Background: Drebrin binds to F-actin and CXCR4 in T cells. Thus, it is a potential candidate for the modulation of HIV-1 infection.
Results: Drebrin and CXCR4 accumulate at viral attachment areas. Drebrin knockdown decreases F-actin polymerization, and increases local profilin accumulation and HIV-1 infection.
Conclusion: Drebrin inhibits HIV-1 entry by stabilizing HIV-1-triggered F-actin polymerization.
Significance: Modulation of actin dynamics differentially regulates each viral step for an effective viral infection.
Keywords: Actin, Cxcr4, Cytoskeleton, HIV-1, Virus Entry
Abstract
HIV-1 contact with target cells triggers F-actin rearrangements that are essential for several steps of the viral cycle. Successful HIV entry into CD4+ T cells requires actin reorganization induced by the interaction of the cellular receptor/co-receptor complex CD4/CXCR4 with the viral envelope complex gp120/gp41 (Env). In this report, we analyze the role of the actin modulator drebrin in HIV-1 viral infection and cell to cell fusion. We show that drebrin associates with CXCR4 before and during HIV infection. Drebrin is actively recruited toward cell-virus and Env-driven cell to cell contacts. After viral internalization, drebrin clustering is retained in a fraction of the internalized particles. Through a combination of RNAi-based inhibition of endogenous drebrin and GFP-tagged expression of wild-type and mutant forms, we establish drebrin as a negative regulator of HIV entry and HIV-mediated cell fusion. Down-regulation of drebrin expression promotes HIV-1 entry, decreases F-actin polymerization, and enhances profilin local accumulation in response to HIV-1. These data underscore the negative role of drebrin in HIV infection by modulating viral entry, mainly through the control of actin cytoskeleton polymerization in response to HIV-1.
Introduction
Human immunodeficiency virus (HIV)-1 entry requires fusion between the viral envelope and the plasma membrane of the target cell (1). This process is mediated by the viral envelope glycoprotein complex gp120/gp41 (Env),5 which interacts first with CD4 and then with a co-receptor, one of the chemokine receptors CCR5 or CXCR4 (2, 3).
For the viral and cellular membranes to fuse, a critical number of gp120-CD4/coreceptor engagements are needed to establish an energetically productive fusion pore (4). HIV-1 can also be transmitted from infected to uninfected cells through cell to cell contacts, called virological synapses because of their similarities to the immunological synapse (5). As an example, CD4 and CXCR4 recruitment (6–8) and local actin polymerization take place in both processes (6, 9–11). Receptor clustering is regulated by two main factors: 1) insertion into specific membrane domains by lateral membrane receptor interactions (12) such as lipid rafts (13) or tetraspanin-enriched microdomains (14–17); and 2) actin remodeling (6, 18–20). This phenomenon is well illustrated by experiments in which HIV-1 contact induces CD4 and CXCR4 capping at the plasma membrane of target CD4+ T lymphocytes (21, 22). Receptor capping requires the formation of a subcortical structure enriched in F-actin and actin-binding proteins such as moesin and filamin-A (18, 20). Moesin and filamin-A have been shown to interact with CD4/CXCR4 complexes, controlling their connections with the subcortical F-actin cytoskeleton and directly affecting HIV-1 infection (18, 20).
The role of F-actin remodeling during HIV-1 infection is not well understood (23). The actin cytoskeleton is known to be involved in several steps of the HIV-1 cycle, including entry (18, 20, 24), nucleocapsid transport toward the host nucleus (25, 26), and viral assembly and budding (11, 27, 28). Some recent reports suggest that although F-actin polymerization is needed for receptor clustering, it causes a physical restriction for the following nucleocapsid entry (26, 29). In accordance, activation of the actin-severing protein cofilin has been associated with enhanced HIV entry (26). Moreover, both removal of the bundling activity or siRNA knockdown of α-actinin, an actin microfilament cross-linker, increase HIV-1 infection (30). In addition, knockdown of proteins that link the actin cytoskeleton with the inner leaflet of the plasma membrane, such as talin and vinculin, have also been related with higher HIV-1 infection (31).
Another candidate that might regulate actin cytoskeleton engagement to CD4/CXCR4 complexes, and therefore HIV-1 entry into host cells, is drebrin. Drebrin is an F-actin-binding protein essential for neuronal plasticity because it is able to change the properties of actin filaments, thereby modulating dendritic spine morphology (32). Drebrin is known to stabilize actin filaments (33, 34). We have reported that the N-terminal region of drebrin associates with the cytoplasmic region of CXCR4 in CD4+ T cells by three different approaches: mass spectrometry, pulldown assays, and co-immunoprecipitation (35). Despite containing a N-terminal ADF-H (actin depolymerizing factor homology) domain, drebrin only binds to polymerized (F-) actin and has no severing activity (36). Drebrin appears to regulate F-actin by inducing structural changes in the microfilaments (33, 34) and by competition with other actin-binding proteins such as fascin (37), α-actinin, and tropomyosin (36). Moreover, drebrin regulates the recruitment of other actin-regulatory proteins such as myosin, gelsolin, and profilin through direct association with them (38, 39). We previously showed that interaction between drebrin and CXCR4 occurs at the T lymphocyte membrane and this molecular association is enhanced during superantigen presentation at the immunological synapse (35). However, the role of drebrin and its association with CXCR4 in controlling the cytoskeletal reorganization triggered by HIV-1 infection has not been previously described.
In this report, we assessed the role of drebrin as a possible mediator of CD4/CXCR4 clustering and actin rearrangements during HIV infection. Our results indicate that drebrin is recruited toward HIV-1 viral envelope glycoprotein and negatively regulates HIV-1 infection by controlling the dynamic reorganization of the actin cytoskeleton and profilin accumulation, preferentially at the viral entry step.
EXPERIMENTAL PROCEDURES
Cell Lines and Reagents
The Jurkat-derived human T cell line J77 was grown in RPMI 1640 medium supplemented with 10% FBS (Cambrex Bioscience). The CEM-T4 T cell line (courtesy of Dr. Paul Clapman), the HeLa TZM-bl reporter cell line (courtesy of Dr. John C. Kappes, Dr. Xiaoyun Wu, and Tranzyme Inc.), and the HxBc2 Jurkat-derived T cell line (courtesy of Dr. Joseph Sodroski (40)) were obtained from the National Institutes of Health AIDS Research and Reference Reagent Program, and cultured according to National Institutes of Health instructions. HxBc2 Jurkat cells were used 3 days after removing doxicycline from the culture media, when maximum Env expression was observed. The chronically infected MT4-NL4.3-GFP cell line (MT4-HIV-GFP) was generated by transfecting parental MT4 T cells with HIV-GFP DNA and allowing subsequent viral infection of the culture. Peripheral blood lymphocytes from healthy donors were isolated by Ficoll-Hypaque gradient centrifugation and cultured for 2 days in RPMI 1640 medium supplemented with 10% FBS (Cambrex Bioscience) and phytohemagglutinin (5 μg/ml) or Staphylococcus enterotoxin E (1 μg/ml). Then, isolated T lymphoblasts were maintained in culture for 5 days in the presence of IL-2 (50 units/ml).
The biotinylated monoclonal anti-CXCR4 antibody was from BD Pharmingen. Rabbit polyclonal anti-CXCR4, which recognizes the N-terminal region, rabbit polyclonal anti-drebrin, and monoclonal anti-α-tubulin and anti-gelsolin (clone GS-2C4) were from Sigma. Mouse monoclonal anti-drebrin (clone M2F6) was from MBL (Nagoya, Japan). Anti-CD4 antibodies used were biotinylated monoclonal anti-CD4 antibody (BD Pharmingen) and CD4v4-FITC (BD Pharmingen). The anti-CD45 mAb used was clone D3/9 (15) and anti-CD45-FITC both from BD Pharmingen. The polyclonal anti-phospho-Moesin (Thr-558, sc-12895) and mouse monoclonal anti-Profilin-1 (sc-136432) were from Santa Cruz, the polyclonal anti-phospho-Cofilin (Ser-3, clone 77G2) was from Cell Signaling, and the monoclonal anti-Rac-1 was from BD Biosciences. Anti-phosphatidylinositol 4,5-bisphosphate mAb was from Santa Cruz Biotechnology (clone 2C11; Santa Cruz Biotechnology, Santa Cruz, CA). HRP-conjugated secondary antibodies were from Pierce and Alexa-conjugated secondary antibodies and phalloidins were from Invitrogen. The intracellular fluorescent trackers CMAC, Calcein-AM, and CMTMR were from Molecular Probes (Camarillo, CA).
The HIV-1-specific fusion inhibitor T20 (also called Enfuvirtide) was from Roche Diagnostics. Azidothymidine (Zidovudine) was from Sigma.
Cell Transfection, DNA, and siRNA
J77 cells (2 × 107) were electroporated in cold Opti-MEM (Invitrogen) with DNA (20 μg) or siRNA (1.25 μm) using a Bio-Rad GenePulser II electroporator (240 V; 950 microfarads). Peripheral blood lymphocytes (2 × 107) were electroporated twice in a 48-h interval with siRNA (1 μm) using these same conditions. Fluorescent protein expression and siRNA knockdown were tested by flow cytometry (24 h) and Western blot (48 h), respectively. The GFP fusion proteins drebrin-GFP, Dreb(1–366)-GFP and Dreb(319–707)-GFP were described previously (41). Cell transfection efficiency was 30–70% GFP+ cells. Overexpression of drebrin constructions displayed a GFP/endogenous drebrin ratio of 1.8, 2.0, and 1.5 for drebrin-GFP, Dreb(1–366)-GFP, and Dreb(319–707)-GFP, respectively. Negative control siRNA was from Eurogentec and the specific siRNA against drebrin (mixture of four sequences) was from Dharmacon (Rockford, IL). siRNA against the non-translated (3′ UTR) region of drebrin mRNA was purchased from Dharmacon. This sequence does not interfere with the expression of exogenous drebrin and was employed as an additional control for siRNA specificity.
HIV-1 Viral Preparation, Viral Production, Viral Attachment/Entry, and Viral Infectivity
Preparation of HIV-1 NL4.3 and measurement of viral replication were performed as described (42). Fluorescent virus-like particles (VLPs: Gag-GFP and Gag-Cherry) were produced at the laboratory of Dr. Martinez-Picado (IrsiCaixa, Barcelona, Spain) (43) by co-transfection of the HIV Gag-eGFP/Cherry plasmid plus the pHXB2 envelope plasmid. For VLPs without HIV envelope, cells were only transfected with the HIV Gag-eGFP plasmid.
For p24 production, T cells were infected with 100 ng of HIV-1 NL4.3 per million cells for 2 h at 37 °C, and then extensively washed with medium to remove non-attached viral particles. Infected cells were kept at 37 °C for 6 days. Supernatants were harvested at days 3 and 6, and the p24 concentration was measured by enzyme-linked immunosorbent assay (Innotest HIV-1 antigen mAb; Innogenetic, Ghent, Belgium).
For HIV attachment and entry measurements, T cells were infected with 20 ng of HIV-1 NL4.3 per million cells for 2 h at 4 (attachment) or 37 °C (entry), then extensively washed with medium to remove viral input, and lysed with RIPA buffer (50 mm Tris-HCl, pH 8, 150 mm NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS). Viral attachment (4 °C) corresponds to the p24 amount measured in samples kept at 4 °C and viral entry to the difference between p24 from samples kept at 37 °C and the samples at 4 °C.
For viral infectivity assays, supernatants containing new released viral particles were harvested at day 3 after infection of control or drebrin-depleted J77 cells, and titrated by p24-ELISA. Equivalent p24 amounts of each supernatant were used to infect the TZM-bl reporter cell line for 24 h at 37 °C. Then, the supernatants were removed and cells were lysed with a Steady Glo luciferase assay system (Promega Corp.) and a 1450 Microbeta Luminescence Counter (Walax, Trilux). Replication-deficient luciferase-HIV-1 viral particles (X4-Luc and VSV-Luc) were kindly provided by Suryaram Gummuluru (Boston University, Boston, MA) and were described previously (15). Briefly, virus stocks were generated by PolyFect transient transfection of HEK293T cells (44). Two days after transfection, cell-free virus-containing supernatants were clarified of cell debris and concentrated by centrifugation (16,000 × g for 1 h at 4 °C) and stored at −80 °C until required. HIV-1 virus preparations were titrated by p24-ELISA.
High titers of HIV VSV-G-pseudotyped recombinant virus stocks (VSV-G-HIV) were produced in 293T cells by co-transfection of pNL4–3.Luc.R-E- (AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, National Institutes of Health: pNL4–3.Luc.R-E- from Dr. Nathaniel Landau) together with the pcDNA3-VSV plasmid encoding the vesicular stomatitis virus G-protein using the calcium phosphate transfection system (AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, National Institutes of Health: pHEF-VSV-G from Dr. Lung-Ji Chang). Supernatants containing virus stocks were harvested 48 h post-transfection, centrifuged to remove cell debris, and stored at −80 °C until use. Cell-free viral stock was tested using an enzyme-linked immunoassay for antigen HIV-p24 detection (Innogenetics N.V., Belgium).
Luciferase Virus Assay
Untreated or transfected T cells were infected with 200 ng/million cells of luciferase-based virus with X4-tropic HIV-1 envelope or VSV envelope for 2 h at 37 °C. Virus was removed by washing infected cells. After 48 h, luciferase activity was determined with a luciferase assay kit (Promega Corp.) and a 1450 Microbeta Luminescence Counter (Walax, Trilux). Protein contents were measured by the bicinchoninic acid method (BCA protein assay kit from Pierce).
Env-induced Cell Fusion Assay
A dual-fluorescence cell-fusion assay was performed as described previously (15). Briefly, CMTMR-loaded Env+ Jurkat-Hxbc2 cells were mixed with calcein-AM-loaded parental or GFP-transfected CEM-T4 cells for 16 h at 37 °C. The double-labeled cells were detected by flow cytometry.
Immunoprecipitation
J77 T cells (4 × 107) were incubated with free HIV particles for 1 h at 37 °C, washed and lysed with 1% Nonidet P-40 in TBS supplemented with a protease/inhibitor mixture. Lysates were centrifuged at 11,000 × g for 10 min at 4 °C, and cell lysate supernatants were incubated for 2 h at 4 °C with cyanogen bromide (CNBr) beads (Amersham Biosciences) blocked with BSA, then incubated with the indicated antibody covalently coupled to CNBr beads. Pellets were washed with lysis buffer and resuspended in reducing Laemmli buffer. Samples were separated by SDS-PAGE and transferred to nitrocellulose membranes and incubated with the indicated antibodies.
Immunofluorescence and Confocal Microscopy
T cells were either incubated with HIV-1-wt or VLPs for 30 min at 37 °C, with Env+ cells (HxBc2) for 2 h at 37 °C, or with HIV-1 infected cells (MT4-HIV-GFP) for 1 h at 37 °C. Cells were then seeded on 50 μg/ml of poly-l-lysine for 30 min at 37 °C and fixed with 3% paraformaldehyde. When necessary, samples were permeabilized during 5 min with TBS, 0.5% Triton X-100, stained, and mounted with Prolong (Invitrogen). Images were obtained with a photomicroscope (DMR; Leica, Germany) fitted with an HCX PL APO 63/1.32–210.6 oil objective (Leica) and coupled to a COHU 4912–5010 charge-coupled device camera. Confocal images were obtained with a Leica TCS-SP5 confocal scanning laser microscope fitted with either an HCX PL APO λ blue ×63/1.4 oil immersion objective or an HCX PL APO λ blue ×100/1.4 oil immersion objective and analyzed with Image J.
For drebrin/CD45 recruitment to HIV-1-wt-induced capping areas, accumulation of drebrin or CD45 staining at CD4 or CXCR4 caps was quantified in cells infected with HIV-1-wt and in cell-cell contacts between uninfected cells and MT4-HIV-infected cells. For drebrin clustering quantification, drebrin staining was transformed with the pseudocolor-intensity look-up tables and yellow-white patches at fluorescent VLP locations were considered positive for clustering. Pearson co-localization factors for Drebrin/CXCR4 or GFP/CXCR4 were obtained by analyzing double-stained or simple-stained confocal images respectively, at the maximal intercellular/VLP-cell contact plane with the “Colocalization Threshold” Image J plug-in.
For F-actin and profilin accumulation at intercellular contacts, the Image J plug-in “Synapsemeasure” was used as described previously (45). Briefly, by selecting regions of interest of the same area for all measurements, the fluorescence intensity at the cell to cell contact area (V), an area of the infected cell not in contact with the target cell (I), an area of the target T cell not in contact with the infected cell (T), and the background (Bg) were quantified. Background signal was subtracted from all other measurements, and then the ratio of the fluorescence intensity accumulated at the cellular contact relative to the rest of the target T cell surface was analyzed according to the formula (V-I)/T.
Western Blot Assays
J77 T cells were lysed in TBS, 1% Nonidet P-40 supplemented with a mixture of protease and phosphatase inhibitors. Lysates were centrifuged at 11,000 × g for 10 min at 4 °C, and supernatants were mixed with reducing Laemmli buffer and boiled for 5 min. Lysates were separated by SDS-PAGE and immunoblotted with specific antibodies. Protein bands were analyzed using the LAS-1000 CCD system and Image Gauge 3.4 (Fuji Photo Film Co., Tokyo, Japan).
Flow Cytometry
Cells were incubated with TBS, 5% BSA for 30 min at 4 °C and then with primary monoclonal antibody (30 min at 4 °C). After washing, cells were incubated with a FITC-conjugated secondary antibody and analyzed in a FACScalibur flow cytometer (BD Biosciences). Data were analyzed with Cell-Quest Pro (BD Biosciences).
F-actin Quantification
Cells were stimulated with HIV NL4.3-free virions (100 ng/million cells) for different times at 37 °C, fixed with 4% formaldehyde, permeabilized with TBS, 0.5% Triton X-100 (5 min) and stained with phalloidin/Alexa 488 or phalloidin/Alexa 647 (Invitrogen). Mean fluorescence intensity of F-actin staining was analyzed in a FACScalibur cytometer (BD Biosciences).
Quantitative PCR Analysis
Cells were spin-infected with 200 ng/106 with VSV-G-HIV at 1200 × g during 1 h at 37 °C with a further incubation period of 1 h at 37 °C, washed, incubated 24 or 48 h at 37 °C, and lysed in 0.2% Nonidet P-40. Total genomic DNA was extracted using QiAamp DNA mini kit (Qiagen) and amplified using Power SYBR Green PCR master mixture (Applied Biosystems): forward primer, 5′-CAGGATTCTTGCCTGGAGCTG-3′ and reverse primer, 5′-GGAGCAGCAGGAAGCACTATG-3′ for early reverse transcription products, and forward primer, 5′-TGTGTGCCCGTCTGTTGTGT-3′ and reverse primer 5′-CGAGTCCTGCGTCGAGAGAT-3′ for late reverse transcription products. The β-actin gene was amplified to measure DNA concentration and used for normalization. Each reaction was performed in triplicate. Azidothymidine (Zidovudine) was added in each experiment as a negative control, inhibiting between 70 and 90% the amount of early and late reverse transcription products.
Statistical Analysis
Statistical significance was calculated on the raw data using paired Student's t test or the parametric one-way analysis of variance with Bonferroni's post hoc multiple comparison test. Significant differences are labeled *, p < 0.05; **, p < 0.01; and ***, p < 0.001.
RESULTS
Drebrin Interacts with CXCR4 during HIV-1 Infection and Is Recruited by the Viral Envelope
We previously reported that the N-terminal region of drebrin interacts with the C-terminal cytoplasmic tail of CXCR4 in primary T lymphoblasts and T cell lines, and that this interaction is increased by antigen presentation (35). To assess whether this interaction also occurs during HIV-1 infection, where CXCR4 is the viral co-receptor, we incubated J77 CD4+ target T cells with free HIV-1 viral particles (NL4.3 strain). Immunoblotting of drebrin immunoprecipitates revealed that the association between drebrin and CXCR4 is maintained in HIV-1-infected T cells (Fig. 1A).
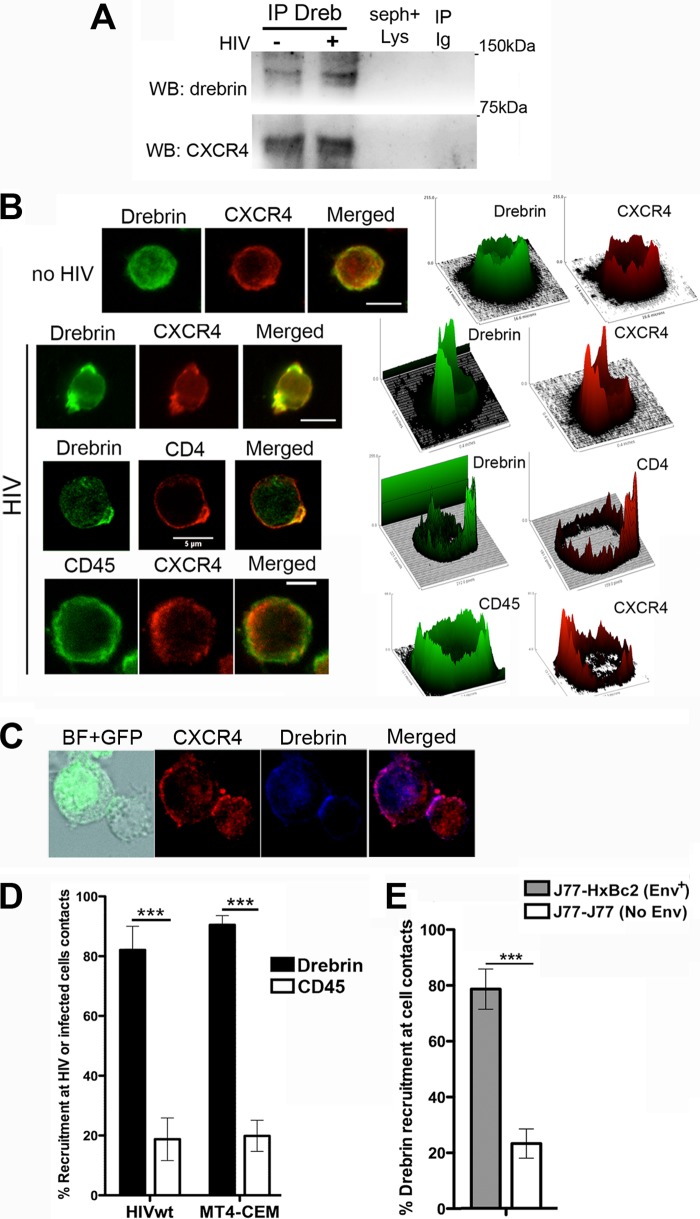
FIGURE 1.
Drebrin interacts and localizes with CXCR4 during HIV infection of CD4+ T cells. A, T cells incubated with HIV-1 viral particles were lysed, immunoprecipitated with anti-drebrin M2F6 mAb (lanes IP drebrin), and blotted with CXCR4 pAb. Western blot of drebrin is shown as a loading control. IP Ig, immunoprecipitation with the isotype control Ab; seph+lys, lysate immunoprecipitated with beads uncoupled to antibody. B, CEM-T4 cells incubated for 30 min at 37 °C with HIV-1 viral particles (lower panels) or virus-free medium (upper panel) were fixed and stained for drebrin (M2F6 mAb), CXCR4 (BD biotinylated mAb), CD4 (BD biotinylated mAb), or CD45 (BD FITC-mAb). Summatory projections of confocal stack images are shown. Bars, 5 μm. Surface plots beside the corresponding images represent fluorescence distribution versus intensity. C, target T cells were conjugated for 2 h at 37 °C with HIV-1-infected T cells (MT4-HIV-GFP) and stained for drebrin (M2F6 mAb) and CXCR4 (N-t pAb). Summatory projections of confocal stack images are shown. Bar, 5 μm. D, quantification of drebrin (black bars) and CD45 (white bars) recruitment at the capping areas (HIV-wt) or at target-MT4-HIV intercellular contacts (MT4-CEM) are depicted (mean ± S.D.). More than 100 particles or contacts were quantified for each category in two independent experiments. E, quantification of drebrin recruitment toward J77-Env+ HxBc2 cells contacts (gray bar) or J77-J77 contacts (white bar). More than 100 contacts were quantified for each category in two independent experiments.
Next, we assessed whether free viral particles were able to induce the recruitment of drebrin to viral attachment sites: CD4+ T cells (CEM-T4) were incubated with free HIV-1 viral particles, which promote CD4 and CXCR4 polarization (“capping”) (21, 22, 46). Staining for drebrin and either CXCR4 or CD4 revealed that HIV-1 viruses triggered the recruitment of drebrin to the CD4/CXCR4 capping areas (Fig. 1, B and D, for quantification), whereas in the absence of HIV virus drebrin displayed a diffuse subcortical pattern (Fig. 1B, upper panel). CD45, included as a control, did not show specific accumulation at HIV-1-triggered CD4/CXCR4 capping areas (Fig. 1, B and D, for quantification). Localization of drebrin-CXCR4 complexes during HIV-induced cell to cell contact areas was also determined by incubating CEM-T4 cells with T cells infected with HIV-GFP (MT4-HIV). Staining for drebrin and CXCR4 showed that both proteins were accumulated at cell to cell contact areas, where viral synapses are established (Fig. 1, C and D, for quantification). CD45, as a control, did not display specific enrichment at MT4-HIV-target cell contacts (Fig. 1D). To assess whether HIV-1 Env was sufficient for drebrin recruitment, the assay was repeated using target Jurkat cells (clone J77) and a Jurkat clone expressing Env at the plasma membrane (Env+ Jurkat-HxBc2 cells). As before, drebrin and CXCR4 were specifically enriched at Env-driven cell to cell contacts (Fig. 1E). Remarkably, no specific drebrin recruitment was observed in cellular contacts between two target cells, where there is no HIV-1 Env (Fig. 1E). Accumulation of drebrin at Env-induced capping areas and Env-induced cellular contacts, but not at unspecific cell to cell contacts, illustrates the specificity of drebrin recruitment by HIV-1 Env.
Drebrin Accumulates at Viral Attachment Sites and Partially Co-localizes with Viral Particles after Internalization
To ascertain whether HIV Env was enhancing drebrin-CXCR4 association, the co-localization of these two proteins was measured. T cells were incubated with Env+ fluorescent virus-like particles (VLPs), which allow visualization of individual particles (data not shown). Co-localization of drebrin-CXCR4 was enhanced at regions of VLP contacts compared with regions where no VLPs were attached to the cell membrane (Fig. 2A).
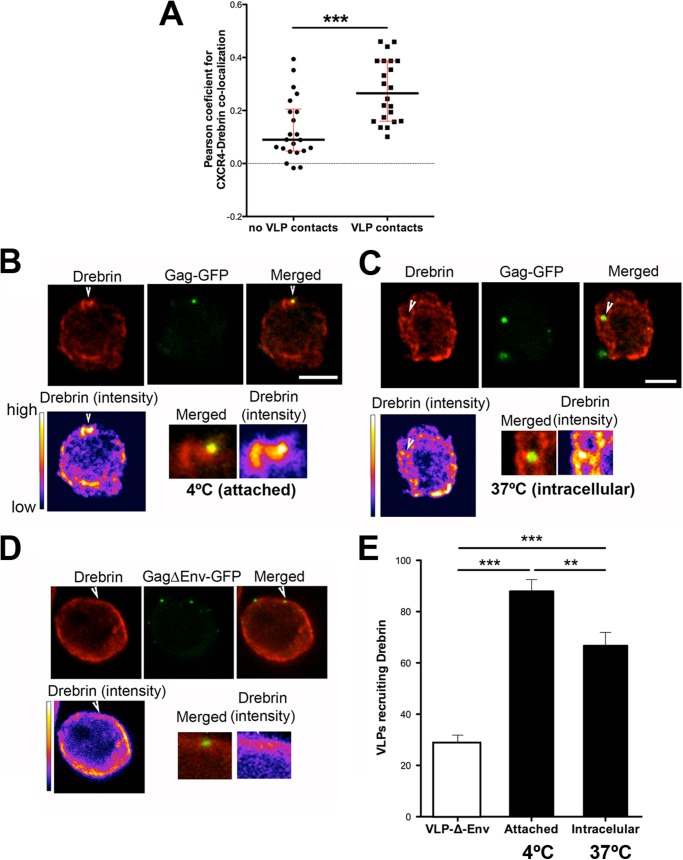
FIGURE 2.
Drebrin recruitment at different steps of HIV-1 infection with VLPs. A, measurement of Pearson's coefficient for CXCR4-drebrin co-localization at VLPs contact sites or other membrane areas. Each point represents the Pearson's coefficient for one VLP contact site/membrane area. The medians and the interquartile ranges are also depicted on the graph. More than 20 areas were quantified for each category in two independent experiments. B, confocal images showing the recruitment of endogenous drebrin toward attached VLPs (Gag-GFP) at 4 °C. Images are projections of the planes in which each individual VLP is observed. Bar, 5 μm. Drebrin staining is shown in pseudocolor intensity-coding format below the confocal image. The small lower panels are zoomed views of VLP contacts from the merged immunostaining and pseudocolor drebrin images (×3). Note that the maximum intensity of drebrin staining, in white at the pseudocolor scale images, fits with the VLP attachment site. C, confocal images showing the endogenous drebrin distribution in a target T cell with internalized VLPs (at 37 °C). Images are projections of the planes in which each individual VLP is observed. Bar, 5 μm. Drebrin staining is shown in pseudocolor intensity-coding format below the confocal image. Lower panels as described in B. D, confocal images showing the absence of recruitment of endogenous drebrin when cells were incubated with VLPs without Env (VLPΔEnv-GFP) (at 37 °C). Images are projections of the planes in which each individual VLP is observed. Bar, 5 μm. Drebrin staining is shown in pseudocolor intensity-coding format below the confocal image. Lower panels as described in B. E, quantification of VLPs that cluster drebrin at the plasma membrane of target cells (4 °C, attached particles), or at the cytoplasm (37 °C, intracellular particles). VLPs not covered with a viral envelope (VLP-Δ-Env) induce basal drebrin accumulation (white bar). More than 100 particles were quantified for each category in three independent experiments.
We next investigated drebrin accumulation at virus attachment sites and upon viral internalization. VLPs were incubated with CEM-T4 cells for 30 min at 4 or 37 °C, fixed, and immunostained for drebrin. At 4 °C, a temperature that does not allow membrane fusion, thus arresting VLPs at the attachment step, drebrin was enriched at 87.95% of VLP contacts (Fig. 2, B and E, for quantification). In this analysis, only the confocal planes where the VLP is observed (usually 1–2 planes separated by 0.25 μm) were analyzed to avoid counting false positives due to fortuitous superposition. Moreover, the accumulation was considered positive only if the intensity of drebrin labeling at the VLP contact area was depicted in white-yellow when using the pseudocolor intensity look-up tables. Incubation at the permissive temperature (37 °C) revealed that 66.71% of intracellular VLPs still localized with drebrin clusters (Fig. 2, C and E, for quantification). Most importantly, accumulation of drebrin at the VLP contact sites was dependent on HIV-1 envelope, because VLPs not covered with a viral envelope (VLP-Δ-Env) were able to induce drebrin recruitment in only 28.59% of the cases (Fig. 2, D and E, for quantification).
Our results indicate that drebrin is recruited together with CD4 and CXCR4 during HIV-1 attachment at the plasma membrane in an Env-dependent manner. This localization is partly maintained after virus internalization, suggesting that drebrin may form part of the actin complex associated with the viral nucleocapsid during its internalization.
Drebrin Regulates HIV-1-induced Cell Fusion and Viral Production
To assess the specific role of drebrin during HIV-1 infection, target T cells were transfected with GFP-tagged drebrin or two different truncated forms: Dreb(1–366)-GFP, which contains only the first 366 amino acids and lacks the Pro-rich motives needed for the interaction with profilin and other molecules; and Dreb(319–707)-GFP, which on the contrary lacks the actin-binding domain and keeps the Pro-rich region (Fig. 3A) (35, 39, 41). In the absence of HIV-1 viral particles, drebrin-GFP and Dreb(1–366)-GFP distributed similarly to their endogenous counterparts (Fig. 3B, left panels). Equal distribution of Dreb(1–366)-GFP is consistent with its ability to interact with CXCR4 and F-actin through its intact N-terminal region. Dreb(319–707)-GFP had a delocalized cytoplasmic distribution in the absence or presence of HIV viral particles, as expected because it can neither bind F-actin nor CXCR4 (Fig. 3B) (35). Both drebrin-GFP and Dreb(1–366)-GFP were recruited together with CXCR4 to the capping areas induced by HIV-1 particles (Fig. 3B, right panels). Drebrin-GFP and Dreb(1–366)-GFP both co-localize with CXCR4 in the absence of viral particles, whereas much lower co-localization was observed for Dreb(319–707)-GFP and CXCR4 (Fig. 3C). CXCR4 co-localization with drebrin-GFP and Dreb(1–366)-GFP was significantly enhanced in VLPs membrane contact areas, whereas no increase was observed for Dreb(319–707)-GFP/CXCR4 co-localization (Fig. 3C). Similar results were obtained at Env-driven cell to cell contacts (Fig. 3D and data not shown).
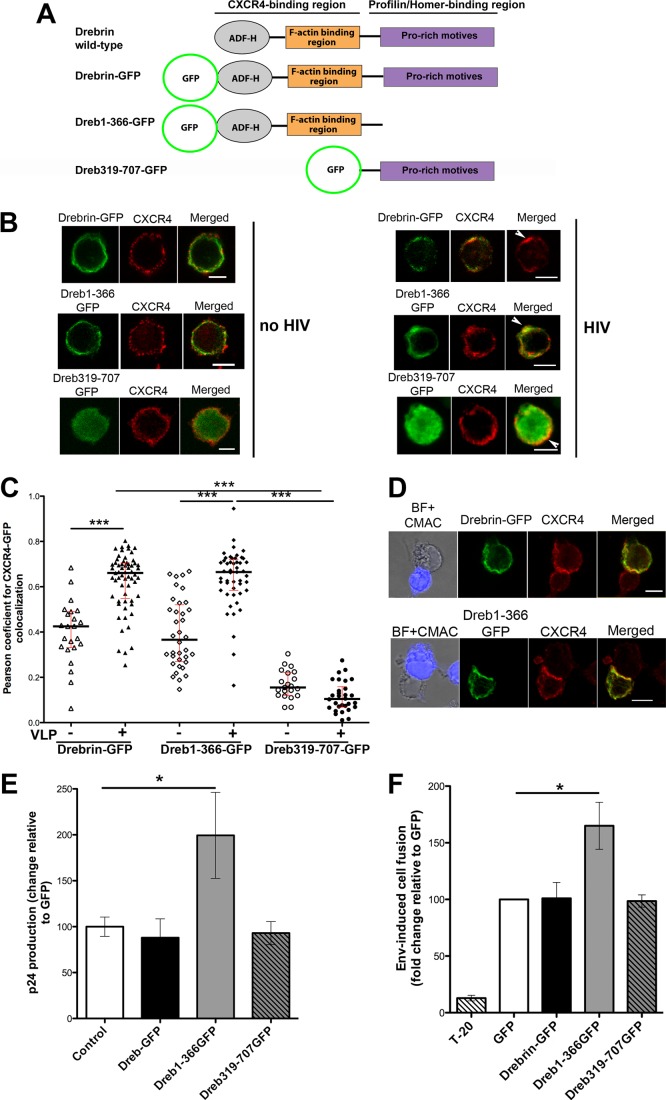
FIGURE 3.
Effect of the overexpression of drebrin wild-type or the deletion mutants, Dreb(1–366)-GFP and Dreb(319–707)-GFP, on HIV infection. A, scheme showing drebrin domains and the different GFP-fused constructs (drebrin-GFP, Dreb(1–366)-GFP, and Dreb(319–707)-GFP). ADF-H stands for actin-depolymerizing factor-homology domain. B, CEM-T4 cells overexpressing drebrin-GFP, Dreb(1–366)-GFP, or Dreb(319–707)-GFP were incubated for 30 min at 37 °C with HIV-1 viral particles (right panel) or virus-free medium (left panel), fixed and stained for CXCR4 (BD mAb). Summatory projections of confocal stack images are shown. Bars, 5 μm. C, measurement of Pearson's coefficient for CXCR4/GFP co-localization at cells overexpressing drebrin-GFP, Dreb(1–366)-GFP, or Dreb(319–707)-GFP in the absence or presence of VLPs. Each point represents the Pearson's coefficient for one VLP contact. The medians and the interquartile ranges are also depicted on the graph. 30–60 VLPs contacts were quantified in each category from three independent experiments. No statistically significant variation was observed between drebrin-GFP and Dreb(1–366)-GFP in the absence of VLPs; drebrin-GFP and Dreb(1–366)-GFP in the presence of VLPs; or Dreb(319–707)-GFP in the absence or presence of VLPs. D, CEM-T4 cells overexpressing drebrin-GFP or Dreb(1–366)-GFP were conjugated with Env+ HxBc2 cells (in blue) for 2 h at 37 °C, fixed, and stained for CXCR4 (BD mAb). Summatory projections of confocal stack images are shown. Bars, 5 μm. E, HIV-1 infection was enhanced by Dreb(1–366)-GFP overexpression. Infection was measured as viral production, quantified by ELISA as the p24 viral protein present in supernatants at day 3 after infection at 37 °C. Data are the fold-induction relative to control cells transfected with GFP alone (mean ± S.D. of four experiments performed in triplicate). F, cell fusion triggered by HIV-1 Env+ HxBc2 cells was increased in cells overexpressing Dreb(1–366)-GFP. Cells were incubated for 16 h at 37 °C and syncytia formation was quantified by flow cytometry. Data are presented as the fold-induction relative to GFP-transfected cells (mean ± S.D. of four experiments performed in duplicate).
The functional effect of drebrin overexpression on HIV-1 infection was assessed by infecting target cells transfected with GFP alone, drebrin-GFP, Dreb(1–366)-GFP, or Dreb(319–707)-GFP. Viral production was measured 3 days after infection by p24 ELISA. As shown in Fig. 3E, overexpression of drebrin-GFP or Dreb(319–707)-GFP did not affect viral production, whereas Dreb(1–366)-GFP expression increased it.
Because drebrin was recruited at the plasma membrane by free HIV-1 viral particles and at Env-driven cell contacts, the step enhanced by Dreb(1–366)-GFP could be HIV-1-induced membrane fusion. We next measured Env-mediated cell fusion by flow cytometry using Env+ HxBc2 cells and target CEM-T4 cells. Env+ cells were marked with a red intracellular tracker (CMTMR) and incubated overnight with target T cells transfected with GFP alone or each of the GFP-tagged drebrin constructs. Env-driven cell fusion leads to syncytia formation, detected as double fluorescence positive events of greater size and complexity. As a control of specificity, syncytia formation was blocked by the HIV-1 fusion inhibitor T20. Quantification of these events yielded results comparable with the viral production experiments: overexpression of wild-type drebrin or Dreb(319–707)-GFP did not affect Env-mediated cell fusion, whereas Dreb(1–366)-GFP increased it (Fig. 3F), thus suggesting that the N-terminal region of drebrin is needed for the inhibition of HIV-1-induced membrane fusion.
Down-regulation of Drebrin Expression Increases HIV-1 Syncytia Formation and Viral Entry
To confirm the regulatory role of drebrin during HIV-1 infection, we knocked down its expression using siRNA. Down-regulation was assessed for each experiment by Western blot (Fig. 4A, representative blot). Control target cells were transfected with a siRNA sequence that does not hybridize with any eukaryotic mRNA. FACS analysis confirmed that CD4 and CXCR4 expression at the plasma membrane of target cells was not affected by drebrin silencing (data not shown). As expected, CXCR4 co-localization with drebrin at VLPs contacts or Env-driven cell to cell contacts was reduced in drebrin-depleted cells (Fig. 4B and data not shown).
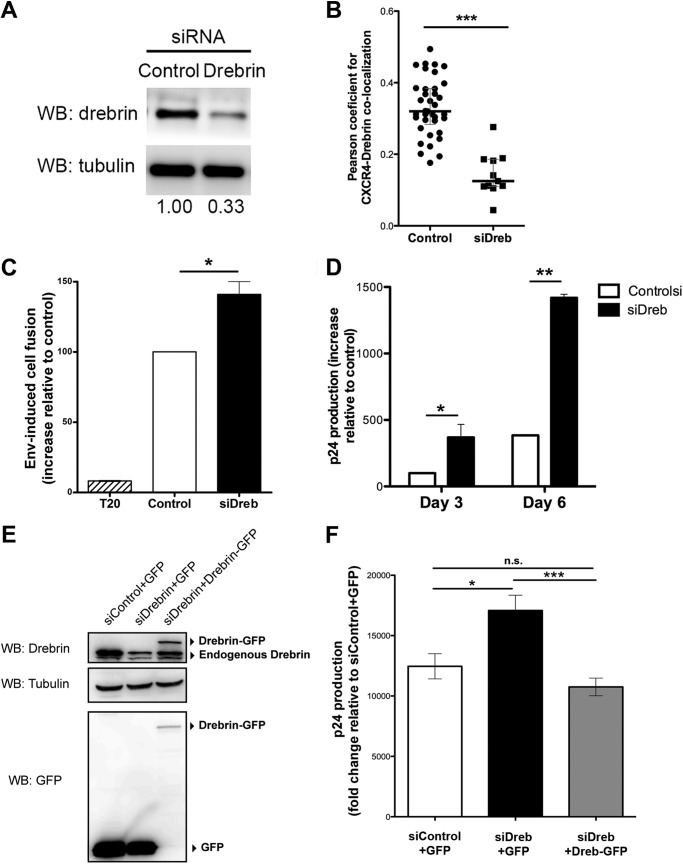
FIGURE 4.
Knockdown of drebrin expression increases HIV-1 infection and Env-induced cell fusion. A, representative Western blot showing silencing of drebrin endogenous expression 48 h after siRNA transfection of the negative oligonucleotide (Control) or siRNA against drebrin (Drebrin) in Jurkat T cells. Tubulin was used as loading control. Silencing is quantified below as the band intensity ratio with respect to tubulin. B, measurement of Pearson's coefficient for CXCR4/drebrin co-localization at VLPs contact areas in control or drebrin-depleted cells. Each point represents the Pearson's coefficient for one VLP contact. The medians and the interquartile ranges are also depicted on the graph. Two independent experiments were quantified. C, drebrin silencing in target T cells increased Env+-induced cell fusion. Cells were incubated for 16 h at 37 °C and syncytia formation was quantified by flow cytometry. Data are presented as fold-induction relative to cells transfected with control siRNA (Control). Results are mean ± S.D. of four independent experiments performed in duplicate. D, effect of drebrin silencing on HIV-1 infection, as measured as viral production after 3 and 6 days at 37 °C. Results were quantified as p24 viral protein content in supernatants by ELISA. Data are the mean fold-induction relative to control cells ± S.D. of four experiments performed in triplicate. E, representative Western blot (WB) of Jurkat T cells co-transfected with siRNA against the non-coding sequence of drebrin mRNA (siDrebrin) or control siRNA, and GFP or drebrin-GFP. Cell lysates were immunoblotted for drebrin and GFP. Tubulin was used as loading control. F, control or drebrin-depleted T cells overexpressing either GFP or drebrin-GFP were infected with HIV NL4.3 strain virus for 2 h at 37 °C. Supernatants were harvested at day 3 after infection and p24 was measured by ELISA. Results are the mean ± S.E. of three experiments performed in triplicates.
To assess Env-driven cell fusion, silenced T cells marked with a green cell tracker were incubated overnight with Env+ HxBc2 cells loaded with CMTMR. Drebrin-silenced cells exhibited enhanced cell fusion with Env+ cells compared with control cells (Fig. 4C). In parallel, control and silenced cells were infected with HIV-1 viral particles and viral production was measured by p24 ELISA 3 and 6 days after infection. Drebrin-silenced cells produced higher titers of HIV than control cells in both time points (Fig. 4D), indicating that drebrin negatively regulates HIV-1 infection. Additionally, possible nonspecific side effects of siRNA were ruled out by using a siRNA against the non-coding sequence of drebrin mRNA in cells overexpressing GFP or drebrin-GFP (Fig. 4E). Cells with endogenous drebrin knocked down and GFP expression produced higher titers of HIV than cells transfected with control oligonucleotide and GFP (Fig. 4F). Drebrin-GFP overexpression in cells with endogenous drebrin knocked down rescued the normal levels of infection (Fig. 4F).
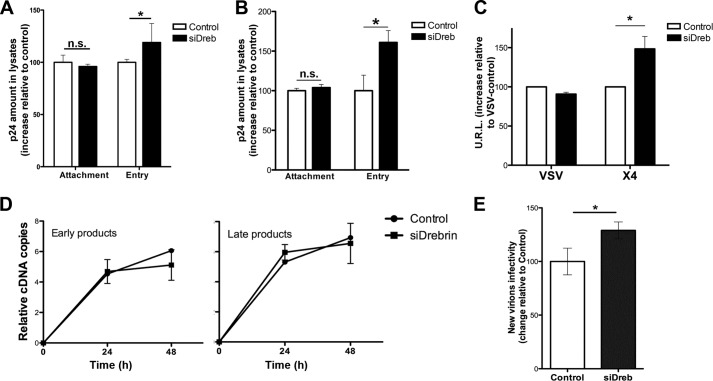
To dissect the precise involvement of drebrin in the first steps of HIV infection, drebrin-silenced or control T cells were incubated at 4 or 37 °C for 2 h with HIV-1 particles at a low infection rate (avoiding superinfection and cooperative entry of virions), washed, and lysed. The amount of p24 in the lysates was measured as an estimate of HIV attachment (4 °C) or HIV entry (subtraction of values of 4 °C from the values of 37 °C). The p24 amount was equal in control or drebrin-silenced cells incubated with HIV-1 at 4 °C. However, it was increased in silenced cells at 37 °C (and therefore at 37 °C less 4 °C), suggesting that HIV entry is specifically enhanced by drebrin silencing (Fig. 5A). Similar results were obtained when this experiment was repeated using primary T lymphoblasts (Fig. 5B). To confirm these observations, we infected target cells with one-cycle luciferase virus pseudotyped with an HIV-1 envelope (X4-Luc), or VSV (vesicular stomatitis virus) envelope (VSV-Luc) as a control. Higher luciferase levels were found in X4-Luc-infected drebrin-silenced cells than in controls, confirming that drebrin knockdown specifically increases viral entry (Fig. 5C). VSV infection was similar in silenced and control cells, demonstrating that the effect of drebrin silencing is HIV-1 Env-dependent (Fig. 5C). These results indicate that drebrin negatively regulates HIV-1 entry without affecting viral attachment.
FIGURE 5.
Drebrin silencing increases HIV-1 entry and infectivity of the viral progeny. A and B, effect of in drebrin silencing on HIV-1 attachment (4 °C) and entry (37- 4 °C) into target T cells (A, J77 cell line; and B, primary human T lymphoblasts). Data are the fold-induction relative to control cells; mean ± S.D. of three and four experiments performed in triplicate, respectively. C, HIV-1 entry in drebrin-silenced cells was measured by infection with one-cycle viral particles carrying a luciferase reporter gene. Infection was performed at 37 °C. Data represent the mean fold-induction of luciferase-HIV-1-envelope virus relative to control cells and normalized with respect to luciferase-VSV-envelope virus (mean ± S.D.). Results are from four independent experiments performed in triplicate. D, time course of viral reverse transcription as measured by quantitative PCR analysis of control and drebrin-silenced T cells infected with VSV-G-HIV at 37 °C. Data represent the mean fold-induction of early or late HIV products normalized with respect to β-actin transcription (mean ± S.D.). Results are from three independent experiments performed in triplicate. E, effect of drebrin silencing on new virions infectivity. Equal p24 amounts of HIV-1 produced by control or drebrin-depleted cells were used to infect the reporter cell line TZM-bl for 24 h at 37 °C. Data represent the mean fold-induction relative to HIV-1 virus produced by control cells (mean ± S.D.). Results are from four independent experiments performed in triplicate.
To further analyze whether drebrin was affecting other steps after viral entry, we assessed HIV-1 reverse transcription. To measure possible changes in this post-entry step without the observed effect of drebrin silencing in HIV-1 entry, we used HIV-1 pseudotyped with VSV-G envelope glycoprotein (VSV-G-HIV) instead of HIV-1 WT, avoiding the increase observed in HIV-1 entry by drebrin knock-down masked any specific effect on reverse transcription. Quantitative PCR for early and late viral reverse transcription products was performed in control or drebrin-silenced cells after infection with VSV-G-HIV. Drebrin silencing did not alter the amount of either early or late viral products (Fig. 5D), suggesting that drebrin is not affecting HIV-1 reverse transcription step.
To test the consequences of drebrin silencing in longer kinetics, the infectivity of the following viral generation was measured. New virions released by either infected control cells or drebrin-depleted cells were harvested and quantified by p24 ELISA. Equivalent p24 amounts of virus produced by control or drebrin-depleted cells were used to infect the TZM-bl reporter cell line (Fig. 5E). Interestingly, viral particles released from drebrin-depleted cells were more infectious than the ones released from control cells, suggesting that drebrin also has a limiting role in the infectivity of the following viral progeny.
Drebrin Regulates Actin Polymerization Induced by HIV-1 Contact
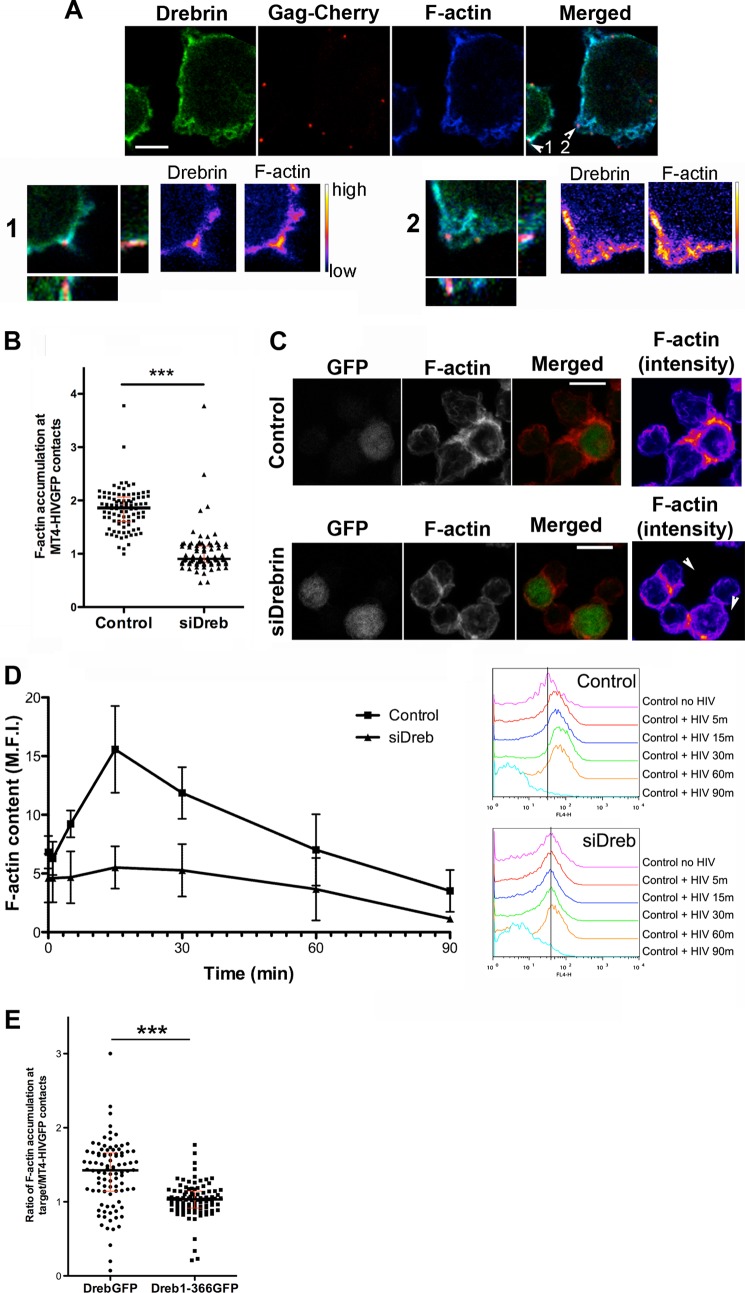
HIV-1 infection is known to trigger actin remodeling, which is essential for CD4/CXCR4 clustering and effective virus entry and viral synapse formation (6, 19). Drebrin is also an important mediator of actin polymerization in neurons (32) and T cells in response to antigen presentation (35). Drebrin and F-actin have been shown to bind and/or co-localize in keratinocytes, neurons, and T cells (35, 41, 47). To study F-actin distribution during HIV-1 infection, we detected F-actin and drebrin during VLP-target cell contacts using confocal microscopy. Staining patterns of drebrin and F-actin were almost identical throughout the plasma membrane, with both showing accumulation at VLP attachment sites (Fig. 6A).
FIGURE 6.
Actin polymerization is reduced in drebrin-silenced cells. A, Jurkat J77 T cells were incubated with fluorescent VLPs (Gag-Cherry) for 30 min at 37 °C, fixed, and stained for drebrin (M2F6 mAb) and F-actin (Phalloidin). Summatory projections of confocal stack images are shown. Bar, 5 μm. Lower panels show zoomed images of selected confocal planes in which individual VLPs are observed (numbered arrows in the merged image), together with the respective orthogonal sections for each particle site, and drebrin and F-actin stainings in pseudocolor intensity-coding format. B, quantification of F-actin accumulation at contacts between control or drebrin-silenced cells and MT4-HIV-GFP-infected cells. Each point represents F-actin accumulation intensity ratio at one contact. More than 100 events were counted in each condition. The medians and the interquartile ranges are also depicted on the graph. C, control or drebrin knockdown target cells were incubated with MT4-HIV-GFP cells (green) for 2 h at 37 °C, fixed, permeabilized, and stained for F-actin (red). Summatory projections of representative confocal stack images are shown, as well as merged and F-actin staining in pseudocolor intensity-coding format on the right panels. Arrowheads indicate contacts between HIV-1-infected and target T cells. Bar, 10 μm. D, time profile of F-actin content upon HIV-1 contact for different times at 37 °C, measured by flow cytometry in drebrin-silenced and control cells. Data are the mean ± S.D. of three independent experiments. p < 0.01. Right panels show flow cytometry histograms of a representative experiment of F-actin fluorescence intensity upon HIV-1 contact at the different time points analyzed in drebrin-silenced and control cells. E, quantification of F-actin accumulation at contacts between cells overexpressing Dreb-GFP or Dreb(1–366)-GFP and MT4-HIV-GFP-infected cells labeled with the cell tracker CMAC. Each point represents the F-actin accumulation intensity ratio at one contact. More than 100 events were counted in each condition. The medians and the interquartile ranges are also depicted on the graph.
Quantification of F-actin staining intensity at contacts between target control or drebrin-silenced cells and infected cells (MT4-HIV-GFP) showed a decrease in F-actin accumulation at cell to cell contacts, with respect to the rest of the cell membrane (Fig. 6, B and C). We also assessed the progression of actin polymerization after HIV-1 contact in drebrin-silenced T cells. HIV-1 induced a marked increase in F-actin content in control T cells, and this increase was impaired by drebrin knockdown (Fig. 6D). This indicates that drebrin is needed for the actin polymerization triggered by HIV-1 attachment to the plasma membrane. Decreased actin polymerization in target T cells caused by RNAi-mediated down-regulation of drebrin correlates with higher HIV-1 entry. In addition, F-actin staining intensity at contacts between target cells overexpressing wild-type drebrin-GFP or Dreb(1–366)-GFP, and infected cells (MT4-HIV-GFP) showed a decrease in F-actin accumulation in cells overexpressing Dreb(1–366)-GFP truncated mutant (Fig. 6E).
Therefore, drebrin knockdown reduces F-actin polymerization triggered by HIV-1 viral particles or HIV-1 Env at cell to cell contacts, in correlation with higher HIV-1 entry. Overexpression of the truncated form Dreb(1–366)-GFP also decreases F-actin accumulation at cell to cell contacts with infected cells.
Drebrin Regulates Profilin Accumulation at Intercellular Contacts with Infected MT4-HIV-GFP Cells
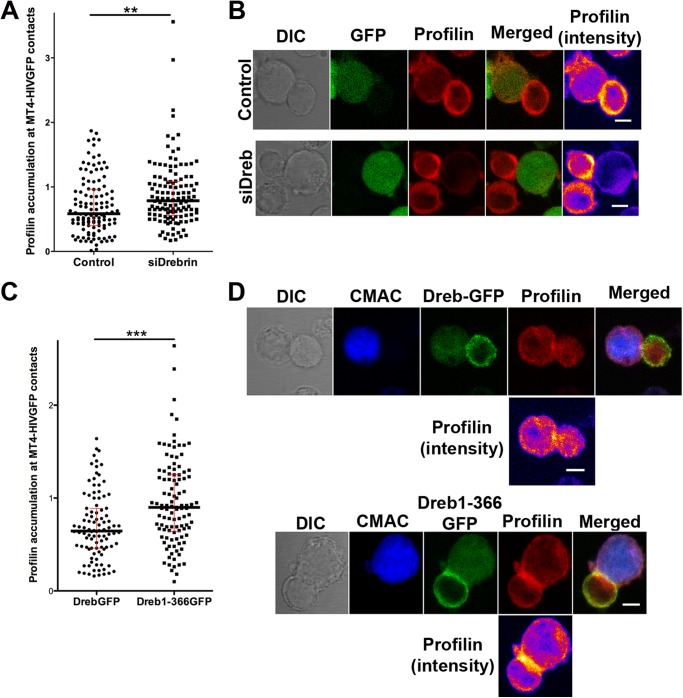
To further assess the mechanism of the drebrin limiting role in HIV-1 infection and its relationship with F-actin remodeling, the distribution of other F-actin regulatory proteins was analyzed. The distribution of phospho-Moesin, phospho-Cofilin, gelsolin, Rac, and phosphatidylinositol 4,5-bisphosphate at VLPs or Env-driven cell to cell contacts were not affected by drebrin knockdown or Dreb(1–366)-GFP overexpression (data not shown). Importantly, profilin accumulation at cell-cell contacts between drebrin-depleted cells and infected cells (MT4-HIV-GFP) was significantly increased in comparison to control cells (Fig. 7, A and B). In addition, cells overexpressing the truncated mutant Dreb(1–366)-GFP displayed a similar profile, with higher profilin local accumulation in comparison with cells overexpressing drebrin-GFP (Fig. 7, C and D). This enhancement in profilin relocalization toward the infected cells was not observed in cells overexpressing GFP alone or the mutant Dreb(319–707)-GFP, which showed comparable profilin distribution as cells overexpressing drebrin-GFP (data not shown). Taken together, our results indicate that both drebrin knockdown and overexpression of the Dreb(1–366)-GFP-truncated protein enhance profilin local accumulation at contacts with infected cells, which might lead to a reduction of F-actin polymerization.
FIGURE 7.
Profilin recruitment at contacts with HIV-1-infected cells is increased in drebrin-depleted and Dreb(1–366)-GFP overexpressing cells. A, quantification of profilin accumulation at cellular contacts between control or drebrin-depleted cells and MT4-HIV-1-GFP-infected cells. Each point represents profilin accumulation intensity ratio at one intercellular contact. More than 100 events were counted for each condition in three independent experiments. The medians and the interquartile ranges are also depicted on the graph. B, control or drebrin-depleted cells were incubated with MT4-HIV-GFP-infected cells for 1 h at 37 °C, fixed, and stained for profilin. Summatory projections of representative confocal stack images are shown. Profilin staining is also shown in pseudocolor intensity-coding format on the right panels. Bar, 5 μm. C, quantification of profilin accumulation at cellular contacts between cells overexpressing drebrin-GFP or Dreb(1–366)-GFP and MT4-HIV-1-GFP-infected cells. Each point represents the profilin accumulation intensity ratio at one intercellular contact. More than 100 events were counted for each condition in three independent experiments. The medians and the interquartile ranges are also depicted on the graph. D, cells overexpressing drebrin-GFP or Dreb(1–366)-GFP were incubated with MT4-HIV-GFP-infected cells (previously labeled with the blue cell tracker CMAC) for 1 h at 37 °C, fixed, and stained for profilin. Summatory projections of representative confocal stack images are shown. Profilin staining is also shown in pseudocolor intensity-coding format below each series. Note that in the green channel both cytoplasmic GFP of MT4-HIV-GFP cells and drebrin-GFP or Dreb(1–366)-GFP cortical distribution are observed. Bar, 5 μm.
DISCUSSION
In this study, we show that drebrin associates with CXCR4 before and during HIV-1 infection, and both are co-localized and specifically enriched at viral and Env-driven cell to cell contacts. Drebrin regulates HIV-1 infection, modulating viral triggered actin polymerization and profilin local accumulation.
Classical HIV-1 entry into its target cells requires fusion of the viral envelope with the plasma membrane (48, 49), although recent reports show productive infection via endocytosis (50–52). At any rate, the plasma membrane is what the virus particle encounters first. Thus, the spatial organization of transmembrane proteins at the cell surface and the physical state of the lipid bilayer are critical regulators of HIV-1 entry (11, 14, 15, 18, 53–55). Supporting this statement, there is an increasing number of studies showing that cellular proteins affecting either the clustering of viral receptors (such as tetraspanins, EWI-2, moesin, and filamin-A) or the subcortical actin cytoskeleton (such as syntenin-1, α-actinin, talin, vinculin, cofilin, profilin, WASp, WAVE-2, diaphanous-2, and Arp2/3) alter HIV-1 infection effectiveness (15, 18, 20, 24, 26, 30, 31, 56–60). Initial HIV-1 binding to the target cell surface induces local actin polymerization, which probably facilitates receptor clustering at the plasma membrane (6, 19, 61). Clustering of CD4/CXCR4 has been suggested to depend on their incorporation into tetraspanin-enriched microdomains (14–16, 62) and to require cholesterol-enriched domains (46, 54, 55), attesting the importance of membrane microdomains for HIV-1 entry. Tetraspanin-enriched microdomains are linked to actin cytoskeleton through actin-binding proteins ERMs and α-actinin (30, 63). ERMs and filamin-A, another actin-binding protein, are known to facilitate CD4/CXCR4 clustering during HIV-1 attachment and promote viral infection (18, 20). Our data regarding the effect of drebrin in Env-mediated cell fusion and HIV attachment/entry support drebrin involvement during the HIV-1 entry step. Therefore, drebrin, by dynamically reorganizing the actin cytoskeleton, could regulate actin-mediated clustering of CD4/CXCR4 in a different fashion to ERMs and filamin-A (18, 20). However, the distribution of phosphorylated moesin in contacts between MT4-HIV-GFP and target drebrin-silenced cells was not modified, and receptor clustering was apparently not impaired, suggesting that the role of drebrin was not related with receptor clustering.
Recent data support a negative role for the actin cytoskeleton after viral-induced membrane fusion. The dense actin structure that favors CD4/CXCR4 clustering during attachment might subsequently act as a physical barrier that impairs introduction of the viral nucleocapsid. Therefore, a functional but loose actin cytoskeleton seems to be favorable for HIV-1 entry into the host cell (26, 57). We have previously reported that silencing syntenin-1, a scaffold protein that binds to the cytoplasmic region of CD4, reduced F-actin polymerization in response to HIV-1 and this promoted HIV-1 entry (57). Here we show that drebrin also plays a role in HIV-triggered actin polymerization. Reduction of the F-actin structure may render cells more permissive to subsequent HIV-1 nucleocapsid entry, accounting for the negative role of drebrin and syntenin-1 in the HIV-1 entry step (57). Although syntenin-1 silencing increases PIP-2 formation at Env-mediated contacts, drebrin does not. Drebrin regulation of actin polymerization seems to be related to local profilin accumulation. Profilin and drebrin have opposite effects regarding actin: whereas profilin enhances actin dynamics, drebrin stabilizes actin filaments (33, 64). Profilin is known to behave in different ways depending on its concentration. At high concentrations, profilin prevents actin polymerization, whereas at low concentrations it enhances actin polymerization (65). In this scenario, both drebrin knockdown and overexpression of its truncated form Dreb(1–366)-GFP enhance profilin recruitment toward HIV-1 entry sites, and consequently its capacity to prevent actin polymerization. High profilin concentrations at cell contacts between drebrin-depleted cells and HIV-infected cells are in accordance with the observed reduction of polymerized actin in response to HIV-1 in these cells. In this sense, profilin inhibition has been reported to suppress viral production in HIV-infected monocytes (58). Enhancement in HIV-1 entry in drebrin-depleted cells could then be a consequence of the observed profilin local increase and its effect on actin remodeling, together with the reduction in the stabilization of F-actin due to the absence of drebrin (33).
Overexpression of different drebrin constructs suggests how drebrin regions are involved in HIV-1 infection. Overexpression of wild-type drebrin-GFP or Dreb(319–707)-GFP does not affect HIV-1 infection, whereas overexpression of Dreb(1–366)-GFP renders similar effects as drebrin knocking down. The absence of effect on HIV-1 infection when overexpressing wild-type drebrin-GFP could be the consequence of high endogenous expression levels, which might be already saturating F-actin and CXCR4 drebrin-binding sites. In agreement with this hypothesis, HIV-1 infection of target cells knocked down for endogenous drebrin and overexpressing wild-type drebrin-GFP displayed similar infection levels as control cells. Concerning the absence of effect on HIV-1 infection when overexpressing Dreb(319–707)-GFP, we have previously shown that this mutant does not bind to CXCR4 (35). Drebrin binding to CXCR4 seems then to be necessary for the drebrin negative role on HIV-1 infection. Dreb(1–366)-GFP, a drebrin construct able to bind F-actin and CXCR4 but unable to recruit profilin, behaves as a dominant-negative of drebrin regarding HIV-1 infection. Indeed, enhancement in profilin accumulation at contacts with infected cells is observed when this mutant, with no drebrin profilin-binding region, is overexpressed in target cells. Profilin accumulation at these areas might be the consequence of profilin interaction with G-actin and/or with other proteins known as profilin partners (39). Therefore, drebrin limits profilin local concentration and stabilizes actin polymerization. The role of drebrin in the stabilization of plasma membrane domains enriched in F-actin structures has been previously shown in neurons and other cell types (32).
Alternatively, drebrin might regulate F-actin in a subsequent step, after virus-promoted local clearance of F-actin. Our results showing that drebrin is not involved in HIV-1 reverse transcription do not rule out its involvement in viral RNA and/or DNA transport to the nucleus. In this regard, part of the internalized VLPs was still associated with drebrin clusters at the cytoplasm. Interestingly, new viral particles released from drebrin-depleted cells are more infective, suggesting not only that drebrin-depleted cells are more easily infected but also that new virions released are more infective, explaining why at day 6 the effect of drebrin silencing was amplified. In this sense, previous reports have shown that other host proteins that get incorporated into the viral particles, affect both viral entry and infectivity (15, 66, 67). In summary, our results identify a regulatory role for the CXCR4-binding protein drebrin during HIV-1 entry into target T cells, by modulating profilin accumulation and actin polymerization at Env-driven contacts.
Acknowledgments
We thank José Román Cabrero for helpful biochemical advice, Manuel Perez-Martinez for confocal assistance, Soraya López-Martín for technical support, Laura Diaz (Research Support Contract (CA11/00290) from the Instituto de Salud Carlos III; Instituto de Investigación Sanitaria Gregorio Marañón, Hospital General Universitario Gregorio Marañón) for technical assistance at the Flow Cytometry Unit, and Miguel Vicente-Manzanares for critical reading of the manuscript. Simon Bartlett (CNIC) provided English editing.
This work was supported by Grant SAF2011-25834 from the Spanish Ministry of Science and Innovation, Grant INDISNET-S2011/BMD-2332 from the Comunidad de Madrid, and Grant FIPSE 36658/07 from the FIPSE Foundation (to F. S.-M.).
- Env
- envelope
- VLP
- virus-like particles
- VSV
- vesicular stomatitis virus
- ERM(s)
- Ezrin-Radixin-Moesin proteins.
REFERENCES
- 1. Dimitrov D. S. (2004) Virus entry. Molecular mechanisms and biomedical applications. Nat. Rev. Microbiol. 2, 109–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Duncan C. J., Sattentau Q. J. (2011) Viral determinants of HIV-1 macrophage tropism. Viruses 3, 2255–2279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Goodenow M. M., Collman R. G. (2006) HIV-1 coreceptor preference is distinct from target cell tropism. A dual-parameter nomenclature to define viral phenotypes. J. Leukocyte Biol. 80, 965–972 [DOI] [PubMed] [Google Scholar]
- 4. Doms R. W. (2000) Beyond receptor expression: the influence of receptor conformation, density, and affinity in HIV-1 infection. Virology 276, 229–237 [DOI] [PubMed] [Google Scholar]
- 5. Piguet V., Sattentau Q. (2004) Dangerous liaisons at the virological synapse. J. Clin. Invest. 114, 605–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jolly C., Kashefi K., Hollinshead M., Sattentau Q. J. (2004) HIV-1 cell to cell transfer across an Env-induced, actin-dependent synapse. J. Exp. Med. 199, 283–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kao H., Lin J., Littman D. R., Shaw A. S., Allen P. M. (2008) Regulated movement of CD4 in and out of the immunological synapse. J. Immunol. 181, 8248–8257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kumar A., Humphreys T. D., Kremer K. N., Bramati P. S., Bradfield L., Edgar C. E., Hedin K. E. (2006) CXCR4 physically associates with the T cell receptor to signal in T cells. Immunity 25, 213–224 [DOI] [PubMed] [Google Scholar]
- 9. Gordon-Alonso M., Veiga E., Sanchez-Madrid F. (2010) Actin dynamics at the immunological synapse. Cell Health Cytoskelet. 2, 33–47 [Google Scholar]
- 10. Haller C., Tibroni N., Rudolph J. M., Grosse R., Fackler O. T. (2011) Nef does not inhibit F-actin remodelling and HIV-1 cell-cell transmission at the T lymphocyte virological synapse. Eur. J. Cell Biol. 90, 913–921 [DOI] [PubMed] [Google Scholar]
- 11. Jolly C., Mitar I., Sattentau Q. J. (2007) Requirement for an intact T-cell actin and tubulin cytoskeleton for efficient assembly and spread of human immunodeficiency virus type 1. J. Virol. 81, 5547–5560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Finnegan C. M., Rawat S. S., Cho E. H., Guiffre D. L., Lockett S., Merrill A. H., Jr., Blumenthal R. (2007) Sphingomyelinase restricts the lateral diffusion of CD4 and inhibits human immunodeficiency virus fusion. J. Virol. 81, 5294–5304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kamiyama H., Yoshii H., Tanaka Y., Sato H., Yamamoto N., Kubo Y. (2009) Raft localization of CXCR4 is primarily required for X4-tropic human immunodeficiency virus type 1 infection. Virology 386, 23–31 [DOI] [PubMed] [Google Scholar]
- 14. Fournier M., Peyrou M., Bourgoin L., Maeder C., Tchou I., Foti M. (2010) CD4 dimerization requires two cysteines in the cytoplasmic domain of the molecule and occurs in microdomains distinct from lipid rafts. Mol. Immunol. 47, 2594–2603 [DOI] [PubMed] [Google Scholar]
- 15. Gordón-Alonso M., Yañez-Mó M., Barreiro O., Alvarez S., Muñoz-Fernández M. A., Valenzuela-Fernández A., Sánchez-Madrid F. (2006) Tetraspanins CD9 and CD81 modulate HIV-1-induced membrane fusion. J. Immunol. 177, 5129–5137 [DOI] [PubMed] [Google Scholar]
- 16. Weng J., Krementsov D. N., Khurana S., Roy N. H., Thali M. (2009) Formation of syncytia is repressed by tetraspanins in human immunodeficiency virus type 1-producing cells. J. Virol. 83, 7467–7474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yáñez-Mó M., Barreiro O., Gordon-Alonso M., Sala-Valdés M., Sánchez-Madrid F. (2009) Tetraspanin-enriched microdomains. A functional unit in cell plasma membranes. Trends Cell Biol. 19, 434–446 [DOI] [PubMed] [Google Scholar]
- 18. Barrero-Villar M., Cabrero J. R., Gordón-Alonso M., Barroso-González J., Alvarez-Losada S., Muñoz-Fernández M. A., Sánchez-Madrid F., Valenzuela-Fernández A. (2009) Moesin is required for HIV-1-induced CD4-CXCR4 interaction, F-actin redistribution, membrane fusion and viral infection in lymphocytes. J. Cell Sci. 122, 103–113 [DOI] [PubMed] [Google Scholar]
- 19. Iyengar S., Hildreth J. E., Schwartz D. H. (1998) Actin-dependent receptor colocalization required for human immunodeficiency virus entry into host cells. J. Virol. 72, 5251–5255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jiménez-Baranda S., Gómez-Moutón C., Rojas A., Martínez-Prats L., Mira E., Ana Lacalle R., Valencia A., Dimitrov D. S., Viola A., Delgado R., Martínez-A C., Mañes S. (2007) Filamin-A regulates actin-dependent clustering of HIV receptors. Nat. Cell Biol. 9, 838–846 [DOI] [PubMed] [Google Scholar]
- 21. Dianzani U., Bragardo M., Buonfiglio D., Redoglia V., Funaro A., Portoles P., Rojo J., Malavasi F., Pileri A. (1995) Modulation of CD4 lateral interaction with lymphocyte surface molecules induced by HIV-1 gp120. Eur. J. Immunol. 25, 1306–1311 [DOI] [PubMed] [Google Scholar]
- 22. Lyengar S., Schwartz D. H. (2004) How do cell-free HIV virions avoid infecting dead-end host cells and cell fragments? AIDS Rev. 6, 155–160 [PubMed] [Google Scholar]
- 23. Liu Y., Belkina N. V., Shaw S. (2009) HIV infection of T cells. Actin-in and actin-out. Sci. Signal. 2, pe23. [DOI] [PubMed] [Google Scholar]
- 24. Harmon B., Campbell N., Ratner L. (2010) Role of Abl kinase and the Wave2 signaling complex in HIV-1 entry at a post-hemifusion step. PLoS Pathog. 6, e1000956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Warrilow D., Harrich D. (2007) HIV-1 replication from after cell entry to the nuclear periphery. Curr. HIV Res. 5, 293–299 [DOI] [PubMed] [Google Scholar]
- 26. Yoder A., Yu D., Dong L., Iyer S. R., Xu X., Kelly J., Liu J., Wang W., Vorster P. J., Agulto L., Stephany D. A., Cooper J. N., Marsh J. W., Wu Y. (2008) HIV envelope-CXCR4 signaling activates cofilin to overcome cortical actin restriction in resting CD4 T cells. Cell 134, 782–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cooper J., Liu L., Woodruff E. A., Taylor H. E., Goodwin J. S., D'Aquila R. T., Spearman P., Hildreth J. E., Dong X. (2011) Filamin A protein interacts with human immunodeficiency virus type 1 Gag protein and contributes to productive particle assembly. J. Biol. Chem. 286, 28498–28510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gladnikoff M., Shimoni E., Gov N. S., Rousso I. (2009) Retroviral assembly and budding occur through an actin-driven mechanism. Biophys. J. 97, 2419–2428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Vasiliver-Shamis G., Cho M. W., Hioe C. E., Dustin M. L. (2009) Human immunodeficiency virus type 1 envelope gp120-induced partial T-cell receptor signaling creates an F-actin-depleted zone in the virological synapse. J. Virol. 83, 11341–11355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gordón-Alonso M., Sala-Valdés M., Rocha-Perugini V., Pérez-Hernández D., López-Martín S., Ursa A., Alvarez S., Kolesnikova T. V., Vázquez J., Sánchez-Madrid F., Yáñez-Mó M. (2012) EWI-2 association with α-actinin regulates T cell immune synapses and HIV viral infection. J. Immunol. 189, 689–700 [DOI] [PubMed] [Google Scholar]
- 31. Brown C., Morham S. G., Walsh D., Naghavi M. H. (2011) Focal adhesion proteins talin-1 and vinculin negatively affect paxillin phosphorylation and limit retroviral infection. J. Mol. Biol. 410, 761–777 [DOI] [PubMed] [Google Scholar]
- 32. Majoul I., Shirao T., Sekino Y., Duden R. (2007) Many faces of drebrin. From building dendritic spines and stabilizing gap junctions to shaping neurite-like cell processes. Histochem. Cell Biol. 127, 355–361 [DOI] [PubMed] [Google Scholar]
- 33. Mikati M. A., Grintsevich E. E., Reisler E. (2013) Drebrin-induced stabilization of actin filaments. J. Biol. Chem. 288, 19926–19938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sharma S., Grintsevich E. E., Phillips M. L., Reisler E., Gimzewski J. K. (2011) Atomic force microscopy reveals drebrin induced remodeling of F-actin with subnanometer resolution. Nano. Lett. 11, 825–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pérez-Martínez M., Gordón-Alonso M., Cabrero J. R., Barrero-Villar M., Rey M., Mittelbrunn M., Lamana A., Morlino G., Calabia C., Yamazaki H., Shirao T., Vázquez J., González-Amaro R., Veiga E., Sánchez-Madrid F. (2010) F-actin-binding protein drebrin regulates CXCR4 recruitment to the immune synapse. J. Cell Sci. 123, 1160–1170 [DOI] [PubMed] [Google Scholar]
- 36. Ishikawa R., Hayashi K., Shirao T., Xue Y., Takagi T., Sasaki Y., Kohama K. (1994) Drebrin, a development-associated brain protein from rat embryo, causes the dissociation of tropomyosin from actin filaments. J. Biol. Chem. 269, 29928–29933 [PubMed] [Google Scholar]
- 37. Sasaki Y., Hayashi K., Shirao T., Ishikawa R., Kohama K. (1996) Inhibition by drebrin of the actin-bundling activity of brain fascin, a protein localized in filopodia of growth cones. J. Neurochem. 66, 980–988 [DOI] [PubMed] [Google Scholar]
- 38. Hayashi K., Ishikawa R., Ye L. H., He X. L., Takata K., Kohama K., Shirao T. (1996) Modulatory role of drebrin on the cytoskeleton within dendritic spines in the rat cerebral cortex. J. Neurosci. 16, 7161–7170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mammoto A., Sasaki T., Asakura T., Hotta I., Imamura H., Takahashi K., Matsuura Y., Shirao T., Takai Y. (1998) Interactions of drebrin and gephyrin with profilin. Biochem. Biophys. Res. Commun. 243, 86–89 [DOI] [PubMed] [Google Scholar]
- 40. Cao J., Park I. W., Cooper A., Sodroski J. (1996) Molecular determinants of acute single-cell lysis by human immunodeficiency virus type 1. J. Virol. 70, 1340–1354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hayashi K., Ishikawa R., Kawai-Hirai R., Takagi T., Taketomi A., Shirao T. (1999) Domain analysis of the actin-binding and actin-remodeling activities of drebrin. Exp. Cell Res. 253, 673–680 [DOI] [PubMed] [Google Scholar]
- 42. Valenzuela-Fernández A., Alvarez S., Gordon-Alonso M., Barrero M., Ursa A., Cabrero J. R., Fernández G., Naranjo-Suárez S., Yáñez-Mo M., Serrador J. M., Muñoz-Fernández M. A., Sánchez-Madrid F. (2005) Histone deacetylase 6 regulates human immunodeficiency virus type 1 infection. Mol. Biol. Cell 16, 5445–5454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Izquierdo-Useros N., Naranjo-Gómez M., Archer J., Hatch S. C., Erkizia I., Blanco J., Borràs F. E., Puertas M. C., Connor J. H., Fernández-Figueras M. T., Moore L., Clotet B., Gummuluru S., Martinez-Picado J. (2009) Capture and transfer of HIV-1 particles by mature dendritic cells converges with the exosome-dissemination pathway. Blood 113, 2732–2741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gummuluru S., KewalRamani V. N., Emerman M. (2002) Dendritic cell-mediated viral transfer to T cells is required for human immunodeficiency virus type 1 persistence in the face of rapid cell turnover. J. Virol. 76, 10692–10701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Calabia-Linares C., Robles-Valero J., de la Fuente H., Perez-Martinez M., Martín-Cofreces N., Alfonso-Pérez M., Gutierrez-Vázquez C., Mittelbrunn M., Ibiza S., Urbano-Olmos F. R., Aguado-Ballano C., Sánchez-Sorzano C. O., Sanchez-Madrid F., Veiga E. (2011) Endosomal clathrin drives actin accumulation at the immunological synapse. J. Cell Sci. 124, 820–830 [DOI] [PubMed] [Google Scholar]
- 46. Nguyen D. H., Giri B., Collins G., Taub D. D. (2005) Dynamic reorganization of chemokine receptors, cholesterol, lipid rafts, and adhesion molecules to sites of CD4 engagement. Exp. Cell Res. 304, 559–569 [DOI] [PubMed] [Google Scholar]
- 47. Peitsch W. K., Hofmann I., Bulkescher J., Hergt M., Spring H., Bleyl U., Goerdt S., Franke W. W. (2005) Drebrin, an actin-binding, cell-type characteristic protein. Induction and localization in epithelial skin tumors and cultured keratinocytes. J. Invest Dermatol. 125, 761–774 [DOI] [PubMed] [Google Scholar]
- 48. Doms R. W., Trono D. (2000) The plasma membrane as a combat zone in the HIV battlefield. Genes Dev. 14, 2677–2688 [DOI] [PubMed] [Google Scholar]
- 49. Gallo S. A., Finnegan C. M., Viard M., Raviv Y., Dimitrov A., Rawat S. S., Puri A., Durell S., Blumenthal R. (2003) The HIV Env-mediated fusion reaction. Biochim. Biophys. Acta 1614, 36–50 [DOI] [PubMed] [Google Scholar]
- 50. Carter G. C., Bernstone L., Baskaran D., James W. (2011) HIV-1 infects macrophages by exploiting an endocytic route dependent on dynamin, Rac1 and Pak1. Virology 409, 234–250 [DOI] [PubMed] [Google Scholar]
- 51. Hübner W., McNerney G. P., Chen P., Dale B. M., Gordon R. E., Chuang F. Y., Li X. D., Asmuth D. M., Huser T., Chen B. K. (2009) Quantitative 3D video microscopy of HIV transfer across T cell virological synapses. Science 323, 1743–1747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Miyauchi K., Kim Y., Latinovic O., Morozov V., Melikyan G. B. (2009) HIV enters cells via endocytosis and dynamin-dependent fusion with endosomes. Cell 137, 433–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Jolly C., Mitar I., Sattentau Q. J. (2007) Adhesion molecule interactions facilitate human immunodeficiency virus type 1-induced virological synapse formation between T cells. J. Virol. 81, 13916–13921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Liao Z., Cimakasky L. M., Hampton R., Nguyen D. H., Hildreth J. E. (2001) Lipid rafts and HIV pathogenesis: host membrane cholesterol is required for infection by HIV type 1. AIDS Res. Hum Retroviruses 17, 1009–1019 [DOI] [PubMed] [Google Scholar]
- 55. Popik W., Alce T. M., Au W. C. (2002) Human immunodeficiency virus type 1 uses lipid raft-colocalized CD4 and chemokine receptors for productive entry into CD4+ T cells. J. Virol. 76, 4709–4722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Aggarwal A., Iemma T. L., Shih I., Newsome T. P., McAllery S., Cunningham A. L., Turville S. G. (2012) Mobilization of HIV spread by diaphanous 2-dependent filopodia in infected dendritic cells. PLoS Pathog. 8, e1002762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Gordón-Alonso M., Rocha-Perugini V., Álvarez S., Moreno-Gonzalo O., Ursa A., López-Martín S., Izquierdo-Useros N., Martínez-Picado J., Muñoz-Fernández M. Á., Yáñez-Mó M., Sánchez-Madrid F. (2012) The PDZ-adaptor protein syntenin-1 regulates HIV-1 entry. Mol. Biol. Cell 23, 2253–2263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kadiu I., Ricardo-Dukelow M., Ciborowski P., Gendelman H. E. (2007) Cytoskeletal protein transformation in HIV-1-infected macrophage giant cells. J. Immunol. 178, 6404–6415 [DOI] [PubMed] [Google Scholar]
- 59. Komano J., Miyauchi K., Matsuda Z., Yamamoto N. (2004) Inhibiting the Arp2/3 complex limits infection of both intracellular mature vaccinia virus and primate lentiviruses. Mol. Biol. Cell 15, 5197–5207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Prasad A., Kuzontkoski P. M., Shrivastava A., Zhu W., Li D. Y., Groopman J. E. (2012) Slit2N/Robo1 inhibit HIV-gp120-induced migration and podosome formation in immature dendritic cells by sequestering LSP1 and WASp. PloS One 7, e48854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Pontow S. E., Heyden N. V., Wei S., Ratner L. (2004) Actin cytoskeletal reorganizations and coreceptor-mediated activation of rac during human immunodeficiency virus-induced cell fusion. J. Virol. 78, 7138–7147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Krementsov D. N., Weng J., Lambelé M., Roy N. H., Thali M. (2009) Tetraspanins regulate cell-to-cell transmission of HIV-1. Retrovirology 6, 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Sala-Valdés M., Ursa A., Charrin S., Rubinstein E., Hemler M. E., Sánchez-Madrid F., Yáñez-Mó M. (2006) EWI-2 and EWI-F link the tetraspanin web to the actin cytoskeleton through their direct association with ezrin-radixin-moesin proteins. J. Biol. Chem. 281, 19665–19675 [DOI] [PubMed] [Google Scholar]
- 64. Yarmola E. G., Bubb M. R. (2006) Profilin. Emerging concepts and lingering misconceptions. Trends Biochem. Sci. 31, 197–205 [DOI] [PubMed] [Google Scholar]
- 65. Bubb M. R., Yarmola E. G., Gibson B. G., Southwick F. S. (2003) Depolymerization of actin filaments by profilin. Effects of profilin on capping protein function. J. Biol. Chem. 278, 24629–24635 [DOI] [PubMed] [Google Scholar]
- 66. Grigorov B., Attuil-Audenis V., Perugi F., Nedelec M., Watson S., Pique C., Darlix J. L., Conjeaud H., Muriaux D. (2009) A role for CD81 on the late steps of HIV-1 replication in a chronically infected T cell line. Retrovirology 6, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Sato K., Aoki J., Misawa N., Daikoku E., Sano K., Tanaka Y., Koyanagi Y. (2008) Modulation of human immunodeficiency virus type 1 infectivity through incorporation of tetraspanin proteins. J. Virol. 82, 1021–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]