Abstract
Kinase inhibitors have emerged as effective cancer therapeutics in a variety of human cancers. Glioblastoma (GBM), the most common malignant brain tumor in adults, represents a compelling disease for kinase inhibitor therapy because the majority of these tumors harbor genetic alterations that result in aberrant activation of growth factor signaling pathways. Attempts to target the Ras—Phosphatidylinositol 3-kinase (PI3K)—mammalian Target of Rapamycin (mTOR) axis in GBM with first generation receptor tyrosine kinase (RTK) inhibitors and rapalogs have been disappointing. However, there is reason for renewed optimism given the now very detailed knowledge of the cancer genome in GBM and a wealth of novel compounds entering the clinic, including next generation RTK inhibitors, class I PI3K inhibitors, mTOR kinase inhibitors (TORKinibs), and dual PI3(K)/mTOR inhibitors. This chapter reviews common genetic alterations in growth factor signaling pathways in GBM, their validation as therapeutic targets in this disease, and strategies for future clinical development of kinase inhibitors for high grade glioma.
1 Introduction
Gliomas represent a spectrum of primary brain tumors which are classified by the World Health Organization (WHO) into low grade and high grade tumors based on the degree of tumor cell proliferation, cellular atypia, and microvascular proliferation (Louis et al. 2007). The median survival for patients with GBM has remained below 2 years despite multimodality therapy, including surgery, radiation, chemotherapy (Stupp et al. 2005), and most recently the anti-VEGF antibody bevacizumab (Friedman et al. 2009; Kreisl et al. 2009a). The term “low-grade” glioma (WHO grade II) refers to a group of tumors with histopathologically less aggressive features. However, many patients with these tumors also succumb to their disease within 3–10 years due to tumor “transformation” to an anaplastic glioma (WHO grade III) or GBM (WHO grade IV). GBMs that have evolved from a clinically overt, low-grade precursor lesion are referred to as “secondary” GBMs in contrast to de novo or “primary” GBMs. Primary and secondary GBMs differ substantially in their molecular pathogenesis (Lai et al. 2011; Ohgaki and Kleihues 2007).
The histopathological appearance of GBM is particularly diverse and has earned it the moniker “multi-forme” (multiformis [Latin]: many shapes) (Louis et al. 2007). This morphological heterogeneity of GBM is often viewed as a reflection of the exceptional genetic heterogeneity of this cancer. Recent genomic studies provide a perhaps more encouraging view of GBM with a finite number of highly recurrent gene copy number alterations (Beroukhim et al. 2009) and missense mutations (TCGA 2005; Parsons et al. 2008). Genome wide RNA expression profiling identifies distinct disease subgroups (Phillips et al. 2006) each of which is enriched for particular mutations (Verhaak et al. 2010).
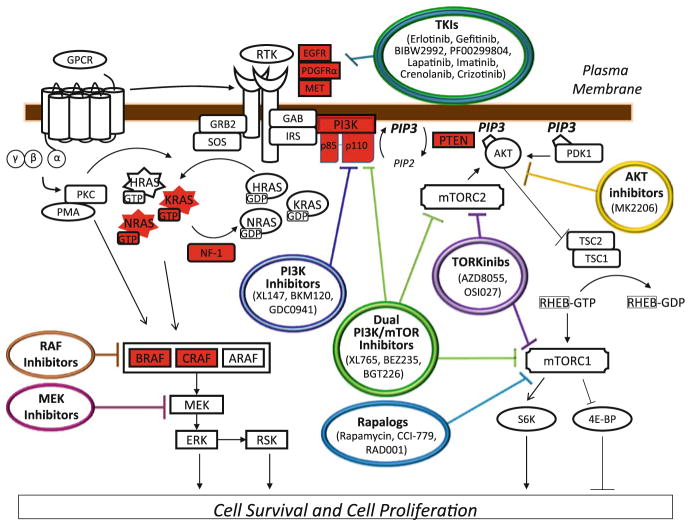
One key result of the extensive profiling of human glioma samples (Beroukhim et al. 2007; Kotliarov et al. 2006; McLendon et al. 2008; Misra et al. 2005; Parsons et al. 2008) is the re-appreciation that nearly all human GBMs harbor genetic alterations in three “core” pathways, namely the RTK/RAS/PI3K signaling axis, the p53-ARF-MDM2/MDM4 pathway, and the RB-CDK4-INK4A pathway. Many of the genetic lesions most consistently found in human tumors have been shown to cooperate in glioma formation in mice (Chow et al. 2011; Reilly et al. 2000; Zheng et al. 2008; Zhu et al. 2005a) and represent the currently most “actionable” drug targets (Fig. 1). This chapter highlights genetic alterations in growth factor signaling pathways in GBM and discusses new directions to develop kinase inhibitors as glioma therapeutics. Our comments largely focus on the adult patient population as genetic alterations (Bax et al. 2010; Paugh et al. 2010) and considerations regarding clinical drug development differ considerably for pediatric glioma patients.
Fig. 1.
The Ras-PI(3)K-mTOR pathway in GBM. Pathway members highlighted in red are mutated in human GBM tumor samples. Pathway inhibitors that have been or will be explored as therapeutics for GBM are indicated
2 Mutations in Growth Factor Receptors
Receptor tyrosine kinases (RTKs) are proteins which transmit signals from the cell surface to the nucleus and participate in most fundamental aspects of cell growth, survival, differentiation, and metabolism. Signaling through RTKs is initiated by ligand binding and terminated by receptor internalization from the cell surface, dissociation of the receptor-ligand complex, receptor dephosphorylation, and degradation of the receptor protein (Lemmon and Schlessinger 2010). The RTK family of proteins includes the epidermal growth factor receptor family (EGFR, HER2, ERBB3, and ERBB4), the platelet-derived growth factor receptor family (PDGFR-α and PDGFR-β), the MET receptor tyrosine kinase, the Vascular-Endothelial Growth Factor Receptor family (VEGFR1/FLT1, VEGFR2/KDR/ FLK1, and VEGFR3/FLT4), and others. Many human cancers harbor mutations in RTKs which relieve auto-inhibitory constraints on the kinase activity or impair the downregulation of the ligand-activated receptor protein (Blume-Jensen and Hunter 2001). Within the RTK family, mutations in the genes encoding EGFR, PDGFR-α, and MET are the most common in high grade gliomas (Table 1 and Fig. 2).
Table 1.
Mutations in Growth Factor Signaling Pathways in GBM (adult patients)
| Gene | Alteration | Frequency in GBM % |
|---|---|---|
| EGFR | Amplification | 35–40 |
| EGFR-variant III (EGFRvIII) | ~20 | |
| Nonsynonymous Mutations | 10–15 | |
| other in-frame deletions/truncations | 5–10 | |
| PDGFRA | Amplification | 5–15 |
| delta-8,9 truncation | 2–5 | |
| Nonsynonymous Mutations | <2 | |
| MET | Amplification | 2–5 |
| Nonsynonymous Mutations | <2 | |
| KRAS | Nonsynonymous Mutations | <2 |
| NRAS | Nonsynonymous Mutations | <2 |
| NF1 | Homozygous Deletion | 2–5 |
| Hemizygous Deletion | 5–10 | |
| Nonsynonymous Mutations | ~15 | |
| BRAF | V600E BRAF | <2 (2–5 in PA) |
| KIAA1549-BRAF fusions | n.d. (60–70 in PA) | |
| p.A598_T599insT BRAF | n.d. (1–3 in PA) | |
| FAM131B-BRAF fusion | n.d. (1–3 in PA) | |
| CRAF | SRGAP3-CRAF fusion | n.d. (1–3 in PA) |
| PIK3CA | Nonsynonymous Mutations | 5–10 |
| Amplification | ~2 | |
| PIK3R1 | Nonsynonymous Mutations | 5–10 |
| PTEN | Homozygous Deletion | 5–10 |
| Hemizygous Deletion | ~70 | |
| Nonsynonymous Mutations | 15–25 |
Please see text for references
n.d not detected, PA Pilocytic Astrocytoma
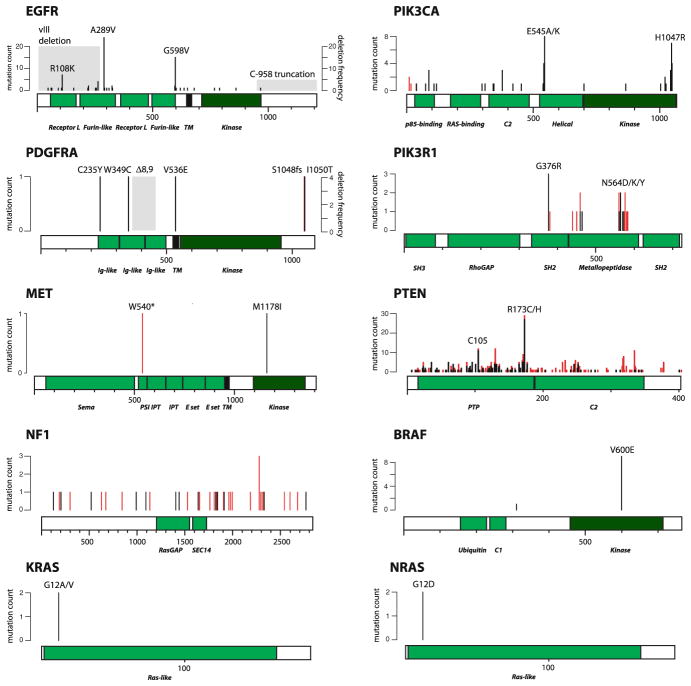
Fig. 2.
Mutations in the Ras-PI(3)K-mTOR axis in WHO grade IV astrocytoma. Shown are all confirmed somatic or previously reported mutations that have been listed in COSMIC v54 under astrocytoma grade IV. The y-axis indicates the number of times a particular mutant has been observed in a primary tumor sample, xenograft tumor, or cell line. Missense mutations are shown in black; nonsense and frameshift mutations as well as small in-frame insertions and deletions are shown in red. Recurrent deletions/truncations in the coding regions of EGFR and PDGFRA are indicated in shaded gray and their estimated frequency is shown as percent of all GBMs (right y-axis)
2.1 Epidermal Growth Factor Receptor (EGFR)
Genetic alterations that result in uncontrolled EGFR kinase activity were amongst the first to be associated with human cancer (Gschwind et al. 2004). A number of alterations involving the EGFR gene have been described in GBM. These include: (a) EGFR gene amplification in ~40% of primary GBMs (Libermann et al. 1985; Wong et al. 1987); extra gene copies reside on double-minutes and are easily detected by fluorescence-in situ hybridization (FISH) (Jansen et al. 2010); (b) In-frame deletions affecting the 5′ end of the EGFR gene (Malden et al. 1988; Yamazaki et al. 1988); these are found mostly, but not exclusively, in tumors with EGFR gene amplification. The most common EGFR variant IIII (or EGFRvIII) is a deletion of exons 2–7, resulting in an 801 amino acid in-frame deletion within the EGFR extracellular domain (Sugawa et al. 1990). The EGFRvIII mutant does not bind the ligands EGF or TGF-α, but is constitutively active (Ekstrand et al. 1994); (c) truncations affecting the C-terminus of the EGFR protein. These alterations are seen in 15–25% of high grade gliomas with EGFR gene amplification (Ekstrand et al. 1992; Eley et al. 1998; Frederick et al. 2000). The EGFR C-terminus encodes receptor portions that are required for ligand-induced receptor internalization (Chen et al. 1989; Decker et al. 1992) and (d) missense mutations in the EGFR extracellular domain in about 10% of primary GBMs (Lee et al. 2006b).
Most EGFR alterations found in human GBM have been shown to represent gain-of-function events. Expression of a truncated EGF receptor lacking the extracellular ligand binding domain induces transformation of immortalized rodent fibroblasts (Haley et al. 1989). Expression of the EGFRvIII mutant enhances the tumorigenicity of GBM cells (Nishikawa et al. 1994) and is able to transform mouse NIH-3T3 fibroblasts in the absence of ligand (Batra et al. 1995). In mice, low grade oligodendrogliomas develop when the retroviral oncogene v-erb-B, encoding a truncated version of EGFR (Schatzman et al. 1986), is expressed under the control of the S100β promoter (Weiss et al. 2003). Expression of the EGFRvIII mutant is not sufficient to induce glioma formation in mice but cooperates with mutant H-RAS or inactivation of CDKN2A in glioma formation (Bachoo et al. 2002; Ding et al. 2003; Holland et al. 1998; Zhu et al. 2009). A C-terminal EGFR truncation mutant has been shown to confer anchorage-independent growth and tumorigenicity to NIH-3T3 cells (Wells et al. 1990; Masui et al. 1991). Several of the extracellular EGFR missense mutations (R108K, T263P, A289V, G598V) have been shown to transform NIH3T3 cells and confer tumorigenicity (Lee et al. 2006b). In contrast to the EGFR mutants described above, overexpression of wild-type EGFR does not transform mouse NIH3T3 in the absence of exogenous EGF (Di Fiore et al. 1987). In mice, overexpression of wild-type EGFR induces glioma formation only in the presence of CDKN2A deletion and, even then, with very low efficiency (Zhu et al. 2009).
The role of mutant EGFR for tumor maintenance needs to be defined more extensively, but current data suggests that at least a subset of EGFR mutant gliomas require EGFR signals for maintenance of the malignant phenotype (Eller et al. 2002; Martens et al. 2008; Sarkaria et al. 2007).
2.2 Platelet-Derived Growth Factor Receptor (PDGFR)
Platelet-derived growth factor (PDGF) is a potent mitogen for glia-derived cells (Richardson et al. 1988) and consists of five dimeric isoforms. These include homodimers of A-, B-, C-, and D-polypeptide chains (i.e., PDGF-AA, -BB, -CC, and –DD) and a PDGF-AB heterodimer. PDGF dimers bind to the RTKs PDGFR-α and PDGFR-β and activate the receptors by inducing receptor dimerization. Different types of receptor dimers are induced by different ligands: A-, B-, and C-chains of PDGF bind to PDGFR-α, whereas B-and D-chains bind to PDGFR-β. PDGF ligands and receptors are frequently co-expressed in human gliomas, perhaps reflecting the presence of an autocrine signaling loop (Hermanson et al. 1992; Lokker et al. 2002).
Mutations in genes encoding PDGF ligands or receptors have been found in a variety of human cancers, including Dermatofibrosarcoma Protuberans (PDGFB), Chronic Myelomonocytic Leukemia (PDGFR-β), and Gastrointestinal Stromal Tumors (PDGFR-α). Many of these cancers respond to PDGFR kinase inhibitors such as imatinib (Ostman and Heldin 2007). Kumabe et al. first reported amplification of the PDGFR-α gene locus in 1/9 (11%) human gliomas. Interestingly, the one case with PDGFR-α gene amplification also harbored an in-frame deletion of exons 8 and 9 of PDGFR-α (Kumabe et al. 1992). Subsequent studies reported PDGFR-α gene amplification in 8–11% of primary human GBMs (Beroukhim et al. 2009; Fleming et al. 1992). Functional characterization of the PDGFR-α-Δ8,9 mutant by Peter Dirk’s laboratory showed that the 243 base pairs deletion results in a constitutively active receptor with greater transforming and tumorigenic ability than wild-type PDGFR-α (Clarke and Dirks 2003). Other PDGFR-α mutations reported in GBM include a two basepair deletion in exon 23, resulting in a truncation of the C-terminus of the receptor (Rand et al. 2005) and an oncogenic gene fusion between the 5′ end of the kinase insert domain receptor (KDR) and the kinase domain and 3′ portion of PDGFR-α (Ozawa et al. 2010) (Table 1).
The contribution of PDGF signaling toward glioma formation in mice has been well documented. Injection of a PDGF-B chain-encoding retrovirus into the brain of newborn C57B16 mice induced brain tumor formation in 40% of animals (Uhrbom et al. 1998). This result has been confirmed by somatic cell type-specific gene transfer experiments in which targeted expression of PDGF-B in nestin-expressing neural progenitors or GFAP-expressing astrocytes induced gliomas in 60 and 40% of mice, respectively (Dai et al. 2001). Retroviral infection of adult white matter progenitor cells similarly resulted in glioma formation (Assanah et al. 2006) and expression of doxycycline-regulated PDGF-B in the spinal cord produced mixed oligoastrocytomas (Hitoshi et al. 2008).
2.3 MET Tyrosine Kinase Receptor
The MET tyrosine kinase is the cell surface receptor for hepatocyte growth factor (HGF). Aberrant activation of MET in human cancers results from amplification of the MET gene (e.g., gastric/esophageal carcinoma, medulloblastoma), missense mutations in the MET gene (e.g., papillary renal cancer), and transcriptional upregulation of MET and its ligand HGF (Comoglio et al. 2008; Koochekpour et al. 1997). While missense mutations in MET are rare in human GBM, focal amplification of the MET gene occurs in about 5% of primary human GBMs (Beroukhim et al. 2009). Amplification of MET has been linked with increased sensitivity to MET kinase inhibition in a panel of human cancer cell lines (McDermott et al. 2007) and radiographic regression of a MET-amplified GBM was recently reported in a patient treated with the MET/ALK kinase inhibitor crizotinib (PF-02341066) (Chi et al. 2011).
3 Mutations in the Ras-Raf Axis
The family of RAS GTPases (HRAS, NRAS, and KRAS) are proteins which cycle between a GTP-bound active form and a GDP-bound inactive form. Guanine nucleotide exchange factors (GEFs) promote formation of GTP-bound RAS, whereas GTPase-activating proteins (GAPs) stimulate the hydrolysis of GTP on RAS. The first critical effector of Ras to be identified in mammalian cells was the RAF-MEK-ERK pathway. Serine/threonine kinases of the RAF family (C-Raf or Raf-1, A-Raf, and B-Raf) bind to RAS-GTP and then relocalize to the plasma membrane where they are phosphorylated. Once activated, they phosphorylate and activate mitogen-activated protein kinase kinase (MAPKK or MEK) that, in turn, phosphorylates and activates extracellular signal-regulated kinases 1 and 2 (ERK1/2) (Castellano and Downward 2011; Shaw and Cantley 2006). The RAS-RAF axis is activated in glioma through several mechanisms, including mutations in NRAS and KRAS, inactivating mutations in the neurofibromatosis gene (NF1), and mutations involving BRAF and CRAF (Table 1 and Fig. 2).
3.1 Mutations in KRAS, NRAS, HRAS
Mutations in KRAS, and NRAS are rare in human gliomas and particularly rare in WHO grade III and IV gliomas in adult patients. Sequencing of 94 high grade gliomas for mutations in NRAS (entire coding region), KRAS (entire coding region) and the first exon of HRAS identified 2 cases (2/94 = 2%) with G12D-NRAS mutation (Knobbe et al. 2004). Another study examined 93 gliomas for “hotspot” mutations in HRAS (exon 2/3), KRAS (exon 2/3), and NRAS (exon 2/3) and reported one NRAS mutation (G10E-NRAS) (Jeuken et al. 2007). Mutations in KRAS have been reported in 5–10% of pediatric gliomas (Cin et al. 2011; Forshew et al. 2009; Janzarik et al. 2007; Maltzman et al. 1997; Schiffman et al. 2010; Sharma et al. 2005). Based on the effects of mutant RAS on glioma formation and maintenance in genetically engineered mouse models (de Vries et al. 2010; Ding et al. 2001; Holland et al. 2000; Holmen and Williams 2005; Marumoto et al. 2009; Uhrbom et al. 2002), Ras mutations are likely relevant for the biology and drug response of the (rare) gliomas in which they are found.
3.2 Mutations in NF1
Neurofibromin, the protein product of the neurofibromatosis gene (NF1) gene, is a p21ras-GTPase activating protein (RasGAP) which critically regulates Ras signal output (Buday and Downward 2008; Shaw and Cantley 2006). Somatic missense mutations in the NF1 gene were first reported in astrocytoma many years ago (Li et al. 1992; Thiel et al. 1995). More complete sequencing of the NF1 coding sequence in a larger number of tumors has uncovered missense mutations in NF1 in ~15% of GBM patients (McLendon et al. 2008; Parsons et al. 2008). Other mechanisms of NF1 silencing in GBM include heterozygous or homozygous NF1 copy loss and posttranslational modifications that result in destabilization of the NF1 protein (McGillicuddy et al. 2009). There is strong evidence for a role of NF1 in gliomagenesis. Patients with neurofibromatosis type I, a genetic disorder caused by germline mutations in NF1, are at increased risk to develop high grade gliomas (Listernick et al. 1999; Rodriguez et al. 2008) and these gliomas often show inactivation of the second NF1 allele (Gutmann et al. 2003). Mice with targeted disruption of the NF1 locus develop astrocytosis and NF1 inactivation cooperates with TP53 inactivation and PTEN inactivation to produce high grade gliomas with rapid onset and high penetrance (Alcantara Llaguno et al. 2009; Bajenaru et al. 2003; Reilly et al. 2000; Zhu et al. 2005a, b).
3.3 Mutations in BRAF
The frequency and type of BRAF alterations in glioma varies substantially between adults and children and between distinct glioma subtypes. V600E-BRAF or V600M-BRAF mutations are rare in WHO grade III/IV gliomas in adults, ranging from 0 to 3% in most studies (Basto et al. 2005; El-Habr et al. 2010; Hagemann et al. 2009; Jeuken et al. 2007; Knobbe et al. 2004; Schindler et al. 2011). The frequency of V600E-BRAF mutations is substantially higher in pediatric gliomas. One series reported V600E-BRAF in 23% (7/31) of WHO grade II–IV pediatric astrocytomas (Schiffman et al. 2010); another series observed V600E-BRAF mutations in 9% (4/42) of WHO grade III/IV pediatric gliomas (Schindler et al. 2011). V600E BRAF mutations are particularly common in the two glioma subtypes pleomorphic xanthoastrocytoma (60–70%) and gangliogliomas (20–60%) (Dougherty et al. 2010; MacConaill et al. 2009; Schindler et al. 2011).
The majority (50–70%) of pilocytic astrocytomas (PAs), the most common central nervous system tumor in children, show low-level copy gain at the BRAF gene locus at 7q34 (Bar et al. 2008; Deshmukh et al. 2008; Pfister et al. 2008). Detailed characterization of this genomic alteration in 44 human PAs by Peter Collins’ group uncovered a tandem duplication that produces fusion proteins between KIAA1549 and BRAF in 66% (29/44) of pilocytic astrocytomas. The most common event fuses KIAA1549 exon 16 and BRAF exon 9. KIAA1549-BRAF fusions generate proteins that lack the BRAF autoregulatory domain and exhibit enhanced BRAF kinase activity (Jones et al. 2008). Further examination of PAs without 7q34 duplication identified alternative mechanisms for activation of the RAF-MEK axis. These included NF1 inactivation (family history of neurofibromatosis type I) (3/44 cases), V600E BRAF mutation (2/44 cases), a tandem duplication at 3p25 fusing SLIT-ROBO Rho GTPase Activating Protein 3 (SRGAP3) and CRAF/RAF1 (1/44 cases), and a 3 bp insertion at codon 598 in BRAF (1/44 cases). The SRGAP3-RAF1 fusion protein and the p.A598_T599insT BRAF mutant both increased the kinase activity of RAF1 and BRAF, respectively. Altogether, 82% (36/44) of pilocytic astrocytomas in this series showed mutational activation of the RAS/RAF axis (Jones et al. 2008, 2009).
Profiling of human pilocytic astrocytomas by other groups confirmed near-universal activation of the RAS/RAF/MEK signaling axis in this disease through KIAA1549-BRAF fusions (~70%), V600E BRAF mutations (5–10%), BRAF codon 598 insertions (1–3%), NF1 inactivation (1–3%), KRAS mutations (1–3%), the SRGAP3-RAF1 fusion (1–3%), and a recently described fusion involving BRAF and Family with sequence similarity 131, member B (FAM131B) (2%) (Cin et al. 2011; Eisenhardt et al. 2010; Forshew et al. 2009; Jacob et al. 2009; Pfister et al. 2008; Schindler et al. 2011; Sievert et al. 2009; Yu et al. 2009). Functional studies support a role of glioma-related BRAF mutants and activated Raf-1 in transformation and gliomagenesis, in particular in combination with CDKN2A inactivation (Gronych et al. 2011; Jones et al. 2008, 2009; Lyustikman et al. 2008; Robinson et al. 2010, 2011).
4 Mutations in PI3K and PTEN
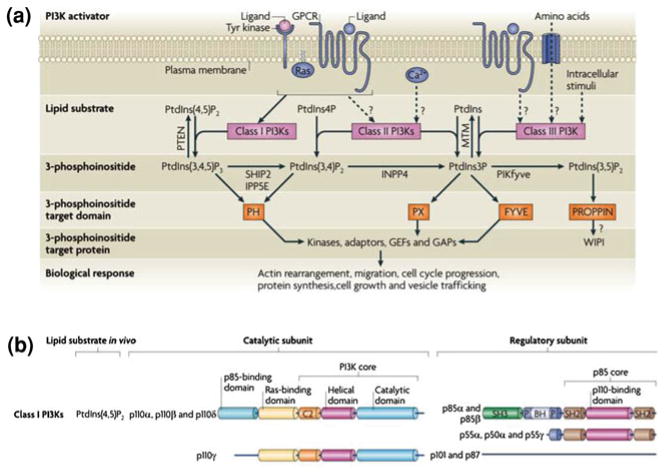
Phosphatidyloinositide 3-kinases (PI3K) belong to a family of lipid kinases that phosphorylate the 3-hydroxyl group of the inositol ring of phosphatidylinositol (PtdIns), PtdIns4P, and PtdIns (4, 5)P2. PI3Ks have been assigned to different classes based on substrate preference and structural features. Class I PI(3)Ks are activated by receptor tyrosine kinases and G-protein coupled receptors and use PtdIns(4, 5)P2 as substrate to generate phosphoinositide 3,4,5 trisphosphate (PIP3). Other classes are class II and class II PI3Ks (Fig. 3a) and the family of PI3K-related protein kinases (PIKKs) which include mammalian Target of Rapamycin (mTOR), ATM, ATR, DNA-PK, and hSMG1 (Vanhaesebroeck et al. 2010).
Fig. 3.
The 3-phosphoinositide lipid network. (a) Following activation by upstream agonists, phosphoinositide 3-kinases (PI3Ks) generate phosphatidylinositol-3,4,5-trisphosphate (PtdIns(3,4,5)P3), PtdIns-3,4-bisphosphate (PtdIns(3,4)P2) and PtdIns-3-phosphate (PtdIns3P). These lipids interact with lipid binding domains in PI3K effector proteins and change their localization and/or activity. Lipid phosphatases degrade or interconvert 3-phosphoinositides. These include lipid phosphatases for PtdIns(3,4,5)P3, such as phosphatase and tensin homologue deleted on chromosome 10 (PTEN), inositol polyphosphate-5-phosphatase E (IPP5E), and SH2 domain-containing inositol 5-phosphatase type 2 (SHIP2). GAP, GTPase-activating protein; GEF, guanine nucleotide exchange factor; GPCR, G protein-coupled receptor. (b) Classification and domain structure of mammalian Class I PI3Ks. All PI3K catalytic subunits have a PI3K core structure consisting of a C2 domain, a helical domain and a catalytic domain. Class I PI3Ks exist in complex with a regulatory subunit, either a p85 isoform (for p110α, p110β and p110δ) or p101 or p87 (for p110γ). All p85 isoforms have two Src homology 2 (SH2) domains and are encoded by either PIK3R1 (which encodes p85α, p55α and p50α), PIK3R2 (which encodes p85β) and PIK3R3 (which encodes p55γ). Figure modified from (Vanhaesebroeck et al. 2010)
PI(3)Ks are composed of a catalytic subunit and a regulatory subunit (Fig. 3b). Catalytic subunits include p110α (encoded by PIK3CA), p110β (encoded by PIK3CB), p110γ (encoded by PIK3CG), and p110δ (encoded by PIK3CD). Regulatory subunits include p85α, p55α, and p50α (all encoded by PIK3R1), p85β (encoded by PIK3R2), p55γ (encoded by PIK3R3), p101 and p87. p110 subunits share a five-domain structure, which includes an N-terminal adaptor binding domain (ABD domain), a Ras binding domain (RBD domain), a C2 (Protein kinase C homology-2) domain, a helical domain, and a catalytic domain. All p85 isoforms (p85α, p55α, p50α, p85β, p55γ) have two Src homology domains 2 (SH2) domains and an intervening domain (iSH2) that binds to the adapter binding domain in p110 (Vanhaesebroeck et al. 2010). P85 isoforms provide at least three functions to p110 proteins: (i) they stabilize the intrinsically unstable p110 protein, (ii) they recruit p110 proteins to pTyr residues in receptor and adaptor molecules (through the SH2 domains of p85 isoforms) upon activation, and (iii) they restrain the kinase activity of p110 proteins in their un-activated state.
PI3K were first linked to cancer through the study of oncogenic viruses. Their critical role in the pathogenesis of human cancer was fully recognized following the discovery that many human tumors harbor mutations in genes whose gene products regulate levels of the lipid molecule PIP3 (Shaw and Cantley 2006; Vogt et al. 2009). These include the phosphatase and tensin homolog deleted on chromosome 10 (PTEN) (Li et al. 1997; Steck et al. 1997), the regulatory subunit p85 (PIK3R1) (Philp et al. 2001), and the catalytic subunit p110α (PIK3CA) (Samuels et al. 2004), and PIK3R2 (Cheung et al. 2011). Almost 100 mutations have been found throughout the PIK3CA coding region. The Ras binding domain (RBD) appears to be spared from mutations and mutation “hot-spots” include the p85-binding domain, the helical domain, and the kinase domain (Vanhaesebroeck et al. 2010; Vogt et al. 2009). Many studies have documented the biochemical effects and transforming ability of PTEN-inactivation (Salmena et al. 2008) and various PIK3CA (Vogt et al. 2009) and PIK3R1 mutations (Huang et al. 2007; Jaiswal et al. 2009; Philp et al. 2001; Sun et al. 2010; Wu et al. 2009). Mutations have not been found in genes encoding other PI3K catalytic subunits (PIK3CB, PIK3CG, PIK3CD). In contrast to p110α, however, which only shows transforming ability it its mutant form, overexpression of wild-type p110β, p110γ, and p110δ can transform chicken fibroblasts (Kang et al. 2006) and these PI3K family members may represent critical oncogenic units in certain genetic contexts (Berenjeno and Vanhaesebroeck 2009). In GBM, mutations in the PI(3)K-mTOR axis are most frequently found in the genes encoding the catalytic and regulatory subunit of PI(3)K (PIK3CA and PIK3R1, respectively) and in PTEN (Table 1 and Fig. 2).
4.1 Mutations in PIK3CA
The initial discovery of PIK3CA mutations in cancer reported a mutation prevalence of 26.7% (4/15) for GBM (Samuels et al. 2004). Subsequent studies in larger numbers of tumors suggest that these mutations occur less frequently in primary GBMs. Sequening of exons 9 and 20 of PIK3CA in a diverse panel of primary brain tumors found mutations in 5/105 (5%) GBMs, 3/21 (14%) anaplastic oligodendrogliomas (WHO grade III), 1/31 (3%) anaplastic astrocytomas (WHO grade III), 0/24 (0%) WHO grade II astrocytomas, 4/78 (5%) medulloblastomas, and 0/26 (0%) ependymomas (Broderick et al. 2004). Other studies screened the entire PIK3CA coding region and reported mutations in 0/30 (0%) (Mueller et al. 2005) and 5/70 (7%) GBMs (Hartmann et al. 2005). Direct sequencing of a large part of the PIK3CA coding sequence (exons 1, 2, 4, 5, 7, 9, 12, 13, 18, 20) in 38 primary human GBMs identified mutations in 3/14 (21%) pediatric and 4/24 (17%) adult GBMs (Gallia et al. 2006). Another study focused on exons 9 and 20 of PIK3CA and reported mutations in 5/107 (5%) de novo GBMs and in 1/32 (3%) secondary GBMs (Kita et al. 2007). Differences between studies in the reported frequency of PIK3CA mutations are likely due to a combination of factors, including the sensitivity of the mutation detection assay (single strand conformational polymorphism versus direct bidirectional sequencing), gene coverage (mutation hot-spots versus entire coding sequence), glioma subtype, sample size, and inclusion of cell lines.
Amplification of the PIK3CA gene locus was first reported as oncogenic event in ovarian cancer (Shayesteh et al. 1999) and later in other cancers. No evidence for PIK3CA gene copy gain (>5-fold) or RNA overexpression was found in an analysis of 145 primary brain tumors (including 50 GBMs and 35 WHO grade III gliomas) (Broderick et al. 2004). Using a slighty less stringent cutoff for PIK3CA gene copy gain (>3-fold), Kita et al. reported PIK3CA amplifications in 14/107 (13%) de novo and 3/32 (9%) secondary GBMs (Kita et al. 2007).
4.2 Mutations in PIK3R1
A truncating mutation in PIK3R1 was first reported in a GBM in 2004 (Mizoguchi et al. 2004). More recent studies reported mutations in PIK3R1 in~10% of GBMs (McLendon et al. 2008; Parsons et al. 2008). These mutations clustered in the N-terminal SH2-domain and the inter-SH2 domain of the PIK3R1 gene, regions that interact with the C2 and helical domains of p110α. Detailed characterization of selected p85 mutants showed that they were unable to negatively regulate p110α, p110β, and p110δ activity, despite retaining their ability to bind and stabilize them (Huang et al. 2007; Jaiswal et al. 2009; Wu et al. 2009). Peter Vogt’s group expressed the full panel of GBM p85 mutants (G376R, E439del, KS459delN, D560Y, DKRMNS560del, N564K, R574fs, T576del, W583del) in chicken fibroblasts. All mutants induced a transformed phenotype, retained the ability to interact with p110α, stabilized the endogenous p110α protein, and stimulated phosphorylation of Akt and 4EBP-1. Efficiencies of transformation varied between mutants and did not clearly correlate with their biochemical effects on Akt and 4EBP1 (Sun et al. 2010).
4.3 Mutations in PTEN
Allelic loss of chromosome 10 has long been known to be common in high grade human glioma (James et al. 1988). After the identification of PTEN as tumor suppressor on chromosome 10 (Li et al. 1997; Steck et al. 1997), missense mutations in PTEN were reported in 17/63 (27%) of GBMs (Liu et al. 1997). Examination of the entire PTEN coding sequence in 331 gliomas reported PTEN missense mutations in 20/142 (14%) GBMs; homozygous PTEN deletions were observed in 7/142 (5%) GBMs (Duerr et al. 1998). Another study reported PTEN point mutations and homozygous deletions in 32/110 (29%) and 11/110 (10%) of GBMs (Smith et al. 2001). Later studies reported PTEN mutations in 14 (Hartmann et al. 2005) to 24% (Ohgaki et al. 2004). PTEN expression is silenced in additional GBMs through promoter methylation, micro-RNAs, and posttranslational modifications.
Genetically engineered mouse models (GEMMs) support a prominent tumor suppressor function of PTEN during glioma progression. Loss of PTEN in various cell types within the brain does not result in tumorigenesis (Fraser et al. 2004; Groszer et al. 2001; Kwon et al. 2001). Inactivation of PTEN in glioma-prone mice, in which oncogenic V12H-Ras is expressed from the GFAP promoter, greatly accelerated malignant glioma progression (Wei et al. 2006). In the NF1/p53 astrocytoma model, haploinsufficiency of PTEN accelerated formation of WHO grade III astrocytomas, whereas loss of PTEN heterozygosity coincided with progression into WHO grade IV tumors (Kwon et al. 2008). Combined deletion of PTEN and TP53 in neural stem cells of the subventricular zone is sufficient to induce glioma formation (Jacques et al. 2010; Zheng et al. 2008). Combined inactivation of PTEN and TP53 in mature astrocytes in the adult brain induced high grade gliomas that showed striking similarity to human GBMs in terms of secondary gene copy number alterations and genome wide RNA expression changes (Chow et al. 2011).
5 Experience with First-Generation RTK Inhibitors
The success with first-generation EGFR kinase inhibitors (gefitinib, erlotinib) in lung cancer and the high frequency of oncogenic EGFR alterations in GBM (~40%) raised expectations that these agents will show activity against GBM. This expectation has largely not been fulfilled. While most studies reported individual patients with tumor regressions in response to erlotinib or gefitinib, the frequency of these response is substantially lower (<5%) than the frequency of oncogenic EGFR alterations in GBM (Mellinghoff et al. 2011). The experience with imatinib, an ATP-site competitive inhibitor of the PDGFR, KIT and ABL-kinases has been similarly disappointing. A Phase II study of imatinib in 112 patients with recurrent gliomas reported a partial response for only 5 patients (Raymond et al. 2008). The radiographic response rates for the subgroup of GBM patients was 3/51 (6%) in this study. In a Phase I/II study of the North American Brain Tumor Consortium 3/57 (5%) GBM patients showed partial radiographic responses and no responses were observed for other histologic subgroups (Wen et al. 2006).
In contrast to the landmark clinical trials with imatinib in chronic myeloid leukemia (Druker et al. 2001) and with the HER2 antibody trastuzumab in breast cancer (Slamon et al. 2001), clinical trials with first-generation EGFR and PDGFR kinase inhibitors in GBM were not enriched for patients whose tumors harbored mutations in EGFR and PDGFR-α, respectively. This lack of molecular preselection likely contributed to the disappointing results in the clinic. Studies to identify determinants of EGFR kinase inhibitor response in GBM have associated tumor regressions with the presence of oncogenic EGFR alterations (EGFR gene amplification or EGFRvIII) and expression of the PTEN tumor suppressor protein or low p-AKT levels (Haas-Kogan et al. 2005; Mellinghoff et al. 2005). Other studies have questioned the relationship between oncogenic EGFR and EGFR kinase inhibitor response in GBM (van den Bent et al. 2009). This discrepancy may, at least in part, be attributable to differences in the determination of EGFRvIII status in tumor tissue.
Several lines of evidence support that PTEN inactivation mediates EGFR kinase inhibitor resistance: (1) PTEN restoration sensitizes EGFR amplified/PTEN deleted cancer cells to cell death induction by EGFR kinase inhibitors (Bianco et al. 2003); (2) PTEN knockdown is sufficient to render EGFR mutant cancer cells resistant to cell death induction by EGFR kinase inhibitors (Vivanco et al. 2010); and (3) PTEN emerged as most consistent resistance factor from a shRNA library screen performed in breast cancer cells to identify mechanisms of resistance to trastuzumab (which targets the EGFR co-receptor HER2) (Berns et al. 2007), and (4) GBMs with intact PTEN expression showed enhanced responsiveness to combination therapy of erlotinib plus temozolomide (Prados et al. 2009).
How does PTEN status influence EGFR kinase inhibitor response? By dephosphorylation the second messenger PIP3, PTEN plays an important role in signal termination downstream of many RTKs. Even if one RTK is effectively inhibited, PTEN loss may allow the accumulation of sufficient PIP3 to activate PIP3 effector molecules, such as the serine-threonine kinase Akt. In other words, PTEN deficient cells may be able to better compensate for single RTK inhibition than cells with intact PTEN. This concept of redundant RTK activation is supported by the biochemical evidence for RTK coactivation in primary GBM tumor samples and the experimental observation that inhibition of multiple RTK, but not a single RTK, induces cell death in certain GBM lines (Stommel et al. 2007). We recently made the surprising discovery that PTEN inactivation raises EGFR protein levels and EGFR kinase activity by interfering with the CBL-mediated downregulation of the activated EGF receptor. Of note, PTEN knockdown did not confer “absolute” EGFR kinase inhibitor resistance but instead right-shifted the cell-death response to EGFR kinase inhibition toward drug concentrations which are difficult to achieve in the central nervous system with currently available EGFR kinase inhibitors (Vivanco et al. 2010). Further studies are needed to determine whether more complete EGFR kinase inhibition, simultaneous inhibition of multiple RTKs, or both are required to overcome EGFR kinase inhibitor resistance in GBM.
In many ways, the current experience with EGFR and PDGFR inhibitors is reminiscent of the experience in melanoma where the disappointing clinical activity of RAF and MEK inhibitors questioned the role of mutant BRAF for the maintenance of these tumors (Smalley and Sondak 2010). The vast majority of clinical trials with RTK inhibitors in neurooncology did not include tumor biopsies during treatment and it is hence unknown to what extent a negative clinical trial result might be attributable to poor drug penetration into the brain tumor. The available data indeed suggests that first-generation RTK inhibitors, given at standard daily doses, result in only weak (if any) pathway inhibition in GBM. Fresh operative samples from three GBM patients who received erlotinib or gefitinib prior to surgery and for whom a frozen pretreatment sample was available showed inconsistent drug effects on phosphorylation of EGFR, Erk, and Akt (Lassman et al. 2005). A more extensive analysis of multiple candidate EGFR effector molecules (p-AKT, p-GSK-3α/β, p-NFκB p65, p-STAT3, p-ERK1/2, p-MEK1, p-p38MAPK, p-p90RSK, p-p70S6 Kinase, p-S6 ribosomal Protein, p-PDGFR-B, and p-SRC) in GBM patients treated with gefitinib similarly showed poor EGFR pathway inhibition (Hegi et al. 2011).
Conclusions regarding target inhibition in GBM have to be viewed as preliminary because tumors with the most informative genotype(s) and strong basal pathway activation were generally underpresented in these studies and because of difficulties to assemble a sufficiently large number of drug-naive “control” tumor samples. Genotype-enriched clinical trials with a surgical arm cohort (Cloughesy et al. 2008) could address this important issue and may be instrumental to “weed out” compounds that fail to achieve sufficient kinase inhibition in GBM tumor tissue and therefore do not warrant further clinical testing in GBM.
6 Clinical Experience with Rapalogs
The PI(3)K pathway represents a very active area of drug development in cancer. Strategies to target members of this pathway are of particular interest in GBM because the majority of these tumors harbor mutations in one or more genes that regulate this pathway (McLendon et al. 2008), including EGFR, PDGFRA, MET, PTEN, NF1, PIK3CA and PIK3R1 (Fig. 1). Biochemical evidence for frequent PI(3)K pathway activation in GBM is provided by immunohistochemical studies which showed phosphorylation of the downstream pathway member S6 ribosomal protein in the majority of these tumors (Choe et al. 2003). The optimal strategy to target the PI(3)K pathway in glioma and other human cancers is currently unclear. Options include inhibitors of PI(3)K, the serine-threonine kinase Akt, the mammalian target of rapamycin (mTOR), and a combination thereof (Workman et al. 2010).
The first member of the PI(3)K pathway for which a clinical grade inhibitor became available was mTOR which exists as a member of two distinct protein complexes called complex I (mTORC1) and complex 2 (mTORC2). mTORC1 responds to a variety of stimuli (growth factors, changes in amino acid availability, energy status, oxygen levels, DNA damage) and promotes protein translation by phosphorylating p70 ribosomal S6 kinase (S6K) (Threonine 389) and eukaryotic initiation factor 4E-binding protein (4EBP) (Threonine 37/46). mTORC2 functions are incompletely understood and include phosphorylation of Akt (Serine 473), serum glucocorticoid-induced kinase (SGK), and specific Protein Kinase C isoforms (Mendoza et al. 2011; Sengupta et al. 2010). The natural product rapamycin inhibits mTORC1 functions allosterically by binding with high affinity to the immunophilin FK506-binding protein-12 (FKBP12).
The potent antiproliferative activity of rapamycin against PTEN-deficient tumor models (Neshat et al. 2001; Podsypanina et al. 2001) motivated a clinical trial with single-agent rapamycin for patients with PTEN-deficient, recurrent GBM. Screening for PTEN protein expression was performed by immunohistochemistry on the specimen from the initial tumor resection and rapamycin was given for 1–2 weeks prior to resection of recurrent tumor. Drug levels of rapamycin were measured in blood and tumor tissue and inhibition of mTOR was evaluated with phosphosite-specific antibodies against the S6K substrate S6 Ribosomal Protein and 40 kDa proline-rich AKT substrate (PRAS40). Inhibition of tumor cell proliferation by rapamycin correlated with mTOR pathway inhibition. About half the tumors showed increased phosphorylation of the Akt substrate PRAS40 during rapamycin treatment and PRAS40 hyperphosphorylation was associated with poor clinical outcome (Cloughesy et al. 2008). Akt activation during treatment with rapalogs has been observed in other cancers and has been attributed to de-inhibition of a negative feeback loop between the mTORC1 substrate S6K1 and adaptor protein insulin receptor substrate (IRS-1) (O’Reilly et al. 2006; Sun et al. 2005). Reactivation of the PI3-K pathway, and also the MAPK pathway (Carracedo et al. 2008), may contribute to the overall disappointing clinical activity of rapalogs in high grade glioma (Chang et al. 2005; Cloughesy et al. 2008; Galanis et al. 2005) and other human cancers (Dancey 2010).
Since PTEN-loss has been associated with EGFR kinase inhibitor resistance and mTOR represents an important effector protein downstream of PI(3)K, combined blockade of EGFR andmTOR might overcome this resistance. In GBM cell lines and other preclinical models, rapamycin indeed showed synergism with the EGFR kinase inhibitor erlotinib (Buck et al. 2006; Wang et al. 2006). However, the combination of rapalogs and first-generation EGFR kinase inhibitors in patients with recurrent GBM was complicated by toxicity requiring substantial dose reductions and failed to show compelling clinical activity (Kreisl et al. 2009b; Reardon et al. 2010).
7 New Approaches to Target the PI(3)K-mTOR Axis
The observed PI(3)K pathway activation in response to rapalogs suggests that inhibitors of PI(3)K (e.g., XL147, GDC-0941, GSK1059615, ZSTK474, PX866) or dual PI(3)K/mTOR inhibitors (e.g., XL765, SF1126, BEZ235, GSK1059615) might accomplish superior pathway blockade and biological activity. Many such inhibitors have been synthesized and have shown broad antiproliferative activity in preclinical GBM models (Fan et al. 2006; Guillard et al. 2009; Shuttleworth et al. 2011). The majority of first generation PI(3)K inhibitors entering the clinic block all members of the class I PI(3) K family and appear to be surprisingly well tolerated (Shuttleworth et al. 2011) despite the central role of individual class I PI(3)Ks in glucose metabolism and immune function (Okkenhaug et al. 2002; Sasaki et al. 2000). Determination of their clinical activity in GBM is eagerly awaited.
Whether more selective inhibition of individual class I PI(3)Ks will widen the therapeutic window of these agents without compromising their activity (Foukas et al. 2010), is an open question. A screen of six GBM cell lines with a panel of chemotypically diverse and isoform-selective inhibitors of the PI(3)K family demonstrated that inhibitors of p110α or p110β were able to inhibit phosphorylation of Akt, but only p110α inhibitors induced proliferative arrest. The PI3K isoforms p110δ and p110γ were not expressed in these cells and inhibitors of p110β and p110δ had no effects on proliferation (Fan et al. 2006). More recent studies identified a critical role for p110β in certain PTEN-deficient malignancies (Jia et al. 2008; Wee et al. 2008). Further work is needed to determine which PI3K isoforms are most critical for tumor maintenance in GBM as many of these tumors harbor multiple lesions within the RTK/Ras/PI(3)-signaling axis (e.g., EGFR mutation and PTEN loss).
The only modest activity of rapalogs against many human cancers may, at least in part, be due to the fact that these drugs do not effectively block many functions of mTORC2 and mTORC1. This shortcoming could be overcome by a new class of TOR kinase domain inhibitors (also called TORKinibs). These “second-generation” mTOR inhibitors have shown compelling antiproliferative activity in a range of preclinical cancer models and have also advanced to clinical testing in GBM and other cancer types (Feldman and Shokat 2010; Liu et al. 2009a).
8 Leveraging the Cancer Genome Atlas for Clinical Drug Development
The clinical experience with kinase inhibitors as cancer therapeutics suggests that these drugs are most effective in patients whose tumors harbor gain-of-function mutations in the targeted kinase. Examples include BCR-ABL mutant leukemia, KIT- or PDGFRA-mutant sarcoma, EGFR or ALK mutant lung cancer, and BRAF or KIT mutant melanoma (Sawyers 2009). While the presence of an oncogenic mutation clearly does not guarantee response to the corresponding kinase inhibitor, the relationship between tumor genotype and drug response is compelling enough that mutational profiling is now routinely performed in the clinic for several human cancers (e.g., lung cancer, melanoma, colorectal cancer) and results incorporated into treatment decisions (Gerber and Minna 2010). A link between tumor genotype and drug response still remains to be proven for GBM, but many centers have begun to prospectively profile gliomas for the most commonly found alterations (e.g., EGFR gene amplification, MGMT methylation, IDH1/2 mutations, 1p19q deletion) (Jansen et al. 2010; Riemenschneider et al. 2010). The extent of profiling is currently driven by institutional expertise, assay cost, and tissue availability but is likely to increase in the near future as newer profiling platforms yield robust results from formalin-fixed and paraffin-embedded clinical specimen.
While the impact of tumor profiling on disease classification and treatment in GBM remains to be firmly established, currently its greatest utility may be for clinical drug development. A logistic obstacle toward genotype-focused clinical development in GBM is the low absolute number of patients with tumors of a particular tumor genotype. As discussed above, many presumed “driver” mutations in GBM (e.g., PDGFRA, PIK3R1, PIK3CA, BRAF, MET) occur in only a small subset of patients and a substantial number of these patients will be ineligible for clinical trial participation due to disease-related morbidity. By determining the distribution of mutations within a large number of patients with primary GBMs, TCGA (2005) has provided valuable information for clinical trial planning. For example, mutations in EGFR and PDGFRA are rarely found in the same tumor (Fig. 4) and one could envision using prospective genotyping information to assign patients to clinical trials targeting either of these lesions. Genotype-enriched trials for other presumed “driver” mutations, such as MET gene amplification, PIK3CA mutations, or PIK3R1 mutations will require screening of a substantially larger number of patients as these mutations are even less common. Based on the distribution of mutations throughout the coding sequence of various pathway members (Fig. 2), perhaps a combination of phosphoproteomic “pathway activation” measurements (Solit and Mellinghoff 2010) and selected genotyping for mutation hotspots (Jansen et al. 2010) may represent the most cost-effective screening approach until predictive biomarkers have been properly validated.
Fig. 4.

Potential strategy for genotype-enriched clinical trials in GBM. Tumor tissue collected during the initial surgery (“GBM Diagnosis”) is profiled to identify candidate “driver” mutations and clinical trial participation at the time of tumor recurrence (i.e., following standard “Upfront Therapy”) is guided by the molecular profiling results. Shown is the distribution of mutations in 138 GBMs which were profiled by the TCGA (2005) and for whom complete Sanger Sequencing and Gene Copy Number data is available
9 Future Perspective
The clinical development of PI(3)Kinhibitors for GBM would be greatly accelerated by enrichment of clinical trials for patients whose tumors that are more likely to respond. While several ATP-site competitive pan-class I PI(3)K inhibitors have shown antiproliferative activity against a broad panel of human GBM cell lines (Fan et al. 2006; Guillard et al. 2009; Koul et al. 2010; Liu et al. 2009b; Prasad et al. 2011), it is not clear whether any disease subgroup might exhibit a degree of PI(3)K pathway “addiction” associated with tumor cell death and tumor regressions in other cancers. In breast cancer, for example, such PI(3)K pathway addiction appears to exist for tumors with PIK3CA mutation and HER2 gene amplification as evidenced by cell death induction in response to pan-class I PI(3)K inhibitors (Ihle et al. 2009; Junttila et al. 2009; Mallon et al. 2010; O’Brien et al. 2010; Serra et al. 2008), Akt inhibitors (She et al. 2008), and the emerging class of mTOR kinase domain inhibitors (Weigelt et al. 2011). Lung cancer cell lines harboring EGFR kinase mutations, on the other hand, do not appear to depend on PI(3)K signals for survival (Faber et al. 2009). More extensive testing of novel compounds in genetically faithful glioma models (Hambardzumyan et al. 2011) and cell lines (Lee et al. 2006a) will provide the foundation to formulate hypotheses for genotype-enriched clinical trials.
Different dosing schedules (e.g., intermittent or “pulsatile” dosing) (Shah et al. 2008) and isoform-specific (e.g., PI3K) or mutant-specific (e.g., BRAF) compounds may increase the therapeutic window of individual kinase inhibitors. Ultimately, however, a combination of agents may be required to achieve clinically meaningful tumor regressions. Such therapies may include: (i) a combination of multiple selective RTK inhibitors, (ii) the simultaneous inhibition of multiple members within the same signaling pathway (e.g., PI3K and mTOR), (iii) a multipronged attack on key oncoproteins through distinct pharmacological approaches (e.g., RTK antibody plus RTK kinase inhibitor or HSP90 inhibitor plus RTK inhibitor), or (iv) a combination of agents based on synthetic lethal relationships between signaling pathways (Kaelin 2005). These strategies warrant rigorous testing in genetically faithful preclinical models.
One example for a rational combination therapy is the combination of autophagy inhibitors with PI(3)K-mTOR inhibitors. Autophagy is a process of “self-degradation” of cellular components which can provide substrates for energy production during periods of low extracellular nutrients. mTORC1 suppresses autophagy by blocking autophagosome initiation via phosphorylation of ATG13 and ULK1 (Zoncu et al. 2011). Conversely, inhibition of mTORC1 by rapamycin (Takeuchi et al. 2005) or dual pan-class I PI(3)K/mTOR inhibitors (Guillard et al. 2009; Liu et al. 2009b) induces autophagy in GBMcells. Induction of autophagy may represent an important survival mechanism during mTOR inhibitor therapy and explain why inhibitors of PI3K and mTOR induce growth arrest without cell death. Consistent with that model, recent studies show that inhibitors of autophagy can synergize with PI3K-mTOR inhibitors to induce apoptosis (Fan et al. 2010; Xu et al. 2011), representing an opportunity for mechanism-based combination therapy.
GBM has long been known as a disease with mutations in growth factor signaling pathways. The full extent of these alterations and their relationship to each other has emerged more clearly through The Cancer Genome Atlas. Despite discouraging clinical results with first-generation RTK inhibitors and rapalogs, there is considerable optimism that new RTK inhibitors, “second-generation” mTOR inhibitors, and compounds inhibiting class I PI(3)Ks will be better suited to effectively shut down critical signaling nodes in GBM. As the number of clinical grade inhibitors continues to grow, it will become increasingly important to develop an approach to clinical drug development which fully leverages our knowledge of the GBM cancer genome, lessons from kinase inhibitor therapy in other human cancers, and the ability to extract robust molecular information from small amounts of routinely collected human tumor tissue.
Acknowledgments
We thank Ms. Farzeen Aslam for secretarial assistance. This work was supported through NIH U54 CA143798 and R21 CA137896 (IKM), NIH/NS 73831 (PSM), the Leon Levy Foundation (IKM), the Doris Duke Charitable Foundation (IKM), the Sontag Foundation (IKM), the James S. McDonnell Foundation (IKM), and an Advanced Clinical Research Award in Glioma from the American Society of Clinical Oncology (IKM). TFC acknowledges funding support by Art of the Brain, the Ziering Family Foundation in memory of Sigi Ziering, the Singleton Family Foundation, the Clarence Klein Fund for Neuro-Oncology, and the John W. Carson Foundation.
Contributor Information
Ingo K. Mellinghoff, Email: mellingi@mskcc.org, Human Oncology and Pathogenesis Program, Department and Neurology, Memorial Sloan-Kettering Cancer Center, New York, NY, USA
Nikolaus Schultz, Computational Biology Program, Memorial Sloan-Kettering Cancer Center, New York, NY, USA.
Paul S. Mischel, Department of Pathology, University of California Los Angeles, Los Angeles, CA, USA
Timothy F. Cloughesy, Department of Neurology, University of California Los Angeles, Los Angeles, CA, USA
References
- Alcantara Llaguno S, Chen J, Kwon CH, Jackson EL, Li Y, Burns DK, Alvarez-Buylla A, Parada LF. Malignant astrocytomas originate from neural stem/progenitor cells in a somatic tumor suppressor mouse model. Cancer Cell. 2009;15:45–56. doi: 10.1016/j.ccr.2008.12.006. S1535-6108(08)00409-[pii] 10.1016/ j.ccr.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assanah M, Lochhead R, Ogden A, Bruce J, Goldman J, Canoll P. Glial progenitors in adult white matter are driven to form malignant gliomas by platelet-derived growth factor-expressing retroviruses. J Neurosci. 2006;26:6781–6790. doi: 10.1523/JNEUROSCI.0514-06.2006. 26/25/6781[pii] 10.1523/JNEUROSCI. 0514-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachoo RM, Maher EA, Ligon KL, Sharpless NE, Chan SS, You MJ, Tang Y, DeFrances J, Stover E, Weissleder R, Rowitch DH, Louis DN, DePinho RA. Epidermal growth factor receptor and Ink4a/Arf: convergent mechanisms governing terminal differentiation and transformation along the neural stem cell to astrocyte axis. Cancer Cell. 2002;1:269–277. doi: 10.1016/s1535-6108(02)00046-6. [DOI] [PubMed] [Google Scholar]
- Bajenaru ML, Hernandez MR, Perry A, Zhu Y, Parada LF, Garbow JR, Gutmann DH. Optic nerve glioma in mice requires astrocyte Nf1 gene inactivation and Nf1 brain heterozygosity. Cancer Res. 2003;63:8573–8577. [PubMed] [Google Scholar]
- Bar EE, Lin A, Tihan T, Burger PC, Eberhart CG. Frequent gains at chromosome 7q34 involving BRAF in pilocytic astrocytoma. J Neuropathol Exp Neurol. 2008;67:878–887. doi: 10.1097/NEN.0b013e3181845622. 10.1097/ NEN.0b013e3181845622. [DOI] [PubMed] [Google Scholar]
- Basto D, Trovisco V, Lopes JM, Martins A, Pardal F, Soares P, Reis RM. Mutation analysis of B-RAF gene in human gliomas. Acta Neuropathol. 2005;109:207–210. doi: 10.1007/s00401-004-0936-x. 10.1007/ s00401-004-0936-x. [DOI] [PubMed] [Google Scholar]
- Batra SK, Castelino-Prabhu S, Wikstrand CJ, Zhu X, Humphrey PA, Friedman HS, Bigner DD. Epidermal growth factor ligand-independent, unregulated, cell-transforming potential of a naturally occurring human mutant EGFRvIII gene. Cell Growth Differ. 1995;6:1251–1259. [PubMed] [Google Scholar]
- Bax DA, Mackay A, Little SE, Carvalho D, Viana-Pereira M, Tamber N, Grigoriadis AE, Ashworth A, Reis RM, Ellison DW, Al-Sarraj S, Hargrave D, Jones C. A distinct spectrum of copy number aberrations in pediatric high-grade gliomas. Clin Cancer Res. 2010;16:3368–3377. doi: 10.1158/1078-0432.CCR-10-0438. 1078-0432.CCR-10-0438[pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berenjeno IM, Vanhaesebroeck B. PI3K regulatory subunits lose control in cancer. Cancer Cell. 2009;16:449–450. doi: 10.1016/j.ccr.2009.11.017. S1535-6108(09)00393-6[pii] [DOI] [PubMed] [Google Scholar]
- Berns K, Horlings HM, Hennessy BT, Madiredjo M, Hijmans EM, Beelen K, Linn SC, Gonzalez-Angulo AM, Stemke-Hale K, Hauptmann M, Beijersbergen RL, Mills GB, van de Vijver MJ, Bernards R. A functional genetic approach identifies the PI3K pathway as a major determinant of trastuzumab resistance in breast cancer. Cancer Cell. 2007;12:395–402. doi: 10.1016/j.ccr.2007.08.030. S1535-6108(07)00262-0[pii] [DOI] [PubMed] [Google Scholar]
- Beroukhim R, Getz G, Nghiemphu L, Barretina J, Hsueh T, Linhart D, Vivanco I, Lee JC, Huang JH, Alexander S, Du J, Kau T, Thomas RK, Shah K, Soto H, Perner S, Prensner J, Debiasi RM, Demichelis F, Hatton C, Rubin MA, Garraway LA, Nelson SF, Liau L, Mischel PS, Cloughesy TF, Meyerson M, Golub TA, Lander ES, Mellinghoff IK, Sellers WR. Assessing the significance of chromosomal aberrations in cancer: methodology and application to glioma. Proc Natl Acad Sci U S A. 2007;104:20007–20012. doi: 10.1073/pnas.0710052104. 0710052104[-pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beroukhim R, Getz G, Mellinghoff IK. Genomic Identification of Significant Targets in Cancer (GISTIC): Methodology and Application to Glioma and Other Cancers. In: Van Meir E, editor. CNS Cancer. Models, Markers, Prognostic Factors, Targets, and Therapeutic Approaches. Springer; Berlin: 2009. [Google Scholar]
- Bianco R, Shin I, Ritter CA, Yakes FM, Basso A, Rosen N, Tsurutani J, Dennis PA, Mills GB, Arteaga CL. Loss of PTEN/MMAC1/TEP in EGF receptor-expressing tumor cells counteracts the antitumor action of EGFR tyrosine kinase inhibitors. Oncogene. 2003;22:2812–2822. doi: 10.1038/sj.onc.1206388. [DOI] [PubMed] [Google Scholar]
- Blume-Jensen P, Hunter T. Oncogenic kinase signalling. Nature. 2001;411:355–365. doi: 10.1038/35077225. [DOI] [PubMed] [Google Scholar]
- Broderick DK, Di C, Parrett TJ, Samuels YR, Cummins JM, McLendon RE, Fults DW, Velculescu VE, Bigner DD, Yan H. Mutations of PIK3CA in anaplastic oligodendrogliomas, high-grade astrocytomas, and medulloblastomas. Cancer Res. 2004;64:5048–5050. doi: 10.1158/0008-5472.CAN-04-1170. [DOI] [PubMed] [Google Scholar]
- Buck E, Eyzaguirre A, Brown E, Petti F, McCormack S, Haley JD, Iwata KK, Gibson NW, Griffin G. Rapamycin synergizes with the epidermal growth factor receptor inhibitor erlotinib in non-small-cell lung, pancreatic, colon, and breast tumors. Mol Cancer Ther. 2006;5:2676–2684. doi: 10.1158/1535-7163.MCT-06-0166. 5/11/2676[pii] [DOI] [PubMed] [Google Scholar]
- Buday L, Downward J. Many faces of Ras activation. Biochim Biophys Acta. 2008 doi: 10.1016/j.bbcan.2008.05.001. S0304-419X(08)00024-3[pii] [DOI] [PubMed] [Google Scholar]
- Carracedo A, Ma L, Teruya-Feldstein J, Rojo F, Salmena L, Alimonti A, Egia A, Sasaki AT, Thomas G, Kozma SC, Papa A, Nardella C, Cantley LC, Baselga J, Pandolfi PP. Inhibition of mTORC1 leads to MAPK pathway activation through a PI3K-dependent feedback loop in human cancer. J Clin Invest. 2008;118:3065–3074. doi: 10.1172/JCI34739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellano E, Downward J. RASInteractionwith PI3K:More Than Just Another Effector Pathway. Genes Cancer. 2011;2:261–274. doi: 10.1177/194760191140807910.1177_1947601911408079[pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang SM, Wen P, Cloughesy T, Greenberg H, Schiff D, Conrad C, Fink K, Robins HI, De Angelis L, Raizer J, Hess K, Aldape K, Lamborn KR, Kuhn J, Dancey J, Prados MD. Phase II study of CCI-779 in patients with recurrent glioblastoma multiforme. Invest New Drugs. 2005;23:357–361. doi: 10.1007/s10637-005-1444-0. [DOI] [PubMed] [Google Scholar]
- Chen WS, Lazar CS, Lund KA, Welsh JB, Chang CP, Walton GM, Der CJ, Wiley HS, Gill GN, Rosenfeld MG. Functional independence of the epidermal growth factor receptor from a domain required for ligand-induced internalization and calcium regulation. Cell. 1989;59(1):33–43. doi: 10.1016/0092-8674(89)90867-2. [DOI] [PubMed] [Google Scholar]
- Cheung LWT, Hennessy BT, Li J. High Frequency of PIK3R1 and PIK3R2 Mutations in Endometrial Cancer Elucidates a Novel Mechansim for Regulation of PTEN Protein Stability. Cancer Discovery. 2011;1:170–185. doi: 10.1158/2159-8290.CD-11-0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi AS, Kwak EL, Clark JW, Wang DL, Louis DN, Iafrate AJ, Batchelor T. Clinical improvement and rapid radiographic regression induced by a MET inhibitor in a patient with MET-amplified glioblastoma. J Clin Oncol. 2011;29:2011. doi: 10.1200/JCO.2011.38.4586. (suppl; abstr 2072) [DOI] [PubMed] [Google Scholar]
- Choe G, Horvath S, Cloughesy TF, Crosby K, Seligson D, Palotie A, Inge L, Smith BL, Sawyers CL, Mischel PS. Analysis of the phosphatidylinositol 3′-kinase signaling pathway in glioblastoma patients in vivo. Cancer Res. 2003;63:2742–2746. [PubMed] [Google Scholar]
- Chow LM, Endersby R, Zhu X, Rankin S, Qu C, Zhang J, Broniscer A, Ellison DW, Baker SJ. Cooperativity within and among Pten, p53, and Rb pathways induces high-grade astrocytoma in adult brain. Cancer Cell. 2011;19:305–316. doi: 10.1016/j.ccr.2011.01.039. S1535-6108(11)00051-1[pii] 10.1016/j.ccr.2011. 01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cin H, Meyer C, Herr R, Janzarik WG, Lambert S, Jones DT, Jacob K, Benner A, Witt H, Remke M, Bender S, Falkenstein F, Van Anh TN, Olbrich H, von Deimling A, Pekrun A, Kulozik AE, Gnekow A, Scheurlen W, Witt O, Omran H, Jabado N, Collins VP, Brummer T, Marschalek R, Lichter P, Korshunov A, Pfister SM. Oncogenic FAM131B-BRAF fusion resulting from 7q34 deletion comprises an alternative mechanism of MAPK pathway activation in pilocytic astrocytoma. Acta Neuropathol. 2011;121:763–774. doi: 10.1007/s00401-011-0817-z. [DOI] [PubMed] [Google Scholar]
- Clarke ID, Dirks PB. A human brain tumor-derived PDGFR-alpha deletion mutant is transforming. Oncogene. 2003;22:722–733. doi: 10.1038/sj.onc.12061601206160[pii]. [DOI] [PubMed] [Google Scholar]
- Cloughesy TF, Yoshimoto K, Nghiemphu P, Brown K, Dang J, Zhu S, Hsueh T, Chen Y, Wang W, Youngkin D, Liau L, Martin N, Becker D, Bergsneider M, Lai A, Green R, Oglesby T, Koleto M, Trent J, Horvath S, Mischel PS, Mellinghoff IK, Sawyers CL. Antitumor activity of rapamycin in a Phase I trial for patients with recurrent PTEN-deficient glioblastoma. PLoS Med. 2008;5:e8. doi: 10.1371/journal.pmed.0050008. 07-PLME-RA-0191[pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comoglio PM, Giordano S, Trusolino L. Drug development of MET inhibitors: targeting oncogene addiction and expedience. Nat Rev Drug Discov. 2008;7:504–516. doi: 10.1038/nrd2530. nrd2530[pii] 10.1038/ nrd2530. [DOI] [PubMed] [Google Scholar]
- Dai C, Celestino JC, Okada Y, Louis DN, Fuller GN, Holland EC. PDGF autocrine stimulation dedifferentiates cultured astrocytes and induces oligodendrogliomas and oligoastrocytomas from neural progenitors and astrocytes in vivo. Genes Dev. 2001;15:1913–1925. doi: 10.1101/gad.903001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dancey J. mTOR signaling and drug development in cancer. Nat Rev Clin Oncol. 2010;7:209–219. doi: 10.1038/nrclinonc.2010.21. nrclinonc.2010.21[pii] [DOI] [PubMed] [Google Scholar]
- de Vries NA, Bruggeman SW, Hulsman D, de Vries HI, Zevenhoven J, Buckle T, Hamans BC, Leenders WP, Beijnen JH, van Lohuizen M, Berns AJ, van Tellingen O. Rapid and robust transgenic high-grade glioma mouse models for therapy intervention studies. Clin Cancer Res. 2010;16:3431–3441. doi: 10.1158/1078-0432.CCR-09-3414. 1078-0432.CCR-09-3414[pii] [DOI] [PubMed] [Google Scholar]
- Decker SJ, Alexander C, Habib T. Epidermal growth factor (EGF)-stimulated tyrosine phosphorylation and EGF receptor degradation in cells expressing EGF receptors truncated at residue 973. J Biol Chem. 1992;267:1104–1108. [PubMed] [Google Scholar]
- Deshmukh H, Yeh TH, Yu J, Sharma MK, Perry A, Leonard JR, Watson MA, Gutmann DH, Nagarajan R. High-resolution, dual-platform aCGH analysis reveals frequent HIPK2 amplification and increased expression in pilocytic astrocytomas. Oncogene. 2008;27:4745–4751. doi: 10.1038/onc.2008.110. onc2008110[pii] [DOI] [PubMed] [Google Scholar]
- Di Fiore PP, Pierce JH, Fleming TP, Hazan R, Ullrich A, King CR, Schlessinger J, Aaronson SA. Overexpression of the human EGF receptor confers an EGF-dependent transformed phenotype to NIH 3T3 cells. Cell. 1987;51:1063–1070. doi: 10.1016/0092-8674(87)90592-7. [DOI] [PubMed] [Google Scholar]
- Ding H, Roncari L, Shannon P, Wu X, Lau N, Karaskova J, Gutmann DH, Squire JA, Nagy A, Guha A. Astrocyte-specific expression of activated p21-ras results in malignant astrocytoma formation in a transgenic mouse model of human gliomas. Cancer Res. 2001;61:3826–3836. [PubMed] [Google Scholar]
- Ding H, Shannon P, Lau N, Wu X, Roncari L, Baldwin RL, Takebayashi H, Nagy A, Gutmann DH, Guha A. Oligodendrogliomas result from the expression of an activated mutant epidermal growth factor receptor in a RAS transgenic mouse astrocytoma model. Cancer Res. 2003;63:1106–1113. [PubMed] [Google Scholar]
- Dougherty MJ, Santi M, Brose MS, Ma C, Resnick AC, Sievert AJ, Storm PB, Biegel JA. Activating mutations in BRAF characterize a spectrum of pediatric low-grade gliomas. Neuro Oncol. 2010 doi: 10.1093/neuonc/noq007. noq007[pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Druker BJ, Talpaz M, Resta DJ, Peng B, Buchdunger E, Ford JM, Lydon NB, Kantarjian H, Capdeville R, Ohno-Jones S, Sawyers CL. Efficacy and safety of a specific inhibitor of the BCR-ABL tyrosine kinase in chronic myeloid leukemia. N Engl J Med. 2001;344:1031–1037. doi: 10.1056/NEJM200104053441401. [DOI] [PubMed] [Google Scholar]
- Duerr EM, Rollbrocker B, Hayashi Y, Peters N, Meyer-Puttlitz B, Louis DN, Schramm J, Wiestler OD, Parsons R, Eng C, von Deimling A. PTEN mutations in gliomas and glioneuronal tumors. Oncogene. 1998;16:2259–2264. doi: 10.1038/sj.onc.1201756. [DOI] [PubMed] [Google Scholar]
- Eisenhardt AE, Olbrich H, Roring M, Janzarik W, Van Anh TN, Cin H, Remke M, Witt H, Korshunov A, Pfister SM, Omran H, Brummer T. Functional characterization of a BRAF insertion mutant associated with pilocytic astrocytoma. Int J Cancer. 2010 doi: 10.1002/ijc.25893. 10.1002/ ijc.25893. [DOI] [PubMed] [Google Scholar]
- Ekstrand AJ, Sugawa N, James CD, Collins VP. Amplified and rearranged epidermal growth factor receptor genes in human glioblastomas reveal deletions of sequences encoding portions of the N- and/or C-terminal tails. Proc Natl Acad Sci U S A. 1992;89:4309–4313. doi: 10.1073/pnas.89.10.4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekstrand AJ, Longo N, Hamid ML, Olson JJ, Liu L, Collins VP, James CD. Functional characterization of an EGF receptor with a truncated extracellular domain expressed in glioblastomas with EGFR gene amplification. Oncogene. 1994;9:2313–2320. [PubMed] [Google Scholar]
- Eley G, Frederick L, Wang XY, Smith DI, James CD. 3′ end structure and rearrangements of EGFR in glioblastomas. Genes Chromosomes Cancer. 1998;23:248–254. doi: 10.1002/(sici)1098-2264(199811)23:3<248::aid-gcc7>3.0.co;2-1. 10.1002/ (SICI)1098-2264(199811)23:3<248:AID-GCC7>3.0.CO;2-1. [pii] [DOI] [PubMed] [Google Scholar]
- El-Habr EA, Tsiorva P, Theodorou M, Levidou G, Korkolopoulou P, Vretakos G, Petraki L, Michalopoulos NV, Patsouris E, Saetta AA. Analysis of PIK3CA and B-RAF gene mutations in human astrocytomas: association with activation of ERK and AKT. Clin Neuropathol. 2010;29:239–245. doi: 10.5414/npp29239. 7714[pii] [DOI] [PubMed] [Google Scholar]
- Eller JL, Longo SL, Hicklin DJ, Canute GW. Activity of anti-epidermal growth factor receptor monoclonal antibody C225 against glioblastoma multiforme. Neurosurgery. 2002;51:1005–1013 d. doi: 10.1097/00006123-200210000-00028. iscussion 1013–4. [DOI] [PubMed] [Google Scholar]
- Faber AC, Li D, Song Y, Liang MC, Yeap BY, Bronson RT, Lifshits E, Chen Z, Maira SM, Garcia-Echeverria C, Wong KK, Engelman JA. Differential induction of apoptosis in HER2 and EGFR addicted cancers following PI3K inhibition. Proc Natl Acad Sci U S A. 2009;106:19503–19508. doi: 10.1073/pnas.0905056106. 0905056106[pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan QW, Knight ZA, Goldenberg DD, Yu W, Mostov KE, Stokoe D, Shokat KM, Weiss WA. A dual PI3 kinase/mTOR inhibitor reveals emergent efficacy in glioma. Cancer Cell. 2006;9:341–349. doi: 10.1016/j.ccr.2006.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan QW, Cheng C, Hackett C, Feldman M, Houseman BT, Nicolaides T, Haas-Kogan D, James CD, Oakes SA, Debnath J, Shokat KM, Weiss WA. Akt and autophagy cooperate to promote survival of drug-resistant glioma. Sci Signal. 2010;3:ra81. doi: 10.1126/scisignal.2001017. 3/147/ ra81[pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman ME, Shokat KM. New inhibitors of the PI3K-Akt-mTOR pathway: insights into mTOR signaling from a new generation of Tor Kinase Domain Inhibitors (TORKinibs) Curr Top Microbiol Immunol. 2010;347:241–262. doi: 10.1007/82_2010_64. [DOI] [PubMed] [Google Scholar]
- Fleming TP, Saxena A, Clark WC, Robertson JT, Oldfield EH, Aaronson SA, Ali IU. Amplification and/or overexpression of platelet-derived growth factor receptors and epidermal growth factor receptor in human glial tumors. Cancer Res. 1992;52:4550–4553. [PubMed] [Google Scholar]
- Forshew T, Tatevossian RG, Lawson AR, Ma J, Neale G, Ogunkolade BW, Jones TA, Aarum J, Dalton J, Bailey S, Chaplin T, Carter RL, Gajjar A, Broniscer A, Young BD, Ellison DW, Sheer D. Activation of the ERK/MAPK pathway: a signature genetic defect in posterior fossa pilocytic astrocytomas. J Pathol. 2009;218:172–181. doi: 10.1002/path.2558. [DOI] [PubMed] [Google Scholar]
- Foukas LC, Berenjeno IM, Gray A, Khwaja A, Vanhaesebroeck B. Activity of any class IA PI3K isoform can sustain cell proliferation and survival. Proc Natl Acad Sci U S A. 2010;107:11381–11386. doi: 10.1073/pnas.0906461107. 0906461107[pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser MM, Zhu X, Kwon CH, Uhlmann EJ, Gutmann DH, Baker SJ. Pten loss causes hypertrophy and increased proliferation of astrocytes in vivo. Cancer Res. 2004;64:7773–7779. doi: 10.1158/0008-5472.CAN-04-2487. 64/21/7773[pii] [DOI] [PubMed] [Google Scholar]
- Frederick L, Wang XY, Eley G, James CD. Diversity and frequency of epidermal growth factor receptor mutations in human glioblastomas. Cancer Res. 2000;60:1383–1387. [PubMed] [Google Scholar]
- Friedman HS, Prados MD, Wen PY, Mikkelsen T, Schiff D, Abrey LE, Yung WK, Paleologos N, Nicholas MK, Jensen R, Vredenburgh J, Huang J, Zheng M, Cloughesy T. Bevacizumab alone and in combination with irinotecan in recurrent glioblastoma. J Clin Oncol. 2009;27:4733–4740. doi: 10.1200/JCO.2008.19.8721. JCO.2008.19.8721[pii] [DOI] [PubMed] [Google Scholar]
- Galanis E, Buckner JC, Maurer MJ, Kreisberg JI, Ballman K, Boni J, Peralba JM, Jenkins RB, Dakhil SR, Morton RF, Jaeckle KA, Scheithauer BW, Dancey J, Hidalgo M, Walsh DJ. Phase II trial of temsirolimus (CCI-779) in recurrent glioblastoma multiforme: a North Central Cancer Treatment Group Study. J Clin Oncol. 2005;23:5294–5304. doi: 10.1200/JCO.2005.23.622. [DOI] [PubMed] [Google Scholar]
- Gallia GL, Rand V, Siu IM, Eberhart CG, James CD, Marie SK, Oba-Shinjo SM, Carlotti CG, Caballero OL, Simpson AJ, Brock MV, Massion PP, Carson BS, Sr, Riggins GJ. PIK3CA gene mutations in pediatric and adult glioblastoma multiforme. Mol Cancer Res. 2006;4:709–714. doi: 10.1158/1541-7786.MCR-06-0172. 4/10/709[pii] [DOI] [PubMed] [Google Scholar]
- Gerber DE, Minna JD. ALK inhibition for non-small cell lung cancer: from discovery to therapy in record time. Cancer Cell. 2010;18:548–551. doi: 10.1016/j.ccr.2010.11.033. S1535-6108(10)00491-5[pii] 10.1016/ j.ccr.2010.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gronych J, Korshunov A, Bageritz J, Milde T, Jugold M, Hambardzumyan D, Remke M, Hartmann C, Witt H, Jones DT, Witt O, Heiland S, Bendszus M, Holland EC, Pfister S, Lichter P. An activated mutant BRAF kinase domain is sufficient to induce pilocytic astrocytoma in mice. J Clin Invest. 2011;121:1344–1348. doi: 10.1172/JCI44656. 44656[pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groszer M, Erickson R, Scripture-Adams DD, Lesche R, Trumpp A, Zack JA, Kornblum HI, Liu X, Wu H. Negative regulation of neural stem/progenitor cell proliferation by the Pten tumor suppressor gene in vivo. Science. 2001;294:2186–2189. doi: 10.1126/science.1065518. [DOI] [PubMed] [Google Scholar]
- Gschwind A, Fischer OM, Ullrich A. The discovery of receptor tyrosine kinases: targets for cancer therapy. Nat Rev Cancer. 2004;4:361–370. doi: 10.1038/nrc1360. [DOI] [PubMed] [Google Scholar]
- Guillard S, Clarke PA, Te Poele R, Mohri Z, Bjerke L, Valenti M, Raynaud F, Eccles SA, Workman P. Molecular pharmacology of phosphatidylinositol 3-kinase inhibition in human glioma. Cell Cycle. 2009;8:443–453. doi: 10.4161/cc.8.3.7643. 7643[pii] [DOI] [PubMed] [Google Scholar]
- Gutmann DH, James CD, Poyhonen M, Louis DN, Ferner R, Guha A, Hariharan S, Viskochil D, Perry A. Molecular analysis of astrocytomas presenting after age 10 in individuals with NF1. Neurology. 2003;61:1397–1400. doi: 10.1212/wnl.61.10.1397. [DOI] [PubMed] [Google Scholar]
- Haas-Kogan DA, Prados MD, Tihan T, Eberhard DA, Jelluma N, Arvold ND, Baumber R, Lamborn KR, Kapadia A, Malec M, Berger MS, Stokoe D. Epidermal growth factor receptor, protein kinase B/Akt, and glioma response to erlotinib. J Natl Cancer Inst. 2005;97:880–887. doi: 10.1093/jnci/dji161. [DOI] [PubMed] [Google Scholar]
- Hagemann C, Gloger J, Anacker J, Said HM, Gerngras S, Kuhnel S, Meyer C, Rapp UR, Kammerer U, Vordermark D, Flentje M, Roosen K, Vince GH. RAF expression in human astrocytic tumors. Int J Mol Med. 2009;23:17–31. [PubMed] [Google Scholar]
- Haley JD, Hsuan JJ, Waterfield MD. Analysis of mammalian fibroblast transformation by normal and mutated human EGF receptors. Oncogene. 1989;4:273–283. [PubMed] [Google Scholar]
- Hambardzumyan D, Parada LF, Holland EC, Charest A. Genetic modeling of gliomas in mice: new tools to tackle old problems. Glia. 2011;59:1155–1168. doi: 10.1002/glia.21142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann C, Bartels G, Gehlhaar C, Holtkamp N, von Deimling A. PIK3CA mutations in glioblastoma multiforme. Acta Neuropathol. 2005;109:639–642. doi: 10.1007/s00401-005-1000-1. [DOI] [PubMed] [Google Scholar]
- Hegi ME, Diserens AC, Bady P, Kamoshima Y, Kouwenhoven MC, Delorenzi M, Lambiv WL, Hamou MF, Matter MS, Koch A, Heppner FL, Yonekawa Y, Merlo A, Frei K, Mariani L, Hofer S. Pathway Analysis of Glioblastoma Tissue after Preoperative Treatment with the EGFR Tyrosine Kinase Inhibitor Gefitinib–A Phase II Trial. Mol Cancer Ther. 2011;10:1102–1112. doi: 10.1158/1535-7163.MCT-11-0048. 1535-7163.MCT-11-0048[pii] [DOI] [PubMed] [Google Scholar]
- Hermanson M, Funa K, Hartman M, Claesson-Welsh L, Heldin CH, Westermark B, Nister M. Platelet-derived growth factor and its receptors in human glioma tissue: expression of messenger RNA and protein suggests the presence of autocrine and paracrine loops. Cancer Res. 1992;52:3213–3219. [PubMed] [Google Scholar]
- Hitoshi Y, Harris BT, Liu H, Popko B, Israel MA. Spinal glioma: platelet-derived growth factor B-mediated oncogenesis in the spinal cord. Cancer Res. 2008;68:8507–8515. doi: 10.1158/0008-5472.CAN-08-1063. 68/20/ 8507[pii] [DOI] [PubMed] [Google Scholar]
- Holland EC, Hively WP, DePinho RA, Varmus HE. A constitutively active epidermal growth factor receptor cooperates with disruption of G1 cell-cycle arrest pathways to induce glioma-like lesions in mice. Genes Dev. 1998;12:3675–3685. doi: 10.1101/gad.12.23.3675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland EC, Celestino J, Dai C, Schaefer L, Sawaya RE, Fuller GN. Combined activation of Ras and Akt in neural progenitors induces glioblastoma formation in mice. Nat Genet. 2000;25:55–57. doi: 10.1038/75596. [DOI] [PubMed] [Google Scholar]
- Holmen SL, Williams BO. Essential role for Ras signaling in glioblastoma maintenance. Cancer Res. 2005;65:8250–8255. doi: 10.1158/0008-5472.CAN-05-1173. 65/18/8250[pii] [DOI] [PubMed] [Google Scholar]
- Huang CH, Mandelker D, Schmidt-Kittler O, Samuels Y, Velculescu VE, Kinzler KW, Vogelstein B, Gabelli SB, Amzel LM. The structure of a human p110alpha/p85alpha complex elucidates the effects of oncogenic PI3Kalpha mutations. Science. 2007;318:1744–1748. doi: 10.1126/science.1150799. 318/5857/1744[pii] [DOI] [PubMed] [Google Scholar]
- Ihle NT, Lemos R, Jr, Wipf P, Yacoub A, Mitchell C, Siwak D, Mills GB, Dent P, Kirkpatrick DL, Powis G. Mutations in the phosphatidylinositol-3-kinase pathway predict for antitumor activity of the inhibitor PX-866 whereas oncogenic Ras is a dominant predictor for resistance. Cancer Res. 2009;69:143–150. doi: 10.1158/0008-5472.CAN-07-6656. 69/1/143[pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob K, Albrecht S, Sollier C, Faury D, Sader E, Montpetit A, Serre D, Hauser P, Garami M, Bognar L, Hanzely Z, Montes JL, Atkinson J, Farmer JP, Bouffet E, Hawkins C, Tabori U, Jabado N. Duplication of 7q34 is specific to juvenile pilocytic astrocytomas and a hallmark of cerebellar and optic pathway tumours. Br J Cancer. 2009;101:722–733. doi: 10.1038/sj.bjc.6605179. 6605179[-pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacques TS, Swales A, Brzozowski MJ, Henriquez NV, Linehan JM, Mirzadeh Z, OMC, Naumann H, Alvarez-Buylla A, Brandner S. Combinations of genetic mutations in the adult neural stem cell compartment determine brain tumour phenotypes. EMBO J. 2010;29:222–235. doi: 10.1038/emboj.2009.327. emboj2009327[pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaiswal BS, Janakiraman V, Kljavin NM, Chaudhuri S, Stern HM, Wang W, Kan Z, Dbouk HA, Peters BA, Waring P, Dela Vega T, Kenski DM, Bowman KK, Lorenzo M, Li H, Wu J, Modrusan Z, Stinson J, Eby M, Yue P, Kaminker JS, de Sauvage FJ, Backer JM, Seshagiri S. Somatic mutations in p85alpha promote tumorigenesis through class IA PI3K activation. Cancer Cell. 2009;16:463–474. doi: 10.1016/j.ccr.2009.10.016. S1535-6108(09)00385-7[pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- James CD, Carlbom E, Dumanski JP, Hansen M, Nordenskjold M, Collins VP, Cavenee WK. Clonal genomic alterations in glioma malignancy stages. Cancer Res. 1988;48:5546–5551. [PubMed] [Google Scholar]
- Jansen M, Yip S, Louis DN. Molecular pathology in adult gliomas: diagnostic, prognostic, and predictive markers. Lancet Neurol. 2010;9:717–726. doi: 10.1016/S1474-4422(10)70105-8. S1474-4422(10)70105-8[pii] 10.1016/ S1474-4422(10)70105-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janzarik WG, Kratz CP, Loges NT, Olbrich H, Klein C, Schafer T, Scheurlen W, Roggendorf W, Weiller C, Niemeyer C, Korinthenberg R, Pfister S, Omran H. Further evidence for a somatic KRAS mutation in a pilocytic astrocytoma. Neuropediatrics. 2007;38:61–63. doi: 10.1055/s-2007-984451. [DOI] [PubMed] [Google Scholar]
- Jeuken J, van den Broecke C, Gijsen S, Boots-Sprenger S, Wesseling P. RAS/RAF pathway activation in gliomas: the result of copy number gains rather than activating mutations. Acta Neuropathol. 2007;114:121–133. doi: 10.1007/s00401-007-0239-0. [DOI] [PubMed] [Google Scholar]
- Jia S, Liu Z, Zhang S, Liu P, Zhang L, Lee SH, Zhang J, Signoretti S, Loda M, Roberts TM, Zhao JJ. Essential roles of PI(3)K-p110beta in cell growth, metabolism and tumorigenesis. Nature. 2008;454:776–779. doi: 10.1038/nature07091. nature07091[pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DT, Kocialkowski S, Liu L, Pearson DM, Backlund LM, Ichimura K, Collins VP. Tandem duplication producing a novel oncogenic BRAF fusion gene defines the majority of pilocytic astrocytomas. Cancer Res. 2008;68:8673–8677. doi: 10.1158/0008-5472.CAN-08-2097. 68/21/8673[pii] 10.1158/0008-5472. CAN-08-2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DT, Kocialkowski S, Liu L, Pearson DM, Ichimura K, Collins VP. Oncogenic RAF1 rearrangement and a novel BRAF mutation as alternatives to KIAA1549:BRAF fusion in activating the MAPK pathway in pilocytic astrocytoma. Oncogene. 2009;28:2119–2123. doi: 10.1038/onc.2009.73. onc200973 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junttila TT, Akita RW, Parsons K, Fields C, Lewis Phillips GD, Friedman LS, Sampath D, Sliwkowski MX. Ligand-independent HER2/HER3/PI3K complex is disrupted by trastuzumab and is effectively inhibited by the PI3K inhibitor GDC-0941. Cancer Cell. 2009;15:429–440. doi: 10.1016/j.ccr.2009.03.020. S1535-6108(09)00109-3[pii] [DOI] [PubMed] [Google Scholar]
- Kaelin WG., Jr The concept of synthetic lethality in the context of anticancer therapy. Nat Rev Cancer. 2005;5:689–698. doi: 10.1038/nrc1691. nrc1691[pii] [DOI] [PubMed] [Google Scholar]
- Kang S, Denley A, Vanhaesebroeck B, Vogt PK. Oncogenic transformation induced by the p110beta, -gamma, and -delta isoforms of class I phosphoinositide 3-kinase. Proc Natl Acad Sci U S A. 2006;103:1289–1294. doi: 10.1073/pnas.0510772103. 0510772103[pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kita D, Yonekawa Y, Weller M, Ohgaki H. PIK3CA alterations in primary (de novo) and secondary glioblastomas. Acta Neuropathol. 2007;113:295–302. doi: 10.1007/s00401-006-0186-1. [DOI] [PubMed] [Google Scholar]
- Knobbe CB, Reifenberger J, Reifenberger G. Mutation analysis of the Ras pathway genes NRAS, HRAS, KRAS and BRAF in glioblastomas. Acta Neuropathol. 2004;108:467–470. doi: 10.1007/s00401-004-0929-9. [DOI] [PubMed] [Google Scholar]
- Koochekpour S, Jeffers M, Rulong S, Taylor G, Klineberg E, Hudson EA, Resau JH, Vande Woude GF. Met and hepatocyte growth factor/scatter factor expression in human gliomas. Cancer Res. 1997;57:5391–5398. [PubMed] [Google Scholar]
- Kotliarov Y, Steed ME, Christopher N, Walling J, Su Q, Center A, Heiss J, Rosenblum M, Mikkelsen T, Zenklusen JC, Fine HA. High-resolution global genomic survey of 178 gliomas reveals novel regions of copy number alteration and allelic imbalances. Cancer Res. 2006;66:9428–9436. doi: 10.1158/0008-5472.CAN-06-1691. 66/19/9428[pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koul D, Shen R, Kim YW, Kondo Y, Lu Y, Bankson J, Ronen SM, Kirkpatrick DL, Powis G, Yung WK. Cellular and in vivo activity of a novel PI3K inhibitor, PX-866, against human glioblastoma. Neuro Oncol. 2010;12:559–569. doi: 10.1093/neuonc/nop058. nop058[pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreisl TN, Kim L, Moore K, Duic P, Royce C, Stroud I, Garren N, Mackey M, Butman JA, Camphausen K, Park J, Albert PS, Fine HA. Phase II trial of single-agent bevacizumab followed by bevacizumab plus irinotecan at tumor progression in recurrent glioblastoma. J Clin Oncol. 2009a;27:740–745. doi: 10.1200/JCO.2008.16.3055. JCO.2008.16.3055[pii] 10.1200/JCO.2008. 16.3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreisl TN, Lassman AB, Mischel PS, Rosen N, Scher HI, Teruya-Feldstein J, Shaffer D, Lis E, Abrey LE. A pilot study of everolimus and gefitinib in the treatment of recurrent glioblastoma (GBM) J Neurooncol. 2009b;92:99–105. doi: 10.1007/s11060-008-9741-z. [DOI] [PubMed] [Google Scholar]
- Kumabe T, Sohma Y, Kayama T, Yoshimoto T, Yamamoto T. Amplification of alpha-platelet- derived growth factor receptor gene lacking an exon coding for a portion of the extracellular region in a primary brain tumor of glial origin. Oncogene. 1992;7:627–633. [PubMed] [Google Scholar]
- Kwon CH, Zhu X, Zhang J, Knoop LL, Tharp R, Smeyne RJ, Eberhart CG, Burger PC, Baker SJ. Pten regulates neuronal soma size: a mouse model of Lhermitte-Duclos disease. Nat Genet. 2001;29:404–411. doi: 10.1038/ng781ng781[pii]. [DOI] [PubMed] [Google Scholar]
- Kwon CH, Zhao D, Chen J, Alcantara S, Li Y, Burns DK, Mason RP, Lee EY, Wu H, Parada LF. Pten haploinsufficiency accelerates formation of high-grade astrocytomas. Cancer Res. 2008;68:3286–3294. doi: 10.1158/0008-5472.CAN-07-6867. 68/9/3286[pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai A, Kharbanda S, Pope WB, et al. Evidence for sequenced molecular evolution of IDH1 mutant glioblastoma from a distinct cell of origin. Journal of Clinical Oncology. 2011 doi: 10.1200/JCO.2010.33.8715. [in press] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassman AB, Rossi MR, Raizer JJ, Abrey LE, Lieberman FS, Grefe CN, Lamborn K, Pao W, Shih AH, Kuhn JG, Wilson R, Nowak NJ, Cowell JK, DeAngelis LM, Wen P, Gilbert MR, Chang S, Yung WA, Prados M, Holland EC. Molecular study of malignant gliomas treated with epidermal growth factor receptor inhibitors: tissue analysis from North American Brain Tumor Consortium Trials 01–03 and 00–01. Clin Cancer Res. 2005;11:7841–7850. doi: 10.1158/1078-0432.CCR-05-0421. 11/21/ 7841[pii] [DOI] [PubMed] [Google Scholar]
- Lee J, Kotliarova S, Kotliarov Y, Li A, Su Q, Donin NM, Pastorino S, Purow BW, Christopher N, Zhang W, Park JK, Fine HA. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell. 2006a;9:391–403. doi: 10.1016/j.ccr.2006.03.030. S1535-6108(06)00117-6[pii] 10.1016/ j.ccr.2006.03.030. [DOI] [PubMed] [Google Scholar]
- Lee JC, Vivanco I, Beroukhim R, Huang JH, Feng WL, DeBiasi RM, Yoshimoto K, King JC, Nghiemphu P, Yuza Y, Xu Q, Greulich H, Thomas RK, Paez JG, Peck TC, Linhart DJ, Glatt KA, Getz G, Onofrio R, Ziaugra L, Levine RL, Gabriel S, Kawaguchi T, O’Neill K, Khan H, Liau LM, Nelson SF, Rao PN, Mischel P, Pieper RO, Cloughesy T, Leahy DJ, Sellers WR, Sawyers CL, Meyerson M, Mellinghoff IK. Epidermal growth factor receptor activation in glioblastoma through novel missense mutations in the extracellular domain. PLoS Med. 2006b;3:e485. doi: 10.1371/journal.pmed.0030485. 06-PLME-RA-0223R2[pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon MA, Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2010;141:1117–1134. doi: 10.1016/j.cell.2010.06.011. S0092-8674(10)00665-3[pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Bollag G, Clark R, Stevens J, Conroy L, Fults D, Ward K, Friedman E, Samowitz W, Robertson M, et al. Somatic mutations in the neurofibromatosis 1 gene in human tumors. Cell. 1992;69:275–281. doi: 10.1016/0092-8674(92)90408-5. 0092-8674(92)90408-5[pii] [DOI] [PubMed] [Google Scholar]
- Li J, Yen C, Liaw D, Podsypanina K, Bose S, Wang SI, Puc J, Miliaresis C, Rodgers L, McCombie R, Bigner SH, Giovanella BC, Ittmann M, Tycko B, Hibshoosh H, Wigler MH, Parsons R. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science. 1997;275:1943–1947. doi: 10.1126/science.275.5308.1943. [DOI] [PubMed] [Google Scholar]
- Libermann TA, Nusbaum HR, Razon N, Kris R, Lax I, Soreq H, Whittle N, Waterfield MD, Ullrich A, Schlessinger J. Amplification, enhanced expression and possible rearrangement of EGF receptor gene in primary human brain tumours of glial origin. Nature. 1985;313:144–147. doi: 10.1038/313144a0. [DOI] [PubMed] [Google Scholar]
- Listernick R, Charrow J, Gutmann DH. Intracranial gliomas in neurofibromatosis type 1. Am J Med Genet. 1999;89:38–44. doi: 10.1002/(SICI)1096-8628(19990326)89:1<38:AID-AJMG8>3.0.CO;2-M[pii]. [DOI] [PubMed] [Google Scholar]
- Liu W, James CD, Frederick L, Alderete BE, Jenkins RB. PTEN/MMAC1 mutations and EGFR amplification in glioblastomas. Cancer Res. 1997;57:5254–5257. [PubMed] [Google Scholar]
- Liu Q, Thoreen C, Wang J, Sabatini D, Gray NS. mTOR Mediated Anti-Cancer Drug Discovery. Drug Discov Today Ther Strateg. 2009a;6:47–55. doi: 10.1016/j.ddstr.2009.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu TJ, Koul D, LaFortune T, Tiao N, Shen RJ, Maira SM, Garcia-Echevrria C, Yung WK. NVP-BEZ235, a novel dual phosphatidylinositol 3-kinase/mammalian target of rapamycin inhibitor, elicits multifaceted antitumor activities in human gliomas. Mol Cancer Ther. 2009b;8:2204–2210. doi: 10.1158/1535-7163.MCT-09-0160. 1535-7163.MCT-09-0160[pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lokker NA, Sullivan CM, Hollenbach SJ, Israel MA, Giese NA. Platelet-derived growth factor (PDGF) autocrine signaling regulates survival and mitogenic pathways in glioblastoma cells: evidence that the novel PDGF-C and PDGF-D ligands may play a role in the development of brain tumors. Cancer Res. 2002;62:3729–3735. [PubMed] [Google Scholar]
- Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, Scheithauer BW, Kleihues P. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114:97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyustikman Y, Momota H, Pao W, Holland EC. Constitutive activation of Raf-1 induces glioma formation in mice. Neoplasia. 2008;10:501–510. doi: 10.1593/neo.08206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacConaill LE, Campbell CD, Kehoe SM, Bass AJ, Hatton C, Niu L, Davis M, Yao K, Hanna M, Mondal C, Luongo L, Emery CM, Baker AC, Philips J, Goff DJ, Fiorentino M, Rubin MA, Polyak K, Chan J, Wang Y, Fletcher JA, Santagata S, Corso G, Roviello F, Shivdasani R, Kieran MW, Ligon KL, Stiles CD, Hahn WC, Meyerson ML, Garraway LA. Profiling critical cancer gene mutations in clinical tumor samples. PLoS ONE. 2009;4:e7887. doi: 10.1371/journal.pone.0007887. 10.1371/ journal.pone.0007887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malden LT, Novak U, Kaye AH, Burgess AW. Selective amplification of the cytoplasmic domain of the epidermal growth factor receptor gene in glioblastoma multiforme. Cancer Res. 1988;48:2711–2714. [PubMed] [Google Scholar]
- Mallon R, Hollander I, Feldberg L, Lucas J, Soloveva V, Venkatesan A, Dehnhardt C, Delos Santos E, Chen Z, Dos Santos O, Ayral-Kaloustian S, Gibbons J. Antitumor efficacy profile of PKI-402, a dual phosphatidylinositol 3-kinase/mammalian target of rapamycin inhibitor. Mol Cancer Ther. 2010;9:976–984. doi: 10.1158/1535-7163.MCT-09-0954. 1535-7163.MCT-09-0954[pii] [DOI] [PubMed] [Google Scholar]
- Maltzman TH, Mueller BA, Schroeder J, Rutledge JC, Patterson K, Preston-Martin S, Faustman EM. Ras oncogene mutations in childhood brain tumors. Cancer Epidemiol Biomarkers Prev. 1997;6:239–243. [PubMed] [Google Scholar]
- Martens T, Laabs Y, Gunther HS, Kemming D, Zhu Z, Witte L, Hagel C, Westphal M, Lamszus K. Inhibition of glioblastoma growth in a highly invasive nude mouse model can be achieved by targeting epidermal growth factor receptor but not vascular endothelial growth factor receptor-2. Clin Cancer Res. 2008;14:5447–5458. doi: 10.1158/1078-0432.CCR-08-0147. 14/17/5447[pii] [DOI] [PubMed] [Google Scholar]
- Marumoto T, Tashiro A, Friedmann-Morvinski D, Scadeng M, Soda Y, Gage FH, Verma IM. Development of a novel mouse glioma model using lentiviral vectors. Nat Med. 2009;15:110–116. doi: 10.1038/nm.1863. nm.1863[pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masui H, Wells A, Lazar CS, Rosenfeld MG, Gill GN. Enhanced tumorigenesis of NR6 cells which express non-down-regulating epidermal growth factor receptors. Cancer Res. 1991;51(22):6170–6175. [PubMed] [Google Scholar]
- McDermott U, Sharma SV, Dowell L, Greninger P, Montagut C, Lamb J, Archibald H, Raudales R, Tam A, Lee D, Rothenberg SM, Supko JG, Sordella R, Ulkus LE, Iafrate AJ, Maheswaran S, Njauw CN, Tsao H, Drew L, Hanke JH, Ma XJ, Erlander MG, Gray NS, Haber DA, Settleman J. Identification of genotype-correlated sensitivity to selective kinase inhibitors by using high-throughput tumor cell line profiling. Proc Natl Acad Sci U S A. 2007;104:19936–19941. doi: 10.1073/pnas.0707498104. 0707498104[pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGillicuddy LT, Fromm JA, Hollstein PE, Kubek S, Beroukhim R, De Raedt T, Johnson BW, Williams SM, Nghiemphu P, Liau LM, Cloughesy TF, Mischel PS, Parret A, Seiler J, Moldenhauer G, Scheffzek K, Stemmer-Rachamimov AO, Sawyers CL, Brennan C, Messiaen L, Mellinghoff IK, Cichowski K. Proteasomal and genetic inactivation of the NF1 tumor suppressor in gliomagenesis. Cancer Cell. 2009;16:44–54. doi: 10.1016/j.ccr.2009.05.009. S1535-6108(09) 00175-5[pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLendon R, Friedman A, Bigner D, Van Meir EG, Brat DJ, Mastrogianakis M, Olson JJ, Mikkelsen T, Lehman N, Aldape K, Alfred Yung WK, Bogler O, Vandenberg S, Berger M, Prados M, Muzny D, Morgan M, Scherer S, Sabo A, Nazareth L, Lewis L, Hall O, Zhu Y, Ren Y, Alvi O, Yao J, Hawes A, Jhangiani S, Fowler G, San Lucas A, Kovar C, Cree A, Dinh H, Santibanez J, Joshi V, Gonzalez-Garay ML, Miller CA, Milosavljevic A, Donehower L, Wheeler DA, Gibbs RA, Cibulskis K, Sougnez C, Fennell T, Mahan S, Wilkinson J, Ziaugra L, Onofrio R, Bloom T, Nicol R, Ardlie K, Baldwin J, Gabriel S, Lander ES, Ding L, Fulton RS, McLellan MD, Wallis J, Larson DE, Shi X, Abbott R, Fulton L, Chen K, Koboldt DC, Wendl MC, Meyer R, Tang Y, Lin L, Osborne JR, Dunford-Shore BH, Miner TL, Delehaunty K, Markovic C, Swift G, Courtney W, Pohl C, Abbott S, Hawkins A, Leong S, Haipek C, Schmidt H, Wiechert M, Vickery T, Scott S, Dooling DJ, Chinwalla A, Weinstock GM, Mardis ER, Wilson RK, Getz G, Winckler W, Verhaak RG, Lawrence MS, O’Kelly M, Robinson J, Alexe G, Beroukhim R, Carter S, Chiang D, Gould J, et al. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellinghoff IK, Wang MY, Vivanco I, Haas-Kogan DA, Zhu S, Dia EQ, Lu KV, Yoshimoto K, Huang JH, Chute DJ, Riggs BL, Horvath S, Liau LM, Cavenee WK, Rao PN, Beroukhim R, Peck TC, Lee JC, Sellers WR, Stokoe D, Prados M, Cloughesy TF, Sawyers CL, Mischel PS. Molecular determinants of the response of glioblastomas to EGFR kinase inhibitors. N Engl J Med. 2005;353:2012–2024. doi: 10.1056/NEJMoa051918. [DOI] [PubMed] [Google Scholar]
- Mellinghoff IK, Lassman AB, Wen PY. Signal transduction inhibitors and antiangiogenic therapies for malignant glioma. Glia. 2011 doi: 10.1002/glia.21137. [DOI] [PubMed] [Google Scholar]
- Mendoza MC, Er EE, Blenis J. The Ras-ERK and PI3K-mTOR pathways: cross-talk and compensation. Trends Biochem Sci. 2011;36:320–328. doi: 10.1016/j.tibs.2011.03.006. S0968-0004(11)00050-8[pii] 10.1016/ j.tibs.2011.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra A, Pellarin M, Nigro J, Smirnov I, Moore D, Lamborn KR, Pinkel D, Albertson DG, Feuerstein BG. Array comparative genomic hybridization identifies genetic subgroups in grade 4 human astrocytoma. Clin Cancer Res. 2005;11:2907–2918. doi: 10.1158/1078-0432.CCR-04-0708. 11/8/2907[pii] 10.1158/ 1078-0432.CCR-04-0708. [DOI] [PubMed] [Google Scholar]
- Mizoguchi M, Nutt CL, Mohapatra G, Louis DN. Genetic alterations of phosphoinositide 3-kinase subunit genes in human glioblastomas. Brain Pathol. 2004;14:372–377. doi: 10.1111/j.1750-3639.2004.tb00080.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller W, Mizoguchi M, Silen E, D’Amore K, Nutt CL, Louis DN. Mutations of the PIK3CA gene are rare in human glioblastoma. Acta Neuropathol. 2005;109:654–655. doi: 10.1007/s00401-005-1001-0. 10.1007/ s00401-005-1001-0. [DOI] [PubMed] [Google Scholar]
- Neshat MS, Mellinghoff IK, Tran C, Stiles B, Thomas G, Petersen R, Frost P, Gibbons JJ, Wu H, Sawyers CL. Enhanced sensitivity of PTEN-deficient tumors to inhibition of FRAP/mTOR. Proc Natl Acad Sci U S A. 2001;98:10314–10319. doi: 10.1073/pnas.171076798171076798[pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa R, Ji XD, Harmon RC, Lazar CS, Gill GN, Cavenee WK, Huang HJ. A mutant epidermal growth factor receptor common in human glioma confers enhanced tumorigenicity. Proc Natl Acad Sci U S A. 1994;91:7727–7731. doi: 10.1073/pnas.91.16.7727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien C, Wallin JJ, Sampath D, GuhaThakurta D, Savage H, Punnoose EA, Guan J, Berry L, Prior WW, Amler LC, Belvin M, Friedman LS, Lackner MR. Predictive biomarkers of sensitivity to the phosphatidylinositol 3′ kinase inhibitor GDC-0941 in breast cancer preclinical models. Clin Cancer Res. 2010;16:3670–3683. doi: 10.1158/1078-0432.CCR-09-2828. 1078-0432.CCR-09-2828[pii] 10.1158/1078-0432. CCR-09-2828. [DOI] [PubMed] [Google Scholar]
- Ohgaki H, Kleihues P. Genetic pathways to primary and secondary glioblastoma. Am J Pathol. 2007;170:1445–1453. doi: 10.2353/ajpath.2007.070011. S0002-9440(10)61358-2[pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohgaki H, Dessen P, Jourde B, Horstmann S, Nishikawa T, Di Patre PL, Burkhard C, Schuler D, Probst-Hensch NM, Maiorka PC, Baeza N, Pisani P, Yonekawa Y, Yasargil MG, Lutolf UM, Kleihues P. Genetic pathways to glioblastoma: a population-based study. Cancer Res. 2004;64:6892–6899. doi: 10.1158/0008-5472.CAN-04-1337. 64/19/6892[pii] [DOI] [PubMed] [Google Scholar]
- Okkenhaug K, Bilancio A, Farjot G, Priddle H, Sancho S, Peskett E, Pearce W, Meek SE, Salpekar A, Waterfield MD, Smith AJ, Vanhaesebroeck B. Impaired B and T cell antigen receptor signaling in p110delta PI 3-kinase mutant mice. Science. 2002;297:1031–1034. doi: 10.1126/science.10735601073560[pii]. [DOI] [PubMed] [Google Scholar]
- O’Reilly KE, Rojo F, She QB, Solit D, Mills GB, Smith D, Lane H, Hofmann F, Hicklin DJ, Ludwig DL, Baselga J, Rosen N. mTOR inhibition induces upstream receptor tyrosine kinase signaling and activates Akt. Cancer Res. 2006;66:1500–1508. doi: 10.1158/0008-5472.CAN-05-2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostman A, Heldin CH. PDGF receptors as targets in tumor treatment. Adv Cancer Res. 2007;97:247–274. doi: 10.1016/S0065-230X(06)97011-0. S0065-230X(06)97011-0[pii] [DOI] [PubMed] [Google Scholar]
- Ozawa T, Brennan CW, Wang L, Squatrito M, Sasayama T, Nakada M, Huse JT, Pedraza A, Utsuki S, Yasui Y, Tandon A, Fomchenko EI, Oka H, Levine RL, Fujii K, Ladanyi M, Holland EC. PDGFRA gene rearrangements are frequent genetic events in PDGFRA-amplified glioblastomas. Genes Dev. 2010;24:2205–2218. doi: 10.1101/gad.1972310. 24/19/2205[pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Siu IM, Gallia GL, Olivi A, McLendon R, Rasheed BA, Keir S, Nikolskaya T, Nikolsky Y, Busam DA, Tekleab H, Diaz LA, Jr, Hartigan J, Smith DR, Strausberg RL, Marie SK, Shinjo SM, Yan H, Riggins GJ, Bigner DD, Karchin R, Papadopoulos N, Parmigiani G, Vogelstein B, Velculescu VE, Kinzler KW. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. 1164382[pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paugh BS, Qu C, Jones C, Liu Z, Adamowicz-Brice M, Zhang J, Bax DA, Coyle B, Barrow J, Hargrave D, Lowe J, Gajjar A, Zhao W, Broniscer A, Ellison DW, Grundy RG, Baker SJ. Integrated molecular genetic profiling of pediatric high-grade gliomas reveals key differences with the adult disease. J Clin Oncol. 2010;28:3061–3068. doi: 10.1200/JCO.2009.26.7252. JCO.2009.26.7252[-pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfister S, Janzarik WG, Remke M, Ernst A, Werft W, Becker N, Toedt G, Wittmann A, Kratz C, Olbrich H, Ahmadi R, Thieme B, Joos S, Radlwimmer B, Kulozik A, Pietsch T, Herold-Mende C, Gnekow A, Reifenberger G, Korshunov A, Scheurlen W, Omran H, Lichter P. BRAF gene duplication constitutes a mechanism of MAPK pathway activation in low-grade astrocytomas. J Clin Invest. 2008;118:1739–1749. doi: 10.1172/JCI33656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips HS, Kharbanda S, Chen R, Forrest WF, Soriano RH, Wu TD, Misra A, Nigro JM, Colman H, Soroceanu L, Williams PM, Modrusan Z, Feuerstein BG, Aldape K. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9:157–173. doi: 10.1016/j.ccr.2006.02.019. S1535-6108(06)00056-0[pii] [DOI] [PubMed] [Google Scholar]
- Philp AJ, Campbell IG, Leet C, Vincan E, Rockman SP, Whitehead RH, Thomas RJ, Phillips WA. The phosphatidylinositol 3′-kinase p85alpha gene is an oncogene in human ovarian and colon tumors. Cancer Res. 2001;61:7426–7429. [PubMed] [Google Scholar]
- Podsypanina K, Lee RT, Politis C, Hennessy I, Crane A, Puc J, Neshat M, Wang H, Yang L, Gibbons J, Frost P, Dreisbach V, Blenis J, Gaciong Z, Fisher P, Sawyers C, Hedrick-Ellenson L, Parsons R. An inhibitor of mTOR reduces neoplasia and normalizes p70/S6 kinase activity in Pten ± mice. Proc Natl Acad Sci U S A. 2001;98:10320–10325. doi: 10.1073/pnas.171060098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prados MD, Chang SM, Butowski N, DeBoer R, Parvataneni R, Carliner H, Kabuubi P, Ayers-Ringler J, Rabbitt J, Page M, Fedoroff A, Sneed PK, Berger MS, McDermott MW, Parsa AT, Vandenberg S, James CD, Lamborn KR, Stokoe D, Haas-Kogan DA. Phase II study of erlotinib plus temozolomide during and after radiation therapy in patients with newly diagnosed glioblastoma multiforme or gliosarcoma. J Clin Oncol. 2009;27:579–584. doi: 10.1200/JCO.2008.18.9639. J-CO. 2008.18.9639[pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad G, Sottero T, Yang X, Mueller S, James CD, Weiss WA, Polley MY, Ozawa T, Berger MS, Aftab DT, Prados MD, Haas-Kogan DA. Inhibition of PI3K/mTOR pathways in glioblastoma and implications for combination therapy with temozolomide. Neuro Oncol. 2011;13:384–392. doi: 10.1093/neuonc/noq193. noq193[pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rand V, Huang J, Stockwell T, Ferriera S, Buzko O, Levy S, Busam D, Li K, Edwards JB, Eberhart C, Murphy KM, Tsiamouri A, Beeson K, Simpson AJ, Venter JC, Riggins GJ, Strausberg RL. Sequence survey of receptor tyrosine kinases reveals mutations in glioblastomas. Proc Natl Acad Sci U S A. 2005;102:14344–14349. doi: 10.1073/pnas.0507200102. 0507200102[pii] 10.1073/ pnas.0507200102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond E, Brandes AA, Dittrich C, Fumoleau P, Coudert B, Clement PM, Frenay M, Rampling R, Stupp R, Kros JM, Heinrich MC, Gorlia T, Lacombe D, van den Bent MJ. Phase II study of imatinib in patients with recurrent gliomas of various histologies: a European Organisation for Research and Treatment of Cancer Brain Tumor Group Study. J Clin Oncol. 2008;26:4659–4665. doi: 10.1200/JCO.2008.16.9235. 26/28/4659[pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reardon DA, Desjardins A, Vredenburgh JJ, Gururangan S, Friedman AH, Herndon JE, 2nd, Marcello J, Norfleet JA, McLendon RE, Sampson JH, Friedman HS. Phase 2 trial of erlotinib plus sirolimus in adults with recurrent glioblastoma. J Neurooncol. 2010;96:219–230. doi: 10.1007/s11060-009-9950-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reilly KM, Loisel DA, Bronson RT, McLaughlin ME, Jacks T. Nf1;Trp53 mutant mice develop glioblastoma with evidence of strain-specific effects. Nat Genet. 2000;26:109–113. doi: 10.1038/79075. [DOI] [PubMed] [Google Scholar]
- Richardson WD, Pringle N, Mosley MJ, Westermark B, Dubois-Dalcq M. A role for platelet-derived growth factor in normal gliogenesis in the central nervous system. Cell. 1988;53:309–319. doi: 10.1016/0092-8674(88)90392-3. 0092-8674(88)90392-3[pii] [DOI] [PubMed] [Google Scholar]
- Riemenschneider MJ, Jeuken JW, Wesseling P, Reifenberger G. Molecular diagnostics of gliomas: state of the art. Acta Neuropathol. 2010;120:567–584. doi: 10.1007/s00401-010-0736-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JP, VanBrocklin MW, Guilbeault AR, Signorelli DL, Brandner S, Holmen SL. Activated BRAF induces gliomas in mice when combined with Ink4a/Arf loss or Akt activation. Oncogene. 2010;29:335–344. doi: 10.1038/onc.2009.333. onc2009333[pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JP, Vanbrocklin MW, Lastwika KJ, McKinney AJ, Brandner S, Holmen SL. Activated MEK cooperates with Ink4a/Arf loss or Akt activation to induce gliomas in vivo. Oncogene. 2011;30:1341–1350. doi: 10.1038/onc.2010.513. onc2010513[pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez FJ, Perry A, Gutmann DH, O’Neill BP, Leonard J, Bryant S, Giannini C. Gliomas in neurofibromatosis type 1: a clinicopathologic study of 100 patients. J Neuropathol Exp Neurol. 2008;67:240–249. doi: 10.1097/NEN.0b013e318165eb7500005072-200803000-00007[pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmena L, Carracedo A, Pandolfi PP. Tenets of PTEN tumor suppression. Cell. 2008;133:403–414. doi: 10.1016/j.cell.2008.04.013. S0092-8674(08)00504-7[pii] [DOI] [PubMed] [Google Scholar]
- Samuels Y, Wang Z, Bardelli A, Silliman N, Ptak J, Szabo S, Yan H, Gazdar A, Powell SM, Riggins GJ, Willson JK, Markowitz S, Kinzler KW, Vogelstein B, Velculescu VE. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304:554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- Sarkaria JN, Yang L, Grogan PT, Kitange GJ, Carlson BL, Schroeder MA, Galanis E, Giannini C, Wu W, Dinca EB, James CD. Identification of molecular characteristics correlated with glioblastoma sensitivity to EGFR kinase inhibition through use of an intracranial xenograft test panel. Mol Cancer Ther. 2007;6:1167–1174. doi: 10.1158/1535-7163.MCT-06-0691. 6/3/1167[pii] [DOI] [PubMed] [Google Scholar]
- Sasaki T, Irie-Sasaki J, Jones RG, Oliveira-dos-Santos AJ, Stanford WL, Bolon B, Wakeham A, Itie A, Bouchard D, Kozieradzki I, Joza N, Mak TW, Ohashi PS, Suzuki A, Penninger JM. Function of PI3Kgamma in thymocyte development, T cell activation, and neutrophil migration. Science. 2000;287:1040–1046. doi: 10.1126/science.287.5455.1040. 8264[pii] [DOI] [PubMed] [Google Scholar]
- Sawyers CL. Shifting paradigms: the seeds of oncogene addiction. Nat Med. 2009;15:1158–1161. doi: 10.1038/nm1009-1158. nm1009-1158[pii] [DOI] [PubMed] [Google Scholar]
- Schatzman RC, Evan GI, Privalsky ML, Bishop JM. Orientation of the v-erb-B gene product in the plasma membrane. Mol Cell Biol. 1986;6:1329–1333. doi: 10.1128/mcb.6.4.1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiffman JD, Hodgson JG, VandenBerg SR, Flaherty P, Polley MY, Yu M, Fisher PG, Rowitch DH, Ford JM, Berger MS, Ji H, Gutmann DH, James CD. Oncogenic BRAF mutation with CDKN2A inactivation is characteristic of a subset of pediatric malignant astrocytomas. Cancer Res. 2010;70:512–519. doi: 10.1158/0008-5472.CAN-09-1851. 0008-5472.CAN-09-1851[pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler G, Capper D, Meyer J, Janzarik W, Omran H, Herold-Mende C, Schmieder K, Wesseling P, Mawrin C, Hasselblatt M, Louis DN, Korshunov A, Pfister S, Hartmann C, Paulus W, Reifenberger G, von Deimling A. Analysis of BRAF V600E mutation in 1, 320 nervous system tumors reveals high mutation frequencies in pleomorphic xanthoastrocytoma, ganglioglioma and extra-cerebellar pilocytic astrocytoma. Acta Neuropathol. 2011;121:397–405. doi: 10.1007/s00401-011-0802-6. [DOI] [PubMed] [Google Scholar]
- Sengupta S, Peterson TR, Sabatini DM. Regulation of the mTOR complex 1 pathway by nutrients, growth factors, and stress. Mol Cell. 2010;40:310–322. doi: 10.1016/j.molcel.2010.09.026. S1097-2765(10)00754-9[pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serra V, Markman B, Scaltriti M, Eichhorn PJ, Valero V, Guzman M, Botero ML, Llonch E, Atzori F, Di Cosimo S, Maira M, Garcia-Echeverria C, Parra JL, Arribas J, Baselga J. NVP-BEZ235, a dual PI3K/mTOR inhibitor, prevents PI3K signaling and inhibits the growth of cancer cells with activating PI3K mutations. Cancer Res. 2008;68:8022–8030. doi: 10.1158/0008-5472.CAN-08-1385. 68/19/ 8022[pii] [DOI] [PubMed] [Google Scholar]
- Shah NP, Kasap C, Weier C, Balbas M, Nicoll JM, Bleickardt E, Nicaise C, Sawyers CL. Transient potent BCR-ABL inhibition is sufficient to commit chronic myeloid leukemia cells irreversibly to apoptosis. Cancer Cell. 2008;14:485–493. doi: 10.1016/j.ccr.2008.11.001. S1535-6108(08)00368-1[pii] 10.1016/ j.ccr.2008.11.001. [DOI] [PubMed] [Google Scholar]
- Sharma MK, Zehnbauer BA, Watson MA, Gutmann DH. RAS pathway activation and an oncogenic RAS mutation in sporadic pilocytic astrocytoma. Neurology. 2005;65:1335–1336. doi: 10.1212/01.wnl.0000180409.78098.d7. 65/ 8/1335[pii] [DOI] [PubMed] [Google Scholar]
- Shaw RJ, Cantley LC. Ras, PI(3)K and mTOR signalling controls tumour cell growth. Nature. 2006;441:424–430. doi: 10.1038/nature04869. [DOI] [PubMed] [Google Scholar]
- Shayesteh L, Lu Y, Kuo WL, Baldocchi R, Godfrey T, Collins C, Pinkel D, Powell B, Mills GB, Gray JW. PIK3CA is implicated as an oncogene in ovarian cancer. Nat Genet. 1999;21:99–102. doi: 10.1038/5042. [DOI] [PubMed] [Google Scholar]
- She QB, Chandarlapaty S, Ye Q, Lobo J, Haskell KM, Leander KR, DeFeo-Jones D, Huber HE, Rosen N. Breast tumor cells with PI3K mutation or HER2 amplification are selectively addicted to Akt signaling. PLoS ONE. 2008;3:e3065. doi: 10.1371/journal.pone.0003065. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Shuttleworth SJ, Silva FA, Cecil AR, Tomassi CD, Hill TJ, Raynaud FI, Clarke PA, Workman P. Progress in the Preclinical Discovery and Clinical Development of Class I and Dual Class I/IV Phosphoinositide 3-Kinase (PI3K) Inhibitors. Curr Med Chem. 2011;18:2686–2714. doi: 10.2174/092986711796011229. BSP/CMC/E-Pub/2011/181[pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievert AJ, Jackson EM, Gai X, Hakonarson H, Judkins AR, Resnick AC, Sutton LN, Storm PB, Shaikh TH, Biegel JA. Duplication of 7q34 in pediatric low-grade astrocytomas detected by high-density single-nucleotide polymorphism-based genotype arrays results in a novel BRAF fusion gene. Brain Pathol. 2009;19:449–458. doi: 10.1111/j.1750-3639.2008.00225.x. BPA225[pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slamon DJ, Leyland-Jones B, Shak S, Fuchs H, Paton V, Bajamonde A, Fleming T, Eiermann W, Wolter J, Pegram M, Baselga J, Norton L. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N Engl J Med. 2001;344:783–792. doi: 10.1056/NEJM200103153441101. [DOI] [PubMed] [Google Scholar]
- Smalley KS, Sondak VK. Melanoma–an unlikely poster child for personalized cancer therapy. N Engl J Med. 2010;363:876–878. doi: 10.1056/NEJMe1005370. [DOI] [PubMed] [Google Scholar]
- Smith JS, Tachibana I, Passe SM, Huntley BK, Borell TJ, Iturria N, O’Fallon JR, Schaefer PL, Scheithauer BW, James CD, Buckner JC, Jenkins RB. PTEN mutation, EGFR amplification, and outcome in patients with anaplastic astrocytoma and glioblastoma multiforme. J Natl Cancer Inst. 2001;93:1246–1256. doi: 10.1093/jnci/93.16.1246. [DOI] [PubMed] [Google Scholar]
- Solit DB, Mellinghoff IK. Tracing cancer networks with phosphoproteomics. Nat Biotechnol. 2010;28:1028–1029. doi: 10.1038/nbt1010-1028. nbt1010-1028[pii] [DOI] [PubMed] [Google Scholar]
- Steck PA, Pershouse MA, Jasser SA, Yung WK, Lin H, Ligon AH, Langford LA, Baumgard ML, Hattier T, Davis T, Frye C, Hu R, Swedlund B, Teng DH, Tavtigian SV. Identification of a candidate tumour suppressor gene, MMAC1, at chromosome 10q23.3 that is mutated in multiple advanced cancers. Nat Genet. 1997;15:356–362. doi: 10.1038/ng0497-356. [DOI] [PubMed] [Google Scholar]
- Stommel JM, Kimmelman AC, Ying H, Nabioullin R, Ponugoti AH, Wiedemeyer R, Stegh AH, Bradner JE, Ligon KL, Brennan C, Chin L, DePinho RA. Coactivation of receptor tyrosine kinases affects the response of tumor cells to targeted therapies. Science. 2007;318:287–290. doi: 10.1126/science.1142946. 1142946[pii] [DOI] [PubMed] [Google Scholar]
- Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn U, Curschmann J, Janzer RC, Ludwin SK, Gorlia T, Allgeier A, Lacombe D, Cairncross JG, Eisenhauer E, Mirimanoff RO. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- Sugawa N, Ekstrand AJ, James CD, Collins VP. Identical splicing of aberrant epidermal growth factor receptor transcripts from amplified rearranged genes in human glioblastomas. Proc Natl Acad Sci U S A. 1990;87:8602–8606. doi: 10.1073/pnas.87.21.8602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun SY, Rosenberg LM, Wang X, Zhou Z, Yue P, Fu H, Khuri FR. Activation of Akt and eIF4E survival pathways by rapamycin-mediated mammalian target of rapamycin inhibition. Cancer Res. 2005;65:7052–7058. doi: 10.1158/0008-5472.CAN-05-0917. [DOI] [PubMed] [Google Scholar]
- Sun M, Hillmann P, Hofmann BT, Hart JR, Vogt PK. Cancer-derived mutations in the regulatory subunit p85alpha of phosphoinositide 3-kinase function through the catalytic subunit p110alpha. Proc Natl Acad Sci U S A. 2010;107:15547–15552. doi: 10.1073/pnas.1009652107. 1009652107[-pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi H, Kondo Y, Fujiwara K, Kanzawa T, Aoki H, Mills GB, Kondo S. Synergistic augmentation of rapamycin-induced autophagy in malignant glioma cells by phosphatidyl-inositol 3-kinase/protein kinase B inhibitors. Cancer Res. 2005;65:3336–3346. doi: 10.1158/0008-5472.CAN-04-3640. 65/8/ 3336[pii] [DOI] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Research Network (TCGA) (2005)
- Thiel G, Marczinek K, Neumann R, Witkowski R, Marchuk DA, Nurnberg P. Somatic mutations in the neurofibromatosis 1 gene in gliomas and primitive neuroectodermal tumours. Anticancer Res. 1995;15:2495–2499. [PubMed] [Google Scholar]
- Uhrbom L, Hesselager G, Nister M, Westermark B. Induction of brain tumors in mice using a recombinant platelet-derived growth factor B-chain retrovirus. Cancer Res. 1998;58:5275–5279. [PubMed] [Google Scholar]
- Uhrbom L, Dai C, Celestino JC, Rosenblum MK, Fuller GN, Holland EC. Ink4a-Arf loss cooperates with KRas activation in astrocytes and neural progenitors to generate glioblastomas of various morphologies depending on activated Akt. Cancer Res. 2002;62:5551–5558. [PubMed] [Google Scholar]
- van den Bent MJ, Brandes AA, Rampling R, Kouwenhoven MC, Kros JM, Carpentier AF, Clement PM, Frenay M, Campone M, Baurain JF, Armand JP, Taphoorn MJ, Tosoni A, Kletzl H, Klughammer B, Lacombe D, Gorlia T. Randomized phase II trial of erlotinib versus temozolomide or carmustine in recurrent glioblastoma: EORTC brain tumor group study 26034. J Clin Oncol. 2009;27:1268–1274. doi: 10.1200/JCO.2008.17.5984. JCO.2008.17.5984[pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanhaesebroeck B, Guillermet-Guibert J, Graupera M, Bilanges B. The emerging mechanisms of isoform-specific PI3K signalling. Nat Rev Mol Cell Biol. 2010;11:329–341. doi: 10.1038/nrm2882. nrm2882[pii] [DOI] [PubMed] [Google Scholar]
- Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, Miller CR, Ding L, Golub T, Mesirov JP, Alexe G, Lawrence M, O’Kelly M, Tamayo P, Weir BA, Gabriel S, Winckler W, Gupta S, Jakkula L, Feiler HS, Hodgson JG, James CD, Sarkaria JN, Brennan C, Kahn A, Spellman PT, Wilson RK, Speed TP, Gray JW, Meyerson M, Getz G, Perou CM, Hayes DN. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. S1535-6108(09)00432-2[pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivanco I, Rohle D, Versele M, Iwanami A, Kuga D, Oldrini B, Tanaka K, Dang J, Kubek S, Palaskas N, Hsueh T, Evans M, Mulholland D, Wolle D, Rajasekaran S, Rajasekaran A, Liau LM, Cloughesy TF, Dikic I, Brennan C, Wu H, Mischel PS, Perera T, Mellinghoff IK. The phosphatase and tensin homolog regulates epidermal growth factor receptor (EGFR) inhibitor response by targeting EGFR for degradation. Proc Natl Acad Sci U S A. 2010;107:6459–6464. doi: 10.1073/pnas.0911188107. 0911188107[pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt PK, Gymnopoulos M, Hart JR. PI 3-kinase and cancer: changing accents. Curr Opin Genet Dev. 2009;19:12–17. doi: 10.1016/j.gde.2008.11.011. S0959-437X(08)00188-3[pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang MY, Lu KV, Zhu S, Dia EQ, Vivanco I, Shackleford GM, Cavenee WK, Mellinghoff IK, Cloughesy TF, Sawyers CL, Mischel PS. Mammalian target of rapamycin inhibition promotes response to epidermal growth factor receptor kinase inhibitors in PTEN-deficient and PTEN-intact glioblastoma cells. Cancer Res. 2006;66:7864–7869. doi: 10.1158/0008-5472.CAN-04-4392. 66/16/7864[pii] 10.1158/ 0008-5472.CAN-04-4392. [DOI] [PubMed] [Google Scholar]
- Wee S, Wiederschain D, Maira SM, Loo A, Miller C, deBeaumont R, Stegmeier F, Yao YM, Lengauer C. PTEN-deficient cancers depend on PIK3CB. Proc Natl Acad Sci U S A. 2008;105:13057–13062. doi: 10.1073/pnas.0802655105. 0802655105[pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Q, Clarke L, Scheidenhelm DK, Qian B, Tong A, Sabha N, Karim Z, Bock NA, Reti R, Swoboda R, Purev E, Lavoie JF, Bajenaru ML, Shannon P, Herlyn D, Kaplan D, Henkelman RM, Gutmann DH, Guha A. High-grade glioma formation results from postnatal pten loss or mutant epidermal growth factor receptor expression in a transgenic mouse glioma model. Cancer Res. 2006;66:7429–7437. doi: 10.1158/0008-5472.CAN-06-0712. 66/15/7429[pii] [DOI] [PubMed] [Google Scholar]
- Weigelt B, Warne PH, Downward J. PIK3CA mutation, but not PTEN loss of function, determines the sensitivity of breast cancer cells to mTOR inhibitory drugs. Oncogene. 2011;30:3222–3233. doi: 10.1038/onc.2011.42. onc201142[pii] [DOI] [PubMed] [Google Scholar]
- Weiss WA, Burns MJ, Hackett C, Aldape K, Hill JR, Kuriyama H, Kuriyama N, Milshteyn N, Roberts T, Wendland MF, DePinho R, Israel MA. Genetic determinants of malignancy in a mouse model for oligodendroglioma. Cancer Res. 2003;63:1589–1595. [PubMed] [Google Scholar]
- Wells A, Welsh JB, Lazar CS, Wiley HS, Gill GN, Rosenfeld MG. Ligand-induced transformation by a noninternalizing epidermal growth factor receptor. Science. 1990;247(4945):962–964. doi: 10.1126/science.2305263. [DOI] [PubMed] [Google Scholar]
- Wen PY, Yung WK, Lamborn KR, Dahia PL, Wang Y, Peng B, Abrey LE, Raizer J, Cloughesy TF, Fink K, Gilbert M, Chang S, Junck L, Schiff D, Lieberman F, Fine HA, Mehta M, Robins HI, DeAngelis LM, Groves MD, Puduvalli VK, Levin V, Conrad C, Maher EA, Aldape K, Hayes M, Letvak L, Egorin MJ, Capdeville R, Kaplan R, Murgo AJ, Stiles C, Prados MD. Phase I/II study of imatinib mesylate for recurrent malignant gliomas: North American Brain Tumor Consortium Study 99–08. Clin Cancer Res. 2006;12:4899–4907. doi: 10.1158/1078-0432.CCR-06-0773. 12/16/4899[pii] [DOI] [PubMed] [Google Scholar]
- Wong AJ, Bigner SH, Bigner DD, Kinzler KW, Hamilton SR, Vogelstein B. Increased expression of the epidermal growth factor receptor gene in malignant gliomas is invariably associated with gene amplification. Proc Natl Acad Sci U S A. 1987;84:6899–6903. doi: 10.1073/pnas.84.19.6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Workman P, Clarke PA, Raynaud FI, van Montfort RL. Drugging the PI3 kinome: from chemical tools to drugs in the clinic. Cancer Res. 2010;70:2146–2157. doi: 10.1158/0008-5472.CAN-09-4355. 0008-5472.CAN-09-4355[pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Shekar SC, Flinn RJ, El-Sibai M, Jaiswal BS, Sen KI, Janakiraman V, Seshagiri S, Gerfen GJ, Girvin ME, Backer JM. Regulation of Class IA PI 3-kinases: C2 domain-iSH2 domain contacts inhibit p85/p110alpha and are disrupted in oncogenic p85 mutants. Proc Natl Acad Sci U S A. 2009;106:20258–20263. doi: 10.1073/pnas.0902369106. 0902369106[pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu CX, Zhao L, Yue P, Fang G, Tao H, Owonikoko TK, Ramalingam SS, Khuri FR, Sun SY. Augmentation of NVP-BEZ235's anticancer activity against human lung cancer cells by blockage of autophagy. Cancer Biol Ther. 2011;12 doi: 10.4161/cbt.12.6.16397. 16397 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki H, Fukui Y, Ueyama Y, Tamaoki N, Kawamoto T, Taniguchi S, Shibuya M. Amplification of the structurally and functionally altered epidermal growth factor receptor gene (c-erbB) in human brain tumors. Mol Cell Biol. 1988;8:1816–1820. doi: 10.1128/mcb.8.4.1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Deshmukh H, Gutmann RJ, Emnett RJ, Rodriguez FJ, Watson MA, Nagarajan R, Gutmann DH. Alterations of BRAF and HIPK2 loci predominate in sporadic pilocytic astrocytoma. Neurology. 2009;73:1526–1531. doi: 10.1212/WNL.0b013e3181c0664a. WNL.0b013e3181c0664a[pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H, Ying H, Yan H, Kimmelman AC, Hiller DJ, Chen AJ, Perry SR, Tonon G, Chu GC, Ding Z, Stommel JM, Dunn KL, Wiedemeyer R, You MJ, Brennan C, Wang YA, Ligon KL, Wong WH, Chin L, DePinho RA. p53 and Pten control neural and glioma stem/ progenitor cell renewal and differentiation. Nature. 2008;455:1129–1133. doi: 10.1038/nature07443. nature07443[-pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Guignard F, Zhao D, Liu L, Burns DK, Mason RP, Messing A, Parada LF. Early inactivation of p53 tumor suppressor gene cooperating with NF1 loss induces malignant astrocytoma. Cancer Cell. 2005a;8:119–130. doi: 10.1016/j.ccr.2005.07.004. S1535-6108(05)00228-X[pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Harada T, Liu L, Lush ME, Guignard F, Harada C, Burns DK, Bajenaru ML, Gutmann DH, Parada LF. Inactivation of NF1 in CNS causes increased glial progenitor proliferation and optic glioma formation. Development. 2005b;132:5577–5588. doi: 10.1242/dev.02162. 132/24/5577[pii] 10.1242/ dev.02162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Acquaviva J, Ramachandran P, Boskovitz A, Woolfenden S, Pfannl R, Bronson RT, Chen JW, Weissleder R, Housman DE, Charest A. Oncogenic EGFR signaling cooperates with loss of tumor suppressor gene functions in gliomagenesis. Proc Natl Acad Sci U S A. 2009;106:2712–2716. doi: 10.1073/pnas.0813314106. 0813314106[pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoncu R, Efeyan A, Sabatini DM. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol. 2011;12:21–35. doi: 10.1038/nrm3025. nrm3025[pii] [DOI] [PMC free article] [PubMed] [Google Scholar]