Background: The function of many proteins is regulated through post-translational methylation.
Results: METTL21A was identified as a human protein methyltransferase targeting Hsp70 proteins, thereby altering their ability to interact with client proteins.
Conclusion: METTL21A is a specific methyltransferase modulating the function of Hsp70 proteins.
Significance: The activity of a human protein-modifying enzyme is unraveled, and the modification is demonstrated to have functional consequences.
Keywords: Enzyme Catalysis, Enzymes, Heat Shock Protein, Molecular Chaperone, Protein Methylation
Abstract
Hsp70 proteins constitute an evolutionarily conserved protein family of ATP-dependent molecular chaperones involved in a wide range of biological processes. Mammalian Hsp70 proteins are subject to various post-translational modifications, including methylation, but for most of these, a functional role has not been attributed. In this study, we identified the methyltransferase METTL21A as the enzyme responsible for trimethylation of a conserved lysine residue found in several human Hsp70 (HSPA) proteins. This enzyme, denoted by us as HSPA lysine (K) methyltransferase (HSPA-KMT), was found to catalyze trimethylation of various Hsp70 family members both in vitro and in vivo, and the reaction was stimulated by ATP. Furthermore, we show that HSPA-KMT exclusively methylates 70-kDa proteins in mammalian protein extracts, demonstrating that it is a highly specific enzyme. Finally, we show that trimethylation of HSPA8 (Hsc70) has functional consequences, as it alters the affinity of the chaperone for both the monomeric and fibrillar forms of the Parkinson disease-associated protein α-synuclein.
Introduction
Many cellular proteins are post-translationally methylated. These modifications are introduced by protein methyltransferases (MTases),3 usually on amine groups on the side chains of lysine and arginine residues (1). The functional role of protein methylation has been most thoroughly studied in the case of histones, where methylation is described as a key element in the “histone code,” which is a strong determinant of chromatin state and gene transcription status (2). Lysine can accept up to three methyl groups, and many of the so-called “readers” of the histone code represent protein domains that specifically interact with a lysine of a certain methylation state, leading to the recruitment of transcriptional modulators and/or chromatin-modifying enzymes (3, 4). In addition to the frequent lysine methylations on histone proteins, many non-histone proteins are also subject to such methylation (1).
Besides methylating proteins, MTases are responsible for methylation of a wide variety of cellular macromolecules and metabolites (5, 6). Bioinformatics analysis has predicted that MTases constitute ∼1% of all human protein-coding genes, and most of the >200 corresponding enzymes remain uncharacterized (7). MTases are subdivided into five different classes based on their three-dimensional structure. Of these, the “seven-beta strand” (7BS) MTases are the most abundant, comprising 126 enzymes in humans (7). The 7BS-MTases adopt a characteristic Rossmann fold-like twisted β-sheet and contain conserved sequence motifs involved in the binding of the methyl donor S-adenosylmethionine (SAM), facilitating their identification by sequence homology searches (8). The second largest class of MTases is constituted by the SET domain proteins, which are lysine-specific protein MTases responsible for methylations on the histone tails, as well as several methylations on non-histone substrates (9). However, some 7BS-MTases are also lysine-specific, and so far, three such human enzymes have been characterized. These are the histone-specific MTase DOT1L, which methylates Lys-79 in the globular part of histone H3; CaM-KMT, which trimethylates Lys-115 in calmodulin; and VCP-KMT, which trimethylates Lys-315 in the molecular chaperone valosin-containing protein (VCP) (see below) (10–12).
Hsp70 (heat shock protein of ∼70 kDa) proteins represent ATP-dependent molecular chaperones present in all three domains of life and constitute one of the most evolutionarily conserved protein families (13). Humans have 13 genes encoding Hsp70s (HSPA proteins), and whereas some of these proteins are induced by heat shock and other types of stress, others are constitutively expressed (14). Although Hsp70s have been extensively studied, several remain largely uncharacterized. Some human Hsp70s are associated with specific subcellular compartments, such as the endoplasmic reticulum resident HSPA5 (BiP/Grp78), mitochondrial HSPA9 (mortalin), cytosolic constitutively expressed HSPA8 (Hsc70), and cytosolic/nuclear classical stress-inducible HSPA1 (Hsp70). Other members are testis-specific HSPA2 and cytosolic HSPA6, which has been reported to chaperone p53 (13). Hsp70s share a domain architecture consisting of an N-terminal nucleotide-binding domain (NBD), followed by a substrate-binding domain, which can be further subdivided into a peptide-binding domain, which binds stretches of hydrophobic residues, and a C-terminal “lid” domain, which can fold back onto the peptide-binding domain and lock client proteins to Hsp70 (15). This process, and thus the affinity for unfolded substrates and the binding/release cycles of the chaperone, is allosterically regulated by ATP hydrolysis catalyzed by the NBD, which in turn is stimulated by co-chaperones from the Hsp40/DNAJ family and by nucleotide exchange factors. Together, these proteins are often referred to as the “Hsp70 chaperone machinery” (16).
In a recent study (12), we reported that the previously uncharacterized human putative MTase METTL21D was responsible for trimethylating Lys-315 in the abundant and essential molecular chaperone VCP, and we denoted this enzyme VCP-KMT. Moreover, our results indicated that VCP-KMT and its close relatives METLL21A, METTL21B, and METTL21C, together with calmodulin (CaM-KMT) and five uncharacterized MTases, constitute a family of human protein MTases (12). In this work, we report the function of METTL21A and show by a combination of enzymatic and cellular studies that it catalyzes the trimethylation of a conserved Lys residue found in the lid domain of several human Hsp70s. Moreover, we show that trimethylation of HSPA8 modulates its chaperone activity, manifested as a reduced ability to sequester the Parkinson disease-associated protein α-synuclein (α-Syn) in a soluble state and to bind to preformed α-Syn fibrils.
EXPERIMENTAL PROCEDURES
Bioinformatics Analyses
NCBI BLAST was used for identification of proteins homologous to human Hsp70s and METTL21A (17). Multiple protein sequence alignments were performed using the Muscle algorithm embedded in Jalview v2.8 (18). The secondary structure of proteins was predicted from the primary sequence using the PSIPRED algorithm (19).
Cloning and Mutagenesis
All plasmids used in this study and the detailed cloning strategy used to generate them are listed in supplemental Table S1. Briefly, ORFs were amplified with Phusion DNA polymerase (Thermo Scientific) using primers that generated PCR products with restriction sites flanking the ORF. The PCR product was then digested with the indicated restriction enzymes (New England Biolabs) and cloned into pNTAPa (Agilent Technologies), pGEX-6P (GE Healthcare), or pET28a (Novagen). To generate the pcDNA5/FRT/TO-METL21A-CBP-SBP construct, METTL21A was first cloned into pNTAPa and then subcloned into pcDNA5/FRT/TO (Invitrogen) using ApaI and NotI. Mutagenesis was performed using the QuikChange site-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions. The identity of all constructs was verified by DNA sequencing.
Generation and Cultivation of Human Cell Lines
A cell line for inducible overexpression of calmodulin-binding peptide (CBP)- and streptavidin-binding peptide (SBP)-tagged METTL21A was generated using the Flp-InTM T-RExTM HEK-293 system (Invitrogen) according to the manufacturer's instructions. Briefly, Flp-In T-REx HEK-293 cells were cotransfected with pcDNA5/FRT/TO-METTL21A-CBP-SBP and pOG44 (Invitrogen) using Lipofectamine® 2000 (Invitrogen). Successful incorporation of the METTL21A construct was selected for with 200 μg/ml hygromycin B (Invitrogen). Cells were thereafter maintained in high-glucose DMEM (Lonza) supplemented with tetracycline-reduced 10% (v/v) FBS (AH diagnostics), l-glutamine (Lonza), 5 μg/ml blasticidin S (Invitrogen), 10 μg/ml streptomycin (Invitrogen), and 10 units/ml penicillin (Invitrogen).
Tandem Affinity Purification
Tandem affinity purification (TAP) was performed using the InterPlay mammalian TAP system (Agilent) according to the manufacturer's instructions with a few exceptions. Briefly, the Flp-In T-REx HEK-293 METTL21A-CBP-SBP cell line was expanded onto 20 T175 flasks. Overexpression of the bait was induced in 10 flasks with 1 μg/ml doxycycline (Sigma-Aldrich D9891) for 24 h, whereas the remaining flasks were untreated to serve as a negative control. As our prior experience indicated that S-adenosyl-l-homocysteine may stabilize 7BS-MTase·substrate complexes, all buffers were supplemented with 100 μm S-adenosyl-l-homocysteine, and 1 mm PMSF (Sigma-Aldrich) and protease inhibitor mixture (Sigma-Aldrich) were added to promote protein stability throughout the purification. Proteins were eluted by boiling in 2× NuPAGE gel loading buffer (Invitrogen) and stored at −20 °C until analyzed.
Recombinant Protein Purification
pET28a- and pGEX-6P-derived plasmids harboring recombinant His6- and GST-tagged proteins, respectively, were transfected into the Escherichia coli BL21-CodonPlus (DE3)-RIPL expression strain (Agilent). Cells were cultured with appropriate antibiotics at 37 °C and shaking at 250 rpm until the absorbance at 600 nm reached 0.5. The temperature was then reduced to 18 °C, and the expression of His6- or GST-tagged proteins was induced by 100 or 500 μm isopropyl β-d-thiogalactopyranoside, respectively, and allowed to proceed for 16 h. Cells were thereafter harvested by centrifugation. For purification of His6-tagged proteins, cell pellets were resuspended in lysis buffer (50 mm Tris (pH 8.0), 500 mm NaCl, 10% (w/v) glycerol, 0.5% (w/v) Nonidet P-40, 30 mm imidazole, 3 mm 2-mercaptoethanol, Complete protease inhibitor mixture (Roche Applied Science), 0.5 mg/ml lysozyme (Sigma-Aldrich), and 25 units/ml Benzonase (Sigma-Aldrich)). Imidazole was omitted for GST-tagged constructs. Cell lysates were cleared by centrifugation, and recombinant proteins were allowed to bind to nickel-nitrilotriacetic acid-agarose (His6-tagged proteins; Qiagen) or glutathione-Sepharose 4B (GST-tagged proteins; GE Healthcare) at 4 °C for 16 h. Glutathione-Sepharose 4B affinity resins were extensively washed with wash buffer (50 mm Tris (pH 7.5), 500 mm NaCl, and 10% (w/v) glycerol), whereas nickel-nitrilotriacetic acid-agarose resins were washed with wash buffer supplemented with 30 mm imidazole. Recombinant proteins were eluted in elution buffer (50 mm Tris-HCl (pH 8.0) and 500 mm NaCl) supplemented with either 300 mm imidazole (His6-tagged proteins) or 15 mm reduced glutathione (GST-tagged proteins). Eluates were then buffer-exchanged to storage buffer (100 mm NaCl, 20 mm Tris (pH 6.8), and 1 mm DTT) by sequential concentration and dilution using Vivaspin 20 ultrafiltration columns with a molecular mass cutoff of 10 or 50 kDa (Sartorius AG). Proteins were aliquoted and stored at −80 °C, and concentrations were determined using the BCA protein assay kit (Pierce).
For tag removal, GST-fused HSPA proteins were incubated with PreScission protease (GE Healthcare) at 4 °C for 16 h to allow proteolytic cleavage. Free GST, GST-fused HSPA proteins, and protease were removed with glutathione-Sepharose according to the manufacturer's instructions, and untagged HSPA proteins were buffer-exchanged to storage buffer as described above.
Measurement of in Vitro Methyltransferase Activity
Methyltransferase reactions were performed in 50-μl volumes for 1 h at 37 °C in methyltransferase reaction buffer (50 mm Tris (pH 7.8), 50 mm KCl, 5 mm MgCl2, 1 mm ATP, 13 μm [3H]SAM (2 μCi; PerkinElmer Life Sciences), 4 μm substrate, and 2 μm METTL21A) unless stated otherwise. For mass spectrometric analysis, radiolabeled SAM was replaced with 1.2 mm nonradioactive SAM (New England Biolabs). Reactions were stopped by precipitating proteins with TCA for liquid scintillation counting of radioactivity or by denaturation in NuPAGE sample buffer for SDS-PAGE analysis (followed by MS or fluorographic analysis).
For fluorography, reactions were separated on NuPAGE 4–12% gradient Bis-Tris gels (Invitrogen), and proteins were transferred to PVDF membranes using the XCell SureLockTM system (Invitrogen). Membranes were then stained with Ponceau S, treated with the scintillation enhancer EN3HANCE (PerkinElmer Life Sciences), and exposed to Carestream Kodak BioMax MS film (Sigma-Aldrich) for 24–72 h at −80 °C.
For liquid scintillation counting, proteins were precipitated in 10% (w/v) TCA at 4 °C for 1 h. Acid-insoluble material was retained on glass fiber filters (Whatman GF/C) by vacuum filtration. Filters were washed with 10% (w/v) TCA and EtOH and placed in scintillation fluid (Ultima GoldTM XR, PerkinElmer Life Sciences), and incorporated radioactivity was measured by scintillation counting.
Mass Spectrometric Analysis
LC-MS/MS of proteolytic peptides was performed using a Hypercarb 5-μm particle size column (G&T Septech AS) for sample concentration and a C18 column (GlycproSIL C18–80Å, Glycpromass) for analytical separation of peptides. Samples were washed with a mobile phase (0.1% (v/v) formic acid and 2.5% (v/v) acetonitrile) and eluted with a binary gradient of increasing acetonitrile up to 60% (v/v). The LC setup was coupled to a LTQ Orbitrap XL mass spectrometer (Thermo Scientific) via nanoelectrospray. In-gel proteolytic digestion was performed with Asp-N (Roche Applied Science) or trypsin (Sigma-Aldrich). Peptide samples were analyzed with a collision-induced dissociation fragmentation method, acquiring one Orbitrap survey scan in the mass range of m/z 200–2000, followed by MS/MS of the most intense ions in the Orbitrap.
Mass spectrometric data were analyzed with searches using Proteome Discoverer SEQUESTTM (Thermo Scientific) against in-house maintained databases of either HSPA proteins or the human proteome. The mass tolerances for fragment ions and parent ions were set to 0.5 Da and 10 ppm, respectively. Methionine oxidation and lysine mono-, di-, and trimethylation and acetylation were selected as variable modifications. MS/MS spectra of relevant peptides were manually extracted using Qual Browser (v2.0.7).
Chromatograms representing various methylation states of peptides were generated by gating for m/z ratios corresponding to un-, mono-, di-, and trimethylated species of the predominant charge state using the BOXCAR algorithm and a sensitivity of 10 ppm in Qual Browser (v2.0.7). To achieve comparable chromatograms, the y axis was normalized with respect to signal intensity for the different methylation states. Relative quantification of peptides was performed by integrating area under curves using Qual Browser (v2.0.7).
Antibody Generation and Western Blotting
The following primary antibodies were used (dilution in Western blotting is indicated): anti-HSPA1 (Abcam ab79852; 1:10,000), anti-HSPA8 (Abcam EP1531Y/ab51052; 1:1000), anti-GAPDH (Ambion AM4300; 1:4000), and anti-CBP (Millipore 07-482; 1:4000). For detection of HSPA1-K561me3 (HSPA1(me3)), a custom antibody was used at a dilution of 1:10,000 in Western blots. The antibody was obtained by affinity purification of serum from rabbits immunized with a synthetic peptide (acetyl-CDEGLKGK(me3)ISEA-amide; New England Peptide).
Transfer of samples to PVDF membrane was performed as described above. Membranes were blocked with 5% (w/v) BSA in TBS containing 0.05% (w/v) Tween 20 and incubated with the appropriate HRP-conjugated secondary antibodies. Blots were thereafter incubated with SuperSignal West Dura (Thermo Scientific), and chemiluminescence signals were detected with a Image Station 4000R Pro system (Kodak).
siRNA-mediated Knockdown
For knockdown assays, Flp-In T-REx HEK-293 cells were transfected with either a pool of siRNAs versus the METTL21A transcript (Sigma-Aldrich SASI_Hs01_00047371, SASI_Hs01_00047373, and SASI_Hs01_00047374) or a scrambled sequence as a negative control (Sigma-Aldrich SIC001), or RNA was omitted. Lipofectamine RNAiMAX (Invitrogen) was used as the transfection agent according to the manufacturer's instructions, and knockdown was assayed by Western blotting and MS after 72 h.
Measurement of ATPase Activity
Reactions were performed in 50-μl volumes in a 96-well format in assay buffer (25 mm HEPES (pH 7.4), 1 mm DTT, 5 mm MgCl2, and 50 mm KCl) supplemented with HSPA1 (1 μm), HSPA8 (1 μm), DNAJB1 (1 μm), HSPA-KMT (0.5 μm), SAM (1 mm), and ATP (100 μm) as indicated. After incubation of the plate for 60 min at 37 °C in a humidity chamber, 100 μl of BIOMOL Green reagent (Enzo Life Sciences) was added. After additional incubation of the plate at room temperature for 30 min, the absorbance at 620 nm was measured. The concentration of liberated free phosphate was determined from a standard curve.
α-Syn Purification and Assembly into Fibrils
Recombinant α-Syn was purified as described previously (20). The α-Syn concentration was determined spectrophotometrically using an extinction coefficient of 5960 m−1 cm−1 at 280 nm. Pure α-Syn (0.5 mm) in 50 mm Tris-HCl (pH 7.5) and 150 mm KCl was passed through sterile 0.22-μm filters and stored at −80 °C.
Assembly of α-Syn fibrils was performed as described previously (21). Briefly, soluble α-Syn was incubated with continuous shaking in an Eppendorf Thermomixer set at 600 rotations/min at 37 °C. At regular time intervals, aliquots were withdrawn, and the assembly process was monitored using thioflavin T binding by averaging the emission signal at 480 nm for an excitation wavelength of 440 nm over 30 s using a Cary Eclipse spectrofluorometer (Varian Inc., Palo Alto, CA). The morphology and structure of α-Syn assemblies were assessed using a Jeol 1400 transmission electron microscope following adsorption of the samples onto carbon-coated 200-mesh grids and negative staining with 1% (w/v) uranyl acetate. The images were recorded with a Gatan ORIUS CCD camera.
RESULTS
Bioinformatics Analysis of METTL21A
Human METTL21A belongs, together with METTL21B, METTL21C, and VCP-KMT (METTL21D), to a subgroup of four closely related enzymes within MTase family 16 (pfam PF10294) (12). To identify putative METTL21A orthologs in other organisms, BLAST searches were performed to identify proteins with substantial sequence similarity to METTL21A (expect value < 10−15). Next, these candidate proteins were used as queries in reciprocal BLAST searches versus human sequences, and those that yielded METTL21A, rather than its paralogs, as the best human hit were categorized as putative orthologs. According to this analysis, METTL21A orthologs are found in a wide range of eukaryotes, including all vertebrates and several invertebrates, but appear to be absent in bacteria (Table 1). However, METTL21A shows a rather scattered distribution among eukaryotes; for example, METTL21A orthologs are absent in the common model organisms Arabidopsis thaliana, Caenorhabditis elegans, and Schizosaccharomyces pombe but are present in other members of the respective organism groups, i.e. plants, nematodes, and fungi (Table 1).
TABLE 1.
Presence of METTL21A-like sequences in various organisms
| Closest METTL21A homolog |
Reciprocal BLAST versus human (expect value) |
|||||
|---|---|---|---|---|---|---|
| Organism | Expect value | Accession no. | METTL21A (NP_660323.3) | METTL21B (NP_056248.2) | METTL21C (NP_001010977.1) | METTL21D (NP_078834.2) |
| Vertebrates | ||||||
| H. sapiens (human) | NP_660323.3 | 1E-161a | 1E-48 | 6E-23 | 5E-28 | |
| Mus musculus (mouse) | 3E-133 | NP_080240.1 | 5E-145 | 3E-47 | 2E-24 | 3E-25 |
| D. rerio (zebrafish) | 1E-80 | NP_001013584.1 | 2E-92 | 4E-49 | 5E-26 | 3E-24 |
| Gallus gallus (chicken) | 2E-94 | XP_421949.1 | 3E-106 | 4E-43 | 2E-28 | 5E-26 |
| Insects | ||||||
| Acyrthosiphon pisum (pea aphid)b | 5E-20 | XP_001945214.1 | 5E-22 | 5E-25 | 4E-15 | 2E-38 |
| Drosophila melanogaster (fruit fly) | 4E-07 | NP_573368.2 | 9E-9 | 5E-11 | 0.00002 | 0.024 |
| Nematodes | ||||||
| A. suum (pig roundworm)b | 5E-31 | ADY46645.1 | 5E-33 | 1E-22 | 3E-11 | 1E-10 |
| C. elegans | 6E-10 | NP_001122759.1 | 7E-12 | 9E-21 | 7E-13 | 5E-29 |
| Plants | ||||||
| Z. mays (maize)b | 1E-25 | NP_001142219.1 | 2E-27 | 2E-20 | 3E-18 | 9E-25 |
| A. thaliana (thale cress) | 1E-21 | AAU94386.1 | 2E-23 | 2E-24 | 3E-18 | 4E-24 |
| Fungi | ||||||
| Serpula lacrymans (dry rot fungus)b | 7E-29 | EGO01576 | 1E-30 | 3E-18 | 2E-14 | 4E-28 |
| S. cerevisiae (budding yeast) | 1E-17 | NP_014374.1 | 1E-19 | 5E-13 | 2E-11 | 2E-16 |
| S. pombe (fission yeast) | 9E-11 | NP_593178.1 | 2E-12 | 6E-11 | 4E-8 | 1E-14 |
| Othersc | ||||||
| Crassostrea gigas (Pacific oyster) | 7E-69 | EKC35706.1 | 9E-71 | 3E-44 | 4E-22 | 4E-24 |
| N. vectensis (starlet sea anemone) | 7E-64 | XP_001623378.1 | 8E-66 | 2E-34 | 1E-22 | 4E-30 |
| Strongylocentrotus purpuratus (sea urchin) | 4E-51 | XP_781765.3 | 4E-58 | 7E-31 | 1E-22 | 1E-17 |
a Boldface indicates best human hit in a reciprocal search.
b Best hit within a group (insect, nematode, etc.).
c Best hits among invertebrates.
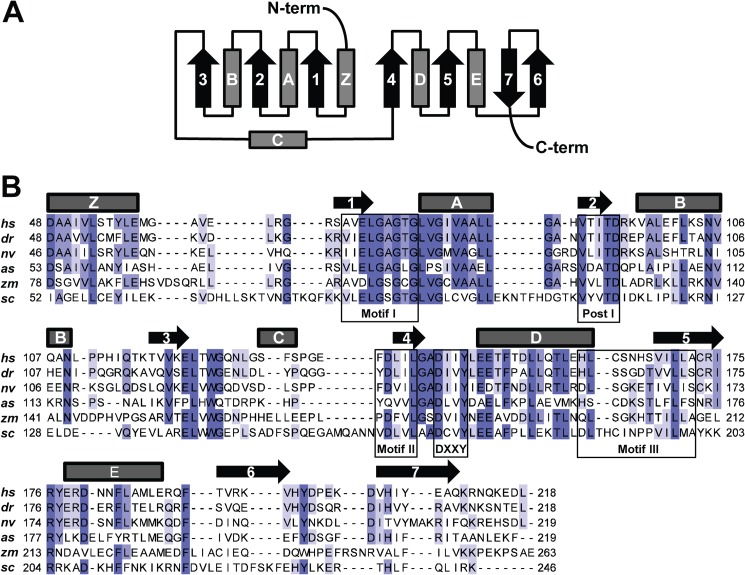
METTL21A was predicted to be a 7BS-MTase in a recent bioinformatics study aimed at uncovering the human methyltransferasome (7). Accordingly, a sequence alignment of putative METTL21A orthologs from various organisms revealed the presence of conserved sequence motifs corresponding to motifs typical of the 7BS-MTases, denoted Motif 1, Post 1, Motif 2, and Motif 3, as well as a hallmark (D/E)XX(Y/F) motif found in members of MTase family 16 (Fig. 1) (8, 12). Furthermore, secondary structure prediction demonstrated that the expected 7BS topology and the conserved motifs are localized at the expected positions relative to the respective β-strands. In conclusion, this initial bioinformatics analysis established METTL21A as an archetypical 7BS-MTase with putative orthologs in a wide range of eukaryotes.
FIGURE 1.
Human METTL21A harbors motifs characteristic of a 7BS-MTase. A, topology diagram of the canonical 7BS-MTase fold. α-Helices and β-sheets are depicted as barrels and arrows, respectively. B, protein sequence alignment of Homo sapiens METTL21A (hs; Q8WXB1, NP_660323.3) and putative orthologs from Danio rerio (dr; NP_001013584.1), Nematostella vectensis (nv; XP_001623378.1), Ascaris suum (as; F1L941_ASCSU), Zea mays (zm; NP_001142219.1), and S. cerevisiae (sc; NP_014374.1). Boxes indicate motifs characteristic for 7BS-MTases (Motif 1, Post 1, Motif 2, and Motif 3), as well as the (D/E)XX(Y/F) motif typical of MTase family 16 (8, 12). The predicted secondary structure for human METTL21A is indicated above the sequence alignment.
METTL21A Interacts with and Methylates Hsp70 Proteins
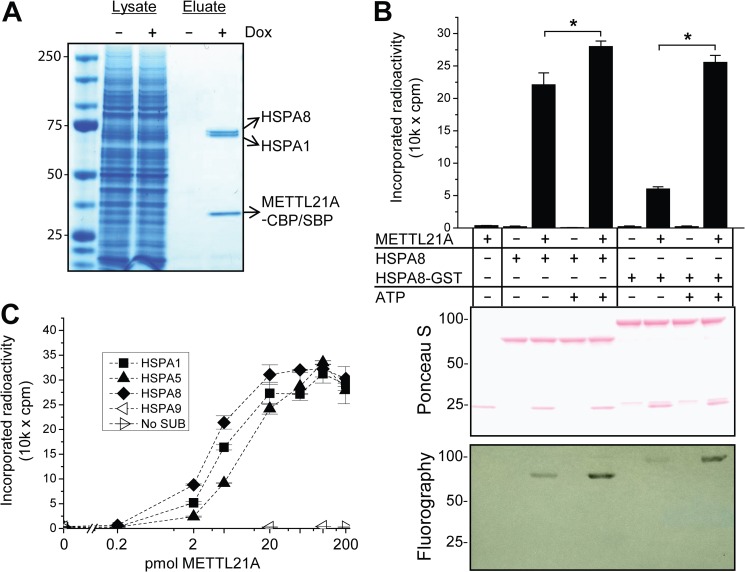
Reasoning that METTL21A substrate(s) could be found among its interaction partners, we applied several established techniques to explore the interactome of the protein, i.e. (i) yeast two-hybrid screen, (ii) pulldown assays with recombinant proteins, and (iii) TAP. The proteins ARPC4 (P59998) and prohibitin (PHB; P35232) were identified as interactants by yeast two-hybrid screen and pulldown experiments, respectively. However, no MTase activity was observed towards these proteins (data not shown), and they are not annotated as being methylated (22). Furthermore, no MTase activity was detected towards DNA, RNA, recombinant histones, or BSA (data not shown). For TAP, a HEK-293-derived cell line with doxycycline-inducible overexpression of CBP- and SBP-tagged METTL21A was generated. As expected, the cell line displayed selective induction of tagged bait protein upon addition of the antibiotic, and the bait was selectively enriched by TAP (data not shown). SDS-PAGE analysis of the proteins obtained by TAP revealed that two ∼70-kDa proteins co-purified with METTL21A. Peptide mass fingerprinting identified these as two members of the Hsp70 protein family: stress-inducible HSPA1 (Hsp70) and constitutively expressed HSPA8 (Hsc70) (Fig. 2A).
FIGURE 2.
METTL21A interacts with and methylates members of the human Hsp70 protein family. A, members of the Hsp70 family co-purify with METTL21A in TAP. Shown is a Coomassie Blue-stained SDS-polyacrylamide gel of lysates and eluates from TAP from a Flp-In T-REx HEK-293-derived cell line treated, where indicated, with doxycycline (Dox) to induce expression of SBP- and CBP-tagged METTL21A. The identity of bait and co-purifying proteins was determined by peptide mass fingerprinting and is indicted to the right. Molecular mass standards (in kilodaltons) are indicated to the left. B, METTL21A methylates HSPA8. 200 pmol of untagged or GST-tagged HSPA8 was incubated with [3H]SAM in the presence or absence of 1 mm ATP and 100 pmol of METTL21A. Upper panel, quantification of incorporated TCA-precipitable radioactivity by liquid scintillation counting. Error bars indicate S.D. (n = 3). *, significant effects of ATP addition (Student's two-tailed t test; alpha level of 0.05, assuming unequal variance). Middle and lower panels, Ponceau S staining and fluorography, respectively, of membranes containing SDS-PAGE-separated methyltransferase reactions corresponding to those in the upper panel. C, activity of METTL21A on various human Hsp70 proteins. The indicated Hsp70 proteins (200 pmol) were incubated with 1 mm ATP and varying amounts of METTL21A in the presence of [3H]SAM. Error bars represent the range of duplicate experiments. No SUB, no substrate.
We then tested whether recombinant HSPA8, expressed and purified as a GST fusion protein, is subject to METTL21A-catalyzed methylation in vitro. In these experiments, substrate and enzyme were incubated in the presence of [3H]SAM, and the incorporation of methyl groups into proteins was measured as TCA-insoluble radioactivity and by fluorography of proteins separated by SDS-PAGE. Hsp70s can exist in two main conformations depending on nucleotide binding to the NBD: an ATP-bound open conformation with low affinity for client proteins and an ADP-bound state associated with a closed conformation and high affinity for clients (15). On this basis, methylation reactions were conducted in both the presence and absence of ATP. Our results show that GST-tagged HSPA8, as well as HSPA8 from which the tag had been proteolytically removed, was subject to METTL21A-mediated methylation. The methylation reaction was significantly stimulated by ATP, a stimulatory effect that was more pronounced with the tagged substrate (Fig. 2B), and we decided to include ATP in future methylation reactions. Furthermore, we also tested other human Hsp70s as potential substrates for METTL21A, i.e. endoplasmic reticulum-associated HSPA5 (BiP/Grp78), stress-inducible HSPA1 (Hsp70), and mitochondrial HSPA9 (mortalin) (Fig. 2C). Interestingly, both HSPA1 and HSPA5 were also METTL21A substrates and were methylated with similar efficiency as HSPA8, whereas the more distantly related protein HSPA9 was not a substrate for methylation. On the basis of the above results, we henceforth refer to METTL21A as Hsp70 lysine methyltransferase, denoted HSPA-KMT.
HSPA-KMT Methylates a Conserved Lysine Found in a Subgroup of Human Hsp70 Proteins
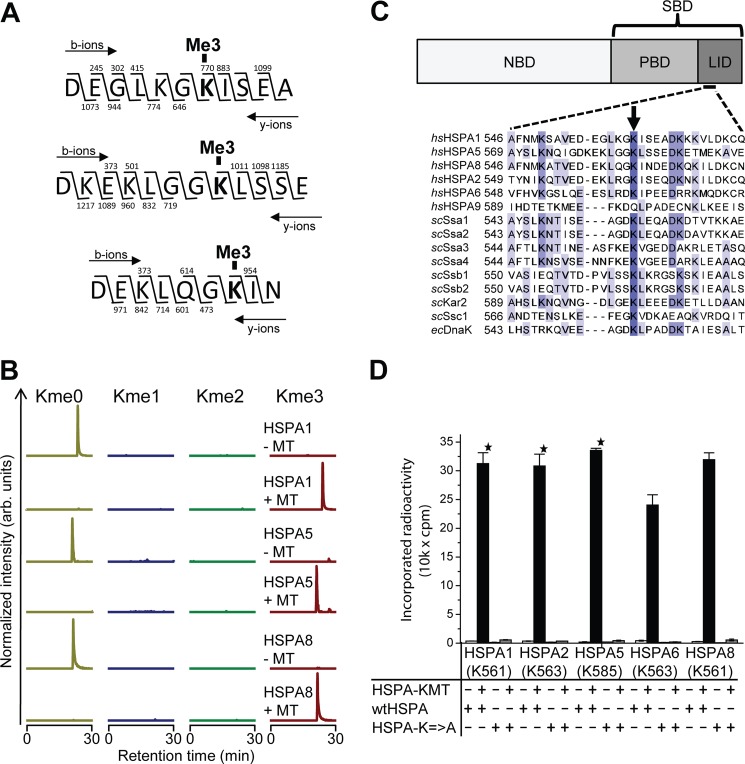
To identify the methylation site(s), HSPA1, HSPA5, and HSPA8 were incubated with HSPA-KMT in the presence of nonradioactive SAM, followed by digestion with the endoprotease Asp-N and MS analysis of the resulting peptides. Interestingly, unique peptides harboring mass shifts corresponding to three added methyl groups were detected in all three test substrates upon incubation with HSPA-KMT (Fig. 3A). MS/MS analysis revealed trimethylation of a single lysine residue, i.e. Lys-561 in HSPA1, Lys-585 in HSPA5, and Lys-561 in HSPA8. Complete trimethylation of these lysine residues was observed after HSPA-KMT treatment, and conversely, the corresponding peptides from control reactions lacking HSPA-KMT were exclusively found in an unmethylated state (Fig. 3B).
FIGURE 3.
HSPA-KMT trimethylates an evolutionarily conserved lysine in human Hsp70s. A, HSPA-KMT trimethylates HSPA1, HSPA5, and HSPA8. Shown are MS/MS fragmentation patterns of proteolytic trimethylated peptides generated by Asp-N digestion of HSPA-KMT-treated proteins. b- and y-ions supporting trimethylation of HSPA1 Lys-561 (top), HSPA5 Lys-585 (middle), and HSPA8 Lys-561 (bottom) are indicated above and below each peptide sequence, respectively. B, MS chromatograms of Asp-N-generated peptides from Hsp70 proteins incubated with HSPA-KMT. Each chromatogram is gated for the un-, mono-, di-, or trimethylated forms of peptides covering Asp-555–Ala-565 of HSPA1, Asp-578–Glu-589 of HSPA5, or Asp-555–Asn-563 of HSPA8. Samples not treated with HSPA-KMT (MT) are shown as controls. arb., arbitrary. C, HSPA domain organization and protein sequence alignment of selected Hsp70 proteins. Upper panel, schematic representation of Hsp70 domain structure illustrating the N-terminal NBD and the C-terminal substrate-binding domain (SBD), constituted by a β-stranded peptide-binding domain (PBD) and an α-helical lid domain. Lower panel, protein sequence alignment of various Hsp70 proteins from H. sapiens (hs), S. cerevisiae (sc), and E. coli (ec) showing the region surrounding the Lys targeted by HSPA-KMT (arrow). The following proteins are shown: human HSPA1 (P08107), HSPA5 (P11021), HSPA8 (P11142), HSPA2 (P54652), HSPA6 (P17066), and HSPA9 (P38646); S. cerevisiae Ssa1 (P10591), Ssa2 (P10592), Ssa3 (P09435), Ssa4 (P22202), Ssb1 (P11484), Ssb2 (P40150), Kar2 (P16474), and Ssc1 (P0CS90); and E. coli DnaK (P0A6Y8). D, mutational analysis of HSPA-KMT substrates. Wild-type human Hsp70 proteins or mutants (200 pmol) in which the conserved lysine in C had been mutated to alanine were incubated with or without 100 pmol of HSPA-KMT. Error bars indicate S.D. (n = 3) or range of duplicate (star) experiments.
Interestingly, sequence alignment of various Hsp70s revealed that methylation occurred at the analogous lysine residue in all three substrates (Fig. 3C). Although found in the relatively poorly conserved lid domain, this residue has been well conserved throughout evolution and is found, for example, in DnaK from E. coli and the Ssa and Ssb proteins from Saccharomyces cerevisiae. Conspicuously, the more distantly related human HSPA9 protein, which was not a substrate for methylation, does not have a lysine residue at this position. To investigate the possibility that HSPA-KMT can methylate sites other than the identified lysine, this residue was mutated to alanine in HSPA1, HSPA5, and HSPA8, as well as in HSPA2 and HSPA6, which also have lysine at this position. Notably, none of these mutant proteins could be methylated, whereas all of the corresponding wild-type proteins were efficiently methylated (Fig. 3D), firmly demonstrating that HSPA-KMT targets a single lysine residue in Hsp70s.
HSPA-KMT Methylates Hsp70 Proteins in Vivo
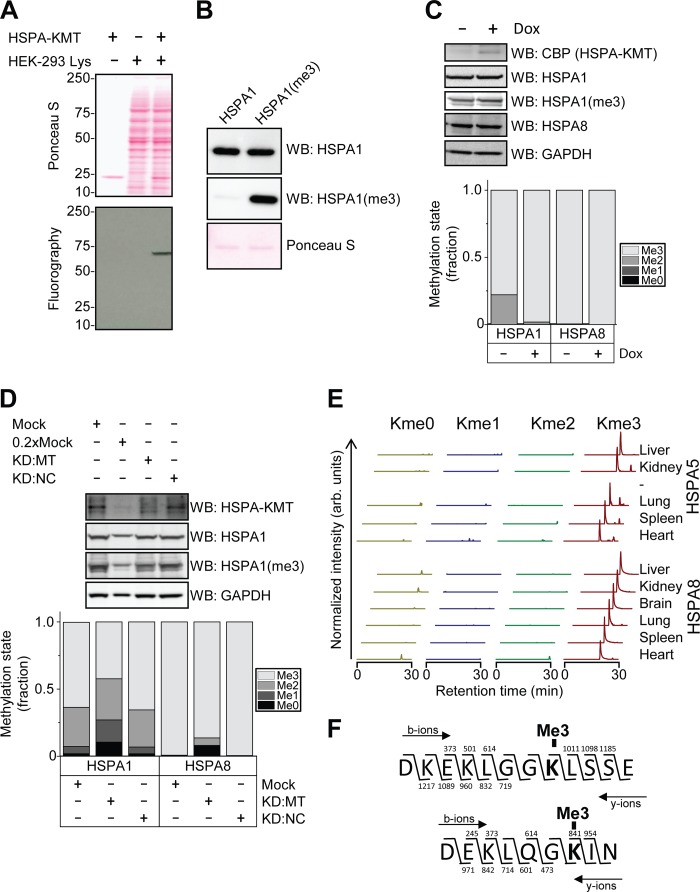
To investigate the possibility that proteins other than Hsp70s are substrates for HSPA-KMT, a protein extract from HEK-293 cells was subjected to in vitro methylation with recombinant enzyme, and methylated proteins were detected by fluorography. In these experiments, only a single band of ∼70 kDa was observed, corresponding to the molecular mass of Hsp70s, demonstrating that HSPA-KMT is a highly specific enzyme, for which the Hsp70s are likely to be the only substrates (Fig. 4A).
FIGURE 4.
HSPA-KMT methylates mammalian Hsp70 proteins in vivo. A, HSPA-KMT-mediated methylation of a 70-kDa protein in human cell extracts. 20 μg of protein extract was methylated with 100 pmol of HSPA-KMT. Upper panel, Ponceau S-stained membrane containing SDS-PAGE-separated methylation reactions. Lower panel, fluorography of membrane. B, specificity of the anti-HSPA1-K561me3 antibody. Western blotting (WB) was performed using an antibody recognizing total HSPA1 or the custom-made antibody targeting HSPA1-K561me3 (HSPA1(me3)) against 100 ng of untreated or HSPA-KMT-treated HSPA1. C, overexpression of HSPA-KMT increases methylation levels. Upper panel, Western blots against CBP (HSPA-KMT), HSPA1, HSPA1-K561me3, HSPA8, and GAPDH in cell extracts from Flp-In T-REx HEK-293 METTL21A-CBP-SBP cells incubated in the presence or absence of doxycycline (Dox). Lower panel, methylation state of Hsp70 proteins determined by MS quantification of Asp-N-generated peptides described in the legend to Fig. 3. D, siRNA-mediated knockdown (KD) of HSPA-KMT decreases methylation levels. Upper panel, Western blots against HSPA1, HSPA1-K561me3, and GAPDH in cell extracts from wild-type Flp-In T-REx HEK-293 cells treated with transfection agent alone (Mock), a pool of siRNAs versus the HSPA-KMT transcript (KD:MT), or siRNA with a randomized sequence as a negative control (KD:NC). 0.2×Mock represents the same sample as the mock but with one-fifth of the amount loaded. Cells were lysed 72 h post-transfection. Lower panel, methylation status of Hsp70 proteins from siRNA-treated cells determined by MS. E, methylation status of murine HSPA5 and HSPA8 in a panel of mouse organs. Chromatograms were gated for different methylation states of Asp-N-generated peptides corresponding to amino acids 579–590 in mouse HSPA5 (P20029) and amino acids 555–563 in mouse HSPA8 (P63017). arb., arbitrary. F, MS/MS fragmentation patterns supporting lysine trimethylation of Hsp70s in mouse liver. The fragmentation patterns corresponding to the major peaks observed in mouse liver in E supporting trimethylation of Lys-586 in mouse HSPA5 (upper panel) and Lys-561 in mouse HSPA8 (lower) are shown.
To assess the methylation state of Hsp70s in cells, we analyzed Asp-N-generated peptides encompassing the methylation site (shown in Fig. 3) by MS, produced by in-gel digestion of cellular proteins separated by SDS-PAGE. For HSPA1, we also generated an antibody against a peptide representing the trimethylated lysine and surrounding residues. This antibody efficiently recognized HSPA1 trimethylated at Lys-561 (HSPA1(me3)), but not the unmethylated counterpart (Fig. 4B). To address whether HSPA-KMT is responsible for Hsp70 methylation in vivo, we first investigated the methylation status of various Hsp70s in the HEK-293 cell line used for TAP, which carries an HSPA-KMT-encoding gene under the control of an inducible promoter. In non-induced cells, the relevant Lys residue in HSPA8 was found to be completely trimethylated, whereas HSPA1 displayed, in addition to the dominating trimethylated state, substantial amounts of dimethylation (Fig. 4C). When overexpression of HSPA-KMT was induced, the fraction of trimethylated HSPA1 shifted from ∼0.80 to ∼0.98 as measured by MS, indicating that overexpressed HSPA-KMT is functional and capable of performing HSPA1 methylation in vivo. When the HSPA1(me3)-specific antibody was used to assess cellular HSPA1 methylation, a slightly increased signal was observed after HSPA-KMT overexpression, in good agreement with the MS data.
Conversely, when HSPA-KMT protein levels were knocked down by transfecting cells with siRNA oligonucleotides, a substantial reduction in methylation level was observed both for HSPA1 and HSPA8 (Fig. 4D). The un- and monomethylated species of HSPA1 were increased by factors of ∼5 and ∼3, respectively, whereas the trimethylated species was reduced by a factor of ∼1.5. Accordingly, the signal obtained with the HSPA1(me3)-specific antibody also showed a substantial reduction. For HSPA8, which was exclusively trimethylated in untreated cells, siRNA treatment led to the appearance of all methylation states, and the trimethylated fraction decreased to below 0.90.
Having established that HSPA-KMT-mediated methylation occurs in mammalian cell culture, we next investigated the methylation status of Hsp70s in various mouse tissues. Protein extracts were prepared from a panel of mouse organs, and the methylation status of HSPA5 and HSPA8 was assessed by MS. Of note, HSPA1 was not detected in this analysis, presumably due to its low expression levels in healthy unstressed animals (23). In all analyzed mouse organs, the peptides covering HSPA8 Lys-561 and HSPA5 Lys-586 were exclusively detected in the trimethylated state (Fig. 4, E and F). Interestingly, previous studies have reported that peptides covering Lys-585 in human HSPA5 (corresponding to Lys-586 in mouse HSPA5) are acetylated (24, 25). Notably, trimethylation and acetylation give very similar changes in molecular mass, +42.0106 Da, and +42.0470 Da, respectively, and cannot be distinguished by low-resolution MS. However, the Orbitrap instrument used for our analyses has the resolution required to distinguish between these modifications, and our results clearly support the presence of trimethyl rather than acetyl modification at this site (data not shown). Our analysis of the methylation status of Hsp70s in cells and tissues clearly demonstrates that the HSPA-KMT activity observed in vitro mirrors a reaction that also occurs in vivo and indicates that the Lys residues targeted by HSPA-KMT exist primarily in a trimethylated state in mammals.
Functional Role of Hsp70 Methylation
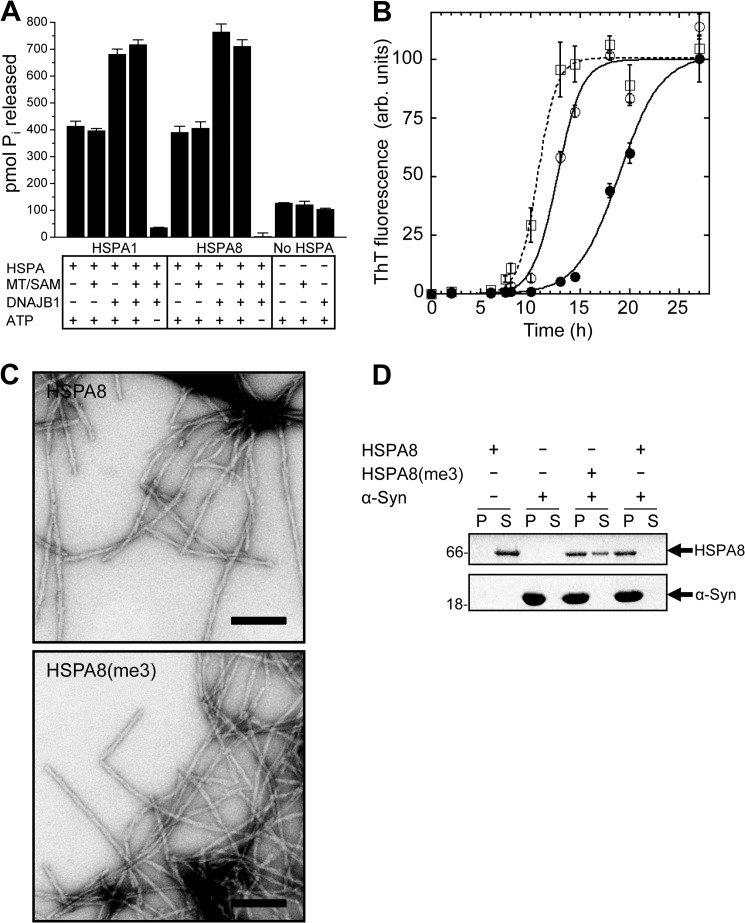
The affinity of Hsp70s for their client proteins is determined by the conformation of the lid domain, which in turn is regulated by nucleotide binding to the NBD. ATP hydrolysis has been shown to be stimulated by DNAJ proteins, whereas the exchange of ADP for ATP is regulated by nucleotide exchange factors. To assess whether HSPA-KMT has a functional role in regulating the ATP hydrolysis rate, ATPase activity was assayed for HSPA1 and HSPA8 alone or in the presence of DNAJB1 and/or HSPA-KMT/SAM. As reported previously, DNAJB1 stimulated HSPA-mediated ATP hydrolysis of both HSPA1 (26) and HSPA8 (21) (Fig. 5A). Notably, neither basal nor DNAJB1-stimulated ATP hydrolysis was affected by adding HSPA-KMT and SAM in amounts sufficient to obtain full trimethylation, indicating that methylation is not involved in regulating or modulating ATP hydrolysis.
FIGURE 5.
Functional effect of HSPA-KMT-mediated methylation. A, stimulation of basal ATPase activity. Shown is the ATPase activity of HSPA1 and HSPA8 in the absence or presence of DNAJB1 and/or HSPA-KMT (MT) and SAM. ATP and HSPA proteins were omitted in relevant control reactions. ATP hydrolysis was indirectly quantified by the amount of liberated Pi after 1 h at 37 °C. Bars represent the mean, and error bars indicate S.D. (n = 3). B, effect of HSPA-KMT-mediated methylation of HSPA8 on α-Syn assembly. Time courses of α-Syn fibrillation in the absence of HSPA8 (□) or in the presence of unmethylated (●) or Lys-561-trimethylated (○) HSPA8. Data are presented as means ± S.D. (n = 3). A sigmoidal curve (line) was fitted to the experimental data (symbols). ThT, thioflavin T; arb., arbitrary. C, electron micrographs of fibrils formed in the presence of HSPA8 and HSPA8-K561me3 (HSPA8(me3)). Scale bars = 200 nm. D, HSPA-KMT-mediated methylation alters the affinity of HSPA8 for fibrillar α-Syn. Shown are the results from SDS-PAGE analysis of the distribution of HSPA8-K561me3 and HSPA8 in the supernatant (S) and pellet (P) fractions following incubation with preformed fibrillar α-Syn (1 h) and centrifugation at 40,000 × g for 20 min at 20 °C. Control reactions containing only HSPA8 or α-Syn are shown. Molecular mass markers (in kilodaltons) are shown to the left.
Redeker et al. (27) recently investigated the interaction interface between HSPA8 and α-Syn, whose aggregation is associated with Parkinson disease. It was shown that (i) recombinant HSPA8 can suppress fibrillation of α-Syn in vitro; (ii) HSPA8 binds to preformed fibrils and reduces their toxicity; and (iii) Lys-561, the residue targeted by HSPA-KMT, can be cross-linked to α-Syn in vitro (27).
To test whether HSPA-KMT-mediated methylation influences the interaction between HSPA8 and α-Syn, the ability of methylated or unmethylated HSPA8 to affect α-Syn assembly was assessed. Interestingly, methylated HSPA8 showed reduced ability to delay α-Syn fibrillation kinetics compared with the unmodified counterpart (Fig. 5B). This indicates that the methylated form of HSPA8 has reduced affinity for soluble α-Syn. Although methylation reduced the ability of HSPA8 to delay fibril assembly, the morphology and branching pattern of the resulting fibrils appeared similar by electron microscopy (Fig. 5C).
To investigate whether the affinity for fibrillar α-Syn also was affected, co-sedimentation assays with preformed α-Syn fibrils were carried out. Interestingly, compared with its unmethylated counterpart, methylated HSPA8 co-sedimented less efficiently with fibrillar α-Syn (Fig. 5D), suggesting that methylation also reduces its affinity for α-Syn fibrils. Taken together, our data demonstrate that HSPA-KMT-mediated methylation does not affect basal or DNAJB1-stimulated ATP hydrolysis of Hsp70s but that it does reduce the affinity of HSPA8 for the pathologically relevant client protein α-Syn.
DISCUSSION
Recently, we reported that the uncharacterized human enzyme METTL21A belongs to a family of putative and established protein MTases, the so-called methyltransferase family 16, encompassing 10 enzymes in humans (12). Here, we firmly established METTL21A as a protein MTase, demonstrating its ability to trimethylate, both in vitro and in vivo, a conserved Lys residue found in Hsp70s, and METTL21A was therefore renamed HSPA-KMT. Furthermore, we have shown that HSPA-KMT-mediated methylation modulates the affinity of HSPA8 for α-Syn, whose aggregation is tightly linked to Parkinson disease onset.
Although numerous studies have addressed the basic functions of Hsp70s, none have pin-pointed a specific function for the lysine residue targeted by HSPA-KMT or for the surrounding region. However, when the x-ray structure of the substrate-binding domain of the E. coli Hsp70 protein DnaK was solved, this residue (Lys-556) was highlighted as one of only three conserved surface-exposed residues found in the lid domain (28). This underscores the importance of this lysine residue and suggests that further improvement of Hsp70 function may be achieved by post-translational modification at this site. Although canonical Hsp70s with a potential HSPA-KMT modification site are apparently present in all eukaryotes (29), putative HSPA-KMT orthologs show a scattered distribution within this kingdom (Table 1). This indicates that HSPA-KMT-mediated methylation is not essential for the basal functions of Hsp70s, and accordingly, such methylation did not affect the ATPase activity of HSPA1 and HSPA8. Interestingly, HSPA1, HSPA5, and HSPA8 were all found to be predominantly or completely trimethylated in cells and/or tissues, and these proteins show important differences with respect to cellular localization (13) and regulation (inducible versus constitutive expression) (30). On this basis, we consider it less likely that HSPA-KMT-mediated methylation serves a regulatory purpose but rather believe that lysine trimethylation of the various Hsp70s represents a protein-editing event optimizing the general chaperone function.
We found that methylation of HSPA8 diminished its interaction with the Parkinson disease-associated protein α-Syn. However, it appears counterintuitive that a reduced ability to chaperone α-Syn, a pathologically relevant client protein, should represent the evolutionary force driving the emergence of the HSPA-KMT function. We rather favor the notion that methylation has yielded human Hsp70s that display improved functionality with respect to the full cellular complement of client proteins, possibly through fine-tuning the binding and release cycles of the chaperone, a pivotal and tightly regulated feature of Hsp70s (16).
Many previous studies addressing client binding of mammalian Hsp70s in vitro were performed with recombinant Hsp70s expressed in E. coli, which, according to our results, are unmethylated. Given the observed effect of methylation on the HSPA8/α-Syn interaction, future studies of mammalian Hsp70s should be performed also with methylated proteins.
Human Hsp70s have been reported to be post-translationally modified by, for example, acetylation, phosphorylation, ubiquitination, and methylation (22, 31). However, the site occupancy and specific function of the individual modifications remain largely uncharacterized and poorly understood with a few exceptions. Phosphorylation of Tyr-525 has been shown to correlate with nuclear accumulation of HSPA1 after heat stress (32). The E3 ubiquitin ligase CHIP (C terminus of Hsp70-interacting protein), which also targets Hsp70 clients for degradation, can ubiquitinate several Lys residues in HSPA1 in vitro (33), and overexpression of CHIP in murine fibroblasts reduces steady-state levels of HSPA1 (34). Finally, phosphorylation at the C terminus of HSPA1 reduces its affinity for CHIP and increases its affinity for HOP (Hsp70/Hsp90-organizing protein), skewing the degradation/refolding balance of the chaperone machinery toward refolding (35). Although numerous studies have been performed on Hsp70s, our current study is, to the best of our knowledge, the first demonstration of how a post-translational modification directly modulates the affinity of an Hsp70 for a client protein.
In a very recent study, Cloutier et al. (36) confirmed our finding that METTL21D is a VCP-specific MTase and also showed that its close relative METTL21A methylates human Hsp70s in vitro. In this work, we have substantially expanded these findings by performing a comparative analysis of the activity of HSPA-KMT on a wide range of substrates, and we have shown that the methylation reaction is stimulated by ATP. Also, we showed that the enzyme methylates only 70-kDa proteins in human cell extracts, demonstrating that the MTase is highly specific for Hsp70s. Furthermore, we showed that Hsp70s are methylated in a wide range of mammalian cells and organs and that HSPA-KMT is the responsible enzyme, and importantly, we demonstrated that methylation affects HSPA8 activity.
In agreement with our results, Cho et al. (37) recently reported that HSPA1 (Hsp70) is methylated at Lys-561, but their study focused on the dimethylated form, K561me2, which they found to be present in ectopically expressed HSPA1. However, the authors concluded that SETD1A was the responsible MTase because altering the level of this MTase by siRNAs or overexpression caused corresponding changes in the K561me2 levels. SETD1A is already well established as a histone MTase that introduces gene-activating methyl groups on Lys-4 in histone H3, and no in vitro activity of SETD1A on HSPA1 was demonstrated (37, 38). In contrast, we have shown here that HSPA-KMT can methylate Lys-561 both in vitro and in vivo. Thus, we consider it likely that the observed effect of SETD1A on HSPA1-K561me2 methylation is indirect, e.g. mediated through altered expression of HSPA-KMT or of a lysine-specific demethylase, several of which have been identified in mammals in recent years.
HSPA1 is expressed at very low levels in healthy mammals (mouse) (23), but its expression is often increased during disease, such as Behçet's disease (39) and various cancers (40). Indeed, elevated HSPA1 levels have already been reported as a biomarker for these diseases (41), and assessing the levels of the individual methylated forms (me0, me1, me2, and me3) will yield additional information reflecting chaperone functionality, which is likely to improve the performance of HSPA1 as a biomarker. As Cho et al. recently detected HSPA1-K561me2 in a wide range of cancers and suggested its use as a cancer biomarker, it will be interesting to also specifically investigate the remaining three Lys-561 methylation states in this respect.
Acknowledgments
We thank Markus Fusser and Arne Klungland (Oslo University Hospital) for providing protein extracts from mouse organs and Erna Davydova, Angela Ho, and Jedrzej Malecki (University of Oslo) for critically reading the manuscript and for stimulating discussions throughout the project.
The work performed in the Falnes laboratory was supported by the University of Oslo and by grants from the Norwegian Cancer Society and the Research Council of Norway.

This article contains supplemental Table S1.
- MTase
- methyltransferase
- 7BS
- seven-beta strand
- SAM
- S-adenosylmethionine
- VCP
- valosin-containing protein
- NBD
- nucleotide-binding domain
- α-Syn
- α-synuclein
- CBP
- calmodulin-binding peptide
- SBP
- streptavidin-binding peptide
- TAP
- tandem affinity purification
- Bis-Tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol.
REFERENCES
- 1. Clarke S. G. (2013) Protein methylation at the surface and buried deep: thinking outside the histone box. Trends Biochem. Sci. 38, 243–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Greer E. L., Shi Y. (2012) Histone methylation: a dynamic mark in health, disease and inheritance. Nat. Rev. Genet. 13, 343–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Margueron R., Trojer P., Reinberg D. (2005) The key to development: interpreting the histone code? Curr. Opin. Genet. Dev. 15, 163–176 [DOI] [PubMed] [Google Scholar]
- 4. Taverna S. D., Li H., Ruthenburg A. J., Allis C. D., Patel D. J. (2007) How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat. Struct. Mol. Biol. 14, 1025–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Schubert H. L., Blumenthal R. M., Cheng X. (2003) Many paths to methyltransfer: a chronicle of convergence. Trends Biochem. Sci. 28, 329–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bestor T. H. (2000) The DNA methyltransferases of mammals. Hum. Mol. Genet 9, 2395–2402 [DOI] [PubMed] [Google Scholar]
- 7. Petrossian T. C., Clarke S. G. (2011) Uncovering the human methyltransferasome. Mol. Cell. Proteomics 10, 10.1074/mcp.M110.000976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Katz J. E., Dlakić M., Clarke S. (2003) Automated identification of putative methyltransferases from genomic open reading frames. Mol. Cell. Proteomics 2, 525–540 [DOI] [PubMed] [Google Scholar]
- 9. Del Rizzo P. A., Trievel R. C. (2011) Substrate and product specificities of SET domain methyltransferases. Epigenetics 6, 1059–1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Feng Q., Wang H., Ng H. H., Erdjument-Bromage H., Tempst P., Struhl K., Zhang Y. (2002) Methylation of H3-lysine 79 is mediated by a new family of HMTases without a SET domain. Curr. Biol. 12, 1052–1058 [DOI] [PubMed] [Google Scholar]
- 11. Magnani R., Dirk L. M., Trievel R. C., Houtz R. L. (2010) Calmodulin methyltransferase is an evolutionarily conserved enzyme that trimethylates Lys-115 in calmodulin. Nat. Commun. 1, 43. [DOI] [PubMed] [Google Scholar]
- 12. Kernstock S., Davydova E., Jakobsson M., Moen A., Pettersen S., Mælandsmo G. M., Egge-Jacobsen W., Falnes P. Ø. (2012) Lysine methylation of VCP by a member of a novel human protein methyltransferase family. Nat. Commun. 3, 1038. [DOI] [PubMed] [Google Scholar]
- 13. Hageman J., van Waarde M. A., Zylicz A., Walerych D., Kampinga H. H. (2011) The diverse members of the mammalian HSP70 machine show distinct chaperone-like activities. Biochem. J. 435, 127–142 [DOI] [PubMed] [Google Scholar]
- 14. Kampinga H. H., Hageman J., Vos M. J., Kubota H., Tanguay R. M., Bruford E. A., Cheetham M. E., Chen B., Hightower L. E. (2009) Guidelines for the nomenclature of the human heat shock proteins. Cell Stress Chaperones 14, 105–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hartl F. U., Bracher A., Hayer-Hartl M. (2011) Molecular chaperones in protein folding and proteostasis. Nature 475, 324–332 [DOI] [PubMed] [Google Scholar]
- 16. Kampinga H. H., Craig E. A. (2010) The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat. Rev. Mol. Cell Biol. 11, 579–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. McGinnis S., Madden T. L. (2004) BLAST: at the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Res. 32, W20–W25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Waterhouse A. M., Procter J. B., Martin D. M., Clamp M., Barton G. J. (2009) Jalview Version 2–a multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. McGuffin L. J., Bryson K., Jones D. T. (2000) The PSIPRED protein structure prediction server. Bioinformatics 16, 404–405 [DOI] [PubMed] [Google Scholar]
- 20. Ghee M., Melki R., Michot N., Mallet J. (2005) PA700, the regulatory complex of the 26S proteasome, interferes with α-synuclein assembly. FEBS J. 272, 4023–4033 [DOI] [PubMed] [Google Scholar]
- 21. Pemberton S., Madiona K., Pieri L., Kabani M., Bousset L., Melki R. (2011) Hsc70 protein interaction with soluble and fibrillar α-synuclein. J. Biol. Chem. 286, 34690–34699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hornbeck P. V., Kornhauser J. M., Tkachev S., Zhang B., Skrzypek E., Murray B., Latham V., Sullivan M. (2012) PhosphoSitePlus: a comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. Nucleic Acids Res. 40, D261–D270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Huang L., Mivechi N. F., Moskophidis D. (2001) Insights into regulation and function of the major stress-induced hsp70 molecular chaperone in vivo: analysis of mice with targeted gene disruption of the hsp70.1 or hsp70.3 gene. Mol. Cell. Biol. 21, 8575–8591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pehar M., Lehnus M., Karst A., Puglielli L. (2012) Proteomic assessment shows that many endoplasmic reticulum (ER)-resident proteins are targeted by Nϵ-lysine acetylation in the lumen of the organelle and predicts broad biological impact. J. Biol. Chem. 287, 22436–22440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kahali S., Sarcar B., Fang B., Williams E. S., Koomen J. M., Tofilon P. J., Chinnaiyan P. (2010) Activation of the unfolded protein response contributes toward the antitumor activity of vorinostat. Neoplasia 12, 80–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gao X. C., Zhou C. J., Zhou Z. R., Wu M., Cao C. Y., Hu H. Y. (2012) The C-terminal helices of heat shock protein 70 are essential for J-domain binding and ATPase activation. J. Biol. Chem. 287, 6044–6052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Redeker V., Pemberton S., Bienvenut W., Bousset L., Melki R. (2012) Identification of protein interfaces between α-synuclein, the principal component of Lewy bodies in Parkinson disease, and the molecular chaperones human Hsc70 and the yeast Ssa1p. J. Biol. Chem. 287, 32630–32639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhu X., Zhao X., Burkholder W. F., Gragerov A., Ogata C. M., Gottesman M. E., Hendrickson W. A. (1996) Structural analysis of substrate binding by the molecular chaperone DnaK. Science 272, 1606–1614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gribaldo S., Lumia V., Creti R., Conway de Macario E., Sanangelantoni A., Cammarano P. (1999) Discontinuous occurrence of the hsp70 (dnaK) gene among Archaea and sequence features of HSP70 suggest a novel outlook on phylogenies inferred from this protein. J. Bacteriol. 181, 434–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hageman J., Kampinga H. H. (2009) Computational analysis of the human HSPH/HSPA/DNAJ family and cloning of a human HSPH/HSPA/DNAJ expression library. Cell Stress Chaperones 14, 1–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wang C., Lazarides E., O'Connor C. M., Clarke S. (1982) Methylation of chicken fibroblast heat shock proteins at lysyl and arginyl residues. J. Biol. Chem. 257, 8356–8362 [PubMed] [Google Scholar]
- 32. Knowlton A. A., Grenier M., Kirchhoff S. R., Salfity M. (2000) Phosphorylation at tyrosine-524 influences nuclear accumulation of HSP72 with heat stress. Am. J. Physiol. Heart Circ. Physiol. 278, H2143–H2149 [DOI] [PubMed] [Google Scholar]
- 33. Kundrat L., Regan L. (2010) Identification of residues on Hsp70 and Hsp90 ubiquitinated by the cochaperone CHIP. J. Mol. Biol. 395, 587–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Dai Q., Zhang C., Wu Y., McDonough H., Whaley R. A., Godfrey V., Li H. H., Madamanchi N., Xu W., Neckers L., Cyr D., Patterson C. (2003) CHIP activates HSF1 and confers protection against apoptosis and cellular stress. EMBO J. 22, 5446–5458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Muller P., Ruckova E., Halada P., Coates P. J., Hrstka R., Lane D. P., Vojtesek B. (2013) C-terminal phosphorylation of Hsp70 and Hsp90 regulates alternate binding to co-chaperones CHIP and HOP to determine cellular protein folding/degradation balances. Oncogene 32, 3101–3110 [DOI] [PubMed] [Google Scholar]
- 36. Cloutier P., Lavallée-Adam M., Faubert D., Blanchette M., Coulombe B. (2013) A newly uncovered group of distantly related lysine methyltransferases preferentially interact with molecular chaperones to regulate their activity. PLoS Genet. 9, e1003210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cho H. S., Shimazu T., Toyokawa G., Daigo Y., Maehara Y., Hayami S., Ito A., Masuda K., Ikawa N., Field H. I., Tsuchiya E., Ohnuma S., Ponder B. A., Yoshida M., Nakamura Y., Hamamoto R. (2012) Enhanced HSP70 lysine methylation promotes proliferation of cancer cells through activation of Aurora kinase B. Nat. Commun. 3, 1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dillon S. C., Zhang X., Trievel R. C., Cheng X. (2005) The SET-domain protein superfamily: protein lysine methyltransferases. Genome. Biol. 6, 227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Birtas-Atesoglu E., Inanc N., Yavuz S., Ergun T., Direskeneli H. (2008) Serum levels of free heat shock protein 70 and anti-HSP70 are elevated in Behçet's disease. Clin. Exp. Rheumatol. 26, S96–S98 [PubMed] [Google Scholar]
- 40. Dakappagari N., Neely L., Tangri S., Lundgren K., Hipolito L., Estrellado A., Burrows F., Zhang H. (2010) An investigation into the potential use of serum Hsp70 as a novel tumour biomarker for Hsp90 inhibitors. Biomarkers 15, 31–38 [DOI] [PubMed] [Google Scholar]
- 41. Abe M., Manola J. B., Oh W. K., Parslow D. L., George D. J., Austin C. L., Kantoff P. W. (2004) Plasma levels of heat shock protein 70 in patients with prostate cancer: a potential biomarker for prostate cancer. Clin. Prostate Cancer 3, 49–53 [DOI] [PubMed] [Google Scholar]