Abstract
The absence of resistance genes against biotic stresses like Tobacco streak virus (TSV) within compatible peanut germplasm necessitates the deployment of genetic engineering strategy to develop transgenic resistance. Transgenic resistance in peanut (Arachis hypogaea L.) to peanut stem necrosis disease caused by TSV was obtained by transferring coat protein (CP) gene of TSV through Agrobacterium-mediated transformation of de-embryonated cotyledons and immature leaves of peanut cultivars Kadiri 6 (K6) and Kadiri 134 (K134). Integration of the transgene in T1, T2 and T3 generations were confirmed by PCR with gene-specific primers. On the basis of segregation analysis of the PCR amplicons, homozygosity was confirmed in progeny from five transgenic lines. Six transgenic plants from three different single copy transgenic lines homozygous for the transgene were selected for challenge inoculation in T3 generations. The transgenic lines remained symptomless throughout and showed traces or no systemic accumulation of virus indicating the tolerance/resistance to the TSV infection. CP gene expression was observed in transgenic lines by RT-PCR, real-time PCR and ELISA. The findings provide an effective strategy for developing peanut with resistance to peanut stem necrosis disease.
Electronic supplementary material
The online version of this article (doi:10.1007/s13337-013-0157-9) contains supplementary material, which is available to authorized users.
Keywords: Arachis hypogaea, Coat protein-mediated transgenic resistance, Peanut stem necrosis disease, Agrobacterium-mediated genetic transformation
Introduction
Peanut (Arachis hypogaea L.) is one of the important commercial crops of the world, and is a major source of protein and oil. Reduction in yield caused by the various diseases including those caused by viruses is a major production constraint in peanut cultivation. Millions of smallholder farmers in the rainfed semi-arid tropics of the Indian subcontinent depend on peanut farming for their livelihood. A new threat to peanut emerged in 2000 in the form of a virus that causes lethal necrosis. This disease, peanut stem necrosis disease (PSND), led to losses over $64 million in 2000 in the district of Anantapur alone, a major peanut producing area in India [16]. Tobacco streak virus (TSV) was found associated with the disease [18], which was reported for the first time in peanut from India.
TSV belongs to the genus Ilarvirus of the family Bromoviridae, spreads mainly through the weeds of crop species which are alternate host to the virus. TSV can survive on cowpea, black gram, green gram, marigold and sunflower. Parthenium hysterophorous, a wide spread weed acts as a symptomless carrier and virus is spread through pollen grains by the three species of thrips namely Frankliniella schultzei, Scirtothrips dorsalis and Megalothrips usitatus. In case of peanut, Frankliniella schultzei acts as the viral vector [18].
Despite several years efforts, still confirmed sources of genetic resistance/tolerance to TSV could not be identified in the gene pool of cultivated peanut for their use in the breeding programmes, and hence, so far, no cultivar resistant to this disease has been developed. Genetically engineered resistance has been actively investigated in recent years as an alternative to cope up with this type of situations [11].
Coat protein-mediated resistance, a form of pathogen-derived resistance, where the degree of protection ranges from a delay in symptom expression to absence of disease symptoms and virus accumulation, has been established as an effective means of protection against viral infection and the prevention of crop loss [2, 3]. Coat protein (CP) genes have been shown to confer partial or complete resistance as was observed for TSV in tobacco [23], cucumber mosaic virus in cucumber [14], and potato virus Y in potato plants [7].
Thus, considering the economic importance of PSND in the peanut cultivation, we have resorted to the transgenic approach using CP gene to develop virus tolerant genotypes in cultivated peanut. This work report successful deployment of the CP gene of TSV in peanut, achieved through Agrobacterium mediated genetic transformation.
Materials and methods
Plasmid constructs and Agrobacterium strain
The gene construct was prepared by inserting the 717 bp CP gene (GenBank Acc. No. AF400664) of TSV, downstream of an enhanced double 35S promoter [10] into the binary vector pCAMBIA 1305.1. The TSV-CP gene sequences of all the reported isolates in NCBI databank are highly conserved and AF400664 had only 0–2 % diversity in nucleotides and 0–4 % in amino acids [1]. The CP gene cloned in pGEM-T Easy vector (Promega, USA), was released by restriction digesting using NcoI and BstEII and gel purified using Promega gel purification kit (Promega, Madison, WI, USA). The GUS gene from the binary vector pCAMBIA 1305.1 was removed by NcoI and BstEII digestions and the vector was gel purified. The TSV-CP gene was ligated in pCAMBIA1305.1 in place of GUS, between the CaMV 35S promoter and the NOS terminator (Fig. 1). The ligated product was transformed into E. coli (strain DH5α) using standard molecular biology protocols [20]. The putative clones were initially screened by PCR and subsequently confirmed by restriction digestion with NcoI and BstEII. TSV-CP gene was mobilized into the Agrobacterium (strain LBA4404) by using freeze and thaw method [9]. The putative colonies were screened by colony PCR and the confirmed clones were maintained on Luria agar plate containing kanamycin (50 μg/mL) and rifampicin (50 μg/mL). The modified binary vector carrying the selectable marker gene hygromycin phosphotransferase (hptII) was used for co-cultivation of explants in plant tissue culture.
Fig. 1.
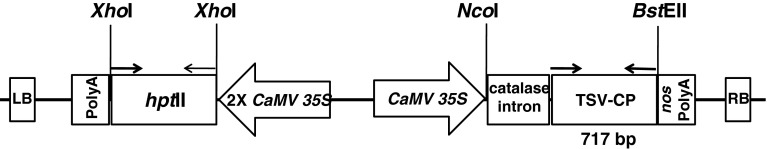
Schematic representation of the T-DNA region of pCAMBIA1305.1 binary plasmid used for transformation of de-embryonated cotyledons and immature leaves with Agrobacterium tumefaciens strain LBA4404. The position of the primers used in PCR assays are shown by arrows on top the TSV-CP gene. LB left border, RB right border, hptII (kan+) hygromycin resistance gene, NOS polyA nopaline synthase terminator
Plant materials and transformation
The mature seeds of the commercial cultivars of peanut, K6 and K134, which are cultivated mainly in the areas where the PSND was epidemic in India, were used in the study. The seeds were obtained from the Genetic Resources Section of the Directorate of Groundnut Research (DGR). Agrobacterium-mediated transformation and regeneration were performed as described by Radhakrishnan et al. [17]. The de-embryonated cotyledons and immature leaves explants from mature pre-soaked seeds were co-cultivated with A. tumefaciens strain LBA4404 harboring the binary plasmid pCAMBIA1305.1:TSV-CP gene. The regeneration frequency was calculated on the number of explants regenerated over the number of explants co-cultured. The transformation frequency was worked out on the final number of confirmed transgenics produced over the number of explants regenerated.
Molecular analysis of putative transformants
PCR analysis
Initial screening of the putative transgenic plants was done by PCR for presence of the transgene. Genomic DNA was extracted from fresh terminal leaves of the glasshouse grown plants by following the protocol described by Radhakrishnan et al. [17]. The PCR reaction was performed with 25 μl of a total reaction mixture containing 100 ng of genomic DNA, 2.5 μl of 10× PCR buffer (containing 15 mM MgCl2), 1.6 μl of 2 mM dNTP mix, 1 μl of 25 pM each of the forward and reverse gene-specific primer, and 2U of Taq DNA polymerase. A control devoid of the template DNA was used in each reaction. DNA from transgenic tobacco and/or the plasmid were used as positive controls. The thermal cycles comprised an initial denaturing at 94 °C for 4 min, followed by 30 cycles of 94 °C for 30 s, Ta °C (depending upon the annealing temperature of the gene-specific primers; Table 1) for 45 s, 72 °C for 1 min and a final extension of 10 min at 72 °C. The amplification products were resolved on 1.2 % agarose gel, stained with EtBr, scanned and documented using a Fuji FLA5200 imaging system.
Table 1.
Sequence of gene-specific primers used for PCR analysis of putative transgenics
| Primer | Sequence | Tm (°C) |
|---|---|---|
| TSV Forward | 5′ CCA TGG ATG AAT ACT TTG ATC CAA GG 3′ | 55.8 |
| TSV Reverse | 5′ GGT NAC CTC AGT CTT GAT TCA CCA G 3′ | 58.0 |
| hptII Forward | 5′ AAA ACT GAT CGA AAA ATA CCG CTG C 3′ | 60.2 |
| hptII Reverse | 5′ TCC CCA GTA AGT CAA AAA ATA GCT C 3′ | 57.2 |
Southern analysis
To ascertain the integration pattern and the number of copies of the inserted DNA, genomic Southern hybridization analysis was performed with 10 μg of genomic DNA of the putative transgenics (T0 generation) which was separately digested with the restriction enzyme PstI or BamHI or BstPI, or NotI or NcoI or EcoRI. The restriction product was resolved on a 1.2 % agarose gel and transferred to Hybond™ –N+ nylon membrane (GE Health Care). For developing the filters and probing with biotin labeled probes prepared from nucleotide sequences of the TSV-CP gene used in the transformation using the DNA labeling kit of Fermentas (Cat. No. K0651) and the biotin chromogenic detection kit of Fermentas (Cat. No. K0661 and K0662) following the manufacturer’s instructions.
Gene segregation analysis
Five selected T0 plants which produced sufficient number of seeds were selected for segregation analysis. The T1 progeny of the five plants were grown in pots under controlled conditions in a glass house. The plantlets at 2–4 leaf stage were used for PCR analysis using gene-specific primers to score the amplicons. On the basis of expected and observed frequencies the χ2 test was conducted in the progeny from all the five transgenic lines.
Reverse transcriptase (RT)-PCR
Total mRNA was isolated from randomly selected transformed and non-transformed plants using Trizol LS (Invitrogen) as per the instructions provided by the kit manufacturer and estimated using ND-1000 spectrophotometer (NanoDrop Technologies Inc., USA) followed by DNase I (Invitrogen) treatment. The first strand DNA was synthesized using the first strand cDNA synthesis kit, Fermentas, USA (K1612) following the protocol manual supplied. This cDNA was used for amplification of the second strand by normal PCR reactions using the gene-specific primers. As a control (-RT), RNA samples without reverse transcriptase were included to detect the possible contamination with genomic DNA.
Quantitative (real-time) PCR analysis
The transcript for TSV-CP was quantified by SYBR Green chemistry. The real-time PCR was carried out using an ABI StepOne real-time PCR machine. The relative quantification of TSV-CP transcript was normalized with respect to the internal (housekeeping gene) control, 18S rRNA (GenBank Acc. No. AF156675.2).
The primers used in the reactions were designed using the software Primer Express™ (Applied Biosystems). The primers for TSV-CP, (Forward) 5′-GAATGACCGCACCAATTCCT-3′ and (Reverse) 5′-TGGGTAGCTTCAACGATGTCTTC-3′ and the primers for 18S rRNA (Forward) 5′- ATTCCTAGTAAGCGCGAGTCATCAG-3′ and (Reverse) 5′-CAATGATCCTTCCGCAGGTTCAC -3′ which produced amplicons sizes 69 bp and 226 bp respectively were used. Each reaction was performed in 25 μl (total volume) and consisted of SYBR Green Master mix (Qiagen QuantiFast SYBR Green PCR Kit; Cat No. 204054), 10 pM of each primer and 1/10-fold diluted cDNA template. Each experiment included triplicate reactions with the same cDNA stock. The 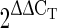 method was used to calculate the comparative fold expression of the TSV-CP [12]. The ∆CT was determined by subtracting 18S rRNA CT from TSV-CP CT in a given sample. The ∆∆CT value was determined by subtracting the lowest expressing plant (calibrator) ∆CT from the ∆CT of each plant.
method was used to calculate the comparative fold expression of the TSV-CP [12]. The ∆CT was determined by subtracting 18S rRNA CT from TSV-CP CT in a given sample. The ∆∆CT value was determined by subtracting the lowest expressing plant (calibrator) ∆CT from the ∆CT of each plant.
Challenge inoculation with TSV
TSV-infected leaves from cowpea plants showing primary symptoms were selected as source of inoculum. The leaves were macerated to fine homogenate using chilled inoculation buffer. For every 100 mg of leaf tissue 0.1 M phosphate buffer, pH 7.0 was used (1:10 w/v). The inoculum was used immediately.
Healthy leaves of the transgenic plants selected for inoculation were rubbed gently with the dust abrasive carborundum and subsequently the inoculum was applied using a cotton swab. The leaves were rinsed with distilled water immediately after inoculation and the plants were covered with polythene sheets for overnight. Along with transgenics the untransformed plants of the cultivar K6 were also inoculated in the similar manner. Using the same inoculums the cowpea plants were also inoculated as a control. Inoculated plants were observed daily for 15–20 days for the development of symptoms and compared with the controls. Plants that did not develop any symptoms were checked by back inoculation tests to detect latent infection, if any.
ELISA analyses
Direct-antigen-coating enzyme-linked immunosorbent assay (DAC-ELISA) was done with 500 mg leaf samples of infected transgenic, non-transgenic and un-inoculated healthy plants as described by Hobbs et al. [8]. Microtiter plates were coated with leaf extracts prepared in 50 mM sodium carbonate buffer, pH 9.6. Polyclonal antiserum raised against CP of TSV-New Delhi isolate was diluted to 1:10,000 or 1:50,000 with extracts of healthy peanut leaves and cross absorbed. Goat antirabbit immunoglobulin conjugated to alkaline phosphatase (Sigma-Aldrich, St. Louis) were used at a 1:5,000 dilution of the commercial stock. P-nitrophenyl phosphate at 1 mg/mL was used as substrate. Absorbance was recorded at 405 nm after incubation for 1 h at room temperature following the addition of the substrate using a BioTek Make ELISA Plate Reader (Model Epoch).
Results
Transformation, selection and multiplication
In vitro regeneration from the de-embryonated cotyledons and immature leaf explants of Arachis hypogaea L. cultivars K6 and K134 resulted in direct shoot initiation after 100–105 days. All transgenic lines appeared normal in morphology and development (Fig. 2).
Fig. 2.
Genetic transformation and regeneration of peanut from de-embryonated cotyledons (1–4 shoot buds initiating from co-cultured immature leaves, 5–6 shoot buds initiating from co-cultured de-embryonated cotyledons, 7–9 shoot buds in antibiotic selection medium, 10 shoots developing roots in the rooting medium
In the case of cultivar K6, 1229 and 746 shoots regenerated from de-embryonated cotyledon and immature leaf explants respectively, survived the antibiotic selection pressure, whereas in the case of cultivar K134, it was 484 and 82 shoots. In the present study, from de-embryonated cotyledon explants overall regeneration frequencies were 60.4 and 84.3 % respectively for the cv. K6 and K134. The shoot regeneration frequencies from the immature leaf explants of the cv. K6 and K134 were 77.4 and 76.9 % respectively. The transformation efficiency of de-embryonated cotyledon was 34 and 16.5 %; and from immature leaf 11.3 and 2.2 % respectively for the two cultivars cv. K6 and K134 respectively (Table 2).
Table 2.
Genetic transformation and regeneration of peanut explants from the cultivars K6 and K134
| No. of co-cultivations | Total explants co-cultured | Explants regenerated | Shoots produced | Shoots passed antibiotic selection | Shoots produced roots | Plantlets hardened | Plants survived in glasshouse | Final recovery of putative transgenics |
|---|---|---|---|---|---|---|---|---|
| cv. K6 | ||||||||
| 10 (DC) | 409 | 247 | 3,938 | 1,229 | 1,038 | 1,010 | 919 | 84 (34.0) |
| 6 (IL) | 1,028 | 796 | 2,263 | 746 | 538 | 368 | 301 | 9 (0.9) |
| cv. K134 | ||||||||
| 5 (DC) | 108 | 91 | 932 | 484 | 280 | 238 | 205 | 15 (16.5) |
| 3 (IL) | 459 | 353 | 273 | 82 | 65 | 51 | 41 | 8 (2.2) |
Values in parenthesis are percentage over explants regenerated
DC De-embryonated cotyledons, IL immature leaves
PCR and Southern analysis of T0 transgenic plants
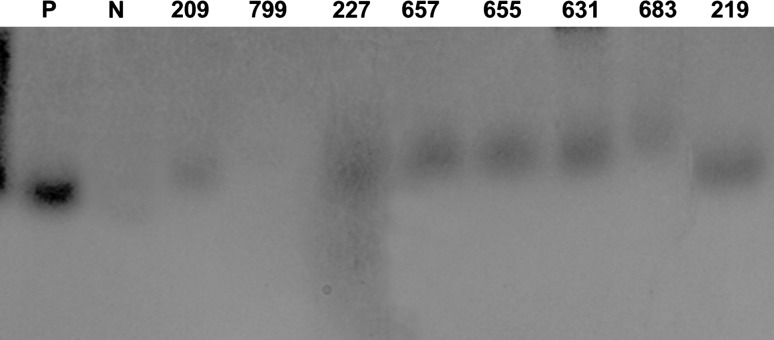
In order to confirm the presence of the transgene in the plants selected in antibiotic containing medium, 1466 PCRs were done (Table 3). The 919 putative transgenic plants developed from the de-embryonated cotyledons of the cultivar K6 and 205 plants from the cultivar K134 were tested for the integration of TSV-CP gene using gene-specific primers. Amplicon of the expected size (717 bp) were produced in 99 T0 plants (Fig. 3).
Table 3.
PCR and Southern analysis of the peanut transgenic lines to confirm the integration of the TSV-CP transgene
| Variety | Total | ||||
|---|---|---|---|---|---|
| K6 | K134 | ||||
| DC | IL | DC | IL | ||
| Number of plants PCR tested | 919 | 301 | 205 | 41 | 1,466 |
| Number of PCR positives | 84 | 9 | 15 | 8 | 116 |
| Number of plants screened by Southern blot assay | 53 | 6 | 5 | 1 | 65 |
DC De-embryonated cotyledon, IL Immature leaves
Fig. 3.
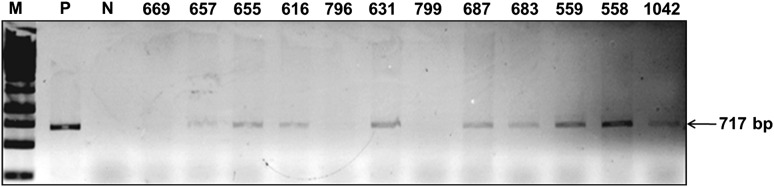
PCR analysis of TSV-CP transgenic peanut plants. TSV-CP gene-specific primers were used for PCR amplification and the size of expected product was 717 bp as that of the full-length TSV-CP cds. Lane M 1 kb ladder, lane P positive control (pCAMBIA1305.1 TSV-CP plasmid), lane N untransformed peanut plant, and lane 4–15 transgenic peanut lines represented by their unique plant identity no
The 301 putative transgenic plants developed form the immature leaves of the cultivar K6 and 41 from the cultivar K134 were also tested for presence of TSV-CP gene with gene-specific primers and 17 T0 plants had amplicons of the expected size.
Sixty-five randomly selected T0 plants were tested by Southern hybridization and results indicate the transgene was inserted into the transgenic plant genome. Molecular evidence for stable transgene integration and expression in a few selected T0 transgenic lines is shown in Fig. 4. Transgenic plants that were hardened and raised in the glasshouse were morphologically comparable to the non-transgenic peanut plants.
Fig. 4.
Southern blot analysis of the T0 transgenic plants. Genomic DNA (10 μg) was digested with BstEII, size fractionated on 1.2 % agarose gel, transferred to a nylon membrane and hybridized with a biotin labeled probes of TSV-CP probe. Lane P positive control (pCAMBIA1305.1 TSV-CP plasmid), lane N untransformed peanut plant, and lane 3–10 transgenic peanut lines
In total, the PCR analyses of the 1466 putative transgenics developed, 116 (7.9 %) revealed the integration of the TSV-CP by producing the amplicon of the expected size (717 bp).
Detection of transcriptional activity in the transgenic plants
The confirmed transgenic lines having single copy insertion were analyzed for expression of TSV-CP gene through reverse transcription; presented active transcriptional activity that corresponded with the recombinant CP fragment, whereas no amplification was observed in untransformed plants (Fig. 5). Direct PCR was performed on the RNA preparations, to rule out the possibility that contaminant DNA was amplified in the samples. No amplified DNA fragments could be detected, confirming the RNA specificity of the reaction.
Fig. 5.
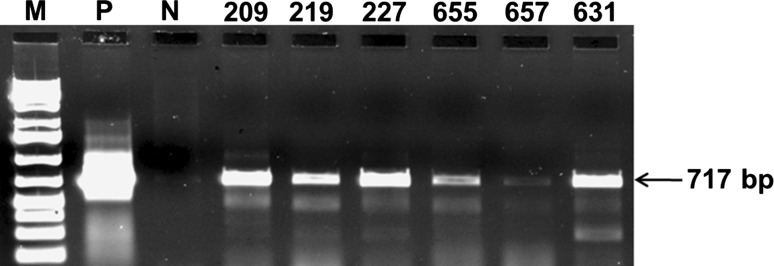
Confirmation of the expression of the TSV-CP transgene in transgenic peanut plants. The total mRNA from the transgenic plants were extracted and the first strand cDNA was prepared. Gene-specific primers were used to amplify the transcripts of the TSV-CP transgene. Lane P positive control (pCAMBIA1305.1 TSV-CP plasmid), lane N untransformed peanut plants and lane 4–9 transgenic peanut lines
Transgene inheritance analysis
Plants from five T1 generation transgenic lines were used for segregation analysis of the transgene. PCR analysis of the plants at 2–4 leaf stage was done using gene-specific primers to score the presence/absence of amplicons. On the basis of expected and observed frequencies of the gene-specific amplicons the χ2 test was conducted with all five transgenic lines (Table 4). The values of χ2 in all five lines were found non-significant (p = 0.05) indicating close agreement between the observed and expected frequencies. The segregation of the transgene followed 3:1 Mendelian ratio.
Table 4.
Segregation analysis of the transgene in the T1 progeny of TSV-CP containing peanut plants
| Transgenic lines | Total no. of plants | Transgene present | Transgene absent | Observed ratio | Expected ratio | χ2 value |
|---|---|---|---|---|---|---|
| TSV.22 | 17 | 12 | 5 | 2.4:1 | 3:1 | 0.093 |
| TSV.209 | 12 | 9 | 3 | 3:1 | 3:1 | 0.000 |
| TSV.219 | 16 | 12 | 4 | 3:1 | 3:1 | 0.000 |
| TSV.224 | 16 | 14 | 4 | 3.5:1 | 3:1 | 0.285 |
| TSV.227 | 26 | 19 | 7 | 2.7:1 | 3:1 | 0.050 |
(df = 1, p = 0.05)
Quantitative analysis by real-time PCR
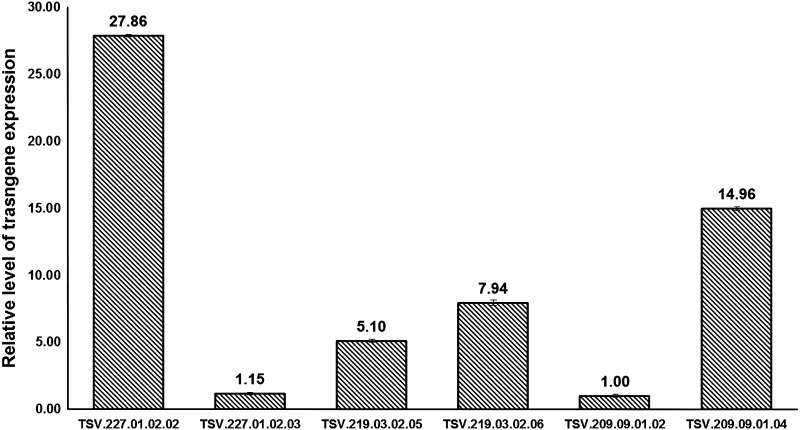
The representative homozygous T3 transgenic lines were analysed for the expression of the TSV-CP gene using real-time PCR. The  method was used to calculate relative changes in expression. Amplicon abundance was monitored in real-time by measuring SYBR® Green fluorescence. The level of TSV-CP transcript in the T3 plants was normalized with reference to 18S rRNA taken as an internal control (Fig. 6 and Supplemental Table 1). The bar in the figure denotes the fold expression as compared to the lowest expressing plant TSV.209.09.01.02 which is taken as the calibrator. The transgene expression in the plant TSV.227.01.02.03 was almost equal to the calibrator. The transgenic line TSV.219.03.02.05 showed about fivefold transgene expressions; while the transgenic line TSV.219.03.02.06, had eightfolds expression of the transgene relative to that of the calibrator. More than 25 fold expression was observed in TSV.227.01.02.02 that was highest followed by TSV.209.09.01.04 (with >14 fold).
method was used to calculate relative changes in expression. Amplicon abundance was monitored in real-time by measuring SYBR® Green fluorescence. The level of TSV-CP transcript in the T3 plants was normalized with reference to 18S rRNA taken as an internal control (Fig. 6 and Supplemental Table 1). The bar in the figure denotes the fold expression as compared to the lowest expressing plant TSV.209.09.01.02 which is taken as the calibrator. The transgene expression in the plant TSV.227.01.02.03 was almost equal to the calibrator. The transgenic line TSV.219.03.02.05 showed about fivefold transgene expressions; while the transgenic line TSV.219.03.02.06, had eightfolds expression of the transgene relative to that of the calibrator. More than 25 fold expression was observed in TSV.227.01.02.02 that was highest followed by TSV.209.09.01.04 (with >14 fold).
Fig. 6.
Quantification of the expression of TSV-CP transcript in transgenic peanut plants. The transcripts were analyzed by quantitative PCR. The level of TSV-CP transcript in T3 plants was normalized with reference to 18S rRNA taken as an internal control. Bars denote fold expression as compared to the lowest expressing plant TSV.209.09.01.02. The control (negative RT) showed no transcript
Bioassay of transgenic plants
Six plants from three independent single copy integration lines were selected for challenge inoculation in T3 generation. Challenge inoculation of 4–6 leaf stage plants was done with crude extract inoculum prepared from 100 mg of cowpea leaves, infected with homologous strain of TSV, macerated in 1 mL 0.1 M phosphate buffer, pH 7.0. After inoculation, necrotic lesions and veinal necrosis in young leaves were observed within 3–4 days on the untransformed control peanut plants. The five transgenic plants which expressed the CP (as observed in reverse transcriptase-PCR) did not show symptoms on the inoculated leaf 1 week after inoculation. Transformed lines were back inoculated with crude extract of TSV infected cowpea to check latent infection, if any. But all transgenic lines remained symptom-less whereas untransformed peanut showed complete stem necrosis and died (Fig. 7) within a month. DAC-ELISA conducted with polyclonal antiserum against TSV (New Delhi isolate), on the upper leaves in all transgenic lines showed reduced virus accumulation as compared to control with the highest reduction observed (approximately three to five times lower than the diseased control) in the transgenic lines TSV.22, TSV.209, TSV.219, TSV.227 and TSV.224 (Table 5). Failure to produce symptoms through back inoculation tests further confirmed resistance in the transgenic lines.
Fig. 7.
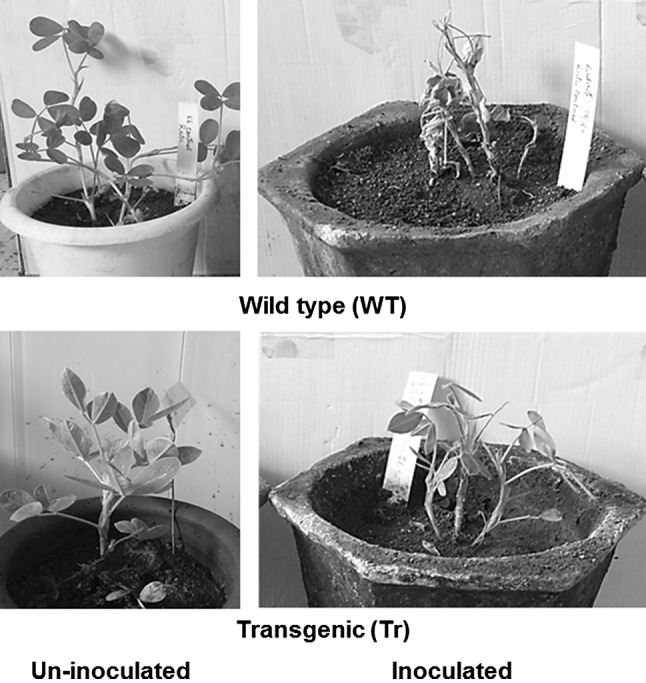
Response of 15 days old transgenic peanut lines (Tr) to 2 weeks of challenge inoculation with homologous strain of TSV as compared to non-transgenic (WT) controls
Table 5.
Reaction of the TSV infected transgenic peanut and control plants estimated by DAC-ELISA
| Sr. No. | Detail of sample | ELISA reading |
|---|---|---|
| 1 | Coating buffer | 0.00 |
| 2 | WT peanut inoculated | 0.24 |
| 3 | WT peanut un-inoculated | 0.00 |
| 4 | TSV.227.01.03.01 un-inoculated | 0.98 |
| 5 | TSV.209.09.01.02 inoculated | 0.09 |
| 6 | TSV.209.09.01.04 inoculated | 0.13 |
| 7 | TSV.219.03.02.05 inoculated | 0.07 |
| 8 | TSV.219.03.02.06 inoculated | 0.09 |
| 9 | TSV.227.01.02.02 inoculated | 0.13 |
| 10 | TSV.227.01.02.03 inoculated | 0.10 |
| 11 | Cowpea un-inoculated | 0.13 |
| 12 | Cowpea inoculated | 0.51 |
Discussion
The main objective of this study was to determine whether the CP-mediated resistance strategy could be applied for developing resistant transgenic peanut utilizing the CP gene of the TSV strain New Delhi isolate. The transformation protocol employed could give a reasonably good success rate. The transgene followed the Mendelian pattern of inheritance as expected.
CP genes have been shown to be effective in preventing the viral infection or reducing disease caused by homologous and closely related viruses [6]. More than 116 transgenic lines were generated using Agrobacterium-mediated transformation. Though the overall regeneration frequency was much different between the de-embryonated cotyledon and the immature leaf explants, the recovery of transgenics was much more in de-embryonated cotyledons. Further, each regenerating de-embryonated cotyledons explant had more than 10 shoots and this had contributed to the higher frequency of recovery of transgenic plants.
The transformed peanut plants analysed by PCR, Southern, RT-PCR, quantitative (real-time) PCR and DAC-ELISA confirmed that the TSV-CP gene had successfully integrated into the plant genome and was being transcribed. On selfing of the transgenic lines in T1 and T2 generation, the expected Mendelian segregation ratio of 3:1 was observed. Evaluation of resistance in different transgenic plants to TSV infection in the T3 generations, raised under glasshouse conditions, as determined by virus accumulation levels indicated total resistance to the expression of disease symptoms. Similar finding have been reported earlier by Srivastav and Raj [21], while delayed symptoms to none were reported in challenge inoculations by Nakajima et al. [13, 14]. Srivastav and Raj [21] has also reported resistance in transgenic plants that remained symptomless throughout their life cycle, although they accumulated virus at high levels in their upper leaves (but less than the control) in conformation to the earlier reports of Powell-Abel et al. [15]. In this study, the resistance shown by transgenic plants to TSV is likely to be protein-mediated as CP was detected by ELISA in the leaves of transgenic plants. It may be due to the interactions of TSV-CP in the transgenic plant and viruses in the challenge inoculums [4, 15]. Higher CP levels in inoculated transgenic plants have presumably affected the uncoating of the virus particle at later stages. According to Reimann-Philipp [19], a reduced rate of virus accumulation in inoculated leaves and slower systemic spread are frequently observed in transgenic CP-accumulating plants owing to slower replication rates or interference with local or systemic virus transport. Higher accumulation of TSV in inoculated leaves without systemic spread may be due to interference with either entry into the phloem or vascular long-distance transport as suggested earlier by Taliansky and Garcia-Arenal [22].
Evaluation of the transgenic peanut plants up to the third generation showed that resistance could be achieved against PSND. Our findings demonstrate the potential of using CP-based transgenic expression systems as effective strategy against PSND viruses in groundnut. Further investigations will be taken up in future for determining the efficacy and performance of the identified transgenic lines at field level.
Electronic supplementary material
Acknowledgments
The Department of Biotechnology, Government of India is gratefully acknowledged for financial support to this project.
References
- 1.Bag S, Singh RS, Jain RK. Further analysis of coat protein gene sequences of Tobacco streak virus isolates from diverse locations and hosts in India. Indian Phytopathol. 2008;61:118–123. [Google Scholar]
- 2.Baulcombe DC. Mechanisms of pathogen-derived resistance to viruses in transgenic plants. Plant Cell. 1996;8:1833–1844. doi: 10.1105/tpc.8.10.1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beachy RN. Mechanisms and applications of pathogen-derived resistance in transgenic plants. Curr Opin Biotechnol. 1997;8:215–220. doi: 10.1016/S0958-1669(97)80105-X. [DOI] [PubMed] [Google Scholar]
- 4.Bendahmane M, Chen I, Asurmendi S, Bazzini AA, Szecsi J, Beachy RN. Coat protein-mediated resistance to TMV infection of Nicotiana tabacum involves multiple modes of interference by coat protein. Virology. 2007;366:107–116. doi: 10.1016/j.virol.2007.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gielen J, Ultzen T, Bontems S, Loots W, Schepen A, Westerbock A, Haan P, van Grinsven M. Coat protein mediated protection of Cucumber mosaic virus infection in cultivated tomato. Euphytica. 1996;88:139–149. doi: 10.1007/BF00032445. [DOI] [Google Scholar]
- 6.Gonsalves D, Slightom JL. Coat-protein mediated protection: analysis of transgenic plants for resistance in a variety of crops. Semin Virol. 1993;4:397–406. doi: 10.1006/smvy.1993.1039. [DOI] [Google Scholar]
- 7.Hefferon KL, Khalilian H, AbouHaidar MG. Expression of the PVY(O) coat protein (CP) under the control of the PVX CP gene leader sequence: protection under greenhouse and field conditions against PVY(O) and PVY(N) infection in three potato cultivars. Theor Appl Genet. 1997;94(2):287–292. doi: 10.1007/s001220050412. [DOI] [Google Scholar]
- 8.Hobbs HA, Reddy DVR, Rajeswari R, Reddy AS. Use of direct antigen coating and protein A coating ELISA procedures for detection of three peanut viruses. Plant Dis. 1987;71:747–749. doi: 10.1094/PD-71-0747. [DOI] [Google Scholar]
- 9.Höfgen R, Willmitzer L. Storage of competent cells for Agrobacterium transformation. Nucleic Acids Res. 1988;16:9877. doi: 10.1093/nar/16.20.9877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kay R, Chan A, Daly M, McPherson J. Duplication of CaMV 35S promoter sequences creates a strong enhancer for plant genes. Science. 1987;236:1299–1302. doi: 10.1126/science.236.4806.1299. [DOI] [PubMed] [Google Scholar]
- 11.Li ZJ, Jarret RL, Demski JW. Engineered resistance to tomato spotted wilt virus in transgenic peanut expressing the viral nucleocapsid gene. Transgenic Res. 1997;6:297–305. doi: 10.1023/A:1018462729127. [DOI] [Google Scholar]
-
12.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR & the
 method. Methods. 2001;25:402–8. [DOI] [PubMed]
method. Methods. 2001;25:402–8. [DOI] [PubMed] - 13.Nakajima M, Hayakawa T, Nakamura I, Suzuki M. Protection against cucumber mosaic virus (CMV) strains O and Y and chrysanthemum mild mottle virus in transgenic tobacco plants expressing CMV-O coat protein. J Gen Virol. 1993;74:319–322. doi: 10.1099/0022-1317-74-2-319. [DOI] [PubMed] [Google Scholar]
- 14.Nishibayashi S, Hayakawa T, Nakajima T, Suzuki M, Kaneko H. CMV protection in transgenic cucumber plants with an introduced CMV-O cp gene. Theor Appl Genet. 1996;93:672–678. doi: 10.1007/BF00224061. [DOI] [PubMed] [Google Scholar]
- 15.Powell-Abel P, Nelson RS, De B, Hoffmann N, Rogers SG, Fraley RT, Beachy RN. Delay of disease development in transgenic plants that express the tobacco mosaic virus coat protein gene. Science. 1986;232:738–743. doi: 10.1126/science.3457472. [DOI] [PubMed] [Google Scholar]
- 16.Prasada Rao RDVJ, Reddy AS, Chander Rao AS, Varaprasad KS, Thirumala Devi K, Nagaraju Muniyappa V, Reddy DVR. Tobacco streak ilarvirus as a causal agent of sunflower necrosis disease in India. J Oilseeds Res. 2000;17:400–401. [Google Scholar]
- 17.Radhakrishnan T, Murthy TGK, Chandran K, Bandyopadhyay A. Micropropagation in peanut (A. hypogaea L.) Biol Plant. 2000;43:447–450. doi: 10.1023/A:1026743822546. [DOI] [Google Scholar]
- 18.Reddy AS, Prasad Rao RDVJ, Thirimala Devi K, Reddy SV, Maya MA, Roberts I, Satyanarayana T, Subramaniam K, Reddy DVR. Occurrence of Tobacco streak virus on peanut (Arachis hypogaea) in India. Plant Dis. 2002;86:173–178. doi: 10.1094/PDIS.2002.86.2.173. [DOI] [PubMed] [Google Scholar]
- 19.Reimann-Phillipp U. Mechanism of resistance: expression of coat protein. In: Foster GD, Taylor SC, editors. Methods in molecular biology, plant virology protocols: from virus isolation to transgenic resistance. Humana Press: New Jersey; 1998. pp. 521–532. [Google Scholar]
- 20.Sambrook J, Russell DW. Molecular cloning: a laboratory manual. New York: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 21.Srivastava A, Raj SK. Coat protein-mediated resistance against an Indian isolate of the Cucumber mosaic virus subgroup IB in Nicotiana benthamiana. J Biosci. 2008;33:249–257. doi: 10.1007/s12038-008-0042-7. [DOI] [PubMed] [Google Scholar]
- 22.Taliansky ME, Garcia AF. Role of Cucumovirus capsid protein in long-distance movement within the infected plants. J Virol. 1995;69:916–922. doi: 10.1128/jvi.69.2.916-922.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Dun CM, Overduin B, van Vloten-Doting L, Bol JL. Transgenic tobacco expressing tobacco streak virus or mutated alfalfa mosaic virus coat protein does not cross protect against alfalfa mosaic virus infection. Virology. 1988;164:383–389. doi: 10.1016/0042-6822(88)90551-X. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.