Background: YidC is required for LacY folding, but the details of this process are not clear.
Results: YidC binds multiple LacY TM domains and is involved in LacY folding but not insertion.
Conclusion: YidC binding to LacY directs the proper helix-helix interactions of LacY.
Significance: This is the first detailed in vivo analysis of LacY insertion and YidC-LacY interactions.
Keywords: Chaperone, Chaperonin, Membrane Biogenesis, Membrane Enzymes, Membrane Proteins, Protein Folding, LacY, YidC, Membrane Insertion
Abstract
To understand how YidC and SecYEG function together in membrane protein topogenesis, insertion and folding of the lactose permease of Escherichia coli (LacY), a 12-transmembrane helix protein LacY that catalyzes symport of a galactoside and an H+, was studied. Although both the SecYEG machinery and signal recognition particle are required for insertion of LacY into the membrane, YidC is not required for translocation of the six periplasmic loops in LacY. Rather, YidC acts as a chaperone, facilitating LacY folding. Upon YidC depletion, the conformation of LacY is perturbed, as judged by monoclonal antibody binding studies and by in vivo cross-linking between introduced Cys pairs. Disulfide cross-linking also demonstrates that YidC interacts with multiple transmembrane segments of LacY during membrane biogenesis. Moreover, YidC is strictly required for insertion of M13 procoat protein fused into the middle cytoplasmic loop of LacY. In contrast, the loops preceding and following the inserted procoat domain are dependent on SecYEG for insertion. These studies demonstrate close cooperation between the two complexes in membrane biogenesis and that YidC functions primarily as a foldase for LacY.
Introduction
In bacteria, membrane proteins are inserted into the membrane by two evolutionarily conserved pathways, the SecYEG and the YidC pathways (for reviews, see Refs. 1–4). In the Sec pathway, the Sec translocase is the principal translocase and plays a critical role in membrane insertion of proteins into the plasma membrane in bacteria and archaea, the endoplasmic reticulum in eukaryotes, and the chloroplast thylakoid membrane in plants (5). In the YidC pathway, the YidC insertase (known as Oxa1 in mitochondria and Alb3 in chloroplasts) catalyzes the insertion and folding of proteins into the bacterial plasma (cytoplasmic) membrane as well as the inner membrane of mitochondria and the thylakoid membrane in chloroplasts (6, 7).
The Sec translocase is comprised of the protein-conducting SecYEG channel and the trimeric SecDF-YajC complex in addition to the peripheral protein subunit SecA. The SecYEG channel functions to translocate the periplasmic loops of membrane proteins across the membrane and to integrate hydrophobic segments into the lipid bilayer (8). In some cases, SecA, the motor ATPase, is required for translocating periplasmic loops of membrane proteins (9–11). SecA uses the energy from ATP hydrolysis to promote translocation of the polypeptide chain through the SecYEG channel in step sizes of 20–30 amino acid residues (12). Finally, SecDF enhances protein translocation and membrane insertion (13, 14) by using the electrochemical proton gradient to drive conformational changes within the SecDF P1 preprotein-interacting domain (15).
Escherichia coli YidC is a 60-kDa protein with six transmembrane (TM)2 helices that may function as a hydrophobic platform to promote insertion of membrane proteins into the lipid bilayer (16, 17). The YidC insertase can autonomously insert phage coat proteins (18–20), subunit c of ATP synthase (21–24), and the N-tail of CyoA (25–27) and MscL (28) into the E. coli inner membrane. YidC interacts with the hydrophobic region of membrane protein substrates during insertion via residues in TM1, TM3, TM4, and TM5 (29, 30).
In addition to acting alone, YidC can function in concert with the Sec translocase to mediate membrane protein insertion and folding (31, 32). Sec-dependent substrates that also require YidC include subunit a of ATP synthase (21, 22), C-terminal domain CyoA (25, 26), LacY (32), MalF (33), and TatC (34). Cross-linking studies show that YidC contacts the transmembrane segments of membrane protein substrates as they insert into the membrane (35–37). Beck et al. (38) proposed, with mannitol permease, that YidC may act as an assembly site for the folding of α-helical bundles in membrane proteins, which may be why YidC is required for the folding and stability of polytopic membrane proteins such as LacY and MalF (32, 33). Interestingly, the translocase itself is very dynamic. Boyd and Koch (39) showed that the SecYEG translocase forms a stable complex with YidC in the presence of mannitol permease but not when the secretory ProOmpA is trapped in the SecYEG channel.
Previously, LacY has been shown to require the signal recognition particle (SRP)/FtsY components and the Sec translocase for membrane insertion (40–43). In addition, the involvement of YidC in the folding of LacY rather than insertion has been shown (32). However, it is not clear whether one or more of the periplasmic loops of LacY require YidC for translocation or whether the helix packing of LacY is perturbed by YidC depletion.
To define a precise role of YidC in the insertion and folding of the galactoside/H+ symporter LacY, we examined the translocation of each of the six periplasmic loops of LacY utilizing a Cys-based alkylation method (44). The findings demonstrate that YidC is not required for translocation of the periplasmic loops of Sec-dependent LacY but is necessary for proper folding. YidC can be disulfide-cross-linked to LacY, indicating that YidC makes contact with LacY during membrane biogenesis. Furthermore, YidC has the capacity to translocate a domain added to the middle cytoplasmic loop of a LacY chimera with an M13 procoat insertion. Membrane insertion of the procoat domain is strictly YidC-dependent, whereas the domains preceding and following the procoat domain are SecYEG-dependent. It is concluded that YidC and SecYEG translocases cooperate in the membrane insertion process and that YidC functions as a foldase for native LacY but can also function as an insertase for an internal loop of the LacY chimera.
EXPERIMENTAL PROCEDURES
Strains, Plasmids, and Materials
E. coli strains JS7131, CM124, and WAM121 were from our collection. E. coli FTL85 was a gift from Tracy Palmer. Lysozyme, amino acids, and Mal-PEG were purchased from Sigma. Trans-[35S]label, a mixture of 85% [35S]Met and 15% [35S]Cys, 1000 Ci/mmol, was from PerkinElmer Life Sciences. AMS (4-acetamido-4′-maleimidylstilbene-2,2′-disulfonic acid) was purchased from Invitrogen. Factor Xa and proteinase K solution were from New England Biolabs. N-ethylmaleimide (NEM) and Immunopure immobilized protein A AffinityPak columns were from Pierce. Monoclonal anti-Penta-His antibody was purchased from Qiagen. The pRSF-1b vector was from Novagen.
The pT7-5 expression vector containing the lacY cassette encoding Cys-less LacY (with a His tag at the C terminus) under the control of the T7/Lac promoter was from the Kaback collection. The heat-inducible T7 RNA polymerase expression vector pGP1–2 (KanR, p15A origin) was purchased from the ATCC. Transformation of pT7-5-LacY (AmpR, ColE1 origin) and pGP1–2 into the YidC depletion E. coli strain JS7131 allows expression of LacY. The construction of pLZ2-LacY, harboring both LacY with a His tag and the T7 RNA polymerase genes, is described in supplemental Fig. S1A and Methods.
The LacY cassette containing the lacY gene and T7/lac tandem promoter and the T7 RNA polymerase cassette were subcloned from pT7-5-LacY and pGP1–2, respectively, into the pRSF-1b vector (KanR, RSF origin), yielding the expression vector pLZ2-LacY. Transformation of the expression vector pLZ2-LacY (KanR, RSF origin) into the SecE depletion strain CM124 allows expression of LacY.
The T7 RNA polymerase expression cassette was subcloned from pGP1–2 to pACYC184, producing pLZ-t7 (CmR, p15A origin) as described in supplemental Fig. S1B and Methods. Transformation of WAM121 (45, 46) with pLZ-t7 and pT7-5-LacY allows expression of LacY.
pMS119 (AmpR, ColE1) harboring lacIq and the Tac promoter was used to express the procoat-leader peptidase fusion protein (PClep) WT and H5, in which the cytoplasmic region is extended by residues 223–323 of the P2 domain of leader peptidase (14). The genes encoding the procoat (WT) and procoat (H5) domains were PCR-amplified from pMS119 PClep (WT) and pMS119 PClep (H5) and used to make the LacY-procoat chimeras pT7-5-LacY-PC (WT) and pT7-5-LacY-PC (H5). The insertion point of the M13 phage gene 8 that encodes the procoat was 3′ of the Leu-210 codon of lacY in the pT7-5-LacY vector.
Growth Conditions
The YidC depletion strain JS7131 containing the pGP1–2 and pT7-5-LacY plasmids was grown at 30 °C in LB medium supplemented with 0.2% arabinose. To deplete YidC, overnight cultures of JS7131 cells were washed twice with fresh LB medium and then diluted 1:100 into LB medium with 0.2% glucose and grown for ∼5 h. The cells were then switched into M9 minimal medium supplemented with 0.5% fructose and 19 amino acids, excluding methionine (Met), and grown for an additional 30 min at 30 °C. Next, the cells were heat-shocked at 42 °C for 15 min to express T7 RNA polymerase and treated with 0.2 mg/ml rifampicin and 0.4 mm IPTG for another 15 min to suppress bacterial protein synthesis by inhibiting E. coli RNA polymerase activity. Cells expressing LacY were then pulse-labeled with trans-[35S]Met for 5 min at 30 °C.
The Ffh depletion strain WAM121 possessing the pT7-5-LacY and pLZ-t7 vectors was cultured by growing cells in LB medium containing 0.2% arabinose at 30 °C. Overnight cultures of WAM121 were diluted 1:100 in LB medium supplemented with 0.2% glucose or 0.2% arabinose. The cells were grown to mid-log phase at 30 °C and then switched to prewarmed M9 minimal medium. The cells were heat-shocked at 42 °C, treated with rifampicin and IPTG, and radiolabeled as described above for JS7131 cells. The SecE depletion strain CM124 bearing pLZ2-LacY was grown overnight at 30 °C in M9 medium containing 0.4% glucose and 0.2% arabinose. The overnight culture of CM124 cells was then diluted 1:100 and grown for 10–12 h at 30 °C in M9 medium containing 0.4% glucose plus 19 amino acids minus Met and supplemented with or without 0.2% arabinose. The cells were radiolabeled by using the rifampicin blocking technique as described above for YidC depletion.
The signal peptidase 1 (SP1) (also known as leader peptidase) depletion strain FTL85 (47) harboring pLZ-t7 and pT7-5-LacY-PC (WT) was grown in LB medium supplemented with 0.2% arabinose. The culture, grown overnight, was washed with fresh LB medium and diluted 1:100 in LB medium supplemented with 0.2% arabinose or 0.2% glucose. Cells were grown at 30 °C for 5 h and analyzed for LacY expression as described in the YidC depletion study. Where appropriate, ampicillin (100 μg/ml final concentration), kanamycin (50 μg/ml final concentration), and chloramphenicol (50 μg/ml final concentration) were added to the medium.
Protease Accessibility Studies
JS7131 cells containing pMS119 encoding PClep (WT) or (H5) were grown to mid-log phase under arabinose or glucose conditions. PClep was induced with 1 mm IPTG (final concentration) for 3 min before pulse-labeling for 1 min with trans-[35S]Met (0.2 mCi/ml final concentration). The radiolabeled cells were converted to spheroplasts, treated with 0.75 mg/ml proteinase K for 1 h, and then incubated with 5 mm PMSF (final concentration) to quench the protease (48). After quenching, the samples were immunoprecipitated with antibodies against leader peptidase (Lep) to precipitate PClep and against outer membrane protein A (OmpA). OmpA is digested in spheroplasts but not in intact cells and serves as a positive control for spheroplasts formation. The samples were then analyzed by SDS-PAGE using a 15% polyacrylamide gel and visualized by phosphorimaging.
Single and Double-Cys Mutants
The single-Cys mutants encoded by the plasmid pT7-5-LacY were constructed by site-directed mutagenesis using the Cys-less LacY expression vector pT7-5-LacY. Unless indicated otherwise, the Cys-less LacY expression vector pKR35 encoding LacY with a Factor Xa protease site in the cytoplasmic loop between TM6 and TM7 was used for the cross-linking studies (49). The double-Cys mutants were constructed using this pKR35 vector.
Site-directed Alkylation of Single-Cys Mutants of LacY
The radiolabeled cells expressing the LacY mutants with single Cys residues were split into four aliquots and pelleted, washed with 1 ml PBS (0.1 m sodium phosphate, 0.15 m sodium chloride (pH7.2)), and resuspended in 200 μl of PBS. Tube a was treated with 5 mm AMS (final concentration, dissolved in N,N-dimethyl formamide) to modify extracellular Cys residues. Tube b was treated with 20 mm NEM (final concentration) to modify intracellular and extracellular Cys residues. Tube c (negative control) and tube d (positive control) were left untreated. Tubes a and b were incubated at room temperature for 30 min. Cells were pelleted, washed twice with PBS, and resuspended in 25 mm Tris HCl (pH7.5). For each aliquot, crude membranes were isolated by centrifugation at 150,000 × g for 1 h after the cells were sonified. Membrane pellets were solubilized in 30 μl of 1% dodecyl-β-d-maltopyranoside (DDM)/20 mm Tris HCl (pH7.4). Tubes a, b, and d were incubated with 5 mm Mal-PEG (molecular weight 5000, Sigma) at 37 °C for 30 min. Tube c was also incubated with PEG5000 at 37 °C for 30 min as a mock treatment. All samples were mixed with 5× SDS gel loading dye, and the proteins were separated by SDS-PAGE followed by phosphorimaging.
Site-directed Cross-linking
Trans-[35S]Met-labeled cells expressing the double-Cys LacY with a Factor Xa protease site (unless indicated otherwise) in the middle of the large cytoplasmic loop were divided into five aliquots, and right side-out membrane vesicles were prepared by lysozyme-EDTA treatment and osmotic lysis (50, 51). The vesicles were resuspended in 50 mm potassium Pi (pH 6.6). For BMH cross-linking, the membrane vesicles were treated with 0.5 mm BMH (final concentration; dissolved in DMSO) at 25 °C for 10 min, and the reaction was stopped by addition of 10 mm DTT. For o-PDM and p-PDM, the reaction was carried out by adding the cross-linking reagent at a concentration of 0.5 mm to the membrane vesicles. The samples were incubated at 25 °C for 30 min, and the reactions were quenched by addition of 10 mm DTT. For disulfide cross-linking, the membranes were treated with 1 mm copper 1,10-phenanthroline (CuP) for 10 min at 25 °C. The 50 mm CuP stock solution was freshly prepared by mixing 225 mm 1,10-phenanthroline monohydrate in ethanol and 150 mm CuSO4 in water in a 2:1 ratio (v/v). The disulfide cross-linking reaction was terminated by the addition of 10 mm EDTA and 10 mm NEM. Following cross-linking, the membranes were isolated and solubilized in 30 μl of DDM buffer (20 mm Tris HCl (pH7.4), 100 mm NaCl, 2.0 mm CaCl2, and 1% DDM), and treated with Factor Xa protease (1 μg) overnight at 4 °C. The samples were mixed with 5× SDS gel loading dye and subjected to SDS-PAGE. The dried gels were scanned using a Typhoon PhosphorImager. The bands were then quantitated by using the public domain ImageJ software developed at the National Institutes of Health..
Immunoprecipitation of in Vivo-synthesized LacY
Monoclonal antibodies against LacY were purified by using Immunopure immobilized protein A AffinityPak columns and concentrated using Ambion Ultra columns (50 kDa molecular weight cutoff membrane, Millipore) as described (32). After radiolabeling, JS7131 cells expressing LacY were converted to spheroplasts as described previously. The spheroplasts were lysed in 50 mm Tris HCl (pH7.5), 150 mm NaCl, 1.0 mm EDTA, and 2% DDM and incubated overnight at 4 °C with a given monoclonal antibody. Supernatants isolated after ultracentrifugation were treated with protein A-Sepharose beads for 90 min at 4 °C. The beads were pelleted by a brief centrifugation, treated four times with wash buffer (50 mm Tris HCl (pH7.5), 150 mm NaCl, 1 mm EDTA, and 0.2% DDM) and resuspended in 1% DDM and 1.0 mm DDT. After addition of gel loading dye, the supernatant obtained from centrifugation was analyzed by SDS-PAGE and phosphorimaging.
Disulfide Cross-linking between YidC and LacY
Given combinations of YidC and LacY single-Cys mutants were analyzed by cotransforming the expression vectors pACYC184-YidC and pLZ2-LacY into the YidC depletion E. coli strain JS7131. The overnight culture was grown in LB medium supplemented with 0.2% glucose, 50 μg/ml kanamycin, and 50 μg/ml chloramphenicol at 30 °C. The overnight culture was then diluted into fresh medium and grown to an A600 of ∼0.6. The cells were switched into fresh M9 minimal medium and incubated at 30 °C for 30 min prior to rifampicin treatment, as described above. The cells were pulse-labeled for 5 min with trans-[35S]Met with or without 1 mm CuP treatment for 10 min at 30 °C. 10 mm NEM and 10 mm EDTA were added to stop the cross-linking reaction. Pulse-labeled cells were chased with 50 mg/ml non-radioactive Met for 5 min. A portion of the protein samples was treated with 200 mm DTT to reverse the disulfide cross-linking. Membrane samples harvested by high-speed centrifugation were then solubilized by DDM buffer (25 mm Tris HCl (pH7.5) and 1% DDM). Immunoprecipitation of LacY or YidC was performed using anti-Penta-His or anti-YidC antibody. Protein samples were resolved by using 10% SDS-PAGE.
RESULTS
Structure and Expression of LacY in Vivo
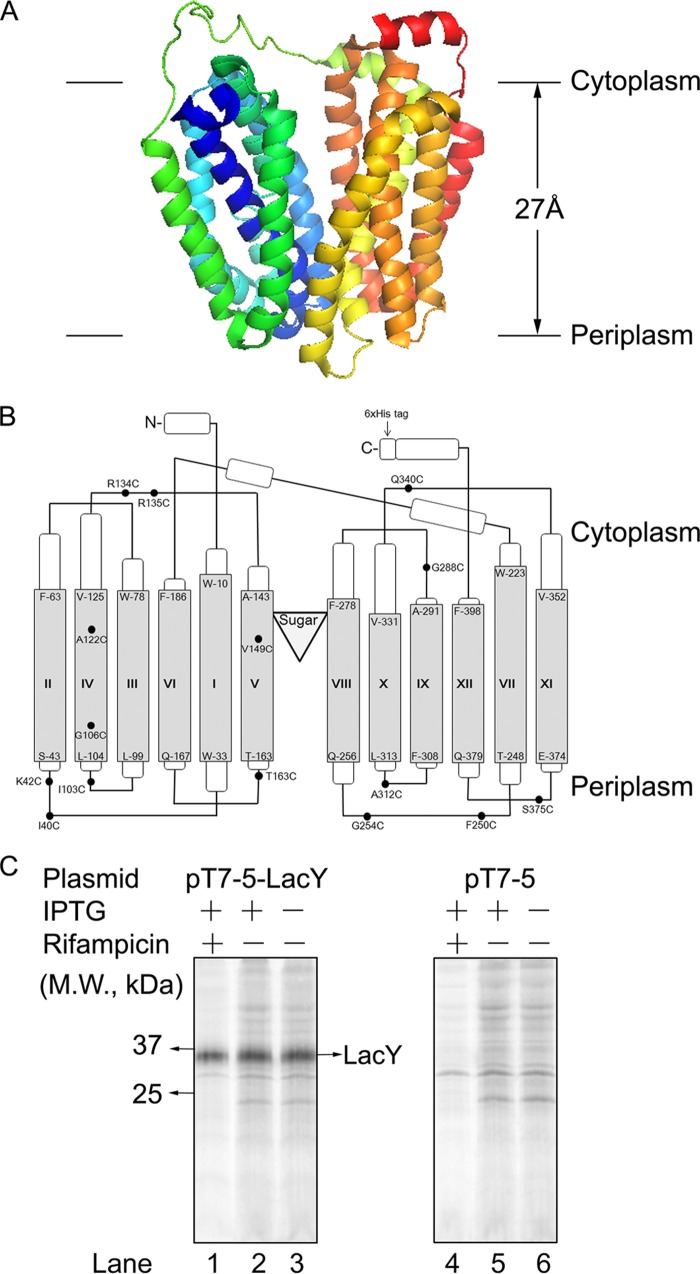
LacY is 86% helical and spans the membrane 12 times, with the N and C termini facing the cytoplasm (Fig. 1, A and B). The x-ray structure of LacY in the inward conformation exhibits two pseudosymmetrical α-helical bundles comprised of the TM1–6 and TM7–12 segments, respectively. The bundles surround a deep hydrophilic cavity with the sugar- and H+-binding sites at the apex (52) (Fig. 1A).
FIGURE 1.
Structure and expression of LacY. A, the structure of LacY in the inward conformation viewed across the membrane plane. The lactose homolog β-d-galactopyranosyl-1-thio-β-d-galactopyranoside is not shown in the structure. B, the membrane topology and secondary structure model of LacY on the basis of the x-ray structure. The loops depict the connectivity. The Cys residues used in the alkylation and cross-linking studies are indicated in the LacY schematic. We also show the position where PC(H5) is inserted in LacY in the LacY-PC chimera (arrow). C, selective [35S]Met-labeling of LacY in the membrane by rifampicin treatment. Membranes were isolated from JS7131 cells containing the pT7-5-LacY vector (left panel) or empty vector pT7-5 (right panel) that were [35S]Met-labeled before or after IPTG induction. Lanes 1 and 4 represent membranes that are derived from rifampicin-treated cells. M.W., molecular weight.
To probe LacY biogenesis in vivo, the Tabor and Richardson method (53) was used to radiolabel LacY in vivo (54). Using this approach, cells are transformed with two plasmids, pT7-5 encoding LacY under the control of the T7/lac promoter and pGP1–2 encoding a heat-inducible T7 RNA polymerase. Rifampicin is added to inhibit the bacterial RNA polymerase, which suppresses synthesis of endogenous E. coli proteins. However, T7 polymerase is insensitive to rifampicin, and synthesis of LacY, regulated by the T7 promoter, is not suppressed. Cells are induced with IPTG in the presence of rifampicin, and [35S]Met is added to radiolabel the proteins, followed by isolation of the membranes. Fig. 1C shows radiolabeling of LacY with this technique. LacY is detected in the membrane fraction when the cells are treated with IPTG and rifampicin (Fig. 1C, left panel, lane 1), and expression is partially induced by IPTG (compare lanes 2 and 3), whereas rifampicin efficiently suppresses endogenous protein synthesis (compare lanes 1 and 2). Importantly, the prominent band at 35 kDa is not observed with the empty pT7-5 vector (Fig. 1C, right panel).
Gel Shift Assay Monitors Translocation of Periplasmic Loops
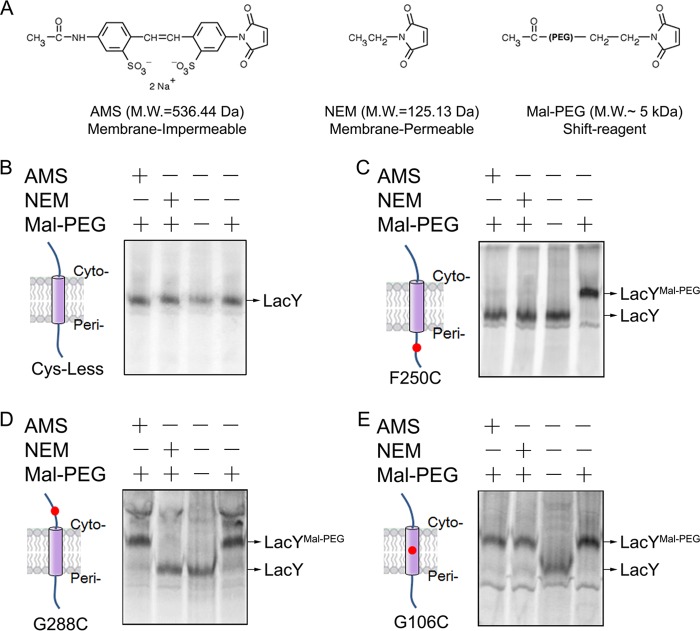
Site-directed alkylation (55) was utilized to test translocation of the periplasmic loops and to assess the topology of LacY (44). Single Cys residues were introduced into the periplasmic TM or cytoplasmic region of a functional LacY mutant devoid of the native Cys residues (56). After radiolabeling the Cys-less mutant in the presence of rifampicin, cells were treated for a specific time with chemical reagents that pass across the outer membrane. Cells treated with the inner membrane-impermeant AMS or the membrane-permeant NEM were then washed and subsequently treated with Mal-PEG as described under “Experimental Procedures” (Fig. 2A). No change in mobility was observed, particularly with Mal-PEG added to the Cys-less LacY (Fig. 2B). In contrast, addition of Mal-PEG to the single-Cys mutant LacY F250C without pretreatment by AMS or NEM yields a higher molecular weight band (Fig. 2C). Furthermore, this effect is blocked by pretreatment with AMS or NEM, showing that F250C in periplasmic loop VII/VIII is translocated to the periplasm.
FIGURE 2.
The gel shift assay to assess the topology of LacY. A, Cys-reactive reagents that were used to probe the topology of LacY. The gel shift assay for the Cys-less (B), F250C (periplasm (Peri)) (C), G288C (cytoplasm (Cyto)) (D), and G106C (transmembrane) LacY proteins (E) is shown. M.W., molecular weight. Rifampicin-treated radiolabeled cells expressing the indicated LacY mutants were treated with or without membrane-impermeable AMS or with membrane-permeable NEM. The cells were then washed with PBS buffer to remove the unreactive chemical reagent and pelleted. Membranes were isolated, and LacY was solubilized in detergent buffer. Where indicated, the samples were treated with Mal-PEG, which modifies unreacted Cys residues, and then analyzed by SDS-PAGE and phosphorimaging.
To provide further evidence that the gel shift assay with Mal-PEG effectively reflects LacY topology, it is shown that membrane-impermeable AMS does not prevent mutant G288C with a single Cys in the cytoplasmic extension of TM VIII (loop VIII/IX) from being modified by Mal-PEG, whereas the reaction with membrane-permeant NEM prevents Mal-PEG from modifying G288C (Fig. 2D). In contrast, G106C LacY (Fig. 2E) with a Cys in TM IV near the periplasmic border is modified with Mal-PEG regardless of whether cells were pretreated with AMS or NEM. Membrane-permeable NEM does not react with a Cys at position 106, either because the thiol group is protonated because of location in a hydrophobic environment or because the position is sterically blocked in the folded protein (Fig. 2E). Mal-PEG can react with this membrane Cys when LacY is solubilized in detergent.
LacY Insertion via the SRP/Sec Pathway
We first wanted to confirm, using our gel shift assays, that LacY requires the Sec machinery for membrane insertion, as shown previously (32, 42). The Sec dependence of LacY insertion was assayed using the SecE depletion strain CM124, where the secE gene is under the control of the araBAD promoter (57). SecE depletion causes a reduction of SecY, and proteins that require the SecYEG machinery are strongly inhibited when SecE and SecY are depleted, whereas Sec-independent proteins are unaffected. Membrane insertion was studied under SecE depletion conditions by growing the bacteria in M9 growth medium supplemented with glucose. For this study, a Cys was introduced into periplasmic loop P4 (VII/VIII) of LacY at position 250 to test for translocation by site-directed alkylation. CM124 cells expressing LacY F250C were grown under SecE depletion or expression conditions and then analyzed for translocation as described under “Experimental Procedures.” Fig. 3A shows that LacY F250C in CM124 is inserted under SecE expression conditions. Growth of the cell under SecE depletion conditions results in accumulation of the periplasmic loop of LacY F250C in the cytoplasm, but LacY still associates with the membrane, most likely peripherally. Thus, the membrane-permeant NEM, but not the membrane-impermeant AMS, prevents the shift caused by Mal-PEG. Western blot analyses support the conclusion that SecE is indeed depleted in CM124 (Fig. 3B). SecE is required to stabilize SecY (58), and the full-length SecY is observed only in cells expressing SecE.
FIGURE 3.
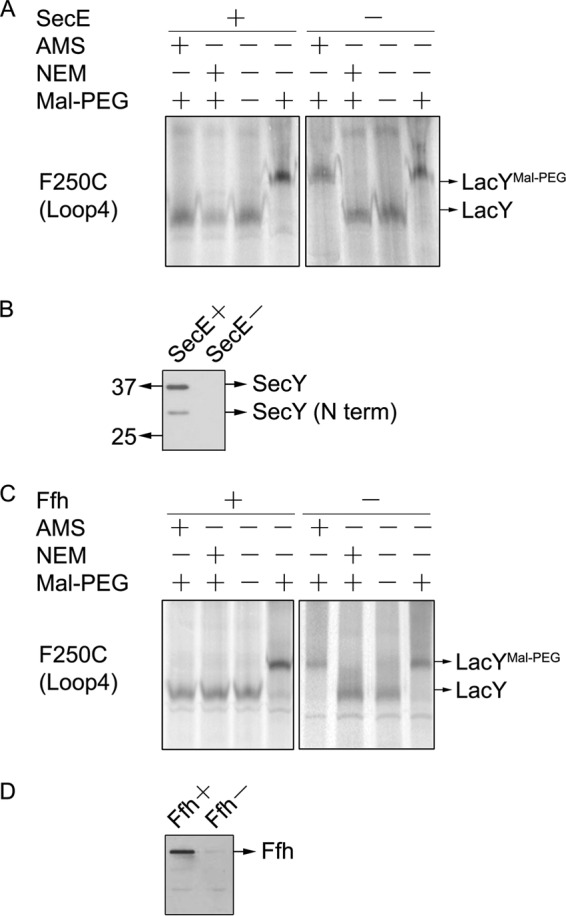
Membrane insertion of LacY is dependent on SRP and SecYEG. A, translocation of the fourth periplasmic loop of LacYF250C under SecE expression (+) and SecE depletion conditions (−). E. coli strain CM124 bearing pT7-5 encoding LacY F250C was grown with arabinose or glucose. Cells were treated with rifampicin and IPTG and labeled with [35S]Met, and the isolated membranes were analyzed by gel shift assay as described in Fig. 2. B, Western blot analysis confirms that SecY is depleted when SecE is depleted from the membrane by growth of CM124 under glucose conditions. N term, N-terminal. C, translocation of loop 4 (F250C) in LacY under Ffh expression (+) and Ffh depletion (−) conditions. E. coli WAM121 grown in the presence of arabinose or glucose was analyzed exactly as described in A. D, Western blot analysis showing that Ffh is depleted from the membrane by growth of WAM121 in glucose medium (see “Experimental Procedures” for details).
In addition, our Cys-based alkylation studies confirmed that LacY requires SRP for membrane targeting and insertion (40, 41). E. coli WAM121, an Ffh depletion strain, was used to test SRP dependence in vivo. In bacteria, SRP is composed of the protein component Ffh and a 4.5 S RNA. WAM121 contains ffh under the control of the araBAD promoter, as described for the YidC depletion strain. Therefore, E. coli WAM121 grown under glucose condition is depleted of Ffh. As shown in Fig. 3C, addition of membrane-impermeant AMS does not block the shift of LacY F250C caused by Mal-PEG under Ffh depletion conditions. In contrast, the membrane-impermeant NEM prevents the shift by Mal-PEG. Thus, membrane insertion of LacY F250C is blocked when Ffh is depleted (Fig. 3C). Western blot analyses verify that Ffh is depleted from the membrane of WAM121 cells grown in glucose (Fig. 3D). The findings confirm that LacY is targeted to the membrane by the SRP pathway and requires the SecYEG machinery for translocation.
YidC Does Not Function in Translocation of Periplasmic Loops of LacY
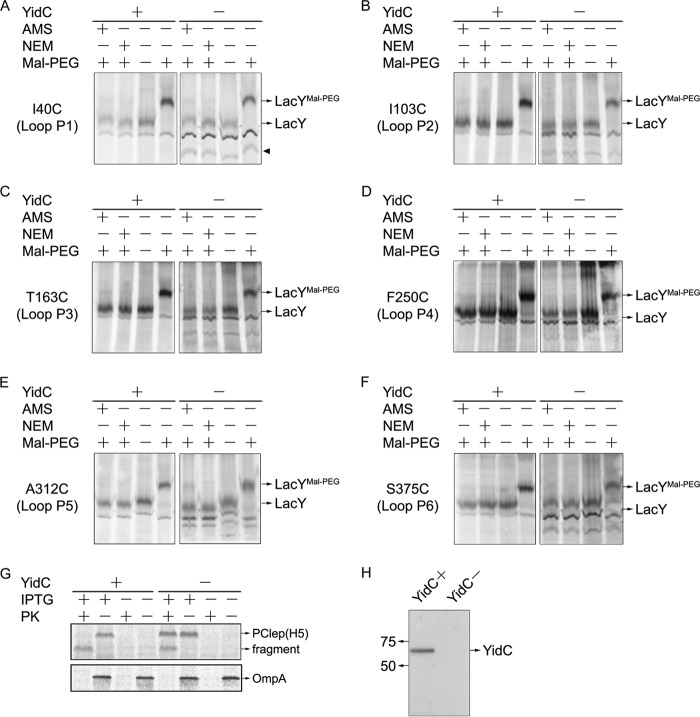
To examine whether YidC functions as an insertase for LacY, we tested whether YidC is required for translocation of the periplasmic loops in LacY. We used E. coli JS7131, a YidC depletion strain containing YidC under the control of the araBAD promoter, similar to the other depletion strains described, and tested whether YidC depletion leads to a defect in insertion of the first periplasmic loop (I/II) of YidC by studying the accessibility/reactivity of a Cys introduced in place of Ile40 (Fig. 4A). Both AMS and NEM prevent the Mal-PEG-induced increase in LacY molecular weight after reaction, regardless of whether YidC is depleted or not. Therefore, the first periplasmic loop of LacY is translocated independently of YidC. It is also noteworthy that expression under the YidC depletion condition leads to a new band at a lower Mr (Fig. 4A, arrowhead), which may be the induced phage shock protein (59, 60).
FIGURE 4.
Translocation of the periplasmic loops of LacY does not require YidC. Translocation of the periplasmic loops of LacY under YidC expression (+) and YidC depletion (−) conditions. E. coli strain JS7131 bearing pT7-5 lacY I40C (A), I103C (B), T163C (C), F250C (D), A312C (E), and S375C (F) were grown, labeled with [35S]Met, and analyzed by the gel shift method as described in Fig. 3B. Note that there is a YidC depletion-induced band at the lower molecular weight position in A–F (arrowhead, see A). G, membrane insertion of procoat-Lep H5 (S13C) under YidC expression (+) and YidC depletion conditions (−). JS7131 containing pMS119 procoat-Lep H5 (S13C) was [35S]Met-labeled without rifampicin treatment and analyzed by protease accessibility assay. OmpA served as a positive control for proteolysis. Samples were analyzed by SDS-PAGE and phosphorimaging. H, a representative Western blot confirms that YidC is depleted by growth of JS7131 bearing the pGP1–2 and pT7-5-LacY plasmids in LB medium with glucose added for 5 h at 30 °C.
Having shown that YidC is not involved in translocating loop P1 (I/II) of LacY, a Cys was introduced at position 103, 163, 250, 312, or 375 as shown in Fig. 1B to assay translocation of periplasmic loop P2 (III/IV), P3 (V/VI), P4 (VII/VIII), P5 (IX/X), or P6 (XI/XII). The data presented in Fig. 4, B–F, indicate that Mal-PEG reactivity is blocked with each single-Cys mutant by AMS as well as NEM, regardless of whether or not YidC is depleted. Therefore, periplasmic loops 2, 3, 4, 5, and 6 are translocated independently of YidC, thereby indicating that YidC does not function as an insertase for the translocation of periplasmic loops of LacY.
By using YidC-dependent Procoat-Lep (H5), which is not cleaved by signal peptidase 1 (SP1) (66), it was confirmed that YidC was depleted under the experimental conditions used (Fig. 4G). It is well established that YidC has to be significantly depleted for the insertion of procoat to be inhibited (19). Although Procoat-Lep (H5) is completely digested by externally added proteinase K and converted to a lower Mr band under YidC expression conditions, it is mostly resistant to proteolysis after YidC depletion. Moreover, Western blotting as well as alkylation of PClep (H5) S13C (supplemental Fig. S2B) provide direct confirmation that YidC is depleted when E. coli JS7131 is grown in glucose medium (Fig. 4H).
YidC Functions as a Foldase
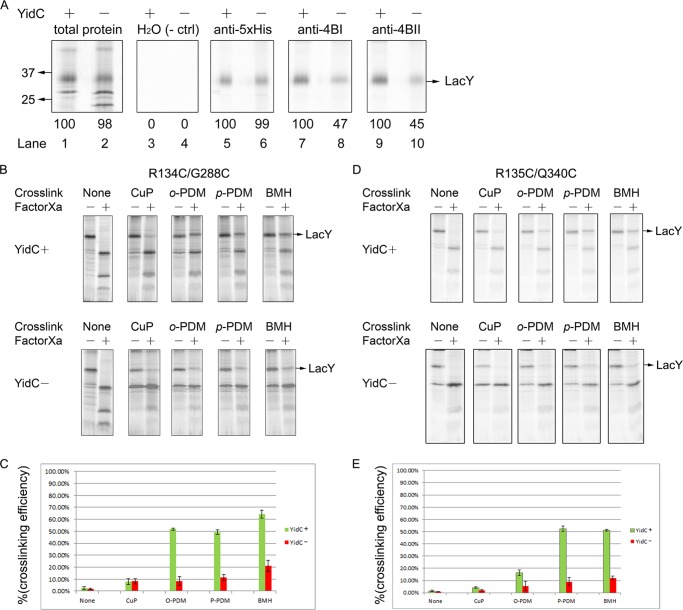
In vitro studies showed previously that the conformation of LacY is perturbed upon YidC depletion (32). Here, two methods were used to determine whether the conformation of LacY is perturbed in vivo when YidC was depleted. First, we utilized two monoclonal antibodies to confirm previous in vitro results, indicating that YidC is important for folding of LacY (32). mAb 4B1 recognizes folded periplasmic loop P4 (VII/VIII) (61), and mAb 4B11 recognizes a folded epitope that includes cytoplasmic loops C4 (VIII/IX) and C5 (X/XI) (62). JS7131 expressing LacY with a C-terminal His tag was grown under YidC depletion or expression conditions and analyzed by [35S]Met labeling in the presence of rifampicin. As positive controls, total 35S-labeled proteins and anti-Penta-His antibody-treated samples were analyzed (Fig. 5A) to assess the level of radiolabeled LacY (lanes 1 and 2), and immunoprecipitated LacY (lanes 5 and 6) was the same under YidC plus and YidC minus conditions. In contrast, the binding of mAb 4B1 (Fig. 5A, lanes 7 and 8) or mAb 4B11 (lanes 9 and 10) to LacY is decreased by ∼50% under YidC depletion conditions. Thus, the conformations of at least half of the 4B1 and 4B11 epitopes are perturbed upon YidC depletion. As a negative control, mock immunoprecipitation with water was performed to show that immunoprecipitation is specific (Fig. 5A, lanes 3 and 4). These in vivo results provide confirmation of previous studies carried out in vitro (32, 63).
FIGURE 5.
YidC is required for proper helix packing of LacY. A, the binding of monoclonal antibodies 4B1 and 4B11 to LacY is impaired under YidC depletion conditions. Expression of LacY under YidC expression (+) and YidC depletion (−) conditions was performed in JS7131 as described in Fig. 4A. Radiolabeled cells in either YidC (+) or YidC (−) conditions were split into five aliquots and treated equally. Radioactive bands were visualized using a Typhoon PhosphorImager. The total 35S-labeled proteins in the membrane before immunoprecipitation are shown in lanes 1 and 2. Mock treatment by immunoprecipitation against water was used as a negative control (ctrl, lanes 3 and 4). Penta-His antibody (Qiagen) recognizing the His-tagged LacY was used as a positive control (lanes 5 and 6). Immunoprecipitation using monoclonal antibodies against the 4B1 and 4B11 epitopes was used to assay the folding of LacY at periplasmic loop 4 (VII/VIII) (lanes 7 and 8) and cytoplasmic loops VIII/IX and X/XI (lanes 9 and 10), respectively. The induced band in the total protein in the membrane that appears slightly below the 25-kDa molecular mass upon YidC depletion is most likely the PspA. B–E, LacY folding is impaired under YidC depletion conditions, as determined by intramolecular cross-linking between Cys pairs of LacY. B, the E. coli strain JS7131 bearing pKR35-R134C/G288C was grown in the presence of arabinose (YidC+) and glucose (YidC−), radiolabeled using the rifampicin blocking technique as described in Fig. 4, and then membranes were isolated. Where indicated, the right-side-out vesicles were treated with BMH, p-PDM, o-PDM, and CuP. Thereafter, solubilized membrane protein samples were treated with Factor Xa protease (NEB Co.). Protein samples were then analyzed by SDS-PAGE and phosphorimaging. D, JS7131 cells harboring pKR35-R135C/Q340C were grown in arabinose and glucose conditions and radiolabeled using the rifampicin technique. The LacY R135C/Q340C in the membrane fraction derived from cells grown under YidC (+) and YidC (−) conditions was analyzed for cross-linking as described in B and subjected to SDS-PAGE and phosphorimaging. The Cys pairs were introduced into LacY L6XB, having a Factor Xa-biotin acceptor domain between TM6 and TM7 (75). The efficiency of LacY cross-linking under YidC (+) and YidC (−) conditions between the R134C/G288C (C) and R135C/Q340C pairs (E) was determined by quantification of the band density of the full-length LacY using ImageJ software. The error bars indicate the S.D. in triplicate measurements.
As a second method, site-directed Cys cross-linking was employed to probe the effect of YidC depletion on the conformation of LacY. Cross-linking between paired Cys residues in a Cys-less background was monitored by proteolysis of an engineered Factor Xa protease site in the middle cytoplasmic loop of LacY between the two Cys residues (49, 64). Cross-linking between the Cys residues is readily assayed after proteolysis because the two LacY fragments remain covalently bound in a single complex with an Mr of ∼35, like the intact molecule. First, cross-linking between Cys-134 (R134C) in cytoplasmic loop C2 (IV/V) and Cys-288 (G288C) in cytoplasmic loop C4 (VIII/IX) was tested with the following cross-linking agents: CuP, which induces formation of a disulfide bond; BMH, ∼16 Å; p-PDM, 10 Å; and o-PDM, 6 Å. In the absence of cross-linking, Factor Xa protease completely cleaves full-length LacY with YidC expression or depletion (Fig. 5B). In the YidC expression condition, BMH, p-PDM, or o-PDM cross-links 50–60% of the LacY, but cross-linking is reduced to 10–20% when YidC is depleted (Fig. 5, B and C). Similar differences in cross-linking efficiency with BMH and p-PDM are observed with respect to YidC depletion and expression, where Cys residues are at positions 135 in C2 (loop IV/V, R135C) and 340 in C5 (loop X/XI, Q340C) (Fig. 5, D and E). In contrast, cross-linking of Cys pairs K42C/G254C (supplemental Fig. S4, A and C) and A122C/V149C (supplemental Fig. S4, B and D) is not influenced significantly by YidC depletion.
Contact between YidC and LacY
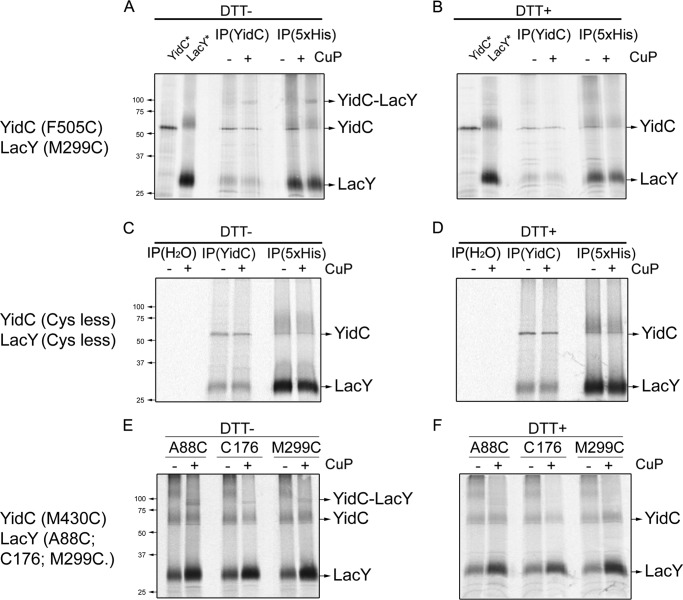
The above results show that YidC is important for LacY to obtain its proper conformation, supporting the notion that YidC is a molecular chaperone involved in folding. To act as a chaperone, YidC needs to make contact with LacY during membrane protein biogenesis. Cys residues were introduced into TM9 (M299C) of LacY and into TM5 of YidC (F505C in the proposed YidC topology (65)) to determine whether YidC interacts with LacY during membrane insertion. The F505C variant has been shown previously (29) to cross-link efficiently to Pf3 coat protein. The YidC depletion strain JS7131 expressing LacY (M299C) and YidC (F505C) was grown in glucose to deplete endogenous YidC and analyzed by pulse-labeling in the presence of rifampicin for 5 min in the presence or absence of CuP. The labeled cells were then split into four equal aliquots, and a portion was treated with the reducing agent DTT. Samples were first immunoprecipitated with anti-YidC (Fig. 6A, lane 1) or anti-Penta-His antibody (lane 2) to indicate the positions of the respective proteins that were immunoprecipitated. In the CuP-treated samples, a higher molecular weight band (corresponding to cross-linked YidC-LacY) between Mr 75–100 is detected (YidC-LacY arrow) with YidC or anti-His antibodies (Fig. 6A, compare lanes 3 and 4 and lanes 5 and 6, respectively). As expected, these bands are not observed when DTT is added after treatment with CuP (Fig. 6B). The cross-linked bands were also not detected when the experiment was performed with Cys-less LacY and YidC derivatives (Fig. 6C) nor with Cys-less YidC and LacY M299C (supplemental Fig. S5B) or YidC F505C and Cys-less LacY present (supplemental Fig. S5A). Notably, the Cys-less LacY and YidC, which are not covalently cross-linked, interact in detergent and can be copurified as a complex (Fig. 6, C and D). On the SDS-PAGE gel, LacY is detected after immunoprecipitation under native conditions using antiserum against YidC. Similarly, YidC is detected when LacY is immunoprecipitated with anti-Penta His antibody. Neither protein is detected in mock immunoprecipitation with water.
FIGURE 6.
YidC contacts LacY during membrane protein biogenesis. Cross-linking between the single-Cys YidC and single-Cys LacY mutants was performed under conditions where the chromosomal YidC was depleted. A and B, to deplete the chromosomally encoded YidC, E. coli JS7131 harboring the YidC and LacY expression vectors pACYC184-YidC (F505C) and pLZ2-LacY (M299C) were grown in the presence of 0.2% (m/v) glucose at 30 °C. The cells were expressed using the rifampicin technique as described in Fig. 2. JS7131 cells were pulse-labeled with trans-[35S]Met for 5 min with or without treatment of 1 mm CuP for 10 min at 30 °C (see “Experimental Procedures”). The total membrane protein sample was immunoprecipitated (IP) with antibody to either YidC or Penta-His (recognizing the 6× His tag at the LacY C terminus). Protein samples with (B) or without (A) adding reducing agent (200 mm DTT), were analyzed using a 10% (m/v) SDS-PAGE and by phosphorimaging. The LacY (lane 2) or YidC (lane 1) protein controls were obtained as follows. E. coli BL21 cells bearing pET28b-YidC (WT) were pulse-labeled for 1 min with trans-[35S]Met, and total protein was immunoprecipitated using anti-YidC serum. E. coli T184 cells cotransformed with pGP1–2 and pT7-5-LacY were pulse-labeled for 5 min after treating with rifampicin, and total membrane proteins were immunoprecipitated using Penta-His antibody. C and D, mock cross-linking between YidC and LacY was performed with E. coli JS7131, with both of the expressed Cys-less proteins, using pACYC184-YidC (Cys-less) and pLZ2-LacY (Cys-less). In addition to YidC and Penta-His immunoprecipitation, a mock immunoprecipitation was performed with water. After CuP treatment, samples were treated with (D) or without (C) DTT. E and F, disulfide cross-linking between YidC (M430C) and LacY single-Cys mutants (A88C, C176, or M299C) was performed as described in A and B. Penta-His antiserum was used to pull down LacY with a 6× His tag.
YidC with a Cys in TM3 (M430C) and LacY single-Cys variants at position 88, 176, or 299 in TM3, TM6, and TM9, respectively, were constructed, and interactions were tested by using disulfide cross-linking (Fig. 6E). New higher Mr bands (YidC-LacY arrows) are observed after CuP treatment (Fig. 6E) but not after subsequent exposure to DTT (F). Thus, YidC makes contact with multiple TM segments of LacY during biogenesis.
YidC Can also Function as an Insertase
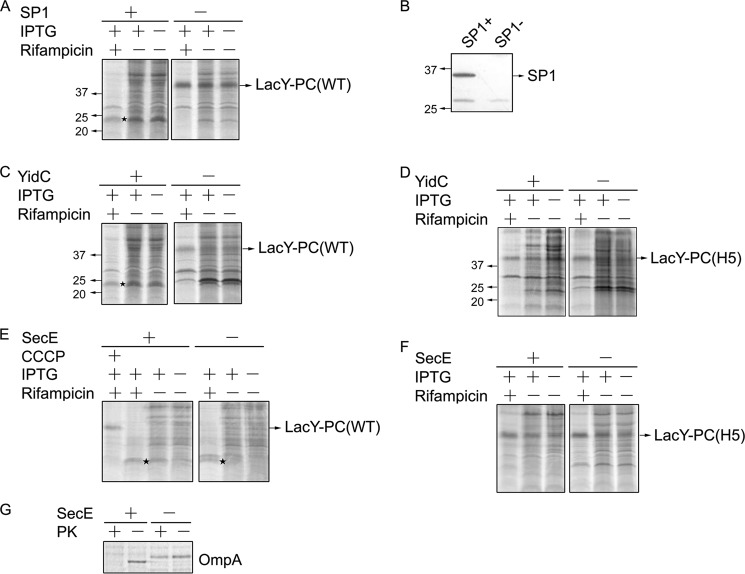
Our data here show YidC is a molecular chaperone that binds LacY and is involved in its folding. Still, it is conceivable that YidC does have the capacity to insert and translocate internal loops of multispanning membrane proteins across the membrane. To test this idea, a chimera was constructed in which the YidC-dependent M13 phage WT procoat (PC), which is cleaved by signal peptidase 1 (SP1), was fused into the middle cytoplasmic loop of LacY. By using an SP1 depletion strain in which lepB (the signal peptidase gene) is under the control of the araBAD promoter (47), we show that the full-length LacY-PC (WT) is detected when SP1 is depleted (SP1-) but not when SP1 is expressed (SP1+) (Fig. 7A). In contrast, a prominent band ∼24 kDa in size is observed (★), indicating the position of the cleaved LacY-PC fragments under SP1-expressed conditions (Fig. 7A, SP1+). This is expected because SP1 cleaves LacY-PC to produce two approximately equal-size fragments (26 and 28 kDa). Thus, LacY-PC (WT) is cleaved by SP1 during membrane protein biogenesis, showing that the procoat domain inserts across the membrane. As shown by the Western blot analyses in Fig. 7B, SP1 was indeed depleted from FTL85 cells grown in the presence of glucose.
FIGURE 7.
YidC can function to insert the procoat domain fused within LacY. A, E. coli FTL85 bearing pLZ-t7 and pT7-5-LacY-PC (WT) was grown under SP1 expression (+) and SP1 depletion (−) conditions. As indicated, cells were treated with rifampicin and/or IPTG prior to radiolabeling. B, Western blot analysis confirms that SP1 is depleted by growth of FTL85 in LB medium plus 0.2% glucose at 30 °C for 5 h (SP1-). C, E. coli JS7131 bearing pT7-5-LacY-PC (WT) was grown under YidC expression (+) and YidC depletion conditions (−). Where indicated, cells were treated with rifampicin and/or IPTG prior to radiolabeling. D, JS7131 harboring pT7-5-LacY-PC (H5) was analyzed as described in panel C. For A and C, ★ depicts the SP1-cleaved fragments of LacY-PC. E. coli CM124 cells bearing pLZ2 encoding LacY-PC (WT) (E) or LacY-PC (H5) (F) were grown in LB medium with arabinose or glucose to express or deplete SecE, respectively, and then treated, where indicated, with rifampicin and IPTG prior to radiolabeling. A portion of CM124 cells grown under arabinose conditions was treated with 50 μm (final concentration) carbonyl cyanide m-chlorophenylhydrazone (CCCP) for 45 s after incubation with rifampicin/IPTG prior to pulse-labeling with trans-[35S]Met for 5 min (E, lane 1). G, A portion of CM124 cells bearing pLZ2-LacY-PC (WT) was grown under arabinose or glucose conditions, switched into fresh M9 minimal medium, and incubated at 30 °C for 30 min. The CM124 cells were pulse-labeled with [35S]Met without rifampicin treatment and analyzed by protease accessibility assay. Sec-dependent OmpA protein was immunoprecipitated using anti-OmpA serum, and the protein samples were analyzed by SDS-PAGE and phosphorimaging as described in Fig. 4G. PK, proteinase K.
To establish whether YidC is required for the PC domain insertion, we tested the YidC dependence of membrane insertion of the PC domain within the PC-LacY chimera. Although, under YidC+ conditions, full-length LacY-PC (WT) was not detected because it is cleaved by SP1 (Fig. 7C, YidC+), under YidC depletion conditions, full-length LacY-PC (WT) chimera is detected (YidC− and IPTG+/Rifampicin+). This shows that, under YidC depletion conditions, the procoat domain is not inserting across the membrane and cleaved by SP1 (Fig. 7C, YidC−). Under YidC+ conditions, we observe the 24-kDa band (Fig. 7C, ★). The results indicate that the procoat domain within chimeric LacY can insert into the membrane when YidC is present but is inhibited when YidC is depleted. We also constructed a chimera in which the YidC-dependent H5 procoat that is not processed by SP1 (66) was fused into the middle cytoplasmic loop of LacY. During insertion of the LacY-PC chimera, the H5 procoat domain is not be cleaved by SP1. Thus, the full-length LacY PC (H5) was detected under YidC expression (+) and YidC depletion (−) conditions (Fig. 7D, IPTG+/Rifampicin+).
To further probe the insertion mechanism of the procoat domain of the LacY-PC chimera, we also examined Sec dependence. First we show that the full-length LacY-PC (H5), which is not cleaved by SP1, is detected in CM124 under SecE+ and SecE− conditions (Fig. 7F). In contrast, the full-length cleavable LacY-PC (WT), under SecE expression or SecE depletion conditions (Fig. 7E, lanes 2 and 5), is only detected if the insertion is prevented by carbonyl cyanide m-chlorophenylhydrazone (lane 1), supporting that the internal PC domain is cleaved by SP1 and that its insertion occurs independently of the SecYEG machinery. The insertion of LacY-PC requires YidC (Fig. 7C), and it has been established previously that the procoat inserts into the inner membrane in a protonmotive force-dependent manner (67). The addition of carbonyl cyanide m-chlorophenylhydrazone prior to [35S]Met labeling of cells for 5 min results in a band at the expected size for the full-length LacY-PC when the protonmotive force is dissipated because the procoat domain requires the protonmotive force for insertion. To ensure SecE depletion in the presence of glucose, a portion of CM124 cells (bearing the LacY-PC (WT) expression vector) grown in glucose medium was pulse-labeled, and the Sec-dependent OmpA protein was immunoprecipitated using anti-OmpA antibody. As shown in Fig. 7G, the precursor form of OmpA accumulates when the cells are grown in glucose medium (under SecE-depletion conditions) and is protected by spheroplasts from proteinase K digestion because the Sec-dependent proOmpA is not translocated across the membrane, whereas mature OmpA is digested by proteinase K as it inserts under SecE expression conditions. These data confirm that SecE has been depleted in the CM124 grown in glucose. The results presented in Fig. 7 show that the procoat domain within LacY inserts into the membrane by a YidC-only mechanism.
YidC and Sec Translocase Work Together in the Membrane Topogenesis of a LacY Chimera
Because the procoat domain inserts by a Sec-independent, YidC-dependent mechanism, we wanted to determine whether the loops before and after the procoat domain (loops P3 and P4 of LacY, respectively) are still inserted in a YidC-independent, Sec-dependent manner. A Cys was introduced into loop P3 (V/VI) at position 163 or loop P4 (VII/VIII) at position 250 of the LacY chimera, respectively. Insertion was then examined using the alkylation method, as described under “Experimental Procedures.” The addition of AMS prevented the shift in LacY-PC (Fig. 8A, H5, T163C*) and LacY-PC (8C, H5, F250C*) by Mal-PEG both under YidC+ and YidC− conditions. This shows that these loops are inserted by a YidC-independent mechanism. In contrast, AMS does not prevent the shift by Mal-PEG under YidC depletion conditions where a Cys is added to the PC domain, showing that the insertion of the procoat domain of the PC-LacY chimera (where a Cys is added to the PC domain) is YidC-dependent (Fig. 8B). We also confirmed, using the SecE-depletion strain CM124, that the loops before and after the procoat domain are not translocated under SecE depletion conditions using an alkylation assay (supplemental Fig. S6). The combined data indicate that the LacY loops before and after the procoat domain are inserted in a YidC-independent manner, showing that the Sec machinery is sufficient to insert these loops, whereas the PC domain is inserted in a strict YidC-dependent, Sec-independent mechanism. We conclude that YidC is capable of sensing the procoat domain within the middle of a LacY chimera and can act as an insertase to translocate an internal loop within the multispanning membrane protein LacY.
FIGURE 8.
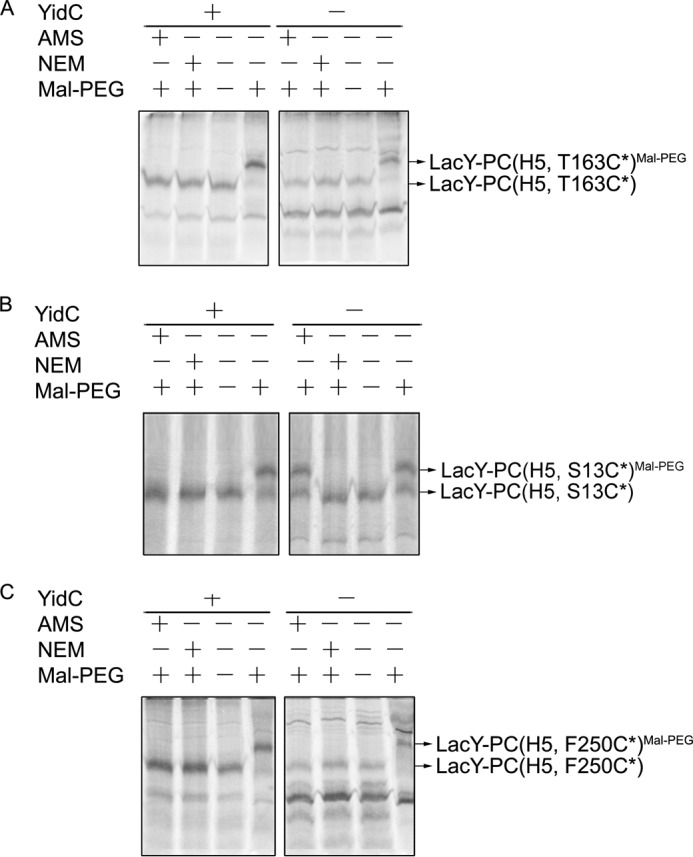
Different YidC translocase requirements for insertion of the PC domain and the flanking hydrophilic loops of LacY-PC. LacY-PC (H5) with a Cys in the loop preceding the PC domain (T163C*) (A), in the PC domain at position 13 (PC S13C*) (B), or in a loop immediately after the PC domain (F250C*) (C) were analyzed for membrane insertion under YidC expression (+) and YidC depletion conditions (−). JS7131 cells bearing pGP1–2 and pT7-5 encoding the respective LacY-PC (H5) derivatives were grown in medium supplemented with arabinose or glucose. Cells were treated with rifampicin and IPTG, radiolabeled with trans-[35S]Met, and analyzed using cysteine modification reagents as described in Fig. 2. The isolated membrane protein samples were treated with Mal-PEG and analyzed by SDS-PAGE and phosphorimaging.
DISCUSSION
In this report, we examine the role of YidC in the insertion and folding of the polytopic membrane protein LacY in vivo. The folding defect because of YidC depletion described by Nagamori et al. (32) was possibly due to one or more TM helices not inserting into the membrane, leading to a membrane topology alteration or a direct effect on helix-helix packing. Therefore, we determined whether YidC is required for translocation of the periplasmic loops in LacY or for the α-helical TM segments to interact, pack, and assume a correct tertiary fold after insertion takes place.
To address the function of YidC in membrane insertion of LacY, we monitored the translocation of the six periplasmic loops of LacY individually under YidC depletion conditions. To do so, we used a gel shift alkylation assay (44), which has an advantage over protease accessibility assays because chemical modification can monitor translocation of individual loops of multispanning membrane proteins regardless of loop size. Moreover, these studies and others were facilitated using rifampicin in the labeling reaction to inhibit RNA polymerase and do not affect membrane insertion of LacY under YidC/Sec plus conditions, although they could potentially have unrelated effects because of shutting down endogenous protein synthesis. However, it should be noted that we see the same YidC-independent results for the translocation of loop 4 of LacY when labeling is performed without using rifampicin (supplemental Fig. S3).
First, we showed that YidC does not function as an insertase for the SRP/Sec-dependent LacY because translocation of all six loops does not require YidC (Fig. 4). However, we cannot exclude that YidC is important for the translocation kinetics of the loops because a 5-min pulse was performed, and it takes time at room temperature to derivatize the periplasmic Cys residues. Rather, YidC has a foldase function (Fig. 5) involved in the packing of TM helices, which reinforces the proposal that YidC is a chaperone involved in the folding of LacY (32). This was shown using two monoclonal antibodies directed against conformational epitopes of LacY and by cross-linking studies between Cys pairs (Fig. 5).
The role of YidC as a chaperone in folding LacY is supported by disulfide cross-linking results (Figs. 6). Previously, YidC residues in TM1, TM3, and TM5 were shown to contact the hydrophobic region of the Pf3 coat and MscL during insertion (29, 30). We now show that Cys-430 in TM3 and Cys-505 in TM5 of YidC contacts LacY at Cys-299 in TM9 (Fig. 6, A and E). In the presence of CuP, disulfide cross-linking is observed, which is reversed by DTT. No cross-linking is observed with the Cys-less YidC or LacY variants (Fig. 6C). In addition to contacting TM9 of LacY, cross-linking to TM3 and TM6 of LacY (Fig. 6E) is shown, demonstrating that YidC contacts multiple TM segments of LacY as it inserts into and assembles in the membrane.
How is membrane protein folding catalyzed? By protein chaperones, other cofactors or by the intrinsic property of a protein chain to fold into its proper conformation? Dowhan and co-workers (68, 69) have shown that the membrane topology of LacY undergoes a phospholipid-dependent rearrangement so that the N-terminal half of LacY adopts a reversed topology in the absence of phosphatidylethanolamine. It was reasoned that the nature of the phospholipids in the membrane is the determinant for membrane protein folding and that no accessory proteins are necessarily required. It was then reported that folding of two monoclonal antibody epitopes in LacY are perturbed by YidC depletion in vitro (32). It is shown here that both folding of the monoclonal antibody epitopes and helix packing of LacY are disrupted when YidC is depleted in vivo. These data, along with YidC contacting LacY during membrane biogenesis (Fig. 6), strengthen the proposal that YidC plays the role of a foldase for LacY and suggests that misfolding of LacY is a direct effect of YidC depletion.
What is the driving force for the interaction of YidC with LacY and what mechanism does YidC employ to facilitate folding of membrane proteins? Generally, ATP hydrolysis is the energy source that is important to promote folding of soluble proteins (70, 71). Most molecular chaperones, typified by GroEL/GroES and DnaK, assist in an ATP-dependent manner in the non-covalent folding and assembly of macromolecules or in the unfolding and disassembly of protein complexes (72, 73). However, unlike these molecular chaperones, YidC promotes protein folding in the membrane and does not have Walker A or B motifs (17). We hypothesize that hydrophobic interactions promote the YidC/LacY interaction that is important for folding. YidC possibly acts as an insulator between the newly synthesized unfolded protein and the lipid bilayer, where it releases the protein upon maturation into its final conformation. This “chamber” reduces energetically unfavorable contacts between the hydrophobic lipid bilayer and hydrophilic parts of the unfolded protein, which is critical for decreasing the energy barriers for folding.
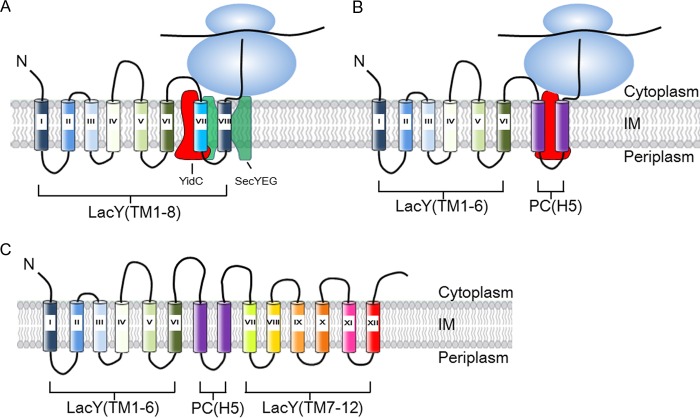
Remarkably, even though YidC does not function as an insertase for the native LacY, it has the capacity to translocate a loop within a multispanning membrane protein like LacY (Figs. 7 and 8). As shown previously, M13 procoat protein requires YidC for membrane insertion (18). We found that YidC is absolutely required for insertion of the procoat domain added to the middle cytoplasmic loop of a LacY chimera (LacY-PC). The results are intriguing because the Sec machinery inserts the first six TM segments of the LacY chimera but does not insert the flanking C-terminal procoat domain. Rather, it needs YidC for insertion of the procoat domain. Clearly, after insertion of the procoat domain, the Sec machinery resumes function and inserts the remaining six TM segments of the LacY chimera (Fig. 8). This indicates that SecYEG and YidC cooperate in the topogenesis of the LacY chimera in a dynamic manner. Each membrane insertion event in the biogenesis of a multispanning protein determines whether it requires SecYEG or YidC. The procoat domain within LacY requires YidC for membrane insertion, and SecYEG alone cannot accomplish this. Most likely, during insertion and assembly of the helical bundle domains of LacY, the SecYEG machinery functions as a translocase to insert the TM regions of LacY across the membrane, whereas YidC binds to LacY TM segments as they leave the lateral gate and helps fold LacY (Fig. 9A). In the case of the LacY-PC chimera, the Sec machinery inserts TM segments 1–6 of LacY across the membrane. YidC then takes over to insert the procoat domain into the membrane, at which point SecYEG resumes its translocase function and, subsequently, inserts the remaining TM7–12 of LacY (Fig. 9B).
FIGURE 9.
Proposed model for cooperative insertion of LacY-PC by SecYEG and YidC. A, the periplasmic loops of LacY are inserted by the SecYEG machinery, whereas YidC binds the TM segments and helps fold the protein. N, designates the N terminus. B, the procoat domain of LacY-PC is inserted by the YidC-only pathway, whereas all the LacY periplasmic loops are inserted by the SecYEG machinery. In this latter pathway, YidC presumably plays a role in the folding of LacY as well. It should be noted that it is not known for LacY when the TM segments are released from YidC and integrated into the lipid bilayer. IM, inner membrane. C, the topology of LacY-PC (H5) on the basis of this study. The PC (H5) region is depicted in purple.
Cooperation of the two inner membrane translocation machineries has been observed previously for membrane insertion of E. coli inner membrane proteins. For example, insertion of bacterial CyoA, which spans the membrane twice, requires the cooperation of YidC and SecYEG/SecA for insertion. CyoA is synthesized in a precursor form (preCyoA) with a cleavable signal peptide and is processed by lipoprotein signal peptidase. The N-terminal region of preCyoA is inserted by the YidC-only pathway, whereas the large C-terminal region is inserted by the SecYEG/SecA pathway (25, 26). Also, close cooperation between SecYEG and YidC in biogenesis was observed with the ABC sugar transporter mannitol permease (MtlA) (38) as well as MalF (33). In vitro photo-cross-linking of insertion intermediates of MtlA revealed that the TM segments inserted initially in a SecYEG environment and then moved to where they contacts only YidC (38). This led to the proposal that YidC constitutes a site for TM domain assembly prior to integration into the lipid bilayer (38). In mitochondria, the ABC transporter Mdl1, which spans the membrane six times, has two machineries that cooperate in the inner membrane insertion process (74). Mdl1 is imported into the mitochondria from the cytoplasm and inserts into the inner membrane via the TIM23 complex. After engaging the TIM23 complex, the amino-terminal domain is integrated into the inner membrane by a stop transfer mechanism where it is released from the TIM23 machinery into the membrane. In contrast, the central loop is translocated completely across the inner membrane into the mitochondrial matrix and is reinserted into the inner membrane by the Oxa1 complex (the mitochondrial YidC homolog). Conversely, the C-terminal domain is inserted into the inner membrane using the TIM23-stop transfer mechanism similar to the insertion of the Mdl1 amino-terminal domain.
Our results fit nicely with Nagamori et al. (75), who showed that the amino-terminal six TM segments of LacY remain in a proteinaceous environment before LacY is fully translated on the ribosome and folded into a quasi-native conformation after insertion into the bilayer. YidC may act as an insulator for newly synthesized LacY. YidC-independent and Sec-dependent LacY is cross-linked to YidC in vivo, indicating that YidC is in close proximity to the protein even if membrane insertion is independent of YidC (Fig. 6). Interestingly, the interruption of the N6 and C6 helical bundles of LacY by an internal M13 procoat domain does not alter the insertion requirements for either LacY halves or the M13 procoat domain. This finding is in agreement with the translocase requirement being dictated by the amino acid composition of the TM segment and polar domains of membrane proteins (48). Taken together, YidC may be broadly described as an insertase for protein translocation and as a chaperone for folding of proteins that utilize the SecYEG machinery.
This work was supported, in whole or in part, by National Institutes of Health Grants DK51131, DK069463, and GM073210 (to H. R. K.). This work was also supported by National Science Foundation Grants MCB-1052033 (to R. E. D.) and MCB-1129551 (to H. R. K.).

This article contains supplemental Figs. S1–S6 and Methods.
- TM
- transmembrane
- SRP
- signal recognition particle
- DDM
- dodecyl-β-d-maltopyranoside
- Mal-PEG
- methoxypolyethylene glycol maleimide
- AMS
- 4-acetamido-4′-maleimidylstilbene-2,2′-disulfonic acid
- NEM
- N-ethylmaleimide
- IPTG
- isopropyl 1-thio-β-d-galactopyranoside
- Lep
- leader peptidase
- BMH
- 1,6-bis(maleimido)hexane
- o-PDM
- N,N′-o-phenylenedimaleimide
- p-PDM
- N,N′-p-phenylenedimaleimide
- CuP
- copper 1,10-phenanthroline
- PC
- procoat.
REFERENCES
- 1. Xie K., Dalbey R. E. (2008) Inserting proteins into the bacterial cytoplasmic membrane using the Sec and YidC translocases. Nat. Rev. Microbiol. 6, 234–244 [DOI] [PubMed] [Google Scholar]
- 2. Driessen A. J., Nouwen N. (2008) Protein translocation across the bacterial cytoplasmic membrane. Annu. Rev. Biochem. 77, 643–667 [DOI] [PubMed] [Google Scholar]
- 3. Dalbey R. E., Wang P., Kuhn A. (2011) Assembly of bacterial inner membrane proteins. Annu. Rev. Biochem. 80, 161–187 [DOI] [PubMed] [Google Scholar]
- 4. Luirink J., Yu Z., Wagner S., de Gier J. W. (2012) Biogenesis of inner membrane proteins in Escherichia coli. Biochim. Biophys. Acta 1817, 965–976 [DOI] [PubMed] [Google Scholar]
- 5. Stephenson K. (2005) Sec-dependent protein translocation across biological membranes. Evolutionary conservation of an essential protein transport pathway (review). Mol. Membr. Biol. 22, 17–28 [DOI] [PubMed] [Google Scholar]
- 6. Wang P., Dalbey R. E. (2011) Inserting membrane proteins. The YidC/Oxa1/Alb3 machinery in bacteria, mitochondria, and chloroplasts. Biochim. Biophys. Acta 1808, 866–875 [DOI] [PubMed] [Google Scholar]
- 7. Funes S., Kauff F., van der Sluis E. O., Ott M., Herrmann J. M. (2011) Evolution of YidC/Oxa1/Alb3 insertases. Three independent gene duplications followed by functional specialization in bacteria, mitochondria and chloroplasts. Biol. Chem. 392, 13–19 [DOI] [PubMed] [Google Scholar]
- 8. Van den Berg B., Clemons W. M., Jr., Collinson I., Modis Y., Hartmann E., Harrison S. C., Rapoport T.A. (2004) X-ray structure of a protein-conducting channel. Nature 427, 36–44 [DOI] [PubMed] [Google Scholar]
- 9. Kuhn A. (1988) Alterations in the extracellular domain of M13 procoat protein make its membrane insertion dependent on SecA and SecY. Eur. J. Biochem. 177, 267–271 [DOI] [PubMed] [Google Scholar]
- 10. Deitermann S., Sprie G. S., Koch H. G. (2005) A dual function for SecA in the assembly of single spanning membrane proteins in Escherichia coli. J. Biol. Chem. 280, 39077–39085 [DOI] [PubMed] [Google Scholar]
- 11. Andersson H., von Heijne G. (1993) Sec dependent and sec independent assembly of E. coli inner membrane proteins. The topological rules depend on chain length. EMBO J. 12, 683–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Economou A., Pogliano J. A., Beckwith J., Oliver D. B., Wickner W. (1995) SecA membrane cycling at SecYEG is driven by distinct ATP binding and hydrolysis events and is regulated by SecD and SecF. Cell 83, 1171–1181 [DOI] [PubMed] [Google Scholar]
- 13. Pogliano J. A., Beckwith J. (1994) SecD and SecF facilitate protein export in Escherichia coli. EMBO J. 13, 554–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chen M., Xie K., Yuan J., Yi L., Facey S. J., Pradel N., Wu L. F., Kuhn A., Dalbey R. E. (2005) Involvement of SecDF and YidC in the membrane insertion of M13 procoat mutants. Biochemistry 44, 10741–10749 [DOI] [PubMed] [Google Scholar]
- 15. Tsukazaki T., Mori H., Echizen Y., Ishitani R., Fukai S., Tanaka T., Perederina A., Vassylyev D. G., Kohno T., Maturana A. D., Ito K., Nureki O. (2011) Structure and function of a membrane component SecDF that enhances protein export. Nature 474, 235–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jiang F., Chen M., Yi L., de Gier J. W., Kuhn A., Dalbey R. E. (2003) Defining the regions of Escherichia coli YidC that contribute to activity. J. Biol. Chem. 278, 48965–48972 [DOI] [PubMed] [Google Scholar]
- 17. Dalbey R. E., Kuhn A. (2004) YidC family members are involved in the membrane insertion, lateral integration, folding, and assembly of membrane proteins. J. Cell Biol. 166, 769–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Samuelson J. C., Chen M., Jiang F., Möller I., Wiedmann M., Kuhn A., Phillips G. J., Dalbey R. E. (2000) YidC mediates membrane protein insertion in bacteria. Nature 406, 637–641 [DOI] [PubMed] [Google Scholar]
- 19. Samuelson J. C., Jiang F., Yi L., Chen M., de Gier J. W. (2001) Function of YidC for the insertion of M13 procoat protein in E. coli. Translocation of mutants that show differences in their membrane potential dependence and Sec requirement. J. Biol. Chem. 276, 34847–34852 [DOI] [PubMed] [Google Scholar]
- 20. Chen M., Samuelson J. C., Jiang F., Muller M., Kuhn A., Dalbey R. E. (2002) Direct interaction of YidC with the Sec-independent Pf3 coat protein during its membrane protein insertion. J. Biol. Chem. 277, 7670–7675 [DOI] [PubMed] [Google Scholar]
- 21. Yi L., Jiang F., Chen M., Cain B., Bolhuis A., Dalbey R. E. (2003) YidC is strictly required for membrane insertion of subunits a and c of the F(1)F(0)ATP synthase and SecE of the SecYEG translocase. Biochemistry 42, 10537–10544 [DOI] [PubMed] [Google Scholar]
- 22. Yi L., Celebi N., Chen M., Dalbey R. E. (2004) Sec/SRP requirements and energetics of membrane insertion of subunits a, b, and c of the Escherichia coli F1F0 ATP synthase. J. Biol. Chem. 279, 39260–39267 [DOI] [PubMed] [Google Scholar]
- 23. van der Laan M., Bechtluft P., Kol S., Nouwen N., Driessen A. J. (2004) F1F0 ATP synthase subunit c is a substrate of the novel YidC pathway for membrane protein biogenesis. J. Cell Biol. 165, 213–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. van Bloois E., Jan Haan G., de Gier J. W., Oudega B., Luirink J. (2004) F(1)F(0) ATP synthase subunit c is targeted by the SRP to YidC in the E. coli inner membrane. FEBS Lett. 576, 97–100 [DOI] [PubMed] [Google Scholar]
- 25. Celebi N., Yi L., Facey S. J., Kuhn A., Dalbey R. E. (2006) Membrane biogenesis of subunit II of cytochrome bo oxidase. Contrasting requirements for insertion of N-terminal and C-terminal domains. J. Mol. Biol. 357, 1428–1436 [DOI] [PubMed] [Google Scholar]
- 26. van Bloois E., Haan G. J., de Gier J. W., Oudega B., Luirink J. (2006) Distinct requirements for translocation of the N-tail and C-tail of the Escherichia coli inner membrane protein CyoA. J. Biol. Chem. 281, 10002–10009 [DOI] [PubMed] [Google Scholar]
- 27. du Plessis D. J., Nouwen N., Driessen A. J. (2006) Subunit a of cytochrome o oxidase requires both YidC and SecYEG for membrane insertion. J. Biol. Chem. 281, 12248–12252 [DOI] [PubMed] [Google Scholar]
- 28. Facey S. J., Neugebauer S. A., Krauss S., Kuhn A. (2007) The mechanosensitive channel protein MscL is targeted by the SRP to the novel YidC membrane insertion pathway of Escherichia coli. J. Mol. Biol. 365, 995–1004 [DOI] [PubMed] [Google Scholar]
- 29. Klenner C., Kuhn A. (2012) Dynamic disulfide scanning of the membrane-inserting Pf3 coat protein reveals multiple YidC substrate contacts. J. Biol. Chem. 287, 3769–3776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Neugebauer S. A., Baulig A., Kuhn A., Facey S. J. (2012) Membrane protein insertion of variant MscL proteins occurs at YidC and SecYEG of Escherichia coli. J. Mol. Biol. 417, 375–386 [DOI] [PubMed] [Google Scholar]
- 31. Scotti P. A., Urbanus M. L., Brunner J., de Gier J. W., von Heijne G., van der Does C., Driessen A. J., Oudega B., Luirink J. (2000) YidC, the Escherichia coli homologue of mitochondrial Oxa1p, is a component of the Sec translocase. EMBO J. 19, 542–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nagamori S., Smirnova I. N., Kaback H. R. (2004) Role of YidC in folding of polytopic membrane proteins. J. Cell Biol. 165, 53–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wagner S., Pop O. I., Pop O., Haan G. J., Baars L., Koningstein G., Klepsch M. M., Genevaux P., Luirink J., de Gier J. W. (2008) Biogenesis of MalF and the MalFGK(2) maltose transport complex in Escherichia coli requires YidC. J. Biol. Chem. 283, 17881–17890 [DOI] [PubMed] [Google Scholar]
- 34. Zhu L., Klenner C., Kuhn A., Dalbey R. E. (2012) Both YidC and SecYEG are required for translocation of the periplasmic loops 1 and 2 of the multispanning membrane protein TatC. J. Mol. Biol. 424, 354–367 [DOI] [PubMed] [Google Scholar]
- 35. Urbanus M. L., Scotti P. A., Froderberg L., Saaf A., de Gier J. W., Brunner J., Samuelson J. C., Dalbey R. E., Oudega B., Luirink J. (2001) Sec-dependent membrane protein insertion. Sequential interaction of nascent FtsQ with SecY and YidC. EMBO Rep. 2, 524–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Houben E. N., Urbanus M. L., Van Der Laan M., Ten Hagen-Jongman C. M., Driessen A. J., Brunner J., Oudega B., Luirink J. (2002) YidC and SecY mediate membrane insertion of a type I transmembrane domain. J. Biol. Chem. 277, 35880–35886 [DOI] [PubMed] [Google Scholar]
- 37. Klenner C., Yuan J., Dalbey R. E., Kuhn A. (2008) The Pf3 coat protein contacts TM1 and TM3 of YidC during membrane biogenesis. FEBS Lett. 582, 3967–3972 [DOI] [PubMed] [Google Scholar]
- 38. Beck K., Eisner G., Trescher D., Dalbey R. E., Brunner J., Müller M. (2001) YidC, an assembly site for polytopic Escherichia coli membrane proteins located in immediate proximity to the SecYE translocon and lipids. EMBO Rep. 2, 709–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Boy D., Koch H. G. (2009) Visualization of distinct entities of the SecYEG translocon during translocation and integration of bacterial proteins. Mol. Biol. Cell 20, 1804–1815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Macfarlane J., Müller M. (1995) The functional integration of a polytopic membrane protein of Escherichia coli is dependent on the bacterial signal-recognition particle. Eur. J. Biochem. 233, 766–771 [DOI] [PubMed] [Google Scholar]
- 41. Seluanov A., Bibi E. (1997) FtsY, the prokaryotic signal recognition particle receptor homologue, is essential for biogenesis of membrane proteins. J. Biol. Chem. 272, 2053–2055 [DOI] [PubMed] [Google Scholar]
- 42. Ito K., Akiyama Y. (1991) In vivo analysis of integration of membrane proteins in Escherichia coli. Mol. Microbiol. 5, 2243–2253 [DOI] [PubMed] [Google Scholar]
- 43. Shimohata N., Nagamori S., Akiyama Y., Kaback H. R., Ito K. (2007) SecY alterations that impair membrane protein folding and generate a membrane stress. J. Cell Biol. 176, 307–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Neale E. J., Rong H., Cockcroft C. J., Sivaprasadarao A. (2007) Mapping the membrane-aqueous border for the voltage-sensing domain of a potassium channel. J. Biol. Chem. 282, 37597–37604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. de Gier J. W., Mansournia P., Valent Q. A., Phillips G. J., Luirink J., von Heijne G. (1996) Assembly of a cytoplasmic membrane protein in Escherichia coli is dependent on the signal recognition particle. FEBS Lett. 399, 307–309 [DOI] [PubMed] [Google Scholar]
- 46. Phillips G. J., Silhavy T. J. (1992) The E. coli ffh gene is necessary for viability and efficient protein export. Nature 359, 744–746 [DOI] [PubMed] [Google Scholar]
- 47. Lüke I., Handford J. I., Palmer T., Sargent F. (2009) Proteolytic processing of Escherichia coli twin-arginine signal peptides by LepB. Arch. Microbiol. 191, 919–925 [DOI] [PubMed] [Google Scholar]
- 48. Zhu L., Wasey A., White S. H., Dalbey R. E. (2013) Charge-composition features of model single-span membrane proteins that determine selection of YidC and SecYEG translocase pathways in Escherichia coli. J. Biol. Chem. 288, 7704–7716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Consler T. G., Persson B. L., Jung H., Zen K. H., Jung K., Privé G. G., Verner G. E., Kaback H. R. (1993) Properties and purification of an active biotinylated lactose permease from Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 90, 6934–6938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kaback H. R. (1971) Bacterial Membranes. Methods Enzymol. 22, 99–120 [Google Scholar]
- 51. Short S. A., Kaback H. R., Kohn L. D. (1975) Localization of D-lactate dehydrogenase in native and reconstituted Escherichia coli membrane vesicles. J. Biol. Chem. 250, 4291–4296 [PubMed] [Google Scholar]
- 52. Guan L., Kaback H. R. (2006) Lessons from lactose permease. Annu. Rev. Biophys. Biomol. Struct. 35, 67–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Tabor S., Richardson C. C. (1985) A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc. Natl. Acad. Sci. U.S.A. 82, 1074–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bibi E., Kaback H. R. (1990) In vivo expression of the lacY gene in two segments leads to functional lac permease. Proc. Natl. Acad. Sci. U.S.A. 87, 4325–4329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kaback H. R., Dunten R., Frillingos S., Venkatesan P., Kwaw I., Zhang W., Ermolova N. (2007) Site-directed alkylation and the alternating access model for LacY. Proc. Natl. Acad. Sci. U.S.A. 104, 491–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. van Iwaarden P. R., Pastore J. C., Konings W. N., Kaback H. R. (1991) Construction of a functional lactose permease devoid of cysteine residues. Biochemistry 30, 9595–9600 [DOI] [PubMed] [Google Scholar]
- 57. Traxler B., Murphy C. (1996) Insertion of the polytopic membrane protein MalF is dependent on the bacterial secretion machinery. J. Biol. Chem. 271, 12394–12400 [DOI] [PubMed] [Google Scholar]
- 58. Matsuyama S., Akimaru J., Mizushima S. (1990) SecE-dependent overproduction of SecY in Escherichia coli. Evidence for interaction between two components of the secretory machinery. FEBS Lett. 269, 96–100 [DOI] [PubMed] [Google Scholar]
- 59. Wang P., Kuhn A., Dalbey R. E. (2010) Global change of gene expression and cell physiology in YidC-depleted Escherichia coli. J. Bacteriol. 192, 2193–2209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. van der Laan M., Urbanus M. L., Ten Hagen-Jongman C. M., Nouwen N., Oudega B., Harms N., Driessen A. J., Luirink J. (2003) A conserved function of YidC in the biogenesis of respiratory chain complexes. Proc. Natl. Acad. Sci. U.S.A. 100, 5801–5806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Sun J., Wu J., Carrasco N., Kaback H. R. (1996) Identification of the epitope for monoclonal antibody 4B1 which uncouples lactose and proton translocation in the lactose permease of Escherichia coli. Biochemistry 35, 990–998 [DOI] [PubMed] [Google Scholar]
- 62. Sun J., Li J., Carrasco N., Kaback H. R. (1997) The last two cytoplasmic loops in the lactose permease of Escherichia coli comprise a discontinuous epitope for a monoclonal antibody. Biochemistry 36, 274–280 [DOI] [PubMed] [Google Scholar]
- 63. van Bloois E., Nagamori S., Koningstein G., Ullers R. S., Preuss M., Oudega B., Harms N., Kaback H. R., Herrmann J. M., Luirink J. (2005) The Sec-independent function of Escherichia coli YidC is evolutionary-conserved and essential. J. Biol. Chem. 280, 12996–13003 [DOI] [PubMed] [Google Scholar]
- 64. Sahin-Tóth M., Dunten R. L., Kaback H. R. (1995) Design of a membrane protein for site-specific proteolysis. Properties of engineered factor Xa protease sites in the lactose permease of Escherichia coli. Biochemistry 34, 1107–1112 [DOI] [PubMed] [Google Scholar]
- 65. Sääf A., Monné M., de Gier J. W., von Heijne G. (1998) Membrane topology of the 60-kDa Oxa1p homologue from Escherichia coli. J. Biol. Chem. 273, 30415–30418 [DOI] [PubMed] [Google Scholar]
- 66. Kuhn A., Wickner W. (1985) Conserved residues of the leader peptide are essential for cleavage by leader peptidase. J. Biol. Chem. 260, 15914–15918 [PubMed] [Google Scholar]
- 67. Date T., Goodman J. M., Wickner W. T. (1980) Procoat, the precursor of M13 coat protein, requires an electrochemical potential for membrane insertion. Proc. Natl. Acad. Sci. U.S.A. 77, 4669–4673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Dowhan W., Bogdanov M. (2009) Lipid-dependent membrane protein topogenesis. Annu. Rev. Biochem. 78, 515–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Vitrac H., Bogdanov M., Dowhan W. (2013) In vitro reconstitution of lipid-dependent dual topology and postassembly topological switching of a membrane protein. Proc. Natl. Acad. Sci. U.S.A. 110, 9338–9343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Gething M. J., Sambrook J. (1992) Protein folding in the cell. Nature 355, 33–45 [DOI] [PubMed] [Google Scholar]
- 71. Bukau B., Weissman J., Horwich A. (2006) Molecular chaperones and protein quality control. Cell 125, 443–451 [DOI] [PubMed] [Google Scholar]
- 72. Horwich A. L., Farr G. W., Fenton W. A. (2006) GroEL-GroES-mediated protein folding. Chem. Rev. 106, 1917–1930 [DOI] [PubMed] [Google Scholar]
- 73. Young J. C., Agashe V. R., Siegers K., Hartl F. U. (2004) Pathways of chaperone-mediated protein folding in the cytosol. Nat. Rev. Mol. Cell Biol. 5, 781–791 [DOI] [PubMed] [Google Scholar]
- 74. Bohnert M., Rehling P., Guiard B., Herrmann J. M., Pfanner N., van der Laan M. (2010) Cooperation of stop-transfer and conservative sorting mechanisms in mitochondrial protein transport. Curr. Biol. 20, 1227–1232 [DOI] [PubMed] [Google Scholar]
- 75. Nagamori S., Vázquez-Ibar J. L., Weinglass A. B., Kaback H. R. (2003) In vitro synthesis of lactose permease to probe the mechanism of membrane insertion and folding. J. Biol. Chem. 278, 14820–14826 [DOI] [PubMed] [Google Scholar]