Abstract
Some adults fail to adapt to chronic stress, developing symptoms of depression and anxiety. In this issue of Neuron, Uchida and colleagues link maladaptive stress responses to GDNF through a comprehensive investigation of the neurotrophic factor’s regulation. Further, this study is an excellent example for investigators interested in neuroepigenetics research.
Major depressive disorder affects nearly 10% of the adult population in the US and is the country’s leading cause of disability. Many do not respond to treatment and those that do experience a high rate of recurrence. A great deal of attention is focused on developing effective treatments for this debilitating disorder. However, an additionally important goal is prevention (Holtzheimer and Nemeroff, 2006; Avenevoli and Merikangas, 2006). This seemingly simple goal requires unraveling the complexities that underlie the development of depression and the associated risk factors. Early-life stress can predispose individuals to major depressive disorder in adulthood through a variety of mechanisms, including lasting epigenetic modifications (Meaney and Szyf, 2005). As the term suggests, “epigenetics” refers to persisting changes made above the genome. But in addition to early-life stress, chronic stress in adulthood also appears to precipitate depression in some individuals. As we are all too aware, chronic stress is a common experience for adults and has a number of deleterious effects. These range from weakening the strength of our immune system to damaging our mental health (McEwen, 2000). An impressive number of mechanisms have been identified in relation to the development of depression, including epigenetic regulation of the growth factor brain-derived neurotrophic factor (BDNF) (Krishnan and Nestler, 2008), and the field is beginning to understand the contribution of stress through interactions between corticotrophin releasing factor (CRF) and serotonin receptors (Magalhaes et al., 2010). But the glaring question remains: why does stress precipitate depression in some adults, while others are seemingly protected?
In this issue of Neuron, Uchida and colleagues explored the specific contribution of growth factors to stress-induced susceptibility to depression (Uchida et al., 2011). As a model of differential stress response, the authors used the clever approach of comparing two strains of mice known to have different baseline levels of anxiety-like behaviors. Relative to C57BL/6J (B6) mice, BALB/cJ (BALB) mice display high measures of anxiety when tested for things like exploration of the center of an open field and time spent in the arms of an elevated plus maze that lacks walls. Uchida et al. (2011) subjected “low anxiety” B6 and “high anxiety” BALB mice to 6 weeks of mild, daily stress and employed tests designed to assess behaviors associated with symptoms of depression: anxiety (novelty suppressed feeding), despair (forced swim test), anhedonia (sucrose preference test), and avoidance of social situations (social interaction test). Results of the behavior tests indicated that B6 mice adapted well to the chronic stress. BALB mice, on the other hand, experienced an exacerbation of their anxiety-like behaviors and developed depression-like behaviors.
To identify potential growth factors contributing to this differential stress response, the authors next compared transcript levels of nine different neurotrophic factors (e.g., BDNF, GDNF, IGF, etc.) in five different brain regions (e.g., hippocampus, prefrontal cortex, etc.) of the BALB mice with and without chronic stress. GDNF expression in the nucleus accumbens (NAc) emerged as a factor of particular interest. Following chronic stress, GDNF’s transcript and protein levels were decreased in BALB mice but increased in B6. Importantly, the BALB behavioral deficits that correlated with GDNF levels were corrected by GDNF overexpression in the NAc. Based on this convincing data for GDNF’s important role in developing an adaptive stress response, the authors then embarked on a heroic endeavor directed at identifying the mechanism(s) of GDNF misregulation. It almost seems criminal to summarize some of their months-long experiments with a single sentence. Nevertheless, I will do just that—with the goal of clearly conveying the group’s exciting findings.
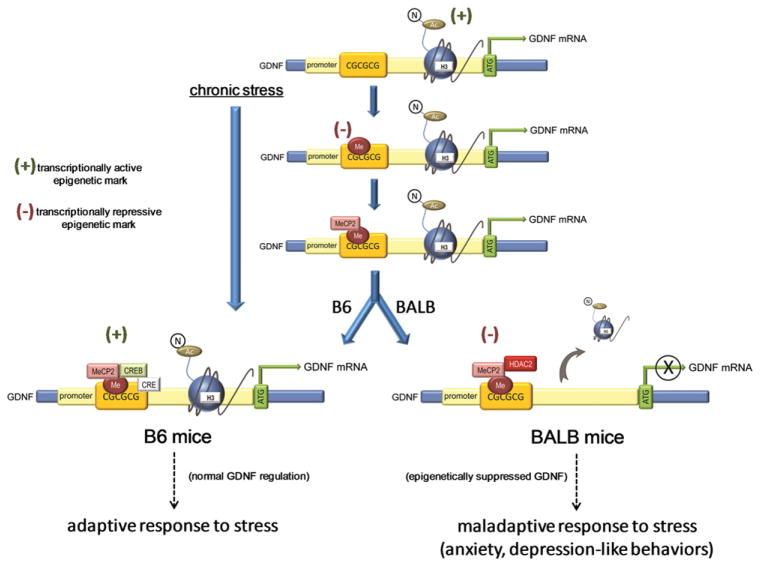
Resequence analysis of the GDNF promoter revealed no differences between strains, so the authors focused their efforts on epigenetic-induced differences in GDNF regulation (Figure 1). Epigenetic mechanisms consist of a set of posttranslational modifications (PTMs) of DNA and histone proteins that produce lasting alterations in chromatin structure and gene expression. Highly basic histone proteins are the major component of chromatin and in its native state, transcription is repressed through tight binding of histones to DNA. This binding prevents the necessary RNA polymerase II enzyme interaction. Therefore, chromatin’s tightly bound structure must be disrupted in order for transcription to occur. This can be achieved through acetylation and phosphorylation of histone tails, as these PTMs are associated with transcriptional activation. The covalent modification of DNA, on the other hand, induces long-term suppression of gene expression through direct interference with transcription factor binding and recruitment of chromatin remodeling enzymes via the action of methyl-CpG binding proteins (MBDs), such as MeCP2. While an oversimplification of the process, cytosines located next to guanines (CpGs) are preferentially methylated. This is particularly true in CpG-rich regions known as CpG islands.
Figure 1. Epigenetic Regulation of GDNF in the “Low Anxiety” B6 and “High Anxiety” BALB Mouse Strains following Chronic Stress.
Chronic stress induces DNA methylation of GDNF’s promoter and recruitment of MeCP2 in the NAc of both low-anxiety B6 mice and high-anxiety BALB mice. The differential transcription rates of GDNF may be accounted for by MeCP2’s recruiting partner. In BALB mice, MeCP2 recruits HDAC2, which presumably deacetylates GDNF’s promoter, leading to the gene’s transcriptional repression. However, in B6 mice, MeCP2 recruits CREB, creating a complex demonstrated to support transcriptional activation. Together with as-yet-unknown mechanisms, epigenetic repression of GDNF leads to maladaptive stress responses in the BALB mice, including anxiety and depression-like behaviors.
The authors began by measuring association between GDNF’s promoter region and the transcriptionally active mark of acetylation on histone H3 (H3Ac). Following stress, H3Ac-GDNF association was decreased in BALB mice but increased in B6 mice. This was consistent with the GDNF transcript levels measured in the previous experiment. Acetyl groups are actively removed from histones by histone deacetylase enzymes (HDACs). Covering all of the class I, II, and IV HDACs, the authors next examined HDACs 1–11 in the NAc and found that only HDAC2 was altered by stress. The HDAC2 increase was associated with GDNF’s promoter and specific to stressed BALB mice, further developing a model where GDNF is actively suppressed with stress by removal of H3Ac from GDNF’s promoter. Histone acetylation can be pharmacologically elevated by treatment with various HDAC inhibitors (HDACi). The HDACi SAHA targets class I HDACs, including HDAC2 (Kilgore et al., 2010). The authors demonstrated that both systemic treatment with SAHA and local viral-mediated knockdown of HDAC2 in the NAc normalized GDNF levels, as well as the anxiety and depression-like behaviors in stressed BALB mice. Conversely, overexpression of HDAC2 further exacerbated the BALBs’ maladaptive behavioral and transcriptional responses to stress.
Histone acetylation can work with DNA methylation to regulate gene transcription and behavior (Miller et al., 2008). Therefore, the authors used bisulfite mapping to detail the specific sites of cytosine methylation within GDNF’s promoter and just downstream of the transcription start site. One promoter CpG site (CpG 2) was hypermethylated in the NAc of both strains of mice with stress, while another was specifically hypermethylated in stressed BALB mice (CpG 3). Stress also increased levels of DNMT 1 and 3a, iso-forms of the enzymes responsible for methylating DNA. Continuous delivery of a DNMT inhibitor into the NAc via an osmotic pump reversed the GDNF hypermethylation, reduction in GDNF mRNA and malapdaptive behavioral stress responses in the BALB mice.
MBDs, such as MeCP2, bind to methylated DNA and repress transcription. Unexpectedly, MeCP2 binding to the GDNF promoter was elevated in both strains of mice following stress. The authors have now observed two putative negative regulators of transcription in both strains with stress. To understand this apparent paradox in epigenetic tagging, the authors next explored the specific proteins that complexed with MeCP2 on GDNF’s promoter. Association of a MeCP2-HDAC2 complex with GDNF’s promoter was specifically increased in stressed BALB mice. As demonstrated in Figure 1, this set of epigenetic changes is consistent with the transcriptional repression of GDNF observed in the stressed BALB mice. On the other hand, in stressed B6 mice, MeCP2-CREB complexed with GDNF’s promoter. The authors’ clever examination of a MeCP2-CREB complex arose from the finding that MeCP2-CREB binding to methylated DNA can have the unexpected effect of activating transcription (Chahrour et al., 2008). In support of this possibility, there is a predicted CRE site adjacent to the CpG that was hypermethylated in both strains (CpG 2). Therefore, this complex is consistent with the transcriptional activation of GDNF measured in stressed B6 mice (Figure 1).
As with any compelling study, the findings of Uchida and colleagues (2011) raise important questions. The authors clearly demonstrate the beneficial behavioral effects of GDNF upregulation in the NAc of stressed BALB mice. Further, the authors recognize that GDNF-HDAC2 interactions are unlikely to be the only factor involved in the BALB’s maladaptive response to stress. Along these lines, it would be very instructive to see if the beneficial effects of SAHA or viral-mediated knockdown of HDAC2 are negated by concomitant knockdown of GDNF. If not, a microarray or deep-sequencing approach could be used to identify additional transcriptional targets regulated by SAHA. In light of the developing case for HDACi treatment of depression (Covington et al., 2009; Grayson et al., 2010), this question of SAHA’s targets under stressful conditions is of particular interest. Along these same lines, the authors examined the effect of SAHA in stressed BALB mice and found that it normalized their social inhibition, anhedonia, and anxiety. It would be interesting to know what effect SAHA would have under the same conditions of stress in B6 mice. Drawing a parallel between the adaptive B6 mice and the human condition introduces a question: what if an individual that is properly coping with daily stress were mistakenly prescribed an HDACi? Could the drug have the potential to shift behavior to the point of removing adaptive inhibitions (e.g., in social situations)?
The work by Uchida and colleagues (2011) also raises a cautionary point with regards to methodology that is relevant to any researcher interested in investigating DNA methylation. While laborious, the authors used the most detailed method of DNA methylation analysis, sodium bisulfite mapping. Unfortunately, a recent discovery has revealed a challenge that all epigeneticists, but particularly those studying the brain, must grapple with. Bisulfite modification, the critical step in sodium bisulfite mapping, protects both 5-methylcytosine (5mC) and a relatively new player, 5-hydroxy-methylcytosine (5hmC). This means that bisulfite mapping cannot distinguish between 5mC and the “sixth base,” 5hmC. This is a particularly serious complication for neuroscientists to consider going forward because the highest levels of 5hmC are found in the brain and its exact function is still unclear (Globisch et al., 2010). Fortunately, new methods of detection have been published in the past few months that will slowly begin to be incorporated into the already complicated toolbox for epigenetic detection.
The findings of Uchida and colleagues (2011) further suggest the intriguing possibility that GDNF serum levels may be predictive of an individual’s coping ability. Interestingly, GDNF serum levels are reported to be lower in patients with major depression and bipolar disorder (Takebayashi et al., 2006), and a positive response to electroconvulsive therapy in patients with pharmacologic-resistant depression has been associated with increased GDNF serum levels (Zhang et al., 2009). Perhaps individuals with a family history of depression may someday benefit from a test of their stress-induced GDNF response and subsequent pharmacologic intervention.
References
- Avenevoli S, Merikangas KR. Am J Prev Med. 2006;31(Suppl 1):S126–S135. doi: 10.1016/j.amepre.2006.07.003. [DOI] [PubMed] [Google Scholar]
- Chahrour M, Jung SY, Shaw C, Zhou X, Wong ST, Qin J, Zoghbi HY. Science. 2008;320:1224–1229. doi: 10.1126/science.1153252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covington HE, 3rd, Maze I, LaPlant QC, Vialou VF, Ohnishi YN, Berton O, Fass DM, Renthal W, Rush AJ, 3rd, Wu EY, et al. J Neurosci. 2009;29:11451–11460. doi: 10.1523/JNEUROSCI.1758-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Globisch D, Münzel M, Müller M, Michalakis S, Wagner M, Koch S, Brückl T, Biel M, Carell T. PLoS ONE. 2010;5:e15367. doi: 10.1371/journal.pone.0015367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grayson DR, Kundakovic M, Sharma RP. Mol Pharmacol. 2010;77:126–135. doi: 10.1124/mol.109.061333. [DOI] [PubMed] [Google Scholar]
- Holtzheimer PE, 3rd, Nemeroff CB. Dialogues Clin Neurosci. 2006;8:175–189. doi: 10.31887/DCNS.2006.8.2/pholtzheimer. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilgore M, Miller CA, Fass DM, Hennig KM, Haggarty SJ, Sweatt JD, Rumbaugh G. Neuropsychopharmacology. 2010;35:870–880. doi: 10.1038/npp.2009.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan V, Nestler EJ. Nature. 2008;455:894–902. doi: 10.1038/nature07455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magalhaes AC, Holmes KD, Dale LB, Comps-Agrar L, Lee D, Yadav PN, Drysdale L, Poulter MO, Roth BL, Pin JP, et al. Nat Neurosci. 2010;13:622–629. doi: 10.1038/nn.2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwen BS. Brain Res. 2000;886:172–189. doi: 10.1016/s0006-8993(00)02950-4. [DOI] [PubMed] [Google Scholar]
- Meaney MJ, Szyf M. Dialogues Clin Neurosci. 2005;7:103–123. doi: 10.31887/DCNS.2005.7.2/mmeaney. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CA, Campbell SL, Sweatt JD. Neurobiol Learn Mem. 2008;89:599–603. doi: 10.1016/j.nlm.2007.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takebayashi M, Hisaoka K, Nishida A, Tsuchioka M, Miyoshi I, Kozuru T, Hikasa S, Okamoto Y, Shinno H, Morinobu S, Yamawaki S. Int J Neuropsychopharmacol. 2006;9:607–612. doi: 10.1017/S1461145705006085. [DOI] [PubMed] [Google Scholar]
- Uchida S, Hara K, Kobayashi A, Otsuki K, Yamagata H, Hobara T, Suzuki T, Miyata N, Watanabe Y. Neuron. 2011;69:359–372. doi: 10.1016/j.neuron.2010.12.023. this issue. [DOI] [PubMed] [Google Scholar]
- Zhang X, Zhang Z, Sha W, Xie C, Xi G, Zhou H, Zhang Y. Psychiatry Res. 2009;170:273–275. doi: 10.1016/j.psychres.2009.01.011. [DOI] [PubMed] [Google Scholar]