Abstract
Plus-strand RNA virus replication occurs in tight association with cytoplasmic host cell membranes. Both, viral and cellular factors cooperatively generate distinct organelle-like structures, designated viral replication factories. This compartmentalization allows coordination of the different steps of the viral replication cycle, highly efficient genome replication and protection of the viral RNA from cellular defense mechanisms. Electron tomography studies conducted during the last couple of years revealed the three dimensional structure of numerous plus-strand RNA virus replication compartments and highlight morphological analogies between different virus families. Based on the morphology of virus-induced membrane rearrangements, we propose two separate subclasses: the invaginated vesicle/spherule type and the double membrane vesicle type. This review discusses common themes and distinct differences in the architecture of plus-strand RNA virus-induced membrane alterations and summarizes recent progress that has been made in understanding the complex interplay between viral and co-opted cellular factors in biogenesis and maintenance of plus-strand RNA virus replication factories.
Keywords: Viral replication factory, Viral replication complex, Plus-strand RNA virus, Membrane remodeling, Virus-host interaction, Alphavirus, Enterovirus, Coronavirus, Flavivirus, Hepatitis C virus
INTRODUCTION
As obligate intracellular parasites, all viruses depend on the host cell biosynthetic machinery in order to replicate their genome and generate progeny virus particles. A common feature among many different viruses is the induction of specialized membranous compartments, often forming organelle-like structures, within the cytoplasm of an infected cell. The unique features of those structures that facilitate virus propagation are best expressed by the commonly accepted term “viral replication factories”. This review focuses on animal plus-strand RNA virus induced replication compartments. However, the concept of viral replication factories also applies to plant plus-strand RNA viruses as well as to other animal RNA viruses or some large DNA viruses such as vaccinia virus. These virus groups are covered by other reviews and will not be discussed here[1-3]. By analogy to an industrial manufacturing unit, virus induced replication factories serve multiple purposes.
To increase the local concentration of all necessary factors thus ensuring high efficacy
Local enrichment of the viral replication factors and co-opted cellular proteins, together with all required metabolites, is the basis for highly efficient genome replication. This is achieved by reducing the three-dimensional (3-D) diffusion in the cytosol to two-dimensional diffusion on the surface of a membrane, thus increasing local reaction efficacy presumably by several orders of magnitude[4]. In fact, many replication enzymes encoded by plus-strand RNA viruses are membrane anchored as exemplified by the hepatitis C virus (HCV)[5]. In addition, also substrates such as ribonucleotides and metabolites providing energy for biosynthetic processes are enriched in replication compartments. Along these lines a recent study convincingly demonstrated locally elevated ATP levels at HCV replication sites[6].
To spatially coordinate different processes of the replication cycle such as RNA translation, replication and assembly
Compartmentalization allows the spatial separation and regulation of RNA translation, replication and packaging of the viral genome, thereby preventing an interference between these processes. Hence, ribosomes responsible for RNA translation and RNA-binding assembly factors are excluded from replication sites, thus avoiding interference with the replication machinery. Indeed, replication factories of all plus-strand RNA viruses are built up of ribosome-free membranes[7-11], and for all Flaviviridae (excess) amounts of core/capsid protein, binding RNA with high affinity, are frequently targeted to other cellular organelles such as lipid droplets (LDs) in the case of HCV and dengue virus (DENV)[12-14], or to the nucleus as observed for DENV[15,16] Japanese encephalitis virus[17] and West Nile virus (WNV)[18].
To create a protected environment shielding viral RNA and eventually also proteins from a hostile degradative environment
The generation of specialized membranous replication compartments protects viral replicase complexes and genomic RNA from degradation by cellular proteases or nucleases, respectively and “hides” the viral RNA genome from cytoplasmic sensors of the innate immune response. The RigI-like receptors efficiently recognize 5’ triphosphorylated RNAs as well as double-stranded RNA (dsRNA) in a length-dependent manner[19,20], leading to mitochondrial antiviral signaling-mediated induction of interferons and nuclear factor κB-mediated inflammation[21]. Minimizing the exposure of stimuli to the innate immune surveillance, by the induction of innate sensor-protected organelle-like replication factories, is therefore an important evolutionary conserved feature of plus-strand RNA virus infection.
In the following we will summarize recent insights into the 3-D ultrastructure of plus-strand RNA virus-induced membrane rearrangements and discuss possible mechanisms of their biogenesis. Furthermore, viral subversion of host cell membrane biology, by interference with signaling pathways and recruitment of host cell factors contributing to biogenesis and maintenance of viral replication factories are highlighted.
MORPHOLOGY OF PLUS-STRAND RNA VIRUS REPLICATION FACTORIES
In the last few years, electron tomography has been instrumental to decipher the 3-D architecture of viral replication factories (for technical review see[22,23]). This accounts for evolutionary diverse plus-strand RNA viruses such as flock-house virus (FHV)[24], rubella virus (RUBV)[8], the two enteroviruses coxsackievirus B3 (CVB3)[25] and poliovirus (PV)[26], severe acute respiratory syndrome coronavirus (SARS-CoV)[11], equine arterivirus (EAV)[27], the two flaviviruses DENV[10] and WNV[9] and HCV[28]. Despite many differences in host range, virion morphology, genome organization, or donor membrane usage (Table 1), these analyses revealed that plus-strand RNA viruses appear to induce one of two different membrane alterations: the invaginated vesicle (InV) or spherule type and the double membrane vesicle (DMV) type. These morphologies that will be used in this review to group plus-strand RNA viruses might reflect the use of different host cell pathways and factors exploited by these viruses to establish the membranous replication compartment.
Table 1.
Overview of plus-strand RNA viruses and induced replication factories
| Invaginated vesicle/spherule type | |||||||
| Superfamily/order | Alphavirus like | Alphavirus like | Alphavirus like | Alphavirus like | |||
| Family | Togaviridae | Togaviridae | Togaviridae | Bromoviridae | Nodaviridae | Flaviviridae | Flaviviridae |
| Genus | Alphavirus | Alphavirus | Rubivirus | Bromovirus | Alphanodavirus | Flavivirus | Flavivirus |
| Species | SFV | SINV | RUBV | BMV | FHV | DENV | WNV |
| Genome | ss(+) RNA | ss(+) RNA | ss(+) RNA | tripartite ss(+) RNA | bipartite ss(+) RNA | ss(+) RNA | ss(+) RNA |
| Gene order | NS-S | NS-S | NS-S | NS-S | NS-S | S-NS | S-NS |
| Genome size (bases) | 13000 | 12000 | 10000 | 8300 | 4500 | 10000 | 10000 |
| Virion size (nm) | 70 | 70 | 70 | 27 | 30 | 50 | 50 |
| Envelope | Yes | Yes | Yes | No | No | Yes | Yes |
| Host | Mosquitoes, humans | Mosquitoes, humans | Humans | Plants, yeast | Insects | Mosquitoes, humans | Mosquitoes, mammals |
| Disease | Encephalitis | Sinbis fever | German measels | Plant disease | Unknown | Dengue fever | Mostly asymtomatic |
| Type of replication factories | Spherules at PM and CPVs | Spherules at PM and CPVs | Invaginations at CPVs | Spherules at the ER | Spherules at mitochondria | Invaginated vesicles at the ER | Invaginated vesicles at the ER |
| Diameter of invaginations (nm) | 50 | 50 | 80-500 | 50-70 | 50 | 90 | 50-150 |
| Primary membrane source | PM, endosomes | PM, endosomes | Endosomes | ER | Outer mitochondrial membrane | ER | ER |
| Proposed sites of replication | Inside spherules | Inside spherules | Inside invaginated vesicles | Inside spherules | Inside spherules | Inside invaginated vesicles | Inside invaginated vesicles |
| Double membrane vesicle type | |||||||
| Superfamily/order | Picornavirales | Picornavirales | Nidovirales | Nidovirales | Nidovirales | ||
| Family | Flaviviridae | Picornaviridae | Picornaviridae | Coronaviridae | Coronaviridae | Arteriviridae | |
| Genus | Hepacivirus | Enterovirus | Enterovirus | Coronavirus | Coronavirus | Arterivirus | |
| Species | HCV | PV | CVB3 | SARS CoV | MHV | EAV | |
| Genome | ss(+) RNA | ss(+) RNA | ss(+) RNA | ss(+) RNA | ss(+) RNA | ss(+) RNA | |
| Gene order | S-NS | S-NS | S-NS | S-NS | S-NS | S-NS | |
| Genome size (bases) | 9600 | 8000 | 8000 | 30000 | 30000 | 13000 | |
| Virion size (nm) | 50 | 30 | 30 | 80-160 | 80-160 | 40-60 | |
| Envelope | Yes | No | No | Yes | Yes | Yes | |
| Host | Humans | Humans | Humans | Humans | Mice | Horses, donkeys | |
| Disease | Hepatitis | Poliomyelitis | HFM disease | SARS | HMM illness | Haemorragic fever | |
| Type of replication factories | DMVs | SMTs, DMVs | SMTs, DMVs | DMVs, VPs | DMVs | DMVs | |
| Length/diameter of replication factories | 150 nm | 100-200 nm, 100-300 nm | 650 nm, 150 nm | 150 nm, 1-5 μm | 200-350 nm | 90 nm | |
| Primary membrane source | ER | Golgi, ER | Golgi, ER | ER | ER | ER | |
| Proposed sites of replication | Possible role of DMVs but largely unknown | On SMT/DMV outer membrane | On SMT/DMV outer membrane | Inside DMVs | Inside DMVs | Inside DMVs | |
NS: Genes encoding nonstructural proteins; S: Genes encoding structural proteins; ER: Endoplasmic reticulum; PM: Plasma membrane; HFM: Hand-foot-and mouth; SARS-CoV: Severe acute respiratory syndrome coronavirus; HMM: High mortality murine; DMVs: Double membrane vesicles; SMTs: Single membrane tubular vesicles; VPs: Vesicle packets; PV: Poliovirus; HCV: Hepatitis C virus; CVB3: Coxsackievirus B3; MHV: Mouse hepatitis virus; EAV: Equine arterivirus; CPV: Cytoplasmic vacuoles; RUBV: Rubella virus; SFV: Semliki Forrest virus; FHV: Flock-house virus; DENV: Dengue virus; WNV: West Nile virus; BMV: Brome mosaic virus; SINV: Sindbis virus.
Architecture of replication factories corresponding to the InV/spherule type
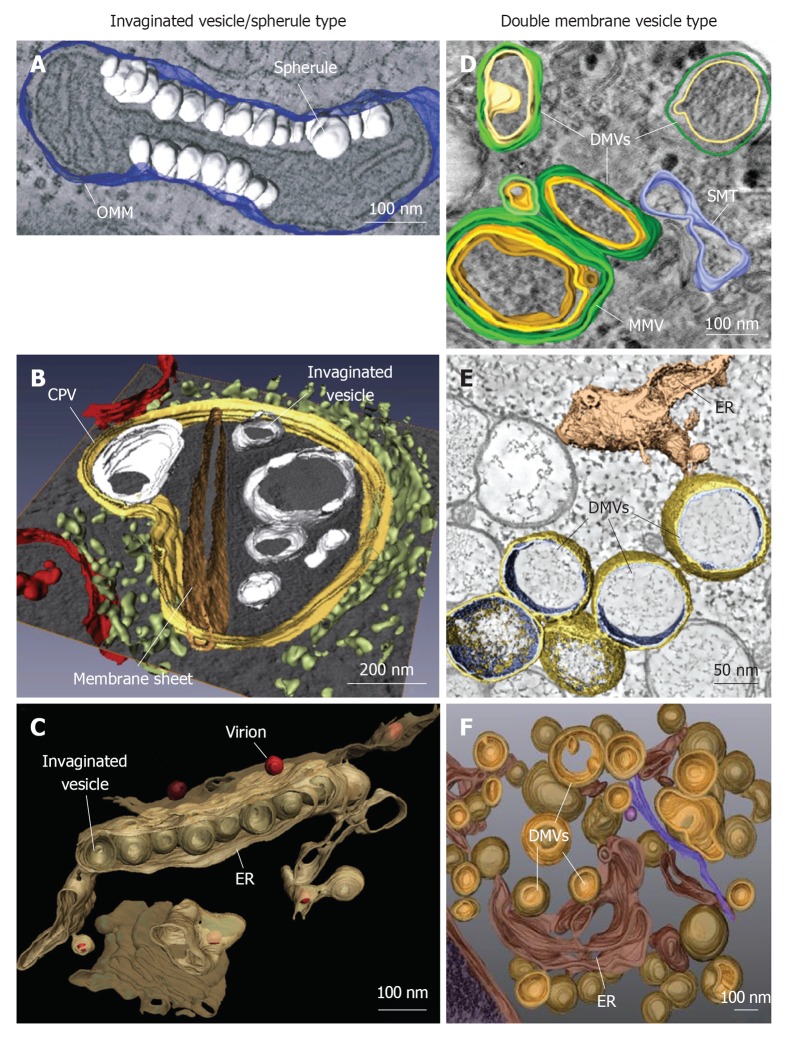
Viral replication factories of the InV/spherule type are induced by alphaviruses such as Semliki Forrest virus (SFV)[29,30] and Sindbis virus[31], by FHV[24], RUBV[8], DENV[10] and WNV[9]. Although no 3-D reconstruction of alphavirus replication factories has been published yet, pioneering classical electron microscopy (EM) studies from Grimley et al[29] describing SFV replication sites at modified membranous structures, date back to the 1960s. Alphavirus infection induces so called cytoplasmic vacuoles (CPVs) (600-2000 nm in size), containing small invaginations called “spherules” with an average diameter of approximately 50 nm[29,32-34]. Surprisingly, at early time points after alphavirus infection, spherules are frequently found at the plasma membrane[30,31]. These spherules are subsequently internalized and become part of the endo-lysosomal membrane system, giving rise to CPVs. The single membrane invagination of spherules is continuous with its donor membrane and an approximately 8 nm small opening connects its interior with the cytoplasm[7]. Viral replicase proteins nsp1 to nsp4 as well as newly synthesized viral RNA localize to spherules[29,30]. Interestingly, the spherules themselves are devoid of ribosomes and viral capsid protein, which are frequently found juxtaposed to the spherule openings[7]. The first 3-D reconstruction of a plus-strand RNA virus replication factory was published by Kopek et al[24]. Electron tomography of FHV-infected cells revealed InVs on the outer mitochondrial membrane (OMM) (Figure 1A). Similar to alphavirus spherules, InVs found in FHV-infected cells are approximately 50 nm in diameter[35]. The single membrane building up the spherule is continuous with the OMM and a membranous neck with an interior diameter of approximately 10 nm connects the spherule lumen to the cytoplasm[24]. The sole viral replicase factor protein A as well as nascent viral RNA localize to spherules. By using high resolution electron tomography in combination with biochemical analyses, Kopek et al[24] determined that each spherule contains approximately 100 copies of protein A, covering the complete interior surface of the spherule and one or two copies of minus-strand RNA, the replication intermediate. RUBV induces replication factories with similar architecture. Replicase proteins p150 and p90, as well as nascent viral RNA localize to remodeled endosomes/lysosomes, termed cytopathic vacuoles[36-39]. Fontana et al[8] conducted electron tomography studies of RUBV replicon cells and observed spherule-like membrane invaginations. In addition, the authors detected bigger vacuolar invaginations and rigid straight membrane sheets, which are continuous with the outer membrane of the CPV and whose interior is connected to the cytoplasm (Figure 1B). Mitochondria localized in close proximity to CPVs and were probably engaged in supplying energy for viral replication. Importantly, CPV membranes were free of ribosomes, but frequently surrounded by rough ER sheets and Golgi stacks, thus facilitating translation and encapsidation of viral RNA, respectively[8,40]. In contrast to the replication compartments of these plus-strand RNA viruses, replication factories of Flaviviridae, exemplified by WNV and DENV, are most likely derived from the ER. Apart from the rather undefined convoluted membranes (CMs) found in DENV-infected cells, our laboratory revealed by means of electron tomography vesicular invaginations of approximately 90 nm diameter in the rough ER, which are connected by approximately 10 nm diameter necked channel openings to the cytoplasm[10] (Figure 1C). Likewise, Gillespie et al[9] observed in WNV replicon-containing cells, InVs in the rough ER, which were connected via neck-like membranous pores to the cytoplasmic space. For both viruses, replicase proteins and dsRNA, assumed to represent a replication intermediate, localized to InVs[10,41], whereas ribosomes were excluded from the InVs, but localized in close proximity on ER membranes[9,10]. Interestingly, DENV virion budding was observed frequently close to or directly opposite of pore-like vesicle openings[10], highlighting the spatial orchestration of replication and assembly steps in the DENV replication cycle.
Figure 1.
Classification and morphologies of plus-strand RNA virus-induced membrane alterations as revealed by electron tomography and three-dimensional reconstruction. Morphological similarities of membrane structures belonging to the invaginated vesicle (InV)/spherule type (A-C) or the DMV type (D-F). A: Flock-house virus (FHV); B: Rubella virus (RUBV); C: Dengue virus (DENV); D: Poliovirus (PV); E: Severe acute respiratory syndrome coronavirus (SARS-CoV); F: Hepatitis C virus (HCV). A: FHV-induced spherules (white) in the outer mitochondrial membrane (OMM; blue); B: InVs (white) and rigid membrane sheets (dark brown) in a modified endosome (the cytopathic vacuole, CPV) (yellow) in cells replicating RUBV. The rough endoplasmic reticulum (ER) is shown in light-green and mitochondria in red; C: Spherule-like invaginations (InVs) in the endoplasmic reticulum (ER) (brown) observed in DENV-infected cells. Newly formed progeny virions are shown in red; D: PV infection induces single membrane tubules (SMT) (blue), double membrane vesicles (DMVs) and multi membrane vesicles (MMVs). The DMV outer membrane is shown in green, the second membrane in yellow and a third membrane in case of MMVs in orange. These membranes are primarily derived from the Golgi; E: An interconnected network of ER-derived DMVs is found in SARS-CoV infected cells. The ER is colored in light-brown, interconnected outer membranes of DMVs in yellow and DMV inner membranes in blue; F: HCV-induced DMVs protruding from the ER (dark-brown). Outer membranes of DMVs are shown in light-brown and inner membranes in orange. Images are modified with permission from ©2007 Kopek et al[24] Plos Biol (A); ©2010 Elsevier Inc.[8] (B); ©2009 Elsevier Inc.[10] (C); ©2012 American Society for Microbiology[26] (D); ©2008 Knoops et al[11] Plos Biol (E); ©2012 Romero-Brey et al[28] Plos Pathog (F).
Architecture of DMV-like replication factories
DMV-like replication factories are formed by the enteroviruses CVB3[25] and PV[26], SARS-CoV[11], EAV[27] and HCV[28]. For PV, probably the best studied plus-strand RNA virus, structural alterations of cellular membranes were observed already more than 50 years ago. They were first described as membranous vesicles of heterogeneous size and termed clear vacuoles and U-bodies[42]. Recent electron tomography studies of PV-infected cells by Belov et al[26] revealed apparently empty and heterogeneous single membrane-branching tubular structures (100-200 nm diameter) that were detected at very early time points after infection. As infection progresses, these tubular structures developed into DMVs (100-300 nm in diameter), filled with presumably cytoplasmic material (Figure 1D). The very early membrane alterations were derived from the Golgi[26], but PV-induced vesicles were earlier shown to “bud” from the ER in a COPII-dependent manner and to contain lysosomal markers[43,44]. Replicase proteins 2C and 3A as well as nascent viral RNA localize to the outside of both single and double membrane structures[26,44]. A similar 3-D architecture of virus-induced membrane alterations was recently described for CVB3, another member of the Picornaviridae family. By using electron tomography, Limpens et al[25] observed interconnected single membrane tubular structures (approximately 650 nm in length and approximately 80 nm in diameter) at the onset of infection. These structures transformed into DMVs (approximately 160 nm in diameter) and multimembranous structures by an enwrapping process. Immunofluorescence analyses showed co-localization of CVB3 replicase proteins 3A and 3D with dsRNA[25] during the log phase of viral replication, when predominantly single membrane tubules were present in infected cells. However, the exact localization of viral proteins and RNA with respect to the rearranged membranes remains to be determined. In contrast to picornavirus-infected cells, Knoops et al[11] revealed that early after infection SARS-CoV replication factories appear as presumably ER-derived CMs and interconnected DMVs (250 nm in diameter) (Figure 1E), which subsequently merge into vesicle packets (VPs) (1-5 μm in size), most likely by fusion of the DMV outer membranes. Electron tomography analysis showed that SARS-CoV-induced DMVs do not exist as isolated structures, but their outer membrane is continuous to other DMVs, CMs, or the rER, thus explaining the transition from DMVs to VPs[11]. Viral replicase proteins nsp3, 5 and 8 were sporadically found in DMVs, whereas the majority localized to CMs. In contrast, dsRNA a marker for viral replication intermediates, was predominantly found inside DMVs or inside the vesicles of VPs[11]. The DMV interior was devoid of ribosomes, which were found on the outer membranes of DMVs or rER sheets in close proximity to DMVs. Virus budding was detected at the outer membrane of VPs, arguing for spatial organization of the different steps of the SARS-CoV replication cycle[11]. A comparable architecture of virus-induced membrane rearrangements was observed for EAV, a SARS-CoV related virus that also belongs to the order Nidovirales. By means of electron tomography, Knoops et al[27] unraveled a reticulovesicular network of interconnected DMVs in EAV-infected cells. However, in contrast to SARS-CoV induced DMVs, those observed for EAV were smaller in diameter (approximately 90 nm), but also exhibited continuous outer DMV membranes. Replicase proteins nsp3 and 9 associated with DMVs and surrounding membranes and as in case of SARS-CoV, dsRNA was prominently found inside DMVs[27]. In addition to the reticulovesicular DMV network, membrane tubules with approximately 43 nm diameter were associated with the EAV capsid protein N and frequently found in close proximity to DMVs[27], thus providing another example for the spatial coordination of replication and assembly steps. Although belonging to the Flaviviridae that includes DENV and WNV, HCV-induced viral replication factories are very different from the InV/spherule type, but more comparable to those of unrelated Picornaviridae or Coronaviridae. Electron tomography of HCV-infected cells conducted in our laboratory, revealed ER-derived DMVs as the predominant structures, which partially develop into MMVs at later stages of infection[28] (Figure 1F). HCV-induced DMVs have an average diameter of approximately 150 nm. Around 50% of DMVs were linked to the ER via the outer membrane giving rise to a neck-like structure, but later on DMVs seem to detach from their donor membrane. Only a small subset (approximately 10%) exhibited a pore-like opening to the cytoplasm, whereas the vast majority showed “sealed” inner and outer membranes.
By using correlative-light-EM, DMVs were shown to contain the viral replicase protein non-structural protein (NS)5A, which co-localizes with dsRNA as shown by fluorescence microscopy[28]. However, direct detection of the HCV replication site at the ultrastructural level was not successful. DMVs localized in close proximity to the rER and LDs, an important organelle for HCV assembly[13], arguing for a compartmentalization of viral RNA translation, replication and assembly steps[28].
TOPOLOGY OF PLUS-STRAND RNA VIRUS REPLICATION SITES
In order to identify viral replication sites, various methods have been applied to detect viral RNA at the ultrastructural level. Initial studies employed metabolic radiolabeling of nascent viral RNA with 3H-labeled uridine or adenosine and subsequent EM-based detection by autoradiography using film emulsions[29,33]. Others applied in situ hybridization methods by using nucleic acid probes and detection via colloidal gold particles[45], labeling of dsRNA replication intermediates by using a dsRNA-specific antibody[10,11,27,41], or antibody-based detection of bromouridine, incorporated into nascent viral RNA[24,26,30]. Importantly, for identification of viral replication sites the detection of metabolically labeled viral RNA is the only reliable method; hybridization techniques do not discriminate between replicating RNA or RNA used e.g., for translation or virion formation. The same applies to detection of viral RNA with the dsRNA-specific antibody that can bind to RNA structures present in genomes of plus-strand RNA viruses, thus excluding specificity for detection of active viral RNA replication. Indeed, by using the incorporation of ethynyl-uridine (EU) into actively replicating coronavirus RNA and subsequent detection via click-chemistry, Hagemeijer et al[46] observed a loss of overlap of the EU- signal with dsRNA labeling during the course of infection. For replication factories of the InV/spherule type it is commonly accepted that viral replicase complexes reside on the invaginated membrane and thus, RNA replication takes place in the spherule lumen[9,10,24,30,36,41,47] (Figure 2A). The neck-like connection to the cytoplasm allows import of all required metabolites (e.g., ribonucleotides) and export of newly synthesized RNA destined for translation or packaging into the capsid. By using bromouridine labeling of nascent viral RNA, this assumption has been confirmed for RUBV[36] alphaviruses[30], FHV[24] and brome mosaic virus (BMV)[47]. However, identification of flavivirus replication sites (DENV and WNV) up to now relies only on the immuno-detection of dsRNA[10,41]. DMV-type replication factories exhibit a topologically more complex situation, as both membranes of the DMV are often sealed and no connection to the cytosol is obvious[11,25-28]. Hence, for this group of plus-strand RNA viruses different topologies of replication sites are currently proposed (Figure 2B). In case of enteroviruses, replication is thought to occur on the cytoplasmic side of single and double-membrane structures[26,48]. However, the fact that during the log phase of viral replication predominantly single membrane tubules are found in PV-or CVB3-infected cells[25,26] suggests that active enteroviral replicase complexes primarily reside on these membrane structures. In this case one might speculate that RNA replication ceases after membrane enwrapping and DMV sealing, but this remains to be determined. In case of SARS-CoV and EAV, the bulk of viral RNA is found inside the lumen of DMVs as determined by dsRNA labeling[11,27]. Moreover, nascent EAV and mouse hepatitis virus (MHV) RNA is associated with DMV membranes as shown by bromouridine labeling[49,50] arguing that DMVs are sites of viral replication. However, if replication takes place inside the DMV lumen, it is unclear how import of metabolites and export of viral RNA occurs. Even with high resolution electron tomography, neck-like membrane discontinuities of approximately 10 nm, as observed for InV/spherule like replication factories, have not been found[11,27]. Thus, proteinaceous channels or transporters might be responsible for linking DMV interior to the cytosol. In fact, proteins such as the ER translocon complex or the mitochondrial transporter outer membrane and transporter inner membrane complexes mediating transport across one or two membranes, respectively (reviewed in[51,52]), have not been visualized by electron tomography so far. Thus, an up to now unidentified enzymatic function of viral proteins containing multiple transmembrane passages and/or recruitment of a cellular factor executing this transport function is in principle possible.
Figure 2.
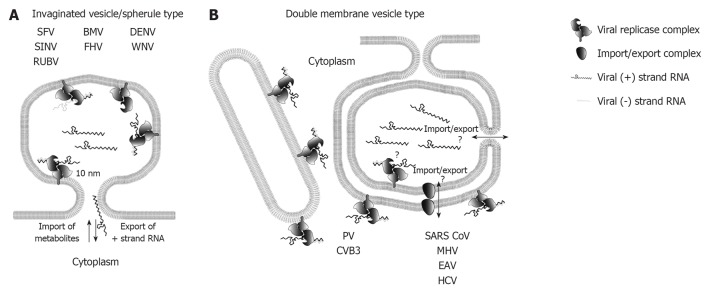
Topology of plus-strand RNA virus replication sites. A: RNA replication of plus-strand RNA viruses inducing invaginated vesicles/spherules is thought to occur in the spherule lumen. Examples of viruses inducing this type of remodeled membranes are denoted. The neck like opening to the cytoplasm allows export of progeny RNA destined for translation or packaging, as well as import of metabolites required for RNA replication; B: Possible topologies of RNA replication site for double membrane vesicle (DMV)-type factories. Enterovirus replication is thought to take place on the outer membrane of single membrane tubules and DMVs, whereas the exact replication site of nidovirales and hepatitis C virus (HCV) is still unclear as indicated by the question marks. Replication in the DMV lumen would require import and export across two membranes, which could be mediated via a proteinacous transport complex, or via an opening to the cytosol of not yet fully closed DMVs (see main text for details). SFV: Semliki Forrest virus; SINV: Sindbis virus; RUBV: Rubella virus; BMV: Brome mosaic virus; FHV: Flock-house virus; DENV: Dengue virus; WNV: West Nile virus; PV: Poliovirus; CVB3: Coxsackievirus B3; SARS-CoV: Severe acute respiratory syndrome coronavirus; MHV: Mouse hepatitis virus; EAV: Equine arterivirus.
Another hypothesis is discussed for HCV-induced DMVs, which accumulate during the log phase of viral RNA replication[28]. Only a small fraction of DMVs has an opening to the cytoplasm, whereas the majority exhibits entirely closed membranes. It is speculated that replication might occur in the interior of DMVs as long as they are linked to the cytosol, but upon membrane sealing DMVs would contain dead end replication complexes that are no longer active. Apart from this possibility, the other models described above also apply to HCV, but further studies are required to define the exact site of RNA replication.
MECHANISMS OF MEMBRANE ALTERATIONS INDUCED BY VIRAL PROTEINS
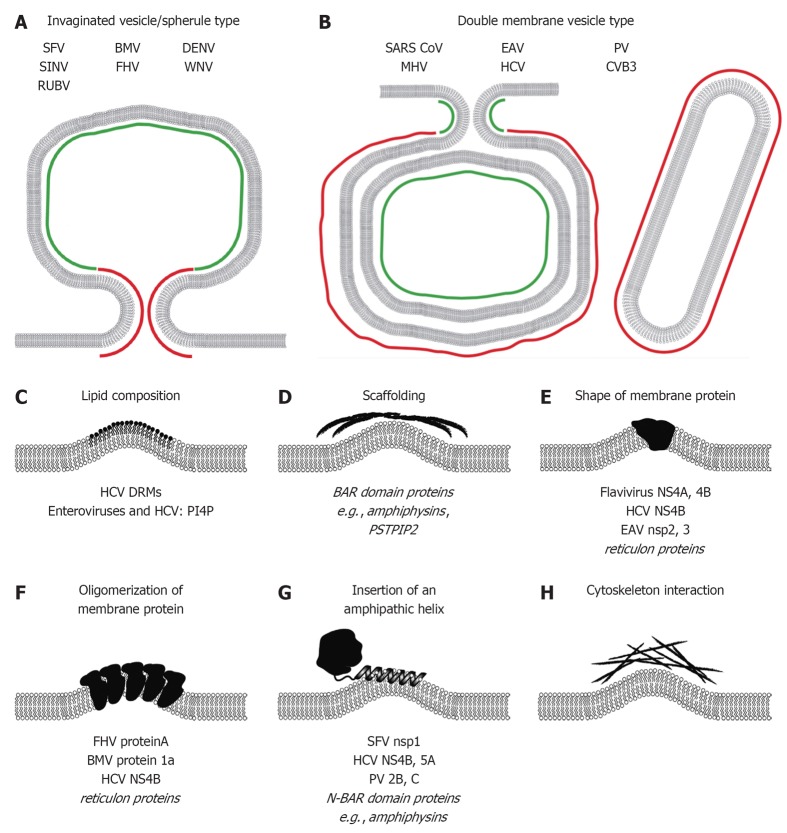
Given the highly complex architecture of plus-strand RNA virus replication factories on one hand and the small genetic coding capacity of these viruses on the other hand, it is obvious that membrane alterations are induced by the concerted action of viral and cellular factors. In this section we will discuss the intrinsic membrane-active properties of viral proteins; host cell factors contributing to formation of replication factories are summarized in section 6 of this article. In principle, membrane bending can be achieved by (1) local alterations of membrane lipid composition; or (2) asymmetric interaction of proteins with membranes (Figure 3). The latter includes scaffolding of the membrane by peripheral proteins, insertion of asymmetric proteins or protein complexes into the membrane, insertion of amphipathic helices into one leaflet of the lipid bilayer or interactions of the membrane with the cytoskeleton (reviewed in[53-55]) (Figure 3C-H). Negative membrane curvature is predominant for the InV/spherule like replication factories (Figure 3A), with spherules bending the membrane away from the cytoplasm, whereas DMV-like replication factories exhibit positive membrane curvature, i.e., the outer DMV membrane is bent towards the cytosol (Figure 3B). However, regions of positive membrane curvature are also present in neck like openings of spherules (Figure 3A) and the inner membranes of DMVs exhibit negative membrane curvature (Figure 3B), respectively, highlighting the complexity of membrane bending events in the induction of plus-strand RNA virus replication factories.
Figure 3.
Mechanisms responsible for induction of membrane curvature likely contributing to formation of plus-strand RNA virus replication factories. Positive and negative membrane curvature for invaginated vesicle/spherule type (A) and double membrane vesicle type (B) replication factories is indicated in red and green, respectively. Mechanisms of membrane modifications/bending are schematically depicted in (C-H). Examples of viral and cellular (in italics) proteins are given at the bottom of each panel (see main text for details). SFV: Semliki Forrest virus; SINV: Sindbis virus; RUBV: Rubella virus; BMV: Brome mosaic virus; FHV: Flock-house virus; DENV: Dengue virus; WNV: West Nile virus; SARS-CoV: Severe acute respiratory syndrome coronavirus; MHV: Mouse hepatitis virus; EAV: Equine arterivirus; HCV: Hepatitis C virus; PV: Poliovirus; CVB3: Coxsackievirus B3; DRM: Detergent-resistant-membrane; BAR: Bin-Amphiphysin-Rvs; PSTPIP2: Proline-serine-threonine phosphatase interacting protein 2; NS4A: Non-structural protein 4A; NS4B: Non-structural protein 4B; N-BAR: N-terminal amphipathic helix containing BAR; PI4P: Phosphatidylinositol-4-phosphate.
Several viral proteins were identified that play crucial roles in the induction of membrane rearrangements. For instance, the membrane-associated protein nsp1 of alphaviruses interacts with anionic phospholipids via an amphipathic helix (AH) (Figure 3G)[56] and is additionally tethered to membranes via a palmitic acid residue in the carboxy-terminal region of the protein[57]. AH-mediated nsp1 membrane association proved crucial for SFV replication[58], whereas palmitoylation mutants could be rescued by second site compensatory muations in vitro[59] but are attenuated in vivo[60]. Nonetheless, only expression of the P123 protein precursor induced spherule-like structures as seen in SFV-infected cells[61], demonstrating the need of nsp1 to recruit and interact with other viral and/or cellular proteins to induce spherule-like membrane rearrangements. The FHV replicase factor protein A exhibits binding affinity to specific anionic phospholipids, namely phosphatidic acid, phosphatidylglycerol and cardiolipin[62], which are enriched in mitochondrial membranes. Protein A is inserted into the OMM by an N-terminal mitochondria-targeting transmembrane sequence[63]. Self-interaction of protein A is required for FHV replication and mediated by multiple domains (Figure 3F)[64]. Nevertheless, protein A is not sufficient to induce spherule like membrane invaginations in the OMM, which requires in addition a replication-competent template RNA and protein A’s polymerase activity[65]. Interestingly, these requirements do not apply to the FHV-related plant virus BMV, for which expression of protein 1a is sufficient to induce spherule-like membrane invaginations[47]. All flaviviral proteins are associated directly or indirectly with membranes either by amphipathic helices or transmembrane domains or both (Figure 3E-G). Fully processed DENV and WNV NS4A is sufficient to induce rearrangements of the ER membrane[66,67], although membrane structures induced by sole NS4A expression are morphologically distinct from those induced in infected cells. Studies of the membrane topology of DENV NS4A revealed an ER luminal helix in the central region of the protein, lying in plane in the luminal membrane leaflet[66]. This topology is compatible with the negative curvature of InVs, arguing that NS4A increases the surface area of the ER membrane on the luminal side. However, the role of NS4A self-interaction and a possible interplay with other viral (e.g., NS4B) and cellular factors, in membrane remodeling remain to be determined. Also for HCV that in contrast to DENV and WNV induces DMV-type replication factories, membrane remodeling activity has been attributed to viral proteins, most prominently to NS4B. It has been proposed that expression of this protein is sufficient to induce the membranous HCV replication compartment (the “membranous web”)[68]. NS4B is a highly hydrophobic protein that contains at least four transmembrane passages[69]. It is palmitoylated at its C-terminus[70] and in addition contains multiple AHs in its N-and C-terminal domains[71-73] (Figure 3E-G). The membrane-associated C-terminal domain of NS4B or the AH within it have been reported to alter membrane integrity (Figure 3G)[74,75], corroborating membrane remodeling activity of NS4B. In addition, NS4B self-interacts in a homo- and heterotypic fashion[76], which is required for HCV RNA replication[76,77] (Figure 3F). Interestingly, mutations in NS4B that inhibit HCV RNA replication also alter NS4B self-interaction and morphology of HCV-induced DMVs[77], implying a functional relationship between DMV morphology and HCV RNA replication. Inspite of the membrane remodeling activity of NS4B, we recently found that only an NS3-5A polyprotein fragment is sufficient to induce DMV structures with morphological similarity to those observed in HCV-infected cells[28]. In fact, MMVs and a small number of DMVs were observed in cells over-expressing just NS5A, which is a dimeric and eventually oligomeric replicase factor with an N-terminal AH[78-82] (Figure 3G). Based on these observations we assume that a concerted action of HCV replicase factors is required for the biogenesis of the membranous web. Similar observations have been made for Nidovirales. For instance, coronavirus nsp3, 4 and 6 contain multiple membrane-spanning domains[83-85]. These proteins appear to play a central role in membrane remodeling (Figure 3E), because mutations e.g., in nsp4 exhibit impaired viral replication and altered DMV morphology[86]. Nsp3 of the related EAV also contains multiple transmembrane segments and is another key player in the induction of DMVs. Expression of nsp2-3 suffices to induce DMVs[87] and their morphology is dramatically altered by nsp3 mutations affecting an ER luminal loop[88].
In case of enteroviruses, proteins 2B, 2C or 3A are membrane-associated via transmembrane passages or AHs[89-91] and the expression of 2BC either alone[92] or in concert with 3A[93] induces structures similar to those in infected cells. Interestingly, expression of only 2C induces more dramatic membrane remodeling including tubular membrane swirls inside a highly dilated ER[92], implying a regulatory role of 2B in 2C membrane remodeling. Taken together these data suggest that membrane-associated proteins of plus-strand RNA viruses have intrinsic membrane remodeling properties. However, in most cases more than one viral factor, eventually in concert with recruited cellular proteins are involved in generation of viral replication factories. Further studies using e.g., recombinant viral proteins and model membranes are required to elucidate the intrinsic membrane remodeling properties of these viral proteins.
POSSIBLE ROLE OF AUTOPHAGY IN THE FORMATION OF DMV LIKE REPLICATION FACTORIES
Autophagy is an evolutionary conserved catabolic mechanism for degradation of long-lived organelles and cytoplasmic material and is crucial for cell homeostasis (reviewed in[94]). Virus-host co-evolution has shaped multiple mechanisms involving autophagy that either promote or restrict viral replication (reviewed in[95]). Due to morphological similarities between DMV-like replication factories and the also double-membrane nature of autophagosomes, it has been suggested that autophagy plays a role in biogenesis of viral replication compartments of this type as induced by enteroviruses[44,96,97], coronaviruses[98,99] and HCV[100,101]. Indeed, lipidation of microtubule - associated protein 1 light chain 3 (LC3), a key event in the induction of autophagy, was observed after enterovirus infection[96,97,102] or over-expression of 2BC[102]. Interestingly, knock-down of central autophagy components or pharmacological inhibition of the pathway decreased viral replication only slightly, whereas generation of progeny virus was clearly reduced arguing that autophagy contributes primarily to virus assembly and release[103]. In case of PV, electron tomography studies of infected cells revealed primarily single membrane structures during the log phase of viral replication, which subsequently developed into DMVs by collapsing and/or enwrapping events[26]. These data argue against a central role of autophagy per se in PV replication, although some factors of this pathway might contribute to heterogeneity of vesicular structures at later stages of infection. Similar observations have been made for the Coronaviridae. MHV infection induces lipidation of LC3 that co-localizes with nsp2/3 in infected cells[99]. Moreover, the non-lipidated form of LC3 is prominently found on isolated membrane fractions from MHV-infected cells, together with components (e.g., EDEM1) of the ER-associated degradation pathway[99]. However, functional studies based on knockouts of central autophagy factors led to contradictory results. In one study, ATG5 knockout decreased virus propagation dramatically[98], whereas others found that MHV replication is independent of ATG5[104] or ATG7[99]. Hence, a central role of the complete autophagy pathway in generation of coronavirus DMVs seems unlikely, although some single factors such as LC3 might add to it. Conflicting observations concerning a role of autophagy in virus propagation have also been made in case of HCV. One study claims that autophagy is implicated in translation of incoming viral RNA, but is dispensable for RNA replication[105]. Other studies suggest that autophagosomes are sites of active replication[101] or promote assembly and release of progeny virus[106]. In addition, a recent study showed an autophagy-mediated down-regulation of innate immune response[107]. In this case, knock-down of autophagy components reduces HCV RNA replication due to a stronger innate immune response. The reasons for these discrepant results are not known, but might be due, at least in part, to the use of different Huh7 cell clones, which are known to differ in their capacity to mount innate antiviral defenses. Although LC3 lipidation is observed upon HCV infection[105] and the protein is associated with HCV membrane fractions[100], EM-based studies of membrane remodeling events argues for a role of autophagy in formation of MMVs eventually as part of a cellular stress response induced by massive membrane alterations[28]. Taken together, the role of autophagy in biogenesis of DMV like replication factories remains rather elusive. Single components of the conventional cellular autophagy are possibly involved, whereas a direct contribution of the complete pathway in generation of DMV replication compartments is rather unlikely. Finally, autophagy might be an epiphenomenom, being activated as a cellular stress response to tremendous amounts of virus induced cytoplasmic membrane alterations, engaged in cell homeostasis and survival during viral infection.
PLUS-STRAND RNA VIRUS SUBVERSION OF CELLULAR MEMBRANE BIOLOGY
Since plus-strand RNA viruses induce massive remodeling of cytoplasmic membranes, but most often have very limited genetic coding capacity it is not surprising that these viruses utilize membrane-active host cell factors and exploit cellular pathways involved in membrane homeostasis.
Viral utilization of co-opted membrane-active proteins of the host cell
Enteroviruses and the plant virus BMV co-opt cellular reticulon proteins, which are required for membrane remodeling and viral RNA replication[108,109]. The evolutionary conserved reticulon protein family is characterized by a common reticulon homology domain (RHD) involved in shaping the ER by inducing and stabilizing highly curved ER tubules[110,111]. Morphogenic properties can be attributed to elongated hydrophobic, partially membrane-spanning hairpin structures within the RHD[112], which in concert with its oligomerization properties[113] increases the surface on the cytoplasmic membrane leaflet, thereby inducing positive curvature (Figure 3E and F). Reticulon 3 has been shown to directly interact with the enterovirus 2C protein[108] and is thus likely engaged in induction and/or stabilization of positive membrane curvature of enterovirus replication factories. BMV replicase protein 1a directly binds to and recruits reticulon proteins to spherule-like membrane invaginations and might stabilize positive membrane curvature in neck-like openings to the cytoplasm or facilitate expansion of the spherule volume by partially neutralizing overall negative membrane curvature[109]. It remains to be determined whether reticulon proteins are co-opted also by other plus-strand RNA viruses, especially those deriving their replication compartments from the ER. Another example of cellular membrane-shaping proteins are amphiphysins that are involved in formation of alphavirus replication factories[114]. These Bin-Amphiphysin-Rvs (BAR) domain containing proteins play pivotal roles in endocytosis and intracellular membrane trafficking (reviewed in[115]). Structural analyses revealed dimerization of the BAR domain mediated by helical coiled-coil interactions[116], giving rise to a concave banana-shaped structure. This complex has a positively charged inner surface, which interacts with negatively charged membrane phospholipids (reviewed in[117]). Thus, BAR domain-containing proteins sense and stabilize membrane curvature by scaffolding mechanisms, to which in case of N-terminal amphipathic helix containing BAR proteins membrane insertion of N-terminal amphipathic helices contributes (Figure 3D and G). Alphavirus nsp3 binds to SH3 domains in amphiphysin via a proline-rich sequence and mediates the recruitment to viral replication factories. Knock-down experiments proved an important role of amphiphysins in alphavirus replication[114]. Although their contribution to membrane remodeling during alphavirus infection remains to be discovered, one could envisage a similar mechanism as proposed for BMV and reticulon proteins. Proline-serine-threonine phosphatase interacting protein 2 (PSTPIP2) also belongs to the BAR protein superfamily (Figure 3D) and was recently shown to be involved in membrane alterations induced by HCV[118]. PSTPIP2 binds to NS4B and NS5A and thereby is recruited to the membranous replication compartment[118]. In fact, knockdown experiments showed the crucial role of PSTPIP2 in HCV membrane remodeling and RNA replication[118]. Interestingly, upon over-expression PSTPIP2 induces cytoplasmic tubular membranes[118], highlighting its ability to induce positive membrane curvature. Hence, PSTPIP2 is probably engaged in inducing and/or stabilizing positive membrane curvature of HCV replication factories. Another mechanism by which viruses can rearrange intracellular membranes has become evident for enteroviruses. PV appears to hijack components of the cellular secretory pathway, explaining why PV replication is sensitive to brefeldin A (BFA) treatment[119]. Viral proteins 3A and 3CD recruit the ADP ribosylation factor (Arf)-GEFs (GTP exchange factors) Golgi-specific BFA-resistance guanine nucleotide-exchange factor 1 (GBF1) and BFA-inhibited guanine nucleotide-exchange protein 1/2 (BIG1/2) to replication sites, leading to elevated levels of activated Arf-GTP in infected cells[120]. Arf proteins are central regulators of membrane dynamics and vesicle budding in the secretory pathway. Upon activation by Arf-GEFs, the GTP-bound form exerts its function on target membranes by recruitment of effector proteins such as lipid modifying enzymes and coat complexes, thus facilitating vesicle budding (reviewed in[121,122]). GBF1-mediated Arf activation is crucial for PV replication, although typical membrane alterations can be observed in BFA-treated cells that over-express viral proteins[123]. Nevertheless, PV replication factories of BFA-treated cells were shown to be inactive, implying a functional difference of remodeled membranes independent from morphological abnormalities[123]. Hence, PV subversion of cellular Arf-GEFs is important for maintenance of functional replication factories, rather than primary membrane remodeling. In summary, plus-strand RNA viruses recruit and utilize membrane-active host cell proteins in order to generate and/or maintain replication factories. Donor membrane usage defines engaged cellular factors, as enteroviruses employ Arf-GEFs from the Golgi, alphaviruses subvert amphiphysins localized at the plasma membrane and enteroviruses and BMV utilize ER-resident reticulon proteins (Figure 4).
Figure 4.
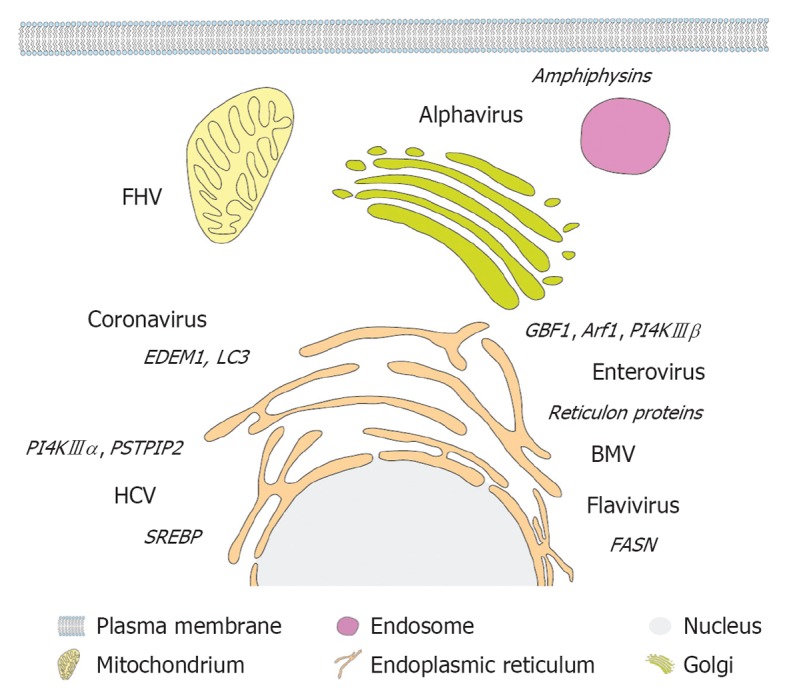
Primary membrane source and host cell factors subverted by different plus-strand RNA viruses. A eukaryotic cell and (endo)membrane organelles are depicted schematically as indicated on the bottom. Viruses are displayed next to their primary membrane source organelle and recruited host cell factors involved in lipogenesis and membrane remodeling are denoted in italics adjacent to each virus name. FHV: Flock-house virus; PSTPIP2: Proline-serine-threonine phosphatase interacting protein 2; HCV: Hepatitis C virus; SREBP: Sterol regulatory element-binding protein; PI4K: Phosphatidylinositol-4-kinase; GBF1: Golgi-specific BFA-resistance guanine nucleotide-exchange factor 1; BMV: Brome mosaic virus; FASN: Fatty acid synthase.
Viral exploitation of pathways regulating homeostasis of cellular membranes
Apart from remodeling existing intracellular membranes, virus infection often induces de novo lipid and membrane biosynthesis in order to increase membrane surface area, which is required for the formation of viral replication factories. Indeed, alteration of cellular lipid homeostasis and virus-induced lipogenesis has been reported for a broad range of plus-strand RNA viruses, including FHV[124], BMV[125], RUBV[126], DENV[127,128], WNV[129], enteroviruses[130] and HCV[131-133]. FHV mainly stimulates glycerophospholipid metabolism[124] and inhibition of phospholipid synthesis leads to destabilization of protein A and decreases viral replication[134]. In case of DENV, high-throughput analysis revealed multiple changes in the cellular lipidome of infected cells, including specific up or down regulation of main structural lipids species, highlighting the link to virus-induced membrane remodeling[128]. Additionally, DENV NS3 recruits fatty acid synthase (FASN), which catalyzes the rate limiting step in lipid biosynthesis at the sites of viral replication[127]. Thus, DENV appears to subvert FASN for de novo lipid synthesis in order to generate new membranes for the formation of viral replication factories. This feature is also observed for the closely related flavivirus WNV[129], stressing the impact of this particular virus host interaction to generate flavivirus replication factories.
HCV induces lipogenesis via the sterol regulatory element-binding protein (SREBP) pathway[131]. SREBPs are major transcription factors for expression of genes required for lipid biosynthesis. SREBPs reside as inactive membrane-bound precursors in the ER, which upon stimulation traffic to the Golgi. There they are proteolytically activated by site 1 protease (S1P) and S2P and subsequently stimulate gene transcription (reviewed in[135]). Proteolytic cleavage of SREBP and transactivating phosphorylation has been observed in HCV-infected cells or upon over-expression of NS4B, leading to elevated levels of transcripts involved in lipogenesis such as FASN[131,136]. However, by using S1P-specific inhibitors SREBP-mediated lipogenesis was found to be dispensable for HCV replication but required for assembly and release of progeny virus[137]. In addition, the HCV replicase complex was shown to reside in detergent-resistant membranes[138]. These membrane micro domains, designated lipid rafts, are enriched for cholesterol, sphingolipids and certain proteins and they form nanoscale-ordered protein-lipid assemblies (reviewed in[139]). Sphingolipid synthesis is stimulated upon and required for HCV replication[140] and it was shown that the NS5B RdRP is activated by sphingomyelin in a genotype-specific manner[141]. A shared feature of plus-strand RNA viruses inducing DMV-like replication factories is their dependence on members of the phosphatidylinositol-4-kinase (PI4K) family and their product, phosphatidylinositol-4-phosphate (PI4P). Both enteroviruses and HCV rely on PI4P for functional replication factories, which is generated by recruitment of PI4KIIIβ in case of enteroviruses[130] and PI4KIIIα in case of HCV[142]. In non-infected cells PI4P localizes to the Golgi and the inner leaflet of the plasma membrane, where it fulfills important functions by providing “signatures” to distinct membrane compartments and by recruiting multiple factors involved in vesicle budding and lipid biosynthesis[143,144]. Subversion of PI4KIIIβ by enteroviruses is executed via 3A-GBF1 interaction, activating Arf that in turn recruits PI4KIIIβ to viral replication factories[130]. Locally elevated PI4P levels allow specific binding of the viral 3D RdRp to the membrane favoring viral replication. Knock-down as well as pharmacological inhibition of PI4KIIIβ dramatically decreases enteroviral replication[130].
In case of HCV, the replicase proteins NS5A and NS5B directly interact with PI4KIIIα, which is thereby recruited to HCV replication sites[142]. Importantly, in HCV-infected cells PI4P that is usually enriched at the Golgi and the plasma membrane, is prominently enriched at the ER-derived sites of HCV replication[142,145]. Numerous siRNA screens identified PI4KIIIα as a major host dependency factor for HCV replication[142,146-149], and pharmacological inhibition of PI4KIIIα activity efficiently blocks viral replication[145]. In the absence of PI4KIIIα, expression of HCV proteins NS3-5B still induces DMVs, but these vesicles are smaller in diameter, very homogeneous and tend to cluster[142]. This morphological change of the overall structure of the membranous web correlates with impaired replication although DMV morphology per se is only moderately affected. This phenomenon is reminiscent to what has been shown for BFA-treated cells over-expressing PV proteins[123]. These findings argue for a role of PI4P in recruiting viral factors such as PV 3D RdRp or so far unidentified cellular proteins, rather than acting as a structural lipid building up membranous replication factories. Importantly, the closely HCV-related flaviviruses DENV and WNV, which form replication factories of the InV/sperule type, do not depend on PI4KIIIα/β or PI4P[129,142], highlighting a functional relationship between PI4P and DMV-like replication factories. Therefore it would be very interesting to investigate the dependence of EAV and coronavirus replication on PI4P. We note that for the latter PI4KIIIβ was shown to be required for virus entry[150].
Taken together, the strong dependence of positive strand RNA viruses on cellular membrane-active proteins and on pathways implicated in cellular membrane homeostasis, renders those host cells factors very attractive targets for future antiviral drug development.
CONCLUSION
Although important discoveries on the 3D architecture of plus-strand RNA virus replication factories have been made, current knowledge is largely descriptive and important information about mechanisms is missing. For instance, the exact topology of RNA replication sites for DMV-type replication factories is elusive. Identification of these sites will require novel experimental techniques such as metabolic labeling of nascent viral RNA and its visualization by using microscopy methods with high resolution and specificity. Likewise, membrane remodeling events responsible for the biogenesis of replication factories are mostly unknown. They are probably mediated by a complex interplay of viral and cellular factors, but precise contributions of individual factors and their temporal and spatial coordination remain to be discovered. Studying the impact of single proteins or combinations thereof on model membranes in vitro or on membranes in cellulo by using correlative light-EM based methods as described recently for studies of membrane remodeling events during endocytosis[151] are possible ways to address this topic. Furthermore, determining the proteome and lipidome of purified viral replication factories will shed light on viral and host cell factors involved in biogenesis and activity of these membranous compartments. This approach has been used with great success for small intracellular vesicles such as COPI vesicles or neuronal transport vesicles[152,153] providing insight into individual membranous structures with unprecedented detail. Another emerging field is the specific in-membrane interaction of proteins with certain lipids, as recently shown for COPI machinery protein p24 and the sphingolipid SM 18 being implicated in regulation of COPI vesicle budding[154]. The tight membrane association of proteins of plus-strand RNA viruses suggests that such specific protein-lipid interactions also occur for this large virus group. Finally, time-resolved (imaging) techniques might shed light onto coupling of viral RNA translation and replication as recently shown for HCV[155] and furthermore onto transport processes of viral and cellular components inside and outside replication factories. These studies will be instrumental to integrate their functional role into the complete viral replication cycle.
ACKNOWLEDGMENTS
We want to express our gratitude to Ahlquist P, Belov G, Risco C, Romero-Brey I, Snijder E and Welsch S for providing EM reconstruction images and apologize to all colleagues whose work could not be cited due to space limitations.
Footnotes
Supported by The DFG, SFB638, TP A5 and SFB/TRR83, TP 13
P- Reviewers Chi-Ho C, Shih WL S- Editor Gou SX L- Editor A E- Editor Zheng XM
References
- 1.Laliberté JF, Sanfaçon H. Cellular remodeling during plant virus infection. Annu Rev Phytopathol. 2010;48:69–91. doi: 10.1146/annurev-phyto-073009-114239. [DOI] [PubMed] [Google Scholar]
- 2.Novoa RR, Calderita G, Arranz R, Fontana J, Granzow H, Risco C. Virus factories: associations of cell organelles for viral replication and morphogenesis. Biol Cell. 2005;97:147–172. doi: 10.1042/BC20040058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Netherton C, Moffat K, Brooks E, Wileman T. A guide to viral inclusions, membrane rearrangements, factories, and viroplasm produced during virus replication. Adv Virus Res. 2007;70:101–182. doi: 10.1016/S0065-3527(07)70004-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Monine MI, Haugh JM. Reactions on cell membranes: comparison of continuum theory and Brownian dynamics simulations. J Chem Phys. 2005;123:074908. doi: 10.1063/1.2000236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moradpour D, Penin F, Rice CM. Replication of hepatitis C virus. Nat Rev Microbiol. 2007;5:453–463. doi: 10.1038/nrmicro1645. [DOI] [PubMed] [Google Scholar]
- 6.Ando T, Imamura H, Suzuki R, Aizaki H, Watanabe T, Wakita T, Suzuki T. Visualization and measurement of ATP levels in living cells replicating hepatitis C virus genome RNA. PLoS Pathog. 2012;8:e1002561. doi: 10.1371/journal.ppat.1002561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Froshauer S, Kartenbeck J, Helenius A. Alphavirus RNA replicase is located on the cytoplasmic surface of endosomes and lysosomes. J Cell Biol. 1988;107:2075–2086. doi: 10.1083/jcb.107.6.2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fontana J, López-Iglesias C, Tzeng WP, Frey TK, Fernández JJ, Risco C. Three-dimensional structure of Rubella virus factories. Virology. 2010;405:579–591. doi: 10.1016/j.virol.2010.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gillespie LK, Hoenen A, Morgan G, Mackenzie JM. The endoplasmic reticulum provides the membrane platform for biogenesis of the flavivirus replication complex. J Virol. 2010;84:10438–10447. doi: 10.1128/JVI.00986-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Welsch S, Miller S, Romero-Brey I, Merz A, Bleck CK, Walther P, Fuller SD, Antony C, Krijnse-Locker J, Bartenschlager R. Composition and three-dimensional architecture of the dengue virus replication and assembly sites. Cell Host Microbe. 2009;5:365–375. doi: 10.1016/j.chom.2009.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Knoops K, Kikkert M, Worm SH, Zevenhoven-Dobbe JC, van der Meer Y, Koster AJ, Mommaas AM, Snijder EJ. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol. 2008;6:e226. doi: 10.1371/journal.pbio.0060226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barba G, Harper F, Harada T, Kohara M, Goulinet S, Matsuura Y, Eder G, Schaff Z, Chapman MJ, Miyamura T, et al. Hepatitis C virus core protein shows a cytoplasmic localization and associates to cellular lipid storage droplets. Proc Natl Acad Sci USA. 1997;94:1200–1205. doi: 10.1073/pnas.94.4.1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miyanari Y, Atsuzawa K, Usuda N, Watashi K, Hishiki T, Zayas M, Bartenschlager R, Wakita T, Hijikata M, Shimotohno K. The lipid droplet is an important organelle for hepatitis C virus production. Nat Cell Biol. 2007;9:1089–1097. doi: 10.1038/ncb1631. [DOI] [PubMed] [Google Scholar]
- 14.Samsa MM, Mondotte JA, Iglesias NG, Assunção-Miranda I, Barbosa-Lima G, Da Poian AT, Bozza PT, Gamarnik AV. Dengue virus capsid protein usurps lipid droplets for viral particle formation. PLoS Pathog. 2009;5:e1000632. doi: 10.1371/journal.ppat.1000632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tadano M, Makino Y, Fukunaga T, Okuno Y, Fukai K. Detection of dengue 4 virus core protein in the nucleus. I. A monoclonal antibody to dengue 4 virus reacts with the antigen in the nucleus and cytoplasm. J Gen Virol. 1989;70(Pt 6):1409–1415. doi: 10.1099/0022-1317-70-6-1409. [DOI] [PubMed] [Google Scholar]
- 16.Bulich R, Aaskov JG. Nuclear localization of dengue 2 virus core protein detected with monoclonal antibodies. J Gen Virol. 1992;73(Pt 11):2999–3003. doi: 10.1099/0022-1317-73-11-2999. [DOI] [PubMed] [Google Scholar]
- 17.Mori Y, Okabayashi T, Yamashita T, Zhao Z, Wakita T, Yasui K, Hasebe F, Tadano M, Konishi E, Moriishi K, et al. Nuclear localization of Japanese encephalitis virus core protein enhances viral replication. J Virol. 2005;79:3448–3458. doi: 10.1128/JVI.79.6.3448-3458.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oh W, Yang MR, Lee EW, Park KM, Pyo S, Yang JS, Lee HW, Song J. Jab1 mediates cytoplasmic localization and degradation of West Nile virus capsid protein. J Biol Chem. 2006;281:30166–30174. doi: 10.1074/jbc.M602651200. [DOI] [PubMed] [Google Scholar]
- 19.Kato H, Takahasi K, Fujita T. RIG-I-like receptors: cytoplasmic sensors for non-self RNA. Immunol Rev. 2011;243:91–98. doi: 10.1111/j.1600-065X.2011.01052.x. [DOI] [PubMed] [Google Scholar]
- 20.Binder M, Eberle F, Seitz S, Mücke N, Hüber CM, Kiani N, Kaderali L, Lohmann V, Dalpke A, Bartenschlager R. Molecular mechanism of signal perception and integration by the innate immune sensor retinoic acid-inducible gene-I (RIG-I) J Biol Chem. 2011;286:27278–27287. doi: 10.1074/jbc.M111.256974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Belgnaoui SM, Paz S, Hiscott J. Orchestrating the interferon antiviral response through the mitochondrial antiviral signaling (MAVS) adapter. Curr Opin Immunol. 2011;23:564–572. doi: 10.1016/j.coi.2011.08.001. [DOI] [PubMed] [Google Scholar]
- 22.Koster AJ, Grimm R, Typke D, Hegerl R, Stoschek A, Walz J, Baumeister W. Perspectives of molecular and cellular electron tomography. J Struct Biol. 1997;120:276–308. doi: 10.1006/jsbi.1997.3933. [DOI] [PubMed] [Google Scholar]
- 23.Frey TG, Perkins GA, Ellisman MH. Electron tomography of membrane-bound cellular organelles. Annu Rev Biophys Biomol Struct. 2006;35:199–224. doi: 10.1146/annurev.biophys.35.040405.102039. [DOI] [PubMed] [Google Scholar]
- 24.Kopek BG, Perkins G, Miller DJ, Ellisman MH, Ahlquist P. Three-dimensional analysis of a viral RNA replication complex reveals a virus-induced mini-organelle. PLoS Biol. 2007;5:e220. doi: 10.1371/journal.pbio.0050220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Limpens RW, van der Schaar HM, Kumar D, Koster AJ, Snijder EJ, van Kuppeveld FJ, Bárcena M. The transformation of enterovirus replication structures: a three-dimensional study of single- and double-membrane compartments. MBio. 2011;2:e00166–11. doi: 10.1128/mBio.00166-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Belov GA, Nair V, Hansen BT, Hoyt FH, Fischer ER, Ehrenfeld E. Complex dynamic development of poliovirus membranous replication complexes. J Virol. 2012;86:302–312. doi: 10.1128/JVI.05937-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Knoops K, Bárcena M, Limpens RW, Koster AJ, Mommaas AM, Snijder EJ. Ultrastructural characterization of arterivirus replication structures: reshaping the endoplasmic reticulum to accommodate viral RNA synthesis. J Virol. 2012;86:2474–2487. doi: 10.1128/JVI.06677-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Romero-Brey I, Merz A, Chiramel A, Lee JY, Chlanda P, Haselman U, Santarella-Mellwig R, Habermann A, Hoppe S, Kallis S, et al. Three-dimensional architecture and biogenesis of membrane structures associated with hepatitis C virus replication. PLoS Pathog. 2012;8:e1003056. doi: 10.1371/journal.ppat.1003056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grimley PM, Berezesky IK, Friedman RM. Cytoplasmic structures associated with an arbovirus infection: loci of viral ribonucleic acid synthesis. J Virol. 1968;2:1326–1338. doi: 10.1128/jvi.2.11.1326-1338.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kujala P, Ikäheimonen A, Ehsani N, Vihinen H, Auvinen P, Kääriäinen L. Biogenesis of the Semliki Forest virus RNA replication complex. J Virol. 2001;75:3873–3884. doi: 10.1128/JVI.75.8.3873-3884.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Frolova EI, Gorchakov R, Pereboeva L, Atasheva S, Frolov I. Functional Sindbis virus replicative complexes are formed at the plasma membrane. J Virol. 2010;84:11679–11695. doi: 10.1128/JVI.01441-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Acheson NH, Tamm I. Replication of Semliki Forest virus: an electron microscopic study. Virology. 1967;32:128–143. doi: 10.1016/0042-6822(67)90261-9. [DOI] [PubMed] [Google Scholar]
- 33.Friedman RM, Levin JG, Grimley PM, Berezesky IK. Membrane-associated replication complex in arbovirus infection. J Virol. 1972;10:504–515. doi: 10.1128/jvi.10.3.504-515.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grimley PM, Levin JG, Berezesky IK, Friedman RM. Specific membranous structures associated with the replication of group A arboviruses. J Virol. 1972;10:492–503. doi: 10.1128/jvi.10.3.492-503.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miller DJ, Schwartz MD, Ahlquist P. Flock house virus RNA replicates on outer mitochondrial membranes in Drosophila cells. J Virol. 2001;75:11664–11676. doi: 10.1128/JVI.75.23.11664-11676.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kujala P, Ahola T, Ehsani N, Auvinen P, Vihinen H, Kääriäinen L. Intracellular distribution of rubella virus nonstructural protein P150. J Virol. 1999;73:7805–7811. doi: 10.1128/jvi.73.9.7805-7811.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee JY, Marshall JA, Bowden DS. Characterization of rubella virus replication complexes using antibodies to double-stranded RNA. Virology. 1994;200:307–312. doi: 10.1006/viro.1994.1192. [DOI] [PubMed] [Google Scholar]
- 38.Magliano D, Marshall JA, Bowden DS, Vardaxis N, Meanger J, Lee JY. Rubella virus replication complexes are virus-modified lysosomes. Virology. 1998;240:57–63. doi: 10.1006/viro.1997.8906. [DOI] [PubMed] [Google Scholar]
- 39.Fontana J, Tzeng WP, Calderita G, Fraile-Ramos A, Frey TK, Risco C. Novel replication complex architecture in rubella replicon-transfected cells. Cell Microbiol. 2007;9:875–890. doi: 10.1111/j.1462-5822.2006.00837.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Risco C, Carrascosa JL, Frey TK. Structural maturation of rubella virus in the Golgi complex. Virology. 2003;312:261–269. doi: 10.1016/S0042-6822(03)00384-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Westaway EG, Mackenzie JM, Kenney MT, Jones MK, Khromykh AA. Ultrastructure of Kunjin virus-infected cells: colocalization of NS1 and NS3 with double-stranded RNA, and of NS2B with NS3, in virus-induced membrane structures. J Virol. 1997;71:6650–6661. doi: 10.1128/jvi.71.9.6650-6661.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kallman F, Williams RC, Dulbecco R, Vogt M. Fine structure of changes produced in cultured cells sampled at specified intervals during a single growth cycle of polio virus. J Biophys Biochem Cytol. 1958;4:301–308. doi: 10.1083/jcb.4.3.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Egger D, Bienz K. Intracellular location and translocation of silent and active poliovirus replication complexes. J Gen Virol. 2005;86:707–718. doi: 10.1099/vir.0.80442-0. [DOI] [PubMed] [Google Scholar]
- 44.Schlegel A, Giddings TH, Ladinsky MS, Kirkegaard K. Cellular origin and ultrastructure of membranes induced during poliovirus infection. J Virol. 1996;70:6576–6588. doi: 10.1128/jvi.70.10.6576-6588.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Troxler M, Egger D, Pfister T, Bienz K. Intracellular localization of poliovirus RNA by in situ hybridization at the ultrastructural level using single-stranded riboprobes. Virology. 1992;191:687–697. doi: 10.1016/0042-6822(92)90244-j. [DOI] [PubMed] [Google Scholar]
- 46.Hagemeijer MC, Vonk AM, Monastyrska I, Rottier PJ, de Haan CA. Visualizing coronavirus RNA synthesis in time by using click chemistry. J Virol. 2012;86:5808–5816. doi: 10.1128/JVI.07207-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schwartz M, Chen J, Janda M, Sullivan M, den Boon J, Ahlquist P. A positive-strand RNA virus replication complex parallels form and function of retrovirus capsids. Mol Cell. 2002;9:505–514. doi: 10.1016/s1097-2765(02)00474-4. [DOI] [PubMed] [Google Scholar]
- 48.Bienz K, Egger D, Pasamontes L. Association of polioviral proteins of the P2 genomic region with the viral replication complex and virus-induced membrane synthesis as visualized by electron microscopic immunocytochemistry and autoradiography. Virology. 1987;160:220–226. doi: 10.1016/0042-6822(87)90063-8. [DOI] [PubMed] [Google Scholar]
- 49.Pedersen KW, van der Meer Y, Roos N, Snijder EJ. Open reading frame 1a-encoded subunits of the arterivirus replicase induce endoplasmic reticulum-derived double-membrane vesicles which carry the viral replication complex. J Virol. 1999;73:2016–2026. doi: 10.1128/jvi.73.3.2016-2026.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gosert R, Kanjanahaluethai A, Egger D, Bienz K, Baker SC. RNA replication of mouse hepatitis virus takes place at double-membrane vesicles. J Virol. 2002;76:3697–3708. doi: 10.1128/JVI.76.8.3697-3708.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Johnson AE, van Waes MA. The translocon: a dynamic gateway at the ER membrane. Annu Rev Cell Dev Biol. 1999;15:799–842. doi: 10.1146/annurev.cellbio.15.1.799. [DOI] [PubMed] [Google Scholar]
- 52.Endo T, Yamamoto H, Esaki M. Functional cooperation and separation of translocators in protein import into mitochondria, the double-membrane bounded organelles. J Cell Sci. 2003;116:3259–3267. doi: 10.1242/jcs.00667. [DOI] [PubMed] [Google Scholar]
- 53.McMahon HT, Gallop JL. Membrane curvature and mechanisms of dynamic cell membrane remodelling. Nature. 2005;438:590–596. doi: 10.1038/nature04396. [DOI] [PubMed] [Google Scholar]
- 54.Zimmerberg J, Kozlov MM. How proteins produce cellular membrane curvature. Nat Rev Mol Cell Biol. 2006;7:9–19. doi: 10.1038/nrm1784. [DOI] [PubMed] [Google Scholar]
- 55.Graham TR, Kozlov MM. Interplay of proteins and lipids in generating membrane curvature. Curr Opin Cell Biol. 2010;22:430–436. doi: 10.1016/j.ceb.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ahola T, Lampio A, Auvinen P, Kääriäinen L. Semliki Forest virus mRNA capping enzyme requires association with anionic membrane phospholipids for activity. EMBO J. 1999;18:3164–3172. doi: 10.1093/emboj/18.11.3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Laakkonen P, Ahola T, Kääriäinen L. The effects of palmitoylation on membrane association of Semliki forest virus RNA capping enzyme. J Biol Chem. 1996;271:28567–28571. doi: 10.1074/jbc.271.45.28567. [DOI] [PubMed] [Google Scholar]
- 58.Spuul P, Salonen A, Merits A, Jokitalo E, Kääriäinen L, Ahola T. Role of the amphipathic peptide of Semliki forest virus replicase protein nsP1 in membrane association and virus replication. J Virol. 2007;81:872–883. doi: 10.1128/JVI.01785-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zusinaite E, Tints K, Kiiver K, Spuul P, Karo-Astover L, Merits A, Sarand I. Mutations at the palmitoylation site of non-structural protein nsP1 of Semliki Forest virus attenuate virus replication and cause accumulation of compensatory mutations. J Gen Virol. 2007;88:1977–1985. doi: 10.1099/vir.0.82865-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ahola T, Kujala P, Tuittila M, Blom T, Laakkonen P, Hinkkanen A, Auvinen P. Effects of palmitoylation of replicase protein nsP1 on alphavirus infection. J Virol. 2000;74:6725–6733. doi: 10.1128/jvi.74.15.6725-6733.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Salonen A, Vasiljeva L, Merits A, Magden J, Jokitalo E, Kääriäinen L. Properly folded nonstructural polyprotein directs the semliki forest virus replication complex to the endosomal compartment. J Virol. 2003;77:1691–1702. doi: 10.1128/JVI.77.3.1691-1702.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stapleford KA, Rapaport D, Miller DJ. Mitochondrion-enriched anionic phospholipids facilitate flock house virus RNA polymerase membrane association. J Virol. 2009;83:4498–4507. doi: 10.1128/JVI.00040-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Miller DJ, Ahlquist P. Flock house virus RNA polymerase is a transmembrane protein with amino-terminal sequences sufficient for mitochondrial localization and membrane insertion. J Virol. 2002;76:9856–9867. doi: 10.1128/JVI.76.19.9856-9867.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dye BT, Miller DJ, Ahlquist P. In vivo self-interaction of nodavirus RNA replicase protein a revealed by fluorescence resonance energy transfer. J Virol. 2005;79:8909–8919. doi: 10.1128/JVI.79.14.8909-8919.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kopek BG, Settles EW, Friesen PD, Ahlquist P. Nodavirus-induced membrane rearrangement in replication complex assembly requires replicase protein a, RNA templates, and polymerase activity. J Virol. 2010;84:12492–12503. doi: 10.1128/JVI.01495-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Miller S, Kastner S, Krijnse-Locker J, Bühler S, Bartenschlager R. The non-structural protein 4A of dengue virus is an integral membrane protein inducing membrane alterations in a 2K-regulated manner. J Biol Chem. 2007;282:8873–8882. doi: 10.1074/jbc.M609919200. [DOI] [PubMed] [Google Scholar]
- 67.Roosendaal J, Westaway EG, Khromykh A, Mackenzie JM. Regulated cleavages at the West Nile virus NS4A-2K-NS4B junctions play a major role in rearranging cytoplasmic membranes and Golgi trafficking of the NS4A protein. J Virol. 2006;80:4623–4632. doi: 10.1128/JVI.80.9.4623-4632.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Egger D, Wölk B, Gosert R, Bianchi L, Blum HE, Moradpour D, Bienz K. Expression of hepatitis C virus proteins induces distinct membrane alterations including a candidate viral replication complex. J Virol. 2002;76:5974–5984. doi: 10.1128/JVI.76.12.5974-5984.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lundin M, Monné M, Widell A, Von Heijne G, Persson MA. Topology of the membrane-associated hepatitis C virus protein NS4B. J Virol. 2003;77:5428–5438. doi: 10.1128/JVI.77.9.5428-5438.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yu GY, Lee KJ, Gao L, Lai MM. Palmitoylation and polymerization of hepatitis C virus NS4B protein. J Virol. 2006;80:6013–6023. doi: 10.1128/JVI.00053-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Elazar M, Liu P, Rice CM, Glenn JS. An N-terminal amphipathic helix in hepatitis C virus (HCV) NS4B mediates membrane association, correct localization of replication complex proteins, and HCV RNA replication. J Virol. 2004;78:11393–11400. doi: 10.1128/JVI.78.20.11393-11400.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gouttenoire J, Castet V, Montserret R, Arora N, Raussens V, Ruysschaert JM, Diesis E, Blum HE, Penin F, Moradpour D. Identification of a novel determinant for membrane association in hepatitis C virus nonstructural protein 4B. J Virol. 2009;83:6257–6268. doi: 10.1128/JVI.02663-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gouttenoire J, Montserret R, Kennel A, Penin F, Moradpour D. An amphipathic alpha-helix at the C terminus of hepatitis C virus nonstructural protein 4B mediates membrane association. J Virol. 2009;83:11378–11384. doi: 10.1128/JVI.01122-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Guillén J, González-Alvarez A, Villalaín J. A membranotropic region in the C-terminal domain of hepatitis C virus protein NS4B interaction with membranes. Biochim Biophys Acta. 2010;1798:327–337. doi: 10.1016/j.bbamem.2009.07.011. [DOI] [PubMed] [Google Scholar]
- 75.Palomares-Jerez MF, Nemesio H, Villalaín J. Interaction with membranes of the full C-terminal domain of protein NS4B from hepatitis C virus. Biochim Biophys Acta. 2012;1818:2536–2549. doi: 10.1016/j.bbamem.2012.06.012. [DOI] [PubMed] [Google Scholar]
- 76.Gouttenoire J, Roingeard P, Penin F, Moradpour D. Amphipathic alpha-helix AH2 is a major determinant for the oligomerization of hepatitis C virus nonstructural protein 4B. J Virol. 2010;84:12529–12537. doi: 10.1128/JVI.01798-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Paul D, Romero-Brey I, Gouttenoire J, Stoitsova S, Krijnse-Locker J, Moradpour D, Bartenschlager R. NS4B self-interaction through conserved C-terminal elements is required for the establishment of functional hepatitis C virus replication complexes. J Virol. 2011;85:6963–6976. doi: 10.1128/JVI.00502-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Brass V, Bieck E, Montserret R, Wölk B, Hellings JA, Blum HE, Penin F, Moradpour D. An amino-terminal amphipathic alpha-helix mediates membrane association of the hepatitis C virus nonstructural protein 5A. J Biol Chem. 2002;277:8130–8139. doi: 10.1074/jbc.M111289200. [DOI] [PubMed] [Google Scholar]
- 79.Penin F, Brass V, Appel N, Ramboarina S, Montserret R, Ficheux D, Blum HE, Bartenschlager R, Moradpour D. Structure and function of the membrane anchor domain of hepatitis C virus nonstructural protein 5A. J Biol Chem. 2004;279:40835–40843. doi: 10.1074/jbc.M404761200. [DOI] [PubMed] [Google Scholar]
- 80.Tellinghuisen TL, Marcotrigiano J, Rice CM. Structure of the zinc-binding domain of an essential component of the hepatitis C virus replicase. Nature. 2005;435:374–379. doi: 10.1038/nature03580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Love RA, Brodsky O, Hickey MJ, Wells PA, Cronin CN. Crystal structure of a novel dimeric form of NS5A domain I protein from hepatitis C virus. J Virol. 2009;83:4395–4403. doi: 10.1128/JVI.02352-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lim PJ, Chatterji U, Cordek D, Sharma SD, Garcia-Rivera JA, Cameron CE, Lin K, Targett-Adams P, Gallay PA. Correlation between NS5A dimerization and hepatitis C virus replication. J Biol Chem. 2012;287:30861–30873. doi: 10.1074/jbc.M112.376822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kanjanahaluethai A, Chen Z, Jukneliene D, Baker SC. Membrane topology of murine coronavirus replicase nonstructural protein 3. Virology. 2007;361:391–401. doi: 10.1016/j.virol.2006.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Oostra M, te Lintelo EG, Deijs M, Verheije MH, Rottier PJ, de Haan CA. Localization and membrane topology of coronavirus nonstructural protein 4: involvement of the early secretory pathway in replication. J Virol. 2007;81:12323–12336. doi: 10.1128/JVI.01506-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Oostra M, Hagemeijer MC, van Gent M, Bekker CP, te Lintelo EG, Rottier PJ, de Haan CA. Topology and membrane anchoring of the coronavirus replication complex: not all hydrophobic domains of nsp3 and nsp6 are membrane spanning. J Virol. 2008;82:12392–12405. doi: 10.1128/JVI.01219-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gadlage MJ, Sparks JS, Beachboard DC, Cox RG, Doyle JD, Stobart CC, Denison MR. Murine hepatitis virus nonstructural protein 4 regulates virus-induced membrane modifications and replication complex function. J Virol. 2010;84:280–290. doi: 10.1128/JVI.01772-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Snijder EJ, van Tol H, Roos N, Pedersen KW. Non-structural proteins 2 and 3 interact to modify host cell membranes during the formation of the arterivirus replication complex. J Gen Virol. 2001;82:985–994. doi: 10.1099/0022-1317-82-5-985. [DOI] [PubMed] [Google Scholar]
- 88.Posthuma CC, Pedersen KW, Lu Z, Joosten RG, Roos N, Zevenhoven-Dobbe JC, Snijder EJ. Formation of the arterivirus replication/transcription complex: a key role for nonstructural protein 3 in the remodeling of intracellular membranes. J Virol. 2008;82:4480–4491. doi: 10.1128/JVI.02756-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.van Kuppeveld FJ, Galama JM, Zoll J, van den Hurk PJ, Melchers WJ. Coxsackie B3 virus protein 2B contains cationic amphipathic helix that is required for viral RNA replication. J Virol. 1996;70:3876–3886. doi: 10.1128/jvi.70.6.3876-3886.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Echeverri AC, Dasgupta A. Amino terminal regions of poliovirus 2C protein mediate membrane binding. Virology. 1995;208:540–553. doi: 10.1006/viro.1995.1185. [DOI] [PubMed] [Google Scholar]
- 91.Towner JS, Ho TV, Semler BL. Determinants of membrane association for poliovirus protein 3AB. J Biol Chem. 1996;271:26810–26818. doi: 10.1074/jbc.271.43.26810. [DOI] [PubMed] [Google Scholar]
- 92.Cho MW, Teterina N, Egger D, Bienz K, Ehrenfeld E. Membrane rearrangement and vesicle induction by recombinant poliovirus 2C and 2BC in human cells. Virology. 1994;202:129–145. doi: 10.1006/viro.1994.1329. [DOI] [PubMed] [Google Scholar]
- 93.Suhy DA, Giddings TH, Kirkegaard K. Remodeling the endoplasmic reticulum by poliovirus infection and by individual viral proteins: an autophagy-like origin for virus-induced vesicles. J Virol. 2000;74:8953–8965. doi: 10.1128/jvi.74.19.8953-8965.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yang Z, Klionsky DJ. Mammalian autophagy: core molecular machinery and signaling regulation. Curr Opin Cell Biol. 2010;22:124–131. doi: 10.1016/j.ceb.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Chiramel AI, Brady NR, Bartenschlager R. Divergent roles of autophagy in virus infection. Cells. 2013;2:83–104. doi: 10.3390/cells2010083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Huang SC, Chang CL, Wang PS, Tsai Y, Liu HS. Enterovirus 71-induced autophagy detected in vitro and in vivo promotes viral replication. J Med Virol. 2009;81:1241–1252. doi: 10.1002/jmv.21502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kemball CC, Alirezaei M, Flynn CT, Wood MR, Harkins S, Kiosses WB, Whitton JL. Coxsackievirus infection induces autophagy-like vesicles and megaphagosomes in pancreatic acinar cells in vivo. J Virol. 2010;84:12110–12124. doi: 10.1128/JVI.01417-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Prentice E, Jerome WG, Yoshimori T, Mizushima N, Denison MR. Coronavirus replication complex formation utilizes components of cellular autophagy. J Biol Chem. 2004;279:10136–10141. doi: 10.1074/jbc.M306124200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Reggiori F, Monastyrska I, Verheije MH, Calì T, Ulasli M, Bianchi S, Bernasconi R, de Haan CA, Molinari M. Coronaviruses Hijack the LC3-I-positive EDEMosomes, ER-derived vesicles exporting short-lived ERAD regulators, for replication. Cell Host Microbe. 2010;7:500–508. doi: 10.1016/j.chom.2010.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ferraris P, Blanchard E, Roingeard P. Ultrastructural and biochemical analyses of hepatitis C virus-associated host cell membranes. J Gen Virol. 2010;91:2230–2237. doi: 10.1099/vir.0.022186-0. [DOI] [PubMed] [Google Scholar]
- 101.Sir D, Kuo CF, Tian Y, Liu HM, Huang EJ, Jung JU, Machida K, Ou JH. Replication of hepatitis C virus RNA on autophagosomal membranes. J Biol Chem. 2012;287:18036–18043. doi: 10.1074/jbc.M111.320085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Taylor MP, Kirkegaard K. Modification of cellular autophagy protein LC3 by poliovirus. J Virol. 2007;81:12543–12553. doi: 10.1128/JVI.00755-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Jackson WT, Giddings TH, Taylor MP, Mulinyawe S, Rabinovitch M, Kopito RR, Kirkegaard K. Subversion of cellular autophagosomal machinery by RNA viruses. PLoS Biol. 2005;3:e156. doi: 10.1371/journal.pbio.0030156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zhao Z, Thackray LB, Miller BC, Lynn TM, Becker MM, Ward E, Mizushima NN, Denison MR, Virgin HW. Coronavirus replication does not require the autophagy gene ATG5. Autophagy. 2007;3:581–585. doi: 10.4161/auto.4782. [DOI] [PubMed] [Google Scholar]
- 105.Dreux M, Gastaminza P, Wieland SF, Chisari FV. The autophagy machinery is required to initiate hepatitis C virus replication. Proc Natl Acad Sci USA. 2009;106:14046–14051. doi: 10.1073/pnas.0907344106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Tanida I, Fukasawa M, Ueno T, Kominami E, Wakita T, Hanada K. Knockdown of autophagy-related gene decreases the production of infectious hepatitis C virus particles. Autophagy. 2009;5:937–945. doi: 10.4161/auto.5.7.9243. [DOI] [PubMed] [Google Scholar]
- 107.Ke PY, Chen SS. Activation of the unfolded protein response and autophagy after hepatitis C virus infection suppresses innate antiviral immunity in vitro. J Clin Invest. 2011;121:37–56. doi: 10.1172/JCI41474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Tang WF, Yang SY, Wu BW, Jheng JR, Chen YL, Shih CH, Lin KH, Lai HC, Tang P, Horng JT. Reticulon 3 binds the 2C protein of enterovirus 71 and is required for viral replication. J Biol Chem. 2007;282:5888–5898. doi: 10.1074/jbc.M611145200. [DOI] [PubMed] [Google Scholar]
- 109.Diaz A, Wang X, Ahlquist P. Membrane-shaping host reticulon proteins play crucial roles in viral RNA replication compartment formation and function. Proc Natl Acad Sci USA. 2010;107:16291–16296. doi: 10.1073/pnas.1011105107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Voeltz GK, Prinz WA, Shibata Y, Rist JM, Rapoport TA. A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell. 2006;124:573–586. doi: 10.1016/j.cell.2005.11.047. [DOI] [PubMed] [Google Scholar]
- 111.Hu J, Shibata Y, Voss C, Shemesh T, Li Z, Coughlin M, Kozlov MM, Rapoport TA, Prinz WA. Membrane proteins of the endoplasmic reticulum induce high-curvature tubules. Science. 2008;319:1247–1250. doi: 10.1126/science.1153634. [DOI] [PubMed] [Google Scholar]
- 112.Zurek N, Sparks L, Voeltz G. Reticulon short hairpin transmembrane domains are used to shape ER tubules. Traffic. 2011;12:28–41. doi: 10.1111/j.1600-0854.2010.01134.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Shibata Y, Voss C, Rist JM, Hu J, Rapoport TA, Prinz WA, Voeltz GK. The reticulon and DP1/Yop1p proteins form immobile oligomers in the tubular endoplasmic reticulum. J Biol Chem. 2008;283:18892–18904. doi: 10.1074/jbc.M800986200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Neuvonen M, Kazlauskas A, Martikainen M, Hinkkanen A, Ahola T, Saksela K. SH3 domain-mediated recruitment of host cell amphiphysins by alphavirus nsP3 promotes viral RNA replication. PLoS Pathog. 2011;7:e1002383. doi: 10.1371/journal.ppat.1002383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Qualmann B, Koch D, Kessels MM. Let‘s go bananas: revisiting the endocytic BAR code. EMBO J. 2011;30:3501–3515. doi: 10.1038/emboj.2011.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Peter BJ, Kent HM, Mills IG, Vallis Y, Butler PJ, Evans PR, McMahon HT. BAR domains as sensors of membrane curvature: the amphiphysin BAR structure. Science. 2004;303:495–499. doi: 10.1126/science.1092586. [DOI] [PubMed] [Google Scholar]
- 117.Rao Y, Haucke V. Membrane shaping by the Bin/amphiphysin/Rvs (BAR) domain protein superfamily. Cell Mol Life Sci. 2011;68:3983–3993. doi: 10.1007/s00018-011-0768-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Chao TC, Su WC, Huang JY, Chen YC, Jeng KS, Wang HD, Lai MM. Proline-serine-threonine phosphatase-interacting protein 2 (PSTPIP2), a host membrane-deforming protein, is critical for membranous web formation in hepatitis C virus replication. J Virol. 2012;86:1739–1749. doi: 10.1128/JVI.06001-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Maynell LA, Kirkegaard K, Klymkowsky MW. Inhibition of poliovirus RNA synthesis by brefeldin A. J Virol. 1992;66:1985–1994. doi: 10.1128/jvi.66.4.1985-1994.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Belov GA, Altan-Bonnet N, Kovtunovych G, Jackson CL, Lippincott-Schwartz J, Ehrenfeld E. Hijacking components of the cellular secretory pathway for replication of poliovirus RNA. J Virol. 2007;81:558–567. doi: 10.1128/JVI.01820-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kahn RA. Toward a model for Arf GTPases as regulators of traffic at the Golgi. FEBS Lett. 2009;583:3872–3879. doi: 10.1016/j.febslet.2009.10.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Nie Z, Hirsch DS, Randazzo PA. Arf and its many interactors. Curr Opin Cell Biol. 2003;15:396–404. doi: 10.1016/s0955-0674(03)00071-1. [DOI] [PubMed] [Google Scholar]
- 123.Belov GA, Feng Q, Nikovics K, Jackson CL, Ehrenfeld E. A critical role of a cellular membrane traffic protein in poliovirus RNA replication. PLoS Pathog. 2008;4:e1000216. doi: 10.1371/journal.ppat.1000216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Castorena KM, Stapleford KA, Miller DJ. Complementary transcriptomic, lipidomic, and targeted functional genetic analyses in cultured Drosophila cells highlight the role of glycerophospholipid metabolism in Flock House virus RNA replication. BMC Genomics. 2010;11:183. doi: 10.1186/1471-2164-11-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Lee WM, Ahlquist P. Membrane synthesis, specific lipid requirements, and localized lipid composition changes associated with a positive-strand RNA virus RNA replication protein. J Virol. 2003;77:12819–12828. doi: 10.1128/JVI.77.23.12819-12828.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Williams LL, Lew HM, Davidorf FH, Pelok SG, Singley CT, Wolinsky JS. Altered membrane fatty acids of cultured human retinal pigment epithelium persistently infected with rubella virus may affect secondary cellular function. Arch Virol. 1994;134:379–392. doi: 10.1007/BF01310575. [DOI] [PubMed] [Google Scholar]
- 127.Heaton NS, Perera R, Berger KL, Khadka S, Lacount DJ, Kuhn RJ, Randall G. Dengue virus nonstructural protein 3 redistributes fatty acid synthase to sites of viral replication and increases cellular fatty acid synthesis. Proc Natl Acad Sci USA. 2010;107:17345–17350. doi: 10.1073/pnas.1010811107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Perera R, Riley C, Isaac G, Hopf-Jannasch AS, Moore RJ, Weitz KW, Pasa-Tolic L, Metz TO, Adamec J, Kuhn RJ. Dengue virus infection perturbs lipid homeostasis in infected mosquito cells. PLoS Pathog. 2012;8:e1002584. doi: 10.1371/journal.ppat.1002584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Martín-Acebes MA, Blázquez AB, Jiménez de Oya N, Escribano-Romero E, Saiz JC. West Nile virus replication requires fatty acid synthesis but is independent on phosphatidylinositol-4-phosphate lipids. PLoS One. 2011;6:e24970. doi: 10.1371/journal.pone.0024970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Hsu NY, Ilnytska O, Belov G, Santiana M, Chen YH, Takvorian PM, Pau C, van der Schaar H, Kaushik-Basu N, Balla T, et al. Viral reorganization of the secretory pathway generates distinct organelles for RNA replication. Cell. 2010;141:799–811. doi: 10.1016/j.cell.2010.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Waris G, Felmlee DJ, Negro F, Siddiqui A. Hepatitis C virus induces proteolytic cleavage of sterol regulatory element binding proteins and stimulates their phosphorylation via oxidative stress. J Virol. 2007;81:8122–8130. doi: 10.1128/JVI.00125-07. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 132.Su AI, Pezacki JP, Wodicka L, Brideau AD, Supekova L, Thimme R, Wieland S, Bukh J, Purcell RH, Schultz PG, et al. Genomic analysis of the host response to hepatitis C virus infection. Proc Natl Acad Sci USA. 2002;99:15669–15674. doi: 10.1073/pnas.202608199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Diamond DL, Syder AJ, Jacobs JM, Sorensen CM, Walters KA, Proll SC, McDermott JE, Gritsenko MA, Zhang Q, Zhao R, et al. Temporal proteome and lipidome profiles reveal hepatitis C virus-associated reprogramming of hepatocellular metabolism and bioenergetics. PLoS Pathog. 2010;6:e1000719. doi: 10.1371/journal.ppat.1000719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Sharma M, Sasvari Z, Nagy PD. Inhibition of phospholipid biosynthesis decreases the activity of the tombusvirus replicase and alters the subcellular localization of replication proteins. Virology. 2011;415:141–152. doi: 10.1016/j.virol.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Eberlé D, Hegarty B, Bossard P, Ferré P, Foufelle F. SREBP transcription factors: master regulators of lipid homeostasis. Biochimie. 2004;86:839–848. doi: 10.1016/j.biochi.2004.09.018. [DOI] [PubMed] [Google Scholar]
- 136.Park CY, Jun HJ, Wakita T, Cheong JH, Hwang SB. Hepatitis C virus nonstructural 4B protein modulates sterol regulatory element-binding protein signaling via the AKT pathway. J Biol Chem. 2009;284:9237–9246. doi: 10.1074/jbc.M808773200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Olmstead AD, Knecht W, Lazarov I, Dixit SB, Jean F. Human subtilase SKI-1/S1P is a master regulator of the HCV Lifecycle and a potential host cell target for developing indirect-acting antiviral agents. PLoS Pathog. 2012;8:e1002468. doi: 10.1371/journal.ppat.1002468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Shi ST, Lee KJ, Aizaki H, Hwang SB, Lai MM. Hepatitis C virus RNA replication occurs on a detergent-resistant membrane that cofractionates with caveolin-2. J Virol. 2003;77:4160–4168. doi: 10.1128/JVI.77.7.4160-4168.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Simons K, Sampaio JL. Membrane organization and lipid rafts. Cold Spring Harb Perspect Biol. 2011;3:a004697. doi: 10.1101/cshperspect.a004697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Hirata Y, Ikeda K, Sudoh M, Tokunaga Y, Suzuki A, Weng L, Ohta M, Tobita Y, Okano K, Ozeki K, et al. Self-enhancement of hepatitis C virus replication by promotion of specific sphingolipid biosynthesis. PLoS Pathog. 2012;8:e1002860. doi: 10.1371/journal.ppat.1002860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Weng L, Hirata Y, Arai M, Kohara M, Wakita T, Watashi K, Shimotohno K, He Y, Zhong J, Toyoda T. Sphingomyelin activates hepatitis C virus RNA polymerase in a genotype-specific manner. J Virol. 2010;84:11761–11770. doi: 10.1128/JVI.00638-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Reiss S, Rebhan I, Backes P, Romero-Brey I, Erfle H, Matula P, Kaderali L, Poenisch M, Blankenburg H, Hiet MS, et al. Recruitment and activation of a lipid kinase by hepatitis C virus NS5A is essential for integrity of the membranous replication compartment. Cell Host Microbe. 2011;9:32–45. doi: 10.1016/j.chom.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Graham TR, Burd CG. Coordination of Golgi functions by phosphatidylinositol 4-kinases. Trends Cell Biol. 2011;21:113–121. doi: 10.1016/j.tcb.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Hammond GR, Fischer MJ, Anderson KE, Holdich J, Koteci A, Balla T, Irvine RF. PI4P and PI(4,5)P2 are essential but independent lipid determinants of membrane identity. Science. 2012;337:727–730. doi: 10.1126/science.1222483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Bianco A, Reghellin V, Donnici L, Fenu S, Alvarez R, Baruffa C, Peri F, Pagani M, Abrignani S, Neddermann P, et al. Metabolism of phosphatidylinositol 4-kinase IIIα-dependent PI4P Is subverted by HCV and is targeted by a 4-anilino quinazoline with antiviral activity. PLoS Pathog. 2012;8:e1002576. doi: 10.1371/journal.ppat.1002576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Li Q, Brass AL, Ng A, Hu Z, Xavier RJ, Liang TJ, Elledge SJ. A genome-wide genetic screen for host factors required for hepatitis C virus propagation. Proc Natl Acad Sci USA. 2009;106:16410–16415. doi: 10.1073/pnas.0907439106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Tai AW, Benita Y, Peng LF, Kim SS, Sakamoto N, Xavier RJ, Chung RT. A functional genomic screen identifies cellular cofactors of hepatitis C virus replication. Cell Host Microbe. 2009;5:298–307. doi: 10.1016/j.chom.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Trotard M, Lepère-Douard C, Régeard M, Piquet-Pellorce C, Lavillette D, Cosset FL, Gripon P, Le Seyec J. Kinases required in hepatitis C virus entry and replication highlighted by small interference RNA screening. FASEB J. 2009;23:3780–3789. doi: 10.1096/fj.09-131920. [DOI] [PubMed] [Google Scholar]
- 149.Berger KL, Cooper JD, Heaton NS, Yoon R, Oakland TE, Jordan TX, Mateu G, Grakoui A, Randall G. Roles for endocytic trafficking and phosphatidylinositol 4-kinase III alpha in hepatitis C virus replication. Proc Natl Acad Sci USA. 2009;106:7577–7582. doi: 10.1073/pnas.0902693106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Yang N, Ma P, Lang J, Zhang Y, Deng J, Ju X, Zhang G, Jiang C. Phosphatidylinositol 4-kinase IIIβ is required for severe acute respiratory syndrome coronavirus spike-mediated cell entry. J Biol Chem. 2012;287:8457–8467. doi: 10.1074/jbc.M111.312561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Kukulski W, Schorb M, Kaksonen M, Briggs JA. Plasma membrane reshaping during endocytosis is revealed by time-resolved electron tomography. Cell. 2012;150:508–520. doi: 10.1016/j.cell.2012.05.046. [DOI] [PubMed] [Google Scholar]
- 152.Brügger B, Sandhoff R, Wegehingel S, Gorgas K, Malsam J, Helms JB, Lehmann WD, Nickel W, Wieland FT. Evidence for segregation of sphingomyelin and cholesterol during formation of COPI-coated vesicles. J Cell Biol. 2000;151:507–518. doi: 10.1083/jcb.151.3.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Takamori S, Holt M, Stenius K, Lemke EA, Grønborg M, Riedel D, Urlaub H, Schenck S, Brügger B, Ringler P, et al. Molecular anatomy of a trafficking organelle. Cell. 2006;127:831–846. doi: 10.1016/j.cell.2006.10.030. [DOI] [PubMed] [Google Scholar]
- 154.Contreras FX, Ernst AM, Haberkant P, Björkholm P, Lindahl E, Gönen B, Tischer C, Elofsson A, von Heijne G, Thiele C, et al. Molecular recognition of a single sphingolipid species by a protein’s transmembrane domain. Nature. 2012;481:525–529. doi: 10.1038/nature10742. [DOI] [PubMed] [Google Scholar]
- 155.Liu HM, Aizaki H, Machida K, Ou JH, Lai MM. Hepatitis C virus translation preferentially depends on active RNA replication. PLoS One. 2012;7:e43600. doi: 10.1371/journal.pone.0043600. [DOI] [PMC free article] [PubMed] [Google Scholar]