Background: Aberrant expression of histone deacetylases (HDACs) has been found in several kinds of cancer.
Results: HDAC10, a class IIb HDAC, can suppress cervical cancer metastasis.
Conclusion: Unlike most other HDACs, which promote carcinogenesis, HDAC10 serves as a tumor suppressor in cervical cancer.
Significance: Isoform-specific HDAC inhibitors could be indicated in the clinical treatment of cervical cancer.
Keywords: Cell Invasion, Cell Migration, Histone Deacetylase, Matrix Metalloproteinase (MMP), Metastasis
Abstract
Aberrant expression of histone deacetylases (HDACs) is associated with carcinogenesis. Some HDAC inhibitors are widely considered as promising anticancer therapeutics. A major obstacle for development of HDAC inhibitors as highly safe and effective anticancer therapeutics is that our current knowledge on the contributions of different HDACs in various cancer types remains scant. Here we report that the expression level of HDAC10 was significantly lower in patients exhibiting lymph node metastasis compared with that in patients lacking lymph node metastasis in human cervical squamous cell carcinoma. Forced expression of HDAC10 in cervical cancer cells significantly inhibited cell motility and invasiveness in vitro and metastasis in vivo. Mechanistically, HDAC10 suppresses expression of matrix metalloproteinase (MMP) 2 and 9 genes, which are known to be critical for cancer cell invasion and metastasis. At the molecular level, HDAC10 binds to MMP2 and -9 promoter regions, reduces the histone acetylation level, and inhibits the binding of RNA polymerase II to these regions. Furthermore, an HDAC10 mutant lacking histone deacetylase activity failed to mimic the functions of full-length protein. These results identify a critical role of HDAC10 in suppression of cervical cancer metastasis, underscoring the importance of developing isoform-specific HDAC inhibitors for treatment of certain cancer types such as cervical squamous cell carcinoma.
Introduction
Cervical cancer is a common gynecologic malignant neoplasm. The occurrence of invasion and distant metastasis are important events that affect the prognosis and treatment of cervical cancer. For the earliest stage of cervical cancer, more than 90% of women survive at least 5 years after diagnosis. However, later stages of cervical cancer with invasion or metastasis have a significantly worse outlook. Fewer than 20% of women with stage IV cervical cancer survive for 5 or more years (1). Some of the cervical cancer cells can metastasize through lymph nodes or blood circulation and form a secondary cancer. They most often spread to the liver, lungs, and bones. Prognosis drops deeply in patient with metastasis as treatment of local lesions is generally more effective than whole body treatments (2). Thus, understanding the molecular and cellular mechanisms of cell invasion and metastasis is critical for developing effective cervical cancer therapies and improving patient survival rate.
Matrix metalloproteinases (MMPs)3 are closely associated with the pathogenesis and progression of cancer. They are zinc-dependent endopeptidases, which are capable of degrading the extracellular matrix (3). They play important roles in many cell behaviors such as cell proliferation, migration, adhesion, differentiation, angiogenesis, and apoptosis. MMP2 and -9 are two widely studied matrix metalloproteinases that can regulate cell migration and invasion of cancer cells and are linked to the process of metastasis (4–6). Their expression levels are significantly increased in many human tumors (7–10) and are regarded as important prognostic factors in cervical cancer as well as other diseases (11, 12). The MMPs are inhibited by specific endogenous tissue inhibitor of metalloproteinases (TIMPs), which form tight 1:1 complexes with MMPs and block their function. MMP2 is specifically inhibited by TIMP2, whereas MMP9 is specifically inhibited by TIMP1 (13). Clinical data show that patients with high expression levels of MMP2 and -9 and low expression levels of TIMP1 and -2 are more likely to exhibit high invasion and metastasis in cervical cancer. However, how expression of MMP2 and -9 is regulated remains unclear.
Aberrant expression of HDACs is often associated with carcinogenesis and cancer clinical prognosis (14). The main function of HDACs is to remove acetyl groups from the N-acetyllysines on histone and thus modify the chromatin structure and modulate gene transcription (15). HDACs can regulate the expression of cancer-relevant genes, so they are targets of a promising class of anticancer drugs. Several HDAC inhibitors (e.g. suberoylanilide hydroxamic acid, valproic acid, and trichostatin A) have been used for cancer treatment clinically or are under clinical trials (14). They inhibit carcinogenesis through multiple mechanisms including inhibiting cancer cell proliferation (16), migration (17), and angiogenesis (18, 19). The HDAC family contains 18 proteins, which are grouped into classes I–IV based on their structure and homology. Classes I, II, and IV contain 11 family members, which are called classical HDACs, whereas the seven class III family members are referred to as sirtuins (20). HDAC10 is a class IIb HDAC, which, unlike the other class IIb HDAC HDAC6 that has two tandem deacetylase domains, has one deacetylase (DAC) domain and one additional catalytically inactive leucine-rich domain (LRD) (21). It has been reported that HDAC10 can relieve repression on the melanogenic program (22), suppress the accumulation of reactive oxygen species (23), and play an important role in homologous recombination (24). However, compared with many other HDACs, the function of HDAC10 in cancer is largely unknown.
In this study, we show that HDAC10 is inversely related to lymph node metastasis in human patients with cervical squamous cell carcinoma. Furthermore, we demonstrate that HDAC10 inhibits cervical cancer cell migration and invasion in vitro and metastasis in vivo. Additionally, we found that HDAC10 binds to the promoter regions of MMP2 and -9, deacetylates histones H3 and H4 in these regions, blocks the binding of RNA polymerase II, and consequently down-regulates MMP2 and -9 expression. Our studies demonstrate for the first time that HDAC10 acts as a suppressor of cancer metastasis. Below, we present our data leading to the findings and discuss the significance and implication of our study on the design of HDAC inhibitor-based anticancer therapy.
EXPERIMENTAL PROCEDURES
Cell Culture
The human cervical carcinoma cell line HeLa-S3 and human embryonic kidney (HEK) 293T cells were purchased from the American Type Culture Collection (ATCC). The human cervical carcinoma cell line Caski was purchased from the Cell Bank at the Shanghai Institutes for Biological Sciences of Chinese Academy of Sciences. HeLa-S3 and 293T cells were cultured in Dulbecco's modified Eagle's medium (DMEM; Invitrogen). Caski cells were cultured in RPMI 1640 medium. All media were supplemented with 10% fetal bovine serum, 100 units/ml streptomycin, and 100 units/ml penicillin. All cells were maintained at 37 °C with 5% CO2.
Plasmids, Transfections, and Virus Generation
To express FLAG-tagged HDAC10, the coding region of HDAC10 (a gift from Wen-Ming Yang; Ref. 22) was cloned into pFLAG-CMV-6c and fugw vectors. Deletion mutations of HDAC10 were made in this laboratory using restriction enzymes and PCR. HeLa cells and HEK293T cells were transfected with X-tremeGENE HP (Roche Applied Science) or ExcellGene DNA transfection reagent. To generate lentivirus particles, lentiviral vector fugw, which contains the target gene, was co-transfected with a helper plasmid encoding gag, pol, and env into HEK293T cells, and virus was obtained 48 h after transfection.
Transwell Assay
HeLa cells were transfected with DNA vectors or siRNA duplexes. 48 h after transfection, cells were collected, and cell migration ability was analyzed using Transwell chambers (Corning catalog number 3422). For migration, HeLa cells were suspended in DMEM with 1% FBS and added to the upper chambers (5 × 104cells/well). Then the chambers were incubated at 37 °C for 16 h. After that, cells on the upper surface of the membrane were removed. The membranes were fixed with 4% paraformaldehyde, and cells on the undersurface were stained with Hoechst 33342. The chambers were observed under a fluorescence microscope, and cells from five randomly fields were counted. For invasion, the chambers were precoated with matrigel (BD Biosciences; 50 mg/ml; 1:8) at 37 °C for 4 h. After cells were added (1 × 105cells/well), the chambers were incubated at 37 °C for 20 h. The experimental conditions for Caski cells were as follows: 1 × 105 cells/well, 16 h for migration and 1 × 105 cells/well, 24 h for invasion.
Antibodies and Western Blotting
Anti-HDAC10 (H3413), anti-MMP9 (HPA001238), and anti-FLAG (F7425) antibodies were purchased from Sigma. Anti-GAPDH (sc-47724) and anti-p65 (sc-372) antibodies were purchased from Santa Cruz Biotechnology. Anti-AP1 (9165) antibody was purchased from Cell Signaling Technology. Anti-TIMP1 (BS1697), anti-TIMP2 (BS1366), and anti-MMP2 (BS1236) antibodies were purchased from Bioworld. For Western blotting, cells were lysed with 1× SDS-PAGE loading buffer. The lysates were sonicated and centrifuged. Then an equal amount of protein was loaded for 10% SDS-PAGE. Signals were developed using enhanced chemiluminescence (ECL).
RNA Interference
siRNAs recognizing HDAC10 were purchased from Sigma. The sequences of these two siRNAs are as follows: CGGAGUCAGUGUGCAUGACAGUACA and UCACUGCACUUGGGAAGCUCCUGUA. To generate virus, shRNA against the second site was cloned into pLKO.1 lentivirus vector.
Quantitative Real Time PCR
Total RNA of the cells was extracted using RNAiso Plus (TaKaRa). 1 μg of total RNA was reverse transcribed with TaKaRa PrimeScript RT reagent kit according to the manufacturer's instruction. Real time PCR was performed using SYBR Premix Ex Taq (TaKaRa) on a Stratagene Mx3000P (Stratagene). Data were collected and analyzed. The expression level of each gene was normalized to actin expression and further normalized to the control group. The primer sequences were as follows: for MMP2, 5′-CCGTCGCCCATCATCAAGTT-3′ and 5′-CTGTCTGGGGCAGTCCAAAG-3′; for MMP9, 5′-GGGACGCAGACATCGTCATC-3′ and 5′-TCGTCATCGTCGAAATGGGC-3′; for TIMP1, 5′-GGGACACCAGAAGTCAACCA-3′ and 5′-GGCTTGGAACCCTTTATACATC-3′; for TIMP2, 5′-AAAGCGGTCAGTGAGAAGGA-3′ and 5′-CTTCTTTCCTCCAACGTCCA-3′; and for β-actin, 5′-GACCTGTACGCCAACACAG-3′ and 5′-CTCAGGAGGAGCAATGATC-3′.
Tissue Microarray and Evaluation of Immunostaining
Cervical cancer tissue microarrays were purchased from the National Engineering Center for BioChips in Shanghai, China. Institutional Review Board permission for the use of samples was obtained. The evaluation of expression was made blindly by two independent observers simultaneously, and a consensus score was recorded. MMP2 (sc-10736) and MMP9 (sc-21733) antibodies used in staining were purchased from Santa Cruz Biotechnology. The staining was scored according to the staining intensity and the percentage of cells stained. Staining intensity was scored as 0 (negative), 1 (weakly positive), 2 (strongly positive), and 3 (very strongly positive). The percentages of cells stained were scored into five categories: 0 (0%), 1 (1–25%), 2 (26–50%), 3 (51–75%), and 4 (76–100%). The final staining scores were calculated by staining intensity × percentage of stained cells.
Chromatin Immunoprecipitation
The chromatin immunoprecipitation (ChIP) assay was carried out according to a published procedure (25). Briefly, the non-treated and HDAC10-transfected HeLa cells were sonicated and fixed with 1% paraformaldehyde. Then the chromatin fragments were immunoprecipitated with antibodies specific to HDAC10 (Sigma), acetyl-H3 (Millipore), acetyl-H4 (Millipore), RNA polymerase II (Millipore), or control rabbit IgG (Millipore) at 4 °C overnight. After DNAs were dissociated from immunoprecipitated chromatin, the amounts of the specific DNA fragments were quantified by real time PCR and normalized against the genomic DNA preparation from the same cells.
Experimental Metastasis Assay
Ten 6–8-week-old male nonobese diabetic-severe combined immunodeficient (NOD-SCID) mice were injected with 5 × 106 control or HDAC10-overexpressng HeLa cells (five mice per group). Cells were washed and resuspended in PBS and injected through the lateral tail vein of the mice (200 μl/mouse). 4 weeks after injection, mice were euthanized, and lungs were isolated and photographed. The number of surface metastases per lung was counted and recorded. All experiments were carried out at the Laboratory Animal Center of Tongji University with Institutional Animal Care and Use Committee approval in accordance with institutional guidelines.
Lymph Node Metastasis Model
To establish this model, 30 BALB/c nude mice (15 per group) were anesthetized, and their right hind limb foot pads were slowly injected with 50 μl of control or HDAC10-overexpressing HeLa cell suspension at a concentration of 8 × 104/μl. 4, 6, and 8 weeks after injection, mice were euthanized (n = 5), and their right popliteal, inguinal, and common iliac lymph node tissues were fixed and analyzed by H&E staining.
Co-immunoprecipitation
Plasmids encoding FLAG-HDAC10 was transfected into HEK293T cells. 48 h after transfection, cells were collected and lysed with lysis buffer (1% Triton X-100 in 50 mm Tris-HCl, pH 7.4 containing 150 mm NaCl, 2 mm Na3VO4, 100 mm NaF, and protease inhibitors). Then the cell lysates were incubated with anti-FLAG mAb M2-agarose beads (Sigma) for 4 h. The beads were washed with lysis buffer three times, and the immunoprecipitates were analyzed by Western blotting with antibodies as specified.
RESULTS
HDAC10 Expression Inversely Correlates with Lymph Node Metastasis in Cervical Cancer
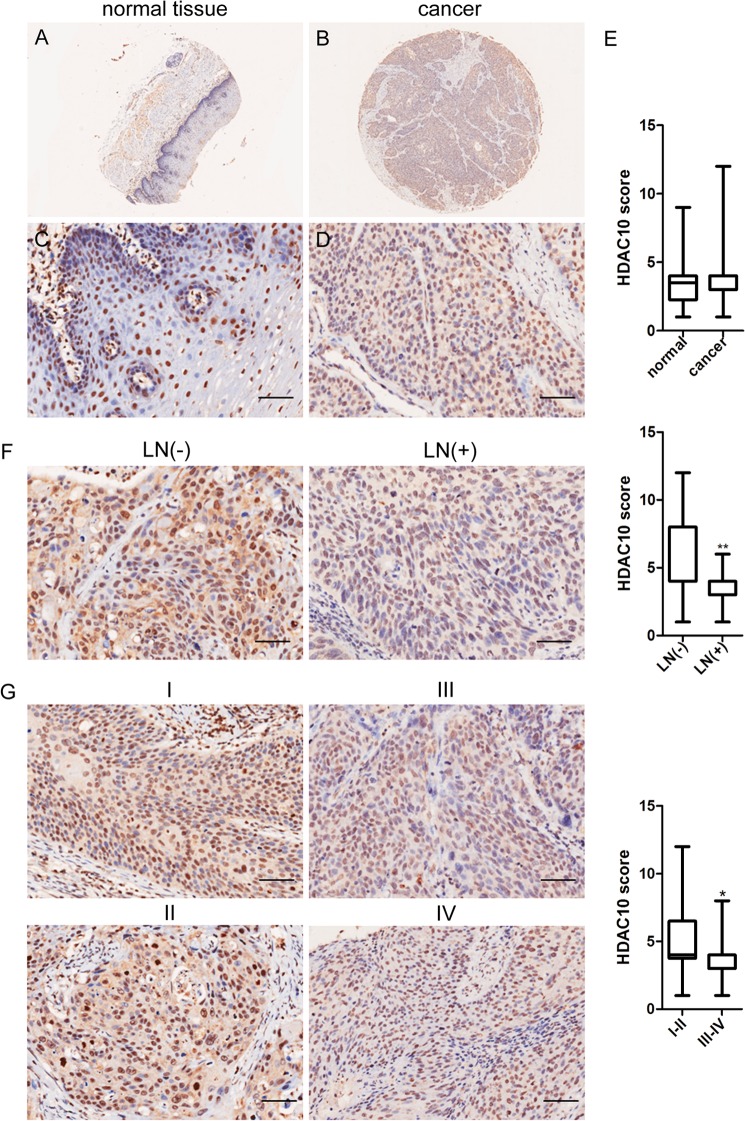
To evaluate the clinical relevance of HDAC10 in cervical cancer patients, we analyzed cervical cancer microarrays with HDAC10 antibody. The cervical cancer microarrays that we used contain 60 patients. A sample of cancer tissue and a corresponding normal tissue from each patient were included. The clinical information of these patients is shown in Table 1. HDAC10 expression was detected in normal cervical tissues as well as cervical squamous cell carcinoma (Fig. 1, A–D). Furthermore, HDAC10 was expressed in both the cell nucleus and cytoplasm. There was no difference in the level of HDAC10 between normal and carcinoma tissues (Fig. 1E). However, we found that in carcinoma tissues the level of HDAC10 in patients with lymph node metastasis was significantly lower than those lacking lymph node metastasis (Fig. 1F). Furthermore, the HDAC10 expression level was significantly higher in patients with TNM stages I and II than those with stages III and IV (Fig. 1G, I and II versus III and IV). These results suggest that the expression level of HDAC10 is inversely correlated with lymph node metastasis and TNM stage in cervical cancer.
TABLE 1.
Clinical and pathologic characteristics of patient with cervical cancer (n = 60)
p values less than 0.05, indicating that the difference of these two groups is significant, are shown in bold. The minimum and maximum ages in each group are shown in parentheses. 60 patients were divided into two groups according to their age, tumor size, lymph node metastasis status, and TNM stage, respectively. The numbers of patients in each group are shown in the n = 60 column.
| Characteristics | n = 60 | HDAC10 score | p value |
|---|---|---|---|
| Age | |||
| <40 years | 16 (28–38 years) | 4.313 | 0.814 |
| ≥40 years | 44 (40–73 years) | 4.182 | |
| Tumor size | |||
| <4 cm | 44 | 4.136 | 0.5879 |
| ≥4 cm | 16 | 4.438 | |
| Lymph node metastasis | |||
| Negative | 23 | 5.087 | 0.038 |
| Positive | 37 | 3.676 | |
| TNM stage | |||
| I-II | 22 | 4.955 | 0.0193 |
| III-IV | 38 | 3.789 | |
FIGURE 1.
HDAC10 expression inversely correlates with lymph node metastasis in cervical cancer. A–D, HDAC10 levels in representative normal and carcinoma tissues. Immunohistochemistry using rabbit anti-HDAC10 antibody was performed on normal cervical tissues (A and C) and cervical squamous cell carcinoma tissues (B and D). E, plot representation of scores according to immunohistochemical expression of HDAC10 in normal tissues and the counterpart tumor tissues. A total of 60 specimens were analyzed. The scores were calculated by intensity × percentage of stained cells. F, correlation between HDAC10 expression levels and lymph node (LN) metastasis status in carcinoma tissues. G, correlation between HDAC10 expression levels and TNM stage in carcinoma tissues. All data were analyzed by Student's t test. *, p < 0.05; **, p < 0.01. Scale bars, 100 μm. Error bars represent S.D.
HDAC10 Inhibits Cervical Cancer Cell Migration and Invasion but Not Proliferation or Survival
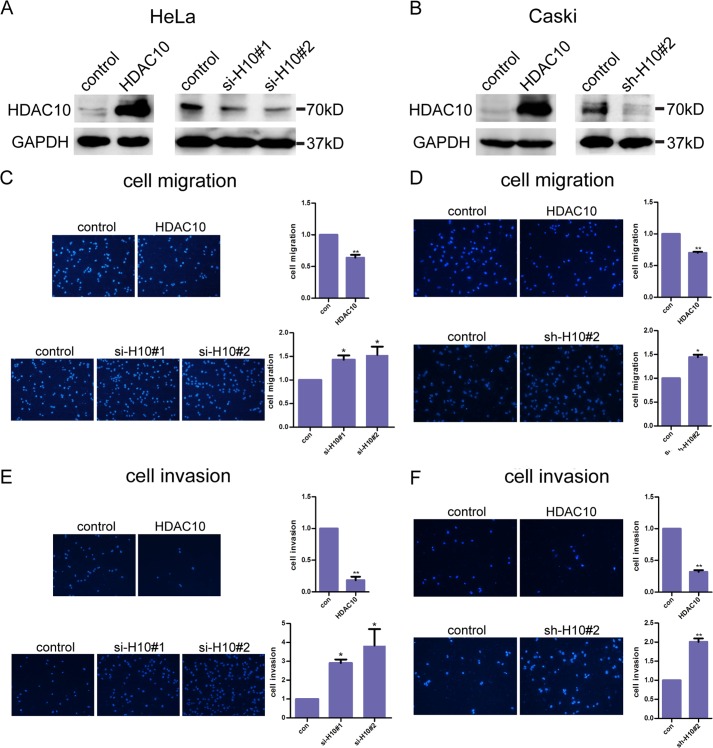
The inverse relationship between HDAC10 and lymph node metastasis that we observed in human cervical cancer patients prompted us to test whether HDAC10 plays a role in regulation of cervical cancer cell migration and invasion, processes that are critical for metastasis. To do this, we transiently transfected human HeLa cervical cancer cells with plasmids containing HDAC10 cDNA and two different siRNAs targeting HDAC10, respectively. Western blotting confirmed that the level of HDAC10 was increased in cells transfected with HDAC10 cDNA expression vector but reduced in cells transfected with HDAC10 siRNAs (Fig. 2A). As the second siRNA is more efficient, we designed an shRNA according to this target sequence and cloned it into a pLKO.1 lentivirus vector. Lentivirus particles containing HDAC10 cDNA or this shRNA were used to infect the human cervical cancer cell line Caski, which has low transient transfection efficiency. The infection efficiency is shown in Fig. 2B. Next, we tested whether cell migration was altered in response to a change in HDAC10 expression with a Transwell assay. The results showed that overexpression of HDAC10 significantly reduced the migration of these two cervical cancer cell lines (Fig. 2, C and D, upper panels). In contrast, depletion of HDAC10 significantly increased cell migration (Fig. 2, C and D, lower panels).
FIGURE 2.
HDAC10 inhibits cervical cancer cell migration and invasion. A and B, HDAC10 overexpression and knockdown. A, HeLa cells were transfected with empty vectors or vectors encoding HDAC10 (left panel) and control siRNA or two siRNAs targeting HDAC10 (right panel). The cell lysates were analyzed by Western blotting with anti-HDAC10 antibodies. B, Caski cells were infected with lentivirus encoding HDAC10 (left panel) or short hairpin RNAs targeting HDAC10 (right panel). The expression of HDAC10 was analyzed by Western blotting with anti-HDAC10 antibodies. C and D, cell migration. HeLa cells (C) or Caski cells (D) of the control (con), HDAC10 overexpression, or HDAC10 knockdown group were collected and added to Transwell inserts. 16 h later, cells on the undersurface were fixed, stained, photographed, and counted. E and F, cell invasion. The upper surface of the inserts was precoated with Matrigel for 4 h. Control, HDAC10-overexpressing, and HDAC10 knockdown HeLa cells (E) and Caski cells (F) were added to the precoated Transwell chambers. Several hours later, cells were detected. Photos are shown on the left, and the dots are nuclei of the cells. Student's t test was performed, and the statistical analysis is shown on the right. Every experiment was repeated three times. *, p < 0.05; **, p < 0.01. Error bars represent S.D.
We next tested the effect of HDAC10 on cervical cancer cell invasion. To do this, Transwell chambers were precoated with matrigel. The detailed steps are described under “Experimental Procedures.” The results showed that cell invasion of these two cell lines was dramatically inhibited by HDAC10 overexpression. Conversely, HDAC10-deficient cells exhibited a much stronger invasive activity than control cells (Fig. 2, E and F).
Next, we analyzed the proliferation rate of HeLa cells expressing different levels of HDAC10. Depletion or overexpression of HDAC10 does not significantly alter cervical cancer cell proliferation. Consistent with this, fluorescence-activated cell sorting (FACS) analysis showed that the proportion of the cells in G1, S, and G2 phases was not influenced by the level of HDAC10 (data not shown). Similarly, neither elevating nor reducing the expression level of HDAC10 induced cancer cell apoptosis (data not shown). Collectively, these results suggest that HDAC10 plays an important and specific role in regulation of cervical cell migration and invasion but not proliferation or apoptosis.
HDAC10 Down-regulates the Expression Level of MMP2 and -9
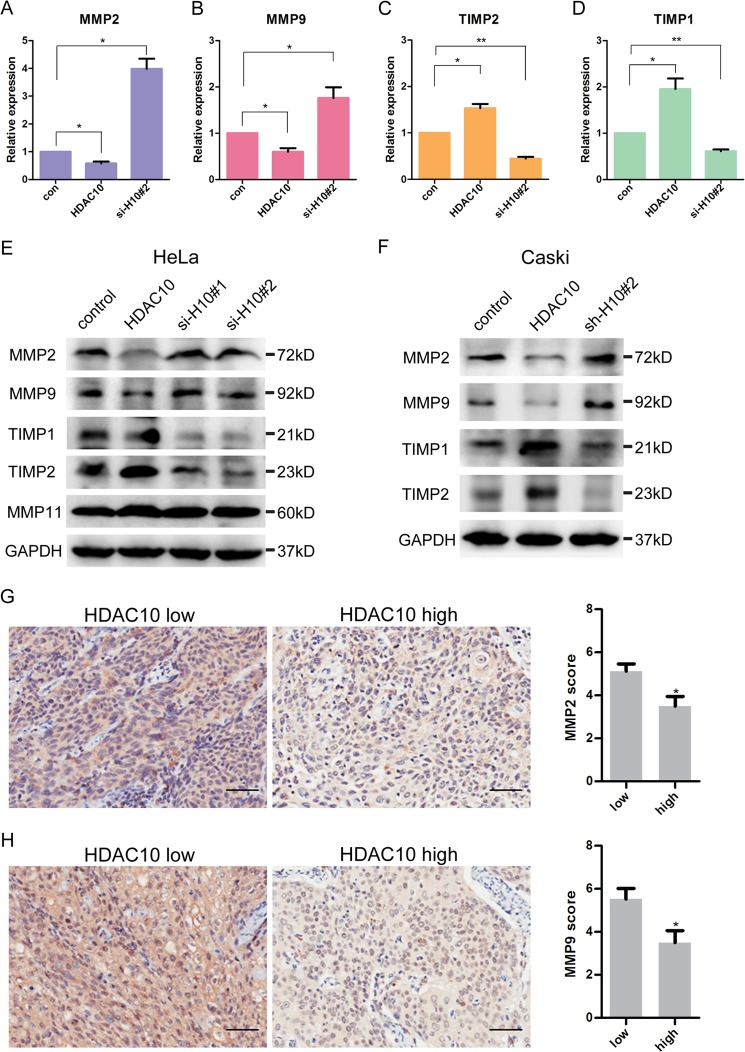
Previous studies have shown that MMP2 and -9 are pivotal for cervical cancer invasion (26). To test whether HDAC10, which suppresses cervical cancer cell migration and invasion (Fig. 2), plays a role in regulation of MMP2 and -9, we analyzed the effects of HDAC10 on MMP2 and -9 expression levels. In HeLa cells, both the mRNA level (Fig. 3, A and B) and protein level (Fig. 3E) of MMP2 and -9 were decreased in response to increased HDAC10 expression. Conversely, depletion of HDAC10 significantly increased the mRNA and protein levels of MMP2 and -9 (Fig. 3, A, B, and E). Although alteration of HDAC10 expression regulates the expression of MMP2 and MMP9, it does not affect MMP11 (Fig. 3E).
FIGURE 3.
HDAC10 inhibits MMP2 and -9 expression. A–D, HDAC10 down-regulates MMP2 and -9 expression and up-regulates TIMP1 and -2 expression. HeLa cells were transiently transfected with empty vector, HDAC10-encoding vector, and HDAC10 siRNA, respectively. The mRNA levels of these genes were detected by quantitative RT-PCR analysis in control (con) cells, HDAC10-overexpressing, and HDAC10 knockdown cells. *, p < 0.05; **, p < 0.01 versus the control group. E and F, Western blotting was used to analyze the expression of MMP2, -9, -11, TIMP1, and -2 in the control, HDAC10 overexpression, and HDAC10 knockdown groups in HeLa cells (E) and Caski cells (F). GAPDH serves as a loading control. G and H, the expression levels of MMP2 (G) and MMP9 (H) in carcinoma tissues were detected in HDAC10 low and HDAC10 high groups by tissue microarray (HDAC10 low, score <6; HDAC10 high, score ≥6). Data were analyzed by Student's t test. *, p < 0.05; **, p < 0.01. Scale bars, 100 μm. Error bars represent S.D.
Previous studies have shown that MMP2 can interact with TIMP2 and be specifically inhibited by it, whereas MMP9 can be specifically inhibited by TIMP1 (13). Thus, we analyzed the expression levels of TIMP1 and TIMP2 in response to alterations of HDAC10 expression in HeLa cells. The results showed that increased expression of HDAC10 up-regulated whereas depletion of HDAC10 down-regulated the levels of TIMP1 and TIMP2 (Fig. 3, C–E). To confirm the function of HDAC10 in regulating these genes, we replicated these experiments in the cervical cancer cell line Caski and obtained similar results (Fig. 3F).
Next, we further investigated the relationship between the expression level of HDAC10 and MMP2 and -9 in clinical cervical cancer patients. Microarrays shown in Fig. 1 were analyzed with MMP2 and MMP9 antibodies. Patients were divided into two groups according to HDAC10 score. The results of microarrays reveal that there is an inverse correlation between the expression of HDAC10 and MMP2 or MMP9 (Fig. 3, G and H) in cervical cancer patients.
Histone Deacetylase Activity of HDAC10 Is Required for Down-regulation of MMP2 and -9 Expression
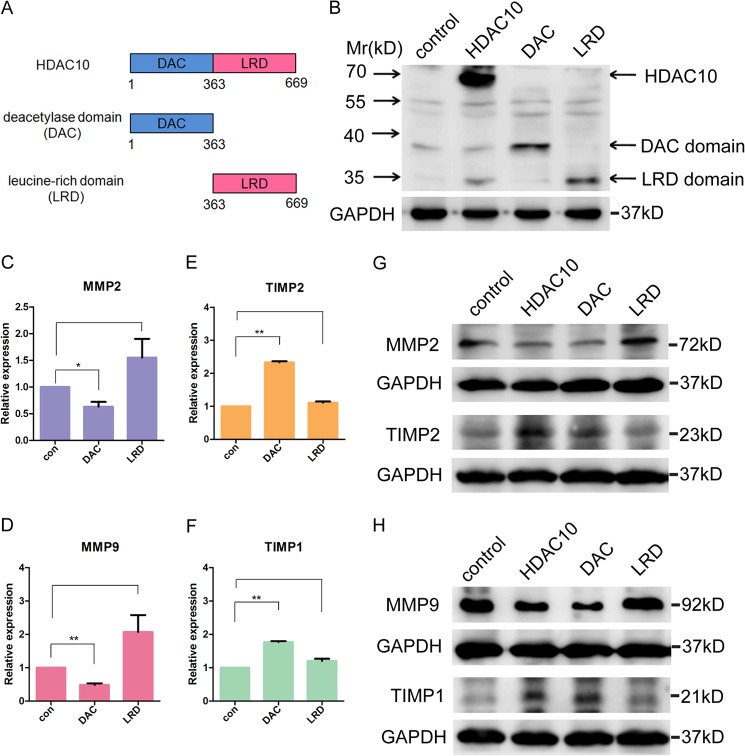
HDAC10 contains one DAC domain in the N-terminal region and one catalytically inactive LRD in the C-terminal region (Fig. 4A) (21). To investigate whether the histone deacetylase activity is required for its regulation of MMP2 and -9 expression, we used PCR to generate two plasmids, each of which encodes either the DAC domain or the LRD (Fig. 4B). Real time PCR and Western blotting showed that the DAC domain, but not the LRD, can mimic the effects of full-length HDAC10 on regulating the mRNA level of MMP2, MMP9, TIMP1, and TIMP2 (Fig. 4, C–F) and the protein level of these genes (Fig. 4, G and H), suggesting that the histone deacetylase activity of HDAC10 is required in this process.
FIGURE 4.
Histone deacetylase activity is required for down-regulation of MMP2 and -9 expression. A, domain structure of HDAC10. The N-terminal region contains a DAC domain, and the C-terminal region contains an LRD. B, plasmids encoding FLAG-tagged full-length HDAC10, DAC domain, and LRD were constructed. The expression of the gene and its fragments was confirmed by Western blotting with an anti-FLAG antibody. C–F, quantitative real time PCR was used to detect the mRNA level of MMP2 and -9 and TIMP1 and -2 in control HeLa cells and cells transfected with plasmids encoding DAC domain or LRD. *, p < 0.05; **, p < 0.01 versus the control group (con). G and H, the change in MMP2, -9, TIMP1, and -2 expression levels after overexpressing full-length HDAC10, DAC domain, and LRD. The protein levels were analyzed by Western blotting. GAPDH was used as a loading control. Error bars represent S.D.
HDAC10 Binds to the Promoter Regions of MMP2 and -9 and Deacetylates Histones H3 and H4 in These Regions
Because the HDAC10-mediated suppression of MMP2 and -9 transcription requires its histone deacetylase domain (Fig. 4, C and D), we hypothesized that HDAC10 might interact with the promoter regions of MMP2 and -9 and alter the acetylation level of histone in these regions, resulting in suppression of MMP2 and -9 expression. To test this hypothesis, we first used a ChIP assay to investigate whether HDAC10 binds to the MMP2 and MMP9 promoter regions. The primers used in the ChIP assay were designed according to the binding site of transcriptional factors in these regions (27–34). The results indicate that HDAC10 can indeed bind to the promoter regions of MMP2 and MMP9 (Fig. 5, A and B). To normalize the efficiency of PCR primers, the products were compared with input DNA amplified by the same primers. Then we immunoprecipitated the DNA using antibodies recognizing acetylated histones H3 and H4 and compared the acetylation levels between the control group and HDAC10 overexpression group in the transcriptional factor binding sites mentioned above. As shown in Fig. 5C, a reduction of histone acetylation was observed after HDAC10 induction in the MMP2 promoter region, and the strongest effect was observed at the AP1 binding site. In the MMP9 promoter region, which is shown in Fig. 5D, the decline of histone acetylation was mainly observed at the second locus containing NF-κB and sp1 binding sites.
FIGURE 5.
HDAC10 binds to the promoter regions of MMP2 and -9 and deacetylates histones H3 and H4 in these regions. A–J, results of ChIP assay. A and B, HDAC10 binds to MMP2 and -9 promoter regions. Top, diagrams of MMP2 and -9 promoter regions. The transcriptional factor binding sites and the primers designed are shown. Bottom, the amount of DNA precipitated by either control IgG or anti-HDAC10 antibody was expressed as a percentage of the total input genomic DNA. C and D, HDAC10 decreases acetylation of histones H3 and H4 in MMP2 and -9 promoter regions. Acetylated histone H3 and H4 antibodies were used to precipitate DNA in ChIP assay. Primers associated with the transcription factor binding sites in MMP2 and -9 promoter regions were used for quantitative real time PCR. The data are from three independent experiments. E and F, reduced acetylation of H3 and H4 blocks the association of RNA polymerase II to the transcription factors binding sites. DNA of HeLa cells in the control group and HDAC10 overexpression group was precipitated by anti-RNA polymerase II (pol II) antibody in a ChIP assay. G and H, HeLa cells overexpressing full-length HDAC10, DAC domain, and LRD were collected and analyzed using a ChIP assay. DNA was precipitated by acetylated histone H3 and H4 antibodies. Primers recognizing MMP2 (−1298 to −1073) (G) and MMP9 (−670 to −486) (H) were used to analyze the amount of DNA precipitated. Columns, results of triplicates; error bars, S.D. *, p < 0.05; **, p < 0.01 versus the control group. I and J, co-immunoprecipitation (IP) assay. Cell lysates of the control and HDAC10 overexpression groups were incubated with anti-FLAG-agarose beads. The immunoprecipitates were analyzed by Western blotting with antibodies recognizing FLAG, AP1, or p65. CREB, cAMP-response element-binding protein. IB, immunoblot.
Previous studies suggest that reduced histone acetylation could lead to DNA in these regions being condensed, which may prevent RNA polymerase II binding to these regions (15). To test whether this occurs in HDAC10-mediated regulation of MMP2 and -9 expression, antibody against RNA polymerase II was used in a ChIP assay. Consistent with reduced acetylation of histones H3 and H4, the binding of RNA polymerase II in the promoter regions of MMP2 and -9, especially at the second locus, was significantly reduced in response to increased expression of HDAC10 (Fig. 5, E and F). To further test this, we overexpressed DAC domain and LRD, respectively, and analyzed histone acetylation in the second locus of MMP2 and -9 where the acetylation level was reduced most dramatically in response to an elevation of HDAC10. As we expected, LRD, which lacks histone deacetylase activity, cannot mimic the effect of the full-length HDAC10 and DAC domain (Fig. 5, G and H).
HDACs lack a DNA-binding motif, so they are recruited to their chromatin targets by interacting with other proteins. In previous studies, HDACs were found as subunits of several complexes and could interact with various transcription factors (35). Based on these studies, we further investigated the interaction between HDAC10 and transcription factors in the promoter regions of MMP2 and MMP9. Considering the strongest effect of histone deacetylation was detected in the AP1 binding site of MMP2 promoter (Fig. 5C) and NF-κB binding site of MMP9 promoter (Fig. 5D), we used a co-immunoprecipitation assay to test whether HDAC10 could interact with these transcription factors. Interaction between HDAC10 and AP1 was not observed (Fig. 5I). However, we detected that HDAC10 could interact with p65 (Fig. 5J), an important component of NF-κB complex.
We also tested whether HDAC10 can interact with the promoter regions of TIMP1 and TIMP2. The results of the ChIP assay showed that, in contrast to MMP2 and -9 promoter regions, the promoter regions of TIMP1 and TIMP2 do not interact with HDAC10 (data not shown). Thus, HDAC10 likely up-regulates TIMP1 and TIMP2 expression (Fig. 3, C–F) through an indirect mechanism.
HDAC10 Inhibits Cervical Cancer Cell Migration and Invasion by Down-regulating MMP2 and -9 Expression
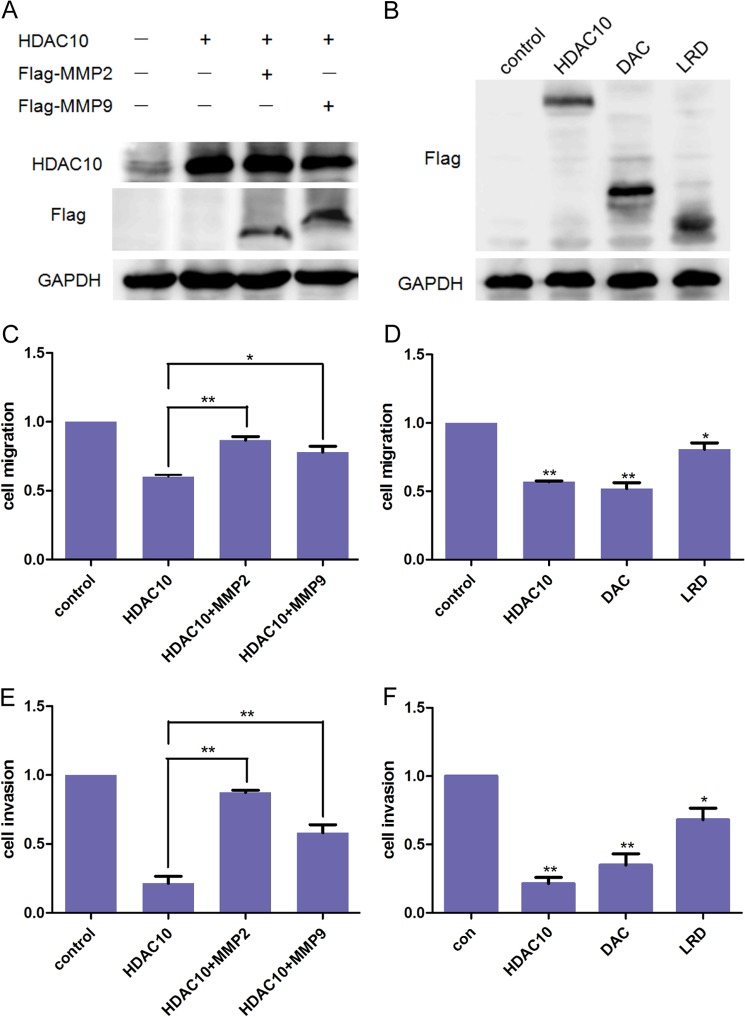
To demonstrate that the alteration of MMP2 and -9 expression is critical in HDAC10-mediated suppression of cervical cancer cell migration and invasion, we generated vectors encoding human MMP2 and -9 and co-transfected them into HeLa cells. Overexpression of MMP2 and -9 was confirmed by Western blotting (Fig. 6A). The results of the Transwell assay showed that HDAC10-meidated suppression of HeLa cell migration and invasion was reversed in part by the expression of MMP9 and to a great extent by the expression of MMP2 (Fig. 6, C and E). Thus, HDAC10 suppresses HeLa cell migration and invasion through inhibition of MMP2 and to a lesser extent MMP9 expression. Consistent with the results described earlier, both the full-length HDAC10 and the DAC domain, but not the LRD, dramatically inhibited cancer cell migration and invasion (Fig. 6, B, D, and F).
FIGURE 6.
HDAC10 inhibits HeLa cell migration and invasion by down-regulating MMP2 and -9 expression. A and B, Western blotting. A, HeLa cells were transfected with HDAC10 expression vector alone or co-transfected with HDAC10 and MMP2 or MMP9 expression vectors. B, plasmids encoding full-length HDAC10, DAC domain, or LRD were transfected into HeLa cells. The effect of overexpression was detected 48 h after transfection. C and D, cell migration. Cells of the groups described above were collected and added to the Transwell chambers at a density of 5 × 104 cells/well. 16 h later, cells on the undersurface of the membrane were detected. E and F, cell invasion. Transwell chambers were precoated, and 1 × 105 HeLa cells were added to each well. The results were analyzed 20 h later. The results are from three independent experiments. The data were analyzed by Student's t test. *, p < 0.05; **, p < 0.01 versus the control group (con). Error bars represent S.D.
HDAC10 Suppresses Cervical Cancer Cell Metastasis in Vivo
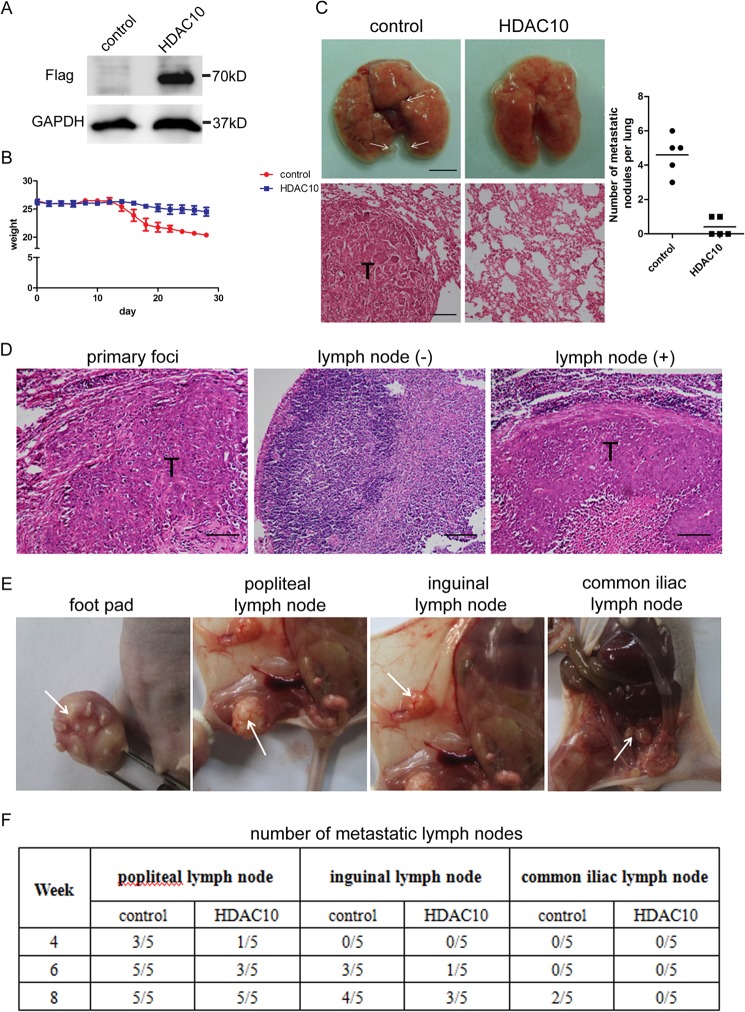
To test the ability of HDAC10 to inhibit cervical cancer cell lung colonization in vivo, we developed an experimental metastasis model in NOD-SCID mice. HeLa cells were infected with lentivirus encoding HDAC10. The increased HDAC10 expression was confirmed by Western blotting (Fig. 7A). Next, we injected equal numbers of HeLa cells overexpressing HDAC10 or those expressing a normal level of HDAC10 as a control into the tail vein of the NOD-SCID mice. After injection, the weights of the mice were measured once every other day. 4 weeks later, the mice of the control group were more morbid, and their weights were lower than those of the HDAC10 overexpression group (Fig. 7B). The mice were then killed, and the metastatic nodules on each lung were counted and recorded. The numbers of nodules in the HDAC10 overexpression group were significantly smaller than those of the control group (Fig. 7C).
FIGURE 7.
HDAC10 inhibits cell metastasis in vivo. A, HDAC10 overexpression. HeLa cells were infected with lentivirus encoding FLAG-tagged HDAC10 or control virus. 72 h after infection, cells were collected, and cell lysate was analyzed by Western blotting with anti-FLAG antibody. GAPDH was used as a loading control. B, weights of the mice. Weights of the mice in each group were measured and recorded every other day after tail vein injection until the mice were killed. Data were calculated, and the average and S.D. of weights in the control group and HDAC10 overexpression group are shown in the diagram. C, 4 weeks after tail vein injection, lungs were isolated and examined. Left, upper panel, photos of lungs. Scale bar, 5 mm. Lower panel, H&E staining of lung metastatic tumors. Scale bar, 100 μm. Right, the number of metastatic nodules on each lung in the control and HDAC10 overexpression groups were counted, and the results are shown (n = 5). D, H&E staining results of primary foci on the foot pad and negative and positive lymph nodes. Scale bar, 100 μm. E, images of primary tumor and metastatic popliteal, inguinal, and common iliac lymph nodes were taken 8 weeks after foot pad injection. F, 4, 6, and 8 weeks after injection, the numbers of positive lymph nodes were determined and recorded (n = 5). Error bars represent S.D.
To further investigate the effects of HDAC10 on lymph node metastasis, we established a model in nude mice. HeLa cells were injected into the right hind limb foot pad of the mice. Several weeks after injection, the cervical cancer cells can metastasize to the lymph nodes nearby. The lymph node metastasis ability of cervical cancer cells can be evaluated by analyzing the numbers of positive lymph nodes in each group. The metastasis status of lymph nodes was determined by H&E staining. The staining results of primary foci, positive lymph node, and negative lymph node are shown in Fig. 7D. 4 weeks after injection, metastasis presented in popliteal lymph node, the first echelon node of the foot pad. Then cancer cells spread to inguinal and common iliac lymph nodes, the second and third echelon lymph nodes. The images of foot pads and lymph nodes are shown in Fig. 7E. Although HDAC10-overexpressing cells still have the ability to metastasize, the number of positive lymph nodes was decreased (Fig. 7F). These results demonstrate that HDAC10 can impair cervical cancer lymph node metastasis in vivo.
To test whether HDAC10 affects HeLa cell proliferation in vivo, we developed a tumor growth model using nude mice. We injected HDAC10-expressing and control HeLa cells subcutaneously into the mice. Consistent with the in vitro proliferation assays, neither the weights nor the volumes of the xenografts were affected by the increased expression of HDAC10. There was also no difference in the weight of the mice between the two groups (data not shown). Collectively, these results suggest that increased expression of HDAC10 effectively suppresses cervical cancer cell metastasis but not tumor growth in vivo.
DISCUSSION
It has been well established that many members of the HDAC family are key factors in promoting carcinogenesis. The histone acetylation levels in the promoter regions of some tumor suppressor genes are aberrant in cancer (36). Class I HDACs can promote cell cycles and cancer cell proliferation (35, 37). HDAC6, a member of the class II HDACs, can regulate the cytoskeleton and alter cell motility (38). It alters the acetylation level of cortactin (39) and microtubules (40) to increase cell migration. Thus, HDACs are promising targets for anticancer therapeutics. However, in certain cases, HDACs may act as tumor suppressors. For example, elevated class I HDAC expression is associated with improved prognosis in cutaneous T-cell lymphoma (14). Class II HDAC expression decreases in some tumors, and high expression of these isoforms in some entities such as non-small cell lung carcinoma (41) and breast carcinoma (14) predicted better patient outcome. Thus, to successfully use HDAC inhibitors in combating cancer, we must fully understand the role of each specific HDAC in the genesis and progression of cancer. In this study, we have obtained several lines of evidence suggesting that HDAC10 acts as a metastasis suppressor in cervical cancer. First, clinically HDAC10 expression is significantly lower in cervical cancer tissues of patients with lymph node metastasis. Second, HDAC10 impairs migration and invasion of cervical cancer cells in culture and metastasis in vivo. Third, HDAC10 suppresses expression of MMP2 and -9, proteins that are known to promote cancer cell invasion and metastasis.
HDACs can regulate gene expression through several mechanisms (15), and frequently they lead to transcription suppression (42). For example, they can regulate the binding of transcription factors to DNA, change the structure of chromatin (43), and serve as a signal to regulate the expression of downstream genes (44). Using a ChIP assay, we demonstrate in this study that HDAC10 binds to the promoter regions of MMP2 and -9, deacetylates histones H3 and H4 in these regions, and reduces the binding of RNA polymerase II. Using a co-immunoprecipitation assay, we detected an interaction between HDAC10 and p65. Based on these findings, we propose that HDAC10 may be recruited to the promoter region by interacting with transcription factors such as p65 in the MMP9 promoter region. However, we have not been able to detect an interaction between HDAC10 and AP1, which is known to be a major transcription factor regulating MMP2 expression. One possible explanation is that HDAC10 may interact with other transcription factors nearby. Alternatively, the reaction might be transient and thus was not detected under the condition used in our experiments. After the binding of HDAC10 to these promoter regions, it may change the structure of chromatin in the promoter regions by deacetylating the histone and making the DNAs more condensed, resulting in reduced binding of RNA polymerase II. Consistent with this, our experiments with the DAC domain and LRD of HDAC10 suggest that the enzymatic activity of HDAC10 is essential for its function. However, it does not mean that LRD is totally nonfunctional, and the function of HDAC10 cannot be split into two parts simply. Although LRD does not have enzymatic activity and cannot change the expression of MMP2 and -9, it may still retain some function of full-length HDAC10. It may have the ability to interact with other proteins such as some components of the transcription complex; this needs further research. By the same token, the DAC domain we used in the experiment probably contains other functions beyond its deacetylation activity. Interestingly, although HDAC10 does not bind to the promoter regions of TIMP1 and -2 it does increase the expression levels of TIMP1 and -2. Thus, HDAC10 likely regulates TIMP1 and -2 expression through an indirect mechanism. For example, HDAC10 might down-regulate genes that suppress TIMP1 and -2 expression. Clearly, the mechanism through which HDAC10 promotes TIMP1 and -2 expression requires further investigation in future studies.
Cervical cancer is the second most common and the fifth deadliest cancer in women in the world (45). Prognosis drops dramatically in patients with invasive cervical cancer. The in situ carcinoma can be successfully treated with surgery, radiation, or chemotherapy. However, the metastatic tumor cannot be removed thoroughly by surgery. In radiation treatment, metastasis can only be removed by increasing the area of treatment, which leads to undesirable side effects. According to statistics, 35% of patients with invasive cervical cancer have persistent or recurrent disease after treatment (2). Thus, suppression of cancer metastasis is critical for treatment of cervical cancer. Clinical studies have demonstrated an association of MMP2 and -9 expression during progression of cervical cancer (46–48). Several agents that regulate the expression of MMP2 and -9 in cervical cancer including epigallocatechin gallate, actinomycin D, cycloheximide, retinoic acid, and dexamethasone (10) have been investigated. However, how the expression of MMP2 and -9 is regulated was not clear. In this study, we have identified HDAC10 as a key suppressor of MMP2 and -9 expression in cervical cancer. Furthermore, we have demonstrated that HDAC10 suppresses MMP2 and -9 expression by interacting with the promoter regions of MMP2 and -9 and deacetylating histones H3 and H4, revealing a new epigenetic regulatory mechanism of MMP2 and -9 expression.
HDAC inhibitors are widely considered as promising anticancer therapeutics. One key consideration of using HDAC inhibitors for treatment of cancers is the balance between safety and efficacy. The studies described in this report together with other studies clearly demonstrate that the functions of HDACs are diverse and isoform-dependent. Although many HDACs, in particular class I HDACs, clearly promote tumor growth and therefore inhibitors to these HDACs are attractive anticancer therapeutic targets, certain HDACs (e.g. HDAC10 described in this study) play important roles in suppressing cancer progression. Thus, developing isoform-specific HDAC inhibitors that do not target metastasis-suppressing HDACs (e.g. HDAC10) will be an important consideration for treatment of cervical squamous cell carcinoma and perhaps other cancers (e.g. non-small cell lung carcinoma (41)).
Acknowledgment
We thank Yizeng Tu (University of Pittsburgh) for technical assistance during the initial stage of this study.
This work was supported, in whole or in part, by National Institutes of Health Grant GM65188 (to C. W.). This work was also supported by Ministry of Science and Technology Grants 2011CB965100, 2010CB944900, 2011CBA01100, 2011DFA30480, 2010CB945000, and 2012CB966603; National Natural Science Foundation of China Grants 91219305, 31171432, 31101061, 31210103905, 31071306, 31000378, and 81170499; Science and Technology Commission of Shanghai Municipality Grants 11ZR1438500 and 11XD1405300; and Ministry of Education Grants IRT1168 and 20110072110039.
- MMP
- matrix metalloproteinase
- HDAC
- histone deacetylase
- TIMP
- tissue inhibitor of metalloproteinase
- DAC
- deacetylase
- LRD
- leucine-rich domain
- NOD-SCID
- nonobese diabetic-severe combined immunodeficient
- TNM
- tumor node metastasis.
REFERENCES
- 1. The Merck Manual Home Health Handbook (2013) Cervical Cancer, www.merckmanuals.com/home/womens_health_issues/cancers_of_the_female_reproductive_system/cervical_cancer.html
- 2. Armenian Medical Network (2013) Cervical Cancer, www.health.am/cr/cervical-cancer/
- 3. Boothello R. (2011) Matrix Metalloproteinases: Its Implications in Cardiovascular Disorders, pharmaxchange.info/press/2011/11/matrix-metalloproteinases-its-implications-in-cardiovascular-disorders/
- 4. Wang X. X., Cheng Q., Zhang S. N., Qian H. Y., Wu J. X., Tian H., Pei D. S., Zheng J. N. (May 22, 2013) PAK5-Egr1-MMP2 signaling controls the migration and invasion in breast cancer cell. Tumour Biol. 10.1007/s13277-013-0824-x [DOI] [PubMed] [Google Scholar]
- 5. Yu G., Li H., Wang X., Wu T., Zhu J., Huang S., Wan Y., Tang J. (2013) MicroRNA-19a targets tissue factor to inhibit colon cancer cells migration and invasion. Mol. Cell. Biochem. 380, 239–247 [DOI] [PubMed] [Google Scholar]
- 6. Yang Y., Jiao L., Hou J., Xu C., Wang L., Yu Y., Li Y., Yang C., Wang X., Sun Y. (2013) Dishevelled-2 silencing reduces androgen-dependent prostate tumor cell proliferation and migration and expression of Wnt-3a and matrix metalloproteinases. Mol. Biol. Rep. 40, 4241–4250 [DOI] [PubMed] [Google Scholar]
- 7. Bérubé M., Deschambeault A., Boucher M., Germain L., Petitclerc E., Guérin S. L. (2005) MMP-2 expression in uveal melanoma: differential activation status dictated by the cellular environment. Mol. Vis. 11, 1101–1111 [PubMed] [Google Scholar]
- 8. Di Nezza L. A., Misajon A., Zhang J., Jobling T., Quinn M. A., Ostör A. G., Nie G., Lopata A., Salamonsen L. A. (2002) Presence of active gelatinases in endometrial carcinoma and correlation of matrix metalloproteinase expression with increasing tumor grade and invasion. Cancer 94, 1466–1475 [DOI] [PubMed] [Google Scholar]
- 9. Sato T., Sakai T., Noguchi Y., Takita M., Hirakawa S., Ito A. (2004) Tumor-stromal cell contact promotes invasion of human uterine cervical carcinoma cells by augmenting the expression and activation of stromal matrix metalloproteinases. Gynecol. Oncol. 92, 47–56 [DOI] [PubMed] [Google Scholar]
- 10. Roomi M. W., Monterrey J. C., Kalinovsky T., Rath M., Niedzwiecki A. (2010) In vitro modulation of MMP-2 and MMP-9 in human cervical and ovarian cancer cell lines by cytokines, inducers and inhibitors. Oncol. Rep. 23, 605–614 [DOI] [PubMed] [Google Scholar]
- 11. Boeker K. H., Haberkorn C. I., Michels D., Flemming P., Manns M. P., Lichtinghagen R. (2002) Diagnostic potential of circulating TIMP-1 and MMP-2 as markers of liver fibrosis in patients with chronic hepatitis C. Clin. Chim. Acta 316, 71–81 [DOI] [PubMed] [Google Scholar]
- 12. Branca M., Ciotti M., Giorgi C., Santini D., Di Bonito L., Costa S., Benedetto A., Bonifacio D., Di Bonito P., Paba P., Accardi L., Syrjänen S., Favalli C., Syrjänen K., and HPV-PathogenISS Study Group (2006) Matrix metalloproteinase-2 (MMP-2) and its tissue inhibitor (TIMP-2) are prognostic factors in cervical cancer, related to invasive disease but not to high-risk human papillomavirus (HPV) or virus persistence after treatment of CIN. Anticancer Res. 26, 1543–1556 [PubMed] [Google Scholar]
- 13. Vasala K. (2008) Matrix metalloproteinase MMP-2 and MMP-9 and their inhibitors TIMP-1 And TIMP-2 in bladder carcinoma. Acta Univ. Oul. D 983 [Google Scholar]
- 14. Weichert W. (2009) HDAC expression and clinical prognosis in human malignancies. Cancer Lett. 280, 168–176 [DOI] [PubMed] [Google Scholar]
- 15. Sengupta N., Seto E. (2004) Regulation of histone deacetylase activities. J. Cell. Biochem. 93, 57–67 [DOI] [PubMed] [Google Scholar]
- 16. Venkataramani V., Rossner C., Iffland L., Schweyer S., Tamboli I. Y., Walter J., Wirths O., Bayer T. A. (2010) Histone deacetylase inhibitor valproic acid inhibits cancer cell proliferation via down-regulation of the Alzheimer amyloid precursor protein. J. Biol. Chem. 285, 10678–10689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhang L., Wang G., Wang L., Song C., Leng Y., Wang X., Kang J. (2012) VPA inhibits breast cancer cell migration by specifically targeting HDAC2 and down-regulating Survivin. Mol. Cell. Biochem. 361, 39–45 [DOI] [PubMed] [Google Scholar]
- 18. Ellis L., Hammers H., Pili R. (2009) Targeting tumor angiogenesis with histone deacetylase inhibitors. Cancer Lett. 280, 145–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Michaelis M., Michaelis U. R., Fleming I., Suhan T., Cinatl J., Blaheta R. A., Hoffmann K., Kotchetkov R., Busse R., Nau H., Cinatl J., Jr. (2004) Valproic acid inhibits angiogenesis in vitro and in vivo. Mol. Pharmacol. 65, 520–527 [DOI] [PubMed] [Google Scholar]
- 20. Witt O., Deubzer H. E., Milde T., Oehme I. (2009) HDAC family: what are the cancer relevant targets? Cancer Lett. 277, 8–21 [DOI] [PubMed] [Google Scholar]
- 21. Tong J. J., Liu J., Bertos N. R., Yang X. J. (2002) Identification of HDAC10, a novel class II human histone deacetylase containing a leucine-rich domain. Nucleic Acids Res. 30, 1114–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lai I. L., Lin T. P., Yao Y. L., Lin C. Y., Hsieh M. J., Yang W. M. (2010) Histone deacetylase 10 relieves repression on the melanogenic program by maintaining the deacetylation status of repressors. J. Biol. Chem. 285, 7187–7196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lee J. H., Jeong E. G., Choi M. C., Kim S. H., Park J. H., Song S. H., Park J., Bang Y. J., Kim T. Y. (2010) Inhibition of histone deacetylase 10 induces thioredoxin-interacting protein and causes accumulation of reactive oxygen species in SNU-620 human gastric cancer cells. Mol. Cells 30, 107–112 [DOI] [PubMed] [Google Scholar]
- 24. Kotian S., Liyanarachchi S., Zelent A., Parvin J. D. (2011) Histone deacetylases 9 and 10 are required for homologous recombination. J. Biol. Chem. 286, 7722–7726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Im H., Grass J. A., Johnson K. D., Boyer M. E., Wu J., Bresnick E. H. (2004) Measurement of protein-DNA interactions in vivo by chromatin immunoprecipitation. Methods Mol. Biol. 284, 129–146 [DOI] [PubMed] [Google Scholar]
- 26. Kato Y., Yamashita T., Ishikawa M. (2002) Relationship between expression of matrix metalloproteinase-2 and matrix metalloproteinase-9 and invasion ability of cervical cancer cells. Oncol. Rep. 9, 565–569 [PubMed] [Google Scholar]
- 27. Chernov A. V., Sounni N. E., Remacle A. G., Strongin A. Y. (2009) Epigenetic control of the invasion-promoting MT1-MMP/MMP-2/TIMP-2 axis in cancer cells. J. Biol. Chem. 284, 12727–12734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kim Y., Jeoung D. (2009) The cancer/testis antigen CAGE induces MMP-2 through the activation of NF-κB and AP-1. BMB Rep. 42, 758–763 [DOI] [PubMed] [Google Scholar]
- 29. Lee C. W., Lin C. C., Lin W. N., Liang K. C., Luo S. F., Wu C. B., Wang S. W., Yang C. M. (2007) TNF-α induces MMP-9 expression via activation of Src/EGFR, PDGFR/PI3K/Akt cascade and promotion of NF-κB/p300 binding in human tracheal smooth muscle cells. Am. J. Physiol. Lung Cell. Mol. Physiol. 292, L799–L812 [DOI] [PubMed] [Google Scholar]
- 30. Lin L. F., Chuang C. H., Li C. F., Liao C. C., Cheng C. P., Cheng T. L., Shen M. R., Tseng J. T., Chang W. C., Lee W. H. (2010) ZBRK1 acts as a metastatic suppressor by directly regulating MMP9 in cervical cancer. Cancer Res. 70, 192–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Satpathy M., Shao M., Emerson R., Donner D. B., Matei D. (2009) Tissue transglutaminase regulates matrix metalloproteinase-2 in ovarian cancer by modulating cAMP-response element-binding protein activity. J. Biol. Chem. 284, 15390–15399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Song H., Ki S. H., Kim S. G., Moon A. (2006) Activating transcription factor 2 mediates matrix metalloproteinase-2 transcriptional activation induced by p38 in breast epithelial cells. Cancer Res. 66, 10487–10496 [DOI] [PubMed] [Google Scholar]
- 33. Takahra T., Smart D. E., Oakley F., Mann D. A. (2004) Induction of myofibroblast MMP-9 transcription in three-dimensional collagen I gel cultures: regulation by NF-κB, AP-1 and Sp1. Int. J. Biochem. Cell Biol. 36, 353–363 [DOI] [PubMed] [Google Scholar]
- 34. Yoshizaki T., Sato H., Furukawa M., Pagano J. S. (1998) The expression of matrix metalloproteinase 9 is enhanced by Epstein-Barr virus latent membrane protein 1. Proc. Natl. Acad. Sci. U.S.A. 95, 3621–3626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Reichert N., Choukrallah M. A., Matthias P. (2012) Multiple roles of class I HDACs in proliferation, differentiation, and development. Cell. Mol. Life Sci. 69, 2173–2187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hellebrekers D. M., Griffioen A. W., van Engeland M. (2007) Dual targeting of epigenetic therapy in cancer. Biochim. Biophys. Acta 1775, 76–91 [DOI] [PubMed] [Google Scholar]
- 37. Glaser K. B., Li J., Staver M. J., Wei R. Q., Albert D. H., Davidsen S. K. (2003) Role of class I and class II histone deacetylases in carcinoma cells using siRNA. Biochem. Biophys. Res. Commun. 310, 529–536 [DOI] [PubMed] [Google Scholar]
- 38. Valenzuela-Fernández A., Cabrero J. R., Serrador J. M., Sánchez-Madrid F. (2008) HDAC6: a key regulator of cytoskeleton, cell migration and cell-cell interactions. Trends Cell Biol. 18, 291–297 [DOI] [PubMed] [Google Scholar]
- 39. Zhang X., Yuan Z., Zhang Y., Yong S., Salas-Burgos A., Koomen J., Olashaw N., Parsons J. T., Yang X. J., Dent S. R., Yao T. P., Lane W. S., Seto E. (2007) HDAC6 modulates cell motility by altering the acetylation level of cortactin. Mol. Cell 27, 197–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hubbert C., Guardiola A., Shao R., Kawaguchi Y., Ito A., Nixon A., Yoshida M., Wang X. F., Yao T. P. (2002) HDAC6 is a microtubule-associated deacetylase. Nature 417, 455–458 [DOI] [PubMed] [Google Scholar]
- 41. Osada H., Tatematsu Y., Saito H., Yatabe Y., Mitsudomi T., Takahashi T. (2004) Reduced expression of class II histone deacetylase genes is associated with poor prognosis in lung cancer patients. Int. J. Cancer 112, 26–32 [DOI] [PubMed] [Google Scholar]
- 42. Kristensen L. S., Nielsen H. M., Hansen L. L. (2009) Epigenetics and cancer treatment. Eur. J. Pharmacol. 625, 131–142 [DOI] [PubMed] [Google Scholar]
- 43. Cress W. D., Seto E. (2000) Histone deacetylases, transcriptional control, and cancer. J. Cell. Physiol. 184, 1–16 [DOI] [PubMed] [Google Scholar]
- 44. Luger K., Mäder A. W., Richmond R. K., Sargent D. F., Richmond T. J. (1997) Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 389, 251–260 [DOI] [PubMed] [Google Scholar]
- 45. Armstrong E. P. (2010) Prophylaxis of cervical cancer and related cervical disease: a review of the cost-effectiveness of vaccination against oncogenic HPV types. J. Manag. Care Pharm. 16, 217–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Asha Nair S., Karunagaran D., Nair M. B., Sudhakaran P. R. (2003) Changes in matrix metalloproteinases and their endogenous inhibitors during tumor progression in the uterine cervix. J. Cancer Res. Clin. Oncol. 129, 123–131 [DOI] [PubMed] [Google Scholar]
- 47. Zhou C. Y, Yao J. F., Chen X. D. (2002) Expression of matrix metalloproteinase-2, 9 and their inhibitor-TIMP 1, 2 in human squamous cell carcinoma of uterine cervix. Ai Zheng 21, 735–739 [PubMed] [Google Scholar]
- 48. Xu P., Cai H., Wang D. (2009) The significance of matrix metalloproteinase-2 (MMP-2) and matrix metalloproteinase-9 (MMP-9) in the course of development of cervical carcinoma. Med. J. West China 21, 2040–2042 [Google Scholar]