Abstract
In this study, we considered some competitive learning methods including hard competitive learning and soft competitive learning with/without fixed network dimensionality for reliability analysis in microarrays. In order to have a more extensive view, and keeping in mind that competitive learning methods aim at error minimization or entropy maximization (different kinds of function optimization), we decided to investigate the abilities of mixture decomposition schemes. Therefore, we assert that this study covers the algorithms based on function optimization with particular insistence on different competitive learning methods. The destination is finding the most powerful method according to a pre-specified criterion determined with numerical methods and matrix similarity measures. Furthermore, we should provide an indication showing the intrinsic ability of the dataset to form clusters before we apply a clustering algorithm. Therefore, we proposed Hopkins statistic as a method for finding the intrinsic ability of a data to be clustered. The results show the remarkable ability of Rayleigh mixture model in comparison with other methods in reliability analysis task.
Keywords: Clustering, cluster validity, microarrays, reliability analysis
INTRODUCTION
In DNA microarray technology, we may face the microarray hybridization experiments where a small fraction of the genes will be expressed. The low intensities (unreliable genes that will not appear) may cause higher false positive results in forthcoming researches. Asyali[1] has proposed two classification methods (FCM and NMM) for discrimination of reliable and unreliable data points and compared the results of both approaches against the reference sets constructed by them. The overall agreement between the results of two approaches and their execution times were also reported. Many similar researches have already been conducted.
Wang et al.[2] introduced two classification models for tumor classification and marker gene prediction for microarray data. The noisy gene expression profiles were first summarized into self-organizing maps with optimally selected map units, followed by tumor sample classification using fuzzy c-means (FCM) clustering. The prediction of marker genes of each type of tumor class is then performed by either manual feature selection (model one) or automatic feature selection (model two) using the pair-wise Fisher linear discriminant.
Seo et al.[3] first employed three methods – common scale, location transformation, and the lowest normalization – to normalize both simulated and microarray data sets. The experimental result revealed that the lowest normalization is more robust for clustering of genes than the other two methods for handling noisy data. In the next phase, they used the FCM algorithm to find the groups of genes with similar expression patterns. The experimental result demonstrated that the FCM clustering perform better than hard clustering methods for the same normalized data sets.
Asyali and Alci[1] employed two classification methods – FCM and normal mixture modeling – to analyze the reliability of microarray data. In the normal mixture modeling classification method, the probability density function of microarray data with two bi-variate normal probability density functions was modeled. Before using the expectation-maximization (EM) algorithm for estimation of the mixture model parameters, the parameters were initialized using the k-means algorithm to find an acceptable local maximum and to reduce computational cost. Once the parameters are estimated, the posterior probability that any data point belongs to a certain class was calculated. Finally, the Bayesian decision theory is used to obtain the optimal decision boundary based on the estimated class posterior probabilities.
Gasch and Eisen[4] proposed a modified FCM clustering method that incorporated hierarchical clustering and principle component analysis (PCA) to handle gene expression data that followed the response of yeast cells to environmental changes. In that method, PCA was used to identify the seeding prototype centroids on a random sample of the data; hierarchical clustering was used to identify the number of clusters, and FCM clustering was used to identify conditionally co-expressed genes and to identify regulatory motifs common to the promoters of similarly expressed genes.
Fu and Medico[5] proposed a fuzzy clustering algorithm called FLAME. The algorithm improved the partitioning of genes based on their expression profile. The key features of FLAME include defining neighborhood relations and defining fuzzy membership assignment by local approximation.
Sanguinetti et al.[6] presented a modified PCA method based on a latent variable model known as probabilistic PCA. The method could automatically determine the correct number of principal components in order to select the relevant genes. In that method, an extended probabilistic PCA model with a non-spherical noise distribution that is not independent and identically distributed was proposed. An EM algorithm was used to estimate the parameters of their model. Prominent capabilities of the method included reducing the importance of the genes with large associated variance in the downstream analysis and automatically implementing a cut-off by down-weighting genes with high associated variance.
In this study, we considered some competitive learning methods including hard competitive learning (HCL) and soft competitive learning (SCL) with/without fixed network dimensionality for reliability analysis in microarrays. In order to have a more extensive view, and keeping in mind that competitive learning methods aim at error minimization or entropy maximization (different kinds of function optimization), we decided to investigate the abilities of mixture decomposition schemes. Therefore, we assert that this study covers the algorithms based on function optimization, with particular insistence on different competitive learning methods. The destination is finding the most powerful method according to a pre-specified criterion determined with numerical methods and matrix similarity measures. Furthermore, we should provide an indication showing the intrinsic ability of the dataset to form clusters before we apply a clustering algorithm. Therefore, we proposed Hopkins statistic as a method for finding the intrinsic ability of a data to be clustered.
MATERIALS AND METHODS
Experimental Data
We used three datasets from Asyali[1] that were achieved from three independent experiments of microarray gene expression from the same cell system (monocytic leukemia cell line, THP-1, induced by the endotoxin, LPS). The CDNA was used and contained about 2000 CDNA distinct probes and a total of about 4000 elements. The data consist of Cy3 (green) and Cy5 (red) channel fluorescence signal intensities. More detail about the data set may be found in a previous study.[1]
Comparison of Clustering Methods
In order to compare different clustering methods, we use a hypothesis test.[7,8] In a good clustering, one expects to find a kind of structure, far from a random partitioning. Therefore, we considered our clustering methods (C) in one side and a random partitioning (P) method in other side. Then, we tested how similar was our clustering to a random partitioning.
The Numerical Methods
In order to make a detailed comparison between the proposed method (C) and a random partitioning (P), we selected two samples in our dataset[9] and affixed a label for this pair. The pairs were labeled SS, DD, SD, and Ds, according to a set of clearly formulated rules:
SS: If both samples are located in identical clusters in both clustering methods
DD: If both samples are located in different clusters in both clustering methods
SD: If the samples were located in identical clusters in our clustering methods, but in different clusters in random mode
DS: If the samples were located in identical clusters in random clustering methods, but in different clusters in our method.
Next, we selected the entire possible pairs in the dataset and calculated the described four labels for each pair. The next step is summing up the produced labels in the dataset to calculate:
A = number of SSs
B = number of SDs
C = number of DSs
D = number of DDs
M = A + B + C + D (1)
Then, we should define indexes representing the similarity/dissimilarity of our clustering method to a random partitioning. There is no doubt that the lower correlation shows higher reliability of our method.

where, m1 indicates the number classmates in our clustering and m2 shows the number of classmates in random partitioning. It is obvious that lower indexes represent lower correlation (between the proposed method and a random partitioning) and, consequently, a more reliable clustering method.
The Matrix Similarity Measure
In this section, we have defined another system of measuring the similarity of the proposed method (C) and a random partitioning (P) based on traditional definition of correlation between two systems. For this purpose, we defined two matrixes:
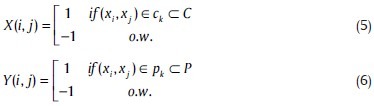
where, ck and pk represent the collection of classmate pairs in the proposed method (C) and in a random partitioning (P), respectively. If we assume that N stands for the number of points in our clustering problem, 1 ≤ i ≤ N, 1 ≤ j ≤ N. The correlation matrix is then formulated as:
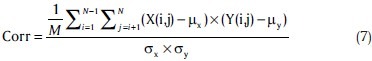
Where, M=N2 and
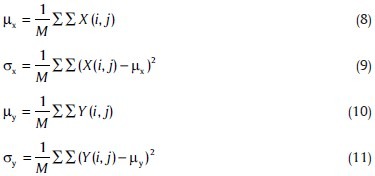
There is no doubt that similar to numerical methods, the lower correlation (between the proposed method and a random partitioning) is an index of more credible clustering algorithm.
Clustering Methods
Our study is mostly based on different competitive learning methods that may be categorized to HCL methods and SCL methods with/without fixed network dimensionality. In order to have a comparison between these methods, we examined the “c-means algorithm” as a representative of HCL methods, the “Neural Gas” and “Neural Gas plus Competitive Hebbian Learning” as an indicator of SCL methods without fixed network dimensionality, and the “Self organizing Map” as a member of SCL methods with fixed network dimensionality. We are aware that the goal of competitive learning methods is error minimization or entropy maximization. Such a goal can introduce competitive learning methods as a subclass in broad category of “clustering schemes based on function optimization.” Other subdivision of function optimization algorithms are mixture models, Fuzzy clustering Algorithms and Maximum Entropy methods. The Normal Mixture Model (NMM) and FCM approach were considered by Asyali and Alci[1] in analyzing the reliability of microarray data. In order to have a more extensive view, we checked Rayleigh Mixture Model and Maximum Entropy methods and we asserted that this study covers the algorithms based on function optimization, with particular insistence on different competitive learning methods.
Competitive Learning Methods
In the area of competitive learning, a rather large number of models exist that have similar goals, but differ considerably in the way they work. A common goal of those algorithms is to distribute a certain number of vectors in a possibly high-dimensional space. The distribution of these vectors should reflect (in one of several possible ways) the probability distribution of the input signals, which in general is not given explicitly but only through sample vectors.
HCL methods
HCL (winner-take-all learning) comprises methods where each input signal only determines the adaptation of one unit, the winner.
The c-means algorithm
The c-means or Isodata Algorithm is one of the most popular and well-known clustering algorithms[10,11] among HCL methods. The squared Euclidean distance was adopted to measure the dissimilarity between vectors and cluster representatives.
The c-means procedure can be described in this algorithm:
Start an initialization and choose two centers (w1, w2). The number of clusters is two in our application since we wanted to discriminate reliable and unreliable points.
-
Choose one sample at random (x) and find the winner center (s):
Winner: s = argmin∥x–wi∥ (12)
-
Update only the winner:
Δws=ε(x–ws) (13)
Stop in the case of little change in centers, otherwise go to 2.
The parameter ε may be selected to have a decreasing exponential form like (18). This method only updates the winner point and, thus, is considered to be a HCL method.
SCL Methods Without Fixed Network Dimensionality
SCL without fixed network dimensionality comprises methods where each input signal determines the adaptation of more than one unit, and no topology of a fixed dimensionality is imposed on the network.
Neural gas clustering
In this method of SCL without fixed network dimensionality, there is no topology at all. In simple words, this algorithm sorts for each input signal the units of the network according to the distance of their reference vector to the input. Based on this “rank order,” a certain number of units are adapted. Both the number of adapted units and the adaptation strength are decreased according to a fixed schedule.
Neural Gas[12,13] gets its principal idea from dynamic of Gas Theory and, similar to K-means clustering, runs with a predetermined number of clusters (chosen to be two in this application), located in arbitrary points of space [wi, i = 1, 2…, N = 4 (number of clusters)]. Then a member of sample space (rth for example) should be chosen randomly and be indexed to the winner cluster which is determined by:
R = argmin∥x–wi∥ (14)
Being known as a soft competitive clustering method, Neural Gas not only updates the cluster center of the winner, but also changes the centers of all other clusters based on their proximity to the randomly selected member of sample space. Therefore, the most approximate wi gets the index k = 0, the second gets the index k = 1, …, and the last winner (loser) gets the index N–1.
The update procedure then can be performed according to the amount of k (a sign of proximity) for each cluster center (wi):
Δwi = ε(t).hτ(ki).(x–wi) (15)
Where,
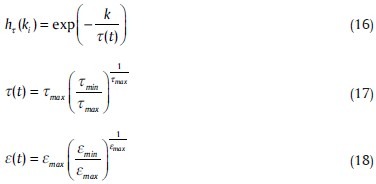
where, t shows the time passing from the start of algorithm, and, according to the formulas, every increment in t, makes a decrease in τ and ε.
Neural gas plus competitive hebbian learning
This method is a straight forward superposition of neural gas and competitive hebbian learning.[14] It is sometimes denoted as “topology-representing networks.” This term, however, is rather general and would also apply to the growing neural gas model. At each adaptation step, a connection between the winner and the second-nearest unit is created (this is competitive hebbian learning). Since the reference vectors are adapted according to the neural gas method, a mechanism is needed to remove edges that are not valid anymore. This is done by a local edge aging mechanism. The complete neural gas which is competitive hebbian learning is as follows:
Initialize
Choose two centers (w)
Using Neural gas (NG) find the index of the winner and the relative indexes for other samples, showing their proximity (i0, i1, i2, i3,…)
Update the centers like NG
If there is no connection between i0, i1: make a connection between these two points: c = c ∪{i0, i1}
Define an age for each connection: age(i0, i1) and increase the age of connections which contain i0: Age(i0, i1) = Age(i0, i1)+1
Remove old connections with age >T(t)
Stop in the case of little change in centers, otherwise go to 2
SCL Methods with Fixed Network Dimensionality
SCL with fixed network dimensionality comprises methods where each input signal determines the adaptation of more than one unit and has a network of a fixed dimensionality like k, which has to be chosen in advance. One advantage of a fixed network dimensionality is that such a network defines a mapping from the n-dimensional input space (with n being arbitrarily large) to a k-dimensional structure. This makes it possible to get a low-dimensional representation of that data, which may be used for visualization purposes.
Self-organizing map
The model is similar to the (much later developed) neural gas model. Since a decaying neighborhood range and adaptation strength are used. An important difference, however, is the topology that is constrained to be a two-dimensional grid and does not change during self organization. The distance on this grid is used to determine how strongly a unit is adapted when the other unit is the winner.
So far, we have implicitly assumed that the representatives wj are not interrelated. We will now remove this assumption. Specifically, for each representative wj, we defined a neighborhood of representatives Qj(t) centered at wj.[15] The neighborhood is defined with respect to the indice j, and it is independent of the distances between representatives in the vector space. If wj wins the current input vector x, all representatives in Qj (t) will be updated. This is the well-known Kohonen self-organizing mapping (SOM) scheme. In its simplest form, SOM may be viewed as a special case of the generalized competitive learning scheme (GCLS). The update formula will be like as follows:
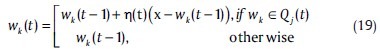
where, η(t) is a variable learning rate. After convergence, the representatives wi are topographically ordered and in a way representative of the distribution of the data. That is, neighboring representatives also lie “close” in terms of their distance in the vector space.
Other Clustering Schemes Based on Function Optimization
One of the most commonly used families of clustering schemes relies on the optimization of a cost function J using differential calculus techniques. The cost J is a function of the vectors of the data set X and it is parameterized in terms of an unknown parameter vector, θ. For most of the schemes of the family, the number of clusters m is assumed to be known. The goal is the estimation of θ that characterizes best the clusters underlying X. Such a goal can introduce competitive learning methods as a subclass in broad category of “clustering schemes based on function optimization.” Other subdivision of function optimization algorithms are mixture models, Fuzzy clustering Algorithms and Maximum Entropy methods.
Rayleigh mixture method
Mixture models are good clustering techniques in cases of known number of clusters and comprise important allotment of “clustering schemes based on function optimization.” The basic reasoning behind this algorithmic family springs from our familiar Bayesian philosophy. Asyali[1] used a Gaussian mixture model in the same data set, but we propose a similar algorithm with Rayleigh distribution. The idea arises from the reality that our dataset consists of two channels of intensities and the proposed mixture model on a two-dimensional space will lie on a distance-based space. If we assume – as proposed by Asyali – that the logarithmic measurement of intensities make them more suitable for Gaussian distribution, there is no doubt that the distances between points will follow a Rayleigh distribution. This method is very similar to NMM method except in the basic formula:
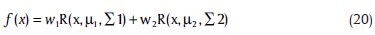
where, R represents the rayleight distribution. We neglected the details of algorithm, which is fully described by Asyali.[1,16] The initial parameters are estimated by k-means clustering and the iterated Expectation and Maximization steps are continued until reaching a change of 0.0001 in parameters or passing from 300 iterations.
Maximum entropy
As mentioned above, we test Maximum Entropy as another subgroup of “clustering schemes based on function optimization.” This method is similar to the c-means algorithm except in the third step (13) of updating the winners.[10] This step should be modified to:
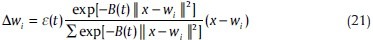
The important difference, as it can be seen in (21), is that Maximum Entropy updates the weights corresponding to all of the points; however, the c means algorithm only updates the weight of the winner. Therefore this algorithm (Maximum Entropy) is categorized as soft clustering.
Clustering Tendency
A common distinguishing quality between the majorities of the clustering algorithms, as discussed in the previous sections, is that they impose a clustering structure on the data set X-even though X may not possess intrinsically any sub-group. In a compact case of X that has a low tendency to be subgrouped, the produced results after the application of a clustering algorithm are not real sub-structures of the data. The problem of verifying whether X inherit a grouping inclination (clustering structure), without identifying it explicitly, is known as clustering tendency.
Test for spatial randomness
We used a test based on Nearest Neighbor Distance and we selected the Hopkins test from this category. Let X’ = {yi, i = 1… M}, M << N, be a set of vectors that are randomly distributed in the sampling window, following the uniform distribution. Also, let X1∈ X be a set of M randomly chosen vectors of X. Let dj be the distance from yj∈ X’ to its closest vector in X1, denoted by xj, and djbe the distance from xj to its closest vector in X1 – {xj}. Then the Hopkins statistic[10,17] involves the l th powers of dj and dj and it is defined as:

This statistic compares the nearest neighbor distribution of the points in X1 with that from the points in X’. When X contains clusters, the distances between nearest neighbor points in X1 are expected to be small, on the average, and, thus, large values of h are expected. Therefore, large values of h indicate the presence of a clustering structure in X.
RESULTS
The implementation of predefined methods is performed under Matlab™ (The math Works Inc., Natick, MA), on a Dell-E6400 Notebook with 4 GB of RAM, running under Windows XP™ operating system.
Comparison of Results for Different Clustering Methods
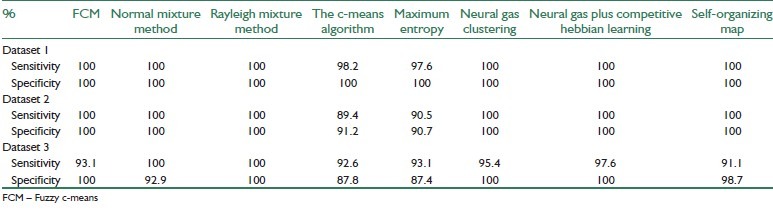
Tables 1 and 2 are represent the classification performance of different methods in comparison with the reference sets. To compare different methods, three criteria were defined as Total Accuracy (TA), sensitivity, and specificity.
Table 1.
The classification performance of different methods in comparison with the reference sets

Table 2.
The classification performance of different methods in comparison with the reference sets
The first criterion is defined as Total Accuracy (TA), in which, the desired method can distinguish reliable genes (in all of three datasets) from unreliable ones. We proposed the below formula for this purpose.
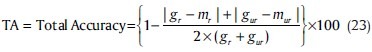
Where, gr and gur represent number of reliable and unreliable genes in target (gold), while mr and mur are similar numbers for each clustering method.
For better demonstration of the results, the sensitivity and specificity criteria, defined below, are applied on the results which can be seen in Table 2.
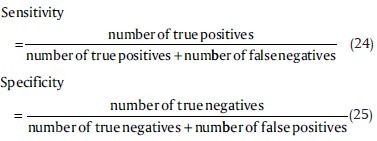
The results reveal that Rayleigh mixture model and Neural Gas plus Competitive Hebbian Learning can surpass the other methods, in accuracy, specificity, and sensitivity.
Comparison of Different Clustering Methods
In this section, we compared the ability of methods by the hypothesis test as described in Comparison of Clustering Methods section. Furthermore, in order to have a control study to prove the validity of hypothesis test in comparison of clustering methods, we used the gene expression data published by Yeoh et al.[18] in 2002.
Numerical methods
The indexes of R, J, and FM are compared in Table 3. Note that the best method should have the lowest amount of mentioned indexes. Table 3is showing one minus the indexes and the interpretation of the results should be based on the higher values. As it is vividly seen in this table, Rayleigh mixture model and Neural Gas plus Competitive Hebbian Learning exceed the other methods. However, one should consider other advantages of the remaining methods, like simplicity of algorithm, speed, and reproducibility to rigorously select the best winner.
Table 3.
Indexes of R, J, and FM
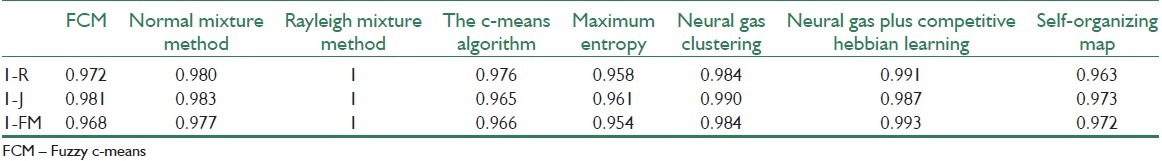
The matrix similarity measure
The correlation can be computed using matrix similarity measure and the results for different methods may be seen in Table 4. Similar to other proposed comparison methods, the results of Table 4demonstrate the exceeding ability of Rayleigh mixture model and Neural Gas plus Competitive Hebbian Learning as compared to other procedures.
Table 4.
Correlation for different methods

Proving the validity of proposed methods
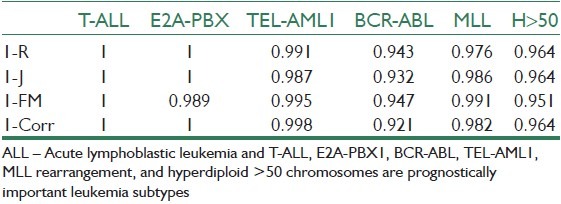
In order to have a control study to prove the validity of hypothesis test in comparison of clustering methods, we used the gene expression data published by Yeoh et al.[18] in 2002. In this dataset, the data samples are labeled to known classes and, consequently, we expect the proposed indexes (R, J, FM, Corr.) take low values when we calculate them on truly classified samples. Table 5 shows the completely low values of indexes for a correct classification that can be considered as a point for justifying the mentioned method for quality assessment of a clustering method.
Table 5.
Indexes of R, J, FM, and correlation on classified results of Yeoh et al
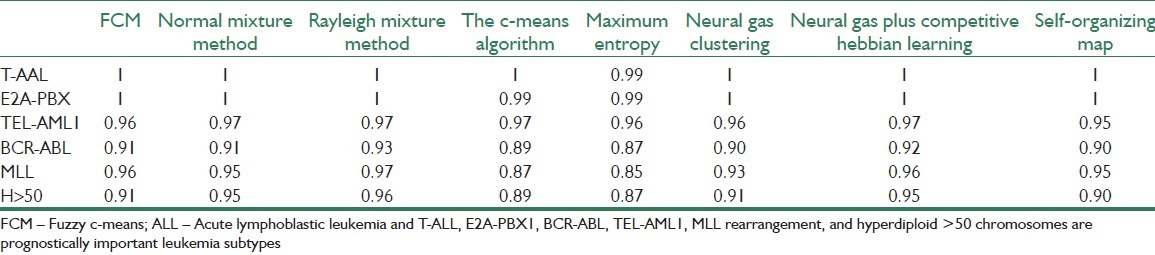
We also tested the proposed clustering algorithms on datasets of Yeoh et al. that were normalized using the z-score method, and the “best” few genes were chosen using Chi Sq gene selection methods. Table 6 demonstrates the J index for comparing different clustering methods.
Table 6.
Index 1-J showing the ability of proposed clustering methods on different dataset of Yeoh et al.
Clustering Tendency
As described in The Matrix Similarity Measure section, we should calculate the measure for ability of a dataset to be clustered. This index is low for very complicated datasets and small sections of set that have no tendency to be classified. On the other hand, it should be high for datasets that have intrinsic ability for being partitioned.
For this purpose, we calculated the clustering tendency index for each of datasets. Furthermore, we tried the index on clustered partitions of each dataset to show if the clustering was able to produce data parts, which are intrinsically compact or not. The result may be found in Table 7.
Table 7.
Clustering tendency index for each of datasets

Table 7 shows that Dataset3 had lower tendency to be clustered (a complicated data population) and this is fully compatible with the weak performance of all clustering methods on this dataset. The next part of this table is another evidence for clustering ability of different method. To clear up, we can see the points clustered to be reliable with Rayleigh mixture method has a very low tendency to be clustered again. While the other methods (sorted with their ability) have a partition that may be clustered again, and this shows the weaker ability of such algorithms.
DISCUSSION AND CONCLUSION
We assert that this study covers the algorithms based on function optimization, with particular insistence on different competitive learning methods. In this study, we considered clustering methods for reliability analysis like Rayleigh mixture method, c-Means Algorithm, Maximum Entropy, Neural Gas clustering, Neural Gas plus Competitive Hebbian Learning, and Self-organizing map. To have a measurement on abilities of different clustering methods, we used numerical- and matrix-based correlation between the proposed methods and a random partitioning and declared the remarkable ability of Rayleigh mixture model in comparison to other methods in reliability analysis task. Furthermore, we calculated an indication of the intrinsic ability of datasets to form clusters using Hopkins statistic; however, as it could be found in results of this paper, Neural Gas plus Competitive Hebbian Learning is the leading competitor of Rayleigh mixture model, according to correlation analysis. Since no statistically meaningful predominance of Rayleigh mixture model is available, we may find both of these methods as the best winners of our proposed platform.
We tried to prove the ability of correlation calculation through numerical- and matrix-based comparison of a clustering method with a random partitioning. Therefore, we used the well-known dataset of Yeoh et al.[18] in two distinct phases. In the first step, we showed that the results of correct classification reveal low values of correlation with a random partitioning. In the second step, we demonstrated the ability of proposed clustering algorithms on datasets of dataset of Yeoh et al. and could prove that Rayleigh mixture model and Neural Gas plus Competitive Hebbian Learning can surpass the other methods, even in this particular dataset.
Lots of other efforts may be suggested to continue the structure of this research. The most important one is evaluation of method on newer and more comprehensive dataset with a more reliable labeling. In addition, the time and computation complexity of methods may be applied as a penalty to their clustering ability and, as a result, finding the winner that can provide the best performance in the least possible time. Besides, the reproducibility of the proposed algorithms can be considered similar to what is fully described in[19].
According to the presented results, we may draw the conclusion that clustering methods that mostly rely on a structural background (like Neural Gas plus Competitive Hebbian Learning) and which are designed based on intrinsic statistical models of the investigated datasets (like Rayleigh mixture model in this application) can surpass the other methods in general.
BIOGRAPHIES

Rahele Kafieh received the BS degree from Sahand University of Technology, Iran, in 2005 and the MS degree from the Isfahan University of Medical Sciences, Iran, in 2008, both in biomedical engineering. She is currently working toward the PhD degree in the Department of Biomedical Engineering at Isfahan University of Medical Sciences. Her research interests are in biomedical image processing, computer vision, graph algorithms, and sparse transforms. She is a student member of the IEEE and the IEEE Signal Processing Society
E-mail: r_kafieh@yahoo.com

Alireza Mehridehnavi was born in Isfahan province at 1961. He had educated in Electronic Engineering at Isfahan University of Technology at 1988. He had finished Master of Engineering in Measurement and Instrumentation at Indian Institute of Technology Roorkee (IIT Roorkee) in India at 1992. He has finished his PhD in Medical Engineering at Liverpool University in UK at 1996. He is an Associate Professor of Medical Engineering at Medical Physics and Engineering Department in Medical School of Isfahan University of Medical Sciences. He is currently visiting at School of Optometry and Visual Science at University of Waterloo in Canada. His research interests are medical optics, devices and signal processing
Footnotes
Source of Support: Nil
Conflict of Interest: None declared.
REFERENCES
- 1.Asyali MH, Alci M. Reliability analysis of microarray data using fuzzy c-means and normal mixture modeling based classification methods. Bioinformatics. 2005;21:644–9. doi: 10.1093/bioinformatics/bti036. [DOI] [PubMed] [Google Scholar]
- 2.Wang J, Bo TH, Jonassen I, Myklebost O, Hovig E. Tumor classification and marker gene prediction by feature selection and fuzzy c-means clustering using microarray data. BMC Bioinformatics. 2003;4:60. doi: 10.1186/1471-2105-4-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim SY, Lee JW, Bae JS. Effect of data normalization on fuzzy clustering of DNA microarray data. BMC Bioinformatics. 2006;7:134. doi: 10.1186/1471-2105-7-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gasch AP, Eisen MB. Exploring the conditional coregulation of yeast gene expression through fuzzy k-means clustering. Genome Biol. 2002;3:1–22. doi: 10.1186/gb-2002-3-11-research0059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fu L, Medico E. A novel fuzzy clustering method for the analysis of DNA micro-array data. BMC Bioinformatics. 2007;8:3. doi: 10.1186/1471-2105-8-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sanguinetti G, Milo M, Rattray M, Lawrence ND. Accounting for probe-level noise in principal component analysis of microarray data. Bioinformatics. 2005;21:3748–54. doi: 10.1093/bioinformatics/bti617. [DOI] [PubMed] [Google Scholar]
- 7.Meila M, Heckerman D. An experimental comparison of model-based clustering methods. Mach Learn. 2001;42:9–29. [Google Scholar]
- 8.Hearst MA, Pedersen JO. Reexamining the cluster hypothesis: Scatter/gather on retrieval results. Proceedings of the 19th annual international ACM SIGIR conference on Research and development in information retrieval. 1996:76–84. [Google Scholar]
- 9.Li G, Ma Q, Liu B, Tang H, Paterson AH, Xu Y. A close-to optimum bi-clustering algorithm for microarray gene expression data. Proc LSS Comput Syst Bioinform. 2009;8:139–49. [Google Scholar]
- 10.Theodoridis S, Koutroumbas K. United States: Academic Press; 2006. Pattern recognition; pp. 572–4. [Google Scholar]
- 11.Duda RO, Hart PE, Stork DG. 2nd ed. New York: John Wiley; 2001. Pattern classification. [Google Scholar]
- 12.Martinetz T, Schulten K. Artificial Neural Networks. Amsterdam: Elsevier; 1991. A neural gas network learns topologies; pp. 397–402. [Google Scholar]
- 13.Canales F, Chacón M. Progress in Pattern Recognition, Image Analysis and Applications. Springer Berlin Heidelberg; 2007. Modification of the growing neural gas algorithm for cluster analysis; pp. 684–93. [Google Scholar]
- 14.Martinetz TM, Schulten KJ. Topology representing networks. Neural Netw. 1994;7:522. [Google Scholar]
- 15.Kohonen T. The self-organizing map. Neurocomputing. 1998;21:1–6. [Google Scholar]
- 16.Dempster AP, Laird NM, Rubin DB. Maximum likelihood from incomplete data via the EM algorithm. J R Stat Soc Series B Stat Methodol. 1977;39:1–38. [Google Scholar]
- 17.Banerjee A, Dave RN. Validating clusters using the Hopkins statistic. The Proceedings of the IEEE International Conference on Fuzzy Systems (FUZZ-IEEE 2004) 2004:149–53. [Google Scholar]
- 18.Yeoh EJ, Ross ME, Shurtleff SA, Williams WK, Patel D, Mahfouz R, et al. Classification, subtype discovery, and prediction of outcome in pediatric acute lymphoblastic leukemia by gene expression profiling. Cancer Cell. 2002;1:133–43. doi: 10.1016/s1535-6108(02)00032-6. [DOI] [PubMed] [Google Scholar]
- 19.Zhang M, Zhang L, Zou J, Yao C, Xiao H, Liu Q, et al. Evaluating reproducibility of differential expression discoveries in microarray studies by considering correlated molecular changes. Bioinformatics. 2009;25:1662–8. doi: 10.1093/bioinformatics/btp295. [DOI] [PMC free article] [PubMed] [Google Scholar]


