Background: Protease resistance and aggregation state, two fundamental properties of abnormal prion protein (PrPSc), remain significantly unexplored.
Results: Prion isolates show differences in PrPSc PK digestion profile that are only partially explained by differences in aggregate size.
Conclusion: Molecular factors, likely acting on aggregate stability, significantly influence PrPSc PK sensitivity.
Significance: The study provides insight into prion strain variability with respect to PrPSc PK sensitivity and aggregation state.
Keywords: Neurodegenerative Diseases, Prions, Protease, Protein Aggregation, Protein Conformation, Protein Purification, Creutzfeldt-Jacob Disease
Abstract
Prion diseases are characterized by tissue accumulation of a misfolded, β-sheet-enriched isoform (scrapie prion protein (PrPSc)) of the cellular prion protein (PrPC). At variance with PrPC, PrPSc shows a partial resistance to protease digestion and forms highly aggregated and detergent-insoluble polymers, two properties that have been consistently used to distinguish the two proteins. In recent years, however, the idea that PrPSc itself comprises heterogeneous species has grown. Most importantly, a putative proteinase K (PK)-sensitive form of PrPSc (sPrPSc) is being increasingly investigated for its possible role in prion infectivity, neurotoxicity, and strain variability. The study of sPrPSc, however, remains technically challenging because of the need of separating it from PrPC without using proteases. In this study, we have systematically analyzed both PK resistance and the aggregation state of purified PrPSc across the whole spectrum of the currently characterized human prion strains. The results show that PrPSc isolates manifest significant strain-specific differences in their PK digestion profile that are only partially explained by differences in the size of aggregates, suggesting that other factors, likely acting on PrPSc aggregate stability, determine its resistance to proteolysis. Fully protease-sensitive low molecular weight aggregates were detected in all isolates but in a limited proportion of the overall PrPSc (i.e. <10%), arguing against a significant role of slowly sedimenting PK-sensitive PrPSc in the biogenesis of prion strains. Finally, we highlight the limitations of current operational definitions of sPrPSc and of the quantitative analytical measurements that are not based on the isolation of a fully PK-sensitive PrPSc form.
Introduction
Prion diseases or transmissible spongiform encephalopathies (TSEs)2 are a phenotypically heterogeneous group of rare neurodegenerative disorders affecting humans and other mammals. These diseases are characterized by tissue deposition of a misfolded form of the endogenous cellular prion protein (PrPC), a membrane glycosylphosphatidylinositol-anchored glycoprotein of 231 amino acids comprising three glycoforms (i.e. diglycosylated, monoglycosylated, and unglycosylated) that are generated by the non-obligatory addition of N-linked glycan chains at residues Asn-181 and Asn-197 (1). Human PrPC is composed of an unstructured N-terminal domain followed by a well characterized globular domain that comprises two β-sheets and three α-helices (2). Although it is well established that PrPC conversion into its misfolded form, PrPSc, involves consistent changes in the secondary structure with part of the α-helical structure turning into a β-sheet (3, 4), a further structural characterization of PrPSc has been hampered to date by the propensity of the misfolded protein to form highly aggregated and detergent-insoluble polymers.
Human prion diseases are characterized by a number of clinicopathological types or variants. Different strains of the TSE agent or prion are believed to be the main cause of this phenotypic diversity (5). In addition, the host variability in the gene encoding PrP (PRNP) as determined in humans by polymorphisms or mutations also modulates the disease phenotype (6–8). In sporadic Creutzfeldt-Jakob disease (sCJD), the most common human TSE, six major disease phenotypes have been characterized that strongly correlate at the molecular level with the genotype at the polymorphic codon 129 (methionine (M) or valine (V)) of PRNP and two PrPSc profiles or types (type 1 and type 2) comprising distinctive physicochemical properties such as size after protease treatment and glycoform ratio (9, 10). The six sCJD types differ among each other in several clinical and histopathological features, and at least three of them (MM1, VV2, and MM2-thalamic (MM 2T)) propagate in animal models as distinct prion strains (11–14). Besides sCJD, a novel form of human sporadic prion disease designated variably protease-sensitive prionopathy (VPSPr) has been recently identified. When compared with sCJD, VPSPr displays “atypical” histopathologic and clinical features and shows a quite distinctive Western blot profile of abnormal PrP comprising truncated fragments in the 8–17 molecular weight range that are not seen in classic CJD (15). Finally, variant CJD (vCJD), an acquired form of the disease that originated from a bovine prion strain, also displays a very distinctive CJD phenotype (16, 17). PrPSc extracted from vCJD brains is biochemically distinguishable from the type 2 found in sCJD due to a relatively high representation of the diglycosylated PrPSc glycoform (18, 19). To date, only individuals that are methionine homozygotes in PRNP codon 129 have been affected by this CJD phenotype (20).
A central issue in prion biology concerns the molecular basis of prion strains. It is currently widely believed that prion strain diversity is encoded in the PrPSc conformation (1). Support for this idea comes from several studies showing that PrPSc exists in a number of molecular types that differ in one or more of the following properties: fragment size after protease digestion, glycoform profile, aggregate size, and stability in chaotropes (for a review, see Ref. 21). However, as this evidence is largely indirect, the structural basis of this heterogeneity remains to be established.
Although resistance to proteinase K (PK) and detergent insolubility have been traditionally used to operationally define PrPSc and to distinguish it from PrPC, the recent characterization of a PK-sensitive form of PrPSc (sPrPSc) has highlighted a more complex picture and added a new perspective to the study of PrPSc properties and their relationship to prion strains (22–25).
Following the observation that in a minority of subjects affected by Gerstmann-Sträussler-Scheinker disease linked to the P102L mutation detergent-insoluble full-length PrPSc is easily digested at a relatively low PK concentration (26), similar PK-sensitive forms of PrPSc have been found in other rare, atypical variants of both human and animal prion diseases (15, 27–30).
However, by showing that the denaturation-dependent immunoreactivity enhancement effect that is attributed to PrPSc is considerably weakened by a pretreatment with PK, Safar et al. (24, 31) have proposed that sPrPSc does not only characterize atypical prions but rather constitutes an invariable and quantitatively significant component of prions, contributing up to 90% of the whole PrPSc signal even in TSEs such as sCJD and classical scrapie. With this novel approach, called conformation-dependent immunoassay (CDI), a correlation between the relative amount of sPrPSc and strain-specific properties such as the incubation period after inoculation and the clinical duration of the disease has also been reported (24, 25, 32). These findings have on one hand called into question the rationale on which TSE diagnosis is currently based (i.e. demonstration of rPrPSc) (24, 33, 34) and on the other supported the idea that sPrPSc contributes to the molecular basis of phenotypic variability and strain diversity in both animal and human prion diseases.
Concerning the physical nature of sPrPSc with respect to its “resistant” counterpart, the isolation and characterization of a sPrPSc fraction consisting of aggregates of shorter size with respect to rPrPSc (22, 23) has suggested that the resistance of PrPSc to proteolysis may, at least in part, depend on its quaternary structure. However, these PK-sensitive aggregates with low sedimenting properties are unlikely to reflect the whole sPrPSc signal defined by CDI because low sedimenting PrPC-rich fractions contain little or no conformation-dependent immunoreactive material (22, 23), thus leaving the structural basis of the CDI-related sPrPSc signal unexplored. Furthermore, PrPSc associated with atypical animal and human prion diseases such as Nor98 or subtypes of Gerstmann-Sträussler-Scheinker disease did not exhibit a higher proportion of slowly sedimenting species when compared with classic strains (29, 35), arguing that, at least in these atypical prionopathies, the pronounced PK sensitivity of PrPSc is not due to low size aggregates but rather to its tertiary structure or other factors.
To further address these issues and gain insights into the molecular basis of phenotypic variability and disease pathogenesis in human prion disease, in the present study we have characterized the detergent-insoluble PrPSc fraction in relation to both PK resistance and the aggregation state across the whole spectrum of the currently characterized human prion strains. In particular, for each strain or disease subtype we have searched for and quantified the low sedimenting sPrPSc fraction, defined the sedimenting velocity profile of PrPSc, studied the overall resistance of PrPSc to PK digestion, and finally compared the resistance of PrPSc to proteolysis between strains in fractions with similarly sized aggregates.
EXPERIMENTAL PROCEDURES
Patients and Tissues
We have studied brain tissues from 41 cases of sCJD, three cases of vCJD, and two cases of VPSPr, the latter carrying, respectively, MM and MV at PRNP codon 129. sCJD tissues comprised the whole spectrum of pure phenotypic variants described by Parchi et al. (9) and included eight MM1, eight VV1, seven MV 2K, eight VV2, six MM2-cortical (MM 2C), and four MM 2T cases (9). In addition, three brains lacking significant histopathological changes were used as prion negative controls. Brain tissues were obtained at autopsy and kept frozen at −80 °C until use. All samples used in this study were taken from the frontal cerebral cortex.
Preparation of Total Brain Homogenates (THs)
50 mg of gray matter were homogenized (10%, w/v) in LB100, a lysis buffer with high buffer capacity (100 mm Tris, 100 mm NaCl, 10 mm EDTA, 0,5% Nonidet P-40, 0,5% sodium deoxycholate) at pH 6.9 (36). Total protein concentration was measured using a standard colorimetric method based on bicinchoninic acid (Pierce).
PrPSc Purification
Purified PrPSc was obtained from about 500 mg of gray matter after three cycles of Sarkosyl extraction and differential centrifugation steps (adapted from Ref. 37). Briefly, tissues were homogenized in 1.75 ml of 2× TEND (20 mm Tris, pH 8.3, 1 mm EDTA, 130 mm NaCl, 1 mm dithiothreitol) and 1.75 ml of 20% Sarkosyl and cleared by ultracentrifugation at 22,500 × g (Beckman rotor SW55Ti) for 25 min at 4 °C. Supernatants (S1) were then underlaid with a sucrose cushion (1 ml of 20% sucrose in H20) and ultracentrifuged at 150,000 × g for 143 min at 4 °C. After discarding the supernatants (S2), the pellets (P2) were incubated overnight at 4 °C in 100 μl of TBS and then resuspended in 2.5 ml 1× TEND, 10% NaCl, and 1% Sarkosyl through three cycles of freezing and thawing after a short and standardized sonication. Samples were then again laid over a sucrose cushion and ultracentrifuged at 150,000 × g for 143 min at 18 °C. After removal of the supernatant (S3), pellets (P3) were resuspended in 400 μl of TNS (10 mm Tris, 150 mm NaCl, 1% Sarkosyl), pH 7.4; incubated overnight; and then dissolved through three cycles of freezing and thawing after a short and standardized sonication.
Sucrose Gradient Centrifugation Assay
Total protein concentration in P3s was adjusted to a final value of 300 μg/ml. S2s and P3s (400 μl) were loaded on top of a 10–60% sucrose step gradient (e.g. 500 μl of 10, 20, 30% 40, 50, and 60% solutions of sucrose in TNS, pH 7.4 going from the top to the bottom of the tube) and ultracentrifuged at 200,000 × g for 1 h at 4 °C. After centrifugation, 12 fractions of 280 μl were collected from the top to the bottom of the tube.
PK Titration Assays
P3s were adjusted to a total protein concentration of 150 μg/ml and digested using serial dilutions of PK activity ranging from 0.004 to 20 units/ml. Because of the higher protein content, the total protein concentration of THs was adjusted to 6 mg/ml, whereas the PK activity used ranged from 0.03 to 256 units/ml.
Finally, the selected fractions (6, 9, and 12) from MM1 and VV2 cases were adjusted to a total protein concentration of 50 μg/ml and a sucrose content of 20%, whereas the range of PK dilutions varied from 0.016 to 40 units/ml. PK digestion was performed for 1 h at 37 °C unless otherwise specified.
PrP Deglycosylation
N-Linked glycans were removed by using a peptide-N-glycosidase F kit (New England Biolabs, Ipswich, MA) according to the manufacturer's instructions.
Western Blot and Quantitative Analysis of Protein Signal
After the addition of sample buffer (final concentration, 3% SDS, 4% β-mercaptoethanol, 10% glycerol, 2 mm EDTA, 62.5 mm Tris, pH 6.8) and boiling for 6 min at 100 °C, samples were run on 13 or 15% (only for PK digestion experiments on THs) acrylamide gels (Criterion, Bio-Rad) and transferred to Immobilon-P membranes (Millipore Corp., Billerica, MA). After blocking in 10% nonfat milk in Tween-Tris-buffered saline, membranes were incubated with mAb SAF 60 (working dilution, 1:2000), which maps on residues 157–161 of the prion protein or with mAb 3F4 (Signet Laboratories, Dedham, MA; working dilution, 1:30,000), which maps on residues 106–110.
After four washings in Tween-Tris-buffered saline, membranes were incubated for 1 h with an anti-mouse secondary antibody conjugated to horseradish peroxidase (GE Healthcare; working dilution, 1:4000) and washed again four times in Tween-Tris-buffered saline. The immunoreactive signal was detected by enhanced chemiluminescence (ECL; GE Healthcare) on an LAS 3000 camera (Fujifilm Corp., Tokyo, Japan) or (only for the PK digestion experiments on THs) on Kodak Biomax Light films (Eastman Kodak Co.).
To quantify PrP content, the Western blot signal of each sample was compared with a standard curve obtained by loading in each gel the same serial dilution (n = 6) of a chosen sCJD sample. Western blot signals were measured by densitometry using the software AIDA (Image Data Analyzer v.4.15, Raytest, Isotopenmessgeraete GmbH, Straubenhardt, Germany).
For each PK titration assay on P3s, semilogarithmic curves were obtained by plotting the percentage of protein remaining after digestion (with respect to the undigested sample) against the corresponding PK concentration. The ED50 (e.g. the PK concentration needed to digest 50% of PrPSc) for each sample was calculated by means of the equation of the straight line that best fitted the linear portion of the curve (R2 ≥ 0.95).
For each sucrose gradient centrifugation assay, the relative amount of each undigested fraction was calculated as a percentage of the sum of all fractions. The aggregation ratio was determined as the ratio between the relative amount of the last six fractions and that of the first six. Finally, the percentage of PrP remaining after digestion in the PK titration curves on THs was not calculated with respect to the undigested sample but rather with respect to the PrP amount detected after digestion at a 0.25 unit/ml PK activity because it was assumed that lower PK dilutions do not completely digest PrPC.
Statistical Analyses
All statistical analyses were performed with SigmaStat 3.5 (Systat Software Inc., Chicago, IL). One-way analysis of variance (ANOVA) followed by all pairwise multiple comparison procedures were used to detect statistically significant differences among CJD variants in the ED50, aggregation ratio, or PrPSc content in grouped fractions of the sucrose gradient assay. Student's t test was used to detect statistically significant differences between MM1 and VV2 in the amount of PrPSc in fractions 6, 9, and 12 for each PK dilution used in the titration curve and among the relative amounts of each of the three differently glycosylated PrPSc bands after digestion at two PK concentrations for each CJD subtype.
RESULTS
Analysis of PrP Species in Fractions Obtained during PrPSc Purification
It is well established that, in both CJD and non-CJD brains, human PrP encompasses truncated fragments in addition to the full-length protein. The known truncated species associated with the non-CJD brain include a major 18-kDa C-terminal fragment, known as C1, which is generated by the cleavage of full-length PrPC between residues 111 and 112, and a minor ∼20-kDa C-terminal fragment (CTF) (38).
Besides the C1 and the ∼20-kDa CTF, the CJD brain accumulates variable amounts of the PK-resistant and detergent-insoluble core of PrPSc, the so-called PrP27–30 (also known as C2), which is generated in vivo by the preferential cleavage of the protein at either residue 82 (type 1, 21 kDa) or 97 (type 2, 19 kDa) (39). Moreover, PK-resistant CTFs of about 12–13 kDa are seen, particularly in the disease variants associated with PrPSc type 1 (40, 41).
In the present study, we have assessed PrPSc protease resistance and aggregation state across the spectrum of human prions using purified PrPSc preparations. The critical separation of PrPC from PrPSc has been obtained through cycles of ultracentrifugation in the presence of detergents, taking advantage of their divergent solubility. To monitor the efficiency of separation of the two PrP species during the purification protocol, we have measured the relative content of the above mentioned truncated fragments in most of the fractions (S1, S2, and P3) obtained. S3 always contained a barely detectable PrP signal, indicating that a complete separation of PrPC from PrPSc in the supernatant is already achieved after the second ultracentrifugation step (S2).
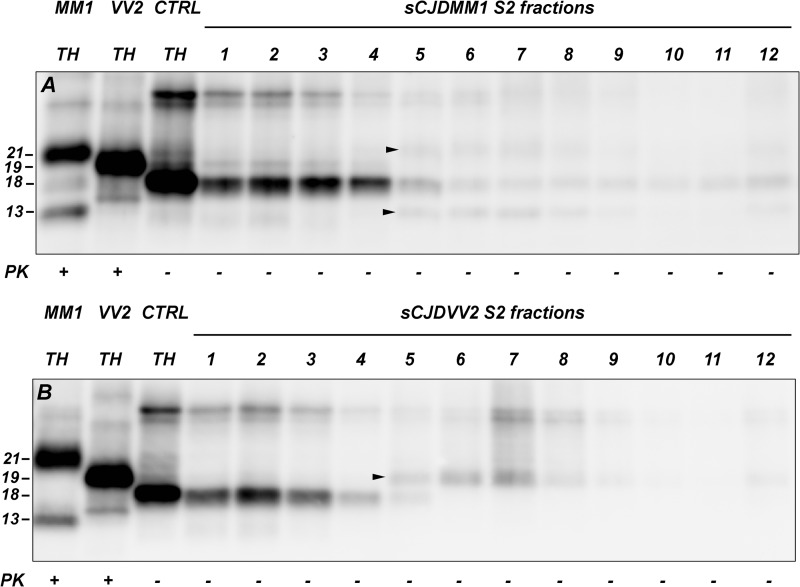
To monitor the degree of PrPSc enrichment in the pellet fractions during the purification procedure, we measured the relative amount of truncated PrP fragments in S1 and S2. In control brains, S1 and S2 showed a very similar C1/20-kDa CTF ratio (C1 comprised 66.4–86.0% of the truncated PrP species in S1 and 69.5–84.1% in S2) and a similar overall amount of both full-length and C1 PrPC, further indicating that virtually all PrPC is recovered in the S2 fraction (data not shown). At variance with control brains, the S1 fraction from sCJD brains revealed a more significant variability of the relative percentage of C1 (from 34.6 to 74.5% of all truncated species) due to the presence of variable amounts of C2. In the S2, however, the C1/C2 ratio fell within the range of the C1/20-kDa CTF ratio of control brains (C1 ranged from 67.9 to 83.6%), thus excluding the presence of a significant amount of PrPSc in S2. The sucrose gradient centrifugation assay of the S2 fraction more specifically showed that virtually all sPrPSc is recovered in P2 (Fig. 1). In particular, all PrPSc-associated truncated fragments (i.e. the 19- or 21-kDa PrPSc cores and the 13-kDa CTF) appeared from the fifth fraction forward, whereas the C1 fragment was the only truncated PrP species in the first four fractions. Thus, because sPrPSc (as it is defined in the present work) is expected to sediment in fractions 1–4 (see below), the result makes it very hard to hypothesize a quantitatively relevant sPrPSc retention in S2.
FIGURE 1.
PrP truncated species in fractionated S2 samples after sucrose gradient centrifugation. In both MM1 (A) and VV2 (B) sCJD cases, the C1 fragment is clearly detected until the fifth fraction, whereas the 21- and 13-kDa (A) or the 19-kDa (B) PrPSc fragments are seen from the fifth fraction onward (arrowheads). Fractions 7–12 have been concentrated 4 times before loading with respect to fractions 1–6. Membranes were probed with mAb SAF 60. Approximate molecular masses are in kDa. CTRL, control.
PrPSc Associated with Distinct Human Prion Strains Shows Variable Resistance to Protease Digestion
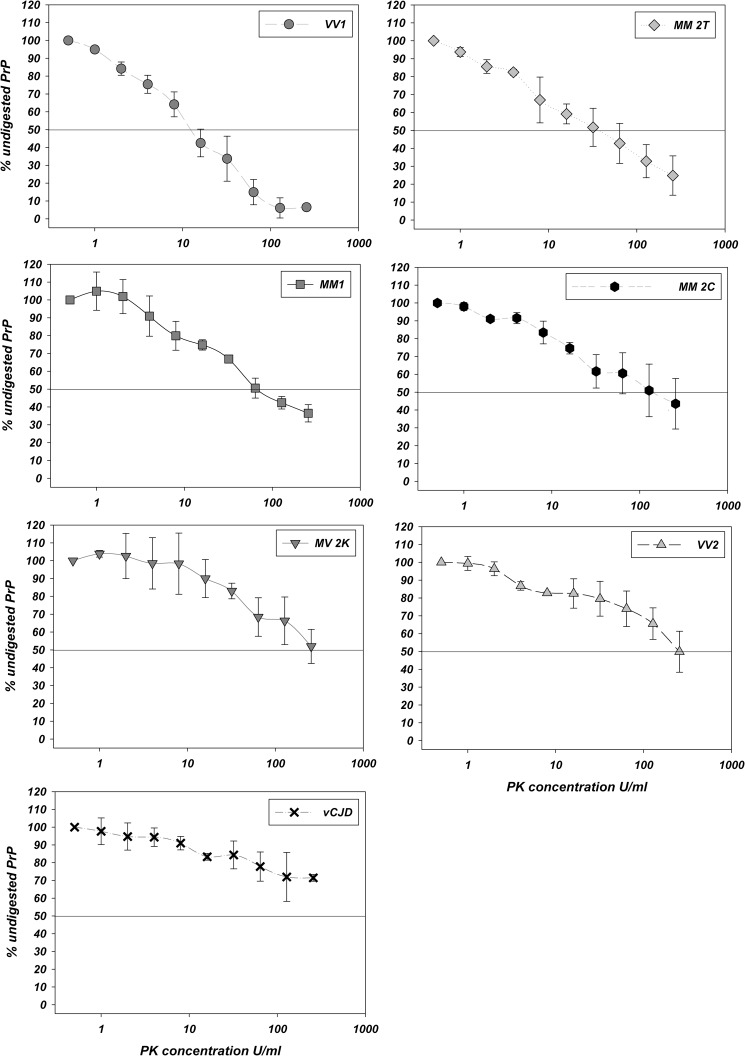
PK titration curves for each sCJD type, VPSPr, and vCJD performed on both purified PrPSc (P3) and THs are shown in Figs. 2, 3, and 4. In P3, the calculated PrPSc ED50 (Table 1) was lowest for VV1 and VPSPr and highest for MV 2K/VV2 and vCJD with MM1, MM 2T, and MM 2C showing intermediate values. A similar trend across the spectrum of human prion strains was also obtained by calculating the percentage of detectable PrPSc signal at the highest tested PK activity rather than the ED50 (Table 2). Results obtained with THs were highly consistent with those obtained with P3s with the only exception being MM 2T-PrPSc, which was slightly more PK-sensitive than MM1-PrPSc in the THs (Fig. 4), whereas the opposite was true in P3. Overall, the type 1-linked variants were less PK-resistant than those associated with type 2, although differences were also observed within groups associated with the same PrPSc type.
FIGURE 2.
Analyses of protease resistance of purified PrPSc by PK titration assay. Purified PrPSc from the six sCJD types, VPSPr, and vCJD was run either untreated (first lane) or after digestion with increasing amounts of PK for 1 or 2 h (last lane only) at 37 °C. Membranes were probed with mAb SAF 60. Approximate molecular masses are in kilodaltons.
FIGURE 3.
PK digestion profiles of purified PrPSc. The plots represent the amount (mean ± S.D. (error bars)) of PrPSc detected after PK digestion expressed as a percentage of the undigested sample for each sCJD group, VPSPr, and vCJD. The PK concentrations used (x axis) are on a logarithmic scale.
FIGURE 4.
PK digestion profiles of total PrP. The plots represent the amount (mean ± S.D. (error bars)) of PrP detected after PK digestion expressed as a percentage of the sample digested with 0.25 unit/ml PK for each sCJD group, VPSPr, and vCJD. The PK concentrations used (x axis) are on a logarithmic scale.
TABLE 1.
ED50 in PK titration assays of PrPSc purified from each CJD type tested
ED50 is expressed as mean ± S.D. Statistical significance was evaluated by means of Kruskal-Wallis one-way ANOVA on ranks followed by pairwise multiple comparisons (Q value estimated with Dunn's method; Q critical value = 3.124 for p < 0.05). Statistically significant comparisons included vCJD versus VV1 (Q = 3.811), VV2 versus VV1 (Q = 4.346), VV2 versus MM1 (Q = 3.309), and MV 2K versus VV1 (Q = 3.642).
| CJD type | Sample size (n) | ED50 |
|---|---|---|
| units/ml | ||
| VV1 | 6 | 0.034 ± 0.028 |
| VPSPr | 2 | 0.069 ± 0.010 |
| MM1 | 6 | 0.093 ± 0.068 |
| MM 2T | 4 | 0.134 ± 0.089 |
| MM 2C | 6 | 0.276 ± 0.210 |
| MV 2K | 7 | 1.407 ± 0.892 |
| VV2 | 6 | 4.137 ± 3.562 |
| vCJD | 3 | 5.192 ± 2.378 |
TABLE 2.
PK dilution values corresponding to the lowest detectable purified PrPSc signal on Western blot
Values are expressed as mean ± S.D. and were calculated as the percentage of PrPSc remaining after digestion with respect to the undigested sample. For each PK dilution, digestion was carried out for 1 h at 37 °C unless otherwise specified.
| CJD type | PK dilution |
|||
|---|---|---|---|---|
| 5 units/ml | 10 units/ml | 20 units/ml | 20 units/ml, 2 h | |
| % | ||||
| VPSPr | 9.01 ± 2.24 | |||
| VV1 | 4.40 ± 1.71 | |||
| MM1 | 8.17 ± 4.65 | |||
| MM 2T | 18.16 ±3.34 | 9.39 ± 4.79 | 5.45 ± 4.58 | |
| MM 2C | 9.62 ± 5.56 | 4.56 ± 3.75 | 2.33 ± 1.69 | |
| MV 2K | 21.95 ± 5.75 | 10.04 ± 3.57 | ||
| VV2 | 27.87 ± 11.58 | 13.75 ± 3.93 | ||
| vCJD | 30.33 ± 0.85 | 17.56 ± 0.97 | ||
Comparison of the PrPSc glycoform ratio obtained after digestion of the protein at different PK concentrations (Table 3) indicated similar kinetics of digestion for each of three glycoforms with only a minor trend toward a preferential digestion of the diglycosylated form, which was most evident in VV1. Interestingly, the PrPSc ED50 positively correlated with the relative percentage of diglycosylated PrPSc in both “type 1” (VV1 and MM1) and “type 2” (MM 2T, MM 2C, MV 2K, VV2, and vCJD) CJD groups (Table 3). Furthermore, the PrPSc ED50 appeared to be inversely correlated with the relative amount of the 13-kDa truncated C-terminal fragment generated by PK digestion (Fig. 2).
TABLE 3.
Effect of PK activity on PrPSc glycoform ratio
Values are expressed as relative percentage ± S.D. of diglycosylated (D), monoglycosylated (M), and unglycosylated (U) PrPSc based on densitometric analyses of immunoblots probed with mAb 3F4. Statistically significant comparisons (Student's t test or Mann-Whitney rank sum test) included diglycosylated PrPSc in VV1 (p < 0.01).
| CJD type | Lowest tested PK activitya |
PK activity at ED50b |
||||
|---|---|---|---|---|---|---|
| D | M | U | D | M | U | |
| VV1 | 21.0 ± 1.6 | 40.0 ± 4.0 | 39.0 ± 2.4 | 17.5 ± 1.0 | 40.7 ± 3.1 | 41.8 ± 3.4 |
| MM1 | 26.1 ± 1.5 | 49.0 ± 1.6 | 24.9 ± 2.3 | 25.9 ± 1.1 | 47.5 ± 2.4 | 26.6 ± 1.3 |
| MM 2T | 22.0 ± 0.4 | 44.5 ± 2.3 | 33.5 ± 1.9 | 20.3 ± 0.8 | 44.0 ± 0.5 | 35.7 ± 0.4 |
| MM 2C | 25.4 ± 2.0 | 42.0 ± 0.5 | 32.6 ± 1.9 | 24.6 ± 1.3 | 43.3 ± 2.4 | 32.1 ± 2.8 |
| MV 2K | 27.9 ± 1.9 | 40.6 ± 2.0 | 31.6 ± 0.5 | 27.5 ± 2.0 | 39.1 ± 0.8 | 33.4 ± 1.5 |
| VV2 | 39.1 ± 5.2 | 41.9 ± 1.5 | 19.0 ± 5.1 | 38.7 ± 3.9 | 41.2 ± 1.8 | 20.1 ± 3.5 |
| vCJD | 49.1 ± 0.7 | 34.5 ± 2.3 | 16.4 ± 1.6 | 47.0 ± 1.6 | 35.4 ± 1.9 | 17.7 ± 0.4 |
a Activity was 0.004 unit/ml for VV1 and MM 2T and 0.0315 unit/ml for all other subtypes.
b Activity was 0.0625 unit/ml for VV1, 0.125 unit/ml for MM1, 0.25 unit/ml for MM 2T and MM 2C, 2 units/ml for MV 2K, 5 units/ml for VV2, and 10 units/ml for vCJD.
In P3, the analyses of the titration curves showed additional differences among disease subtypes. For example, PrPSc-MM 2C had a higher ED50 than PrPSc-MM 2T (Table 1) despite the latter maintaining about 10% of the signal up to a higher PK concentration (10 versus 5 units/ml) (Table 2). Finally, it is worth noting that the ED50 of PrPSc purified from VPSPr carrying the MM or MV genotype was within the range of those observed among the classic sCJD variants.
PrPSc Associated with Distinct Human Prion Strains Shows Differences in Aggregation State
The analyses of PrPSc distribution among the fractions collected after PrPSc purification and sucrose gradient ultracentrifugation showed that PrPSc preferentially localizes in the intermediate and late fractions irrespective of the disease subtype (Fig. 5, A–C). Nevertheless, the fraction containing the highest amount of PrPSc correlated to some extent with the CJD subtype (i.e. fraction 8 in VPSPr and VV1; fraction 9 in MM1, MV 2K, VV2, and MM 2T; fraction 10 in vCJD; and fraction 12 in MM 2C). Furthermore, the distribution of the largest PrPSc aggregates (i.e. fractions 9–12) was significantly influenced by both the type of PrPSc and the codon 129 genotype with both type 2 and methionine at codon 129 favoring the distribution of the abnormal protein in the late fractions (Fig. 5D).
FIGURE 5.

PrPSc aggregate distribution among fractions after sucrose gradient centrifugation assay. A–C, for each CJD type and VPSPr, the amount (mean ± S.D. (error bars)) of PrPSc in each fraction collected after sucrose gradient centrifugation is expressed as a percentage of all fractions. D, the PrPSc amount of combined fractions 1–4, 5–8, and 9–12 is shown. Note that the PrPSc content of fractions 1–4, containing the least aggregated and fully PK-sensitive PrPSc, does not exceed 10% in all groups. Nevertheless, the PrPSc content in these slowly sedimenting fractions is significantly higher in MM 2C than in VV2, MV 2K, MM 2T, MM1 (p < 0.001), VV1, and VPSPr (p < 0.002) (one-way ANOVA test followed by all pairwise multiple comparisons with the Holm-Sidak method). Intermediate (5–8) and bottom fractions (9–12) contain partially PK-resistant PrPSc aggregates that also vary in relative proportion among CJD types. The PrPSc amount in fractions 9–12 was statistically different in the following pairs: (i) VV1 versus vCJD, MM 2C, MM 2T, and MV 2K (p < 0.001); (ii) VPSPr versus vCJD, MM 2T, MM 2C (p < 0.001), MV 2K (p = 0.001), and VV2 (p = 0.002); and (iii) MM1 versus vCJD (p < 0.001) (one-way ANOVA test followed by Holm-Sidak method for all pairwise multiple comparisons).
To obtain a semiquantitative index of the overall state of PrPSc aggregation for each CJD type, we calculated the ratio between the amounts of PrPSc in fractions 7–12 and those in fractions 1–6. Statistical analyses were significant between groups at the very extreme of the range (Table 4). Thus, VPSPr and VV1, the groups with the lowest aggregation ratio, showed significant differences with respect to those with the highest values such as VV2 and MV 2K or vCJD. In addition, PrPSc from MM1 significantly differed from that of vCJD PrPSc. Finally, the analyses of the PrPSc aggregation ratio also distinguished MM 2C from MM 2T, two rare sCJD variants sharing the PrP primary sequence and many PrPSc properties including the fragment size after PK digestion, the degree of protease resistance, and the propensity to mainly form aggregates of relatively large size (i.e. fractions 9–12).
TABLE 4.
PrPSc aggregation ratio for each CJD type
The aggregation ratio is expressed as mean ± S.D. Statistical significance was evaluated by means of one-way ANOVA followed by pairwise multiple comparisons carried out by the Holm-Sidak method. Statistically significant comparisons included VPSPr versus MM 2T, versus vCJD, versus VV2 (p < 0.001), and versus MV 2K (p = 0.001); VV1 versus MM 2T, versus vCJD (p < 0.001), and versus VV2 (p = 0.001); MM 2C versus MM 2T and versus vCJD (p < 0.001); and MM1 versus MM 2T and versus vCJD (p < 0.001).
| CJD type | Sample size (n) | Aggregation ratio |
|---|---|---|
| VPSPr | 2 | 3.76 ± 0.28 |
| VV1 | 6 | 5.47 ± 0.46 |
| MM 2C | 5 | 5.68 ± 0.99 |
| MM1 | 6 | 6.43 ± 2.15 |
| MV 2K | 6 | 8.38 ± 1.93 |
| VV2 | 6 | 8.68 ± 1.22 |
| MM 2T | 4 | 10.92 ± 1.44 |
| vCJD | 3 | 11.65 ± 3.35 |
To assess the relationship between PrPSc aggregation and PK resistance for each strain, we then compared the PrPSc “aggregation ratio” with the ED50. The results of this analysis (Tables 1 and 4) showed a positive correlation between the two variables with the only exception being MM 2T-PrPSc, which showed an intermediate degree of PK resistance despite the very high aggregation ratio with the latter being comparable with that of vCJD-PrPSc, the most aggregated and most PK-resistant PrPSc type.
Detection of sPrPSc
Digestion of the P3 fractions obtained from the sucrose gradient centrifugation assay revealed a totally PK-sensitive component in each type. Fully PK-sensitive PrPSc distributed exclusively among fractions 1–4 (Fig. 6) irrespective of the CJD type, indicating that this sPrPSc component is less aggregated than rPrPSc. Overall, sPrPSc accounted for less than 10% of the whole PrPSc with MM 2C showing the highest and VV2 showing the lowest relative amount (Fig. 5D). Thus, the amount of this sPrPSc fraction seems to be unrelated to the overall degree of PrPSc PK resistance even if it is contributing to the aggregation ratio.
FIGURE 6.
Detection of fully sensitive PrPSc in slowly sedimenting fractions. Western blot analysis of PrPSc distribution among fractions collected after sucrose gradient centrifugation in MM 2C is shown. A, PrPSc distribution among the 12 fractions before PK digestion. Serial dilutions of a sample with known PrPSc content have been used as a standard curve. B, the PrPSc amount in the same fractions after PK digestion (PK dilution used, 0.25 unit/ml). Note that the signal in fractions 1–4 has completely disappeared, whereas the standard curve shows a comparable intensity of signal. C, overexposure of A showing that in fractions 1–4 (i) PrP dimers, typical of PrPSc and absent in PrPC, are well represented (arrowhead) and (ii) the PrP glycosylation pattern is typical of PrPSc and different from that of PrPC. D, overexposure of B demonstrating the complete disappearance of sPrPSc signal after PK digestion. Membranes were probed with mAb SAF 60. Approximate molecular masses are in kilodaltons.
The PK sensitivity of the PrPSc recovered in fractions 1–4 was comparable with that of PrPC because after adjusting for the total protein concentration it was almost completely digested by the minimal PK concentration required for PrPC digestion (data not shown). Thus, it seems that the aggregate size represents a main determinant of PrPSc resistance to PK digestion. Accordingly, changes in rPrPSc aggregate size could potentially affect sPrPSc content. Indeed, prolonged sonication after the ultracentrifugation steps during PrPSc purification resulted in an increase of sPrPSc amount in fractions 1–4 (data not shown).
rPrPSc PK Resistance in the Same Fraction from Different Types: MM1 Versus VV2
To determine whether CJD type-specific rPrPSc preparations with similar aggregation size maintain or not their difference in PK resistance, we performed PK titration curves on selected fractions of the sucrose gradient assay. For this experiment, we chose to compare fractions 6, 9, and 12 between MM1 and VV2, two common human sCJD variants, which showed statistically significant differences in their PK resistance (e.g. ED50) but not in their aggregation ratio (Fig. 7). By showing that VV2-rPrPSc is significantly more resistant than MM1-rPrPSc in each of the fractions analyzed, the results replicated the differences observed for the PK titration curves of the whole purified PrPSc (Fig. 8), indicating that this fundamental property of prions is also determined by structural PrPSc properties that are independent from the relative size of the protein aggregates.
FIGURE 7.
Analysis of protease resistance of purified PrPSc fractions with similar aggregation size: comparison between MM1 and VV2. For each sample, PK titration curves have been performed on fractions 6, 9, and 12 collected after subjecting purified PrPSc to sucrose gradient centrifugation. Samples were run either untreated (first lane) or after digestion with increasing amounts of PK for 1 or 2 h (last lane only) at 37 °C. Membranes were probed with mAb SAF 60. Approximate molecular masses are in kilodaltons.
FIGURE 8.
PK digestion profiles of purified PrPSc fractions with similar aggregation size: comparison between MM1 and VV2. The plots represent the amount (mean ± S.D. (error bars)) of PrPSc detected after PK digestion in fractions 6, 9, and 12 expressed as a percentage of the undigested sample. The PK concentrations used (x axis) are in a logarithmic scale. An asterisk (*) indicates statistically significant differences between MM1 and VV2 (Student's t test or Mann-Whitney rank sum test).
DISCUSSION
In the present study, we have carried out the first systematic analysis of PrPSc protease resistance and aggregation state across the spectrum of human sporadic and vCJD prions. The results show significant strain-specific differences in the PK digestion profile of human PrPSc that are only partially explained by differences in aggregate size, indicating that other factors also determine this fundamental property of prions.
Overall, CJD variants linked to PrPSc type 1 (VV1 and MM1 subtypes) were less PK-resistant than those associated with PrPSc type 2 (MM 2T, MM 2C, MV 2K, VV2, and vCJD), although clear differences were also seen among CJD prions sharing the same PrPSc type. In particular, VV1 PrPSc was by far the least resistant, whereas MV 2K/VV2- and vCJD-associated PrPSc showed the highest resistance to PK digestion. Interestingly, the PrPSc ED50 also appeared to be inversely correlated with the relative amount of the 13-kDa truncated C-terminal fragment generated by PK digestion (41), suggesting that the protease resistance of PrPSc also depends on the accessibility of PK to the PrPSc C terminus.
By showing that purified PrPSc forming low molecular mass aggregates is digested completely at a PK activity comparable with the lowest activity that is needed to fully digest PrPC, our data confirm that a minimal aggregation size for the PrPSc molecules is required to make the protein resistant to proteolysis and that this property, at least partially, depends on its quaternary structure. Furthermore, our data confirm the existence of a bona fide sPrPSc across the whole spectrum of human prions. However, our semiquantitative analyses clearly show that this slowly sedimenting sPrPSc represents only a relatively minor component of abnormal PrP not exceeding 10% of total detergent-insoluble PrPSc in any of the human sporadic TSE subtypes. Thus, previous concerns raised about the common practice of diagnosing human prion diseases by means of rPrPSc detection and characterization (24) should be, at least for classic sCJD and vCJD, significantly reduced. Moreover, our finding of a comparable relative amount of sPrPSc across the whole spectrum of CJD prions comprising six distinct strains an including VPSPr linked to MM or MV at codon 129 argues against the idea that sPrPSc, or at least its slowly sedimenting component, plays a major role in prion strain determination in human sporadic prion diseases.
The significant difference between our estimate of sPrPSc among sCJD isolates and that of Safar et al. (24) may depend on methodological aspects or data interpretation. First, the two approaches differ in the means by which they distinguish between PrPC and PrPSc. Indeed, although our method completely separated PrPC in a verifiable manner (Ref. 26 and present study), CDI maintains PrPC in the sample and estimates the amount of sPrPSc assuming that the epitope unmasking induced by guanidine hydrochloride is specific for PrPSc and as a consequence that PrPC would not contribute to the increase of the PrP signal after denaturation.
Second, although our estimate of sPrPSc has been obtained after the physical separation of a fully PK-sensitive PrPSc fraction forming aggregates of relatively low size, CDI measures sPrPSc as a part of the whole PrPSc without defining any actual structural or physical difference between the two (s versus r) PrPSc species, raising the issue of whether the sPrPSc signal detected by CDI reflects the properties of a distinct PrPSc molecule, is an indivisible part of the features of the PrPSc molecule, or a mixture of both.
Furthermore, it is noteworthy that PrPSc was originally defined as an abnormal conformer that is only partially resistant to protease digestion, a property that is also well illustrated in the present study. Thus, the CDI estimate of the amount of sPrPSc may also comprise this phenomenon. However, it is well known that the experimental conditions in which the protease digestion is performed (i.e. pH, temperature, and protein concentration) may dramatically vary the resistance of a given substrate (36). In addition, there are significant differences in the degree of protease resistance of PrPSc that depend on the type of prion isolate as shown in the present and a previous study from our group (42) for human PrPSc. Altogether, these considerations underline the difficulty and limits of creating a distinction between PrPSc isoforms (s versus r as it is increasingly reported in the literature) only based on a single digestion condition and without any definition of the actual structural or physical differences between the two PrP species. Furthermore, the strict dependence on the experimental conditions used would make any interlaboratory quantitative comparison between the putative “sensitive” and resistant components extremely difficult.
Our detailed PK digestion curves on the whole purified PrPSc, by showing the progression of the loss of PrPSc signal at increasing PK activity for each of the sCJD prions and vCJD, exemplify some of the issues discussed above and show the heterogeneity of this additional potential “sPrPSc-like” component according to the CJD subtype at a given PK activity. When the percentage of PrPSc signal loss is considered at the lowest PK activity we have used for the most resistant strains, which was still capable of trimming the N-terminal portion of PrPSc, generating PrP27–30, it was 1.9% in vCJD, 22.3% in VV2, and 53.4% in VV1. Nevertheless, it is noteworthy that even in the most sensitive prion isolate tested here, the VV1, the disappearance of the large majority of the PrPSc signal (85%) was only obtained in stringent experimental conditions (PK activity of 200 μg/ml, pH 7.4 and a very low protein concentration of 150 μg/ml). Therefore, it appears very unlikely that in less stringent experimental conditions such as those used in the CDI the loss of PrPSc after PK digestion would reach the amount estimated by CDI (24) in any of the CJD subtypes.
To further examine the relationship between aggregation size and PK sensitivity, we calculated the aggregation ratio, which measures the proportion of smaller PrP aggregates with respect to larger aggregates. We found a trend of values among the CJD strains reproducing, to some extent, those obtained with the PK digestion profiles. In particular, there was a positive correlation between the aggregation ratio and both PrPSc type 2 and codon 129 MM genotype with the vCJD group showing the highest value. Similarly, the relative amount of PrPSc among the fastest sedimenting fractions (9–12) also showed a statistically significant increase for both Met polymorphism and type 2, although in this case the value was highest for MM 2C rather than vCJD. Overall, these data are in agreement with the results of the study of Kobayashi et al. (43) suggesting that sCJD MM2 brains contain larger PrPSc aggregates than MM1 brains and with the work of Pham et al. (44) demonstrating that human recombinant PrP is more prone to aggregate when carrying Met at codon 129 instead of Val.
According to our findings, however, the positive correlation between the aggregation ratio and the ED50 expressing the resistance to proteolysis is not complete given, for example, that MM1 cases were significantly different from VV2s in their PK resistance despite the similar aggregation ratio. This is also in keeping with a previous observation obtained in Gerstmann-Sträussler-Scheinker disease or atypical scrapie where PrPSc forms relatively large aggregates that are easily digested by PK (26, 28–30, 45). To further address this issue, we also compared the PK resistance of single fractions containing aggregates of similar sizes obtained from different CJD strains. By showing that VV2 prions remain more PK-resistant than MM1 prions even when the influence of the size of the aggregates is eliminated, our findings strongly suggest that PrPSc sensitivity is also caused by other strain-specific factors. Given that this strain-specific difference in PK sensitivity becomes significant only when the PK activity is relatively high and that PrPSc is completely digested under a minimal aggregation size, indicating that PrPSc aggregation is a necessary condition for PK resistance to occur, we propose that this second factor comprises the stability of PrPSc aggregates. In other words, the most PK-resistant prions would be characterized not only by PrPSc aggregates of larger size but also kept together by stronger interactions. According to Masel and Jansen (46), PK digestion is biphasic: during the first phase, PK would attack single monomers (or exposed sites of aggregated polymers), whereas during the second phase, polymers must disaggregate to be digested. As a consequence, in this latter phase the digestion rate would exclusively depend on the rate of PrPSc disaggregation. Thus, if PrPSc associated with different CJD subtypes has indeed a heterogeneous propensity to fragmentation, then PK digestion will occur at different rates even in equally aggregated samples. This scenario (Fig. 9) would not only fit with our findings but also accommodate previous observations, particularly in Gerstmann-Sträussler-Scheinker disease or atypical scrapie where PrPSc forms relatively large aggregates that are easily digested by PK (26, 28–30, 35, 45). These could instead be explained by postulating that PrPSc forms less stable aggregates compared with classic CJD or scrapie rather than smaller aggregates. It is noteworthy that differences in aggregate frangibility among strains have already been demonstrated for yeast prions (47–49).
FIGURE 9.
PrPSc aggregate stability: a model to explain the heterogeneity of PrPSc resistance to proteolysis among prion strains. While strain A shows a compact PrPSc core with low tendency to crumble into monomers, strain B is characterized by a more unstable core that is more prone to break up into separate monomers. In the presence of a relatively low PK activity, both strains A and B produce a few monomers that are easily digested together with the exposed portions of the whole core. In the presence of a relatively higher PK activity, the fragmentation into new monomers is significantly accelerated only for the PrPSc core of strain B, whereas the more compact PrPSc core of strain A is less affected.
Interestingly, this scenario would implicate that PrPSc resistance to protease digestion could be reduced by lowering its aggregation size, although proportionally to the strength that keeps the PrPSc particles together. Previous studies have demonstrated that sonication reduces PrPSc aggregate size (50–52), and our preliminary studies suggest that a brief sonication cycle reduces PrPSc resistance to digestion in VV1 and MM1 subtypes but not in MV 2K or VV2s. The reduction in size of PrPSc aggregates would likely make them more susceptible to endogenous protease activity and may increase their clearance, which would have a significant therapeutic implication. To this aim, it would be important to understand the factors modulating PrPSc aggregate stability. Given that the two sCJD subgroups with the most significant difference in aggregate stability (VV1 and VV2) have identical PRNP sequence, it is likely that factors other than primary PrP sequence are involved. Several studies have pointed to the glycosaminoglycans as major players in protein aggregation related to neurodegeneration (53) and as prion disease therapeutic targets as well (54). Increasing evidence suggests that carbohydrate-involving interactions play a role in protein aggregation and aggregate stabilization. These would be promoted, for example, by glycation related to oxidative stress, which occurs between lateral amino acidic chains of the protein and carbonyl-containing groups, causing the formation of cross-links that stabilize protein aggregates (55). PrP contains a glycosaminoglycan-binding motif at its N terminus (56) and can be glycated in the same region (57). Furthermore, PrP contains two glycosylation sites (Asn-181 and Asn-197) that can carry N-linked oligosaccharides. The nature of these carbohydrates is still poorly understood, and it is not clear whether they are qualitatively different among sCJD subtypes. Nevertheless, host PrP glycosylation patterns can affect disease progression, alter the infectious properties of a specific prion strain, and even protect against disease onset (58–60). Our finding of a positive correlation between the relative percentage of diglycosylated PrPSc and the overall degree of PrPSc resistance to PK digestion also supports the notion that N-linked glycans influence protein aggregate stability. On the other hand, the observation that the kinetics of digestion of the three PrPSc glycoforms is fairly similar is only in apparent contrast with the idea formulated above because evidence indicates that PrPSc particles or aggregates always contain a mixture of glycoforms (61). Thus, the possibility that N-linked oligosaccharides modulate stability by themselves or contribute together with other protein-linked carbohydrate structures to determine the strain-specific PrPSc aggregate stability remains a scenario that should be considered and further explored.
In summary, in the present study we have documented that human prion strains differ in PrPSc PK sensitivity over a 100-fold range of PK concentration and that the differences stem from both aggregate stability and size. By providing new insight into prion strain variability with respect to PrPSc PK sensitivity and aggregation state, our findings could also serve as a starting point for understanding the properties of other disease-associated misfolded proteins. Indeed, increasing evidence argues that other protein aggregates (i.e. tau, α-synuclein, amyloid-β, and huntingtin aggregates), like prions, can seed the misfolding of their normal conformers (62) and even show a “strain” specificity in their PK digestion profile and in vivo cross-seeding ability as was recently shown for α-synuclein (63).
Acknowledgments
Prof. James Ironside at the National CJD Surveillance Unit in the United Kingdom kindly provided the tissues of variant CJD used for this study. We thank Dr. Barbara Polischi for excellent technical assistance.
This work was supported by Italian Ministry of Health Grant RF-2009-1474624, University of Bologna Grants RFO 2008–2010, and the Gino Galletti Foundation.
- TSE
- transmissible spongiform encephalopathy
- PrPC
- cellular prion protein
- PrPSc
- scrapie prion protein
- PrP
- prion protein
- CJD
- Creutzfeldt-Jacob disease
- sCJD
- sporadic Creutzfeldt-Jacob disease
- VPSPr
- variably protease-sensitive prionopathy
- vCJD
- variant CJD
- PK
- proteinase K
- sPrPSc
- PK-sensitive form of PrPSc
- CDI
- conformation-dependent immunoassay
- rPrPSc
- PK-resistant PrPSc
- TH
- total brain homogenate
- S1
- supernatant obtained after the first step of ultracentrifugation
- S2
- supernatant obtained after the second step of ultracentrifugation
- P2
- pellet obtained after the first step of ultracentrifugation
- S3
- supernatant obtained after the third step of ultracentrifugation
- P3
- pellet obtained after the third step of ultracentrifugation
- ANOVA
- analysis of variance
- C1
- 18-kDa C-terminal PrPC fragment associated with non-CJD brains
- C2 or PrP27–30
- 19/21-kDa PK-resistant PrPSc core associated with CJD brains
- CTF
- C-terminal fragment
- PRNP
- gene encoding PrP
- C
- cortical
- T
- thalamic.
REFERENCES
- 1. Prusiner S. B. (1998) Prions. Proc. Natl. Acad. Sci. U.S.A. 95, 13363–13383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zahn R., Liu A., Lührs T., Riek R., von Schroetter C., López García F., Billeter M., Calzolai L., Wider G., Wüthrich K. (2000) NMR solution structure of the human prion protein. Proc. Natl. Acad. Sci. U.S.A. 97, 145–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Caughey B. W., Dong A., Bhat K. S., Ernst D., Hayes S. F., Caughey W. S. (1991) Secondary structure analysis of the scrapie-associated protein PrP 27–30 in water by infrared spectroscopy. Biochemistry 30, 7672–7680 [DOI] [PubMed] [Google Scholar]
- 4. Pan K. M., Baldwin M., Nguyen J., Gasset M., Serban A., Groth D., Mehlhorn I., Huang Z., Fletterick R. J., Cohen F. E. (1993) Conversion of α-helices into β-sheets features in the formation of the scrapie prion proteins. Proc. Natl. Acad. Sci. U.S.A. 90, 10962–10966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Aguzzi A., Heikenwalder M., Polymenidou M. (2007) Insights into prion strains and neurotoxicity. Nat. Rev. Mol. Cell Biol. 8, 552–561 [DOI] [PubMed] [Google Scholar]
- 6. Gambetti P., Kong Q., Zou W., Parchi P., Chen S. G. (2003) Sporadic and familial CJD: classification and characterisation. Br. Med. Bull. 66, 213–239 [DOI] [PubMed] [Google Scholar]
- 7. Ghetti B., Tagliavini F., Takao M., Bugiani O., Piccardo P. (2003) Hereditary prion protein amyloidoses. Clin. Lab. Med. 23, 65–85 [DOI] [PubMed] [Google Scholar]
- 8. Capellari S., Strammiello R., Saverioni D., Kretzschmar H., Parchi P. (2011) Genetic Creutzfeldt-Jakob disease and fatal familial insomnia: insights into phenotypic variability and disease pathogenesis. Acta Neuropathol. 121, 21–37 [DOI] [PubMed] [Google Scholar]
- 9. Parchi P., Giese A., Capellari S., Brown P., Schulz-Schaeffer W., Windl O., Zerr I., Budka H., Kopp N., Piccardo P., Poser S., Rojiani A., Streichemberger N., Julien J., Vital C., Ghetti B., Gambetti P., Kretzschmar H. (1999) Classification of sporadic Creutzfeldt-Jakob disease based on molecular and phenotypic analysis of 300 subjects. Ann. Neurol. 46, 224–233 [PubMed] [Google Scholar]
- 10. Parchi P., Strammiello R., Notari S., Giese A., Langeveld J. P., Ladogana A., Zerr I., Roncaroli F., Cras P., Ghetti B., Pocchiari M., Kretzschmar H., Capellari S. (2009) Incidence and spectrum of sporadic Creutzfeldt-Jakob disease variants with mixed phenotype and co-occurrence of PrPSc types: an updated classification. Acta Neuropathol. 118, 659–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Parchi P., Cescatti M., Notari S., Schulz-Schaeffer W. J., Capellari S., Giese A., Zou W.-Q., Kretzschmar H., Ghetti B., Brown P. (2010) Agent strain variation in human prion disease: insights from a molecular and pathological review of the National Institutes of Health series of experimentally transmitted disease. Brain 133, 3030–3042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bishop M. T., Will R. G., Manson J. C. (2010) Defining sporadic Creutzfeldt-Jakob disease strains and their transmission properties. Proc. Natl. Acad. Sci. U.S.A. 107, 12005–12010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mastrianni J. A., Nixon R., Layzer R., Telling G. C., Han D., DeArmond S. J., Prusiner S. B. (1999) Prion protein conformation in a patient with sporadic fatal insomnia. N. Engl. J. Med. 340, 1630–1638 [DOI] [PubMed] [Google Scholar]
- 14. Moda F., Suardi S., Di Fede G., Indaco A., Limido L., Vimercati C., Ruggerone M., Campagnani I., Langeveld J., Terruzzi A., Brambilla A., Zerbi P., Fociani P., Bishop M. T., Will R. G., Manson J. C., Giaccone G., Tagliavini F. (2012) MM2-thalamic Creutzfeldt-Jakob disease: neuropathological, biochemical and transmission studies identify a distinctive prion strain. Brain Pathol. 22, 662–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zou W.-Q., Puoti G., Xiao X., Yuan J., Qing L., Cali I., Shimoji M., Langeveld J. P., Castellani R., Notari S., Crain B., Schmidt R. E., Geschwind M., Dearmond S. J., Cairns N. J., Dickson D., Honig L., Torres J. M., Mastrianni J., Capellari S., Giaccone G., Belay E. D., Schonberger L. B., Cohen M., Perry G., Kong Q., Parchi P., Tagliavini F., Gambetti P. (2010) Variably protease-sensitive prionopathy: a new sporadic disease of the prion protein. Ann. Neurol. 68, 162–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Will R. G., Ironside J. W., Zeidler M., Cousens S. N., Estibeiro K., Alperovitch A., Poser S., Pocchiari M., Hofman A., Smith P. G. (1996) A new variant of Creutzfeldt-Jakob disease in the UK. Lancet 347, 921–925 [DOI] [PubMed] [Google Scholar]
- 17. Bruce M. E., Will R. G., Ironside J. W., McConnell I., Drummond D., Suttie A., McCardle L., Chree A., Hope J., Birkett C., Cousens S., Fraser H., Bostock C. J. (1997) Transmissions to mice indicate that “new variant” CJD is caused by the BSE agent. Nature 389, 498–501 [DOI] [PubMed] [Google Scholar]
- 18. Collinge J., Sidle K. C., Meads J., Ironside J., Hill A. F. (1996) Molecular analysis of prion strain variation and the aetiology of “new variant” CJD. Nature 383, 685–690 [DOI] [PubMed] [Google Scholar]
- 19. Parchi P., Capellari S., Chen S. G., Petersen R. B., Gambetti P., Kopp N., Brown P., Kitamoto T., Tateishi J., Giese A., Kretzschmar H. (1997) Typing prion isoforms. Nature 386, 232–234 [DOI] [PubMed] [Google Scholar]
- 20. Ironside J. W. (2010) Variant Creutzfeldt-Jakob disease. Haemophilia 16, Suppl. 5, 175–180 [DOI] [PubMed] [Google Scholar]
- 21. Cobb N. J., Surewicz W. K. (2009) Prion diseases and their biochemical mechanisms. Biochemistry 48, 2574–2585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tzaban S., Friedlander G., Schonberger O., Horonchik L., Yedidia Y., Shaked G., Gabizon R., Taraboulos A. (2002) Protease-sensitive scrapie prion protein in aggregates of heterogeneous sizes. Biochemistry 41, 12868–12875 [DOI] [PubMed] [Google Scholar]
- 23. Pastrana M. A., Sajnani G., Onisko B., Castilla J., Morales R., Soto C., Requena J. R. (2006) Isolation and characterization of a proteinase K-sensitive PrPSc fraction. Biochemistry 45, 15710–15717 [DOI] [PubMed] [Google Scholar]
- 24. Safar J. G., Geschwind M. D., Deering C., Didorenko S., Sattavat M., Sanchez H., Serban A., Vey M., Baron H., Giles K., Miller B. L., Dearmond S. J., Prusiner S. B. (2005) Diagnosis of human prion disease. Proc. Natl. Acad. Sci. U.S.A. 102, 3501–3506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kim C., Haldiman T., Cohen Y., Chen W., Blevins J., Sy M.-S., Cohen M., Safar J. G. (2011) Protease-sensitive conformers in broad spectrum of distinct PrPSc structures in sporadic Creutzfeldt-Jakob disease are indicator of progression rate. PLoS Pathog. 7, e1002242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Parchi P., Chen S. G., Brown P., Zou W., Capellari S., Budka H., Hainfellner J., Reyes P. F., Golden G. T., Hauw J. J., Gajdusek D. C., Gambetti P. (1998) Different patterns of truncated prion protein fragments correlate with distinct phenotypes in P102L Gerstmann-Sträussler-Scheinker disease. Proc. Natl. Acad. Sci. U.S.A. 95, 8322–8327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Benestad S. L., Arsac J.-N., Goldmann W., Nöremark M. (2008) Atypical/Nor98 scrapie: properties of the agent, genetics, and epidemiology. Vet. Res. 39, 19. [DOI] [PubMed] [Google Scholar]
- 28. Polymenidou M., Prokop S., Jung H. H., Hewer E., Peretz D., Moos R., Tolnay M., Aguzzi A. (2011) Atypical prion protein conformation in familial prion disease with PRNP P105T mutation. Brain Pathol. 21, 209–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Monaco S., Fiorini M., Farinazzo A., Ferrari S., Gelati M., Piccardo P., Zanusso G., Ghetti B. (2012) Allelic origin of protease-sensitive and protease-resistant prion protein isoforms in Gerstmann-Sträussler-Scheinker disease with the P102L mutation. PLoS One 7, e32382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jansen C., Parchi P., Capellari S., Strammiello R., Dopper E. G., van Swieten J. C., Kamphorst W., Rozemuller A. J. (2011) A second case of Gerstmann-Sträussler-Scheinker disease linked to the G131V mutation in the prion protein gene in a Dutch patient. J. Neuropathol. Exp. Neurol. 70, 698–702 [DOI] [PubMed] [Google Scholar]
- 31. Safar J., Wille H., Itri V., Groth D., Serban H., Torchia M., Cohen F. E., Prusiner S. B. (1998) Eight prion strains have PrPSc molecules with different conformations. Nat. Med. 4, 1157–1165 [DOI] [PubMed] [Google Scholar]
- 32. Kim C., Haldiman T., Surewicz K., Cohen Y., Chen W., Blevins J., Sy M.-S., Cohen M., Kong Q., Telling G. C., Surewicz W. K., Safar J. G. (2012) Small protease sensitive oligomers of PrPSc in distinct human prions determine conversion rate of PrPC. PLoS Pathog. 8, e1002835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cronier S., Gros N., Tattum M. H., Jackson G. S., Clarke A. R., Collinge J., Wadsworth J. D. (2008) Detection and characterization of proteinase K-sensitive disease-related prion protein with thermolysin. Biochem. J. 416, 297–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Thackray A. M., Hopkins L., Klein M. A., Bujdoso R. (2007) Mouse-adapted ovine scrapie prion strains are characterized by different conformers of PrPSc. J. Virol. 81, 12119–12127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tixador P., Herzog L., Reine F., Jaumain E., Chapuis J., Le Dur A., Laude H., Béringue V. (2010) The physical relationship between infectivity and prion protein aggregates is strain-dependent. PLoS Pathog. 6, e1000859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Notari S., Capellari S., Giese A., Westner I., Baruzzi A., Ghetti B., Gambetti P., Kretzschmar H. A., Parchi P. (2004) Effects of different experimental conditions on the PrPSc core generated by protease digestion: implications for strain typing and molecular classification of CJD. J. Biol. Chem. 279, 16797–16804 [DOI] [PubMed] [Google Scholar]
- 37. Chen S. G., Parchi P., Brown P., Capellari S., Zou W., Cochran E. J., Vnencak-Jones C. L., Julien J., Vital C., Mikol J., Lugaresi E., Autilio-Gambetti L., Gambetti P. (1997) Allelic origin of the abnormal prion protein isoform in familial prion diseases. Nat. Med. 3, 1009–1015 [DOI] [PubMed] [Google Scholar]
- 38. Chen S. G., Teplow D. B., Parchi P., Teller J. K., Gambetti P., Autilio-Gambetti L. (1995) Truncated forms of the human prion protein in normal brain and in prion diseases. J. Biol. Chem. 270, 19173–19180 [DOI] [PubMed] [Google Scholar]
- 39. Parchi P., Zou W., Wang W., Brown P., Capellari S., Ghetti B., Kopp N., Schulz-Schaeffer W. J., Kretzschmar H. A., Head M. W., Ironside J. W., Gambetti P., Chen S. G. (2000) Genetic influence on the structural variations of the abnormal prion protein. Proc. Natl. Acad. Sci. U.S.A. 97, 10168–10172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zou W.-Q., Capellari S., Parchi P., Sy M.-S., Gambetti P., Chen S. G. (2003) Identification of novel proteinase K-resistant C-terminal fragments of PrP in Creutzfeldt-Jakob disease. J. Biol. Chem. 278, 40429–40436 [DOI] [PubMed] [Google Scholar]
- 41. Notari S., Strammiello R., Capellari S., Giese A., Cescatti M., Grassi J., Ghetti B., Langeveld J. P., Zou W.-Q., Gambetti P., Kretzschmar H. A., Parchi P. (2008) Characterization of truncated forms of abnormal prion protein in Creutzfeldt-Jakob disease. J. Biol. Chem. 283, 30557–30565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Notari S., Capellari S., Langeveld J., Giese A., Strammiello R., Gambetti P., Kretzschmar H. A., Parchi P. (2007) A refined method for molecular typing reveals that co-occurrence of PrPSc types in Creutzfeldt-Jakob disease is not the rule. Lab. Invest. 87, 1103–1112 [DOI] [PubMed] [Google Scholar]
- 43. Kobayashi A., Satoh S., Ironside J. W., Mohri S., Kitamoto T. (2005) Type 1 and type 2 human PrPSc have different aggregation sizes in methionine homozygotes with sporadic, iatrogenic and variant Creutzfeldt-Jakob disease. J. Gen. Virol. 86, 237–240 [DOI] [PubMed] [Google Scholar]
- 44. Pham N., Yin S., Yu S., Wong P., Kang S.-C., Li C., Sy M.-S. (2008) Normal cellular prion protein with a methionine at position 129 has a more exposed helix 1 and is more prone to aggregate. Biochem. Biophys. Res. Commun. 368, 875–881 [DOI] [PubMed] [Google Scholar]
- 45. Nazor K. E., Kuhn F., Seward T., Green M., Zwald D., Pürro M., Schmid J., Biffiger K., Power A. M., Oesch B., Raeber A. J., Telling G. C. (2005) Immunodetection of disease-associated mutant PrP, which accelerates disease in GSS transgenic mice. EMBO J. 24, 2472–2480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Masel J., Jansen V. A. (1999) The kinetics of proteinase K digestion of linear prion polymers. Proc. Biol. Sci. 266, 1927–1931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tanaka M., Collins S. R., Toyama B. H., Weissman J. S. (2006) The physical basis of how prion conformations determine strain phenotypes. Nature 442, 585–589 [DOI] [PubMed] [Google Scholar]
- 48. Immel F., Jiang Y., Wang Y.-Q., Marchal C., Maillet L., Perrett S., Cullin C. (2007) In vitro analysis of SpUre2p, a prion-related protein, exemplifies the relationship between amyloid and prion. J. Biol. Chem. 282, 7912–7920 [DOI] [PubMed] [Google Scholar]
- 49. Falsig J., Nilsson K. P., Knowles T. P., Aguzzi A. (2008) Chemical and biophysical insights into the propagation of prion strains. HFSP J. 2, 332–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhou X. B., Gao J. M., Gao C., Zhang B. Y., Yuan Y. K., Dong X. P. (2004) Influence of ultrasonic processing on the aggregation of PrP-Sc in the brain extracts of the scrapie-infected hamsters. Zhonghua Shi Yan He Lin Chuang Bing Du Xue Za Zhi 18, 118–121 [PubMed] [Google Scholar]
- 51. Weber P., Giese A., Piening N., Mitteregger G., Thomzig A., Beekes M., Kretzschmar H. A. (2006) Cell-free formation of misfolded prion protein with authentic prion infectivity. Proc. Natl. Acad. Sci. U.S.A. 103, 15818–15823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Weber P., Reznicek L., Mitteregger G., Kretzschmar H., Giese A. (2008) Differential effects of prion particle size on infectivity in vivo and in vitro. Biochem. Biophys. Res. Commun. 369, 924–928 [DOI] [PubMed] [Google Scholar]
- 53. Papy-Garcia D., Christophe M., Huynh M. B., Fernando S., Ludmilla S., Sepulveda-Diaz J. E., Raisman-Vozari R. (2011) Glycosaminoglycans, protein aggregation and neurodegeneration. Curr. Protein Pept. Sci. 12, 258–268 [DOI] [PubMed] [Google Scholar]
- 54. Zsila F., Gedeon G. (2006) Binding of anti-prion agents to glycosaminoglycans: evidence from electronic absorption and circular dichroism spectroscopy. Biochem. Biophys. Res. Commun. 346, 1267–1274 [DOI] [PubMed] [Google Scholar]
- 55. Vicente Miranda H., Outeiro T. F. (2010) The sour side of neurodegenerative disorders: the effects of protein glycation. J. Pathol. 221, 13–25 [DOI] [PubMed] [Google Scholar]
- 56. Yin S., Pham N., Yu S., Li C., Wong P., Chang B., Kang S.-C., Biasini E., Tien P., Harris D. A., Sy M.-S. (2007) Human prion proteins with pathogenic mutations share common conformational changes resulting in enhanced binding to glycosaminoglycans. Proc. Natl. Acad. Sci. U.S.A. 104, 7546–7551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Choi Y.-G., Kim J.-I., Jeon Y.-C., Park S.-J., Choi E.-K., Rubenstein R., Kascsak R. J., Carp R. I., Kim Y.-S. (2004) Nonenzymatic glycation at the N terminus of pathogenic prion protein in transmissible spongiform encephalopathies. J. Biol. Chem. 279, 30402–30409 [DOI] [PubMed] [Google Scholar]
- 58. Tuzi N. L., Cancellotti E., Baybutt H., Blackford L., Bradford B., Plinston C., Coghill A., Hart P., Piccardo P., Barron R. M., Manson J. C. (2008) Host PrP glycosylation: a major factor determining the outcome of prion infection. PLoS Biol. 6, e100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Cancellotti E., Bradford B. M., Tuzi N. L., Hickey R. D., Brown D., Brown K. L., Barron R. M., Kisielewski D., Piccardo P., Manson J. C. (2010) Glycosylation of PrPC determines timing of neuroinvasion and targeting in the brain following transmissible spongiform encephalopathy infection by a peripheral route. J. Virol. 84, 3464–3475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Cancellotti E., Mahal S. P., Somerville R., Diack A., Brown D., Piccardo P., Weissmann C., Manson J. C. (2013) Post-translational changes to PrP alter transmissible spongiform encephalopathy strain properties. EMBO J. 32, 756–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Khalili-Shirazi A., Summers L., Linehan J., Mallinson G., Anstee D., Hawke S., Jackson G. S., Collinge J. (2005) PrP glycoforms are associated in a strain-specific ratio in native PrPSc. J. Gen. Virol. 86, 2635–2644 [DOI] [PubMed] [Google Scholar]
- 62. Prusiner S. B. (2012) Cell biology. A unifying role for prions in neurodegenerative diseases. Science 336, 1511–1513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Guo J. L., Covell D. J., Daniels J. P., Iba M., Stieber A., Zhang B., Riddle D. M., Kwong L. K., Xu Y., Trojanowski J. Q., Lee V. M. (2013) Distinct α-synuclein strains differentially promote tau inclusions in neurons. Cell 154, 103–117 [DOI] [PMC free article] [PubMed] [Google Scholar]