Abstract
Genetic transfer of neutralizing antibodies has been shown to confer strong and persistent protection against bacterial and viral infectious agents. While it is well established that for many exogenous neutralizing antibodies increased antigen affinity correlates with protection, the effect of antigen affinity on antibodies produced in situ following adenoviral gene transfer has not been examined. The mouse IgG2b monoclonal antibody 2C12.4 recognizes the Yersinia pestis Type III secretion apparatus protein LcrV (V antigen) and confers protection in mice when administered as an IgG intraperitoneally or, following genetic immunization with engineered, replication-defective serotype 5 human adenovirus (Ad) 1. 2C12.4 was expressed as a scFv fragment in E. coli and was shown to display a KD=3.5 nM by surface plasmon resonance (SPR) analysis. The 2C12.4 scFv was subjected to random mutagenesis and variants with increased affinity were isolated by flow cytometry using the Anchored Periplasmic Expression (APEx) bacterial display system. After a single round of mutagenesis, variants displaying up to 35-fold lower KD values (H8, KD=100 pM) were isolated. The variable domains of the H8 scFv were used to replace those of the parental 2C12.4 IgG encoded in the Ad vector, AdαV giving rise to AdαV.H8. The two adenoviral vectors resulted in similar titers of anti-V antigen antibodies 3 days post-immunization with 109, 1010 or 1011 particle units. Following intranasal challenge with 363 LD50Y. pestis CO92, 54% of the mice immunized with 1010 pu of AdαV.H8 survived at the 14 day end point compared to only 15% survivors for the group immunized with AdαV expressing the lower affinity 2C12.4 (P<0.04, AdαV versus AdαV.H8). These results indicate that affinity maturation of a neutralizing antibody delivered by genetic transfer may confer increased protection not only for Y. pestis challenge but possibly for other pathogens.
Keywords: antibody, affinity maturation, Yersinia pestis, adenovirus gene transfer, protection
Introduction
Yersinia pestis is the etiologic agent of the plague and has been responsible for pandemic outbreaks occurring throughout the course of history. Although advances in our current living conditions, public health practices, and antibiotic therapies make future pandemics unlikely, outbreaks of plague resulting from biological warfare are a real threat. The features of Y. pestis that make it an attractive option for use as a biological weapon include availability of the organism, capacity for aerosol dissemination, potential for spread of secondary cases, and the high fatality rate of the pneumonic form of plague. In endemic regions of the world, the bacterium survives by causing chronic disease in animal reservoirs. It is spread among these animals and occasionally to humans predominantly through a flea vector, such as Xenopsylla cheopis2,3. Without prompt antibiotic therapy, approximately 50% of bubonic plague infections are fatal and can progress to the more dangerous pneumonic plague 2. Respiratory droplets from a pneumonic infected individual promote rapid spread through a susceptible population. Symptoms develop in 1 to 6 days post-infection and the disease progresses rapidly from a flu-like illness to severe pneumonia with cough, chest pain, and bloody sputum. To be effective, antibiotic therapy must be administered early. If treatment is delayed more than 24 hours following the onset of symptoms, the fatality rate is high 4. Additionally, the presence of antibiotic-resistant strains of Y. pestis renders antibiotic therapy unreliable. For these reasons, Y. pestis is a likely agent to be used as a biological weapon since aerosolized bacteria can confer widespread pneumonic plague 2.
Of the 11 Yersinia species, only Y. pestis, Y. enterocolitica, and Y. pseudotuberculosis are human pathogens. Y. pestis is a gram-negative, non-motile, non-spore-forming bacterium that replicates intracellularly during the early stages of infection and grows predominantly extracellularly at later stages of the infectious cycle 2. At present, no plague vaccine has been approved for use in the US. Passive immunization with antibodies specific for the LcrV protein (V antigen) is an attractive alternative to vaccines and have been shown to be effective against lethal challenges with Y. pestis1,5-7. The V antigen plays a central role in plague pathogenesis. It activates the type III secretion system and thus mediates translocation of effector proteins (Yops) into host macrophages. It is also released from the bacteria and has immunosuppressive functions manifested by increasing levels of the anti-inflammatory cytokine interleukin 10 and decreasing levels of TNF-α 2. A recently developed anti-V antigen monoclonal antibody (mAb) 2C12.4 has been shown to confer protection against lethal challenge with intranasally administered Y. pestis in a dose-dependent manner 1.
For several neutralizing antibodies the degree of protection against challenge with pathogen correlates with antigen binding affinity 8-11. For example, while monoclonal antibodies and antibody fragments to the Protective Antigen (PA) of Bacillus anthracis with a KD=11 nM fail to confer protection against challenge with the holotoxin or with intranasally administered spores, engineered antibody variants displaying 40- to 200-fold higher affinities were protective in different animal models 8,12. Notably, protection appeared to be mediated by blocking the ability of PA to bind to its receptor since PEGylated antibody fragments exhibiting a KD=35 pM but lacking an Fc domain, and hence incapable of engaging innate immunity mechanisms of pathogen clearance, were protective 12. Engineering antibodies with high affinity has been shown to improve protection for other protein toxins and viruses including Botulism, human immunodeficiency virus (HIV), and human respiratory syncytial virus (RSV) and have increased efficacy when targeting inflammatory cytokines such as TNF-α 8-11,13,14.
In this study, we evaluated whether Ad-mediated delivery of an engineered 2C12.4 IgG exhibiting markedly increased affinity directed towards the V antigen can improve protection against Y. pestis challenge in mice 1. Improvement would be demonstrated by conferring a higher level of protection through immunization with an equivalent dose of a recombinant adenovirus encoding the higher affinity neutralizing antibody. However, the mutations needed to confer higher affinity could have unexpected effects on protein expression from the Ad delivered IgG DNA or biodistribution of the IgG, thus making it difficult to predict how protection to pathogen challenge might be affected. A scFv fragment incorporating the VH and VL domains of the 2C12.4 IgG was constructed and subjected to affinity maturation by screening a library generated by random mutagenesis. The latter was performed using the E. coli display technique Anchored Periplasmic Expression (APEx) coupled with fluorescent activated cell sorting (FACS) 15. In APEx, the protein library is tethered to the periplasmic side of the inner membrane of the bacterium via fusion to the 6 N-terminus amino acids of the native lipoprotein NlpA. The resulting NlpA-scFv fusion is lipidated in vivo and thus becomes attached to the inner membrane. Following permeabilization of the E. coli outer membrane by chemical and enzymatic means, the cells are labeled with fluorescent antigen and the brightest cells are isolated by FACS. A single round of random mutagenesis and APEx screening resulted in the isolation of a clone, H8, displaying a KD=100 pM which is 35-fold higher affinity than the parental 2C12.4. The H8 VH and VL domains were used to exchange those of 2C12.4 IgG in the adenoviral vector AdαV giving rise to AdαV.H8. We show that delivery of the two vectors in vivo resulted in approximately the same Ab serum titers and that mice immunized with 1010 pu AdαV.H8 conferred a statistically significant increase in protection relative to the parental AdαV.
Materials and methods
Bacterial strains and plasmid vectors
Escherichia coli Jude-1 (E. coli DH10B F-mcrA Δ(mrr-hsdRMS-mcrBC) φ80dlacZ ΔM15 ΔlacX74 recA1 araD139 Δ(ara leu)7697 galU galK rpsL endA1 nupG harboring an F′ from E. coli XL1-blue introduced by conjugation) were used for all antibody engineering experiments. Plasmid pMoPac16 was used for soluble single-chain antibody fragment (scAb) expression and has been previously described 16. Plasmids pAPEx1 was used for E. coli display of soluble single-chain variable fragment antibodies (scFv) 15. pFLAG-APEx was constructed by replacing the polyhistidine and c-myc sequences in pAPEx1 via BamHI and NotI restriction sites with the FLAG peptide epitope sequence amplified by PCR using primers TVB100 (5′-GTCGCTGCGGCCGCAGATTACAAAGACGACGATGACAAGTAGTGATATCGCAAGCTTGACC-3′) and TVB101 (5′-CAGCGAGGATCCGTGACGCAGTAGCGGTAAACGGC-3′).
Protein expression and purification
Recombinant V antigen was expressed and purified as previously described 1,17. Monomeric scAbs were expressed and purified according to published procedures 15,16. Briefly, E. coli Jude-1 containing pMoPac16 derivatives encoding the desired scAb genes were streaked from frozen stocks onto agar plates containing Luria-Burtani Miller (LB) medium (Becton Dickinson Difco™, Sparks, MD) supplemented with 2% (wt/vol) glucose (2% glc) and 200 μg/ml ampicillin (Amp200) (20 h, 25 °C). Individual colonies were cultured in 2 ml of Terrific Broth (TB) medium (Becton Dickinson Difco™, Sparks, MD) + 2% glc + Amp200 (8 h, 250 rpm, 30 °C) then 1 ml was used to inoculate 500 ml of TB medium + 2% glc + Amp200 (250 rpm, 30 °C). After overnight growth, cells were pelleted by centrifugation (15 min, 4,400 × g, 4 °C) and resuspended in 500 ml of TB medium + Amp200 + 1 mM Isopropyl-β-d-thiogalactopyranoside (IPTG) (Sigma-Aldrich, St. Louis, MO) to induce protein expression. After 4 h of incubation at 25 °C, the cells were pelleted by centrifugation (15 min, 4,400 × g, 4 °C), and the periplasmic fraction from the cell pellet was isolated by osmotic shock according to published procedures 16. scAb proteins were purified from the shockate by immobilized metal affinity chromatography (IMAC) using Ni-NTA Agarose based on the manufacturer's protocol (Qiagen, Hilden, Germany), and monomeric scAbs were further isolated by size-exclusion FPLC (SEC) on a Superdex™ 200 column (GE Healthcare, Uppsala, Sweden) using HEPES buffered saline with surfactant P20 (HBS-P) (GE Healthcare, Uppsala, Sweden). Proteins were quantified based on the A280 measured using a NanoDrop™ 1000 (Thermo Fisher Scientific, Wilmington, DE) and the appropriate extinction coefficients calculated using Protein Calculator (http://www.scripps.edu/∼cdputnam/protcalc.html, Putnam Lab at The Scripps Research Institute, La Jolla, CA). Relative concentrations were verified and purity determined by SDS-PAGE using a 4-20% gel (NuSep, Lawrenceville, GA) stained with GelCode Blue (Thermo Fisher Scientific, Rockford, IL). All scAbs used in this work were at least 90% pure.
Anti-V antigen antibody fragment cloning, expression and analysis
Genes encoding the variable heavy chain (VH) and the variable light chain (VK) domains of anti-V antigen 2C12.4 IgG were amplified by PCR from the previously described adenovirus vector plasmid DNA 1 using primers TVB102 (5′-GCAGCGAGGCCCAGCCGGCCATGGCGcaggtaactctgaaagagtctg-3′) and TVB103 (5′-agaGCCGCC GCCGCCgctaCCaCCaCCaCCagaaCCaCCaCCaCCtgaggagactgtgagagtggtg-3′) for VH and TVB104 (5′-GGtGGtGGtGGtagcGGCGGCGGCGGCtctGGCGGCGGCGGCtccgacattgtgctgacac agtcg-3′) and TVB105 (5′-CGAATTCGGCCCCCGAGGCccgttttacttccagcttggtc-3′) for VK1. The resulting PCR products were combined by overlap extension PCR and cloned into pMoPac16 via the non-compatible SfiI restriction sites for soluble expression as a scAb. The resulting ligation product was transformed into E. coli Jude-1, and scAb expression was performed as previously described 16,18. Briefly, individual colonies were cultured overnight in 2 ml of TB medium + 2% glc + Amp200 at 30 °C. After overnight growth, cells were pelleted by centrifugation (10 min at 4,600 × g, 4 °C) and resuspended in 2 ml of TB medium + Amp200 + 1 mM IPTG to induce protein expression. After 4 h incubation at 25 °C, 1 ml of cells were pelleted by centrifugation (10 min at 4,600 × g, 4 °C), and the cell pellet was resuspended in 1 ml of lysis buffer which consisted of BugBuster™ HT Protein Extraction Reagent (Novagen, Madison, WI) diluted 1:4 in phosphate buffered saline (PBS) 19. The cells were incubated in lysis buffer while rotating on an inverter (30 min, 60 RPM, 25 °C) then the insoluble fraction was removed by centrifugation (1 min, 10,000 × g, 4 °C).
The soluble fraction was analyzed for the presence of full-length scAb specific for the V antigen by Western blot analysis and enzyme-linked immunosorbent assay (ELISA), respectively. Western blot analysis with anti-polyhistidine (anti-His) peroxidase (HRP) conjugate (Sigma-Aldrich, St. Louis, MO) was performed as previously described 20. ELISA was performed by coating Costar® high binding 96-well EIA/RIA plates (Corning, Corning, NY) overnight at 4° C with 50 μl of 5 μg/ml recombinant V antigen or bovine serum albumin (BSA) in PBS. The plates were washed three times with PBS followed by blocking with 400 μl PBS supplemented with 2% milk for 4 h at room temperature. Samples were diluted 1:1 in PBS with 2% milk and incubated for 1 h at room temperature. The plates were washed three times with PBS then an additional three times with PBS containing 0.1% Tween-20. Immunocomplexes were detected using an anti-human kappa light chain (anti-HuCκ) polyclonal serum HRP conjugate (Sigma, St. Louis, MO) applied at a 1:10,000 dilution and incubated for 30 min at room temperature. The plates were washed as described above and developed using the chromogenic HRP substrate TMB+ (Dako, Glostrup, Denmark) as described by the manufacturer. The A450 was measured with a mircroplate reader (BioTek, Winooski, VT).
Isolation of high affinity variants of the 2C12.4 scFv
The anti-V antigen 2C12.4 scFv and the anti-digoxin 26.10 scFv were cloned into pAPEx1 and pFLAG-APEx via the non-compatible SfiI restriction sites 15. Purified recombinant V antigen was conjugated to BODIPY® using 6-((4,4-difluoro-5,7-dimethyl-4-bora-3a,4a-diaza-s-indacene-3-propionyl)amino)hexanoic acid succinimidyl ester (Invitrogen, Carlsbad, CA) at a molar ratio of 1:5 as described by the manufacturer. Free BODIPY FL-X SE was removed using a NAP-10 gel filtration column as described by the manufacturer (GE Healthcare, Uppsala, Sweden). The extent of conjugation was determined based on the ratio of A280 and A504 measured using a NanoDrop™ 1000 and the appropriate extinction coefficients and correction factors as recommended by the manufacturer (Invitrogen, Carlsbad, CA; http://www.scripps.edu/∼cdputnam/protcalc.html, Putnam Lab at The Scripps Research Institute, La Jolla, CA).
The anti-V antigen 2C12.4 scFv was subjected to random mutagenesis by error-prone PCR 21, the amplified DNA was cloned into pFLAG-APEx via the non-compatible SfiI restriction sites, and the ligation product was transformed into electrocompetent E. coli Jude-1 15. Cells were cultured on agar plates containing LB medium + 2% glc + 30 μg/ml chloramphenicol (Cm30) (22 h, 25 °C). Cells were transferred to LB medium + 2% glc + 15% glycerol (v/v) to A600 ∼10 then stored at -80 °C. Frozen cell stocks were thawed on ice and subcultured to A600 0.1 in TB medium + 2% glc + Cm30 at 37 °C, grown to A600 ∼0.8-1.2, cooled for 30 min to 25 °C, and protein expression was induced with the addition of 1mM IPTG. After 4h induction at 25 °C, 1 ml of cells at A600 5.0 were converted to spheroplasts using Tris/Sucrose/EDTA/lysozyme as previously described 15,19. The spheroplasted cells were first labeled with 5 μg/ml PhycoLink® anti-Flag® R-Phycoerythrin (anti-Flag PE) (Prozyme, San Leandro, CA) in PBS supplemented with 1% BSA (PBSB) (30 min, 150 rpm, 25 °C). The spheroplasts were washed with PBSB and incubated with 500 nM V antigen conjugated to BODIPY (V antigen-BODIPY) in PBSB (1 h, 150 rpm, 25 °C). Labeled spheroplasts were washed with PBSB and analyzed on a FACSAria (Becton Dickinson Biosciences, San Jose, CA) droplet deflection flow cytometer by exciting with a 488-nm laser and measuring the fluorescence emission spectrum of BODIPY and PE with 530/40 and 570/40 band-pass filters, respectively. Fluorescence compensation was performed using the APEx controls as previously described 19. The library population was gated to avoid aggregates as determined by the forward scatter (FSC) and side scatter (SSC) parameters, and the brightest 1-2% of the population based on both the BODIPY and PE emission spectrums was collected 15,19. The collected spheroplasts were immediately resorted, and the scFv genes in the resort solution were amplified by PCR using primers BRH06 (5′-GCGGATAACAATTTCACACAGG-3′) and AHX89 (5′-CGCAGTAGCGGTAAACGGC-3′) 15. The amplified DNA was cloned into pFLAG-APEx via the non-compatible SfiI restriction sites, and the ligation mixture was transformed into electrocompetent E. coli Jude-1. At least 10-fold excess of colonies relative to the number of events in the resort were obtained. Colonies were scraped from the agar plates into liquid media as above and subjected to an additional two rounds of sorting exactly as described above.
The scFv genes from the third round of sorting were subcloned into pMoPac16, and the ligation product was transformed into E. coli Jude-1 for expression of soluble scAb similar to above 15. In addition, E. coli Jude-1 containing pMoPac16 encoding 2C12.4 scAb was streaked from a frozen stock onto an agar plate containing LB medium + 2% glc + Amp200. Further, the anti-digoxin 26.10 scFv was cloned into pMoPac16 via the non-compatible SfiI restriction sites, and the ligation product was transformed into E. coli Jude-1. 72 individual colonies from round III, 8 colonies of 2C12.4 scAb and 4 colonies of 26.10 scAb were picked and cultured overnight in sterile Costar® round bottom 96-well microtiter plates (Corning, Corning, NY) containing 200 μl of TB medium + 2% glc + Amp200 on a microtiter plate shaker. After overnight growth (150 rpm, 30 °C), 4 μl aliquots were transferred to a master microtiter plate well containing 156 μl of TB medium + 2% glc + Amp200, cultured on a plate shaker (8 h, 150 rpm, 30 °C) and stored at 4 °C for up to two weeks. The remaining cells were pelleted by centrifugation (10 min at 4,600 × g, 4 °C) and resuspended in 200 μl of TB medium + Amp200 + 1 mM IPTG to induce protein expression. After 4 h induction at 25 °C, cells were pelleted by centrifugation (10 min at 4,600 × g, 4 °C), the cell pellet was resuspended in 1 ml of lysis buffer for 30 min at 25 °C, and the insoluble fraction was removed by centrifugation (20 min at 4,600 × g, 4 °C). The soluble fraction was further clarified using 96-well MultiScreen® HTS NA clearing filter plates (Millipore, Billerica, MA) as described by the manufacturer. Filtered lysates were transferred to Costar® non-binding 96-well plates (Corning, Corning, NY) and covered with microplate foil (GE Healthcare, Uppsala, Sweden). Relative expression levels of scAb proteins in filtered lysate were obtained using a Minifold® I Dot Blot System (Whatman, GE Healthcare, Uppsala, Sweden) by transferring 10 μl of filtered lysate diluted with 90 μl of PBS to a nitrocellulose membrane following the manufacturer guidelines. Immunological detection of the polyhistidine tagged scAb proteins immobilized on the nitrocellulose membrane was performed with anti-His HRP conjugate as previously described 20. The scAbs in filtered lysate were analyzed by SPR as described below.
Surface Plasmon resonance (SPR)
Antigen binding kinetics of scAb proteins in filtered lysate were analyzed via SPR using a Biacore™ 3000 (GE Healthcare, Uppsala, Sweden). Approximately 200 response units (RU) of V antigen in 10 mM sodium acetate, pH 5.0 were coupled to a CM5 chip (GE Healthcare, Uppsala, Sweden) by using 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide/N-hydroxy succinimide chemistry. BSA was similarly coupled to the chip and used for in-line subtraction. Kinetic analysis was performed in HBS-P (GE Healthcare, Uppsala, Sweden) at a flow rate of 25 μl/min at 25 °C unless indicated. Samples were injected for 3 min and dissociation was monitored for 5 min. The surface was regenerated with two 12 sec injections of 50 mM phosphoric acid. The antigen binding kinetics of purified monomeric scAb were determined in HBS-P at a flow rate of 50 μl/min. Samples were injected for 2 min and dissociation was monitored for 10 min. The surface was regenerated with two 12 sec injections of 50 mM phosphoric acid. Four 2-fold dilutions of each antibody beginning at 160 nM were performed in triplicate, double referenced with a blank, and globally fit to a 1:1 Langmuir binding model for the calculation of both kon and koff using BIAevaluation software (GE Healthcare, Uppsala, Sweden) 22.
Ad vectors
AdαV is a replication-defective human adenovirus (Ad) serotype 5, E1-E3- gene transfer vector constructed to direct the expression of the heavy and light chains of the anti-V antigen monoclonal antibody 2C12.4 from a single promoter 1. The expression cassette contains (5′ to 3′) the cytomegalovirus (CMV) immediate early promoter/enhancer, the antibody IgG2b heavy chain coding sequence, a 4 amino acid furin cleavage site, the 24 amino acid self-cleaving 2A peptide, the kappa light chain coding sequence, and the simian virus (SV) 40 polyadenylation signal. AdαV.H8 was constructed by assembling the genes encoding the variable heavy chain (VH) and variable light chain (VK) domains of the high affinity anti-V antigen 2C12.4 variant, H8, by overlap PCR using the AdαV IgG2b expression cassette as a template and the resulting fragment was used to replace the 2C12.4 coding sequence in AdαV. AdαPA is a similarly constructed gene transfer vector encoding an unrelated antibody (14B7-1H) against Bacillus anthracis protective antigen (PA) and was used as a negative control 23. AdαV, AdαV.H8, and AdαPA were produced in human embryonic kidney 293 cells (American Type Culture Collection, Manassas, VA) and purified by centrifugation twice through a CsCl gradient, as described 24. The titer of each recombinant Ad preparation was determined spectrophotometrically and expressed as particle units (pu) as described 25.
Assessment of AdαV.H8 In Vivo
Male C57BL/6 mice (n=5/group) (The Jackson Laboratory, Bar Harbor, ME) were housed under specific-pathogen-free conditions and used at 6 to 8 weeks of age. Mice were administered AdαV or AdαV.H8 (109, 1010, or 1011 pu) via the intravenous route. Naïve mice or mice injected with AdαPA (1011 pu) were used as negative controls. Ad vectors were diluted with saline to the specified dose. Serum was collected via the tail vein three days following vector administration, centrifuged at 8000× g for 20 min, and stored at -20 °C. Anti-V antigen serum titers were assessed by ELISA using flat bottomed 96-well EIA/RIA plates (Corning, New York, NY) coated with 100 μl 5 μg/ml recombinant V antigen in 0.05 M carbonate buffer, pH 7.4 overnight at 4 °C. The plates were washed three times with PBS and blocked with 200 μl PBS supplemented with 5% milk for 1 hr at 23 °C. Serial serum dilutions were added to each well and incubated for 1 hr at 23°C. The plates were washed three times with PBS containing 0.05% Tween-20 (PBST). Immunocomplexes were detected using a sheep anti-mouse IgG HRP conjugate (Sigma, St. Louis, MO) applied at a 1:10,000 dilution in PBS supplemented with 1% milk incubated for 1 hr at 23 °C. The plates were washed four times with PBST and once with PBS, developed using 100 μl of peroxidase substrate (Bio-Rad, Hercules, CA) incubated for 15 min at 23 °C, and the reaction quenched by the addition of 100 μl 2% oxalic acid. The A415 was measured with a microplate reader (Bio-Rad, Hercules, CA). Antibody titers were calculated based on log(optical density)-log(dilution) normalized with purified anti-V antigen antibodies to account for affinity differences. Mice challenge studies were conducted at The Public Health Research Institute (PHRI) at the International Center for Public Health (Newark, NJ) under biosafety level 3 (BSL3) conditions. Y. pestis CO92 was grown aerobically in Heart Infusion Broth (Becton Dickinson Difco™, Sparks, MD) at 30 °C and diluted in saline solution for a challenge dose of 2 × 104 cfu which corresponds to 363 LD50. Twenty-five μl of bacterial suspension was used for intranasal infection of mice; bacterial dose was controlled by plating on Yersinia Selective Agar (YSA) (Oxoid, Hampshire, UK) and counting colonies for colony-forming units (cfu) determination. Survival was monitored daily for 14 days.
Statistical Analyses
The data are presented as mean ± standard error of the mean. Statistical analyses were performed using the non-paired two-tailed Student's t-test, assuming equal variance. Survival evaluation was carried out using Kaplan-Meier analysis. Statistical significance was determined at p<0.1.
Results
Engineering high affinity anti-V antigen antibodies
The genes encoding the variable heavy chain (VH) and light chain (VK) domainof the Y. pestis neutralizing mAb 2C12.4 were used to create a single chain variable fragment antibody (scFv). This scFv gene was cloned into pMoPac16 for soluble expression of the antibody fragment in the single chain antibody (scAb) format 16. The scAb format is comprised of a scFv in which the C termini of the variable light chain domain is fused to the human kappa constant domain, resulting in improved expression in E. coli without altering the antibody binding affinity 26. Lysates from cells expressing the 2C12.4 scAb were confirmed to express full length protein via Western blotting and displayed binding activity as determined via ELISA using purified Y. pestis V antigen.
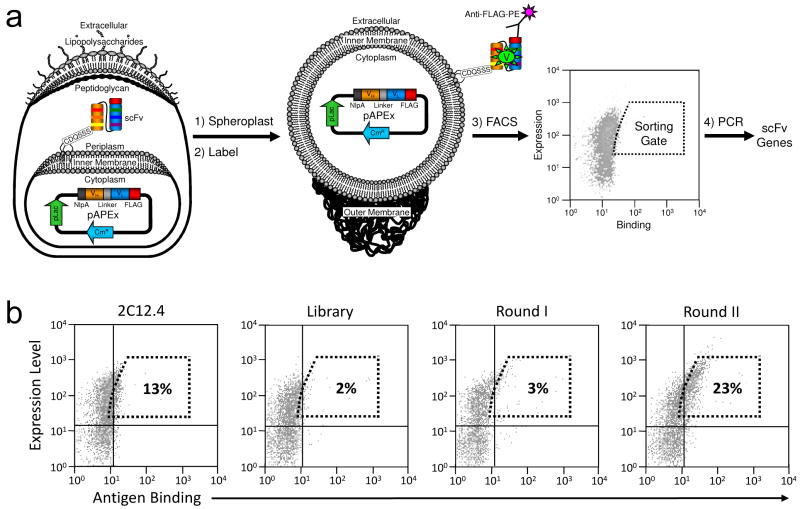
The gene encoding the scFv version of 2C12.4 was subjected to one round of random mutagenesis by error-prone PCR. Amplified DNA was cloned into pFLAG-APEx and the ligation product was transformed into E. coli Jude-1 cells yielding >2 × 106 independent transformants. DNA sequencing of 10 clones selected at random revealed an average of 1.75% nucleotide substitutions with a standard deviation of 0.75%. Cells were grown in liquid culture, protein expression was induced, and 4 hours later, the cells were harvested and converted into spheroplasts. The spheroplasted cells were then labeled with anti-Flag-PE and V antigen-BODIPY to determine full-length scFv expression levels and antigen binding, respectively (Figure 1a). A total of 8 × 107 cells (40-fold library coverage) were subjected to high-throughput fluorescence activated cell sorting (FACS) and the top 2% events in terms of PE and BODIPY fluorescence emissions were collected (Figure 1b). The collected spheroplasts were immediately resorted as above without additional labeling to maximize the isolation of antibodies with slow dissociation rate constants toward V antigen 15. The scFv genes in the spheroplasts collected during the resort were amplified by PCR, and the DNA was subcloned back into the pFLAG-APEx vector. Following transformation, the cells were subjected to two additional rounds of sorting using increasingly stringent collection criteria, namely a higher fluorescence threshold. After the third round of sorting, the number of events falling within the window used for the 1st round increased from 2% to 23% (Figure 1b).
Figure 1.
Affinity maturation of the anti-V antigen scFv 2C12.4 using APEx. (a) Schematic diagram of anchored periplasmic expression (APEx) (b) FACS analysis of E. coli spheroplasts expressing 2C12.4 scFv, the library of random mutants and the populations recovered following two rounds of FACS. Spheroplasts were labeled with 500 nM V antigen-BODIPY and 5 μg/ml Anti-FLAG-PE. The dashed box represents the gate used to isolate cells by FACS from the original library. The percentage of the spheroplast in this gate for each population is indicated.
The scFv gene pool from the third round of sorting was amplified by PCR and cloned into pMoPac16 for expression in the scAb format. SPR was used to rank order the dissociation rate constants of individual clones (Supplementary Figure 1). Of the 72 colonies tested, 41 (57%) expressed scAbs that displayed significant binding towards V antigen immobilized on the Biacore chip. Of these 41 clones, four clones exhibited dissociation rate constants at least 10-fold slower than those expressing the parental antibody (Supplementary Figure 1e and 1f).
These four scAbs, as well as the 2C12.4 and 26.10 scAbs, were expressed at the 500 ml scale in shake flasks, the periplasmic fraction was isolated by osmotic shock, and monomeric scAb proteins were purified by immobilized metal affinity chromatography (IMAC) and size exclusion FPLC. Consistent with the isolation of the respective clones based on increased PE fluorescence, the protein yields for all four scAbs was higher than that of the parental 2C12.4 scAb (Table 1). All antibodies exhibited at least 10-fold lower equilibrium dissociation constants (KD) relative to the parental 2C12.4 scAb (Table 1) which is consistent with the measurements obtained using crude cell lysates (Supp. Fig 1F). The highest affinity clone, H8, exhibited an affinity of 100 pM that translates to a 35-fold improvement compared to the parental antibody 2C12.4. The improved affinity for all clones was almost exclusively the result of a decrease in the dissociation rate constant whereas the association rate constant remained essentially unchanged. The four variants, H8, E4, F4 and G8, contained either 7 or 9 amino acid mutations compared to the parental antibody (Table 2) of which two were common: L49S in framework 2 of the VH, which is immediately prior to complementary determining region 2 (CDR2); and Y32F in CDR1 of the light chain 27. Further, three of the variants contained an additional four common mutations (D1N, T5S, R24K, T97R) in the light chain.
Table 1. Kinetic analysis of anti-V antigen antibody fragments.
| Antibody | kon (M-1s-1) | koff (s-1) | KD (pM) | Affinity Improvementa | Expression Improvementb |
|---|---|---|---|---|---|
| 2C12.4 | 9.2 × 104 | 3.2 × 10-4 | 3500 | 1x | 1x |
| H8 | 1.2 × 105 | 1.2 × 10-5 | 100 | 35x | 4x |
| E4 | 1.0 × 105 | 1.8 × 10-5 | 180 | 20x | 5x |
| F4 | 1.2 × 105 | 2.6 × 10-5 | 220 | 16x | 3x |
| G8 | 9.8 × 104 | 2.4 × 10-5 | 250 | 14x | 2x |
Affinity improvement relative to the parental antibody 2C12.4
Yield of purified scAb (mg/L A600) compared to 2C12.4
Table 2. Sequence analysis of anti-V antigen antibody fragments.
| Variable Heavy Chaina | Variable Light Chaina | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| FW1 | CDR1 | FW2 | CDR2 | FW3 | FW1 | CDR1 | FW2 | CDR2 | FW3 | CDR3 | ||||||||
| Clone | 3 | 11 | 32 | 45 | 49 | 51 | 65 | 67 | 82 | 1 | 5 | 24 | 32 | 39 | 53 | 72 | 84 | 97 |
|
| ||||||||||||||||||
| 2C12.4 | T | I | S | L | L | I | S | L | I | D | T | R | Y | K | N | T | A | T |
|
| ||||||||||||||||||
| H8 | S | V | N | S | K | F | R | |||||||||||
| E4 | V | P | S | F | R | S | T | |||||||||||
| F4 | A | P | S | V | N | S | K | F | R | |||||||||
| G8 | S | G | T | N | S | K | F | A | R | |||||||||
Amino acid substitutions based on Kabat numbering. FW, framework; CDR, complementary determining region
Genetic immunization using IgG encoding adenovirus vectors and challenge with Y. pestis
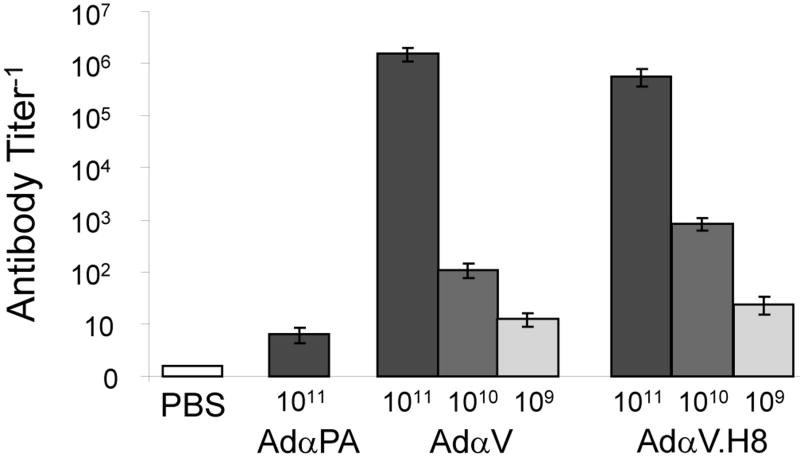
The VH and VK domains of the 2C12.4 mouse IgG2b antibody were replaced with those from the highest affinity antibody variant, H8, and engineered into a replication-defective adenovirus serotype 5 E1-E3- gene transfer vector to create AdαV.H8 1. Mice were immunized with varying doses of AdαV and AdαVH8 [109 – 1011 particle units (pu)], or, as controls, the highest dose (1011 pu) of AdαPA, an adenovirus vector encoding a mouse IgG mAb specific for the B. anthracis PA or PBS. Serum anti-V antigen antibody titers were measured via ELISA three days post immunizations (Figure 2). A similar dose dependent response was observed for both AdαV and AdαV.H8. The titers for equivalent immunization levels were within 10-fold for both AdαV and AdαV.H8. Both AdαPA and PBS controls did not result in any measurable anti-V antibody titers. Three days after immunization, the mice were challenged intranasally with 363 LD50 of of the fully virulent Y. pestis CO92 and survival monitored for two weeks (Figure 3). All mice immunized with the highest dosage (1011 pu) of AdαV and AdαV.H8 survived (Figure 4). However, at a 10-fold lower dose (1010 pu), only 15% of the mice immunized with AdαV survived while 54% of the mice immunized with this dose of AdαV.H8 survived (P < 0.04, AdαV versus AdαV.H8). In addition, 15% of the mice immunized with an additional 10-fold lower dose (109 pu) of AdαV.H8 survived, but none survived with the same dosage level of AdαV.H8 (P < 0.1, AdαV versus AdαV.H8).
Figure 2.
Anti-V antigen IgG titers in mice following immunization with Ad vectors. C57BL/6 mice (n = 13/group for AdαV and AdαV.H8; n = 1/group for AdαPA and PBS) were administered intravenously with the indicated Ad vectors or PBS. Serum anti-V antigen antibody titers were measured three days post immunization via ELISA. PBS, negative control; AdαPA, vector encoding an IgG specific for the B. anthracis PA; AdαV, vector encoding the 2C12.4 IgG; AdαV.H8, vector encoding the 2C12.4 IgG mutant H8.
Figure 3.
Survival of mice challenged with Y. pestis following prophylactic administration of Ad vectors. Three days following intravenous administration of Ad vectors or PBS, C57BL/6 mice (n = 13/group for AdαV and AdαV.H8; n = 13/group for AdαPA and PBS) were challenged with a lethal dose of Y. pestis intranasally. Survival of the mice was monitored for 14 days following challenge. (a) Immunization with AdαV compared to AdαPA and PBS. (b) Immunization with AdαV.H8 compared to AdαPA and PBS.
Figure 4.
Net survival of mice challenged with Y. pestis following prophylactic administration of Ad vectors. Percentage of C57BL/6 mice (n = 13/group for AdαV and AdαV.H8; n = 13/group for AdαPA and PBS) surviving 14 days following intranasal challenge with a lethal dose of Y. pestis.
Discussion
Passive immunization with engineered antibodies displaying enhanced antigen affinity has been shown to increase the neutralization efficacy of a number of bacterial or viral pathogens and toxins 8-11,13,14. However, to the best of our knowledge, neither the effect of antigen affinity for antibodies expressed in situ following viral transfer of the antibody gene, nor the relationship between V antigen affinity and protection to the potential biowarfare agent Y. pestis, have been examined. Here we found that increasing the affinity of the Y. pestis neutralizing antibody 2C12.4 from 3.5 nM to 100 pM (35-fold) could confer significant protection against a lethal challenge with Y. pestis at a 10-fold lower dose of a recombinant adenovirus encoding the neutralizing antibody.
The affinities of antibodies generated by the natural immune system are constrained by the kinetics of in vivo selection 28. Most high affinity mouse monoclonal antibodies display nanomolar affinities and this is the case for the anti-V antigen antibody 2C12.4 which displays a KD=3.5 nM by surface Plasmon resonance (SPR). Combinatorial mutagenesis and library screening by phage display or other high throughput techniques has been employed for affinity maturation of the variable domains of monoclonal antibodies 8-11,13,14. In earlier studies, we used random mutagenesis and screening of E. coli displayed libraries by APEx to isolate picomolar affinity antibodies to the B. anthracis PA which in turn proved to have markedly improved neutralization potency in vitro and in various animal models 8,12,29. Key advantages of this strategy are the ease with which libraries of random mutants can be constructed in E. coli and screened by FACS. FACS is a high-throughput screening technique that enables real-time quantitative multi-parameter analysis on a cell by cell basis. Therefore, multiple properties can be monitored simultaneously to specifically isolate antibodies with desired characteristics. In this work antibody fragments were isolated from a library on the basis of both antigen affinity and expression level (Figure 1). Subsequently, clones isolated from the last round of FACS were rank-ordered by comparing the dissociation rate constants in crude lysates (Supplementary Figure 1). This approach significantly expedited the analysis of the clones obtained after screening, a process that often represents the rate limiting step in antibody affinity maturation. Thus, a single round of mutagenesis followed by three rounds of FACS screening resulted in the isolation of antibodies displaying affinities in the 100 pM range and also higher expression yields in E. coli (Figure 1b, Table 1). In addition, the conversion of these monovalent antibody fragments to full-length antibodies can further enhance their potency due to the avidity associated with these bivalent molecules leading to an even higher apparent affinity.
Genetic delivery of full-length antibodies in vivo has been carried out using adenovirus, adeno-associated virus, and vaccinia virus vectors 30-32 and is an attractive alternative to the administration of antibody protein which requires production, purification, and formulation prior to injection into the patient 31,33. The administration of antibodies as injectable therapeutics is complicated by bioavailability issues, and by the potential for increased immunogenicity resulting from large dosages required for therapeutic purposes and the ensuing aggregation issues 34. A common side effect is infusion reactions with symptoms ranging from mild to life-threatening and typically occur near the time of administration 35. Following infusions, complicated pharmacokinetics result in a continual decrease in serum concentrations which effects the bioavailability and the pharmacodynamic properties of the therapeutic antibody 34,36. However, genetic delivery can provide immediate and sustained serum levels for several months from a single treatment with both adenovirus (Ad) and adeno-associated virus (AAV) vectors encoding the same monoclonal antibody 37. This approach has been demonstrated with a neutralizing antibody and offered almost immediate protection against anthrax toxin challenge in vivo which was sustained for 6 months 23. This approach circumvents many of the issues associated with conventional methods and has the additional benefits of increasing patient quality of life and protecting soldiers entering combat for extended periods.
Here we demonstrate that the affinity of antibodies produced in situ following adenoviral gene transfer correlates with protection. Although human Ad serotype 5(Ad5) has been effectively used in a variety of animal models for genetic delivery, it has limited use because approximately 35 to 50% of humans have preexisting neutralizing antibodies against it 38. However, several non-human primate adenovirus vectors for which no natural immunity exists in humans have been described and can be deployed to circumvent the neutralization or clearance of the vector 37. There are many pathogens to which an effective vaccine has yet to be produced and neutralizing antibodies remain the only means for prophylaxis and therapy. The potency of neutralizing antibodies can benefit from engineering their affinity, stability and neutralization potency (for instance through mutations that increase antibody-dependent cell-mediated cytotoxicity 39). The in situ synthesis of optimized neutralizing antibodies by low immunogenicity genetic delivery vectors could represent a promising alternative for the treatment of a number of infectious diseases.
Supplementary Material
Supplementary Figure 1. Determination of antibody dissociation rate constants by SPR of cell lysates. (a) Flow chart describing the method. Antibodies are expressed in the scAb format and all steps are performed using 96-well microtiter plates. (b & c) Demonstration of the methods reproducibility. Four colonies each expressing 2C12.4 scAb were individually picked and processed according to the method described here. The dissociation rate constant (koff) for each sample was compared to the actual value obtained using purified 2C12.4 scAb monomers. Accurate values for H8 were not determined because the calculated koff were lower than the reported sensitivity of the Biacore™ 3000 40 (d) Individual sensograms from 72 clones isolated by APEx following three rounds of FACS, 8 replicates of 2C12.4 scAb (reference) and 4 replicates of 26.10 scAb (negative control). (e & f) Analysis of the four clones with the lowest dissociation rate constants (koff) as compared to 2C12.4.
Acknowledgments
This work was supported by a grant from the Clayton Foundation (to GG), a NIH grant U54 AI057158 (to RGC), and a gift from Robert A. Belfer to Support Development of an Antibioterrorism Vaccine (to RGC). We would like to thank Mridula Rani for generating the sequencing encoding the glycine-serine linker for the antibody fragments. We would also like to thank the staff at the Public Health Research Institute for conducting the Y. pestis challenge studies.
Footnotes
Conflict of interest: The authors declare no conflict of interest.
Supplementary Information is available at Gene Therapy's website (http://www.nature.com/gt)
References
- 1.Sofer-Podesta C, et al. Adenovirus-mediated Delivery of an Anti-V Antigen Monoclonal Antibody Protects Mice Against a Lethal Yersinia pestis Challenge. Infect Immun. 2009;77:1561–1568. doi: 10.1128/IAI.00856-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smiley ST. Immune defense against pneumonic plague. Immunol Rev. 2008;225:256–271. doi: 10.1111/j.1600-065X.2008.00674.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brouqui P, Raoult D. Arthropod-borne diseases in homeless. Ann N Y Acad Sci. 2006;1078:223–235. doi: 10.1196/annals.1374.041. [DOI] [PubMed] [Google Scholar]
- 4.Zhou D, Han Y, Yang R. Molecular and physiological insights into plague transmission, virulence and etiology. Microbes Infect. 2006;8:273–284. doi: 10.1016/j.micinf.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 5.Hill J, et al. Administration of antibody to the lung protects mice against pneumonic plague. Infect Immun. 2006;74:3068–3070. doi: 10.1128/IAI.74.5.3068-3070.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hill J, et al. Synergistic protection of mice against plague with monoclonal antibodies specific for the F1 and V antigens of Yersinia pestis. Infect Immun. 2003;71:2234–2238. doi: 10.1128/IAI.71.4.2234-2238.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hill J, et al. Regions of Yersinia pestis V antigen that contribute to protection against plague identified by passive and active immunization. Infect Immun. 1997;65:4476–4482. doi: 10.1128/iai.65.11.4476-4482.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maynard JA, et al. Protection against anthrax toxin by recombinant antibody fragments correlates with antigen affinity. Nat Biotechnol. 2002;20:597–601. doi: 10.1038/nbt0602-597. [DOI] [PubMed] [Google Scholar]
- 9.Wu H, et al. Development of motavizumab, an ultra-potent antibody for the prevention of respiratory syncytial virus infection in the upper and lower respiratory tract. J Mol Biol. 2007;368:652–665. doi: 10.1016/j.jmb.2007.02.024. [DOI] [PubMed] [Google Scholar]
- 10.Marks JD. Deciphering antibody properties that lead to potent botulinum neurotoxin neutralization. Mov Disord. 2004;19(Suppl 8):S101–108. doi: 10.1002/mds.20023. [DOI] [PubMed] [Google Scholar]
- 11.Barbas CF, 3rd, et al. In vitro evolution of a neutralizing human antibody to human immunodeficiency virus type 1 to enhance affinity and broaden strain cross-reactivity. Proc Natl Acad Sci U S A. 1994;91:3809–3813. doi: 10.1073/pnas.91.9.3809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mabry R, et al. Passive protection against anthrax by using a high-affinity antitoxin antibody fragment lacking an Fc region. Infect Immun. 2005;73:8362–8368. doi: 10.1128/IAI.73.12.8362-8368.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Riano-Umbarila L, et al. A strategy for the generation of specific human antibodies by directed evolution and phage display. An example of a single-chain antibody fragment that neutralizes a major component of scorpion venom. Febs J. 2005;272:2591–2601. doi: 10.1111/j.1742-4658.2005.04687.x. [DOI] [PubMed] [Google Scholar]
- 14.Rajpal A, et al. A general method for greatly improving the affinity of antibodies by using combinatorial libraries. Proc Natl Acad Sci U S A. 2005;102:8466–8471. doi: 10.1073/pnas.0503543102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harvey BR, et al. Anchored periplasmic expression, a versatile technology for the isolation of high-affinity antibodies from Escherichia coli-expressed libraries. Proc Natl Acad Sci U S A. 2004;101:9193–9198. doi: 10.1073/pnas.0400187101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hayhurst A, et al. Isolation and expression of recombinant antibody fragments to the biological warfare pathogen Brucella melitensis. Journal of Immunological Methods. 2003;276:185–196. doi: 10.1016/s0022-1759(03)00100-5. [DOI] [PubMed] [Google Scholar]
- 17.Chiuchiolo MJ, et al. Protective immunity against respiratory tract challenge with Yersinia pestis in mice immunized with an adenovirus-based vaccine vector expressing V antigen. J Infect Dis. 2006;194:1249–1257. doi: 10.1086/507644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen G, et al. Isolation of high-affinity ligand-binding proteins by periplasmic expression with cytometric screening (PECS) Nat Biotechnol. 2001;19:537–542. doi: 10.1038/89281. [DOI] [PubMed] [Google Scholar]
- 19.Mazor Y, Van Blarcom T, Iverson BL, Georgiou G. E-clonal antibodies: selection of full-length IgG antibodies using bacterial periplasmic display. Nat Protoc. 2008;3:1766–1777. doi: 10.1038/nprot.2008.176. [DOI] [PubMed] [Google Scholar]
- 20.Ribnicky B, Van Blarcom T, Georgiou G. A scFv antibody mutant isolated in a genetic screen for improved export via the twin arginine transporter pathway exhibits faster folding. J Mol Biol. 2007;369:631–639. doi: 10.1016/j.jmb.2007.03.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fromant M, Blanquet S, Plateau P. Direct random mutagenesis of gene-sized DNA fragments using polymerase chain reaction. Anal Biochem. 1995;224:347–353. doi: 10.1006/abio.1995.1050. [DOI] [PubMed] [Google Scholar]
- 22.Myszka DG. Improving biosensor analysis. J Mol Recognit. 1999;12:279–284. doi: 10.1002/(SICI)1099-1352(199909/10)12:5<279::AID-JMR473>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 23.De BP, Hackett NR, Crystal RG, Boyer JL. Rapid/sustained anti-anthrax passive immunity mediated by co-administration of Ad/AAV. Mol Ther. 2008;16:203–209. doi: 10.1038/sj.mt.6300344. [DOI] [PubMed] [Google Scholar]
- 24.Rosenfeld MA, et al. In vivo transfer of the human cystic fibrosis transmembrane conductance regulator gene to the airway epithelium. Cell. 1992;68:143–155. doi: 10.1016/0092-8674(92)90213-v. [DOI] [PubMed] [Google Scholar]
- 25.Mittereder N, March KL, Trapnell BC. Evaluation of the concentration and bioactivity of adenovirus vectors for gene therapy. J Virol. 1996;70:7498–7509. doi: 10.1128/jvi.70.11.7498-7509.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hayhurst A. Improved expression characteristics of single-chain Fv fragments when fused downstream of the Escherichia coli maltose-binding protein or upstream of a single immunoglobulin-constant domain. Protein Expr Purif. 2000;18:1–10. doi: 10.1006/prep.1999.1164. [DOI] [PubMed] [Google Scholar]
- 27.Martin AC. Accessing the Kabat antibody sequence database by computer. Proteins. 1996;25:130–133. doi: 10.1002/(SICI)1097-0134(199605)25:1<130::AID-PROT11>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 28.Foote J, Eisen HN. Kinetic and affinity limits on antibodies produced during immune responses. Proc Natl Acad Sci U S A. 1995;92:1254–1256. doi: 10.1073/pnas.92.5.1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mohamed N, et al. A high-affinity monoclonal antibody to anthrax protective antigen passively protects rabbits before and after aerosolized Bacillus anthracis spore challenge. Infect Immun. 2005;73:795–802. doi: 10.1128/IAI.73.2.795-802.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fang J, et al. Stable antibody expression at therapeutic levels using the 2A peptide. Nat Biotechnol. 2005;23:584–590. doi: 10.1038/nbt1087. [DOI] [PubMed] [Google Scholar]
- 31.Jiang M, et al. Gene therapy using adenovirus-mediated full-length anti-HER-2 antibody for HER-2 overexpression cancers. Clin Cancer Res. 2006;12:6179–6185. doi: 10.1158/1078-0432.CCR-06-0746. [DOI] [PubMed] [Google Scholar]
- 32.BenAmmar-Ceccoli S, et al. Recombinant vaccinia viruses expressing immunoglobulin variable regions efficiently and selectively protect mice against tumoral B-cell growth. Cancer Gene Ther. 2001;8:815–826. doi: 10.1038/sj.cgt.7700376. [DOI] [PubMed] [Google Scholar]
- 33.Daugherty AL, Mrsny RJ. Formulation and delivery issues for monoclonal antibody therapeutics. Adv Drug Deliv Rev. 2006;58:686–706. doi: 10.1016/j.addr.2006.03.011. [DOI] [PubMed] [Google Scholar]
- 34.Samaranayake H, et al. Challenges in monoclonal antibody-based therapies. Ann Med. 2009:1–10. doi: 10.1080/07853890802698842. [DOI] [PubMed] [Google Scholar]
- 35.Chung CH. Managing premedications and the risk for reactions to infusional monoclonal antibody therapy. Oncologist. 2008;13:725–732. doi: 10.1634/theoncologist.2008-0012. [DOI] [PubMed] [Google Scholar]
- 36.Joshi A, et al. An overview of the pharmacokinetics and pharmacodynamics of efalizumab: a monoclonal antibody approved for use in psoriasis. J Clin Pharmacol. 2006;46:10–20. doi: 10.1177/0091270005283282. [DOI] [PubMed] [Google Scholar]
- 37.Boyer JL, Kobinger G, Wilson JM, Crystal RG. Adenovirus-based genetic vaccines for biodefense. Hum Gene Ther. 2005;16:157–168. doi: 10.1089/hum.2005.16.157. [DOI] [PubMed] [Google Scholar]
- 38.Hashimoto M, et al. Induction of protective immunity to anthrax lethal toxin with a nonhuman primate adenovirus-based vaccine in the presence of preexisting anti-human adenovirus immunity. Infect Immun. 2005;73:6885–6891. doi: 10.1128/IAI.73.10.6885-6891.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lazar GA, et al. Engineered antibody Fc variants with enhanced effector function. Proc Natl Acad Sci U S A. 2006;103:4005–4010. doi: 10.1073/pnas.0508123103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu H, et al. Ultra-potent antibodies against respiratory syncytial virus: effects of binding kinetics and binding valence on viral neutralization. J Mol Biol. 2005;350:126–144. doi: 10.1016/j.jmb.2005.04.049. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Determination of antibody dissociation rate constants by SPR of cell lysates. (a) Flow chart describing the method. Antibodies are expressed in the scAb format and all steps are performed using 96-well microtiter plates. (b & c) Demonstration of the methods reproducibility. Four colonies each expressing 2C12.4 scAb were individually picked and processed according to the method described here. The dissociation rate constant (koff) for each sample was compared to the actual value obtained using purified 2C12.4 scAb monomers. Accurate values for H8 were not determined because the calculated koff were lower than the reported sensitivity of the Biacore™ 3000 40 (d) Individual sensograms from 72 clones isolated by APEx following three rounds of FACS, 8 replicates of 2C12.4 scAb (reference) and 4 replicates of 26.10 scAb (negative control). (e & f) Analysis of the four clones with the lowest dissociation rate constants (koff) as compared to 2C12.4.