Background: The mechanism of botulinum neurotoxin D light chain (LC/D) substrate recognition is not well defined.
Results: A dual recognition strategy employed by LC/D was revealed, in which one site of VAMP-2 was recognized by two independent, functionally similar LC/D sites that were complementary to each other.
Conclusion: LC/D utilizes a unique mechanism for substrate recognition.
Significance: This study provides insights for LC/D engineering and antitoxin development.
Keywords: Bacterial Toxins, Enzyme Catalysis, Enzyme Kinetics, Metalloprotease, Neurotoxin
Abstract
Botulinum neurotoxins are the most potent protein toxins in nature. Despite the potential to block neurotransmitter release at the neuromuscular junction and cause human botulism, they are widely used in protein therapies. Among the seven botulinum neurotoxin serotypes, mechanisms of substrate recognition and specificity are known to a certain extent in the A, B, E, and F light chains, but not in the D light chain (LC/D). In this study, we addressed the unique substrate recognition mechanism of LC/D and showed that this serotype underwent hydrophobic interactions with VAMP-2 at its V1 motif. The LC/D B3, B4, and B5 binding sites specifically recognize the hydrophobic residues in the V1 motif of VAMP-2. Interestingly, we identified a novel dual recognition mechanism employed by LC/D in recognition of VAMP-2 sites at both the active site and distal binding sites, in which one site of VAMP-2 was recognized by two independent, but functionally similar LC/D sites that were complementary to each other. The dual recognition strategy increases the tolerance of LC/D to mutations and renders it a good candidate for engineering to improve its therapeutic properties. In conclusion, in this study, we identified a unique multistep substrate recognition mechanism by LC/D and provide insights for LC/D engineering and antitoxin development.
Introduction
The seven existing botulinum neurotoxin (BoNT)2 serotypes, A–G, are the most potent protein toxins. BoNT causes human and animal botulism, a flaccid paralysis caused by the blocking of neurotransmitter (usually acetylcholine) release at the neuromuscular junction. Among the seven different serotypes, BoNT/A, BoNT/B, BoNT/E, and BoNT/F are involved in human botulism, whereas BoNT/D is responsible mainly for animal botulism (1–3).
BoNTs are zinc-dependent proteases and contain a His-Glu-X-X-His zinc-binding motif of metalloendopeptidases in the central region of their light chains (4). BoNTs are 150-kDa dichain proteins with typical A (active)-B (binding) structure-function. BoNTs are activated by proteolysis to generate dichain organization (2). There are three domains found for BoNTs, including an N-terminal catalytic domain (light chain (LC)), an internal translocation domain (heavy chain), and a C-terminal receptor-binding domain (heavy chain) (5). The SNARE complex, formed by the vesicle SNARE (VAMP-2 (vesicle-associated membrane protein-2)) and the plasma membrane SNAREs (SNAP25 and syntaxin 1a), is the driving force of vesicle fusion during the mammalian neuronal exocytosis process (6). These SNARE proteins are the targets of the seven BoNTs: serotypes B, D, F, and G cleave VAMP-2; serotypes A and E cleave SNAP25; and serotype C cleaves both SNAP25 and syntaxin 1a (2).
Due to their extreme toxicity and ease of production, handling, and delivery through aerosol or liquid route, BoNTs represent a potential biological warfare agent and have been classified as a category A agent by the Center for Disease Control and Prevention in the United States (7, 8). However, another key feature of BoNTs is that their intoxication can be reversed by the replacement of affected nerves with new ones (5, 9, 10), thus making BoNTs effective agents for the therapies of a range of neuromuscular disorders such as strabismus (11, 12). A better understanding of the mechanism of action and substrate recognition of BoNTs will enable us to develop an antidote for BoNT intoxication and novel therapies to extend its clinical applications. Recently, the mechanisms of BoNT/A, BoNT/B, BoNT/E, and BoNT/F have been thoroughly studied, with results indicating that an extended region (exosite) is necessary for substrate binding and cleavage (13–16). In 2006, Arndt et al. (17) reported the structure of the light chain of BoNT/D and proposed the recognition of the hydrophobic SNARE V1 motif through structural comparison with LC/F. Sikorra et al. (18) reported the contribution of VAMP-2 residues to LC/D substrate recognition and found that LC/D requires a relatively short sequence for optimal substrate cleavage compared with other serotypes such as LC/F and the tetanus neurotoxin LC. However, the mechanism underlying the effective recognition and cleavage of VAMP-2 by LC/D is still unclear, which prompted us in this study to depict the mechanism of substrate recognition and specificity by LC/D.
Our data show that, similar to other serotypes, LC/D recognition of VAMP-2 occurs though multistep binding and, in particular, recognition of VAMP-2 sites by LC/D substrate recognition pockets. Interestingly, in contrast to all other BoNT LCs and metalloproteinases, LC/D employs a novel dual recognition mechanism, in which one VAMP-2 site is recognized by two independent LC/D sites that are complementary to each other. The dual recognition strategy increases the tolerance of LC/D to mutations and makes it a good candidate for engineering to improve its pharmacological properties.
EXPERIMENTAL PROCEDURES
Molecular Modeling
The structure of the LC/D·VAMP-2 complex was modeled and analyzed using SWISS-MODEL and refined using PyMOL as described previously (15).
Plasmid Construction and Protein Expression
Codon-optimized DNA encoding the LC domain of BoNT/D (residues 1–430) was synthesized by EZBiolab (Westfield, IN). The PCR product was then subcloned into the pET-15b vector and transformed into Escherichia coli BL21(DE3)-RIL cells (Stratagene). Protein expression and purification were achieved as described previously (14, 15, 19). VAMP-2(1–97) was constructed by PCR amplification of the cDNA clone purchased from American Type Culture Collection (GenBankTM accession number NM_014232) and subcloning into pGEX-2T. The plasmid containing the VAMP-2(1–97) fragment was transformed into E. coli BL21(DE3) cells, and expression and purification of VAMP-2 were achieved as described previously (14, 15, 19).
VAMP-2 and LC/D Mutagenesis
The introduction of point mutations into LC/D and VAMP-2 genes was also performed using the QuikChange® protocol as described previously (15, 16). Plasmids were sequenced to confirm the mutations and that additional mutations were not present within the open reading frame of VAMP-2 and LC/D. Mutant proteins were produced and purified as described previously (15, 16).
Linear Velocity and Kinetic Constants
Determination of the linear velocity and kinetic constants of LC/D and its derivatives was performed as described previously (14, 15, 19). Briefly, 10 μm VAMP-2 or the indicated VAMP-2 derivatives were incubated with various concentrations of LC/D or its derivatives in 10 μl of reaction buffer (10 mm Tris-HCl (pH 7.6) and 20 mm NaCl) at 37 °C for 20 min. The reactions were stopped by adding an equal volume of SDS-PAGE sample buffer, boiled at 100 °C for 5 min, and analyzed for the relative abundance of the substrate and cleaved product by SDS-PAGE (12% polyacrylamide gels). The amount of cleaved VAMP-2 was determined by densitometry. Km and kcat determinations were performed using the same assay, in which VAMP-2 concentrations were adjusted to between 1 and 20 μm to achieve ∼10% cleavage by LC/D or its derivatives. The amount of cleaved VAMP-2 substrate was determined by densitometry, and the velocity was determined by dividing the amount of substrate cleavage by the reaction time. The reaction velocity against the substrate concentration was fitted to the Michaelis-Menten equation, and kinetic constants were derived using GraphPad. At least five independent assays were performed to determine the kinetic constants for each protein.
Trypsin Digestion of LC/D and Its Derivatives
10 μm LC/D and its derivatives were incubated with 2 mm trypsin in a 20-μl reaction volume at 37 °C for 30 min. The reactions were stopped by adding SDS-PAGE sample buffer, subjected to SDS-PAGE, and stained to visualize the partial trypsin digestion profiles.
Far-UV Circular Dichroism Analysis
LC/D and its derivatives were subjected to far-UV CD analysis. CD spectroscopy was performed at a wavelength range of 200–250 nm at room temperature with a JASCO J-810 spectropolarimeter. Far-UV CD data were obtained with a 10-mm path length quartz cuvette containing 500 μl of protein solution (0.1–0.4 mg/ml protein in 10 mm Tris-HCl (pH 7.9) and 20 mm NaCl) at a scanning speed of 50 nm/min and a 2-s response time. Each sample was measured in triplicate, the CD data were converted to molar ellipticity, and the spectrum was generated using GraphPad.
RESULTS
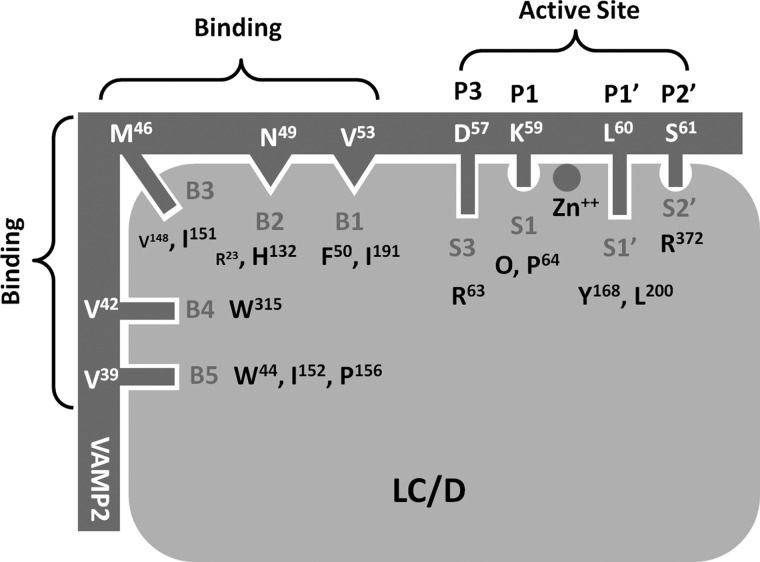
A previous study has shown that several residues of VAMP-2 contribute to substrate cleavage by LC/D, including Val39, Val42, Met46, Val49, Asp53, Lys59, Leu60, and Ser61 (18). To better quantify the degree of contribution of these residues to LC/D substrate cleavage, linear velocity assays were performed to depict the impact of alanine mutagenesis of these residues. The results show that amino acid changes in VAMP-2, including V39A, V42A, M46A, V49A, D53A, D57A, K59A, L60A, and S61A, caused ∼25-, 10-, 125-, 20-, 25-, 10-, 20-, 25-, and 20-fold reductions in LC/D cleavage, respectively. These data confirm that the residues in VAMP-2 contribute significantly to LC/D substrate recognition and cleavage. Analysis of the modeled structure of the LC/D·VAMP-2 complex identified three putative substrate recognition pockets in the active site of LC/D, S2′, S1′, and S3, which may specifically recognize the VAMP-2 P2′ (Ser61), P1′ (Leu60), and P3 (Asp53) sites, respectively (Fig. 1). In addition, we also predicted several other substrate-binding pockets distal to the active site of LC/D, including the B1–B5 binding sites (Fig. 1). To characterize the substrate recognition pockets and to confirm the specific recognition of VAMP-2 sites, the LC/D residues constituting the substrate recognition pockets were mutated to different amino acid residues to test the effects of these changes on substrate recognition. To exclude the possibility that the effect was due to the overall conformational changes as a result of the mutations, we performed partial trypsin digestion on different mutant proteins, and the result indicated that all of the LC/D mutants had an identical digestion profile to WT-LC/D, suggesting that the mutations did not cause any conformational change in LC/D (data not shown). In addition, far-UV CD analysis of LC/D and its derivatives indicated that LC/D(I151D) had a slightly different spectrum compared with WT-LC/D, whereas all other LC/D derivatives had the same far-UV CD spectrum as WT-LC/D (Fig. 2). The curve vertexes of LC/D and its derivatives at ∼240 nm looked different, which is probably due to the high degree of flexibility of LC/D owing to its relatively high number of turns and random coils. Therefore, the different curves at ∼240 nm did not reflect the conformational changes of LC/D derivatives (Fig. 2). The CD spectra of the other LC/D derivatives were similar to that of WT-LC/D (data not shown).
FIGURE 1.
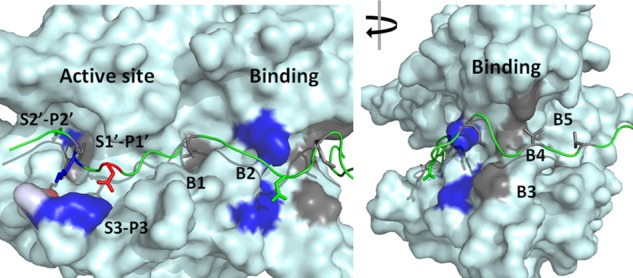
Overall view of the modeled LC/D·VAMP-2 complex structure. Left panel, view of the active site side; right panel, view after a 90° clockwise turn. LC/D is shown as a surface structure, and VAMP-2 is shown as a ribbon structure. The active site recognition and binding site interactions are highlighted. Negatively charged residues are shown in red, positively charged residues are shown in blue, hydrophobic residues are shown in gray, and polar residues are shown in green.
FIGURE 2.
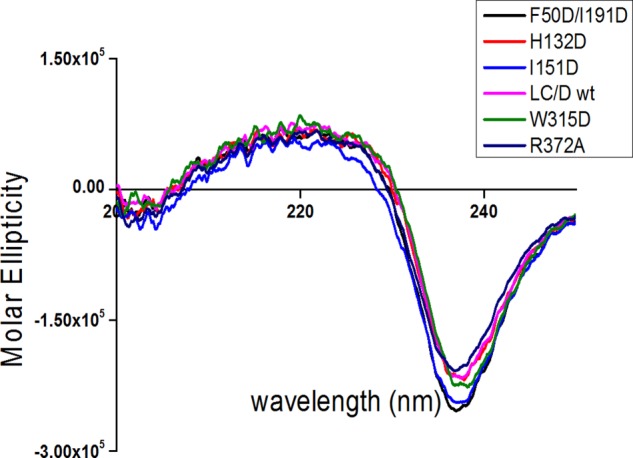
CD spectroscopy analysis of LC/D and its derivatives. Far-UV CD (200–250 nm) data were obtained for LC/D and its derivatives with a JASCO J-810 spectropolarimeter at room temperature. The data for the most representative LC/D derivatives are shown in different colors.
Recognition of the P2′ Site (Ser61) of VAMP-2 by the S2′ Pocket of LC/D
The P2′ site (Ser61) of VAMP-2 plays a certain role in LC/D substrate recognition, as the VAMP-2(S61A) mutation reduced LC/D substrate hydrolysis by ∼20-fold. A S2′ pocket in LC/D that recognized the P2′ site of VAMP-2 at Ser61 was identified through analysis of the modeled structure of the complex of LC/D and VAMP-2. The S2′ pocket is composed of Arg372, and the R372A mutation resulted in an ∼40-fold reduction of LC/D activity, with almost the same Km and an ∼40-fold lower kcat compared with WT-LC/D (Fig. 3a and Table 1). These data suggest that the S2′ pocket (Arg372) of LC/D may recognize the P2′ site of VAMP-2 by forming a hydrogen bond between these two residues.
FIGURE 3.
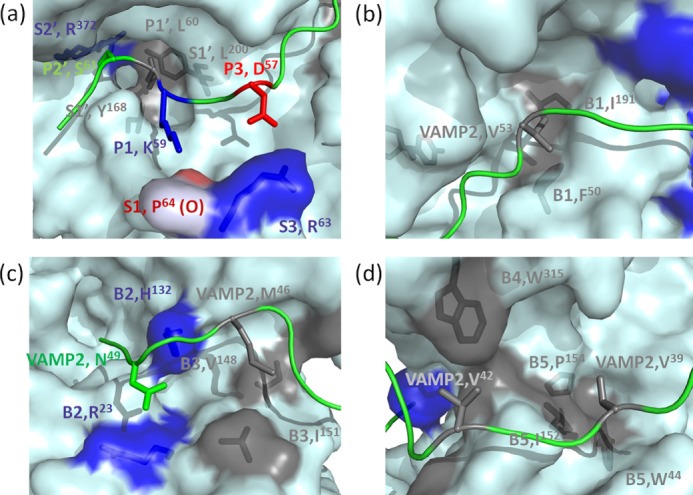
Specific recognition of VAMP-2 by LC/D pockets. Shown are surface representations of the recognition of different P sites of VAMP-2 by the S pockets and recognition of VAMP-2-binding sites by the B1–B5 binding sites of LC/D. Negatively charged residues are shown in red, positively charged residues are shown in blue, hydrophobic residues are shown in gray, and polar residues are shown in green. a, recognition of the P sites of VAMP-2 by the active site pockets of LC/D. The P2′ site (Ser61) of VAMP-2 is recognized by the S2′ pocket (Arg372). The P1′ site (Leu60) of VAMP-2 is recognized by the S1′ pocket (Tyr168 and Leu200) of LC/D. The P1 site (Lys59) of VAMP-2 interacts with the oxygen atom of Pro64 of LC/D. The P3 site (Asp57) of VAMP-2 is recognized by the S3 pocket (Arg63) of LC/D. b, recognition of Val53 by the B1 binding site (Phe50 and Ile191) of LC/D. c, recognition of Asn49 of VAMP-2 by the B2 binding site (Arg23 and His132) of LC/D and recognition of Met46 of VAMP-2 by the B3 binding site (Val148 and Ile151) of LC/D. d, recognition of Val42 by the B4 binding site (Trp315) of LC/D and recognition of Val39 by the B5 binding site (Trp44, Ile152, and Pro154) of LC/D.
TABLE 1.
Efficiency of VAMP-2 hydrolysis and kinetic constants of LC/D and its derivatives
| LC/D pockets | VAMP-2 site recognition | LC/D derivatives | Activity reductiona | Km | kcat | kcat/Km |
|---|---|---|---|---|---|---|
| -fold | μm | s−1 | s−1 μm−1 | |||
| WT- LC/D | 1 | 2.91 (0.74)b | 6.88 | 2.36 | ||
| AS-S2′c | P2′ (Ser61) | R372A | 40 | 2.09 (0.23) | 0.17 | 8.1 × 10−2 |
| AS-S1′ | P1′ (Leu60) | Y168A | 1 | —d | — | — |
| Y168D | 2 | — | — | — | ||
| L200A | 2 | — | — | — | ||
| L200D | 8 | 2.40 (0.36) | 0.81 | 3.4 × 10−1 | ||
| Y168A/L200A | 2 | — | — | — | ||
| Y168D/L200D | 60 | 3.33 (0.66) | 0.12 | 3.6 × 10−2 | ||
| AS-S3 | P3 (Asp57) | R63A | 10 | 2.19 (0.24) | 0.40 | 1.8 × 10−1 |
| R63E | 50 | 2.26 (0.35) | 0.15 | 6.6 × 10−2 | ||
| Binding B1 | Val53 | F50A | 1 | — | — | — |
| I191A | 1 | — | — | — | ||
| F50D | 4 | 3.24 (0.12) | 1.60 | 4.9 × 10−1 | ||
| I191D | 4 | 3.56 (0.21) | 1.40 | 3.9 × 10−1 | ||
| F50A/I191A | 60 | 4.81 (0.64) | 0.28 | 5.8 × 10−2 | ||
| F50D/I191D | 400 | 4.46 (0.57) | 0.04 | 8.9 × 10−3 | ||
| Binding B2 | Asn49 | R23A | 1 | — | — | — |
| H132A | 1 | — | — | — | ||
| H132D | NDd | NDb | ND | ND | ||
| H132Q | 100 | 35.56 (3.48) | 0.86 | 2.4 × 10−2 | ||
| R23D/H132A | 25 | 32.87 (5.25) | 2.38 | 7.2 × 10−2 | ||
| Binding B3 | Met46 | V148A | 1 | — | — | — |
| V148D | 1 | — | — | — | ||
| I151A | 2 | — | — | — | ||
| I151D | 1000 | 7.96 (1.284) | 0.02 | 2.5 × 10−3 | ||
| V148/I151A | 15 | |||||
| Binding B4 | Val42 | W315A | 20 | 9.90 (1.51) | 1.52 | 1.5 × 10−1 |
| W315D | 40 | 47.63 (15.12) | 3.92 | 8.2 × 10−2 | ||
| Binding B5 | Val39 | W44D | 2 | — | — | — |
| I152D | 2 | — | — | — | ||
| P154D | 4 | 11.86 (3.46) | 6.35 | 5.4 × 10−1 | ||
| W44A/I152A/P154A | 20 | 32.45 (8.87) | 5.86 | 1.8 × 10−1 |
a WT-VAMP-2 hydrolysis was measured as the ratio of the amount of LC/D derivatives needed to cleave 50% of WT-VAMP-2 to the amount of WT-LC/D needed to cleave 50% of WT-VAMP-2.
b The numbers in parentheses are the S.E. of at least five independent experiments.
c AS, active site; ND, not detectable. The mutant was too inactive to determine its kinetic constants in our experiments.
d —, kinetic constants were not determined.
Dual Recognition of the VAMP-2 P1′ Site by the S1′ Pocket of LC/D
The P1′ site of VAMP-2 was shown to be important for LC/D substrate recognition, as seen in other serotypes of BoNT (14, 15, 20, 21). Mutation of the VAMP-2 P1′ residue (L60A) reduced LC/D substrate hydrolysis by ∼25-fold. A corresponding S1′ pocket composed of two hydrophobic residues, Tyr168 and Leu200, was identified in LC/D (Fig. 3a). The LC/D(Y168A) mutation had no effect on LC/D substrate hydrolysis, and LC/D(Y168D) affected substrate hydrolysis by only ∼2-fold. The LC/D(L200A) mutation resulted in an ∼2-fold reduction of LC/D substrate hydrolysis, whereas LC/D(L200D) affected substrate hydrolysis by ∼8-fold (Table 1). The complementary effect of Tyr168 and Leu200 was also examined, and we found that although LC/D(Y168A/L200A) resulted in only an ∼2-fold reduction of substrate hydrolysis, LC/D(Y168D/L200D) reduced substrate hydrolysis by 60-fold, with no effect on Km and an ∼60-fold reduction of kcat. These data suggest that a hydrophobic S1′ pocket is necessary to maintain the full recognition of VAMP-2 Leu60 and that both hydrophobic residues in the S1′ pocket, Tyr168 and Leu200, play a complementary role in Leu60 recognition.
The S3 Pocket Residue of LC/D Interacts with the P3 Residue (Asp57) of VAMP-2
Asp57 at the P3 site of VAMP-2 plays a certain role in LC/D substrate recognition, as the D57A mutation reduced LC/D substrate hydrolysis by ∼20-fold. The S3 pocket of LC/D that specifically recognized the P3 site residue (Asp57) of VAMP-2 contains Arg63. The R63A mutation had almost no effect on Km, but reduced substrate catalysis by ∼13-fold (Fig. 3a and Table 1). The charge reversal mutation LC/D(R63E) caused an ∼50-fold reduction of LC/D substrate hydrolysis, with no effect on Km and an ∼50-fold reduction of kcat. These data suggest that a salt bridge or a side chain hydrogen bond is important for recognition of Asp57 of VAMP-2 by Arg63 of LC/D.
The Main Chain Oxygen Atom of LC/D Pro64 Interacts with the VAMP-2 P1 Site Residue (Lys59)
There was no obvious residue or pocket that showed an interaction with the P1 site residue (Lys59) of VAMP-2. The VAMP-2(K59A) mutation caused an ∼20-fold reduction of LC/D substrate hydrolysis, suggesting a role for Lys59 in LC/D substrate recognition. Structural analysis indicated that Lys59 could potentially interact with the oxygen atom of LC/D Pro64 through formation of a hydrogen bond. This interaction could not be tested through mutational analysis (Fig. 3a).
Substrate Binding and Recognition Distal to the Active Site of LC/D
Recognition of VAMP-2 Val53 by the LC/D B1 Binding Site
The VAMP-2(V53A) mutation caused an ∼25-fold reduction of LC/D hydrolysis. The B1 binding site of LC/D, which is formed by two residues, Phe50 and Ile191, may interact with Val53 (Fig. 3b). The LC/D(F50A) and LC/D(I191A) mutations had no effect on LC/D substrate hydrolysis, whereas LC/D(F50D) and LC/D(I191D) caused an ∼4-fold reduction of VAMP-2 hydrolysis. To test for the complementary effect of these two residues on Val53 recognition, the effects of double mutations were tested. The double mutation LC/D(F50A/I191A) resulted in an ∼60-fold reduction of substrate hydrolysis, with an ∼2-fold increase in Km and an ∼25-fold decrease in kcat, whereas LC/D(F50D/I191D) reduced substrate hydrolysis by ∼400-fold, with an ∼2-fold increase in Km and an ∼200-fold decrease in kcat (Table 1). These data suggest that Phe50 and Ile191 have independent but complementary effects on Val53 substrate recognition. The recognition of Val53 might contribute mainly to the fine orientation of VAMP-2 for optimal substrate recognition in the active site of LC/D because this recognition site contributed mainly to substrate catalysis (kcat), but not substrate binding (Km).
Recognition of VAMP-2 Asn49 by the LC/D B2 Binding Site
The VAMP-2(N49A) mutation was associated with an ∼20-fold reduction in cleavage efficiency of LC/D. The B2 binding site, which is composed of Arg23 and His132 of LC/D and may interact with Asn49, was revealed though analysis of the structure of the LC/D·VAMP-2 complex (Fig. 3c). The LC/D(R23A) and LC/D(H132A) mutations had no effect on LC/D substrate hydrolysis (Table 1). Surprisingly, the charge reversal mutation LC/D(R23D) still maintained the full activity on VAMP-2 as did WT-LC/D. The charge reversal mutation LC/D(H132D) became inactive in cleaving VAMP-2, whereas LC/D(H132Q) reduced substrate hydrolysis by ∼100-fold (Table 1), suggesting that the formation of a hydrogen bond between LC/D His132 and VAMP-2 Asn49 may contribute to this recognition and that a negatively charged residue at position 132 may impair this interaction. However, the minimal effect of the LC/D(H132A) mutation on LC/D substrate hydrolysis may be due to the complementary effect of Arg23. To test this hypothesis, the effect of R23D and H132A double mutations on LC/D substrate hydrolysis was tested. LC/D(R23D/H132A) reduced Km by ∼11-fold and kcat by ∼3-fold (Table 1), suggesting that both Arg23 and His132 play a role in LC/D substrate catalysis and that His132 plays a dominant role in Asn49 recognition. This substrate recognition contributes significant to substrate binding.
Recognition of VAMP-2 Met46 by the LC/D B3 Binding Site
Compared with Asn49, Met46 of VAMP-2 plays a more important role in VAMP-2 recognition by LC/D. Consistent with previous data (18), under our assay conditions, the VAMP-2(M46A) mutation reduced LC/D substrate hydrolysis by ∼125-fold. Structural analysis also identified the B3 binding site, which is composed of two residues, Ile151 and Val148, and which may interact with Met46 (Fig. 3c). The point mutations associated with the LC/D(V148A) and LC/D(I151A) alterations did not show any effect on VAMP-2 hydrolysis. LC/D(V148D) also did not have any impact on VAMP-2 hydrolysis, but LC/D(I151D) reduced LC/D substrate hydrolysis by ∼1000-fold (Table 1). The dramatic effect of the LC/D(I151D) mutation may be partially due to its conformational change based on our far-UV CD analysis. Similar to Asn49 recognition, the minimal effect of the LC/D(I151A) mutation may be related to the complementary effect of Val148. To test this hypothesis, the effect of Ile151 and Val148 double mutations on LC/D substrate hydrolysis was tested. LC/D(I151A/V148D) reduced Km by ∼10-fold and kcat by ∼2-fold (Table 1), suggesting that both Ile151 and Val148 play a role in LC/D substrate catalysis, with Ile151 playing a dominant role in substrate recognition. This substrate recognition contributes significantly to substrate binding.
Recognition of VAMP-2 Val42 by the LC/D B4 Binding Site
Val42 of VAMP-2 is also important for LC/D substrate hydrolysis, and the VAMP-2(V42A) mutation affected LC/D cleavage of VAMP-2 by ∼10-fold. Based on the modeled structure of the LC/D·VAMP-2 complex, LC/D Trp315 in the B4 binding site was predicted to have a direct interaction with Val42 (Fig. 3d). The LC/D(W315A) mutation reduced VAMP-2 hydrolysis by ∼20-fold, with an ∼4-fold increase in Km and an ∼5-fold decrease in kcat, whereas LC/D(W315D) reduced VAMP-2 hydrolysis by ∼40-fold, with an ∼20-fold increase in Km and an ∼2-fold decrease in kcat. These data suggest that LC/D Trp315 and VAMP-2 Val42 substrate recognition contributes significantly to substrate binding.
Recognition of VAMP-2 Val39 by the LC/D B5 Binding Site
Another hydrophobic residue in VAMP-2 that contributes to LC/D substrate hydrolysis is Val39. The mutation VAMP-2(V39A) resulted in an ∼25-fold reduction of LC/D hydrolysis. A hydrophobic pocket in LC/D, the B5 binding site, which is formed by Trp44, Ile152, and Pro154, was identified through analysis of the structure of the LC/D·VAMP-2 complex (Fig. 3d). The W44D, P152D, and P154D mutations were associated with ∼2-, 2-, and 4-fold reductions of substrate hydrolysis, respectively (Table 1). These mutations had no effect on kcat, but caused a 4-fold increase in Km, suggesting a role for this pocket in substrate binding. Interestingly, the triple mutation LC/D(W44A/I152A/P154A) resulted in an ∼20-fold increase in Km, but had no effect on kcat (Table 1), suggesting a complementary effect of these three residues on Val39 recognition and VAMP-2 binding.
DISCUSSION
It was proposed that botulinum neurotoxins recognize their substrates through two separate regions, one that contains the scissile bond and the other distal to the scissile bond and containing the SNARE motif (22). A two-region substrate recognition model has been demonstrated in LC/A, LC/B, LC/E, LC/F, and the tetanus neurotoxin LC (14, 15, 20, 21). The significance of the SNARE motif was not consistently proven in these toxins. However, for LC/D, the SNARE V1 motif (38QVDEVVDIMR47) was shown to be important for substrate recognition and hydrolysis. Three conservative hydrophobic residues in the V1 motif of VAMP-2, Val39, Val42, and Met47, are critical for efficient LC/D substrate hydrolysis. In addition, LC/D also utilizes other hydrophobic interactions to recognize the VAMP-2 substrate, such as the recognition of the P1′ site (Leu60) of VAMP-2 by the hydrophobic S1′ pocket of LC/D and the recognition of Val53 of VAMP-2 by the hydrophobic B1 binding site of LC/D. In contrast to the substrate binding contributed by the SNARE V1 motif (B3, B4, and B5), the B1 and B2 binding sites contribute more to LC/D substrate catalysis than substrate binding. The data suggest that the V1 motif plays a significant role in LC/D substrate binding, whereas the B1 and B2 binding sites may help more in fine-tuning the orientation of the substrate for specific recognition by the active site of LC/D rather than direct substrate binding.
In this study, we revealed the mechanism of LC/D substrate recognition and specificity. After internalization to the cytoplasm of neuronal cells, LC/D attacks the free form of VAMP-2 through interaction with and recognition of hydrophobic residues in the V1 motif of VAMP-2, including Val39, Val42, and Met46, by the substrate-binding regions B5, B4, and B3 of LC/D on the substrate-binding cleft, respectively. In particular, binding of Met46 of VAMP-2 to the LC/D B3 binding site was suggested to be very important for LC/D substrate recognition. This binding facilitates further binding of VAMP-2 Asn49 and Val53 to the B2 and B1 binding sites located at the active site surface of LC/D. The recognition of VAMP-2 Val53 by the LC/D Phe50/Ile191 pocket further orientates and stabilizes VAMP-2 for subsequent recognition of its different P sites by the corresponding S pockets in the active site of LC/D. Active P site recognition includes the formation of a salt bridge between P3 (Asp57) of VAMP-2 and S3 (Arg63) of LC/D, a hydrogen bond interaction between P1 (Lys58) of VAMP-2 and the main chain oxygen atom of Pro64, recognition of P1′ (Leu60) of VAMP-2 by the S1′ pocket (Tyr168 and Leu200) of LC/D, and finally a hydrogen bond interaction between P2′ (Ser61) of VAMP-2 and the S2′ pocket (Arg372) of LC/D. The anchoring of VAMP-2 P sites to different S pockets in the active site of LC/D aligns the VAMP-2 scissile bond close enough to the active site zinc ion to facilitate peptide bond cleavage (Fig. 4).
FIGURE 4.
Mechanism of substrate recognition by LC/D. After internalization to the cytoplasm of neuronal cells, LC/D attacks the free form of VAMP-2 through interaction with and recognition of hydrophobic residues in the V1 motif of VAMP-2, including Val39, Val42, and Met46, by the substrate-binding regions B5, B4, and B3 of LC/D on the substrate-binding cleft, respectively. In particular, binding of Met46 of VAMP-2 to the LC/D B3 binding site was suggested to be very important for LC/D substrate recognition. This binding facilitates further binding of VAMP-2 Asn49 and Val53 to the B2 and B1 binding sites located at the active site surface of LC/D. The recognition of VAMP-2 Val53 by the LC/D Phe50/Ile191 pocket further orientates and stabilizes VAMP-2 for subsequent recognition of different P sites of VAMP-2 by the corresponding S pockets in the active site of LC/D. Active P site recognition includes the formation of a salt bridge between P3 (Asp57) of VAMP-2 and S3 (Arg63) of LC/D, a hydrogen bond interaction between P1 (Lys58) of VAMP-2 and the main chain oxygen atom of Pro64, recognition of P1′ (Leu60) of VAMP-2 by the S1′ pocket (Tyr168 and Leu200) of LC/D, and finally a hydrogen bond interaction between P2′ (Ser61) of VAMP-2 and the S2′ pocket (Arg372) of LC/D. The anchoring of VAMP-2 P sites to different S pockets in the active site of LC/D aligns the VAMP-2 scissile bond close enough to the active site zinc ion to facilitate peptide bond cleavage.
Compared with substrate recognition by other serotypes of BoNT, LC/D possesses unique features of substrate recognition (14, 15, 20, 21). First, hydrophobic interaction between LC/D and VAMP-2 plays an important role in substrate recognition. The interaction between VAMP-2 Met46 and LC/D Ile151 seems to be critical for LC/D substrate recognition. This may be the first step in substrate recognition, which may facilitate the conformational change in VAMP-2 from a double helix to a free loop confirmation, favoring the subsequent substrate binding and catalysis by different regions of LC/D. Further research may be needed to test this hypothesis. However, far-UV CD analysis showed that I151D displays a slightly different conformation compared with WT-LC/D, suggesting that the significant effect of I151D substrate recognition may be partially due to the conformational change in the whole protein, but not the loss of the Ile151 site recognition. Second, in contrast to the recognition of one site of the substrate by one pocket of the LC for other serotypes of BoNT, LC/D utilizes two functionally similar residues to recognize one site of VAMP-2, such as the S1′ pocket (Tyr168–Leu200) of LC/D for recognition of the P1′ site (Leu60) of VAMP-2 in the active site of LC/D. In addition, dual recognition is also commonly employed at the substrate-binding regions. VAMP-2 Val53 is recognized by the LC/D pocket formed by Phe50 and Ile191. Mutation to each residue did not have much effect (maximum of 4-fold) on substrate hydrolysis, whereas mutation to both residues resulted in a dramatic reduction (400-fold) of substrate hydrolysis. VAMP-2 Asn49 and Met46 are also recognized by pockets with the dual recognition mechanism. The pocket that recognizes Asn49 of VAMP-2 is formed by Arg23 and His132 of LC/D. Although His132 plays a dominant role in Asn49 recognition, the H132A mutation can be complemented by Arg23. Similar to Asn49, the pocket that recognizes Met46 of VAMP-2 is formed by Val148 and Ile151 of LC/D. Ile151 plays a dominant role in Met46 recognition, whereas Val148 of VAMP-2 can play a complementary role when Ile151 is mutated to alanine. Finally, the pocket that recognizes Val39 of VAMP-2 is formed by three hydrophobic residues (Trp44, Ile152, and Pro154) of LC/D. Mutation of each residue to alanine or asparagine had no effect or only a minor effect on substrate hydrolysis, whereas triple mutations to alanine resulted in a much stronger reduction of substrate hydrolysis, highlighting the complementary effects of the three residues forming this pocket. The presence of two or more functionally similar residues in the same substrate recognition pocket enables LC/D to tolerate mutations. This property of LC/D makes it a good candidate for further protein engineering.
Unlike BoNT/A, which is the most toxic botulinum neurotoxin and is implicated in human botulism, BoNT/D is responsible mainly for animal botulism, such as cattle botulism. However, our data indicate that LC/D and LC/A exhibit a similar potency in hydrolyzing their substrates under in vitro conditions (16). The role of BoNT/D as a human therapy or bioterrorism weapon remains to be investigated. Our data provide insights into the development of novel BoNT-based therapies and BoNT/D antitoxins.
Acknowledgments
We thank Edward Chan and Xiaodong Lin for critical reading of the manuscript and members of the Sheng laboratory for helpful discussions.
This work was supported by Grant B-Q25N from the Hong Kong Research Grant Council (to S. C.).
- BoNT
- botulinum neurotoxin
- LC
- light chain.
REFERENCES
- 1. Montecucco C., Schiavo G. (1993) Tetanus and botulism neurotoxins: a new group of zinc proteases. Trends Biochem. Sci. 18, 324–327 [DOI] [PubMed] [Google Scholar]
- 2. Montecucco C., Schiavo G. (1994) Mechanism of action of tetanus and botulinum neurotoxins. Mol. Microbiol. 13, 1–8 [DOI] [PubMed] [Google Scholar]
- 3. Jankovic J., Brin M. F. (1991) Therapeutic uses of botulinum toxin. N. Engl. J. Med. 324, 1186–1194 [DOI] [PubMed] [Google Scholar]
- 4. Schiavo G., Rossetto O., Benfenati F., Poulain B., Montecucco C. (1994) Tetanus and botulinum neurotoxins are zinc proteases specific for components of the neuroexocytosis apparatus. Ann. N.Y. Acad. Sci. 710, 65–75 [DOI] [PubMed] [Google Scholar]
- 5. Davletov B., Bajohrs M., Binz T. (2005) Beyond BOTOX: advantages and limitations of individual botulinum neurotoxins. Trends Neurosci. 28, 446–452 [DOI] [PubMed] [Google Scholar]
- 6. Brunger A. T. (2005) Structure and function of SNARE and SNARE-interacting proteins. Q. Rev. Biophys. 38, 1–47 [DOI] [PubMed] [Google Scholar]
- 7. Bossi P., Bricaire F. (2003) Botulism toxin, bioterrorist weapon. Presse Med. 32, 463–465 [PubMed] [Google Scholar]
- 8. Arnon S. S., Schechter R., Inglesby T. V., Henderson D. A., Bartlett J. G., Ascher M. S., Eitzen E., Fine A. D., Hauer J., Layton M., Lillibridge S., Osterholm M. T., O'Toole T., Parker G., Perl T. M., Russell P. K., Swerdlow D. L., Tonat K. (2001) Botulinum toxin as a biological weapon: medical and public health management. JAMA 285, 1059–1070 [DOI] [PubMed] [Google Scholar]
- 9. Klein A. W. (2004) The therapeutic potential of botulinum toxin. Dermatol. Surg. 30, 452–455 [DOI] [PubMed] [Google Scholar]
- 10. Pearce L. B., First E. R., MacCallum R. D., Gupta A. (1997) Pharmacologic characterization of botulinum toxin for basic science and medicine. Toxicon 35, 1373–1412 [DOI] [PubMed] [Google Scholar]
- 11. Glogau R. G. (2002) Review of the use of botulinum toxin for hyperhidrosis and cosmetic purposes. Clin. J. Pain 18, S191–197 [DOI] [PubMed] [Google Scholar]
- 12. Cheng C. M., Chen J. S., Patel R. P. (2006) Unlabeled uses of botulinum toxins: a review, part 2. Am. J. Health Syst. Pharm. 63, 225–232 [DOI] [PubMed] [Google Scholar]
- 13. Breidenbach M. A., Brunger A. T. (2004) Substrate recognition strategy for botulinum neurotoxin serotype A. Nature 432, 925–929 [DOI] [PubMed] [Google Scholar]
- 14. Chen S., Kim J. J., Barbieri J. T. (2007) Mechanism of substrate recognition by botulinum neurotoxin serotype A. J. Biol. Chem. 282, 9621–9627 [DOI] [PubMed] [Google Scholar]
- 15. Chen S., Barbieri J. T. (2007) Multiple pocket recognition of SNAP25 by botulinum neurotoxin serotype E. J. Biol. Chem. 282, 25540–25547 [DOI] [PubMed] [Google Scholar]
- 16. Chen S., Barbieri J. T. (2006) Unique substrate recognition by botulinum neurotoxins serotypes A and E. J. Biol. Chem. 281, 10906–10911 [DOI] [PubMed] [Google Scholar]
- 17. Arndt J. W., Chai Q., Christian T., Stevens R. C. (2006) Structure of botulinum neurotoxin type D light chain at 1.65 Å resolution: repercussions for VAMP-2 substrate specificity. Biochemistry 45, 3255–3262 [DOI] [PubMed] [Google Scholar]
- 18. Sikorra S., Henke T., Galli T., Binz T. (2008) Substrate recognition mechanism of VAMP/synaptobrevin-cleaving clostridial neurotoxins. J. Biol. Chem. 283, 21145–21152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chen S., Hall C., Barbieri J. T. (2008) Substrate recognition of VAMP-2 by botulinum neurotoxin B and tetanus neurotoxin. J. Biol. Chem. 283, 21153–21159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chen S., Karalewitz A. P., Barbieri J. T. (2012) Insights into the different catalytic activities of Clostridium neurotoxins. Biochemistry 51, 3941–3947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chen S., Wan H. Y. (2011) Molecular mechanisms of substrate recognition and specificity of botulinum neurotoxin serotype F. Biochem. J. 433, 277–284 [DOI] [PubMed] [Google Scholar]
- 22. Rossetto O., Schiavo G., Montecucco C., Poulain B., Deloye F., Lozzi L., Shone C. C. (1994) SNARE motif and neurotoxins. Nature 372, 415–416 [DOI] [PubMed] [Google Scholar]