Abstract
Small ubiquitin-like modifier (SUMO)ylation is a key post-translational modification mechanism that controls the function of a plethora of proteins and biological processes. Given its central regulatory role, it is not surprising that it is widely exploited by viruses. A number of viral proteins are known to modify and/or be modified by the SUMOylation system to exert their function, to create a cellular environment more favorable for virus survival and propagation, and to prevent host antiviral responses. Since the SUMO pathway is a multi-step cascade, viral proteins engage with it at many levels, to advance and favor each stage of a typical infection cycle: replication, viral assembly and immune evasion. Here we review the current knowledge on the interplay between the host SUMO system and viral lifecycle.
Keywords: Virus, Small ubiquitin-like modifier, Small ubiquitin-like modifier-ylation, Exploitation, Virus assembly, Immune evasion, Innate immunity
INTRODUCTION
Pathogenic organisms possess the remarkable ability to hijack the cellular machinery of host cells to their advantage. Viruses in particular manipulate several physiological cellular pathways to prevent antiviral responses and to create an environment that facilitates their survival and propagation.
A common strategy to create a more conducive milieu to viral development consists in exploiting cellular post-translational modifications (PTMs) mechanisms.
Between the numerous PTMs occurring in cells, small ubiquitin-like modifier (SUMO)ylation is emerging as a key PTM that controls the function of a plethora of proteins and biological processes. Hence, given its central regulatory role, the SUMOylation pathway is widely exploited by viruses, whose proteins can either modify and/or be modified by the SUMOylation system with various consequences.
The aim of this review is therefore focused on the mechanisms through which viruses exploit the SUMOylation pathway and the implications for viral infections and diseases. First, we will describe the general characteristics of the SUMO cycle and the enzymes involved in this pathway. Next, we will give a comprehensive overview on the latest findings on viral proteins and SUMOylation interplay, focusing in particular on the mechanisms that can promote viral infections by altering the SUMO system.
SUMO PATHWAY
The SUMO is a member of the ubiquitin-like proteins (Ubls) family. The common feature of Ubls is that they are attached to a target protein amino group of a lysine residue through similar but distinct enzymatic cascades[1]. After conjugation, Ubls reversibly alter protein functions, without the need for new protein synthesis, thus providing cells with a rapid and versatile mechanism to quickly respond to changes in the surrounding environment.
SUMOylation was identified as a reversible PTM in 1996[2,3]. There are four different genes in the human genome coding for the different SUMO modifiers: SUMO-1, -2, -3 and -4. SUMO-2 and -3 are nearly identical in sequence, differing from each other by only three N-terminal residues, and are therefore collectively referred to as SUMO-2/3[4]. On the contrary, SUMO-2/3 significantly diverge from SUMO-1, sharing only about 45% similarity with its sequence[5]. Finally, a SUMO-4 isoform has been described[6], which shares 86% homology with SUMO-2. In humans, while SUMO-1 and SUMO-2/3 are ubiquitous, SUMO-4 expression seems to be restricted to kidneys, lymph nodes and spleen[6].
SUMO-1 conjugation has been implicated in the regulation of physiological processes because it is virtually all bound to target proteins, while SUMO-2/3 appears to be more widely expressed as a pool of free non-conjugated proteins, readily available for stress responses[4]. SUMO-4 is probably not conjugated under normal conditions and its biological role is still unclear[7]. Moreover, the different SUMO paralogs do preferentially conjugate some substrates[4,8,9], although other proteins can be equally modified by SUMO-1 or SUMO-2/3[8,10,11].
SUMO attachment to target proteins is mediated by enzymatic reactions (schematized in Figure 1) that catalyze the formation of an isopeptide bond between the SUMO C-terminus and the ε-amino group of an internal lysine in the target, generally but not necessarily found within a SUMO modification consensus motif, ψKxE[12,13] (where ψ is a bulky aliphatic residue, X is any residue).
Figure 1.
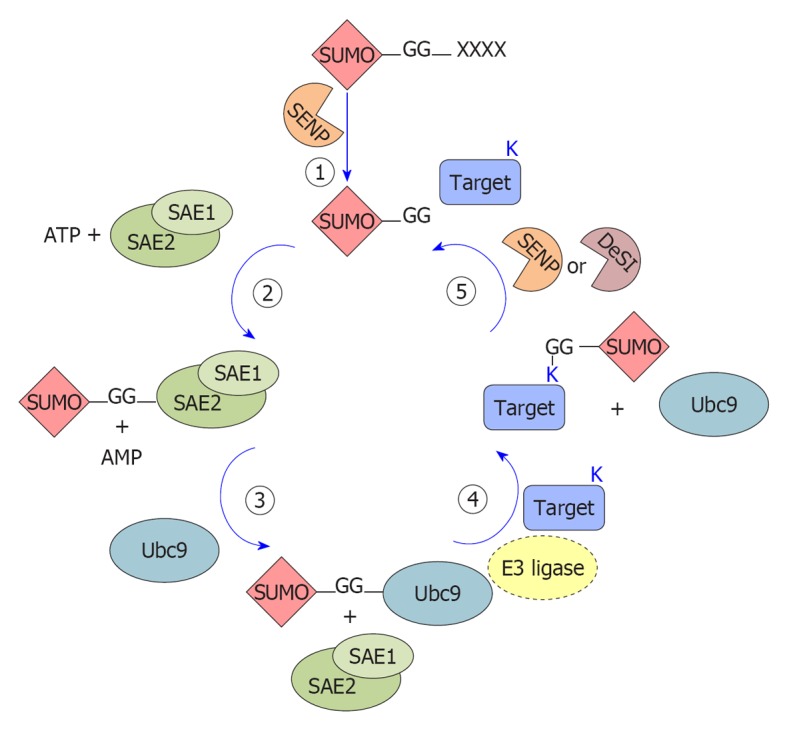
Schematic representation of the small ubiquitin-like modifier conjugation enzymatic cascade. The cartoon schematically represents the enzymatic steps of small ubiquitin-like modifier (SUMO) protein conjugation on lysines of substrate proteins. 1: SUMO protein precursors are processed by SUMO proteases (SENP) that remove the C-terminal tetrapeptide (X) and free the diglycine motif (-GG); 2: The mature form of SUMO is activated by adenylation at the C-terminal diglycine motif by the E1 enzyme (the SUMO activating enzyme, SAE1-SAE2 or AOS1-UBA2) promoting a thioester bond with a conserved Cys of the E1 enzyme; 3: SUMO is then transferred to a Cys on the E2 conjugating enzyme (Ubc9) forming an E2-SUMO thioester; 4: An isopeptide bond is formed between the diglycine motif of SUMO and a lysine (K) residue in the substrate. E3 ligases are dispensable in vitro but most likely required in vivo; 5: SUMO proteins are removed from substrates by the action of SUMO proteases (SENPs) or DeSumoylating-isopeptidase (DeSI) and free SUMO proteins are available for another cycle of conjugation.
Interestingly, SUMO-2/3 also bear the ψKxE motif and therefore can be SUMOylated, forming chains on substrate proteins through their internal lysine residue[14]. Although the formation of SUMO-1 chains has also been observed both in vitro and in vivo via non canonical consensus sites[15-17], usually SUMO-1 acts as terminator of SUMO-2/3 polymeric chains[15]. Although target proteins are predominantly conjugated to monomeric SUMO, SUMO chains also play roles in replication, turnover of SUMO targets, mitosis and meiosis[18].
SUMO proteins are 11 kDa and, similarly to most other Ubls, are synthesized as inactive precursor proteins carrying an extension of variable length (ranging from 2 to 11 amino acids). These primary translated products undergo a C-terminal cleavage to expose the diglycine motif that will be linked to the target proteins. Removal of this C-terminal end is mediated by a specific protease belonging to the sentrin-specific proteases (SENPs) family[19]. In addition to its role in SUMO processing, SENP activity is also required for SUMO depolymerization and deconjugation from its substrates[19], as detailed below.
The mature form of SUMO is conjugated to the target proteins by a three-step enzymatic cascade, very similar to the ubiquitin pathway but involving different enzymes: E1 activating enzyme, E2 conjugating enzyme and E3 ligases (Figure 1).
SUMO E1 is a 110 kDa protein, composed of a heterodimer of SUMO-activating enzyme subunit (SAE) 1/2 subunits (also known as AOS1-UBA2[20,21]). During each conjugation cycle, SAE1/2 activate SUMO proteins[20] through the formation of a high-energy thioester bond between SAE2 and the C-terminal portion of SUMO[22]. Activated SUMO is then transferred[22] to the E2 enzyme ubiquitin-conjugating 9 (Ubc9). Opposite to the ubiquitin pathway, where numerous conjugating enzymes have been described, Ubc9 is the only known SUMO-conjugating enzyme[23,24] and is essential for viability in most eukaryotes[25]. Although Ubc9 itself can transfer SUMO to targets[26], specific SUMO E3 ligases are required for efficient modification.
SUMO E3 ligases can be classified into three groups on the basis of their similarity to the ubiquitin E3 ligases and in their mechanism of action, but they share the ability to act as a bridge between the Ubc9-SUMO complex and the target protein, functioning as substrate recognizers[27]. The first group encompasses members of the protein inhibitor of activated STAT (PIAS) family (PIAS1, PIAS3, PIASxα, PIASxβ and PIASy, reviewed in[28]). In addition to the PIAS proteins, other secretory protein (SP)-RING domain-containing proteins function as SUMO E3 ligases (TOPORS[29], MUL1[30] and MMS21[31]). All these members contain a RING domain (SP, Siz/PIAS-RING) similar to the one found in ubiquitin E3 ligases. The second group is represented exclusively by the nucleoporin RanBP2 that seems to act as a composite E3 ligase in the RanBP2/RanGAP1*SUMO1/Ubc9 complex[32]. The third group comprises E3 ligases lacking the RING-domain such as the polycomb member Pc2[33], histone deacetylase (HDAC)4[34], HDAC7[35], the G-protein Rhes[36], the RNA-binding protein translocated in liposarcoma[37] and tumor-necrosis-factor-associated protein 7[38]. Moreover, members of the diverse tripartite motif (TRIM) family have been very recently discovered as a new group of SUMO E3 ligases, requiring TRIM (defined by a RING domain, one or two zinc-binding domains and a coiled-coil dimerization region) to stimulate the conjugation of both SUMO-1 and SUMO-2/3 to target proteins[39,40].
SUMOylation is a reversible process, governed by SUMO-specific proteases belonging to the SENP family and by the recently found DeSumoylating-isopeptidase (DeSI) proteins. Six true human SENP proteins have been described so far (SENP1, 2, 3, 5, 6, 7), differing in their cellular distribution, selectivity for SUMO maturation and deconjugation towards different SUMO paralogs[41]. SENP1 and SENP2 are specific for both SUMO-1 and SUMO-2/3 processing and deconjugation, while SENP3 and SENP5 act preferentially on SUMO2/3. SENP6 and SENP7 seem involved mainly in deconjugating SUMO2/3 chains (see[41] and citations therein). Finally, SENP8 shows substrate specificity to another Ubl, NEDD8[42]. All the SENPs localize to the nucleus or nucleus-associated structures; on the contrary, DeSI (-1 and -2) proteins localize also in the cytoplasm and show deSUMOylating but not processing activity for SUMO1 and for both monomeric and polymeric SUMO2/3 chains[43].
Most cellular SUMO targets are transcription factors and usually SUMOylation exerts an inhibitory effect on their transactivating activity[44], by sequestering the transcription factor in ProMyelocyticLeukemia nuclear bodies (PML-NBs)[45], a nuclear domain whose assembly requires an active and efficient SUMOylation pathway[46].
Usually, after undergoing SUMOylation, the substrate protein is recognized by a binding partner containing a SUMO-interaction motif (SIM)[47]. This interplay can lead to an altered binding with interacting proteins or DNA, promotes the recruitment of another SIM-containing effector, and affects the stability, localization or enzymatic activity of the SUMOylated protein. Through these mechanisms, SUMOylation regulates a number of cellular processes, such as transcriptional regulation, mRNA maturation, meiosis, mitosis, chromatin remodeling, ion channel activity, cell growth and apoptosis (reviewed in[48]).
Therefore, because of SUMOylation marked involvement in the regulation of cell functions, it is easy to understand why viruses have evolved a variety of mechanisms to exploit this system to their advantage.
MANIPULATION OF THE SUMO PATHWAY: A VERSATILE SWITCH TO PROMOTE VIRAL LIFESPAN
Many different viral proteins have been characterized for their ability to interact with the SUMO pathway. Since the SUMO pathway is a multi-step cascade, viral proteins can interact with and exploit it at many levels, in order to promote each stage of a typical infection cycle (Table 1). Indeed, viruses can utilize the SUMO cellular machinery to support viral persistence and replication, assembly of the virus, and to avoid the host immune system.
Table 1.
Viral life cycle and small ubiquitin-like modifier pathway
| Virus | Protein | Interaction with SUMO | Proposed impact on viral or cellular activity | Ref. |
| Chicken embryo lethal orphan Adenovirus | Gam1 | Overall SUMOylation decrease through SAE1/2 degradation and reduced Ubc9 expression | Cellular transcriptional activation | [49,50] |
| PML-NBs dispersion | [51] | |||
| Interference with HDAC1 SUMOylation | Cellular transcriptional activation | [51] | ||
| Geminivirus | AL1 | Promotes SUMOylation of selected host factors | Viral replication | [57,58] |
| Adenovirus | E1B-55K | E3 ligase SUMO-1 specific | Regulator of cell cycle and apoptosis | [59,60] |
| SUMOylated | Cellular transformation | [63] | ||
| Interaction with PML-NBs | [62,64] | |||
| Kaposi’s sarcoma-associated herpesvirus | K-bZip | E3 ligase SUMO-2/3 specific | Regulator of cell cycle and apoptosis | [66] |
| SUMOylated | Cellular transcriptional repression | [65,66] | ||
| K-Rta | E3 ubiquitin ligase activity against SUMOylated proteins | PML-NBs dispersion | [119] | |
| Vaccinia | E3 | SUMOylated | Regulator of apoptosis | [69] |
| A40R | SUMOylated | Viral replication | [74] | |
| Bovine papillomavirus | E1 | SUMOylated | Viral replication | [71,72] |
| Human papillomavirus | L2 | SUMOylated | Viral capsid assembly | [93] |
| Increase in cellular SUMO 2/3 conjugation | [93] | |||
| Moloney murine leukemia | CA | SUMOylated | Viral replication | [75] |
| Influenza A | NS1 | SUMOylated | Viral replication | [78] |
| Cytomegalovirus | IE1 | SUMOylated | Viral replication | [80,81] |
| Interaction with selected host factors | PML dispersion | [123] | ||
| SUMOylated | ||||
| IE2 | Interaction with SUMOylated proteins | Viral replication | [82] | |
| SUMOylated | Viral replication | [83] | ||
| UL44 | Interaction with Ubc9 | Viral replication | [84] | |
| Epstein-Barr | BZLF1 | SUMOylated | Reactivation of latent infections | [85,86,88] |
| Rta | SUMOylated | Reactivation of latent infections | [89] | |
| BGLF4 | Interaction with SUMOylated proteins | Viral replication and reactivation of latent infections | [91] | |
| PML-NBs dispersion | ||||
| Hantaan | NP | Interaction with Ubc9 and SUMO-1 | Virus assembly | [95,96] |
| Mason-Pfizer; human immunodeficiency | Gag | Interaction with Ubc9 | Virus assembly | [97] |
| [98-100] | ||||
| Influenza A | M1 | SUMOylated | Virus assembly | [102,103] |
| Ebola Zaire | VP35 | Promotes SUMOylation of selected host factors | IFN inhibition | [109,111] |
| Herpes simplex type-1 | ICP0 | E3 ubiquitin ligase activity against SUMOylated proteins | PML-NBs dispersion | [115-117] |
| Varicella zoster | ORF61 | Interaction with SUMOylated proteins | PML-NBs dispersion | [120,121] |
| Encephalomyocarditis | 3C | Interaction with selected host factors | PML degradation | [122] |
The table schematizes known viral proteins interacting with the small ubiquitin-like modifier (SUMO) pathway, with a brief description of their relationship with the SUMOylation machinery and the proposed biological outcome. References are also reported (see text for further details). SAE: SUMO-activating enzyme subunit; HDAC: Histone deacetylase; PML-NBs: ProMyelocyticLeukaemia nuclear bodies; IFN: Interferon.
In the following sections we will detail some examples of the current knowledge about the interplay between SUMO dysregulation and viral lifecycle.
SUMOylation and viral replication
Viruses lack some of the components required to replicate their genetic material and therefore need to redirect cell activities to promote their own reproduction. A large body of evidence supports a role for SUMOylation in replication of many viruses. In particular, viruses can subvert the transcriptional profile or the proliferation activity of the host cell, dysregulate the host cell cycle or interfere with the apoptotic process, or exploit SUMOylation to regulate the transcriptional activity of viral proteins involved in virus replication (Figure 2). Here we will describe only some illustrative examples of the strategies used by viruses to promote their own replication using the SUMOylation machinery. In fact, there are many other viral proteins known to interact with and/or modify the SUMO pathway and in many cases the biological significance of this interplay is still obscure.
Figure 2.
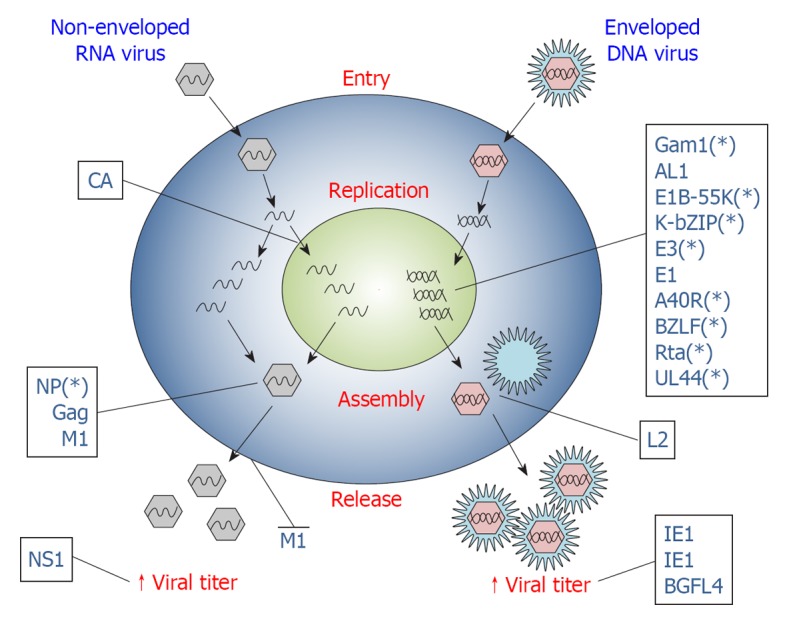
Viral proteins exploit small ubiquitin-like modifier at different steps of virus lifecycle. Scheme representing the different stages of viral infection (entry, replication, assembly, release) in host cells. Viral proteins interact with the small ubiquitin-like modifier (SUMO) machinery to promote different steps of viral life cycle, as represented. Viral proteins are designed with their acronym. The asterisk (*) indicates that the marked viral protein has not been formally shown to directly influence viral life cycle by exploiting SUMOylation, but indications of a mechanistic link are known. See text for further details on exploitation of the SUMO machinery by single viral proteins.
A hallmark of viral infection is the increase of host transcription to sustain viral replication. Gam1 is an early gene expressed by chicken embryo lethal orphan (CELO) Adenovirus that has the remarkable ability to inhibit global SUMOylation. Work from our group demonstrated that Gam1 decreases the overall SUMOylation by interaction and consequent degradation of the E1 heterodimer[49,50]. Specifically, Gam1 recruits both SAE1 and SAE2 into Cul2/5-EloB/C-Roc1 ubiquitin ligase complexes and subsequently targets SAE1 for ubiquitylation and degradation. SAE2 depletion is not tightly related to Gam1, but is rather an effect of SAE1 disappearance[50].
We also observed that Ubc9 levels are reduced upon Gam1 expression, by a yet undefined mechanism[49]. Furthermore, Gam1 disperses PML-NBs concomitant to a strong loss of SUMO-1 from the nucleus[51]. As SUMO conjugation to many transcription factors represses their activities[48], the overall decrease in SUMO conjugation caused by Gam1 could increase cellular transcriptional activity, which in turn could facilitate viral replication. Gam1 also interferes with SUMOylation of endogenous proteins such as HDAC1[51]. HDAC1 SUMOylation has an impact on the transcriptional repressive potential of the deacetylase[52,53]. Moreover, HDAC1-containing chromatin remodeling complexes are known to be exploited by viruses to regulate the progression of their infection[54]. Interestingly, a replication deficient ∆Gam1 CELO virus[55] can be rescued by HDAC inhibitors treatment[56], suggesting the existence of a cross talk between cellular SUMOylation and acetylation that can be subverted and exploited by Gam1, an essential gene for CELO replication[55].
While Gam1 promotes broad changes in the global SUMOylation pattern of the host cell, AL1 protein encoded by the plant pathogen Geminivirus alters the SUMOylation status of only selected proteins[57]. AL1 is the only plant pathogen protein described so far as interacting with the SUMO pathway[58], by associating with the plant E2 conjugating enzyme SCE1. As mentioned, AL1-SCE1 complex in plants does not produce an overall alteration of host proteins, but seems to modulate the SUMOylation level of selected host factors to create an environment suitable for viral infection.
E1B-55K is an Adenoviral early protein that functions as an E3 SUMO ligase that specifically conjugates SUMO-1 but not SUMO-2/3 to p53, inhibiting its transcriptional activity[59]. Indeed, expression of E1B-55K protein induces p53 SUMOylation[60] and E1KB-55K/p53 co-localization to PML-NBs, thus restricting p53 nuclear mobility in living cells[59]. p53 sequestration in PML-NBs seems to be a prerequisite to the altered p53 localization and activity observed in Adenovirus infected cells, preceding and addressing its ubiquitin-proteasome dependent degradation in cytoplasmic aggresomes[61]. Notably, E1B-55K associates with PML-NBs at early times after infection[62], as does p53[61]. Hence, since p53 is one of the most recognized regulators of cell cycle arrest and apoptosis, the p53-SUMO-1 conjugation could be a key event in the oncogenic transformation of primary cells induced by Adenoviruses. Interestingly, E1B-55K is also itself SUMOylated by all SUMO paralogs in a phosphorylation-dependent mechanism[63] and both SUMOylation[64] and phosphorylation[63] are required for its activity. In addition, the recent findings that E1B-55K itself interacts with Ubc9 strongly highlight that this viral protein extensively cooperates with the SUMO pathway to promote Adenovirus lifespan[63].
The K-bZIP protein encoded by Kaposi’s sarcoma-associated herpes virus (KSHV) is another viral protein that utilizes the SUMO pathway to alter the host cell cycle. K-bZIP is a strong transcriptional repressor whose activity, similarly to E1-55K, depends on SUMOylation[65], catalyzed by K-bZip itself[66]. Other similarities to the adenoviral protein include the PML-NBs localization[65] and the ability to recruit p53 to PML-NBs[67]. Finally, K-bZIP also exhibits E3 SUMO ligase activity but, unlike E1-55K, shows preferential selectivity towards SUMO-2/3 paralogs[66]. Notably, p53 SUMO-2/3 conjugation catalyzed by K-bZIP enhances p53 transcription factor ability, suggesting a p53-mediated growth arrest by prolongation of the G1 phase of the cell cycle[66]. Growth arrest is a common outcome of herpes viruses infection[68], that poses the cell in a specific phase of the cell cycle, encouraging viral replication and protecting the host cells from undergoing apoptosis.
An additional viral protein that takes advantage of the SUMO pathway to regulate the cellular apoptotic process is the E3 protein encoded by Vaccinia virus. Indeed, recent findings[69] demonstrate that E3 SUMO-1 or SUMO-2 modification has a negative effect on E3 transcriptional transactivation of the p53-upregulated modulator of apoptosis and APAF-1 genes. Therefore, these results could indicate that SUMO conjugation is a negative regulator of the transcriptional activation of p53 by E3.
Also, bovine papillomavirus (BPV) E1, the major initiator protein for BPV replication[70], is SUMO modified but, opposite to Vaccinia virus E3, only by the SUMO-1 paralog[71]. This covalent modification is required for E1 intranuclear localization and influences viral replication activity[72].
A40R is another Vaccinia virus protein that interacts with the SUMO system to accomplish its function. Vaccinia are unique among DNA viruses because DNA replication occurs entirely in discrete cytoplasmic structures enveloped by endoplasmic reticulum (mini-nuclei) membrane, rather than in the nucleus of the infected host cell[73]. A40R gene product is SUMO-1 modified, but unlike what has been described so far, this modification appears to be very stable and not subjected to SENP deconjugating activity[74]. Consistently, all other viral proteins SUMO-1 modified are localized into the nucleus, while A40R-SUMO-1 expression has been found in the cytosolic side of endoplasmic reticulum, the same membranes that wrap the virus replication sites. The specific localization of A40R strongly suggests a role for SUMOylation in Vaccinia replication[74].
Also, Moloney murine leukemia retrovirus capsid protein (CA) utilizes a similar mechanism. In fact, this protein interacts simultaneously with both Ubc9 and E3 ligase PIASy[75], resulting in covalent transfer of SUMO-1 to CA. Surprisingly, suppression of SUMO-1 attachment by CA mutations at Ubc9 or PIASy binding sites blocks virus replication in vivo, but does not affect late stages of viral gene expression or virion assembly[75]. On the contrary, Rous sarcoma virus (RSV) CA-Ubc9 interaction and SUMO-1 conjugation does not influence RSV replication[76].
Nonstructural protein 1 (NS1) is one of the major factors involved in Influenza A virus replication[77]. NS1 is able to interact with human Ubc9 and is preferentially modified by SUMO-1[78]. This characteristic seems to be conserved among most Influenza virus strains, underlining the importance of SUMO modification in Influenza virus infection. SUMO-1 modification enhances the stability of NS1 and its ability to suppress host protein expression causing an acceleration in viral replication rate[78].
Cytomegalovirus (CMV) immediate early 1 (IE1) is a viral protein that acts as a key regulator of early events in virus infection cycle together with IE2. While IE2 activates a wide range of viral and cellular promoters, IE1 only modestly promotes both cellular and viral transcription[79]. However, SUMO modification of IE1 contributes to efficient CMV replication by enhancing the expression of IE2 mRNA derived from the same transcription unit, by a yet unidentified post-translational mechanism[80,81]. Furthermore, IE2 is also SUMOylated by both non-covalent and covalent SUMO-modification[82]. IE2 SUMOylation is necessary for the function of this viral transcription factor and for human CMV replication[82], opposite to the activities of most transcription factors that are regulated in a negative manner by SUMO attachment[44]. Importantly, IE2 also contains a SIM motif to interact with other SUMOylated partners, such as TAF12, a component of the transcription factor IID complex[83]. This interaction enhances the transactivation activity of IE2, playing a further role in the progression of the CMV lytic cycle[83].
Recently, Loregian’s group reported the first evidence of SUMOylation of a viral DNA-polymerase processivity factor: the UL44 protein from human CMV[84]. UL44 strongly binds to cellular Ubc9 and is widely SUMOylated during CMV infection, with accumulation at a later time post-infection. Interestingly, UL44 SUMOylation is dependent on its correct dimerization and proper DNA binding. CMV infection in cells overexpressing SUMO1 protein results in increased viral replication and viral titer, as well as a faster relocalization of UL44 from replicative foci, suggesting that UL44 SUMOylation could perhaps support later functions important for viral propagation[84].
Finally, other interesting examples of viral proteins SUMO-modified are Epstein-Barr virus (EBV), BZLF1 (also known as Zta), Rta and BGLF4 proteins.
EBV is usually maintained under latent conditions in B lymphocytes and to proliferate it must enter the lytic cycle driven by BZLF1 and Rta. BZLF1 is post-translationally modified by both SUMO-1[85] and SUMO-2/3[86]. BZLF1 is a transcriptional activator involved in the reactivation of EBV[87], allowing its switch from latent to lytic stage, characteristic of the EBV infection cycle. SUMOylation of BLZF1 plays a key role in this mechanism, negatively affecting its transcriptional activity. In fact, SUMOylated BLZF1 associates with HDAC3 and this association allocates HDAC3 to BLZF1-responsive promoters, repressing the transcription of BLZF1-induced genes[88]. Furthermore, the SUMO-mediated repression of BLZF1 is reverted by the action of a specific protein kinase (EBV-PK) that, by inhibiting BLZF1 SUMO-conjugation, promotes the transcription of BLZF1 target genes and replication of the viral genome[86].
Similarly, also Rta SUMOylation, mediated by the adaptor cellular protein RanBPM, enhances its transactivation activity and promotes viral replication of the latent EBV virus[89].
BGLF4 is a protein kinase that phosphorylates both viral and host proteins[90], strongly contributing to the EBV infection cycle. BGLF4 carries SIM motifs responsible for its binding to SUMO-2 conjugated proteins. The SUMO binding function of BGLF4, among others, is also required to enhance the production of extracellular virus during EBV lytic replication and to disperse PML-NBs[91]. Indeed, BGLF4 seems to inhibit SUMOylation, thus promoting activation of the EBV BZLF1 protein (see above), probably by SIM-mediated recruitment and phosphorylation on SUMOylated BZLF1[91].
SUMO and virus assembly
Virus assembly is the result of a series of protein-protein and protein-lipid interactions that permits localization of different viral components at sites of virus budding. Although specific for each virus strain, virus assembly typically involves the expression of late genes that direct capsid assembling and enveloping. Besides its key role in the activity of the early expressed viral proteins that drive viral replication, SUMOylation also plays fundamental roles in viral assembly processes (Figure 2).
L2, together with L1, is a structural protein of the human papillomavirus (HPV) capsid critical for the generation of infectious viral particles as well as in early events of HPV infection[92]. L2 is preferentially modified by SUMO2/3, affecting its stability[93]. In fact, SUMOylated L2 has an increased half-life compared to the non-SUMOylated mutant. Moreover, the effect of SUMOylation negatively affects L2 capacity to interact with its physiological interactor L1, suggesting a mechanism by which capsid assembly may be modulated in HPV infected cells[93]. Moreover, L2 also increases the overall SUMO-2/3 conjugation of host proteins[93].
Nucleocapsid protein (NP) of Hantaan virus (HTNV) is a structural protein that, through its oligomerization and ability to bind RNA[94], is involved in viral assembly in the infected cell. Ubc9 and SUMO-1 interaction with NP[95] determines its localization at the perinuclear region where viral replication occurs[96] and, therefore, could regulate the assembly of the HTNV. Notably, Ubc9 was also identified as a cellular protein that interacts with the Gag protein of Mason-Pfizer monkey[97] and Human Immunodeficiency viruses[98-100], regulating viral assembly, trafficking and Env incorporation. However, these activities are not dependent on Ubc9 conjugation activity, indicating that SUMOylation may not be strictly required for assembly of these viruses[97,99].
A large body of evidence shows that Influenza A virus M1 protein is essential for viral assembly and budding[101]. M1, together with other viral proteins are SUMOylated during Influenza virus infection[102,103]. Moreover, abolishment of M1 SUMOylation resulted in dramatic reduction of the virus titer in the culture fluid, accompanied by accumulation of intracellular viral proteins and viral RNA, indicating that SUMOylation of M1 modulates the assembly of Influenza A virus. On the other hand, other steps of the viral life cycle, such as virus entry, RNA replication and translation, are not affected by M1 SUMOylation[103].
SUMOylation and viral host immune evasion
Multicellular organisms normally fight infections via their immune system. The immune system recognizes and combats invading foreign agents through two main canonical pathways, the innate and adaptive immunity. In addition, intrinsic cellular defenses are also employed by the host to clear viral infections[104]. Intrinsic resistance represents the first line of intracellular antiviral defense that employs the classical pathogen recognition receptors (PRRs), shared by the innate immunity response, to sense viruses and to rapidly produce antiviral molecules in order to limit the initial stages of infection. Consequently and not surprisingly, viruses have evolved a variety of mechanisms to overcome cellular defenses and SUMO represents one of the most exploited pathways to this end (Figure 3).
Figure 3.
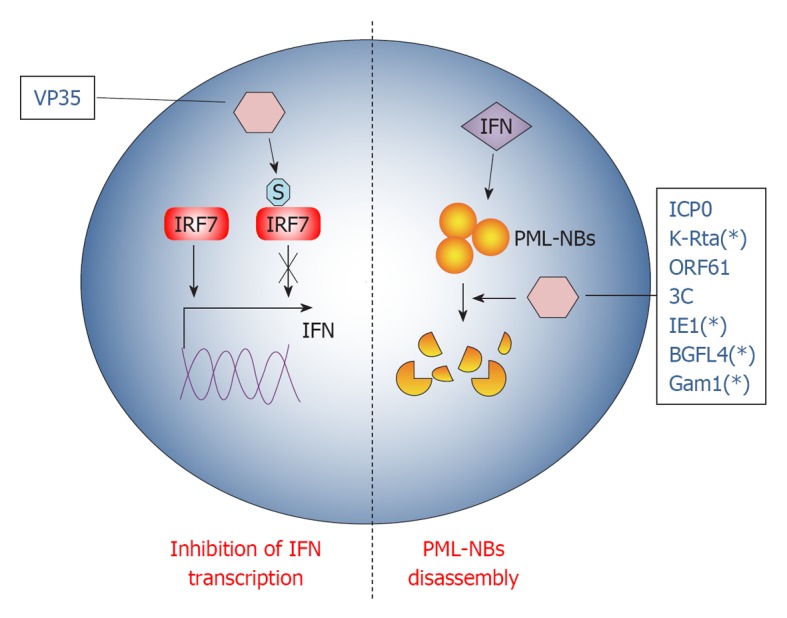
Viral proteins exploit small ubiquitin-like modifier to promote immune escape from innate and intrinsic responses. Schematic representation of the strategies used by viral proteins to counteract host innate and intrinsic responses through small ubiquitin-like modifier (SUMO). Viral proteins are designated with their acronym. S stands for SUMO. The asterisk (*) indicates that the marked viral protein has not been formally shown to directly influence cellular antiviral activity by exploiting SUMOylation, but indications of a mechanistic link are known. See text for further details on exploitation of the SUMO machinery by single viral proteins. IFN: Interferon; IRF: IFN regulatory factor; PML-NBs: ProMyelocyticLeukemia nuclear bodies.
The intrinsic and innate immune responses are primed by the activation of PRRs, such as the toll-like receptors and retinoic acid-inducible gene I which, in turn, lead to interferons (IFNs) production by IFN regulatory factor (IRF)-dependent mechanisms[105].
Ebola Zaire virus (EBOV) is a human pathogen that infects initially dendritic cells and macrophages[106], inhibiting the production of the pro-inflammatory IFN type I (IFN-I)[107]. Namely, EBOV VP35 protein potently inhibits IFN-I transcription[108] using the cellular SUMO machinery in dendritic cells. Indeed, VP35 increases SUMOylation of IRF7[109], the principal cellular factor required for IFN-I transcription[110], in a PIAS1 dependent manner. VP35 forms a complex with the SUMO ligase PIAS1 and IRF7, thus increasing PIAS1-mediated IRF7 SUMO-1 and SUMO-3 conjugation[109]. Interestingly, IRFs SUMOylation appears to be a physiological process orchestrating INFs production after viral infection[111], allowing clearing of the infecting virus. Therefore, VP35 exploits SUMO to turn off IFN-I production by dendritic cells, probably worsening the maturation of these cells[112] and weakening the host innate immunity against EBOV infection.
Taken together, these reports strongly suggest the existence of a correlation between SUMO pathway exploitation by viruses and escape from the host innate immune system. However, viruses also possess a large number of mechanisms to escape the intrinsic immune system. This is not surprising, considering that the intrinsic response is the first host defense to fight viral infections.
PML-NBs are nuclear inclusions rich of SUMOylated proteins, known to be crucial organelles involved in intrinsic anti-viral response. In fact, PML-NBs seem to be implicated in the downstream effect of INF-mediated antiviral action[113]. Notably, PML-NBs are disassembled during most viral infections at very early stages, indicating that targeting PML-NBs could be an efficient viral strategy to evade IFN action[113]. Therefore, most of the mechanisms developed by DNA viruses to overcome cellular defense disperse PML-NBs: one rapid way to achieve this goal is by hijacking the SUMOylation pathway.
Herpes simplex virus type-1 (HSV-1) protein ICP0 structure encompasses a RING finger domain that acts as an E3 ubiquitin ligase, redirecting specific cellular proteins for proteasome-dependent degradation[114]. Earlier during infection, PML-NBs components are quickly recruited at sites closely associated with the viral genome in a SUMO-dependent manner[115,116], promoting the transcription of anti-viral genes. However, ICP0 counteracts this PML-NBs response targeting SUMOylated proteins for degradation, thanks to its E3 ligase activity[115]. This HSV-response is strictly required for its infection cycle, since in this way ICP0 inhibits cellular mechanisms that would otherwise repress viral transcription[114]. Interestingly, it has been shown that ICP0 falls in the SUMO targeted ubiquitin ligases (STUbLs) family[117], a class of RING finger ubiquitin ligases that contains SIMs[118]. Therefore, through its SIM motifs, ICP0 binds to important SUMOylated transcription factors in PML-NBs that, in turn, are degraded by the E3 ubiquitin ligase activity of its RING motif. This dual action of ICP0 efficiently counteracts intrinsic antiviral resistance to HSV-1 infection[117].
Like ICP0, KHSV K-Rta protein also belongs to the STUbLs family. Indeed, K-Rta contains SIM motifs and conjugates ubiquitin to SUMO and SUMO-chains, disrupting PML-NBs in a ubiquitin ligase dependent fashion[119].
A similar mechanism is also conducted by Varicella Zoster virus protein ORF61. Indeed, ORF61 colocalizes and disperses PML-NBs shortly after virus entry in its target cell[120]. It also contains three SIM motifs through which it counteracts intrinsic SUMO-promoted anti-viral control by PML-NBs[121]. Consistently, ORF61 SIM mutants are unable to disperse PML and the overall degree of virus infection is dramatically impaired when SUMO-conjugation is inhibited[121]. As for ICP0, PML-NBs dispersal by ORF61 is a two-step process accomplished by different protein domains: the ORF61 SIMs that recognize SUMOylated PML protein in PML-NBs and the RING domain that executes their dispersal[121]. However, ORF61 RING domain does not share the E3 ligase activity with ICP0 and is thus not able to degrade PML-NBs. A similar PML-NBs disruption mechanism seems to be also carried out by the already described EBV BGLF4 protein[91].
While in all the examples described above viruses extensively interact with a number of proteins in PML-NBs, encephalomyocarditis virus (EMCV) counteracts antiviral pathway targeting the PML protein alone[122]. In fact, during infection, PML is first transferred by EMCV from the nucleoplasm to the nuclear matrix and then the viral protease 3C induces PML degradation. Both PML delocalization and degradation are a consequence of covalent SUMO-1, -2 and -3 conjugation promoted by EMCV[122].
In addition to the ability to be itself SUMOylated (see above) for its transactivation functions, CMV IE1 also efficiently inhibits the intrinsic antiviral response by preventing the accumulation of SUMOylated forms of PML[123]. In this regard, CMV seems to behave as EMCV, since IE1 does not induce PML degradation.
CONCLUSION
In recent years, SUMOylation has emerged as a major regulator system involved in a variety of cellular processes. SUMO is indeed conjugated to a number of proteins that in turn can interact with many other partners through the SUMO interacting (SIM) motifs. Therefore, the SUMOylation machinery virtually affects and directs most of cellular activities, crucially regulating cellular homeostasis. Thus, exploiting SUMOylation represents a very convenient way to quickly promote and sustain pathogen survival in the host.
Viruses, in particular, exploit SUMOylation in several key steps of their intracellular life and, importantly, they also use the SUMO pathway to subvert the immune response of the host (Table 1). Both DNA and RNA viruses can use SUMOylation to promote viral genes transcription, virus assembly (Figure 2) and immune evasion (Figure 3), using apparently different mechanisms. Some viral proteins (i.e., E3, E1, L2, A40R, CA, IE1, IE2, BLZF1, Rta, BGLF4, M1, E1B-55k, K-bZip, UL44) are modified by SUMO in order to activate their function; alternatively, they can influence the SUMOylation level of a specific target protein (AL1, VP35) or the global SUMOylation status of infected cells (Gam1, ICP0, K-Rta, L2). Finally, other viral products could interact with SUMO components or with host SUMO-containing proteins (NP, Gag, 3C, ORF61), usually through a SIM motif, or mimicking SUMOylation enzymes (K-bZIP, E1B-55K). Remarkably, the same virus (KSHV, CMV, EBV, Vaccinia Virus, Papillomavirus) can exploit the SUMO pathway through various proteins, as well as the same protein (Gam1, IE1, IE2, E1B-55K, K-bZIP, BGLF4) can interact with SUMO using several mechanisms perhaps also to promote different steps of viral infection.
It is interesting to note that the vast majority of viral proteins known to interact with the SUMOylation system are immediate-early or early proteins, suggesting a crucial role for SUMO in counteracting viral infection.
What we can learn from the complex network of interplay between the SUMO pathway and viruses in the virus-host interactions is that the same crucial pathway can be hijacked by different pathogens in very different ways to obtain a common goal, i.e., sustaining viral infection. More studies are required to define the global picture but the findings presented here can strongly indicate the SUMO pathway as a promising target for specific antiviral therapies.
Footnotes
P- Reviewer Brown JC S- Editor Gou SX L- Editor Roemmele A E- Editor Zheng XM
References
- 1.Hochstrasser M. Origin and function of ubiquitin-like proteins. Nature. 2009;458:422–429. doi: 10.1038/nature07958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Matunis MJ, Coutavas E, Blobel G. A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J Cell Biol. 1996;135:1457–1470. doi: 10.1083/jcb.135.6.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mahajan R, Delphin C, Guan T, Gerace L, Melchior F. A small ubiquitin-related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2. Cell. 1997;88:97–107. doi: 10.1016/s0092-8674(00)81862-0. [DOI] [PubMed] [Google Scholar]
- 4.Saitoh H, Hinchey J. Functional heterogeneity of small ubiquitin-related protein modifiers SUMO-1 versus SUMO-2/3. J Biol Chem. 2000;275:6252–6258. doi: 10.1074/jbc.275.9.6252. [DOI] [PubMed] [Google Scholar]
- 5.Lapenta V, Chiurazzi P, van der Spek P, Pizzuti A, Hanaoka F, Brahe C. SMT3A, a human homologue of the S. cerevisiae SMT3 gene, maps to chromosome 21qter and defines a novel gene family. Genomics. 1997;40:362–366. doi: 10.1006/geno.1996.4556. [DOI] [PubMed] [Google Scholar]
- 6.Bohren KM, Nadkarni V, Song JH, Gabbay KH, Owerbach D. A M55V polymorphism in a novel SUMO gene (SUMO-4) differentially activates heat shock transcription factors and is associated with susceptibility to type I diabetes mellitus. J Biol Chem. 2004;279:27233–27238. doi: 10.1074/jbc.M402273200. [DOI] [PubMed] [Google Scholar]
- 7.Owerbach D, McKay EM, Yeh ET, Gabbay KH, Bohren KM. A proline-90 residue unique to SUMO-4 prevents maturation and sumoylation. Biochem Biophys Res Commun. 2005;337:517–520. doi: 10.1016/j.bbrc.2005.09.090. [DOI] [PubMed] [Google Scholar]
- 8.Vertegaal AC, Andersen JS, Ogg SC, Hay RT, Mann M, Lamond AI. Distinct and overlapping sets of SUMO-1 and SUMO-2 target proteins revealed by quantitative proteomics. Mol Cell Proteomics. 2006;5:2298–2310. doi: 10.1074/mcp.M600212-MCP200. [DOI] [PubMed] [Google Scholar]
- 9.Tatham MH, Kim S, Jaffray E, Song J, Chen Y, Hay RT. Unique binding interactions among Ubc9, SUMO and RanBP2 reveal a mechanism for SUMO paralog selection. Nat Struct Mol Biol. 2005;12:67–74. doi: 10.1038/nsmb878. [DOI] [PubMed] [Google Scholar]
- 10.Evdokimov E, Sharma P, Lockett SJ, Lualdi M, Kuehn MR. Loss of SUMO1 in mice affects RanGAP1 localization and formation of PML nuclear bodies, but is not lethal as it can be compensated by SUMO2 or SUMO3. J Cell Sci. 2008;121:4106–4113. doi: 10.1242/jcs.038570. [DOI] [PubMed] [Google Scholar]
- 11.Citro S, Chiocca S. Sumo paralogs: redundancy and divergencies. Front Biosci (Schol Ed) 2013;5:544–553. doi: 10.2741/s388. [DOI] [PubMed] [Google Scholar]
- 12.Rodriguez MS, Dargemont C, Hay RT. SUMO-1 conjugation in vivo requires both a consensus modification motif and nuclear targeting. J Biol Chem. 2001;276:12654–12659. doi: 10.1074/jbc.M009476200. [DOI] [PubMed] [Google Scholar]
- 13.Sampson DA, Wang M, Matunis MJ. The small ubiquitin-like modifier-1 (SUMO-1) consensus sequence mediates Ubc9 binding and is essential for SUMO-1 modification. J Biol Chem. 2001;276:21664–21669. doi: 10.1074/jbc.M100006200. [DOI] [PubMed] [Google Scholar]
- 14.Tatham MH, Jaffray E, Vaughan OA, Desterro JM, Botting CH, Naismith JH, Hay RT. Polymeric chains of SUMO-2 and SUMO-3 are conjugated to protein substrates by SAE1/SAE2 and Ubc9. J Biol Chem. 2001;276:35368–35374. doi: 10.1074/jbc.M104214200. [DOI] [PubMed] [Google Scholar]
- 15.Matic I, van Hagen M, Schimmel J, Macek B, Ogg SC, Tatham MH, Hay RT, Lamond AI, Mann M, Vertegaal AC. In vivo identification of human small ubiquitin-like modifier polymerization sites by high accuracy mass spectrometry and an in vitro to in vivo strategy. Mol Cell Proteomics. 2008;7:132–144. doi: 10.1074/mcp.M700173-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blomster HA, Imanishi SY, Siimes J, Kastu J, Morrice NA, Eriksson JE, Sistonen L. In vivo identification of sumoylation sites by a signature tag and cysteine-targeted affinity purification. J Biol Chem. 2010;285:19324–19329. doi: 10.1074/jbc.M110.106955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Galisson F, Mahrouche L, Courcelles M, Bonneil E, Meloche S, Chelbi-Alix MK, Thibault P. A novel proteomics approach to identify SUMOylated proteins and their modification sites in human cells. Mol Cell Proteomics. 2011;10:M110.004796. doi: 10.1074/mcp.M110.004796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vertegaal AC. SUMO chains: polymeric signals. Biochem Soc Trans. 2010;38:46–49. doi: 10.1042/BST0380046. [DOI] [PubMed] [Google Scholar]
- 19.Hay RT. SUMO-specific proteases: a twist in the tail. Trends Cell Biol. 2007;17:370–376. doi: 10.1016/j.tcb.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 20.Desterro JM, Rodriguez MS, Kemp GD, Hay RT. Identification of the enzyme required for activation of the small ubiquitin-like protein SUMO-1. J Biol Chem. 1999;274:10618–10624. doi: 10.1074/jbc.274.15.10618. [DOI] [PubMed] [Google Scholar]
- 21.Gong L, Li B, Millas S, Yeh ET. Molecular cloning and characterization of human AOS1 and UBA2, components of the sentrin-activating enzyme complex. FEBS Lett. 1999;448:185–189. doi: 10.1016/s0014-5793(99)00367-1. [DOI] [PubMed] [Google Scholar]
- 22.Lois LM, Lima CD. Structures of the SUMO E1 provide mechanistic insights into SUMO activation and E2 recruitment to E1. EMBO J. 2005;24:439–451. doi: 10.1038/sj.emboj.7600552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson ES, Blobel G. Ubc9p is the conjugating enzyme for the ubiquitin-like protein Smt3p. J Biol Chem. 1997;272:26799–26802. doi: 10.1074/jbc.272.43.26799. [DOI] [PubMed] [Google Scholar]
- 24.Desterro JM, Thomson J, Hay RT. Ubch9 conjugates SUMO but not ubiquitin. FEBS Lett. 1997;417:297–300. doi: 10.1016/s0014-5793(97)01305-7. [DOI] [PubMed] [Google Scholar]
- 25.Hayashi T, Seki M, Maeda D, Wang W, Kawabe Y, Seki T, Saitoh H, Fukagawa T, Yagi H, Enomoto T. Ubc9 is essential for viability of higher eukaryotic cells. Exp Cell Res. 2002;280:212–221. doi: 10.1006/excr.2002.5634. [DOI] [PubMed] [Google Scholar]
- 26.Bernier-Villamor V, Sampson DA, Matunis MJ, Lima CD. Structural basis for E2-mediated SUMO conjugation revealed by a complex between ubiquitin-conjugating enzyme Ubc9 and RanGAP1. Cell. 2002;108:345–356. doi: 10.1016/s0092-8674(02)00630-x. [DOI] [PubMed] [Google Scholar]
- 27.Melchior F, Schergaut M, Pichler A. SUMO: ligases, isopeptidases and nuclear pores. Trends Biochem Sci. 2003;28:612–618. doi: 10.1016/j.tibs.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 28.Jackson PK. A new RING for SUMO: wrestling transcriptional responses into nuclear bodies with PIAS family E3 SUMO ligases. Genes Dev. 2001;15:3053–3058. doi: 10.1101/gad.955501. [DOI] [PubMed] [Google Scholar]
- 29.Weger S, Hammer E, Heilbronn R. Topors acts as a SUMO-1 E3 ligase for p53 in vitro and in vivo. FEBS Lett. 2005;579:5007–5012. doi: 10.1016/j.febslet.2005.07.088. [DOI] [PubMed] [Google Scholar]
- 30.Braschi E, Zunino R, McBride HM. MAPL is a new mitochondrial SUMO E3 ligase that regulates mitochondrial fission. EMBO Rep. 2009;10:748–754. doi: 10.1038/embor.2009.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao X, Blobel G. A SUMO ligase is part of a nuclear multiprotein complex that affects DNA repair and chromosomal organization. Proc Natl Acad Sci USA. 2005;102:4777–4782. doi: 10.1073/pnas.0500537102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Werner A, Flotho A, Melchior F. The RanBP2/RanGAP1*SUMO1/Ubc9 complex is a multisubunit SUMO E3 ligase. Mol Cell. 2012;46:287–298. doi: 10.1016/j.molcel.2012.02.017. [DOI] [PubMed] [Google Scholar]
- 33.Kagey MH, Melhuish TA, Wotton D. The polycomb protein Pc2 is a SUMO E3. Cell. 2003;113:127–137. doi: 10.1016/s0092-8674(03)00159-4. [DOI] [PubMed] [Google Scholar]
- 34.Grégoire S, Yang XJ. Association with class IIa histone deacetylases upregulates the sumoylation of MEF2 transcription factors. Mol Cell Biol. 2005;25:2273–2287. doi: 10.1128/MCB.25.6.2273-2287.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gao C, Ho CC, Reineke E, Lam M, Cheng X, Stanya KJ, Liu Y, Chakraborty S, Shih HM, Kao HY. Histone deacetylase 7 promotes PML sumoylation and is essential for PML nuclear body formation. Mol Cell Biol. 2008;28:5658–5667. doi: 10.1128/MCB.00874-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Subramaniam S, Sixt KM, Barrow R, Snyder SH. Rhes, a striatal specific protein, mediates mutant-huntingtin cytotoxicity. Science. 2009;324:1327–1330. doi: 10.1126/science.1172871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oh SM, Liu Z, Okada M, Jang SW, Liu X, Chan CB, Luo H, Ye K. Ebp1 sumoylation, regulated by TLS/FUS E3 ligase, is required for its anti-proliferative activity. Oncogene. 2010;29:1017–1030. doi: 10.1038/onc.2009.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morita Y, Kanei-Ishii C, Nomura T, Ishii S. TRAF7 sequesters c-Myb to the cytoplasm by stimulating its sumoylation. Mol Biol Cell. 2005;16:5433–5444. doi: 10.1091/mbc.E05-08-0731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liang Q, Deng H, Li X, Wu X, Tang Q, Chang TH, Peng H, Rauscher FJ, Ozato K, Zhu F. Tripartite motif-containing protein 28 is a small ubiquitin-related modifier E3 ligase and negative regulator of IFN regulatory factor 7. J Immunol. 2011;187:4754–4763. doi: 10.4049/jimmunol.1101704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chu Y, Yang X. SUMO E3 ligase activity of TRIM proteins. Oncogene. 2011;30:1108–1116. doi: 10.1038/onc.2010.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim JH, Baek SH. Emerging roles of desumoylating enzymes. Biochim Biophys Acta. 2009;1792:155–162. doi: 10.1016/j.bbadis.2008.12.008. [DOI] [PubMed] [Google Scholar]
- 42.Mendoza HM, Shen LN, Botting C, Lewis A, Chen J, Ink B, Hay RT. NEDP1, a highly conserved cysteine protease that deNEDDylates Cullins. J Biol Chem. 2003;278:25637–25643. doi: 10.1074/jbc.M212948200. [DOI] [PubMed] [Google Scholar]
- 43.Shin EJ, Shin HM, Nam E, Kim WS, Kim JH, Oh BH, Yun Y. DeSUMOylating isopeptidase: a second class of SUMO protease. EMBO Rep. 2012;13:339–346. doi: 10.1038/embor.2012.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gill G. Something about SUMO inhibits transcription. Curr Opin Genet Dev. 2005;15:536–541. doi: 10.1016/j.gde.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 45.Seeler JS, Dejean A. SUMO: of branched proteins and nuclear bodies. Oncogene. 2001;20:7243–7249. doi: 10.1038/sj.onc.1204758. [DOI] [PubMed] [Google Scholar]
- 46.Müller S, Matunis MJ, Dejean A. Conjugation with the ubiquitin-related modifier SUMO-1 regulates the partitioning of PML within the nucleus. EMBO J. 1998;17:61–70. doi: 10.1093/emboj/17.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kerscher O. SUMO junction-what’s your function New insights through SUMO-interacting motifs. EMBO Rep. 2007;8:550–555. doi: 10.1038/sj.embor.7400980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wilson VG. SUMO regulation of cellular processes. Dordrecht: Springer; 2009. [Google Scholar]
- 49.Boggio R, Colombo R, Hay RT, Draetta GF, Chiocca S. A mechanism for inhibiting the SUMO pathway. Mol Cell. 2004;16:549–561. doi: 10.1016/j.molcel.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 50.Boggio R, Passafaro A, Chiocca S. Targeting SUMO E1 to ubiquitin ligases: a viral strategy to counteract sumoylation. J Biol Chem. 2007;282:15376–15382. doi: 10.1074/jbc.M700889200. [DOI] [PubMed] [Google Scholar]
- 51.Colombo R, Boggio R, Seiser C, Draetta GF, Chiocca S. The adenovirus protein Gam1 interferes with sumoylation of histone deacetylase 1. EMBO Rep. 2002;3:1062–1068. doi: 10.1093/embo-reports/kvf213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.David G, Neptune MA, DePinho RA. SUMO-1 modification of histone deacetylase 1 (HDAC1) modulates its biological activities. J Biol Chem. 2002;277:23658–23663. doi: 10.1074/jbc.M203690200. [DOI] [PubMed] [Google Scholar]
- 53.Cheng J, Wang D, Wang Z, Yeh ET. SENP1 enhances androgen receptor-dependent transcription through desumoylation of histone deacetylase 1. Mol Cell Biol. 2004;24:6021–6028. doi: 10.1128/MCB.24.13.6021-6028.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Roizman B. The checkpoints of viral gene expression in productive and latent infection: the role of the HDAC/CoREST/LSD1/REST repressor complex. J Virol. 2011;85:7474–7482. doi: 10.1128/JVI.00180-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Glotzer JB, Saltik M, Chiocca S, Michou AI, Moseley P, Cotten M. Activation of heat-shock response by an adenovirus is essential for virus replication. Nature. 2000;407:207–211. doi: 10.1038/35025102. [DOI] [PubMed] [Google Scholar]
- 56.Chiocca S, Kurtev V, Colombo R, Boggio R, Sciurpi MT, Brosch G, Seiser C, Draetta GF, Cotten M. Histone deacetylase 1 inactivation by an adenovirus early gene product. Curr Biol. 2002;12:594–598. doi: 10.1016/s0960-9822(02)00720-0. [DOI] [PubMed] [Google Scholar]
- 57.Sánchez-Durán MA, Dallas MB, Ascencio-Ibañez JT, Reyes MI, Arroyo-Mateos M, Ruiz-Albert J, Hanley-Bowdoin L, Bejarano ER. Interaction between geminivirus replication protein and the SUMO-conjugating enzyme is required for viral infection. J Virol. 2011;85:9789–9800. doi: 10.1128/JVI.02566-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Castillo AG, Kong LJ, Hanley-Bowdoin L, Bejarano ER. Interaction between a geminivirus replication protein and the plant sumoylation system. J Virol. 2004;78:2758–2769. doi: 10.1128/JVI.78.6.2758-2769.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pennella MA, Liu Y, Woo JL, Kim CA, Berk AJ. Adenovirus E1B 55-kilodalton protein is a p53-SUMO1 E3 ligase that represses p53 and stimulates its nuclear export through interactions with promyelocytic leukemia nuclear bodies. J Virol. 2010;84:12210–12225. doi: 10.1128/JVI.01442-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Muller S, Dobner T. The adenovirus E1B-55K oncoprotein induces SUMO modification of p53. Cell Cycle. 2008;7:754–758. doi: 10.4161/cc.7.6.5495. [DOI] [PubMed] [Google Scholar]
- 61.Liu Y, Shevchenko A, Shevchenko A, Berk AJ. Adenovirus exploits the cellular aggresome response to accelerate inactivation of the MRN complex. J Virol. 2005;79:14004–14016. doi: 10.1128/JVI.79.22.14004-14016.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Doucas V, Ishov AM, Romo A, Juguilon H, Weitzman MD, Evans RM, Maul GG. Adenovirus replication is coupled with the dynamic properties of the PML nuclear structure. Genes Dev. 1996;10:196–207. doi: 10.1101/gad.10.2.196. [DOI] [PubMed] [Google Scholar]
- 63.Wimmer P, Blanchette P, Schreiner S, Ching W, Groitl P, Berscheminski J, Branton PE, Will H, Dobner T. Cross-talk between phosphorylation and SUMOylation regulates transforming activities of an adenoviral oncoprotein. Oncogene. 2012:Epub ahead of print. doi: 10.1038/onc.2012.187. [DOI] [PubMed] [Google Scholar]
- 64.Wimmer P, Schreiner S, Everett RD, Sirma H, Groitl P, Dobner T. SUMO modification of E1B-55K oncoprotein regulates isoform-specific binding to the tumour suppressor protein PML. Oncogene. 2010;29:5511–5522. doi: 10.1038/onc.2010.284. [DOI] [PubMed] [Google Scholar]
- 65.Izumiya Y, Ellison TJ, Yeh ET, Jung JU, Luciw PA, Kung HJ. Kaposi’s sarcoma-associated herpesvirus K-bZIP represses gene transcription via SUMO modification. J Virol. 2005;79:9912–9925. doi: 10.1128/JVI.79.15.9912-9925.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chang PC, Izumiya Y, Wu CY, Fitzgerald LD, Campbell M, Ellison TJ, Lam KS, Luciw PA, Kung HJ. Kaposi’s sarcoma-associated herpesvirus (KSHV) encodes a SUMO E3 ligase that is SIM-dependent and SUMO-2/3-specific. J Biol Chem. 2010;285:5266–5273. doi: 10.1074/jbc.M109.088088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Katano H, Ogawa-Goto K, Hasegawa H, Kurata T, Sata T. Human-herpesvirus-8-encoded K8 protein colocalizes with the promyelocytic leukemia protein (PML) bodies and recruits p53 to the PML bodies. Virology. 2001;286:446–455. doi: 10.1006/viro.2001.1005. [DOI] [PubMed] [Google Scholar]
- 68.Flemington EK. Herpesvirus lytic replication and the cell cycle: arresting new developments. J Virol. 2001;75:4475–4481. doi: 10.1128/JVI.75.10.4475-4481.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.González-Santamaría J, Campagna M, García MA, Marcos-Villar L, González D, Gallego P, Lopitz-Otsoa F, Guerra S, Rodríguez MS, Esteban M, et al. Regulation of vaccinia virus E3 protein by small ubiquitin-like modifier proteins. J Virol. 2011;85:12890–12900. doi: 10.1128/JVI.05628-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wilson VG, Ludes-Meyers J. A bovine papillomavirus E1-related protein binds specifically to bovine papillomavirus DNA. J Virol. 1991;65:5314–5322. doi: 10.1128/jvi.65.10.5314-5322.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rangasamy D, Wilson VG. Bovine papillomavirus E1 protein is sumoylated by the host cell Ubc9 protein. J Biol Chem. 2000;275:30487–30495. doi: 10.1074/jbc.M003898200. [DOI] [PubMed] [Google Scholar]
- 72.Rangasamy D, Woytek K, Khan SA, Wilson VG. SUMO-1 modification of bovine papillomavirus E1 protein is required for intranuclear accumulation. J Biol Chem. 2000;275:37999–38004. doi: 10.1074/jbc.M007777200. [DOI] [PubMed] [Google Scholar]
- 73.Roberts KL, Smith GL. Vaccinia virus morphogenesis and dissemination. Trends Microbiol. 2008;16:472–479. doi: 10.1016/j.tim.2008.07.009. [DOI] [PubMed] [Google Scholar]
- 74.Palacios S, Perez LH, Welsch S, Schleich S, Chmielarska K, Melchior F, Locker JK. Quantitative SUMO-1 modification of a vaccinia virus protein is required for its specific localization and prevents its self-association. Mol Biol Cell. 2005;16:2822–2835. doi: 10.1091/mbc.E04-11-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yueh A, Leung J, Bhattacharyya S, Perrone LA, de los Santos K, Pu SY, Goff SP. Interaction of moloney murine leukemia virus capsid with Ubc9 and PIASy mediates SUMO-1 addition required early in infection. J Virol. 2006;80:342–352. doi: 10.1128/JVI.80.1.342-352.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Spidel JL, Wilson CB, Craven RC, Wills JW. Genetic Studies of the beta-hairpin loop of Rous sarcoma virus capsid protein. J Virol. 2007;81:1288–1296. doi: 10.1128/JVI.01551-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hale BG, Randall RE, Ortín J, Jackson D. The multifunctional NS1 protein of influenza A viruses. J Gen Virol. 2008;89:2359–2376. doi: 10.1099/vir.0.2008/004606-0. [DOI] [PubMed] [Google Scholar]
- 78.Xu K, Klenk C, Liu B, Keiner B, Cheng J, Zheng BJ, Li L, Han Q, Wang C, Li T, et al. Modification of nonstructural protein 1 of influenza A virus by SUMO1. J Virol. 2011;85:1086–1098. doi: 10.1128/JVI.00877-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Castillo JP, Kowalik TF. Human cytomegalovirus immediate early proteins and cell growth control. Gene. 2002;290:19–34. doi: 10.1016/s0378-1119(02)00566-8. [DOI] [PubMed] [Google Scholar]
- 80.Spengler ML, Kurapatwinski K, Black AR, Azizkhan-Clifford J. SUMO-1 modification of human cytomegalovirus IE1/IE72. J Virol. 2002;76:2990–2996. doi: 10.1128/JVI.76.6.2990-2996.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nevels M, Brune W, Shenk T. SUMOylation of the human cytomegalovirus 72-kilodalton IE1 protein facilitates expression of the 86-kilodalton IE2 protein and promotes viral replication. J Virol. 2004;78:7803–7812. doi: 10.1128/JVI.78.14.7803-7812.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Berndt A, Hofmann-Winkler H, Tavalai N, Hahn G, Stamminger T. Importance of covalent and noncovalent SUMO interactions with the major human cytomegalovirus transactivator IE2p86 for viral infection. J Virol. 2009;83:12881–12894. doi: 10.1128/JVI.01525-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kim ET, Kim YE, Huh YH, Ahn JH. Role of noncovalent SUMO binding by the human cytomegalovirus IE2 transactivator in lytic growth. J Virol. 2010;84:8111–8123. doi: 10.1128/JVI.00459-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sinigalia E, Alvisi G, Segré CV, Mercorelli B, Muratore G, Winkler M, Hsiao HH, Urlaub H, Ripalti A, Chiocca S, et al. The human cytomegalovirus DNA polymerase processivity factor UL44 is modified by SUMO in a DNA-dependent manner. PLoS One. 2012;7:e49630. doi: 10.1371/journal.pone.0049630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Adamson AL, Kenney S. Epstein-barr virus immediate-early protein BZLF1 is SUMO-1 modified and disrupts promyelocytic leukemia bodies. J Virol. 2001;75:2388–2399. doi: 10.1128/JVI.75.5.2388-2399.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hagemeier SR, Dickerson SJ, Meng Q, Yu X, Mertz JE, Kenney SC. Sumoylation of the Epstein-Barr virus BZLF1 protein inhibits its transcriptional activity and is regulated by the virus-encoded protein kinase. J Virol. 2010;84:4383–4394. doi: 10.1128/JVI.02369-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wen W, Iwakiri D, Yamamoto K, Maruo S, Kanda T, Takada K. Epstein-Barr virus BZLF1 gene, a switch from latency to lytic infection, is expressed as an immediate-early gene after primary infection of B lymphocytes. J Virol. 2007;81:1037–1042. doi: 10.1128/JVI.01416-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Murata T, Hotta N, Toyama S, Nakayama S, Chiba S, Isomura H, Ohshima T, Kanda T, Tsurumi T. Transcriptional repression by sumoylation of Epstein-Barr virus BZLF1 protein correlates with association of histone deacetylase. J Biol Chem. 2010;285:23925–23935. doi: 10.1074/jbc.M109.095356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chang LK, Liu ST, Kuo CW, Wang WH, Chuang JY, Bianchi E, Hong YR. Enhancement of transactivation activity of Rta of Epstein-Barr virus by RanBPM. J Mol Biol. 2008;379:231–242. doi: 10.1016/j.jmb.2008.04.011. [DOI] [PubMed] [Google Scholar]
- 90.Gershburg E, Pagano JS. Conserved herpesvirus protein kinases. Biochim Biophys Acta. 2008;1784:203–212. doi: 10.1016/j.bbapap.2007.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Li R, Wang L, Liao G, Guzzo CM, Matunis MJ, Zhu H, Hayward SD. SUMO binding by the Epstein-Barr virus protein kinase BGLF4 is crucial for BGLF4 function. J Virol. 2012;86:5412–5421. doi: 10.1128/JVI.00314-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Holmgren SC, Patterson NA, Ozbun MA, Lambert PF. The minor capsid protein L2 contributes to two steps in the human papillomavirus type 31 life cycle. J Virol. 2005;79:3938–3948. doi: 10.1128/JVI.79.7.3938-3948.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Marusic MB, Mencin N, Licen M, Banks L, Grm HS. Modification of human papillomavirus minor capsid protein L2 by sumoylation. J Virol. 2010;84:11585–11589. doi: 10.1128/JVI.01269-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gött P, Stohwasser R, Schnitzler P, Darai G, Bautz EK. RNA binding of recombinant nucleocapsid proteins of hantaviruses. Virology. 1993;194:332–337. doi: 10.1006/viro.1993.1263. [DOI] [PubMed] [Google Scholar]
- 95.Lee BH, Yoshimatsu K, Maeda A, Ochiai K, Morimatsu M, Araki K, Ogino M, Morikawa S, Arikawa J. Association of the nucleocapsid protein of the Seoul and Hantaan hantaviruses with small ubiquitin-like modifier-1-related molecules. Virus Res. 2003;98:83–91. doi: 10.1016/j.virusres.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 96.Maeda A, Lee BH, Yoshimatsu K, Saijo M, Kurane I, Arikawa J, Morikawa S. The intracellular association of the nucleocapsid protein (NP) of hantaan virus (HTNV) with small ubiquitin-like modifier-1 (SUMO-1) conjugating enzyme 9 (Ubc9) Virology. 2003;305:288–297. doi: 10.1006/viro.2002.1767. [DOI] [PubMed] [Google Scholar]
- 97.Weldon RA, Sarkar P, Brown SM, Weldon SK. Mason-Pfizer monkey virus Gag proteins interact with the human sumo conjugating enzyme, hUbc9. Virology. 2003;314:62–73. doi: 10.1016/s0042-6822(03)00348-9. [DOI] [PubMed] [Google Scholar]
- 98.Gurer C, Berthoux L, Luban J. Covalent modification of human immunodeficiency virus type 1 p6 by SUMO-1. J Virol. 2005;79:910–917. doi: 10.1128/JVI.79.2.910-917.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Jaber T, Bohl CR, Lewis GL, Wood C, West JT, Weldon RA. Human Ubc9 contributes to production of fully infectious human immunodeficiency virus type 1 virions. J Virol. 2009;83:10448–10459. doi: 10.1128/JVI.00237-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Martinez NW, Xue X, Berro RG, Kreitzer G, Resh MD. Kinesin KIF4 regulates intracellular trafficking and stability of the human immunodeficiency virus type 1 Gag polyprotein. J Virol. 2008;82:9937–9950. doi: 10.1128/JVI.00819-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Rossman JS, Lamb RA. Influenza virus assembly and budding. Virology. 2011;411:229–236. doi: 10.1016/j.virol.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Pal S, Santos A, Rosas JM, Ortiz-Guzman J, Rosas-Acosta G. Influenza A virus interacts extensively with the cellular SUMOylation system during infection. Virus Res. 2011;158:12–27. doi: 10.1016/j.virusres.2011.02.017. [DOI] [PubMed] [Google Scholar]
- 103.Wu CY, Jeng KS, Lai MM. The SUMOylation of matrix protein M1 modulates the assembly and morphogenesis of influenza A virus. J Virol. 2011;85:6618–6628. doi: 10.1128/JVI.02401-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Boo KH, Yang JS. Intrinsic cellular defenses against virus infection by antiviral type I interferon. Yonsei Med J. 2010;51:9–17. doi: 10.3349/ymj.2010.51.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Cavlar T, Ablasser A, Hornung V. Induction of type I IFNs by intracellular DNA-sensing pathways. Immunol Cell Biol. 2012;90:474–482. doi: 10.1038/icb.2012.11. [DOI] [PubMed] [Google Scholar]
- 106.Bray M, Geisbert TW. Ebola virus: the role of macrophages and dendritic cells in the pathogenesis of Ebola hemorrhagic fever. Int J Biochem Cell Biol. 2005;37:1560–1566. doi: 10.1016/j.biocel.2005.02.018. [DOI] [PubMed] [Google Scholar]
- 107.Mohamadzadeh M, Chen L, Schmaljohn AL. How Ebola and Marburg viruses battle the immune system. Nat Rev Immunol. 2007;7:556–567. doi: 10.1038/nri2098. [DOI] [PubMed] [Google Scholar]
- 108.Leung LW, Park MS, Martinez O, Valmas C, López CB, Basler CF. Ebolavirus VP35 suppresses IFN production from conventional but not plasmacytoid dendritic cells. Immunol Cell Biol. 2011;89:792–802. doi: 10.1038/icb.2010.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Chang TH, Kubota T, Matsuoka M, Jones S, Bradfute SB, Bray M, Ozato K. Ebola Zaire virus blocks type I interferon production by exploiting the host SUMO modification machinery. PLoS Pathog. 2009;5:e1000493. doi: 10.1371/journal.ppat.1000493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Honda K, Yanai H, Negishi H, Asagiri M, Sato M, Mizutani T, Shimada N, Ohba Y, Takaoka A, Yoshida N, et al. IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature. 2005;434:772–777. doi: 10.1038/nature03464. [DOI] [PubMed] [Google Scholar]
- 111.Kubota T, Matsuoka M, Chang TH, Tailor P, Sasaki T, Tashiro M, Kato A, Ozato K. Virus infection triggers SUMOylation of IRF3 and IRF7, leading to the negative regulation of type I interferon gene expression. J Biol Chem. 2008;283:25660–25670. doi: 10.1074/jbc.M804479200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Jin H, Yan Z, Prabhakar BS, Feng Z, Ma Y, Verpooten D, Ganesh B, He B. The VP35 protein of Ebola virus impairs dendritic cell maturation induced by virus and lipopolysaccharide. J Gen Virol. 2010;91:352–361. doi: 10.1099/vir.0.017343-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Everett RD, Chelbi-Alix MK. PML and PML nuclear bodies: implications in antiviral defence. Biochimie. 2007;89:819–830. doi: 10.1016/j.biochi.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 114.Boutell C, Sadis S, Everett RD. Herpes simplex virus type 1 immediate-early protein ICP0 and is isolated RING finger domain act as ubiquitin E3 ligases in vitro. J Virol. 2002;76:841–850. doi: 10.1128/JVI.76.2.841-850.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Cuchet-Lourenço D, Boutell C, Lukashchuk V, Grant K, Sykes A, Murray J, Orr A, Everett RD. SUMO pathway dependent recruitment of cellular repressors to herpes simplex virus type 1 genomes. PLoS Pathog. 2011;7:e1002123. doi: 10.1371/journal.ppat.1002123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Cuchet D, Sykes A, Nicolas A, Orr A, Murray J, Sirma H, Heeren J, Bartelt A, Everett RD. PML isoforms I and II participate in PML-dependent restriction of HSV-1 replication. J Cell Sci. 2011;124:280–291. doi: 10.1242/jcs.075390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Boutell C, Cuchet-Lourenço D, Vanni E, Orr A, Glass M, McFarlane S, Everett RD. A viral ubiquitin ligase has substrate preferential SUMO targeted ubiquitin ligase activity that counteracts intrinsic antiviral defence. PLoS Pathog. 2011;7:e1002245. doi: 10.1371/journal.ppat.1002245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Uzunova K, Göttsche K, Miteva M, Weisshaar SR, Glanemann C, Schnellhardt M, Niessen M, Scheel H, Hofmann K, Johnson ES, et al. Ubiquitin-dependent proteolytic control of SUMO conjugates. J Biol Chem. 2007;282:34167–34175. doi: 10.1074/jbc.M706505200. [DOI] [PubMed] [Google Scholar]
- 119.Campbell M, Izumiya Y. Post-Translational Modifications of Kaposi’s Sarcoma-Associated Herpesvirus Regulatory Proteins - SUMO and KSHV. Front Microbiol. 2012;3:31. doi: 10.3389/fmicb.2012.00031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Reichelt M, Brady J, Arvin AM. The replication cycle of varicella-zoster virus: analysis of the kinetics of viral protein expression, genome synthesis, and virion assembly at the single-cell level. J Virol. 2009;83:3904–3918. doi: 10.1128/JVI.02137-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Wang L, Oliver SL, Sommer M, Rajamani J, Reichelt M, Arvin AM. Disruption of PML nuclear bodies is mediated by ORF61 SUMO-interacting motifs and required for varicella-zoster virus pathogenesis in skin. PLoS Pathog. 2011;7:e1002157. doi: 10.1371/journal.ppat.1002157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.El McHichi B, Regad T, Maroui MA, Rodriguez MS, Aminev A, Gerbaud S, Escriou N, Dianoux L, Chelbi-Alix MK. SUMOylation promotes PML degradation during encephalomyocarditis virus infection. J Virol. 2010;84:11634–11645. doi: 10.1128/JVI.01321-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Lee HR, Kim DJ, Lee JM, Choi CY, Ahn BY, Hayward GS, Ahn JH. Ability of the human cytomegalovirus IE1 protein to modulate sumoylation of PML correlates with its functional activities in transcriptional regulation and infectivity in cultured fibroblast cells. J Virol. 2004;78:6527–6542. doi: 10.1128/JVI.78.12.6527-6542.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]


