Background: Inhibition of complex sphingolipid synthesis by inositol starvation activates the PKC-MAPK signaling pathway.
Results: Sir2p-dependent telomeric silencing is activated by interrupting sphingolipid synthesis and requires the MAPK, Slt2p.
Conclusion: Telomeric silencing is regulated by PKC-MAPK signaling in response to interruption of sphingolipid synthesis.
Significance: Sphingolipid metabolism plays an important role in regulating silencing and chronological life span.
Keywords: Inositol Phospholipid, NAD Biosynthesis, Protein Kinase C (PKC), Sirtuins, Sphingolipid
Abstract
Depriving wild type yeast of inositol, a soluble precursor for phospholipid, phosphoinositide, and complex sphingolipid synthesis, activates the protein kinase C (PKC)-MAPK signaling pathway, which plays a key role in the activation of NAD+-dependent telomeric silencing. We now report that triggering PKC-MAPK signaling by inositol deprivation or by blocking inositol-containing sphingolipid synthesis with aureobasidin A results in increased telomeric silencing regulated by the MAPK, Slt2p, and the NAD+-dependent deacetylase, Sir2p. Consistent with the dependence on NAD+ in Sir2p-regulated silencing, we found that inositol depletion induces the expression of BNA2, which is required for the de novo synthesis of NAD+. Moreover, telomeric silencing is greatly reduced in bna2Δ and npt1Δ mutants, which are defective in de novo and salvage pathways for NAD+ synthesis, respectively. Surprisingly, however, omitting nicotinic acid from the growth medium, which reduces cellular NAD+ levels, leads to increased telomeric silencing in the absence of inositol and/or at high temperature. This increase in telomeric silencing in response to inositol starvation is correlated to chronological life span extension but is Sir2p-independent. We conclude that activation of the PKC-MAPK signaling by interruption of inositol sphingolipid synthesis leads to increased Sir2p-dependent silencing and is dependent upon the de novo and salvage pathways for NAD+ synthesis but is not correlated with cellular NAD+ levels.
Introduction
Microarray studies have revealed that the transcript levels of hundreds of genes in yeast are affected by the availability of the phospholipid precursor, inositol (1–4). The INO1 gene, encoding inositol-3-phosphate synthase, and other genes involved in lipid metabolism are repressed by the Opi1p repressor in response to the presence of exogenous inositol (5–10). However, the majority of the genes that are regulated in response to inositol availability in wild type cells are not involved in phospholipid metabolism and are not under the control of the Opi1p repressor (1, 2). Many of genes that are induced in the absence of inositol are targets of stress response pathways that have been shown to be activated in the absence of exogenous inositol, including the unfolded protein response and the protein kinase C (PKC) pathway (1–3, 11–13). Moreover, mutations in these same stress response pathways confer inositol auxotrophy (Ino− phenotype) indicating that signaling through these pathways is essential for survival of cells experiencing stress associated with inositol deprivation (3, 11–14). Thus, growth in the absence of inositol is an inherently stress-activating condition, analogous to growth at elevated temperature, high or low osmolarity, and/or exposure to agents such as tunicamycin, caffeine, or calcofluor white (3, 8, 13). In wild type cells, inositol deprivation also results in dramatic changes in lipid metabolism, including rapid depletion of phosphatidylinositol (15, 16). Phosphatidylinositol serves as precursor to all other inositol-containing lipids in the cell, including important signaling lipids such as the inositol-containing sphingolipids and phosphatidylinositol phosphates (8, 17–19), as well as inositol phosphates and pyrophosphates derived from phosphatidylinositol phosphates (20, 21). In a previous study (13), we demonstrated that the slowing of synthesis of the inositol-containing sphingolipids is responsible for triggering activation of PKC signaling in inositol-depleted wild type cells. Furthermore, the phosphatidylinositol 4-phosphate and phosphatidylinositol 4,5-biphosphate pools on the plasma membrane were shown to be essential to the activation of PKC signaling in the absence of inositol (13).
Strikingly, the PKC pathway has also been shown to play a key role in activation of NAD+-dependent gene silencing (22–24). The Sir complex, consisting of Sir2p, Sir3p, and Sir4p, controls silencing at telomeric regions of chromosomes and at the silent mating type loci HMR and HML in yeast (25–28). Sir3p is a specific phosphorylation target of Slt2p, the MAPK of the PKC pathway (22, 24), and hyper-phosphorylation of Sir3p strengthens telomeric silencing (29). NAD+ is essential to the activity of NAD+-dependent histone/protein deacetylases (Fig. 1), named “sirtuins” after Sir2p (silent information regulator 2 protein), the founding member of this class of proteins (30–32). Saccharomyces cerevisiae has five sirtuins, Sir2p, Hst1p, Hst2p, Hst3p, and Hst4p (33). In the absence of exogenous nicotinic acid (NA)4 (Fig. 1), the Hst1p sirtuin is responsible for activation of BNA2, which encodes the enzyme that catalyzes the first step in the pathway for the de novo synthesis of NAD+ from tryptophan (Fig. 1) (34, 35). However, BNA2 was also found to be up-regulated in microarray studies of cells grown in the absence of inositol (1, 2) and in cells grown at 37 °C (36), a stress condition that, similar to inositol starvation, activates PKC signaling (37).
FIGURE 1.
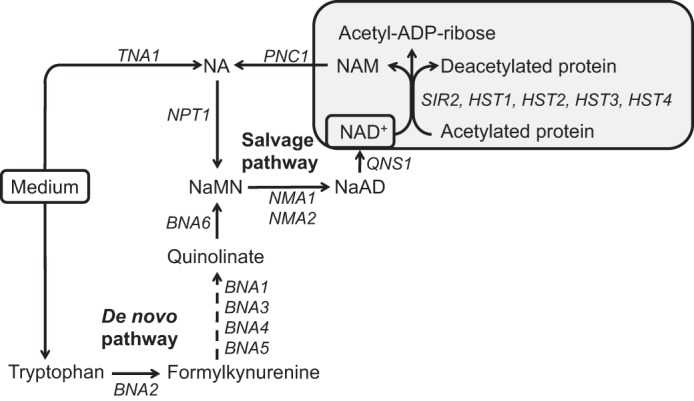
NAD+ in S. cerevisiae is synthesized from NA via the salvage pathway and from tryptophan through the de novo pathway. NA and tryptophan imported from the growth medium serve as precursors of NAD+. A gray box indicates the activity of sirtuins (Sir2p and Hst1p-4p), NAD+-dependent protein deacetylases, which hydrolyze NAD+ into nicotinamide and ADP-ribose, in a reaction coupled with protein/histone deacetylation. The enzymes involved in the salvage pathway for NAD+ synthesis are primarily localized in the nucleus (52, 62), whereas the enzymes in the de novo pathway are distributed in the nucleus and cytoplasm (52). The de novo pathway converts tryptophan to nicotinic acid mononucleotide in a series of reactions catalyzed by Bna1p–6p. Nicotinic acid mononucleotide is the point of convergence for the de novo and salvage pathways for NAD+ biosynthesis. Abbreviations used are as follows: NAD+, nicotinamide adenine dinucleotide; NA, nicotinic acid; NAM, nicotinamide; NaMN, nicotinic acid mononucleotide; NaAD, deamido-NAD.
We now report that wild type cells grown in medium lacking inositol exhibit significant strengthening of Sir2p-dependent telomeric silencing. In contrast, the slt2Δ mutant, defective in PKC signaling, exhibited a loss of silencing relative to wild type under these same growth conditions. Whereas increased telomeric silencing triggered by inositol starvation was dependent on both the de novo and salvage pathways for NAD+ biosynthesis, it was not tightly correlated to overall NAD+ levels. We also confirmed, as reported previously (29, 38), that Sir2p-dependent telomeric silencing is enhanced at 37 °C in wild type cells and that deletion of the SLT2 gene resulted in a loss of silencing at this growth temperature. Previously, we demonstrated that activation of PKC signaling during inositol starvation was specifically triggered by slowing of synthesis of the complex inositol-containing sphingolipids, rather than the build up of sphingoid bases (13). Strikingly, we demonstrate here that blocking sphingolipid synthesis with aureobasidin A (AbA) strengthens Sir2p-dependent telomeric silencing in a fashion similar to the effects observed in cells growing in the absence of inositol and that this response is also mediated by an active PKC pathway. We also report that strengthening of telomeric silencing in wild type cells in the absence of inositol is correlated to increase chronological life span. We conclude that increased Sir2p-dependent telomeric silencing in the absence of inositol requires an active PKC signaling pathway and is dependent on the de novo and salvage pathways for NAD+ biosynthesis but is not correlated with absolute cellular NAD+ levels.
EXPERIMENTAL PROCEDURES
Media and Growth Conditions
Strains were maintained on plates containing 1% yeast extract, 2% bactopeptone, 2% glucose, and 2% agar (YEPD medium). All experiments were carried out using defined synthetic media containing (per liter) the following: 20 g of glucose, 5 g of ammonium sulfate, 1 g of potassium phosphate, 0.5 g of magnesium sulfate, 0.1 g of sodium chloride, 0.1 g of calcium chloride, 0.5 mg of boric acid, 0.04 mg of cupric sulfate, 0.1 mg of potassium iodide, 0.2 mg of ferric chloride, 0.4 mg of manganese sulfate, 0.2 mg of sodium molybdate, 0.4 mg of zinc sulfate, 2 μg of biotin, 400 μg of calcium pantothenate, 2 μg of folic acid, 200 μg of p-aminobenzoic acid, 400 μg of pyridoxine hydrochloride, 200 μg of riboflavin, 400 μg of thiamine, 20 mg of adenine sulfate, 20 mg of arginine, 20 mg of histidine, 60 mg of leucine, 230 mg of lysine, 20 mg of methionine, 300 mg of threonine, 20 mg of tryptophan, and 40 mg of uracil. The medium described above lacks both inositol (I) and nicotinic acid (NA) and is therefore referred to as I−NA− medium. Where indicated, I−NA− medium was supplemented with 75 μm inositol (I+NA− medium) or 3.25 μm nicotinic acid (I−NA+ medium) or both (I+NA+ medium). Plates containing these media also contained 2% agar.
Yeast Strains
All strains (Table 1) were derived from the S288C genetic background and were either purchased from Research Genetics (Invitrogen) or were constructed from strains in the SC288C background. All strains obtained from Research Genetics were crossed and subjected to tetrad analysis to confirm that all phenotypes segregated 2:2.
TABLE 1.
Yeast strains used in this study
| Strain | Genotype | Source or Ref. |
|---|---|---|
| BY4742 | MATα; his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 | Invitrogen |
| SLY44 | MATα; his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 bna2Δ::kanMX | Invitrogen |
| SLY43 | MATα; his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 npt1Δ::kanMX | Invitrogen |
| SLY30 | MATα; his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 sir2Δ::kanMX | Invitrogen |
| SJY804 | MATα; his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 slt2Δ::HIS3 | This study |
| SLY72 | MATα; his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 TELVR::URA3 | This study |
| LGY441 | MATα; his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 bna2Δ::kanMX TELVR::URA3 | This study |
| SLY75 | MATα; his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 sir2Δ::kanMX TELVR::URA3 | This study |
| LGY450 | MATα; his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 slt2Δ::HIS3 TELVR::URA3 | This study |
| SLY73 | MATα; his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 npt1Δ::kanMX TELVR::URA3 | This study |
Measurement of Total Cellular NAD+ Levels
500-ml cultures were grown continuously in the four different growth media (i.e. I+NA+, I−NA+, I+NA−, and I−NA−) to A600 nm = 1.0 at 30 or 37 °C as indicated. Cells were harvested by filtration and stored at −80 °C. Harvested cells were extracted with 250 μl of ice-cold 1 m formic acid saturated with butanol for 30 min. 62.5 μl of trichloroacetic acid (TCA) was added, and samples were precipitated on ice for 15 min. Samples were centrifuged for 5 min, and the acid-soluble supernatants were collected. The pellets were washed with 125 μl of 20% TCA and centrifuged. These supernatants were combined with the supernatants obtained from the 100% TCA extraction (39, 40). 150-μl aliquots of extract were reacted with 10 μl of 5 mg/ml alcohol dehydrogenase and 840 μl of Tris/lysine Buffer (pH 9.7), which contained 0.24% ethanol. This mixture was incubated for 20 min at room temperature to allow NAD+ to be converted into NADH, which was measured as absorbance at 340 nm (40, 41). The absorbance was converted to NAD+ molar concentration using the NADH extinction coefficient (6220 m−1cm−1), and the molar concentration in the extract was calculated using a factor of 6.67 and a haploid cell assumed volume of 7 × 10−14 liters (42). All NAD+ molar concentration values were corrected for recovery efficiency as described by Sporty et al. (43).
Measurement of BNA2 and THI4 Expression; RNA Isolation and RT-PCR Analysis
To measure expression of the BNA2 gene, cells were grown in medium containing inositol with and without NA to mid-logarithmic phase at 30 or 37 °C. At A600 nm = 0.5–0.6, cultures were divided in half. One-half of each culture was filtered, washed with prewarmed medium containing inositol, and resuspended in new media containing inositol with and without NA, maintaining the same temperature. The other half was also filtered, washed with prewarmed medium lacking inositol, and resuspended in new media lacking inositol with and without NA at its original incubation temperature of 30 or 37 °C. Samples were harvested by filtration at 3 h following the media shift, flash-frozen in dry ice, and stored at −80 °C. Total RNA was isolated using RNeasy® mini kit, including DNA digestion with RNase-free DNase set (Qiagen). cDNA was synthesized from 1 μg of total RNA using oligo(dT)12–18 primer (0.5 μg), PCR grade dNTP mix (0.5 μm), First strand buffer (1×), DTT (10 mm), and 100 units of SuperScript® II reverse transcriptase (Invitrogen). Real time PCR was performed on a StepOnePlusTM Real Time PCR system (Applied Biosystems) using TaqMan® universal PCR master mix, No AmpErase® UNG (Applied Biosystems), and the following TaqMan® probes and primers: BNA2, TaqMan® probe, 5′-Fam-ATG GGC TGG ATG TCT-Tamra-3′; forward primer (5′-AAG GTG CGG AGC GTC ATC-3′) and reverse primer (5′-GCC CAA GAT CGT CTC ATC CA-3′); THI4, TaqMan® probe, 5′-Fam-TTC TGC GCC AAG AGA ATC GTC GAC ATT-Tamra-3′; forward primer (5′-CGG TCA TGA TGG TCC ATT TG-3′) and reverse primer (5′-CGC CCA ATT TTT GGT TTT GA-3′). The ACT1 gene was used as an endogenous control for normalization of RNA levels. In brief, the reaction mix in a volume of 25 μl consisted of 0.5 μm primers, 0.2 μm TaqMan® probe, 1× Master Mix, and 5 ng of cDNA. Nontemplate control (5 ng of RNA) and nonreaction control (diethyl pyrocarbonate/water) were routinely performed. The thermal program for the PCR included stage 1 as follows: 95 °C, 10 min; stage 2, 95 °C, 0.5 min, and 60 °C, 1 min for a total of 40 cycles. Relative gene expression levels were calculated as 2−ΔΔCT. ΔΔCT was calculated as follows: (gene CTS − ACT1 CTS) − (gene CTR − ACT1 CTR) and represents the change in mRNA expression after ACT1 normalization relative to mRNA derived from the wild type control. “Gene” represents the mRNA under study (BNA2 or THI4). “S” refers to the strain from which the mRNA to be tested was derived (i.e. wild type), grown under several different growth conditions (I+NA+, I−NA+, I+NA−, or I−NA− media at 30 or 37 °C). In the formula above, “R” refers to the control mRNA derived from the wild type strain grown in I+NA+ medium to mid-logarithmic phase at 30 °C. CT (cycle threshold) is defined as the number of cycles required for the fluorescent signal to cross the threshold (i.e. to exceed background level). Each RT-PCR experiment was performed at least in triplicate.
Silencing Assay Using the URA3 Reporter Gene
The plasmid pV-UCA, which carries the URA3 reporter gene inserted adjacent to the right end of chromosome V (telomere VR on a HindIII fragment) was kindly provided by T. Petes. The EcoRI fragment of pV-UCA derived from this plasmid was transformed into wild type, bna2Δ, npt1Δ, slt2Δ, and sir2Δ cells. Using the strains carrying this construct, silencing was assessed by measurement of expression of the URA3 reporter gene by RT-PCR, using methods described above for BNA2 and THI4, using ACT1 as an endogenous control for normalization of RNA levels. The TaqMan® probes and primers for URA3 were as follows: TaqMan® probe, 5′-Fam-CCG CTA AAG GCA TTA TCC GCC AAG TAC A-Tamra-3′, forward primer (5′-TTC CAT GGA GGG CAC AGT TAA-3′) and reverse primer (5′-GTA TTA CCA ATG TCA GCA AAT TTT CTG-3′); ACT1, TaqMan® probe, 5′-Fam-TGC AAA CCG CTG CTC AAT CTT CTT CAA T-Tamra-3′, forward primer (5′-CGC CTT GGA CTT CGA ACA AG-3′) and reverse primer (5′-GAC CAT CTG GAA CTT CGT AGG ATT-3′).
For RT-PCR analysis, strains carrying the URA3 construct were grown on synthetic medium (I+NA+ or I+NA−) at 30 or 37 °C until mid-logarithmic phase and diluted back to A600 nm = 0.2 while maintaining NA supplementation level and growth temperature constant. Cells precultured in I+NA+ media were shifted to fresh I+NA+ and I−NA+; cells cultured in I+NA− were shifted to fresh I+NA− or I−NA− media in each case maintaining the preculture temperature and allowed to grow until they again reached an A600 nm of 1.0. Where indicated, AbA was added to a final concentration of 2 μg/ml immediately following a shift for 120 min to I+NA+. There was little significant difference in the growth rate of any of these strains in any of the media and/or at either temperature, or in response to the addition of AbA, during the interval specified for each experiment. Total RNA isolation, cDNA synthesis, and RT-PCR were performed as described above.
Chronological Life Span
Yeast longevity can be measured as replicative life span, defined as the number of daughter cells produced by a mother cell (44), and/or chronological life span, defined as the number of days that nondividing cells maintain viability in a saturated culture or while resuspended in water (45). Because Huang et al. (46) recently reported that complex inositol sphingolipids are involved in chronological life span extension in yeast and we had observed that interruption of synthesis of inositol-containing sphingolipids activates PKC signaling, we monitored chronological life span in cells grown in the presence or absence of inositol. Chronological life span of cells incubated in either I+ or I− medium was measured as described previously (45). In brief, one colony from a fresh I+ plate was inoculated in 10 ml of I+ or I− medium. The overnight culture was diluted in 50 ml of I+ or I− medium, and cells were allowed to grow until saturation. Cell survival was measured every 2 days by diluting yeast cultures and plating an appropriate number of cells onto YEPD plates to monitor the colony-forming units (CFUs). The number of CFUs at day 2 was considered to be the initial survival (100%) and was used to determine the age-dependent mortality.
RESULTS
Telomeric Silencing Increases Significantly in Wild Type Cells Grown in the Absence of Inositol and/or at 37 °C
Telomeric silencing was measured by analyzing the expression of a URA3 reporter inserted near the right end of chromosome V (47, 48) as described under “Experimental Procedures.” We found increases in silencing in response to both inositol deprivation and growth at 37 °C, as depicted in Fig. 2, A and B. The data are shown as log2 ratios so that relative changes in expression of URA3, both positive and negative, can be observed and compared in similar scales on the same graph. In this assay, an increase in URA3 expression is indicative of decreased silencing of the reporter gene relative to the wild type control, whereas decreased expression is indicative of increased silencing.
FIGURE 2.
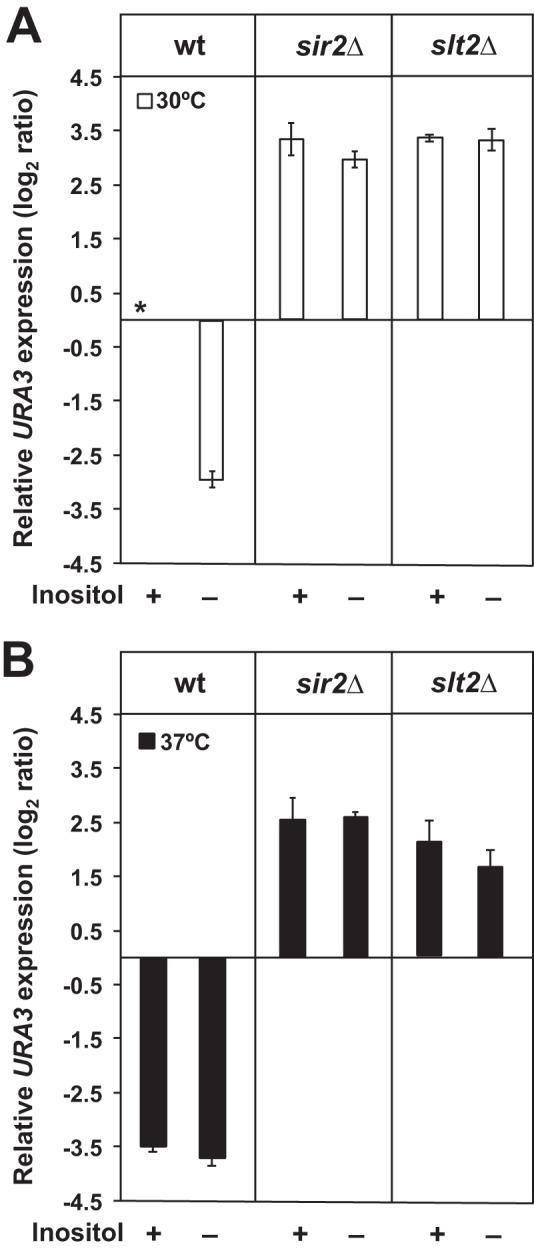
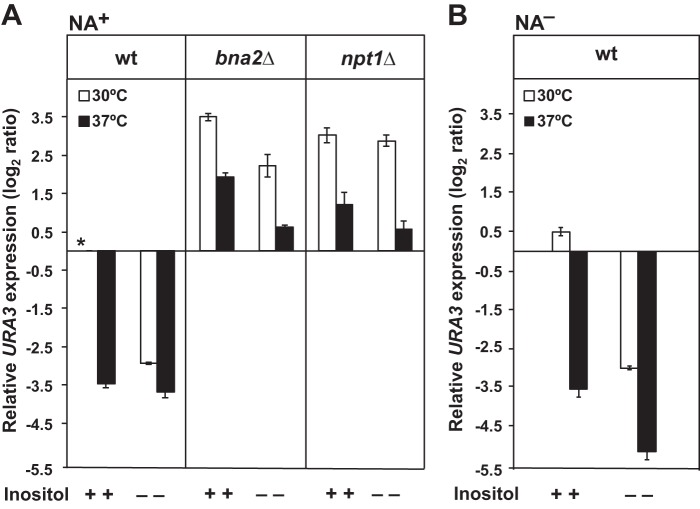
Increased silencing in the absence of inositol and/or at 37 °C is dependent on Sir2p and Slt2p. Wild type, sir2Δ, and slt2Δ strains carrying the TELVR::URA3 construct were grown on I+ medium until mid-logarithmic phase, diluted back to A600 nm = 0.2 in I+ and I− media at 30 or 37 °C, and allowed to grow until an A600 nm of 1.0. Gene expression levels were examined by RT-PCR and calculated using the comparative CT method (ΔΔCT) as described under “Experimental Procedures.” The changes in URA3 expression levels are log2 ratios of the level of expression measured by RT-PCR in a specific strain, under the given growth condition, relative to URA3 expression in the control culture. The asterisk refers to the mRNA derived from the wild type control grown in I+NA+ medium at 30 °C and served as a reference (unit = 1) for measuring relative URA3 expression levels. A indicates the strains grown at 30 °C. B indicates the strains grown at 37 °C. Data represent the mean ± S.E. of three independent experiments, n = 3.
Wild type cells growing in I− medium at 30 °C, exhibited an approximate 3-fold decrease, log2 scale, in URA3 expression as compared with the control growing in I+ medium at 30 °C (Fig. 2A). Growth at 37 °C also resulted in an increase in silencing in wild type cells, regardless of the presence of inositol, as shown by a decrease of ∼3.5-fold, log2 scale, in URA3 expression, in both I+ and I− media (Fig. 2B). This increase in silencing observed at 37 °C is consistent with the reports of Stone and Pillus (29) and Bi et al. (38), who reported that telomeric silencing is strengthened at higher growth temperatures.
Increased Silencing in the Absence of Inositol and/or at 37 °C Is Dependent on Both Sir2p and on Slt2p, the Terminal MAPK of the PKC Stress Response Pathway
The sir2Δ mutant, as expected and consistent with its primary defect in silencing (48, 49), exhibited a significant loss of silencing in comparison with the wild type, as indicated by 2.5–3.5-fold higher expression (log2 scale) of the URA3 reporter. The loss of silencing in sir2Δ was not significantly improved by growth in the absence of inositol (Fig. 2), indicating that the increases in silencing observed in wild type cells in the absence of inositol are Sir2p-dependent. Similar to the sir2Δ mutant, the slt2Δ mutant exhibited elevated expression of the URA3 reporter compared with wild type under all conditions tested (Fig. 2). These results suggest that the increase in silencing in response to inositol deprivation in wild type cells is also dependent on Slt2p. However, at 37 °C, in comparison with 30 °C, a slight, but significant, decrease in URA3 expression was observed in the slt2Δ mutant, whether inositol was present or not, suggesting that a minor component of the observed increase in silencing seen at 37 °C in wild type cells may be Slt2p-independent.
Interruption of Synthesis of Inositol-containing Sphingolipids Activates PKC Signaling, Resulting in Increased Silencing
Previously, we reported that the synthesis of complex sphingolipids is substantially reduced in yeast cells grown in the absence of inositol (13). In these experiments, interruption of sphingolipid synthesis whether by inositol deprivation or exposure to AbA was responsible for activation of PKC signaling. Given that increased silencing in the absence of inositol is dependent on Slt2p, we questioned if interruption in the synthesis of inositol-containing sphingolipids was also responsible for the strengthening of silencing that we observed in the absence of inositol. Indeed, inhibition of synthesis of inositol-containing sphingolipids by the addition of AbA to cells growing in the presence of inositol resulted in an increase in silencing in wild type cells relative to untreated control (Fig. 3). This pattern is similar to the strength in silencing that we observed in the absence of inositol (Figs. 2A and 3). In contrast to wild type, the presence of AbA did not lead to an increase in silencing in the slt2Δ strain. In the slt2Δ strain, the high level of URA3 expression observed in both I+ and I− medium remained unchanged when cells were grown in the presence of AbA in I+ medium, indicating no restoration of silencing when sphingolipid synthesis is blocked. These results indicate that activation of Slt2p-dependent PKC signaling resulting from a reduction in the synthesis of inositol-containing sphingolipids is responsible for the increase in silencing in wild type cells grown in the absence of inositol.
FIGURE 3.
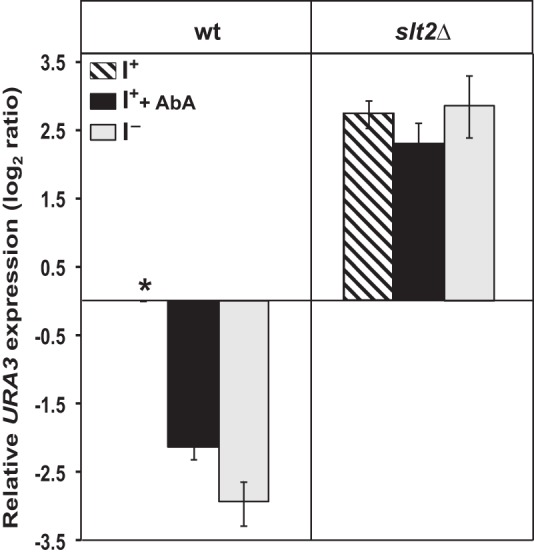
Inhibiting complex sphingolipid synthesis results in increased silencing. Wild type and slt2Δ strains carrying the TELVR::URA3 construct were grown on I+ medium until mid-logarithmic phase, diluted back to A600 nm = 0.2 in I+ and I− media at 30 °C, and allowed to grow until an A600 nm of 1.0. An aliquot of the cultures growing in I+ medium were treated with AbA to final concentration of 2 μg/ml, incubated at 30 °C for 120 min, and harvested. Gene expression levels were examined by RT-PCR and calculated using the comparative CT method (ΔΔCT) as described under “Experimental Procedures.” The changes in URA3 expression levels are log2 ratios of the level of expression measured by RT-PCR in a specific strain, under the given growth condition, relative to URA3 expression in the control culture. The asterisk refers to the mRNA derived from the wild type control grown in I+NA+ medium at 30 °C and served as a reference (unit = 1) for measuring relative URA3 expression levels. Data represent the mean ± S.E.) of three independent experiments, n = 3.
Sir2p-dependent Telomeric Silencing Is Lost in npt1Δ and bna2Δ Mutants under All Conditions Tested
To determine whether NAD+ synthesis through the de novo and/or salvage pathways for NAD+ biosynthesis (Fig. 1) is required to support the increased Sir2p-dependent telomeric silencing observed, we tested the expression of the telomere-associated reporter URA3 gene in the npt1Δ and bna2Δ strains in NA-containing medium at both growth temperatures, with and without inositol. At 30 °C in I+ medium, the bna2Δ and npt1Δ mutants both exhibited increases of 3.5- to 3.0-fold log2 ratio in URA3 expression relative to wild type (Fig. 4A), indicative of decreases in silencing similar in magnitude to those seen in the sir2Δ mutant (Fig. 2). At 30 °C, lack of inositol supplementation had little effect on the loss of silencing in the npt1Δ strain, although the bna2Δ mutant exhibited a significant decrease in URA3 expression when inositol was absent (Fig. 4A). However, at 37 °C, URA3 expression levels were significantly reduced in both strains. In bna2Δ, the reduction in URA3 expression at 37 °C was most pronounced in the absence of inositol, and the effects of inositol deprivation and growth at 37 °C on the reduction in URA3 expression appeared to be additive (Fig. 4A). In the npt1Δ strain, the reduction in URA3 expression at 37 °C was greatest in medium lacking inositol, despite the lack of any apparent effect of inositol deprivation on URA3 expression at 30 °C (Fig. 4A).
FIGURE 4.
Influence of the de novo and salvage pathways for NAD+ biosynthesis on Sir2-dependent telomeric silencing. Wild type, bna2Δ, and npt1Δ strains carrying the TELVR::URA3 construct were grown on I+ medium until mid-logarithmic phase, diluted back to A600 nm = 0.2 in I+ and I− media at 30 or 37 °C, and allowed to grow until an A600 nm of 1.0. As a control experiment, wild type cells were grown under the same conditions with the exception that NA was omitted from the medium. Gene expression levels were examined by RT-PCR and calculated using the comparative CT method (ΔΔCT) as described under “Experimental Procedures.” The changes in URA3 expression levels are log2 ratios of the level of expression measured by RT-PCR in a specific strain, under the given growth condition, relative to URA3 expression in the control culture. The asterisk refers to the mRNA derived from the wild type control grown in I+NA+ medium at 30 °C and served as a reference (unit = 1) for measuring relative URA3 expression levels. White bars indicate the strains grown at 30 °C, and black bars indicate the strains grown at 37 °C. A indicates the strains grown in the presence of NA. B indicates the strain grown in the absence of NA. Data represent the mean ± S.E. of three independent experiments, n = 3.
Previously, a loss of silencing was reported in wild type cells growing in the absence of NA, an important precursor in the synthesis of NAD+ (41). Consistent with these prior results, we found decreased silencing in wild type cells grown in medium lacking NA but containing inositol (I+NA− medium) at 30 °C (Fig. 4B). However, surprisingly wild type cells growing at 30 °C in medium lacking both inositol and NA exhibited increased silencing (Fig. 4B). Growth at 37 °C also resulted in increased silencing in wild type cells growing in the absence of NA (Fig. 4B). Importantly, when both inositol and NA (I−NA− medium) were omitted from the medium at 37 °C, a further increase in silencing occurred. Overall, in I−NA− medium at 37 °C, URA3 expression was about 5-fold lower, log2 scale, than in the I+NA+ control at 30 °C indicative of the highest level of silencing observed in these experiments (Fig. 4B). Thus, both inositol starvation and increased growth temperature resulted in increased silencing independent of NA supplementation.
BNA2 Expression in Wild Type Cells Is Increased in an Additive Fashion in Response to Inositol Deprivation and Growth at 37 °C in the Presence or Absence of NA
The decrease in silencing in the bna2Δ mutant suggested that the expression of this gene is important for satisfying the cellular demands for NAD+ generated by the observed increase in telomeric silencing in the absence of inositol and/or growth at 37 °C. To analyze BNA2 expression, we grew wild type cells on medium containing inositol with and without NA to mid-logarithmic phase at 30 or 37 °C. The cultures were then divided in half and shifted to new medium containing or lacking inositol as described under “Experimental Procedures.” In wild type cells grown in I+NA+ medium at 37 °C, BNA2 expression was elevated about 5-fold in comparison with cells grown in the same medium at 30 °C (Fig. 5A). In addition and consistent with the report of Bedalov et al. (35), we confirmed that BNA2 expression was about 40-fold higher in cells grown in I+NA− relative to cells grown in I+NA+ medium. Moreover, the effects of the higher growth temperature and the lack of NA on BNA2 expression were additive. Thus, in the wild type strain, the overall increase in expression of BNA2 in I+NA− medium at 37 °C, in comparison with its expression in I+NA+ medium at 30 °C, was over 50-fold (Fig. 5, A and B).
FIGURE 5.
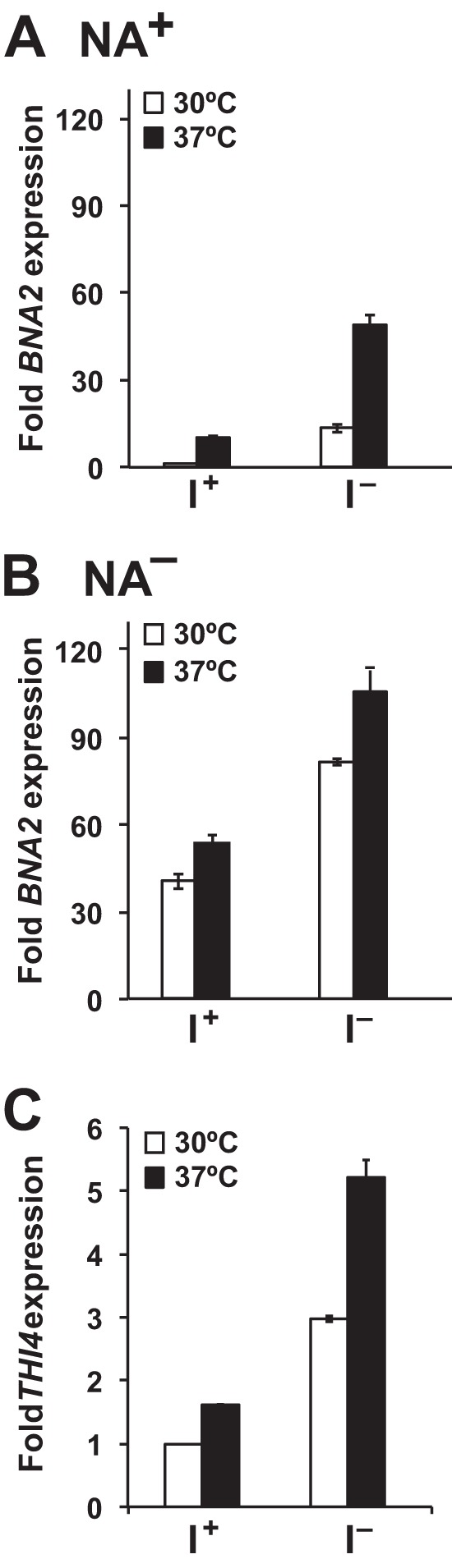
Inositol availability affects expression of the BNA2 and THI4 genes in wild type cells. Gene expression levels were determined by quantitative RT-PCR and calculated using the comparative CT method (ΔΔCT) as described under “Experimental Procedures.” Cells were grown in medium containing inositol (I+NA+ and I+NA−) to mid-logarithmic phase. The cultures were divided in half. One-half was shifted to inositol-free medium (I−NA+ and I−NA−), while maintaining constant temperature at 30 or 37 °C. The remaining culture was shifted to medium containing inositol (I+NA+ and I+NA−) and was also incubated at 30 or 37 °C. RNA samples were collected at 3 h following the media shift. The ACT1 gene served as an endogenous control. Data represent the mean ± S.E. of three independent experiments, n = 3. A, BNA2 expression levels in cells grown in NA+. B, BNA2 expression levels in cells grown in NA−. C, THI4 expression levels in cells grown in NA+. The growth conditions indicated are: 30 °C (white bars) and 37 °C (black bars).
Following a shift of wild type cells to I− medium at 30 °C in the presence of NA, consistent with our earlier microarray studies (1, 2), we observed an almost 10-fold increase in BNA2 expression (Fig. 5A). In cells shifted to I−NA+ at 37 °C, BNA2 expression reached a level 5-fold higher than the cells shifted to fresh I+NA+ medium at 37 °C. Overall, BNA2 expression was some 50-fold higher in the cells shifted to I−NA+ medium at 37 °C than in the I+NA+ cultures at 30 °C (Fig. 5A). Thus, the effects of inositol deprivation and growth at 37 °C on BNA2 expression appeared to be additive when NA was present.
An increase in BNA2 expression was also observed when wild type cells growing in I+NA− medium were shifted to I−NA− medium at 30 or 37 °C (Fig. 5B). Under these conditions, the shift to I− NA− medium resulted in BNA2 expression levels about 80-fold higher than the I+NA+ cultures at 30 °C (Fig. 5, A and B). Cells shifted to I−NA− medium at 37 °C also underwent a further increase of about 2-fold in BNA2 expression, reaching levels some 100-fold higher than those observed in wild type cells grown at 30 °C in I+NA+ medium (Fig. 5B). Thus, shifting wild type cells to inositol-free medium resulted in increased BNA2 expression at both 30 and 37 °C, whether NA was present or not. Moreover, the effects of higher growth temperature and the absence of NA and inositol on BNA2 expression appeared to be additive (Fig. 5, A and B).
The Hst1p sirtuin, which was shown by Bedalov et al. (35) to regulate BNA2 expression in response to NA, also regulates THI4 and other genes in thiamine biosynthesis in response to NA (50). To confirm that the pattern of BNA2 regulation we observed in response to inositol and temperature was also characteristic of Hst1p-regulated genes, we examined THI4 expression in wild type cells under these same growth conditions. Up-regulation of THI4 expression in response to both inositol deprivation and elevated growth temperature (Fig. 5C) was similar in pattern, although less dramatic in overall fold changes, than BNA2 (Fig. 5A). These observations are consistent with the model of coordinate regulation of genes in thiamine and NAD+ biosynthesis by the Hst1p sirtuin (50).
Cellular NAD+ Levels Increase in the Wild Type Strain in the Absence of Exogenous Inositol When NA Is Present
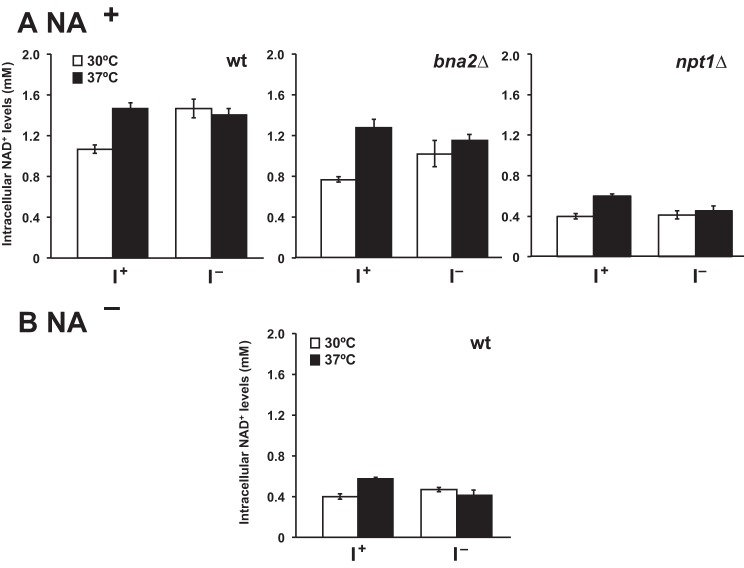
As described in the Introduction, telomeric silencing and regulation of BNA2 expression are processes dependent on the activity of two different NAD+-dependent sirtuins, Sir2p and Hst1p, respectively. Reduced NAD+ levels in wild type cells grown in the absence of NA were found to be associated with increased expression of BNA2 and other genes under the control of the Hst1p sirtuin (35, 51). Consistent with these previous reports, we found that total cellular NAD+ levels in cells grown in NA+ medium, at both 30 and 37 °C, were at least 50% lower in wild type cells grown in NA− medium as compared with NA+ medium whether inositol was present or not (Fig. 6, A and B).
FIGURE 6.
Exogenous inositol influences NAD+ levels in wild type cells grown in the presence of NA. NAD+ levels were measured as described under “Experimental Procedures” using the alcohol dehydrogenase assay. A, wild type, bna2Δ, and npt1Δ strains were grown continuously up to A600 nm = 1 in I+NA+ and I−NA+ medium at 30 and 37 °C. B, wild type cells were grown in I+NA− and I−NA− at 30 and 37 °C and were harvested at A600 nm = 1. Data represent the mean ± S.E. of at least three independent experiments, n = 3 or 4. The growth conditions indicated are: 30 °C (white bars) and 37 °C (black bars).
However, at 30 °C cellular NAD+ levels were elevated about 25% in wild type cells grown in the absence of inositol (I−NA+ medium), in comparison with the same cells grown in I+NA+ medium (Fig. 6A). Growth temperature also affected NAD+ levels in wild type cells, but again only when NA was present. In I+NA+ medium at 37 °C, wild type cells exhibited NAD+ levels about 25% higher than the levels observed in cells grown in the same medium at 30 °C (Fig. 6A). The level of cellular NAD+ observed in wild type cells growing in I+NA+ medium at 37 °C was almost identical to the level seen in the same cells growing in the absence of inositol (i.e. in I−NA+ medium) at 30 °C (Fig. 6A). However, NAD+ levels in cells growing in I−NA+ 37 °C were not significantly different from the levels seen in the same cells growing at 37 °C in I+NA+ medium (Fig. 6A). Thus, the effects of inositol deprivation and growth at 37 °C on NAD+ levels in wild type cells growing in NA+ medium were not additive.
To determine whether the increased NAD+ levels observed in wild type cells grown at 37 °C and/or in the absence of inositol were dependent on up-regulation of the de novo NAD+ and/or the salvage biosynthetic pathways, we examined the NAD+ content of bna2Δ and npt1Δ mutant strains. Because the bna2Δ strain is auxotrophic for NA, it could only be tested in medium containing NA. We found that the NAD+ levels in bna2Δ were slightly but significantly lower than in wild type cells under all growth conditions tested, with the possible exception of cells growing in I+NA+ medium at 37 °C (Fig. 6A). In general, the bna2Δ mutant, similar to wild type, exhibited nonadditive increases in cellular NAD+ levels in response to growth at 37 °C and inositol deprivation, indicating that the increased NAD+ levels observed in wild type cells under these conditions are, at least in part, independent of induction of enzymes of de novo NAD+ biosynthesis. The data do, however, indicate that Bna2p contributes to maintenance of the overall NAD+ levels achieved in wild type cells when NA is present.
In contrast to the de novo pathway, the salvage pathway for NAD+ biosynthesis clearly plays a more important role, because NAD+ levels in the npt1Δ strain were severely reduced in comparison with both the wild type and bna2Δ strains and were not influenced greatly by either temperature or inositol (Fig. 6A). Indeed, as reported by Bedalov et al. (35) the overall levels of NAD+ in the npt1Δ strain (Fig. 6A), with and without inositol supplementation at both temperatures, resembled those observed in wild type cells growing in the absence of NA (Fig. 6B). Thus, although both pathways are involved in maintaining NAD+ levels in wild type cells, it appears that the salvage pathway plays the more important role under all the conditions we examined. This is consistent with the results reported by Sandmeier et al. (52) in synthetic complete medium at 30 °C.
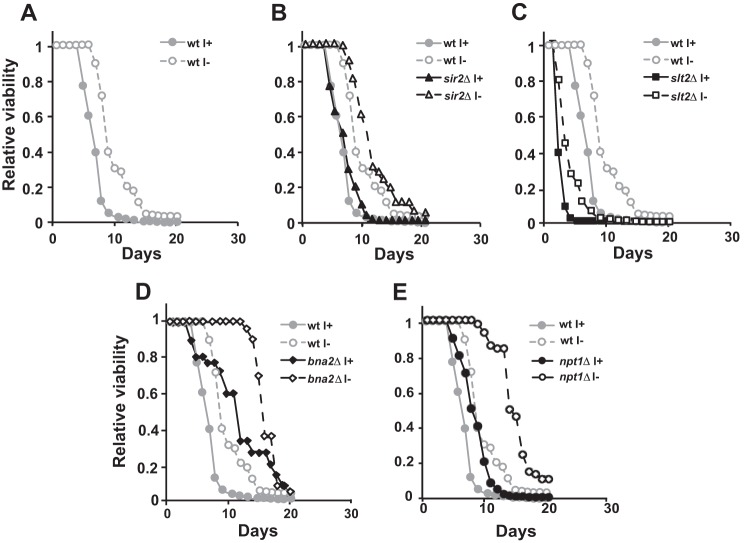
Chronological Life Span Is Extended in Wild Type Cells in the Absence of Inositol
The effect of the inositol deprivation in wild type cells on telomeric silencing suggested that inositol might also influence the extent of chronological life span in yeast cells. Accordingly, we tested the effect of inositol supplementation on chronological life span in wild type, sir2Δ, slt2Δ, bna2Δ, and npt1Δ strains. Strikingly, wild type cells growing in the absence of inositol exhibited an extended chronological life span, as compared with cells growing in medium containing inositol (Fig. 7A). However, no significant difference in chronological life span was observed between the wild type control and the sir2Δ mutant, with or without inositol supplementation (Fig. 7B). Although the slt2Δ, bna2Δ and npt1Δ strains exhibited significant decreases in telomeric silencing in comparison with wild type (Figs. 2 and 4), the effects of these mutations on chronological life span were distinctively different (Fig. 7). The slt2Δ strain exhibited a shorter chronological life span in comparison with wild type, whether inositol was present or not (Fig. 7C), whereas the bna2Δ and npt1Δ mutants exhibited chronological life span extension relative to wild type (Fig. 7, D and E). However, similar to wild type, the slt2Δ, bna2Δ, and npt1Δ strains had a relatively shorter chronological life span when grown in the presence of inositol as compared with growth in its absence (Fig. 7, C–E). Thus, the increase in chronological life span observed in wild type cells in the absence of inositol appears to be at least partially independent of the SLT2, BNA2, and NPT1 gene products.
FIGURE 7.
Absence of inositol causes an increase in yeast chronological life span. Cells were inoculated into I+ or I− medium and allowed to grow into late stationary phase. Aliquots from the cultures were diluted in water and plated into YEPD plates such that single colonies could be counted (CFU). Two dilutions per culture were used to reduce the fluctuations due to manual error. CFU were counted after 2 days. The number of CFUs at day 2 was considered to be the initial survival (100% survival) and was used to determine the age-dependent percent survival.
DISCUSSION
It has long been known that inositol-containing sphingolipid synthesis is reduced during inositol starvation (13, 53–55). It has also been shown that treating cells with AbA (56) inhibits inositol phosphorylceramide synthase activity leading to a reduction in inositol-containing sphingolipid synthesis (57). Previous work from our laboratory established that reduction in the synthesis of sphingolipids in the absence of inositol or by treatment with either AbA or myriocin triggers the activation of Slt2p, the terminal MAPK of the PKC pathway (13). However, PKC activation resulting from interruption of sphingolipid synthesis by treatment with myriocin was shown to be significantly attenuated by supplementation of dihydroxysphingosine and phytosphingosine, yet these sphingoid bases did not restore PKC signaling in cells treated with AbA (13). Moreover, supplementation of untreated wild type cells with exogenous dihydroxysphingosine or phytosphingosine did not lead to activation of PKC signaling. Together, these prior results strongly supported the conclusion that activation of PKC signaling by inhibition of sphingolipid synthesis, whether by inositol deprivation or treatment with myriocin or AbA, is due to the reduced synthesis of inositol-containing sphingolipids and not by increased levels of sphingoid bases (13).
In this study we have presented evidence for the first time that growth of wild type cells in medium lacking inositol, or treatment with AbA, also leads to increased telomeric silencing. This process is dependent on both Sir2p and Slt2p (Figs. 2 and 3). These results indicate that PKC activation induced by a reduction in the synthesis of inositol-containing sphingolipids promotes Sir2p-dependent telomeric silencing in wild type cells. Growth at 37 °C, as reported previously by Stone and Pillus (29) and Bi et al. (38), also resulted in increased Sir2p-dependent telomeric silencing (Fig. 2). Increased silencing in response to both inositol deprivation and growth at 37 °C is dependent on the activity of the de novo and salvage pathways for NAD+ biosynthesis (Fig. 4) but is not correlated to absolute cellular NAD+ levels (Fig. 6). The strengthening of telomeric silencing in wild type cells in the absence of inositol is correlated with chronological life span extension under this growth condition (Fig. 7). These observations are discussed below in the context of the effects related to activation of PKC signaling in response to inositol starvation.
Absence of Inositol Triggers Sir2p-dependent Telomeric Silencing
We have shown that depleting wild type cells of inositol or treating them with AbA promote Sir2p-dependent telomeric silencing (Figs. 2 and 3). We also point out that inositol deprivation capable of triggering this response is expected to occur in any study using synthetic complete media based on yeast nitrogen base for continuous growth. Hanscho et al. (58) reported that the concentration of inositol (10 μm) in yeast nitrogen base is too low to sustain fast proliferation of yeast until glucose is exhausted. Their findings are consistent with the report of Hirsch and Henry (59) that INO1 is derepressed by mid-logarithmic phase in wild type cells grown in yeast nitrogen base media containing 10 μm inositol. To avoid the exhaustion of inositol in the media, 75–100 μm inositol should be used to avoid the derepression of INO1 as well as activation of hundreds of genes controlled by stress response pathways, including PKC, which are induced by inositol deprivation (1, 2, 13)
Earlier studies have shown that Slt2p phosphorylates Sir3p (22, 24), a key component of the Sir complex containing Sir2p (25, 26, 28). Moreover, phosphorylation of Sir3p is associated with strengthening of silencing (29). Thus, the loss of silencing that we observed in the slt2Δ mutant is consistent with its reported role of activation of Sir3p by phosphorylation. Thus, our data are consistent with a mechanism by which inositol deprivation activates Slt2p, as shown by Jesch et al. 2010 (13), which in turn is expected to result in phosphorylation of Sir3p, as reported by Stone and Pillus (29), resulting in an increase in silencing. However, the slight decrease in telomeric silencing at 37 °C that we observed in the slt2Δ strain suggests that a secondary mechanism such as super-coiling of DNA, independent of Sir3p phosphorylation as reported previously by Bi et al. (38), could also be involved.
Increased Sir2p-dependent Telomeric Silencing in the Absence of Inositol and at 37 °C Is Dependent on Ongoing Synthesis of NAD+ but Is Not Correlated to Absolute Cellular NAD+ Levels
In yeast, cellular levels of NAD+ are maintained by a variety of mechanisms, including regeneration of NAD+ from nicotinamide via the salvage pathway, as well as activity of the de novo pathway for synthesis of NAD+ when exogenous NA is unavailable (Fig. 1). The activity of Sir2p and its function in silencing are sensitive to cellular NAD+ concentration (31, 60). Accordingly, we found that the activities of both the de novo and salvage pathways for NAD+ biosynthesis were required to maintain high levels of Sir2p-dependent telomeric silencing in response to high growth temperature as well as a lack of inositol (Figs. 4 and 5). Consistent with previous microarray studies (1, 2, 36), we observed that expression of the BNA2 gene increased in response to growth at 37 °C and to the absence of inositol, even in the presence of NA (Fig. 5). Given the well established mechanism of regulation of BNA2 by the Hst1p sirtuin (35), these results suggest that wild type cells experience higher NAD+ demand under these conditions.
Indeed, the highest level of BNA2 induction occurred in cells growing at 37 °C in medium lacking both NA and inositol (Fig. 5), a condition under which we observed low cellular levels of NAD+ (Fig. 6), consistent with the report of Bedalov et al. (35). However, given that NAD+ levels were reported to regulate the enzymatic activity of Sir2p (61), we were surprised that the highest level of Sir2p-dependent telomeric silencing also occurred under this growth condition (Fig. 4). However, it has been shown that calorie restriction, a condition that enhances Sir2p activity, does not result in increased cellular NAD+ levels (62–64). Another study supports the idea that alterations in the NAD+/NADH ratio regulate Sir2p activity (65). Our findings indicate that even though Sir2p-dependent telomeric silencing is strengthened in the absence of both NA and inositol in cells growing at 37 °C, the cellular pool of NAD+ does not increase. Despite these observations, we found that the de novo and salvage NAD+ biosynthetic pathways are both required to sustain the high level of telomeric silencing seen in wild type cells both at high growth temperature and under inositol starvation.
Growth in the Absence of Inositol Leads to Chronological Life Span Extension in Wild Type
The absence of inositol is a stress-activating condition (3, 8, 13), analogous to growth at elevated temperature, high or low osmolarity, and calorie or amino acid restriction. In yeast, many of these stresses are associated with chronological life span extension (63, 66). Consistent with the view that inositol deprivation is a stress provoking growth condition in yeast, we found that wild type cells growing in medium lacking inositol exhibit increased chronological life span (Fig. 7A). Yet, this increase in chronological life span is not dependent on Sir2p, because no significant difference in chronological life span was observed when the wild type control and the sir2Δ mutant were compared (Fig. 7B). Similarly, Sir2p is not required for calorie restriction-mediated chronological life span extension in yeast (66, 67). Instead, our results are consistent with other studies that found that blocking sphingolipid synthesis leads to chronological life span extension. For example, yeast strains lacking mannosyl-di-inositol phosphoceramide, including single and double deletions mutants in IPT1 and/or SKN1, exhibit increased chronological life span (68). Likewise, down-regulation of sphingolipid synthesis by myriocin treatment or by using the tetO7-LCB1 or tetO7-LCB2 strains has been reported to prolong yeast chronological life span (46). Together, those results and the results reported here suggest that the decrease in complex sphingolipid synthesis in the absence of inositol (13) could be mediating the chronological life span extension. Future studies are necessary to understand how sphingolipid metabolism mediates the increase in chronological life span in wild type yeast and how life span extension is influenced by various forms of cellular stress.
Acknowledgments
We thank all the members of the Henry laboratory for discussions throughout the course of this work. We are indebted to Dr. Thomas D. Petes for plasmids used in the telomeric silencing assays in this study.
This work was supported, in whole or in part, by National Institutes of Health Grant GM-19629 (to S. A. H.). This study is taken in part from the Ph.D. thesis of Sojin Lee, Cornell University, 2009.
- NA
- nicotinic acid
- AbA
- aureobasidin A
- Fam
- 6-carboxyfluorescein
- Tamra
- tetramethylrhodamine.
REFERENCES
- 1. Jesch S. A., Liu P., Zhao X., Wells M. T., Henry S. A. (2006) Multiple endoplasmic reticulum-to-nucleus signaling pathways coordinate phospholipid metabolism with gene expression by distinct mechanisms. J. Biol. Chem. 281, 24070–24083 [DOI] [PubMed] [Google Scholar]
- 2. Jesch S. A., Zhao X., Wells M. T., Henry S. A. (2005) Genome-wide analysis reveals inositol, not choline, as the major effector of Ino2p-Ino4p and unfolded protein response target gene expression in yeast. J. Biol. Chem. 280, 9106–9118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nunez L. R., Jesch S. A., Gaspar M. L., Almaguer C., Villa-Garcia M., Ruiz-Noriega M., Patton-Vogt J., Henry S. A. (2008) The cell wall integrity-map kinase pathway is essential for lipid homeostasis. J. Biol. Chem. 283, 34204–34217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Santiago T. C., Mamoun C. B. (2003) Genome expression analysis in yeast reveals novel transcriptional regulation by inositol and choline and new regulatory functions for Opi1p, Ino2p, and Ino4p. J. Biol. Chem. 278, 38723–38730 [DOI] [PubMed] [Google Scholar]
- 5. Carman G. M., Han G. S. (2009) Regulation of phospholipid synthesis in yeast. J. Lipid Res. 50, S69–S73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Carman G. M., Henry S. A. (1989) Phospholipid biosynthesis in yeast. Annu. Rev. Biochem. 58, 635–669 [DOI] [PubMed] [Google Scholar]
- 7. Carman G. M., Henry S. A. (1999) Phospholipid biosynthesis in the yeast Saccharomyces cerevisiae and interrelationship with other metabolic processes. Prog. Lipid Res. 38, 361–399 [DOI] [PubMed] [Google Scholar]
- 8. Henry S. A., Kohlwein S. D., Carman G. M. (2012) Metabolism and regulation of glycerolipids in the yeast Saccharomyces cerevisiae. Genetics 190, 317–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Henry S. A., Patton-Vogt J. L. (1998) Genetic regulation of phospholipid metabolism: yeast as a model eukaryote. Prog. Nucleic Acids Res. Mol. Biol. 61, 133–179 [DOI] [PubMed] [Google Scholar]
- 10. Loewen C. J., Gaspar M. L., Jesch S. A., Delon C., Ktistakis N. T., Henry S. A., Levine T. P. (2004) Phospholipid metabolism regulated by a transcription factor sensing phosphatidic acid. Science 304, 1644–1647 [DOI] [PubMed] [Google Scholar]
- 11. Chang H. J., Jones E. W., Henry S. A. (2002) Role of the unfolded protein response pathway in regulation of INO1 and in the sec14 bypass mechanism in Saccharomyces cerevisiae. Genetics 162, 29–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cox J. S., Chapman R. E., Walter P. (1997) The unfolded protein response coordinates the production of endoplasmic reticulum protein and endoplasmic reticulum membrane. Mol. Biol. Cell 8, 1805–1814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jesch S. A., Gaspar M. L., Stefan C. J., Aregullin M. A., Henry S. A. (2010) Interruption of inositol sphingolipid synthesis triggers Stt4p-dependent protein kinase C signaling. J. Biol. Chem. 285, 41947–41960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Villa-García M. J., Choi M. S., Hinz F. I., Gaspar M. L., Jesch S. A., Henry S. A. (2011) Genome-wide screen for inositol auxotrophy in Saccharomyces cerevisiae implicates lipid metabolism in stress response signaling. Mol. Genet. Genomics 285, 125–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gaspar M. L., Aregullin M. A., Jesch S. A., Henry S. A. (2006) Inositol induces a profound alteration in the pattern and rate of synthesis and turnover of membrane lipids in Saccharomyces cerevisiae. J. Biol. Chem. 281, 22773–22785 [DOI] [PubMed] [Google Scholar]
- 16. Gaspar M. L., Hofbauer H. F., Kohlwein S. D., Henry S. A. (2011) Coordination of storage lipid synthesis and membrane biogenesis: evidence for cross-talk between triacylglycerol metabolism and phosphatidylinositol synthesis. J. Biol. Chem. 286, 1696–1708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dickson R. C. (2008) Thematic review series: sphingolipids. New insights into sphingolipid metabolism and function in budding yeast. J. Lipid Res. 49, 909–921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Strahl T., Thorner J. (2007) Synthesis and function of membrane phosphoinositides in budding yeast, Saccharomyces cerevisiae. Biochim. Biophys. Acta 1771, 353–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Balla T. (2013) Phosphoinositides: Tiny lipids with giant impact on cell regulation. Physiol. Rev. 93, 1019–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shears S. B., Ganapathi S. B., Gokhale N. A., Schenk T. M., Wang H., Weaver J. D., Zaremba A., Zhou Y. (2012) Defining signal transduction by inositol phosphates. Subcell. Biochem. 59, 389–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ye C., Bandara W. M., Greenberg M. L. (2013) Regulation of inositol metabolism is fine-tuned by inositol pyrophosphates in Saccharomyces cerevisiae. J. Biol. Chem. 288, 24898–24908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ai W., Bertram P. G., Tsang C. K., Chan T. F., Zheng X. F. (2002) Regulation of subtelomeric silencing during stress response. Mol. Cell 10, 1295–1305 [DOI] [PubMed] [Google Scholar]
- 23. Kamada Y., Jung U. S., Piotrowski J., Levin D. E. (1995) The protein kinase C-activated MAP kinase pathway of Saccharomyces cerevisiae mediates a novel aspect of the heat shock response. Genes Dev. 9, 1559–1571 [DOI] [PubMed] [Google Scholar]
- 24. Ray A., Hector R. E., Roy N., Song J. H., Berkner K. L., Runge K. W. (2003) Sir3p phosphorylation by the Slt2p pathway effects redistribution of silencing function and shortened life span. Nat. Genet. 33, 522–526 [DOI] [PubMed] [Google Scholar]
- 25. Hecht A., Strahl-Bolsinger S., Grunstein M. (1996) Spreading of transcriptional repressor SIR3 from telomeric heterochromatin. Nature 383, 92–96 [DOI] [PubMed] [Google Scholar]
- 26. Moazed D., Johnson D. (1996) A deubiquitinating enzyme interacts with SIR4 and regulates silencing in S. cerevisiae. Cell 86, 667–677 [DOI] [PubMed] [Google Scholar]
- 27. Rine J., Herskowitz I. (1987) Four genes responsible for a position effect on expression from HML and HMR in Saccharomyces cerevisiae. Genetics 116, 9–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Strahl-Bolsinger S., Hecht A., Luo K., Grunstein M. (1997) SIR2 and SIR4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes Dev. 11, 83–93 [DOI] [PubMed] [Google Scholar]
- 29. Stone E. M., Pillus L. (1996) Activation of an MAP kinase cascade leads to Sir3p hyperphosphorylation and strengthens transcriptional silencing. J. Cell Biol. 135, 571–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Blander G., Guarente L. (2004) The Sir2 family of protein deacetylases. Annu. Rev. Biochem. 73, 417–435 [DOI] [PubMed] [Google Scholar]
- 31. Imai S., Armstrong C. M., Kaeberlein M., Guarente L. (2000) Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 403, 795–800 [DOI] [PubMed] [Google Scholar]
- 32. Rusche L. N., Kirchmaier A. L., Rine J. (2003) The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Annu. Rev. Biochem. 72, 481–516 [DOI] [PubMed] [Google Scholar]
- 33. Frye R. A. (2000) Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochem. Biophys. Res. Commun. 273, 793–798 [DOI] [PubMed] [Google Scholar]
- 34. Panozzo C., Nawara M., Suski C., Kucharczyka R., Skoneczny M., Bécam A. M., Rytka J., Herbert C. J. (2002) Aerobic and anaerobic NAD+ metabolism in Saccharomyces cerevisiae. FEBS Lett. 517, 97–102 [DOI] [PubMed] [Google Scholar]
- 35. Bedalov A., Hirao M., Posakony J., Nelson M., Simon J. A. (2003) NAD+-dependent deacetylase Hst1p controls biosynthesis and cellular NAD+ levels in Saccharomyces cerevisiae. Mol. Cell. Biol. 23, 7044–7054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gasch A. P., Spellman P. T., Kao C. M., Carmel-Harel O., Eisen M. B., Storz G., Botstein D., Brown P. O. (2000) Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 11, 4241–4257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Levin D. E. (2011) Regulation of cell wall biogenesis in Saccharomyces cerevisiae: the cell wall integrity signaling pathway. Genetics 189, 1145–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bi X., Yu Q., Sandmeier J. J., Elizondo S. (2004) Regulation of transcriptional silencing in yeast by growth temperature. J. Mol. Biol. 344, 893–905 [DOI] [PubMed] [Google Scholar]
- 39. Cornell N. W., Veech R. L. (1983) Enzymatic measurement of ethanol or NAD in acid extracts of biological samples. Anal. Biochem. 132, 418–423 [DOI] [PubMed] [Google Scholar]
- 40. Smith J. S., Avalos J., Celic I., Muhammad S., Wolberger C., Boeke J. D. (2002) SIR2 family of NAD+-dependent protein deacetylases. Methods Enzymol. 353, 282–300 [DOI] [PubMed] [Google Scholar]
- 41. Belenky P., Racette F. G., Bogan K. L., McClure J. M., Smith J. S., Brenner C. (2007) Nicotinamide riboside promotes Sir2 silencing and extends life span via Nrk and Urh1/Pnp1/Meu1 pathways to NAD+. Cell 129, 473–484 [DOI] [PubMed] [Google Scholar]
- 42. Sherman F. (2002) Getting started with yeast. Methods Enzymol. 350, 3–41 [DOI] [PubMed] [Google Scholar]
- 43. Sporty J. L., Kabir M. M., Turteltaub K. W., Ognibene T., Lin S. J., Bench G. (2008) Single sample extraction protocol for the quantification of NAD and NADH redox states in Saccharomyces cerevisiae. J. Sep. Sci. 31, 3202–3211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mortimer R. K., Johnston J. R. (1959) Life span of individual yeast cells. Nature 183, 1751–1752 [DOI] [PubMed] [Google Scholar]
- 45. Fabrizio P., Longo V. D. (2003) The chronological life span of Saccharomyces cerevisiae. Aging Cell 2, 73–81 [DOI] [PubMed] [Google Scholar]
- 46. Huang X., Liu J., Dickson R. C. (2012) Down-regulating sphingolipid synthesis increases yeast life span. PLoS Genet. 8, e1002493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Craven R. J., Petes T. D. (2000) Involvement of the checkpoint protein Mec1p in silencing of gene expression at telomeres in Saccharomyces cerevisiae. Mol. Cell. Biol. 20, 2378–2384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Gottschling D. E., Aparicio O. M., Billington B. L., Zakian V. A. (1990) Position effect at S. cerevisiae telomeres: reversible repression of Pol II transcription. Cell 63, 751–762 [DOI] [PubMed] [Google Scholar]
- 49. Smith J. S., Boeke J. D. (1997) An unusual form of transcriptional silencing in yeast ribosomal DNA. Genes Dev. 11, 241–254 [DOI] [PubMed] [Google Scholar]
- 50. Li M., Petteys B. J., McClure J. M., Valsakumar V., Bekiranov S., Frank E. L., Smith J. S. (2010) Thiamine biosynthesis in Saccharomyces cerevisiae is regulated by the NAD+-dependent histone deacetylase Hst1. Mol. Cell. Biol. 30, 3329–3341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Evans C., Bogan K. L., Song P., Burant C. F., Kennedy R. T., Brenner C. (2010) NAD+ metabolite levels as a function of vitamins and calorie restriction: evidence for different mechanisms of longevity. BMC Chem. Biol. 10, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sandmeier J. J., Celic I., Boeke J. D., Smith J. S. (2002) Telomeric and rDNA silencing in Saccharomyces cerevisiae are dependent on a nuclear NAD+ salvage pathway. Genetics 160, 877–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Becker G. W., Lester R. L. (1977) Changes in phospholipids of Saccharomyces cerevisiae associated with inositol-less death. J. Biol. Chem. 252, 8684–8691 [PubMed] [Google Scholar]
- 54. Hanson B. A., Lester R. L. (1980) The extraction of inositol-containing phospholipids and phosphatidylcholine from Saccharomyces cerevisiae and Neurospora crassa. J. Lipid Res. 21, 309–315 [PubMed] [Google Scholar]
- 55. Alvarez-Vasquez F., Sims K. J., Cowart L. A., Okamoto Y., Voit E. O., Hannun Y. A. (2005) Simulation and validation of modelled sphingolipid metabolism in Saccharomyces cerevisiae. Nature 433, 425–430 [DOI] [PubMed] [Google Scholar]
- 56. Endo M., Takesako K., Kato I., Yamaguchi H. (1997) Fungicidal action of aureobasidin A, a cyclic depsipeptide antifungal antibiotic, against Saccharomyces cerevisiae. Antimicrob. Agents Chemother. 41, 672–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Nagiec M. M., Nagiec E. E., Baltisberger J. A., Wells G. B., Lester R. L., Dickson R. C. (1997) Sphingolipid synthesis as a target for antifungal drugs. Complementation of the inositol phosphorylceramide synthase defect in a mutant strain of Saccharomyces cerevisiae by the AUR1 gene. J. Biol. Chem. 272, 9809–9817 [DOI] [PubMed] [Google Scholar]
- 58. Hanscho M., Ruckerbauer D. E., Chauhan N., Hofbauer H. F., Krahulec S., Nidetzky B., Kohlwein S. D., Zanghellini J., Natter K. (2012) Nutritional requirements of the BY series of Saccharomyces cerevisiae strains for optimum growth. FEMS Yeast Res. 12, 796–808 [DOI] [PubMed] [Google Scholar]
- 59. Hirsch J. P., Henry S. A. (1986) Expression of the Saccharomyces cerevisiae inositol-1-phosphate synthase (INO1) gene is regulated by factors that affect phospholipid synthesis. Mol. Cell. Biol. 6, 3320–3328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Smith J. S., Brachmann C. B., Celic I., Kenna M. A., Muhammad S., Starai V. J., Avalos J. L., Escalante-Semerena J. C., Grubmeyer C., Wolberger C., Boeke J. D. (2000) A phylogenetically conserved NAD+-dependent protein deacetylase activity in the Sir2 protein family. Proc. Natl. Acad. Sci. U.S.A. 97, 6658–6663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Lin S. J., Guarente L. (2003) Nicotinamide adenine dinucleotide, a metabolic regulator of transcription, longevity and disease. Curr. Opin. Cell Biol. 15, 241–246 [DOI] [PubMed] [Google Scholar]
- 62. Anderson R. M., Bitterman K. J., Wood J. G., Medvedik O., Cohen H., Lin S. S., Manchester J. K., Gordon J. I., Sinclair D. A. (2002) Manipulation of a nuclear NAD+ salvage pathway delays aging without altering steady-state NAD+ levels. J. Biol. Chem. 277, 18881–18890 [DOI] [PubMed] [Google Scholar]
- 63. Anderson R. M., Bitterman K. J., Wood J. G., Medvedik O., Sinclair D. A. (2003) Nicotinamide and PNC1 govern life span extension by calorie restriction in Saccharomyces cerevisiae. Nature 423, 181–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Ashrafi K., Lin S. S., Manchester J. K., Gordon J. I. (2000) Sip2p and its partner snf1p kinase affect aging in S. cerevisiae. Genes Dev. 14, 1872–1885 [PMC free article] [PubMed] [Google Scholar]
- 65. Lin S. J., Ford E., Haigis M., Liszt G., Guarente L. (2004) Calorie restriction extends yeast life span by lowering the level of NADH. Genes Dev. 18, 12–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Smith D. L., Jr., McClure J. M., Matecic M., Smith J. S. (2007) Calorie restriction extends the chronological life span of Saccharomyces cerevisiae independently of the Sirtuins. Aging Cell 6, 649–662 [DOI] [PubMed] [Google Scholar]
- 67. Fabrizio P., Gattazzo C., Battistella L., Wei M., Cheng C., McGrew K., Longo V. D. (2005) Sir2 blocks extreme life-span extension. Cell 123, 655–667 [DOI] [PubMed] [Google Scholar]
- 68. Aerts A. M., François I. E., Bammens L., Cammue B. P., Smets B., Winderickx J., Accardo S., De Vos D. E., Thevissen K. (2006) Level of M(IP)2C sphingolipid affects plant defensin sensitivity, oxidative stress resistance and chronological life-span in yeast. FEBS Lett. 580, 1903–1907 [DOI] [PubMed] [Google Scholar]