Abstract
Autism is a neurodevelopmental disorder associated with social deficits and behavioral abnormalities. Recent evidence suggests that mitochondrial dysfunction and oxidative stress may contribute to the etiology of autism. This is the first study to compare the activities of mitochondrial electron transport chain (ETC) complexes (I–V) and pyruvate dehydrogenase (PDH), as well as mitochondrial DNA (mtDNA) copy number in the frontal cortex tissues from autistic and age-matched control subjects. The activities of complexes I, V and PDH were most affected in autism (n=14) being significantly reduced by 31%, 36% and 35%, respectively. When 99% confidence interval (CI) of control group was taken as a reference range, impaired activities of complexes I, III and V were observed in 43%, 29% and 43% of autistic subjects, respectively. Reduced activities of all five ETC complexes were observed in 14% of autistic cases, and the activities of multiple complexes were decreased in 29% of autistic subjects. These results suggest that defects in complexes I and III (sites of mitochondrial free radical generation) and complex V (adenosine triphosphate synthase) are more prevalent in autism. PDH activity was also reduced in 57% of autistic subjects. The ratios of mtDNA of three mitochondrial genes ND1, ND4 and Cyt B (that encode for subunits of complexes I and III) to nuclear DNA were significantly increased in autism, suggesting a higher mtDNA copy number in autism. Compared with the 95% CI of the control group, 44% of autistic children showed higher copy numbers of all three mitochondrial genes examined. Furthermore, ND4 and Cyt B deletions were observed in 44% and 33% of autistic children, respectively. This study indicates that autism is associated with mitochondrial dysfunction in the brain.
Keywords: autism, electron transport chain, mitochondrial DNA, mitochondrial dysfunction, oxidative stress, pyruvate dehydrogenase
Introduction
Autism is a neurodevelopmental disorder associated with social deficits and behavioral abnormalities. It belongs to a group of disorders known as autism spectrum disorders (ASDs), which also include Asperger's syndrome and pervasive developmental disorder-not otherwise specified. According to the Centers for Disease Control and Prevention, the prevalence of ASDs in the United States is 1 in 88 children.1 Although the cause of autism is elusive, numerous studies suggest that enhanced oxidative stress is a prevalent biochemical condition in autism.2, 3, 4, 5, 6, 7, 8, 9, 10, 11 A recent meta-analysis study showed prevalence of mitochondrial disorder (MD) in the ASD population to be 5%, which is much higher than observed in general population (∼0.01%).12
The main function of mitochondria is to generate energy in the form of adenosine triphosphate (ATP) through oxidative phosphorylation, a process which requires the action of five electron transport chain (ETC) complexes, that is, complex I (NADH dehydrogenase), complex II (succinate dehydrogenase), complex III (cytochrome bc1 complex), complex IV (cytochrome-c oxidase) and complex V (ATP synthase).13, 14 Mitochondrial pyruvate dehydrogenase (PDH) occupies a central position in cellular energy metabolism because it catalyzes the conversion of pyruvate to generate acetyl CoA and NADH, thereby linking the glycolysis pathway to tricarboxylic acid cycle and subsequent oxidative phosphorylation to produce ATP in the mitochondria. NADH and FADH2 formed in the glycolytic and tricarboxylic acid cycle carry electrons to ETC at complexes I and II, respectively. During electron transfer along ETC, the complexes I, III and IV transport protons from mitochondrial matrix to intermembrane space of mitochondria, thus generating proton gradient (membrane potential). This proton gradient is used by ATP synthase (complex V) for phosphorylating adenosine diphosphate (ADP) to produce ATP. The ETC in mitochondria, particularly complex I and III, also serves as a prime mechanism for the generation of free radicals.15, 16 The number of mitochondria per cell is related to the energy demands of the cell, and the copy number of mitochondrial DNA (mtDNA) can vary depending upon the energy needs of a cell17 and oxidative stress conditions.18, 19
The brain has a high demand for energy, and neurons contain a large number of mitochondria. Extensive evidence suggests that mitochondrial dysfunction occurs in the early stages of major neurodegenerative diseases, such as Alzheimer's disease,20, 21, 22, 23 Parkinson's disease (PD),22, 23, 24, 25, 26 Huntington's disease27 and amyotrophic lateral sclerosis.28, 29 In addition, mitochondrial dysfunction in the brain of some individuals with schizophrenia has been reported.30, 31, 32, 33 Recent reviews have suggested that mitochondrial abnormalities may also affect high-energy supply of developing brain and trigger a cascade of events, leading to neurodevelopmental disorders including autism.12, 32, 34, 35, 36
Although a few studies of blood and muscle biopsy samples37, 38, 39, 40, 41, 42 have suggested compromised mitochondrial energy metabolism and defects in ETC complexes in autism, the information on brain mitochondrial dysfunction in autism is very limited. Preliminary magnetic resonance spectroscopy studies showed decreased synthesis of ATP and a disturbance of energy metabolism in the brain of individuals with autism.43, 44 Recently, we reported brain region-specific deficit in the protein expression of ETC complexes in the cerebellum, and cortices from frontal and temporal regions of the children with autism.6 None of the five ETC complexes was affected in the parietal and occipital cortices in the subjects with autism.6 The present study was undertaken to analyze whether brain mitochondrial activities of the ETC complexes and PDH enzyme are affected in autism.
Mitochondrial function is under the dual genetic control of mtDNA and nuclear DNA (nDNA). More than 70 proteins are components of mitochondrial ETC complexes. The mtDNA contains 37 genes that code for 13 subunits of ETC complexes I, III, IV and V.45 Mitochondrial NADH dehydrogenase genes, that is, ND1, ND2, ND3, ND4, ND4L and ND6, encode seven subunits of complex I. Cytochrome-b is the mitochondrial Cyt B-encoded complex III subunit. Mitochondrial cytochrome-c oxidase genes, that is, CO1, CO2 and CO3, encode three subunits of complex IV, and the mitochondrial ATP6 and ATP8 genes encode two subunits of complex V. On the other hand, complex II is exclusively coded by nuclear genome. Mitochondria disorders can be caused by abnormal ETC structure and/or function, and defects of mtDNA or nDNA.
Deletions and duplications of a chromosomal segment, known as copy number variation (CNVs), are now emerging as important factors in the etiology of neuropsychiatric disorders, including autism,46, 47, 48, 49, 50, 51, 52, 53, 54 bipolar disorder55, 56, 57 and schizophrenia.47, 57, 58, 59, 60 Pons et al.61 reported mtDNA mutations or mtDNA deletion in five cases with autism. Recently, increased copy numbers of three mitochondrial genes (Cyt B, ND1 and ND4) because of mtDNA overreplication was reported in the lymphocytes from 5 out of 10 subjects with autism.38 Although overreplication of mitochondrial genes in the peripheral tissues of subjects with autism was an important observation, such study has not been done in the brains of autistic subjects. Considering the importance of ETC complexes I and III in the generation of free radicals, and of oxidative stress in autism, we also compared the copy number and deletions of Cyt B, ND1 and ND4 mtDNA genes for complexes I and III in the frontal cortex of subjects with autism and age-matched controls.
Materials and methods
Materials
Samples of frozen postmortem frontal cortices of brain from autistic and age-matched control subjects were obtained from the National Institute of Child Health and Human Development Brain and Tissue Bank for Developmental Disorders at the University of Maryland, Baltimore (UMB). Demographics of autistic and control subjects, including age, postmortem interval (PMI), medications and cause of death are summarized in Table 1. Donors with autism fit the diagnostic criteria of the Diagnostic and Statistical Manual-IV, as confirmed by the Autism Diagnostic Interview-Revised. The age (mean±s.e.) for autistic subjects was 10.36±1.46 years (N=14) and for control subjects was 10.83±1.72 years (N=12). Brain samples were stored at −70 °C. This study was approved by the Institutional Review Board of the New York State Institute for Basic Research in Developmental Disabilities.
Table 1. Case history of autistic and control subjects.
| UMB# | Diagnosis | Age (y) | Sex | PMI (h) | Medications | Cause of death |
|---|---|---|---|---|---|---|
| 4671 | Autism | 4.5 | F | 13 | — | Multiple injuries from fall |
| 5308 | Autism | 4.5 | M | 21 | — | Skull fractures |
| 1349 | Autism | 5.6 | M | 39 | — | Drowning |
| 4849 | Autism | 7.5 | M | 20 | — | Drowning |
| 1174 | Autism | 7.8 | F | 14 | Depakote, Tegretol | Multi-system organ failure |
| 4231 | Autism | 9.9 | F | 24 | Zyprexia, Reminyl | Drowning |
| 4721 | Autism | 8.8 | M | 16 | — | Drowning |
| 797 | Autism | 9.3 | M | 13 | Desipramine | Drowning |
| 1182 | Autism | 8.8 | M | 12 | — | Smoke inhalation due to fire |
| M2004M | Autism | 10.1 | M | 25 | Clonidine | Drowning |
| 144 | Autism | 10.4 | M | 22 | Depakote, Haldol, Imipramine, Luvox, Olanzepine, Paxil, Wellbutrin | Drowning |
| 4899 | Autism | 14.3 | M | 9 | Clonidine, Trileptal, Zoloft, Melatonin | Drowning |
| 4999 | Autism | 20.8 | M | 14 | — | Cardiac arrthymia |
| 5176 | Autism | 22.6 | M | 18 | Risperdal | Subdural hemorrhage |
| 1499 | Control | 4.5 | F | 21 | — | Lymphocytic myocarditis |
| 1185 | Control | 4.7 | M | 17 | — | Drowning |
| 4332 | Control | 5.7 | M | 18 | — | Brochopneumonia |
| 4898 | Control | 7.7 | M | 12 | Concerta, Clonidone | Drowning |
| 1708 | Control | 8.1 | F | 20 | — | Multiple injuries from accident |
| 4337 | Control | 8.3 | M | 16 | — | Blunt force neck injury |
| 1407 | Control | 9.1 | F | 20 | Albuterol, Zirtec, Alegra, Rodact, Flovent, Flonase | Asthma |
| 616 | Control | 11.6 | M | 25 | — | Multiple injuries from accident |
| M3228M | Control | 11.8 | M | 20 | — | Internal bleeding |
| 5163 | Control | 14.9 | M | 12 | — | Drowning |
| 4590 | Control | 20.5 | M | 19 | — | Dilated Cardiomyopathy |
| 5342 | Control | 23.0 | M | 12 | — | Multiple injuries from accident |
Abbreviation: PMI, postmortem interval.
Mitochondria isolation and protein assay
Mitochondria were isolated from frontal cortex tissue samples by a differential centrifugation method following the manufacturer's protocol of mitochondria isolation kit (ab110169; Abcam, Cambridge, MA, USA). Brain tissue samples were homogenized in isolation buffer by Dounce homogenizer (about 30 strokes) on ice. Then, the brain homogenates were transferred to the tubes and centrifuged at 1000 g for 10 min at 4 °C. The supernatants were again centrifuged at 12 000 g for 15 min at 4 °C to isolate the mitochondria (the pellet). Protein concentrations of the mitochondria in samples were assayed by bicinchoninic acid protein assay kit (Pierce, Rockford, IL, USA).
Assay for activities of mitochondrial ETC complexes I, II, III, IV and V
The activities of mitochondrial ETC complexes I, II, III, IV, V were analyzed using microplate assay kits (ab109721, ab109908, ab109905, ab109909 and ab109714, respectively) from Abcam, as described in the Supplementary Information. All the tests were done in duplicate. The absorbance was read by Spetramax M5 microplate reader (Molecular Device, Downingtown, PA, USA). For the activity assays of complex I, II, IV and V, detergent (1/10 volume) was added to the mitochondria to extract transmembrane proteins into the solution. To select the optimal amount of the mitochondria for the activity assays, a dose–effect curve was plotted for each assay.
Assay for mitochondrial PDH enzyme activity
The PDH enzyme activity was analyzed using an Abcam assay kit (ab109902). Intact and functional PDH enzymes were extracted after adding detergent (1/19 volume) to the mitochondria, and 100 μg proteins from each sample were used for this assay. PDH activity was determined by following the reduction of NAD+ to NADH, coupled to the reduction of a reporter dye to yield a yellow-colored reaction product whose concentration was monitored by measuring the increase in absorbance at 450 nm.
Assay for activity of mitochondrial citrate synthase
Mitochondrial citrate synthase is an exclusive marker of the mitochondrial matrix, and the content and activity of citrate synthase depend on the number of mitochondria. To exclude any effect of the mitochondrial number on the activities of mitochondrial ETC complexes and PDH enzyme, their activities were normalized to the activity of citrate synthase. Citrate synthase activity was detected using microplate assay kit (ab119692; Abcam), as described in the Supplementary Information.
Analysis of mtDNA copy number by real-time PCR
Total genomic DNA was extracted from the frontal cortex samples of children with autism (age: 4.5–10.4 years) and age-matched controls (age: 4.5–11.8 years) using DNeasy kit (Qiagen, Gaithersburg, MD, USA), and the concentration of DNA was measured using Nano Drop (Nano Drop, Wilmington, DE, USA). ND1, ND4 and Cyt B genes were used to represent the mtDNA, and pyruvate kinase (PK) gene was used to represent the nDNA. Relative mtDNA copy numbers were assessed after Cyt B, ND1 and ND4 normalization by the single-copy nuclear gene PK. Human primers for PK were: forward 5′-AGCCCAAATGGCCTTGAA-3′ reverse 5′-AGAGACAGAATGCCAGTGAGC-3′ primers for ND1 were: forward 5′-CCCTAAAACCCGCCACATCT-3′ reverse 5′-GAGCGATGGTGAGAGCTAAGGT-3′ primers for ND4 were: forward 5′-CCATTCTCCTCCTATCCCTCAAC-3′ reverse 5′-CACAATCTGATGTTTTGGTTAAACTATATTT-3′ and primers for Cyt B were: forward 5′-CACGATTCTTTACCTTTCACTTCATC-3′ reverse 5′-TGATCCCGTTTCGTGCAAG-3′. These primers were purchased from Sigma (St Louis, MO, USA). All reactions were performed in 96-well plates on a Bio-Rad iCycler IQ system instrument (Hercules, CA, USA). The reaction solution was prepared by combining the SYBR green fluor qPCR Mastermix (Bio-Rad) and 320 nM each of primers, and each sample was analyzed in triplicate. The DNA concentration in each sample was diluted to 20 ng μl−1, and 1 μl of DNA was added to each well as the template. Negative controls without template were run for each gene. Amplification program was 10 min at 95 °C, followed by 40 cycles of 15 s at 95 °C and 60 s at 60 °C. At the end of the amplification process, the amplification specificity of gene was assessed by melting curve between 55 and 95 °C. The corresponding real-time PCR efficiencies for each mitochondrial and nuclear gene amplification were calculated according to the equation: E=10(−1/slope)−1. The efficiency of ND1, ND4, Cyt B and PK gene was 96%, 94%, 106% and 94%, respectively. Relative mtDNA copy number (mtDNA amount/nDNA amount) was calculated by a comparative Ct method, using the following equation: mtDNA/nDNA=2−ΔCt, where ΔCt=Ctmitochondrial−Ctnuclear.
Statistics
The statistical analysis of the data was performed using Graphpad Prism 5.0 software (La Jolla, CA, USA). The difference between autism and control groups was examined by the unpaired Student's t-test (two-tailed), and values of P<0.05 were considered significant. In addition, 95 or 99% confidence interval (CI) of control group was used as the normal range to evaluate whether the data of autistic group were out of the normal range.
Results
Reduced activities of mitochondrial ETC complexes in the frontal cortex of subjects with autism
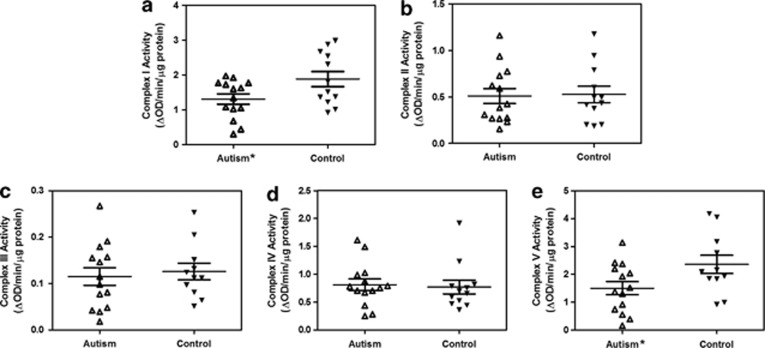
The activities of mitochondrial ETC complexes normalized to the activity of citrate synthase in the frontal cortex of autistic and control subjects are represented in Table 2. Figure 1 shows the scattered plots of the data. Our results showed that the activity of complex I was significantly reduced by 31% (P=0.0343) in autism (mean±s.e.=1.309±0.147) compared with the control group (mean±s.e.=1.884±0.217). The activity of complex V in the subjects with autism (mean±s.e.=1.502±0.236) was also significantly decreased by 36% (P=0.0404) compared with the control group (mean±s.e.=2.356±0.327). If a 99% CI of control group was taken as the reference range, 6 of the 14 (43%) autistic subjects had less complex I and/or V activities than the lower limit of 99% CI of the control group, suggesting that 6 subjects with autism were affected with deficiencies of complex I and/or V activity (Table 2). Although there was no significant difference in the activity of complex III between the autistic and control groups, its lower activity was observed in a subset, that is, 29% (4 out of 14) of the autistic population when compared with the lower limit of the 99% CI of the control group (Table 2). Reduced activities of multiple complexes were observed in 4 of the 14 (29%) autistic subjects, and 2 of the 14 (14%) autistic subjects showed underactivity of all ETC complexes. For complexes II and IV, 2 of the 14 (14%) autistic subjects showed underactivity, whereas other 2 autistic subjects (14%) showed overactivity of one of these complexes. Altogether, there was no significant difference in the activities of these complexes II and IV between autism and control groups. Two autistic subjects (#5 and 9) showed a decrease in both complexe II and III activities. The deficiency of complex III activity in these two autism samples could also be caused by the lower activity of complex II, because the whole mitochondria (including complex II and III) was used, and electrons need to be transferred from complex II to III to produce reduced cytochrome-c in the complex III activity assay. Taken together, the order of deficiency in the activities of ETC complexes was I and V (43%), III (29%), II and IV (14%) in the brain samples from subjects with autism.
Table 2. Activities of mitochondrial ETC complexes and PDH from frontal cortex in autism and control subjects.
| Complex Ia | Complex II | Complex III | Complex IV | Complex Va | Affected complexes | PDHa | |
|---|---|---|---|---|---|---|---|
| Autism | |||||||
| 1 | 1.921 | 1.161b | 0.157 | 0.987 | 2.370 | None | 1.283b |
| 2 | 1.600 | 0.267 | 0.098 | 0.705 | 3.139 | None | 0.624 |
| 3 | 1.080c | 0.271 | 0.048c | 0.873 | 2.038 | I, III | 0.499c |
| 4 | 1.772 | 0.729 | 0.155 | 1.022 | 1.377 | None | 1.041 |
| 5 | 0.305c | 0.234c | 0.039c | 0.280c | 1.301c | I, II, III, IV, V | 0.680 |
| 6 | 1.729 | 0.939b | 0.179 | 0.671 | 0.555c | V | 0.475c |
| 7 | 1.626 | 0.776 | 0.267b | 1.490b | 0.390c | V | 0.540c |
| 8 | 1.773 | 0.384 | 0.191 | 1.610b | 0.742c | V | 0.305c |
| 9 | 1.027c | 0.156c | 0.018c | 0.250c | 0.917c | I, II, III, IV, V | 0.601 |
| 10 | 1.052c | 0.310 | 0.081 | 0.750 | 2.196 | I | 0.325c |
| 11 | 0.683c | 0.277 | 0.042c | 0.704 | 0.158c | I, III, V | 0.329c |
| 12 | 1.974 | 0.624 | 0.115 | 0.790 | 2.411 | None | 0.583c |
| 13 | 1.343 | 0.592 | 0.146 | 0.779 | 1.516 | None | 0.649 |
| 14 | 0.445c | 0.425 | 0.077 | 0.438 | 1.923 | I | 0.375c |
| Mean±s.e. | 1.309±0.147a | 0.510±0.081 | 0.115±0.019 | 0.811±0.104 | 1.502±0.236a | 0.594±0.074a | |
| 99% CI | (0.865–1.753) | (0.268–0.753) | (0.058–0.172) | (0.498–1.123) | (0.792–2.213) | (0.372–0.815) | |
| Affected subjectsc | 43% | 14% | 29% | 14% | 43% | 57% | |
| Control | |||||||
| Mean±s.e. | 1.884±0.217 | 0.529±0.089 | 0.126±0.018 | 0.769±0.123 | 2.356±0.327 | 0.917±0.103 | |
| 99% CI | (1.210–2.557) | (0.253–0.805) | (0.069–0.183) | (0.387–1.151) | (1.320–3.392) | (0.598–1.236) | |
Abbreviations: CI, confidence interval; PDH, pyruvate dehydrogenase.
The activities of ETC complexes and PDH were normalized to citrate synthase activity in order to exclude any effect of mitochondrial number on their activities, and are represented as ΔOD per min per μg protein.
Significant difference between autistic group and control group (P<0.05).
The value was higher than upper 99% CI of control.
Activities were lower than the lower limit of 99% CI of control group.
Figure 1.
Scattered plots of activities of mitochondrial ETC complexes (I, II, III, IV and V) in the frontal cortex of brain from autism and control subjects. Activities of complex I (a) and V (e) were significantly decreased in subjects with autism compared with the control group. Panels b, c and d represent the activities of ETC complexes II, III and IV, respectively. *P<0.05.
Impaired activity of PDH enzyme in the frontal cortex of subjects with autism
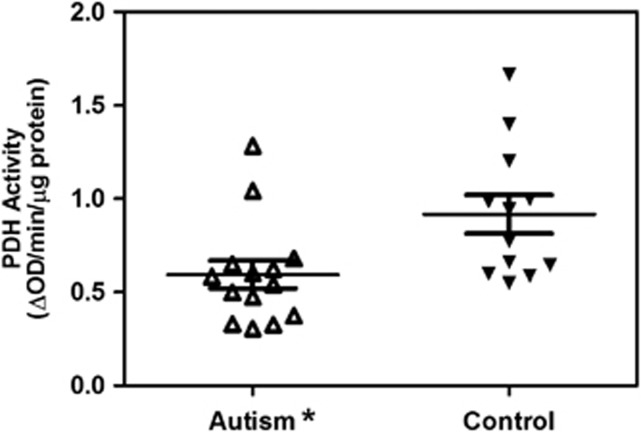
The enzyme activity of PDH normalized to the activity of citrate synthase in autistic and control subjects is represented in Table 2 and Figure 2. PDH enzyme activity was significantly reduced by 35% (P=0.015) in the frontal cortex of subjects with autism (mean±s.e.=0.594±0.074) compared with the control group (mean±s.e.=0.917±0.103). Compared with the lower 99% CI of the control group, 8 of the 14 (57%) autistic samples showed a deficiency of PDH activity (Table 2). Two subjects with autism (#5 and 9) showed all five complexes with defects but without a PDH enzyme defect, one autistic subject (#12) had no ETC complex defect but had a PDH enzyme defect, whereas three subjects with autism (#2, 4 and 13) had neither ETC complex nor PDH enzyme defect.
Figure 2.
Scattered plots of PDH activity in the frontal cortex of brain from autism and control subjects. Activity of PDH was significantly decreased in the subjects with autism compared with the control group. *P<0.05.
Increased mtDNA copy number in the frontal cortex of children with autism
Relative mtDNA copy numbers were assessed in frontal cortex from autism and age-matched control children after normalization of Cyt B, ND1 and ND4 mtDNA genes to a single-copy nuclear gene PK (Table 3). The ratios of mtDNA for all three genes (ND1, Cyt B and ND4) to nDNA (PK) were increased in the frontal cortex from autistic subjects compared with the controls. The ratio of ND1/PK in the autistic group (mean±s.e.=2871±348.5) was significantly higher by 1.65-fold (P=0.0244) than its ratio in the control group (mean±s.e.=1742±291.6). The ratio of ND1/PK in six out of nine autistic samples exceeded the upper limit of 95% CI of the controls, indicating that 67% of autistic subjects had overreplication. Similarly, the ratio of ND4/PK was higher by 1.54-fold in autistic group (mean±s.e.=1981±252.5) than in the control group (mean±s.e.=1285±223.6), and five out of nine autistic subjects were affected, although there was no significant difference between the two groups (P=0.0556). The Cyt B/PK ratio of the autistic group (mean±s.e.=5036±824.7) was significantly higher (P=0.0401) by 1.74-fold than in the control group (mean±s.e.=2893±488.9), and five out of nine autistic subjects (56%) were affected. These results indicate a significant increase of mtDNA copy number in the frontal cortex of brain from the autistic subjects as compared with the control subjects. In four autistic subjects (UMB #4671, 5308, 1349 and 144), all three mtDNA genes showed increased copy number, whereas only ND1 and ND4 showed increased mtDNA copy number in one autistic subject (UMB #4849). In another autistic subject (UMB #4721), there was a slight increase for ND1 and Cyt B.
Table 3. Mitochondrial DNA copy number in the frontal cortex from children with autism and control subjects.
| UMB # | ND1/PKa | ND4/PK | Cyt B/PKa |
|---|---|---|---|
| Autism | |||
| 4671 | 3717b | 2195b | 7089b |
| 5308 | 3030b | 2288b | 6950b |
| 1349 | 2798b | 1978b | 7082b |
| 4849 | 3641b | 3148b | 3304 |
| 4231 | 1252 | 852 | 1795 |
| 4721 | 2504b | 1641 | 4640b |
| 797 | 1808 | 1287 | 2978 |
| 1182 | 2402 | 1479 | 2740 |
| 144 | 4688b | 2964b | 8742b |
| Mean±s.e. | 2871±348.5a | 1981±252.5 | 5036 ±824.7a |
| 95% CI | (2067–3675) | (1399–2564) | (3134–6937) |
| Affected subjects | 67% | 56% | 56% |
| Control | |||
| Mean±s.e. | 1742±291.6 | 1285±223.6 | 2893±488.9 |
| 95% CI | (1070–2414) | (769.7–1801) | (1766–4021) |
Abbreviations: CI, confidence interval; PK, pyruvate kinase.
Age of the children with autism was 4.5–10.4 years, and of control subjects was 4.5–11.8 years. Relative mtDNA copy numbers of ND1, ND4 and Cyt B were assessed by normalization with single-copy nuclear gene of PK.
Significant difference between autistic group and control group (P<0.05).
Values were higher than the upper limit of 95% CI in control group.
Mitochondrial DNA deletion in the frontal cortex of subjects with autism
To evaluate the deletion of Cyt B and ND4 genes, we calculated the ratio of ND4/ND1 and Cyt B/ND1 because the ND1gene is rarely deleted. The results (Table 4) showed that ND4/ND1 ratios in four out of nine autistic subjects (44%) were lower than the 95% CI of the control group (0.674–0.791), suggesting that 44% of the autistic group had ND4 deletion present. For Cyt B/ND1, three out of nine autistic subjects (33%) had lower values than the 95% CI of control group (1.537–1.834), suggesting that three cases with autism had a Cyt B deletion present.
Table 4. Mitochondrial DNA deletion in the frontal cortex from children with autism and control subjects.
| UMB # | ND4/ND1 | Cyt B/ND1 |
|---|---|---|
| Autism | ||
| 4671 | 0.59a | 1.91 |
| 5308 | 0.76 | 2.29 |
| 1349 | 0.71 | 2.53 |
| 4849 | 0.86 | 0.91a |
| 4231 | 0.68 | 1.43a |
| 4721 | 0.66a | 1.85 |
| 797 | 0.71 | 1.65 |
| 1182 | 0.62a | 1.14a |
| 144 | 0.63a | 1.86 |
| Mean±s.e. | 0.691±0.027 | 1.730±0.172 |
| 95% CI | 0.628–0.754 | 1.333–2.127 |
| Affected subjects | 44% | 33% |
| Control | ||
| Mean±s.e. | 0.732±0.025 | 1.686±0.064 |
| 95% CI | (0.674–0.791) | (1.537–1.834) |
Abbreviation: CI, confidence interval.
As compared with the control, values were lower than the lowest limit of 95% CI.
Effects of age, gender and medications on mitochondrial function.
To analyze the effect of age on the activities of mitochondrial ETC complexes and PDH, the autism group was subdivided into group A of children (age: 4.5–10.4 years) and group B of adults (age: 14.3–22.6 years). A significant decrease in the activities of complexes I, V and PDH by 32.2%, 38.7% and 63.7%, respectively, was observed in the autistic group A children (n=11) vs age-matched control subjects (n=9) (Supplementary Figure S1, Supplementary Information). Although a decrease in the activities of these complexes and PDH was also observed in the adult autistic group B compared with the control group (n=3), it was not statistically significant (Supplementary Figure S1). Linear regression analysis between age and the activities of these complexes and PDH, or of mtDNA copy number of ND1, ND4 and Cyt B, did not show a correlation between age and these mitochondrial parameters (data not shown). The comparison of male vs female autistic group did not show any effect of gender on any parameter of mitochondrial function studied (data not shown).
As shown in the case histories of autistic and control subjects (Table 1), 7 of 14 autistic subjects and 2 of 10 control subjects were taking one or more medications. A significant effect of medications was only observed on PDH activity (P=0.029: mean±s.e.=0.439±0.055 (n=7) in autism subjects with medications and 0.748±0.112 (n=7) in autism subjects without medications). There was no effect of medications on the activities of ETC complexes and mtDNA copy numbers of ND1, ND4 and Cyt B (data not shown).
Discussion
Accumulating evidence supports the hypothesis that mitochondrial dysfunction has an important role in the pathophysiology of autism. We recently reported decreased protein levels of mitochondrial ETC complexes in the cerebellum, frontal and temporal cortices of children with autism.6 These alterations were brain region-specific and were not observed in parietal and occipital cortices from autistic subjects.6 The results of the present study suggest that mitochondrial dysfunction observed in autism could also be attributed to the reduction in enzyme activities of ETC complexes and PDH, as well as to the alterations in the copy number of the mitochondrial genes involved in encoding the proteins of ETC. The activities of ETC complexes I and V, as well as PDH enzyme, were significantly reduced in the frontal cortex of subjects with autism compared with the controls. In our study, reduced activities of complexes I and V were the most prevalent ETC defects in the brain tissue of subjects with autism, and this was observed in 43% of the autistic cases, followed by deficiency in activity of complex III in 29% of autism cases. Deficiency in the activities of multiple ETC complexes was observed in 29% of autistic subjects, and 14% of autistic subjects showed underactivity of all ETC complexes. Although this is the first study on the activities of ETC complexes in the brain tissue of autism subjects, other groups have also reported abnormal expression and activities of mitochondrial ETC complexes in blood or muscle biopsy samples from subjects with autism. In muscle biopsy samples, Shoffner et al.41 reported a defect of complex I in 50% and of complex V in 14.3% of 28 subjects with ASDs and MD. Weissman et al.42 performed a retrospective analysis of 25 individuals with autism and reported enzyme- or mutation-defined mitochondrial ETC dysfunction. Complex I activity in the muscle tissue was reduced in 64% of autistic cases, followed by a deficiency in complex III (in 20%), II (in 5%) and IV (in 4%). A population-based survey of children with ASD showed that the most frequent deficiency in muscle tissue was in complexes I, IV and V.40 Giulivi et al.38 also reported more common deficiency in the activities of complex I and V in the lymphocytes from 60% and 40%, respectively, of 10 children with autism, followed by decrease in the activities of complex IV or III in 10% of autism cases. In a recent meta-analysis, Rossignol and Frye12 reported deficiencies of complexes I, III, V, IV and II in 53%, 30%, 23%, 20% and 9%, respectively, of children with ASD/MD. Multiple complex deficiencies were found in 36% of the children with ASD/MD. Altogether, these reports and our study suggest that deficiencies of complex I, III and V are more common in ASDs. Results of this study and previous report on protein levels of these complexes6 suggest that the reduced activities of these complexes could also be due to their decreased protein levels.
In our study, mitochondrial PDH activity was significantly decreased in the frontal cortex of autistic subjects, with 57% of autistic subjects being affected when compared with the control group. Reduced PDH activity will lead to decreased decarboxylation of pyruvate to acetyl CoA, impaired tricarboxylic acid cycle activity, decreased generation of NADH and increased metabolism of pyruvate to lactate and alanine in autism. Based on analysis of blood and/or muscle biopsy samples in the individuals with ASDs, several studies have reported mitochondrial dysfunction associated with high lactate and alanine levels in autism.12, 32, 34, 35, 36 Our results suggest that impairment in PDH activity may be partly responsible for abnormal levels of these metabolites and mitochondrial dysfunction in autism.
Complex I is composed of at least 45 subunits, of which 7 are coded by mtDNA and 38 are coded by nDNA.62 It is the first and largest enzyme of the ETC. Complex I catalyzes the oxidation of NADH to NAD+ and transports two electrons to coenzyme Q10 (also known as ubiquinone), and it generates a proton gradient across the inner mitochondrial membrane.63 Complex III is made of 11 subunits, of which 1 subunit (cytochrome-b) is coded by mtDNA and 10 subunits are coded by nDNA. It transfers two electrons from coenzyme Q10 to cytochrome-c and transports protons across the inner mitochondrial membrane at the same time. These protons in the intermembrane are used by complex V to generate ATP from ADP. Complexes I and III are also the main sites of mitochondrial free radical (superoxide) production.64, 65 Under normal physiological conditions, superoxide is released into mitochondrial matrix or intermembrane where it is converted to hydrogen peroxide. The disturbance in the function of complexes I, III and V can reduce ATP production, increase superoxide levels and disturb calcium homeostasis. Our results suggest that increased oxidative stress observed in the brain of autistic subjects5, 6, 7, 8, 11 could be attributed, in part, to a disturbance of the function of ETC complexes I and III.
The role of mitochondrial dysfunction has also been implicated in many other neuropsychiatric disorders. Although defects in mitochondrial ETC complexes I, III and V seem to be more common in autism, abnormalities in complexes I, II and IV are more prevalent in other brain-related disorders. As observed in autism, deficiency of complex I is also the most frequently encountered cause of childhood-onset mitochondrial disease66 and has been detected in PD,22, 23, 24, 25, 26 bipolar disorder67 and amyotrophic lateral sclerosis.29 In PD, deficiency of complex I has been suggested as a major contributor for increased free radical generation and dopaminergic-neuronal cell death.22, 23, 24, 25, 26 The predominance of complex I deficiency has also been noted in mitochondrial cytopathies.63 The studies conducted with postmortem brain samples from Down's syndrome patients also depicted decreased levels of complex I in the temporal cortex, occipital cortex, caudate nucleus and thalamus, and of complex V β-chain in the frontal cortex.68, 69 Although no significant effect on complex IV was observed in autism in the present study, decreased activity of complex IV is the most common finding in Alzheimer's disease patients and animal models of Alzheimer's disease.22, 23 Reduced activity of complex IV was also reported in the temporal and frontal cortex of schizophrenia subjects.31, 32, 33 In the caudate nucleus of Huntington's disease patients, severe deficiency in the activities of complexes II, III and IV have been reported.27 The importance of deficiencies of mitochondrial ETC complexes in brain-related disorders is also emphasized by the reports that inhibitors of complexes I and II are used in animal models to induce symptoms of PD22, 23, 24, 25, 26 and Huntington's disease,27 respectively. Taken together, our present study and above reports suggest that impaired mitochondrial ETC functions, and as a consequence, impaired cellular energy state, may be one of the mechanisms underlying the pathophysiology of neurodegenerative and neurodevelopmental disorders.
CNVs and copy number polymorphism are the most prevalent type of structural variations in human genome.70, 71 CNVs contribute to genetic heterogeneity because 12–15% of human genome varies in copy number, which has an important role in health and various diseases.72 Recent study in autism has shown CNVs of genes that are important in mitochondrial oxidative phosphorylation.53 In our study with the brain tissues of subjects with autism, we have observed increased copy numbers of three mitochondrial genes, that is, ND1, ND4 and Cyt B that encode for subunits of mitochondrial ETC complexes I and III. Increased mtDNA copy numbers in autism may represent a mechanism to compensate for the lower activity of ETC complexes I and III in autism. Our current study in the brain of autistic subjects supports the study of Giulivi et al.38 who also reported increased copy number in these mitochondrial genes in the lymphocytes of subjects with autism. These findings suggest that alteration of copy number of mitochondrial genes occurs not only in the peripheral tissue but also in the brain of subjects with autism. Our results also suggest mtDNA deletion of ND4 and Cyt B in the frontal cortex of 44% and 33% of autistic subjects, respectively. Pons et al.61 have previously reported mtDNA mutations or mtDNA deletion in the muscle tissue of five autistic cases. Higher ratio of deleted to wild-type mtDNA has also been reported in the cerebral cortex of patients with bipolar disorder as compared with age-matched controls.73, 74
Deficiencies of protein expression and activities of ETC complexes and the PDH enzyme in autism can be a primary etiology or secondary to other causes because mitochondria are vulnerable to several endogenous and exogenous factors, which are linked by excessive production of free radicals. Mitochondria are not only the source of free radicals but also the target of oxidative damage. Oxidative stress has also been shown to increase the mtDNA copy number.18, 19 Extensive evidence from our and other groups showed increased markers of oxidative stress in the brain, urine and blood of subjects with autism.2, 3, 4, 5, 6, 7, 8, 9, 10, 11 Several studies have suggested that mtDNA is more prone than nDNA to oxidative damage because mtDNA does not have protective histones, and mtDNA damage is more extensive and persists longer under oxidative stress conditions.75 Therefore, it is highly likely that increase in copy numbers of Cyt B, ND1 and ND4 in the brain of children with autism may be attributed to increased oxidative stress or defective scavenging of free radicals in autism. Alternatively, increased production of reactive oxygen species by damaged mitochondria could elicit chronic oxidative stress, which may increase mtDNA replication and repair. The higher mtDNA copy number in autism subjects may act as the compensatory mechanism to increase the number of wild-type mtDNA templates so that normal levels of mitochondrial transcripts can be maintained.76 It may also be possible that increased mtDNA copy number could be due to defective replication and/or repair of mtDNA inflicted upon by genetic or environmental factors. Whatever the case may be for increased copy number of Cyt B, ND1 and ND4, it will result in perturbed energy production in the brain and may lead to neuronal dysfunctions in autism.
CNV occurs not only in the mtDNA segments but also in nDNA segments of children with autism.46, 47, 48, 49, 50, 51, 52, 53, 54 Using comparative genomic hybridization technique, Sebat et al.52 reported strong association of de novo copy number mutations in autism. Recently, CNVs including duplications at 1q21.1, 7q11.23, 15q11-q13 and 22q11.21, as well as deletions at 16p11.2, have been suggested as risk factors for autism.47, 50, 51, 54
In conclusion, mitochondria may be having a significant role in the etiology of ASDs. The high-energy demand of the developing brain may trigger a cascade of structural and functional changes, leading to the autistic phenotype if mitochondrial functions are impaired and the energy need of the brain is not fulfilled. Therefore, abnormalities observed in the mtDNA or ETC functions along with oxidative stress may be responsible for some of the core features of autism.
Acknowledgments
Human brain tissues were obtained from the National Institute of Child Health and Human Development Brain and Tissue Bank for Developmental Disorders at the University of Maryland, Baltimore, MD. This work was supported by funds from the Department of Defense Autism Spectrum Disorders Research Program AS073224P2, the Autism Research Institute and the NYS Office for People with Developmental Disabilities.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on the Translational Psychiatry website (http://www.nature.com/tp)
Supplementary Material
References
- Wingate M, Mulvihill B, Kirby RS, Pettygrove S, Cunniff C, Meaney F, et al. Prevalence of autism spectrum disorders—autism and developmental disabilities monitoring network, 14 sites, United States, 2008. MMWR Surveill Summ. 2012;61:1–19. [PubMed] [Google Scholar]
- Adams JB, Audhya T, McDonough-Means S, Rubin RA, Quig D. Nutritional and metabolic status of children with autism vs. neurotypical children, and the association with autism severity. Nutr Metab (Lond) 2011;8:34. doi: 10.1186/1743-7075-8-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan A, Chauhan V, Brown WT, Cohen I. Oxidative stress in autism: increased lipid peroxidation and reduced serum levels of ceruloplasmin and transferrin--the antioxidant proteins. Life Sci. 2004;75:2539–2549. doi: 10.1016/j.lfs.2004.04.038. [DOI] [PubMed] [Google Scholar]
- Chauhan A, Chauhan V. Oxidative stress in autism. Pathophysiology. 2006;13:171–181. doi: 10.1016/j.pathophys.2006.05.007. [DOI] [PubMed] [Google Scholar]
- Chauhan A, Chauhan V, Brown WT.(eds)Autism: oxidative stress, inflammation and immune abnormalities CRC Press: Taylor and Francis group: Florida; 2009 [Google Scholar]
- Chauhan A, Gu F, Essa MM, Wegiel J, Kaur K, Brown WT, et al. Brain region-specific deficit in mitochondrial electron transport chain complexes in children with autism. J Neurochem. 2011;117:209–220. doi: 10.1111/j.1471-4159.2011.07189.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan A, Audhya T, Chauhan V. Brain region-specific glutathione redox imbalance in autism. Neurochem Res. 2012;37:1681–1689. doi: 10.1007/s11064-012-0775-4. [DOI] [PubMed] [Google Scholar]
- Gu F, Chauhan V, Chauhan A. Impaired synthesis and antioxidant defense of glutathione in the cerebellum of autistic subjects: alternations in the activities and protein expression of glutathione-related enzymes. Free Radic Biol Med. 2013;65:488–496. doi: 10.1016/j.freeradbiomed.2013.07.021. [DOI] [PubMed] [Google Scholar]
- James SJ, Melnyk S, Jernigan S, Cleves MA, Halsted CH, Wong DH, et al. Metabolic endophenotype and related genotypes are associated with oxidative stress in children with autism. Am J Med Genet B Neuropsychiatr Genet. 2006;141B:947–956. doi: 10.1002/ajmg.b.30366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ming X, Stein TP, Brimacombe M, Johnson WG, Lambert GH, Wagner GC. Increased excretion of a lipid peroxidation biomarker in autism. Prostaglandins Leukot Essent Fatty Acids. 2005;73:379–384. doi: 10.1016/j.plefa.2005.06.002. [DOI] [PubMed] [Google Scholar]
- Sajdel-Sulkowska EM, Xu M, McGinnis W, Koibuchi N. Brain region-specific changes in oxidative stress and neurotrophin levels in autism spectrum disorders (ASD) Cerebellum. 2011;10:43–48. doi: 10.1007/s12311-010-0223-4. [DOI] [PubMed] [Google Scholar]
- Rossignol DA, Frye RE. Mitochondrial dysfunction in autism spectrum disorders: a systematic review and meta-analysis. Mol Psychiatry. 2012;17:290–314. doi: 10.1038/mp.2010.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram R, Gram Pedersen M, Luciani DS, Sherman A. A simplified model for mitochondrial ATP production. J Theor Biol. 2006;243:575–586. doi: 10.1016/j.jtbi.2006.07.019. [DOI] [PubMed] [Google Scholar]
- Boekema EJ, Braun HP. Supramolecular structure of the mitochondrial oxidative phosphorylation system. J Biol Chem. 2007;282:1–4. doi: 10.1074/jbc.R600031200. [DOI] [PubMed] [Google Scholar]
- Cadenas E, Davies KJ. Mitochondrial free radical generation, oxidative stress, and aging. Free Radic Biol Med. 2000;29:222–230. doi: 10.1016/s0891-5849(00)00317-8. [DOI] [PubMed] [Google Scholar]
- Lenaz G. The mitochondrial production of reactive oxygen species: mechanisms and implications in human pathology. IUBMB Life. 2001;52:159–164. doi: 10.1080/15216540152845957. [DOI] [PubMed] [Google Scholar]
- Shay JW, Pierce DJ, Werbin H. Mitochondrial DNA copy number is proportional to total cell DNA under a variety of growth conditions. J Biol Chem. 1990;265:14802–14807. [PubMed] [Google Scholar]
- Liu CS, Tsai CS, Kuo CL, Chen HW, Lii CK, Ma YS, et al. Oxidative stress-related alteration of the copy number of mitochondrial DNA in human leukocytes. Free Radic Res. 2003;37:1307–1317. doi: 10.1080/10715760310001621342. [DOI] [PubMed] [Google Scholar]
- Lee HC, Wei YH. Mitochondrial biogenesis and mitochondrial DNA maintenance of mammalian cells under oxidative stress. Int J Biochem Cell Biol. 2005;37:822–834. doi: 10.1016/j.biocel.2004.09.010. [DOI] [PubMed] [Google Scholar]
- Wang X, Su B, Zheng L, Perry G, Smith MA, Zhu X. The role of abnormal mitochondrial dynamics in the pathogenesis of Alzheimer's disease. J Neurochem. 2009;109 (Suppl 1:153–159. doi: 10.1111/j.1471-4159.2009.05867.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aliev G, Palacios HH, Walrafen B, Lipsitt AE, Obrenovich ME, Morales L. Brain mitochondria as a primary target in the development of treatment strategies for Alzheimer disease. Int J Biochem Cell Biol. 2009;41:1989–2004. doi: 10.1016/j.biocel.2009.03.015. [DOI] [PubMed] [Google Scholar]
- Morais VA, De Strooper B. Mitochondria dysfunction and neurodegenerative disorders: cause or consequence. J Alzheimers Dis. 2010;20 (Suppl 2:S255–S263. doi: 10.3233/JAD-2010-100345. [DOI] [PubMed] [Google Scholar]
- Moran M, Moreno-Lastres D, Marin-Buera L, Arenas J, Martin MA, Ugalde C. Mitochondrial respiratory chain dysfunction: implications in neurodegeneration. Free Radic Biol Med. 2012;53:595–609. doi: 10.1016/j.freeradbiomed.2012.05.009. [DOI] [PubMed] [Google Scholar]
- Sterky FH, Hoffman AF, Milenkovic D, Bao B, Paganelli A, Edgar D, et al. Altered dopamine metabolism and increased vulnerability to MPTP in mice with partial deficiency of mitochondrial complex I in dopamine neurons. Hum Mol Genet. 2012;21:1078–1089. doi: 10.1093/hmg/ddr537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie W, Wan OW, Chung KK. New insights into the role of mitochondrial dysfunction and protein aggregation in Parkinson's disease. Biochim Biophys Acta. 2010;1802:935–941. doi: 10.1016/j.bbadis.2010.07.014. [DOI] [PubMed] [Google Scholar]
- Zhu J, Chu CT. Mitochondrial dysfunction in Parkinson's disease. J Alzheimers Dis. 2010;20 (Suppl 2:S325–S334. doi: 10.3233/JAD-2010-100363. [DOI] [PubMed] [Google Scholar]
- Gu M, Gash MT, Mann VM, Javoy-Agid F, Cooper JM, Schapira AH. Mitochondrial defect in Huntington's disease caudate nucleus. Ann Neurol. 1996;39:385–389. doi: 10.1002/ana.410390317. [DOI] [PubMed] [Google Scholar]
- Wiedemann FR, Manfredi G, Mawrin C, Beal MF, Schon EA. Mitochondrial DNA and respiratory chain function in spinal cords of ALS patients. J Neurochem. 2002;80:616–625. doi: 10.1046/j.0022-3042.2001.00731.x. [DOI] [PubMed] [Google Scholar]
- Ghiasi P, Hosseinkhani S, Noori A, Nafissi S, Khajeh K. Mitochondrial complex I deficiency and ATP/ADP ratio in lymphocytes of amyotrophic lateral sclerosis patients. Neurol Res. 2012;34:297–303. doi: 10.1179/1743132812Y.0000000012. [DOI] [PubMed] [Google Scholar]
- Bubber P, Tang J, Haroutunian V, Xu H, Davis KL, Blass JP, et al. Mitochondrial enzymes in schizophrenia. J Mol Neurosci. 2004;24:315–321. doi: 10.1385/JMN:24:2:315. [DOI] [PubMed] [Google Scholar]
- Cavelier L, Jazin EE, Eriksson I, Prince J, Bave U, Oreland L, et al. Decreased cytochrome-c oxidase activity and lack of age-related accumulation of mitochondrial DNA deletions in the brains of schizophrenics. Genomics. 1995;29:217–224. doi: 10.1006/geno.1995.1234. [DOI] [PubMed] [Google Scholar]
- Chauhan A, Gu F, Chauhan V. Mitochondrial respiratory chain defects in autism and other neurodevelopmental disorders. J Pediatr Biochem. 2012;2:213–223. [Google Scholar]
- Maurer I, Zierz S, Moller H. Evidence for a mitochondrial oxidative phosphorylation defect in brains from patients with schizophrenia. Schizophr Res. 2001;48:125–136. doi: 10.1016/s0920-9964(00)00075-x. [DOI] [PubMed] [Google Scholar]
- Haas RH. Autism and mitochondrial disease. Dev Disabil Res Rev. 2010;16:144–153. doi: 10.1002/ddrr.112. [DOI] [PubMed] [Google Scholar]
- Gargus JJ, Imtiaz F. Mitochondrial energy-deficient endophenotype in autism. Am J Biochem Biotech. 2008;4:198–207. [Google Scholar]
- Palmieri L, Persico AM. Mitochondrial dysfunction in autism spectrum disorders: cause or effect. Biochim Biophys Acta. 2010;1797:1130–1137. doi: 10.1016/j.bbabio.2010.04.018. [DOI] [PubMed] [Google Scholar]
- Correia C, Coutinho AM, Diogo L, Grazina M, Marques C, Miguel T, et al. Brief report: High frequency of biochemical markers for mitochondrial dysfunction in autism: no association with the mitochondrial aspartate/glutamate carrier SLC25A12 gene. J Autism Dev Disord. 2006;36:1137–1140. doi: 10.1007/s10803-006-0138-6. [DOI] [PubMed] [Google Scholar]
- Giulivi C, Zhang YF, Omanska-Klusek A, Ross-Inta C, Wong S, Hertz-Picciotto I, et al. Mitochondrial dysfunction in autism. JAMA. 2010;304:2389–2396. doi: 10.1001/jama.2010.1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guevara-Campos J, Gonzalez-Guevara L, Briones P, Lopez-Gallardo E, Bulan N, Ruiz-Pesini E, et al. Autism associated to a deficiency of complexes III and IV of the mitochondrial respiratory chain. Invest Clin. 2010;51:423–431. [PubMed] [Google Scholar]
- Oliveira G, Diogo L, Grazina M, Garcia P, Ataide A, Marques C, et al. Mitochondrial dysfunction in autism spectrum disorders: a population-based study. Dev Med Child Neurol. 2005;47:185–189. doi: 10.1017/s0012162205000332. [DOI] [PubMed] [Google Scholar]
- Shoffner J, Hyams L, Langley GN, Cossette S, Mylacraine L, Dale J, et al. Fever plus mitochondrial disease could be risk factors for autistic regression. J Child Neurol. 2010;25:429–434. doi: 10.1177/0883073809342128. [DOI] [PubMed] [Google Scholar]
- Weissman JR, Kelley RI, Bauman ML, Cohen BH, Murray KF, Mitchell RL, et al. Mitochondrial disease in autism spectrum disorder patients: a cohort analysis. PLoS One. 2008;3:e3815. doi: 10.1371/journal.pone.0003815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chugani DC, Sundram BS, Behen MLML, Moore GJ. Evidence of altered energy metabolism in autistic children. Prog Neuropsychopharmacol Biol Psychiatry. 1999;23:635–641. doi: 10.1016/s0278-5846(99)00022-6. [DOI] [PubMed] [Google Scholar]
- Minshew NJ, Goldstein G, Dombrowski SM, Panchalingam K, Pettegrew JW. A preliminary 31P MRS study of autism: evidence for undersynthesis and increased degradation of brain membranes. Biol Psychiatry. 1993;33:762–773. doi: 10.1016/0006-3223(93)90017-8. [DOI] [PubMed] [Google Scholar]
- Cotter D, Guda P, Fahy E, Subramaniam S. MitoProteome: mitochondrial protein sequence database and annotation system. Nucleic Acids Res. 2004;32:D463–D467. doi: 10.1093/nar/gkh048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho SC, Yim SH, Yoo HK, Kim MY, Jung GY, Shin GW, et al. Copy number variations associated with idiopathic autism identified by whole-genome microarray-based comparative genomic hybridization. Psychiatr Genet. 2009;19:177–185. doi: 10.1097/YPG.0b013e32832bdafa. [DOI] [PubMed] [Google Scholar]
- Crespi BJ, Crofts HJ. Association testing of copy number variants in schizophrenia and autism spectrum disorders. J Neurodev Disord. 2012;4:15. doi: 10.1186/1866-1955-4-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin B, Scherer SW. Genetic architecture in autism spectrum disorder. Curr Opin Genet Dev. 2012;22:229–237. doi: 10.1016/j.gde.2012.03.002. [DOI] [PubMed] [Google Scholar]
- Griswold AJ, Ma D, Cukier HN, Nations LD, Schmidt MA, Chung RH, et al. Evaluation of copy number variations reveals novel candidate genes in autism spectrum disorder-associated pathways. Hum Mol Genet. 2012;21:3513–3523. doi: 10.1093/hmg/dds164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller DT, Shen Y, Weiss LA, Korn J, Anselm I, Bridgemohan C, et al. Microdeletion/duplication at 15q13.2q13.3 among individuals with features of autism and other neuropsychiatric disorders. J Med Genet. 2009;46:242–248. doi: 10.1136/jmg.2008.059907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders SJ, Ercan-Sencicek AG, Hus V, Luo R, Murtha MT, Moreno-De-Luca D, et al. Multiple recurrent de novo CNVs, including duplications of the 7q11.23 Williams syndrome region, are strongly associated with autism. Neuron. 2011;70:863–885. doi: 10.1016/j.neuron.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebat J, Lakshmi B, Malhotra D, Troge J, Lese-Martin C, Walsh T, et al. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M, Flodman PL, Gargus JJ, Simon MT, Verrell K, Haas R, et al. Mitochondrial and ion channel gene alterations in autism. Biochim Biophys Acta. 2012;1817:1796–1802. doi: 10.1016/j.bbabio.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss LA, Shen Y, Korn JM, Arking DE, Miller DT, Fossdal R, et al. Association between microdeletion and microduplication at 16p11.2 and autism. N Engl J Med. 2008;358:667–675. doi: 10.1056/NEJMoa075974. [DOI] [PubMed] [Google Scholar]
- Girirajan S, Eichler EE. De novo CNVs in bipolar disorder: recurrent themes or new directions. Neuron. 2011;72:885–887. doi: 10.1016/j.neuron.2011.12.008. [DOI] [PubMed] [Google Scholar]
- Priebe L, Degenhardt FA, Herms S, Haenisch B, Mattheisen M, Nieratschker V, et al. Genome-wide survey implicates the influence of copy number variants (CNVs) in the development of early-onset bipolar disorder. Mol Psychiatry. 2012;17:421–432. doi: 10.1038/mp.2011.8. [DOI] [PubMed] [Google Scholar]
- Malhotra D, McCarthy S, Michaelson JJ, Vacic V, Burdick KE, Yoon S, et al. High frequencies of de novo CNVs in bipolar disorder and schizophrenia. Neuron. 2012;72:951–963. doi: 10.1016/j.neuron.2011.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosak L, Silhan P, Hosakova J. Genomic copy number variations: A breakthrough in our knowledge on schizophrenia etiology. Neuro Endocrinol Lett. 2012;33:183–190. [PubMed] [Google Scholar]
- Kirov G, Grozeva D, Norton N, Ivanov D, Mantripragada KK, Holmans P, et al. Support for the involvement of large copy number variants in the pathogenesis of schizophrenia. Hum Mol Genet. 2009;18:1497–1503. doi: 10.1093/hmg/ddp043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye T, Lipska BK, Tao R, Hyde TM, Wang L, Li C, et al. Analysis of copy number variations in brain DNA from patients with schizophrenia and other psychiatric disorders. Biol Psychiatry. 2012;72:651–654. doi: 10.1016/j.biopsych.2012.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pons R, Andreu AL, Checcarelli N, Vila MR, Engelstad K, Sue CM, et al. Mitochondrial DNA abnormalities and autistic spectrum disorders. J Pediatr. 2004;144:81–85. doi: 10.1016/j.jpeds.2003.10.023. [DOI] [PubMed] [Google Scholar]
- Carroll J, Fearnley IM, Shannon RJ, Hirst J, Walker JE. Analysis of the subunit composition of complex I from bovine heart mitochondria. Mol Cell Proteomics. 2003;2:117–126. doi: 10.1074/mcp.M300014-MCP200. [DOI] [PubMed] [Google Scholar]
- Janssen RJ, Nijtmans LG, van den Heuvel LP, Smeitink JA. Mitochondrial complex I: structure, function and pathology. J Inherit Metab Dis. 2006;29:499–515. doi: 10.1007/s10545-006-0362-4. [DOI] [PubMed] [Google Scholar]
- Barja G. Mitochondrial oxygen radical generation and leak: sites of production in states 4 and 3, organ specificity, and relation to aging and longevity. J Bioenerg Biomembr. 1999;31:347–366. doi: 10.1023/a:1005427919188. [DOI] [PubMed] [Google Scholar]
- Muller FL, Liu Y, Van Remmen H. Complex III releases superoxide to both sides of the inner mitochondrial membrane. J Biol Chem. 2004;279:49064–49073. doi: 10.1074/jbc.M407715200. [DOI] [PubMed] [Google Scholar]
- Kirby DM, Crawford M, Cleary MA, Dahl HH, Dennett X, Thorburn DR. Respiratory chain complex I deficiency: an underdiagnosed energy generation disorder. Neurology. 1999;52:1255–1264. doi: 10.1212/wnl.52.6.1255. [DOI] [PubMed] [Google Scholar]
- Andreazza AC, Shao L, Wang JF, Young LT. Mitochondrial complex I activity and oxidative damage to mitochondrial proteins in the prefrontal cortex of patients with bipolar disorder. Arch Gen Psychiatry. 2010;67:360–368. doi: 10.1001/archgenpsychiatry.2010.22. [DOI] [PubMed] [Google Scholar]
- Kim SH, Vlkolinsky R, Cairns N, Fountoulakis M, Lubec G. The reduction of NADH ubiquinone oxidoreductase 24- and 75-kDa subunits in brains of patients with Down syndrome and Alzheimer's disease. Life Sci. 2001;68:2741–2750. doi: 10.1016/s0024-3205(01)01074-8. [DOI] [PubMed] [Google Scholar]
- Kim SH, Vlkolinsky R, Cairns N, Lubec G. Decreased levels of complex III core protein 1 and complex V beta chain in brains from patients with Alzheimer's disease and Down syndrome. Cell Mol Life Sci. 2000;57:1810–1816. doi: 10.1007/PL00000661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iafrate AJ, Feuk L, Rivera MN, Listewnik ML, Donahoe PK, Qi Y, et al. Detection of large-scale variation in the human genome. Nat Genet. 2004;36:949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, Andrews TD, et al. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almal SH, Padh H. Implications of gene copy-number variation in health and diseases. J Hum Genet. 2012;57:6–13. doi: 10.1038/jhg.2011.108. [DOI] [PubMed] [Google Scholar]
- Kato T, Inubushi T, Kato N. Magnetic resonance spectroscopy in affective disorders. J Neuropsychiatry Clin Neurosci. 1998;10:133–147. doi: 10.1176/jnp.10.2.133. [DOI] [PubMed] [Google Scholar]
- Rezin GT, Amboni G, Zugno AI, Quevedo J, Streck EL. Mitochondrial dysfunction and psychiatric disorders. Neurochem Res. 2009;34:1021–1029. doi: 10.1007/s11064-008-9865-8. [DOI] [PubMed] [Google Scholar]
- Yakes FM, Van Houten B. Mitochondrial DNA damage is more extensive and persists longer than nuclear DNA damage in human cells following oxidative stress. Proc Natl Acad Sci USA. 1997;94:514–519. doi: 10.1073/pnas.94.2.514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RS. Mitochondrial gene expression in mammalian striated muscle. Evidence that variation in gene dosage is the major regulatory event. J Biol Chem. 1986;261:12390–12394. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.