Abstract
Hepatocellular Carcinoma (HCC) ranks among the deadliest of cancers and has a complex etiology. Proteomics analysis using iTRAQ provides a direct way to analyze perturbations in protein expression during HCC progression from early- to late-stage but suffers from consistency and coverage issues. Appropriate use of network-based analytical methods can help to overcome these issues.
We built an integrated and comprehensive protein-protein interaction network (PPIN) by merging several major databases. Additionally, the network was filtered for GO coherent edges. Significantly differential genes (seeds) were selected from iTRAQ data and mapped onto this network. Undetected proteins linked to seeds (linked proteins) were identified and functionally characterized.
The process of network cleaning provides a list of higher quality linked proteins, which are highly enriched for similar biological process Gene Ontology terms. Linked proteins are also enriched for known cancer genes and are linked to many well-established cancer processes such as apoptosis and immune response. We found that there is an increased propensity for known cancer genes to be found in highly linked proteins. Three highly-linked proteins were identified that may play an important role in driving HCC progression—the G-protein coupled receptor signaling proteins, ARRB1/2 and the structural protein beta-actin, ACTB. Interestingly, both ARRB proteins evaded detection in the iTRAQ screen. ACTB was not detected in the original dataset derived from Mascot but was found to be strongly supported when we re-ran analysis using another protein detection database (Paragon).
Identification of linked proteins helps to partially overcome the coverage issue in shotgun proteomics analysis. The set of linked proteins are found to be enriched for cancer-specific processes, and more likely so if they are more highly linked. Additionally, a higher quality linked set is derived if network-cleaning is performed prior. This form of network-based analysis complements the cluster-based approach, and can provide a larger list of proteins on which to perform functional analysis, as well as for biomarker identification.
Keywords: Biological Networks, PPINs, MaxLink, Liver Cancer, HCC, Hepatitis B, Proteomics Expansion Pipeline
1 Introduction
Hepatocellular Carcinoma (HCC) ranks among the deadliest cancers(9). Its risk factors are varied—and include viral infection, germline mutations and alcohol-induction(34). Additionally, this cancer type can be histologically classified into poor, moderate and well-differentiated stages. Generally, the more poorly differentiated, the more advanced the cancer. However, histological characterization is limited, and may have poor accuracy in properly staging the cancer patient. It also provides limited insight into the molecular interactions underlying the disease.
On one hand, high-throughput methods such as microarrays and RNA sequencing have been very useful in enhancing our molecular understanding of HCC. However, they only measure RNA levels, not protein level. Thus, the evidences provided are indirect. On the other hand, there are many difficulties associated with high-throughput protein analyses or proteomics. Recent improvements in mass spectrometry (MS)-based technologies, however, have greatly increased coverage and detection range. In particular, isobaric Tag relative and absolute quantitation (iTRAQ)-based technologies have recently gained widespread popularity for their higher detection limits and ability to multiplex up to 8 samples simultaneously (33; 8). Despite these improvements, proteomics still suffer from coverage and consistency issues. The coverage issue—that is, the ability to cover the entire proteome—arises in part due to the limited detection range of MS instruments, as well as due to inherent sample complexity. The consistency issue—that is, whether the same results are produced in repeated runs—arises in the seemingly random data acquisition and mechanics of current MS instruments, caused by dominance of the proteome by a few highly abundant proteins which leads to the oversampling of high abundance peptide ions (16).
These two problems make it difficult to analyze MS data in a comprehensive way. However, it is possible to partially overcome these issues by taking advantage of the fact that proteins tend to work in groups rather than as singular entities. In our previous work, we proposed a powerful complex prediction algorithm termed the Proteomics Expansion Pipeline (PEP) (12). PEP first identifies the group of high-confidence proteins or “seeds” from the proteomic screen—i.e., proteins that are consistently found in patients with significant over- or under-expression. These seeds are then mapped to nodes in a large integrated protein-protein interaction network (PPIN). An expanded subnetwork is then extracted from the PPIN by taking the immediate neighbors of the seeds in the PPIN. The subnetwork is then clustered using CFinder (1). Each cluster is then ranked based on the average expression value of the proteins it contains. This includes the expression values of non-seeds as well. Proteins (in high-ranking clusters) not found in the proteomics screen are then screened against the original mass spectra for evidence of existence.
PEP uses a very comprehensive PPIN comprising data from HPRD (23), BioGRID (31), IntAct (3), and DIP (36), as well as data from literature (32; 25). Although combining PPINs improves coverage of the protein interactome, it also compounds the noise present in them (35). So, PEP uses the iterated Czekanowski-Dice distance (CD-distance) technique from CMC (15) to identify and eliminate potential noise edges from the integrated PPIN. The CD-distance technique is very effective—it produced a cleaned integrated PPIN having a significantly higher level of functional and localization coherence, after eliminating about 50% of the edges from the original integrated PPIN.
We applied PEP to a group of 12 hepatocellular carcinoma (HCC) patients, of whom 5 were clinically diagnosed to be in the moderate (mod) and 7 in the poor stage. In our analysis, we found that most of the detected mod-stage proteins were also found in poor-stage patients. In terms of pathway enrichments, mod-stage patients appeared to exhibit signs of immune response not observed in poor-stage patients, while poor-stage patients exhibited widespread metabolic deregulations. From the network-based PEP analysis, we uncovered several interesting clusters which might be crucial in driving mod-stage cancer to poor stage. Of these, the cluster comprising of PRKDC, WRN, XRCC5/6 and PCNA appeared most interesting.
The PEP approach is largely focused on cluster discovery and analysis, as well as recovery of low abundance and low confidence proteins. However, there are other network-based approaches which can be used on the cleaned PPIN. This may produce results that can augment our existing findings. More interestingly, it may reveal insights that have been missed. One useful approach may be Maxlink, introduced by Ostlund et al (21). It is a method for identifying novel cancer genes based on a given set of identified oncogenes. Maxlink first requires a set of oncogenes (seeds) to be identified based on literature search and the Cancer Gene Census (11). It then produces a ranked list of new candidate genes based on the number of links they have in the FunCoup PPIN database (2) to the seed set. The higher the number of connections to seeds, and the lower the number of connections to non-seeds, the higher the rank. This approach relies on two reasonable hypotheses. The first hypothesis is that a protein should participate in the same biological processes, biological functions, or protein complexes that are over-represented among its interaction partners (28; 14). The second hypothesis is that proteins in the same complex should have more interactions between themselves than with proteins outside the complex (6).
Maxlink has not yet been explicitly tested on proteomics data. In this work, we apply a Maxlink-type approach on our HCC proteomics data.
2 Methods
2.1 Experimental setup
The experimental setup is described briefly here; details are given in supplementary methods. Liver tissues were obtained from 12 male patients diagnosed with HCC and suffered from cirrhosis with chronic Hepatitis B virus (HBV) infection. There was no metastasis at the point of surgery. Tissues collected were grouped according to histology report; 5 had moderately differentiated HCC (mod) and 7 had poorly differentiated HCC (poor). Paired tissues were obtained from each patient, one from the adjacent non-tumor region (normal) and the other from the tumor region of the resected liver. Mixed protein lysate from each patient was put through an initial phase of iTRAQ followed by 2D liquid chromatography. Finally, the resultant spectrum was resolved by peptide database search using Mascot.
2.2 Selection of seed proteins
A seed is defined as meeting the following requirements: Support by at least 4 poor patients, and with a combined differential score ≥ 1.2. The combined differential score is calculated as the average score of the protein ratios (tumor over self non-tumor). If the ratio is below 1 (under-expressed), the reciprocal is used.
2.3 Network integration and cleaning
An integrated PPIN was built comprising of data from HPRD (23), BioGRID(31), IntAct (3), and DIP (36), as well as data from literature (32; 25). The various IDs were mapped using BioMart to gene names. This network was then filtered using the iterated CD-distance method from CMC (15), and the top 90% of the highest non-zero scoring edges are kept. The resultant combined network displayed the properties of a typical PPIN such as a power-law distribution of the degrees, disassortativity (hubs less likely to be linked to each other) and small-world (small diameter) (data not shown).
2.4 Identification of linked proteins
The code was written in PERL. Let the network G be comprised of nodes V and edges E. From the set of seeds X ⊆ V, the set of non-seeds Y is derived (Y = V − X). The set of linked proteins L are those proteins in Y that have at least 1 connection to proteins in X. That is, L = {y ∈ Y ∣ 1 ≤ ∣ {x ∈ X ∣ (x, y) ∈ E}∣}.
2.5 Gene-Ontology (GO)-based characterization and coherence measurement
Annotation and the GO tree (ver 1.2 OBO) files for Homo Sapiens were downloaded from geneontology.org (dated 23 April 2011). UniProtKB accessions were mapped to Ensembl Gene IDs and gene names via Biomart. Informative biological process terms were extracted from the GO OBO file; as in (37), a term is considered informative if it is annotated to at least 30 genes and no direct descendent of the term is annotated to at least 30 genes. Significance testing for each cluster was performed using the hypergeometric test with Bonferroni correction (p ≤ 0.05).
To evaluate the quality of linked proteins derived from the cleaned and uncleaned integrated network, we measured Gene Ontology biological process (BP), cellular component (CC) and molecular function (MF) term coherence for every edge—i.e., a seed protein connected to a linked protein—derived from the cleaned and uncleaned network. Edge coherence is calculated by counting the number of shared GO terms in each category for every GO-annotated edge divided by the total number of considered edges.
3 Results
3.1 Identification of linked proteins and the important effects of network cleaning
235 seeds were returned from the dataset. From the cleaned dataset, 288 linked proteins were found to share at least one other connection with a seed. From the uncleaned dataset, 902 linked proteins were returned.
We then built two sets of derived networks from cleaned and uncleaned networks, obtaining edges formed between seed and linked proteins (seed + linked) from the reference integrated PPIN, and checked the extent of GO term sharing. It is observed that the cleaned network boasts much higher quality edges where the joined nodes tend to have deep sharing of GO terms. Hence, the linked proteins derived from the cleaned network is likely to be more biologically relevant. The improvement in quality as a result of the cleaning step is observed to be at least two folds; see Table 1.
Table 1. GO term coherence of linked proteins derived from cleaned and uncleaned networks.
It can be observed that the cleaned network boasts higher quality edges where the joined nodes tend to have deep sharing of GO terms. Hence, the linked proteins derived from the cleaned network is likely to be more biologically relevant (BP: biological process, MF: molecular function and CC: cellular localization)
| Network | BP | MF | CC |
|---|---|---|---|
| Cleaned | 0.180 | 0.376 | 0.755 |
| Uncleaned | 0.035 | 0.121 | 0.300 |
To see whether the improvement in the 3 GO categories (biological process - BP, molecular function - MF and cellular localization - CC) is greater than the network generally, we calculated the log odds ratio. That is, seed+linked from cleaned network/ seed+linked from uncleaned network divided over total cleaned/total uncleaned network. Interestingly, there was a 1.5X enhancement for BP terms whereas there were no improvements for MF (1.02X) and CC terms (1.02X). This indicates that the cleaned seed+linked protein network is highly enriched for proteins in shared biological processes. The significant enhancement of GO term coherence in the cleaned network indicates that the cleaning step is important. It also improves analytical results in combination with the Maxlink approach.
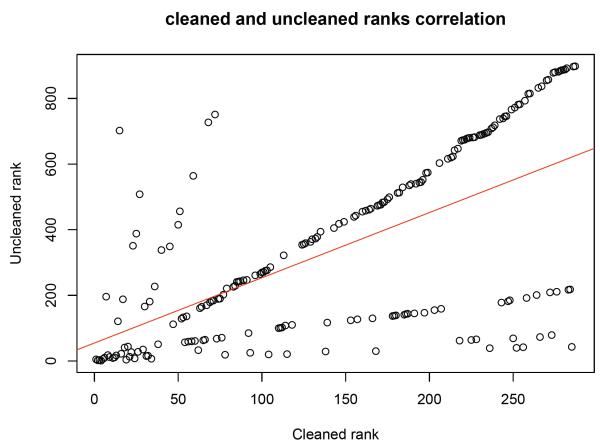
There appears to be a strong linear correlation between the ranks of the linked proteins (sorted in descending order by the number of connections to seeds) in the cleaned and uncleaned networks; Figure 1. This is evident from the string of points forming a near perfect diagonal and is not particularly surprising. However, it can also be seen that a large number of points are ranked below the diagonal. This means that they are ranked relatively higher than they would actually be after the cleaning step. It also means that the cleaned linked proteins is enriched for linked proteins with high ranks from the poor network. Although this is less direct evidence than measuring GO coherence as above, it does demonstrate the efficacy and relevance of the cleaning procedure.
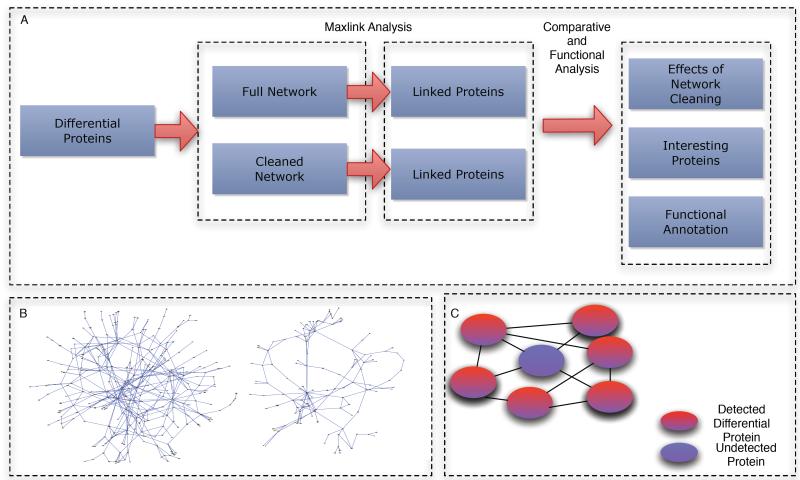
Figure 1.
A) Overview of analytical pipeline. Two sets of networks are used; a cleaned and uncleaned network, to discover linked proteins undetected by the MS screen. The results are then compared using GO coherence measurements and ranks correlation analysis. The set of interesting linked proteins are then functionally annotated using GO terms. B) A cleaned (left) and uncleaned network (right). Note that this is for illustration purposes. The actual networks are too complex to visualize. C) An example of a Maxlinked protein (blue) A Maxlinked protein is one that is highly connected to detected differentially expressed proteins.
We then turned to informative GO term enrichment for the linked proteins in the cleaned network. This produced 87 significant GO BP terms (for 288 proteins). To find out whether these terms are closely associated in the GO tree, we calculated the shortest-path lengths between all terms, and returned the average path length. A null distribution is then generated by picking a number of proteins equal to the linked proteins from the reference network, calculating the significant informative GO terms, and calculating the average GO term path length. This is repeated 1000 times. For linked proteins, we find that the GO terms are significantly more closely associated (Z-score = −3.98, p = 0.000034).
3.2 Properties of the most highly linked proteins: ACTB and the ARRB1/2
In the cleaned dataset, 34 proteins share at least 3 connections to the seeds; Supplementary Table 1. This set includes known oncogenes such as NFKB2, RAF1, REL, TP53 and VHL.
Of these 34 highly linked proteins, ARRB1/2 and ACTB were found to be most connected to the seeds. Interestingly, these 3 proteins were not found in the set of detected non-seed proteins either. Hence, it is possible that these proteins were not picked up by MS.
To verify this, we turned to another MS-protein identification algorithm, Paragon. In Goh et al (12), we found that there was good correlation between the reported ranks and ratios of Mascot and Paragon. However, Paragon reported many more proteins than Mascot even although these extra proteins were found to originate from lower quality MS/MS spectra. We generated a Paragon excess list comprising the read outs from all 12 patients not found in Mascot. Here, ACTB was found to be supported in all 12 patients. It was also found to be very confidently predicted in Paragon with a normalized average rank of 0.028 (out of 1). The omission of ACTB in the set of detected proteins could be due to a variety of factors — e.g. Mascot’s filtering parameters, incomplete coverage in its database or differences in peptide matching algorithms (29). ARBB1 and 2 however, were not found to be predicted in Paragon.
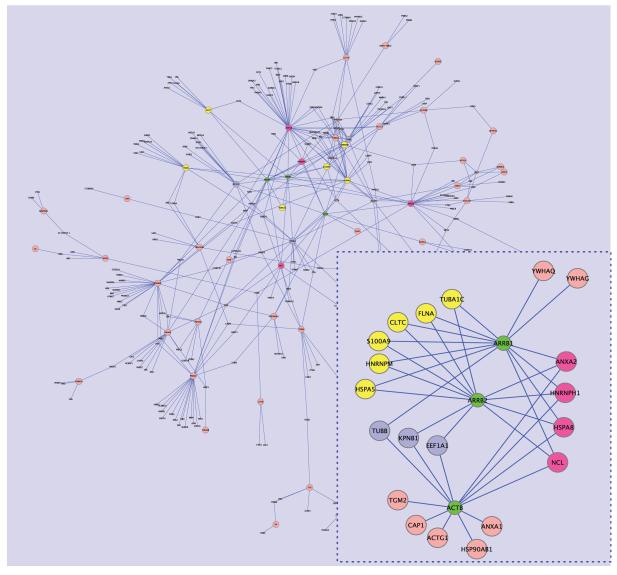
The inter-connections of linked proteins to the seeds are shown in Figure 2. This network appears to be quite sparse, and is probably not suitable for performing cluster analysis. ARRB1 and ARRB2 share many seeds (Figure 2 inset). This includes HSPA8, HNRNPM, HSPA5, TUBA1C, HNRNPH1, FLNA, CLTC, S100A9, NCL and ANXA2. This set of proteins appear to be important in vesicle-mediated transport (p = 0.0016), as well as actin cytoskeleton reorganization (p = 0.00885).
Figure 2. Ranks correlations between uncleaned and cleaned networks.
There is a strong linear ranks correlation between the linked proteins found in the cleaned and uncleaned networks. The trendline is approximated with a gradient of 2 and y-intercept of 55 (adjusted R-squared = 0.39, p ≤ 2.2e – 16).
The subnet formed by ARRB1/2 and ACTB (Figure 2 inset) shows that ACTB is less strongly connected to ARRB1/2. Here, proteins linked only to ARRB1 and 2 are colored yellow; ARRB1 and ACTB in light blue; all 3 in purple; proteins not shared are in pink. GO term analysis of the 20 connected seeds does not reveal any term typically associated with cancers. Instead, many of the terms are more akin to functionalities associated with the liver, e.g. vesicle-mediated transport and transport. However, stress responses and wound healing is represented by half of the proteins, and it does agree with our previous observations where many of the significant clusters were also enriched for stress responses.
ARRB1/2 are signaling proteins of G protein-coupled receptors (GPCRs). They are known to play an important role in tumor tissue invasion and metastasis. Rosano et al (24) showed that in ovarian cancer, silencing of both ARRB1 and 2 inhibited endothelin-A (ET(A)R) receptor-driven silencing, resulting in SRC suppression, mitogen-activated protein kinase (MAPK), AKT activation, EGFR transactivation and, most importantly, complete inhibition of ET-1-induced beta-catenin/TCF transcriptional activity and cell invasion. In colorectal cancer, it was reported that the association of ARRB1 with SRC is critical for carcinoma cell migration as well as metastatic spread of cancer to liver in vivo (4). This association is stimulated by the expression of prostagladin E, and may act by activation of the EGFR controlled pathways. Like Rosano et al (24), this study also implied a functional role for ARRB1 as an important mediator of tumor invasion and metastasis. Interestingly, to our best knowledge, ARRB1/2 have not been reported as a crucial factor in driving oncogenic progression in HCC from mod to poor. However, the fact that it is linked to the most number of our MS-detected dysregulated proteins, coupled to its enrichment in other metastatic tumors suggests a potentially important role in driving HCC progression.
Actins are highly conserved proteins that are involved in cell motility, structure, and integrity. ACTB or beta-action is a major constituent of the contractile apparatus and one of two nonmuscle cytoskeletal actins. Because it is a housekeeping protein, it is commonly used for normalization in gene expression studies. However, here, we found that ACTB is connected to a disproportionate number of dysregulated proteins in the cleaned network (as well as in uncleaned), and could possibly be involved in driving HCC progression. Indeed, several studies have shown that ACTB is differentially expressed in cancer. This includes differential expression of ACTB in N1S1 rat hepatoma (5), colon carcinoma/colorectal cancer (CRC) (27), and blood cancers such as chronic myelogenous leukemia (CML), chronic lymphocytic leukemia (CLL), acute myelogenous leukemia (AML) (17) . In human colon adenocarcinoma (20), hepatoma (22) and melanoma (13), there is a tendency for ACTB to be dysregulated in cells with greater metastatic capacity.
Of the 34 most highly linked proteins, there is an enrichment for significant GO terms commonly associated with cancer. For example, apoptosis and programmed cell death (n = 16, p = 5.19e – 09), activation of immune responses (n = 7, p = 6.15e – 08), response to stress (n = 18, p = 1.40e – 06), positive regulation of NF-kappaB transcription factor activity (n = 5, p = 4.30e – 05), and negative regulation of cell proliferation (n = 8, p = 7.33e – 05). Comparing the highly linked proteins (at least 3 connections to seeds) and lower linked proteins against the Cancer Gene Census (11), we found 1.86X more enrichment for known cancer genes among the highly linked proteins. This reinforces the notion that increased connectivity to seed proteins is likely to imply potential oncogenic function.
4 Discussions
4.1 How the Maxlink approach complements PEP
Both Maxlink (21) and PEP (12) addresses to an extent incomplete coverage issues in proteomics. However, they do this differently. Maxlink identifies additional proteins based on the number of connections to seed proteins whereas PEP identifies significantly differentially expressed submodules formed by the neighbors of the seeds. Functional analysis reveals a common enrichment of terms such as apoptosis and stress responses. However, Maxlink picked up immune responses which we did not observe in PEP. One likely possibility is that these proteins are poorly connected in the reference PPIN and therefore did not qualify as clusters. Since Maxlink only considers proteins connected to seeds regardless of their inter-connectivity, it provides an additional dimension to the results from cluster analysis. The Maxlink approach is also dependent on the quality of the reference network. We show here that the process of network cleaning greatly reduces the number of linked proteins from 902 to 288 but the latter set is enriched for coherent terms as well as highly ranked linked proteins in the former.
However, both methods are not able to deal adequately with the consistency issue in proteomics—viz., the unrepeatability of results from the same samples. Furthermore, the dependence on identifying seeds from the iTRAQ screen filters off a large amount of the limited available information, because only proteins supported by the majority of samples and clearly differentially expressed are primarily considered.
Hence, there is still avenue for further development of methods that can deal with these shortfalls.
4.2 The role of ARRB1/2 proteins and ACTB in driving HCC progression
It is interesting that the most linked proteins in HCC to seeds turned out to be non-classical oncogenes. This reinforces the notion of how complex cancer is, and how limited current knowledge is. There is limited literature evidence on the roles of ARRB1/2 and even beta-actin in driving metastasis. Although many of the reported literature documents other cancer types, especially more aggressive cancers, it is possible that dysregulation of these proteins can also have similar effects in driving HCC progression.
Furthermore, although found in more aggressive tumors, GO term analysis of the shared neighbors by these 3 proteins revealed no significant cancer associated term, aside from wound healing and stress response. This could be also be due to the limited annotations in GO, as well as due to the limited scope in analyzing only PPIN information. ARRB1/2 proteins are GPCR signaling proteins and may drive invasiveness and metastasis via several different pathways ranging from ET(A)R, SRC and EGFR, AKT and MAPK (24; 4).
The role of beta-actin is more interesting given that it is a well-known housekeeping protein with widespread expression. It is typically used, alongside GADPH, as a marker for normalization of gene expression experiments. Its functional role in cancer is not particularly well-characterized despite literature evidence indicating its dysregulation in more aggressive cancer types (26; 22; 20).
In our derived network, we noted that ARRB1/2’s shared neighbors were enriched for the GO term, actin cytoskeleton reorganization (p = 0.00885). This is effected by FLNA and S100A9, which are shared between ARRB1/2. FLNA or filamin-A, an actin-binding protein, that is widely expressed and regulates re-organization of the actin cytoskeleton by interacting with integrins, transmembrane receptor complexes and second messengers. S100A9 is a calcium binding protein, and may be implicated in leukemia (7). Furthermore, we found significant crosstalk with actin via shared neighbors. Indeed, by looking at the shared neighbors they seemed to converge on mRNA fate. NCL or nucleolin forms part of mRNP complex which decides on mRNA localization, translation and turnover (19). EEF1A1, the translation elongation factor on the other hand is required for binding of aminoacyl-tRNA to the ribosome during translation. The perturbed mRNA dynamics could be the result of 1) interaction of host cell with HBV and/or 2) dysregulated protein synthesis in malignant neoplastic transformation to poorly-differentiated HCC to promote tumor growth.
In the process of dedifferentiation from small, well differentiated to moderately differentiated and finally poorly differentiated HCC tumors, the vasculature remodels substantially and abnormally (30). This vasculogenic and angiogenic switch is critical for tumor growth. Endothelial cell motility drives the formation and maintenance of blood vessels and to do so, actin dynamics is required. Of note, HBV upregulates and stabilises HIF-α, and subsequently stimulate the cascade of signalling events that lead to angiogenesis (18). Similarly, HBVx activates RhoA, a small GTPase that regulates actin (10). Together, our results support active angiogenesis and vasculogenesis as important molecular events that occur in the progression of HBV-induced HCC which requires the participation of actin. At the same time, tubulins A and B and CAP1 are connected to ARRB and actin seeds, suggesting the importance of the regulation of cytoskeletal events in multiple cellular responses during oncogenesis.
5 Conclusion
Identification of linked proteins helps to partially overcome the coverage issue in proteomics analysis. The set of linked proteins are found to be enriched for cancer-specific processes, and more likely so if they are highly linked. Additionally, a higher quality linked set is derived if network-cleaning is performed prior. Here, the most linked proteins (ARBB1/2 and ACTB) turned out to be non-classical cancer genes which have been evidenced to play important roles in metastasis and invasiveness although the mechanisms appear to be very complex. To the best of our knowledge, there is not much known about the role these proteins play in HCC progression.
The Maxlink form of network-based analysis complements cluster-based approaches such as PEP, because it concentrates on seed connections rather than inter-connectivity between seeds and their neighbors. It can therefore add on to the list of proteins on which to perform functional analysis, as well as for biomarker identification. In addition, we find that cleaning the network prior to performing Maxlink provides a higher quality set of linked proteins.
Supplementary Material
Included are supplementary methods and tables.
The cleaned network comprising data pooled from various literature sources and cleaned using the CMC/filterNadd program, which implements the iterated CD-distance method.
iTRAQ data obtained from the 5 moderate stage patients.
iTRAQ data obtained from the 7 poor stage patients.
The PERL code for generating the linked proteins from the seed set. It also outputs an edge list comprising links between linked and seed proteins.
The set of linked genes derived from the data set.
Figure 3. Background: the inter-connections of linked proteins to the seeds; Inset: the connections between ARBB1,2 and ACTB.
Background: The network comprising seeds and linked proteins is topologically sparse. Inset: Here, proteins linked only to ARRB1 and 2 are labelled yellow; ARRB1 and ACTB in light blue; connections to all 3 in purple; proteins not shared are in pink.
Table 2.
List of most highly connected proteins to the seed set (sorted in descending order)
| Protein | Links |
|---|---|
| ARRB1 | 13 |
| ACTB | 12 |
| ARRB2 | 12 |
| TRAF6 | 9 |
| PPP2R2B | 8 |
| MCC | 7 |
| TBK1 | 6 |
| TNFRSF1B | 6 |
| TP53 | 6 |
| CFTR | 5 |
| IKBKG | 5 |
| NFKB2 | 5 |
| RIPK1 | 5 |
| FN1 | 4 |
| PTMA | 4 |
| REL | 4 |
| SMAD2 | 4 |
| SMAD3 | 4 |
| VHL | 4 |
| ARF6 | 3 |
| CASP3 | 3 |
| CBL | 3 |
| CTSB | 3 |
| DYNLL1 | 3 |
| EIF1B | 3 |
| EIF6 | 3 |
| LARP1 | 3 |
| MAP3K7IP1 | 3 |
| PLG | 3 |
| PRKCD | 3 |
| RAF1 | 3 |
| RELB | 3 |
| S100A1 | 3 |
| SUMO4 | 3 |
Acknowledgements
WWBG is supported by a Wellcome Trust Scholarship (83701/Z/07/Z). LW is supported in part by a Singapore National Research Foundation grant NRF-G-CRP-2007-04-082(d). The iTRAQ work is supported by the Singapore Cancer Syndicate.
Biographical notes
Wilson Wen Bin Goh is currently pursuing his PhD in Imperial College London. His research interests include cancer proteomics and miRNA-networks.
Contributor Information
Wilson Wen Bin Goh, Department of Computing, Imperial College London, UK.
Yie Hou Lee, Singapore-MIT Alliance for Research and Technology, Singapore yie.hou@smart.mit.edu.
Zubaidah M. Ramdzan, Rosalind and Morris Goodman Cancer Centre, McGill University, Canada zubaidah.mohamedramdzan@mcgill.ca
Maxey C.M. Chung, Department of Biological Sciences and Department of Biochemistry, National University of Singapore, Singapore maxey_chung@nuhs.edu.sg
Limsoon Wong, Department of Computer Science and Department of Pathology, National University of Singapore, Singapore wongls@comp.nus.edu.sg.
Marek J. Sergot, Department of Computing, Imperial College London, UK, m.sergot@imperial.ac.uk
References
- [1].Adamcsek Balázs, Palla Gergely, Farkas Illés J, Derényi Imre, Vicsek Tamás. Cfinder: locating cliques and overlapping modules in biological networks. Bioinformatics. 2006 Apr;22(8):1021–3. doi: 10.1093/bioinformatics/btl039. [DOI] [PubMed] [Google Scholar]
- [2].Alexeyenko Andrey, Sonnhammer Erik L L. Global networks of functional coupling in eukaryotes from comprehensive data integration. Genome Res. 2009 Jun;19(6):1107–16. doi: 10.1101/gr.087528.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Aranda B, Achuthan P, Alam-Faruque Y, Armean I, Bridge A, Derow C, Feuermann M, Ghanbarian AT, Kerrien S, Khadake J, Kerssemakers J, Leroy C, Menden M, Michaut M, Montecchi-Palazzi L, Neuhauser SN, Orchard S, Perreau V, Roechert B, van Eijk K, Hermjakob H. The intact molecular interaction database in 2010. Nucleic Acids Res. 2010 Jan;38(Database issue):D525–31. doi: 10.1093/nar/gkp878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Buchanan F Gregory, Lee Gorden D, Matta Pranathi, Shi Qiong, Matrisian Lynn M, DuBois Raymond N. Role of beta-arrestin 1 in the metastatic progression of colorectal cancer. Proc Natl Acad Sci U S A. 2006 Jan;103(5):1492–7. doi: 10.1073/pnas.0510562103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Chang TJ, Juan CC, Yin PH, Chi CW, Tsay HJ. Up-regulation of beta-actin, cyclophilin and gapdh in n1s1 rat hepatoma. Oncol Rep. 1998;5(2):469–71. doi: 10.3892/or.5.2.469. [DOI] [PubMed] [Google Scholar]
- [6].Chen Jingchun, Yuan Bo. Detecting functional modules in the yeast protein-protein interaction network. Bioinformatics. 2006 Sep;22(18):2283–90. doi: 10.1093/bioinformatics/btl370. [DOI] [PubMed] [Google Scholar]
- [7].Cheok Meyling H, Yang Wenjian, Pui Ching-Hon, Downing James R, Cheng Cheng, Naeve Clayton W, Relling Mary V, Evans William E. Treatment-specific changes in gene expression discriminate in vivo drug response in human leukemia cells. Nat Genet. 2003 May;34(1):85–90. doi: 10.1038/ng1151. [DOI] [PubMed] [Google Scholar]
- [8].Choe Leila, D’Ascenzo Mark, Relkin Norman R, Pappin Darryl, Ross Philip, Williamson Brian, Guertin Steven, Pribil Patrick, Lee Kelvin H. 8-plex quantitation of changes in cerebrospinal fluid protein expression in subjects undergoing intravenous immunoglobulin treatment for alzheimer’s disease. Proteomics. 2007 Oct;7(20):3651–60. doi: 10.1002/pmic.200700316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].El-Serag Hashem B. Hepatocellular carcinoma: recent trends in the united states. Gastroenterology. 2004 Nov;127(5 Suppl 1):S27–34. doi: 10.1053/j.gastro.2004.09.013. [DOI] [PubMed] [Google Scholar]
- [10].Fukui Koji, Tamura Shinji, Wada Akira, Kamada Yoshihiro, Sawai Yoshiyuki, Imanaka Kazuho, Kudara Takahiko, Shimomura Iichiro, Hayashi Norio. Expression and prognostic role of rhoa gtpases in hepatocellular carcinoma. J Cancer Res Clin Oncol. 2006 Oct;132(10):627–33. doi: 10.1007/s00432-006-0107-7. [DOI] [PubMed] [Google Scholar]
- [11].Futreal P Andrew, Coin Lachlan, Marshall Mhairi, Down Thomas, Hubbard Timothy, Wooster Richard, Rahman Nazneen, Stratton Michael R. A census of human cancer genes. Nat Rev Cancer. 2004 Mar;4(3):177–83. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Goh Wilson Wen Bin, Lee Yie Hou, Zubaidah Ramdzan M, Jin Jingjing, Dong Difeng, Lin Qingsong, Chung Maxey C M, Wong Limsoon. Network-based pipeline for analyzing ms data: an application toward liver cancer. J Proteome Res. 2011 May;10(5):2261–72. doi: 10.1021/pr1010845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Goidin D, Mamessier A, Staquet MJ, Schmitt D, Berthier-Vergnes O. Ribosomal 18s rna prevails over glyceraldehyde-3-phosphate dehydrogenase and beta-actin genes as internal standard for quantitative comparison of mrna levels in invasive and noninvasive human melanoma cell subpopulations. Anal Biochem. 2001 Aug;295(1):17–21. doi: 10.1006/abio.2001.5171. [DOI] [PubMed] [Google Scholar]
- [14].Hishigaki H, Nakai K, Ono T, Tanigami A, Takagi T. Assessment of prediction accuracy of protein function from protein–protein interaction data. Yeast. 2001 Apr;18(6):523–31. doi: 10.1002/yea.706. [DOI] [PubMed] [Google Scholar]
- [15].Liu Guimei, Wong Limsoon, Chua Hon Nian. Complex discovery from weighted ppi networks. Bioinformatics. 2009 Aug;25(15):1891–7. doi: 10.1093/bioinformatics/btp311. [DOI] [PubMed] [Google Scholar]
- [16].Liu Hongbin, Sadygov Rovshan G, Yates John R., 3rd A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal Chem. 2004 Jul;76(14):4193–201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- [17].Lupberger J, Kreuzer KA, Baskaynak G, Peters UR, le Coutre P, Schmidt CA. Quantitative analysis of beta-actin, beta-2-microglobulin and porphobilinogen deaminase mrna and their comparison as control transcripts for rt-pcr. Mol Cell Probes. 2002 Feb;16(1):25–30. doi: 10.1006/mcpr.2001.0392. [DOI] [PubMed] [Google Scholar]
- [18].Moon Eun-Joung, Jeong Chul-Ho, Jeong Joo-Won, Kim Kwang Rok, Yu Dae-Yeul, Murakami Seishi, Kim Chul Woo, Kim Kyu-Won. Hepatitis b virus x protein induces angiogenesis by stabilizing hypoxia-inducible factor-1alpha. FASEB J. 2004 Feb;18(2):382–4. doi: 10.1096/fj.03-0153fje. [DOI] [PubMed] [Google Scholar]
- [19].Moore Melissa J. From birth to death: the complex lives of eukaryotic mrnas. Science. 2005 Sep;309(5740):1514–8. doi: 10.1126/science.1111443. [DOI] [PubMed] [Google Scholar]
- [20].Nowak Dorota, Skwarek-Maruszewska Aneta, Zemanek-Zboch Magdalena, Malicka-Błaszkiewicz Maria. Beta-actin in human colon adenocarcinoma cell lines with different metastatic potential. Acta Biochim Pol. 2005;52(2):461–8. [PubMed] [Google Scholar]
- [21].Ostlund Gabriel, Lindskog Mats, Sonnhammer Erik L L. Network-based identification of novel cancer genes. Mol Cell Proteomics. 2010 Apr;9(4):648–55. doi: 10.1074/mcp.M900227-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Popow A, Nowak D, Malicka-Błaszkiewicz M. Actin cytoskeleton and beta-actin expression in correlation with higher invasiveness of selected hepatoma morris 5123 cells. J Physiol Pharmacol. 2006 Nov;57(Suppl 7):111–23. [PubMed] [Google Scholar]
- [23].Prasad T S Keshava, Kandasamy Kumaran, Pandey Akhilesh. Human protein reference database and human proteinpedia as discovery tools for systems biology. Methods Mol Biol. 2009;577:67–79. doi: 10.1007/978-1-60761-232-2_6. [DOI] [PubMed] [Google Scholar]
- [24].Rosanò Laura, Cianfrocca Roberta, Masi Stefano, Spinella Francesca, Di Castro Valeriana, Biroccio Annamaria, Salvati Erica, Nicotra Maria Rita, Natali Pier Giorgio, Bagnato Anna. Beta-arrestin links endothelin a receptor to beta-catenin signaling to induce ovarian cancer cell invasion and metastasis. Proc Natl Acad Sci U S A. 2009 Feb;106(8):2806–11. doi: 10.1073/pnas.0807158106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Rual Jean-François, Venkatesan Kavitha, Hao Tong, Hirozane-Kishikawa Tomoko, Dricot Amélie, Li Ning, Berriz Gabriel F, Gibbons Francis D, Dreze Matija, Ayivi-Guedehoussou Nono, Klitgord Niels, Simon Christophe, Boxem Mike, Milstein Stuart, Rosenberg Jennifer, Goldberg Debra S, Zhang Lan V, Wong Sharyl L, Franklin Giovanni, Li Siming, Albala Joanna S, Lim Janghoo, Fraughton Carlene, Llamosas Estelle, Cevik Sebiha, Bex Camille, Lamesch Philippe, Sikorski Robert S, Vandenhaute Jean, Zoghbi Huda Y, Smolyar Alex, Bosak Stephanie, Sequerra Reynaldo, Doucette-Stamm Lynn, Cusick Michael E, Hill David E, Roth Frederick P, Vidal Marc. Towards a proteome-scale map of the human protein-protein interaction network. Nature. 2005 Oct;437(7062):1173–8. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- [26].Ruan Wenjing, Lai Maode. Actin, a reliable marker of internal control? Clin Chim Acta. 2007 Oct;385(1-2):1–5. doi: 10.1016/j.cca.2007.07.003. [DOI] [PubMed] [Google Scholar]
- [27].Sagynaliev Emil, Steinert Ralf, Nestler Gerd, Lippert Hans, Knoch Manfred, Reymond Marc-André. Web-based data warehouse on gene expression in human colorectal cancer. Proteomics. 2005 Aug;5(12):3066–78. doi: 10.1002/pmic.200402107. [DOI] [PubMed] [Google Scholar]
- [28].Schwikowski B, Uetz P, Fields S. A network of protein-protein interactions in yeast. Nat Biotechnol. 2000 Dec;18(12):1257–61. doi: 10.1038/82360. [DOI] [PubMed] [Google Scholar]
- [29].Shilov Ignat V, Seymour Sean L, Patel Alpesh A, Loboda Alex, Tang Wilfred H, Keating Sean P, Hunter Christie L, Nuwaysir Lydia M, Schaeffer Daniel A. The paragon algorithm, a next generation search engine that uses sequence temperature values and feature probabilities to identify peptides from tandem mass spectra. Mol Cell Proteomics. 2007 Sep;6(9):1638–55. doi: 10.1074/mcp.T600050-MCP200. [DOI] [PubMed] [Google Scholar]
- [30].Sonoda T, Shirabe K, Takenaka K, Kanematsu T, Yasumori K, Sugimachi K. Angiographically undetected small hepatocellular carcinoma: clinicopathological characteristics, follow-up and treatment. Hepatology. 1989 Dec;10(6):1003–7. doi: 10.1002/hep.1840100619. [DOI] [PubMed] [Google Scholar]
- [31].Stark Chris, Breitkreutz Bobby-Joe, Reguly Teresa, Boucher Lorrie, Breitkreutz Ashton, Tyers Mike. Biogrid: a general repository for interaction datasets. Nucleic Acids Res. 2006 Jan;34(Database issue):D535–9. doi: 10.1093/nar/gkj109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Stelzl Ulrich, Worm Uwe, Lalowski Maciej, Haenig Christian, Brembeck Felix H, Goehler Heike, Stroedicke Martin, Zenkner Martina, Schoenherr Anke, Koeppen Susanne, Timm Jan, Mintzlaff Sascha, Abraham Claudia, Bock Nicole, Kietzmann Silvia, Goedde Astrid, Toksöz Engin, Droege Anja, Krobitsch Sylvia, Korn Bernhard, Birchmeier Walter, Lehrach Hans, Wanker Erich E. A human protein-protein interaction network: a resource for annotating the proteome. Cell. 2005 Sep;122(6):957–68. doi: 10.1016/j.cell.2005.08.029. [DOI] [PubMed] [Google Scholar]
- [33].Tan Hwee Tong, Tan Sandra, Lin Qingsong, Lim Teck Kwang, Hew Choy Leong, Chung Maxey C M. Quantitative and temporal proteome analysis of butyrate-treated colorectal cancer cells. Mol Cell Proteomics. 2008 Jun;7(6):1174–85. doi: 10.1074/mcp.M700483-MCP200. [DOI] [PubMed] [Google Scholar]
- [34].Villanueva Augusto, Minguez Beatriz, Forner Alejandro, Reig Maria, Llovet Josep M. Hepatocellular carcinoma: novel molecular approaches for diagnosis, prognosis, and therapy. Annu Rev Med. 2010;61:317–28. doi: 10.1146/annurev.med.080608.100623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].von Mering Christian, Krause Roland, Snel Berend, Cornell Michael, Oliver Stephen G, Fields Stanley, Bork Peer. Comparative assessment of large-scale data sets of protein-protein interactions. Nature. 2002 May;417(6887):399–403. doi: 10.1038/nature750. [DOI] [PubMed] [Google Scholar]
- [36].Xenarios Ioannis, Salwínski Lukasz, Duan Xiaoqun Joyce, Higney Patrick, Kim Sul-Min, Eisenberg David. Dip, the database of interacting proteins: a research tool for studying cellular networks of protein interactions. Nucleic Acids Res. 2002 Jan;30(1):303–5. doi: 10.1093/nar/30.1.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Zhou Xianghong, Kao Ming-Chih J, Wong Wing Hung. Transitive functional annotation by shortest-path analysis of gene expression data. Proc Natl Acad Sci U S A. 2002 Oct;99(20):12783–8. doi: 10.1073/pnas.192159399. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Included are supplementary methods and tables.
The cleaned network comprising data pooled from various literature sources and cleaned using the CMC/filterNadd program, which implements the iterated CD-distance method.
iTRAQ data obtained from the 5 moderate stage patients.
iTRAQ data obtained from the 7 poor stage patients.
The PERL code for generating the linked proteins from the seed set. It also outputs an edge list comprising links between linked and seed proteins.
The set of linked genes derived from the data set.