Background: Activation of the relaxin receptor RXFP1 is driven by the LDLa module at the RXFP1 N terminus.
Results: LDLa residues Leu-7, Tyr-9, and Lys-17 all contribute to receptor activation via interactions involving their hydrophobic side chains.
Conclusion: These interactions induce the active receptor conformation, suggesting a novel mode of GPCR activation.
Significance: This novel mechanism of GPCR activation may lead to new drug development.
Keywords: G Protein-coupled Receptors (GPCR), NMR, Peptide Hormones, Peptides, Protein Structure, RXFP1, Relaxin
Abstract
The peptide hormone relaxin is showing potential as a treatment for acute heart failure. Although it is known that relaxin mediates its actions through the G protein-coupled receptor relaxin family peptide receptor 1 (RXFP1), little is known about the molecular mechanisms by which relaxin binding results in receptor activation. Previous studies have highlighted that the unique N-terminal low density lipoprotein class A (LDLa) module of RXFP1 is essential for receptor activation, and it has been hypothesized that this module is the true “ligand” of the receptor that directs the conformational changes necessary for G protein coupling. In this study, we confirmed that an RXFP1 receptor lacking the LDLa module binds ligand normally but cannot signal through any characterized G protein-coupled receptor signaling pathway. Furthermore, we comprehensively examined the contributions of amino acids in the LDLa module to RXFP1 activity using both gain-of-function and loss-of-function mutational analysis together with NMR structural analysis of recombinant LDLa modules. Gain-of-function studies with an inactive RXFP1 chimera containing the LDLa module of the human LDL receptor (LB2) demonstrated two key N-terminal regions of the module that were able to rescue receptor signaling. Loss-of-function mutations of residues in these regions demonstrated that Leu-7, Tyr-9, and Lys-17 all contributed to the ability of the LDLa module to drive receptor activation, and judicious amino acid substitutions suggested this involves hydrophobic interactions. Our results demonstrate that these key residues contribute to interactions driving the active receptor conformation, providing further evidence of a unique mode of G protein-coupled receptor activation.
Introduction
Relaxin belongs to the seven-member relaxin peptide family, which is believed to have evolved from insulin early in the evolution of vertebrates (1). There are three relaxin peptides in human, relaxin-1, -2, and -3, named so because of the presence of the well characterized relaxin binding cassette, Arg-X3-Arg-X2-Ile/Val (where X represents any amino acid), in the peptide B-chain. The RLN35 gene was discovered in 2002 (2) and encodes a highly conserved neuropeptide that is the ancestral peptide of the relaxin family (1, 3). Humans and other higher primates possess an RLN1 and an RLN2 gene, whereas other mammals have only an RLN1 gene (1, 4). The function of the RLN1 gene in humans and higher primates is unknown, and indeed no peptide product of this gene has ever been isolated. The human RLN2 gene product, H2 relaxin, is secreted into the blood and is the functional orthologue of the RLN1 gene product, simply called relaxin, found in other non-primate mammalian species (5). Relaxin has long been recognized as a reproductive hormone with important roles during pregnancy (for a review, see Ref. 6). However, it is the recent understanding of its roles in the cardiovascular system as a vasodilator (7–9), cardioprotective agent (10), and angiogenic factor (11) that is generating much interest. Indeed, recombinant H2 relaxin (serelaxin) has been demonstrated to be an effective treatment for acute heart failure in a recently completed Phase III clinical trial (12).
In 2002, the cognate receptor for relaxin was discovered to be the leucine-rich repeat-containing G protein-coupled receptor (GPCR) 7 (13), now known as the relaxin family peptide receptor 1 (RXFP1) (14). Typical of leucine-rich repeat-containing GPCRs, RXFP1 is a multidomain receptor with a classic GPCR seven-transmembrane helical region and a large extracellular domain containing the characteristic leucine-rich repeats formed by α-helices and β-sheets. Similar to other leucine-rich repeat-containing GPCRs, the leucine-rich repeat domain is essential for ligand binding in both RXFP1 (15) and the closely related RXFP2 (16, 17), which is the receptor for insulin-like peptide 3. However, RXFP1 and RXFP2 are unique in both the leucine-rich repeat-containing GPCR and GPCR family in that they have a low density lipoprotein receptor (LDLr) class A (LDLa) module at their N termini. Importantly, this module is conserved across evolution and has been demonstrated to be essential for ligand-stimulated signaling (18, 19). No cAMP responses were observed when RXFP1 and RXFP2 constructs lacking an LDLa module were stimulated with ligand despite the constructs exhibiting ligand binding properties comparable with wild type receptors (18). Later, it was learned that LDLa structural integrity is essential for the function of RXFP1. Structural perturbation via mutation of the Ca2+-ligating Asp-36 in the RXFP1 LDLa (20), abolishment of the disulfide bond at the N (19) or C terminus (20) of the module, or mutation of Phe-10, which is important for residue packing within the module including those involved in Ca2+ ligation (19), resulted in mutants unable to elicit any cAMP response upon stimulation. In addition to a correctly folded module, there is also evidence to suggest that specific amino acid side chains are necessary for the activity of the LDLa module. When the LDLa module of human RXFP1 was swapped with the second ligand binding domain (LB2) of the LDLr with which it shares high structural but relatively low sequence similarity (Fig. 1), the resulting chimera was unable to signal, although it was expressed on the cell surface and bound H2 relaxin like RXFP1 (19). Further characterization of the RXFP1 LDLa module via mutation alongside NMR structural analyses of proteins of equivalent mutant LDLa modules identified two potential LDLa signaling residues, Leu-7 and Tyr-9 (19). Although the study did not conclusively verify the properties of Leu-7 or Tyr-9 essential for activation, it provided compelling evidence to suggest that specific LDLa residues with specific signaling roles exist.
FIGURE 1.
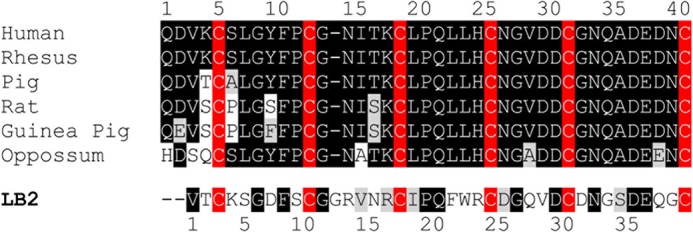
Boxshade alignment of LDLa module sequences from RXFP1 receptors from various mammalian species in comparison with the sequence of LB2 of the human LDLr. The conserved cysteine residues are highlighted in red, conserved amino acids are highlighted in black, and conservative amino acid substitutions are shaded.
It is clear that an obvious gap in the understanding of RXFP1 activation exists as the molecular mechanics surrounding the role of the LDLa module remain elusive. Therefore, this study aimed to advance our understanding of the molecular mechanisms of LDLa-mediated activation of RXFP1. First, we clarified whether the LDLa module is essential for any G protein-mediated signaling. We then adopted a gain-of-function and loss-of-function approach to identify LDLa signaling residues. Subsequently, we sought to understand the molecular mechanism involved in the LDLa action by characterizing the side chain properties engaged in potential protein-protein interactions that form the active receptor.
EXPERIMENTAL PROCEDURES
Generation of HEK293T Cell Lines Stably Expressing RXFP1 without the LDLa Module
HEK293T cells (ATCC number CRL-1573, American Type Tissue Culture Collection) stably expressing RXFP1 without the LDLa module (RXFP1-short) were generated via retrovirus transduction (21). Simply, the receptor-coding DNA sequence was first subcloned in-frame via a 5′ BamHI and a 3′ XhoI restriction site into the MSCV-IRES-GFP retroviral expression vector (a kind gift from Associate Prof. Ross Hannan, Peter MacCallum Cancer Centre, Melbourne, Australia) in the 5′–3′ direction between the 5′ long terminal repeats and the internal ribosomal entry site (IRES) sequence. Green fluorescent protein (GFP) follows the IRES. Following cloning and sequencing, the MSCV-RXFP1-short-IRES-GFP construct was utilized for retrovirus production by co-transfecting HEK293T cells on a 10-cm cell culture dish (Nunc) with the construct together with the Ampho packaging plasmid (a kind gift from Associate Prof. Ross Hannan) for 48 h. Retrovirus secreted into the medium was collected and passed through a 30-mm-diameter 0.45-μm Durapore PVDF syringe filter (Millipore) to remove any cell particulate prior to storage at −80 °C.
HEK293T cells seeded on a 10-cm cell culture dish were transduced with the harvested retrovirus by replacing the cell culture medium with 10 ml of the retrovirus-containing medium mixed with Polybrene (Millipore) to increase the transduction efficiency. Two rounds of 24-h transduction were performed before the cells were allowed to recover for 48 h in Dulbecco's modified Eagle's medium (DMEM) (Multicel) supplemented with 10% heat-inactivated fetal bovine serum (FBS), 1% penicillin/streptomycin, and 1% l-glutamine (referred to as complete DMEM). The cells were then transferred and grown to confluence in a 175-cm2 flask. Transduced cells were sorted into three distinct populations based on the amount of GFP fluorescence using fluorescence-activated cell sorting (FACS) on a MoFloTM XDP cell sorter (Beckman Coulter): high, medium, and low GFP (hence receptor) expression. These cells were then grown to confluence before assessing receptor expression on the cell surface via the cell surface expression assay as outlined below.
Secreted Alkaline Phosphatase (SEAP) Reporter Gene Assay
The SEAP reporter gene assay was performed as described (22) using reporter gene constructs (Clontech) that contained the SEAP gene under the control of specific transcription factor consensus sequences (cAMP response element (CRE), glucocorticoid response element, nuclear factor of κB cells (NFκB), activator protein 1, serum response element, nuclear factor of activated T cells, E-box DNA binding element (Myc), and heat shock element). Briefly, HEK293T cells stably expressing RXFP1 or RXFP1-short seeded in 12-well plates (Nunc) were first transfected with the desired plasmids for 30 h at a 1:1 ratio for a reporter gene of interest and the mCherry plasmid. Cells were then partially serum-starved for 18 h in DMEM supplemented with 0.5% FBS, 1% penicillin/streptomycin, and 1% l-glutamine prior to stimulation using 100 nm H2 relaxin. Stimulation was allowed to occur for 18 h (a time point chosen as responses began to peak at 12 h and were maintained at 24 h (22)) after which medium samples were collected, frozen at −20 °C, and had their SEAP protein content quantified using the BD Great EscAPeTM SEAP Fluorescence Detection kit (Clontech). The transfection efficiency of the reporter gene constructs could affect SEAP protein expression and hence bias the results; therefore, the mCherry plasmid, which constitutively expressed a red fluorescent monomer, served as a control to measure transfection efficiency between and within experiments. Upon medium sample collection, cells in each well were lysed in 500 μl of lysis buffer (50 mm Tris base, 150 mm NaCl, 1 mm EDTA, 1% Triton X-100, pH 7.4) and transferred to a 96-well OptiPlate with white opaque walls. mCherry fluorescence was then measured using a Victor Multilabel Plate Reader with 587 and 610 nm excitation and emission wavelengths, respectively.
Data analysis was performed as described (22) using GraphPad Prism 5. Briefly, discrepancies in cell transfection efficiency were first corrected by standardizing the SEAP protein measurement using the fluorescence measurement of the mCherry protein in each individual well. The transformed SEAP readings were then expressed as -fold change in response to peptide stimulation compared with the effect of vehicle alone. Data from three independent assays were pooled and are displayed as mean -fold change ±S.E. Unpaired two-tailed t tests were performed on the transfection-standardized and background-corrected data, comparing the peptide- and vehicle-treated cells for each reporter gene construct.
Generation of RXFP1-LB2
Human RXFP1 with its LDLa module substituted with the LB2 module was cloned into pcDNA3.1/Zeo (RXFP1-LB2) (19). As the LB2 module was inserted via two artificially engineered EcoRI restriction sites flanking the LDLa sequence, site-directed mutagenesis was used to delete the two additional amino acids (Glu-Phe) at each end of the LDLa module encoded by the EcoRI restriction sites.
Site-directed Mutagenesis
Site-directed mutagenesis was carried out via the QuikChange method as described (23). Polymerase chain reactions were performed with the RXFP1 or RXFP1-LB2 plasmids as templates to generate whole receptor mutants. The pGEV-LB2 plasmid expresses the LB2 module fused at its N terminus to B1 immunoglobulin binding domain of streptococcal protein G (GB1). Mutagenesis was performed on this plasmid to generate mutant LB2 modules for recombinant expression in Escherichia coli. Mutagenic primers were designed as detailed (24). The constructs together with the primers incorporating the mutations of interest are listed in Table 1. The complete sequence of each generated plasmid was verified via fluorescence-based cycle DNA sequencing.
TABLE 1.
RXFP1 variants and their corresponding mutation as well as the template DNA and primers used in QuikChange site-directed mutagenesis
| Receptor construct | Mutation introduced | Template DNA | Sense mutagenic primer (5′–3′ DNA sequence) | Antisense mutagenic primer (5′–3′ DNA sequence) |
|---|---|---|---|---|
| Gain-of-function mutants | ||||
| Single site | ||||
| G12del-RXFP1-LB2 | G12del | RXFP1-LB2 | gctgtggccgtgtcaaccgctgc | ggccacagctgaagtccccggatttgc |
| S34A-RXFP1-LB2 | S34A | RXFP1-LB2 | ggcgcagacgagcaaggctgtggagac | ctcgtctgcgccgttgtcgcagtccacttg |
| SLGYFP-RXFP1-LB2 | K4S, S5L, D7Y, S9P | RXFP1-LB2 | ctgcagtttagggtacttcccctgtgggggccgtgtcaac | cacaggggaagtaccctaaactgcaggtgacggatccacg |
| NITK-RXFP1-LB2 | G12del, R13N, V14I, N15T, R16K | RXFP1-LB2 | ggacttcagctgtgggaacatcacaaagtgcattcctcagttctgg | ccagaactgaggaatgcactttgtgatgttcccacagctgaagtcc |
| LLH-RXFP1-LB2 | F21L, W22L, R23H | RXFP1-LB2 | cgctgcattcctcagctcctgcactgcgatggccaagtgg | ccacttggccatcgcagtgcaggagctgaggaatgcagcg |
| NGVD-RXFP1-LB2 | D25N, Q27V, V28D | RXFP1-LB2 | cctcagttctggaggtgcaacggcgtggacgactgcgacaacggc | gccgttgtcgcagtcgtccacgccgttgcacctccagaactgagg |
| Double site | ||||
| SLGYFP G12del-RXFP1-LB2 | K4S, S5L, D7Y, S9P, G12del | SLGYFP-RXFP1-LB2 | cctgtggccgtgtcaaccgct | cggccacaggggaagtaccctaaactgc |
| SLGYFP NITK-RXFP1-LB2 | K4S, S5L, D7Y, S9P, G12del, R13N, V14I, N15T, R16K | SLGYFP-RXFP1-LB2 | gggaacatcacaaagtgcattcctcagttctggaggtgc | gcactttgtgatgttcccacaggggaagtaccctaaactg |
| Loss-of-function mutants | ||||
| Single residue | ||||
| L7A-RXFP1 | L7A | RXFP1 | ctccgccggctatttcccctgtgggaacatc | gccggcggagcacttgacatcctggtcg |
| L7K-RXFP1 | L7K | RXFP1 | ctccaaaggctatttcccctgtgggaacatc | gcctttggagcacttgacatcctggtcg |
| Y9F-RXFP1 | Y9F | RXFP1 | ggctttttcccctgtgggaacatcacaaag | gaaaaagccaagggagcacttgacatcc |
| Y9M-RXFP1 | Y9M | RXFP1 | ggcatgttcccctgtgggaacatcacaaag | gaacatgccaagggagcacttgacatcc |
| I15A-RXFP1 | I15A | RXFP1 | gaacgccacaaagtgcttgcctcagctc | gtggcgttcccacaggggaaatagccaag |
| T16A-RXFP1 | T16A | RXFP1 | catcgcaaagtgcttgcctcagctcctg | ctttgcgatgttcccacaggggaaatagccaag |
| K17A-RXFP1 | K17A | RXFP1 | cacagcgtgcttgcctcagctcctg | cacgctgtgatgttcccacaggggaaatag |
| K17M-RXFP1 | K17M | RXFP1 | caatgtgcttgcctcagctcctgc | gataaaggggacacccttgtagtgttacac |
| L19A-RXFP1 | L19A | RXFP1 | gtgcgcgcctcagctcctgcactgtaacg | gctgaggcgcgcactttgtgatgttcccacaggg |
| Double residue | ||||
| L7A/L22A-RXFP1 | L7A, L22A | L7A-RXFP1 | caggccctgcactgtaacggtgtggacg | cagggcctgaggcaagcactttgtgatgttcc |
| L7K/L22K-RXFP1 | L7K, L22K | L7K-RXFP1 | cagaaactgcactgtaacggtgtggacg | cagtttctgaggcaagcactttgtgatgttcc |
| L7K/Y9M-RXFP1 | L7K, Y9M | Y9M-RXFP1 | ctccaaaggcatgttcccctgtgggaac | gcctttggagcacttgacatcctggtcg |
| L19A/P20A-RXFP1 | L19A, P20A | RXFP1 | gtgcgcggcacagctcctgcactgtaacggtg | gctgtgccgcgcactttgtgatgttcccacaggg |
| Triple residue | ||||
| L7K/Y9M/K17A-RXFP1 | L7K, Y9M, K17A | L7K/Y9M-RXFP1 | cacagcgtgcttgcctcagctcctg | gcacgctgtgatgttcccacaggggaac |
Generation of SLGYFP NITK C-term-RXFP1-LB2
The entire RXFP1 LDLa C-terminal region beginning from Asn-26 was transposed into the SLGYFP NITK-RXFP1-LB2 construct using a cloning strategy because more RXFP1 LDLa residues were to be introduced into the LB2 module than was viable using single mutagenesis PCR. Hence, the DNA sequence encoding the LB2 module with introduced RXFP1 LDLa residues and flanking EcoRI restriction sites was purchased from GenScript and cloned into the original pcDNA3.1/Zeo construct (19). Site-directed mutagenesis was then used to delete the additional sequences encoded by the EcoRI restriction sites prior to use.
cAMP Activity Assay
cAMP activity assays were performed as described previously (18, 23). Briefly, HEK293T cells seeded on 96-well plates (Nunc) were transfected with plasmids of the receptor construct of interest and the CRE β-galactosidase reporter in a 1:1 ratio for 18 h. Afterward, cells were incubated with 100 μl of H2 relaxin in increasing concentrations made up in complete DMEM. Positive and negative control stimulations using 100 μl of 5 μm forskolin (Sigma-Aldrich) and 100 μl of complete DMEM, respectively, were also performed. After 6 h, the medium was aspirated, and cells were frozen overnight at −80 °C. Measurement of cAMP-driven β-galactosidase expression then followed based on its interaction with its substrate, chlorophenol red β-d-galactopyranoside (Roche Diagnostics). Data were analyzed using GraphPad Prism 5 and are presented as the mean percentage of 5 μm forskolin response ±S.E. These were then standardized for the cell surface expression of each construct. Significance was determined using one-way analysis of variance followed by a Tukey-Kramer multiple comparison post-test.
Cell Surface Expression Assay
All constructs were fused with an N-terminal FLAG epitope tag to enable monitoring of receptor expression using an anti-FLAG antibody. Adhering to the 1:1 ratio, HEK293T cells on 24-well plates (Nunc) were transfected with the plasmid of the receptor construct of interest and empty pcDNA3.1/Zeo for 18 h. Receptor expression was then assessed in triplicates as described (23) using an anti-FLAG mouse monoclonal primary antibody (Sigma-Aldrich) and a goat anti-mouse Alexa Fluor 488 secondary antibody. Data were analyzed using GraphPad Prism 5 and are displayed as the mean percentage of the wild type receptor expression ±S.E. Unpaired two-tailed t test was performed to gauge the significance of the expression of each receptor of interest above the background as well as compared with RXFP1.
Whole Cell Competition Binding Assay Using 125I-Labeled H2 Relaxin (125I-H2 Relaxin)
HEK293T cells on 48-well plates (Nunc) were transfected with the plasmid of the receptor construct of interest for 18 h. Ligand binding was then assessed in triplicates as described (23) using increasing concentrations of H2 relaxin to compete with 125I-H2 relaxin (kindly provided by Mohsin Sarwar and Prof. Roger Summers, Monash Institute of Pharmaceutical Sciences, Melbourne, Australia). Data were analyzed using GraphPad Prism 5 and are presented as the mean percentage of specific binding ±S.E. Significance was determined using one-way analysis of variance followed by a Tukey-Kramer multiple comparison post-test.
Expression and Purification of SLYFP NITK-LB2
SLGYFP NITK-LB2 was expressed using the pGEV-LB2 plasmids appropriately mutated to incorporate RXFP1 sequences of interest. Protein expression and purification were performed as described (19). Briefly, plasmids were transformed into BL21(DE3)trxB (Novagen) for expression of the GB1-LB2 mutant protein. Freshly transformed cells were used for all protein expression cultures. All cultures were grown and protein expression was induced at 37 °C. Proteins were 15N- or 13C,15N-labeled by growing cultures in a 2-liter Braun Biostat fermenter containing 1 liter of minimal medium with 15NH4Cl and d-[13C]glucose as the sole nitrogen and carbon sources. Fermentation was conducted as described by Cai et al. (25). Protein expression was induced by the addition of 1 mm isopropyl 1-thio-β-d-galactopyranoside for 2.5 h after which cells were harvested, pelleted, and stored at −20 °C.
Fusion protein was purified using IgG-Sepharose (GE Healthcare) via the manufacturer's instructions. The eluted protein was buffer-exchanged into 50 mm Tris-HCl, 150 mm NaCl, pH 7.5 and concentrated to ∼100 μg/ml using a 3-kDa-cutoff Vivaspin centrifugal concentrator (Sartorius). Protein concentration was adjusted to 100–300 μg/ml in refolding buffer (3 mm reduced glutathione, 0.3 mm oxidized glutathione, 50 mm Tris-HCl, 150 mm NaCl, 2.5 mm CaCl2, pH 7.5). The sample was incubated overnight at 4 °C with stirring to allow the complete formation of disulfide bonds.
Postoxidation, the GB1 fusion protein was cleaved from the LDLa module overnight by incubation with 10 units of thrombin protease (GE Healthcare)/mg of fusion protein. The cleaved GB1 was separated from the LDLa module by passing the sample over IgG-Sepharose. The unbound LDLa module was further purified by reverse phase HPLC on a Jupiter Proteo 4-μm 90-Å column with a 15–60% gradient (Buffer A, 1% TFA; Buffer B, 100% acetonitrile, 1% TFA). The eluted protein was lyophilized and stored at −20 °C.
NMR Analysis and Structure Determination of SLGYFP NITK-LB2
Lyophilized 13C,15N-labeled SLGYFP NITK-LB2 was diluted to 1 mm with 50 mm imidazole, pH 6.0, 10 mm CaCl2, 10% D2O in a 5-mm Shigemi tube for NMR studies. Three APSY (26) NMR experiments (five-dimensional APSY-CBCACONH, five-dimensional APSY-HACACONH, and four-dimensional APSY-HACANH) were acquired on 500-MHz spectrometer equipped with a cryoprobe. Automated backbone assignments (NH, N, Hα, Cα, and C′) including Cβ were obtained from UNIO-MATCH v.2.0.2 (27) using the input of the three APSY peak lists. Three NOESY spectra with a mixing time of 150 ms (three-dimensional 1H,1H NOESY-15N HSQC, three-dimensional 1H,1H NOESY-13C(aliphatic) HSQC, and three-dimensional 1H,1H NOESY-13C(aromatic) HSQC) were acquired on an AVANCE II 800-MHz spectrometer equipped with a cryoprobe. The backbone assignments after UNIO-MATCH were manually checked with three-dimensional 1H,1H NOESY-15N HSQC and three-dimensional 1H,1H NOESY-13C(aliphatic) HSQC spectra and then provided to UNIO-ATNOS/ASCAN (28) for automated side chain assignments. Finally, all the assignments were interactively checked and extended using CARA. All the NMR experiments were recorded at 25 °C.
Automated structure calculation was performed using UNIO-ATNOS/CANDID (29) with CYANA3.0 (30). Based on the homologous crystal structure of LB5 (Protein Data Bank code 1AJJ) (31), the Ca2+ ion coordinate was assigned with residues Trp-25(O), Gln-30(O), Asp-28(Oδ1), Asp-32(Oδ2), Asp-38(Oδ2), and Glu-39(Oϵ2) and introduced in the structure calculation using a “pseudo-link” from the C terminus of the sequence.
Structure Analysis
All relevant structures were visualized and aligned using MacPyMOL (The PyMOL Molecular Graphics System, Version 1.5.0.4, Schrödinger, LLC) and MOLMOL (32).
RESULTS
SEAP Reporter Gene Assays
Reporter gene assays were utilized to determine whether RXFP1 in the absence of the LDLa module was able to signal via other G protein-implicated pathways. To ensure a constant level of receptor expression at the cell surface, cell lines stably expressing each receptor of interest were used. An RXFP1 stable cell line was already available (33), and an RXFP1-short cell line was generated using retrovirus to transduce HEK293T cells. Transduced cells were sorted into three populations based on the amount of GFP fluorescence: high, medium, and low GFP (hence receptor) expression. Receptor expression and ligand binding assays demonstrated that the high expression cell line did indeed have high expression of the RXFP1-short receptor at levels similar to RXFP1 cells (data not shown); hence, this cell line was used for the SEAP reporter assays.
RXFP1 cells were stimulated with 100 nm H2 relaxin to screen for GPCR-relevant pathways associated with RXFP1 activation. Of all reporter genes tested, three showed changes in the SEAP level present in the cell culture medium compared with the vehicle-treated cells (Fig. 2A). At the CRE reporter gene, secretion of SEAP into the cell culture medium increased significantly upon stimulation to 3.25 ± 0.27-fold of vehicle treatment (n = 3, p < 0.01). SEAP secretion also rose significantly at the glucocorticoid response element reporter gene (2.19 ± 0.11-fold of vehicle treatment, n = 3, p < 0.01), although the response was of a smaller magnitude compared with that seen at the CRE reporter gene. On the other hand, secretion of SEAP into the medium was inhibited at the NFκB reporter gene whereby it decreased significantly to 0.52 ± 0.07-fold of vehicle treatment (n = 3, p < 0.01). No significant change in SEAP secretion compared with vehicle treatment was detected at the heat shock element, serum response element, activator protein 1, nuclear factor of activated T cells, and Myc reporter genes upon receptor stimulation.
FIGURE 2.
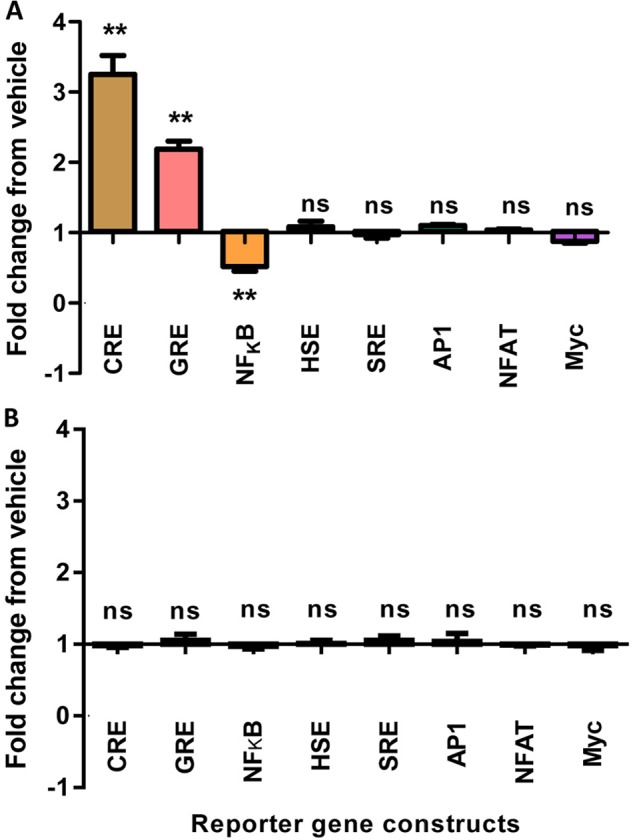
Reporter gene responses upon stimulation of RXFP1 (A) or RXFP1-short (B) stably expressed in HEK293T cells using 100 nm H2 relaxin. Data are -fold change of response from vehicle. Symbols represent means, and vertical bars represent S.E. of triplicate determinations from three independent experiments. GRE, glucocorticoid response element; AP1, activator protein 1; SRE, serum response element; NFAT, nuclear factor of activated T cells; HSE, heat shock element.
In contrast, stimulation of RXFP1-short stable cells with 100 nm H2 relaxin did not induce any change to the transcriptional activity of all reporter genes tested; hence, SEAP levels in the cell culture medium were comparable between the peptide-stimulated and vehicle-treated cells (Fig. 2B). Importantly, a positive control construct used in every experiment produced a transfection-standardized SEAP response that was significantly higher than that of the negative control construct (data not shown).
Substitution of the RXFP1 LDLa with the LB2 Module
We have demonstrated previously that conserving the RXFP1 receptor topology by substituting the LDLa module with the structurally similar but functionally and sequentially distinct LB2 module (Fig. 1) resulted in a non-signaling chimera despite its unchanged affinity for H2 relaxin (19). This suggests the presence of specific amino acids within the module whose side chains are engaged in protein-protein interactions that form the active receptor. Because of the conserved receptor topology, RXFP1-LB2 provided a suitable scaffold for identifying LDLa residues implicated in RXFP1 activation via a systematic introduction of RXFP1 LDLa residues into corresponding positions in RXFP1-LB2 to rescue its activity. However, it is important to note that the original RXFP1-LB2 construct (19) contains additional Glu-Phe residues flanking both ends of the LB2 module encoded by the artificially engineered EcoRI restriction sites used for cloning. To avoid any potential confounding effects, these residues were first deleted so that the RXFP1-LB2 N-terminal configuration completely mimicked that of RXFP1. Like the original RXFP1-LB2, the re-engineered RXFP1-LB2 could not signal, although it bound H2 relaxin normally and was expressed on the cell surface at levels similar to RXFP1 (Fig. 3A and Table 2).
FIGURE 3.
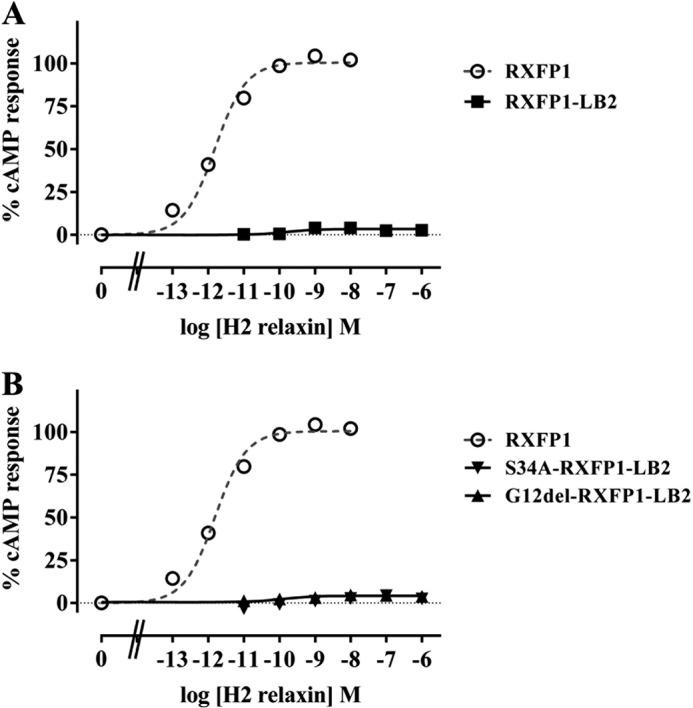
H2 relaxin-induced cAMP response of RXFP1-LB2 (A) and G12del-RXFP1-LB2 and S34A-RXFP1-LB2 (B) compared with RXFP1. cAMP activity is expressed as the percentage of the 5 μm forskolin-stimulated response for each receptor and has been normalized for cell surface expression. Symbols represent means and vertical bars (not visible) represent S.E. of triplicate determinations from at least three independent experiments.
TABLE 2.
Pooled activity data for RXFP1 mutants compared with wild type receptor
Competition binding data showing the pIC50 values, receptor expression data showing the cell surface expression, and cAMP activity data showing the pEC50 values as well as percent forskolin-stimulated maximum responses of RXFP1 and all its variants used in this study. NA, no activity.
| Receptor construct | Binding pIC50 | n | Cell surface expression | n | cAMP activity |
n | |
|---|---|---|---|---|---|---|---|
| Maximum response | pEC50 | ||||||
| % of RXFP1 | |||||||
| RXFP1 | 8.79 ± 0.07 | 5 | 100.10 ± 1.76 | 15 | 102.40 ± 3.52 | 11.72 ± 0.03 | 15 |
| RXFP1-LB2 | 8.69 ± 0.13 | 3 | 102.68 ± 7.58 | 5 | NA | NA | 5 |
| Single site gain-of-function mutants | |||||||
| G12del-RXFP1-LB2 | 8.50 ± 0.10 | 3 | 100.62 ± 9.13 | 5 | NA | NA | 6 |
| S34A-RXFP1-LB2 | 8.61 ± 0.08 | 3 | 101.66 ± 7.84 | 3 | NA | NA | 4 |
| SLGYFP-RXFP1-LB2 | 8.49 ± 0.05 | 3 | 103.22 ± 4.49 | 6 | 12.86 ± 1.44 | 7.70 ± 0.15 | 9 |
| NITK-RXFP1-LB2 | 8.56 ± 0.07 | 3 | 106.79 ± 8.18 | 6 | 32.58 ± 1.32 | 7.86 ± 0.07 | 8 |
| LLH-RXFP1-LB2 | 8.80 ± 0.07 | 3 | 100.90 ± 6.66 | 5 | NA | NA | 3 |
| NGVD-RXFP1-LB2 | 8.73 ± 0.08 | 3 | 104.98 ± 8.55 | 4 | NA | NA | 3 |
| Double site gain-of-function mutants | |||||||
| SLGYFP G12del-RXFP1-LB2 | 8.49 ± 0.10 | 3 | 98.77 ± 5.82 | 3 | 9.31 ± 0.45 | 7.59 ± 0.24 | 3 |
| SLGYFP NITK-RXFP1-LB2 | 8.61 ± 0.09 | 3 | 98.14 ± 5.96 | 4 | 44.07 ± 2.38 | 7.56 ± 0.05 | 6 |
| Triple site gain-of-function mutants | |||||||
| SLGYFP NITK C-term-RXFP1-LB2 | 8.66 ± 0.18 | 3 | 104.05 ± 7.05 | 8 | 41.62 ± 2.90 | 7.84 ± 0.09 | 4 |
| Loss-of-function mutants | |||||||
| L7A-RXFP1 | 8.68 ± 0.12 | 3 | 103.61 ± 9.78 | 3 | 107.6 ± 6.86 | 11.00 ± 0.17 | 7 |
| L7K-RXFP1 | 8.99 ± 0.06 | 3 | 103.00 ± 5.69 | 6 | 97.62 ± 2.25 | 10.09 ± 0.08 | 5 |
| L7A/L22A-RXFP1 | 8.68 ± 0.12 | 3 | 102.08 ± 8.88 | 3 | 99.95 ± 3.26 | 11.09 ± 0.20 | 11 |
| L7K/L22K-RXFP1 | 8.66 ± 0.09 | 3 | 103.87 ± 10.18 | 3 | 99.35 ± 2.01 | 10.29 ± 0.04 | 3 |
| Y9F-RXFP1 | 8.55 ± 0.13 | 3 | 97.68 ± 10.64 | 3 | 101.5 ± 1.89 | 11.82 ± 0.08 | 4 |
| Y9M-RXFP1 | 8.52 ± 0.06 | 3 | 93.74 ± 10.15 | 3 | 96.27 ± 4.77 | 10.46 ± 0.03 | 3 |
| L7K/Y9M-RXFP1 | 8.82 ± 0.10 | 3 | 99.88 ± 9.34 | 3 | 85.44 ± 3.73 | 10.28 ± 0.20 | 7 |
| I15A-RXFP1 | 8.88 ± 0.12 | 3 | 95.80 ± 9.07 | 3 | 96.57 ± 2.65 | 11.71 ± 0.12 | 3 |
| T16A-RXFP1 | 8.89 ± 0.11 | 3 | 97.18 ± 10.06 | 4 | 94.49 ± 2.32 | 11.60 ± 0.09 | 3 |
| K17A-RXFP1 | 8.61 ± 0.08 | 3 | 99.76 ± 9.43 | 5 | 103.3 ± 3.04 | 10.41 ± 0.12 | 4 |
| K17M-RXFP1 | 8.61 ± 0.10 | 3 | 98.35 ± 6.24 | 4 | 102.5 ± 1.53 | 11.37 ± 0.05 | 6 |
| L7K/Y9M/K17A-RXFP1 | 8.57 ± 0.14 | 3 | 99.10 ± 6.93 | 6 | 58.28 ± 3.79 | 9.36 ± 0.07 | 8 |
Characterization of potential RXFP1 LDLa signaling residues was attempted via a two-prong strategy that combined gain-of-function studies on the RXFP1-LB2 chimera and loss-of-function studies on RXFP1. In the gain-of-function studies, residues across the RXFP1 LDLa module were systematically introduced into the non-signaling RXFP1-LB2 that bound H2 relaxin normally to rescue receptor activity. This scanned the entire RXFP1 LDLa module for potential signaling regions. Upon identifying such regions, mutational analysis was performed on RXFP1 to dissect the importance of individual residues in each region in the complementary loss-of-function studies. Importantly, all receptor chimeras and mutants were expressed at the cell surface at levels similar to RXFP1 (Table 2). Additionally, the H2 relaxin binding ability of all the receptor chimeras and mutants was unaffected by any of the mutations introduced (Table 2). This reflected our previous studies whereby H2 relaxin binding was unaffected by the deletion of the RXFP1 LDLa module (18) or by mutations that perturbed receptor signaling (19, 34).
Gain-of-function Studies Utilizing RXFP1-LB2
Alignment of the LB2 and RXFP1 LDLa modules reveals that the LB2 possesses one residue more than the RXFP1 LDLa (Gly-12 as depicted in Fig. 1). Consequently, Gly-12 was deleted to test whether it was responsible for the non-signaling of RXFP1-LB2. G12del-RXFP1-LB2 did not elicit any cAMP response upon H2 relaxin stimulation but was able to bind H2 relaxin normally (Fig. 3B and Table 2). The LB2 module also contains a potential glycosylation site at Asn-32 based on the consensus N-glycosylation sequon (Asn-X-Ser where X represents any amino acid except Pro). The RXFP1 LDLa module also contains an N-glycosylation site that has been demonstrated to be utilized (23) but is in a different position (Asn-14) on the RXFP1 LDLa module. Hence, the influence of Asn-32 glycosylation on the non-signaling phenotype of RXFP1-LB2 was explored by mutating Ser-34 to an Ala to disrupt the glycosylation sequon. S34A-RXFP1-LB2 also did not signal, although it was expressed on the cell surface and bound H2 relaxin like RXFP1 (Fig. 3B and Table 2). These observations suggest that these structural factors are unlikely to be responsible for the non-signaling phenotypes of RXFP1-LB2, supporting its suitability as a scaffold for use in gain-of-function studies.
Subsequently, short stretches of conserved human RXFP1 LDLa residues as represented in Fig. 1 were systematically introduced into RXFP1-LB2 to generate SLGYFP-RXFP1-LB2, NITK-RXFP1-LB2, LLH-RXFP1-LB2, and NGVD-RXFP1-LB2. Importantly, SLGYFP-RXFP1-LB2 and NITK-RXFP1-LB2 showed a significant rescue of signaling, whereas the other mutants were inactive (Fig. 4A). SLGYFP-RXFP1-LB2 and NITK-RXFP1-LB2 demonstrated only partial rescue of activity with both having similar decreased H2 relaxin potency (p < 0.001 versus RXFP1; Table 2) and much lower maximal forskolin-induced cAMP activity (p < 0.01 versus RXFP1; Table 2) compared with RXFP1. Interestingly, the maximal activity of NITK-RXFP1-LB2 was higher compared with SLGYFP-RXFP1-LB2 (32.58 ± 1.32 and 12.86 ± 1.44%, respectively; p < 0.01). These results suggest that residues within these regions are important for RXFP1 activity as each region is able to give a partial gain of function. As evident from Fig. 1, the additional Gly-12 in the LB2 module is three residues to the C-terminal end of the mutation in SLGYFP-RXFP1-LB2. Although the deletion of Gly-12 had no effect on the activity of the non-signaling RXFP1-LB2, in the signaling SLGYFP-RXFP1-LB2, Gly-12 could displace the introduced Ser-4, Leu-5, Tyr-7, and Pro-9 in the LB2 structure as compared with their arrangement in the RXFP1 LDLa. The equivalent Gly-12 was thus deleted in SLGYFP-RXFP1-LB2. The dose-response curve of SLGYFP G12del-RXFP1-LB2 was unchanged compared with SLGYFP-RXFP1-LB2, suggesting that the additional Gly-12 does not affect the arrangement of the introduced residues within the LB2 structure (Table 2).
FIGURE 4.
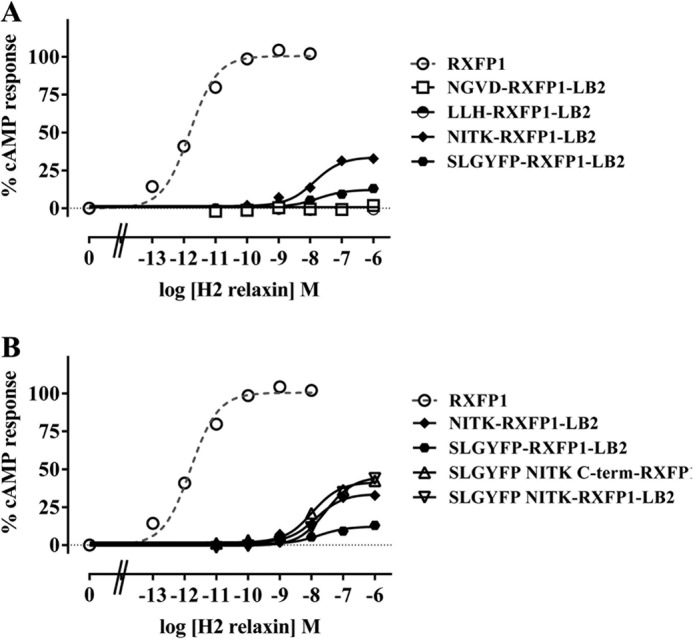
H2 relaxin-induced cAMP response of SLGYFP-RXFP1-LB2, NITK-RXFP1-LB2, LLH-RXFP1-LB2, and NGVD-RXFP1-LB2 (A) and SLGYFP-RXFP1-LB2, NITK-RXFP1-LB2, SLGYFP NITK-RXFP1-LB2, and SLGYFP NITK C-term-RXFP1-LB2 (B) compared with RXFP1. cAMP activity is expressed as the percentage of the 5 μm forskolin-stimulated response for each receptor and has been normalized for cell surface expression. Symbols represent means and vertical bars (not visible) represent S.E. of triplicate determinations from at least three independent experiments.
The mutations in SLGYFP-RXFP1-LB2 and NITK-RXFP1-LB2 were then combined to test for an additive effect on receptor activity. SLGYFP NITK-RXFP1-LB2 responded to H2 relaxin stimulation with a maximum response of 44.07 ± 2.38% forskolin-induced cAMP activity that was moderately but significantly higher than when either mutation was present alone (p < 0.001 compared with SLGYFP-RXFP1-LB2 and p < 0.01 compared with NITK-RXFP1-LB2) (Fig. 4B and Table 2). Although its pEC50 value was still unchanged compared with those of SLGYFP-RXFP1-LB2 and NITK-RXFP1-LB2, the improved cAMP response is evidence that both regions are involved concurrently in the formation of the active RXFP1 conformation.
The sequence at the C-terminal region of the RXFP1 LDLa module is highly similar to the LDLa modules of other proteins due to the important role of the C-terminal region in chelating Ca2+ (19, 34). Consequently, it seems unlikely that this region contains residues important for signaling. Nonetheless, the possibility was still tested. There are four acidic residues in the C-terminal region of the RXFP1 LDLa (19) and the LB2 (35, 36) modules whose ligation of a Ca2+ is crucial for maintaining structural integrity. To avoid disrupting Ca2+ ligation and hence the structure, the entire RXFP1 LDLa C-terminal region beginning from Asn-26 (three residues away from the first Ca2+-ligating residue in both the LDLa and LB2 modules) was transposed into SLGYFP NITK-RXFP1-LB2. The three-residue distance served as a “buffer” in an attempt to maintain the side chain orientation of the Ca2+-ligating residues whose packing in the module might be affected by the side chain of residues adjacent to it. Although SLGYFP NITK C-term-RXFP1-LB2 responded to H2 relaxin stimulation, its dose-response curve was similar to that of SLGYFP NITK-RXFP1-LB2, suggesting that the RXFP1 LDLa C-terminal region does not contain important signaling residues (Fig. 4B and Table 2).
RXFP1 Loss-of-function Mutants
The gain-of-function studies identified two regions in the RXFP1 LDLa module important for signaling. As short stretches of LDLa residues were introduced each time, a complementary loss-of-function was used to dissect the importance of individual residues and their properties essential for RXFP1 signaling.
The first signaling region identified, SLGYFP, consists of the RXFP1-specific Ser-6, Leu-7, Tyr-9, and Pro-11. Position 6 is highly variable across known and predicted mammalian RXFP1 sequences; hence, Ser-6 is unlikely to be important. The three other residues on the other hand are highly conserved across mammalian species. Although Pro-11 is completely conserved, its contribution to signaling was not explored as Pro residues are typically involved in maintaining protein structure due to the rigid side chain. Similarly, Phe-10 was not investigated in this study as it is conserved throughout LDLa modules, and a previous F10A mutation (19) disrupted the ability of the module to fold and ligate Ca2+, confirming its structural importance. As such, only Leu-7 and Tyr-9 were investigated, and indeed, they had been shown previously to be important for RXFP1 signaling (19).
Leu-7 was mutated first to an Ala to confirm the previously observed loss of activity (19) followed by a Lys to test the effect of a positive charge. The L7K mutation retains a similar length of side chain while introducing a positive charge. As expected, L7A-RXFP1 yielded a decrease in ligand potency compared with RXFP1 with no change in maximum response (pEC50 of 11.00 ± 0.17, p < 0.01) (Fig. 5A and Table 2). The introduction of a positive charge also did not affect the maximum response but yielded an even greater rightward shift of the L7K-RXFP1 dose-response curve (pEC50 of 10.09 ± 0.08), which was significantly different from the L7A-RXFP1 curve (p < 0.01). Therefore, these results highlight the importance of the Leu-7 long aliphatic side chain as well as the considerable unfavorableness of having a positive charge at this position.
FIGURE 5.
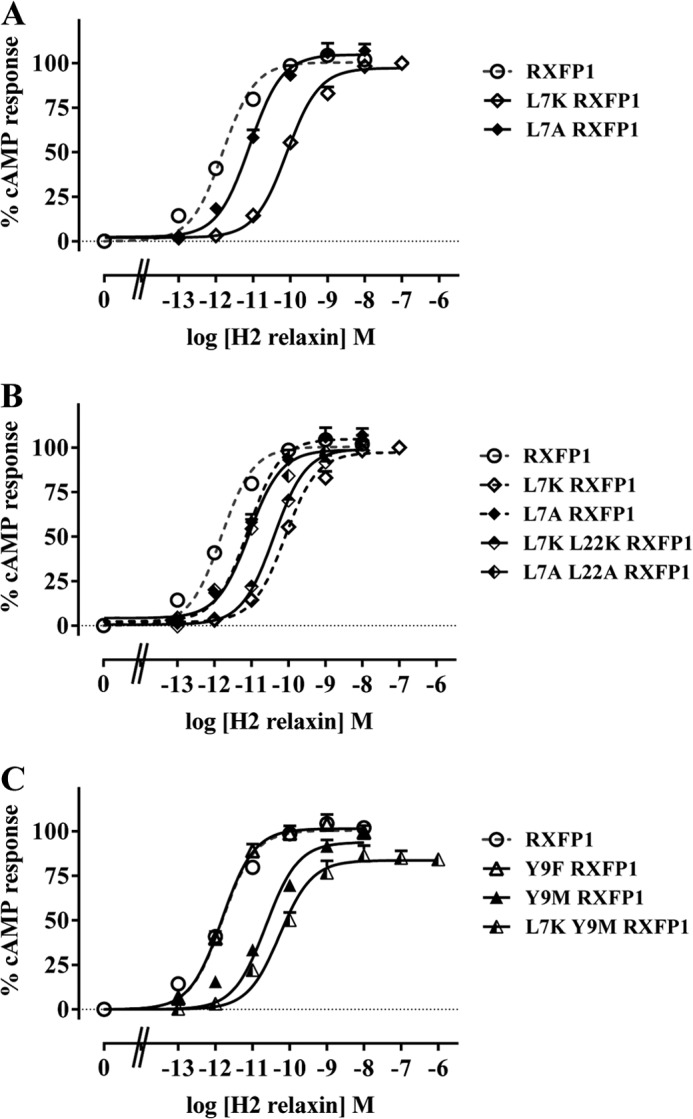
The H2 relaxin-induced cAMP response of L7K-RXFP1 and L7A-RXFP1 (A) and L7K-RXFP1, L7A-RXFP1, L7K/L22K-RXFP1, and L7A/L22A-RXFP1 (B) is shown. C, Y9F-RXFP1, Y9M-RXFP1, and L7K/Y9M-RXFP1 compared with RXFP1. cAMP activity is expressed as the percentage of the 5 μm forskolin-stimulated response for each receptor and has been normalized for cell surface expression. Symbols represent means and vertical bars represent S.E. of triplicate determinations from at least three independent experiments.
We previously mutated Leu-22 to explore its role in RXFP1 signaling, and whereas L22A-RXFP1 produced a rightward-shifted dose-response curve, the shift was not statistically significant (19). We revisited this residue by mutating Leu-22 together with Leu-7 to test its potential as an auxiliary signaling residue. In one mutant, the L7A mutation was combined with a L22A mutation, whereas in another, the L7K mutation was combined with a L22K mutation. The dose-response curves of L7A/L22A-RXFP1 and L7K/L22K-RXFP1 were similar to that of L7A-RXFP1 and L7K-RXFP1, respectively, conclusively showing that Leu-22 has no role in RXFP1 signaling (Fig. 5B and Table 2).
Next, the importance of Tyr-9 was investigated. We previously demonstrated that its mutation to alanine affected the structure of the LDLa module, causing a loss of signaling (19). As the removal of the entire side chain disrupted the structure of the LDLa module, less drastic substitutions were tested to investigate the role of this residue. Hence, Tyr-9 was first mutated to the structurally related Phe to explore the importance of the tyrosine OH group. The dose-response curve of Y9F-RXFP1 resembled that of RXFP1, suggesting that the OH group has no signaling role (Fig. 5C and Table 2). Subsequently, Tyr-9 was mutated to a Met to remove the benzene ring while conserving the size of the side chain. Compared with RXFP1, the dose-response curve of Y9M-RXFP1 was significantly rightward-shifted (pEC50 of 10.46 ± 0.03, n = 3, p < 0.001) whereas the maximum response remained unchanged, implicating the benzene moiety in RXFP1 signaling (Fig. 6B and Table 2). The hydrophobicity of the benzene ring suggests that the active RXFP1 conformation also engages the Tyr benzene ring in hydrophobic interactions.
FIGURE 6.
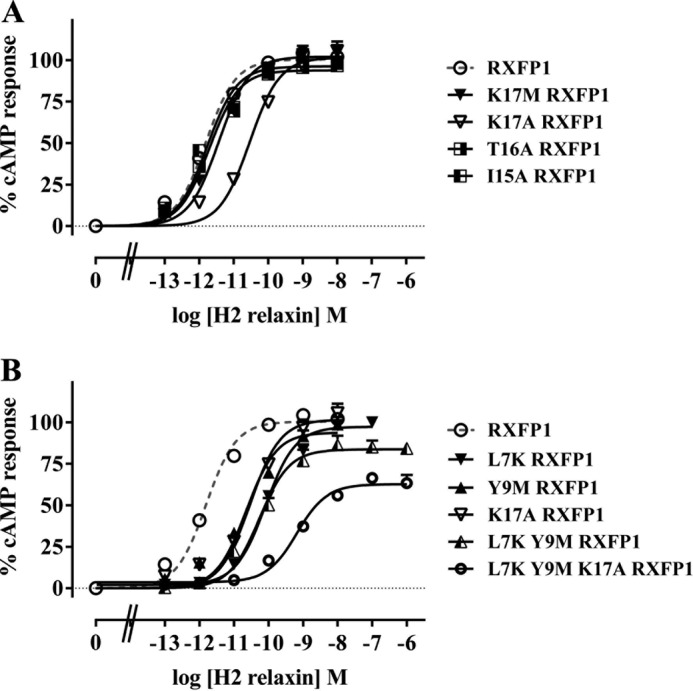
H2 relaxin-induced cAMP response of I15A-RXFP1, T16A-RXFP1, K17A-RXFP1, and K17M-RXFP1 (A) and L7K-RXFP1, Y9M-RXFP1, K17A-RXFP1, L7K/Y9M-RXFP1, and L7K/Y9M/K17A-RXFP1 (B) compared with RXFP1. cAMP activity is expressed as the percentage of the 5 μm forskolin-stimulated response for each receptor and has been normalized for cell surface expression. Symbols represent means and vertical bars represent S.E. of triplicate determinations from at least three independent experiments.
The L7K and Y9M mutations were combined next to test whether the hydrophobicity of both residues was engaged together in interactions that conferred the active receptor conformation. Although the pEC50 value of L7K/Y9M-RXFP1 was not significantly different from those of L7K-RXFP1 and Y9M-RXFP1, its maximum cAMP response of 85.44 ± 3.73% was significantly lower than that of either L7K-RXFP1, Y9M-RXFP1, or RXFP1 (p < 0.05) (Fig. 5C and Table 2). Because this decrease was not associated with a decrease in cell surface expression, it appears that the hydrophobicity of both Leu-7 and Tyr-9 is engaged together in the formation of the active RXFP1 conformation.
Next, the second RXFP1 LDLa region, NITK, consisting of Asn-14, Ile-15, Thr-16, and Lys-17 was examined. All four residues are highly to completely conserved across all mammalian species (Fig. 1); however, only Ile-15, Thr-16, and Lys-17 were studied as Asn-14 has been extensively studied in two previous studies (18, 23). The mutation of Ile-15 to Ala resulted in no change in the dose-response curve of I15A-RXFP1 in comparison with RXFP1, suggesting that Ile-15 has no signaling role (Fig. 6A and Table 2). The same was deduced for Thr-16 as T16A-RXFP1 also produced a dose-response curve similar to that of RXFP1 (Fig. 6A and Table 2). However, when Lys-17 was mutated to an Ala, its dose-response curve was significantly rightward-shifted (pEC50 = 10.41 ± 0.12, p < 0.001), whereas its maximum response remained unchanged compared with that of RXFP1. To determine the side chain property essential for signaling, Lys-17 was mutated to a Met to remove the positive charge while conserving the size of the side chain. K17M-RXFP1 yielded a dose-response curve similar to that of RXFP1, suggesting that the charge is not important for signaling (Fig. 6A and Table 2). It follows that Lys-17 contributes its long aliphatic side chain for hydrophobic interactions that form the active RXFP1 conformation.
Upon detecting this third potential signaling residue, a K17A mutation was introduced into L7K/Y9M-RXFP1 to test whether all three residues act together to engage the necessary interactions for RXFP1 activation. Indeed, L7K/Y9M/K17A-RXFP1 responded to H2 relaxin treatment with a significantly reduced pEC50 value of 9.36 ± 0.07 compared with when either mutation was present alone or when the L7K and Y9M mutations were combined (p < 0.05 compared with L7K-RXFP1, p < 0.01 compared with Y9M-RXFP1, p < 0.001 compared with K17A-RXFP1, and p < 0.001 compared with L7K/Y9M-RXFP1). Similarly, its maximum response of only 58.28 ± 3.79% forskolin-induced cAMP activity was also significantly decreased (p < 0.001 compared with L7K-RXFP1, p < 0.01 compared with Y9M-RXFP1, p < 0.001 compared with K17A-RXFP1, and p < 0.05 compared with L7K/Y9M-RXFP1) (Fig. 6B and Table 2). Thus, it appears that reducing the hydrophobicity of Leu-7, Tyr-9, and Lys-17 concurrently reduced the maximum response and efficacy of H2 relaxin.
Structural Analysis of SLGYFP NITK-LB2
Our previous structural studies of the LDLa module showed that it is sensitive to mutagenesis and that decreases in the maximal response were indicative of either structural perturbation or loss of Ca2+ ligation. As the SLGYFP NITK-LB2 chimera showed only 44.07 ± 2.38% of rescued cAMP activity, the structure of this module was solved and compared with that of RXFP1.
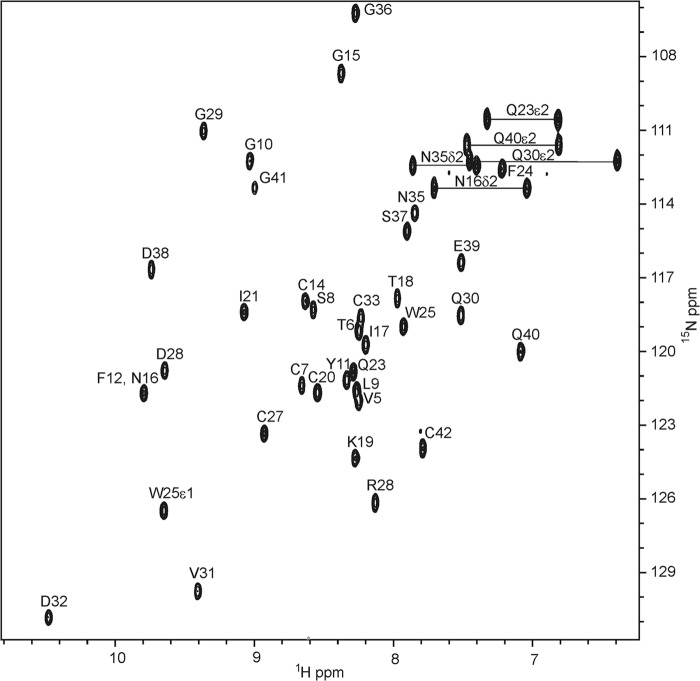
The 1H,15N HSQC spectra of SLYFP NITK-LB2 shows good dispersion of resonances, indicating that the module is folded as a globular domain as a consequence of correct disulfide bond formation and Ca2+ ligation (Fig. 7). As the resonances were significantly shifted from those of both the native LB2 and RXFP1 LDLa modules, reassignment was required. Assignment and structure determination were accomplished rapidly by acquiring four-dimensional and five-dimensional APSY spectra and 15N-edited and 13C-edited NOESY. From the APSY data, complete assignment of the peptide N, NH, C′, Cα, Hα, and Cβ resonances for all residues except the NH group of Asp-34 (numbering based on chimera, Protein Data Bank code 2M7P; equivalent to Asp-31 in LB2; Fig. 1) were made. From the NOESY data, a total of 665 NOEs were assigned (Table 3) and used to define the structure with an overall backbone r.m.s.d. of 0.35 Å between residues 6 and 42. The 20 lowest energy structures are presented in Fig. 8A.
FIGURE 7.
1H,15N HSQC of 1 mm13C,15N-labeled SLGYFP NITK-LB2 in 50 mm imidazole, 10 mm CaCl2, 10% D2O, pH 6. 0 and 25 °C acquired at 800 MHz. Assignments of observed peptide and side chain NH resonances are indicated. Numbering is based on the chimeric LB2 module in Protein Data Bank code 2M7P.
TABLE 3.
Input for the structure calculation and characterization of the bundle of 20 CYANA conformers of SLYFP NITK-LB2 (residues 1–42)
ali, aliphatic; aro, aromatic.
| Quantity | Valuea |
|---|---|
| NOE upper distance constraints | 665 |
| Intraresidual | 135 |
| Short range | 190 |
| Medium range | 159 |
| Long range | 181 |
| Number of long range/residue (6–42) | 5 |
| Number of assigned NOE peaks | |
| Three-dimensional 1H,1H NOESY-15N HSQC (%) | 80.3 |
| Three-dimensional 1H,1H NOESY-13C(ali) HSQC (%) | 86.6 |
| Three-dimensional 1H,1H NOESY-13C(aro) HSQC (%) | 87.6 |
| Residual NOE violations number ≥0.2 Å | 2 ± 1 |
| Residual target function value (Å2) | 1.33 ± 0.09 |
| Deviations from idealized geometry | |
| Bond length (Å) | 0.0128 ± 0.00 |
| Bond angle (°) | 1.009 ± 0.00 |
| r.m.s.d. from mean coordinates (Å) | |
| Backbone (6–42) | 0.35 ± 0.11 |
| All heavy atoms (6–42) | 0.70 ± 0.09 |
| Ramachandran plot statisticsb (6–42) | |
| Most favored and additional allowed regions (%) | 100 |
| Generously allowed regions (%) | 0.0 |
| Disallowed regions (%) | 0.0 |
a Except for the top nine entries, average values and standard deviations for the 20 CYANA conformers are given. The nine entries represent the output generated in the seventh cycle of the UNIO-ATNOS/CANDID with CYANA3.0 calculation. The numbers in parentheses indicate the residues for which the values were calculated.
b As determined by PROCHECK.
FIGURE 8.
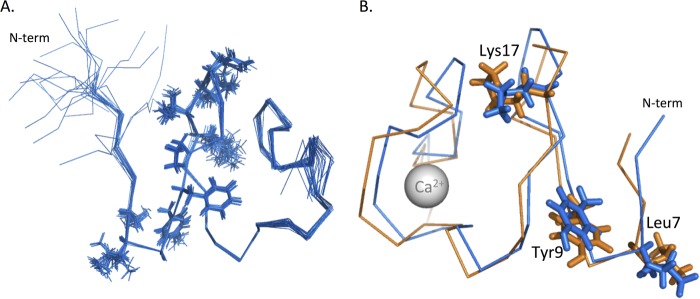
A, ensemble of the 20 lowest energy structures of SLYFP NITK-LB2. Structures are represented as an overlay of the backbone structures in MOLMOL that overlay with an r.m.s.d. of 0.35 Å between residues 6 and 42. The side chains from the two “add-back” regions SLYFP and NITK have been represented as lines. B, overlay of SLYFP NITK-LB2 (blue) onto the structure of RXFP1 LDLa (orange) (Protein Data Bank code 2JM4). The side chains of Leu-7, Tyr-9, and Lys-17 overlay with good agreement. The overall r.m.s.d. of the two structures between residues 6 and 42 is 1.68 Å.
The structure of the SLGYFP NITK-LB2 module overlays well with the RXFP1 LDLa (Protein Data Bank code 2JM4) with a backbone r.m.s.d. of 1.68 Å over residues 7–41. Importantly, the three key signaling residues, Leu-7, Tyr-9, and Lys-17, are in excellent alignment with each other despite the additional Gly in the LB2 scaffold (Fig. 8B). The structure of SLGYFP NITK-LB2 suggests that we successfully recreated the arrangement of the RXFP1 LDLa module within the N-terminal region as the backbone r.m.s.d. between residues 7 and 21 is 1.08 Å, whereas the C-terminal region (residues 21–41) overlays with a backbone r.m.s.d. of 1.2 Å. Interestingly, comparison of the SLGYFP NITK-LB2 structure with the native LB2 suggests that our mutations have made the fold more “RXFP1-like” as the structure overlays with an r.m.s.d. of 1.9 Å to LB2 (Protein Data Bank code 1LDR). The C-terminal region of the mutant shows a distinct difference in the orientation of the Asn residue that is equivalent to position 35 in RXFP1. In the LB2 structure, this Asn (Asn-32) is oriented toward the solvent, whereas in RXFP1 LDLa and SLGYFP NITK-LB2, the Asn is oriented toward the core of the structure (Fig. 9). Taken together, analysis of the SLGYFP NITK-LB2 demonstrates that we have chosen an appropriate scaffold to generate the chimera for the gain-of-function studies and that we have recreated the orientation of the side chains of the LDLa module that contribute to RXFP1 receptor activation.
FIGURE 9.
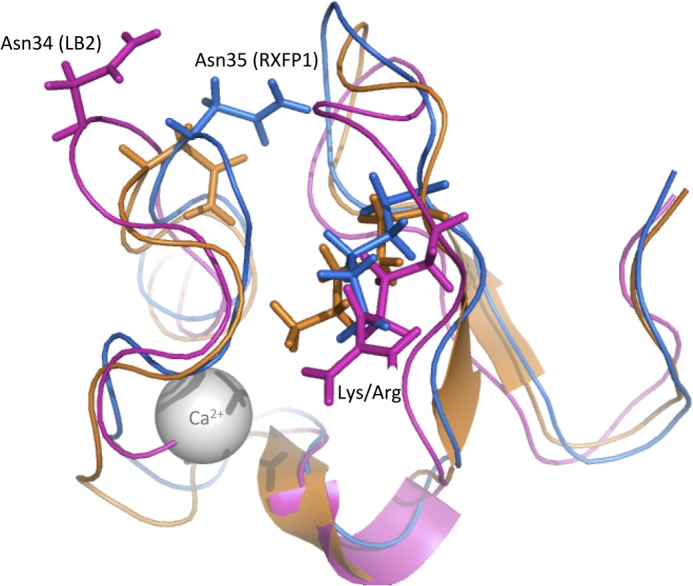
Overlay of the mean structure of SLYFP NITK-LB2 (blue), RXFP1 LDLa (orange), and LB2 (pink) represented as a cartoon model. The side chains of Lys-17 and equivalent Arg in LB2 are represented as sticks in addition to Asn-33 (RXFP1) and the equivalent Asn-32 from LB2. The overlay demonstrates that the chimera is more “RXFP1”-like despite the LB2 scaffold.
DISCUSSION
Several studies have highlighted that the LDLa module is crucial in conferring the active RXFP1 conformation that leads to signal activation as truncation of the module or point mutations that disrupted the structure of the module abolished cAMP related signaling (18–20, 37). Because RXFP1 and RXFP2 are the only mammalian GPCRs with an LDLa module, this represents a unique mode of GPCR signaling. We hypothesize that the LDLa module interacts with other receptor domains to induce the active receptor conformation, and although we have yet to identify the site of the receptor at which the LDLa module binds, we predict that it may interact with the transmembrane domain to induce signaling (38). We previously identified specific residues on the LDLa module that are associated with signaling (19). However, the study was limited as only an alanine scan was performed on the residues that contributed to the hydrophobic surface of the module and hence could have overlooked signaling residues present in other parts of the module. Consequently, in this study, a comprehensive examination of the RXFP1 LDLa module was performed to identify signaling residues.
Additionally, the previous studies that measured signaling in the absence of the LDLa module utilized cAMP-related signaling assays, either direct cAMP measurement (19, 20, 37) or cAMP activity assays using a CRE β-galactosidase reporter (18). It is possible that the RXFP1 mutant lacking the LDLa module, RXFP1-short, which is still able to bind relaxin, is signaling through an alternative GPCR signaling pathway. Therefore, a panel of reporter genes known to be associated with GPCR signaling was used to test whether RXFP1 was able to induce alternative signaling in the absence of the LDLa module. We previously used this system to study signaling at RXFP1 and demonstrated that relaxin stimulation of RXFP1 activated the CRE and glucocorticoid response element reporters but inhibited the NFκB reporter gene (22). Importantly, we were able to confirm these results while at the same time demonstrating that the RXFP1-short receptor is unable to signal through any signaling pathway. These studies also gave us the confidence to utilize the CRE reporter gene to monitor the ability of receptors to signal in subsequent LDLa mutation studies. Moreover, this pathway produced the most robust SEAP response and hence represented the most sensitive measure of receptor activation.
All mutants in this study had cell surface expression and H2 relaxin binding comparable with RXFP1. The unaffected cell surface expression agrees with our previous data whereby even receptors with structurally disrupted LDLa modules were expressed normally on the cell surface (19). The uncompromised H2 relaxin binding was also expected as the LDLa module has been established to have no role in ligand binding (18, 19). As such, these factors clearly did not contribute to the signaling changes observed with a number of the mutants used in this study.
The gain-of-function approach identified two regions of the RXFP1 LDLa module that are key to signal activation. Introduction of the first region consisting of residues Ser-6, Leu-7, Tyr-9, and Pro-11 and the second region consisting of Asn-14, Ile-15, Thr-16, and Lys-17 resulted in an increase in maximal cAMP signaling compared with the RXFP1-LB2, which was unable to signal. Although the introduction of both these regions into RXFP1-LB2 appeared to have an improved effect, it did not completely restore signaling or significantly increase the pEC50 value. In contrast, the introduction of Leu-22, Leu-23, and His-24 did not rescue signaling. The lack of activity of these residues matches our previous work, which demonstrated that Leu-23 was potentially important for correct folding of the module and His-24 may also play an important role in maintaining the integrity of the module (19). Additionally, substitution of Leu-22 in this study and previously (19) highlighted that it is not important to structure or function.
The rescued activity from the reintroduction of the SLGYFP and NITK regions clearly implicates residues within these regions as making important contacts. Our previous work highlighted that Leu-7 made a specific side chain interaction involved in receptor activation. Additionally, mutation of Tyr-9 suggested that it has a dual role in maintaining structural integrity in addition to contributing to the signal activation. Therefore, the regained activity from this region conferred with our previous hypothesis that the loop before the β-hairpin is important. We had not previously considered the residues within the NITK loop as important for signaling because the residues form the motif that leads to N-linked glycosylation of Asn-14. Mutation of Asn-14 to inhibit glycosylation did not affect RXFP1 signaling (18, 23), so the initial observation that this region makes significant contributions to the signaling surface was unexpected. To further dissect the contribution of individual residues within these regions, we made single substitutions of the side chains. However, we opted for subtle changes to probe the importance of the properties of the side chain rather than side chain deletions by alanine substitution, which in general do not appear to be well tolerated by the structure of the module.
To this end, we revealed that it is the hydrophobicity of the Leu-7 aliphatic side chain and the Tyr-9 benzene ring that is engaged in interactions that confer RXFP1 activation. Mutation of these residues individually to conservative non-hydrophobic residues led to significant shifts in the concentration-response curves with relaxin, and the combination of the two resulted in an additive shift. Examination of the second signaling region implicated Lys-17 in RXFP1 signaling. Although Lys-17 is positively charged, mutational investigation showed that RXFP1 activation engages the Lys-17 aliphatic side chain rather than its charge, paralleling the importance of the hydrophobicity of Leu-7 and Tyr-9. Asn-14 is another residue of interest in this region as it forms part of an N-glycosylation sequon (Asn-X-Thr where X represents any amino acid except Pro) and is likely to be glycosylated (18, 23). Mutation of Thr-16 to Ala disrupted the glycosylation sequon but had no effect on signaling, H2 relaxin binding, or receptor cell surface expression, suggesting that Thr-16 has no role in signaling. Additionally, this also suggests that Asn-14 glycosylation is not important for RXFP1 function. This is in agreement with several findings where an N14Q mutation had no (18) or a minor effect on signaling (23) but contradicted another (20) whereby the same N14Q mutation led to decreased cell surface expression and cAMP signaling. Because of the conflicting results, it is unlikely that Asn-14 or its glycosylation represents an important factor in RXFP1 signaling because if either were important substituting the residue and/or changing its glycosylation status should yield reproducible effects.
Subsequently, the K17A mutation was combined with the L7K and Y9M mutations. The dose-response curve of L7K/Y9M/K17A-RXFP1 shifted further to the right compared with when either mutation was present alone or when the L7K and Y9M mutations were combined. Again, the H2 relaxin binding affinity remained unchanged, suggesting that all three residues are engaged in interactions that form the active RXFP1 conformation. With the sequential mutation of each signaling residue, the receptor becomes increasingly less able to signal as signified by progressive decreases in either the maximum cAMP response or both maximum cAMP response and H2 relaxin efficacy.
Identification of Leu-7, Tyr-9, and Lys-17 needed to form the active RXFP1 conformation reveals a signaling surface on the LDLa module as illustrated in Fig. 10. The position of Leu-19 and Pro-20 on this surface may suggest signaling roles, but their significant conservation across all LDLa modules is more indicative of structural importance. Indeed, it is reasonable to predict that mutation of Pro with its rigid side chain could disrupt the fold of the module. As such, Leu-19 and Pro-20 were not further investigated.
FIGURE 10.
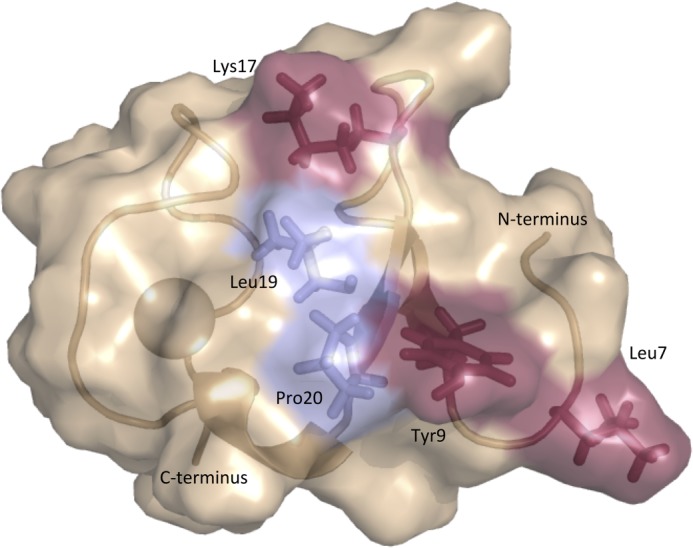
Proposed surface of the RXFP1 LDLa module involved in receptor activation. Mutagenesis studies confirm the importance of residues Leu-7, Tyr-9, and Lys-17. The positions of Leu-19 and Pro-20 suggest that they could contribute to the surface; however, the conservation of these residues throughout the LDLa module family suggests roles in structural maintenance.
Although the introduction of the SLGYFP and NITK regions of RXFP1 onto the RXFP1-LB2 scaffold rescued the cAMP response, the pEC50 value for H2 relaxin was significantly lower in comparison with RXFP1, and the maximum response was only 44.07 ± 2.38% of the maximal cAMP response of RXFP1. It would seem reasonable to assume that if these were the only sites on the module involved then presentation of the key residues on the LB2 scaffold would rescue full activation. Indeed, analysis of the NMR structure of the SLGYFP NITK-LB2 protein chimera revealed that the module retained its fold and that Leu-7, Tyr-9, and Lys-17, the residues our loss-of-function mutants highlighted as having the largest contribution to signaling, were positioned equivalently to those in the native RXFP1 LDLa module. It seems unlikely that the remaining activity lies in other residues from the LDLa module as we have exhaustively examined the ordered regions of the module. It is possible that although the SLGYFP NITK chimera presents the key signaling residues of the module well the effect of the subtle differences within the C-terminal region (residues 21–41) including the 310 helix between the segments cannot be discounted. Such structural complexity is not unusual and has been demonstrated previously by the interaction between reelin and the first LDLa module of apoE receptor-2. Although the key binding residues have been identified, their grafting onto the fourth LDLa module did not rescue binding to reelin (39).
This investigation and our previous studies support the presence of key signaling residues within the N-terminal region of the LDLa module. This is in contrast to the “binding fingerprint” of other LDLa modules from the LDLr family that bind their ligands using a surprisingly small surface area compared with most protein-protein interactions and utilize the acidic residues within the C-terminal region that also contribute to the ligation of the Ca2+ ion. Examples include the structures of the third and fourth ligand binding repeats of the LDLr in complex with the receptor-associated protein whereby the Ca2+-ligating Asp residues are in contact with receptor-associated protein (40), the structure of LA1 from apoE receptor-2 in complex with reelin whose small binding interface encompasses primarily the Ca2+-ligating residues (39), and the recent NMR study of gentamicin binding to complement type repeat 10 from Megalin that shows ligand interactions with the Ca2+-ligating Asp residues (41).
RXFP1 and the related RXFP2 stand out as unique and complex GPCRs as they are the only mammalian GPCRs to contain an LDLa module essential for receptor activation. The mechanism of activation whereby ligand binding alone is unable to induce an active receptor conformation but requires distinct side chain-driven interactions involving the N-terminal LDLa module presents a novel paradigm in GPCR signal activation. Based on the discovery that RXFP1 and RXFP2 form constitutive homodimers (42, 43), we postulate that ligand binding at one monomer of a dimer may position the LDLa module for interaction with the transmembrane domain of its dimerization partner to form the active receptor complex capable of signaling (5). Although hypothetical, this model highlights the complex signaling and binding mechanism of the RXFP1 receptor. We have previously demonstrated the complexity of ligand binding to RXFP1 and RXFP2; it requires coordination of ligand binding to the ectodomain and the transmembrane domain. Although we were able to fully reconstitute the binding of the related relaxin family peptide insulin-like peptide 3 to an RXFP1 ectodomain-only construct, we could not completely reconstitute binding in full-length RXFP1 mutants (17). Therefore, it is likely that our inability to reconstitute 100% of the cAMP response within our chimeric LDLa receptors also lies within this complexity.
Irrespective of the exact mechanism of LDLa-mediated receptor signaling, this study has successfully identified a signaling surface composed of Leu-7, Tyr-9, and Lys-17 on the RXFP1 LDLa N-terminal region whose side chains are involved in hydrophobic protein-protein interactions that direct the active conformation. The unique nature of this activation mechanism and our understanding of these key signaling residues thus present the prospect for new classes of agonists and/or antagonists targeting RXFP1 based on the LDLa-mediated activation mechanism.
Acknowledgments
We thank Sharon Layfield and Tania Ferraro for technical assistance, Corthera for provision of recombinant H2 relaxin, and Mohsin Sarwar and Prof. Roger Summers, Monash Institute of Pharmaceutical Sciences, for the kind gift of 125I-H2 relaxin. We also acknowledge the State of Victoria, Rowden White Foundation, and Australian Research Council-Linkage Infrastructure, Equipment and Facilities for NMR instrumentation funding.
This work was supported in part by National Health and Medical Research Council of Australia Project Grants 628427 and 1043750 (to R. A. D. B. and P. R. G.) and by the Victorian Government Operational Infrastructure Support Program.
The 1H,13C,15N chemical shifts have been deposited in the BioMagResBank under BMRB 19200.
The atomic coordinates and structure factors (code 2M7P) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- RLN
- relaxin
- GPCR
- G protein-coupled receptor
- LDLa
- low density lipoprotein class A
- RXFP
- relaxin family peptide
- LDLr
- low density lipoprotein receptor
- LB2
- second ligand binding domain
- IRES
- internal ribosomal entry site
- SEAP
- secreted alkaline phosphatase
- GB1
- B1 immunoglobulin binding domain of streptococcal protein G
- APSY
- automated projection spectroscopy
- HSQC
- heteronuclear single quantum correlation
- r.m.s.d.
- root mean square deviation
- CRE
- cAMP response element.
REFERENCES
- 1. Wilkinson T. N., Speed T. P., Tregear G. W., Bathgate R. A. (2005) Evolution of the relaxin-like peptide family. BMC Evol. Biol. 5, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bathgate R. A., Samuel C. S., Burazin T. C., Layfield S., Claasz A. A., Reytomas I. G., Dawson N. F., Zhao C., Bond C., Summers R. J., Parry L. J., Wade J. D., Tregear G. W. (2002) Human relaxin gene 3 (H3) and the equivalent mouse relaxin (M3) gene. Novel members of the relaxin peptide family. J. Biol. Chem. 277, 1148–1157 [DOI] [PubMed] [Google Scholar]
- 3. Hsu S. Y. (2003) New insights into the evolution of the relaxin-LGR signaling system. Trends Endocrinol. Metab. 14, 303–309 [DOI] [PubMed] [Google Scholar]
- 4. Evans B. A., Fu P., Tregear G. W. (1994) Characterization of two relaxin genes in the chimpanzee. J. Endocrinol. 140, 385–392 [DOI] [PubMed] [Google Scholar]
- 5. Bathgate R. A., Halls M. L., van der Westhuizen E. T., Callander G. E., Kocan M., Summers R. J. (2013) Relaxin family peptides and their receptors. Physiol. Rev. 93, 405–480 [DOI] [PubMed] [Google Scholar]
- 6. Bathgate R. A. D., Hsueh A. J., Sherwood O. D. (2006) in Physiology of Reproduction (Neill J. D., ed) 3rd Ed., pp. 679–770, Elsevier, San Diego, CA [Google Scholar]
- 7. Bani-Sacchi T., Bigazzi M., Bani D., Mannaioni P. F., Masini E. (1995) Relaxin-induced increased coronary flow through stimulation of nitric oxide production. Br. J. Pharmacol. 116, 1589–1594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bani D., Failli P., Bello M. G., Thiemermann C., Bani Sacchi T., Bigazzi M., Masini E. (1998) Relaxin activates the L-arginine-nitric oxide pathway in vascular smooth muscle cells in culture. Hypertension 31, 1240–1247 [DOI] [PubMed] [Google Scholar]
- 9. Dschietzig T., Bartsch C., Richter C., Laule M., Baumann G., Stangl K. (2003) Relaxin, a pregnancy hormone, is a functional endothelin-1 antagonist: attenuation of endothelin-1-mediated vasoconstriction by stimulation of endothelin type-B receptor expression via ERK-1/2 and nuclear factor-κB. Circ. Res. 92, 32–40 [DOI] [PubMed] [Google Scholar]
- 10. Dschietzig T., Richter C., Bartsch C., Laule M., Armbruster F. P., Baumann G., Stangl K. (2001) The pregnancy hormone relaxin is a player in human heart failure. FASEB J. 15, 2187–2195 [DOI] [PubMed] [Google Scholar]
- 11. Segal M. S., Sautina L., Li S., Diao Y., Agoulnik A. I., Kielczewski J., McGuane J. T., Grant M. B., Conrad K. P. (2012) Relaxin increases human endothelial progenitor cell NO and migration and vasculogenesis in mice. Blood 119, 629–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Teerlink J. R., Cotter G., Davison B. A., Felker G. M., Filippatos G., Greenberg B. H., Ponikowski P., Unemori E., Voors A. A., Adams K. F., Jr., Dorobantu M. I., Grinfeld L. R., Jondeau G., Marmor A., Masip J., Pang P. S., Werdan K., Teichman S. L., Trapani A., Bush C. A., Saini R., Schumacher C., Severin T. M., Metra M. (2013) Serelaxin, recombinant human relaxin-2, for treatment of acute heart failure (RELAX-AHF): a randomised, placebo-controlled trial. Lancet 381, 29–39 [DOI] [PubMed] [Google Scholar]
- 13. Hsu S. Y., Nakabayashi K., Nishi S., Kumagai J., Kudo M., Sherwood O. D., Hsueh A. J. (2002) Activation of orphan receptors by the hormone relaxin. Science 295, 671–674 [DOI] [PubMed] [Google Scholar]
- 14. Bathgate R. A., Ivell R., Sanborn B. M., Sherwood O. D., Summers R. J. (2006) International Union of Pharmacology LVII: recommendations for the nomenclature of receptors for relaxin family peptides. Pharmacol. Rev. 58, 7–31 [DOI] [PubMed] [Google Scholar]
- 15. Büllesbach E. E., Schwabe C. (2005) The trap-like relaxin-binding site of the leucine-rich G-protein-coupled receptor 7. J. Biol. Chem. 280, 14051–14056 [DOI] [PubMed] [Google Scholar]
- 16. Scott D. J., Wilkinson T. N., Zhang S., Ferraro T., Wade J. D., Tregear G. W., Bathgate R. A. (2007) Defining the LGR8 residues involved in binding insulin-like peptide 3. Mol. Endocrinol. 21, 1699–1712 [DOI] [PubMed] [Google Scholar]
- 17. Scott D. J., Rosengren K. J., Bathgate R. A. (2012) The different ligand-binding modes of relaxin family peptide receptors RXFP1 and RXFP2. Mol. Endocrinol. 26, 1896–1906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Scott D. J., Layfield S., Yan Y., Sudo S., Hsueh A. J., Tregear G. W., Bathgate R. A. (2006) Characterization of novel splice variants of LGR7 and LGR8 reveals that receptor signaling is mediated by their unique low density lipoprotein class A modules. J. Biol. Chem. 281, 34942–34954 [DOI] [PubMed] [Google Scholar]
- 19. Hopkins E. J., Layfield S., Ferraro T., Bathgate R. A., Gooley P. R. (2007) The NMR solution structure of the relaxin (RXFP1) receptor lipoprotein receptor class A module and identification of key residues in the N-terminal region of the module that mediate receptor activation. J. Biol. Chem. 282, 4172–4184 [DOI] [PubMed] [Google Scholar]
- 20. Kern A., Agoulnik A. I., Bryant-Greenwood G. D. (2007) The low-density lipoprotein class A module of the relaxin receptor (leucine-rich repeat containing G-protein coupled receptor 7): its role in signaling and trafficking to the cell membrane. Endocrinology 148, 1181–1194 [DOI] [PubMed] [Google Scholar]
- 21. Cepko C., Pear W. (2001) Overview of the retrovirus transduction system. Curr. Protoc. Mol. Biol. Chapter 9, Unit 9.9 [DOI] [PubMed] [Google Scholar]
- 22. Halls M. L., Bathgate R. A., Summers R. J. (2007) Comparison of signaling pathways activated by the relaxin family peptide receptors, RXFP1 and RXFP2, using reporter genes. J. Pharmacol. Exp. Ther. 320, 281–290 [DOI] [PubMed] [Google Scholar]
- 23. Yan Y., Scott D. J., Wilkinson T. N., Ji J., Tregear G. W., Bathgate R. A. (2008) Identification of the N-linked glycosylation sites of the human relaxin receptor and effect of glycosylation on receptor function. Biochemistry 47, 6953–6968 [DOI] [PubMed] [Google Scholar]
- 24. Zheng L., Baumann U., Reymond J.-L. (2004) An efficient one-step site-directed and site-saturation mutagenesis protocol. Nucleic Acids Res. 32, e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cai M., Huang Y., Sakaguchi K., Clore G. M., Gronenborn A. M., Craigie R. (1998) An efficient and cost-effective isotope labeling protocol for proteins expressed in Escherichia coli. J. Biomol. NMR 11, 97–102 [DOI] [PubMed] [Google Scholar]
- 26. Hiller S., Fiorito F., Wüthrich K., Wider G. (2005) Automated projection spectroscopy (APSY). Proc. Natl. Acad. Sci. U.S.A. 102, 10876–10881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Volk J., Herrmann T., Wüthrich K. (2008) Automated sequence-specific protein NMR assignment using the memetic algorithm MATCH. J. Biomol. NMR 41, 127–138 [DOI] [PubMed] [Google Scholar]
- 28. Fiorito F., Herrmann T., Damberger F. F., Wüthrich K. (2008) Automated amino acid side-chain NMR assignment of proteins using 13C- and 15N-resolved 3D [1H, 1H]-NOESY. J. Biomol. NMR 42, 23–33 [DOI] [PubMed] [Google Scholar]
- 29. Herrmann T., Güntert P., Wüthrich K. (2002) Protein NMR structure determination with automated NOE assignment using the new software CANDID and the torsion angle dynamics algorithm DYANA. J. Mol. Biol. 319, 209–227 [DOI] [PubMed] [Google Scholar]
- 30. Güntert P., Mumenthaler C., Wüthrich K. (1997) Torsion angle dynamics for NMR structure calculation with the new program DYANA. J. Mol. Biol. 273, 283–298 [DOI] [PubMed] [Google Scholar]
- 31. Fass D., Blacklow S., Kim P. S., Berger J. M. (1997) Molecular basis of familial hypercholesterolaemia from structure of LDL receptor module. Nature 388, 691–693 [DOI] [PubMed] [Google Scholar]
- 32. Koradi R., Billeter M., Wüthrich K. (1996) MOLMOL: a program for display and analysis of macromolecular structures. J. Mol. Graph. 14, 51–55 [DOI] [PubMed] [Google Scholar]
- 33. Bathgate R. A., Lin F., Hanson N. F., Otvos L., Jr., Guidolin A., Giannakis C., Bastiras S., Layfield S. L., Ferraro T., Ma S., Zhao C., Gundlach A. L., Samuel C. S., Tregear G. W., Wade J. D. (2006) Relaxin-3: improved synthesis strategy and demonstration of its high-affinity interaction with the relaxin receptor LGR7 both in vitro and in vivo. Biochemistry 45, 1043–1053 [DOI] [PubMed] [Google Scholar]
- 34. Hopkins E. J., Bathgate R. A., Gooley P. R. (2005) The human LGR7 low-density lipoprotein class A module requires calcium for structure. Ann. N.Y. Acad. Sci. 1041, 27–34 [DOI] [PubMed] [Google Scholar]
- 35. Bieri S., Atkins A. R., Lee H. T., Winzor D. J., Smith R., Kroon P. A. (1998) Folding, calcium binding, and structural characterization of a concatemer of the first and second ligand-binding modules of the low-density lipoprotein receptor. Biochemistry 37, 10994–11002 [DOI] [PubMed] [Google Scholar]
- 36. Daly N. L., Djordjevic J. T., Kroon P. A., Smith R. (1995) Three-dimensional structure of the second cysteine-rich repeat from the human low-density lipoprotein receptor. Biochemistry 34, 14474–14481 [DOI] [PubMed] [Google Scholar]
- 37. Bogatcheva N. V., Ferlin A., Feng S., Truong A., Gianesello L., Foresta C., Agoulnik A. I. (2007) T222P mutation of the insulin-like 3 hormone receptor LGR8 is associated with testicular maldescent and hinders receptor expression on the cell surface membrane. Am. J. Physiol. Endocrinol. Metab. 292, E138–E144 [DOI] [PubMed] [Google Scholar]
- 38. Hartley B. J., Scott D. J., Callander G. E., Wilkinson T. N., Ganella D. E., Kong C. K., Layfield S., Ferraro T., Petrie E. J., Bathgate R. A. D. (2009) Resolving the unconventional mechanisms underlying RXFP1 and RXFP2 receptor function Ann. N.Y. Acad. Sci. 1160, 67–73 [DOI] [PubMed] [Google Scholar]
- 39. Yasui N., Nogi T., Takagi J. (2010) Structural basis for specific recognition of reelin by its receptors. Structure 18, 320–331 [DOI] [PubMed] [Google Scholar]
- 40. Fisher C., Beglova N., Blacklow S. C. (2006) Structure of an LDLR-RAP complex reveals a general mode for ligand recognition by lipoprotein receptors. Mol. Cell 22, 277–283 [DOI] [PubMed] [Google Scholar]
- 41. Dagil R., O'Shea C., Nykjær A., Bonvin A. M., Kragelund B. B. (2013) Gentamicin binds to the megalin receptor as a competitive inhibitor using the common ligand binding motif of complement type repeats: insight from the NMR structure of the 10th complement type repeat domain alone and in complex with gentamicin. J. Biol. Chem. 288, 4424–4435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Svendsen A. M., Zalesko A., Kønig J., Vrecl M., Heding A., Kristensen J. B., Wade J. D., Bathgate R. A., De Meyts P., Nøhr J. (2008) Negative cooperativity in H2 relaxin binding to a dimeric relaxin family peptide receptor 1. Mol. Cell. Endocrinol. 296, 10–17 [DOI] [PubMed] [Google Scholar]
- 43. Svendsen A. M., Vrecl M., Ellis T. M., Heding A., Kristensen J. B., Wade J. D., Bathgate R. A., De Meyts P., Nøhr J. (2008) Cooperative binding of insulin-like Peptide 3 to a dimeric relaxin family peptide receptor 2. Endocrinology 149, 1113–1120 [DOI] [PubMed] [Google Scholar]