Abstract
Recoverin, a member of the neuronal calcium sensor (NCS) branch of the calmodulin superfamily, is expressed in retinal photoreceptor cells and serves as a calcium sensor in vision. Ca2+-induced conformational changes in recoverin cause extrusion of its covalently attached myristate (termed Ca2+-myristoyl switch) that promote translocation of recoverin to disk membranes during phototransduction in retinal rod cells. Here we report double electron-electron resonance (DEER) experiments on recoverin that probe Ca2+-induced changes in distance as measured by the dipolar coupling between spin labels strategically positioned at engineered cysteine residues on the protein surface. The DEER distance between nitroxide spin-labels attached at C39 and N120C is 2.5 ±0.1 nm for Ca2+-free recoverin and 3.7 ±0.1 nm for Ca2+-bound recoverin. An additional DEER distance (5 - 6 nm) observed for Ca2+-bound recoverin may represent an intermolecular distance between C39 and N120. 15N NMR relaxation analysis and CW-EPR experiments both confirm that Ca2+-bound recoverin forms a dimer at protein concentrations above 100 μM, whereas Ca2+-free recoverin is monomeric. We propose that Ca2+-induced dimerization of recoverin at the disk membrane surface may play a role in regulating Ca2+-dependent phosphorylation of dimeric rhodopsin. The DEER approach will be useful for elucidating dimeric structures of NCS proteins in general for which Ca2+-induced dimerization is functionally important but not well understood.
Keywords: Recoverin, NMR, EPR, calcium, Ca2+-myristoyl switch, DEER, vision, phototransduction, EF-hand
Recoverin, a 23 kDa Ca2+-binding protein and member of the calmodulin superfamily, serves as a Ca2+ sensor for regulating phototransduction in retinal rod cells (1-5). Recoverin prolongs the lifetime of photoexcited rhodopsin by inhibiting rhodopsin kinase only at high Ca2+ levels (2, 6-9). Hence, recoverin makes the desensitization of rhodopsin responsive to Ca2+, and the shortened lifetime of photoexcited rhodopsin at low Ca2+ levels may promote visual recovery and contribute to the adaptation to background light. Recoverin was also found localized in the rod inner segment and may serve as a Ca2+ sensor in synaptic termini (10). Upon light activation (at low Ca2+ levels), 98% of recoverin is translocated to the rod inner segment, whereas recoverin is transported to the outer segment in the dark (high Ca2+ levels) (11). This light-dependent protein translocation is likely facilitated by Ca2+-induced conformational changes, termed Ca2+-myristoyl switch (12, 13) that may play a role in light-adaptation.
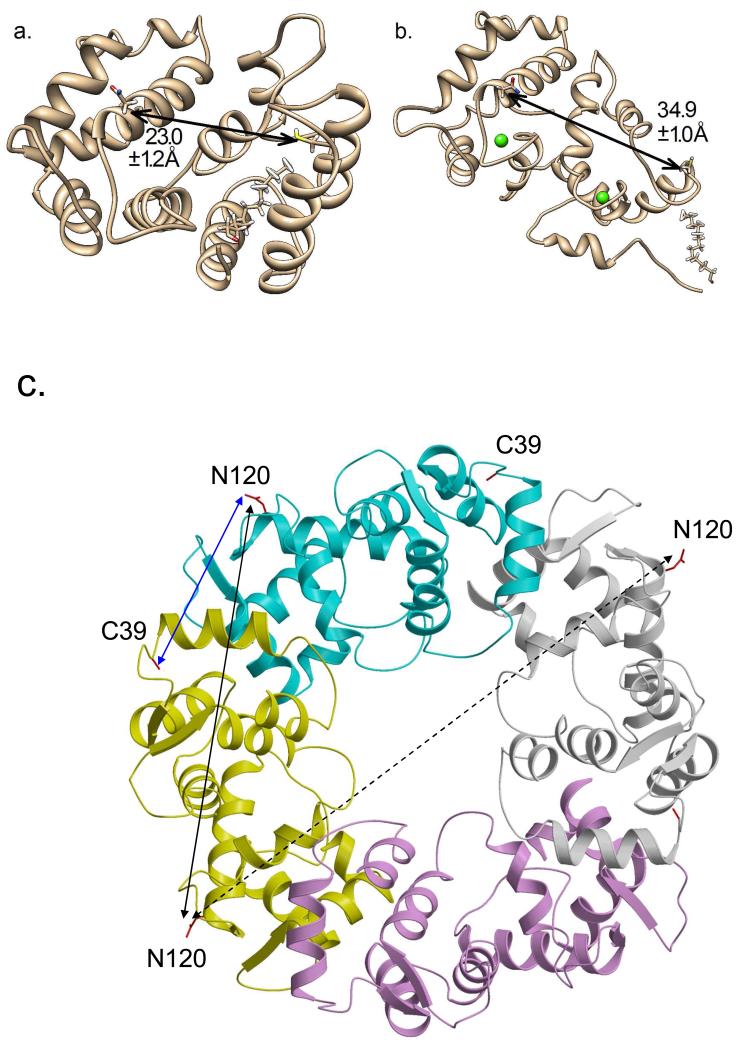
Recoverin belongs to a family of neuronal calcium sensor (NCS) proteins (14, 15), including NCS-1 (16), neurocalcin (17), and hippocalcin (18). The common structural features of NCS proteins are an approximately 200-residue chain containing four EF-hand motifs and amino-terminal myristoylation consensus sequence. Recoverin binds to two Ca2+ at the second and third EF-hands (EF2 and EF3) (19) that induces the binding of myristoylated, but not unmyristoylated, recoverin to rod outer segment disc membranes (12, 13). The three-dimensional structures of myristoylated recoverin with 0 and 2 Ca2+ bound have been determined by NMR spectroscopy (20-22). In the Ca2+-free state, the myristoyl group is sequestered in a deep hydrophobic cavity inside the protein, forming a compact structure (Fig. 1A). The binding of two Ca2+ causes a large protein conformational change that leads to extrusion of the N-terminal myristoyl group (Fig. 1B). The Ca2+-induced exposure of the myristoyl group (Ca2+-myristoyl switch) enables recoverin to bind to membranes only at high Ca2+ (23, 24).
Figure 1.
NMR structures of myristoylated recoverin in (A) Ca2+-free state (1IKU) and (B) Ca2+-bound state (1JSA). Distances between the alpha carbon atoms of C39 and N120 are indicated as the mean and standard deviation of all PDB substructures. (C) Crystal structure of Ca2+-bound non-myristoylated recoverin (2HET) that forms a 4-fold symmetric, crystallographic tetramer. The four tetramer subunits are colored cyan (chain A), yellow (chain B), magenta (chain C), and white (chain D). The intermolecular distance between alpha carbon atoms for C39 and N120 from adjacent subunits (chains A and B) is 2.9 nm (blue arrow). The intermolecular distance between alpha carbon atoms of N120 from adjacent subunits (chains A and B) is 4.7 nm (black solid arrow). The intermolecular distance between alpha carbon atoms of N120 from opposite subunits (chains B and D) is 6.3 nm (dotted arrow).
We report here double electron-electron resonance (DEER) analysis (25-27) on site-directed nitroxide spin labels in recoverin to probe Ca2+-induced conformational changes and protein dimerization. The DEER data reveal a 1.2 nm increase in the distance between spin labels placed at C39 and N120C in recoverin caused by Ca2+ binding. A distance distribution calculated from the DEER data using a rotamer distribution model indicates a single distance between C39 and N120C centered at 2.5 nm for Ca2+-free recoverin, whereas multiple distances (2.9, 3.7, 4.7 and 5.7 nm) are calculated for Ca2+-bound recoverin. The main distance component at 3.7 nm represents the intramolecular distance between C39 and N120C for Ca2+-bound recoverin. The longer distance components (5 - 6 nm) may represent intermolecular distances in the dimeric Ca2+-bound protein. CW-EPR studies on recoverin containing a nitroxide spin label attached at C39 indicate a Ca2+-induced increase in rotational correlation time for recoverin, consistent with Ca2+-induced dimerization. Also, NMR relaxation and pulsed-field gradient echo experiments confirm that Ca2+-bound recoverin forms a dimer at protein concentrations above 100 μM, whereas Ca2+-free recoverin is monomeric. We propose that Ca2+-induced dimerization of recoverin may occur at the disk membrane surface, which could play a role in regulating Ca2+-dependent phosphorylation of dimeric rhodopsin (28-30).
EXPERIMENTAL PROCEDURES
Mutant construction and protein purification
The mutation N120C was introduced into bovine recoverin as described previously (31) and verified by DNA sequencing. The N120C recoverin mutant (hereafter called recoverinN120C) contains two surface cysteine residues (residues 39 and 120) for spin label attachment below.
Recombinant myristoylated wildtype recoverin and recoverinN120C were expressed in E. coli strain, DH5α, co-transfected with plasmids, pTREC2 and pBB131 as described previously (12). The cells were grown in M9-broth in 90% 2H2O at 32° C until the cell culture reached an optical density of 0.7-0.8 at 600 nm. The myristoyl-CoA transferase expression was induced by adding IPTG (0.5 mM). Recoverin expression was induced 30 min. later by rapidly raising the temperature to 42° C and growing the cells for an additional 4 hrs. The protein purification procedure was similar to that used previously (21, 31). Briefly, the cells suspended in lysis buffer were disrupted using a French Pressure Cell (Spectronic Instruments, Inc. USA) and the extract was clarified by centrifugation (10000 g) for 1.5 hrs at 4° C. The crude extract was treated with CaCl2 (3 mM), adsorbed onto a Phenyl Sepharose CL-4B (Pharmacia Biotech) column at 4° C and washed thoroughly using high Ca2+ buffer. The Ca2+-free protein was eluted using Ca2+-free buffer containing EGTA. The protein was further purified using Q-Sepharose chromatography at room temperature and then concentrated to a final concentration of 10 mg/ml using Centricon YM-10 (Millipore Corp., USA). The overall yields were 10 mg from 1 L cultured cells. Proteins were aliquoted, frozen by liquid nitrogen and stored at −80° C. Protein concentrations in this study were determined by measuring the optical density at 280 nm and using a molar absorption coefficient, ε = 23,950 cm−1 M−1. The final purified recoverin sample was more than 98% pure as judged by SDS-PAGE, and more than 95% myrisoylated as confirmed by reverse-phase HPLC and mass spectrometry analysis.
Spin labeling and EPR sample preparation
RecoverinN120C was labeled with MTSSL (2,5-dihydro-2,2,5,5-tetramethyl-3-[[(methylsulfonyl)thio]methyl]-1H-pyrrol-1-yloxy, (1-oxyl-2,2,5,5-tetramethyl-Δ3-pyrroline-3-methyl) methanethiosulfonate spin label) (Toronto Research Chemicals, North York, Ontario, Canada). Any unreacted and/or free MTSSL was removed by gel filtration using two sequential PD-10 columns (GE healthcare). EPR samples of spin-labeled recoverinN120C were prepared at a final protein concentration of 200 μM by exchange into buffer made from 99% D2O containing 50 mM Tris (pD 7.8), 200 mM KCl, and 30% d8-glycerol (Cambridge Isotopes Laboratory). For each sample, 150 μL was loaded into a 4 mm o.d. quartz tube (Wilmad 706-PQ-9.50) and flash frozen in liquid nitrogen.
DEER data collection and analysis
DEER data were acquired at the CalEPR center at UC Davis on a Bruker-Biospin EleXsys E580 spectrometer at 9.5 GHz modified for DEER. Bridge modifications include use of a HP 8761A SPDT switch to input the pump frequency from a HP 83622A source to the manual pulse forming unit (MPFU) and addition of a variable coaxial attenuator (ARRA Inc.) prior to the stripline pulse forming unit (SPFU). This allowed independent power level settings of pump and probe pulses as inputs to the TWTA. A Bruker MS5 split-ring resonator was used in an Oxford Instuments CF935 liquid helium cooling system, at a temperature of 50 K and a magnetic field of about 340 mT. The DEER pulse sequence (π/2)1 − τ1 − (π)1 − τ1 + t − (π)2 − (τ2-t) − (π)1 − τ2 − [echo] was used with pulse lengths of 16/32/32 ns for the probe pulses (subscript 1) and 20 ns for the pump pulse (subscript 2). Pulse lengths were obtained by minimizing TWT-A input power at the observe frequency, followed by minimizing the pump π-pulse width within the HP source power leveler range (15dBm max). The value of τ1 was adjusted to a 1H suppression tau near a maximum in the 2H nuclear modulation envelope (ca. 412 ns), and t was varied between - 60ns and a τ2 setting for about 2.5 modulation periods of the expected dipolar frequency, in increments of 8 or 20 ns. The pump frequency ν2 was centered in the resonator mode and aligned with the spectral maximum. The probe frequency ν1 was 65 MHz above ν2. A repetition time was found that provided at least 95% unsaturated echo intensity. The DEER data were analyzed and fit with DeerAnalysis2011 (32). Through in silico labeling using the molecular dynamics software MMM (33), DEER experimental data was compared to published recoverin structures. A “cryo” setting of 175K, provided the lower temperature conditions to better-capture the liquid nitrogen freezing process which reduces the contributing rotamers present under “ambient” conditions (34).
15N NMR relaxation analysis
15N NMR longitudinal relaxation rates (R1) and transverse relaxation rates (R2) of Ca2+-bound unmyristoylated recoverin were measured using pulse schemes described previously at 60.81 MHz 15N resonance frequency (35). Sample conditions and procedures for the NMR experiments on Ca2+-bound unmyristoylated recoverin were described previously (19). Relaxation delays for R1 experiments were 0.016, 0.16, 0.32, 0.48, 0.65, 0.8, and 0.96 s. Relaxation delays for R2 experiments were 4.8, 9.6, 19.2, 38.4, 57.6, 76.8, and 105.6 ms. The 180° pulses for Car-Purcell-Meiboom-Gill (CPMG) sequence were applied every 1.0 ms. Uncertainty of R1 and R2 values were determined by Monte-Carlo error simulation using signal intensities that contain experimental noise fluctuation. Model-free parameters including generalized order parameters and correlation times for internal motion (36-38) were determined using the protocol described previously (35).
CW-EPR analysis
CW-EPR measurements were carried out in a JEOL X-band spectrometer fitted with a loop-gap resonator (39). Aliquots (5 μl) of purified myristoylated recoverin (150 μM), spin-labeled only at Cys39, were incubated for 12 h at 4°C in buffer: 10 mM Tris pH 8.0, 1 mM MgCl2 and 2 mM CaCl2 (Ca2+-bound recoverin) or 2 mM EGTA (Ca2+-free recoverin) and were placed in sealed quartz capillaries contained in the resonator. Where indicated, a final concentration of 10 % (w/v) sodium-dodecyl sulfate (SDS) was used for protein unfolding, or 30% sucrose (w/v) to increase sample viscosity. Spectra were acquired at room temperature (20–22°C) from a single 2-min scan over a field of 100 G at a microwave power of 2 mW and a modulation amplitude optimized to the natural line width of the individual spectrum (0.5 - 1.5 G). The obtained spectra were double integrated and normalized. Molecular accessibility of spin-labeled side chains to NiEDDA and O2 was determined using successive power saturation scans as described (40). Π1/2 values (which also were used to calculate the contrast function (Φ)) were calculated using software provided by C. Altenbach.
Least-squares simulation of slow/intermediate motion nitroxide EPR spectra were carried out using a LabView version of the NLSL software (41) developed by C. Altenbach downloadable at https://sites.google.com/site/altenbach/labview-programs/epr-programs/multicomponent. Fitting for the effective rotational correlation times of the spin label was carried-out as previuosly described (42), including the standard MTSL magnetic tensor values (gxx= 2.0078, gyy = 2.0058, gzz=2.0022; Axx= 6.2, Ayy= 5.9, Azz=37.0), with the exception that Azz was fixed to 36.6. Rotational correlation times were calculated from the spherical rotational diffusion tensor from the relation τ = 1/(6×10R) (42). All spectra fit reasonably well assuming a two-component macroscopic order microscopic disorder (MOMD) model that describes anisotropic motion of the nitroxide (41) (i.e., sets of rotational diffusion tensors).
RESULTS AND DISCUSSION
Attaching nitroxide spin-labels at strategic sites in recoverin
Recoverin contains only one native cysteine, C39 (Fig. 1), which was used for attachment of a single nitroxide spin label (per recoverin molecule) in the CW-EPR experiments. In order to perform DEER analysis on recoverin, it is necessary to introduce a second cysteine residue using site-directed mutagenesis. The location of the second cysteine was chosen to be on the protein surface such that it undergoes a large change in distance from C39 upon Ca2+-binding to recoverin. The average distance between the alpha carbon atoms for recoverin residues C39 and N120 is 2.3 nm in the NMR structure of Ca2+-free recoverin (Fig. 1A) that increases to 3.5 nm in the NMR structure of Ca2+-bound recoverin (Fig. 1B). Therefore, a cysteine residue was introduced at residue N120, generating the single-site mutant, recoverinN120C (see Methods). This mutant was spin-labeled at both C39 and N120C to measure the distance between these sites in the DEER experiments below.
RecoverinN120C was verified to remain structurally folded and functionally intact. The 15N-1H HSQC spectrum of recoverinN120C looks similar to that of wildtype recoverin (Fig. S1), indicating that recoverinN120C is structurally similar to wildtype. The apparent dissociation constant for Ca2+-binding to recoverinN120C (Kd ~ 20 μM) is nearly the same as that of wildtype (Fig. S2). Lastly, recoverinN120C has an unfolding melting temperature of 62°C, identical to that of wildtype recoverin. Together, these data show that recoverinN120C is structurally intact and undergoes a functional Ca2+-myristoyl switch.
DEER analysis of recoverin
4-pulse DEER measurements were performed on myristoylated recoverinN120C containing nitroxide spin-labels attached at C39 and N120C (Figs. 2A. and 2C and Supplemental Figure S3). Initial DEER experiments on recoverinN120C in the absence of KCl yielded a two-pulse decay time (Tm) of 180 ns that increased significantly to 1.5 microseconds upon addition of 200mM KCl (Fig. 2). The salt dependence of Tm is perhaps explained by increased intermolecular spin-spin interactions due to partial aggregation of the protein in the absence of salt. Thus, all the distance measurements between C39 and N120C in this study were derived from DEER experiments on recoverinN120C in the presence of 200 mM KCl. Proton modulations in the DEER acquisition were large despite a 1H-suppression tau value (time between first and second pulses of sequence). As seen in Figure 2, proton modulations are still present in the sample prepared in D2O, but decreased significantly from that in H2O (not shown). The different proton modulations in the two data sets did not significantly alter the distance distribution, and therefore, the approach of summing over several tau values was not undertaken (43).
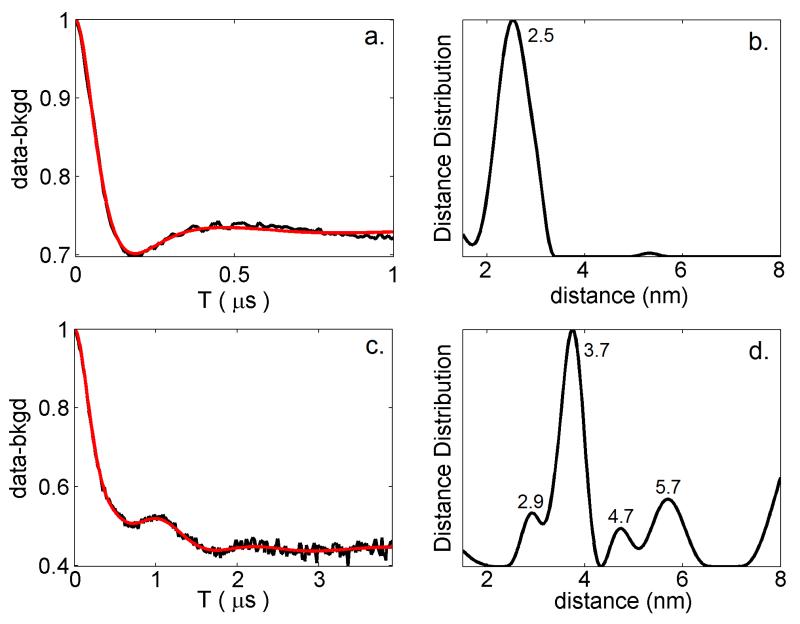
Figure 2.
DEER data (with background subtraction) for Ca2+-free recoverinN120C (A) and Ca2+-bound recoverinN120C (C) collected at 50K, with a rate of 125 Hz. Simulated fits to the DEER data (red solid line) were produced using DeerAnalysis2011 (32). Distance distributions calculated from the DEER simulations are shown for Ca2+-free recoverin (B) and Ca2+-bound recoverin (D).
Distance distributions calculated from the DEER data are shown for Ca2+-free recoverin (Fig. 2B) and Ca2+-bound recoverin (Fig. 2D). Weak peaks in the distance distribution at 5.5 nm (Fig. 2B) and 8 nm (Fig. 2D) are due to background subtraction artifacts. Distribution peaks at 2.5 nm (Fig. 2B) and at 2.9 nm, 3.7 nm, 4.7 nm and 5.7 nm (Fig. 2D) all withstand variations of background subtraction parameters and represent average distances between spin-labels attached at C39 and N120C. For Ca2+-free recoverin, the DEER distance between spin labels attached at C39 and N120C is 2.5 nm (Fig. 2B), which is similar to the intramolecular distance between alpha carbon atoms of C39 and N120 seen in the NMR structure, 1IKU (Fig. 1A). For Ca2+-bound recoverin, the most prominent DEER distance component at 3.7 nm (Fig. 2D) is similar to the intramolecular distance between C39 and N120 seen in the NMR structure, 1JSA (Fig. 1B). Thus, our DEER measurements indicate a 1.2 nm increase in the distance between spin-labels attached at C39 and N120C that occurs upon Ca2+ binding to recoverin. This 1.2 nm increase is in exact agreement with the Ca2+-induced change in the distance between alpha carbon atoms for C39 and N120 as seen in the NMR structures of recoverin (Figs. 1A and 1B). Additional longer distances in the distribution for Ca2+-bound recoverin (5 – 6 nm) may represent intermolecular distances between spin-labels from protein dimers or oligomers. A Ca2+-induced dimerization of recoverin is also supported by a Ca2+-dependent increase in modulation depth, which changes from 0.27 (Ca2+-free recoverin) to 0.55 (Ca2+-bound recoverin), vide infra.
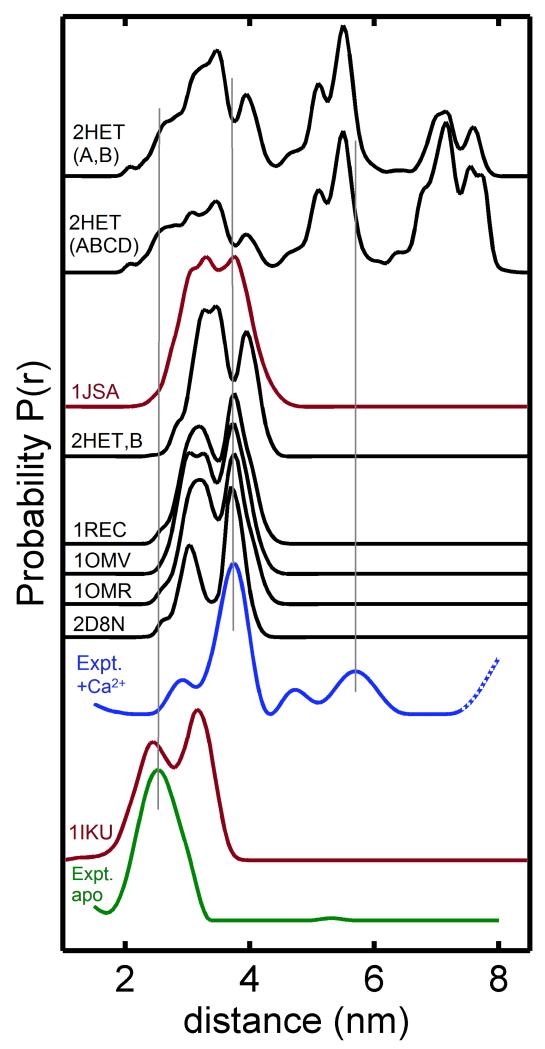
A quantitative comparison of the observed DEER distances with the corresponding distances calculated from known NMR and x-Ray crystal structures of recoverin was performed by in silico spin-labeling using the molecular dynamics software MMM (33). Figure 3 shows distance distributions calculated from the various NMR and x-Ray crystal structures of recoverin (that contain in silico spin labeling at residues 39 and 120) and are compared to the experimental DEER distance distributions. The PDB files for the NMR structures of myristoylated recoverin contain an ensemble of substructures (22 for 1IKU and 24 for 1JSA) that were used to estimate rotamer populations. The NMR structure of myristoylated recoverin with two Ca2+ bound (1JSA) is overall similar to the various crystal structures of non-myristoylated recoverin with one Ca2+-bound (1REC) or two Ca2+-bound (2HET). However, the distance distribution calculated from 1JSA differs slightly from those of the 1REC and 2HET (Fig. 3), and the experimental DEER distance distribution for Ca2+-bound recoverin (Fig. 2D) is most similar to that calculated from 1JSA.
Figure 3.
Distance distributions for spin-labels attached at C39 and N120C were calculated using NMR structures of recoverin (1IKU and 1JSA, highlighted in red) and several x-Ray crystal structures (1REC, 1OMV, 1OMB, 2HET; in black). Experimental distance distructions calculated from the DEER data are shown for Ca2+-free recoverin (green) and Ca2+-bound recoverin (blue).
Distance distributions calculated from the NMR and x-Ray crystal structures predict multiple rotamers that are not seen by the DEER measurements (Fig. 3). For example, the NMR structure of Ca2+-free recoverin (1IKU) calculates two rotamers seen in the distance distribution with resolved peaks centered at 2.45 nm and 3.12 nm. By contrast, the experimental DEER data for Ca2+-free recoverin yields a distance distribution with only a single peak at 2.5 nm. The various structures for Ca2+-bound recoverin (1JSA, 2D8N, 1REC, 1OMV, and 1OMR) each calculate a distance distribution with multiple peaks centered at 3.3 nm and 3.7 nm (Fig. 3). However, the experimental DEER data for Ca2+-bound recoverin generates a distance distribution with a prominent single peak at 3.7 nm and weaker peaks at 2.9 nm, 4.7 nm and 5.7 nm (Fig. 2D). Thus, the various rotamers (predicted from the NMR structures) are not uniformly populated under the conditions of the DEER experiment. Instead, only one main rotamer species is populated for both Ca2+-free recoverin (2.5 nm) and Ca2+-bound recoverin (3.7 nm) under the conditions of the DEER experiment.
Intermolecular DEER distances for Ca2+-bound recoverin
The experimental distance distribution for Ca2+-bound recoverin contains components at 4.7 nm and 5.7 nm that are longer than the length of a single recoverin molecule and may represent an intermolecular distance from a protein oligomer. Indeed, a Ca2+-induced dimerization of recoverin is also consistent with an approximate doubling of modulation depth in the DEER data for Ca2+-bound recoverin (Fig. 2C) compared to that of Ca2+-free recoverin (Fig. 2A). Using the equation Δ = 1−(1−λ)n−1 (44), where Δ is the observed modulation depth, λ is the calculated modulation depth as a function of pump spins, and n is the number of spins contributing, we calculate that n = 4 for Ca2+-bound recoverin versus n = 2 for Ca2+-free recoverin, consistent with Ca2+-induced dimerization.
To better understand the molecular origin of the 4.7 nm and 5.7 nm distances, a quantitative comparison was made to the crystal structure of non-myristoylated recoverin that forms a 4-fold symmetric tetramer in the crystal lattice (2HET in Fig. 1C) (45). The crystallographic tetramer possesses an intramolecular distance of 3.5 nm between the alpha carbon atoms of C39 and N120, which matches the 3.7 nm DEER distance (Fig. 2D) after correcting for a spin-label tether distance of ~0.2 nm. Two adjacent protein subunits in 2HET (chains A and B in Fig. 1C) represent a protein dimer that contains an intermolecular distance between alpha carbon atoms for C39 and N120 at 2.7 nm (blue arrow in Fig. 1C) and a separate intermolecular distance of 4.7 nm between alpha carbons of N120 (see solid black arrow in Fig. 1C). The intermolecular distance between alpha carbon atoms of C39 is also 4.7 nm. These intermolecular distances between adjacent subunits in the crystal structure (2.7 nm and 4.7 nm) are somewhat similar to the observed DEER distances at 2.9 nm and 4.7 nm for Ca2+-bound recoverin (Fig. 2D). This agreement suggests that Ca2+-bound recoverin in solution (measured by DEER) may form a dimer that resembles the dimeric arrangement of adjacent subunits in 2HET (see chains A and B in Fig. 1C and called 2HET(AB) in Fig. 3). Two opposite protein subunits in 2HET (chains B and D in Fig. 1C) have an intermolecular distance of 6.7 nm between alpha carbons of N120 (dotted arrow in Fig. 1C) that is not observed in the DEER data (Fig. 2D).
Q-band DEER traces at longer τ2 times (5 μs for Ca2+-free recoverin and 11 μs for Ca2+-bound recoverin) were acquired to more sensitively look for the long intermolecular distances predicted by 2HET (Fig. S4). For Ca2+-bound recoverin, the longer τ2 time causes detection of a new distance component at 7.4 nm (Fig. S4) that is similar to the intermolecular distance between opposite subunits in the tetramer crystal structure (dotted arrow in Fig. 1C). However, a detailed noise analysis (Fig. S4) indicates that the 7.4 nm component is within the noise level of the Q-band DEER trace and is most likely an artifact.
CW-EPR analysis of Ca2+-induced dimerization of recoverin
Recoverin has one native cysteine residue (C39) located in the loop region between the alpha helices of EF1 with an exposed side-chain in both Ca2+-free and Ca2+-bound recoverin (Fig. 1). This native cysteine was therefore used for attachment of a single nitroxide spin-label per recoverin molecule that was used for CW-EPR analysis. CW-EPR spectra of spin-labeled recoverin showed a relatively high degree of immobilization of the spin-label (shown by spectral broadening in Figure 4). The spectral broadening was even more pronounced for Ca2+-bound recoverin. Motional averaging of the EPR lineshape arises from three different modes: segmental backbone dynamics, reorientation about the bonds connecting the nitroxide ring to the backbone, and global rotational diffusion of the protein. For globular proteins with a mass greater than 30 kDa, the contribution of global rotational diffusion to the line shape becomes minimal. Based on the molecular volume of recoverin, the global tumbling rate should be on the order of 10 ns.
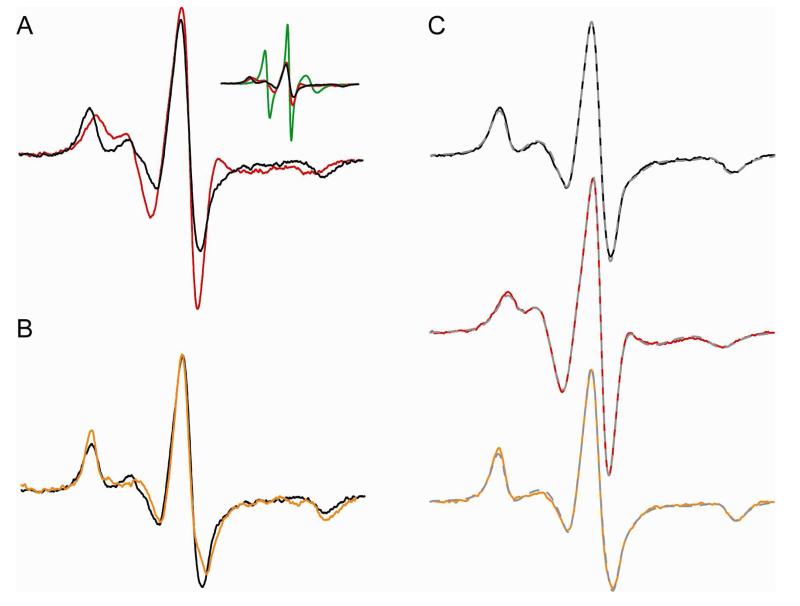
Figure 4.
CW-EPR data show Ca2+-induced dimerization of recoverin. (A) CW-EPR spectra for myristoylated recoverin (110 μM), spin labeled at the native cysteine residue (Cys39) in the Ca2+ free state (red) and Ca2+ bound state (black). Inset shows spectral comparison with the normalized spectrum of denatured protein (10% SDS; green trace). (B) CW-EPR spectra for Ca2+-bound recoverin in the presence of 30% sucrose (orange trace) and without sucrose (black). (C) Simulated EPR spectra (broken gray traces) are overlayed on top of the experimental spectra of recoverin in Ca2+-bound state (black), Ca2+-free state (red) and Ca2+-bound state in the presence of sucrose (orange trace). The simulated spectra were generated by the NLSL program. The rotational correlation times derived from the simulated spectra were 13 ns (Ca2+-free), 29 ns (Ca2+-bound), and 53 ns (Ca2+-bound in the presence of 30% sucrose (w/v)).
It is clear from the narrow spectral line shape for Ca2+-free recoverin that the spin label experiences faster internal dynamics compared to that of Ca2+-bound recoverin. The rotational correlation times for Ca2+-free vs Ca2+-bound recoverin were derived from a line shape analysis of their CW-EPR spectra (Fig. 4C). The EPR spectra were fit to a two-component macroscopic order microscopic disorder (MOMD) model that describes anisotropic motion of the nitroxide (41). The spectral line shape for Ca2+-bound recoverin was dominated (95%) by a single slow component, whereas the lineshape for Ca2+-free recoverin display two components of comparable amplitude. Results of the spectral fitting are given in Table 1. The spectral simulation for Ca2+-free recoverin indicates motion on the order of 6 nsec and a second, slower component of comparable weight with a correlation time of 13 nsec. The EPR line shape for Ca2+-bound recoverin reflects a strongly immobilized spin label, with a correlation time on the order of 29 nsec, consistent with a dimeric protein. The broad spectral lineshape for Ca2+-bound recoverin indicates not only a conformational change due to a more ordered backbone and/or tightly packed side chain, but also a molecular volume with a correlation time longer than 10 nsec. Increasing the viscosity with 30% sucrose (which is expected to obliviate any motional averging by global tumbling for a species >20 kDa (46) increases the effective correlation time to 53 nsec.
Table 1.
Diffusion tensors determined from simulation of EPR spectra.
| Sample | Rxx | Ryy | Rzz | τeff (nsec) | |
|---|---|---|---|---|---|
| −Ca2+, fast component | 7.6 | 6.8 | 7.8 | 7.4 | 6.3 |
| −Ca2+, slow component | 8.0 | 5.0 | 8.3 | 7.1 | 13 |
| +Ca2+ | 7.3 | 4.7 | 8.3 | 6.8 | 29 |
| +Ca2+, 30% sucrose | 7.1 | 4.7 | 7.8 | 6.5 | 53 |
To further probe the nature of the Ca2+-induced conformational change, the collisional frequency of paramagnetic relaxers was determined by power-saturation analysis. By measuring the differential collisional frequency of the diffusible NiEDDA and molecular oxygen, the contrast function (Φ) is calculated from the polar (Π NiEDDA) and apolar (Π O2) accessibility parameters to measure the immediate environment of the spin-labeled side-chain. While the enviroment around the spin label remains polar, the most notable change with Ca2+ is a drop in the NiEDDA accessibility, with a corresponding increase in the hydrophobicity (Φ = −0.69 and −0.57, in the absence and presence of Ca2+, respectively). The lack of a major change in the accessibility suggests that much of the immobilization arises from an ordering of the backbone due to Ca2+ binding, rather than becoming buried within the folded structure.
NMR Analysis of Ca2+-induced dimerization of recoverin
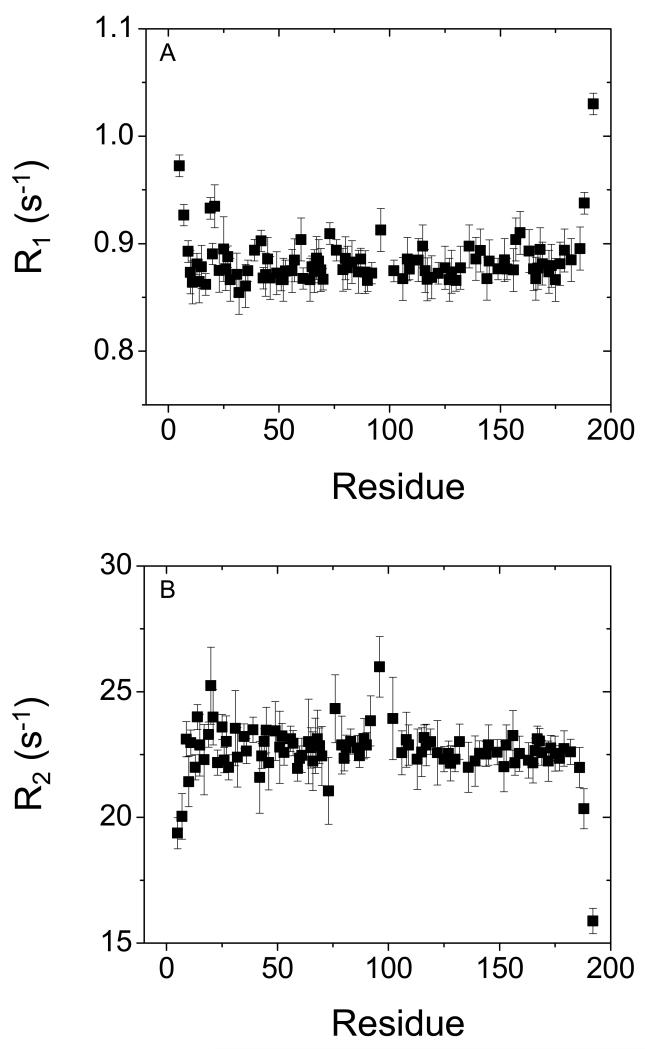
A series of NMR measurements were performed to further confirm that Ca2+-binding to recoverin induces protein dimerization. The rotational correlation time (τc) of recoverin in both Ca2+-free and Ca2+-bound states was measured by 15N NMR relaxation analysis. The longitudinal and transverse 15N NMR relaxation rates (R1 and R2) for each assigned amide resonance of Ca2+-bound recoverin are shown in Fig. 5 and were reported previously for Ca2+-free recoverin (47). Elevated R1 and decreased R2 values are apparent for the first 8 residues from the N-terminus and the last 9 residues at the C-terminus, consistent with these regions being unstructured in Ca2+-bound recoverin (22). For Ca2+-bound recoverin, the average 15N R1 and R2 values from residues in structured regions are 0.87 (± 0.04) s−1 and 22 (± 0.5) s−1, respectively. Assuming isotropic tumbling of recoverin, the average rotational correlation time obtained from R1/R2 ratios of all residues within 1 standard deviation of the average value (48) was calculated to be 25 ±0.5 ns at 310 K, suggesting that Ca2+-bound recoverin is dimeric under NMR conditions (protein concentrations above 100 μM). In addition, NMR pulsed-field gradient diffusion studies (49) on Ca2+-bound recoverin determined a translational diffusion coefficient (D = 1 × 10−10 m2/s) consistent with a molar mass of a protein dimer (~44 kDa). By contrast, Ca2+-free recoverin was shown previously to be monomeric under the same conditions (47).
Figure 5.
15N NMR relaxation data for Ca2+-bound unmyristoylated recoverin. Spin-lattice relaxation rate constants (A) and spin-spin relaxation rate constants (B) are plotted as a function of residue number. All data were measured at 60.81 MHz 15N frequency and 310 K.
Functional implications for Ca2+-induced dimerization of recoverin
The DEER analysis indicates a large Ca2+-induced change in the distance between C39 and N120C (Figs. 1A and 1B) and suggests intermolecular distances consistent with a Ca2+-bound recoverin dimer (Figs. 1C and 2D). Ca2+-induced dimerization of recoverin was further confirmed by CW-EPR (Fig. 4) and NMR relaxation analysis (Fig. 5). The recoverin dimerization observed under NMR and EPR conditions (protein concentration above 100 μM) could not be detected by size-exclusion chromatography, perhaps because recoverin dimerization has a relatively high dissociation constant with fast off-rate kinetics. We suggest that the relatively low affinity for dimerization (and hence fast dissociation rate) might enable recoverin to respond rapidly during phototransduction. Ca2+-dependent dimerization is also functionally important in other NCS proteins, including neurocalcin (17), GCAP2 (50), and DREAM/KChIPs (51, 52).
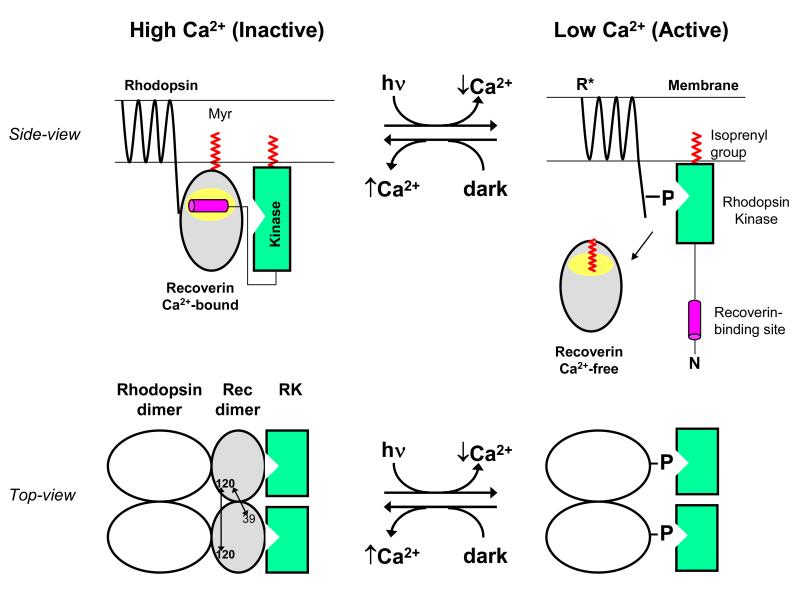
We present a schematic molecular mechanism to explain how Ca2+-induced dimerization of recoverin might regulate desensitization of dimeric rhodopsin (Fig. 6). In the dark, when Ca2+ levels are high, the Ca2+-bound recoverin dimer is at the membrane surface where it interacts with a cognate rhodopsin dimer (28-30) on one side, while the opposite side of recoverin interacts with the N-terminal helix of rhodopsin kinase (RK) (9, 53). In essence, Ca2+-bound recoverin binds to RK on the membrane surface and blocks RK interaction with dark-state rhodopsin (Fig. 6, upper panel). Light activation of retinal rods leads to a lowering of cytosolic Ca2+ (3, 4), resulting in Ca2+ dissociation from recoverin, dissociation of dimeric recoverin into a Ca2+-free monomer, and internal conformational changes in recoverin that cause sequestration of the myristoyl group (22). We suggest that the Ca2+-bound recoverin dimer on the membrane may serve to hold two molecules of RK in close proximity to the rhodopsin dimer (Fig. 6, lower panel) so that RK can rapidly form a 2:2 complex with each rhodopsin dimer once the RK inhibition is removed by light-induced dissociation of recoverin.
Figure 6.
Schematic model showing functional role of Ca2+-induced dimerization of recoverin. A side-view of recoverin on the disk membrane is shown in the upper panel and a top-view of dimeric recoverin is shown in the lower panel. Myristoylation (red) targets Ca2+-bound recoverin to the membrane surface (upper panel). The Ca2+-bound recoverin dimer binds to the N-terminal helix of rhodopsin kinase (magenta), forming a 2:2 complex on the membrane surface that blocks phosphorylation of dimeric rhodopsin (lower panel). Intermolecular DEER distances for the recoverin dimer are indicated by double-headed arrows. Light activation leads to a lowering of cytosolic Ca2+, causing conformational changes in recoverin that promote dimer dissociation, sequester the covalently attached myristoyl group, and disrupt binding to rhodopsin kinase (RK). Ca2+-free monomeric recoverin then dissociates from the membrane surface, allowing RK to phosphorylate the C-terminal tail of light-excited rhodopsin (R*).
Supplementary Material
ACKNOWLEDGMENT
We are grateful to Dr. Jerry Dallas for help with NMR experiments and Frank Delaglio for writing computer software for NMR data processing and analysis.
Footnotes
This work was supported by NIH grants EY012347 (J.B.A.) and RR11973 (UC Davis NMR), and DOE grant DE-FG02-09ER16117 (R.D.B.).
Abbreviations: DEER, double electron-electron resonance; CW-EPR, continuous wave electron paramagnetic resonance; HSQC, heteronuclear single quantum coherence; IPTG, isopropyl β-D-1-thiogalactoside; NMR, nuclear magnetic resonance; SDS PAGE, sodium dodecylsulfate polyacrylamide gel electrophoresis; R1, longitudinal relaxation rate constant; R2, transverse relaxation rate constant; RMSD, root-mean-squared deviation; TM, phase memory time.
SUPPORTING INFORMATION AVAILABLE
Supporting information includes 15N-1H HSQC spectra of Ca2+-free recoverinN120C (Fig. S1); Ca2+-binding data for recoverinN120C (Fig. S2); DEER data and analysis for Ca2+-free and Ca2+-bound recoverinN120C (Fig. S3); and Q-band DEER data recorded at longer τ2 times (Fig. S4). This material will be available free of charge online at http://pubs.acs.org.
REFERENCES
- 1.Dizhoor AM, Ray S, Kumar S, Niemi G, Spencer M, Rrolley D, Walsh KA, Philipov PP, Hurley JB, Stryer L. Recoverin: a calcium sensitive activator of retinal rod guanylate cyclase. Science. 1991;251:915–918. doi: 10.1126/science.1672047. [DOI] [PubMed] [Google Scholar]
- 2.Makino CL, Dodd RL, Chen J, Burn ME, Roca A, Simon MI, Baylor DA. Recoverin regulates light-dependent phosphodiesterase activity in retinal rods. J. Gen. Physiol. 2004;123:729–741. doi: 10.1085/jgp.200308994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Polans A, Baehr W, Palczewski K. Turned on by Ca2+! The physiology and pathology of Ca(2+)-binding proteins in the retina. Trends Neurosci. 1996;19:547–554. doi: 10.1016/s0166-2236(96)10059-x. [DOI] [PubMed] [Google Scholar]
- 4.Palczewski K, Polans AS, Baehr W, Ames JB. Ca(2+)-binding proteins in the retina: structure, function, and the etiology of human visual diseases. Bioessays. 2000;22:337–350. doi: 10.1002/(SICI)1521-1878(200004)22:4<337::AID-BIES4>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 5.Yarfitz S, Hurley JB. Transduction mechanisms of vertebrate and invertebrate photoreceptors. J. Biol. Chem. 1994;269:14329–14332. [PubMed] [Google Scholar]
- 6.Kawamura S. Rhodopsin phosphorylation as a mechanism of cyclic GMP phosphodiesterase regulation by S-modulin. Nature. 1993;362:855–857. doi: 10.1038/362855a0. [DOI] [PubMed] [Google Scholar]
- 7.Gray-Keller MP, Polans AS, Palczewski K, Detwiler PB. The effect of recoverin-like calcium-binding proteins on the photoresponse of retinal rods. Neuron. 1993;10:523–531. doi: 10.1016/0896-6273(93)90339-s. [DOI] [PubMed] [Google Scholar]
- 8.Erickson MA, Lagnado L, Zozulya S, Neubert TA, Stryer L, Baylor DA. The effect of recombinant recoverin on the photoresponse of truncated rod photoreceptors. Proc. Natl. Acad. Sci. USA. 1998;95:6474–9. doi: 10.1073/pnas.95.11.6474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Komolov KE, Senin II, Kovaleva NA, Christoph MP, Churumova VA, Grigoriev II, Akhtar M, Philippov PP, Koch KW. Mechanism of rhodopsin kinase regulation by recoverin. J. Neurochem. 2009;110:72–79. doi: 10.1111/j.1471-4159.2009.06118.x. [DOI] [PubMed] [Google Scholar]
- 10.Sampath AP, Strissel KJ, Elias R, Arshavsky VY, McGinnis JF, Chen J, Kawamura S, Rieke F, Hurley JB. Recoverin improves rod-mediated vision by enhancing signal transmission in the mouse retina. Neuron. 2005;46:413–420. doi: 10.1016/j.neuron.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 11.Strissel KJ, Lishko PV, Trieu LH, Kennedy MJ, Hurley JB, Arshavsky VY. Recoverin undergoes light-dependent intracellular translocation in rod photoreceptors. J Biol Chem. 2005;280:29250–29255. doi: 10.1074/jbc.M501789200. [DOI] [PubMed] [Google Scholar]
- 12.Zozulya S, Stryer L. Calcium-myristoyl protein switch. Proc. Natl. Acad. Sci. USA. 1992;89:11569–73. doi: 10.1073/pnas.89.23.11569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dizhoor AM, Chen CK, Olshevskaya E, Sinelnikova VV, Phillipov P, Hurley JB. Role of the acylated amino terminus of recoverin in Ca(2+)-dependent membrane interaction. Science. 1993;259:829–32. doi: 10.1126/science.8430337. [DOI] [PubMed] [Google Scholar]
- 14.Burgoyne RD, O’Callaghan DW, Hasdemir B, Haynes LP, Tepikin AV. Neuronal Ca2+-sensor proteins: multitalented regulators of neuronal function. Trends Neurosci. 2004;27:203–209. doi: 10.1016/j.tins.2004.01.010. [DOI] [PubMed] [Google Scholar]
- 15.Ames JB, Lim S. Molecular structure and target recognition of neuronal calcium sensor proteins. Biochim. Biophys. Acta. 2012;1820:1205–1213. doi: 10.1016/j.bbagen.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bourne Y, Dannenberg J, Pollmann VV, Marchot P, Pongs O. Immunocytochemical localization and crystal structure of human frequenin (neuronal calcium sensor1) J. Biol. Chem. 2001;276:11949–11955. doi: 10.1074/jbc.M009373200. [DOI] [PubMed] [Google Scholar]
- 17.Vijay-Kumar S, Kumar VD. Crystal structure of recombinant bovine neurocalcin. Nature Struct. Biol. 1999;6:80–88. doi: 10.1038/4956. [DOI] [PubMed] [Google Scholar]
- 18.Tzingounis AV, Kobayashi M, Takamatsu K, Nicoll RA. Hippocalcin gates the calcium activation of the slow afterhyperpolarization in hippocampal pyramidal cells. Neuron. 2007;53:487–493. doi: 10.1016/j.neuron.2007.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ames JB, Porumb T, Tanaka T, Ikura M, Stryer L. Amino-terminal myristoylation induces cooperative calcium binding to recoverin. J Biol Chem. 1995;270:4526–33. doi: 10.1074/jbc.270.9.4526. [DOI] [PubMed] [Google Scholar]
- 20.Tanaka T, Ames JB, Harvey TS, Stryer L, Ikura M. Sequestration of the membrane-targeting myristoyl group of recoverin in the calcium-free state. Nature. 1995;376:444–7. doi: 10.1038/376444a0. [DOI] [PubMed] [Google Scholar]
- 21.Ames JB, Hamasaki N, Molchanova T. Structure and calcium-binding studies of a recoverin mutant (E85Q) in an allosteric intermediate state. Biochemistry. 2002;41:5776–5787. doi: 10.1021/bi012153k. [DOI] [PubMed] [Google Scholar]
- 22.Ames JB, Ishima R, Tanaka T, Gordon JI, Stryer L, Ikura M. Molecular mechanics of calcium-myristoyl switches. Nature. 1997;389:198–202. doi: 10.1038/38310. [DOI] [PubMed] [Google Scholar]
- 23.Valentine K, Mesleh M, Ikura M, Ames JB, Opella S. Structure, topology and dynamics of myristoylated recoverin bound to phospholipid bilayers. Biochemistry. 2003;42:6333–6340. doi: 10.1021/bi0206816. [DOI] [PubMed] [Google Scholar]
- 24.Valentine KG, Peterson RW, Saad JS, Summers MF, Xu X, Ames JB, Wand AJ. Reverse micelle encapsulation of membrane-anchored proteins for solution NMR studies. Structure. 2010;18:9–16. doi: 10.1016/j.str.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Milov AD, Salikhov KM, Shchirov MD. Applications of the double resonance method to electron spin echo in a study of spatial distribution of paramagnetic centers in solids. Sov. Phys. Solid State. 1981;23:565–569. [Google Scholar]
- 26.Larsen RG, Singel DJ. Double electron-electron resonance spin-echo modulation: Spectroscopic measurement of electron spin pair separations in orientationally disordered solids. J. Chem. Phys. 1993;98:5134–5146. [Google Scholar]
- 27.Pannier M, Schädler V, Schöps M, Wiesner U, Jeschke G, Spiess HW. Dead-Time Free Measurement of Dipole-Dipole Interactions between Electron Spins. J. Magn. Reson. 2000;142:331–340. doi: 10.1006/jmre.1999.1944. [DOI] [PubMed] [Google Scholar]
- 28.Knepp AM, Periole X, Marrink SJ, Sakmar TP, Huber T. Rhodopsin forms a dimer with cytoplasmic helix 8 contacts in native membranes. Biochemistry. 2012;51:1819–1821. doi: 10.1021/bi3001598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fotiadis D, Jastrzebska B, Philippsen A, Muller DJ, Palczewski K, Engel A. Structure of the rhodopsin dimer: a working model for G-protein-coupled receptors. Curr. Opin. Struct. Biol. 2006;16:252–259. doi: 10.1016/j.sbi.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 30.Fotiadis D, Liang Y, Filipek S, Saperstein DA, Engel A, Palczewski K. Atomic-force microscopy: Rhodopsin dimers in native disc membranes. Naure. 2003;421:127–128. doi: 10.1038/421127a. [DOI] [PubMed] [Google Scholar]
- 31.Baldwin AN, Ames JB. Core mutations that promote the calcium-induced allosteric transition of bovine recoverin. Biochemistry. 1998;37:17408–19. doi: 10.1021/bi980928s. [DOI] [PubMed] [Google Scholar]
- 32.Jeschke G, Chechik V, Ionita P, Godt A, Zimmermann H, Banham J, Timmel CR, Hilger D, Jung H. DeerAnalysis2006-a comprehensive software package for analyzing pulsed ELDOR data. Appl. Magn. Reson. 2006;30:473–498. [Google Scholar]
- 33.Polyhach Y, Bordignon E, Jeschke G. Rotamer libraries of spin labeled cysteines for protein studies. Phys. Chem. Chem. Phys. 2011;13:2356–2366. doi: 10.1039/c0cp01865a. [DOI] [PubMed] [Google Scholar]
- 34.Georgieva ER, Roy AS, Grigoryants VM, Borbat PP, Earle KA, Scholes CP, Freed JH. Effect of freezing conditions on distances and their distributions derived from Double Electron Electron Resonance (DEER): A study of doubly-spin-labeled T4 lysozyme. J. Magn. Reson. 2012;216:69–77. doi: 10.1016/j.jmr.2012.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Freedberg DI, Ishima R, Jacob J, Wang YX, Kustanovich I, Louis JM, Torchia DA. Rapid structural fluctuations of the free HIV protease flaps in solution: relationship to crystal structures and comparison with predictions of dynamics calculations. Protein Sci. 2002;11:221–232. doi: 10.1110/ps.33202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lipari G, Szabo A. Model-free approach. J. Am. Chem. Soc. 1982;104:4546–4559. [Google Scholar]
- 37.Kay LE, Torchia DA, Bax A. Backbone dynamics of proteins as studied by 15N inverse detected heteronuclear NMR spectroscopy. Biochemistry. 1989;28:8972–8979. doi: 10.1021/bi00449a003. [DOI] [PubMed] [Google Scholar]
- 38.Mandel AM, Akke M, Palmer AG. Backbone dynamics of Escherichia coli ribonuclease HI: correlations with structure and function in an active enzyme. J. Mol. Biol. 1995;246:144–163. doi: 10.1006/jmbi.1994.0073. [DOI] [PubMed] [Google Scholar]
- 39.Hyde JS, Froncisz W. The role of microwave frequency in EPR spectroscopy of copper complexes. Annu. Rev. Biophys. Bioeng. 1982;11:391–417. doi: 10.1146/annurev.bb.11.060182.002135. [DOI] [PubMed] [Google Scholar]
- 40.Oh KJ, Altenbach C, Collier RJ, Hubbell WL. Site-directed spin labeling of proteins. Applications to diphtheria toxin. Methods Mol. Biol. 2000;145:147–169. doi: 10.1385/1-59259-052-7:147. [DOI] [PubMed] [Google Scholar]
- 41.Budil DE, Lee S, Saxena S, Freed JH. Nonlinear-Least-Squares Analysis of Slow-Motion EPR Spectra in One and Two Dimensions Using a Modified Levenberg-Marquardt Algorithm. J. Magn. Reson. 1996;120:155–189. [Google Scholar]
- 42.Fleissner MR, Brustad EM, Kálai T, Altenbach C, Cascio D, Peters FB, Hideg K, Peuker S, Schultz PG, Hubbell WL. Site-directed spin labeling of a genetically encoded unnatural amino acid. Proc. Natl. Acad. Sci. USA. 2009;106:21637–21642. doi: 10.1073/pnas.0912009106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bohme S, Padmavathi PVL, Holterhues J, Ouchni F, Klare JP, Steinhoff HJ. Topology of the amphipathic helices of the colicin A pore-forming domain in E. coli lipid membranes studied by pulse EPR. Phys. Chem. Chem. Phys. 2009;11:6770–6777. doi: 10.1039/b907117m. [DOI] [PubMed] [Google Scholar]
- 44.Bode BE, Margraf D, Plackmeyer J, Durner G, Prisner TF, Schiemann O. Counting the monomers in nanometer-sized oligomers by pulsed electron-electron double resonance. J. Am. Chem. Soc. 2007;129:6736–6745. doi: 10.1021/ja065787t. [DOI] [PubMed] [Google Scholar]
- 45.Weiergraber OH, Senin II, Zernii EY, Churumova VA, Ovaleva NA, Nazipova AA, Pmeryakov EA, Philippov PP, Granzin J, Koch KW. Tuning of a Neuronal Calcium Sensor. J. Biol. Chem. 2006;281:37594–37600. doi: 10.1074/jbc.M603700200. [DOI] [PubMed] [Google Scholar]
- 46.Mchaourab HS, Lietzow MA, Hideg K, Hubbell WL. Motion of spin-labeled side chains in T4 lysozyme. Correlation with protein structure and dynamics. Biochemistry. 1996;35:7692–7704. doi: 10.1021/bi960482k. [DOI] [PubMed] [Google Scholar]
- 47.Xu X, Ishima R, Ames JB. Conformational dynamics of recoverin’s Ca2+-myristoyl switch probed by 15N NMR relaxation dispersion and chemical shift analysis. Proteins. 2011;79:1910–1922. doi: 10.1002/prot.23014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Marion D, Driscoll PC, Kay LE, Wingfield PT, Bax A, Gronenborn AM, Clore GM. Overcoming the overlap problem in the assignment of 1H NMR spectra of larger proteins by use of three-dimensional heteronuclear 1H-15N Hartmann-Hahn-multiple quantum coherence and nuclear Overhauser-multiple quantum coherence spectroscopy: application to interleukin 1 beta. Biochemisty. 1989;28:6150–6156. doi: 10.1021/bi00441a004. [DOI] [PubMed] [Google Scholar]
- 49.Altieri AS, Hinton DP, Byrd RA. Association of biomolecular systems via pulsed field gradient NMR self-diffusion measurements. J. Am. Chem. Soc. 1995;117:7566–7567. [Google Scholar]
- 50.Olshevskaya EV, Ermilov AN, Dizhoor AM. Dimerization of guanylyl cyclase-activating protein. J. Biol. Chem. 1999;274:25583–25587. doi: 10.1074/jbc.274.36.25583. [DOI] [PubMed] [Google Scholar]
- 51.Osawa M, Dace A, Tong KI, Valiveti A, Ikura M, Ames JB. Mg2+ and Ca2+ differentially regulate DNA binding and dimerization of DREAM. J. Biol. Chem. 2005;280:18008–18014. doi: 10.1074/jbc.M500338200. [DOI] [PubMed] [Google Scholar]
- 52.Lusin JD, Vanarotti M, Li C, Valiveti A, Ames JB. NMR Structure of DREAM: Implications for Ca(2+)-Dependent DNA Binding and Protein Dimerization. Biochemistry. 2008;47:2252–2264. doi: 10.1021/bi7017267. [DOI] [PubMed] [Google Scholar]
- 53.Ames JB, Levay K, Wingard JN, Lusin JD, Slepak VZ. Structural basis for calcium-induced inhibition of rhodopsin kinase by recoverin. J Biol Chem. 2006;281:37237–37245. doi: 10.1074/jbc.M606913200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.