Abstract
As a field, lipidomics is in its infancy, yet it has already begun to influence lipid biochemistry in myriad ways. As with other omic technologies, the field is driven by advances in analytical chemistry, particularly by mass spectrometry. At the heart of a renaissance in lipid biochemistry, systems biology is being used to define the cellular lipome, build a comprehensive picture of metabolic interconnections, discover new molecular species and determine how lipids modulate biological functions.
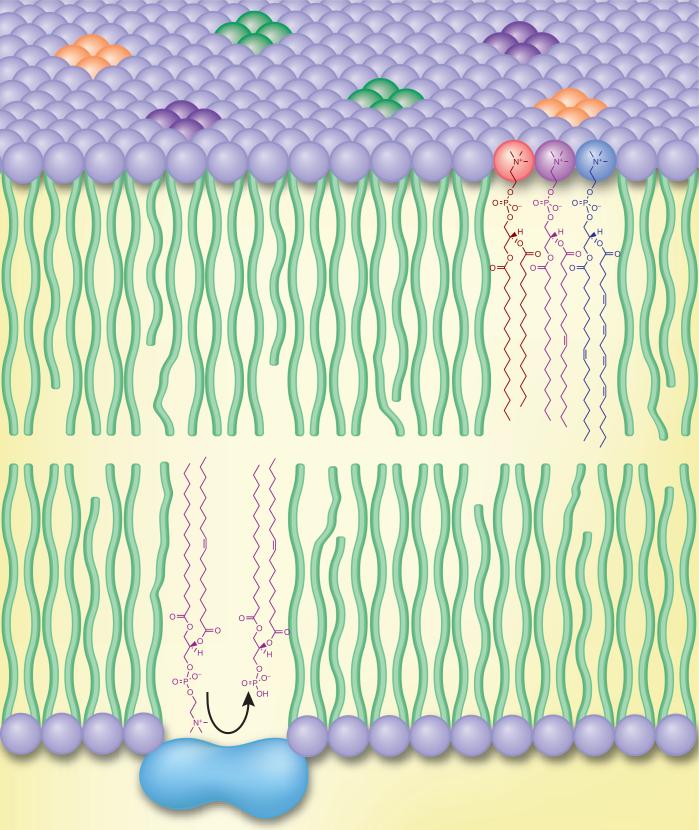
During the past decade, there has been a fervor of activity in lipid biochemistry driven by advances in both biochemistry and analytical methodology, specifically in the area of mass spectrometry and its ancillary techniques. This research has been aimed at enhancing our understanding of the role that lipids play within the cell and biology in general, and it has led to a redefining of lipid substances and an appreciation of the complex biochemistry underlying their synthesis, metabolism, interactions with proteins and roles in regulation of gene expression. The role for lipids even several decades ago was known to be quite broad and diverse. These hydrophobic species include the family of molecules that serve as metabolic and energy storage units and as entities critically involved in membrane structure and scaffolding for membrane proteins. Clearly, the semipermeable membrane resulting from the complex array of phospholipids has played a central role in the development of the life process itself as a result of its ability to separate cells and subcellular organelles from each other (Fig. 1). Other lipids function as signaling molecules that coordinate biochemical events between cells; these include the eicosanoids (prostaglandins and leukotrienes) in mammalian cells1, the estrogens, which are circulating hormones2, and the diacylglycerols, which are important in intracellular signal transduction events3. The variety of lipid utility is illustrated by steroids, which not only serve a physical role in the plasma membrane of cells (cholesterol or ergosterol) but also perform hormonal roles in coordinating biochemical events throughout an entire organism. The extensive diversity in lipid substances from the plant domain is exemplified by the large number of natural products from the isoprenoid and polyketide biosynthetic pathways that have found their way into our pharmacopeia and that have uses in human medicine. Yet within the past few years, our understanding of the roles that lipids play in cell biology has expanded greatly to include repressors and derepressors involved as agonists of nuclear protein receptors such as the peroxisome proliferation–activating receptors (PPARs), the retinoic acid receptors (RXRs) and the liver X receptors (LXRs). Homoserine lactones are now known as messengers in quorum-sensing bacteria4. Polyphosphatidylinositols function as intracellular messengers by modulating the structures of membrane proteins, such as ion channels5, and by providing docking sites for translocation of cytosolic signaling proteins. Thus, those compounds that we call lipids are deeply integrated into the entire fabric and chemistry of cell biology because of their unique properties and structures.
Figure 1.
Biological membranes are often depicted as a sea of homogenous lipid in which membrane proteins reside and cytosolic proteins translocate. In reality, cellular membranes are composed of chemically diverse lipid species that regulate essential biophysical, metabolic and signaling processes.
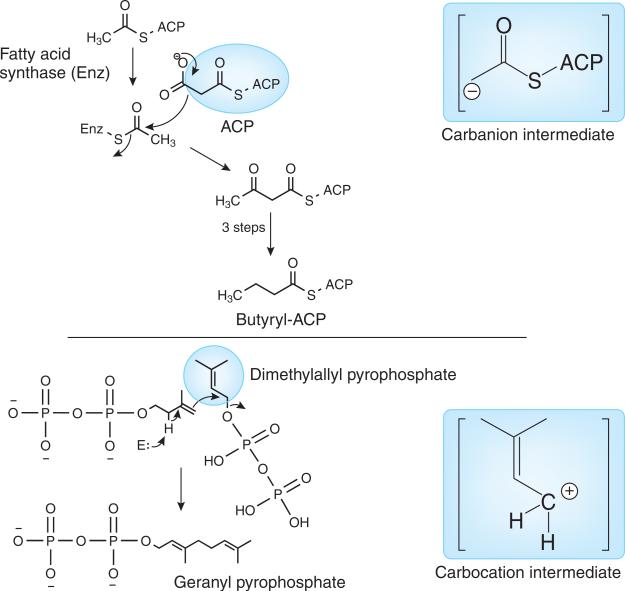
Even at the present time, lipids are defined as molecules soluble in organic solvents or at least extracted from aqueous systems by the use of an immiscible solvent6. There are so many examples of lipid substances that do not obey this simple rule that defining these substances based on their biosynthetic origin is much more descriptive7. We know that lipids in nature are derived from one of two different biosynthetic pathways. The first pathway involves the condensation of acyl carrier protein (ACP) intermediates derived from malonyl-CoA and acetyl-CoA esters and the intermediacy of a carbanion structure (Fig. 2). This mechanism leads to a wide variety of lipids that contain the fatty acyl chain, including fatty acids, phospholipids and glycerolipids. A very similar process, although enzymatically different, is the polyketide biosynthetic pathway in plants8. The second pathway (and perhaps the oldest pathway leading to lipid-like substances on Earth) is the carbocation pathway that involves condensation of branched-chain 5-carbon pyrophosphate intermediates and the intermediacy of a carbocation in the process (Fig. 2). This carbocation pathway leads to all lipids that are present in the Archaea domain and a large number of lipids in the Bacteria and Eukarya domains, such as the steroids, polyisoprenoids and highly pigmented molecules such as the carotenoids9. Thinking of lipids from the standpoint of their biosynthetic mechanism of origin removes many of the technical issues associated with defining them based on organic solvent solubility.
Figure 2.
Known initial biochemical steps responsible for the synthesis of all lipids. Synthesis occurs via either a carbanion intermediate (fatty acyls and polyketides, hence mammalian and plant glycerolipids, sphingolipids, saccharolipids and glycerophospholipids) or a carbocation intermediate (prenols, steroids and archaeal glycerolipids, sphingolipids and glycerophospholipids), according to Fahy et al.7.
Lipid diversity
At the heart of the renaissance in lipid biochemistry is an attempt to answer two very simple questions: how many lipid species are there, and how do they interact with other molecules? The surprising answers are (i) an extraordinarily large number—hundreds of thousands of lipids—and (ii) an ever-expanding list of interactions with proteins and other lipids. This diversity is a reflection of the importance of a unique property—the amphipathicity of lipids with their hydrophobic acyl tails and hydrophilic head groups—involved in the living process and the need to have mechanisms to sustain localized compartments.
The expansion in the numbers and types of identified lipids that are present within a single cell type has been made possible by the application of mass spectrometry and by the development of powerful techniques that can structurally probe these molecules and quantitate their presence within a cell. An important component of lipid analysis in the past has been the use of chromatographic separation, and this approach remains important today. Lipid analysis was quite difficult a century ago, as illustrated by the work of Koch and Woods10 on the estimation of phosphatidylcholines and phosphatidylethanolamines (lecithans) in various tissues. At this early time for biological chemistry, the analysis of phospholipids was largely based on a gravimetric measurement of phosphorus liberated from a specific extract.
The beginning of the modern era for lipid analysis was ushered in by the application of emerging chemical techniques of chromatography such as thin layer chromatography11, and for low-molecular-weight lipids such as fatty acids and fatty acid methyl esters, gas chromatography. Both approaches enabled separation of the complex mixture of lipids that were typically present in organic solvent extracts from biological matrices12. By the 1960s it was possible to also use mass spectrometry to analyze lipids in great molecular detail13, and the emergence of the combined gas chromatograph/mass spectrometer/computer system14 enabled the rapid generation of databases from this tandem analytical instrument that could be searched to detect the elution of unexpected lipids based on chromatographic separation properties and unique mass spectrometric behavior15.
Yet the major advances in the ability to analyze all lipids with the powerful technique of mass spectrometry only came after development of the ionization techniques of electrospray ionization16 and matrix-assisted laser desorption ionization17. Electrospray ionization (ESI) provided a realistic way to approach the analysis of any lipid that could be extracted either by solvent extraction methods or by hydrophobic solid-phase extraction technology. Since ESI could be readily coupled to the effluent of a liquid chromatograph, both normal-phase and reversed-phase HPLC became routinely available for LC/MS direct analysis. The parallel development of mass spectrometric techniques—including collision-induced dissociation of gas-phase ions, tandem mass spectrometry and the strategies for quantitative analysis—enabled mass spectrometry to be applied as a sensitive and specific quantitative tool.
Quickly it became apparent that the world of lipids was one of extreme diversity in structure, number and biochemical utility. For many lipids such as phospholipids, glycerolipids and sphingolipids, thousands of different molecular species were found to be present in tissues and cells. Having the ability to qualitatively define such lipids at the molecular species level led to the next level of questions concerning the absolute quantity of each of these lipid molecular species. Could quantitative information reveal unique biochemical pathways at their intersection? Thus, the field of lipidomics began and was applied to mammalian, plant, fungal and bacterial systems.
Metabolomics and systems biology
Several different approaches have been pursued to glean information about the quantity and identity of lipids that are present within cells or tissues. One of the techniques minimized the use of chromatography and maximized the use of the unique properties of the mass spectrometer and the ionization technique to gather a picture of the lipids that could be detected in an extract. Simultaneous analysis of all lipids either as positive or negative ions became the defining approach. This technique has come to be known as ‘shotgun lipidomics’, and impressive results have been obtained to profile the lipids (including phospholipids, glycerol lipids and sphingolipids) that are present within many different types of biological extracts18,19.
A more traditional approach is to engage the power of the chromatographic separation to divide a complex mixture into major categories, such as neutral lipids, phospholipids and sphingolipids, and analyze each of the lipid subclasses sequentially using electrospray ionization mass spectrometry as well as gas-phase ion chemistry techniques of tandem mass spectrometry to identify specific molecular species. There is important structural information to be found in elution order from HPLC columns that can significantly aid in the characterization of lipids that cannot be readily identified by mass spectrometric analysis alone—for example, the occurrence of ether-linked triglycerides (monoalkyldiacylglycerols) in mixtures of triacylglycerols in lipid bodies20. Chromatographic separation techniques have also enabled investigators to assign the double bond position in fatty acyl groups or the location of fatty acyl groups on the glycerol backbone. In the former case, an interesting approach has been recently described that uses collision-induced dissociation in the presence of ozone and captures the facile chemistry of ozone attack on isolated carbon-carbon double bonds in the formation of unique aldehydes and hydroxyhydroperoxy compounds that indicate the position of double bonds in a particular fatty acyl group21. While the use of the chromatographic strategy and tandem mass spectrometry reduces some of the problems of shotgun lipidomics (such as ion suppression and detection of minor components in complex mixtures), it nonetheless does require considerably more time and effort to implement. At the extreme of this general approach is the analysis of eicosanoids present in a biological sample (eicosanomics), which requires the power of chromatographic separation coupled with mass spectrometric detection to quantitate subpicomolar levels of these arachidonate metabolites generated after cellular activation22. Such analyses would not be possible by the shotgun approach since these lipids are only minor constituents of traditional lipid extracts.
Challenges in quantitation
A major problem in all areas of lipidomics is the availability of both reference standard material and specific lipids that can serve as internal standards for the mass spectrometric quantitation method. Quantitative analysis by mass spectrometry does require having reference compounds to establish a standard curve, yet in most cases there are very few molecular species of a lipid subtype commercially available. At present there are less than 10 reference standard materials available to establish quantitative curves for all of the arachidonate-containing glycerophospholipids, which we now know to exceed 100 in a cell such as the macrophage23. We also know that activation of cell surface receptors results in changes in numerous lipid species rather than just a few specific molecular species24, thus requiring a prohibitively large number of quantitative standard curves to be constructed. Mass spec-trometric analysis is also subject to a number of variables that alter absolute sensitivity that need to be assessed and controlled to render the observed abundance of an ion into a quantitative measure of lipid concentration. Without the availability of true reference standards and the appropriate internal standard (such as a stable isotope-labeled molecule for each target molecular species), a series of compromises is the only avenue open. It is clear that ionization efficiency, stability of ions and relative formation of positive and negative ions are quite dependent on subtle chemical features of the lipid under investigation. Furthermore, slight structural differences—for example, in position of double bonds, position of fatty acylation in the glycerol backbone, carbon chain length, ion adducts, number of double bonds and composite molecular weight—can alter absolute signal levels that are measured as the quantitative index of the lipid. Though compromises are clearly necessary, the results can be sufficiently accurate to reveal answers to biochemical questions. For instance, when comparing samples from parallel lipidomics measurements in which the precision is very high, the lower accuracy in absolute quantity is not that relevant. It is even possible to reveal changes between control and treated samples in a more accurate fashion. Strategies have been developed to label aminophospholipids (glycerophospho-ethanolamine and -serine lipids) with different stable isotope-tagged derivatives that permit very precise comparison of all aminophospholipid molecular species25.
With these limitations in mind, it is important to ask the following question: how precise does one need to be in order to glean important information about lipids present in cells at the molecular species level? Using the tools that are currently available, it is quite likely that one can obtain 20–50% absolute quantitation precision of a lipid in very complex mixtures26. In many cases, this is still below biological variability. Using the isotope-tagging approaches, one can reduce this tenfold in relative quantitation25.
For some biological systems, it remains impossible to absolutely quantitate individual molecular species within a particular class. This is quite apparent with the glycerolipids and wax esters. In large part, this is because of the number of molecules with the same exact molecular weight (isobaric) that are present in the complex mixtures. Molecular ion analysis by mass spectrometry reveals the total number of fatty acyl carbon atoms and the total number of double bonds. For some triglyceride mixtures isolated from mammalian cells, molecular ions can represent mixtures of 6–10 different triacylglycerol molecular species27. Even more complex triacylglycerol mixtures are isolated from fish oil, which contains abundant polyunsaturated fatty acids and isobaric ions corresponding to 20–30 different molecular species of lipids. Thus measurement of a single mass-to-charge ratio does not give a picture of individual molecular species composition, and if one observes a change in ion abundances, that ion does not reveal which one of the ten isobaric species has in fact changed. While chromatographic separations offer some assistance, challenges nonetheless remain.
Dynamics of lipid molecular species
In spite of the power of mass spectrometry and the recognized limitations in quantitative analysis, a major thrust has been to use the information obtained from such studies with informatics techniques to understand lipid pathways and biochemical mechanisms. A tacit assumption is that with sufficient information it should be possible to truly define lipid biochemistry (much like a biblical document made up of a simple character set of lipid identity and absolute lipid concentration) and then apply this information to define normal cellular and pathological processes. Considerable effort has been placed into the analysis of lipids for a number of years, and one of the clear messages emerging is that an individual molecular species concentration is a dynamic variable. Identical quantities of phospholipid and glycerolipid species are difficult to reproduce from laboratory to laboratory and even by the same laboratory at different times, even when there is no attempt to perturb cells. Does this reflect failure to accurately quantify, or is a biological message being revealed? We think the latter is the case. One might entertain the idea that these results reveal that the population of lipids within a cell membrane and within subcellular membranes is very dynamic and that one obtains only a snapshot picture of the lipid molecular species present when one carries out an extraction and stops the lipid remodeling events. For some time it has been appreciated that remodeling of lipids, in particular phospholipids, is an active process and is the result of acyltransferase activity, about which little was known28. However, it is now quite clear that a family of lysophospholipid acyltransferases exist in all cells, and each one has rather specific specificities for both the lysophospholipid polar head group and the fatty acyl-CoAs.
If it is the case that the exact composition and concentrations of lipid species present in a resting cell are dynamically changing, it is highly likely that a cell can sustain critical reactions with many different lipid compositions rather than a single composition. This makes the lipidomic approach to understanding biology and pathology much more complex than that assumed by comparison to the field of genomics.
New frontiers and future directions
A fundamental problem remains in the analysis of lipids at the cellular or even tissue level. Of great importance is knowing where a particular lipid resides within a specific cellular compartment, whether it is at the plasma membrane, Golgi apparatus, mitochondria, nuclear envelope or lipid bodies. Rather traditional techniques of subcellular organelle isolation by differential centrifugation are available to probe intracellular locations of specific lipid molecular species. Yet when one extracts a cell, one is certainly intermixing lipids during the homogenization and extraction process, which would cause information about the specific location of lipids to be lost. When membranes are disrupted, chemistry tells us that they reform, so why would they not re-form as chimeric membranes?
Many advances in cell biology are a direct result of knowing (i) the intracellular sites at which specific proteins reside and (ii) where and how proteins move during cellular activation and during major cellular events such as cell death. Lipids inherently do not have readily accessible chromophores that are sufficiently unique to allow them to be directly observed using very powerful optical microscopic-based techniques. Furthermore, because of their small size, one cannot modify lipids in a way that introduces such a property without disrupting the native chemical character of the lipid. The closest approach is to incorporate radiolabeled tracers, such as tritium-labeled lipids, and use microscopic autoradiography techniques to locate the positions of tritium atoms within a cell. However, the exact chemical nature of the tritium atom remains unknown, and only its beta particle can be detected through silver ion chemistry.
Recently mass spectrometry–based profiling has been used to assign new functions to lipid messengers and to facilitate development of new small-molecule inhibitors. Long et al.29 recently described a role for endogenous 2-arachidonoylglycerol in several behaviors classically associated with cannabinoid pharmacology. These authors used an inhibitor of monoacylglycerol lipase to selectively modify 2-arachidonoylglycerol levels without altering closely related anandamide species. Similarly, Scott et al.30 used mass spectrometry–based profiles to facilitate design of isoform-selective inhibitors of phospholipase D and to explore the role of signaling lipids in metastatic breast cancer models. These approaches illustrate how improvements in analytical methodologies and chemical biology are being applied to important questions in biochemistry and pharmacology.
There are emerging technologies that show considerable promise for addressing this unique void in lipid biochemistry. Mass spectrometers can be used to image tissues and reveal locations of endogenous lipids by abundant secondary ions emitted from the surface either in matrix-assisted laser desorption ionization (MALDI imaging) (Fig. 3)31 or in secondary ion emission (SIMs) techniques. Though SIMs can achieve nanometer-scale lateral resolution of surfaces32, it suffers from the use of high-energy ion beams that tend to decompose fragile lipids into fairly nondescript product ions such as the phosphocho-line ion (m/z 184). Several groups are pushing the limits of both techniques in order to gain information about lipids at the tissue level with eyes on the subcellular region. One of the more attractive advances has been the use of polyatomic ionic particles such as C60+ bucky-balls33. New instruments are being developed that promise to have significantly improved lateral resolution that perhaps can be applied to the analysis of tissues and (more importantly) to lipids in a single cell. Other instruments are being developed that use alternate means to separate ions and thereby improve the analytical power of the mass spectrometric experiment—for example, a drift tube can be introduced between the electrospray ionization source and the mass spectrometer that permits separation based on collisional properties and the shape of a molecule rather than the absolute mass of the ion34.
Figure 3.
Brain PC imaged by MALDI mass spectrometry. Mass spectral imaging of a 10 μm mouse brain slice (mid-sagittal section) for a docosahexaenoic acid–containing phosphatidylcholine (18:0/22:6-PC) as a positive ion at m/z 834.6 using MALDI mass spectrometry. The highest ion intensity (brightest shade) was observed in the cerebellar gray matter (see ref. 31 for methods). c.p.s., counts per second.
Summary
The ability to analyze the large number of lipids that are present within a biological system provides new insights into lipid biochemistry that were previously not possible. It is clear that a great deal of information is embedded in the distribution and concentration of lipids within molecular species. Furthermore, a number of unique lipids are present at very low concentrations in cells and tissues that serve as signals for cellular events. Whereas the majority of biosynthesized lipids are fairly well known for the mammalian organisms, considerably less is known about the lipids present in prokaryotes, Archaea and plants. Fortunately, tools that can address specific challenges and facilitate research in this area are constantly being refined and developed. With continued advances in the sensitivity and throughput of mass spectrometry, we are likely to discover new connections and cross-talk in the metabolism of species in different lipid classes that were previously thought to be disparate and isolated from one another. However, mechanistic insights of such new relationships will not emerge simply by observation of covariance in quantities. Rather, these findings will inspire new questions that must be answered by classical biochemical approaches. Lipidomics does not provide any magical shortcuts to understanding the roles of lipids in biology; rather, this emerging technology provides a view of the gestalt of the biological system. Because of the variety of classes, the enormous diversity of molecular species and the dynamics of remodeling, lipidomics offers a powerful tool for viewing the robust nature of lipid biochemistry with an eye toward a critical examination and eventual understanding of lipid biochemistry in the living process.
ACKNOWLEDGMENTS
This work has been supported in part by a large-scale consortium grant from the US National Institutes of Health (GM 069338).
Contributor Information
H Alex Brown, Department of Pharmacology, Vanderbilt University School of Medicine, Nashville, Tennessee, USA..
Robert C Murphy, Department of Pharmacology, University of Colorado Denver School of Medicine, Aurora, Colorado, USA..
References
- 1.Shimizu T. Annu. Rev. Pharmacol. Toxicol. 2009;49:123–150. doi: 10.1146/annurev.pharmtox.011008.145616. [DOI] [PubMed] [Google Scholar]
- 2.Hewitt SC, Harrell JC, Korach KS. Annu. Rev. Physiol. 2005;67:285–308. doi: 10.1146/annurev.physiol.67.040403.115914. [DOI] [PubMed] [Google Scholar]
- 3.Newton AC. J. Lipid Res. 2009;50:S266–S271. doi: 10.1194/jlr.R800064-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cooley M, Chhabra SR, Williams P. Chem. Biol. 2008;15:1141–1147. doi: 10.1016/j.chembiol.2008.10.010. [DOI] [PubMed] [Google Scholar]
- 5.Suh BC, Hille B. Annu. Rev. Biophys. 2008;37:175–195. doi: 10.1146/annurev.biophys.37.032807.125859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alberts B, et al. Molecular Biology of the Cell. 5th edn Vol. 115. Garland Science; New York: 2008. [Google Scholar]
- 7.Fahy E, et al. J. Lipid Res. 2005;46:839–861. doi: 10.1194/jlr.E400004-JLR200. [DOI] [PubMed] [Google Scholar]
- 8.Austin MB, O'Maille PE, Noel JP. Nat. Chem. Biol. 2008;4:217–222. doi: 10.1038/nchembio0408-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thulasiram HV, Erickson HK, Poulter CD. Science. 2007;316:73–76. doi: 10.1126/science.1137786. [DOI] [PubMed] [Google Scholar]
- 10.Koch W, Woods HS. J. Biol. Chem. 1906;1:203–211. [Google Scholar]
- 11.Fontell K, Holman RT, Lambertsen GJ. Lipid Res. 1960;1:391–404. [PubMed] [Google Scholar]
- 12.James AT, Martin AJ. Biochem. J. 1956;63:144–152. doi: 10.1042/bj0630144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ryhage R, Stenhagen EJ. Lipid Res. 1960;1:361–390. [PubMed] [Google Scholar]
- 14.Hites RA, Biemann K. Anal. Chem. 1968;40:1217–1221. [Google Scholar]
- 15.Murphy RC, Djuricic MV, Markey SP, Biemann K. Science. 1969;165:695–697. doi: 10.1126/science.165.3894.695. [DOI] [PubMed] [Google Scholar]
- 16.Fenn JB, Mann M, Meng CK, Wong SF, Whitehouse CM. Science. 1989;246:64–71. doi: 10.1126/science.2675315. [DOI] [PubMed] [Google Scholar]
- 17.Hillenkamp F, Karas M, Beavis RC, Chait BT. Anal. Chem. 1991;63:1193A–1203A. doi: 10.1021/ac00024a002. [DOI] [PubMed] [Google Scholar]
- 18.Han X, Gross RW. Mass Spectrom. Rev. 2005;24:367–412. doi: 10.1002/mas.20023. [DOI] [PubMed] [Google Scholar]
- 19.Ejsing CS, et al. Proc. Natl. Acad. Sci. USA. 2009;106:2136–2141. doi: 10.1073/pnas.0811700106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hutchins PM, Barkley RM, Murphy RC. J. Lipid Res. 2008;49:804–813. doi: 10.1194/jlr.M700521-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thomas MC, et al. Anal. Chem. 2007;79:5013–5022. doi: 10.1021/ac0702185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murphy RC, et al. Anal. Biochem. 2005;346:1–42. doi: 10.1016/j.ab.2005.04.042. [DOI] [PubMed] [Google Scholar]
- 23.Rouzer CA, Ivanova PT, Byrne MO, Brown HA, Marnett LJ. Biochemistry. 2007;46:6026–6042. doi: 10.1021/bi0621617. [DOI] [PubMed] [Google Scholar]
- 24.Rouzer CA, et al. Biochemistry. 2006;45:14795–14808. doi: 10.1021/bi061723j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zemski Berry K, Murphy RC. Anal. Biochem. 2006;349:118–128. doi: 10.1016/j.ab.2005.11.020. [DOI] [PubMed] [Google Scholar]
- 26.Callender HL, et al. Anal. Chem. 2007;79:263–272. doi: 10.1021/ac061083q. [DOI] [PubMed] [Google Scholar]
- 27.McAnoy AM, Wu CC, Murphy RC. J. Am. Soc. Mass Spectrom. 2005;16:1498–1509. doi: 10.1016/j.jasms.2005.04.017. [DOI] [PubMed] [Google Scholar]
- 28.Shindou H, Shimizu TJ. Biol. Chem. 2009;284:1–5. doi: 10.1074/jbc.R800046200. [DOI] [PubMed] [Google Scholar]
- 29.Long JZ, et al. Nat. Chem. Biol. 2009;5:37–44. doi: 10.1038/nchembio.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scott SA, et al. Nat. Chem. Biol. 2009;5:108–117. doi: 10.1038/nchembio.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hankin JA, Barkley RM, Murphy RC. J. Am. Soc. Mass Spectrom. 2007;18:1646–1652. doi: 10.1016/j.jasms.2007.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ostrowski SG, Van Bell CT, Winograd N, Ewing AG. Science. 2004;305:71–73. doi: 10.1126/science.1099791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carado A, et al. Anal. Chem. 2008;80:7921–7929. doi: 10.1021/ac801712s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McLean JA, Ridenour WB, Caprioli RM. J. Mass Spectrom. 2007;42:1099–1105. doi: 10.1002/jms.1254. [DOI] [PubMed] [Google Scholar]