Abstract
Stem cell technologies provide an exciting avenue to directly access the transcriptome of patients in neuronal-like cell types, which might have more direct relevance to brain research than other peripheral tissues (blood, fibroblasts). Enthusiasm should be tempered by concerns that artifacts and noise might be generated as part of the in vitro process of creating and maintaining these cell type. A solution may be to apply a Convergent Functional Genomics approach, where the data from stem cell-derived neuronal cells are integrated, cross-validated and prioritized using independent lines of evidence from other approaches and platforms (human genetic data, human postmortem brain data, animal model data). I provide a brief overview and an example in support of such an approach.
Keywords: biomarkers, convergent functional genomics, gene expression, GWAS, stem cells
‘Did I request thee, Maker, from my clay To mould me man? Did I solicit thee From darkness to promote me?' —Milton, Paradise Lost
Gene expression and stem cell-derived cells
Gene expression data directly reflect genetic inheritance and acquired genetic mutations, as well as environmental influences. Scientifically, it may also help tie together and unravel epistasis (co-acting gene expression,1 ‘genes that change together work together'), as well as regulatory networks of non-coding single-nucleotide polymorphisms (SNPs),2 epigenetic changes, chromatin modifications, non-coding RNAs and transcription factors responsive to environmental stimuli. Human gene expression studies have been carried out in postmortem brain,3, 4 as well as in peripheral blood,5, 6 fibroblasts,7 olfactory epithelium-derived neurons8 and, more recently, in induced pluripotent stem cell (iPSC)-derived neurons.9, 10 Each particular approach has strengths and limitations (Table 1). The quest for peripheral tissue read-outs and biomarkers is particularly important in psychiatry, as the target organ (brain) is not accessible to biopsies in live humans, for obvious practical and ethical reasons. The integration of genomics with phenomics (for example, quantitative clinical data), in particular the issue of whether a marker reflects state, trait, both, or neither, is important and often overlooked. The ability to correlate peripheral read-outs directly with mental states (for example, symptom severity), or indirectly with mental traits (for example, psychiatric diagnosis), determines what kind of biomarkers can be discovered using different tissues and approaches.
Table 1. Human neuropsychiatric gene expression studies.
| Human postmortem brain | Human blood | Fibroblasts | Olfactory epithelium-derived neurons | iPSC-derived neurons | |
|---|---|---|---|---|---|
| Strengths | Target organ Neuronal cells | Immediate access Non-transformed cells (except when lymphoblastoid cell lines are used) Direct correlation with mental state information | Non- (or less) transformed cells | Neuronal-like cells Non- (or less) transformed cells | Neuronal-like cells |
| Limitations | Postmortem interval artifacts Lack of direct correlation with mental state information | Non-neuronal | Cell-culture artifacts Non-neuronal Lack of direct correlation with mental state information | Cell-culture artifacts Lack of direct correlation with mental state information | Cell-culture artifacts Transformation artifacts Lack of direct correlation with mental state information |
Abbreviation: iPSC, induced pluripotent stem cell.
iPSCs, in addition to future hypothetical organ-building regenerative medicine applications, may be more immediately useful for understanding disease,11 and particularly for drug testing and drug discovery,12 including personalized medicine approaches. However, concerns arise about genetic and gene expression artifacts induced by the in vitro stem cell creation process. Two of the four transcription factors used to create iPSCs (c-Myc and KLF4) are oncogenic. Interestingly, histone deacetylase inhibitors (HDACi), such as the neuropsychiatric agent valproate,13 may provide a safer alternative for helping transform adult cells into iPSCs. Valproate might also expand the pool of neural stem cells in the adult brain.14 The effect of the HDACi per se on the gene expression landscape would have to be factored out in scientific studies. Olfactory epithelium-derived neuronal precursor cells may also have less transformation artifacts,8, 15 although the cell culture and passaging artifacts remain in common with the other cell culture approaches. Finally, all neuronal-like cells derived with these methodologies need to be validated as being indeed reflective of true neurons. Some of the methods used for this are, in the increasing order of relevance, neuronal biochemical marker testing (immunohistochemistry), testing for synapse formation (electrophysiology) and functional integration in vivo.16
The gene expression data obtained from such cells arguably need additional cross-validation for relevance to in vivo functioning and disease states.
Convergent Functional Genomics (CFG)
Genetic and gene expression studies in humans and lower organism model (mice, rats, dogs, zebra fish, Drosophila, Caenorhabditis elegans, yeast) studies of medical disorders are becoming increasingly integrated. Particularly for genomics, the convergence and integration of data across experimental modalities, technical platforms, and species are providing a fit-to-disease way of extracting reproducible and biologically important signal, in contrast to the fit-to-cohort effect and limited reproducibility of human genetic analyses alone. Due to the emerging data from the ENCODE project suggesting that a major portion of the non-coding genome may contain regulatory variants, convergent approaches are going to be important to identify disease-relevant signal from the polymorphic variation present in the population.
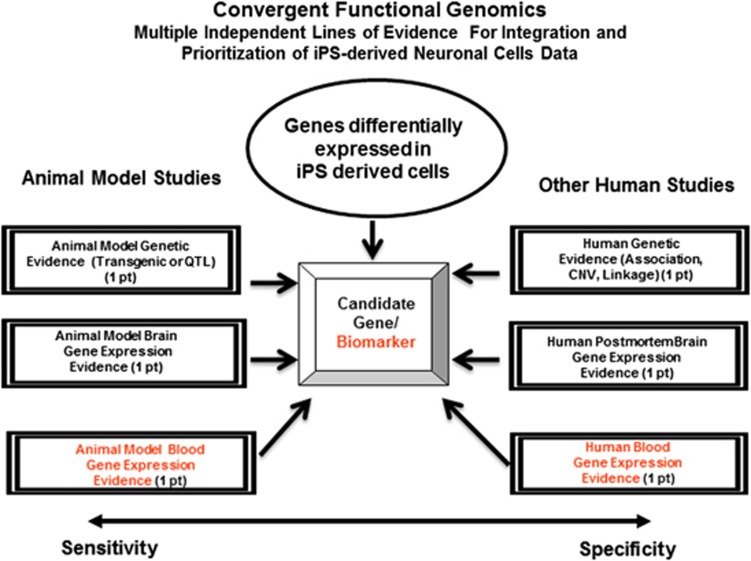
CFG1, 5, 17, 18, 19, 20, 21, 22, 23, 24 is a powerful methodology developed over the past 15 years for extracting signal from noise by gene-level integration of multiple independent lines of evidence from human and lower organisms model studies—genetic, gene expression, proteomics—of brain, peripheral tissues and cell lines (Figure 1). Lower organism model data can provide sensitivity and ability to conduct experimental manipulations not feasible in humans. Human data provide more specificity and relevance to the human disease. Combined, we have an approach that increases our ability to distinguish signal from noise even with limited size cohorts and data sets. CFG helps to identify and prioritize candidate genes for the illness, using a polyevidence score. All these lines of evidence are the result of independent experiments. The virtues of this networked approach are that, even if one or another of the ‘nodes' (lines of evidence) becomes questionable/non-functional upon further evidence in the field, the network is resilient and maintains the functionality. The prioritization of candidates is similar conceptually to the Google PageRank algorithm—the more links (lines of evidence) to a candidate, the higher it will be prioritized. Subsequent biological pathway analyses on these prioritized genes can uncover mechanistic aspects of the disease being studied. More recently, variations and expansions of this approach have been used successfully by other groups as well.25, 26
Figure 1.
Convergent Functional Genomics: multiple independent lines of evidence for integration and prioritization of induced pluripotent stem (iPS)-derived neuronal cells data. CNV, copy number variant; QTL, quantitative trait loci.
Our past work provides evidence for the advantages, reproducibility and consistency of gene-level analyses of data, as opposed to SNP level analyses, pointing to the fundamental issue of genetic heterogeneity at a SNP level.27 In fact, it may be that the more biologically important a gene is for higher mental functions, the more heterogeneity it has at a SNP level and the more evolutionary divergence, for adaptive reasons.28 A similar diversity, for similar adaptive reasons, exists in immune system genes.
On top of the gene-level integration, CFG provides a way to prioritize genes based on disease relevance, not study-specific effects (that is, fit-to-disease as opposed to fit-to-cohort). Reproducibility of findings across different studies, experimental paradigms and technical platforms is deemed more important (and scored as such by CFG) than the strength of finding in an individual study (for example, P-value in a genome-wide association study (GWAS)). This Bayesian-like approach minimizes false positives if one focuses on the top of the distribution, and minimizes false negatives if one goes deeper down the list (Figure 2). Most importantly, the CFG-prioritized genes show reproducibility and predictive ability in independent cohorts, which is the key litmus test for genetic and biomarker studies. Once the genes are identified and prioritized, biological pathway analyses can be conducted and mechanistic models can be constructed.
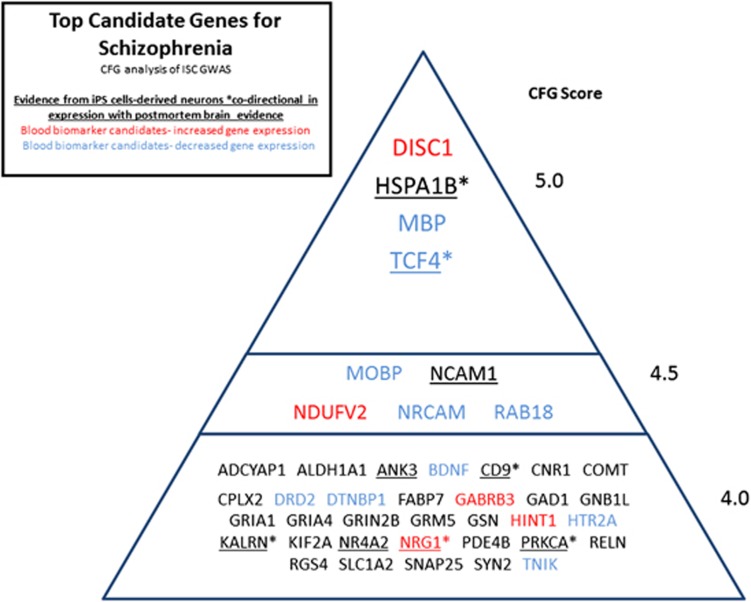
Figure 2.
Top candidate genes for schizophrenia—Convergent Functional Genomics (CFG) analysis of ISC genome-wide association study (GWAS). iPS cell, induced pluripotent stem cell; ISC, International Schizophrenia Consortium.
Using a set of mouse experiments as a driving force,20, 23 or using human blood gene expression5, 6 or GWAS data22, 24, 27 as a driving force, such convergent studies from my group and others have identified and prioritized candidate genes and biomarkers for psychiatric disorders (bipolar disorder,22, 24 schizophrenia,21, 27 anxiety disorders,29 alcoholism19, 30) that show good reproducibility as well as predictive ability in independent cohorts. In essence, the CFG approach is a de facto field-wide collaboration, integrating together the best available evidence at the time the analyses are conducted. Periodic re-analyses as future evidence accumulates in the field can improve and refine the results.
Application of CFG to stem cell-derived data
Data generated from neuronal-like cells derived from iPSCs can be cross-validated and prioritized using a CFG approach with other lines of evidence (Figure 1), or can serve as a line of evidence itself for the cross-validation and prioritization of, for example, GWAS data (Figure 2).
We have used the later approach for schizophrenia.27 Data published by Gage and colleagues from schizophrenia subjects10 in iPSC-derived neuronal-like cells (‘hiPSC neurons') was used as one of the multiple lines of evidence in a convergent approach that incorporated, besides GWAS data,31 human postmortem data, human blood gene expression data6 and animal model pharmacogenomics brain and blood gene expression data (using phencyclidine and clozapine as agonist-antagonist pharmacological agents21). In all, 21% (9 out of 42) of the top schizophrenia candidate genes identified by us in our overall CFG analysis had evidence in the hiPSC neurons study, and in 6 out of 9 of these genes the direction of change in expression in iPS-derived cells was the same as that in postmortem brains from schizophrenics (HSPA1B, TCF4, CD9, KALRN, PRKCA and NRG1) (Figure 2). Given the fact that the ‘hiPSC neurons' data in the original study were derived from only n=4 schizophrenic subjects,10 and there is intra-subject as well as inter-subject variability in cell lines, generating a large ( 596 unique genes) and potentially noisy list of differentially expressed genes, the use of cross-validating approaches such as CFG was essential to pinpoint the most disease-relevant genes.
The case of HSPA1B (heat-shock 70-kDa protein 1B), for example, a previously more obscure gene in terms of involvement in schizophrenia, is illustrative of the utility of a non-hypothesis-driven, convergent approach. HSPA1B, a chaperone involved in stress response, stabilizes existing proteins against aggregation and mediates the folding of newly translated proteins. HSPA1B has some previous genetic evidence for association with schizophrenia.32 It is co-directionally increased in expression in postmortem brains33 and iPSC-derived neurons from schizophrenia patients. HSPA1B is also decreased in expression by antipsychotic treatment with clozapine in the brain and blood of a mouse model, based on our previous work.21 It was also co-directionally increased in the brain and blood in a pharmacogenomic mouse model of anxiety disorders that we have recently described,29 as well as in a stress-reactive genetic mouse model.20 Treatment with the omega-3 fatty acid docosahexaenoic acid reversed the increase in expression of HSPA1B in this stress-reactive genetic mouse model.30 Another closely related molecule, HSPA1A (heat-shock 70-kDa protein 1A), is also present on our list of prioritized candidate genes for schizophrenia, with a lower CFG score of 3.5.27 Heat-shock proteins may be involved in the biological and clinical overlap and interdependence between response to stress,34 anxiety and psychosis.
A CFG approach could also be used in cases where HDACi are used for transformation, to understand which gene expressed in iPSC-derived cells are drug modulated. We have generated in our lab valproate brain and blood gene expression data sets5, 23 from mouse models, which could serve such a role.
Conclusion
Convergent approaches may be important for mining and interpreting gene expression data from pluripotent stem cell-derived cells in psychiatric and non-psychiatric disorders.
Acknowledgments
We thank members of the Niculescu laboratory for their input; Rusty Gage, Alysson Muotri, Akira Sawa and Uli Mueller for stimulating discussions; and our colleagues in the field for the comprehensive work they are doing. This work was supported by an NIH Directors' New Innovator Award (1DP2OD007363) and a VA Merit Award (1I01CX000139-01) to ABN.
The author declares no conflict of interest.
References
- Niculescu AB, 3rd, Segal DS, Kuczenski R, Barrett T, Hauger RL, Kelsoe JR. Identifying a series of candidate genes for mania and psychosis: a convergent functional genomics approach. Physiol Genomics. 2000;4:83–91. doi: 10.1152/physiolgenomics.2000.4.1.83. [DOI] [PubMed] [Google Scholar]
- ENCODE Project Consortium. Bernstein BE, Birney E, Dunham I, Green ED, Gunter C, Snyder M. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banigan MG, Birney E, Dunham I, Green ED, Gunter C, Snyder M. Differential expression of exosomal micrornas in prefrontal cortices of schizophrenia and bipolar disorder patients. PLoS One. 2013;8:e48814. doi: 10.1371/journal.pone.0048814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HJ, Kawasawa YI, Cheng F, Zhu Y, Xu X, Li M. Spatio-temporal transcriptome of the human brain. Nature. 2011;478:483–489. doi: 10.1038/nature10523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le-Niculescu H, Kurian SM, Yehyawi N, Dike C, Patel SD, Edenberg HJ, et al. Identifying blood biomarkers for mood disorders using convergent functional genomics. Mol Psychiatry. 2009;14:156–174. doi: 10.1038/mp.2008.11. [DOI] [PubMed] [Google Scholar]
- Kurian SM, Kurian SM, Yehyawi N, Dike C, Patel SD, Edenberg HJ, et al. Identification of blood biomarkers for psychosis using convergent functional genomics. Mol Psychiatry. 2011;16:37–58. doi: 10.1038/mp.2009.117. [DOI] [PubMed] [Google Scholar]
- Yang S, Van Dongen HP, Wang K, Berrettini W, Bucan M. Assessment of circadian function in fibroblasts of patients with bipolar disorder. Mol Psychiatry. 2009;14:143–155. doi: 10.1038/mp.2008.10. [DOI] [PubMed] [Google Scholar]
- Tajinda K, Ishizuka K, Colantuoni C, Morita M, Winicki J, Le C, et al. Neuronal biomarkers from patients with mental illnesses: a novel method through nasal biopsy combined with laser-captured microdissection. Mol Psychiatry. 2010;15:231–232. doi: 10.1038/mp.2009.73. [DOI] [PubMed] [Google Scholar]
- Lin M, Pedrosa E, Shah A, Hrabovsky A, Maqbool S, Zheng D, et al. RNA-Seq of human neurons derived from iPS cells reveals candidate long non-coding RNAs involved in neurogenesis and neuropsychiatric disorders. PLoS One. 2011;6:e23356. doi: 10.1371/journal.pone.0023356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennand KJ, Simone A, Jou J, Gelboin-Burkhart C, Tran N, Sangar S, et al. Modelling schizophrenia using human induced pluripotent stem cells. Nature. 2011;473:221–225. doi: 10.1038/nature09915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchetto MC, Carromeu C, Acab A, Yu D, Yeo GW, Mu Y, et al. A model for neural development and treatment of Rett syndrome using human induced pluripotent stem cells. Cell. 2010;143:527–539. doi: 10.1016/j.cell.2010.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellin M, Marchetto MC, Gage FH, Mummery CL. Induced pluripotent stem cells: the new patient. Nat Rev Mol Cell Biol. 2012;13:713–726. doi: 10.1038/nrm3448. [DOI] [PubMed] [Google Scholar]
- Huangfu D, Maehr R, Guo W, Eijkelenboom A, Snitow M, Chen AE, et al. Induction of pluripotent stem cells by defined factors is greatly improved by small-molecule compounds. Nat Biotechnol. 2008;26:795–797. doi: 10.1038/nbt1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashi M, Maruta N, Bernstein A, Ikenaka K, Hitoshi S. Mood stabilizing drugs expand the neural stem cell pool in the adult brain through activation of notch signaling. Stem Cells. 2008;26:1758–1767. doi: 10.1634/stemcells.2007-1032. [DOI] [PubMed] [Google Scholar]
- Benitez-King G, Riquelme A, Ortíz-López L, Berlanga C, Rodríguez-Verdugo MS, Romo F, et al. A non-invasive method to isolate the neuronal linage from the nasal epithelium from schizophrenic and bipolar diseases. J Neurosci Methods. 2011;201:35–45. doi: 10.1016/j.jneumeth.2011.07.009. [DOI] [PubMed] [Google Scholar]
- Wernig M, Zhao JP, Pruszak J, Hedlund E, Fu D, Soldner F, et al. Neurons derived from reprogrammed fibroblasts functionally integrate into the fetal brain and improve symptoms of rats with Parkinson's disease. Proc Natl Acad Sci USA. 2008;105:5856–5861. doi: 10.1073/pnas.0801677105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertsch B, Ogden CA, Sidhu K, Le-Niculescu H, Kuczenski R, Niculescu AB, et al. Convergent functional genomics: a Bayesian candidate gene identification approach for complex disorders. Methods. 2005;37:274–279. doi: 10.1016/j.ymeth.2005.03.012. [DOI] [PubMed] [Google Scholar]
- Le-Niculescu H, McFarland MJ, Mamidipalli S, Ogden CA, Kuczenski R, Kurian SM, et al. Convergent Functional Genomics of bipolar disorder: from animal model pharmacogenomics to human genetics and biomarkers. Neurosci Biobehav Rev. 2007;31:897–903. doi: 10.1016/j.neubiorev.2007.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodd ZA, Bertsch BA, Strother WN, Le-Niculescu H, Balaraman Y, Hayden E, et al. Candidate genes, pathways and mechanisms for alcoholism: an expanded convergent functional genomics approach. Pharmacogenomics J. 2007;7:222–256. doi: 10.1038/sj.tpj.6500420. [DOI] [PubMed] [Google Scholar]
- Le-Niculescu H, McFarland MJ, Ogden CA, Balaraman Y, Patel S, Tan J, et al. Phenomic, convergent functional genomic, and biomarker studies in a stress-reactive genetic animal model of bipolar disorder and co-morbid alcoholism. Am J Med Genet B Neuropsychiatr Genet. 2008;147B:134–166. doi: 10.1002/ajmg.b.30707. [DOI] [PubMed] [Google Scholar]
- Le-Niculescu H, Balaraman Y, Patel S, Tan J, Sidhu K, Jerome RE, et al. Towards understanding the schizophrenia code: an expanded convergent functional genomics approach. Am J Med Genet B Neuropsychiatr Genet. 2007;144B:129–158. doi: 10.1002/ajmg.b.30481. [DOI] [PubMed] [Google Scholar]
- Patel SD, Le-Niculescu H, Koller DL, Green SD, Lahiri DK, McMahon FJ, et al. Coming to grips with complex disorders: genetic risk prediction in bipolar disorder using panels of genes identified through convergent functional genomics. Am J Med Genet B Neuropsychiatr Genet. 2010;153B:850–877. doi: 10.1002/ajmg.b.31087. [DOI] [PubMed] [Google Scholar]
- Ogden CA, Rich ME, Schork NJ, Paulus MP, Geyer MA, Lohr JB, et al. Candidate genes, pathways and mechanisms for bipolar (manic-depressive) and related disorders: an expanded convergent functional genomics approach. Mol Psychiatry. 2004;9:1007–1029. doi: 10.1038/sj.mp.4001547. [DOI] [PubMed] [Google Scholar]
- Le-Niculescu H, Patel SD, Bhat M, Kuczenski R, Faraone SV, Tsuang MT, et al. Convergent functional genomics of genome-wide association data for bipolar disorder: comprehensive identification of candidate genes, pathways and mechanisms. Am J Med Genet B Neuropsychiatr Genet. 2009;150B:155–181. doi: 10.1002/ajmg.b.30887. [DOI] [PubMed] [Google Scholar]
- Zhu J, Sova P, Xu Q, Dombek KM, Xu EY, Vu H, et al. Stitching together multiple data dimensions reveals interacting metabolomic and transcriptomic networks that modulate cell regulation. PLoS Biol. 2012;10:e1001301. doi: 10.1371/journal.pbio.1001301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Gaiteri C, Bodea LG, Wang Z, McElwee J, Podtelezhnikov AA, et al. Integrated systems approach identifies genetic nodes and networks in late-onset Alzheimer's disease. Cell. 2013;153:707–720. doi: 10.1016/j.cell.2013.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayalew M, Le-Niculescu H, Levey DF, Jain N, Changala B, Patel SD, et al. Convergent functional genomics of schizophrenia: from comprehensive understanding to genetic risk prediction. Mol Psychiatry. 2012;17:887–905. doi: 10.1038/mp.2012.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niculescu AB, Le-Niculescu H. The P-value illusion: how to improve (psychiatric) genetic studies. Am J Med Genet B Neuropsychiatr Genet. 2010;153B:847–849. doi: 10.1002/ajmg.b.31076. [DOI] [PubMed] [Google Scholar]
- Le-Niculescu H, Balaraman Y, Patel SD, Ayalew M, Gupta J, Kuczenski R, et al. Convergent functional genomics of anxiety disorders: translational identification of genes, biomarkers, pathways and mechanisms. Transl Psychiatry. 2011;1:e9. doi: 10.1038/tp.2011.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le-Niculescu H, Case NJ, Hulvershorn L, Patel SD, Bowker D, Gupta J, et al. Convergent functional genomic studies of omega-3 fatty acids in stress reactivity, bipolar disorder and alcoholism. Transl Psychiatry. 2011;1:e4. doi: 10.1038/tp.2011.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell SM, Wray NR, Stone JL, Visscher PM, O'Donovan MC, Sullivan PF, et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JJ, Mandelli L, Lim S, Lim HK, Kwon OJ, Pae CU, et al. Association analysis of heat shock protein 70 gene polymorphisms in schizophrenia. Eur Arch Psychiatry Clin Neurosci. 2008;258:239–244. doi: 10.1007/s00406-007-0791-6. [DOI] [PubMed] [Google Scholar]
- Arion D, Unger T, Lewis DA, Levitt P, Mirnics K. Molecular evidence for increased expression of genes related to immune and chaperone function in the prefrontal cortex in schizophrenia. Biol Psychiatry. 2007;62:711–721. doi: 10.1016/j.biopsych.2006.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller GE, Chen E, Sze J, Marin T, Arevalo JM, Doll R, et al. A functional genomic fingerprint of chronic stress in humans: blunted glucocorticoid and increased NF-kappaB signaling. Biol Psychiatry. 2008;64:266–272. doi: 10.1016/j.biopsych.2008.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]