Background: Coq7p is a mitochondrial hydroxylase required to synthesize coenzyme Q6 that is regulated by phosphorylation.
Results: Ptc7p is a mitochondrial phosphatase that activates coenzyme Q6 biosynthesis by Coq7p dephosphorylation.
Conclusion: Coq7p is a physiological target of the Ptc7p phosphatase.
Significance: Ptc7p constitutes a new target to increase coenzyme Q10 levels in patients affected by primary or secondary coenzyme Q deficiency.
Keywords: Coenzyme Q, Mitochondrial Metabolism, Phosphatase, Respiratory Chain, Yeast
Abstract
The study of the components of mitochondrial metabolism has potential benefits for health span and lifespan because the maintenance of efficient mitochondrial function and antioxidant capacity is associated with improved health and survival. In yeast, mitochondrial function requires the tight control of several metabolic processes such as coenzyme Q biosynthesis, assuring an appropriate energy supply and antioxidant functions. Many mitochondrial processes are regulated by phosphorylation cycles mediated by protein kinases and phosphatases. In this study, we determined that the mitochondrial phosphatase Ptc7p, a Ser/Thr phosphatase, was required to regulate coenzyme Q6 biosynthesis, which in turn activated aerobic metabolism and enhanced oxidative stress resistance. We showed that Ptc7p phosphatase specifically activated coenzyme Q6 biosynthesis through the dephosphorylation of the demethoxy-Q6 hydroxylase Coq7p. The current findings revealed that Ptc7p is a regulator of mitochondrial metabolism that is essential to maintain proper function of the mitochondria by regulating energy metabolism and oxidative stress resistance.
Introduction
Protein phosphorylation and dephosphorylation cycles mediated by kinases and phosphatases are cell signaling mechanisms found in many biological pathways. Proteomics studies have shown that a large number of proteins are phosphorylated within the mitochondria (1). Protein phosphorylation cycles regulate the activity of mitochondrial proteins involved in many cellular processes such as lipid metabolism, protein import, and regulation of oxidative phosphorylation complexes (2).
Type 2C protein phosphatases (PP2C5; now known as PPMs) are Mg2+- or Mn2+-dependent protein phosphatases distinct from other Ser/Thr phosphatases (3). This enzyme family is found in eukaryotes and contains at least 16 PPM genes in the human genome (3, 4). The conservation throughout phylogeny indicates that the PPM family plays a central role in cell signaling. For instance, human PPM proteins have been shown to regulate cell survival, growth, apoptosis, and stress response (5). In Saccharomyces cerevisiae, the PPM family includes the products of seven genes, PTC1 to PTC7. Proteins Ptc1p to Ptc4p have been characterized in the negative regulation of the high osmolarity glycerol pathway. The phosphatase Ptc5p is responsible for the up-regulation of the pyruvate dehydrogenase complex, and Ptc6p phosphatase is required for mitophagy induction (5, 6).
Yeast Ptc7p is a known mitochondrial protein that conserves the 11 motifs of the PPM family and exhibits phosphatase activity (7). This phosphatase is encoded by PTC7 (YHR079w) gene. PTC7 promoter contains sequences recognized by Hap4p, Yap1p, and Adr1p transcription factors (8, 9). These are transcription factors that lead to the induction of respiratory metabolism and stress resistance genes (10). In agreement with a potential role of Ptc7p in the control of cellular respiration, it has been determined that PTC7 overexpression improves cell growth in low oxygen environments (7). In this report, Jiang et al. (7) determined a significant increase in PTC7 gene expression coinciding with the onset of respiratory metabolism. Interestingly, Juneau et al. (11) determined that PTC7 is spliced to produce two isoforms. The first isoform localizes in the nuclear envelope, whereas the second isoform resides in the mitochondria and is mostly expressed in media with a non-fermentable carbon source, suggesting the involvement of this protein in mitochondrial metabolism (11). The human homolog of Ptc7p has been named T cell-activated protein phosphatase 2C. This protein seems to be involved in T cell activation, a process that requires an enormous amount of energy (12). Unfortunately, the function of the human homolog of Ptc7p has not been thoroughly studied yet.
Coenzyme Q (CoQ; CoQ6 in yeast) is a redox molecule essential for mitochondrial function and antioxidant defenses. CoQ functions mainly as an electron carrier from complex I or II to complex III in the mitochondrial inner membrane. Because of its redox capacity, CoQ also has an important function as an antioxidant in cellular membranes. Additionally, other functions of CoQ have been described in the β-oxidation of fatty acids, opening of the transition pore, and biosynthesis of pyrimidine nucleotides (13). CoQ biosynthesis is a tightly regulated process driven by a multiprotein complex localized in the mitochondria that catalyzes several modifications of the CoQ benzene ring (14). The protein Coq7p/Cat5p (Coq7p) is a hydroxylase that is part of the CoQ6 biosynthesis complex in yeast and catalyzes one of the latest steps required for the conversion of the late intermediate demethoxy-coenzyme Q6 (DMQ6) to CoQ6. This step represents a key regulatory point in the CoQ6 biosynthetic pathway (15). Several proteins involved in CoQ6 biosynthesis such as the O-methyltransferase Coq3p, the C-methyltransferase Coq5p, and Coq7p are known to be phosphorylated (16, 17). We have previously demonstrated that CoQ6 biosynthesis is regulated by the phosphorylation state of Coq7p in yeast. The non-phosphorylated state of Coq7p increased CoQ6 levels, whereas a phosphomimetic version of Coq7p displayed a decreased CoQ6 content, suggesting that a mitochondrial phosphatase regulates the induction of CoQ6 biosynthesis in yeast (16). In this report, we demonstrate that the mitochondrial phosphatase Ptc7p interacts with Coq7p to activate coenzyme Q biosynthesis to promote respiratory metabolism and antioxidant functions.
EXPERIMENTAL PROCEDURES
Yeast Strains and Growth Media
Yeast strains used in this study are listed in Table 1. Growth media for yeast and bacteria were prepared as described previously (16). Yeast were grown at 30 °C with shaking (200 rpm).
TABLE 1.
Strains used
| Strain | Genotype | Source |
|---|---|---|
| ptc7 | BY4742; MATα; his3Δ1; leu2Δ0; lys2Δ0; ura3Δ0; YHR076w::KanMX4; knock-out mutant of the gene YHR076w (PTC7) | Euroscarf |
| atp2 | BY4742; MATα; his3Δ1; leu2Δ0; lys2Δ0; ura3Δ0; YJR121c::KanMX4; knock-out mutant of the gene YJR121c (ATP2) | Euroscarf |
| WT | BY4742; MATα; his3Δ1; leu2Δ0; lys2Δ0; ura3Δ0; wild-type isogenic strain | Euroscarf |
| coq7 | BY4741; MATa; his3Δ1; leu2Δ0; met15Δ0; ura3Δ0; YOR125c::kanMX4; knock-out mutant of the gene YOR125c (COQ7) | Euroscarf |
Plasmid Construction
The vectors and primers used in this study are shown in Tables 2 and 3. GST-Coq7p, GST-Coq7p-AAA, and GST-Ptc7p recombinant proteins were obtained by amplifying the sequences by PCR following ligation in the pGEX4T1 bacterial expression vector. PCR was performed using Fw-GEX and Rw-GEX primers. V5-tagged proteins were obtained by amplifying coding sequences excluding the stop codon with specific V5 primers and cloned using the pYES2.1 TOPO TA Expression kit following the manufacturer's procedures (Invitrogen catalog number K415001). The pCM189-PTC7 version was obtained by amplifying the sequence by PCR with pCM189-PTC7 primers following ligation in the pCM189 yeast expression vector. pmLAAA was cloned by digesting COQ7-AAA from pRS316 with XhoI and HindIII restriction enzymes and ligating in pRS426. DNA sequencing was performed by the MWG-Biotech AG Sequencing Service (Ebergsberg, Germany).
TABLE 2.
Plasmids used
| Name | Description | Specification | Source/Ref. |
|---|---|---|---|
| pGEX-COQ7 | pGEX4T1 including COQ7 WT sequence | GST-Coq7p recombinant protein | 16 |
| pGEX-COQ7-AAA | pGEX4T1 including COQ7- (S20A,S28A,S32A) sequence | GST-Coq7p-AAA recombinant protein | 16 |
| pGEX-PTC7 | pGEX4T1 including PTC7 WT sequence | GST-Ptc7p recombinant protein | This work |
| pCM189 | Yeast expression vector, URA3 auxotrophy | Empty control | 42 |
| pCM189-PTC7 | pCM189 including PTC7 WT sequence | PTC7-null mutant complementation | This work |
| pPTC7-V5 | pYES2.1/V5-His-TOPO including PTC7 WT sequence-V5 tag | Ptc7p tagged with V5 epitope | This work |
| pCOQ7-V5 | pYES2.1/V5-His-TOPO-COQ7 WT sequence-V5 tag | Wild-type Coq7p tagged with V5 epitope | 16 |
| pCOQ7-AAA-V5 | pYES2.1/V5-His-TOPO-COQ7- (S20A,S28A,S32A) sequence-V5 tag | Non-phosphorylatable Coq7p tagged with V5 epitope | 16 |
| pRS426 | Yeast expression vector, URA3 auxotrophy | Empty control | 43 |
| pmCOQ7 | pRS426 including COQ7 WT sequence | COQ7 multicopy complementation | 16 |
| pmCOQ7-AAA | pRS426-COQ7- (S20A,S28A,S32A) sequence | pLAAA-COQ7 multicopy complementation | This work |
TABLE 3.
Primers used
| Name | Sequence |
|---|---|
| Fw-pCM189-PTC7 | 5′-CAGTTAACATGTTTGCAAACGTTGGATTTAGAA-3′ |
| Rw-pCM189-PTC7 | 5′-GCGGCCGCTTAGTCAACTCTCACG-3′ |
| Fw-GEX-COQ7 | 5′-CATAGGATCCATGTTATCCCGTGTTTC-3′ |
| Rw-GEX-COQ7 | 5′-CACTGGATCCTGGTTAAATTCTTTCGGCACTC-3′ |
| Fw-GEX-PTC7 | 5′-CAGGATCCATGTTTGCAAACGTTGGA-3′ |
| Rw-GEX-PTC7 | 5′-CGTCGACTTAGTCAACTCTCACG-3′ |
| Fw-RTPCR-ACT1 | 5′-ATTCTGAGGTTGCTGCTT-3′ |
| Rw-RTPCR-ACT1 | 5′-GGACCACTTTCGTCGTAT-3′ |
| Fw-RTPCR-PTC7 | 5′-GCCGAAAGAAGGGGTAGCAAG-3′ |
| Rw-RTPCR-PTC7 | 5′-CGTTTGTTCTCGCAGCGTTATC-3′ |
| Fw-V5-PTC7 | 5′-AACGACAAAGCCACC-3′ |
| Rw-V5-PTC7 | 5′-GTCAACTCTCACGACAAC-3′ |
| Fw-V5-COQ7 | 5′-AATTCTTTCGGCACTCCATATAGC-3′ |
| Rw-V5-COQ7 | 5′-CATACATATGATGTTATCCCGTGTTTC-3′ |
Quinone Identification and Quantification
Total CoQ6 and DMQ6 quantification in mitochondrial samples was performed by HPLC-electrochemical detection. CoQ9 was used as the internal standard to determine CoQ6 yield. A description of equipment and methods has been published previously (18).
Real Time PCR
Yeast were cultured for 16 h in YPD with 2% (w/v) glucose and then harvested by centrifugation, washed, and resuspended in the same volume of either fresh YPG or YPD and 0.5% (w/v) glucose or buffer (100 mm potassium phosphate, pH 6.2 and 0.2% (w/v) glucose) for stress treatments. The stress agents added were: hydrogen peroxide (2 mm), tert-butyl peroxide (1 mm), linolenic acid (1 mm), cadmium sulfate (25 mm), and sodium chloride (0.6 m). Once resuspended, cells were treated for 0.5 (short term) or 4 h (long term) and stored at −80 °C. Yeast total RNA isolation and quantitative PCR were carried out according to previous reports (18). Yeast total RNA was prepared according to the instructions for the Perfect RNA Eukaryotic Mini kit from Eppendorf after a cell wall digestion with Zymolyase 20T (Seikagaku Corp.). cDNA was synthesized using the iScript cDNA Synthesis kit from Bio-Rad. RT-PCR was performed using iQ SYBR Green Supermix from Bio-Rad. The ACT1 gene was used as a calibrator gene. The expression ratio was calculated according to the 2 − ΔΔCP method. Primers used in this study are listed in Table 3.
Mitochondrial Enzymatic Activities
Fresh mitochondrial samples were used to measure NADH-cytochrome c reductase and succinate-cytochrome c reductase activities. Other respiratory chain activities (NADH-dichlorophenolindophenol reductase, succinate-dichlorophenolindophenol reductase, decylubiquinol-cytochrome c reductase, and cytochrome c oxidase activities) were performed with fresh samples subjected to one freeze-thaw cycle. All respiratory chain activities were determined according to published methods (19).
Determination of Protein Carbonylation
Protein carbonylation in total extracts was quantified using the OxyBlot Protein Oxidation Detection kit according to the manufacturer's instructions (Chemicon International).
Protein Purification
The Escherichia coli strain BL21 pLys was used for expression of recombinant proteins in the pGEX4T1 plasmid (GE Healthcare). Protein purification was performed according to methods published previously (20).
In Vitro Phosphatase Assays
p-Nitrophenyl phosphate, 6,8-difluoro-4-methylumbelliferyl phosphate (DiFMUP), and malachite green assays were performed according to previous reports (21, 22). The para-nitrophenyl phosphate (p-NPP) assay was performed at room temperature in 0.1 m Tris-HCl, pH 8.0, 2 mm dithiothreitol, 2 mm MnCl2, and 1 mm MgCl2 in a 96-well plate in 50-μl reaction mixtures with 1 μg of recombinant proteins and 5 mm p-NPP. After 30 min, the reaction was stopped by addition of 200 μl of 0.5% (w/v) SDS, and the absorbance at 410 nm was recorded with an Eldex microplate reader. For the DiFMUP assay, 5 mm DiFMUP from Molecular Probes, 50 mm imidazole, pH 7.2, 1 mm EGTA, 0.1% (v/v) β-mercaptoethanol, 0.5 mg/ml BSA, and 0.5 μg of recombinant protein were added in a 500-μl final volume. As a standard, 6,8-difluoro-4-methylumbelliferyl from Molecular Probes was used. After 60 min at 37 °C, the reaction was stopped by adding sodium borate, pH 10, and the fluorescence was determined in a florescence spectrophotometer (model SFM-25, Biotek Kontron) with excitation detection at 358 nm and emission detection at 455 nm. For the peptide dephosphorylation assay, the activity of 2 μg of recombinant protein was assayed in the same volume of p-NPP buffer assay with synthetic phosphopeptide at a final concentration of 5 μm. After 15 min, the reaction was stopped by the addition of 100 μl of a malachite green solution (Millipore). After color development for 15 min, the absorbance at 610 nm was determined in a microplate reader. Values were corrected by subtracting the absorbance of blanks, which were reactions without added enzyme.
In Vitro Phosphorylation Assay
Protein phosphorylation was performed with recombinant active PKA (1 μg) (Sigma) from rat brain incubated with 0.1 μCi of [γ-32P]ATP (PerkinElmer Life Sciences), 200 μm ATP (Sigma), and 2 μg of recombinant Coq7p protein in 50 μl of kinase buffer (50 mm Tris-HCl, pH 7.8, 25 mm β-glycerophosphate, 5 mm MgCl2, 5 mm MnCl2, and 1 mm dithiothreitol) and Sigma phosphatase inhibitor mixture 1 (1:100, v/v). Reactions were incubated at 30 °C for 2 h.
In Vitro Dephosphorylation Assay
Two micrograms of recombinant phosphatases or BSA was added to phosphorylated proteins bound to glutathione-Sepharose beads as described above in p-NPP assay buffer. After 2 h at 30 °C, samples were centrifuged (1000 × g for 5 min), and the supernatant was subjected to precipitation by adding 2.5 mg of BSA and 300 μl of 15% (w/v) trichloroacetic acid (TCA) (23). Free phosphate in the supernatant was quantified in a scintillation counter (Beckman LS6500).
In Vivo Protein Phosphorylation Determination
Fresh mitochondria from cells expressing V5-tagged versions of Coq7p (100 μg of protein) were permeabilized in 50 μl of buffer (50 mm Tris-HCl, 25 mm β-glycerophosphate, 5 mm MgCl2, 1 mm MnCl2, 1 mm dithiothreitol, and 0.3 g of 106-μm-diameter glass beads) plus digitonin (1:100, w/v). Samples were then subjected to vortexing for 15 min at 4 °C. After centrifugation at 1000 × g for 5 min at 4 °C, the supernatant was subjected to 12% (w/v) SDS-PAGE or immunoprecipitation. Results were visualized by Pro-Q Diamond gel stain (Molecular Probes), Coomassie Blue, and anti-V5 tag immunodetection (Invitrogen).
Immunoprecipitation and Isoelectric Focusing/SDS-PAGE
Mitochondrial samples (1 mg) from yeast expressing Coq7p-V5 were permeabilized. Immunoprecipitation was performed with anti-V5-agarose gel affinity beads according to the manufacturer's procedure (Sigma). Proteins were subjected to two-dimensional isoelectric focusing/SDS-PAGE as we reported previously (16).
Other Methods
Densitometry analysis was carried out with a GS800 densitometer (Bio-Rad) using ImageJ (v1.41o) software for analysis. Statistical (Student's t test) analyses were carried out using the SigmaStat 3.0 (SPSS) statistical package.
RESULTS
PTC7 Deficiency Compromises CoQ6 Biosynthesis and Mitochondrial Metabolism
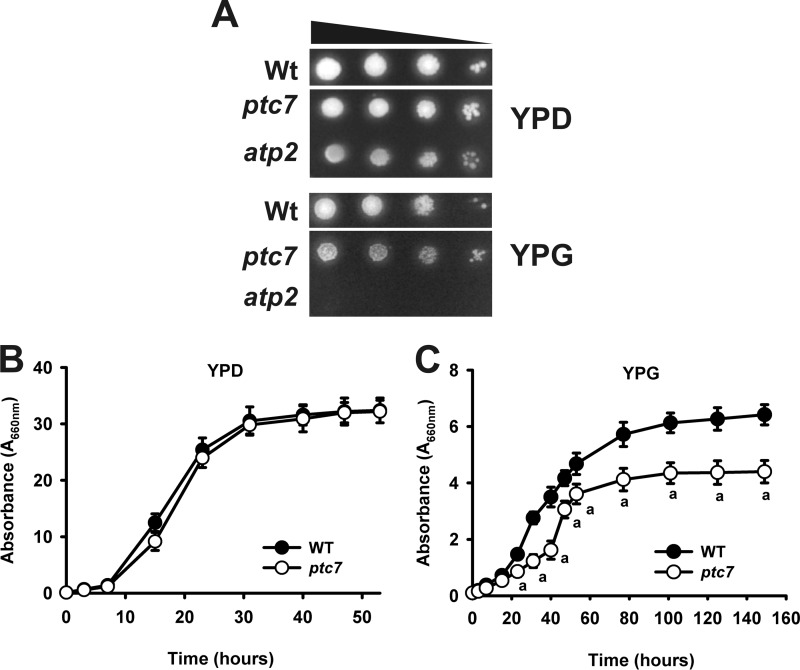
Yeast growth in non-fermentable carbon sources is an initial signal of coenzyme Q deficiency. The yeast ptc7 is a knock-out mutant from the PTC7 gene (ptc7 in the text). We compared the growth rate of mutant ptc7 yeast strain in the presence of different carbon sources both in liquid and solid media. The growth of ptc7 was not affected in the fermentable carbon source medium YPD. Interestingly, in YPG, a non-fermentable carbon source medium, mutant ptc7 yeast strain showed significantly decreased growth (Fig. 1, A–C), supporting a decreased mitochondrial performance in these mutants.
FIGURE 1.
Ptc7p phosphatase is required for normal respiratory growth. A, yeast were grown until stationary phase and spotted by serial dilutions (1:10) from A660 nm = 0.5 onto YPD and YPG plates. Plates were incubated for 3 days at 30 °C and imaged. B and C, WT and mutant ptc7 yeast were inoculated at A660 nm = 0.1. Yeast were harvested at marked time points, and A660 nm was determined. B, YPD medium. C, YPG medium. a, p < 0.05 compared with WT.
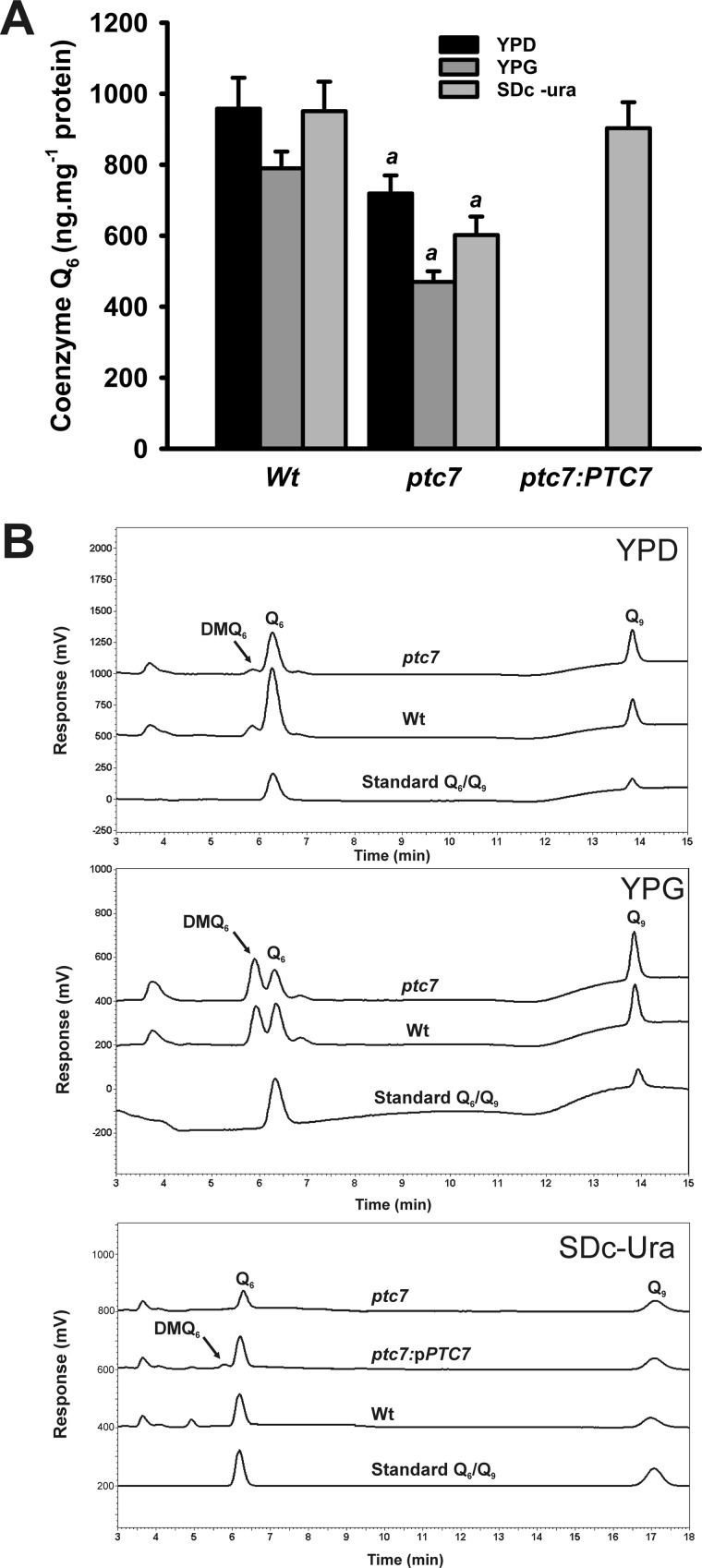
As mentioned previously, CoQ is a redox molecule essential for respiratory growth and antioxidant defenses. Given that PTC7 gene expression is induced in conditions that require respiratory metabolism and increased antioxidant defenses, the same physiological circumstances that induce CoQ6 biosynthesis in yeast (15), we studied the potential role of the Ptc7p phosphatase in the regulation of CoQ6 biosynthesis. We measured the CoQ6 content in the mutant ptc7 yeast grown for 5 days (Fig. 2). CoQ6 levels in wild-type yeast grown on YPD medium were comparable with those reported in other genetic backgrounds (19). Mutant ptc7 yeast strain exhibited decreased levels of CoQ6 in both YPD medium (75%) and YPG (59%) medium, respectively. In accordance with our hypothesis, mutant ptc7 yeast strain complemented with the wild-type PTC7 allele (ptc7:pCM189-PTC7) rescued CoQ6 levels, confirming that this protein is required to synthesize normal CoQ6 levels in yeast.
FIGURE 2.
Mitochondrial coenzyme Q quantification. Mitochondrial samples were subjected to lipid extraction and HPLC-electrochemical detection to quantify quinone in the indicated culture media. A, coenzyme Q6 determination in complemented strains was carried out in SDc-Ura and 2% (w/v) glucose to allow the vector selection by auxotrophy. Error bars represent S.D. a, p < 0.05 compared with WT. n = 3 in all the experiments. B, representative chromatograms. Growth in YPD, growth in YPG, and complementation of ptc7 with pCM189-PTC7 vector on SDc-Ura and 2% (w/v) glucose are shown. Q6, coenzyme Q6; Q9, coenzyme Q9.
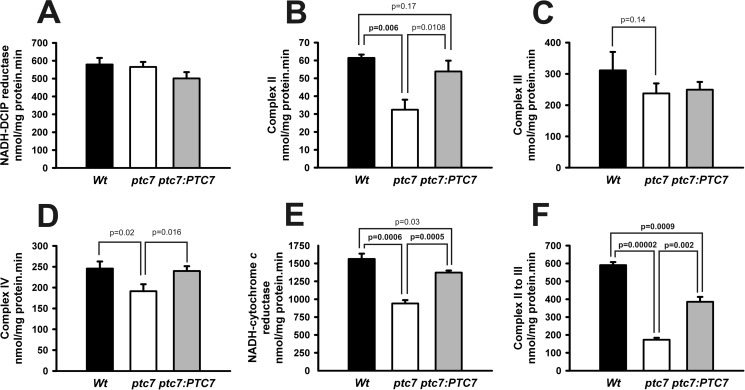
The decrease in CoQ6 content in mutant ptc7 yeast strain is evidence of mitochondrial dysfunction. Thus, we determined mitochondrial activities of the electron transport chain in mutant ptc7 yeast. Mitochondria were purified from wild-type, mutant ptc7, and mutant ptc7 complemented yeast strains (Fig. 3, A–F). CoQ acts as an electron carrier from NADH-coenzyme Q dehydrogenases (NDE1, NDE2, and NDI1) to complex III and from complex II to complex III of the mitochondrial electron transport chain. Yeast NADH dehydrogenases work like the typical complex I from mammals but without proton pumping activity and are composed of a single polypeptide (24). Remarkably, the lack of PTC7 resulted in a severe decrease in complex II, NADH-coenzyme Q dehydrogenase to complex III, and complex II to complex III activities (p < 0.01, two-tailed t test). However, other activities not related directly to CoQ6 such as complex III and complex IV were still affected but to a lesser extent (p = 0.14 and p = 0.02, respectively, two-tailed t test), suggesting that the decrease of CoQ6 or the lack of Ptc7p phosphatase mainly affects CoQ6-dependent enzymatic activities.
FIGURE 3.
Coenzyme Q-dependent activities. A–F, mitochondrial activities were determined in 50 μg of mitochondrial samples. A, NADH-coenzyme Q dehydrogenase, NADH-dichlorophenolindophenol (DCIP) reductase activity. B, complex II, succinate-dichlorophenolindophenol reductase. C, complex III, decylubiquinol-cytochrome c reductase activity. D, complex IV, cytochrome c oxidase activity. E, NADH-coenzyme Q dehydrogenase to complex III, NADH-cytochrome c reductase activity. F, complex II to III, succinate-cytochrome c reductase activity. n = 3 in all the experiments. The statistical analysis is included in the figure when p < 0.05 (in bold when p < 0.01). Errors bars represent S.D.
Antioxidant Defenses Are Impaired in ptc7 Mutants
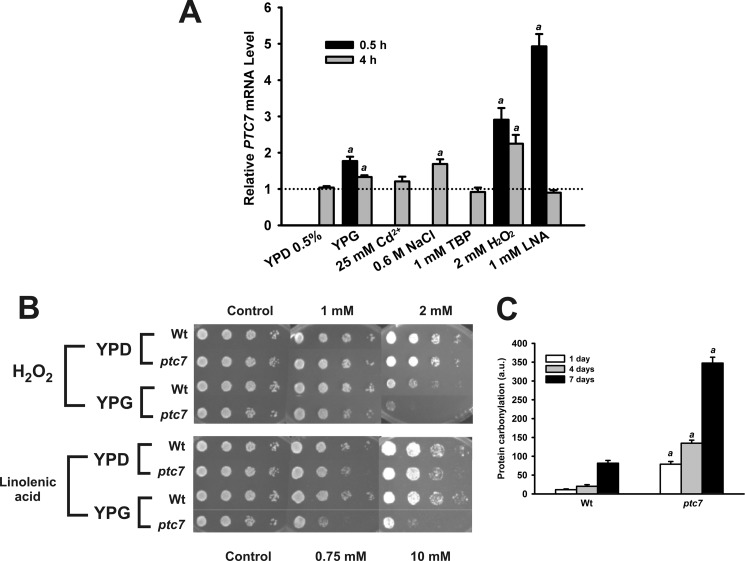
To characterize the function of Ptc7p protein in yeast, we determined the expression of this gene by quantitative PCR in wild-type (WT) yeast grown in several media with different carbon sources and under several stress treatments. PTC7 gene mRNA levels were increased in YPG, a non-fermentable carbon source, reaching 77% in the short term treatment (0.5 h) and 33% in the long term treatment (4 h) (Fig. 4A). PTC7 mRNA levels were increased up to 191% in the short term and up to 125% after long term treatment with hydrogen peroxide. Short term treatment with linolenic acid, a lipophilic oxidative stress agent, resulted in a 493% increase in expression. Interestingly, mRNA levels were not changed in the long term assay, suggesting that the yeast response to acute oxidative stress is a fast and tightly regulated process. When other stress treatments were assayed, we observed only a 69% induction with sodium chloride, which is consistent with previous reports (25). Neither tert-butyl peroxide nor Cd2+ affects the PTC7 gene expression. These results demonstrate that PTC7 gene expression is induced under conditions that require proper respiratory metabolism as well as the induction of antioxidant defenses as was supported by a previously published phosphorylation analysis under respiratory conditions that maintain the Coq7p protein in a poorly phosphorylated state compared with fermentative growth conditions (16).
FIGURE 4.
Knock-out PTC7 mutant presents impaired antioxidant defenses. A, analysis of PTC7 gene expression. Quantitative analysis of PTC7 expression was performed using quantitative RT-PCR in wild-type yeast. Data are expressed as relative expression compared with 2% (w/v) YPD or untreated yeast. a, p < 0.01 significant differences. Error bars represent S.D. Cd2+, cadmium sulfate; TBP, tert-butyl peroxide; LNA, linolenic acid. n = 3 in all experiments. B, cell viability was monitored under induced oxidative stress. Yeast were grown in YPD or YPG. After 5 days, cells were spotted by serial dilutions (1:10) from A660 nm = 0.5 onto YPD plates containing the indicated concentrations of oxidative stress agents. Plates were incubated at 30 °C for 3 days and imaged. C, quantification of protein carbonylation. Yeast were grown for 7 days to monitor the evolution of protein carbonylation content. Fresh yeast extracts (50 μg) were harvested at marked times (days) and analyzed. Carbonylation content is normalized (relative units) to wild-type levels at day 1. Error bars represent S.D. a, p < 0.05 compared with WT. n = 3 in all experiments. a.u., arbitrary units.
Linolenic acid and hydrogen peroxide are well known stressors that cause oxidative damage in yeast (19). These oxidative stress treatments were also able to significantly induce PTC7 expression. To determine the ability to induce antioxidant defenses, we performed oxidative stress treatments (Fig. 4B). Yeast were cultured for 5 days in YPD or YPG and plated in serial dilutions of either hydrogen peroxide or linolenic acid. Both treatments produced a toxic effect on all strains when compared with untreated plates. Hydrogen peroxide treatment showed the highest difference between wild-type and mutant ptc7 yeast strains grown in YPG. Interestingly, linolenic acid, which produces oxidative stress in cellular membranes (26), affected mutant ptc7 yeast survival in both media, revealing an impaired antioxidant defense in this strain. Oxidative stress results in changes to various types of molecules in the cells such as lipids, DNA, and proteins (27). The most distinctive oxidative modification in proteins is carbonylation, which affects their functionality (28). To monitor oxidative damage accumulation in mutant ptc7 yeast strain, we determined the protein carbonylation in yeast extracts (Fig. 4C). In agreement with published data, wild-type yeast showed a low level of protein carbonylation on the 1st day and a progressive accumulation during the length of the treatment (27). The mutant ptc7 strain exhibited severely increased levels of protein carbonylation compared with wild-type yeast, suggesting that antioxidant protection is impaired in yeast lacking PTC7, rendering an increase in oxidative damage accumulation.
Ptc7p Is a PPM Type Phosphatase
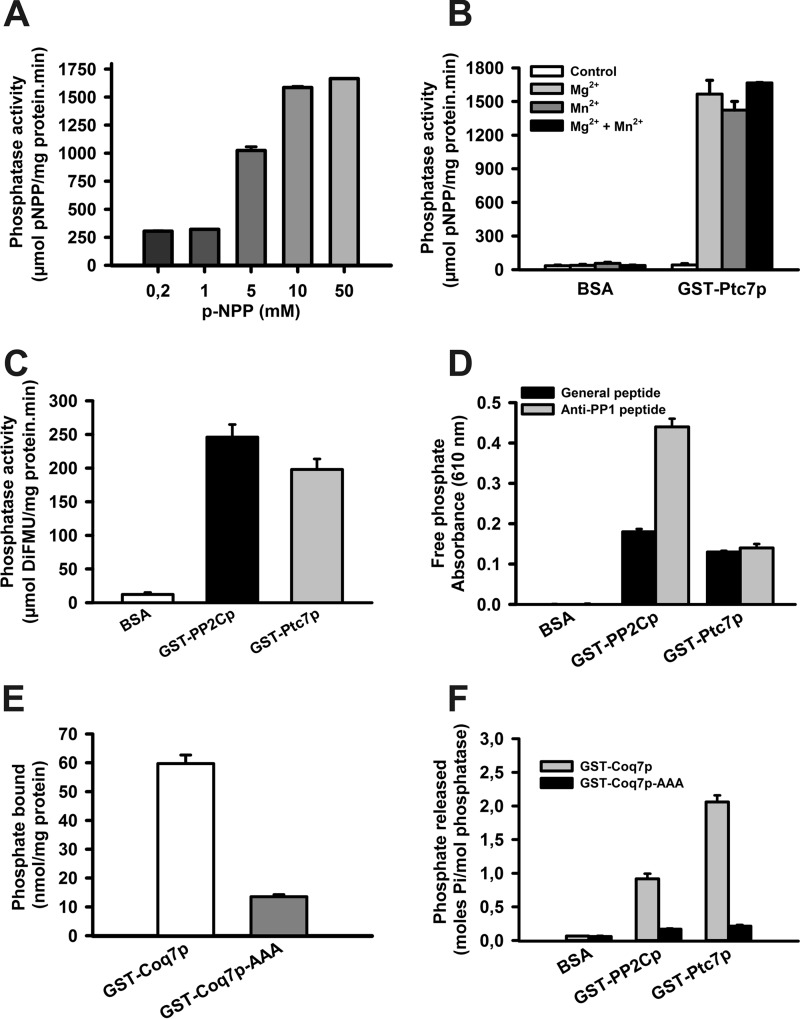
As mentioned previously, Ptc7p protein has been described as a phosphatase (7). We performed in vitro phosphatase assays with GST-Ptc7p recombinant protein. Increased concentrations of p-NPP substrate increased the reaction rate, and the reaction appeared to be saturated at 10–50 mm (Fig. 5A), indicating that the activity is clearly produced by an enzyme. The enzymatic activity of GST-Ptc7p was dependent on the addition of Mn2+ or Mg2+ as described previously for other members of the PPM phosphatase family (Fig. 5B) (3). We also determined GST-Ptc7p activity with DiFMUP as substrate (Fig. 5C) (29). Noticeably, the enzymatic specific activity of GST-Ptc7p was similar to the recombinant human PP2C-α phosphatase (PPM1A), a well known member of the PPM family.
FIGURE 5.
Ptc7p dephosphorylates Coq7p in vitro. A, in vitro phosphatase assay using increasing concentrations of p-NPP. Activity is expressed as μmol of phosphate released/mg of protein·min. Error bars represent S.D. n = 3. B, Ptc7p activity without cofactors. An in vitro phosphatase assay using 40 mm p-NPP was performed to determine the requirement of magnesium and magnesium cofactors for Ptc7 phosphatase activity. 10 mm Mg2+ and 20 mm Mn2+ were added where indicated. BSA was used as a negative control. Activity is expressed as μmol of 6,8-difluoro-4-methylumbelliferyl (DiFMU) produced/mg of protein·min. Error bars represent S.D. n = 3. C, in vitro phosphatase assay using 1 mm DiFMUP as reagent with 0.5 μg of recombinant protein. As a positive control, the human recombinant phosphatase GST-PP2Cp was included. Error bars represent S.D. n = 3. D, phosphopeptide dephosphorylation assays. Data are expressed as increased absorbance at 610 nm using universal and anti-PP1 specific phosphopeptides. As a positive control, the human recombinant phosphatase GST-PP2Cp was included. Error bars represent S.D. n = 3. E, in vitro phosphorylation of versions of Coq7p. Recombinant proteins GST-Coq7p and GST-Coq7p-AAA (2 μg) were phosphorylated with [γ-32P]ATP and PKA. Total phosphorylation was directly determined in a scintillation counter and is expressed as nmol of bound phosphate/mg of protein. Error bars represent S.D. n = 3. F, in vitro Coq7p dephosphorylation. Recombinant proteins GST-Coq7p and GST-Coq7p-AAA were phosphorylated with [32P]ATP and PKA. Dephosphorylation by GST-PP2Cp, GST-Ptc7p, and BSA was determined after TCA precipitation. Activity is expressed as mol of phosphate released/mol of phosphatase. Error bars represent S.D. n = 3 in all experiments.
Once we established the phosphatase activity of GST-Ptc7p with chemical substrates, we examined Ptc7p enzymatic activity using phosphorylated peptides. The assays were performed using the malachite green reagent to quantify the phosphate released from either a universal phosphopeptide (KRpTIRR) that can be dephosphorylated by different phosphatases (30) or a more selective phosphopeptide (RRRRPpTPA) (22), (Fig. 5D) (20). GST-Ptc7p protein phosphatase activity was confirmed with both phosphorylated peptides, demonstrating that Ptc7p is a protein phosphatase with properties similar to PPM1.
Ptc7p Dephosphorylates Coq7p in Vitro
We have previously demonstrated that Coq7 phosphorylation state is a key regulation point in CoQ6 biosynthesis (15, 16). We hypothesized that phosphatase Ptc7p, which is also localized within the mitochondria (11), may dephosphorylate Coq7p hydroxylase to induce CoQ6 biosynthesis in yeast mitochondria. To address this hypothesis, we performed an in vitro phosphorylation assay with two recombinant versions of Coq7p protein, a phosphorylatable Coq7p (GST-Coq7p) and a non-phosphorylatable Coq7p (GST-Coq7p-AAA) (16). Then phosphorylated recombinant proteins bound to glutathione-Sepharose beads were divided into four aliquots that were incubated with 2 μg of GST-PP2Cp, GST-Ptc7p phosphatase, or BSA as a negative control. One aliquot was not treated and directly introduced into a scintillation counter to assess total Coq7p phosphorylation (Fig. 5E). Following incubation, proteins were precipitated with TCA, and the supernatants were used to determine the release of radioactive phosphate (Fig. 5F). The results clearly showed that GST-Ptc7p was able to dephosphorylate GST-Coq7p in an in vitro assay in a similar fashion to human PP2C-α phosphatase, although GST-Ptc7p is more effective than PP2C-α.
The Lack of Ptc7p Affects Coq7p Phosphorylation State in Vivo
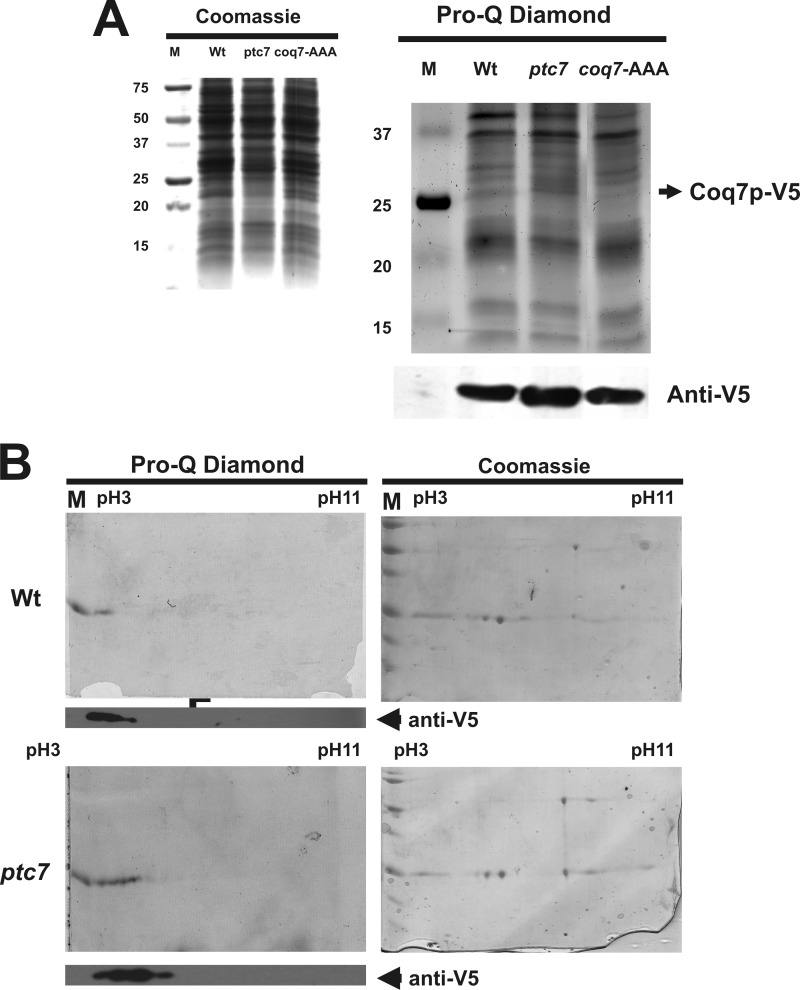
Once we demonstrated the in vitro Coq7p dephosphorylation by Ptc7p, we sought to determine the phosphorylation state of Coq7p in the mutant ptc7 strain. We cloned and expressed the V5-tagged version of wild-type Coq7p (pCOQ7-V5) in both wild-type and mutant ptc7 yeast strains. Moreover, as a negative control for Coq7p phosphorylation, we cloned and expressed a non-phosphorylatable version of Coq7p (pCOQ7-AAA-V5) into the mutant ptc7 strain (16). Yeast were cultured in SDc-Ura and 2% (w/v) galactose for 16 h to induce Coq7p-V5 expression. Then cells were transferred to SDc-Ura and 10% (w/v) glucose for 2 h to promote Coq7p phosphorylation. Subsequently, yeast were resuspended in YPG to induce Coq7p dephosphorylation (16). In accordance with our hypothesis, Coq7p phosphorylation was poorly detected by Pro-Q Diamond in wild-type and Coq7p-AAA compared with mutant ptc7 yeast (Fig. 6A). As further evidence for the differential phosphorylation state of Coq7p-V5 in vivo, the same mitochondrial isolations from mutant ptc7 and wild-type yeast were subjected to immunoprecipitation and two-dimensional separation by isoelectric focusing plus SDS-PAGE (Fig. 6B). Coq7p-V5 migrated at 25 kDa, consistent with its predicted size. Coq7p-V5 protein is detected by Western blot with anti-V5 at acidic pH as a series of protein spots. It is interesting that wild-type yeast strain shows phosphorylation in the more acid pH, whereas mutant ptc7, which shows increased phosphorylation by Pro-Q Diamond staining, ran in a broader extent. In the case of Coq7p protein, we have described previously (16) that it can be phosphorylated at three amino acids (Ser-20, Ser-28, and Thr-32). This means that with the combination of single, double, and full phosphorylation we can obtain three isoforms (four with the non-phosphorylated state) with a different extent of phosphorylation. In the mutant ptc7 strain, we can see several combinations of isoforms, but in wild-type strain, the number of combinations is lower. This could explain the broader spectrum of Coq7p protein migration in mutant ptc7 yeast. Therefore, although phosphorylation levels are clearly reduced in wild-type yeast, minor modifications in the isoelectric focusing could occur. There are other post-translational modifications such as acetylation that could alter the isoelectric point of Coq7p (31).
FIGURE 6.
Ptc7p dephosphorylates Coq7p in vivo. A and B, Coq7p phosphorylation in mutant ptc7 strain. V5-tagged versions of Coq7p were introduced into wild-type and mutant ptc7 yeast. Mitochondrial samples were isolated and analyzed. A, left, Coomassie Blue staining. Right, phosphoprotein determination by Pro-Q Diamond staining. Bottom, Coq7p-V5 immunodetection. M, molecular weight marker. B, mitochondrial samples were analyzed by Coq7p-V5 immunoprecipitation and two-dimensional isoelectric focusing analysis. Left, Pro-Q Diamond staining. Right, Coomassie staining. Bottom, Coq7p-V5 immunodetection.
In accordance with the induced dephosphorylating conditions produced in YPG medium for wild-type yeast, Coq7p-V5 immunoprecipitation showed a slightly phosphorylated Coq7p state but in the spots closest to the acidic pH. Phosphorylation detected in mutant ptc7 mitochondrial extracts is consistent with reduced Coq7p-V5 dephosphorylation in the absence of Ptc7p. In this case, phosphorylation affects all spots detected. These results demonstrate a direct relationship between Ptc7p phosphatase expression and Coq7p phosphorylation state, indicating that Coq7p may be the natural substrate of Ptc7p phosphatase.
Ptc7p Dephosphorylates Coq7p to Activate CoQ6 Biosynthesis
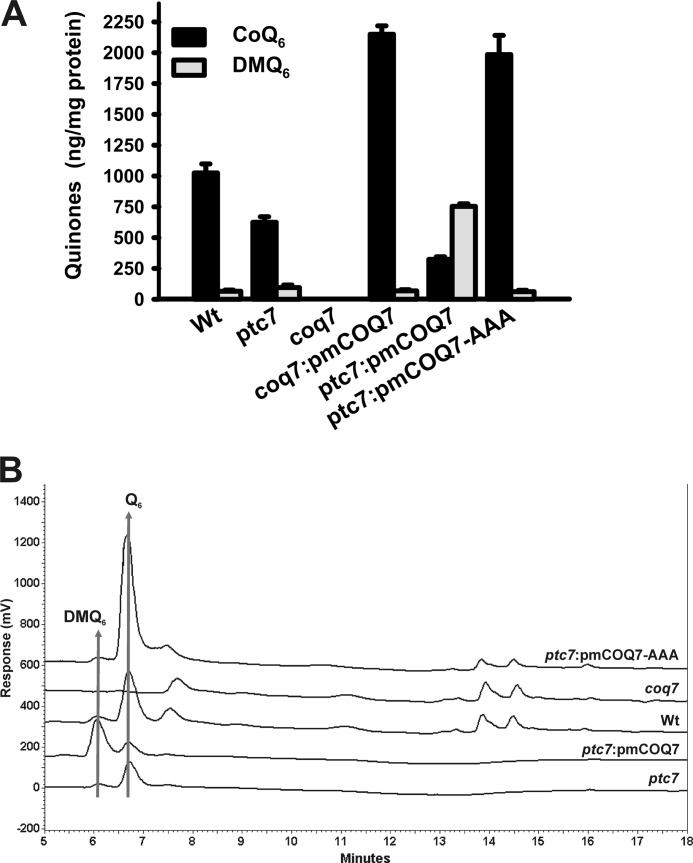
To ascertain the putative Ptc7p phosphatase control of Coq7p hydroxylase activity, we studied whether a constitutively active Coq7p protein could bypass Ptc7p control. Mitochondrial isolations of mutant ptc7 yeast expressing the COQ7 wild-type allele or COQ7-AAA allele, a constitutive dephosphorylated version of Coq7p, were subjected to lipid extraction and quinone determination (Fig. 7, A and B) (16). Interestingly, COQ7 overexpression in the mutant ptc7 strain (ptc7:pmCOQ7) led to a significant increase of DMQ6, the substrate of Coq7p hydroxylase, suggesting that the activation of the conversion of DMQ6 to CoQ6 is driven by a Ptc7p-dependent Coq7p dephosphorylation. In accordance with our hypothesis, the overexpression of the non-phosphorylatable version of Coq7p (COQ7-AAA) in the mutant ptc7 strain (ptc7:pmCOQ7-AAA) severely increased the accumulation of CoQ6, reaching a CoQ6 content similar to what we found previously in a coq7 knock-out mutant strain harboring the same COQ7-AAA allele (16), thereby demonstrating the bypass of the regulatory function of Ptc7p in CoQ6 biosynthesis. Taken together, these data conclusively demonstrate that Ptc7p phosphatase controls CoQ6 biosynthesis through the dephosphorylation of Coq7 hydroxylase in yeast.
FIGURE 7.
Ptc7p phosphatase and Coq7p hydroxylase interact in vivo. A, multicopy plasmids harboring the wild-type COQ7 allele (pmCOQ7) or the non-phosphorylatable COQ7 allele (pmCOQ7-AAA) were introduced in mutant coq7 and ptc7 strains. Yeast were grown in SDc-Ura and 2% (w/v) glucose and subjected to mitochondrial purification. Mitochondrial samples were subjected to lipid extraction and quinone quantification. Error bars represent S.D. n = 3 in all experiments. ND, the wild-type, mutant coq7, and mutant ptc7 strains without the indicated plasmids were transformed with the empty vector pRS426. B, representative chromatograms of mitochondrial quinone determination. Q6, coenzyme Q6.
DISCUSSION
Our study demonstrates that the lack of Ptc7p phosphatase, a mitochondrial phosphatase, results in a metabolically unfavorable state leading to mitochondrial dysfunction, which includes reduced growth in non-fermentable media, compromised mitochondrial activities, and increased oxidative damage accumulation. According to our previous research, coenzyme Q6 biosynthesis can be regulated by phosphorylation/dephosphorylation of the mitochondrial protein Coq7p (16). This regulatory mechanism, initially observed in vitro, was supported by in vivo experiments showing that the Coq7p modifications toward a permanent non-phosphorylated state led to increased amounts of coenzyme Q6. In addition, the growth in non-fermentable sources, when maximal respiratory capacity is required, leads to a reduction of the phosphorylation state of Coq7p.
Several studies point to the idea that coenzyme Q6 biosynthesis is regulated by a phosphorylation-based mechanism. Thirty years ago, Coq3p regulation through cAMP/PKA was suggested (32). Recently, it has been demonstrated that Coq8p, a member of the ADCK2 kinase family (33), is required at least indirectly for the phosphorylation of Coq3p (34) as well as Coq5p and Coq7p (17). Phosphorylation seems to be required for the assembly of the coenzyme Q6 biosynthetic complex (14) because Coq8p overexpression stabilizes this complex in null mutants that do not produce quinone compounds (15). Coq8p overexpression has been used to analyze the accumulation of CoQ6 precursors in COQ-null mutants. This strategy has also been used to bypass coenzyme Q6 biosynthesis in studies using synthetic intermediates (35). Although Coq8p is a promising candidate as the kinase that regulates the complex assembly, unfortunately there is no direct evidence of kinase activity in vitro for Coq8p.
In this study, we have focused on the phosphatase counterpart as the possible positive regulator of CoQ6 biosynthesis (16). Previous research indicated that only six of the 33 phosphatases (36) encoded in the yeast genome are present in the mitochondria based on bioinformatics analysis (37). The phosphatase encoded by the PTC7 gene does not show a specific physiological function, although its mitochondrial localization has been described (11), and it belongs to the MPP phosphatase family (7). We determined that the recombinant Ptc7p has an enzymatic activity that displays Michaelis-Menten kinetics and requires Mg2+ or Mn2+ as was described previously (7). Moreover, we demonstrated that Ptc7p phosphatase activity is extended to other substrates such as DiFMUP and phosphorylated peptides (22).
Knock-out PTC7 mutants (ptc7) show significantly decreased Q6 levels in mitochondria that were completely restored by Ptc7p complementation (Fig. 2). This decrease differs in several culture media but does not compromise the growth in non-fermentable carbon sources (Fig. 1, A–C), although it produces a delayed and decreased maximal growth compared with wild-type controls.
Typical coenzyme Q functions are affected in a similar fashion. Some activities of the mitochondrial respiratory chain such as NADH-cytochrome c reductase, succinate-cytochrome c reductase, and succinate dehydrogenase are decreased in ptc7 mutant strains. These activities are partially restored after PTC7 complementation (p < 0.01, two-tailed t test), although the activities did not reach wild-type levels. Oxidative stress protection is affected in ptc7 mutant strain as was observed after H2O2 and linolenic acid treatment in agar plates (Fig. 4A). This effect is associated with the accumulation of carbonylated proteins during yeast growth (Fig. 4B). These data together with the induction of PTC7 expression under linolenic acid and hydrogen peroxide treatments and the presence of sequences in PTC7 promoter recognized by transcription factors that control mitochondrial metabolism and the antioxidant response such as HAP4, ADR1, and YAP1 (8, 9) support the idea that PTC7 is required to sustain normal respiratory metabolism. Because PTC7 gene expression is induced by conditions that induce the expression of COQ genes (15), it is probable that PTC7 acts as an antioxidant protein mainly regulating Coq7p hydroxylase activity.
At this point, it is reasonable to wonder why coenzyme Q6 biosynthesis is not more severely affected in ptc7 mutants. In addition to PTC7, two other members of the MPP phosphatase family are located in the mitochondria (37). We cannot rule out that PTC5 and PTC6 might partially overlap with PTC7 function in the activation of CoQ6 biosynthesis (38, 39). Additionally, according to our previous research, Coq7p is not completely inactivated by phosphorylation. A basal Coq7p activity remains in versions of Coq7p mimicking the permanent phosphorylated state that allows a decreased but still sufficient accumulation of coenzyme Q6 to maintain yeast metabolism (16).
Mutant ptc7 strains exhibit a high level of phosphorylated Coq7p compared with wild-type control and the non-phosphorylatable version Coq7p-AAA in purified mitochondria. The two-dimensional analysis of immunoprecipitated Coq7p supports the proposed mechanism of action because the phosphorylation and dephosphorylation have been produced in mitochondria without the presence of exogenous kinases and ATP. However, these results are indirect evidence of the relationship between Ptc7p phosphatase and the Coq7p hydroxylase because other proteins, functions, or regulatory mechanisms such as post-translational modifications can be affected by the lack of Ptc7p. Analysis of potential targets for kinases has been useful in the past to elucidate functional interactions between components of the coenzyme Q biosynthetic complex, but so far these analyses are not conclusive (17, 34).
In vitro, recombinant Coq7p phosphorylated by PKA (16) is dephosphorylated by recombinant Ptc7p. These data support the hypothesis of a physical interaction between both proteins. Although there are good examples of physical interactions between phosphatases and their targets based mainly on the existence of protein complexes (40, 41), in this study, we demonstrated this interaction through a functional relationship (Fig. 7). In ptc7 mutant strain, DMQ6 was accumulated after Coq7p overexpression, and coenzyme Q6 content was reduced compared with wild-type control yeast. However, the overexpression in the same strain (ptc7) of the Coq7p-AAA version led to a dramatic increase of coenzyme Q6. The bypass of this regulatory mechanism in this version of Coq7p further indicates the relationship between Coq7p and Ptc7p.
In this report, we propose a new step in the regulation of coenzyme Q6 biosynthesis beyond the initial complex assembly. The presence in humans of Ptc7p phosphatase homologous to the yeast protein (12) allows us to hypothesize the same regulatory mechanism as in yeast and humans and the existence of a new target for therapeutic treatments in patients with coenzyme Q10 deficiency.
The role of the mitochondrial protein phosphatase Ptc7p has not yet been studied. Here we reveal the molecular mechanisms that are regulated by Ptc7p. Our findings indicate that Ptc7p is a key regulator of mitochondrial metabolism that could be related to cases of mitochondrial disease. We determined that Ptc7p orchestrates yeast metabolism to maintain a proper mitochondrial function and antioxidant response. We demonstrated that Ptc7p regulates the activation of energy metabolism and antioxidant responses through the activation of coenzyme Q biosynthesis by Coq7p dephosphorylation.
The work was supported in part by the Spanish Ministerio de Ciencia y Tecnología Grant PI11/00078, Junta de Andalucía Grant P08-CTS-03988, and the International Q10 Association “Phosphorylation based regulation of coenzyme Q biosynthesis in yeast.”
- PP2C
- type 2C protein phosphatase
- PPM
- Mg2+- or Mn2+-dependent protein phosphatase
- CoQ
- coenzyme Q
- DMQ6
- demethoxy-coenzyme Q6
- YPD
- yeast extract-peptone-dextrose
- YPG
- yeast extract-peptone-glycerol
- DiFMUP
- 6,8-difluoro-4-methylumbelliferyl phosphate
- p-NPP
- para-nitrophenyl phosphate
- SDc-Ura
- synthetic complete culture media with glucose and without uracil.
REFERENCES
- 1. Lee J., Xu Y., Chen Y., Sprung R., Kim S. C., Xie S., Zhao Y. (2007) Mitochondrial phosphoproteome revealed by an improved IMAC method and MS/MS/MS. Mol. Cell. Proteomics 6, 669–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Reinders J., Wagner K., Zahedi R. P., Stojanovski D., Eyrich B., van der Laan M., Rehling P., Sickmann A., Pfanner N., Meisinger C. (2007) Profiling phosphoproteins of yeast mitochondria reveals a role of phosphorylation in assembly of the ATP synthase. Mol. Cell. Proteomics 6, 1896–1906 [DOI] [PubMed] [Google Scholar]
- 3. Barford D., Das A. K., Egloff M. P. (1998) The structure and mechanism of protein phosphatases: insights into catalysis and regulation. Annu. Rev. Biophys. Biomol. Struct. 27, 133–164 [DOI] [PubMed] [Google Scholar]
- 4. Shi Y. (2009) Serine/threonine phosphatases: mechanism through structure. Cell 139, 468–484 [DOI] [PubMed] [Google Scholar]
- 5. Lammers T., Lavi S. (2007) Role of type 2C protein phosphatases in growth regulation and in cellular stress signaling. Crit. Rev. Biochem. Mol. Biol. 42, 437–461 [DOI] [PubMed] [Google Scholar]
- 6. Tamura S., Toriumi S., Saito J., Awano K., Kudo T.-A., Kobayashi T. (2006) PP2C family members play key roles in regulation of cell survival and apoptosis. Cancer Sci. 97, 563–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jiang L., Whiteway M., Ramos C. w, Rodriguez-Medina J. R., Shen S.-H. (2002) The YHR076w gene encodes a type 2C protein phosphatase and represents the seventh PP2C gene in budding yeast. FEBS Lett. 527, 323–325 [DOI] [PubMed] [Google Scholar]
- 8. Abdulrehman D., Monteiro P. T., Teixeira M. C., Mira N. P., Lourenço A. B., dos Santos S. C., Cabrito T. R., Francisco A. P., Madeira S. C., Aires R. S., Oliveira A. L., Sá-Correia I., Freitas A. T. (2011) YEASTRACT: providing a programmatic access to curated transcriptional regulatory associations in Saccharomyces cerevisiae through a web services interface. Nucleic Acids Res. 39, D136–D340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ramos C. W., Güldener U., Klein S., Hegemann J. H., Gonzalez S., Rodríguez-Medina J. R. (2000) Molecular analysis of the Saccharomyces cerevisiae YHR076w gene. IUBMB Life 50, 371–377 [DOI] [PubMed] [Google Scholar]
- 10. Forsburg S. L., Guarente L. (1989) Identification and characterization of HAP4: a third component of the CCAAT-bound HAP2/HAP3 heteromer. Genes Dev. 3, 1166–1178 [DOI] [PubMed] [Google Scholar]
- 11. Juneau K., Nislow C., Davis R. W. (2009) Alternative splicing of PTC7 in Saccharomyces cerevisiae determines protein localization. Genetics 183, 185–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mao M., Biery M. C., Kobayashi S. V., Ward T., Schimmack G., Burchard J., Schelter J. M., Dai H., He Y. D., Linsley P. S. (2004) T lymphocyte activation gene identification by coregulated expression on DNA microarrays. Genomics 83, 989–999 [DOI] [PubMed] [Google Scholar]
- 13. Bentinger M., Tekle M., Dallner G. (2010) Coenzyme Q—biosynthesis and functions. Biochem. Biophys. Res. Commun. 396, 74–79 [DOI] [PubMed] [Google Scholar]
- 14. Marbois B., Gin P., Gulmezian M., Clarke C. F. (2009) The yeast Coq4 polypeptide organizes a mitochondrial protein complex essential for coenzyme Q biosynthesis. Biochim. Biophys. Acta 1791, 69–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Padilla S., Tran U. C., Jiménez-Hidalgo M., López-Martín J. M., Martín-Montalvo A., Clarke C. F., Navas P., Santos-Ocaña C. (2009) Hydroxylation of demethoxy-Q6 constitutes a control point in yeast coenzyme Q6 biosynthesis. Cell. Mol. Life Sci. 66, 173–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Martín-Montalvo A., González-Mariscal I., Padilla S., Ballesteros M., Brautigan D. L., Navas P., Santos-Ocaña C. (2011) Respiratory-induced coenzyme Q biosynthesis is regulated by a phosphorylation cycle of Cat5p/Coq7p. Biochem. J. 440, 107–114 [DOI] [PubMed] [Google Scholar]
- 17. Xie L. X., Hsieh E. J., Watanabe S., Allan C. M., Chen J. Y., Tran U. C., Clarke C. F. (2011) Expression of the human atypical kinase ADCK3 rescues coenzyme Q biosynthesis and phosphorylation of Coq polypeptides in yeast coq8 mutants. Biochim. Biophys. Acta 1811, 348–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jiménez-Hidalgo M., Santos-Ocaña C., Padilla S., Villalba J. M., López-Lluch G., Martín-Montalvo A., Minor R. K., Sinclair D. A., de Cabo R., Navas P. (2009) NQR1 controls lifespan by regulating the promotion of respiratory metabolism in yeast. Aging Cell 8, 140–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Padilla S., Jonassen T., Jiménez-Hidalgo M. A., Fernández-Ayala D. J., López-Lluch G., Marbois B., Navas P., Clarke C. F., Santos-Ocaña C. (2004) Demethoxy-Q, an intermediate of coenzyme Q biosynthesis, fails to support respiration in Saccharomyces cerevisiae and lacks antioxidant activity. J. Biol. Chem. 279, 25995–26004 [DOI] [PubMed] [Google Scholar]
- 20. Brautigan D. L., Brown M., Grindrod S., Chinigo G., Kruszewski A., Lukasik S. M., Bushweller J. H., Horal M., Keller S., Tamura S., Heimark D. B., Price J., Larner A. N., Larner J. (2005) Allosteric activation of protein phosphatase 2C by D-chiro-inositol-galactosamine, a putative mediator mimetic of insulin action. Biochemistry 44, 11067–11073 [DOI] [PubMed] [Google Scholar]
- 21. Gee K. R. (1999) Novel fluorogenic substrates for acid phosphatase. Bioorg. Med. Chem. Lett. 9, 1395–1396 [DOI] [PubMed] [Google Scholar]
- 22. Hans S. K., Camara F., Altiti A., Martín-Montalvo A., Brautigan D. L., Heimark D., Larner J., Grindrod S., Brown M. L., Mootoo D. R. (2010) Synthesis of C-glycoside analogues of β-galactosamine-(1→4)-3-O-methyl-D-chiro-inositol and assay as activator of protein phosphatases PDHP and PP2Cα. Bioorg. Med. Chem. Lett. 18, 1103–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Maeda T., Tsai A. Y., Saito H. (1993) Mutations in a protein tyrosine phosphatase gene (PTP2) and a protein serine/threonine phosphatase gene (PTCJ) cause a synthetic growth defect in Saccharomyces cerevisiae. Mol. Cell. Biol. 13, 5408–5417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Joseph-Horne T., Hollomon D. W., Wood P. M. (2001) Fungal respiration: a fusion of standard and alternative components. Biochim. Biophys. Acta 1504, 179–195 [DOI] [PubMed] [Google Scholar]
- 25. Runner V. M., Brewster J. L. (2003) A genetic screen for yeast genes induced by sustained osmotic stress. Yeast 20, 913–920 [DOI] [PubMed] [Google Scholar]
- 26. Colquhoun A., Schumacher R. I. (2001) γ-Linolenic acid and eicosapentaenoic acid induce modifications in mitochondrial metabolism, reactive oxygen species generation, lipid peroxidation and apoptosis in Walker 256 rat carcinosarcoma cells. Biochim. Biophys. Acta 1533, 207–219 [DOI] [PubMed] [Google Scholar]
- 27. Costa V., Moradas-Ferreira P. (2001) Oxidative stress and signal transduction in Saccharomyces cerevisiae: insights into ageing, apoptosis and diseases. Mol. Aspects Med. 22, 217–246 [DOI] [PubMed] [Google Scholar]
- 28. Suzuki Y. J., Carini M., Butterfield D. A. (2010) Protein carbonylation. Antioxid. Redox Signal. 12, 323–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ruan H., Yan Z., Sun H., Jiang L. (2007) The YCR079w gene confers a rapamycin-resistant function and encodes the sixth type 2C protein phosphatase in Saccharomyces cerevisiae. FEMS Yeast Res. 7, 209–215 [DOI] [PubMed] [Google Scholar]
- 30. Rahman A., Brew B. J., Guillemin G. J. (2011) Lead dysregulates serine/threonine protein phosphatases in human neurons. Neurochem. Res. 36, 195–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Henriksen P., Wagner S. A., Weinert B. T., Sharma S., Bacinskaja G., Rehman M., Juffer A. H., Walther T. C., Lisby M., Choudhary C. (2012) Proteome-wide analysis of lysine acetylation suggests its broad regulatory scope in Saccharomyces cerevisiae. Mol. Cell. Proteomics 11, 1510–1522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sippel C. J., Goewert R. R., Slachman F. N., Olson R. E. (1983) The regulation of ubiquinone-6 biosynthesis by Saccharomyces cerevisiae. J. Biol. Chem. 258, 1057–1061 [PubMed] [Google Scholar]
- 33. Leonard C. J., Aravind L., Koonin E. V. (1998) Evolution of the eukaryotic protein kinase superfamily novel families of putative protein kinases in bacteria and archaea: evolution of the eukaryotic protein kinase superfamily. Genome Res. 8, 1038–1047 [DOI] [PubMed] [Google Scholar]
- 34. Tauche A., Krause-Buchholz U., Rödel G. (2008) Ubiquinone biosynthesis in Saccharomyces cerevisiae: the molecular organization of O-methylase Coq3p depends on Abc1p/Coq8p. FEMS Yeast Res. 8, 1263–1275 [DOI] [PubMed] [Google Scholar]
- 35. Xie L. X., Ozeir M., Tang J. Y., Chen J. Y., Jaquinod S.-K., Fontecave M., Clarke C. F., Pierrel F., Kieffer-Jaquinod S. (2012) Overexpression of the Coq8 kinase in Saccharomyces cerevisiae coq null mutants allows for accumulation of diagnostic intermediates of the coenzyme Q6 biosynthetic pathway. J. Biol. Chem. 287, 23571–23581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sakumoto N., Mukai Y., Uchida K., Kouchi T., Kuwajima J., Nakagawa Y., Sugioka S., Yamamoto E., Furuyama T., Mizubuchi H., Ohsugi N., Sakuno T., Kikuchi K., Matsuoka I., Ogawa N., Kaneko Y., Harashima S. (1999) A series of protein phosphatase gene disruptants in Saccharomyces cerevisiae. Yeast 15, 1669–1679 [DOI] [PubMed] [Google Scholar]
- 37. Claros M. G., Vincens P. (1996) Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur. J. Biochem. 241, 779–786 [DOI] [PubMed] [Google Scholar]
- 38. Gey U., Czupalla C., Hoflack B., Rödel G., Krause-Buchholz U.. (2008) Yeast pyruvate dehydrogenase complex is regulated by a concerted activity of two kinases and two phosphatases. J. Biol. Chem. 283, 9759–9767 [DOI] [PubMed] [Google Scholar]
- 39. Krause-Buchholz U., Gey U. (2006) YIL042c and YOR090c encode the kinase and phosphatase of the Saccharomyces cerevisiae pyruvate dehydrogenase complex. FEBS Lett. 580, 2553–2560 [DOI] [PubMed] [Google Scholar]
- 40. Steták A., Csermely P., Ullrich A., Kéri G. (2001) Physical and functional interactions between protein tyrosine phosphatase α, PI 3-kinase, and PKCδ. Biochem. Biophys. Res. Commun. 288, 564–572 [DOI] [PubMed] [Google Scholar]
- 41. Himmelbach A., Hoffmann T., Leube M., Höhener B., Grill E. (2002) Homeodomain protein ATHB6 is a target of the protein phosphatase ABI1 and regulates hormone responses in Arabidopsis. EMBO J. 21, 3029–3038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bellí G., Garí E., Piedrafita L. (1998) An activator/repressor dual system allows tight tetracycline-regulated gene expression in budding yeast. Nucleic Acids Res. 26, 942–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Christianson T. W., Sikorski R. S., Dante M., Shero J. H., Hieter P. (1992) Multifunctional yeast high-copy-number shuttle vectors. Gene 110, 119–122 [DOI] [PubMed] [Google Scholar]