Abstract
BACKGROUND
Prostate cancer (PCa) affects more than 190,000 men each year with ~10% of men diagnosed at ≤ 55 years, i.e., early onset (EO) PCa. Based on historical findings for other cancers, EO PCa likely reflects a stronger underlying genetic etiology.
METHODS
We evaluated the association between EO PCa and previously identified single nucleotide polymorphisms (SNPs) in 754 Caucasian cases from the Michigan Prostate Cancer Genetics Project (mean 49.8 years at diagnosis), 2,713 Caucasian controls from Illumina’s iControlDB database and 1,163 PCa cases diagnosed at >55 years from the Cancer Genetic Markers of Susceptibility Study (CGEMS).
RESULTS
Significant associations existed for 13 of 14 SNPs (rs9364554 on 6q25, rs10486567 on 7p15, rs6465657 on 7q21, rs6983267 on 8q24, rs1447295 on 8q24, rs1571801 on 9q33, rs10993994 on 10q11, rs4962416 on 10q26, rs7931342 on 11q13, rs4430796 on 17q12, rs1859962 on 17q24.3, rs2735839 on 19q13, and rs5945619 on Xp11.22, but not rs2660753 on 3p12). EO PCa cases had a significantly greater cumulative number of risk alleles (mean 12.4) than iControlDB controls (mean 11.2; p=2.1×10−33) or CGEMS cases (mean 11.9; p=1.7 × 10−5). Notably, EO PCa cases had a higher frequency of the risk allele than CGEMS cases at 11 of13 associated SNPs, with significant differences for five SNPs. EO PCa cases diagnosed at <50 (mean 12.8) also had significantly more risk alleles than those diagnosed at 50–55 years (mean 12.1; p = 0.0003).
CONCLUSIONS
These results demonstrate the potential for identifying PCa-associated genetic variants by focusing on the subgroup of men diagnosed with EO disease.
Keywords: Early Onset Prostate Cancer, Polymorphism, Association, Genetics
INTRODUCTION
In 2009 prostate cancer (PCa) was the most commonly diagnosed non-cutaneous cancer among men in the United States with an estimated 192,280 new cases and the second leading cause of cancer-related mortality with an estimated 27,360 PCa-related deaths (1). Although PCa is commonly considered to be a disease of older men, with 63% of men diagnosed over the age of 65, last year over 9% of men diagnosed were ≤ 55 years. The proportion of men diagnosed at younger ages has increased steadily since the introduction of widespread screening with prostate-specific antigen (PSA) and continues to rise despite an apparent stabilization in PCa incidence overall (2,3). This is despite guidelines for PCa early detection that have previously targeted men starting at age 50 except for those perceived to be at increased risk of disease, i.e., those with African American ancestry and/or a family history of prostate cancer, who may begin screening 5–10 years earlier (4,5). PCa in younger men may have different public health implications, since some data suggest that compared to older men with similar clinical features younger men may be more likely to die of their cancer (6), especially those diagnosed with higher grade or locally advanced disease (7).
Early age at diagnosis is a recognized marker of genetic susceptibility for several hereditary cancers including breast (8), colorectal (9), ovarian (10), and endometrial (11). Among hereditary PCa families, risk increases with decreasing age of diagnosis of affected relatives (12) and, on average, hereditary PCa is diagnosed 6–7 years earlier than sporadic PCa (6). Because the lower incidence of PCa at younger ages may indicate the lower overall prevalence of other risk factors for the disease (13), early onset (EO) PCa cases, i.e., PCa cases diagnosed ≤ 55 years of age, may provide an especially rich sub-group of men among whom to search for genes associated with PCa risk.
Several recent genome-wide association studies (GWAS) have provided statistically significant evidence for multiple independent loci associated with PCa (14–21). Efforts to distinguish those loci that may be associated with aggressive PCa, a clinically important form of PCa that is most likely to impact survival, have also been pursued (22–25). In this study, we evaluate the evidence for association between risk of EO PCa and 14 single nucleotide polymorphisms (SNPs) distributed across 10 chromosomes in a sample of 754 unrelated Caucasian American EO PCa cases from the University of Michigan Prostate Cancer Genetics Project (UM-PCGP) who were diagnosed at ≤ 55 years and 2,713 Caucasian controls. These SNPs were selected based on having the strongest evidence in the published literature supporting an association with prostate cancer and, for rs1571801, with aggressive prostate cancer (14, 16, 17, 19, 25, 26). We found significant evidence for an association (p < 0.05) between EO PCa and 13 of the 14 SNPs, with the direction of the association consistent with prior reports. Further, we show that the EO PCa cases had a significantly greater average number of risk alleles across these SNPs than the 1,163 PCa cases from the Cancer Genetic Markers of Susceptibility Study (CGEMS) study who were diagnosed with disease after age 55.
SUBJECTS AND METHODS
Study Subjects
The study population consists of 754 unrelated Caucasian American participants in the UM PCGP diagnosed with histologically confirmed PCa (International Classification of Diseases for Oncology code C61.9) at ≤ 55 years of age. Dates of diagnosis were obtained from the date of diagnostic biopsy for 96.6% of cases, with the date of diagnosis for the remaining cases determined from the date of trans-urethral resection of the prostate, date of radical prostatectomy or physician’s note. The majority of cases (95%) were diagnosed between November, 1993 and February, 2006. Men were aged 27 to 55 years at diagnosis, with average and median ages of 49.8 and 50 years, respectively. Cases completed self-administered questionnaires that collected information on family history of prostate and other cancers, medical history and demographic factors. In addition, detailed clinical information relating to the diagnosis and treatment of PCa, including Gleason score from biopsy, tumor stage, and PSA level at diagnosis, was available from medical records. Peripheral blood samples for preparation of DNA were drawn from all subjects for genotyping. All study procedures have been approved by the University of Michigan Institutional Review Board and were conducted in accordance with Health Insurance Portability and Accountability Act (HIPAA) regulations. Written informed consent was obtained from all study participants prior to participation.
Genotyping EO PCa cases
Fourteen SNPs were selected for genotyping based on published reports identifying them as significantly associated with PCa: rs2660753 on 3p12, rs9364554 on 6q25, rs10486567 on 7p15, rs6465657 on 7q21, rs6983267 and rs1447295 on 8q24, rs1571801 on 9q33, rs10993994 on 10q11, rs4962416 on 10q26, rs7931342 on 11q13, rs4430796 on 17q12, rs1859962 on 17q24.3, rs2735839 on 19q13, and rs5945619 on Xp11.22. Applied Biosystems TaqMan™ SNP assay system was used to genotype individual DNA samples with allelic discrimination performed on an ABI PRISM 7900HT Sequence Detection System. Any SNPs remaining undetermined by the assay were directly sequenced on an Applied Biosystems 3100 Genetic Analyzer using Big Dye version 1.1 chemistries for a final overall average 99.1% call completion, with no individual SNP below 95.6% completion. Quality control included duplicate genotyping of 5% of the samples, distributed evenly among TaqMan™ genotyping batches. Out of 897 duplicate pairs, 16 genotype calls were discrepant, corresponding to 98.2% agreement overall. In total 658 cases had complete genotype data on all 14 SNPs.
iControlDB and CGEMS Study Subjects
An independent set of 2,713 unrelated controls of Caucasian ancestry with available genotype data were obtained from Illumina’s iControlDB database (www.illumina.com/science/icontroldb.ilmn), i.e., iControls, as a comparison group for the UM EO PCa cases. Subjects were anonymous but had information available on age, sex and ancestry. iControls were genotyped with Illumina’s HumanHap550v1 (referred to as V1 subjects; n=1,197) or HumanHap550v3 (referred to as V3 subjects; n=1,516) genome-wide genotyping platforms. The use of iControls in genetic association studies has been documented previously (27,28). Genotype data for all 14 SNPs considered in this study were available in both V1 and V3 iControl samples. For SNP rs5945619, located on the X chromosome, the allele from each male and a single randomly chosen allele from each female in the iControls constituted the iControl sample for this SNP.
Caucasian PCa cases (n=1,163) and screened controls (n=1,113) in the CGEMS GWAS were included as additional comparison groups to our EO PCa cases. PCa cases and controls in the initial CGEMS GWAS were participants in the control arm of the Prostate, Lung, Colorectal and Ovarian (PLCO) Cancer Screening Trial (29). All CGEMS cases and controls were PSA screened; cases were 55–74 years at diagnosis, with an average age at diagnosis > 60 years. CGEMS samples were genotyped using the Illumina HumanHap300 and HumanHap240 chips. Data was available for all 14 SNPs investigated in this report.
Statistical Analyses
The observed genotype distributions for each SNP were tested for consistency with Hardy-Weinberg equilibrium (HWE) expected proportions using 1-degree-of-freedom Pearson chi-square tests in the EO PCa cases and iControls, respectively, using the PLINK software (30) version 1.06 (available from pngu.mgh.harvard.edu/purcell/plink). Unconditional logistic regression models were used to evaluate the association between EO PCa and SNP genotypes assuming a multiplicative, i.e., log-additive, genetic inheritance model. Statistical significance was assessed using 1-degree-of-freedom likelihood-ratio tests. Both 2-sided and 1-sided hypothesis tests were performed, with the direction of the 1-sided test determined from prior published studies (See Table 2 for relevant references). Statistical analyses were performed using SAS version 9.1.3 software package (SAS Institute, Cary, N.C.). Similar analyses were performed using screened controls from the CGEMS GWAS as the comparison group. Finally, allelic-based likelihood ratio tests were used to systematically test for differences in the “risk” allele frequencies between EO PCa cases and PCa cases diagnosed after age 55 years, i.e., CGEMS PCa cases, for each SNP.
Table 2.
Early Onset Prostate Cancer Risk Associated With SNPs When Compared with iControlDB Public Controls.
| SNP | Chr. | Alleles1 | Frequency of Risk Allele | OR (95 %CI)2 | P (two-sided) | P (one-sided)3 | |
|---|---|---|---|---|---|---|---|
| Early-Onset Cases (n=754) | iControls (n=2,713) | ||||||
| rs2660753 (14) | 3p | C/T | 0.127 | 0.140 | 0.90 (0.76,1.07) | 0.22 | 0.89 |
| rs9364554 (14) | 6q | C/T | 0.317 | 0.259 | 1.32 (1.17,1.50) | 1.3×10−05 | 6.3×10−06 |
| rs10486567 (16) | 7p | A/G | 0.806 | 0.761 | 1.30 (1.12,1.49) | 4.0×10−04 | 2.0×10−04 |
| rs6465657 (14) | 7q | T/C | 0.495 | 0.451 | 1.19 (1.06,1.34) | 0.0029 | 0.0015 |
| rs1447295 (26) | 8q | C/A | 0.134 | 0.099 | 1.41 (1.18,1.67) | 1.7×10−4 | 8.7×10−5 |
| rs6983267 (19) | 8q | T/G | 0.606 | 0.499 | 1.55 (1.38,1.75) | 1.9×10−13 | 9.6×10−14 |
| rs1571801 (25) | 9q | G/T | 0.274 | 0.245 | 1.16 (1.02,1.32) | 0.024 | 0.012 |
| rs4962416 (16) | 10q | A/G | 0.326 | 0.295 | 1.15 (1.02,1.30) | 0.023 | 0.012 |
| rs10993994 (16) | 10q | C/T | 0.470 | 0.422 | 1.21 (1.08,1.36) | 0.0011 | 5.5×10−04 |
| rs7931342 (14) | 11q | T/G | 0.578 | 0.544 | 1.15 (1.03,1.30) | 0.017 | 0.0083 |
| rs1859962 (17) | 17q | T/G | 0.534 | 0.474 | 1.26 (1.12,1.41) | 6.4×10−05 | 3.2×10−05 |
| rs4430796 (17) | 17q | C/T | 0.571 | 0.499 | 1.31 (1.17,1.47) | 3.2×10−06 | 1.6×10−06 |
| rs2735839 (14) | 19q | A/G | 0.888 | 0.849 | 1.39 (1.18,1.67) | 1.9×10−04 | 9.3×10−05 |
| rs5945619 (14) | Xp | T/C | 0.432 | 0.364 | 1.33 (1.13,1.57) | 7.9×10−04 | 3.9×10−04 |
Reference allele/allele associated with increased risk of prostate cancer in prior studies.
Odds Ratio for each additional copy of risk allele (as identified in previous studies) assuming a multiplicative model.
One-sided test for direction of alternative hypothesis determined by prior study.
To assess whether the cumulative number of risk alleles across the 13 PCa-associated SNPs was associated with EO PCa and whether EO PCa cases carry more risk alleles on average than older-onset PCa cases, we calculated the total number of risk alleles in each case and each control sample (i.e., EO PCa and CGEMS PCa cases and iControls and CGEMS controls). SNP rs2660753 was not included in these calculations as it was not significantly associated with PCa in the current study, the CGEMS study, nor in a large study of 7,370 PCa cases and 5,742 controls by the PRACTICAL consortium (15). Individual subjects missing genotype data for any of the 13 SNPs used in calculating the sum of risk alleles were excluded from this calculation. Unconditional logistic regression models were used to test whether there were significant differences in the total number of risk alleles between EO PCa cases and iControls and between EO PCa and CGEMS PCa cases. Unconditional logistic regression was also used to evaluate the performance of iControls as a reference group by comparing the distribution of allele frequencies between iControls and CGEMS controls.
Finally, case-only analyses were performed to assess whether SNP genotypes were associated with clinical features observed in our EO PCa cases, specifically, age at diagnosis, biopsy or pathological Gleason score, and serum PSA at diagnosis. Single-SNP case-only analyses were performed using logistic regression (for dichotomous outcomes: family history and aggressive disease) and Spearman’s rank correlation (for continuous outcomes: age, Gleason score, and pre-treatment serum PSA at diagnosis). Analyses were repeated using the cumulative number of risk alleles as a continuous predictor of clinical features of EO PCa.
RESULTS
The majority (63.8%) of EO PCa cases reported a positive family history of PCa, with over 40% having a confirmed, first-degree affected relative (Table 1). Clinically, 76% of cases presented with serum PSA level at diagnosis ≥ 4.0 ng/mL; 8.1% had clinical Gleason scores 8–10. Overall, 29.8% of men with EO disease were diagnosed with aggressive PCa, as defined in Lange et al. (31) All SNPs in EO PCa cases and 13 of 14 SNPs in iControls had genotype frequencies consistent (p > 0.001) with HWE. Among iControls, SNP rs4430796 had an observed genotype distribution inconsistent with HWE (p = 4.4 × 10−4, deficit of heterozygotes compared to expectation). Upon further inspection, 112/1,197 V1 samples had a missing genotype call (compared to 4/1,516 V3 samples) for this SNP. Sample-specific testing for HWE revealed that the genotype distribution for rs4430796 was consistent with HWE among V3 samples, but not among V1 samples (p = 6.1 × 10−4).
Table 1.
Sample Characteristics for 754 Early-Onset Prostate Cancer Cases
| Clinical Features | EO Cases | Range | |
|---|---|---|---|
| n=754 | % | ||
| Age at Diagnosis (years) 1, | 49.8 (3.9) | 50.5 | 34–55 |
| 34 – 45 | 105 | 13.9 | |
| 46 – 50 | 272 | 36.1 | |
| 51 – 55 | 377 | 50.0 | |
| Number of Affected Family Members 1,2 | 1.4 (1.8) | 1 | 0 – 17 |
| 0 | 273 | 36.2 | |
| ≥ 1 | 481 | 63.8 | |
| Family History of Prostate Cancer | |||
| 1st degree relative (confirmed) | 310 | 41.1 | |
| 2nd degree relative (confirmed) | 83 | 11.0 | |
| 1st or 2nd degree relative (unconfirmed) | 73 | 9.7 | |
| 3rd degree relative (unconfirmed) | 15 | 2.0 | |
| Serum PSA (ng/mL) 1,3 | 22.9 (218.7) | 5.4 | 0.3 – 5428 |
| < 4.0 | 172 | 24.0 | |
| 4.0–9.9 | 403 | 56.1 | |
| ≥ 10 | 143 | 19.9 | |
| Gleason Score 1 | 6.4 (0.9) | 6.0 | 3 – 10 |
| 3–6 | 446 | 61.0 | |
| 7 | 227 | 31.1 | |
| 8–10 | 58 | 7.9 | |
| Aggressive Disease 4 | |||
| No | 529 | 70.2 | |
| Yes | 225 | 29.8 | |
For age at diagnosis, number of affected family members, and serum PSA statistics given are: mean (SD), median, and range.
Total numbers of affected family members, not including the proband.
Serum prostate-specific antigen (PSA) measured at diagnosis prior to treatment.
Aggressive disease defined as in Lange et al., 2005. (1)
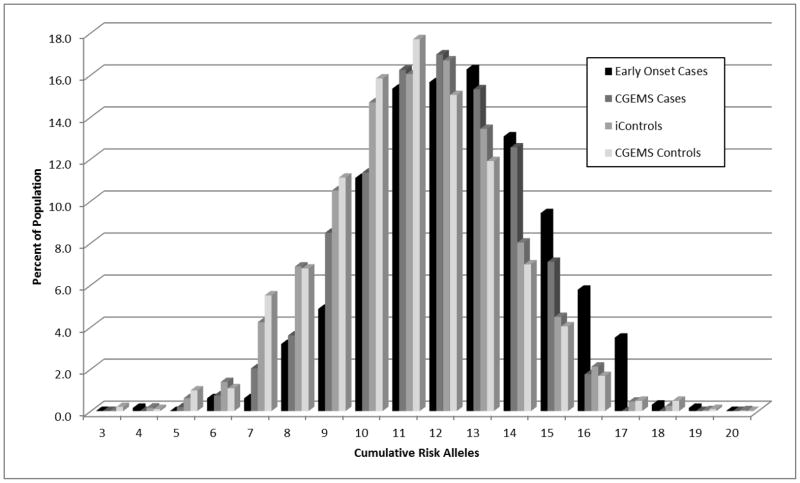
Thirteen of the 14 studied SNPs, excluding rs2660753, demonstrated evidence (p < 0.05) of association with EO PCa. All 13 associated SNPs had a direction of effect consistent with previous reports (Table 2). Ten of the 13 SNPs remained statistically significant after strict application of the Bonferroni correction for multiple testing to the one-sided test results (i.e., pone-sided < 0.0036), while all 13 SNPs remained statistically significant after applying Holm’s less conservative sequential rejection method (i.e., the Holm-Bonferroni method) for multiple testing (33) (data not shown). Similar results were obtained when CGEMS controls were utilized as the reference group (Table 3). SNP rs4430796 was significantly associated with EO PCa when using either V1 (p = 1.8 × 10−4, OR = 1.28) or V3 (p = 3.2 × 10−6, OR = 1.34) iControl samples. No significant evidence for an association was observed between rs2660753 and EO PCa using the combined iControl samples or when restricting the iControl samples to V1 (p = 0.88, OR = 0.99) or V3 (p=0.06, OR = 0.84) samples. In addition, the odds ratio observed between EO PCa and rs2660753 based on V3 control samples was in the opposite direction to the previous report by Eeles et al.(14) Finally, the total number of risk alleles observed in each subject (measured as the sum of risk alleles across 13 SNPs excluding rs2660753) was strongly associated with EO PCa (p = 2.1 × 10−33) (Table 4 and Figure 1).
Table 3.
Distribution of SNP Allele Frequencies in Early Onset Prostate Cancer Cases Compared to CGEMs Cases and Controls.
| SNP | Chr. | Alleles 1 | Early Onset Cases Allele Freq. 2 | CGEMS Controls (n=1,101) | CGEMS Cases (n=1,176) | ||||
|---|---|---|---|---|---|---|---|---|---|
| Allele Freq. 2 | OR (95% CI) 3 | P 4 | Allele Freq. 2 | % Change 5 | P 4 | ||||
| rs2660753 | 3p | C/T | 0.127 | 0.120 | 1.06 (0.87,1.30) | 0.53 | 0.127 | 0.0 % | 0.99 |
| rs9364554 | 6q | C/T | 0.317 | 0.291 | 1.13 (0.98,1.30) | 0.097 | 0.293 | + 8.2 % | 0.13 |
| rs10486567 | 7p | A/G | 0.806 | 0.770 | 1.23 (1.05,1.47) | 0.010 | 0.792 | + 1.8 % | 0.30 |
| rs6465657 | 7q | T/C | 0.495 | 0.460 | 1.14 (1.00,1.30) | 0.046 | 0.472 | + 4.9 % | 0.17 |
| rs1447295 | 8q | C/A | 0.134 | 0.101 | 1.39 (1.13,1.70) | 0.0020 | 0.144 | − 6.9 % | 0.36 |
| rs6983267 | 8q | T/G | 0.606 | 0.489 | 1.60 (1.40,1.84) | 3.7×10−12 | 0.555 | + 9.2 % | 0.0021 |
| rs1571801 | 9q | G/T | 0.274 | 0.242 | 1.18 (1.02,1.37) | 0.029 | 0.285 | − 3.9 % | 0.47 |
| rs4962416 | 10q | A/G | 0.326 | 0.264 | 1.34 (1.16,1.55) | 6.4×10−05 | 0.321 | + 1.6 % | 0.76 |
| rs10993994 | 10q | C/T | 0.470 | 0.368 | 1.49 (1.31,1.70) | 2.0×10−09 | 0.413 | + 13.8 % | 5.6×10−04 |
| rs7931342 | 11q | T/G | 0.578 | 0.498 | 1.37 (1.20,1.56) | 2.5×10−06 | 0.543 | + 6.4 % | 0.033 |
| rs1859962 | 17q | T/G | 0.534 | 0.486 | 1.21 (1.06,1.38) | 0.0045 | 0.532 | + 0.4 % | 0.92 |
| rs4430796 | 17q | C/T | 0.571 | 0.501 | 1.32 (1.15,1.51) | 4.9×10−05 | 0.541 | + 5.5 % | 0.082 |
| rs2735839 | 19q | A/G | 0.888 | 0.844 | 1.45 (1.20,1.79) | 1.4×10−04 | 0.864 | + 2.8 % | 0.027 |
| rs5945619 | Xp | T/C | 0.432 | 0.336 | 1.50 (1.24,1.82) | 3.2×10−05 | 0.413 | + 4.6 % | 1.7×10−04 |
Reference allele/allele associated with increased risk of prostate cancer as identified in prior studies, i.e., the ‘risk’ allele.
Frequency of risk allele.
Odds ratio and 95% confidence interval estimating the risk of prostate cancer associated with each additional copy of a risk allele, assuming a log-additive genetic model.
Two-sided P-value.
Percent change in risk allele frequency (+/− increase or decrease, respectively) in EO compared to CGEMS Cases
Table 4.
Cumulative Number of Risk Alleles for 13 SNPs in EO Cases, iControlDB Controls, CGEMS Cases and CGEMS Controls
| EO Cases1,2 (n=754) | EO Cases 50–55 yrs3 (n=459) | EO Cases < 50 yrs3 (n=295) | iControls1,4 (n=2,713) | CGEMS Cases2 (n=1,1760 | CGEMS Controls4 (n=1,101) | |
|---|---|---|---|---|---|---|
| Mean | 12.39 | 12.13 | 12.81 | 11.16 | 11.92 | 10.97 |
| SD/SE | 2.36/0.092 | 2.42/0.12 | 2.22/0.14 | 2.34/0.047 | 2.30/0.070 | 2.39/0.075 |
EO cases compared to iControls(p = 1.6×10−32).
EO cases compared to CGEMS cases (p = 4.4×10−05).
EO cases diagnosed at < 50 years versus EO cases diagnosed between 50–55 years (p = 0.00030).
iControls versus CGEMS controls (p = 0.033).
Figure 1.
The Distribution of Total Number of Risk Alleles Across 13 SNPs in Early Onset Prostate Cancer Cases Compared to iControlDB Controls, CGEMS Cases and CGEMS Controls.
The frequencies of previously defined risk alleles were higher among the younger EO PCa cases than the older CGEMS cases. Specifically, the risk allele in 11 of the13 SNPs was more common among EO PCa cases (Table 3). This difference was statistically significant (p<0.05) for five of the 11 SNPs. EO PCa cases had significantly more total risk alleles across the 13 SNPs than the PCa cases from the CGEMS study, with 12.42 risk alleles on average compared to 11.92 in CGEMS cases (p = 1.7×10−5) (Table 4). In EO PCa case-only analyses, the frequency of the risk allele at three SNPs, rs1048656 (p = 0.012), rs1099399 (p = 0.0087) and rs1859962 (p = 0.037), was significantly greater in men diagnosed with PCa prior to age 50 (n=295) than in men diagnosed with PCa between the ages of 50–55 years (n=459). Across all 13 associated SNPs, there was significant evidence for more total risk alleles in EO PCa cases diagnosed prior to age 50 (12.81 risk alleles on average) than in men diagnosed with PCa between the ages of 50–55 (12.13 risk alleles on average; p = 0.0003) (Table 4).
There was no significant evidence for any association between individual SNPs or total number of risk alleles measured across the 13 associated SNPs and pre-diagnostic serum PSA. The number of risk alleles at rs2735839, was significantly negatively correlated, after Bonferroni correction, with biopsy Gleason score (Spearman’s correlation = −0.12, p = 0.0016). One SNP was nominally significantly correlated (p < 0.05) with Gleason score (rs1859962, Spearman’s correlation = −0.080, p = 0.033. We found significant evidence for a negative correlation between the cumulative number of risk alleles across the 13 SNPs associated with EO PCa and biopsy Gleason score (Spearman’s correlation = −0.085, p = 0.032)
DISCUSSION
We performed a replication-based genetic association study for 14 SNPs previously reported to be associated with PCa in a sample of 754 Caucasian American EO PCa cases from the UM-PCGP and 2,713 Caucasian American public controls from Illumina’s iControlDB database. We found significant evidence (p < 0.05) for an association between EO PCa and 13 of the14 SNPs, but not rs2660753, with similar direction of effect as in previous reports (14, 16, 17, 19, 25, 26). For 11 of the 13 SNPs, the association observed in the younger EO PCa cases was stronger than those in the existing literature, which reflect older case populations.
To our knowledge our study is the first to report replication for an association between rs1571801 and PCa, although we did not find any significant evidence to support an increased frequency of the risk allele in PCa cases with aggressive disease [data not shown]. The association of rs1571801 with PCa was first identified in a GWAS for aggressive PCa using combined participants from the Cancer of the Prostate in Sweden (CAPS) and CGEMS studies (25). Interestingly, as reported in Duggan et al. (25), the frequency of the risk allele for rs1571801 was greater in non-aggressive than in aggressive cases among CGEMS samples. SNP rs1571801 was one of only two SNPs (the other being rs1447295) with a higher risk allele frequency in CGEMS compared to EO PCa cases. Given that the association between rs1571801 and PCa was previously identified in a study that included CGEMS cases, the higher frequency of the risk allele in CGEMS cases compared to our EO PCa cases may be explained by the winner’s curse phenomenon (34).
SNPs rs4430796 and rs1859962 were first identified by deCODE Genetics from a targeted follow-up study to their original GWAS (26) that was initiated in response to reports of linkage evidence to chromosome 17 in UM-PCGP and John’s Hopkins University PCa pedigrees (17,31). A subset of the UM-PCGP subjects who were included in the reported linkage analysis on chromosome 17 was also included in this current association study. The association of PCa risk with two SNPs at 8q24, rs1447295 and rs6983267, was first reported by deCODE Genetics (26) and the CGEMS GWAS study that expanded on the initial findings at 8q24 (19), respectively. Two SNPs, rs10486567 and rs4962416, were first reported in a follow-up study to the initial GWAS by the CGEMS study (16). We note that our current study includes only those CGEMS PCa cases included in the initial GWAS (19). The remaining seven SNPs (rs2660753, rs9364554, rs6465657, rs10993994, rs7931342, rs2735839 and rs5945619) were first reported in the early-onset and familial PCa GWAS by Eeles et al. (14) and subsequently followed-up in a confirmatory study in 7,370 PCa cases and 5,742 controls by the PRACTICAL consortium (15). In this follow-up study, strong supporting evidence for 6 of the 7 SNPs (excluding rs2660753) was reported.
We used a public control population of Caucasian Americans that have been genotyped in several studies using different genotyping platforms than the platform used to genotype our study cases. The iControls were genotyped on Illumina’s HumanHap550v1 and HumanHap550v3 Beadchip genome-wide SNP platforms compared to the UM-PCGP cases, who were genotyped using Applied Biosystems TaqMan assays. It is possible that the use of different genotyping platforms in cases and controls could have led to a systematic bias in genotyping calls, or a batch effect. Public controls were selected for use in this study based on the limited availability of unrelated controls for genotyping to UM-PCGP investigators and the availability of a large number of reference samples’ control genotypes though Illumina’s iControlDB database. We note that the iControlDB samples have been genotyped using the same Illumina Beadchip technology used in several recent PCa genome-wide studies. While we cannot definitively rule out the possibility of bias resulting from a batch genotyping effect, we note that the direction of the association between EO PCa and 13 SNPs was consistent with previous reports. Further, the allele frequencies for the SNPs were similar between the two independent V1 and V3 iControlDB samples. Prior GWA studies that have included the iControlDB samples have not noted the presence of any major genotype call bias in their reports (27,28). Most importantly, we note that our results did not differ when we used the CGEMS controls’ genotype distribution as the reference group (Table 3) and that our case-only results are not subject to any such possible bias.
EO PCa has been shown to be significantly associated with increased family history of the disease, providing evidence of a stronger underlying genetic etiology of EO disease than for late-onset disease. To date, several multistage GWA studies for PCa have been conducted using a variety of rules for PCa case inclusion. The GWAS based on younger (i.e., ≤ 60 years) PCa cases and cases with a positive family history of PCa by Eeles et al. (14) demonstrated the increased power for detecting SNPs associated with PCa that can be achieved by including cases with enriched genetic susceptibility to the disease. Eeles et al.(14) found significant evidence of association for 7 novel SNPs (p < 1.0 × 10−7) in their stage 1 results in addition to the widely reported PCa susceptibility loci on chromosomes 8q24 and 17q. Although an association with age of diagnosis has been tested and rejected in many prior studies, including the original GWAS reports that identified the SNPs we have tested, the men included in all of these studies were much older than those included in the present study. On average, our EO PCa cases were diagnosed at 49.8 years, ~7 years younger than the PCa cases in the study by Eeles et al. Furthermore, > 60% of our cases had a family history of PCa in 1st or 2nd degree relatives (52.1% confirmed, 9.7% unconfirmed). We found significant evidence (p < 0.05) supporting the association between EO PCa and 13 of 14 SNPs studied, with the same direction of effects as in previous reports. We showed that the frequency of risk alleles, for both individual SNPs and in aggregate across SNPs, is significantly greater in EO PCa cases than in CGEMs cases who were diagnosed at > 55 years. Interestingly, we also found significant evidence that the trend of more risk alleles in younger cases compared to older cases existed among just our EO PCa cases suggesting that the cumulative impact of common genetic risk factors are particularly important in men diagnosed with PCa prior to their 50th birthday.
In summary, our results provide strong evidence that SNPs associated with overall PCa are also likely to be associated with EO PCa and that studies focused on EO PCa could be a particularly powerful resource for future association studies focusing on PCa. From a clinical perspective, these findings suggest that common genetic variants play an increased role in EO PCa, relative to later onset PCa, and that greater emphasis should be placed on measuring the cumulative impact of these variants on EO PCa. It is likely that novel common and rare high-penetrant genetic variants exist and have yet to be identified that will be particularly important in EO and familial PCa. We are in the process of performing a GWAS and high-throughput sequencing efforts that will focus on this important set of patients with EO prostate cancer.
Acknowledgments
Grant Support: NIH Grants RO1 CA79596, RO1 CA136621 and SPORE P50 CA69568.
References
- 1.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer JClin. 2009;59(4):225–249. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 2.Surveillance, Epidemiology, and End Results (SEER) Program (www.seer.cancer.gov) SEER*Stat Database: Incidence - SEER 9 Regs Limited-Use, Nov 2008 Sub (1973–2006) <Katrina/rita Population Adjustment> - Linked to County Attributes - Total U.S., 1969–2006 Counties. National Cancer Institute, DCCPS, Surveillance Research Program, Cancer Statistics Branch, released April 2009, based on the November 2008 submission; 2010.
- 3.Welch HG, Albertsen PC. Prostate cancer diagnosis and treatment after the introduction of prostate-specific antigen screening: 1986–2005. J Natl Cancer Inst. 2009;101(19):1325–1329. doi: 10.1093/jnci/djp278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith RA, von Eschenbach AC, Wender R, Levin B, Byers T, Rothenberger D, Brooks D, Creasman W, Cohen C, Runowicz C, Saslow D, Cokkinides V, Eyre H. American Cancer Society guidelines for the early detection of cancer: update of early detection guidelines for prostate, colorectal, and endometrial cancers. Also: update 2001--testing for early lung cancer detection. CA Cancer J Clin. 2001;51(1):38–75. doi: 10.3322/canjclin.51.1.38. quiz 77–80. [DOI] [PubMed] [Google Scholar]
- 5.(AUA) AUA. Prostate-specific antigen (PSA) best practice policy. Oncology (Williston Park) 2000;14:267–280. [PubMed] [Google Scholar]
- 6.Bratt O, Damber JE, Emanuelsson M, Gronberg H. Hereditary prostate cancer: clinical characteristics and survival. J Urol. 2002;167(6):2423–2426. [PubMed] [Google Scholar]
- 7.Lin DW, Porter M, Montgomery B. Treatment and survival outcomes in young men diagnosed with prostate cancer: a Population-based Cohort Study. Cancer. 2009;115(13):2863–2871. doi: 10.1002/cncr.24324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lynch HT, Guirgis H, Brodkey F, Maloney K, Lynch PM, Rankin L, Lynch J. Early age of onset in familial breast cancer. Genetic and cancer control implications. Arch Surg. 1976;111(2):126–131. doi: 10.1001/archsurg.1976.01360200032006. [DOI] [PubMed] [Google Scholar]
- 9.Lynch HT, de la Chapelle A. Genetic susceptibility to non-polyposis colorectal cancer. J Med Genet. 1999;36(11):801–818. [PMC free article] [PubMed] [Google Scholar]
- 10.Bewtra C, Watson P, Conway T, Read-Hippee C, Lynch HT. Hereditary ovarian cancer: a clinicopathological study. Int J Gynecol Pathol. 1992;11(3):180–187. doi: 10.1097/00004347-199207000-00003. [DOI] [PubMed] [Google Scholar]
- 11.Vasen HF, Watson P, Mecklin JP, Jass JR, Green JS, Nomizu T, Muller H, Lynch HT. The epidemiology of endometrial cancer in hereditary nonpolyposis colorectal cancer. Anticancer Res. 1994;14(4B):1675–1678. [PubMed] [Google Scholar]
- 12.Zeegers MP, Jellema A, Ostrer H. Empiric risk of prostate carcinoma for relatives of patients with prostate carcinoma: a meta-analysis. Cancer. 2003;97(8):1894–1903. doi: 10.1002/cncr.11262. [DOI] [PubMed] [Google Scholar]
- 13.Rothman KJ, Poole C. A strengthening programme for weak associations. Int J Epidemiol. 1988;17(4):955–959. doi: 10.1093/ije/17.4.955. [DOI] [PubMed] [Google Scholar]
- 14.Eeles RA, Kote-Jarai Z, Giles GG, Olama AA, Guy M, Jugurnauth SK, Mulholland S, Leongamornlert DA, Edwards SM, Morrison J, Field HI, Southey MC, Severi G, Donovan JL, Hamdy FC, Dearnaley DP, Muir KR, Smith C, Bagnato M, rdern-Jones AT, Hall AL, O’Brien LT, Gehr-Swain BN, Wilkinson RA, Cox A, Lewis S, Brown PM, Jhavar SG, Tymrakiewicz M, Lophatananon A, Bryant SL, Horwich A, Huddart RA, Khoo VS, Parker CC, Woodhouse CJ, Thompson A, Christmas T, Ogden C, Fisher C, Jamieson C, Cooper CS, English DR, Hopper JL, Neal DE, Easton DF. Multiple newly identified loci associated with prostate cancer susceptibility. Nat Genet. 2008;40(3):316–321. doi: 10.1038/ng.90. [DOI] [PubMed] [Google Scholar]
- 15.Kote-Jarai Z, Easton DF, Stanford JL, Ostrander EA, Schleutker J, Ingles SA, Schaid D, Thibodeau S, Dork T, Neal D, Donovan J, Hamdy F, Cox A, Maier C, Vogel W, Guy M, Muir K, Lophatananon A, Kedda MA, Spurdle A, Steginga S, John EM, Giles G, Hopper J, Chappuis PO, Hutter P, Foulkes WD, Hamel N, Salinas CA, Koopmeiners JS, Karyadi DM, Johanneson B, Wahlfors T, Tammela TL, Stern MC, Corral R, McDonnell SK, Schurmann P, Meyer A, Kuefer R, Leongamornlert DA, Tymrakiewicz M, Liu JF, O’Mara T, Gardiner RA, Aitken J, Joshi AD, Severi G, English DR, Southey M, Edwards SM, Al Olama AA, Eeles RA. Multiple novel prostate cancer predisposition loci confirmed by an international study: the PRACTICAL Consortium. Cancer Epidemiol Biomarkers Prev. 2008;17(8):2052–2061. doi: 10.1158/1055-9965.EPI-08-0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thomas G, Jacobs KB, Yeager M, Kraft P, Wacholder S, Orr N, Yu K, Chatterjee N, Welch R, Hutchinson A, Crenshaw A, Cancel-Tassin G, Staats BJ, Wang Z, Gonzalez-Bosquet J, Fang J, Deng X, Berndt SI, Calle EE, Feigelson HS, Thun MJ, Rodriguez C, Albanes D, Virtamo J, Weinstein S, Schumacher FR, Giovannucci E, Willett WC, Cussenot O, Valeri A, Andriole GL, Crawford ED, Tucker M, Gerhard DS, Fraumeni JF, Jr, Hoover R, Hayes RB, Hunter DJ, Chanock SJ. Multiple loci identified in a genome-wide association study of prostate cancer. Nat Genet. 2008;40(3):310–315. doi: 10.1038/ng.91. [DOI] [PubMed] [Google Scholar]
- 17.Gudmundsson J, Sulem P, Steinthorsdottir V, Bergthorsson JT, Thorleifsson G, Manolescu A, Rafnar T, Gudbjartsson D, Agnarsson BA, Baker A, Sigurdsson A, Benediktsdottir KR, Jakobsdottir M, Blondal T, Stacey SN, Helgason A, Gunnarsdottir S, Olafsdottir A, Kristinsson KT, Birgisdottir B, Ghosh S, Thorlacius S, Magnusdottir D, Stefansdottir G, Kristjansson K, Bagger Y, Wilensky RL, Reilly MP, Morris AD, Kimber CH, Adeyemo A, Chen Y, Zhou J, So WY, Tong PC, Ng MC, Hansen T, Andersen G, Borch-Johnsen K, Jorgensen T, Tres A, Fuertes F, Ruiz-Echarri M, Asin L, Saez B, van BE, Klaver S, Swinkels DW, Aben KK, Graif T, Cashy J, Suarez BK, van Vierssen TO, Frigge ML, Ober C, Hofker MH, Wijmenga C, Christiansen C, Rader DJ, Palmer CN, Rotimi C, Chan JC, Pedersen O, Sigurdsson G, Benediktsson R, Jonsson E, Einarsson GV, Mayordomo JI, Catalona WJ, Kiemeney LA, Barkardottir RB, Gulcher JR, Thorsteinsdottir U, Kong A, Stefansson K. Two variants on chromosome 17 confer prostate cancer risk, and the one in TCF2 protects against type 2 diabetes. Nat Genet. 2007;39(8):977–983. doi: 10.1038/ng2062. [DOI] [PubMed] [Google Scholar]
- 18.Gudmundsson J, Sulem P, Manolescu A, Amundadottir LT, Gudbjartsson D, Helgason A, Rafnar T, Bergthorsson JT, Agnarsson BA, Baker A, Sigurdsson A, Benediktsdottir KR, Jakobsdottir M, Xu J, Blondal T, Kostic J, Sun J, Ghosh S, Stacey SN, Mouy M, Saemundsdottir J, Backman VM, Kristjansson K, Tres A, Partin AW, bers-Akkers MT, Godino-Ivan MJ, Walsh PC, Swinkels DW, Navarrete S, Isaacs SD, Aben KK, Graif T, Cashy J, Ruiz-Echarri M, Wiley KE, Suarez BK, Witjes JA, Frigge M, Ober C, Jonsson E, Einarsson GV, Mayordomo JI, Kiemeney LA, Isaacs WB, Catalona WJ, Barkardottir RB, Gulcher JR, Thorsteinsdottir U, Kong A, Stefansson K. Genome-wide association study identifies a second prostate cancer susceptibility variant at 8q24. Nat Genet. 2007;39(5):631–637. doi: 10.1038/ng1999. [DOI] [PubMed] [Google Scholar]
- 19.Yeager M, Orr N, Hayes RB, Jacobs KB, Kraft P, Wacholder S, Minichiello MJ, Fearnhead P, Yu K, Chatterjee N, Wang Z, Welch R, Staats BJ, Calle EE, Feigelson HS, Thun MJ, Rodriguez C, Albanes D, Virtamo J, Weinstein S, Schumacher FR, Giovannucci E, Willett WC, Cancel-Tassin G, Cussenot O, Valeri A, Andriole GL, Gelmann EP, Tucker M, Gerhard DS, Fraumeni JF, Jr, Hoover R, Hunter DJ, Chanock SJ, Thomas G. Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nat Genet. 2007;39(5):645–649. doi: 10.1038/ng2022. [DOI] [PubMed] [Google Scholar]
- 20.Haiman CA, Patterson N, Freedman ML, Myers SR, Pike MC, Waliszewska A, Neubauer J, Tandon A, Schirmer C, McDonald GJ, Greenway SC, Stram DO, Le ML, Kolonel LN, Frasco M, Wong D, Pooler LC, Ardlie K, Oakley-Girvan I, Whittemore AS, Cooney KA, John EM, Ingles SA, Altshuler D, Henderson BE, Reich D. Multiple regions within 8q24 independently affect risk for prostate cancer. Nat Genet. 2007;39(5):638–644. doi: 10.1038/ng2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Freedman ML, Haiman CA, Patterson N, McDonald GJ, Tandon A, Waliszewska A, Penney K, Steen RG, Ardlie K, John EM, Oakley-Girvan I, Whittemore AS, Cooney KA, Ingles SA, Altshuler D, Henderson BE, Reich D. Admixture mapping identifies 8q24 as a prostate cancer risk locus in African-American men. Proc Natl Acad Sci USA. 2006;103(38):14068–14073. doi: 10.1073/pnas.0605832103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sun J, Zheng SL, Wiklund F, Isaacs SD, Li G, Wiley KE, Kim ST, Zhu Y, Zhang Z, Hsu FC, Turner AR, Stattin P, Liu W, Kim JW, Duggan D, Carpten J, Isaacs W, Gronberg H, Xu J, Chang BL. Sequence variants at 22q13 are associated with prostate cancer risk. Cancer Res. 2009;69(1):10–15. doi: 10.1158/0008-5472.CAN-08-3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cussenot O, Azzouzi AR, Bantsimba-Malanda G, Gaffory C, Mangin P, Cormier L, Fournier G, Valeri A, Jouffe L, Roupret M, Fromont G, Sibony M, Comperat E, Cancel-Tassin G. Effect of genetic variability within 8q24 on aggressiveness patterns at diagnosis and familial status of prostate cancer. Clin Cancer Res. 2008;14(17):5635–5639. doi: 10.1158/1078-0432.CCR-07-4999. [DOI] [PubMed] [Google Scholar]
- 24.Xu J, Isaacs SD, Sun J, Li G, Wiley KE, Zhu Y, Hsu FC, Wiklund F, Turner AR, Adams TS, Liu W, Trock BJ, Partin AW, Chang B, Walsh PC, Gronberg H, Isaacs W, Zheng S. Association of prostate cancer risk variants with clinicopathologic characteristics of the disease. Clin Cancer Res. 2008;14(18):5819–5824. doi: 10.1158/1078-0432.CCR-08-0934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Duggan D, Zheng SL, Knowlton M, Benitez D, Dimitrov L, Wiklund F, Robbins C, Isaacs SD, Cheng Y, Li G, Sun J, Chang BL, Marovich L, Wiley KE, Balter K, Stattin P, Adami HO, Gielzak M, Yan G, Sauvageot J, Liu W, Kim JW, Bleecker ER, Meyers DA, Trock BJ, Partin AW, Walsh PC, Isaacs WB, Gronberg H, Xu J, Carpten JD. Two genome-wide association studies of aggressive prostate cancer implicate putative prostate tumor suppressor gene DAB2IP. J Natl Cancer Inst. 2007;99(24):1836–1844. doi: 10.1093/jnci/djm250. [DOI] [PubMed] [Google Scholar]
- 26.Amundadottir LT, Sulem P, Gudmundsson J, Helgason A, Baker A, Agnarsson BA, Sigurdsson A, Benediktsdottir KR, Cazier JB, Sainz J, Jakobsdottir M, Kostic J, Magnusdottir DN, Ghosh S, Agnarsson K, Birgisdottir B, Le RL, Olafsdottir A, Blondal T, Andresdottir M, Gretarsdottir OS, Bergthorsson JT, Gudbjartsson D, Gylfason A, Thorleifsson G, Manolescu A, Kristjansson K, Geirsson G, Isaksson H, Douglas J, Johansson JE, Balter K, Wiklund F, Montie JE, Yu X, Suarez BK, Ober C, Cooney KA, Gronberg H, Catalona WJ, Einarsson GV, Barkardottir RB, Gulcher JR, Kong A, Thorsteinsdottir U, Stefansson K. A common variant associated with prostate cancer in European and African populations. Nat Genet. 2006;38(6):652–658. doi: 10.1038/ng1808. [DOI] [PubMed] [Google Scholar]
- 27.Silverberg MS, Cho JH, Rioux JD, McGovern DP, Wu J, Annese V, Achkar JP, Goyette P, Scott R, Xu W, Barmada MM, Klei L, Daly MJ, Abraham C, Bayless TM, Bossa F, Griffiths AM, Ippoliti AF, Lahaie RG, Latiano A, Pare P, Proctor DD, Regueiro MD, Steinhart AH, Targan SR, Schumm LP, Kistner EO, Lee AT, Gregersen PK, Rotter JI, Brant SR, Taylor KD, Roeder K, Duerr RH. Ulcerative colitis-risk loci on chromosomes 1p36 and 12q15 found by genome-wide association study. Nat Genet. 2009;41(2):216–220. doi: 10.1038/ng.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wrensch M, Jenkins RB, Chang JS, Yeh RF, Xiao Y, Decker PA, Ballman KV, Berger M, Buckner JC, Chang S, Giannini C, Halder C, Kollmeyer TM, Kosel ML, LaChance DH, McCoy L, O’Neill BP, Patoka J, Pico AR, Prados M, Quesenberry C, Rice T, Rynearson AL, Smirnov I, Tihan T, Wiemels J, Yang P, Wiencke JK. Variants in the CDKN2B and RTEL1 regions are associated with high-grade glioma susceptibility. Nat Genet. 2009;41(8):905–908. doi: 10.1038/ng.408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Prorok PC, Andriole GL, Bresalier RS, Buys SS, Chia D, Crawford ED, Fogel R, Gelmann EP, Gilbert F, Hasson MA, Hayes RB, Johnson CC, Mandel JS, Oberman A, O’Brien B, Oken MM, Rafla S, Reding D, Rutt W, Weissfeld JL, Yokochi L, Gohagan JK. Design of the Prostate, Lung, Colorectal and Ovarian (PLCO) Cancer Screening Trial. Control Clin Trials. 2000;21(6 Suppl):273S–309S. doi: 10.1016/s0197-2456(00)00098-2. [DOI] [PubMed] [Google Scholar]
- 30.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lange EM, Ho LA, Beebe-Dimmer JL, Wang Y, Gillanders EM, Trent JM, Lange LA, Wood DP, Cooney KA. Genome-wide linkage scan for prostate cancer susceptibility genes in men with aggressive disease: significant evidence for linkage at chromosome 15q12. Hum Genet. 2006;119(4):400–407. doi: 10.1007/s00439-006-0149-6. [DOI] [PubMed] [Google Scholar]
- 32.Schaid DJ, McDonnell SK, Zarfas KE, Cunningham JM, Hebbring S, Thibodeau SN, Eeles RA, Easton DF, Foulkes WD, Simard J, Giles GG, Hopper JL, Mahle L, Moller P, Badzioch M, Bishop DT, Evans C, Edwards S, Meitz J, Bullock S, Hope Q, Guy M, Hsieh CL, Halpern J, Balise RR, Oakley-Girvan I, Whittemore AS, Xu J, Dimitrov L, Chang BL, Adams TS, Turner AR, Meyers DA, Friedrichsen DM, Deutsch K, Kolb S, Janer M, Hood L, Ostrander EA, Stanford JL, Ewing CM, Gielzak M, Isaacs SD, Walsh PC, Wiley KE, Isaacs WB, Lange EM, Ho LA, Beebe-Dimmer JL, Wood DP, Cooney KA, Seminara D, Ikonen T, Baffoe-Bonnie A, Fredriksson H, Matikainen MP, Tammela TL, Bailey-Wilson J, Schleutker J, Maier C, Herkommer K, Hoegel JJ, Vogel W, Paiss T, Wiklund F, Emanuelsson M, Stenman E, Jonsson BA, Gronberg H, Camp NJ, Farnham J, Cannon-Albright LA, Catalona WJ, Suarez BK, Roehl KA. Pooled genome linkage scan of aggressive prostate cancer: results from the International Consortium for Prostate Cancer Genetics. Hum Genet. 2006;120(4):471–485. doi: 10.1007/s00439-006-0219-9. [DOI] [PubMed] [Google Scholar]
- 33.Holm S. A simple sequentially rejective multiple test procedure. Scandinavian journal of statistics, theory and applications. 1979;6(2):65–70. [Google Scholar]
- 34.Kraft P. Curses--winner’s and otherwise--in genetic epidemiology. Epidemiology. 2008;19(5):649–651. doi: 10.1097/EDE.0b013e318181b865. [DOI] [PubMed] [Google Scholar]