.
Supplemental Figure 7. Promoter Analysis of Glucosinolate Genes.
Supplemental Figure 8. Conserved MYC and MYB cis-Elements in Arabidopsis lyrata Promoters.
Supplemental Figure 9. Expression of Selected COI1-Dependent Defense Genes.
Supplemental Figure 10. Expression of MYC2, MYC3, and MYC4.
Supplemental Table 1. Microarray Data for Glucosinolate Biosynthesis Genes.
Supplemental Table 2. Glucosinolate Content in Col-0, myc2234, and coi1-1.
Supplemental Table 3. List of GS Genes with Enriched Reads in ChIP-Seq Data.
Supplemental Table 4. List of GS Genes Containing a MYC2 Binding Motif.
Supplemental Table 5. Primers Used in This Study.
Supplemental Table 6. Glucosinolates Analyzed by UHPLC-QTOFMS.
Supplemental Data Set 1. Microarray Data from Col-0 versus myc234 Comparison.
Acknowledgments
We thank Blaise Tissot (University of Lausanne, Switzerland) for maintenance of the plants, and Roland Reist (Syngenta, Stein, Switzerland) for providing S. littoralis eggs. We thank Yunde Zhao (University of California San Diego) for the gift of the cyp79B2 cyp79B3 double mutant, Piero Morandini (University of Milan, Italy) for the myb28 myb29 double mutant, and Jane Glazebrook (University of Minnesota) for the coi1-1 nonglabrous mutant. We thank Hao Yu (National University of Singapore) for jin1-8 pMYC2:MYC2-FLAG seeds. We thank Joe Nery (Salk Institute Genomic Analysis Laboratory) for sequencing operations. Research in R.S.'s laboratory is supported by grants from the Ministry of Science and Innovation (BIO2010-21739, CSD2007-00057-B, and EUI2008-03666). M.Z. was supported by Research Fellowship Za 730/1-1 from the German Research Foundation (Deutsche Forschungsgemeinschaft). M.G.L. was supported by an EU Marie Curie International Outgoing Fellowship (Project 252475) and the National Science Foundation (Grant MCB-1024999). Research in J.R.E.’s laboratory is supported by the National Science Foundation (Grant MCB-1024999), the Howard Hughes Medical Institute, and the Gordon and Betty Moore Foundation. J.R.E. is a Howard Hughes Medical Institute and Gordon and Betty Moore Foundation Investigator. P.R. research is supported by the Swiss National Science Foundation.
AUTHOR CONTRIBUTIONS
F.S., R.S., and P.R. designed the research. F.S., P.F.-C., M.D.-D., S.F., and M.Z. performed the research. G.G. measured GSs. M.G.L. designed and analyzed ChIP-Seq data. R.S., M.G.L., and J.R.E. read and edited the article. F.S. and P.R. wrote the article.
Glossary
- GS
glucosinolate
- JA
jasmonate
- TF
transcription factor
- bHLH
basic helix-loop-helix
- Col-0
Columbia-0
- ChIP-Seq
chromatin immunoprecipitation sequencing
References
- Barth C., Jander G. (2006). Arabidopsis myrosinases TGG1 and TGG2 have redundant function in glucosinolate breakdown and insect defense. Plant J. 46: 549–562 [DOI] [PubMed] [Google Scholar]
- Beekwilder J., et al. (2008). The impact of the absence of aliphatic glucosinolates on insect herbivory in Arabidopsis. PLoS ONE 3: e2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodenhausen N., Reymond P. (2007). Signaling pathways controlling induced resistance to insect herbivores in Arabidopsis. Mol. Plant Microbe Interact. 20: 1406–1420 [DOI] [PubMed] [Google Scholar]
- Bones A.M., Rossiter J.T. (2006). The enzymic and chemically induced decomposition of glucosinolates. Phytochemistry 67: 1053–1067 [DOI] [PubMed] [Google Scholar]
- Browse J. (2009). Jasmonate passes muster: A receptor and targets for the defense hormone. Annu. Rev. Plant Biol. 60: 183–205 [DOI] [PubMed] [Google Scholar]
- Celenza J.L., Quiel J.A., Smolen G.A., Merrikh H., Silvestro A.R., Normanly J., Bender J. (2005). The Arabidopsis ATR1 Myb transcription factor controls indolic glucosinolate homeostasis. Plant Physiol. 137: 253–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chini A., Fonseca S., Chico J.M., Fernández-Calvo P., Solano R. (2009). The ZIM domain mediates homo- and heteromeric interactions between Arabidopsis JAZ proteins. Plant J. 59: 77–87 [DOI] [PubMed] [Google Scholar]
- Chini A., Fonseca S., Fernández G., Adie B.A.T., Chico J.M., Lorenzo O., García-Casado G., López-Vidriero I., Lozano F.M., Ponce M.R., Micol J.L., Solano R. (2007). The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448: 666–671 [DOI] [PubMed] [Google Scholar]
- de Vos M., Kriksunov K.L., Jander G. (2008). Indole-3-acetonitrile production from indole glucosinolates deters oviposition by Pieris rapae. Plant Physiol. 146: 916–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vos M., Van Oosten V.R., Van Poecke R.M.P., Van Pelt J.A., Pozo M.J., Mueller M.J., Buchala A.J., Métraux J.-P., Van Loon L.C., Dicke M., Pieterse C.M. (2005). Signal signature and transcriptome changes of Arabidopsis during pathogen and insect attack. Mol. Plant Microbe Interact. 18: 923–937 [DOI] [PubMed] [Google Scholar]
- Dombrecht B., Xue G.P., Sprague S.J., Kirkegaard J.A., Ross J.J., Reid J.B., Fitt G.P., Sewelam N., Schenk P.M., Manners J.M., Kazan K. (2007). MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis. Plant Cell 19: 2225–2245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlting J., Chowrira S.G., Mattheus N., Aeschliman D.S., Arimura G.-I., Bohlmann J. (2008). Comparative transcriptome analysis of Arabidopsis thaliana infested by diamond back moth (Plutella xylostella) larvae reveals signatures of stress response, secondary metabolism, and signalling. BMC Genomics 9: 154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feller A., Machemer K., Braun E.L., Grotewold E. (2011). Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. Plant J. 66: 94–116 [DOI] [PubMed] [Google Scholar]
- Fernández-Calvo P., et al. (2011). The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell 23: 701–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca S., Chini A., Hamberg M., Adie B.A.T., Porzel A., Kramell R., Miersch O., Wasternack C., Solano R. (2009). (+)-7-iso-Jasmonoyl-L-isoleucine is the endogenous bioactive jasmonate. Nat. Chem. Biol. 5: 344–350 [DOI] [PubMed] [Google Scholar]
- Gendrel A.V., Lippman Z., Yordan C., Colot V., Martienssen R.A. (2002). Dependence of heterochromatic histone H3 methylation patterns on the Arabidopsis gene DDM1. Science 297: 1871–1873 [DOI] [PubMed] [Google Scholar]
- Gigolashvili T., Berger B., Mock H.-P., Müller C., Weisshaar B., Flügge U.-I. (2007a). The transcription factor HIG1/MYB51 regulates indolic glucosinolate biosynthesis in Arabidopsis thaliana. Plant J. 50: 886–901 [DOI] [PubMed] [Google Scholar]
- Gigolashvili T., Engqvist M., Yatusevich R., Müller C., Flügge U.-I. (2008). HAG2/MYB76 and HAG3/MYB29 exert a specific and coordinated control on the regulation of aliphatic glucosinolate biosynthesis in Arabidopsis thaliana. New Phytol. 177: 627–642 [DOI] [PubMed] [Google Scholar]
- Gigolashvili T., Yatusevich R., Berger B., Müller C., Flügge U.-I. (2007b). The R2R3-MYB transcription factor HAG1/MYB28 is a regulator of methionine-derived glucosinolate biosynthesis in Arabidopsis thaliana. Plant J. 51: 247–261 [DOI] [PubMed] [Google Scholar]
- Glauser G., Schweizer F., Turlings T.C.J., Reymond P. (2012). Rapid profiling of intact glucosinolates in Arabidopsis leaves by UHPLC-QTOFMS using a charged surface hybrid column. Phytochem. Anal. 23: 520–528 [DOI] [PubMed] [Google Scholar]
- Godoy M., Franco-Zorrilla J.M., Pérez-Pérez J., Oliveros J.C., Lorenzo O., Solano R. (2011). Improved protein-binding microarrays for the identification of DNA-binding specificities of transcription factors. Plant J. 66: 700–711 [DOI] [PubMed] [Google Scholar]
- Grant C.E., Bailey T.L., Noble W.S. (2011). FIMO: Scanning for occurrences of a given motif. Bioinformatics 27: 1017–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammarström M., Hellgren N., van Den Berg S., Berglund H., Härd T. (2002). Rapid screening for improved solubility of small human proteins produced as fusion proteins in Escherichia coli. Protein Sci. 11: 313–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai M.Y., et al. (2007). Omics-based identification of Arabidopsis Myb transcription factors regulating aliphatic glucosinolate biosynthesis. Proc. Natl. Acad. Sci. USA 104: 6478–6483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong G.-J., Xue X.-Y., Mao Y.-B., Wang L.-J., Chen X.-Y. (2012). Arabidopsis MYC2 interacts with DELLA proteins in regulating sesquiterpene synthase gene expression. Plant Cell 24: 2635–2648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou X., Lee L.Y.C., Xia K., Yan Y., Yu H. (2010). DELLAs modulate jasmonate signaling via competitive binding to JAZs. Dev. Cell 19: 884–894 [DOI] [PubMed] [Google Scholar]
- Howe G.A., Jander G. (2008). Plant immunity to insect herbivores. Annu. Rev. Plant Biol. 59: 41–66 [DOI] [PubMed] [Google Scholar]
- Katsir L., Schilmiller A.L., Staswick P.E., He S.Y., Howe G.A. (2008). COI1 is a critical component of a receptor for jasmonate and the bacterial virulence factor coronatine. Proc. Natl. Acad. Sci. USA 105: 7100–7105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazan K., Manners J.M. (2013). MYC2: The master in action. Mol. Plant 6: 686–703 [DOI] [PubMed] [Google Scholar]
- Kester K., Peterson S., Hanson F., Jackson D., Severson R. (2002). The roles of nicotine and natural enemies in determining larval feeding site distributions of Manduca sexta L. and Manduca quinquemaculata (Haworth) on tobacco. Chemoecology 12: 1–10 [Google Scholar]
- Kliebenstein D.J., Figuth A., Mitchell-Olds T. (2002). Genetic architecture of plastic methyl jasmonate responses in Arabidopsis thaliana. Genetics 161: 1685–1696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebenstein D.J., Kroymann J., Mitchell-Olds T. (2005). The glucosinolate-myrosinase system in an ecological and evolutionary context. Curr. Opin. Plant Biol. 8: 264–271 [DOI] [PubMed] [Google Scholar]
- Klopffleisch K., et al. (2011). Arabidopsis G-protein interactome reveals connections to cell wall carbohydrates and morphogenesis. Mol. Syst. Biol. 7: 532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Salzberg S.L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9: 357–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Zhao Y., McCaig B.C., Wingerd B.A., Wang J., Whalon M.E., Pichersky E., Howe G.A. (2004). The tomato homolog of CORONATINE-INSENSITIVE1 is required for the maternal control of seed maturation, jasmonate-signaled defense responses, and glandular trichome development. Plant Cell 16: 126–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liechti R., et al. (2010). EuroDia: A beta-cell gene expression resource. Database (Oxford) 2010: baq024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mewis I., Tokuhisa J.G., Schultz J.C., Appel H.M., Ulrichs C., Gershenzon J. (2006). Gene expression and glucosinolate accumulation in Arabidopsis thaliana in response to generalist and specialist herbivores of different feeding guilds and the role of defense signaling pathways. Phytochemistry 67: 2450–2462 [DOI] [PubMed] [Google Scholar]
- Mikkelsen M.D., Petersen B.L., Glawischnig E., Jensen A.B., Andreasson E., Halkier B.A. (2003). Modulation of CYP79 genes and glucosinolate profiles in Arabidopsis by defense signaling pathways. Plant Physiol. 131: 298–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller R., de Vos M., Sun J.Y., Sønderby I.E., Halkier B.A., Wittstock U., Jander G. (2010). Differential effects of indole and aliphatic glucosinolates on lepidopteran herbivores. J. Chem. Ecol. 36: 905–913 [DOI] [PubMed] [Google Scholar]
- Niu Y., Figueroa P., Browse J. (2011). Characterization of JAZ-interacting bHLH transcription factors that regulate jasmonate responses in Arabidopsis. J. Exp. Bot. 62: 2143–2154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nour-Eldin H.H., Andersen T.G., Burow M., Madsen S.R., Jørgensen M.E., Olsen C.E., Dreyer I., Hedrich R., Geiger D., Halkier B.A. (2012). NRT/PTR transporters are essential for translocation of glucosinolate defence compounds to seeds. Nature 488: 531–534 [DOI] [PubMed] [Google Scholar]
- Paschold A., Halitschke R., Baldwin I.T. (2007). Co(i)-ordinating defenses: NaCOI1 mediates herbivore- induced resistance in Nicotiana attenuata and reveals the role of herbivore movement in avoiding defenses. Plant J. 51: 79–91 [DOI] [PubMed] [Google Scholar]
- Pauwels L., et al. (2010). NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature 464: 788–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pires N., Dolan L. (2010). Origin and diversification of basic-helix-loop-helix proteins in plants. Mol. Biol. Evol. 27: 862–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi T., Song S., Ren Q., Wu D., Huang H., Chen Y., Fan M., Peng W., Ren C., Xie D. (2011). The Jasmonate-ZIM-domain proteins interact with the WD-Repeat/bHLH/MYB complexes to regulate Jasmonate-mediated anthocyanin accumulation and trichome initiation in Arabidopsis thaliana. Plant Cell 23: 1795–1814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratzka A., Vogel H., Kliebenstein D.J., Mitchell-Olds T., Kroymann J. (2002). Disarming the mustard oil bomb. Proc. Natl. Acad. Sci. USA 99: 11223–11228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renwick J.A.A., Haribal M., Gouinguené S., Städler E. (2006). Isothiocyanates stimulating oviposition by the diamondback moth, Plutella xylostella. J. Chem. Ecol. 32: 755–766 [DOI] [PubMed] [Google Scholar]
- Reymond P., Bodenhausen N., Van Poecke R.M.P., Krishnamurthy V., Dicke M., Farmer E.E. (2004). A conserved transcript pattern in response to a specialist and a generalist herbivore. Plant Cell 16: 3132–3147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymond P., Weber H., Damond M., Farmer E.E. (2000). Differential gene expression in response to mechanical wounding and insect feeding in Arabidopsis. Plant Cell 12: 707–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlaeppi K., Bodenhausen N., Buchala A., Mauch F., Reymond P. (2008). The glutathione-deficient mutant pad2-1 accumulates lower amounts of glucosinolates and is more susceptible to the insect herbivore Spodoptera littoralis. Plant J. 55: 774–786 [DOI] [PubMed] [Google Scholar]
- Schoonhoven, L.M., van Loon, J.J.A., and Dicke, M. (2005). Insect-Plant Biology. (Oxford, UK: Oxford University Press). [Google Scholar]
- Schweizer F., Bodenhausen N., Lassueur S., Masclaux F.G., Reymond P. (2013). Differential contribution of transcription factors to Arabidopsis thaliana defence against Spodoptera littoralis. Front. Plant Sci. 4: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sclep G., Allemeersch J., Liechti R., De Meyer B., Beynon J., Bhalerao R., Moreau Y., Nietfeld W., Renou J.-P., Reymond P., Kuiper M.T., Hilson P. (2007). CATMA, a comprehensive genome-scale resource for silencing and transcript profiling of Arabidopsis genes. BMC Bioinformatics 8: 400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheard L.B., et al. (2010). Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature 468: 400–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J., Heidrich K., Sanchez-Villarreal A., Parker J.E., Davis S.J. (2012). TIME FOR COFFEE represses accumulation of the MYC2 transcription factor to provide time-of-day regulation of jasmonate signaling in Arabidopsis. Plant Cell 24: 2470–2482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shroff R., Vergara F., Muck A., Svatos A., Gershenzon J. (2008). Nonuniform distribution of glucosinolates in Arabidopsis thaliana leaves has important consequences for plant defense. Proc. Natl. Acad. Sci. USA 105: 6196–6201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth G.K. (2004). Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3: e3. [DOI] [PubMed] [Google Scholar]
- Sønderby I.E., Burow M., Rowe H.C., Kliebenstein D.J., Halkier B.A. (2010a). A complex interplay of three R2R3 MYB transcription factors determines the profile of aliphatic glucosinolates in Arabidopsis. Plant Physiol. 153: 348–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sønderby I.E., Geu-Flores F., Halkier B.A. (2010b). Biosynthesis of glucosinolates—Gene discovery and beyond. Trends Plant Sci. 15: 283–290 [DOI] [PubMed] [Google Scholar]
- Sønderby I.E., Hansen B.G., Bjarnholt N., Ticconi C., Halkier B.A., Kliebenstein D.J. (2007). A systems biology approach identifies a R2R3 MYB gene subfamily with distinct and overlapping functions in regulation of aliphatic glucosinolates. PLoS ONE 2: e1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S., Qi T., Huang H., Ren Q., Wu D., Chang C., Peng W., Liu Y., Peng J., Xie D. (2011). The Jasmonate-ZIM domain proteins interact with the R2R3-MYB transcription factors MYB21 and MYB24 to affect Jasmonate-regulated stamen development in Arabidopsis. Plant Cell 23: 1000–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey J.D., Tibshirani R. (2003). Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA 100: 9440–9445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J.Y., Sønderby I.E., Halkier B.A., Jander G., de Vos M. (2009). Non-volatile intact indole glucosinolates are host recognition cues for ovipositing Plutella xylostella. J. Chem. Ecol. 35: 1427–1436 [DOI] [PubMed] [Google Scholar]
- Thines B., Katsir L., Melotto M., Niu Y., Mandaokar A., Liu G., Nomura K., He S.Y., Howe G.A., Browse J. (2007). JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 448: 661–665 [DOI] [PubMed] [Google Scholar]
- Wittstock U., Agerbirk N., Stauber E.J., Olsen C.E., Hippler M., Mitchell-Olds T., Gershenzon J., Vogel H. (2004). Successful herbivore attack due to metabolic diversion of a plant chemical defense. Proc. Natl. Acad. Sci. USA 101: 4859–4864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittstock U., Gershenzon J. (2002). Constitutive plant toxins and their role in defense against herbivores and pathogens. Curr. Opin. Plant Biol. 5: 300–307 [DOI] [PubMed] [Google Scholar]
- Wittstock U., Halkier B.A. (2002). Glucosinolate research in the Arabidopsis era. Trends Plant Sci. 7: 263–270 [DOI] [PubMed] [Google Scholar]
- Xie D.X., Feys B.F., James S., Nieto-Rostro M., Turner J.G. (1998). COI1: An Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280: 1091–1094 [DOI] [PubMed] [Google Scholar]
- Yamashino T., Matsushika A., Fujimori T., Sato S., Kato T., Tabata S., Mizuno T. (2003). A link between circadian-controlled bHLH factors and the APRR1/TOC1 quintet in Arabidopsis thaliana. Plant Cell Physiol. 44: 619–629 [DOI] [PubMed] [Google Scholar]
- Yan Y., Stolz S., Chételat A., Reymond P., Pagni M., Dubugnon L., Farmer E.E. (2007). A downstream mediator in the growth repression limb of the jasmonate pathway. Plant Cell 19: 2470–2483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yatusevich R., Mugford S.G., Matthewman C., Gigolashvili T., Frerigmann H., Delaney S., Koprivova A., Flügge U.-I., Kopriva S. (2010). Genes of primary sulfate assimilation are part of the glucosinolate biosynthetic network in Arabidopsis thaliana. Plant J. 62: 1–11 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W., Liu X.S. (2008). Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9: R137. [DOI] [PMC free article] [PubMed] [Google Scholar]