Background: Glucose metabolism provides acetyl-CoA for histone acetylation.
Results: Inositol polyphosphates (InsPs), produced by the phospholipase C-dependent pathway, are required for degradation of the transcriptional repressor Mth1p, expression of glucose transporters, and normal acetyl-CoA homeostasis.
Conclusion: Defect in InsP synthesis results in global histone hypoacetylation and altered transcriptional regulation.
Significance: InsPs affect synthesis of glucose-derived acetyl-CoA and global histone acetylation.
Keywords: Acetyl-coenzyme A, Chromatin, Chromatin Histone Modification, Histones, Inositol Phosphates, Metabolism, Phospholipase C, Transcription
Abstract
Phospholipase C (Plc1p) is required for the initial step of inositol polyphosphate (InsP) synthesis, and yeast cells with deletion of the PLC1 gene are completely devoid of any InsPs and display aberrations in transcriptional regulation. Here we show that Plc1p is required for a normal level of histone acetylation; plc1Δ cells that do not synthesize any InsPs display decreased acetylation of bulk histones and global hypoacetylation of chromatin histones. In accordance with the role of Plc1p in supporting histone acetylation, plc1Δ mutation is synthetically lethal with mutations in several subunits of SAGA and NuA4 histone acetyltransferase (HAT) complexes. Conversely, the growth rate, sensitivity to multiple stresses, and the transcriptional defects of plc1Δ cells are partially suppressed by deletion of histone deacetylase HDA1. The histone hypoacetylation in plc1Δ cells is due to the defect in degradation of repressor Mth1p, and consequently lower expression of HXT genes and reduced conversion of glucose to acetyl-CoA, a substrate for HATs. The histone acetylation and transcriptional defects can be partially suppressed and the overall fitness improved in plc1Δ cells by increasing the cellular concentration of acetyl-CoA. Together, our data indicate that Plc1p and InsPs are required for normal acetyl-CoA homeostasis, which, in turn, regulates global histone acetylation.
Introduction
Phospholipase C (PLC)3 hydrolyzes phosphatidylinositol 4,5-bisphosphate, yielding two prominent eukaryotic second messengers: 1,2-diacylglycerol and inositol 1,4,5-trisphosphate. In yeast cells, PLC (Plc1p encoded by the PLC1 gene) and four inositol polyphosphate kinases (Ipk2p/Arg82p, Ipk1p, Kcs1p, and Vip1p) constitute a nuclear signaling pathway that is responsible for synthesis of inositol polyphosphates (InsPs) and inositol pyrophosphates (PP-InsPs) (1, 2). InsPs and PP-InsPs play an important role in regulation of DNA and RNA metabolism; they affect transcriptional control (3, 4), export of mRNA from the nucleus (5–8), homologous DNA recombination (9), telomere length (10, 11), and kinetochore function (12, 13). PP-InsPs also bind to the CDI·cyclin·CDK complex Pho81p·Pho80p·Pho85p and promote its inactivation (14, 15), and inhibit Akt signaling (16). Plc1p is required for the initial step of InsPs and PP-InsPs synthesis, and yeast cells with deletion of the PLC1 gene are completely devoid of any InsPs and PP-InsPs (5).
Plc1p and InsPs also regulate recruitment and activity of chromatin remodeling complexes and thus in addition to transcription may affect other chromatin-based processes such as replication, repair, and recombination (17, 18). Eukaryotic DNA is packaged into nucleosomes that represent basic building units of chromatin. The nucleosome structure limits access to DNA and thus the position and modification state of nucleosomes affect many processes in DNA metabolism (19). Chromatin-modifying complexes are classified into two categories. The first category includes ATP-dependent nucleosome-remodeling complexes that noncovalently modify and reposition nucleosomes, such as yeast Swi/Snf and RSC complexes (20–22). The second category includes complexes that post-translationally modify histones by acetylation, methylation, phosphorylation, and ubiquitynation (23–26). Charge neutralization of the histone tails by acetylation of the lysine residues is believed to weaken histone-DNA interactions and alter interactions between neighboring nucleosomes (24, 27–29). In addition, bromodomain-containing proteins such as Swi2p bind acetyl-lysine motifs in the histone tails and facilitate transcription (30). The enzymes responsible for histone acetylation are the histone acetyltransferases (HATs), whereas histone deacetylases (HDACs) remove acetyl groups from histones.
The activity of HATs depends on the concentration of acetyl-CoA in the nucleocytosolic compartment. The acetyl-CoA is produced by intermediary metabolism from glucose; however, the connection between intermediary metabolism and histone acetylation has been appreciated only recently (31–35). In mammalian cells, ATP-citrate lysase (ACL) generates acetyl-CoA in the nucleocytosolic compartment from glucose-derived citrate, and glucose availability affects histone acetylation in an ACL-dependent manner (36). Yeast cells do not have ACL and cytosolic acetyl-CoA is generated from acetate by acetyl-CoA synthetase, encoded by the ACS1 and ACS2 genes. Inactivation of ACS2 impairs global histone acetylation and transcriptional regulation (37). The cellular level of acetyl-CoA is also regulated by acetyl-CoA carboxylase Acc1p that catalyzes carboxylation of acetyl-CoA to malonyl-CoA. Decreased activity of Acc1p results in globally increased histone acetylation (38). In addition, the decreasing concentration of glucose in the medium as cells enter the stationary phase is accompanied by decreased histone acetylation (39, 40), whereas addition of glucose induces histone acetylation by picNuA4 and SAGA HAT complexes (41). The nucleocytosolic acetyl-CoA thus links histone acetylation with the metabolic state of the cell and perhaps provides an additional fine-tuning of the transcriptional regulation (31–33, 41, 42).
The changes in transcriptional control in cells with altered synthesis of InsPs prompted us to further explore the role of InsPs in regulation of the chromatin structutre. We now show that InsPs are required for normal levels of histone acetylation; plc1Δ cells display global histone hypoacetylation. Consequently, plc1Δ mutation is synthetically lethal with mutations in the SAGA and NuA4 complexes, the major yeast HATs. Conversely, deletion of histone deacetylase HDA1 partially suppressed the slow growth phenotype and improved overall fitness of plc1Δ cells. The histone hypoacetylation phenotype of plc1Δ cells is caused by altered synthesis of acetyl-CoA due to the failure to degrade repressor Mth1p. Increasing the cellular level of acetyl-CoA improves growth rate and suppresses the transcriptional defects of plc1Δ cells. Together, our data show that InsPs affect histone acetylation and transcriptional regulation by a mechanism that involves regulation of the acetyl-CoA homeostasis.
EXPERIMENTAL PROCEDURES
Strains and Media
All yeast strains are listed in Table 1. All the strains used in this study are isogenic to W303. Standard genetic techniques were used to manipulate yeast strains and introduce mutations from non-W303 strains into the W303 background (43). Cells were grown in rich medium (YPD; 1% yeast extract, 2% Bacto-peptone, 2% glucose) or under selection in synthetic complete medium containing 2% glucose and, when appropriate, lacking specific nutrients to select for a plasmid or strain with a particular genotype. Meiosis was induced in diploid cells by incubation in 1% potassium acetate.
TABLE 1.
Yeast strains used in this study
| Strain | Genotype | Source/Ref. |
|---|---|---|
| W303-1a | MATa ade2-1 his3-11,15 leu2–3,112 trp1-1 ura3-1 | R. Rothstein |
| ssd1-d2 can1–100 | ||
| W303-1α | MATα ade2-1 his3-11,15 leu2–3,112 trp1-1 ura3-1 | R. Rothstein |
| ssd1-d2 can1–100 | ||
| W303 | MATa/MATα ade2-1/ade2-1 his3-11,15/his3–11,15 | R. Rothstein |
| leu2-3,112/leu2-3,112 trp1-1/trp1-1ura3-1/ura3-1 | ||
| can1–100/ can1–100 | ||
| HL1-1 | W303-1α plc1::URA3 | 12 |
| HL1-3 | W303-1a plc1::URA3 | 103 |
| WPA023 | W303-1a plc1::TRP1 | 103 |
| WPL046 | W303-1α plc1::TRP1 | 103 |
| A0004 | W303-1α ipk1::kanMX | 5 |
| A0003 | W303-1α ipk2::HIS3 | 3 |
| LSY507 | W303-1α kcs1::HIS3 | 11 |
| DY5116 | W303-1a gcn5::TRP1 | 104 |
| FY1292 | MATa gcn5::HIS3 leu2Δ1 lys2–173 trp1Δ63 ura3–52 | 105 |
| ND818 | W303-1a gcn5::HIS3 | This study |
| ND703 | W303-1a spt3::TRP1 | 45 |
| ND713 | W303-1a spt7::LEU2 | 45 |
| DY6707 | W303-1a spt20::HIS3 | 106 |
| YTT2256 | W303-1a yng2::NatMX | 107 |
| YTT3122 | W303-1a htz1::NatMX | 107 |
| YTT2329 | W303-1a eaf1::NatMX | 107 |
| DY5068 | W303-1α hda1::URA3 | 104 |
| LG167 | W303-1a hda1::URA3 | This study |
| LG159 | W303-1a plc1::TRP1 hda1::URA3 | This study |
| CWY1128 | W303-1a grr1::LEU2 RGT1–3HA::KanMX2 | 84 |
| CWY1310 | W303-1a MTH1–6x-myc::KanMX2 | 84 |
| LG442 | W303-1a plc1::URA3 MTH1–6x-myc::KanMX2 | This study |
| LG510 | W303-1a ipk2::HIS3 MTH1–6x-myc::KanMX2 | This study |
| AUY009 | MATa tetO7-ACC1 ura3–52 trp1–63 leu2Δ1::tTA-LEU2 | 82 |
| LG362 | W303-1a tetO7-ACC1 | 38 |
| LG364 | W303-1a tetO7-ACC1 plc1::URA3 | This study |
| YM6266 | MATα his3Δ leu2Δ lys2Δ ura3Δ mth1::kanMX2 | 108 |
| LG471 | W303-1a mth1::kanMX2 | This study |
| LG474 | W303-1a plc1::URA3 mth1::kanMX2 | This study |
Western Blotting
Denatured proteins were separated on 15% denaturing polyacrylamide gels and Western blotting with anti-histone H3 polyclonal antibody (ab1791; Abcam), anti-histone H4 polyclonal antibody (2592; Cell Signaling), anti-acetyl histone H3 (Lys-14) polyclonal antibody (acH3K14; 07-353, Upstate Biotechnology), anti-hyperacetylated histone H4 polyclonal antibody (acH4K5,8,12,16; 06-946; Upstate Biotechnology), anti-Htz1p polyclonal antibody (ab4626; Abcam), anti-acetyl-Htz1p (Lys-14) polyclonal antibody (07-719; Upstate), and anti-myc (A-14) polyclonal antibody (sc-789; Santa Cruz Biotechnology) was carried out as described previously (44). To confirm equivalent amounts of loaded proteins, the membranes were also probed5′ with anti-Pgk1p monoclonal antibody 22C5 (A6457; Invitrogen).
Real-time RT-PCR Analysis
Total RNA was isolated from cultures grown in YPD medium to optical density A600 nm = 1.0 by the hot phenol method, treated with RNase-free DNase (Qiagen), and purified with an RNeasy Mini Kit (Qiagen). Reverse transcription and real-time PCR amplification were performed with the iScript kit (Bio-Rad) using 100 ng of RNA and the following primers: ACT1 (5′-TATGTGTAAAGCCGGTTTTGC-3′ and 5′-GACAATACCGTGTTCAATTGGG-3′), RPS22B (5′-AGCTGATGCTTTGAATGCCA-3′ and 5′-TTCGCCAATGTAACCATGCT-3′), RPL18B (5′-CCACCTGTTTCAGTCTCCAGAAT-3′ and 5′-TTGGGAATTCGAAGATCCTG-3′), HXT1 (5′-CAACTTAAGTGAAAGTCAAGTGCAAC-3′ and 5′-ATGAAACCACCGAAAGCAAC-3′), HXT3 (5′-GCCTTCGAATAGCTCTCAGGTA-3′ and 5′-CACAGTGACATATGCACCTTTACC-3′), HXT4 (5′-TGCCTATCAAGAGGATACAGCAG-3′ and 5′-GTCATCTCTTTCAGCTTTGTTGG-3′), YCR095C (5′-GAGGTCAAGAACCATCCAAGTTT-3′ and 5′-CAGAAGAGCTTTTTACCGGAAC-3′), GIT1 (5′-GGAAGACAAAGATATCACATCGG-3′ and 5′-AGGTTTCAGTACGGGTTGCA-3′), YCR100C (5′-TGTCATCTACGGACATCTGGAT-3′ and 5′-CCTTCCGATAGAATCTTCACGA-3′), YCR106W (5′-CTCGCGATGCCAACAAAATTC-3′ and 5′-TGAATCCATCAGAGTCGTTTGC-3′), HHT1/2 (5′-GAAGCCTCACAGATATAAGCCAG-3′ and 5′-ATCTTGAGCGATTTCTCTGACC-3′), HHF1/2 (5′-CCAAGCGTCACAGAAAGATTCTA-3′ and 5′-ACCAGAAATACGCTTGACACCA-3′), HTA1/2 (5′-CGGTGGTAAAGGTGGTAAAGC-3′ and 5′-TGGAGCACCAGAACCAATTC-3′), HTB1/2 (5′-CAAAGTTTTGAAGCAAACTCACCC-3′ and 5′-GCCAATTTAGAAGCTTCAGTAGC-3′), HTZ1 (5′-CATGGAGGTAAAGGTAAATCCG-3′ and 5′-GTAGCGTGCCTTTTCAGGTAAC-3′). Primer pairs for the canonical histones were designed so that they measure expression of both genes for that particular histone (HTA1 and HTA2, HTB1 and HTB2, HHT1 and HHT2, and HHF1 and HHF2). Gene expression was normalized to ACT1 expression, which is not altered in plc1Δ, ipk2Δ, ipk11Δ, and kcs1Δ cells (44).
Chromatin Immunoprecipitation and Quantitative Real-time PCR Analysis
In vivo chromatin cross-linking and immunoprecipitation were performed as described previously (13, 44–46) with the following antibodies: anti-myc polyclonal antibody A-14 (Santa Cruz Biotechnology), anti-histone H3 polyclonal antibody (ab1791; Abcam), anti-acetyl histone H3 (Lys-14) polyclonal antibody (acH3K14; 07-353, Upstate Biotechnology), and anti-hyperacetylated histone H4 polyclonal antibody (acH4K5,8,12,16; 06-946; Upstate Biotechnology). Total input DNA and coimmunoprecipitated DNA was then analyzed by real-time PCR with the Bio-Rad MyIQ single-color real-time PCR detection system (Bio-Rad). Each immunoprecipitation was performed at least three times using different chromatin samples, and the occupancy was calculated using the POL1 coding sequence as a negative control and corrected for the efficiency of the primers. The results were calculated as fold-increase in occupancy of the particular protein at the particular locus in comparison with the POL1 locus. Primers used for real-time PCR analysis are as follows: PGK1 (5′-CATTGGACGGTAAGAAGATCAC-3′ and 5′-TGAGAAGCCAAGACAACGTATC-3′), ACT1 (5′-CTCTTGTATTCTTCCTTCCCCTTTC-3′ and 5′-ATGGTGCAAGCGCTAGAACATAC-3′), ADH1 (5′-AATCCCACGGTAAGTTGGAATAC-3′ and 5′-AAGCGTGCAAGTCAGTGTGAC-3′), PYK1 (5′-TTGTTGCTGGTTCTGACTTGAG-3′ and 5′-CAATGTTCAAACCAGCCTTTCTC-3′), RPL18B (5′-CGTTACCCGACCTCGTTATTTTAC-3′ and 5′-CCTTTGGTGAAACAGGTAGTTTTG-3′), RPS22B (5′-GCCCATGTGTTGGAGGGAAGG-3′ and 5′-ATCAGCTAAAACGGAAGAGCGAG-3′), PHO5 (5′-CCATTTGGGATAAGGGTAAACATC-3′ and 5′-AGAGATGAAGCCATACTAACCTCG-3′), MDN1 promoter region (5′-GTACCTCTGAGTTAATTTGACATCG-3′ and 5′-GGACATTCTCAGCAGATTATAACGG-3′), MDN1 coding region-middle (5′-ATTGGCCGTTGGTACTTGTTG-3′ and 5′-ATGGAAAACACGTCTACTCTGGG-3′), MDN1 coding region-3′ end (5′-TCTGATGGTATTTGCGAAGACC-3′ and 5′-TGGCTCATGTCTAAGATGGATTC-3′), PMA1 promoter region (5′-TGGTGGGTACCGCTTATGCT-3′ and 5′-TTAG ATGTTAGACGATAATGATAGGACA-3′), PMA1 coding region-middle (5′-GACTTGATGTTGACTGCTTGTTTG-3′ and 5′-TGTACTTGGTCAAAGCGTCCTT-3′), PMA1 coding region-3′ end (5′-TTACTGTCGTCCGTGTCTGGAT-3′ and 5′-CCGTTCATCAATCTGTCAAAGG-3′), YCR095C (5′-GAGGTCAAGAACCATCCAAGTTT-3′ and 5′-CAGAAGAGCTTTTTACCGGAAC-3′), GIT1 (5′-GGAAGACAAAGATATCACATCGG-3′ and 5′-AGGTTTCAGTACGGGTTGCA-3′), YCR100C (5′-TGTCATCTACGGACATCTGGAT-3′ and 5′-CCTTCCGATAGAATCTTCACGA-3′), YCR106W (5′-CTCGCGATGCCAACAAAATTC-3′ and 5′-TGAATCCATCAGAGTCGTTTGC-3′), and POL1 (5′-TCCTGACAAAGAAGGCAATAGAAG-3′ and 5′-TAAAACACCCTGATCCACCTCTG-3′), HMR (5′-GCAGGTACTCCTGGTTTTTGTT-3′ and 5′-TCGCCTACCTTCTTGAACAAGAT-3′), YEF3 (5′-TAACGCCATGCAAGCTGTTG-3′ and 5′-GGAAATCAAAGTTTCGGAAGC-3′), ERG11 (5′-CAGCAGGCTTGAATAGAAACAGA-3′ and 5′-GCCAAGAAATGACTTAAACCAATG-3′).
Acetyl-CoA and Pyruvate Assay
Cells were grown in YPD medium to an optical density of A600 nm = 1.0. Sodium azide was added to a final concentration of 10 mm and 3 × 108 cells were harvested by centrifugation and lysed in 200 μl of 10% perchloric acid with pre-chilled glass beads. The lysate was neutralized with 10 m KOH to pH 7.5. Acetyl-CoA was assayed with a ELISA kit (Cusabio-Antibodies-online GmbH), and pyruvate was assayed with a pyruvate colorimetric assay kit (Abcam).
RESULTS
plc1Δ Cells Display Hypoacetylation of Histones H3 and H4
We have found previously that Plc1p and InsPs are required for recruitment of the HAT complex SAGA to osmoinducible promoters (45). The recruitment was not associated with increased histone acetylation in the corresponding promoters (45), most likely because of the simultaneous recruitment of the Rpd3p HDAC complex (47). However, the role of Plc1p and InsPs in SAGA recruitment prompted us to test whether InsPs affect targeted and/or global histone acetylation. To assess whether plc1Δ mutation affects acetylation of histones H3 and H4, we performed a Western blot analysis of cell lysates prepared from wild-type and plc1Δ cells and found a significant decrease in the levels of both acH3 and acH4 (Fig. 1A). To identify the specific inositol polyphosphate that is required for normal histone acetylation, we also analyzed lysates from ipk2Δ, ipk1Δ, and kcs1Δ strains. Ipk2p converts Plc1p-generated InsP3 into InsP4 and InsP5 (3, 5). Ipk1p converts InsP5 into InsP6, and Kcs1p produces inositol pyrophosphates PP-InsP4 and PP-InsP5 (10, 11, 48). The results show that, similar to plc1Δ cells, the ipk2Δ strain also displays decreased acetylation of histones H3 and H4 (Fig. 1A). Because ipk1Δ and kcs1Δ strains display a wild-type level of histone acetylation, synthesis of InsP4 and InsP5 thus appears to be required for normal histone acetylation.
FIGURE 1.
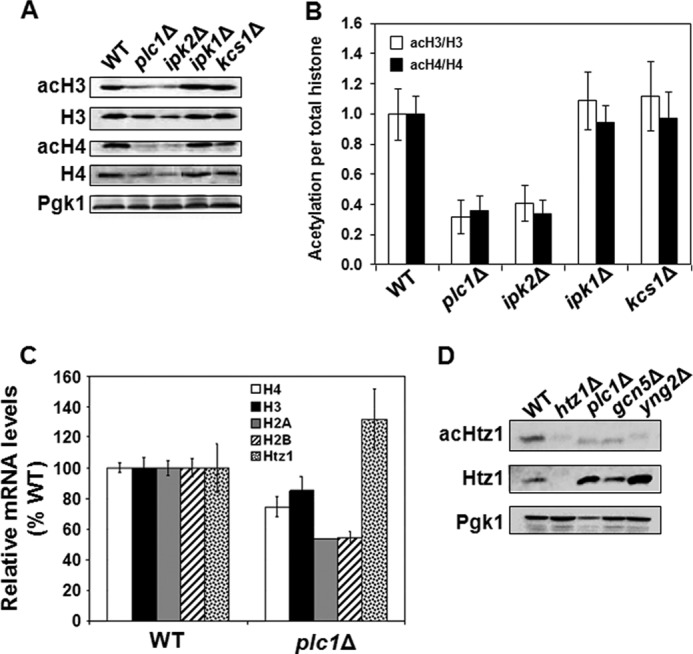
Histones H3 and H4 are hypoacetylated in plc1Δ and ipk2Δ cells. A, plc1Δ and ipk2Δ cells display lower levels of acH3 and acH4. Samples from the indicated strains were analyzed by Western blotting with antibodies against total histone H3 and H4, histone H3 acetylated at lysine 14 (acH3), and hyperacetylated histone H4 (acH4). Even loading of protein samples was confirmed with anti-phosphoglycerate kinase (Pgk1p) antibody. The experiment was performed three times, and representative results are shown. B, quantitative analysis of the Western blots was performed by densitometric analysis of the band intensities using UN-SCAN-IT software (Silk Scientific) and the ratios of acH3 to total histone H3 and acH4 to total histone H4 were plotted. The ratios represent mean ± S.D. from three independent experiments. C, expression of histones H4, H3, H2A, and H2B is reduced and expression of the H2A variant Htz1 is elevated in plc1Δ cells. WT and plc1Δ strains were grown in YPD medium at 30 °C to an A600 of 1.0 and the total RNA was isolated and assayed for H4, H3, H2A, H2B, and HTZ1 transcripts by real-time RT-PCR. The results were normalized to ACT1 RNA and expressed relative to the value for the WT strain. The experiment was repeated three times, and the results are shown as mean ± S.D. D, acetylation of Htz1p is reduced in plc1Δ cells. Samples from the indicated strains were analyzed by Western blotting with antibodies against Htz1p, acetylated Htz1p (acHtz1), and Pgk1. The experiment was performed three times, and representative results are shown.
We also noted that the level of non-acetylated histones H3 and H4 is somewhat reduced in plc1Δ and ipk2Δ cells. To determine, whether the lower abundance of histones H3 and H4 in plc1Δ cells is due to decreased transcription, we determined the mRNA level for histones H2A, H2B, H3, and H4 as well as the histone H2A variant HTZ1 (Fig. 1C). In agreement with the protein levels, the mRNA levels for individual core histones in plc1Δ cells were reduced to 55–85% of the wild-type levels. However, the expression of HTZ1 in plc1Δ cells was increased to ∼130% of the wild-type level. To eliminate the possibility that the lower level of acetylated histones H3 and H4 in plc1Δ cells is due to decreased expression of the histones, we determined the acetylation level of Htz1p. Htz1p is acetylated at Lys-14 by NuA4 and SAGA complexes (49, 50). The level of Htz1p is increased in plc1Δ cells and is thus in agreement with the increased mRNA level (Fig. 1, C and D). However, the level of acK14 Htz1p in plc1Δ cells is lower than in the wild-type cells (Fig. 1D). The results thus suggest that the decreased acetylation of histones H3 and H4 in plc1Δ cells is not due to the lower expression of both histones.
plc1Δ Cells Display Globally Decreased Untargeted Acetylation of Chromatin Histones
To test whether mutation in PLC1 decreases histone acetylation globally or only at specific loci, we evaluated by chromatin immunoprecipitation (ChIP) the occupancy of histone H3 acetylated at lysine 14 (acH3K14) as well as hyperacetylated histone H4 (acH4K5,8,12,16) at the promoter regions of PGK1, ACT1, ADH1, PYK1, and PHO5. We used anti-H3 antibody that recognizes the C-terminal region of H3 that is not post-translationally modified. The ChIP signal obtained with this antibody thus represents total H3 occupancy and can be used to calculate the histone acetylation levels per nucleosome content (51–54).
Histone H3 was 1.1 to 2 times less acetylated and histone H4 was 1.2 to 1.7 times less acetylated in the promoter regions of plc1Δ cells than in the wild-type cells (Fig. 2, A, C, E, G, and I). To account for differences in nucleosome density at the different promoters, we corrected the acH3 and acH4 occupancies for histone H3 content, and generated values that represent acetylation per nucleosome. The acetylation of histones H3 and H4 per nucleosome in the promoter regions was 2.2 to 5.8 and 1.9 to 4.6 times lower in plc1Δ cells than in the wild-type cells, respectively (Fig. 2, B, D, F, H, and J).
FIGURE 2.
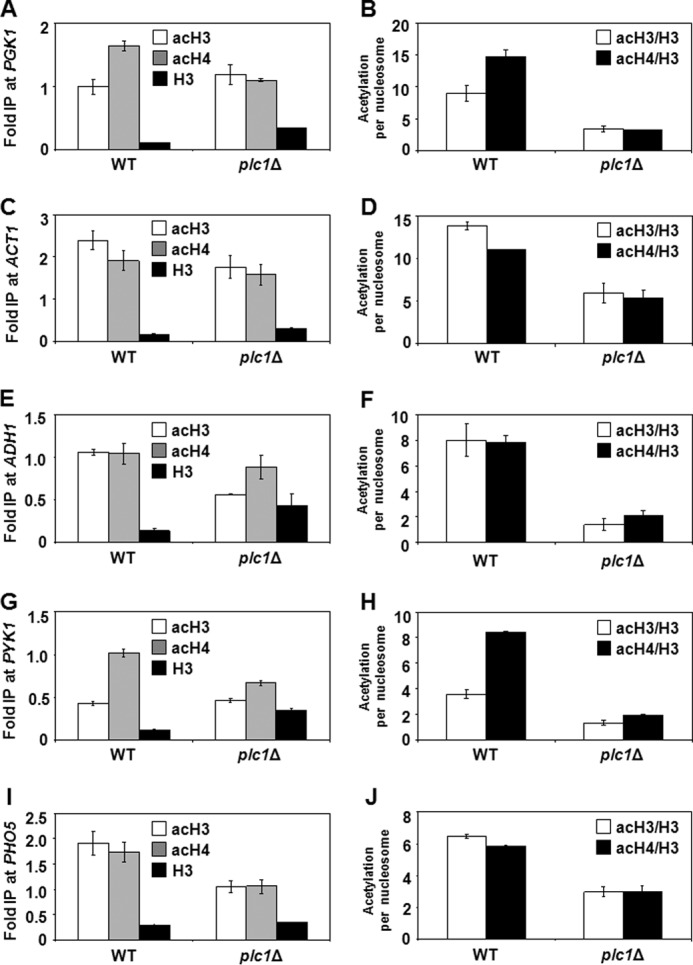
plc1Δ cells display decreased untargeted acetylation of chromatin histones. Wild-type and plc1Δ cells were grown at 30 °C in YPD medium to an A600 = 0.8. ChIP experiments were performed with antibodies against total histone H3 (H3), histone H3 acetylated at lysine 14 (acH3), and hyperacetylated histone H4 (acH4). Occupancies of H3, acH3, and acH4 were determined in the promoter region of PGK1 (A), ACT1 (C), ADH1 (E), PYK1 (G), and PHO5 (I). Acetylation per nucleosome was calculated as ratios of AcH3 to total H3 and acH4 to total H3 (B, D, F, H, and J). The experiments were repeated three times and results are shown as mean ± S.D.
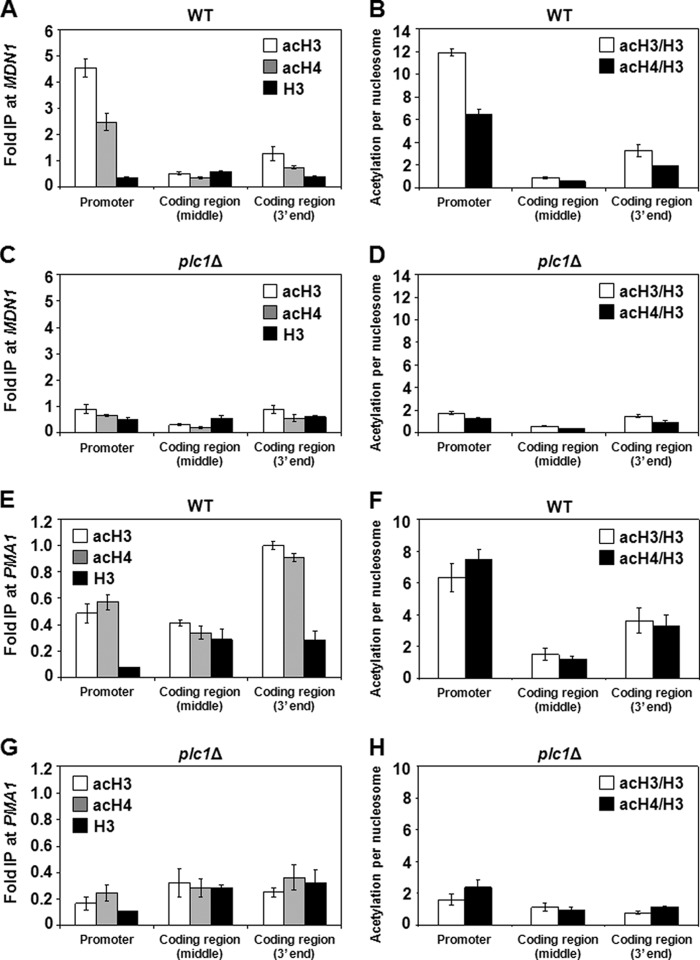
To test whether the decreased acetylation of histones found in the promoter regions of plc1Δ cells is also found in the coding regions, we evaluated the occupancy of acetylated histones in the long coding regions of MDN1 and PMA1. Because the DNA fragments obtained through sonication in the ChIP protocol are randomly generated, the long coding regions of MDN1 (15 kb) and PMA1 (3 kb) allowed us to design 3 sets of primers (promoter, middle of the coding region, and 3′ end of the coding region) far from each other to avoid overlap of the DNA fragments. We found that the total levels of acH3 and acH4 in plc1Δ cells were decreased 1.5 to 5.1 and 1.5 to 3.8 times, respectively, in the MDN1 coding region (Fig. 3, A and C), and decreased 1.3 to 4.0 and 1.2 to 2.5 times in the PMA1 coding region (Fig. 3, E and G), when compared with the wild-type cells. When we corrected the acetylation levels of histones H3 and H4 per nucleosome content, the plc1Δ cells showed a decrease of 1.6 to 6.9 and 1.7 to 5.1 times, respectively, in the MDN1 coding region (Fig. 3, B and D), and a decrease of 1.4 to 4.6 and 1.2 to 3.2 times, respectively, in the PMA1 coding region (Fig. 3, F and H), when compared with the wild-type cells. The decreased acetylation of both histones H3 and H4 at all tested loci suggests that the lack of InsPs in plc1Δ cells results in decreased acetylation of chromatin histones in a global, untargeted manner, and is in agreement with the decreased acetylation of bulk histones, shown by Western blot analysis (Fig. 1A).
FIGURE 3.
plc1Δ cells display decreased histone acetylation at MDN1 and PMA1 promoters and coding regions. Wild-type and plc1Δ cells were grown at 30 °C in YPD medium to an A600 = 0.8. ChIP experiments were performed with antibodies against total histone H3 (H3), histone H3 acetylated at lysine 14 (acH3), and hyperacetylated histone H4 (acH4). Occupancies of H3, acH3, and acH4 were determined in the promoter region, middle of the coding region, and 3′ end of the coding region of MDN1 and PMA1 for wild-type (A and E) and plc1Δ (C and G) cells. Acetylation per nucleosome was calculated as the ratios of acH3 to total H3 and acH4 to total H3 for wild-type (B and F) and plc1Δ (D and H) cells. The experiments were repeated three times and results are shown as mean ± S.D.
Hypoacetylation of Histones in plc1Δ Cells Results in Spread of the SIR Complex and Lower Expression of HMR- and Telomere-proximal Genes
Yeast heterochromatin occupies ribosomal DNA, the silent mating-type loci HMR and HML, and chromatin domains adjacent to telomere ends (55). Silencing at the silent mating loci and telomeres is mediated by Rap1p and the SIR complex that includes Sir1p, Sir2p, Sir3p, and Sir4p. The assembly of heterochromatin involves Sir2p-mediated deacetylation of histone H4 K16, and binding of Sir3p and Sir4p to deacetylated histone tails. The formation of the boundary regions that prevent the spread of heterochromatin into adjoining euchromatin requires acetylation of histone H4 K16 by the HAT SAS (56, 57). This acetylation then allows the incorporation of the histone variant Htz1p (58, 59), acetylation of which is also required for the efficient anti-silencing function of the boundary regions (49). In addition, the anti-silencing function of the boundary regions also requires Gcn5p- and Elp3p-mediated histone H3 acetylation (60). Because the balance between histone acetylation and histone deacetylation demarcates heterochromatin, we wanted to evaluate whether histone hypoacetylation of plc1Δ cells results in the spread of heterochromatin. We analyzed histone acetylation, occupancy of the SIR complex, and expression of genes YCR095C, GIT1, and YCR100C, flanking the HMR silent cassette, and YCR106W, localized close to the telomere of chromosome III (Fig. 4A). As expected, in comparison to the HMR- and telomere-proximal genes, the histone acetylation levels in the HMR locus were significantly lower in both wild-type and plc1Δ cells (Fig. 4B). In plc1Δ cells, the acetylation levels of histones H3 and H4 in the HMR- and telomere-proximal genes were 1.7 to 3.5 and 1.2 to 3.2 times lower than in the wild-type cells, respectively (Fig. 4B). Correcting the acetylation values for nucleosome content also showed a decrease in plc1Δ cells of 1.3 to 2.8 and 1.2 to 2.5 times for histones H3 and H4, respectively, when compared with the wild-type cells (Fig. 4C). The decrease in the histone acetylation level in plc1Δ cells resulted in significantly increased occupancy of Sir3p in HMR- and telomere-proximal genes of chromosome III in comparison with the wild-type cells (Fig. 4D), causing a defective boundary function as reflected by the decreased expression of these genes (Fig. 4E). Interestingly, Sir3p occupancy at the HMR locus was slightly decreased in plc1Δ cells in comparison to the wild-type cells. Our results suggest that histone hypoacetylation in plc1Δ cells affects the spread of Sir3p from heterochromatin regions, and are consistent with a previous study (49) that showed that two main HATs, SAGA and NuA4, are required for proper acetylation of nucleosomes in the HMR and telomere-proximal genes and that mutation in NuA4 severely affects the boundary function.
FIGURE 4.
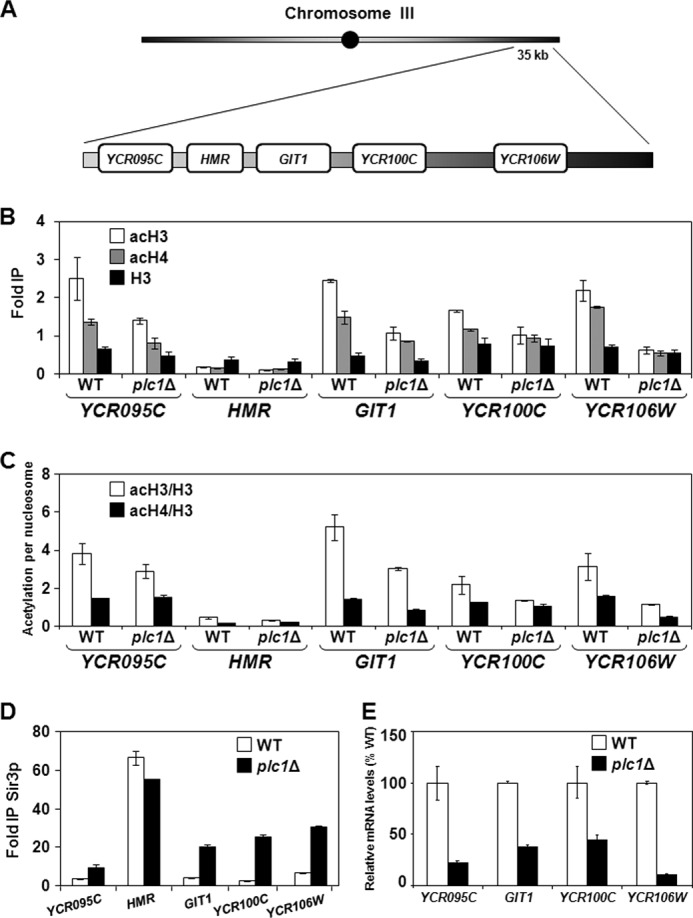
Histone hypoacetylation in plc1Δ cells leads to spread of Sir3p and lower expression of HMR- and telomere-proximal genes. A, diagram of the right arm of chromosome III containing the HMR locus and the flanking genes. B and D, wild-type and plc1Δ cells were grown at 30 °C in YPD medium to an A600 = 0.8 and ChIP experiments were performed with antibodies against total histone H3 (H3), histone H3 acetylated at lysine 14 (acH3), hyperacetylated histone H4 (acH4), and anti-myc (Sir3p). Occupancies of H3, acH3, acH4, and Sir3p were determined in the promoter regions of YCR095C, GIT1, YCR100C, YCR106W, and at the HMR locus. C, acetylation per nucleosome was calculated as ratios of AcH3 to total H3 and acH4 to total H3. The experiments were repeated three times and the results are shown as mean ± S.D. E, expression of YCR095C, GIT1, YCR100C, and YCR106W is reduced in plc1Δ cells. WT and plc1Δ strains were grown in YPD medium at 30 °C to an A600 = 1.0 and the total RNA was isolated and assayed for YCR095C, GIT1, YCR100C, and YCR106W transcripts by real-time RT-PCR. The results were normalized to ACT1 RNA and expressed relative to the value for the WT strain. The experiment was repeated three times and the results are shown as mean ± S.D.
Mutation in Histone Deacetylase HDA1 Improves Growth Rate and Fitness of plc1Δ Cells
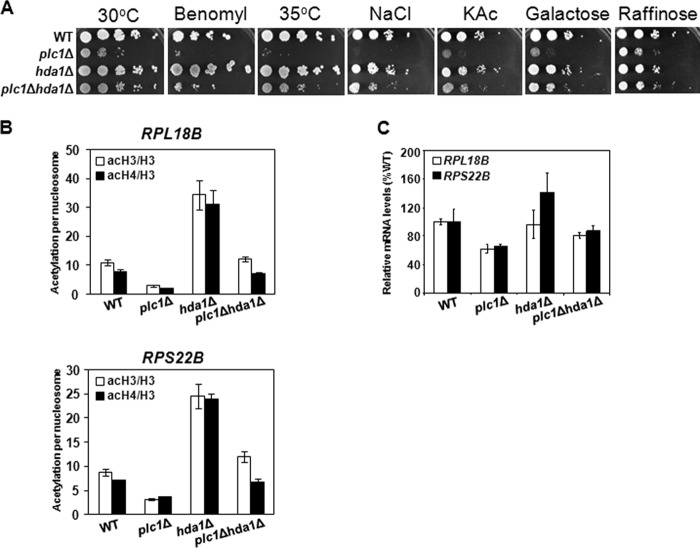
The dynamic balance between histone acetylation and deacetylation, mediated by the activities of HATs and HDACs, is well regulated and required for proper execution of the transcriptional program. The NuA4 and SAGA complexes are the major HAT activities that are counteracted by HDAC activities of the HDA and Rpd3 complexes (61). The NuA4, SAGA, HDA, and Rpd3 complexes provide the bulk control of the dynamic balance of global histone acetylation and deacetylation, with the HDA complex providing the major counterbalancing effect on the HAT activities, indicating that the HDA complex removes the largest amount of acetyl groups (61, 62). Because plc1Δ cells display decreased acetylation of histones H3 and H4 (Figs. 2–4) and number of aberrant phenotypes, including slow growth and temperature sensitivity (63, 64), we tested whether inactivating the HDA complex would improve the growth rate and fitness of plc1Δ cells. Hda1p is the catalytic subunit of the HDA complex that deacetylates H3 and H2B (65, 66). Deletion of HDA1 was shown to reverse the hypoacetylation of H3K9,14 caused by gcn5Δ mutation (61). Introducing the hda1Δ mutation in plc1Δ cells resulted in improved growth rate, as well as increased benomyl resistance, temperature resistance, osmotic resistance, and the improved ability to utilize carbon sources other than glucose (Fig. 5A). These results also suggest that histone hypoacetylation in plc1Δ cells is global and leads to a decreased growth rate and overall fitness of plc1Δ cells. The fact that the rpd3Δ mutation does not suppress the slow growth phenotype of plc1Δ cells (data not shown) can be explained by the role of Rpd3C(S) in regulating transcriptional elongation (67–69). If Plc1p is required for normal acetylation of histones, then one would expect that the plc1Δ mutation would display synthetic genetic interactions with the two major HATs, SAGA and NuA4. In agreement with this prediction, we found that the plc1Δ mutation is synthetically lethal with deletions of several subunits of the SAGA and NuA4 complexes (Table 2).
FIGURE 5.
Mutation in histone deacetylase HDA1 improves growth rate and suppresses defects in expression of ribosomal protein genes in plc1Δ cells. A, slow growth of plc1Δ cells on YPD medium and different carbon sources, as well as sensitivity to benomyl, high temperature, and osmolarity are partially suppressed by hda1Δ mutation. Cells were grown to log phase at 30 °C and 10-fold serial dilutions were spotted onto YPD plates at 30 or 35 °C, with YPD plates containing benomyl (10 μg/ml) or NaCl (0.4 m), or YP plates containing 2% potassium acetate (KAc), 2% galactose, or 2% raffinose, and grown for 2 days at 30 °C. Typical results from three independent experiments are shown. B, histone hypoacetylation in the promoter regions of RPL18B and RPS22B of plc1Δ cells is suppressed by hda1Δ mutation. Occupancies of H3, acH3, and acH4 were determined in the promoter region of RPL18B and RPS22B for wild-type, plc1Δ, hda1Δ, and plc1Δhda1Δ cells. Acetylation per nucleosome was calculated as ratios of acH3 to total H3 and acH4 to total H3 and shown as mean ± S.D. for three independent experiments. C, low expression of ribosomal protein genes in plc1Δ cells is suppressed by hda1Δ mutation. Wild-type, plc1Δ, hda1Δ, and plc1Δhda1Δ cells were grown in YPD medium at 30 °C to an A600 = 1.0 and the total RNA was isolated and assayed for RPL18B and RPS22B transcripts by real-time RT-PCR. The results were normalized to ACT1 RNA and expressed relative to the value for the WT strain. The experiment was repeated three times, and the results are shown as mean ± S.D.
TABLE 2.
Genetic interactions between plc1Δ and HAT mutations
Heterozygous diploids were prepared by standard genetic crosses and the synthetic lethality was deduced from the failure to recover any double mutants after dissecting at least 50 tetrads.
| HAT complex | Mutations synthetically lethal with plc1Δ |
|---|---|
| SAGA | gcn5Δ, spt7Δ, spt3Δ, spt20Δ |
| NuA4 | yng2Δ, eaf1Δ |
The suppression of the slow growth phenotype of plc1Δ cells by the hda1Δ mutation suggests that many loci are hypoacetylated in plc1Δ cells and the corresponding genes have altered expression. The ribosomal protein genes are among the most highly transcribed genes in the yeast genome, and their expression correlates with growth rate (70, 71). Following the general trend that promoters of highly transcribed genes are generally associated with increased histone acetylation (72–74), the acetylation of histones in ribosomal protein gene promoters by NuA4 is known to regulate their transcription (75, 76). Our previous results showed that many ribosomal protein genes have decreased expression in plc1Δ cells (44). Consistently with the notion that plc1Δ cells display global histone hypoacetylation, the acetylation per nucleosome in the promoters of two ribosomal protein genes, RPL18B and RPS22B, was decreased in plc1Δ cells in comparison with wild-type cells, and the decreased acetylation in plc1Δ cells was suppressed by the hda1Δ mutation (Fig. 5B). As expected, the expression of RPL18B and RPS22B genes was reduced in plc1Δ cells to about 60% of the wild-type level and the defect was again partially suppressed by the hda1Δ mutation (Fig. 5C).
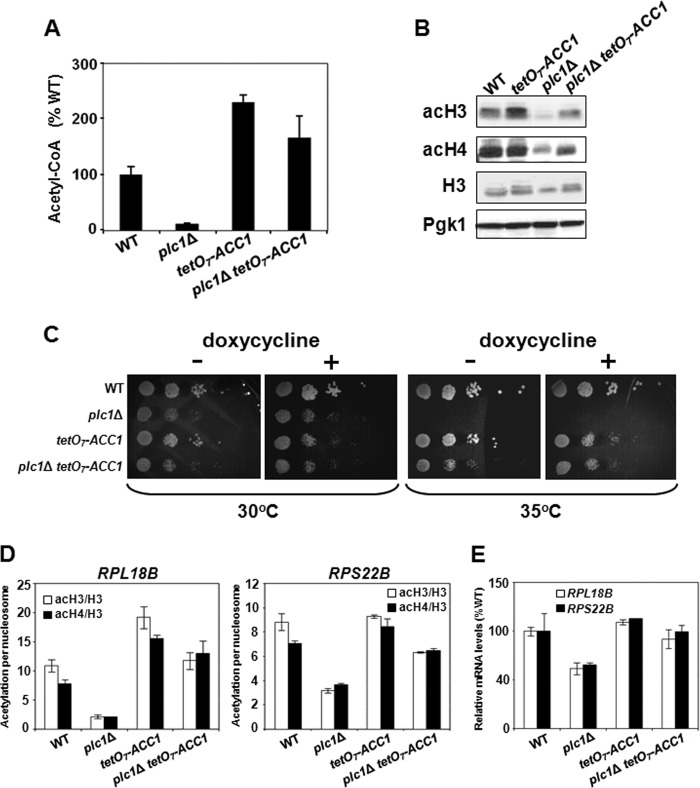
plc1Δ Cells Display Reduced Level of Acetyl-CoA
Nucleocytosolic acetyl-CoA is the common substrate for all HATs, and defect in the acetyl-CoA synthesis results in global untargeted histone hypoacetylation (37). To test the possibility that the histone hypoacetylation phenotype of plc1Δ cells is due to a defect in acetyl-CoA synthesis, we determined the cellular level of acetyl-CoA in wild-type and plc1Δ cells. The acetyl-CoA level in plc1Δ cells was only 12% of the wild-type level (Fig. 6A). In budding yeast, the majority of glycolytically produced pyruvate is converted to acetaldehyde and subsequently into ethanol; only a small fraction of acetaldehyde is converted into acetate (77–79). This acetate is subsequently converted to acetyl-CoA by nucleocytosolic acetyl-CoA synthetase 2 (Acs2p) (37). Because glucose represses tricarboxylic cycle and respiration in Saccharomyces cerevisiae, only a very small fraction of glycolytically produced pyruvate is translocated into mitochondria and converted to acetyl-CoA by the pyruvate dehydrogenase complex (77–79). The mitochondrial pool of acetyl-CoA in glucose-grown cells is very small and because it is biochemically isolated cannot be used for histone acetylation (37). Only the nucleocytosolic acetyl-CoA is available to HATs and its level regulates global histone acetylation (37); however, this acetyl-CoA can be also used for de novo synthesis of fatty acids (80). The first and rate-limiting reaction in de novo synthesis of fatty acids is carboxylation of acetyl-CoA to malonyl-CoA, catalyzed by acetyl-CoA carboxylase (Acc1p) (80). We recently showed that histone acetylation and synthesis of fatty acids compete for the same acetyl-CoA pool and that reduced expression of ACC1 results in globally increased histone acetylation (38). To test the possibility that decreased expression of ACC1 in plc1Δ cells would increase acetyl-CoA and histone acetylation levels, we suppressed expression of the ACC1 gene using the regulatable tetO7 promoter fused to the ACC1 coding region (tetO7-ACC1) (38, 81, 82). Indeed, reduced ACC1 expression rendered by the tetO7-ACC1 allele increased the acetyl-CoA level (Fig. 6A), increased acetylation of bulk histones H3 and H4 (Fig. 6B), and suppressed the temperature sensitivity of plc1Δ cells (Fig. 6C). Suppression of the histone hypoacetylation phenotype of plc1Δ cells by reduced ACC1 expression was also detectable by ChIP analysis at RPL18B and RPS22B promoters (Fig. 6D) and resulted in increased expression of these genes in plc1ΔtetO7-ACC1 cells than in the plc1Δ cells (Fig. 6E).
FIGURE 6.
plc1Δ cells that display reduced levels of acetyl-CoA and histone hypoacetylation in plc1Δ cells can be suppressed by increasing the pool of cytosolic acetyl-CoA. A, low intracellular level of acetyl-CoA in plc1Δ cells is partially suppressed by reduced ACC1 expression. The indicated strains were grown in YPD medium containing 0.05 μg/ml of doxycycline to an A600 = 0.8. The cells were harvested by centrifugation, lysed with glass beads in perchloric acid, and acetyl-CoA was determined in neutralized lysates. The experiment was repeated three times, and the results are shown as mean ± S.D. 100% wild-type levels of acetyl-CoA equals to 1.6 nmol/107 cells. B, histone hypoacetylation in plc1Δ cells is partially suppressed by the tetO7-ACC1 allele. The indicated strains were grown in YPD medium containing 0.05 μg/ml of doxycycline to an A600 = 0.8. Samples were analyzed by Western blotting with antibodies against histone H3 acetylated at lysine 14 (acH3), hyperacetylated histone H4 (acH4), and total histone H3. Even loading of protein samples was confirmed with anti-Pgk1p antibody. The experiment was performed three times, and representative results are shown. C, temperature sensitivity of plc1Δ cells is partially suppressed by reduced ACC1 expression. 10-Fold serial dilutions of the indicated strains were spotted onto YPD plates without doxycycline and YPD plates containing 0.1 μg/ml of doxycycline and grown for 2 days at 30 and 35 °C. Typical results from three independent experiments are shown. D, histone hypoacetylation in the promoter regions of RPL18B and RPS22B of plc1Δ cells is partially suppressed by the tetO7-ACC1 allele. Occupancies of H3, acH3, and acH4 were determined in the promoter regions of RPL18B and RPS22B and the acetylation per nucleosome was calculated as ratios of acH3 to total H3 and acH4 to total H3. The experiment was repeated three times, and the results are shown as mean ± S.D. E, low expression of ribosomal protein genes in plc1Δ cells is suppressed by the tetO7-ACC1 allele. Wild-type, plc1Δ, tetO7-ACC1, and plc1Δ tetO7-ACC1 cells were grown in YPD medium containing 0.05 μg/ml of doxycycline at 30 °C to an A600 = 1.0 and the total RNA was isolated and assayed for RPL18B and RPS22B transcripts by real-time RT-PCR. The results were normalized to ACT1 RNA and expressed relative to the value for the WT strain. The experiment was repeated three times, and the results are shown as mean ± S.D.
Plc1p Is Required for Expression of the HXT Genes
Availability of glucose modulates histone acetylation through glycolysis flux and synthesis of acetyl-CoA (41), which implies that defects in sensing, transport, or catabolism of glucose affect the acetyl-CoA level and global histone acetylation. To test whether plc1Δ cells are able to respond to glucose addition by up-regulating the transcription of glucose transporter genes, we analyzed expression of HXT1, HXT3, and HXT4 genes, which are regulated in response to different glucose concentrations (83, 84). HXT1 is induced in high but not low glucose, HXT3 is efficiently expressed in both high and low glucose, and HXT4 is induced in low but not high glucose (83, 85). Interestingly, all glucose transporters tested were expressed less in plc1Δ cells than in wild-type cells (Fig. 7, A–C). Because the ipk2Δ strain also displays hypoacetylation of histones H3 and H4 (Fig. 1A) and Ipk1p and Kcs1p are required for synthesis of InsP6 and PP-InsP4/PP-InsP5, respectively, we also analyzed expression of HXT1, HXT3, and HXT4 genes in ipk2Δ, ipk1Δ, and kcs1Δ strains. The results show that similarly to plc1Δ cells, the ipk2Δ strain also displays decreased expression of the tested glucose transporters (Fig. 7, A–C). The expression pattern of HXT1, HXT3, and HXT4 genes in ipk1Δ and kcs1Δ strains was similar to the pattern in the wild-type cells. The results suggest that the absence of InsP4 and InsP5 in plc1Δ and ipk2Δ cells hinders expression of glucose transporters. As a negative control we used grr1Δ cells. The F-box protein Grr1p is a component of the Skp1/Cullin/F-box (SCF) E3 ubiquitin ligase complex. SCFGrr1 is required for degradation of Mth1p, a negative regulator of expression of HXT genes (84, 86, 87). Cells lacking Grr1p are unable to degrade Mth1p in response to glucose and therefore cannot express HXT genes (84).
FIGURE 7.
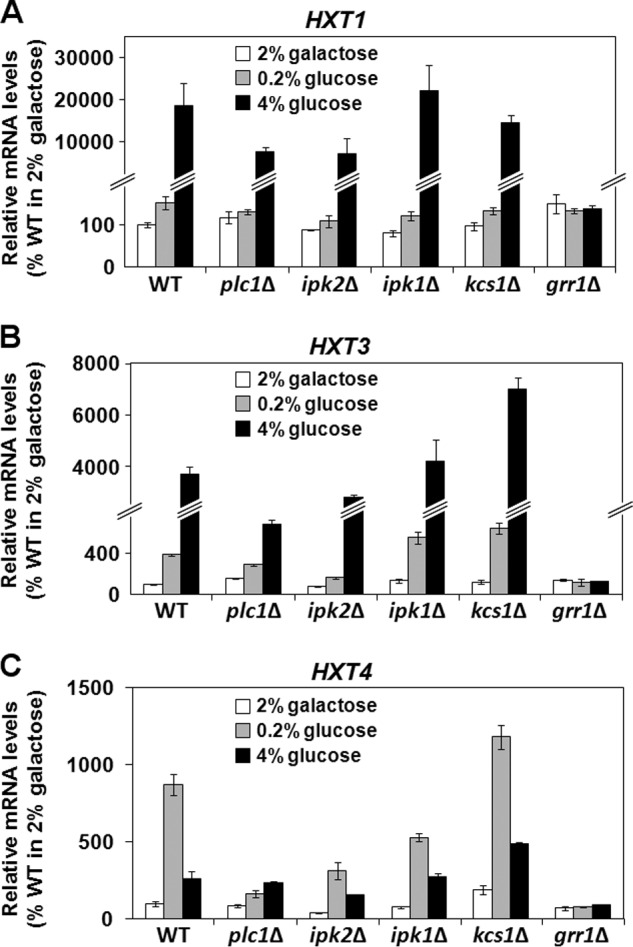
Plc1p and Ipk2p are required for expression of glucose transporters upon glucose induction. Wild-type, plc1Δ, ipk2Δ, ipk1Δ, kcs1Δ, and grr1Δ cells were grown in YP medium containing 2% galactose to an A600 = 0.5 at 30 °C. The culture was split and either left untreated or adjusted to a final concentration of 0.2% or 4% glucose and grown for an additional 2 h. Total RNA was isolated and assayed for HXT1 (A), HXT3 (B), and HXT4 (C) transcripts by real-time RT-PCR. The results were normalized to ACT1 RNA and expressed relative to the value for the WT strain grown in 2% galactose. The experiment was repeated three times, and the results are shown as mean ± S.D.
Plc1p Is Required for Mth1p Degradation and Normal Glucose Metabolism
In S. cerevisiae cells grown under aerobic conditions, glucose is metabolized mainly by glycolysis, generating pyruvate. If glucose sensing, transport, or catabolism is hindered in plc1Δ cells, one would expect a low intracellular level of pyruvate. Indeed, plc1Δ cells contain only 32% of the wild-type pyruvate level (Fig. 8A). This finding is in agreement with the lower level of acetyl-CoA found in plc1Δ cells (Fig. 6A). To elucidate the mechanism responsible for the altered glucose metabolism in plc1Δ cells, we evaluated the kinetics of Mth1p degradation in response to glucose. Transcription of HXT genes is regulated by the repressor Rgt1p and co-repressors Mth1p and Std1p (88). In the absence of glucose, Rgt1p represses transcription of all HXT transporters (89–93). This repression requires co-repressors Mth1p and Std1p (88). The glucose signal results in Grr1p-dependent degradation of Mth1p (84), which exposes Rgt1p to phosphorylation, probably by PKA (94, 95). This phosphorylation results in dissociation of Rgt1p from HXT promoters and alleviates the repressive activity of Rgt1p (Fig. 9). Although Mth1p was rapidly degraded upon addition of glucose in the wild-type cells, Mth1p was still detectable 20 min after addition glucose in plc1Δ and ipk2Δ cells (Fig. 8B). Thus, it seems that Plc1p and Ipk2p are required for proper Mth1p degradation that is needed for efficient expression of the HXT genes. To test whether the defect in Mth1p degradation is responsible for the decreased level of acetyl-CoA and histone hypoacetylation in plc1Δ cells, we introduced the mth1Δ mutation in plc1Δ cells. Indeed, mth1Δ mutation not only increased the cellular level of acetyl-CoA and histone acetylation (Fig. 8, C and D), but also improved the growth rate and partially suppressed temperature sensitivity of plc1Δ cells (Fig. 8E). The notion that proper degradation of the Mth1p repressor and transcription of the glucose transporters is required for histone acetylation is in agreement with the finding that grr1Δ cells also display histone hypoacetylation (Fig. 8F). Cumulatively, our results show that Plc1p and InsPs are important for Mth1p degradation, normal glucose metabolism, and acetyl-CoA synthesis, and highlight the connection between regulation of the intermediary metabolism and global histone acetylation.
FIGURE 8.
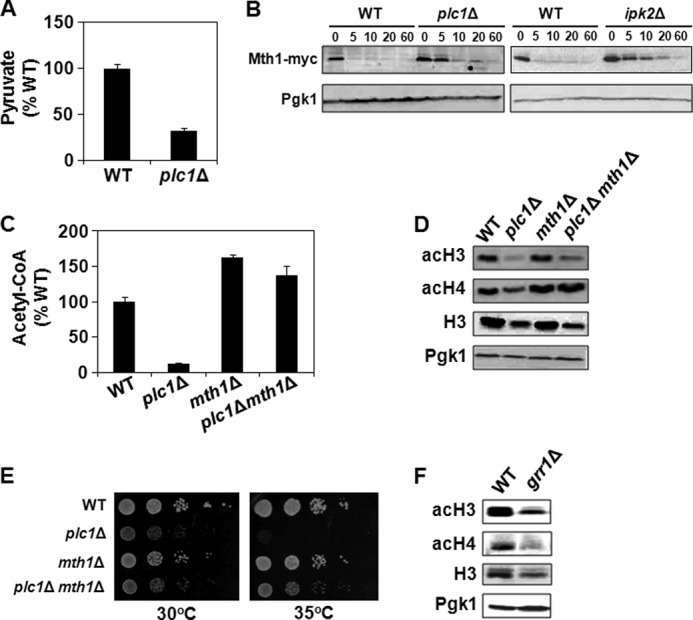
Increased stability of Mth1p is responsible for the decreased level of acetyl-CoA and histone hypoacetylation in plc1Δ cells. A, plc1Δ cells have low intracellular levels of pyruvate. Cells were grown in YPD medium, lysed, and pyruvate was assayed with colorimetric assay kit. The experiment was repeated three times, and the results are shown as mean ± S.D. B, samples from wild-type, plc1Δ, and ipk2Δ cells expressing Mth1-myc were grown in YP medium containing 2% galactose to an A600 = 0.8. Glucose was subsequently added to 4% and samples were taken just before addition of glucose and at the times indicated after the addition of glucose. Cell extracts were analyzed by Western blotting with anti-myc antibodies. Even loading of protein samples was confirmed with anti-Pgk1p antibody. The experiment was performed three times, and representative results are shown. C, low intracellular level of acetyl-CoA in plc1Δ cells is partially suppressed by the mth1Δ mutation. The indicated strains were grown in YPD medium to an A600 = 0.8. The cells were harvested by centrifugation, lysed with glass beads in perchloric acid, and acetyl-CoA was determined in neutralized lysates. The experiment was repeated three times, and the results are shown as mean ± S.D. 100% wild-type levels of acetyl-CoA equals 1.6 nmol/107 cells. D, hypoacetylation of bulk histones in plc1Δ cells is suppressed by mth1Δ mutation. Samples from the indicated strains were analyzed by Western blotting with antibodies against total histone H3, histone H3 acetylated at lysine 14 (acH3), and hyperacetylated histone H4 (acH4). Even loading of protein samples was confirmed with anti-phosphoglycerate kinase (Pgk1p) antibody. The experiment was performed three times, and representative results are shown. E, temperature sensitivity of plc1Δ is partially suppressed by mth1Δ mutation. 10-Fold serial dilutions of the indicated strains were spotted onto YPD plates, and grown for 2 days at 30 and 35 °C. Typical results from three independent experiments are shown. F, grr1Δ cells display global hypoacetylation of histones. Samples from the indicated strains were analyzed by Western blotting with antibodies against total histone H3, histone H3 acetylated at lysine 14 (acH3), and hyperacetylated histone H4 (acH4). Even loading of protein samples was confirmed with anti-phosphoglycerate kinase (Pgk1p) antibody. The experiment was performed three times, and representative results are shown.
FIGURE 9.
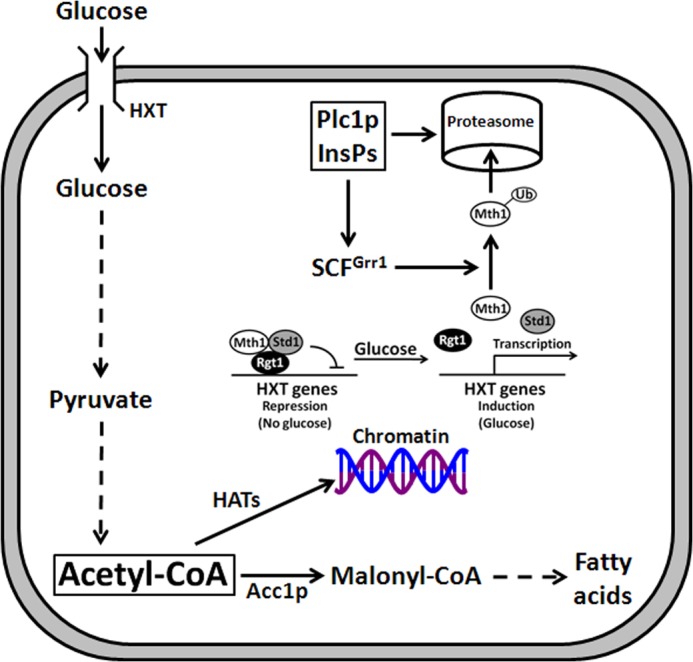
Model for the role of Plc1p and InsPs in regulation of glucose catabolism and histone acetylation. Glucose catabolism requires the expression of the glucose transporters, encoded by the HXT genes, and yields acetyl-CoA, a substrate for HATs. Plc1p and InsPs are important for Mth1p degradation, expression of the glucose transporters, and acetyl-CoA homeostasis. Histone hypoacetylation in plc1Δ cells is caused by a reduced cellular level of acetyl-CoA.
DISCUSSION
Our results show that Plc1p and InsPs are important for the normal level of histone acetylation. This notion is supported by several lines of evidence. First, bulk histones H3 and H4 in total cell lysates are hypoacetylated in plc1Δ cells. Second, ChIP experiments show that different chromosomal loci are hypoacetylated in plc1Δ cells, suggesting that InsPs are important for untargeted, global histone acetylation. Third, genetic interactions strongly suggest that Plc1p and InsPs are important for histone acetylation. Deletions of genes encoding components of both the SAGA complex (gcn5Δ, spt20Δ, and spt7Δ) and NuA4 complex (yng2Δ and eaf1Δ) are synthetically lethal with plc1Δ mutation. Perhaps most importantly, the slow growth phenotype and overall fitness of plc1Δ cells is significantly improved by inactivation of the HDA complex by hda1Δ mutation.
The global untargeted histone hypoacetylation in plc1Δ cells prompted us to test whether this phenotype is a result of a decreased level of nucleocytosolic acetyl-CoA. Indeed, plc1Δ cells display only 12% of the acetyl-CoA level compared with wild-type cells and increasing the level of acetyl-CoA improves the growth rate and partially suppresses the temperature sensitivity of plc1Δ cells (Fig. 6). Our results implicate altered regulation of transcription of the HXT transporters and glucose metabolism as the mechanism responsible for the decreased cellular level of acetyl-CoA and histone hypoacetylation in plc1Δ cells (Fig. 9).
Our results show that plc1Δ cells do not degrade Mth1p efficiently. The defect in HXT transcription as the mechanism responsible for histone hypoacetylation in plc1Δ cells is supported by two findings. First, introducing the mth1Δ mutation in plc1Δ cells partially suppressed histone hypoacetylation, slow growth, and temperature sensitivity of plc1Δ cells (Fig. 8). Second, deletion of GRR1, a component of a Skp1p/Cullin/F box protein (SCF) E3 ubiquitin ligase complex, also resulted in histone hypoacetylation (Fig. 8).
Another mechanism that could account for the role of InsPs in glucose transport and acetyl-CoA homeostasis was suggested by the finding that Plc1p negatively regulates endocytosis of hexose transporters in an Rsp5p-dependent manner (96). Rsp5p, a HECT-type ubiquitin ligase, is involved in ubiquitination of several transporters and permeases in the plasma membrane. The association of Rsp5p with the plasma membrane is likely mediated by its C2 domain, which has a strong affinity for phosphatidylinositol 4,5-bisphosphate. It is possible that the constitutive endocytosis of the hexose transporters in plc1Δ cells is caused by increased recruitment of Rsp5p to the plasma membrane and increased ubiquitination of the hexose transporters (96). However, the ipk2Δ mutation is not expected to affect the level of phosphatidylinositol 4,5-bisphosphate and the association of Rsp5p with the plasma membrane, but it results in reduced transcription of the HXT genes (Fig. 7) and histone hypoacetylation (Fig. 1). Therefore, we believe that the histone hypoacetylation phenotype of plc1Δ and ipk2Δ cells is due to a defect in Mth1p degradation by the ubiquitin/proteasome pathway (84).
How do Plc1p and InsPs affect the ubiquitin/proteasome pathway? There are several indications that Plc1p and InsPs may be involved in regulation of proteasome. First, 26 S proteasome-mediated destruction of C-type cyclin Ume3p/Srb11p/Ssn3p upon oxidative stress requires Plc1p (97). Second, genome-wide identification of protein complexes revealed that Plc1p interacts with Caf130p (98), a component of the Ccr4·Not transcriptional regulatory complex. The Ccr4·Not complex associates with the proteasome (99). One of the subunits of the Ccr4·Not complex is Not4p, an ubiquitin E3 ligase that is required for the activity of the proteasome (100). Alternatively, InsPs may be involved in regulation of Grr1p, a component of the Skp1/Cullin/F-box (SCF) E3 ubiquitin ligase complex. The Arabidopsis homolog of Grr1p, TIR1, is also an F-box protein and a subunit of the SCFTIR1 ubiquitin ligase complex. TIR1 is related to the yeast Grr1p (101) and contains inositol hexakisphosphate (InsP6) as a co-factor (102). Thus, it is possible that Grr1p also binds and is activated by one of the InsPs.
An important conclusion of this work is that Plc1p and InsPs are required for normal acetyl-CoA homeostasis and global histone acetylation. The histone hypoacetylation in plc1Δ cells is due to the defect in Mth1p degradation, and consequently reduced synthesis of glucose-derived acetyl-CoA.
Acknowledgments
We thank Drs. Arndt, Buratowski, Hinnebusch, Nasmyth, Shirra, Stillman, Tsukiyama, Wittenberg, Winston, and York for yeast strains, and members of the Vancura lab and Dr. Vancurova for helpful comments.
This work was supported, in whole or in part, by National Institutes of Health Grant GM087674 (to A. V.).
- PLC
- phospholipase C
- InsPs
- inositol polyphosphates
- PP-InsPs
- inositol pyrophosphates
- HAT
- histone acetyltransferase
- HDAC
- histone deacetylase complex
- ACL
- ATP-citrate lysase.
REFERENCES
- 1. York J. D. (2006) Regulation of nuclear processes by inositol polyphosphates. Biochim. Biophys. Acta 1761, 552–559 [DOI] [PubMed] [Google Scholar]
- 2. Tsui M. M., York J. D. (2010) Roles of inositol phosphates and inositol pyrophosphates in development, cell signaling and nuclear processes. Adv. Enzyme Regul. 50, 324–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Odom A. R., Stahlberg A., Wente S. R., York J. D. (2000) A role for inositol 1,4,5-trisphosphate kinase in transcriptional control. Science 287, 2026–2029 [DOI] [PubMed] [Google Scholar]
- 4. Alcázar-Román A. R., Wente S. R. (2008) Insositol polyphosphates. A new frontier for regulating gene expression. Chromosoma 117, 1–13 [DOI] [PubMed] [Google Scholar]
- 5. York J. D., Odom A. R., Murphy R., Ives E.B., Wente S. R. (1999) A phospholipase C-dependent inositol polyphosphate kinase pathway required for efficient messenger RNA export. Science 285, 96–100 [DOI] [PubMed] [Google Scholar]
- 6. Alcázar-Román A. R., Tran E. J., Guo S., Wente S. R. (2006) Inositol hexakisphosphate and Gle1 activate the DEAD-box protein Dbp5 for nuclear mRNA export. Nat. Cell Biol. 8, 711–716 [DOI] [PubMed] [Google Scholar]
- 7. Weirich C. S., Erzberger J. P., Flick J. S., Berger J. M., Thorner J., Weis K. (2006) Activation of the DExD/H-box protein Dbp5 by the nuclear-pore protein Gle1 and its coactivator InsP6 is required for mRNA export. Nat. Cell Biol. 8, 668–676 [DOI] [PubMed] [Google Scholar]
- 8. Bolger T. A., Folkmann A. W., Tran E. J., Wente S. R. (2008) The mRNA export factor Gle1 and inositol hexakisphosphate regulate distinct stages of translation. Cell 134, 624–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Luo H. R., Saiardi A., Yu H., Nagata E., Ye K., Snyder S. H. (2002) Inositol pyrophosphates are required for DNA hyperrecombination in protein kinase C1 mutant yeast. Biochemistry 41, 2509–2515 [DOI] [PubMed] [Google Scholar]
- 10. Saiardi A., Resnick A. C., Snowman A. M., Wendland B., Snyder S. H. (2005) Inositol pyrophosphates regulate cell death and telomere length through phosphoinositide 3-kinase-related protein kinases. Proc. Natl. Acad. Sci. U.S.A. 102, 1911–1914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. York S. J., Armbruster B. N., Greenwell P., Petes T. D., York J. D. (2005) Inositol diphosphate signaling regulates telomere length. J. Biol. Chem. 280, 4264–4269 [DOI] [PubMed] [Google Scholar]
- 12. Lin H., Choi J. H., Hasek J., DeLillo N., Lou W., Vancura A. (2000) Phospholipase C is involved in kinetochore function in Saccharomyces cerevisiae. Mol. Cell Biol. 20, 3597–3607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Desai P., Guha N., Galdieri L., Hadi S., Vancura A. (2009) Plc1p is required for proper chromatin structure and activity of the kinetochore in Saccharomyces cerevisiae by facilitating recruitment of the RSC complex. Mol. Genet. Genomics 281, 511–523 [DOI] [PubMed] [Google Scholar]
- 14. Lee Y. S., Mulugu S., York J. D., O'Shea E. K. (2007) Regulation of a cyclin-CDK-CDK inhibitor complex by inositol pyrophosphates. Science 316, 109–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. York J. D., Lew D. J. (2008) IP7 guards the CDK gate. Nat. Chem. Biol. 4, 16–17 [DOI] [PubMed] [Google Scholar]
- 16. Chakraborty A., Koldobskiy M. A., Bello N. T., Maxwell M., Potter J. J., Juluri K. R., Maag D., Kim S., Huang A. S., Dailey M. J., Saleh M., Snowman A. M., Moran T. H., Mezey E., Snyder S. H. (2010) Inositol pyrophosphates inhibit Akt signaling, thereby regulating insulin sensitivity and weight gain. Cell 143, 897–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Shen X., Xiao H., Ranallo R., Wu W. H., Wu C. (2003) Modulation of ATP-dependent chromatin-remodeling complexes by inositol polyphosphates. Science 299, 112–114 [DOI] [PubMed] [Google Scholar]
- 18. Steger D. J., Haswell E. S., Miller A. L., Wente S. R., O'Shea E. K. (2003) Regulation of chromatin remodeling by inositol polyphosphates. Science 299, 114–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Felsenfeld G., Groudine M. (2003) Controlling the double helix. Nature 421, 448–453 [DOI] [PubMed] [Google Scholar]
- 20. Vignali M., Hassan A. H., Neely K. E., Workman J. L. (2000) ATP-dependent chromatin-remodeling complexes. Mol. Cell. Biol. 20, 1899–1910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Martens J. A., Winston F. (2003) Recent advances in understanding chromatin remodeling by Swi/Snf complexes. Curr. Opin. Genet. Dev. 13, 136–142 [DOI] [PubMed] [Google Scholar]
- 22. Hogan C., Varga-Weisz P. (2007) The regulation of ATP-dependent nucleosome remodeling factors. Mutat. Res. 618, 41–51 [DOI] [PubMed] [Google Scholar]
- 23. Jenuwein T., Allis C. D. (2001) Translating the histone code. Science 293, 1074–1080 [DOI] [PubMed] [Google Scholar]
- 24. Millar C. B., Grunstein M. (2006) Genome-wide patterns of histone modifications in yeast. Nat. Rev. Mol. Cell Biol. 7, 657–666 [DOI] [PubMed] [Google Scholar]
- 25. Krebs J. E. (2007) Moving marks. Dynamic histone modifications in yeast. Mol. BioSyst. 3, 590–597 [DOI] [PubMed] [Google Scholar]
- 26. Kouzarides T. (2007) Chromatin modifications and their function. Cell 128, 693–705 [DOI] [PubMed] [Google Scholar]
- 27. Roth S. Y., Denu J. M., Allis C. D. (2001) Histone acetyltransferases. Annu. Rev. Biochem. 70, 81–120 [DOI] [PubMed] [Google Scholar]
- 28. Shahbazian M. D., Grunstein M. (2007) Functions of site-specific histone acetylation and deacetylation. Annu. Rev. Biochem. 76, 75–100 [DOI] [PubMed] [Google Scholar]
- 29. Sterner D. E., Berger S. L. (2000) Acetylation of histones and transcription-related factors. Microbiol. Mol. Biol. Rev. 64, 435–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mujtaba S., Zeng L., Zhou M. M. (2007) Structure and acetyl-lysine recognition of the bromodomain. Oncogene 26, 5521–5527 [DOI] [PubMed] [Google Scholar]
- 31. Rathmell J. C., Newgard C. B. (2009) A glucose-to-gene link. Science 324, 1021–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Friis R. M., Schultz M. C. (2009) Untargeted tail acetylation of histones in chromatin. Lessons from yeast. Biochem. Cell Biol. 87, 107–116 [DOI] [PubMed] [Google Scholar]
- 33. Albaugh B. N., Arnold K. M., Denu J. M. (2011) KAT(ching) metabolism by the tail. Insight into the links between lysine acetyltransferases and metabolism. Chembiochem 12, 290–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wellen K. E., Thompson C. B. (2012) A two-way street. Reciprocal regulation of metabolism and signalling. Nat. Rev. Mol. Cell Biol. 13, 270–276 [DOI] [PubMed] [Google Scholar]
- 35. Sassone-Corsi P. (2013) When metabolism and epigenetics converge. Science 339, 148–150 [DOI] [PubMed] [Google Scholar]
- 36. Wellen K. E., Hatzivassiliou G., Sachdeva U. M., Bui T. V., Cross J. R., Thompson C. B. (2009) ATP-citrate lyase links cellular metabolism to histone acetylation. Science 324, 1076–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Takahashi H., McCaffery J. M., Irizarry R. A., Boeke J. D. (2006) Nucleocytosolic acetyl-coenzyme a synthetase is required for histone acetylation and global transcription. Mol. Cell 23, 207–217 [DOI] [PubMed] [Google Scholar]
- 38. Galdieri L., Vancura A. (2012) Acetyl-CoA carboxylase regulates global histone acetylation. J. Biol. Chem. 287, 23865–23876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sandmeier J. J., French S., Osheim Y., Cheung W. L., Gallo C. M., Beyer A. L., Smith J. S. (2002) RPD3 is required for the inactivation of yeast ribosomal DNA genes in stationary phase. EMBO J. 21, 4959–4968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ramaswamy V., Williams J. S., Robinson K. M., Sopko R. L., Schultz M. C. (2003) Global control of histone modification by the anaphase-promoting complex. Mol. Cell. Biol. 23, 9136–9149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Friis R. M., Wu B. P., Reinke S. N., Hockman D. J., Sykes B. D., Schultz M. C. (2009) A glycolytic burst drives glucose induction of global histone acetylation by picNuA4 and SAGA. Nucleic Acids Res. 37, 3969–3980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Cai L., Sutter B. M., Li B., Tu B. P. (2011) Acetyl-CoA induces cell growth and proliferation by promoting the acetylation of histones at growth genes. Mol. Cell 42, 426–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sherman F. (1991) Getting started with yeast. Methods Enzymol. 194, 3–21 [DOI] [PubMed] [Google Scholar]
- 44. Demczuk A., Guha N., Nguyen P. H., Desai P., Chang J., Guzinska K., Rollins J., Ghosh C. C., Goodwin L., Vancura A. (2008) Saccharomyces cerevisiae phospholipase C regulates transcription of Msn2p-dependent stress-responsive genes. Eukaryot. Cell 7, 967–979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Guha N., Desai P., Vancura A. (2007) Plc1p is required for SAGA recruitment and derepression of Sko1p-regulated genes. Mol. Biol. Cell 18, 2419–2428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Galdieri L., Moon J., Vancura A. (2012) Determination of histone acetylation status by chromatin immunoprecipitation. Methods Mol. Biol. 809, 255–265 [DOI] [PubMed] [Google Scholar]
- 47. De Nadal E., Zapater M., Alepuz P. M., Sumoy L., Mas G., Posas F. (2004) The MAPK Hog1 recruits Rpd3 histone deacetylase to activate osmoresponsive genes. Nature 427, 370–374 [DOI] [PubMed] [Google Scholar]
- 48. Mulugu S., Bai W., Fridy P. C., Bastidas R. J., Otto J. C., Dollins D. E., Haystead T. A., Ribeiro A. A., York J. D. (2007) A conserved family of enzymes that phosphorylate inositol hexakisphosphate. Science 316, 106–109 [DOI] [PubMed] [Google Scholar]
- 49. Babiarz J. E., Halley J. E., Rine J. (2006) Telomeric heterochromatin boundaries require NuA4-dependent acetylation of histone variant H2A.Z in Saccharomyces cerevisiae. Genes Dev. 20, 700–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Keogh M. C., Mennella T. A., Sawa C., Berthelet S., Krogan N. J., Wolek A., Podolny V., Carpenter L. R., Greenblatt J. F., Baetz K., Buratowski S. (2006) The Saccharomyces cerevisiae histone H2A variant Htz1 is acetylated by NuA4. Genes Dev. 20, 660–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Erkina T. Y., Erkine A. M. (2006) Displacement of histones at promoters of Saccharomyces cerevisiae heat shock genes is differentially associated with histone H3 acetylation. Mol. Cell. Biol. 26, 7587–7600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Govind C. K., Zhang F., Qiu H., Hofmeyer K., Hinnebusch A. G. (2007) Gcn5 promotes acetylation, eviction, and methylation of nucleosomes in transcribed coding regions. Mol. Cell 25, 31–42 [DOI] [PubMed] [Google Scholar]
- 53. Kristjuhan A., Svejstrup J. Q. (2004) Evidenec for distinct mechanisms facilitating transcript elongation through chromatin in vivo. EMBO J. 23, 4243–4252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lee C. K., Shibata Y., Rao B., Strahl B. D., Lieb J. D. (2004) Evidence for nucleosome depletion at active regulatory regions genome-wide. Nat. Genet. 36, 900–905 [DOI] [PubMed] [Google Scholar]
- 55. Rusche L. N., Kirchmaier A. L., Rine J. (2003) The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Annu. Rev. Biochem. 72, 481–516 [DOI] [PubMed] [Google Scholar]
- 56. Suka N., Luo K., Grunstein M. (2002) Sir2p and Sas2p opposingly regulate acetylation of yeast histone H4 lysine 16 and spreading of heterochromatin. Nat. Genet. 32, 378–383 [DOI] [PubMed] [Google Scholar]
- 57. Kimura A., Umehara T., Horikoshi M. (2002) Chromosomal gradient of histone acetylation established by Sas2p and Sir2p functions as a shield against gene silencing. Nat. Genet. 32, 370–377 [DOI] [PubMed] [Google Scholar]
- 58. Meneghini M. D., Wu M., Madhani H. D. (2003) Conserved histone variant H2A.Z protects euchromatin from the ectopic spread of silent heterochromatin. Cell 112, 725–736 [DOI] [PubMed] [Google Scholar]
- 59. Shia W. J., Li B., Workman J. L. (2006) SAS-mediated acetylation of histone H4 Lys-16 is required for H2A.Z incorporation at subtelomeric regions in Saccharomyces cerevisiae. Genes Dev. 20, 2507–2512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kristjuhan A., Wittschieben B. O., Walker J., Roberts D., Cairns B. R., Svejstrup J. Q. (2003) Spreading of Sir3 protein in cells with severe histone H3 hypoacetylation. Proc. Natl. Acad. Sci. U.S.A. 100, 7551–7556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Lin Y. Y., Qi Y., Lu J. Y., Pan X., Yuan D. S., Zhao Y., Bader J. S., Boeke J. D. (2008) A comprehensive synthetic genetic interaction network governing yeast histone acetylation and deacetylation. Genes Dev. 22, 2062–2074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Vogelauer M., Wu J., Suka N., Grunstein M. (2000) Global histone acetylation and deacetylation in yeast. Nature 408, 495–498 [DOI] [PubMed] [Google Scholar]
- 63. Yoko-o T., Matsui Y., Yagisawa H., Nojima H., Uno I., Toh-e A. (1993) The putative phosphoinositide-specific phospholipase C gene, PLC1, of the yeast Saccharomyces cerevisiae is important for cell growth. Proc. Natl. Acad. Sci. U.S.A. 90, 1804–1808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Flick J. S., Thorner J. (1993) Genetic and biochemical characterization of a phosphatidylinositol-specific phospholipase C in Saccharomyces cerevisiae. Mol. Cell. Biol. 13, 5861–5876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Carmen A. A., Rundlett S. E., Grunstein M. (1996) HDA1 and HDA3 are components of a yeast histone deacetylase (HDA) complex. J. Biol. Chem. 271, 15837–15844 [DOI] [PubMed] [Google Scholar]
- 66. Wu J., Suka N., Carlson M., Grunstein M. (2001) TUP1 utilizes histone H3/H2B-specific HDA1 deacetylase to repress gene activity in yeast. Mol. Cell 7, 117–126 [DOI] [PubMed] [Google Scholar]
- 67. Carrozza M. J., Li B., Florens L., Suganuma T., Swanson S. K., Lee K. K., Shia W. J., Anderson S., Yates J., Washburn M. P., Workman J. L. (2005) Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intrageneic transcription. Cell 123, 581–592 [DOI] [PubMed] [Google Scholar]
- 68. Keogh M. C., Kurdistani S. K., Morris S. A., Ahn S. H., Podolny V., Collins S. R., Schuldiner M., Chin K., Punna T., Thompson N. J., Boone C., Emili A., Weissman J. S., Hughes T. R., Strahl B. D., Grunstein M., Greenblatt J. F., Buratowski S., Krogan N. J. (2005) Cotranscriptional Set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell 123, 593–605 [DOI] [PubMed] [Google Scholar]
- 69. Li B., Gogol M., Carey M., Lee D., Seidel C., Workman J. L. (2007) Combined action of PHD and chromo domains directs the Rpd3S HDAC to transcribed chromatin. Science 316, 1050–1054 [DOI] [PubMed] [Google Scholar]
- 70. Kraakman L. S., Griffioen G., Zerp S., Groeneveld P., Thevelein J. M., Mager W. H., Planta R. J. (1993) Growth related expression of ribosomal protein genes in Saccharomyces cerevisiae. Mol. Gen. Genet. 239, 196–204 [DOI] [PubMed] [Google Scholar]
- 71. Warner J. R. (1999) The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 24, 437–440 [DOI] [PubMed] [Google Scholar]
- 72. Kurdistani S. K., Tavazoie S., Grunstein M. (2004) Mapping global histone acetylation patterns to gene expression. Cell 117, 721–733 [DOI] [PubMed] [Google Scholar]
- 73. Robert F., Pokholok D. K., Hannett N. M., Rinaldi N. J., Chandy M., Rolfe A., Workman J. L., Gifford D. K., Young R. A. (2004) Global position and recruitment of HATs and HDACs in the yeast genome. Mol. Cell 16, 199–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Pokholok D. K., Harbison C.T., Levine S., Cole M., Hannett N. M., Lee T. I., Bell G. W., Walker K., Rolfe P. A., Herbolsheimer E., Zeitlinger J., Lewitter F., Gifford D. K., Young R. A. (2005) Genome-wide map of nucleosome acetylation and methylation in yeast. Cell 122, 517–527 [DOI] [PubMed] [Google Scholar]
- 75. Reid J. L., Iyer V. R., Brown P. O., Struhl K. (2000) Coordinate regulation of yeast ribosomal protein genes is associated with targeted recruitment of Esa1 histone acetylase. Mol. Cell 6, 1297–1307 [DOI] [PubMed] [Google Scholar]
- 76. Rohde J. R., Cardenas M. E. (2003) The Tor pathway regulates genes expression by linking nutrient sensing to histone acetylation. Mol. Cell. Biol. 23, 629–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Gombert A. K., Moreira dos Santos M., Christensen B., Nielsen J. (2001) Network identification and flux quantification in the central metabolism of Saccharomyces cerevisiae under different conditions of glucose repression. J. Bacteriol. 183, 1441–1451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Smedsgaard J., Nielsen J. (2005) Metabolite profiling of fungi and yeast. From phenotype to metabolome by MS and informatics. J. Exp. Bot. 56, 273–286 [DOI] [PubMed] [Google Scholar]
- 79. Heyland J., Fu J., Blank L. M. (2009) Correlation between TCA flux and glucose uptake rate during respire-fermentative growth of Saccharomyces cerevisiae. Microbiology 155, 3827–3837 [DOI] [PubMed] [Google Scholar]
- 80. Tehlivets O., Scheuringer K., Kohlwein S. D. (2007) Fatty acid synthesis and elongation in yeast. Biochim. Biophys. Acta 1771, 255–270 [DOI] [PubMed] [Google Scholar]
- 81. Garí E., Piedrafita L., Aldea M., Herrero E. (1997) A set of vectors with a tetracycline-regulatable promoter system for modulated gene expression in Saccharomyces cerevisiae. Yeast 13, 837–848 [DOI] [PubMed] [Google Scholar]
- 82. Shirra M. K., Patton-Vogt J., Ulrich A., Liuta-Tehlivets O., Kohlwein S. D., Henry S. A., Arndt K. M. (2001) Inhibition of acetyl coenzyme A carboxylase activity restores expression of the INO1 gene in a snf1 mutant strain of Saccharomyces cerevisiae. Mol. Cell. Biol. 21, 5710–5722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Ozcan S., Johnston M. (1995) Three different regulatory mechanisms enable yeast hexose transporter (HXT) genes to be induced by different levels of glucose. Mol. Cell. Biol. 15, 1564–1572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Flick K. M., Spielewoy N., Kalashnikova T. I., Guaderrama M., Zhu Q., Chang H. C., Wittenberg C. (2003) Grr1-dependent inactivation of Mth1 mediates glucose-induced dissociation of Rgt1 from HXT gene promoters. Mol. Biol. Cell 14, 3230–3241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Ozcan S., Johnston M. (1999) Function and regulation of yeast hexose transporters. Microbiol. Mol. Biol. Rev. 63, 554–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Lafuente M. J., Gancedo C., Jauniaux J. C., Gancedo J. M. (2000) Mth1 receives the signal given by the glucose sensors Snf3 and Rgt2 in Saccharomyces cerevisiae. Mol. Microbiol. 35, 161–172 [DOI] [PubMed] [Google Scholar]
- 87. Lakshmanan J., Mosley A. L., Ozcan S. (2003) Repression of transcription by Rgt1 in the absence of glucose requires Std1 and Mth1. Curr. Genet. 44, 19–25 [DOI] [PubMed] [Google Scholar]
- 88. Schmidt M. C., McCartney R. R., Zhang X., Tillman T. S., Solimeo H., Wölfl S., Almonte C., Watkins S. C. (1999) Std1 and Mth1 proteins interact with the glucose sensors to control glucose-regulated gene expression in Saccharomyces cerevisiae. Mol. Cell. Biol. 19, 4561–4571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Ozcan S., Leong T., Johnston M. (1996) Rgt1p of Saccharomyces cerevisiae, a key regulator of glucose-induced genes, is both an activator and a repressor of transcription. Mol. Cell. Biol. 16, 6419–6426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Johnston M., Kim J.-H. (2005) Glucose as a hormone. Receptor-mediated glucose sensing in the yeast Saccharomyces cerevisiae. Biochem. Soc. Trans. 33, 247–252 [DOI] [PubMed] [Google Scholar]
- 91. Santangelo G. M. (2006) Glucose signaling in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 70, 253–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Zaman S., Lippman S. I., Zhao X., Broach J. R. (2008) How Saccharomyces responds to nutrients. Annu. Rev. Genet. 42, 27–81 [DOI] [PubMed] [Google Scholar]
- 93. Gancedo J. M. (2008) The early steps of glucose signaling in yeast. FEMS Microbiol. Rev. 32, 673–704 [DOI] [PubMed] [Google Scholar]
- 94. Kim J. H., Johnston M. (2006) Two glucose-sensing pathways converge on Rgt1 to regulate expression of glucose transporter genes in Saccharomyces cerevisiae. J. Biol. Chem. 281, 26144–26149 [DOI] [PubMed] [Google Scholar]
- 95. Palomino A., Herrero P., Moreno F. (2006) Tpk3 and Snf1 protein kinases regulate Rgt1 association with Saccharomyces cerevisiae HXK2 promoter. Nucleic Acids Res. 34, 1427–1438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Yoshida A., Wei D., Nomura W., Izawa S., Inoue Y. (2012) Reduction of glucose uptake through inhibition of hexose transporters and enhancement of their endocytosis by methylglyoxal in Saccharomyces cerevisiae. J. Biol. Chem. 287, 701–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Cooper K. F., Mallory M. J., Strich R. (1999) Oxidative stress-induced destruction of the yeast C-type cyclin Ume3p requires phosphatidylinositol-specific phospholipase C and the 26 S proteasome. Mol. Cell Biol. 19, 3338–3348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Krogan N. J., Cagney G., Yu H., Zhong G., Guo X., Ignatchenko A., Li J., Pu S., Datta N., Tikuisis A. P., Punna T., Peregrín-Alvarez J. M., Shales M., Zhang X., Davey M., Robinson M. D., Paccanaro A., Bray J. E., Sheung A., Beattie B., Richards D. P., Canadien V., Lalev A., Mena F., Wong P., Starostine A., Canete M. M., Vlasblom J., Wu S., Orsi C., Collins S. R., Chandran S., Haw R., Rilstone J. J., Gandi K., Thompson N. J., Musso G., St Onge P., Ghanny S., Lam M. H., Butland G., Altaf-Ul A. M., Kanaya S., Shilatifard A., O'Shea E., Weissman J. S., Ingles C. J., Hughes T. R., Parkinson J., Gerstein M., Wodak S. J., Emili A., Greenblatt J. F. (2006) Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature 440, 637–643 [DOI] [PubMed] [Google Scholar]
- 99. Laribee R. N., Shibata Y., Mersman D. P., Collins S. R., Kemmeren P., Roguev A., Weissman J. S., Briggs S. D., Krogan N. J., Strahl B. D. (2007) CCR4/NOT complex associates with the proteasome and regulates histone methylation. Proc. Natl. Acad. Sci. U.S.A. 104, 5836–5841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Panasenko O. O., Collart M. A. (2011) Not4 E3 ligase contributes to proteasome assembly and functional integrity in part through Ecm29. Mol. Cell. Biol. 31, 1610–1623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Ruegger M., Dewey E., Gray W. M., Hobbie L., Turner J., Estelle M. (1998) The TIR1 protein of Arabidopsis functions in auxin response and is related to human SKP2 and yeast Grr1p. Genes Dev. 12, 198–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Tan X., Calderon-Villalobos L. I., Sharon M., Zheng C., Robinson C. V., Estelle M., Zheng N. (2007) Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 446, 640–645 [DOI] [PubMed] [Google Scholar]
- 103. De Lillo N., Romero C., Lin H., Vancura A. (2003) Genetic evidence for a role of phospholipase C at the budding yeast kinetochore. Mol. Gen. Genomics 269, 261–270 [DOI] [PubMed] [Google Scholar]
- 104. Yu Y., Eriksson P., Stillman D. J. (2000) Architectural transcription factors and the SAGA complex finction in parallel pathways to activate transcription. Mol. Cell. Biol. 20, 2350–2357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Roberts S. M., Winston F. (1997) Essential functional interactions of SAGA, a Saccharomyces cerevisiae complex of Spt, Ada, and Gcn5 proteins, with the Snf/Swi and Srb/mediator complexes. Genetics 147, 451–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Yu Y., Eriksson P., Bhoite L. T., Stillman D. J. (2003) Regulation of TATA-binding protein binding by the SAGA complex and the Nhp6 high-mobility group protein. Mol. Cell. Biol. 23, 1910–1921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Lindstrom K. C., Vary J. C., Jr., Parthun M. R., Delrow J., Tsukiyama T. (2006) Isw1 functions in parallel with the NuA4 and Swr1 complexes in stress-induced gene repression. Mol. Cell. Biol. 26, 6117–6129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Kim J. H., Brachet V., Moriya H., Johnston M. (2006) Integration of transcriptional and posttranslational regulation in a glucose signal transduction pathway in Saccharomyces cerevisiae. Eukaryot. Cell 5, 167–173 [DOI] [PMC free article] [PubMed] [Google Scholar]