Background: Cellular stress leading to cell death induces the formation of lipid droplets.
Results: Nutrient deprivation induces LD biogenesis and mobilization, fueling fatty acid oxidation to sustain cell viability.
Conclusion: β-Oxidation requires biogenesis and mobilization of LD.
Significance: The role of LD in cell survival and β-oxidation might provide new potential targets for antitumor therapy.
Keywords: Cell Death, Fatty Acid Oxidation, Lipid Droplet, Lipogenesis, Lipolysis, Stress, Starvation
Abstract
Cells exposed to stress of different origins synthesize triacylglycerols and generate lipid droplets (LD), but the physiological relevance of this response is uncertain. Using complete nutrient deprivation of cells in culture as a simple model of stress, we have addressed whether LD biogenesis has a protective role in cells committed to die. Complete nutrient deprivation induced the biogenesis of LD in human LN18 glioblastoma and HeLa cells and also in CHO and rat primary astrocytes. In all cell types, death was associated with LD depletion and was accelerated by blocking LD biogenesis after pharmacological inhibition of Group IVA phospholipase A2 (cPLA2α) or down-regulation of ceramide kinase. Nutrient deprivation also induced β-oxidation of fatty acids that was sensitive to cPLA2α inhibition, and cell survival in these conditions became strictly dependent on fatty acid catabolism. These results show that, during nutrient deprivation, cell viability is sustained by β-oxidation of fatty acids that requires biogenesis and mobilization of LD.
Introduction
Lipid droplets (LD)3 are organelles present virtually in all types of cells (1), made of triacylglycerols (TAG) and cholesteryl esters surrounded by a monolayer of amphipathic lipids with which some proteins interact. Interest in the study of LD has increased during recent years, which have witnessed the shift of a traditional view of LD as inert fat depots to their recognition today as dynamic stores of metabolic fuel and membrane-building blocks, lipid-buffering structures, platforms for protein sorting, or sources of inflammatory mediators and gene transcription regulators (2–6). LD, therefore, appear central in the pathophysiology of metabolic (7, 8) and infectious diseases (9, 10).
Cells in culture generate LD in two different situations from a physiological perspective: when maintained in medium containing free fatty acids and/or lipoproteins or when exposed to a stressor stimulus. LD exhibit different proteomic and lipidomic profiles, depending on the stimulus, and the study of their functional heterogeneity is a challenge (10). Many types of stress leading to cell death induce the formation of LD (11, 12), and in fact LD accumulation can be considered a hallmark of apoptosis (13). LD give rise to NMR signals that increase in response to cell cycle arrest (14), acidic pH (15), or apoptosis (16–18), and these signals are the basis for imaging techniques in cancer diagnosis and therapy (19–21). It is not clear, however, what is the origin and physiological role of LD generated in cells committed to die. Studies on LD biogenesis in stress are often carried out in media containing free fatty acids and/or lipoproteins, making it difficult to assess the origin of their lipid content. In a previous report, we addressed LD biogenesis in response to different types of stress and showed that, in the absence of external lipids, stressors increased the availability of intracellular fatty acids. This effect was mediated by Group VIA phospholipase A2 (iPLA2) and was independent of de novo fatty acid synthesis (22). Further, TAG synthesis correlated closely with LD occurrence (22), showing that phospholipid-linked, preexisting fatty acids were reused for synthesis of TAG and LD biogenesis.
Synthesis of TAG precedes and is required for LD biogenesis, as evidenced by the blunting effect of the acyl-CoA synthetase inhibitor triacsin A (17, 22, 23). Because fatty acid activation by acyl-CoA synthetase is an energy-consuming process, conceivably LD biogenesis in stress may embody an attempt to overcome a metabolic jeopardy. Du et al. (24) reported that neuron survival to starvation is related to the ability to make LD, and Lei et al. (25) have shown that LD attenuate ischemia-induced injury in heart. Similar to ours, buildup of LD in these experiments took place in the absence of exogenous lipids. We therefore hypothesized that recycling fatty acyl moieties of phospholipids into TAG for buildup of LD could be a prosurvival response to stress aimed at supplying catabolic substrates. Should this hypothesis hold, interfering with LD biogenesis might be a potential antitumor strategy.
Here we show that survival of different cell types (CHO, LN18 human glioblastoma, HeLa, or rat astrocytes) under complete nutrient deprivation depends on LD, which confer the capacity to degrade fatty acids through β-oxidation.
EXPERIMENTAL PROCEDURES
Materials
[9,10-3H]Palmitic acid (60 Ci/mmol) was purchased from American Radiolabeled Chemicals, Inc. LipofectamineTM RNAiMAX transfection reagent was from Invitrogen. cPLA2α inhibitor pyrrolidine-2 (py-2, catalogue number 525143) was from Calbiochem; carnitine palmitoyltransferase-1 (CPT1) inhibitor etomoxir (EX), autophagy inhibitor 3-methyladenine (3-MA), sodium oleate, primuline, Nile red, Oil Red O, and propidium iodide (PI) were from Sigma. Rabbit anti-cPLA2α and anti-phospho-Ser505 cPLA2α antibodies were from Cell Signaling; rabbit anti-perilipin 2, anti-perilipin 3, anti-CERK, anti-LC3B, and anti-CPT1 antibodies were from Abcam, and mouse anti-β-actin was from Sigma. 4,4-Difluoro-1,3,5,7,8-pentamethyl-4-bora- 3a,4a-diaza-s-indacene (BODIPY® 493/503) and LysoTracker® Red DND-99 were from Molecular Probes.
Cells
Human glioblastoma LN18 and HeLa cells were cultured in Dulbecco's modified Eagle's medium (DMEM; Sigma), containing 10% fetal bovine serum (FBS; Sigma), 100 units/ml penicillin (Invitrogen), and 100 μg/ml streptomycin (Invitrogen). CHO-K1 cells were cultured in Ham's F-12 medium (Sigma), containing 7.5% FBS, 100 units/ml penicillin, and 100 μg/ml streptomycin. Cell passages were made once a week by trypsinization (Sigma). Primary astrocytes from rat cerebral cortex were cultured as described (26). For experiments, cells were seeded on 6- or 24-well plates at a density of 105 cells/ml and maintained for 24 h in culture medium in the absence of FBS in order to deplete cells of LD before the treatments, as described previously (22, 23). This plating density ensured that cells were far from confluence (60–70%) at the time of treatments, avoiding confluence-induced generation of LD (14, 15, 27). Stress was induced by complete nutrient deprivation, which was achieved washing twice with phosphate-buffered saline (PBS; Sigma) and substituting medium by Krebs-Henseleit (KH) buffer without glucose (116 mm NaCl, 4.7 mm KCl, 25 mm NaHCO3, 1.2 mm MgSO4, 1.2 mm KH2PO4, and 1.3 mm CaCl2, pH 7.4, equilibrated with O2/CO2 (95:5)).
Microscopy
For fluorescence microscopy, cells cultured on glass coverslips were washed with PBS, fixed with 3% paraformaldehyde (Sigma) for 10 min, and washed twice with PBS. The occurrence of LD was routinely checked by microscopic examination of cells stained with Nile red, BODIPY® 493/503, or Oil red-O. For Nile red staining, cells were overlaid with 0.5 ml of a working solution of PBS containing 2 μl/ml Nile red stock solution in acetone (0.2 mg/ml). For BODIPY® 493/503 staining, cells were overlaid with 0.5 ml of a working solution of PBS containing 2.5 μl/ml of a BODIPY® 493/503 stock solution in 98% ethanol (1 mg/ml). Oil Red O staining of LD followed the recommendations of Koopman et al. (28). In most instances, nuclei were counterstained with DAPI. To label acidic compartments, unfixed cells were overlaid with prewarmed culture medium or buffer, containing 75 nm LysoTracker® Red DND-99. Cells were incubated for 30 min and then washed with PBS and fixed with 3% paraformaldehyde. Samples were kept in the dark until photographed in a Leica Qwin 500 microscope with a Leica DFC500 camera, using Leica DCviewer 3.2.0.0 software. Electron microscopy was carried out as described (23).
Flow Cytometry
Indirect quantification of Nile red-stained LD by flow cytometry was performed as described (22, 23, 29) with the only difference that cells were not fixed. Briefly, harvested cells were transferred to tubes, together with their overlaying medium or buffer, to prevent the loss of floating cells. After two washes with PBS, cells were resuspended in 0.5 ml of the Nile red working solution (0.4 μg/ml final concentration). Samples were kept 45 min in the dark to attain equilibrium with the dye. Analysis was carried out with a Cytomics FC 500 (Beckman Coulter) equipped with an argon laser (488 nm), in the FL1 channel (505–545 nm), with the photomultiplier set at 600 V and a gain value of 1. After gating out cellular debris, 10,000 events were acquired. For each assay, we obtained a side scatter versus forward scatter plot (SS/FS plot), a bidimensional representation of each event in terms of structural complexity (SS) and size (FS). Each sample appeared split into two populations differing in FS value. Staining with PI showed that the population with a lower FS value (smaller cells) consisted of dead cells. The shift of dead cells to lower FS values allowed us to quantify separately LD content in viable and dead cells in the same sample, although the Nile red emission spectrum does not allow co-staining with PI. Alternatively, cells were stained with PI and BODIPY® 493/503. To do this, cells were processed as described above and resuspended in 0.5 ml of the BODIPY® 493/503 working solution containing 2.5 μl of the PI stock solution (1 mg/ml in water). The working solution also contained 2.5 μl of RNase (Sigma) stock solution (1 mg/ml in water), to achieve specific DNA staining. Samples were kept in the dark at 4 °C for 20 min before carrying out the analysis in the FL1 and FL3 channels.
Lactate Dehydrogenase (LDH) Assay
To confirm that PI staining monitored cell death accurately, we measured LDH activity released into the medium. Briefly, the medium was collected carefully, centrifuged for 10 min at 13,000 × g, and incubated in a reaction mix containing NADH, pyruvate, and sodium azide. NADH oxidation was followed after the decrease of absorbance at 340 nm, and its rate was referred to total LDH activity, which was estimated after hypotonic lysis of untreated cells.
β-Oxidation
Cells seeded in 24-well plates were maintained for 24 h in medium without FBS and then labeled overnight with about 1 μCi/ml [3H]palmitic acid (10–30 nm, final concentration) in culture medium containing 0.5% delipidated bovine serum albumin (BSA; Sigma). After labeling, cells were washed once with BSA-containing culture medium and twice with PBS and left for 30 min in culture medium. The β-oxidation assay was based on that described by Djouadi et al. (30) and monitored the generation of [3H]water. Treatments were run with quadruplicate determinations and started by substitution of culture medium by 0.5 ml of fresh culture medium or KH buffer without glucose. After treatments, which proceeded for 1–8 h, 0.4 ml were transferred to a 5-ml polypropylene tube containing 1 ml of a 1:2 slurry of Dowex 1 × 2 (chloride form) anionic exchange resin (Sigma) in water. Tubes were vortexed and centrifuged for 10 min at 1,000 rpm. The resin trapped radioactive metabolic intermediates other than [3H]water, and 0.5 ml of the supernatants containing [3H]water were counted for radioactivity. All experiments included a blank control, where [3H]water was determined immediately after the addition of KH buffer without glucose.
Quantification of 3H-Labeled Lipids
Cells seeded in 24-well plates were labeled overnight with 1 μCi/ml [3H]palmitic acid, washed, and treated as explained above. After treatment, lipids were extracted as described (31). To separate the major lipid species, 0.2-ml aliquots of the chloroform phases were evaporated under vacuum, resuspended in 15 μl of chloroform/methanol (3:1, v/v), and spotted onto Silica Gel 60 high performance thin layer chromatography glass plates with concentrating zone (Merck), which were developed in hexane/diethyl ether/acetic acid (70:30:1, v/v/v) and stained with primuline spray (5 mg of primuline in 100 ml of acetone/water (80:20, v/v)). Identification of phospholipids, TAG, and cholesteryl esters was achieved by co-migration with 1-palmitoyl-2-oleoyl-phosphatdylcholine, tripalmitin, and cholesteryl palmitate (Sigma) standards, respectively. To quantify radioactive lipids, the silica gel from regions corresponding to migration of the standards was scraped into scintillation vials.
Immunoblots
Cells were lysed with 62.5 mm Tris-HCl buffer, pH 6.8, containing 2% SDS, 10% glycerol, 50 mm dithiothreitol, and 0.01% bromphenol blue, and around 20 μg of protein were separated by standard SDS-PAGE and transferred to nitrocellulose membranes. Primary (1:1,000) and secondary (1:5,000) antibodies were diluted in 25 mm Tris-HCl buffer, pH 7.4, containing 140 mm NaCl, 0.5% defatted dry milk, bovine serum albumin according to the manufacturer's advice, and 0.1% Tween 20. Membranes were developed using ECL detection reagents from Amersham Biosciences.
siRNA Transfection
Expression of cPLA2α was silenced with a predesigned siRNA (Gene Link) directed against human cPLA2α. CHO cells were transfected at 60% confluence by adding to each 35-mm culture well 1 ml of Opti-MEM (Invitrogen) containing 1.5 μl of the stock siRNA solution (20 μm) and 5 μl of LipofectamineTM RNAiMAX transfection reagent (1 mg/ml). After 5 h, 1 ml of Ham's F-12 medium containing 7.5% FBS was added, and the cells were incubated for 48 h and then changed to serum-free medium for 24 h prior to treatment. Expression of CERK and perilipins 2 and 3 was silenced in human glioblastoma LN18 cells, using predesigned siRNAs from GeneLink (CERK) and Invitrogen (perilipins 2 and 3). Transfection was performed using siPORT NeoFX reagent (Invitrogen).
Statistical Analysis
Data analysis was carried out with Prism software (GraphPad). Values are expressed as mean ± S.E. Differences between mean values were compared using the unpaired Student's one-tailed t test and were considered significant if p was <0.05.
RESULTS
LD Sustain Cell Viability
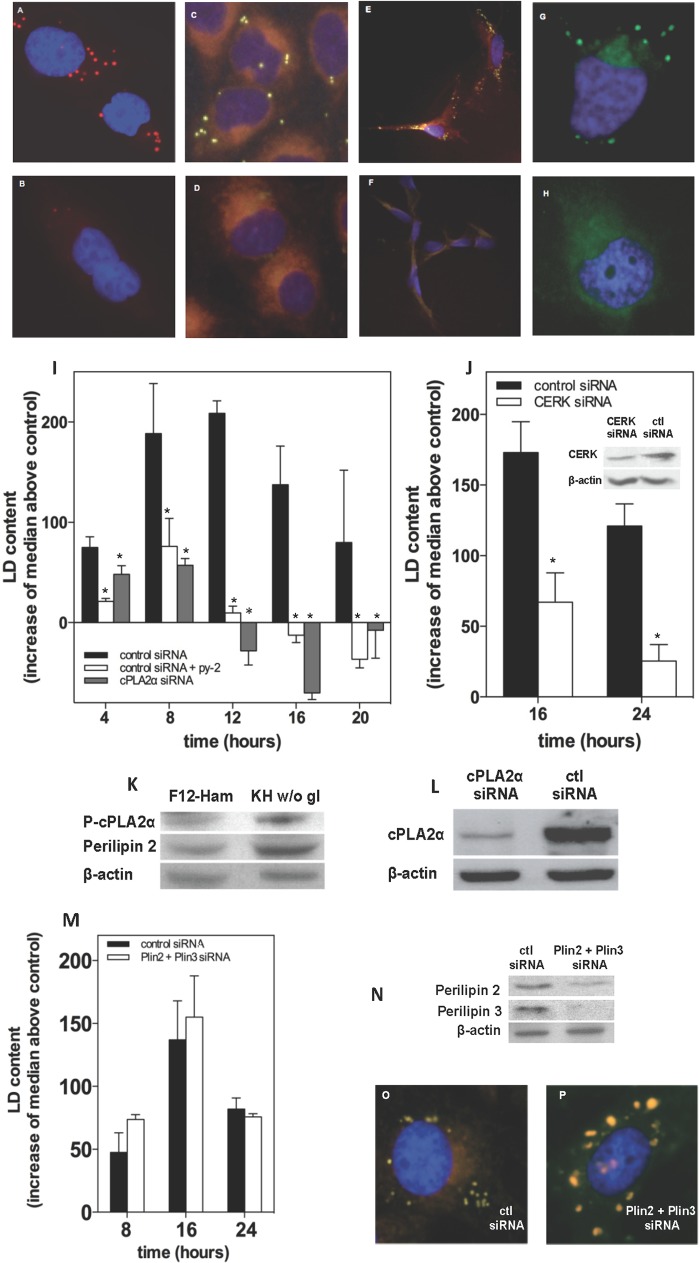
To test the hypothesis that LD generated in stress have a prosurvival role, we used complete nutrient deprivation as a stressor stimulus, because among the different types of stress we previously analyzed, this was the strongest in terms of TAG synthesis and LD occurrence (22). Fig. 1 shows that LN18 (A and B), CHO (C and D), rat astrocytes (E and F), and HeLa cells (G and H) in KH buffer without glucose generated LD. In all cell types, LD occurrence was sensitive to inhibition of cPLA2α with the specific inhibitor (32) py-2. CHO cells under complete nutrient deprivation generated LD that reached a maximum at 8–12 h of treatment and decreased thereafter (Fig. 1I). Nutrient-deprived LN18 and HeLa cells and rat astrocytes also generated LD with a bell-shaped time course (not shown). In agreement with previous results from our laboratory (22, 23, 29) and others (24, 33, 34), LD biogenesis took place after activation of cPLA2α by phosphorylation (Fig. 1K) and was abrogated by py-2 (Fig. 1I) or by down-regulation of the enzyme (Fig. 1, I and L). Similar to cPLA2α, down-regulation of CERK, which participates in cPLA2α activation during the biogenesis of LD (29), also decreased LD occurrence in LN18 cells under nutrient deprivation (Fig. 1J). We also tried to reduce LD levels by down-regulating the LD-associated proteins perilipins 2 and 3. In contrast to CERK or cPLA2α, combined knockdown of perilipins 2 and 3 (Fig. 1N) did not affect the overall LD occurrence induced by nutrient deprivation, as monitored by flow cytometry (Fig. 1M), but LD were clearly larger (Fig. 1O), reflecting a fusion process (35).
FIGURE 1.
Cells maintained in Krebs buffer without glucose form LD in a process that requires cPLA2α. A–H, human LN18 glioblastoma (A and B), hamster CHO-K1 (C and D), rat astrocytes (E and F), and human HeLa cells (G and F) were kept in KH buffer without glucose for 16 h (A, B, E, and F) or 8 h (C, D, G, and H) and in the absence (top panels) or presence (bottom panels) of 1 μm py-2. LD were stained with Oil red O (A and B), Nile red (C–F), or BODIPY 493/503 (G and H). In all cases, treatment with KH without glucose triggered LD biogenesis, which was blocked by py-2. I, CHO cells were transfected with siRNA designed against cPLA2α (gray bars) or with a control siRNA (solid and open bars) and then kept in culture medium without serum for 24 h to deplete LD. Afterward, cells were switched to KH buffer without glucose in the absence (solid and gray bars) or presence of 1 μm py-2 (open bars), harvested at the times indicated, and stained with Nile red. The occurrence of LD was assessed by flow cytometry. Intensities in FL1 were quantified as the median values of the event distributions of each condition. Here, LD content is expressed as the increase of the median values above control (cells kept in FBS-free culture medium), and the results are means ± S.E. (error bars) of 3–5 independent experiments carried out with duplicate determinations. *, significantly different from KH without glucose. Cells maintained in KH buffer without glucose had increased expression of perilipin-2 and phosphorylation of cPLA2α at Ser505 (K). Inhibition of cPLA2α with the specific inhibitor py-2 blocked LD biogenesis (A–I), as did its down-regulation with siRNA (I and L). J, LN18 glioblastoma cells were transfected with siRNA designed against CERK (open bars) or with a control siRNA (solid bars) and switched to KH buffer without glucose after 24 h of FBS depletion. Flow cytometry quantification of LD content showed that down-regulation of CERK reduced LD occurrence. *, significantly different from control siRNA. M, LN18 cells were transfected with siRNA designed against perilipin 2 and perilipin 3 or with a control siRNA. Simultaneous knockdown of both proteins (N) had no effect on LD content as quantified by flow cytometry (M), although LD were bigger than those in control cells (O and P). The results are means ± S.E. of 3–5 independent experiments carried out with duplicate determinations.
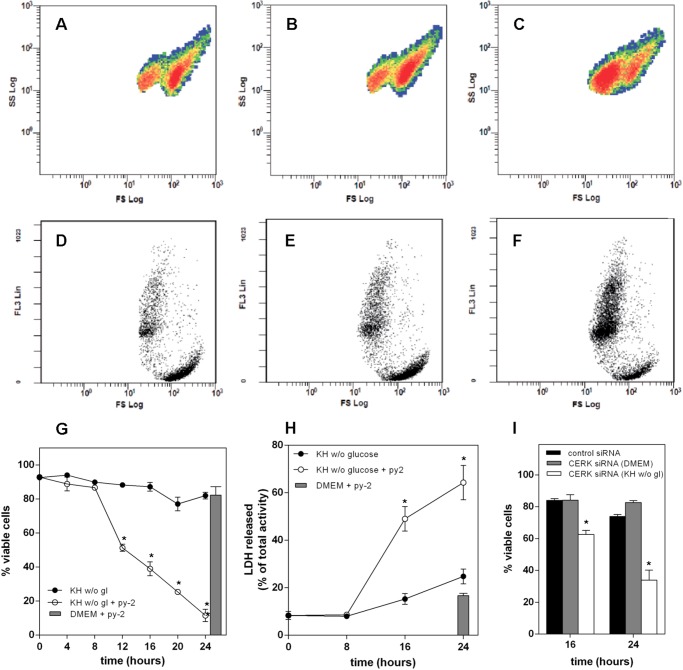
To assess cell death during nutrient deprivation, we used flow cytometry to quantify cell shrinkage, as evidenced by the left shift in SS/FS plots shown in Fig. 2 (A–C). Smaller cells in the low FS values were permeable to PI, as shown by the higher FL3 fluorescence in the low FS population (Fig. 2, D–F) and therefore considered dead, whereas those in the high FS values had a lower FL3 fluorescence and were scored viable. Using this criterion, LN18 cells maintained viability during 24 h of complete nutrient deprivation, as shown in G. However, the same treatment in the presence of 1 μm py-2 induced cell death after a lag of 8 h, leaving only about 10% viable cells after 24 h. This contrasts with the complete lack of toxicity of the cPLA2α inhibitor in cells kept in culture medium. Cell death as monitored by flow cytometry agreed closely with LDH release to the medium, shown in Fig. 2H. Death of starved CHO and HeLa cells proceeded at a faster pace, whereas astrocytes maintained viability longer than LN18 cells. Again, py-2 consistently accelerated death in all cell types under starvation, but it did not affect viability in culture medium (not shown). Similar to pharmacological inhibition of cPLA2α, reduced expression of CERK accelerated death of nutrient-deprived LN18 cells but did not affect the viability of cells in culture medium (Fig. 2I). In contrast, simultaneous knockdown of perilipins 2 and 3 did not alter the death rate of nutrient-deprived cells (not shown).
FIGURE 2.
py-2 induces death of nutrient-deprived cells. A–F, LN18 cells were kept for 16 h in DMEM (A and D) or KH without glucose in the absence (B and E) or presence of 1 μm py-2 (C and F), harvested, stained with PI, and analyzed by flow cytometry. A–C show the distribution of 10,000 events for each condition in terms of shape (SS) and size (FS). In all cases, events were distributed in two distinct populations, characterized by a different FS value. Cells kept in DMEM (A) and KH without glucose (B) appeared mostly in the population with a higher FS value, whereas those treated with KH without glucose + 1 μm py-2 (C) were in the low FS population. D–F show the distribution of the same samples in terms of viability, measured according to the ability of cells to exclude PI, which is expressed as FL3, and size (FS). In all cases, the high FS population excluded PI, as evidenced by the lower FL3 fluorescence. py-2 promoted an increase of the PI-stained population. G, LN18 cells were maintained in KH buffer without glucose in the absence (solid symbols) or presence (open symbols) of 1 μm py-2. At the indicated times, cells were harvested, and percentage viability was determined by flow cytometry. Gray bar, viability of cells in DMEM with 1 μm py-2 during 24 h. Inclusion of 1 μm py-2 accelerated the death of nutrient-deprived cells but was not toxic for cells in culture medium. Results are means ± S.E. (error bars) of six independent experiments carried out with duplicate determinations. H, similar results were obtained by determination of LDH activity released to the medium. Results are means ± S.E. of two independent experiments carried out with quadruplicate determinations. *, significantly different from KH buffer without glucose. I, down-regulation of CERK accelerated death of cells kept in KH buffer without glucose (open bars) as compared with control siRNA (solid bars) but had no effect on cells kept in DMEM (gray bars). Results are means ± S.E. of two independent experiments carried out with triplicate determinations. *, significantly different from control siRNA.
Cell Death Is Associated with LD Depletion
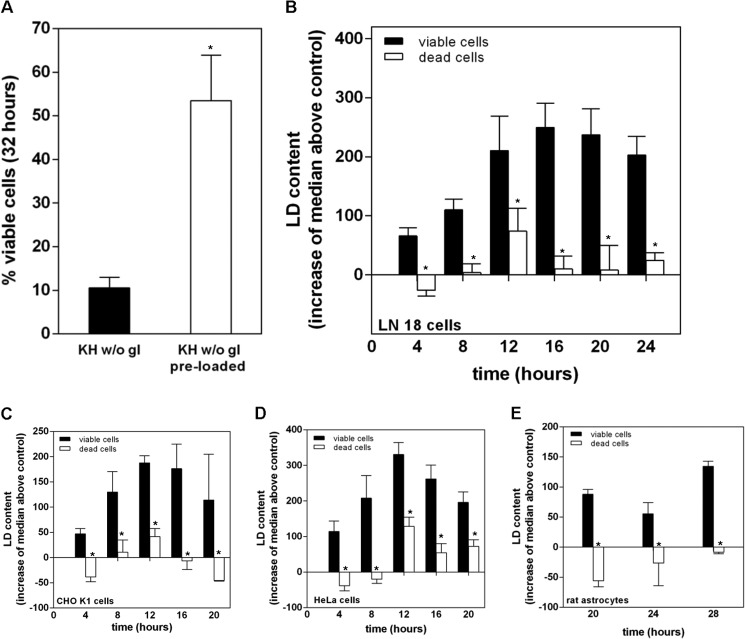
The bell-shaped profile of LD occurrence over time suggests an ongoing process of biogenesis and mobilization that seems relevant for cell survival, given the toxic effects of py-2 and CERK knockdown, and is in keeping with a possible catabolic use of LD generated in response to starvation. As a first approach to test this idea, we loaded LN18 cells with LD using a 16-h treatment with exogenous lipid in DMEM (10% FBS and 100 μm sodium oleate). As expected, LD-preloaded cells survived nutrient deprivation longer than cells that were depleted of LD at the beginning of the treatment, which died rapidly between 24 and 32 h of starvation (Fig. 3A). These results agree with those of Katchadourian and Maysinger (36), who showed that oleic acid preconditioning increases survival of PC12 cells to starvation. If cell viability during starvation is sustained by the catabolic use of LD, conceivably cell death should take place after LD depletion, and this is what we found when analyzing LD content in viable and dead cells. Viable and dead cells can be separated after SS/FS flow cytometry plots, as shown in Fig. 2, and therefore Nile red-stained cells can be gated in two different populations from the same sample to quantify LD occurrence. Fig. 3, B–E, represents the quantification of LD in viable and dead LN18 (B), CHO (C), HeLa (D), and astrocyte (E) cells over time, showing that, regardless of cell type or the time of treatment, dead cells were consistently depleted of LD as compared with viable cells.
FIGURE 3.
Cells surviving nutrient deprivation contain LD, whereas dead cells are devoid of them. A, LN18 cells were maintained overnight in FBS-free DMEM to ensure depletion of LD (filled bar) or DMEM supplemented with 10% FBS and 100 μm sodium oleate (open bar) prior to their treatment with KH without glucose for 32 h. Cells devoid of LD before starvation died faster than those that had been preloaded with LD. B–E, LN18 (B), CHO (C), HeLa (D), or astrocytes (E) were maintained in KH without glucose for the times indicated and then stained with Nile red, and LD content in viable (solid bars) and dead (open bars) cell populations was assessed by flow cytometry. Viable and dead cell populations in Nile red-stained samples were gated in SS/FS plots, and their LD content was determined after the median values of the FL1 event distributions. Results are expressed as FL1 median values above that of control samples devoid of LD, maintained overnight in culture medium without FBS, and are means ± S.E. (error bars) of five (B) or three (C–E) independent experiments carried out with duplicate determinations. *, significantly different from viable cells.
Nutrient Deprivation Induces β-Oxidation of Fatty Acids That Requires LD
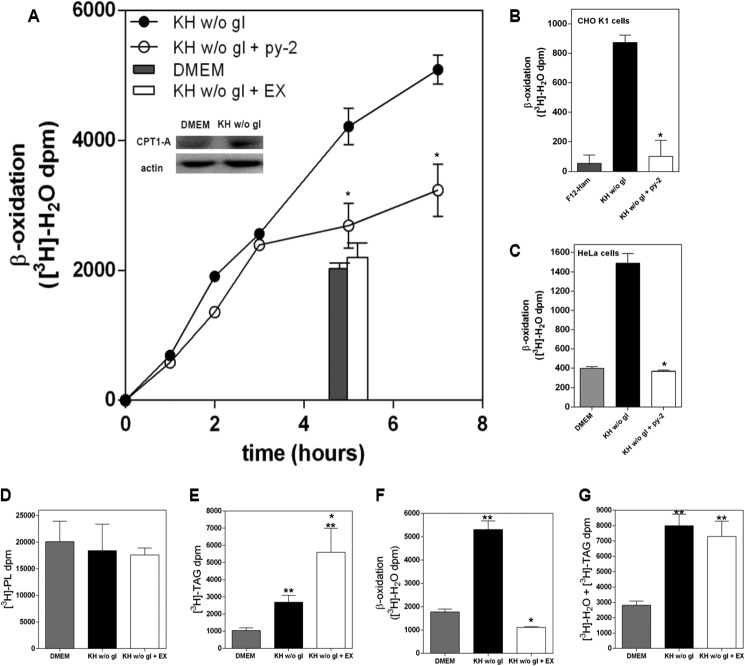
To address whether LD generated in complete nutrient deprivation provide catabolic substrates, we labeled endogenous lipids by incubating LN18 cells overnight in culture medium in the absence of serum and with [3H]palmitic acid at concentrations (10–30 nm) low enough to avoid LD biogenesis (22, 23). After washing out the tracer, we switched the cells to nutrient-deprived conditions and monitored the generation of tritiated water as an index of fatty acid oxidation (30). As shown in Fig. 4A, [3H]palmitate-prelabeled LN18 cells generated [3H]water in a manner that was sensitive to a 30 μm concentration of the CPT1 inhibitor EX, indicating that it was an index of mitochondrial β-oxidation of fatty acids. [3H]Water generation by cells maintained in culture medium was similar to that in cells where β-oxidation was inhibited with EX, showing that nutrient deprivation did induce the oxidation of fatty acids. In agreement with this, fatty acid oxidation was associated with increased expression of CPT1A (Fig. 4A, inset). As with LN18 cells, nutrient deprivation also stimulated fatty acid oxidation in CHO and HeLa cells (Fig. 4, B and C). More importantly, inhibition of LD biogenesis with py-2 in LN18, CHO, and HeLa cells decreased β-oxidation down to a level close to that defined by EX (Fig. 4, A–C) and within a time frame that excluded the possibility that the effect was secondary to cell death. Down-regulation of perilipins 2 and 3, however, did not affect significantly β-oxidation or TAG synthesis (not shown). Analysis of the major lipid species revealed that [3H]palmitate labeling went mainly to phospholipids (Fig. 4D), and nutrient deprivation induced a small but non-significant decrease. Nutrient deprivation induced TAG accumulation that was potentiated after inhibition of β-oxidation with EX (Fig. 4E). Accumulation of cholesteryl esters, the other main lipid contained in LD, was 10-fold lower than that of TAG, and nutrient deprivation induced a 2-fold increase (not shown), probably reflecting increased availability of [3H]palmitate, and it was not affected by EX. Fig. 4G shows the aggregated values of [3H]TAG + [3H]water, the latter shown in Fig. 4F, and illustrates that nutrient deprivation stimulated synthesis of TAG that led to β-oxidation.
FIGURE 4.
Nutrient deprivation induces β-oxidation that requires LD biogenesis. A, LN18 cells were prelabeled overnight with [3H]palmitate prior to their treatment with DMEM (gray bar) or KH without glucose in the absence (solid symbols) or presence of 1 μm py-2 (open symbols) or 30 μm EX (open bar). β-Oxidation was monitored after the generation of [3H]water. The inset shows increased expression of CPT1 after 16 h of starvation. B and C, [3H]palmitate-prelabeled CHO (B) or HeLa (C) cells were treated for 4 h with culture medium (gray bar) or KH without glucose in the absence (solid bar) or presence of 1 μm py-2 (open bar). β-Oxidation was defined as EX-sensitive production of [3H]water. Results are means ± S.E. (error bars) of an experiment carried out with determinations in quadruplicate and are representative of four (A) or three (B and C) independent experiments with similar outcome. *, significantly different from KH without glucose. D–G, [3H]palmitate-prelabeled LN18 cells were maintained in culture medium (gray bars) or KH without glucose in the absence (solid bars) or presence of 30 μm EX (open bars). After 8 h, lipids were extracted and separated by high performance thin layer chromatography, and radioactivity in phospholipids (PL) and TAG was quantified. D, nutrient deprivation did not induce a significant change in phospholipid content. E, nutrient deprivation induced TAG accumulation that was potentiated after inhibition of β-oxidation with EX. F, nutrient deprivation stimulated the generation of [3H]water as an index of β-oxidation, which was inhibited by EX. G, aggregated values of [3H]TAG + [3H]water, illustrating that nutrient deprivation induced a process of lipogenesis that led to the oxidation of fatty acids. Results are means ± S.E. of one experiment with quadruplicate determinations that was repeated once with a similar outcome. *, significantly different from KH without glucose; **, significantly different from DMEM.
β-Oxidation of Fatty Acids Sustains Cell Viability
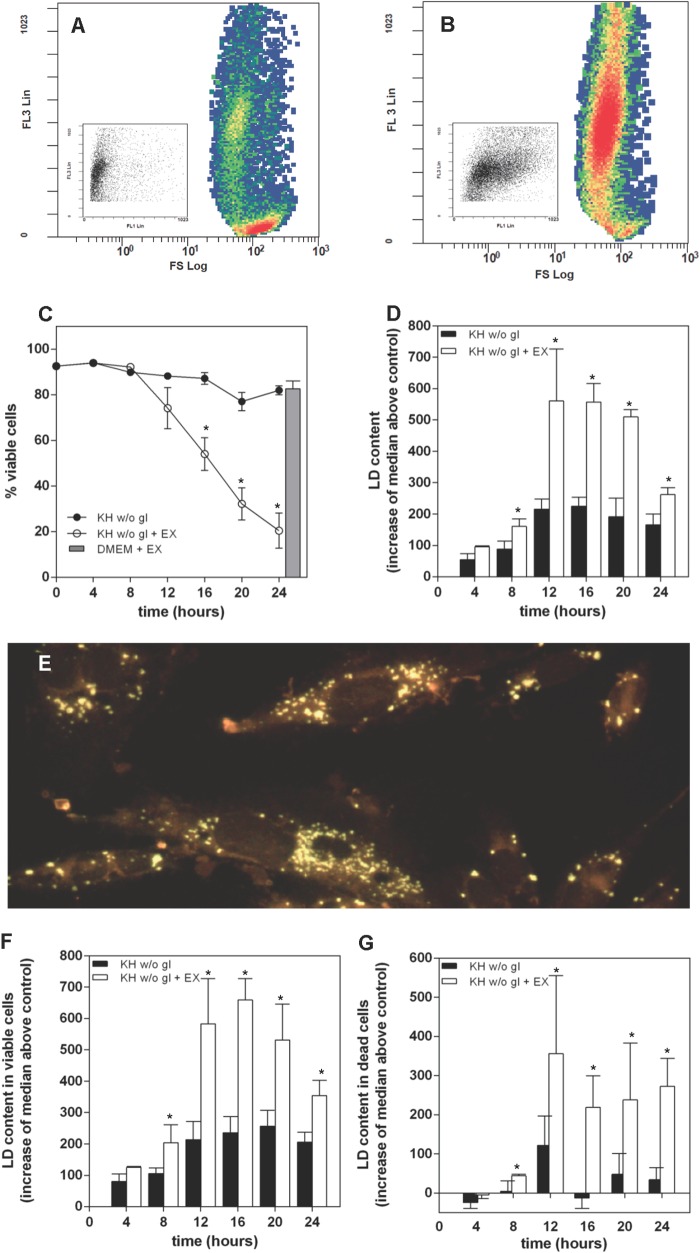
The preceding results are consistent with a necessary role of TAG synthesis and LD biogenesis in fatty acid oxidation during complete nutrient deprivation. Alternatively, because starvation leads ultimately to cell death, LD might accumulate as a consequence of increased fatty acid availability secondary to impaired mitochondrial function, as has been shown in cells undergoing etoposide-induced apoptosis (12). Although this possibility seems unlikely in our case, we analyzed the effect of inhibition of β-oxidation with EX on cell viability and LD content. Fig. 5 (A and B) shows FL3/FS flow cytometry plots of PI- and BODIPY 493/503-stained HeLa cells that had been maintained for 10 h under nutrient-deprived conditions in the absence (A) or presence of 30 μm EX (B). Increased FL3 fluorescence reflects cell permeability to PI and shows that 30 μm EX potentiated starvation-induced cell death. LD content in dead cells was clearly higher in EX-treated samples, as evidenced by the stronger BODIPY FL1 fluorescence, shown as a right shift of FL3/FL1 plots in the panel insets. EX was not toxic to LN18 (Fig. 5C) or CHO cells (not shown) maintained in culture medium (Fig. 5C), which agrees with our data showing no induction of β-oxidation in these conditions (Fig. 4) and is also consistent with the lack of toxicity of py-2 in culture medium (Fig. 2). In contrast, inhibition of β-oxidation accelerated death of nutrient-deprived LN18 cells with a time course similar to that induced by py-2, and the same was true for CHO and HeLa cells (not shown). Moreover, EX increased the overall LD occurrence in LN18 cells induced by nutrient deprivation (Fig. 5, D and E), and this was apparent both in viable and death LN18 cells (Fig. 5, F and G) and also in viable and death CHO cells (not shown). Taken together, the results presented so far show that cell viability during nutrient deprivation is sustained by β-oxidation of fatty acids that requires biogenesis and mobilization of LD.
FIGURE 5.
Under nutrient deprivation, inhibition of β-oxidation accelerates cell death and leads to the accumulation of LD. A and B, HeLa cells were kept for 10 h in KH without glucose and in the absence (A) or presence of 30 μm EX (B). After treatment, they were harvested and stained with PI and BODIPY 493/503 to measure viability and LD content. A and B show the event distribution for each treatment in terms of PI permeability (FL3) and size (FS). Inhibition of β-oxidation (B) induced death of nutrient-deprived cells. Insets show the event distribution of the dead cell population for each treatment in terms of FL3 and LD occurrence (FL1). The increased FL1 signal induced by EX is apparent. C, inhibition of β-oxidation with 30 μm EX accelerated the death of LN18 cells maintained in KH without glucose but had no effect on cells maintained in culture medium (DMEM), as denoted by a gray bar. Treatment with EX led to an overall accumulation of LD (D) that was very apparent in Nile red-stained cells after a 16-h treatment (E). Analysis of viable (F) and dead cells (G) shows increased LD occurrence in EX-treated samples. Results are means ± S.E. (error bars) of six independent experiments carried out with duplicate determinations. *, significantly different from KH without glucose.
Autophagy Does Not Seem to Be Involved in LD Mobilization for β-Oxidation
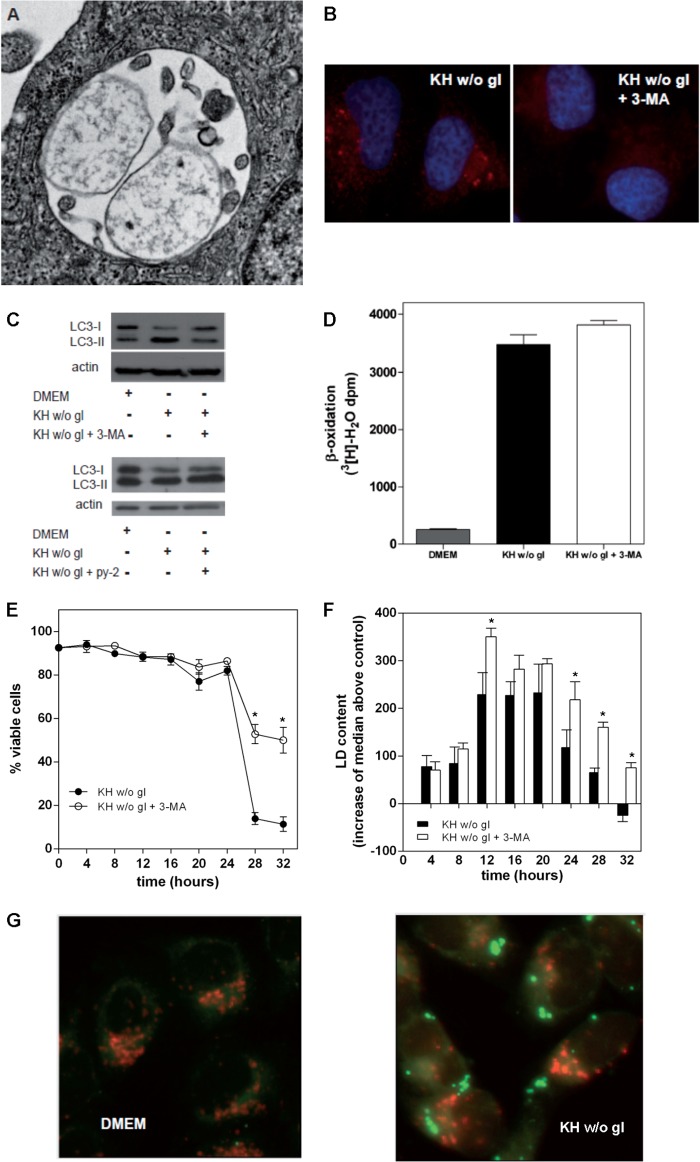
Mobilization of LD by autophagy to fuel fatty acid oxidation has been shown to take place in hepatocytes and liver (37) and also in hypothalamic neurons (38). Autophagy is also involved in the mobilization of LD-stored cholesteryl esters in macrophages for efflux of cholesterol (39), and we addressed whether this mechanism was involved in LD mobilization in our model. As shown in Fig. 6, nutrient deprivation of LN18 cells induced the appearance of autophagosomes (Fig. 6A), microtubule-associated protein light chain 3 (LC3) puncta (Fig. 6B), and the conversion of LC3-I to LC3-II (Fig. 6C), indicative of autophagy. The autophagy inhibitor 3-MA blocked the appearance of LC3 puncta and also conversion of LC3-I to LC3-II (Fig. 6, B and C). In contrast, inhibition of LD biogenesis with py-2 did not affect LC3 conversion (Fig. 6C). Regarding β-oxidation of endogenous fatty acids, 3-MA did not affect the EX-sensitive generation of [3H]water induced by nutrient deprivation (Fig. 6D). Unlike the deleterious effects of pharmacological inhibition of LD biogenesis or β-oxidation, 3-MA did not accelerate LN18 cell death; rather, it increased viability at long treatment times (Fig. 6E), and similar results were found with CHO cells (not shown). These results suggest that the autophagic machinery is involved in cell death, but it does not participate in the mobilization of LD for β-oxidation that is required to sustain cell viability in starvation. We did not find association between LD and acidic compartments (Fig. 6G), as one would expect for an autophagic processing of LD. However, cytometry analysis of Nile red-stained cells revealed that treatment with 3-MA increased LD content in LN18 (Fig. 6F) and CHO cells (not shown), suggesting that, although not involved in LD mobilization for β-oxidation and cell survival, autophagy may account for part of the LD processing activity.
FIGURE 6.
Inhibition of autophagy does not accelerate death of nutrient-deprived cells. Treatment with KH without glucose induced autophagy in LN18 cells. A, electronic microscope image of a late autophagosome after 16 h in KH without glucose. B and C, LN18 cells were treated for 8 h with DMEM or KH without glucose (w/o gl) and with or without 1 μm py-2 or the autophagy inhibitor 3-MA (10 mm). Nutrient deprivation induced autophagy, as monitored by the appearance of LC3 puncta (B, left) or by conversion of LC3-I to LC3-II (C). This process was blocked by 3-MA (B, right; C, top) but not by inhibition of LD biogenesis with py-2 (C, bottom). D, LN18 cells were prelabeled overnight with [3H]palmitate prior to their treatment for 5 h with DMEM or KH without glucose and with or without 3-MA or EX. Inhibition of autophagy with 3-MA had no effect on β-oxidation of fatty acids, which was defined as EX-sensitive production of [3H]water. E, at long treatment times, blocking autophagy had a significant cytoprotective effect. F, flow cytometry analysis of Nile red-stained cells showed an overall accumulation of LD in cells treated with 3-MA. G, LN18 cells were kept 16 h in DMEM (left) or KH without glucose (right); acidic compartments were labeled with LysoTracker® Red DND-99, and LD were stained with BODIPY 493/503. Results in D are means ± S.E. (error bars) of one experiment in quadruplicate that is representative of three experiments with similar results. Results in E and F are means ± S.E. of 4–6 independent experiments carried out with duplicate determinations, and asterisks in these panels denote significance as compared with KH without glucose.
DISCUSSION
This work shows that cells under complete nutrient deprivation synthesize TAG and pack them in LD, which are mobilized to fuel β-oxidation of fatty acids. Biogenesis of LD has been shown in many experimental models of cellular stress and also appears associated with diverse pathological conditions. In order to develop new antitumor or cell-protecting therapeutic strategies, it is of interest to assess their relevance for cell survival. LD have been shown to accumulate in cells undergoing etoposide-induced apoptosis, as a consequence of decreased fatty acid oxidation secondary to impaired mitochondrial function (13). LD formation in tumors, which has long been known to take place in the anoxic core (40–42), might also be secondary to impaired β-oxidation. In these cases, synthesis of TAG and LD biogenesis could buffer excess fatty acids and prevent their toxicity. In this regard, LD formed in response to extracellular fatty acids and ER-linked TAG synthesis appear to relax ER stress (12) and protect cells from lipotoxicity. The potential use of the fatty acid catabolic pathway as a source of new targets for cancer therapy has been highlighted recently (43). Regardless of whether LD in perinecrotic tumor areas develop secondary to mitochondrial dysfunction or, as in our experimental model, they provide catabolic substrates to overcome nutrient starvation, it might be worth pursuing further studies to test whether pharmacological inhibition of cPLA2α with py-2 could prove useful as an adjuvant antitumor therapy to target fatty acid oxidation. Also, LD formed in ischemic heart and neurons appear to have a cell-protective role (24, 25), as lipid-buffering structures and/or as providers of metabolic fuel, and interventions aimed at increasing LD biogenesis could contribute to decrease cell damage.
Although based primarily on the use of py-2 to target LD biogenesis, a relevant finding of this work is that, under complete nutrient deprivation, the main source of fatty acids for β-oxidation is LD. Leaving aside sterol metabolism, the metabolic functions of LD as revealed or confirmed by proteomic studies can be grouped into fatty acid synthesis and activation, TAG biosynthesis, and fatty acid mobilization (44). Fatty acid synthesis does not take place in our glucose-devoid experimental setting, as evidenced by the ability of the fatty acid synthase inhibitors cerulenin and C75 to promote synthesis of TAG and LD biogenesis (22). Because LD are formed in the absence of exogenous lipoproteins or fatty acids, synthesis of TAG required for LD formation is at the expense of preexisting, phospholipid-linked fatty acids by a iPLA2-mediated process (22). This leaves fatty acid activation and TAG synthesis and mobilization as the relevant events involved in cell survival to complete nutrient deprivation. Accumulation of TAG within the ER membranes seems to be associated with LD formation (12, 44). Inhibition of cPLA2α, however, allows the dissociation of the two events, because it precludes LD biogenesis but does not affect synthesis of TAG (22, 23). Activation of cPLA2α for LD biogenesis requires phosphorylation by JNK (29, 33), and this may well account for the leaner phenotype of cPLA2α-defective (47, 48) or JNK1-defective mice (49). Pharmacological inhibition of cPLA2α with py-2 is a useful tool, therefore, to tackle the function of LD as providers of catabolic fuel. Our results show that cell survival to complete nutrient deprivation becomes strictly dependent on the catabolism of fatty acids. In this regard, similar to the inhibition of β-oxidation with EX, precluding the formation of LD with py-2 accelerates the death of starved cells, but it does not affect the viability of cells that do not depend on fatty acid catabolism. The observation that β-oxidation of fatty acids needs the formation of LD implies a dedicated metabolic channeling lipid droplet-mitochondrion, ensuring that fatty acyl-CoA derived from LD mobilization does not mix with other acyl-CoA pools within the cytosol, adding to the complexity of acyl-CoA synthetases in directing fatty acids into different metabolic pathways (50). Otherwise, neither synthesis of TAG after fatty acyl-CoA nor LD biogenesis after TAG would be needed for β-oxidation, and the release of fatty acids from phospholipids by iPLA2 and their activation by acyl-CoA synthetases would meet the requirement to fuel mitochondrial catabolism. Our results, therefore, underline the relevance of LD for fatty acid oxidation, in keeping with the idea that fatty acids liberated from LD may be used more efficiently for some metabolic pathways (45). In this regard, the possibility of direct contacts between LD and mitochondria has been considered (51). Among the LD-associated perilipin proteins, our data confirm that perilipins 2 and 3 shield LD from fusion with each other but do not affect the overall amount of TAG and, importantly, do not affect the rate of fatty acid catabolism, in agreement with the results of Bell et al. (35). LD- and mitochondria-associated perilipin 5 is expressed in highly oxidative tissues (52) and regulates TAG mobilization and flux of fatty acids to the mitochondria by a mechanism involving direct physical contact between the two organelles (53–55). Although we have been unable to detect perilipin 5 in our system, probably due to the unavailability of good antibodies, it is tempting to speculate that the metabolic adaptation to fatty acid oxidation that follows full nutrient deprivation, as described here, could be accompanied by expression of perilipin 5. A similar induction of specific LD-associated proteins may account in part for the high LD content together with a high oxidative capacity found in trained muscle, the so-called athlete's paradox (33, 56). In any case, our model system of total nutrient deprivation, in the absence of extracellular lipoproteins and free fatty acids and where cell viability is tightly dependent on LD mobilization, constitutes a simple setting to dissect the functional coupling between LD and mitochondria.
This work was supported in part by Spanish Ministry of Economy Grants BFU2009-07823 and BFU2012-34987).
- LD
- lipid droplet(s)
- BODIPY® 493/503
- 4,4-difluoro-1,3,5,7,8-pentamethyl-4-bora-3a,4a-diaza-s-indacene
- cPLA2α
- Group IVA phospholipase A2
- CERK
- ceramide kinase
- CPT1
- carnitine palmitoyltransferase-1
- EX
- etomoxir
- KH
- Krebs-Henseleit
- LDH
- lactate dehydrogenase
- LC3
- microtubule-associated protein light chain 3
- 3-MA
- 3-methyladenine
- PI
- propidium iodide
- py-2
- pyrrolidine-2
- SS
- side scatter
- FS
- forward scatter
- TAG
- triacylglycerol(s)
- iPLA2
- Group VIA phospholipase A2.
REFERENCES
- 1. Murphy D. J. (2012) The dynamic roles of intracellular lipid droplets. From archaea to mammals. Protoplasma 249, 541–585 [DOI] [PubMed] [Google Scholar]
- 2. Farese R. V., Jr., Walther T. C. (2009) Lipid droplets finally get a little R-E-S-P-E-C-T. Cell 139, 855–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Greenberg A. S., Coleman R. A. (2011) Expanding roles for lipid droplets. Trends Endocrinol. Metab. 22, 195–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Carman G. M. (2012) Thematic minireview series on the lipid droplet, a dynamic organelle of biomedical and commercial importance. J. Biol. Chem. 287, 2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bozza P. T., Bakker-Abreu I., Navarro-Xavier R. A., Bandeira-Melo C. (2011) Lipid body function in eicosanoid synthesis. An update. Prostaglandins Leukot. Essent. Fatty Acids 85, 205–213 [DOI] [PubMed] [Google Scholar]
- 6. Zechner R., Zimmermann R., Eichmann T. O., Kohlwein S. D., Haemmerle G., Lass A., Madeo F. (2012) Fat Signals. Lipases and lipolysis in lipid metabolism and signaling. Cell Metab. 15, 279–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Le Lay S., Dugail I. (2009) Connecting lipid droplet biology and the metabolic syndrome. Prog. Lipid. Res. 48, 191–195 [DOI] [PubMed] [Google Scholar]
- 8. Greenberg A. S., Coleman R. A., Kraemer F. B., McManaman J. L., Obin M. S., Puri V., Yan Q. W., Miyoshi H., Mashek D. G. (2011) The role of lipid droplets in metabolic disease in rodents and humans. J. Clin. Invest. 121, 2102–2110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Herker E., Ott M. (2011) Unique ties between hepatitis C virus replication and intracellular lipids. Trends Endocrinol. Metab. 22, 241–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Saka H. A., Valdivia R. (2012) Emerging roles for lipid droplets in immunity and host-pathogen interactions. Annu. Rev. Cell Dev. Biol. 28, 411–437 [DOI] [PubMed] [Google Scholar]
- 11. Murphy D. J., Vance J. (1999) Mechanisms of lipid-body formation. Trends Biochem. Sci. 24, 109–115 [DOI] [PubMed] [Google Scholar]
- 12. Hapala I., Marza E., Ferreira T. (2011) Is fat so bad? Modulation of endoplasmic reticulum stress by lipid droplet formation. Biol. Cell 103, 271–285 [DOI] [PubMed] [Google Scholar]
- 13. Boren J., Brindle K. M. (2012) Apoptosis-induced mitochondrial dysfunction causes cytoplasmic lipid droplet formation. Cell Death Differ. 19, 1561–1570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Quintero M., Cabañas M. E., Arús C. (2007) A possible cellular explanation for the NMR-visible mobile lipid (ML) changes in cultured C6 glioma cells with growth. Biochim. Biophys. Acta 1771, 31–44 [DOI] [PubMed] [Google Scholar]
- 15. Barba I., Mann P., Cabañas M.E., Arús C., Gasparovic C. (2001) Mobile lipid production after confluence and pH stress in perfused C6 cells. NMR Biomed. 14, 33–40 [DOI] [PubMed] [Google Scholar]
- 16. Hakumäki J. M., Poptani H., Sandmair A. M., Ylä-Herttuala S., Kauppinen R. A. (1999) 1H MRS detects polyunsaturated fatty acid accumulation during gene therapy of glioma. Implications for the in vivo detection of apoptosis. Nat. Med. 5, 1323–1327 [DOI] [PubMed] [Google Scholar]
- 17. Iorio E., Di Vito M., Spadaro F., Ramoni C., Lococo E., Carnevale R., Lenti L., Strom R., Podo F. (2003) Triacsin C inhibits the formation of 1H NMR-visible mobile lipids and lipid bodies in HuT 78 apoptotic cells. Biochim. Biophys. Acta 1634, 1–14 [DOI] [PubMed] [Google Scholar]
- 18. Schmitz J. E., Kettunen M. I., Hu D.-E., Brindle K. M. (2005) 1H MRS-visible lipids accumulate during apoptosis of lymphoma cells in vitro and in vivo. Magn. Reson. Med. 54, 43–50 [DOI] [PubMed] [Google Scholar]
- 19. Hakumäki J. M., Kauppinen R. A. (2000) 1H NMR visible lipids in the life and death of cells. Trends Biochem. Sci. 25, 357–362 [DOI] [PubMed] [Google Scholar]
- 20. Hakumäki J. M., Brindle K. M. (2003) Techniques. Visualizing apoptosis using nuclear magnetic resonance. Trends Pharmacol. Sci. 24, 146–149 [DOI] [PubMed] [Google Scholar]
- 21. Delikatny E. J., Chawla S., Leung D. J., Poptani H. (2011) MR-visible lipids and the tumor microenvironment. NMR Biomed. 24, 592–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gubern A., Barceló-Torns M., Casas J., Barneda D., Masgrau R., Picatoste F., Balsinde J., Balboa M. A., Claro E. (2009) Lipid droplet biogenesis induced by stress involves triacylglycerol synthesis that depends on group VIA phospholipase A2. J. Biol. Chem. 284, 5697–5708 [DOI] [PubMed] [Google Scholar]
- 23. Gubern A., Casas J., Barceló-Torns M., Barneda D., de la Rosa X., Masgrau R., Picatoste F., Balsinde J., Balboa M. A., Claro E. (2008) Group IVA phospholipase A2 is necessary for the biogenesis of lipid droplets. J. Biol. Chem. 283, 27369–27382 [DOI] [PubMed] [Google Scholar]
- 24. Du L., Hickey R. W., Bayir H., Watkins S. C., Tyurin V. A., Guo F., Kochanek P. M., Jenkins L. W., Ren J., Gibson G., Chu C. T., Kagan V. E., Clark R. S. (2009) Starving neurons show sex difference in autophagy. J. Biol. Chem. 284, 2383–2396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lei P., Baysa A., Nebb H. I., Valen G., Skomedal T., Osnes J. B., Yang Z., Haugen F. (2013) Activation of liver X receptors in the heart leads to accumulation of intracellular lipids and attenuation of ischemia-reperfusion injury. Basic Res. Cardiol. 108, 323–335 [DOI] [PubMed] [Google Scholar]
- 26. Barceló-Torns M., Lewis A. M., Gubern A., Barneda D., Bloor-Young D., Picatoste F., Churchill G. C., Claro E., Masgrau R. (2011) NAADP mediates ATP-induced Ca2+ signals in astrocytes. FEBS Lett. 585, 2300–2306 [DOI] [PubMed] [Google Scholar]
- 27. Barba I., Cabañas I., Arús C. (1999) The relationship between nuclear magnetic resonance-visible lipids, lipid droplets, and cell proliferation in cultured C6 cells. Cancer Res. 59, 1861–1868 [PubMed] [Google Scholar]
- 28. Koopman R., Schaart G., Hesselink M. K. (2001) Optimisation of oil red O staining permits combination with immunofluorescence and automated quantification of lipids. Histochem. Cell Biol. 116, 63–68 [DOI] [PubMed] [Google Scholar]
- 29. Gubern A., Barceló-Torns M., Barneda D., López J. M., Masgrau R., Picatoste F., Chalfant C. E., Balsinde J., Balboa M. A., Claro E. (2009) JNK and ceramide kinase govern the biogenesis of lipid droplets through activation of group IVA phospholipase A2. J. Biol. Chem. 284, 32359–32369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Djouadi F., Bonnefont J.-P., Munnich A., Bastin J. (2003) Characterization of fatty acid oxidation in human muscle mitochondria and myoblasts. Mol. Genet. Metab. 78, 112–118 [DOI] [PubMed] [Google Scholar]
- 31. Bligh E. G., Dyer W. J. (1959) A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37, 911–917 [DOI] [PubMed] [Google Scholar]
- 32. Murakami M., Taketomi Y., Miki Y., Sato H., Hirabayashi T., Yamamoto K. (2011) Recent progress in phospholipase A2 research. From cells to animals to humans. Prog. Lipid Res. 50, 152–192 [DOI] [PubMed] [Google Scholar]
- 33. Guijas C., Pérez-Chacón G., Astudillo A. M., Rubio J. M., Gil-de-Gómez L., Balboa M. A., Balsinde J. (2012) Simultaneous activation of p38 and JNK by arachidonic acid stimulates the cytosolic phospholipase A2-dependent synthesis of lipid droplets in human monocytes. J. Lipid Res. 53, 2343–2354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Menzel N., Fischl W., Hueging K., Bankwitz D., Frentzen A., Haid S., Gentzsch J., Kaderali L., Bartenschlager R., Pietschmann T. (2012) MAP-kinase regulated cytosolic phospholipase A2 activity is essential for production of infectious hepatitis C virus particles. PLoS Pathog. 8, e1002829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bell M., Wang H., Chen H., McLenithan J. C., Gong D. W., Yang R. Z., Yu D., Fried S. K., Quon M. J., Londos C., Sztalryd C. (2008) Consequences of lipid droplet coat protein downregulation in liver cells. Abnormal lipid droplet metabolism and induction of insulin resistance. Diabetes 57, 2037–2045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Khatchadourian A., Maysinger D. (2009) Lipid droplets. Their role in nanoparticle-induced oxidative stress. Mol. Pharm. 6, 1125–1137 [DOI] [PubMed] [Google Scholar]
- 37. Singh R., Kaushik S., Wang Y., Xiang Y., Novak I., Komatsu M., Tanaka K., Cuervo A. M., Czaja M. J. (2009) Autophagy regulates lipid metabolism. Nature 458, 1131–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kaushik S., Rodriguez-Navarro J. A., Arias E., Kiffin R., Sahu S., Schwartz G. J., Cuervo A. M., Singh R. (2011) Autophagy in hypothalamic AgRP neurons regulates food intake and energy balance. Cell Metab. 14, 173–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ouimet M., Franklin V., Mak E., Liao X., Tabas I., Marcel Y. L. (2011) Autophagy regulates cholesterol efflux from macrophage foam cells via lysosomal acid lipase. Cell Metab. 13, 655–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Freitas I., Pontiggia P., Barni S., Bertone V., Parente M., Novarina A., Roveta G., Gerzeli G., Stoward P. (1990) Histochemical probes for the detection of hypoxic tumour cells. Anticancer Res. 10, 613–622 [PubMed] [Google Scholar]
- 41. Freitas I., Bono B., Bertone V., Griffini P., Baronzio G. F., Bonandrini L., Gerzeli G. (1996) Characterization of the metabolism of perinecrotic cells in solid tumors by enzyme histochemistry. Anticancer Res. 16, 1491–1502 [PubMed] [Google Scholar]
- 42. Zoula S., Rijken P. F., Peters J. P., Farion R., Van der Sanden B. P., Van der Kogel A. J., Décorps M., Rémy C. (2003) Pimonidazole binding in C6 rat brain glioma. Relation with lipid droplet detection. Br. J. Cancer 88, 1439–1444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Carracedo A., Cantley L. C., Pandolfi P. P. (2013) Cancer metabolism. Fatty acid oxidation in the limelight. Nat. Rev. Cancer 13, 227–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Goodman J. M. (2009) Demonstrated and inferred metabolism associated with cytosolic lipid droplets. J. Lipid Res. 50, 2148–2156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Fujimoto T., Parton R. G. (2011) Not just fat. The structure and function of the lipid droplet. Cold Spring Harb. Perspect. Biol. 3, a004838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Coen P. M., Goodpaster B. H. (2012) Role of intramyocelluar lipids in human health. Trends Endocrinol. Metab. 23, 391–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ii H., Hatakeyama S., Tsutsumi K., Sato T., Akiba S. (2008) Group IVA phospholipase A2 is associated with the storage of lipids in adipose tissue and liver. Prostaglandins Other Lipid Mediat. 86, 12–17 [DOI] [PubMed] [Google Scholar]
- 48. Ii H., Yokoyama N., Yoshida S., Tsutsumi K., Hatakeyama S., Sato T., Ishihara K., Akiba S. (2009) Alleviation of high-fat diet-induced fatty liver damage in group IVA phospholipase A2-knockout mice. PLoS One 4, e8089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hirosumi J., Tuncman G., Chang L., Görgün C. Z., Uysal K. T., Maeda K., Karin M., Hotamisligil G. S. (2002) A central role for JNK in obesity and insulin resistance. Nature 420, 333–336 [DOI] [PubMed] [Google Scholar]
- 50. Li L. O., Klett E. L., Coleman R. A. (2010) Acyl-CoA synthesis, lipid metabolism and lipotoxicity. Biochim. Biophys. Acta 1801, 246–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Goodman J. M. (2008) The gregarious lipid droplet. J. Biol. Chem. 283, 28005–28009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wang H., Sztalryd C. (2011) Oxidative tissue. Perilipin 5 links storage with the furnace. Trends Endocrinol. Metab. 22, 197–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wang H., Bell M., Sreenivasan U., Hu H., Liu J., Dalen K., Londos C., Yamaguchi T., Rizzo M. A., Coleman R., Gong D., Brasaemle D., Sztalryd C. (2011) Unique regulation of adipose triglyceride lipase (ATGL) by perilipin 5, a lipid droplet-associated protein. J. Biol. Chem. 286, 15707–15715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wang H., Sreenevasan U., Hu H., Saladino A., Polster B. M., Lund L. M., Gong D. W., Stanley W. C., Sztalryd C. (2011) Perilipin 5, a lipid droplet-associated protein, provides physical and metabolic linkage to mitochondria. J. Lipid Res. 52, 2159–2168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Bosma M., Minnaard R., Sparks L. M., Schaart G., Losen M., de Baets M. H., Duimel H., Kersten S., Bickel P. E., Schrauwen P., Hesselink M. K. (2012) The lipid droplet coat protein perilipin 5 also localizes to muscle mitochondria. Histochem. Cell Biol. 137, 205–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Goodpaster B. H., He J., Watkins S., Kelley D. E. (2001) Skeletal muscle lipid content and insulin resistance. Evidence for a paradox in endurance-trained athletes. J. Clin. Endocrinol. Metab. 86, 5755–5761 [DOI] [PubMed] [Google Scholar]