SUMMARY
Numerous cytosolic and nuclear proteins involved in metabolism, DNA maintenance, protein translation, or iron homeostasis depend on iron-sulfur (Fe/S) cofactors, yet their assembly is poorly defined. Here, we identify and characterize human CIA2A (FAM96A), CIA2B (FAM96B), and CIA1 (CIAO1) as components of the cytosolic Fe/S protein assembly (CIA) machinery. CIA1 associates with either CIA2A or CIA2B and the CIA targeting factor MMS19. The CIA2B-CIA1-MMS19 complex binds to and facilitates assembly of most cytosolic-nuclear Fe/S proteins. In contrast, CIA2A specifically matures iron regulatory protein (IRP) 1 which is critical for cellular iron homeostasis. Surprisingly, a second layer of iron regulation involves the stabilization of IRP2 by CIA2A binding or upon depletion of CIA2B or MMS19, even though IRP2 lacks a Fe/S cluster. In summary, CIA2B-CIA1-MMS19 and CIA2A-CIA1 assist different branches of Fe/S protein assembly, and intimately link this process to cellular iron regulation via IRP1 Fe/S cluster maturation and IRP2 stabilization.
INTRODUCTION
Iron-sulfur (Fe/S) clusters are inorganic cofactors of many proteins participating in fundamental biological processes (Ayala-Castro et al., 2008; Lill et al., 2012; Py and Barras, 2010; Rouault, 2012; Sheftel et al., 2010a). They mediate electron transfer (e.g., in respiratory complexes I, II, and III), are part of catalytic centers (mitochondrial aconitase of the TCA cycle), contribute to nucleotide metabolism (glutamine phosphoribosylpyrophosphate amidotransferase, GPAT (Martelli et al., 2007); dihydropyrimidine dehydrogenase, DPYD (Schnackerz et al., 2004)), and are required for essential steps of DNA replication (catalytic subunits of replicative DNA polymerases (Netz et al., 2012b)) and DNA repair (DNA helicases such as Xeroderma pigmentosum complementation group D protein, XPD or Fanconi anemia protein J, FANCJ (Rudolf et al., 2006). In addition, Fe/S proteins such as the bifunctional iron regulatory protein 1 (IRP1) execute sensory functions (Anderson et al., 2012). When IRP1 carries a [4Fe-4S] cluster, it acts as a cytosolic aconitase. In the absence of this cofactor, IRP1 binds to mRNA stem-loop structures called iron-responsive elements (IRE) and post-transcriptionally regulates the expression of various proteins involved in cellular iron homeostasis. A second layer of regulation might include the iron-dependent degradation of IRP1 by the E3 ubiquitin ligase FBXL5, as these proteins were shown to interact (Salahudeen et al., 2009; Vashisht et al., 2009).
The assembly of Fe/S proteins in mitochondria, cytosol and nucleus of eukaryotes depends on the concerted action of complex biogenesis systems (for review see (Lill, 2009; Lill et al., 2012; Rouault, 2012)). The mitochondrial iron-sulfur cluster (ISC) assembly machinery was inherited from bacteria (Johnson et al., 2005), and is required for the maturation of both mitochondrial and extra-mitochondrial Fe/S proteins. The cytosolic iron-sulfur protein assembly (CIA) apparatus is specific for the maturation of cytosolic and nuclear Fe/S proteins. Despite the lack of structural similarities between the ISC and CIA components, the synthesis of Fe/S clusters and their incorporation into apoproteins by these two systems appear to follow similar mechanistic principles. First, a Fe/S cluster is assembled on a scaffold complex requiring the sulfide production by the mitochondrial cysteine desulfurase complex NFS1-ISD11. In a second step, the Fe/S cluster is released from the scaffold, transferred to specific target apoproteins, and incorporated into the polypeptide chains. Each of these steps requires assistance by dedicated ISC or CIA factors.
Initial studies on the CIA machinery were performed in yeast. The cytosolic P-loop NTPases Cfd1 and Nbp35 were characterized as a scaffold on which a transiently bound [4Fe-4S] cluster is assembled (Hausmann et al., 2005; Netz et al., 2012a; Netz et al., 2007; Roy et al., 2003; Sharma et al., 2010). The electron transfer from NADPH to the diflavin oxidoreductase Tah18 and the Fe/S protein Dre2 is essential for the assembly process, and is particularly required for insertion of the N-terminal [4Fe-4S] cluster of Nbp35 (Netz et al., 2010; Zhang et al., 2008). Transfer of the labile Fe/S cluster from the Cfd1-Nbp35 scaffold to target apoproteins requires the Fe-only hydrogenase-like protein Nar1 and the WD40 repeat protein Cia1 (Balk et al., 2005; Balk et al., 2004; Srinivasan et al., 2007). Recently, the HEAT repeat protein Mms19 (also called Met18) has been identified as a CIA component which directly binds to apoproteins and assists Fe/S cluster incorporation into several Fe/S proteins involved in genome stability and DNA maintenance (Gari et al., 2012; Stehling et al., 2012). Mms19 forms a complex with Cia1 and Cia2. The latter protein (encoded by yeast gene YHR122w) contains a DUF59 domain and has been implicated in cytosolic Fe/S protein assembly in a systematic screen for proteins containing reactive cysteine residues (Weerapana et al., 2010). The precise role of Cia2 is unknown.
The eight known CIA components are conserved from yeast to man suggesting similar molecular mechanisms of Fe/S protein assembly in all eukaryotes. So far, only human NBP35, IOP1 (homologue of yeast Nar1; for nomenclature see Table S1) and MMS19 have been functionally investigated, and shown to act similarly to their yeast counterparts (Gari et al., 2012; Seki et al., 2013; Song and Lee, 2008; Stehling et al., 2008; Stehling et al., 2012). Human MMS19 is specifically involved in the maturation of both DPYD and DNA polymerase POLD1, but is dispensable for IRP1 and GPAT assembly, suggesting a target-specific function of MMS19 similar to its yeast counterpart (Stehling et al., 2012). MMS19 interacts with IOP1, CIA1, CIA2B (FAM96B; Table S1) and numerous cytosolic-nuclear Fe/S proteins (Gari et al., 2012; Ito et al., 2010; Luo et al., 2012; Stehling et al., 2012; van Wietmarschen et al., 2012). Hence, the MMS19-CIA1-CIA2B complex was proposed to function as a targeting complex facilitating the specific delivery of the Fe/S clusters to apoproteins. So far, the function of human CIA1 and CIA2B in the maturation of Fe/S proteins has not been studied yet. Intriguingly, and in contrast to yeast, humans possess a second Cia2 homologue designated CIA2A (FAM96A; Table S1) which has not been investigated so far. Notably, gene duplications have also occurred for the mitochondrial ISC component ferredoxin and for cytosolic IOP1, but typically only one of these isoforms participates in Fe/S protein biogenesis (Sheftel et al., 2010b; Song and Lee, 2008).
Here, we used RNAi-mediated protein depletion to investigate the potential involvement of human CIA1, CIA2A and CIA2B in the maturation of cytosolic-nuclear Fe/S proteins. The cell biological studies were complemented by both proteomic analyses and dedicated co-immunoprecipitations to identify the specific protein partners of CIA1, CIA2A and CIA2B. Our results suggest that these proteins form different complexes that act late in the CIA pathway to target different Fe/S proteins. Particularly, the function of CIA2A connects cytosolic Fe/S protein maturation with cellular iron regulation providing a hitherto unknown molecular link between these two important biological processes.
RESULTS
Localization and knock-down of human CIA1, CIA2A, and CIA2B proteins
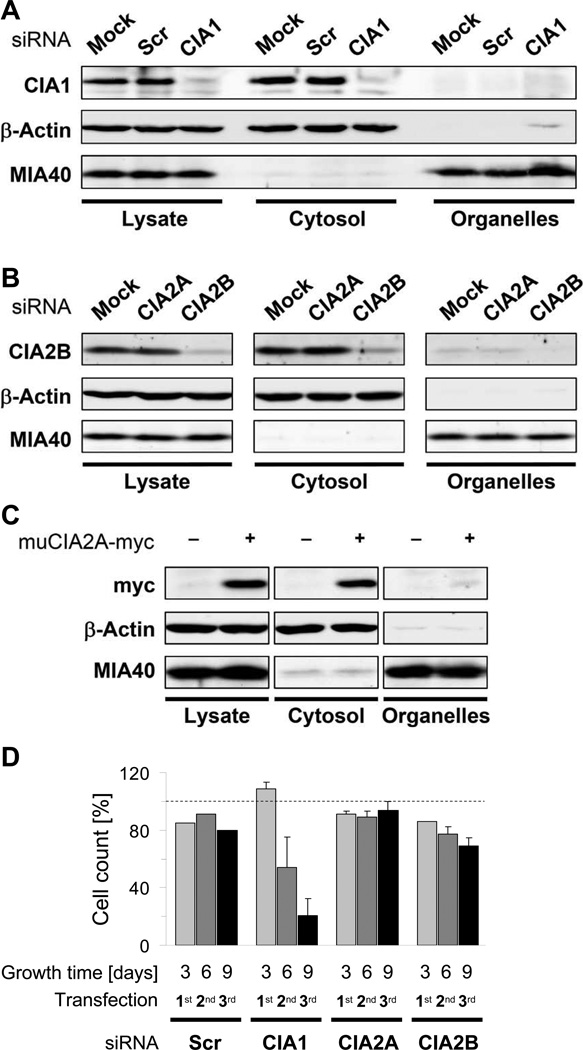
We first determined the subcellular localization of CIA1, CIA2A, and CIA2B by immuno-blotting of cytosolic and organelle-containing fractions derived from digitonin-permeabilised HeLa cells (Stehling et al., 2008). Since, in contrast to CIA1 and CIA2B, no functional CIA2A-directed antibody was available, we analyzed HeLa cells expressing a C-terminally myc-tagged version of murine CIA2A (muCIA2A-myc). CIA1, CIA2B, and muCIA2A-myc were detectable in the cytosolic fraction but nearly absent in the organelle fraction (Fig. 1A–C). Actin and MIA40 served as cytosolic and mitochondrial markers, respectively. RNAi-mediated depletion verified the specificity of the CIA1 and CIA2B immunostaining (Fig. 1A–B). Depletion was achieved by three consecutive transfections at a 3-day interval applying three different siRNAs either individually or as a pool. Particularly for the pooled siRNAs, quantitative real-time PCR and immunostaining showed a strong decrease (>75%) in mRNA and protein levels, respectively (Fig. S1A–B). Individual siRNAs were also highly active, but usually showed lower efficiencies. Scrambled siRNAs did not affect CIA1 and CIA2B levels (Fig. 1A–B). Importantly, CIA2A-directed siRNAs did not diminish protein levels of the related CIA2B (Fig. 1B). Actin and MIA40 levels were not altered by the RNAi treatments. Only depletion of CIA1 resulted in a severe, time-dependent effect on cell growth as measured by cell counting and protein yield (Figs. 1D and S1C–D). Taken together, the digitonin-based cell fractionation approach identified human CIA1, CIA2A, and CIA2B as soluble cytosolic proteins.
Figure 1. see also Figure S1. Subcellular localization of mammalian CIA1, CIA2A, and CIA2B.
A–B) HeLa cells were transfected three times at 3-day intervals either with scrambled siRNAs (SCR) or with pools of three siRNAs directed against CIA1, CIA2A or CIA2B. As a control, cells were mock-transfected. Nine days after the first transfection, cells were fractionated following digitonin treatment (Stehling et al., 2008). Total cell lysates, supernatant (Cytosol) and pellet (Organelles) fractions were analyzed for CIA1 (A), CIA2B (B), β-actin, and mitochondrial MIA40 by immunostaining. C) HeLa cells were transfected three times at 3-day intervals with either a control vector (-) or a vector encoding a muCIA2A-myc fusion protein. Cells were treated and analyzed for localization of the indicated proteins as in parts A-B. D) At each harvest cumulative numbers of siRNA-treated cells were determined. All values are normalized to mock-transfected cells (set to 100%; dashed line) and given as mean ± SD (n=3).
Depletion of CIA2A but not of CIA2B and CIA1 impairs Fe/S cluster maturation of IRP1
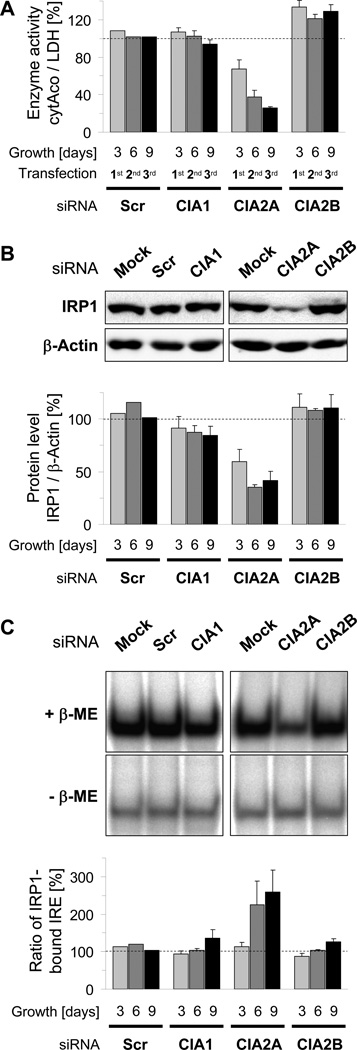
The efficient knock-downs of CIA1, CIA2A, and CIA2B in HeLa cells allowed us to examine the proteins’ putative roles in Fe/S protein assembly. We first tested the effects of their depletion on cytosolic aconitase (cytAco) activity of IRP1 using lactate dehydrogenase activity as a reference (Biederbick et al., 2006; Stehling et al., 2008). Surprisingly, only depletion of CIA2A, but not that of CIA2B or CIA1 strongly decreased the cytAco activity in a depletion time-dependent manner (Fig. 2A). Individual CIA2A-directed siRNAs were also effective (Fig. S2A), while scrambled siRNAs did not elicit any effects. As the apoform of IRP1 lacking the Fe/S cluster is less stable than the holoform (Clarke et al., 2006; Sheftel et al., 2010b; Stehling et al., 2008), we additionally determined the IRP1 protein levels as an indirect measure of Fe/S cluster assembly. In order to account for potential changes in gene expression mRNA levels of IRP1 were also examined. Consistent with the decline in cytAco activity, only RNAi treatment for CIA2A decreased the IRP1 protein amount (Figs. 2B and S2B), while IRP1 mRNA levels remained unchanged (Fig. S2C). In individual experiments, the relative decrease in IRP1 steady-state protein levels was frequently 10% to 30% less pronounced than that in cytAco activity, indicating that the Fe/S cluster assembly defect is preceding IRP1 protein destabilization. Notably, low amounts of CIA2B slightly improved rather than decreased both cytAco activity and protein levels of IRP1 indicating enhanced Fe/S cluster maturation of IRP1 in the absence of CIA2B (Figs. 2A–B and S2A–B). All aconitase effects were highly specific and restricted to the extra-mitochondrial compartment, as both the activities and levels of the mitochondrial Fe/S proteins aconitase (mtAco) and succinate dehydrogenase (SDH) were unchanged upon depletion of CIA1, CIA2A or CIA2B (Fig. S3).
Figure 2. see also Figures S2 and S3. Depletion of CIA2A, but not of CIA2B or CIA1, impairs Fe/S cluster maturation of IRP1.
HeLa cells were depleted of CIA1, CIA2A or CIA2B by RNAi as in Fig. 1 for 3, 6, and 9 days by three consecutive transfections. A) Cells were fractionated as in Fig. 1 and cytosolic aconitase (cytAco) activity of IRP1was determined relative to the activity of the non-Fe/S cluster-containing cytosolic enzyme lactate dehydrogenase (LDH). Ratios were normalized to mock-transfected cells. B) Samples of mock- and siRNA-transfected cells were subjected to immunostaining for IRP1 and β-actin. Upper panel: Representative immunoblots of samples prepared 9 days after the first transfection. Lower panel: IRP1- and β-actin-associated chemiluminescence was quantified and the ratio normalized to mock-transfected cells. C) IRP1 binding activity (IRP2 supershift method) to 32β-labeled IRE of human ferritin mRNA in the presence (+β-ME) or absence (-β-ME) of 1.7% β-mercaptoethanol was analyzed by native gel electrophoresis and subsequent phosphorimaging. Upper panel: Representative autoradiographs of samples prepared 9 days after the first transfection. Lower panel: The ratio of IRP1-bound IRE probe in the absence and presence of β-ME (ratio of IRP1-bound IRE) was normalized to mock-transfected cells (set to 100 %) and represents a relative measure for the Fe/S cluster maturation status of IRP1. All values are given as mean ± SD (n=3); dashed line, mock-transfected cells, SCR; scrambled siRNAs.
We next used a RNA electrophoretic mobility shift assay (REMSA) to test the effects of CIA1, CIA2A and CIA2B depletion on the RNA-binding activity of IRP1 to a [γ-32P]CTP-labeled ferritin IRE probe. IRE binding to IRP1 was distinguished from association with IRP2 by antibody-mediated supershift of IRP2 during native gel electrophoresis (Stehling et al., 2008). To account for changes in IRP1 protein levels as observed in CIA2A-deficient cells, the maximal IRE-binding activity of IRP1 was determined in a second set of samples by pre-treatment with β-mercaptoethanol (β-ME; (Hentze et al., 1989)). The amount of IRP1-associated IRE probe was visualized by autoradiography and quantified by densitometry. The ratio of IRP1-bound probe in the absence (intrinsic binding activity) and presence of β-ME (maximal binding activity) is a measure for the proportion of IRP1 protein being in an IRE-binding conformation thus reflecting the Fe/S cluster maturation status of IRP1. Depletion of CIA2A nearly doubled the proportion of IRP1 being in an IRE-binding state (ratio of IRP1-bound IRE; Figs. 2C and S2F), an effect mainly due to a decrease in IRP1’s maximal IRE-binding activity (maximal IRP1-bound IRE; Fig. S2D) rather than an increase in the protein’s intrinsic binding activity (Fig. S2E). In contrast, RNAi-mediated depletion of CIA1 or CIA2B had only a marginal influence, a result consistent with the minor effects of their depletion on cytAco activity. Taken together, the data suggest that human CIA2A is required for maturation of IRP1 Fe/S protein.
Depletion of CIA1 and CIA2B but not of CIA2A diminishes maturation of DPYD and GPAT
The somewhat surprising specificity of the two human CIA2 isoforms prompted us to examine the requirement of CIA2A, CIA2B, and CIA1 for the biosynthesis of other cytosolic-nuclear Fe/S proteins. A clinically relevant example is dihydropyrimidine dehydrogenase (DPYD), an enzyme involved in the detoxification of pyrimidine derivatives such as the anticancer drug 5-fluorouracil (van Kuilenburg, 2004). DPYD is a homodimer of 220 kDa and catalyzes the reduction of pyrimidines to initiate their degradation. Each DPYD monomer contains one FAD, one FMN, and four Fe/S clusters, the latter being arranged to form two entangled electron conduits that proceed across both subunits to connect the active sites of the enzyme (Schnackerz et al., 2004).
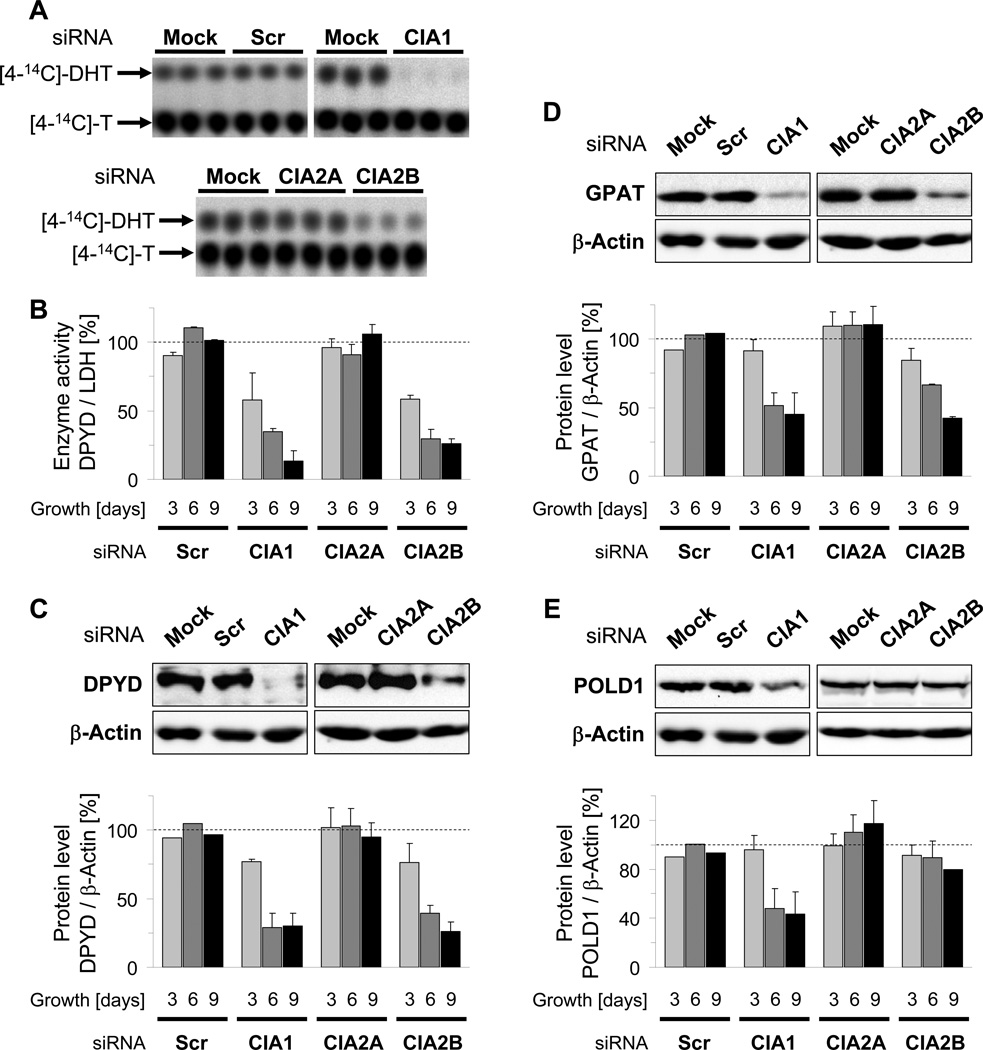
To assess the maturation of DPYD we measured the enzyme’s activity to convert [4-14C]-thymine into [4-14C]-dihydrothymine ([4-14C]-DHT). Substrate and product were separated by thin layer chromatography and quantitated by autoradiography. The assay was validated by analyzing the DPYD activity in HeLa cells depleted for the known ISC and CIA components NFS1, FXN, and NBP35 (Biederbick et al., 2006; Stehling et al., 2004; Stehling et al., 2008). Loss of any one of these established assembly factors resulted in a severe decrease in [4-14C]-DHT-formation compared to control cells, while the activity of LDH was virtually unaffected (Fig. S4). A profound time-dependent decrease (up to 85%) in DPYD activity was also seen for knock-down of both CIA1 and CIA2B, but not for CIA2A (Fig. 3A–B). Application of individual siRNAs directed against CIA1 and CIA2B was nearly as effective as the use of pooled siRNAs demonstrating the specificity of the approach (Fig. S5A–B). Loss of DPYD activity in CIA1 and CIA2B-depleted cells was associated with a diminution of DPYD protein (Figs. 3C and S5C) but not of DPYD mRNA (Fig. S5D). This result is consistent with apoprotein destabilization due to impaired Fe/S cluster assembly. We conclude that the maturation of DPYD depends on CIA2B and CIA1, while CIA2A is dispensable. Thus, CIA2A, CIA2B and CIA1 are bona fide CIA proteins, yet exhibit strikingly different Fe/S target protein specificity.
Figure 3. see also Figures S3 to S5. Depletion of CIA1 and CIA2B, but not of CIA2A, impairs maturation of the cytosolic Fe/S proteins DPYD, GPAT, and POLD1.
CIA1, CIA2A and CIA2B were depleted by RNAi as in Fig. 2, and HeLa cells were analyzed for enzyme activity and steady-state protein levels of DPYD, GPAT, and POLD1. A-B) DPYD-dependent formation of [4-14C]-dihydrothymine ([4-14C]-DHT) from [4-14C]-thymine ([4-14C]-T) was determined by thin layer chromatography and subsequent autoradiography. Representative autoradiographs of replicate samples obtained 9 days after the first transfection are shown in (A). DPYD enzyme activity was calculated from the proportion of [4-14C]-DHT and [4-14C]-T, expressed relative to LDH activity and normalized to mock-transfected cells (B). C-E) DPYD, GPAT, POLD1 and β-actin steady-state protein levels were examined by immunostaining. Upper panel: Representative immunostains of samples prepared 9 days after the first transfection. Lower panel: Protein-associated chemiluminescence was quantified and the protein per β-actin ratio normalized to mock-transfected cells. All values are given as mean ± SD (n=3); dashed line, mock-transfected cells, SCR; scrambled siRNAs.
To examine this target specificity in more detail, we analyzed the maturation of glutamine phosphoribosylpyrophosphate amidotransferase (GPAT) which catalyzes the first step of purine nucleotide synthesis and is translated as an inactive proenzyme. Its activation requires both the incorporation of a [4Fe-4S] cluster and the subsequent removal of an N-terminal undecamer propeptide. Since cluster insertion critically determines protein stability (Martelli et al., 2007; Stehling et al., 2008), the steady-state levels of GPAT measured by immunostaining provide a direct measure of Fe/S cluster assembly. The consequences of CIA2A, CIA2B, and CIA1 depletion on GPAT protein and mRNA levels were similar to those observed for DPYD maturation (Figs. 3D and S5E–F). In conclusion, GPAT maturation requires CIA1 and CIA2B, but not CIA2A, and thus exhibits a similar CIA factor dependency as DPYD.
Depletion of CIA2B but not of CIA2A affects maturation of POLD1
While IRP1, GPAT, and DPYD reside in the cytosol, other Fe/S proteins are present in the nucleus and function in the maintenance of genome integrity. A representative example is POLD1, the catalytic subunit of DNA polymerase δ, and a homologue of yeast Pol3 which was recently shown to carry a [4Fe-4S] cluster (Netz et al., 2012b). Similar to IRP1, GPAT, and DPYD, the steady-state protein level of POLD1 is dependent on efficient Fe/S cluster assembly (Stehling et. al., 2012), and thus provides a suitable measure for the maturation status of the protein. Upon RNAi-mediated depletion of CIA1 the POLD1 protein levels were strongly (>50%) diminished, while knock-downs of CIA2B or CIA2A only slightly decreased or increased POLD1 levels, respectively (Figs. 3E and S5G). In neither case did POLD1 mRNA levels decline (Fig. S5H). Thus, maturation of POLD1 appears to be most dependent on CIA1, and is only mildly affected by depletion of CIA2B. Together, these results indicate that the CIA proteins CIA1, CIA2A and CIA2B exhibit differential target specificity in the assembly of cytosolic-nuclear Fe/S proteins and thus can be viewed as dedicated CIA maturation factors.
CIA2A and CIA2B-MMS19 differentially affect IRE binding and stability of IRP2
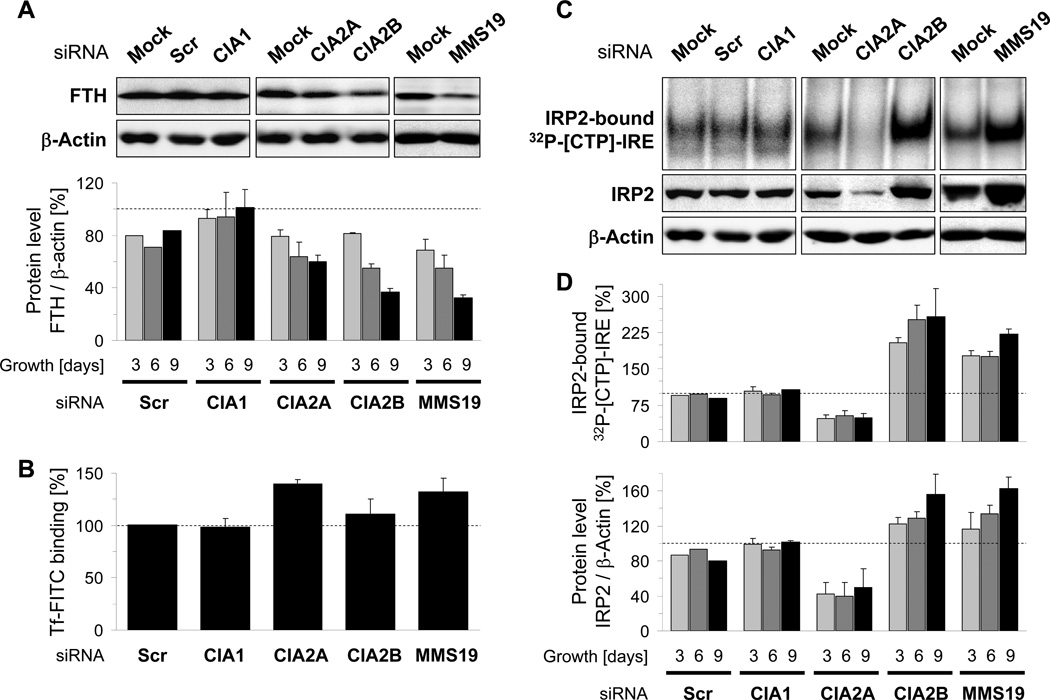
IRP1 is a key protein of mammalian iron homeostasis. Maturation of its Fe/S cluster depends on NBP35, IOP1, and, as shown above, CIA2A but not on CIA2B and CIA1. Thus, we expected that only a deficiency of the former, but not of the latter two CIA factors or of the recently identified CIA component MMS19 (Gari et al., 2012; Stehling et al., 2012) may impact on cellular iron regulation, even though we note that the IRE binding activity of IRP1 hardly changed upon depletion of these CIA proteins (Fig. 2C). To resolve this issue, we depleted CIA1, CIA2A, CIA2B, and MMS19 and measured two indicators of iron homeostasis, i.e. the level of the cytosolic iron storage protein ferritin and the binding of transferrin to its receptor (TfR). While CIA1 depletion did not yield any significant alterations, CIA2B or MMS19 deficiency caused an up to twofold diminution of ferritin protein, even though their mRNA levels remained unchanged (Figs. 4A and S6A–B). In contrast, both TfR-dependent binding of fluorescence labeled holo-transferrin and TfR1 mRNA levels increased 1.3- to 2-fold (Figs. 4B and S6C–D). CIA2A-depleted cells showed similar ferritin and TfR alterations as CIA2B- and MMS19-deficient cells. As a control and in contrast to the changes in TfR1 mRNA levels, mRNA levels of the house-keeping gene glyceraldehyde 3-phosphate dehydrogenase (GAPDH) remained constant (Fig. S6E). Obviously, the depletion of the CIA proteins elicited effects on ferritin and TfR levels that cannot be explained by the function of IRP1.
Figure 4. see also Figure S6. Depletion of CIA2A or CIA2B-MMS19 affects cellular iron metabolism and elicits opposite effects on IRP2.
CIA1, CIA2A, CIA2B, and MMS19 were depleted by RNAi as in Fig. 2 for 9 days, and HeLa cells were analyzed for ferritin, transferrin binding, and IRP2 protein levels or IRP2-IRE binding activity. A) Ferritin heavy chain (FT-H) and β-actin protein levels were examined by immunostaining (upper panel) and quantitation (lower panel) of samples as in Fig. 2B. B) Analysis of fluorescein-labeled transferrin (Tf-FITC) binding as a measure of transferrin receptor levels. Cell-associated fluorescence was corrected for cell density and normalized to mock-transfected cells. C) IRE-binding activity of IRP2 (IRP1 supershift method) was probed by a 32P-labeled IRE of human ferritin mRNA in the presence of 0.3% β-mercaptoethanol and analyzed by native gel electrophoresis and subsequent phosphorimaging as in Fig. 2C (upper panel). Lower panels: IRP2 and β-actin steady-state protein levels examined by immunostaining. D) [32P]-IRE-IRP2 binding, IRP2 and β-actin levels of part C were quantified and normalized to mock-transfected cells. All values are given as mean ± SD (n=4); dashed line, mock-transfected cells, SCR; scrambled siRNAs.
The posttranscriptional regulation of ferritin and TfR expression depends on both the Fe/S protein IRP1 and its non-Fe/S counterpart IRP2 which is known to respond to an iron-rather than Fe/S cluster-dependent signal (Anderson et al., 2012; Salahudeen et al., 2009; Vashisht et al., 2009). We therefore examined to what extent CIA protein depletion affects IRP2. To this end, the IRE binding activity of IRP2 was measured essentially as described above for IRP1, yet this time using an antibody-induced supershift of IRP1. Depletion CIA2B or MMS19 increased both IRE-binding activity and protein levels of IRP2 twofold, an effect consistent with the observed decrease in ferritin and the increase in transferrin binding (Figs. 4C–D and S6F–G). In contrast, CIA2A deficiency lowered both IRE-binding activity and protein levels of IRP2 twofold, which does not explain the lower ferritin levels and higher transferrin binding (Fig. 4). Depletion of CIA1 did not cause any effect, similar as for ferritin and TfR. The amounts of IRP2 mRNA were hardly changed under any depletion condition (Fig. S6H), thereby excluding the possibility that these changes were due to altered IRP2 gene expression. The relative effects of increased transferrin binding of CIA2A-, CIA2B- or MMS19-depleted cells versus control cells were retained even under high or low iron supply in the medium (Fig. S6C). Taken together, the changes in ferritin and TfR levels resulting from CIA2B and MMS19 depletion may be explained by the increases in IRP2 levels. On the contrary, the similar changes of ferritin and TfR levels seen under CIA2A depletion are inconsistent with the expected effects for lowered IRP2 levels. The molecular mechanisms of these new regulatory connections between these CIA components and the iron metabolism remain unclear. Nevertheless, the decrease in IRP2 upon CIA2A depletion may be explained on the basis of the complex formation between the two proteins (see below) which leads to stabilization of IRP2. The complicated modulatory effects of these CIA proteins on cellular iron homeostasis introduce a previously unknown mode of IRP2 regulation.
Mammalian CIA2A and CIA2B form mutually exclusive complexes with CIA1
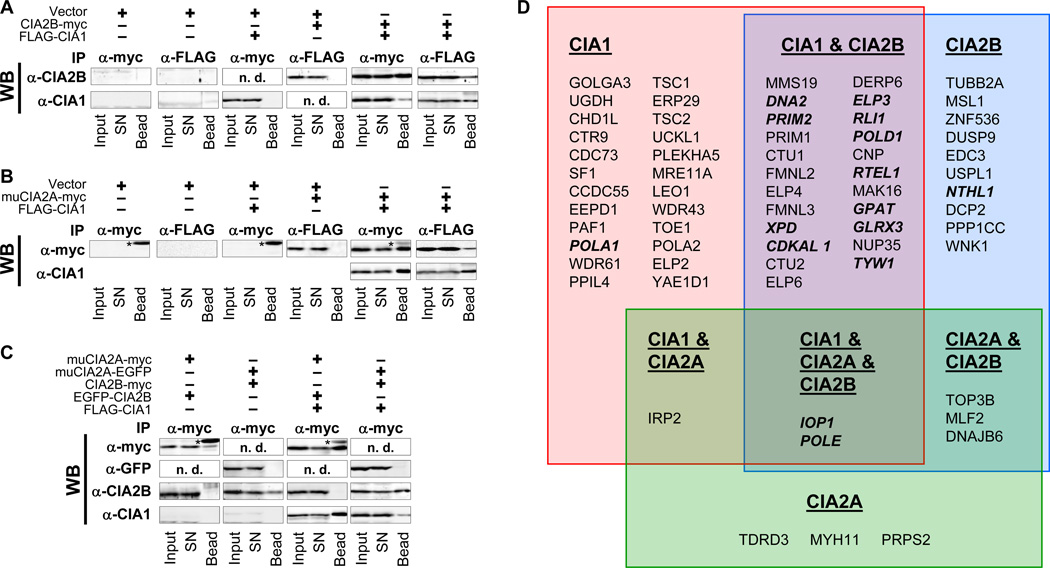
The differential requirement of CIA2A and CIA2B for the maturation of various Fe/S proteins prompted us to examine their specific interaction partners in more detail. First, CIA2B-myc (Fig. 5A) and muCIA2A-myc (Fig. 5B) were co-expressed with N-terminally FLAG-tagged CIA1 (Johnstone et al., 1998). Cleared cell lysates were subjected to anti-FLAG or anti-myc immunoprecipitation, and isolated proteins were analyzed by immunostaining. CIA2B-myc or muCIA2A-myc were each co-isolated with FLAG-CIA1 independently of which tag was used for precipitation (Fig. 5A–B, right parts). In the absence of bait protein no relevant signal was detected (Fig. 5A–B, left parts), confirming the specificity of the interactions.
Figure 5. see also Figure S7 and Table S4. CIA2A and CIA2B form mutually exclusive complexes with CIA1 to allow for selective interaction with different target Fe/S proteins.
A) HeLa cells were transfected with empty vectors (Vector), vectors encoding C-terminally myc-tagged CIA2B, or vectors encoding N-terminally FLAG-tagged CIA1 as indicated. After cell growth for three days, cleared lysates were prepared and subjected to immunoprecipitation (IP) by anti-myc or anti-FLAG-coupled protein A-agarose. Fractions of lysates (Input), and unbound supernatant (SN) or bead-associated material (Bead) were immunostained for CIA2B (α-CIA2B) and CIA1 (α-CIA1). B) HeLa cells were transfected with vectors encoding the indicated proteins, and subjected to immunoprecipitation and immunostaining of muCIA2A (α-myc) and CIA1 (α-CIA1). C) HeLa cells were transfected with vectors encoding the indicated myc-, FLAG- or EGFP-tagged proteins, and subjected to immunoprecipitation and immunostaining. (*), light chains of anti-myc immunoglobulins; n.d., not determined. D) Samples of HEK293T cells expressing N-terminally HA-FLAG-tagged versions of CIA1, CIA2A, or CIA2B were subjected to anti-FLAG affinity purification. Captured proteins were identified by MudPIT-MS and grouped for their association with the respective bait proteins. Known Fe/S proteins are given in bold face.
In order to determine if CIA1, CIA2A, and CIA2B interact in a ternary complex, HeLa cells were transfected with vectors encoding C-terminally EGFP-tagged muCIA2A or N-terminally EGFP-tagged CIA2B fusion proteins and with vectors encoding CIA2B-myc or muCIA2A-myc, respectively. These transfections were done either alone or in combination with a vector encoding FLAG-CIA1. Anti-myc immunoprecipitation revealed that none of the CIA2 bait proteins interacted with the EGFP-tagged CIA2 counterpart (Fig. 5C), irrespective of the presence of FLAG-CIA1. However, both muCIA2A-myc and CIA2B-myc were able to co-precipitate FLAG-CIA1. Thus, muCIA2A and CIA2B do not interact with each other, yet both independently bind to CIA1. These results suggest a mutually exclusive association of muCIA2A or CIA2B with CIA1.
This conclusion was further supported by analyzing the steady-state protein levels of CIA1, CIA2B, and MMS19 when one of these CIA components was depleted by RNAi. In each case, protein levels of the other two binding partners were significantly decreased (Fig. S7A–D). In contrast, knock-down of CIA2A had no effects on CIA1, CIA2B or MMS19, in keeping with its lack of interaction with both CIA2B and MMS19 (Stehling et al., 2012). Apparently, CIA1, CIA2B, and MMS19 stabilize each other by formation of the CIA targeting complex, arguing for a tight association of these proteins.
CIA2A and CIA2B associate with distinct sets of proteins
The mutually exclusive binding of CIA2A or CIA2B to CIA1 and their radically different Fe/S target specificity prompted us to further define their global protein interaction networks. To this end, we used an established affinity purification system to isolate N-terminally HA- FLAG-tagged CIA1, CIA2A, and CIA2B together with associated proteins from doxycycline-inducible HEK293 stable cell lines. Purified proteins were uncovered by Multidimensional Protein Identification Technology (MudPIT)-based proteomic mass spectrometry (Stehling et al., 2012; Wohlschlegel, 2009). Proteins represented by more than 20 peptides and belonging to the 99% percentile of the total sample entity (Table S4) were grouped with respect to their association with one, two, or three of the CIA baits (Fig. 5D). Representative interactions were confirmed by immunostaining (Fig. S7E). Among the top interacting proteins, we found the CIA targeting complex CIA1-CIA2B-MMS19. Its additional interaction with the CIA protein IOP1 may indicate the functional contact of earlier (IOP1) and late parts of the CIA machinery. A similar functional interaction may exist between IOP1 and CIA2A, consistent with the role of IOP1 as a general CIA factor functioning upstream of the two CIA2 isoforms. Strikingly, many of the CIA2B-interacting proteins (Fig. 5D, blue box) are known Fe/S proteins including DNA2, PRIM2, XPD, RLI1, POLD1, RTEL1, GPAT, and NTHL1. As expected, most of these proteins also interacted with both CIA1 (Fig. 5D, red box and Table S4) and MMS19 (Stehling et al., 2012). These results nicely fit with our functional studies showing that the CIA1-CIA2B-MMS19 complex is involved in the maturation of the majority of cytosolic-nuclear Fe/S proteins (Fig. 6).
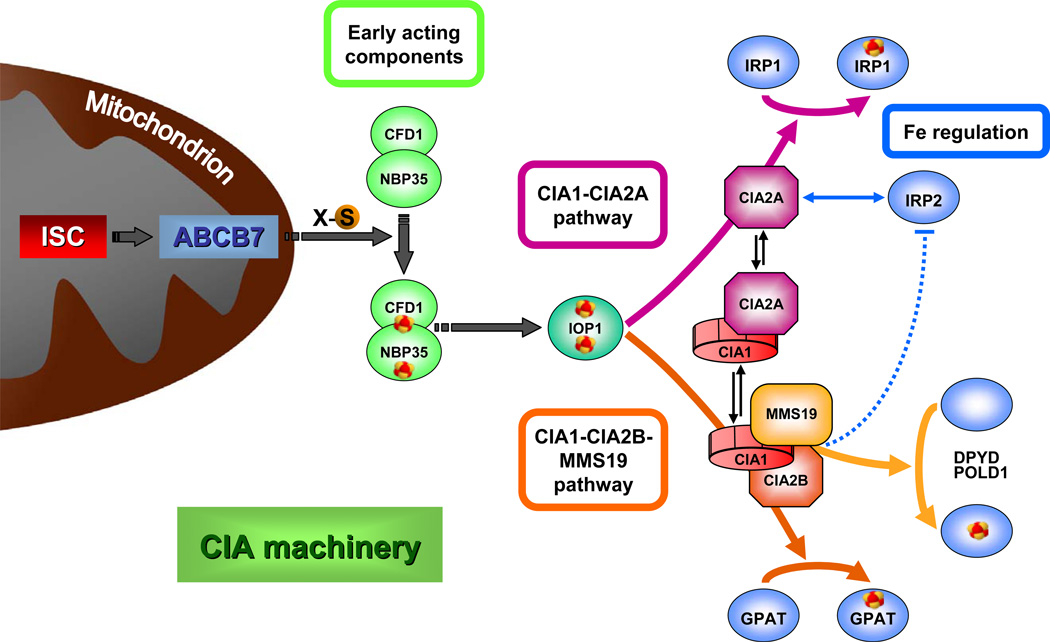
Figure 6. Working model for the maturation of cytosolic-nuclear Fe/S proteins in human cells and the intimate link to cellular iron regulation.
Cytosolic Fe/S protein assembly is a stepwise process and relies on a still unknown sulfur-containing compound (X-S) which is produced by the mitochondrial ISC assembly machinery and transported into the cytosol via the mitochondrial ABC transporter ABCB7 (Lill, 2009). Then, a [4Fe-4S] cluster is transiently assembled on the hetero-tetrameric scaffold complex NBP35-CFD1. The Fe/S cluster is subsequently transferred to different apoproteins by late-acting CIA components, a process facilitated by the action of IOP1 and the two different CIA targeting complexes identified in the present study. The CIA1-CIA2B-MMS19 complex facilitates the specific assembly of the majority of Fe/S proteins including DPYD, GPAT, and DNA polymerases. CIA2A, which also forms a complex with CIA1, supports the specific maturation of IRP1 to cytosolic aconitase, thus directly influencing cellular iron regulation. Hitherto undescribed effects on iron regulation are conferred on IRP2. CIA2A forms a stabilizing complex with IRP2, and hence depletion of CIA2A diminishes IRP2 levels (blue double-arrow). In contrast, deficiency of CIA2B or MMS19 increase IRP2 amounts by an unknown mechanism (dotted blue line).
The large group of proteins exclusively associated with CIA1 included UDP-glucose 6-dehydrogenase, Golgin subfamily A member 3, Chromo-domain-helicase-DNA-binding protein 1-like (Fig. 5D, Table S4). Intriguingly, the majority of these proteins have not been previously linked to Fe/S protein metabolism. They could represent novel CIA or Fe/S proteins, new regulators of CIA1 activity, or relate to so far unknown roles for CIA1 in other biological pathways. CIA2A was not found to associate with IRP1 in these experiments or by dedicated immunoprecipitation, despite its functional relevance for IRP1 Fe/S cluster maturation. However, both CIA2A and CIA1 bound to IRP2 (IREB2) with high specificity, thus shedding light on the mechanism by which CIA2A levels may impact on IRP2 protein amounts. Direct complex formation between IRP2 and CIA2A may lead to stabilization of IRP2. Thus, CIA2A, in addition to its above defined role in IRP1 Fe/S cluster maturation, establishes an unexpected link between Fe/S protein biogenesis and cellular iron metabolism via IRP2 interaction.
DISCUSSION
The importance of cytosolic and nuclear Fe/S proteins in fundamental cellular processes such as DNA replication and repair, chromosome segregation and ribosomal protein translation has only recently become evident. Nevertheless, the mechanism of Fe/S protein maturation in mammalian cells is still poorly defined. Previously, only three cytosolic factors have been implicated in this process, namely NBP35 and IOP1, which act as general CIA biogenesis factors (Song and Lee, 2008; Stehling et al., 2008), and MMS19 which executes a more target-specific function (Gari et al., 2012; Stehling et al., 2012). Here, we used cell biological, biochemical and proteomic approaches to identify and functionally characterize three additional components, CIA1, CIA2A and CIA2B, as bona fide members of the human CIA machinery. In contrast to NBP35 and IOP1, but similarly to MMS19, these proteins directly interact with Fe/S target proteins and are engaged in target-specific maturation steps. CIA1 and CIA2B assemble with MMS19, forming the so-called CIA targeting complex (Stehling et al., 2012) that is involved in the biogenesis of the majority of cytosolic and nuclear Fe/S proteins including GPAT, DPYD, and DNA polymerases such as POLD1 (Fig. 6). Thus, this complex is the functional equivalent of a related entity in yeast. The strong conservation of its components in eukaryotes reflects the importance of cytosolic and nuclear Fe/S proteins. CIA2A on the other hand is present in only few organisms and functions as a dedicated Fe/S cluster maturation factor of IRP1, thereby exerting a direct impact on the regulation of cellular iron homeostasis in mammals (Fig. 6). This situation radically differs from yeast and other lower eukaryotes which harbor only one CIA2 gene and lack IRP1. In yeast, depletion of CIA components has no immediate effects on cellular iron homeostasis. Thus, CIA2A and CIA2B are key components of the mammalian CIA machinery defining the target specificity for Fe/S cluster insertion into distinct sets of Fe/S apoproteins thereby establishing a hitherto undescribed branched pathway for cytosolic Fe/S protein maturation (Fig. 6).
CIA1, CIA2B and MMS19 are known to tightly interact, yet the Fe/S target protein specificity of the individual components may differ. All three CIA proteins were required for maturation of DPYD, but only two of them were crucial for the assembly of POLD1 and GPAT (Fig. 6). RNAi-mediated depletion of CIA2B had only mild effects on POLD1, and functional inactivation of MMS19 left GPAT virtually unaffected. This result is surprising based on the complex formation between the three CIA proteins. Either the CIA targeting complex is dynamic and can dissociate into dimeric sub-complexes with distinct target specificities, or it has multiple binding sites for different Fe/S targets. The existence of dimeric subcomplexes that specifically interact with dedicated Fe/S proteins is corroborated by our current and previous proteomic analyses. For instance, many of the CIA2B-associated proteins identified here have been previously shown to co-purify with MMS19 (Stehling et al., 2012) supporting the existence of a CIA2B-MMS19 dimeric subcomplex. The same might hold true for CIA1. However, our results also suggest that the individual constituents of the CIA targeting complex stabilize each other, as depletion of one component lowered the abundance of the remaining two. This finding is best explained by protein interactions stabilizing the partners of the CIA targeting complex. Importantly, the majority of the cytosolic and nuclear Fe/S proteins are associated with CIA1, CIA2B and MMS19 (Fig. 5D and (Gari et al., 2012; Luo et al., 2012; Stehling et al., 2012; van Wietmarschen et al., 2012)) emphasizing the central importance of the CIA targeting complex for the maturation of the vast majority of cytosolic and nuclear Fe/S proteins.
Our co-immunoprecipitation and proteomic approaches indicate that CIA1 forms mutually exclusive complexes with either CIA2A or CIA2B. Knockdown of CIA2A by RNAi resulted in low cytosolic aconitase activity and diminished protein levels indicative of hampered Fe/S maturation of IRP1 in the absence of CIA2A. Since no direct interaction of these two proteins was detectable, their association may be rather labile. On the contrary, CIA2A was dispensable for GPAT, DPYD, and POLD1 maturation consistent with the lack of interaction with known Fe/S proteins apart from POLE1 which thus might be a second substrate of CIA2A (Fig. 5D). Surprisingly, although CIA2A formed a complex with CIA1, depletion of the latter had no significant effect on IRP1 steady-state protein level and cytAco activity. This is reminiscent of the situation for the CIA1-CIA2B-MMS19 targeting complex in which the individual constituents are not necessarily essential for maturation of distinct targets of the partner CIA proteins.
Our results demonstrate that the late-acting CIA proteins affect cellular iron regulation in previously unknown ways. The direct requirement of the human CIA machinery for IRP1 Fe/S cluster maturation has been documented earlier, but is extended in our current studies by identifying CIA2A as both an IRP1-specific targeting factor and a stabilizing component for IRP2. Another surprising impact of the CIA machinery on iron homeostasis is mediated by CIA2B and MMS19. Their depletion led to the unforeseen increase in cellular levels and IRE binding activity of IRP2, i.e. opposite effects than those seen for the deficiency in CIA2A (this study and (Stehling et al., 2012)). IRP2 is the teammate of IRP1 in the post-transcriptional regulation of iron metabolism, yet is known to exhibit its iron regulatory function independently of a bound Fe/S cluster (Zumbrennen et al., 2009). Instead, IRP2 binding to IRE is critically regulated via its iron-responsive degradation by FBXL5 (Salahudeen et al., 2009; Vashisht et al., 2009), phosphorylation of a critical serine residue (Wallander et al., 2008), and likely an oxoglutarate-dependent dioxygenase (Wang et al., 2004). Although IRP1 and IRP2 were reciprocally affected by knockdown of CIA2A and CIA2B-MMS19, respectively, the physiological effects on ferritin levels and cellular transferrin binding were strikingly similar. In cells lacking CIA2B or MMS19, IRP2 was stabilized resulting in lower levels of ferritin and an increase in transferrin receptor. In contrast, CIA2A depletion lowered the levels of IRP2, yet the impact on ferritin and TfR was similar as for CIA2B and MMS19 depletion, i.e. opposite to the expected situation. The stabilizing effect of CIA2A on IRP2 may be best explained by the physical interaction of IRP2 with CIA2A detected by our proteomic studies. Intriguingly, CIA1 interacts with the CIA2A-IRP2 complex, yet depletion of CIA1 had no detectable consequences on iron metabolism. Thus, the role of this interaction remains to be determined.
The initial discovery of CIA1 and CIA2B implicated them to function in cellular processes other than Fe/S protein assembly. CIA1 was identified in a search for binding partners of Wilms’ tumor protein (Johnstone et al., 1998). The biochemical basis for this connection became evident from recent studies which identified a function of the CIA1 binding partner MMS19 in the dedicated assembly of a subset of Fe/S proteins including many involved in DNA metabolism (Gari et al., 2012; Stehling et al., 2012). Similarly, CIA2B was suggested to perform a function in chromosome segregation (Ito et al., 2010). Intriguingly, the two proteins were found to be part of a large complex including MMS19 and the Fe/S protein XPD, and RNAi-mediated knock-down of CIA2B led to defective chromosome segregation and abnormally shaped nuclei. These tumor-related and chromosome integrity-related functions of CIA1 and CIA2B can now be understood on the basis of our past (Stehling et al., 2012) and present findings that these proteins, together with MMS19, perform a primary function in the maturation of Fe/S proteins involved in DNA-related processes such as the DNA helicases XPD and RTEL1, as well as DNA polymerases.
The mutually exclusive binding of CIA1 to either CIA2A or CIA2B and the opposing effects of CIA2A and CIA2B-MMS19 deficiency on the IRP system propose a branching mechanism for cytosolic-nuclear Fe/S protein assembly, and an intimate connection of this pathway to the regulation of iron homeostasis via CIA2A-dependent IRP1 activation and IRP2 stabilization (Fig. 6). According to this model, knockdown of CIA2B or MMS19 impairs the activity of the CIA targeting complex which might lead to the preferential use of the CIA2A branch thus promoting Fe/S cluster maturation of IRP1 and stabilization of IRP2. Vice versa, binding of CIA2A to CIA1 might negatively affect CIA2B-dependent Fe/S protein maturation implicating a regulatory function of the dynamic CIA2A-CIA1 interaction. Notably, IOP1 interacts with both CIA1-CIA2A and CIA1-CIA2B-MMS19 targeting complexes (Fig. 5D) suggesting that it might be the final general CIA component acting before the branching node within the CIA machinery (Seki et al., 2013). Our present study provides a framework for future investigations on the precise molecular function of the CIA components that assist the assembly of cytosolic-nuclear Fe/S proteins in human cells. Furthermore, our work enables future studies dedicated to the molecular mechanism of IRP2 regulation by late-acting members of the CIA machinery.
EXPERIMENTAL PROCEDURES
Vectors and siRNAs
For nomenclature of human CIA proteins see Table S1. The mammalian expression vector pCMV-FLAG-CIA1 (CIAO1 GeneID: 9391) was kindly provided by R.W. Johnstone (Johnstone et al., 1998). Plasmid pPK368 containing murine CIA2A cDNA (FAM96A GeneID: 68250) was a kind gift of M. Zörnig (Frankfurt, Germany). Cloning vector pOTB7-CIA2B (FAM96B GeneID: 51647) was obtained from imaGenes GmbH (Berlin). For mass spectrometry, plasmids encoding CIA1 (Clone ID: 5219740), CIA2A (Clone ID: 4128736) and CIA2B (Clone ID: 3353705) were purchased from Open Biosystems. Small interfering RNAs (Table S2) were purchased from Ambion. Primer sequences for qRT-PCR are given in Table S3.
Antibodies
Anti-CIA2B antiserum was either raised in rabbits immunized with bacterially expressed, N-terminally His-tagged, and purified full-length protein or was purchased from Proteintech Group. Anti-CIA1 antiserum was either raised in rabbits or obtained from Genway. Further antibodies are listed in the Supplemental Experimental Procedures section.
Cell culture and transfection
Human cervix carcinoma cells (HeLa) were cultured and transfected by electroporation as described (Sheftel et al., 2010b; Stehling et al., 2008; Stehling et al., 2012). A total of 15 µg siRNAs was used, and the amount of plasmids ranged between 5 and 10 µg. In order to prolong the time period of mRNA depletion cells were retransfected twice at three day intervals. Flp-InTM T-RExTM-293 cells were obtained from Invitrogen and cultured as described (Stehling et al., 2012). Stably transfected doxycyclin-inducible (500 ng/mL overnight) 3xHA-3xFLAG-CIA1, 3xHA-3xFLAG-CIA2A, and 3xHA-3xFLAG-CIA2B Flp-InTM T-RExTM-293 cells were generated using the Flp-In system (Invitrogen) according to manufacturer’s instructions. LipofectamineTM 2000 was used in transfections according to the manufacturer’s directions.
Biochemical methods
HeLa cells were harvested, fractionated by digitonin treatment and analyzed as described (Müllner et al., 1989; Sheftel et al., 2010b; Stehling et al., 2008; Stehling et al., 2009; Stehling et al., 2007). Aconitase activity was determined by a coupled aconitase-isocitrate dehydrogenase assay and succinate dehydrogenase activity by the DCIP assay in combination with decyl ubiquinone. Cytochrome-c oxidoreductase, citrate synthase, and lactate dehydrogenase activities were analyzed as published. DPYD enzyme activity was determined by analysis of the conversion of [4-14C]-thymine into [4-14C]-dihydrothymine using thin layer chromatography (TLC) (Stehling et al., 2012). IRE RNA-binding capacities of IRP1 and of IRP2 were examined by REMSA. In order to activate latent IRP2, p-mercaptoethanol was added at a final concentration of 0.3% prior to the addition of the 32P-labeled IRE (Zumbrennen et al., 2009). TfR expression was evaluated by a fluorometric approach (Stehling et al., 2008).
Co-immunoprecipitation
HeLa cells were transfected with vectors encoding tagged fusion proteins and harvested after three days of tissue culture. Cell pellets (corresponding to about 2 mg of total protein) were lysed in TNGT buffer (10 mM Tris-HCl, pH 7.4, 150 mM NaCl, 10% [w/v] glycerol, 0.25% [vol/vol] Triton X-100, 1 mM phenylmethylsulfonyl fluoride) and clarified by centrifugation at 13,000 ×g for 10 min. Protein-A-agarose-conjugated FLAG or myc probe (Santa Cruz biotechnology) was added for 1.5 h under gentle agitation at 4°C. Agarose beads were washed four times with TNGT buffer and resuspended in loading buffer for SDS-PAGE and immunoblotting. About 10% to 30% of the precipitated material was applied in each gel run.
Supplementary Material
Highlights.
CIA1, CIA2A and CIA2B function in cytosolic-nuclear iron-sulfur protein maturation
The CIA1-CIA2B-MMS19 targeting complex assists maturation of dedicated apoproteins
CIA2A is specific for iron-sulfur cluster assembly of IRP1 and stabilization of IRP2
CIA2A and CIA2B integrate iron-sulfur protein assembly and cellular iron regulation
ACKNOWLEDGEMENTS
We thank Dr. S. Molik for assistance with antibody generation, Dr. M. Zörnig for plasmid pPK368, and Dr. H.-P. Elsässer for technical advice. R.L. was supported by Deutsche Forschungsgemeinschaft (SFB 593, GRK1216, and Gottfried-Wilhelm Leibniz Program), von Behring-Röntgen Stiftung, LOEWE program of the state of Hesse, and the Max–Planck Gesellschaft. J.A.W. was supported by grants from the National Institutes of Health (GM089778), University of California Cancer Research Coordinating Committee, and the Jonsson Cancer Center at UCLA.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Anderson CP, Shen M, Eisenstein RS, Leibold EA. Mammalian iron metabolism and its control by iron regulatory proteins. Biochim Biophys Acta. 2012:1468–1483. doi: 10.1016/j.bbamcr.2012.05.010. 1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayala-Castro C, Saini A, Outten FW. Fe-S cluster assembly pathways in bacteria. Microbiol Mol Biol Rev. 2008;72:110–125. doi: 10.1128/MMBR.00034-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balk J, Aguilar Netz DJ, Tepper K, Pierik AJ, Lill R. The essential WD40 protein Cia1 is involved in a late step of cytosolic and nuclear iron-sulfur protein assembly. Mol Cell Biol. 2005;25:10833–10841. doi: 10.1128/MCB.25.24.10833-10841.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balk J, Pierik AJ, Netz DJ, Mühlenhoff U, Lill R. The hydrogenase-like Nar1p is essential for maturation of cytosolic and nuclear iron-sulphur proteins. Embo J. 2004;23:2105–2115. doi: 10.1038/sj.emboj.7600216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biederbick A, Stehling O, Rösser R, Niggemeyer B, Nakai Y, Elsasser HP, Lill R. Role of human mitochondrial Nfs1 in cytosolic iron-sulfur protein biogenesis and iron regulation. Mol Cell Biol. 2006;26:5675–5687. doi: 10.1128/MCB.00112-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke SL, Vasanthakumar A, Anderson SA, Pondarre C, Koh CM, Deck KM, Pitula JS, Epstein CJ, Fleming MD, Eisenstein RS. Iron-responsive degradation of iron-regulatory protein 1 does not require the Fe-S cluster. Embo J. 2006;25:544–553. doi: 10.1038/sj.emboj.7600954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gari K, Leon Ortiz AM, Borel V, Flynn H, Skehel JM, Boulton SJ. MMS19 links cytoplasmic iron-sulfur cluster assembly to DNA metabolism. Science. 2012;337:243–245. doi: 10.1126/science.1219664. [DOI] [PubMed] [Google Scholar]
- Hausmann A, Aguilar Netz DJ, Balk J, Pierik AJ, Mühlenhoff U, Lill R. The eukaryotic P loop NTPase Nbp35: an essential component of the cytosolic and nuclear iron-sulfur protein assembly machinery. Proc Natl Acad Sci U S A. 2005;102:3266–3271. doi: 10.1073/pnas.0406447102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentze MW, Rouault TA, Harford JB, Klausner RD. Oxidation-reduction and the molecular mechanism of a regulatory RNA-protein interaction. Science. 1989;244:357–359. doi: 10.1126/science.2711187. [DOI] [PubMed] [Google Scholar]
- Ito S, Tan LJ, Andoh D, Narita T, Seki M, Hirano Y, Narita K, Kuraoka I, Hiraoka Y, Tanaka K. MMXD, a TFIIH-independent XPD-MMS19 protein complex involved in chromosome segregation. Mol Cell. 2010;39:632–640. doi: 10.1016/j.molcel.2010.07.029. [DOI] [PubMed] [Google Scholar]
- Johnson DC, Dean DR, Smith AD, Johnson MK. Structure, function, and formation of biological iron-sulfur clusters. Annu Rev Biochem. 2005;74:247–281. doi: 10.1146/annurev.biochem.74.082803.133518. [DOI] [PubMed] [Google Scholar]
- Johnstone RW, Wang J, Tommerup N, Vissing H, Roberts T, Shi Y. Ciao 1 is a novel WD40 protein that interacts with the tumor suppressor protein WT1. J Biol Chem. 1998;273:10880–10887. doi: 10.1074/jbc.273.18.10880. [DOI] [PubMed] [Google Scholar]
- Lill R. Function and biogenesis of iron-sulphur proteins. Nature. 2009;460:831–838. doi: 10.1038/nature08301. [DOI] [PubMed] [Google Scholar]
- Lill R, Hoffmann B, Molik S, Pierik AJ, Rietzschel N, Stehling O, Uzarska MA, Webert H, Wilbrecht C, Muhlenhoff U. The role of mitochondria in cellular iron-sulfur protein biogenesis and iron metabolism. Biochim Biophys Acta. 2012:1491–1508. doi: 10.1016/j.bbamcr.2012.05.009. 1823. [DOI] [PubMed] [Google Scholar]
- Luo D, Bernard DG, Balk J, Hai H, Cui X. The DUF59 Family Gene AE7 Acts in the Cytosolic Iron-Sulfur Cluster Assembly Pathway to Maintain Nuclear Genome Integrity in Arabidopsis. The Plant Cell. 2012;24:4135–4148. doi: 10.1105/tpc.112.102608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martelli A, Wattenhofer-Donze M, Schmucker S, Bouvet S, Reutenauer L, Puccio H. Frataxin is essential for extramitochondrial Fe-S cluster proteins in mammalian tissues. Hum Mol Genet. 2007;16:2651–2658. doi: 10.1093/hmg/ddm163. [DOI] [PubMed] [Google Scholar]
- Müllner EW, Neupert B, Kühn LC. A specific mRNA binding factor regulates the iron-dependent stability of cytoplasmic transferrin receptor mRNA. Cell. 1989;58:373–382. doi: 10.1016/0092-8674(89)90851-9. [DOI] [PubMed] [Google Scholar]
- Netz DJ, Pierik AJ, Stumpfig M, Bill E, Sharma AK, Pallesen LJ, Walden WE, Lill R. A bridging [4Fe-4S] cluster and nucleotide binding are essential for function of the Cfd1-Nbp35 complex as a scaffold in iron-sulfur protein maturation. J Biol Chem. 2012a;287:12365–12378. doi: 10.1074/jbc.M111.328914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netz DJ, Pierik AJ, Stümpfig M, Mühlenhoff U, Lill R. The Cfd1-Nbp35 complex acts as a scaffold for iron-sulfur protein assembly in the yeast cytosol. Nat Chem Biol. 2007;3:278–286. doi: 10.1038/nchembio872. [DOI] [PubMed] [Google Scholar]
- Netz DJ, Stith CM, Stumpfig M, Kopf G, Vogel D, Genau HM, Stodola JL, Lill R, Burgers PM, Pierik AJ. Eukaryotic DNA polymerases require an iron-sulfur cluster for the formation of active complexes. Nat Chem Biol. 2012b;8:125–132. doi: 10.1038/nchembio.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netz DJ, Stumpfig M, Dore C, Muhlenhoff U, Pierik AJ, Lill R. Tah18 transfers electrons to Dre2 in cytosolic iron-sulfur protein biogenesis. Nat Chem Biol. 2010;6:758–765. doi: 10.1038/nchembio.432. [DOI] [PubMed] [Google Scholar]
- Py B, Barras F. Building Fe-S proteins: bacterial strategies. Nature Rev. Microbiology. 2010;8:436–446. doi: 10.1038/nrmicro2356. [DOI] [PubMed] [Google Scholar]
- Rouault TA. Biogenesis of iron-sulfur clusters in mammalian cells: new insights and relevance to human disease. Dis Model Mech. 2012;5:155–164. doi: 10.1242/dmm.009019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy A, Solodovnikova N, Nicholson T, Antholine W, Walden WE. A novel eukaryotic factor for cytosolic Fe-S cluster assembly. The EMBO journal. 2003;22:4826–4835. doi: 10.1093/emboj/cdg455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolf J, Makrantoni V, Ingledew WJ, Stark MJ, White MF. The DNA repair helicases XPD and FancJ have essential iron-sulfur domains. Mol Cell. 2006;23:801–808. doi: 10.1016/j.molcel.2006.07.019. [DOI] [PubMed] [Google Scholar]
- Salahudeen AA, Thompson JW, Ruiz JC, Ma HW, Kinch LN, Li Q, Grishin NV, Bruick RK. An E3 ligase possessing an iron-responsive hemerythrin domain is a regulator of iron homeostasis. Science. 2009;326:722–726. doi: 10.1126/science.1176326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnackerz KD, Dobritzsch D, Lindqvist Y, Cook PF. Dihydropyrimidine dehydrogenase: a flavoprotein with four iron-sulfur clusters. Biochim Biophys Acta. 2004:61–74. doi: 10.1016/j.bbapap.2004.06.009. 1701. [DOI] [PubMed] [Google Scholar]
- Seki M, Takeda Y, Iwai K, Tanaka K. IOP1 is an external component of human CIA machinery and functions in MMS19-dependent CIA pathway. J Biol Chem. 2013 doi: 10.1074/jbc.M112.416602. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma AK, Pallesen LJ, Spang RJ, Walden WE. Cytosolic iron-sulfur cluster assembly (CIA) system: factors, mechanism, and relevance to cellular iron regulation. J Biol Chem. 2010;285:26745–26751. doi: 10.1074/jbc.R110.122218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheftel A, Stehling O, Lill R. Iron-sulfur proteins in health and disease. Trends Endocrinol Metab. 2010a;21:302–314. doi: 10.1016/j.tem.2009.12.006. [DOI] [PubMed] [Google Scholar]
- Sheftel AD, Stehling O, Pierik AJ, Elsasser HP, Muhlenhoff U, Webert H, Hobler A, Hannemann F, Bernhardt R, Lill R. Humans possess two mitochondrial ferredoxins, Fdx1 and Fdx2, with distinct roles in steroidogenesis, heme, and Fe/S cluster biosynthesis. Proc Natl Acad Sci U S A. 2010b;107:11775–11780. doi: 10.1073/pnas.1004250107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song D, Lee FS. A role for iron-only hydrogenase like protein 1 in mammalian cytosolic iron-sulfur protein biogenesis. J Biol Chem. 2008;283:9231–9238. doi: 10.1074/jbc.M708077200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan V, Netz DJ, Webert H, Mascarenhas J, Pierik AJ, Michel H, Lill R. Structure of the yeast WD40 domain protein Cia1, a component acting late in iron-sulfur protein biogenesis. Structure. 2007;15:1246–1257. doi: 10.1016/j.str.2007.08.009. [DOI] [PubMed] [Google Scholar]
- Stehling O, Elsässer HP, Bruckel B, Mühlenhoff U, Lill R. Iron-sulfur protein maturation in human cells: evidence for a function of frataxin. Hum Mol Genet. 2004;13:3007–3015. doi: 10.1093/hmg/ddh324. [DOI] [PubMed] [Google Scholar]
- Stehling O, Netz DJ, Niggemeyer B, Rosser R, Eisenstein RS, Puccio H, Pierik AJ, Lill R. Human Nbp35 is essential for both cytosolic iron-sulfur protein assembly and iron homeostasis. Mol Cell Biol. 2008;28:5517–5528. doi: 10.1128/MCB.00545-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stehling O, Sheftel AD, Lill R. Chapter 12 Controlled expression of iron-sulfur cluster assembly components for respiratory chain complexes in mammalian cells. Methods Enzymol. 2009;456:209–231. doi: 10.1016/S0076-6879(08)04412-1. [DOI] [PubMed] [Google Scholar]
- Stehling O, Smith PM, Biederbick A, Balk J, Lill R, Mühlenhoff U. Investigation of iron-sulfur protein maturation in eukaryotes. Methods Mol Biol. 2007;372:325–342. doi: 10.1007/978-1-59745-365-3_24. [DOI] [PubMed] [Google Scholar]
- Stehling O, Vashisht AA, Mascarenhas J, Jonsson ZO, Sharma T, Netz DJ, Pierik AJ, Wohlschlegel JA, Lill R. MMS19 Assembles Iron-Sulfur Proteins Required for DNA Metabolism and Genomic Integrity. Science. 2012;337:195–199. doi: 10.1126/science.1219723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Kuilenburg AB. Dihydropyrimidine dehydrogenase and the efficacy and toxicity of 5-fluorouracil. Eur J Cancer. 2004;40:939–950. doi: 10.1016/j.ejca.2003.12.004. [DOI] [PubMed] [Google Scholar]
- van Wietmarschen N, Moradian A, Morin GB, Lansdorp PM, Uringa EJ. The Mammalian Proteins MMS19, MIP18, and ANT2 Are Involved in Cytoplasmic Iron-Sulfur Cluster Protein Assembly. J Biol Chems. 2012;287:43351–43358. doi: 10.1074/jbc.M112.431270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vashisht AA, Zumbrennen KB, Huang X, Powers DN, Durazo A, Sun D, Bhaskaran N, Persson A, Uhlen M, Sangfelt O, Spruck C, Leibold EA, Wohlschlegel JA. Control of iron homeostasis by an iron-regulated ubiquitin ligase. Science. 2009;326:718–721. doi: 10.1126/science.1176333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallander ML, Zumbrennen KB, Rodansky ES, Romney SJ, Leibold EA. Iron-independent phosphorylation of iron regulatory protein 2 regulates ferritin during the cell cycle. J Biol Chem. 2008;283:23589–23598. doi: 10.1074/jbc.M803005200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Chen G, Muckenthaler M, Galy B, Hentze MW, Pantopoulos K. Iron-mediated degradation of IRP2, an unexpected pathway involving a 2-oxoglutarate-dependent oxygenase activity. Mol Cell Biol. 2004;24:954–965. doi: 10.1128/MCB.24.3.954-965.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weerapana E, Wang C, Simon GM, Richter F, Khare S, Dillon MB, Bachovchin DA, Mowen K, Baker D, Cravatt BF. Quantitative reactivity profiling predicts functional cysteines in proteomes. Nature. 2010;468:790–795. doi: 10.1038/nature09472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlschlegel JA. Identification of SUMO-conjugated proteins and their SUMO attachment sites using proteomic mass spectrometry. Methods Mol Biol. 2009;497:33–49. doi: 10.1007/978-1-59745-566-4_3. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Lyver ER, Nakamaru-Ogiso E, Yoon H, Amutha B, Lee DW, Bi E, Ohnishi T, Daldal F, Pain D, Dancis A. Dre2, a conserved eukaryotic Fe/S cluster protein, functions in cytosolic Fe/S protein biogenesis. Mol Cell Biol. 2008;28:5569–5582. doi: 10.1128/MCB.00642-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zumbrennen KB, Wallander ML, Romney SJ, Leibold EA. Cysteine oxidation regulates the RNA-binding activity of iron regulatory protein 2. Mol Cell Biol. 2009;29:2219–2229. doi: 10.1128/MCB.00004-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.