Background: Identification of SCF1(FBXO25) substrates can be done through in chip ubiquitination on protoarrays.
Results: FBXO25 interacts and mediates ubiquitination and proteasomal degradation of the ELK-1 protooncogene regulator.
Conclusion: The c-Fos regulator ELK-1 is an SCF1(FBXO25) substrate.
Significance: FBXO25 is a potential mitogen pathway regulator through ELK-1 degradation.
Keywords: E3 Ubiquitin Ligase, egr-1, Ets Family Transcription Factor, Proteasome, Protein Degradation
Abstract
FBXO25 is one of the 69 known human F-box proteins that serve as specificity factors for a family of ubiquitin ligases composed of SKP1, Rbx1, Cullin1, and F-box protein (SCF1) that are involved in targeting proteins for degradation across the ubiquitin proteasome system. However, the substrates of most SCF E3 ligases remain unknown. Here, we applied an in chip ubiquitination screen using a human protein microarray to uncover putative substrates for the FBXO25 protein. Among several novel putative targets identified, the c-fos protooncogene regulator ELK-1 was characterized as the first endogenous substrate for SCF1(FBXO25) E3 ligase. FBXO25 interacted with and mediated the ubiquitination and proteasomal degradation of ELK-1 in HEK293T cells. In addition, FBXO25 overexpression suppressed induction of two ELK-1 target genes, c-fos and egr-1, in response to phorbol 12-myristate 13-acetate. Together, our findings show that FBXO25 mediates ELK-1 degradation through the ubiquitin proteasome system and thereby plays a role in regulating the activation of ELK-1 pathway in response to mitogens.
Introduction
Substrates of the ubiquitin proteasome system are eventually degraded following covalent attachment of an ubiquitin chain to the target protein through sequential steps catalyzed by three different enzymes: ubiquitin-activating enzyme (E1), ubiquitin-conjugating enzyme (E2), and ubiquitin-ligase enzyme (E3). The E3s are critical enzymes in this pathway because of their roles in substrate recognition. The polyubiquitinated substrates are directed to 26S proteasome where the ubiquitin is recycled and the protein is degraded (1, 2).
The largest and most well characterized family of E3 ligases is the multisubunit really interesting new gene (RING)5 finger type, known as cullin-RING ligase1 (CRL1) or SCF1 complex. They are composed of a Cullin1 (CUL1) subunit that serves as an organizing scaffold, an E2-recruiting RING-box protein 1 (Roc1/RBX1), an adaptor protein S phase kinase-associated protein 1 (SKP1), and a substrate recognition subunit F-box protein (FBP) (3). In addition to the F-box domain that binds to SKP1, FBP contains distinct protein-protein interaction domains that interact with their substrates. In humans, 69 SCF ligases have been identified to date, each one characterized by a different FBP subunit that provides specificity by recognizing different substrates (4–6). Post-translational modifications, such as protein phosphorylation, prolylhydroxylation, or glycosylation, are important in regulating the interaction of a substrate with the FBP (7).
The F-box protein FBXO25, the closest paralog of atrogin-1 (FBXO32), assembles an active E3 ligase in mammalian cells with SKP1, CUL1, and Roc1 (8–10). FBXO25 localizes to the nucleus as a component of the nuclear body named FANDs (FBXO25-associated nuclear domains), partially colocalizes with CUL1, SKP1, ubiquitinated proteins, and the proteasome. In addition, FANDs recruit polyglutamine-containing proteins and prevent their accumulation in the nucleus (11). Our studies suggested that FANDs are competent sites of polyubiquitination in the nucleus having SCF1(FBXO25) as E3 ligase (8, 9, 11). Interestingly, the inhibition of RNA polymerase I disrupts FANDs, and FBXO25 antibodies interfere with RNA polymerase II transcription in vitro, indicating the role of this FBP in the transcriptional process (9, 11). Previously, we have applied a combination of two proteomic approaches and identified 132 novel FBXO25-interacting partners. The transcriptional factor β-actin was demonstrated to be enriched in and regulates the FANDs dynamics in the nucleus. However, β-actin is not a substrate of SCF1(FBXO25) (9).
Notably, only a small number of the 69 human SCF ubiquitin-ligases have well established substrates, many of which are involved in cell cycle control, cell growth, apoptosis, DNA damage response, and tumorigenesis (12). In the present study, we have applied a powerful and high throughput proteomic approach, known as protoarray, to identify ubiquitinated substrates of SCF1(FBXO25). This technology has been previously validated by identifying targets of purified E3 ligases through in vitro ubiquitination assays (13–16). Here, we identify 75 novel potential SCF1(FBXO25) substrates and validate in vitro and in cellulo the c-fos protooncogene regulator ELK-1 as a substrate for this E3 ubiquitin-ligase. We also report the functional relationship of FBXO25 and two immediate early genes regulated by elk-1, c-fos, and egr-1.
EXPERIMENTAL PROCEDURES
Plasmids and Subcloning
The F-box-deleted (ΔF) fbxo25 gene was subcloned into pcDNA5/FRT/TO plasmid (Invitrogen) using pDEST27-HA-FBXO25-ΔF-box-FLAG described previously (8) as template. The insert was amplified by using the primers ΔF-forward (GAAGCTTATGCCGTTTCTGGG) and ΔF-reverse (CCTCGAGTCAGAACTTGAAG). The products were digested with HindIII and XhoI and subcloned into pcDNA5/FRT/TO. DNA manipulation and transformation procedures were performed according to standard cloning techniques (17). The plasmid encoding elk-1 (ELK-1-FLAG-His6) was kindly provided by Dr. Andrew D. Sharrocks from the University of Manchester. The plasmids encoding the proteins HA-SKP-1, FLAG-CUL1, FLAG-ROC1, GST-HA-FBXO25-ΔF-box-FLAG, and GST-HA-FBXO25-FLAG were used previously (8).
Cells: Culturing, Transient Transfection, and Drug Treatments
HEK293T (CRL-11268, American Type Culture Collection) cells were grown in DMEM (Sigma-Aldrich) supplemented with 10% fetal bovine serum (FBS; Invitrogen) in 5% CO2 atmosphere. The transfections were carried out using FuGENE in accordance with manufacturer (Roche Applied Science) by 48 h. Six hours before lysis, 500 nm epoxomicin proteasome inhibitor (Sigma-Aldrich) was added to cell culture medium. For c-fos and egr-1 expression evaluation, the cells were transfected with fbxo25 or empty vector for 24 h and then submitted to starvation in no-FBS DMEM for an additional 24 h. Thereafter, 100 nm phorbol 12-myristate 13-acetate (PMA) (Invitrogen) was added for the indicated times, and the cell pellets were obtained after 0, 15, and 45 min. The total RNA was extracted, and the c-fos and egr-1 transcript levels were quantified by quantitative PCR.
Purification of SCF1 Complexes
HEK293T cells were transfected with plasmids encoding SCF1 complexes HA-SKP1, CUL1-FLAG, Myc-Roc1, and GST-HA-FBXO25-FLAG or GST-HA-FBXO25-ΔF-box-FLAG. After 48 h, the cells were rinsed in lysis buffer (25 mm Tris-HCl, pH 7.5, 150 mm KCl, and 1% Nonidet P-40) containing protease inhibitor mixture (Sigma-Aldrich) and phosphatase inhibitors (10 mm NaF and 1 mm Na3VO4; Sigma-Aldrich). The SCF1 complex purification was performed by GST pulldown. The lysates were incubated with Sepharose-glutathione resin (GE Healthcare) for 3 h at 4 °C with rocking. After that, the beads were washed with lysis buffer, and the SCF1 complexes were eluted with elution buffer (0.1 m Tris-HCl, pH 7.5, with 0.1 m reduced glutathione). These eluates were dialyzed in ubiquitination buffer and stored at −20 °C until use.
Ubiquitination on Protoarrays
The procedures with ProtoArrays Human Protein Microarrays v4.1 were in according to the manufacturer's instructions (Invitrogen). The protoarray slides were treated in blocking buffer (50 mm HEPES, 200 mm NaCl, 0.08% Triton X-100, 25% glycerol, 20 mm reduced glutathione, 1 mm dithiothreitol (DTT), and 1% bovine serum albumin (BSA) (Invitrogen) for 60 min at 4 °C. The reactions were prepared: purified SCF1(FBXO25) or SCF1(FBXO25-ΔF-box) and 100 ng of E1 + 500 ng of E2 (UbcH5c) or 500 ng of E2DN (dominant negative) + 2.5 μg of ubiquitin N-terminally monobiotinylated + 1 μg of native ubiquitin + ubiquitination buffer (20 mm Tris-HCl, pH 7.6, 20 mm KCl, 5 mm MgCl2, 2 mm ATP, 1 mm DTT, and 10% glycerol). The enzymes E1, E2, and ubiquitins were purchased from BostonBiochem (Boston, MA). 100 μl of the reaction was added to the slide and overlaid with a coverslip followed by incubation for 3 h at 30 °C in humid chamber (Corning Inc.). Slides were washed in assay buffer (50 mm Tris, pH 7.5, 50 mm NaCl, 5 mm MgSO4, 0.1% Tween 20, 1% BSA) (Invitrogen), and the arrays were then incubated with 1.0 ng/μl streptavidin-Alexa Fluor 647 (Invitrogen) for 45 min at 4 °C. Then they were washed five times with assay buffer and once with water. Slides were dried by centrifugation at 1000 × g for 2 min, and the images were acquired immediately.
Data Acquisition and Analyses
The protoarrays were scanned, and the data were acquired with GenePix4000B software (Molecular Devices). Background-subtracted intensities for all spots were normalized among slides centering all intensities on a single reference value. The centering factor for each slide was chosen as the biotin-positive controls average, and they were corrected to match the overall biotin-positive control of all slides. Significant intensity detection was carried out comparing each spot normalized signal with the negative control intensity distribution of the whole slide. For each slide, a background density function was fitted, and a p value was calculated for each spot. The significance cutoff was established in p value < 0.01. Proteins were further considered only if all internal replicates were consistently detected. The final target list was defined as the intersection between two slides with SCF1(FBXO25) minus negative controls, SCF1(FBXO25) with E2 dominant negative and SCF1(FBXO25-ΔF-box).
Pulse Labeling of Cells with [35S]Methionine
Cells seeded in 6-well plates were transfected with empty vector (pcDNA3) or GST-HA-FBXO25-FLAG or GST-HA-FBXO25-ΔF-box-FLAG. After 24 h, the cells were washed with PBS and DMEM without methionine (Sigma-Aldrich) and kept in this medium for 2 h. Then the cells were pulsed for 1 h with 150 μCi/well 35S-radiolabeled methionine (Express labeling mix 35S; PerkinElmer Life Sciences). The time zero was collected immediately and after 12 h and 24 h T1 and T2 were obtained, respectively. The cell pellets were lysed with radioimmune precipitation assay buffer (300 mm NaCl, 2% Nonidet P-40, 0.1% deoxycholate, 0.2% SDS, and 100 mm Tris-HCl, pH 7.5), sonicated, and the supernatants were submitted to immunoprecipitation with rabbit polyclonal anti-ELK-1 antibody (Millipore). The eluted proteins by load buffer were separated by SDS-PAGE, and after the staining with Coomassie Brilliant Blue the gels were dried and exposed to a Phosphor Screen (Amersham Biosciences,). After 24 h at room temperature, the Phosphor Screen was scanned in Molecular Image FX using the software PDQuest (Bio-Rad).
Real-time Quantitative PCR Analysis
Total RNA from HEK293T cells was extracted and isolated using TRIzol reagent (Invitrogen). To synthesize cDNA we used the ImProm II Reverse Transcriptase (Promega) following the manufacturer's protocol. The c-fos and egr-1 transcript levels were quantified relative to GAPDH using the Mastercycler ep realplex real-time PCR system (Eppendorf). The c-fos primer set included forward (5′- GGGGCAAGGTGGAACAGTTATC-3′) and reverse (5′-CTCCGGTTGCGGCATTTGG-3′) primers; the egr-1 set primer was forward (5′-CGCCCACCATGGACAACTAC-3′) and reverse (5′-AGGAAAAGACTCTGCGGTCAG-3′). A gapdh primer set (forward (5′-TGCTGATGCCCCCATGTTCG-3′ and reverse (5′-TGCAGGAGGCATTGCTGATGA-3′)) was included as a sample loading control. The reaction was carried out with Platinum SYBR Green qPCR SuperMix-UDG/ROX (Invitrogen). The experiment was performed on three independent biological replicates. The c-Fos or EGR-1 transcript level was normalized with the endogenous GAPDH control according to the 2−-ΔΔCt method (18). The results were compared by one-way analysis of variance (ANOVA) followed by Tukey's pairwise comparisons testing of significance between selected groups. The differences observed were considered to be statistically significant at p < 0.05.
Construction of Stable Cell Line
Flip-InTM T-RexTM-293 cells contain a single stably integrated FRT site at a transcriptionally active genomic locus. For targeted integration of F-box deleted (ΔF) fbxo25 into the FRT site, we carried out cotransfection of FBXO25-ΔF-box/pcDNA5/FRT/TO vector and Flp recombinase vector pOG44 by using Lipofectamine 2000 (Invitrogen). Flip-InTM T-RexTM-293 cells cotransfected with pcDNA5/FRT/TO empty vector and Flp recombinase were used as control. Forty-eight hours after transfection, 400 μg/ml hygromycin was added to the medium for selection of the cells with integrated sequences coding for FBXO25-ΔF-box. After 2 weeks of antibiotic selection the colonies were picked and expanded to confirm tetracycline-dependent expression of FBXO25-ΔF-box.
In Vitro Ubiquitination of Purified ELK-1
HEK293T cells were transfected with ELK-1-FLAG-His6 and rinsed with lysis buffer. The supernatants were immunoprecipitated by anti-FLAG M2 beads, and the ELK-1-FLAG-His6 was eluted with 100 μg/ml 3×FLAG peptide (Sigma-Aldrich) in Tris-buffered saline (TBS; 50 mm Tris-HCl, pH 7.5, 0.15 m NaCl). The in vitro ubiquitination reactions were developed as follows. Purified SCF1(FBXO25) or SCF1(FBXO25-ΔF-box) + ubiquitin mix (100 ng of E1 + 500 ng of E2 (UbcH5c) + 1 μg of native ubiquitin + 0.5 μg of N-terminal biotin-ubiquitin and ubiquitination buffer) were incubated for 90 min at 30 °C. The proteins were loaded in SDS-PAGE, and the Western blots were probed with anti-polyubiquitin FK2 antibodies (Millipore) and with streptavidin-peroxidase (Thermo Pierce).
In Cellulo Ubiquitination of ELK-1
HEK293T cells were transfected with ELK-1-FLAG-His6 and empty vector (pcDNA3) or GST-HA-FBXO25-FLAG or GST-HA-FBXO25-ΔF-box-FLAG. Six hours before lysis the cells were treated with epoxomicin and lysed with radioimmune precipitation assay buffer, sonicated, and boiled prior to immunoprecipitation. Lysates were cleared by centrifugation and immunoprecipitated with anti-ELK-1 antibodies (Millipore). Then, the resins were washed with radioimmune precipitation assay buffer, and SDS-PAGE load buffer was added. The Western blotting was developed by probing the membranes with anti-polyubiquitin FK2 antibodies (Millipore).
RESULTS
Identification of SCF1(FBXO25) Ubiquitinated Proteins
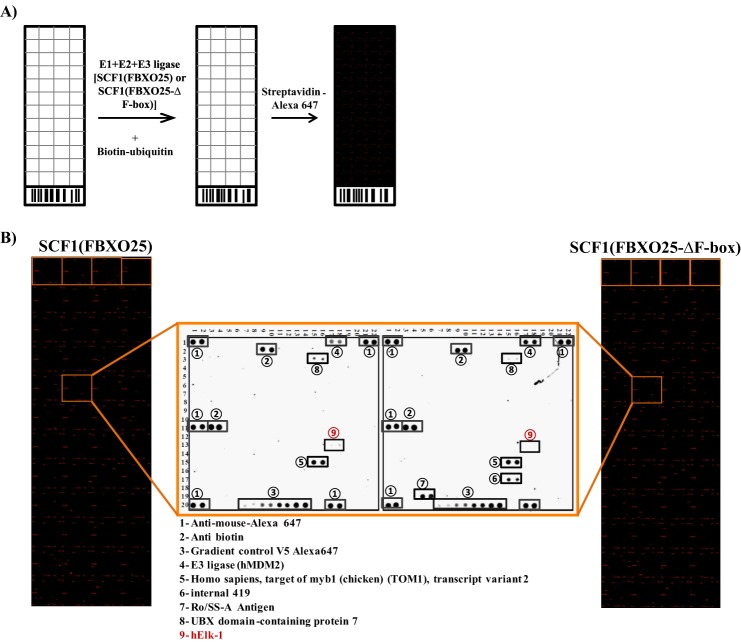
To identify FBXO25 substrate candidates, we have used the human protoarray as target and the purified SCF1(FBXO25) complexes as a source of E3 ligases (Fig. 1A). The protoarrays are based on a robust technology described previously (19) containing >8000 unique full-length proteins spotted in duplicate, covering almost the entire human proteome. We carried out the substrate search using four protoarrays as follows. Two arrays were probed with SCF1 complexes containing FBXO25 wild type (WT) and two negative controls containing SCF1(FBXO25-ΔF-box), depleted of F-box domain (ΔF), or SCF1(FBXO25), with E2DN (dominant negative, unable to transfer ubiquitin). Using the statistical criteria described under “Experimental Procedures,” 75 putative substrates of FBXO25 were identified with high confidence (supplemental Table 1).
FIGURE 1.
The protein ELK-1 was ubiquitinated in the protoarrays by SCF1(FBXO25). A, experimental scheme of the ubiquitination in vitro reaction using the protoarray and SCF1 complexes. The ubiquitin mix (E1+E2 or E2DN+ATP+biotin-ubiquitin+ubiquitin) was added with the SCF1 complexes wild type or mutant in the ubiquitination reaction. Furthermore, the slides were incubated with streptavidin-Alexa Fluor 647 and scanned for visualization of the ubiquitinated proteins. B, protoarray slides ubiquitinated by SCF1(FBXO25) or SCF1(FBXO25-ΔF-box) highlighting a block with proteins differentially ubiquitinated by the wild type SCF1 complex. The protein ELK1, a member of ETS oncogene family, was differentially ubiquitinated by SCF1(FBXO25) (n = 2). The other revealed spots were loading controls or ubiquitin-binding proteins spotted in the slides.
The Protein ELK-1 Is a Potential Target of FBXO25
Because ELK-1 is a nuclear protein that has been reported to be a ubiquitin proteasome system substrate (19), we chose to further explore the possibility of ELK-1 being a FBXO25 substrate. ELK-1 is a ternary complex factor subfamily of ETS domain transcription factors that plays an important role in the induction of immediate early gene expression in response to a extracellular signal (20). ELK-1 was specifically ubiquitinated in the protoarray by SCF1(FBXO25) but not by SCF1(FBXO25-ΔF-box) (Fig. 1B) or SCF1 (FBXO25) with E2DN. This last negative control was used to exclude from the final hits any unspecific target such as those that got a biotin-ubiquitin directly from E1 or were able to keep bound with this molecule and after the washes.
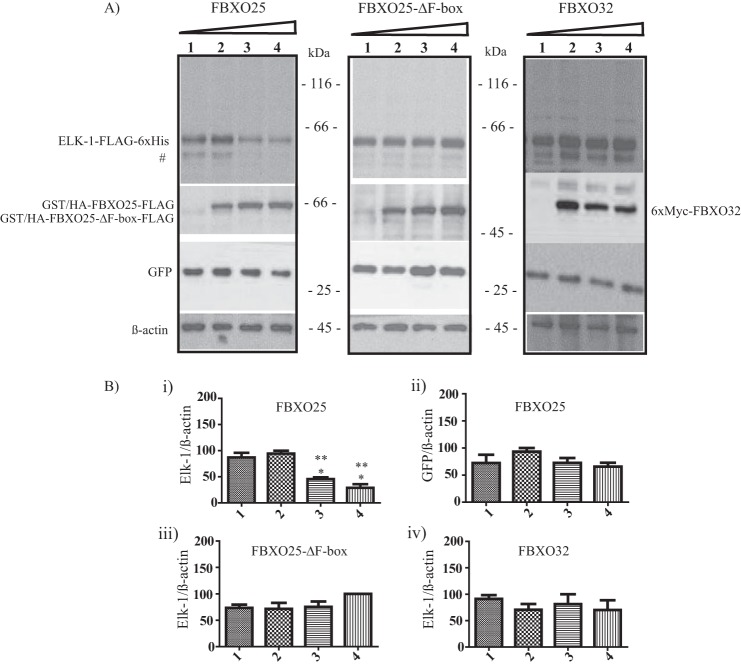
To evaluate the functional relationship of these proteins, we cotransfected HEK293T cells with ELK-1-FLAG-His6 and GFP in combination with increasing amounts of FBXO25 or FBXO25-ΔF-box or FBXO32. A statistically significant reduction of overexpressed ELK-1 protein levels in the presence of FBXO25 was observed in a dose-dependent manner (Fig. 2, left panel and graphic). In contrast, FBXO25-ΔF-box had a weak effect on the levels of ELK-1 protein (Fig. 2, middle panel and graphic), which is in agreement with the inability of the ΔF-box mutant version of FBOX25 to assemble in an active SCF1 complex to promote ubiquitination and degradation of its substrates.
FIGURE 2.
ELK-1 degradation is mediated by FBXO25. A, HEK293T cells were cotransfected with 0.25 μg of ELK-1, 0.50 μg of GFP, in combination with increasing amounts (0, 0.5, 1.0, and 2.0 μg) of FBXO25 or FBXO25-ΔF-box or FBXO32 as indicated. After 48 h the supernatant was obtained and used in Western blotting analyses with the indicated antibodies. For all experiments n = 3. #, this additional band detected by antibodies, possibly represents a smaller isoform of ELK-1 that arises from an internal translation start codon in the elk-1 sequence (41). B, the graphics were obtained from densitometry of ELK-1-FLAG-His6 using β-actin to normalization. *, p < 0.05 compared with lane 1 by one-way ANOVA followed by Tukey's post test; **, p < 0.05 compared with lane 2 by one-way ANOVA followed by Tukey's post test.
We sought to evaluate the specificity of ELK-1 degradation by FBXO25 through its cotransfection with the FBXO25 closest protein family member FBXO32. The overexpression of FBXO32 had no effect on the levels of the ELK-1, confirming the FBXO25 selectivity toward their substrates (Fig. 2, right panel and graphic). Moreover, FBXO25 did not change the steady-state levels of an irrelevant control GFP protein significantly (Fig. 2, right panel and graphic Bii), corroborating the specificity of ELK-1 degradation by FBXO25. Overall, these findings suggested that FBXO25 triggers the ELK-1 degradation, and this effect was dependent on its F-box domain, suggesting that ELK-1 is a substrate of an active SCF1(FBXO25) complex.
FBXO25 Interacts with and Destabilizes ELK-1 by Proteasome-dependent Degradation
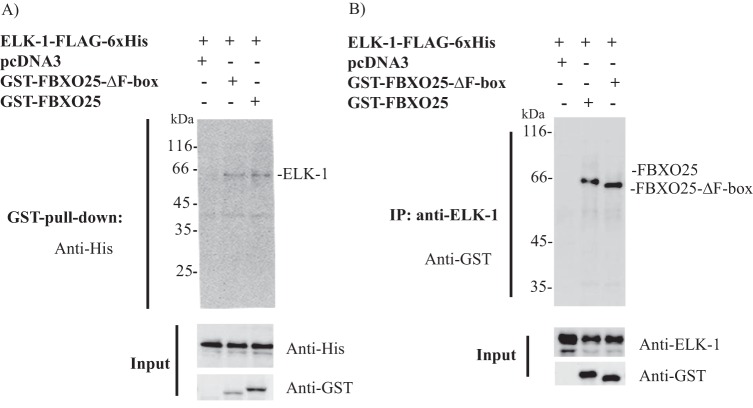
The F-box family of proteins are known to use protein-protein domains to bind tightly and specifically to their protein targets (4, 5). We sought to determine whether FBXO25 interacts with ELK-1 through GST pulldown assays by using cell lysates from HEK293T cotransfected with ELK-1-FLAG-His6 in combination with pcDNA3 or FBXO25 or FBXO25-ΔF-box. GST-FBXO25 was purified using glutathione-Sepharose beads, and ELK-1-FLAG-His6 was detected by Western blotting using anti-His tag antibodies. Consistent with the results shown above, FBXO25 copurified with ELK-1, indicating that they are protein partners (Fig. 3A). To confirm the specificity of this association, we performed a reverse coimmunoprecipitation using HEK293T cell lysates transfected with the same combination of plasmids. Antibodies anti-ELK-1 copurified ELK-1-FLAG-His6 and GST-FBXO25 and FBXO25-ΔF-box (Fig. 3B). It is important to mention that ELK-1 was found associated with GST-FBXO25-ΔF-box in both types of copurification assays, highlighting its possibility to work as a dominant negative by competing with the endogenous FBXO25.
FIGURE 3.
FBXO25 and FBXO25-ΔF-box interacts with ELK-1. A, HEK293T cells transfected with the indicated plasmids. Six hours before cell lysis the proteasome inhibitor epoxomicin was added, and the supernatants were obtained and submitted to GST pulldown. The eluted fraction obtained by reduced glutathione was used in Western blotting analyses with the indicated antibodies. B, immunoprecipitation (IP) of ELK-1 using anti-ELK-1 antibodies from HEK293T cells lysates transfected with indicated plasmids and probed with anti-GST and anti-ELK-1 antibodies.
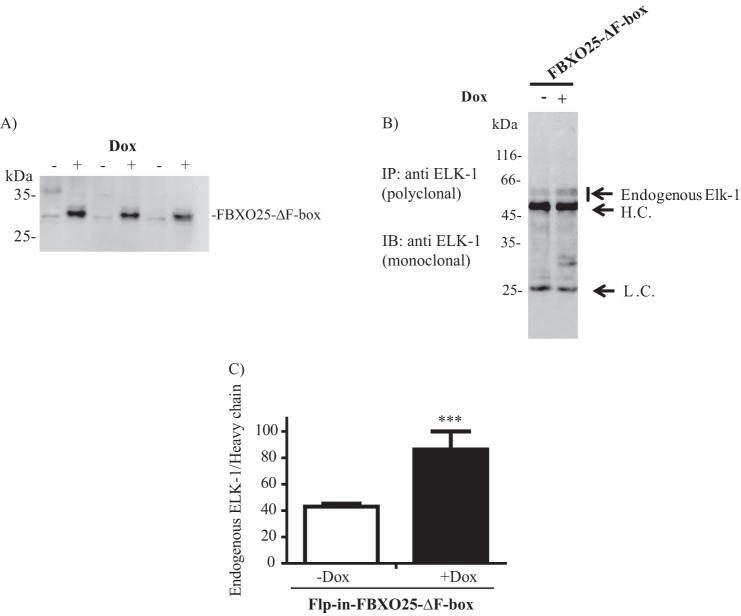
To confirm the suggested dominant negative function of FBXO25-ΔF-box on ELK-1 degradation we generated HEK293T cells expressing stably and inductively this mutant form (Fig. 4A). In this system only clones with integrated genes were selected, avoiding the contamination signal of untransfected cells. The endogenous ELK-1 protein content was evaluated after immunoprecipitation with anti-ELK-1 antibodies before and after induction of the mutant FBXO25 gene with doxycycline. We observed in FBXO25-ΔF-box-induced cells an accumulation of ELK-1, indicating that this mutant was able to protect endogenous ELK-1 from degradation (Fig. 4, B and C).
FIGURE 4.
FBXO25-ΔF-box induction promotes endogenous ELK-1 accumulation in mammalian cells. A, characterization of Flip-InTM T-RexTM-293 cell line (Flp-In-FBXO25-ΔF-box) which stably expresses FBXO25-ΔF-box in a doxycycline-inducible manner. The cells were induced with doxycycline (+Dox) or not (−Dox) for 48 h, and the lysates were submitted to Western blotting (IB) using anti-FBXO25 antibodies. B, protection of endogenous ELK-1 by induced FBXO25-ΔF-box visualized by Western blotting analysis of endogenous ELK-1 immunoprecipitates (IP). C, densitometric graphics analysis of the blot indicated in B. H.C., heavy chain; L.C., light chain. For all experiments n = 3. ***, p < 0.05 compared with untreated by one-way ANOVA followed by Tukey's post test.
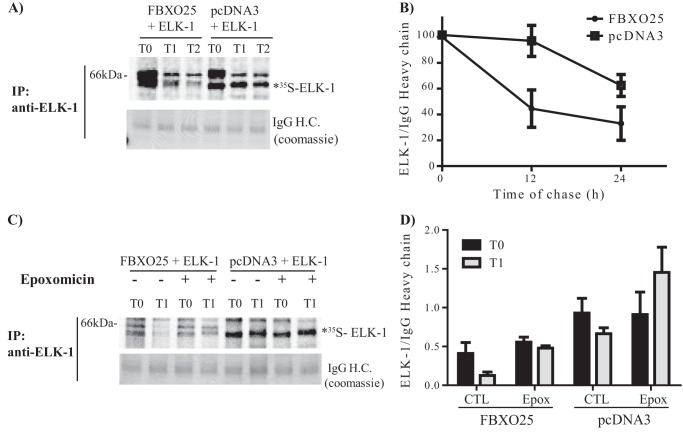
As the F-box proteins catalyze ubiquitination and degradation of their target proteins by the ubiquitin proteasome system, we surmise that the overexpression of FBXO25 would increase the turnover of ELK-1. To evaluate this possibility, we performed methionine chase labeling experiments in cells overexpressing ELK-1-FLAG-His6 either with FBXO25 or pcDNA3. The rate of 35S-labeled ELK-1-FLAG-His6 degradation was increased in cells overexpressing FBXO25 compared with those transfected with empty vector (Fig. 5, A and B). Treatment of the cells with the irreversible, highly specific, proteasome inhibitor epoxomicin blocked ELK-1 proteolysis. These data strongly suggest that the ELK-1 degradation was mediated by FBXO25 and is proteasome-dependent (Fig. 5, C and D). It is interesting to point out that the different levels of immunoprecipitated ELK-1-FALG-His6 comparing FBXO25 and pcDNA3 cotransfection in the T0 should be due to ELK-1 degradation during the 24 h of cotransfection prior to the methionine chase assay.
FIGURE 5.
The degradation of ELK-1 is proteasome-dependent and mediated by FBXO25. The methionine chase labeling experiments were performed using HEK293T cells transfected with the indicated plasmids. After the indicated times, the supernatants were obtained and submitted to immunoprecipitation (IP) with anti-ELK-1 antibody. The immunoprecipitated proteins were visualized by Coomassie Blue, and dried gels were film-exposed to protein visualization. A, FBXO25 and not pcDNA3 mediated ELK-1 degradation. B, 35S-labeled ELK-1-FLAG-His6 is degraded in the presence of FBXO25 after 12 h of transfection but not by pcDNA3 as indicated in densitometry graphics. C, 35S-labeled ELK-1 degradation mediated by FBXO25 is proteasome-dependent. D, the proteasome inhibitor epoxomicin protected 35S-labeled ELK-1-FLAG-His6 of degradation by FBXO25 as indicated in densitometry graphics. For all experiments n = 2. H.C., heavy chain. *, the apparent molecular mass of the radiolabeled band ELK-1-FLAG-His6 on the gels was ascertained by comparison with the mobility of FLAG affinity-purified ELK-1-FLAG-His6 (data not shown).
ELK-1 Is Ubiquitinated by FBXO25
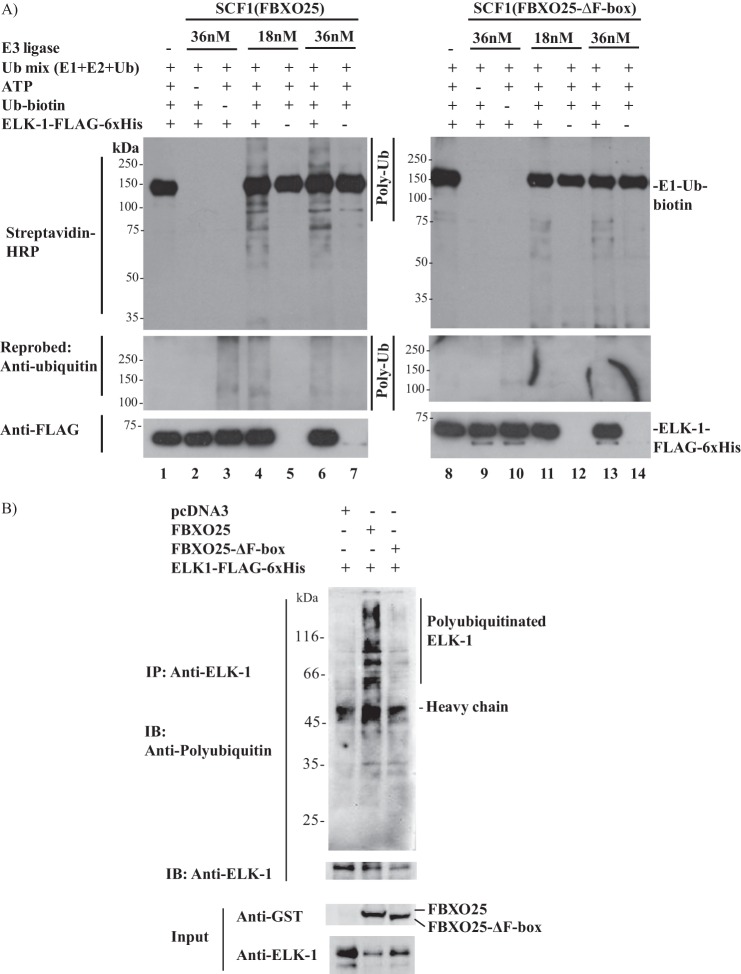
The proteins spotted in the protoarrays are in fusion with GST, and they were purified from insect cells using the baculovirus expression system (Invitrogen information). To eliminate any artifact influence of GST tag or modifications in ELK-1 imposed by this expression system in our in vitro ubiquitination assay, we expressed and purified ELK-1-FLAG-His6 from HEK293T cells, the same cell line used in all experiments (data not shown). The ELK-1 protein purified from HEK293T cells was used as substrate for the in vitro ubiquitination assay. Two different concentrations of SCF1 complexes were used in the presence or absence of ELK-1, ATP, or biotin-ubiquitin (Fig. 6A). The protein ELK-1 was ubiquitinated by SCF1(FBXO25) (Fig. 6A, lanes 4 and 6) but not by ubiquitin mix (lanes 1 and 8) or SCF1(FBXO25-ΔF-box) (lanes 11 and 13). We also observed a more intense smear of polyubiquitinated ELK-1 in the higher concentration of wild type SCF1 complexes (lanes 4 and 6), but no differences were observed in the mutant complexes (lanes 11 and 13). No signal of polyubiquitination was visualized in the absence of ATP (lanes 2 and 9) or biotin-ubiquitin (lanes 3 and 10). We also probed these membranes with anti-ubiquitin antibody, and beyond the aforementioned ubiquitination we were able to see the polyubiquitinated ELK-1 in the reaction without biotin-ubiquitin but with native ubiquitin only in the SCF1(FBXO25) (compare lanes 3 and 10). These results further corroborated the present findings that ELK-1 is ubiquitinated in vitro by SCF1(FBXO25).
FIGURE 6.
In vitro and in cellulo ubiquitination of ELK-1 by SCF1(FBXO25). A, in vitro ubiquitination of ELK-1 by SCF1(FBXO25). Two different concentrations of affinity-purified SCF1(FBXO25) or SCF1(FBXO25-ΔF-box) complexes in combination with purified ELK-1-FLAG-His6 from HEK293T cells, Ub-mix (E1, E2, ubiquitin), biotin-ubiquitin, and ATP, were used in ubiquitination in vitro reactions as indicated. Polyubiquitinated ELK-1 by SCF1(FBXO25) and not by SCF1(FBXO25-ΔF-box) was visualized by Western blotting with streptavidin-HRP and anti-ubiquitin antibodies. B, in cellulo ubiquitination of ELK-1 by FBXO25. HEK293T cells were transfected with the indicated plasmids for 48 h. Six hours before lysis the cells were treated with epoxomicin, and the lysates were boiled to remove any noncovalently bound proteins. The supernatants were used in immunoprecipitation (IP) with polyclonal anti-ELK-1, and polyubiquitinated ELK-1 was visualized by Western blotting (IB) with anti-ubiquitin FK2 antibodies.
In an attempt to endorse the idea that FBXO25 promotes the ubiquitin-dependent degradation of ELK-1, we performed in cellulo ubiquitination experiments in HEK293T cells cotransfected with ELK-1-FLAG-His6 and FBXO25 or FBXO25-ΔF-box or empty vector. To guarantee the accumulation of polyubiquitinated ELK-1, the proteasome was inhibited before cell lysis. The lysates were subjected to ELK-1 immunoprecipitation followed by immunoblotting using the FK2 anti-ubiquitin antibodies. A notably more intense high molecular mass smear of ubiquitin-modified ELK-1 was observed in the presence of FBXO25 compared with the profile obtained with the empty vector or FBXO25-ΔF-box (Fig. 6B), indicating that FBXO25 mediated the in cellulo ubiquitination of ELK-1. Interestingly, the ELK-1 in the input cotransfected with FBXO25 was weaker compared with the empty vector or FBXO25-ΔF-box, indicating that the accumulation of the polyubiquitinated ELK-1 after proteasome inhibition which was evident after immunoprecipitation (Fig. 6B, middle lane).
FBXO25 Suppresses c-fos and egr-1 mRNA Induction
Transcriptions of the protooncogene c-fos or other immediate early genes are stimulated rapidly and transiently by mitogens or serum growth factors (20, 21). This expression is mediated by the ternary complex factors including the transcription factor ELK-1 (22, 23). The mitogen PMA can be used to activate protein kinase C and consequently, enhances the phosphorylation of ERK1/2 MAP kinase. It has been shown that ERK and ELK-1 are downstream targets of the protein kinase Cη in glioblastoma multiform cells stimulated with PMA (24). To uncover the biological significance of FBXO25-mediated degradation of ELK-1, we scrutinized whether overexpression of FBXO25 would affect c-fos and egr-1 gene expression in cells stimulated by PMA. Indeed, HEK293T cells transfected with FBXO25 but not with the empty vector were able to suppress the c-fos and egr-1 expression following PMA stimulation (Fig. 7, A and B). Thus, the present data suggest that the overexpression of FBXO25 led to ubiquitination and proteasomal degradation of ELK-1, which could account for a mechanism of regulation of these two genes expression after PMA stimulation (Fig. 7C).
FIGURE 7.
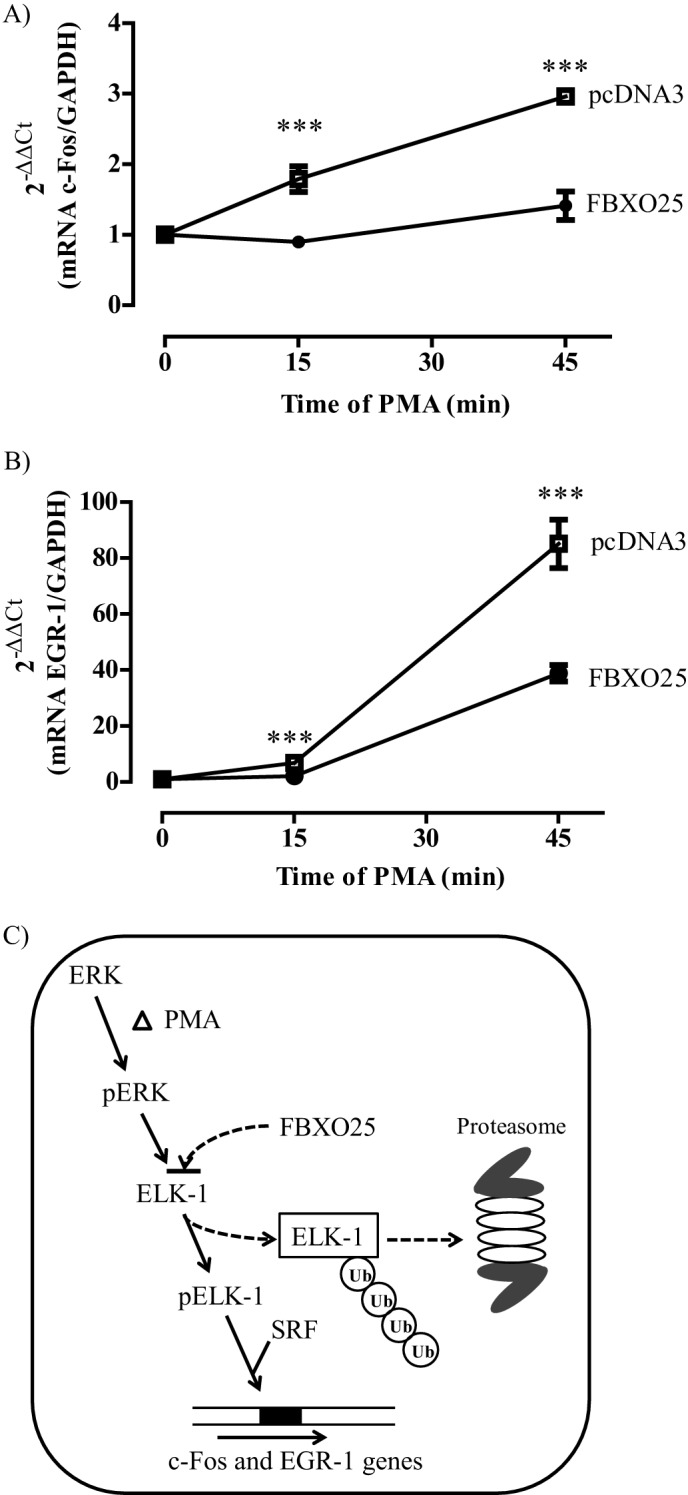
The overexpression of FBXO25 decreased c-fos and egr-1 expression in PMA-treated cells. A and B, HEK293T cells were transfected with pcDNA3 or FBXO25, and after serum starvation of 12 h the cells were stimulated with PMA at the indicated times. The total RNA was extracted at each time, and the expression of c-fos (A) and egr-1 (B) was evaluated by real time quantitative PCR using gapdh as housekeeping gene (n = 3). C, model shows the action of FBXO25 in regulating c-fos gene expression. PMA treatment induces phosphorylation and activation of elk-1 by pERK1/2. The FBXO25 protein promotes ELK-1 ubiquitination and degradation, which could account for the decrease of c-fos and egr-1 gene expression. ***, p < 0.05 compared with empty vector (pCDNA3) by one-way ANOVA followed by Tukey's post test.
DISCUSSION
The growing number of identified FBP substrates has introduced an exquisite cellular mechanism for substrate target selection. It has been shown that the substrate degradation depends on its phosphorylation state (25), cell cycle, and intracellular localization (26). In addition, the functions of some FBPs are cell type-dependent (27–29). In an attempt to bypass these constraints and identify new SCF1(FBXO25) substrates, the ubiquitination in chip approach has been used. These arrays contain most of the human proteome components immobilized directionally by an expression tag (GST) on nitrocellulose-coated glass slides (19), whose 9483 spotted proteins had been expressed and purified from insect cells with their native post-translational modifications, such as phosphorylation state, in accordance with the manufacturer's information. This technology has been used to identify substrates of purified HECT-type E3 ligases, which are composed of a single protein that recruits the E2-ubiquitin enzyme and promotes substrate ubiquitination (13, 15, 30). However, to date there are no reports addressing the use of these arrays for the identification of ubiquitinated substrates by cullin-RING E3 ligases (CRLs or SCF1).
We have identified 75 potential SCF1 (FBXO25) substrates using an active purified complex purified under native conditions by GST pulldown from extract of HEK293T cells cotransfected with SCF1 complex components, as described previously (9). Interestingly, from the ubiquitinated proteins on the arrays, we observed strong ubiquitin-labeled spots of the ubiquitin-binding proteins, such as E1, E2 RING-finger type, and HECT-type E3 ligases, indicating that these spotted proteins remained in their functional native structures. The signals of these ubiquitin-binding proteins were considered false hits, and therefore, they were not included as potential substrates. Ten proteins of the final hit list, which includes PADI4, HIP1, PRKCA, TNIP2, ANKRD13A, C6orf106, PSMD4, FKBP3, UBQLN1, and UBQLN2, were also identified as a SMURF1 targets using the same approach (13). These targets were not FBXO25-specific, but their ubiquitination was SCF1(FBXO25)- or SMURF1-dependent. In fact, some FBPs have been described as responsible for ubiquitination and degradation of the same substrates, such as cyclin D1, which is substrate of five different FBPs or c-Myc by two others, revised by (12). Post-translational modification of the targets as well as cell cycle stage and intracellular localization of the FBPs can be some of the reasons for this apparent promiscuity.
A proteome-wide analysis of endogenous ubiquitinome in HEK293T cells revealed 11,054 endogenous putative ubiquitination sites on 4273 human proteins by measuring the diglycine-modified lysines contents (31). Remarkably, we have found that 33% of the SCF1(FBXO25) ubiquitinated proteins on protoarrays (GTF2B, ODF2, SAMHD1, RPS10, ADRBK1, LUC7L, HP1BP3, TCEAL2, PAK4, DNAJB2, SERBP1, G3BP1, CDK9, PRRG1, CENPB, MAT2B, PPID, EIF4E2, DNAJC8, EGFR, and RABEP2) were found in the aforementioned ubiquitinome. This emphasizes the great potential of in chip ubiquitination assays for revealing substrates of E3 ligases. Surprisingly, the protein ELK-1 was not present in this ubiquitinome as well as other known low expression level FBP substrates. This low level of ELK-1 can explain why we did not find this protein in our FBXO25 interactome (9). Indeed, both ELK-1 protein contents and its phosphorylated state are tightly regulated to maintain the cellular functions such as proliferation, differentiation, and apoptosis in a normal state (32).
From the potential identified targets, we investigated ELK-1 to be validated as a substrate based on its SCF1(FBXO25)-related features, such as nuclear localization-dependent stability and its degradation mediated by proteasome (11, 33). We presented here a combination of two in vitro approaches to show the ubiquitination of ELK-1 by SCF1(FBXO25). First, the protoarrays were used as source of targets, and afterward, we performed the in vitro assay with purified ELK-1 from mammalian cells to guarantee that any modification of the protoarray targets, such as the tags, could not influence the ubiquitination signals. To unveil the relationship of these proteins in vivo, we also showed that SCF1(FBXO25) was able to ubiquitinate ELK-1 in a cellular environment. The ubiquitination of proteins by E3 ligases can be associated with their degradation by proteasome or regulation of their cellular functions. Here, we demonstrated by methionine chase that FBXO25 wild type increased the turnover of overexpressed ELK-1 in HEK293T cells, suggesting that its ubiquitination is related to degradation. In fact, the FBXO25-mediated ELK-1 degradation was completely blocked by epoxomicin, confirming that ELK-1 degradation was proteasome-dependent, as described previously (33).
An F-box deletion mutant of FBXO25, which interacts with ELK-1 but does not promote its ubiquitination, behaved in vivo as a dominant negative. The induction of FBXO25-ΔF-box by doxycycline in selected stable cell lineages increased the steady-state levels of endogenous ELK-1, suggesting a dominant negative effect of this mutant. Dominant negative mutants lacking the F-box domain have been widely used to confirm FBP substrates (34–36). On the other hand, different approaches aiming to reduce the F-box protein levels such as RNAi have been used extensively to evaluate substrate accumulation (37). However, this technology relies on the endogenous protein machinery to remove the existing protein and hence depends largely of its intrinsic stability (37). Interestingly, we have observed by specific RNAi as well as time course experiments by cycloheximide chase that FBXO25 is a very stable protein (data not shown), which affects the analysis of its substrate accumulation in response to mRNA depletion by RNAi. In addition, the use of genetic approaches such as knock-out mouse or mouse embryonic fibroblast cell lineages to evaluate the FBP substrate accumulation has unveiled conflicting results with the literature. The stability of cyclin D1 was not changed either when evaluated in knock-out mouse for each one of the five different FBPs described as responsible for its degradation or in mouse embryonic fibroblast cells from these animals in which a combination of three different FBPs knock-out was inserted (38).
It has been shown that two motifs in ELK-1 determine its premature degradation: the N-terminal region (residues 1–32) in the ETS domain which is responsible for ELK-1 dimerization in the cytosol, and a proline-rich hydrophobic motif (residues 187–216) between the B and S domains that could be the recognition element for E3 ligases (33). Interestingly, the FBXO25 prediction of functional motifs by the ELM (Eukaryotic Linear Motifs) software revealed a proline-rich binding motif in its structure (residues 343–348). As part of the interaction characterization, the mapping of the interaction domains between FBXO25 and ELK-1 remains to be done.
In cells stimulated by mitogen factors ELK-1 is phosphorylated by MAPK and recruited into the serum response factor complex, regulating the transcription of the immediate early genes, such as c-fos, egr-1, egr-2, and mcl-1 by interacting with their serum response element DNA regulatory site (20, 22, 23, 39, 40). We also demonstrated the biological relevance of FBXO25-mediated degradation of ELK-1 through the suppression of the ELK-1 target genes, c-fos and egr-1, after induction by PMA in cells overexpressing FBXO25. The role of FBXO25 in the functional processes mediated by immediate early gene induction regulated by ELK-1, such as cell proliferation, differentiation, and apoptosis, remains to be uncovered. In summary, the in vitro ubiquitination on protoarrays represents a powerful strategy to identify substrates of cullin-RING E3 ligases. We provided in vitro and in cellulo evidence that SCF1(FBXO25) mediates ubiquitination and proteasomal degradation of ELK-1 as well as the functional relevance of this effect in the cells.
Acknowledgments
We thank Dr. Eduardo B. Oliveira (Faculty of Medicine of Ribeirão Preto, University of São Paulo, Brazil) and Dr. Emer S. Ferro (Institute of Biomedical Sciences, University of São Paulo) for helpful discussion in the preparation of the manuscript; Dr. Andrew D. Sharrocks (University of Manchester) for the ELK-1-FLAG-His6 construct; Dr. Heike Laman (University of Cambridge) for support in the ubiquitination in vitro assay; and Cacilda Dias Pereira for excellent technical support. Array scanning was performed in the Dr. Gustavo H. Goldman laboratory (Faculty of Pharmaceutical Sciences of Ribeirao Preto, University of São Paulo) with the technical assistance of Dr. Marcia Eliana da Silva Ferreira Balieiro.
This work was supported by Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) Grant 201013045-4, CNPq Grant GENOPROT 42/2009, Fundação de Apoio ao Ensino, Pequisa e Assistência (FAEPA), and the University of São Paulo through the Núcleo de Apoio à Pesquisa na Interface Proteólise Sinalização Celular (NAPPS).

This article contains supplemental Table 1.
- RING
- really interesting new gene
- ANOVA
- analysis of variance
- CUL1
- Cullin1
- SCF1
- Skp1, Rbx1, Cullin1, and F-box protein
- ETS
- E-twenty-six
- FAND
- FBXO25-associated nuclear domain
- FBP
- F-box protein
- FRT
- Flp recognition target
- PMA
- phorbol 12-myristate 13-acetate
- SKP
- S phase kinase-associated protein.
REFERENCES
- 1. Ciechanover A. (1994) The ubiquitin-proteasome proteolytic pathway. Cell 79, 13–21 [DOI] [PubMed] [Google Scholar]
- 2. Hershko A., Ciechanover A. (1992) The ubiquitin system for protein degradation. Annu. Rev. Biochem. 61, 761–807 [DOI] [PubMed] [Google Scholar]
- 3. Petroski M. D., Deshaies R. J. (2005) Function and regulation of cullin-RING ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 6, 9–20 [DOI] [PubMed] [Google Scholar]
- 4. Cenciarelli C., Chiaur D. S., Guardavaccaro D., Parks W., Vidal M., Pagano M. (1999) Identification of a family of human F-box proteins. Curr. Biol. 9, 1177–1179 [DOI] [PubMed] [Google Scholar]
- 5. Winston J. T., Koepp D. M., Zhu C., Elledge S. J., Harper J. W. (1999) A family of mammalian F-box proteins. Curr. Biol. 9, 1180–1182 [DOI] [PubMed] [Google Scholar]
- 6. Cleveland B. M., Evenhuis J. P. (2010) Molecular characterization of atrogin-1/F-box protein-32 (FBXO32) and F-box protein-25 (FBXO25) in rainbow trout (Oncorhynchus mykiss): expression across tissues in response to feed deprivation. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 157, 248–257 [DOI] [PubMed] [Google Scholar]
- 7. Cardozo T., Pagano M. (2004) The SCF ubiquitin ligase: insights into a molecular machine. Nat. Rev. Mol. Cell Biol. 5, 739–751 [DOI] [PubMed] [Google Scholar]
- 8. Maragno A. L., Baqui M. M., Gomes M. D. (2006) FBXO25, an F-box protein homologue of atrogin-1, is not induced in atrophying muscle. Biochim. Biophys. Acta 1760, 966–972 [DOI] [PubMed] [Google Scholar]
- 9. Teixeira F. R., Yokoo S., Gartner C. A., Manfiolli A. O., Baqui M. M., Assmann E. M., Maragno A. L., Yu H., de Lanerolle P., Kobarg J., Gygi S. P., Gomes M. D. (2010) Identification of FBXO25-interacting proteins using an integrated proteomics approach. Proteomics 10, 2746–2757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gomes M. D., Lecker S. H., Jagoe R. T., Navon A., Goldberg A. L. (2001) Atrogin-1, a muscle-specific F-box protein highly expressed during muscle atrophy. Proc. Natl. Acad. Sci. U.S.A. 98, 14440–14445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Manfiolli A. O., Maragno A. L., Baqui M. M., Yokoo S., Teixeira F. R., Oliveira E. B., Gomes M. D. (2008) FBXO25-associated nuclear domains: a novel subnuclear structure. Mol. Biol. Cell 19, 1848–1861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Skaar J. R., D'Angiolella V., Pagan J. K., Pagano M. (2009) SnapShot: F box proteins II. Cell 137, 1358. [DOI] [PubMed] [Google Scholar]
- 13. Andrews P. S., Schneider S., Yang E., Michaels M., Chen H., Tang J., Emkey R. (2010) Identification of substrates of SMURF1 ubiquitin ligase activity utilizing protein microarrays. Assay Drug Dev. Technol. 8, 471–487 [DOI] [PubMed] [Google Scholar]
- 14. Lu J. Y., Lin Y. Y., Qian J., Tao S. C., Zhu J., Pickart C., Zhu H. (2008) Functional dissection of a HECT ubiquitin E3 ligase. Mol. Cell. Proteomics 7, 35–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Merbl Y., Kirschner M. W. (2009) Large-scale detection of ubiquitination substrates using cell extracts and protein microarrays. Proc. Natl. Acad. Sci. U.S.A. 106, 2543–2548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Persaud A., Alberts P., Amsen E. M., Xiong X., Wasmuth J., Saadon Z., Fladd C., Parkinson J., Rotin D. (2009) Comparison of substrate specificity of the ubiquitin ligases Nedd4 and Nedd4-2 using proteome arrays. Mol. Syst. Biol. 5, 333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sambrook J., Fritsch E. F., Maniatis T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Ed, Cold Spring Harbor Laboratory, Cold Spring Harbor, New York [Google Scholar]
- 18. Livak K. J., Schmittgen T. D. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 19. Zhu H., Bilgin M., Bangham R., Hall D., Casamayor A., Bertone P., Lan N., Jansen R., Bidlingmaier S., Houfek T., Mitchell T., Miller P., Dean R. A., Gerstein M., Snyder M. (2001) Global analysis of protein activities using proteome chips. Science 293, 2101–2105 [DOI] [PubMed] [Google Scholar]
- 20. Gille H., Sharrocks A. D., Shaw P. E. (1992) Phosphorylation of transcription factor p62TCF by MAP kinase stimulates ternary complex formation at c-fos promoter. Nature 358, 414–417 [DOI] [PubMed] [Google Scholar]
- 21. Marais R., Wynne J., Treisman R. (1993) The SRF accessory protein Elk-1 contains a growth factor-regulated transcriptional activation domain. Cell 73, 381–393 [DOI] [PubMed] [Google Scholar]
- 22. Dalton S., Treisman R. (1992) Characterization of SAP-1, a protein recruited by serum response factor to the c-Fos serum response element. Cell 68, 597–612 [DOI] [PubMed] [Google Scholar]
- 23. Hipskind R. A., Rao V. N., Mueller C. G., Reddy E. S., Nordheim A. (1991) Ets-related protein Elk-1 is homologous to the c-Fos regulatory factor p62TCF. Nature 354, 531–534 [DOI] [PubMed] [Google Scholar]
- 24. Uht R. M., Amos S., Martin P. M., Riggan A. E., Hussaini I. M. (2007) The protein kinase Cη isoform induces proliferation in glioblastoma cell lines through an ERK/Elk-1 pathway. Oncogene 26, 2885–2893 [DOI] [PubMed] [Google Scholar]
- 25. Viñas-Castells R., Beltran M., Valls G., Gómez I., García J. M., Montserrat-Sentís B., Baulida J., Bonilla F., de Herreros A. G., Díaz V. M. (2010) The hypoxia-controlled FBXL14 ubiquitin ligase targets SNAIL1 for proteasome degradation. J. Biol. Chem. 285, 3794–3805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kamura T., Hara T., Matsumoto M., Ishida N., Okumura F., Hatakeyama S., Yoshida M., Nakayama K., Nakayama K. I. (2004) Cytoplasmic ubiquitin ligase KPC regulates proteolysis of p27Kip1 at G1 phase. Nat. Cell Biol. 6, 1229–1235 [DOI] [PubMed] [Google Scholar]
- 27. Laman H., Funes J. M., Ye H., Henderson S., Galinanes-Garcia L., Hara E., Knowles P., McDonald N., Boshoff C. (2005) Transforming activity of Fbxo7 is mediated specifically through regulation of cyclin D/cdk6. EMBO J. 24, 3104–3116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lomonosov M., Meziane el K., Ye H., Nelson D. E., Randle S. J., Laman H. (2011) Expression of FBXO7 in haematopoietic progenitor cells cooperates with p53 loss to promote lymphomagenesis. PLoS One 6, e21165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Meziane el K., Randle S. J., Nelson D. E., Lomonosov M., Laman H. (2011) Knockdown of Fbxo7 reveals its regulatory role in proliferation and differentiation of haematopoietic precursor cells. J. Cell Sci. 124, 2175–2186 [DOI] [PubMed] [Google Scholar]
- 30. Phizicky E., Bastiaens P. I., Zhu H., Snyder M., Fields S. (2003) Protein analysis on a proteomic scale. Nature 422, 208–215 [DOI] [PubMed] [Google Scholar]
- 31. Wagner S. A., Beli P., Weinert B. T., Nielsen M. L., Cox J., Mann M., Choudhary C. (2011) A proteome-wide, quantitative survey of in vivo ubiquitylation sites reveals widespread regulatory roles. Mol. Cell. Proteomics 10.1074/mcp.M111.013284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Shao N., Chai Y., Cui J. Q., Wang N., Aysola K., Reddy E. S., Rao V. N. (1998) Induction of apoptosis by Elk-1 and δElk-1 proteins. Oncogene 17, 527–532 [DOI] [PubMed] [Google Scholar]
- 33. Evans E. L., Saxton J., Shelton S. J., Begitt A., Holliday N. D., Hipskind R. A., Shaw P. E. (2011) Dimer formation and conformational flexibility ensure cytoplasmic stability and nuclear accumulation of Elk-1. Nucleic Acids Res. 39, 6390–6402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jin J., Ang X. L., Shirogane T., Wade Harper J. (2005) Identification of substrates for F-box proteins. Methods Enzymol. 399, 287–309 [DOI] [PubMed] [Google Scholar]
- 35. Lin D. I., Barbash O., Kumar K. G., Weber J. D., Harper J. W., Klein-Szanto A. J., Rustgi A., Fuchs S. Y., Diehl J. A. (2006) Phosphorylation-dependent ubiquitination of cyclin D1 by the SCF(FBX4-αB crystallin) complex. Mol. Cell 24, 355–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Carrano A. C., Eytan E., Hershko A., Pagano M. (1999) SKP2 is required for ubiquitin-mediated degradation of the CDK inhibitor p27. Nat. Cell Biol. 1, 193–199 [DOI] [PubMed] [Google Scholar]
- 37. Zhang J., Zheng N., Zhou P. (2003) Exploring the functional complexity of cellular proteins by protein knockout. Proc. Natl. Acad. Sci. U.S.A. 100, 14127–14132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kanie T., Onoyama I., Matsumoto A., Yamada M., Nakatsumi H., Tateishi Y., Yamamura S., Tsunematsu R., Matsumoto M., Nakayama K. I. (2012) Genetic reevaluation of the role of F-box proteins in cyclin D1 degradation. Mol. Cell. Biol. 32, 590–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Townsend K. J., Zhou P., Qian L., Bieszczad C. K., Lowrey C. H., Yen A., Craig R. W. (1999) Regulation of MCL1 through a serum response factor/Elk-1-mediated mechanism links expression of a viability-promoting member of the BCL2 family to the induction of hematopoietic cell differentiation. J. Biol. Chem. 274, 1801–1813 [DOI] [PubMed] [Google Scholar]
- 40. Janknecht R., Ernst W. H., Pingoud V., Nordheim A. (1993) Activation of ternary complex factor Elk-1 by MAP kinases. EMBO J. 12, 5097–5104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Vanhoutte P., Nissen J. L., Brugg B., Gaspera B. D., Besson M. J., Hipskind R. A., Caboche J. (2001) Opposing roles of Elk-1 and its brain-specific isoform, short Elk-1, in nerve growth factor-induced PC12 differentiation. J. Biol. Chem. 276, 5189–5196 [DOI] [PubMed] [Google Scholar]