Abstract
We sought to delineate the molecular regulatory events involved in the energy substrate preference switch from fatty acids to glucose during cardiac hypertrophic growth. α1-adrenergic agonist–induced hypertrophy of cardiac myocytes in culture resulted in a significant decrease in palmitate oxidation rates and a reduction in the expression of the gene encoding muscle carnitine palmitoyltransferase I (M-CPT I), an enzyme involved in mitochondrial fatty acid uptake. Cardiac myocyte transfection studies demonstrated that M-CPT I promoter activity is repressed during cardiac myocyte hypertrophic growth, an effect that mapped to a peroxisome proliferator–activated receptor-α (PPARα) response element. Ventricular pressure overload studies in mice, together with PPARα overexpression studies in cardiac myocytes, demonstrated that, during hypertrophic growth, cardiac PPARα gene expression falls and its activity is altered at the posttranscriptional level via the extracellular signal–regulated kinase mitogen-activated protein kinase pathway. Hypertrophied myocytes exhibited reduced capacity for cellular lipid homeostasis, as evidenced by intracellular fat accumulation in response to oleate loading. These results indicate that during cardiac hypertrophic growth, PPARα is deactivated at several levels, leading to diminished capacity for myocardial lipid and energy homeostasis.
Introduction
Myocardial energy substrate preference is tightly controlled in mammalian organisms during development and in response to diverse dietary, physiologic, and pathophysiologic conditions (1, 2). During the fetal period, glucose and lactate serve as the chief myocardial energy substrates. After birth and during the postnatal period, myocardial energy is derived increasingly from reducing equivalents generated by mitochondrial β-oxidation of long-chain fatty acids (3). In the normal adult heart, mitochondrial fatty acid oxidation (FAO) accounts for the majority of ATP production (1). The importance of the FAO pathway as a source of energy in the postnatal human heart is underscored by the severe clinical manifestations of genetic defects in mitochondrial FAO enzymes, including childhood cardiomyopathy and sudden death, presumably due to the accumulation of myocardial long-chain fatty acid intermediates coupled with depletion of energy stores (4).
The results of studies performed in cell culture and in vivo have established a critical role for members of the nuclear receptor superfamily in the transcriptional control of genes encoding cardiac FAO enzymes (5–7). Peroxisome proliferator–activated receptor-α (PPARα), a lipid-activated nuclear receptor (8), has been shown to regulate basal and fatty acid–induced transcription of FAO enzyme genes, including medium-chain acyl-CoA dehydrogenase (5) and muscle carnitine palmitoyltransferase I (M-CPT I or CPT Iβ) (9, 10). PPARα binds to target DNA elements as a heterodimeric partner with the retinoid X receptor, and is activated by a variety of ligands, including long-chain fatty acids (11). The expression of mitochondrial and peroxisomal FAO enzymes are reduced in postnatal liver and heart of PPARα-null (PPARα–/–) mice (12, 13). Moreover, PPARα–/– mice accumulate myocardial lipid in the context of conditions known to increase FAO rates, such as fasting, indicating that PPARα plays a critical role in the maintenance of cardiac energy and lipid homeostasis by its regulatory influence on cellular fatty acid utilization pathways (14, 15).
During the development of pressure overloadinduced ventricular hypertrophy, myocardial FAO rates decrease and glucose utilization increases, a reversion to the fetal pattern of energy substrate utilization (16–18). The expression of mitochondrial FAO cycle enzymes is downregulated in parallel with fatty acid utilization rates in the rodent and human hypertrophied and failing heart (19, 20). Recently, we showed that nuclear levels of PPARα fall during the development of pressure overload–induced ventricular hypertrophy in mice (20). These results have suggested that reduced activity of PPARα may be responsible for the downregulated expression of cardiac FAO enzyme genes in the hypertrophied heart.
The objective of this study was to delineate the gene regulatory pathway and upstream signaling events responsible for altered capacity for FAO during cardiac hypertrophic growth. We demonstrate that during hypertrophic growth of cardiac myocytes in culture, palmitate oxidation rates are reduced, and the basal and fatty acid–stimulated expression of the M-CPT I gene is repressed, a response that is linked to reduced activity of PPARα. Our results indicate that during hypertrophic growth, PPARα activity is reduced at the level of gene expression as well as by rapid posttranslational effects involving phosphorylation by the extracellular signal–regulated kinase mitogen-activated protein kinase (ERK-MAPK) pathway. Lastly, the myocyte lipid homeostatic response was shown to be defective in the hypertrophied myocyte, consistent with altered PPARα function. We propose that although this molecular regulatory response may be adaptive early in the hypertrophic process, it could lead to myocardial lipid imbalance, causing a predisposition for pathologic remodeling processes such as contractile dysfunction or sudden death, as occurs in humans with inborn errors in FAO enzymes.
Methods
Primary rat neonatal cardiac myocyte culture.
Cardiac myocytes were prepared as described (7, 9). In brief, 1-day-old Sprague-Dawley rats were euthanized by CO2 inhalation. The right and left ventricles were removed and then digested in 0.2% collagenase (Wako Chemicals USA Inc., Richmond, Virginia, USA). The cells were pooled in DMEM (Sigma Chemical Co., St. Louis, Missouri, USA) containing 10% horse serum and 5% FCS (GIBCO BRL, Gaithersburg, Maryland, USA), and then subjected to differential plating for 1 hour to enrich the myocyte fraction. Nonadherent cells (enriched myocyte fraction) were plated to 50% confluence in dishes that had been pretreated with collagen (Sigma Chemical Co.). After 24 hours, the cell medium was switched to serum-free DMEM containing 0.10 mM 5-bromo-2-deoxyuridine, 10 μg/mL insulin, 10 μg/mL transferrin, and 1 mg/mL fatty acid–free BSA (Sigma Chemical Co.). Myocyte hypertrophy was induced by the addition of phenylephrine (PE; Sigma Chemical Co.) to the serum-free medium, to a final concentration of 100 μM. Oleate complexed to BSA (molar ratio 4:1) was added to the serum-free medium to the final concentration listed in Results. In some experiments, eicosatetraynoic acid (ETYA; Biomol Research Laboratories, Plymouth Meeting, Pennsylvania, USA) was added to serum-free medium to a final concentration of 10 μM. The timing and duration of exposure to oleate, PE, and ETYA is described under Results and in the figure legends. The MAPK pathway inhibitors PD98059 (50 μM) and SB202190 (20 μM) (Calbiochem-Novabiochem Corp., San Diego, California, USA) were added upon switching to serum-free medium in a subset of experiments as described in Results. Neutral lipid within cardiac myocytes was detected by oil red O staining of fixed cells on gelatin-coated cover slips as described (14).
Palmitate oxidation studies.
Measurements of cellular palmitate oxidation rates were performed as described previously (5). In brief, cardiac myocytes were prepared as above and then plated in approximately equivalent numbers in T25 flasks. PE or vehicle control (water) exposure began at the switch to serum-free medium. Forty-eight hours later, the cells were given fresh PE- or vehicle-containing medium containing [1-14C]palmitate (American Radiolabeled Chemicals Inc., St.Louis, Missouri, USA), and #1 Whatman filter paper was suspended within each flask. The flasks were sealed, and 24 hours later the cells were lysed with 6 N hydrochloric acid. The 14CO2 collected overnight on the Whatman paper was liberated by alkalization with 2 N sodium hydroxide, and was quantified by scintillation counting.
Plasmid constructs.
The M-CPT I promoter–luciferase gene reporter plasmids MCPT.Luc.781 and MCPT.Luc.781m1, and the TK promoter–luciferase reporter plasmid (FARE-1)2TKLuc have been described (9). (ACO)3TKLuc was generated by cloning tandem copies of the PPARα response element identified in the rat acyl-CoA oxidase promoter (21) into the BamHI site of the TKLuc backbone vector. (UAS)3TKLuc and the PPARα expression plasmid (pCDM.PPAR) have been described (6, 22). The PPAR-Gal4DBD fusion expression vector was created by subcloning a cDNA encoding mouse PPARα into pCMXGal4 (a gift from David D. Moore, Baylor College of Medicine, Houston, Texas, USA).
Cardiac myocyte transfection.
Transient transfection of rat neonatal cardiac myocytes was performed as described using a modified calcium phosphate coprecipitation method (7, 9). Using green fluorescent protein as a marker, we have shown that this approach results in gene transfer in 5–10% of the cultured myocytes (data not shown). For each transfection, 4 μg of reporter plasmid was cotransfected with either 500 ng of SV40–β-Gal (a plasmid containing a β-galactosidase gene downstream of the simian virus promoter), or 100 ng of RSV β-Gal (the β-galactosidase gene downstream of the Rous sarcoma virus promoter), to control for transfection efficiency. Cotransfection experiments were performed with 0.125–1.0 μg of pCDM.PPAR; an appropriate amount of vector backbone was added to normalize the total amount of plasmid DNA per well. Luciferase activities were determined by the standard luciferin-ATP assay (PharMingen, San Diego, California, USA), and β-galactosidase activity was measured by the Galacto-Light chemiluminescence assay (Tropix, Bedford, Massachusetts, USA) in a Monolight 2010 luminometer from Analytical Luminescence Laboratory (San Diego, California, USA).
In vitro kinase assays.
Bacterial expression constructs for mouse PPARα-GST fusion proteins containing an NH2-FLAG epitope were created in pGEX-4T-1 (Amersham Pharmacia Biotech Inc., Piscataway, New Jersey, USA). Site-directed mutagenesis was performed using the QuickChange kit (Stratagene, La Jolla, California, USA). Recombinant wild-type and mutant proteins were produced in the BL21 bacterial strain as described (6). Partially purified proteins were incubated with activated ERK2 (Stratagene) and [γ-32P]ATP for 30 minutes at 30°C. Labeled proteins were resolved on SDS-PAGE, transferred to nitrocellulose, and visualized by autoradiography. Western blot analysis of the same membranes was performed with an anti-FLAG M2 antibody (Sigma Chemical Co.) to control for loading differences.
RNA blot analysis.
Northern blot studies were performed as described (9). Total RNA was isolated from rat neonatal cardiac myocytes in cell culture or from the left ventricles of mice subjected to sham operation or transverse aortic constriction (TAC). The probes were derived from cDNAs encoding rat M-CPT I (9), a human β-actin (9), murine very-long-chain acyl-CoA dehydrogenase (23), murine medium chain acyl-CoA dehydrogenase (MCAD) (7), murine PPARα (20), rat acyl-CoA oxidase (ACO) (14), and rat atrial natriuretic factor (20). Band intensities were quantified by phosphorimaging using a GS 525 Molecular Imager System (Bio-Rad Laboratories, Hercules, California, USA).
Animal studies.
The protocol for the surgical placement of a ligature to produce constriction of the transverse aorta in mice has been described (20, 24). Adult C57BL/6 × SJL/J mice (aged 3–4 months) were used for the transverse aortic constriction TAC experiments shown in Figure 5. All animal experiments and euthanasia protocols were conducted in strict accordance with the National Institutes of Health guidelines for humane treatment of laboratory animals. All animal experiment protocols were reviewed and approved by the Animal Care Committee of Washington University.
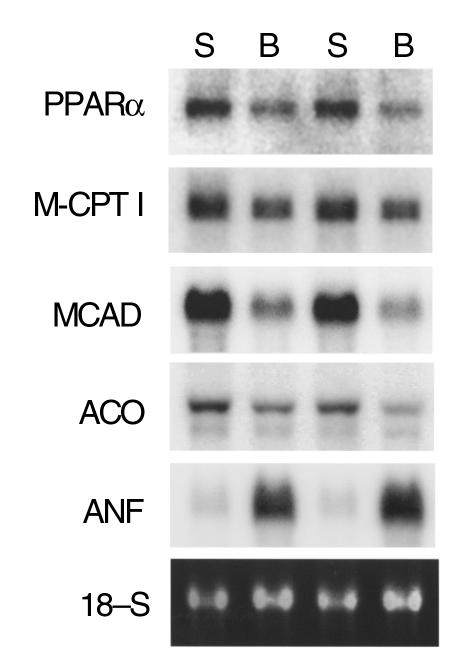
Figure 5.
Downregulation of PPAR-α and FAO enzyme gene expression in the pressure overload–induced hypertrophied mouse heart. Representative autoradiograph of a Northern blot analysis performed with total RNA isolated from the left ventricles of mice 7 days after placement of a constricting band around the transverse aortic arch (B) or sham operation (S). Ten micrograms of RNA was loaded per lane. The blot was sequentially hybridized with the radiolabeled cDNA probes indicated at left (described under Methods). 18S ribosomal RNA stained with ethidium bromide and visualized by UV fluorescence is shown as a loading control. This figure is representative of the results obtained across four pairs of age-matched, banded (n = 7) mice and sham-operated control (n = 7) littermates (aged 3–5 months).
Statistical analysis.
All data are presented as mean ± SEM. Differences between mean values obtained for palmitate oxidation studies, Northern blot studies, and transfection experiments were determined by an unpaired Student’s t test or one-factor ANOVA coupled to the Scheffe test. P < 0.05 was considered significant.
Results
Palmitate oxidation rates and the expression of the M-CPT I gene decrease during α1-adrenergic agonist–induced hypertrophy of rat neonatal cardiac myocytes.
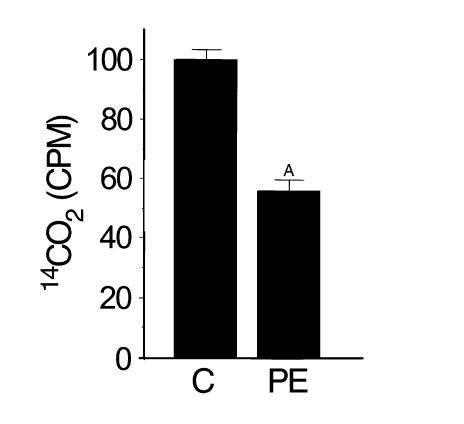
To determine whether hypertrophic growth of cardiac myocytes in culture is associated with a reduced capacity for FAO, we determined the rate of oxidative conversion of [1-14C]palmitate to 14CO2 in primary cultures of rat neonatal cardiac myocytes after exposure to the α1-adrenergic agonist, PE. For these experiments, myocytes were exposed to PE or vehicle for 48 hours in serum-free media (see Methods) before the addition of [1-14C]palmitate. Induction of myocyte hypertrophy by PE was confirmed by an increase in cell size and in atrial natriuretic factor mRNA levels (data not shown). Mean palmitate oxidation rates were 42% lower in PE-treated cells than in control cells (P < 0.001; Figure 1) indicating that, as in the intact heart, FAO rates are reduced in the hypertrophied myocyte in culture. These results also demonstrate that alterations in myocyte lipid utilization occur rapidly after exposure to a stimulus for hypertrophic growth.
Figure 1.
Palmitate oxidation rates decrease during cardiac myocyte hypertrophy. Shown is total 14CO2 (in counts per minute, CPM) elaborated over a 24-hour period by the oxidation of [1-14C]palmitate in cardiac myocytes after exposure to either the α1-adrenergic agonist PE (100 μM) or vehicle (water) control (C) for 72 hours. CPM was determined by scintillation counting (as described in Methods and corrected for cell number), and was normalized to the value obtained in vehicle-treated control cells (= 100%). ASignificantly different (P < 0.001 by Student’s t test) from control. These results represent the mean ± SEM of duplicate conditions in three independent experiments.
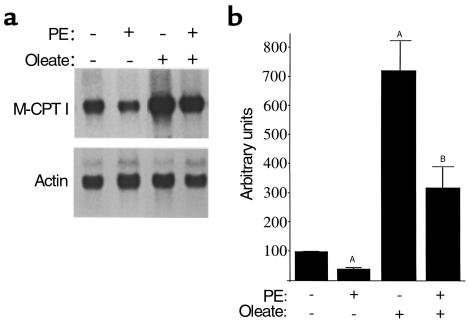
To characterize the gene regulatory events associated with the reduction of FAO rates during cardiac myocyte hypertrophy, we focused on M-CPT I, an enzyme that catalyzes the rate-limiting step in the mitochondrial import of long-chain fatty acids before entering the β-oxidation cycle. CPT I activity has been shown to be a major determinant of mitochondrial FAO flux (25). We and others have demonstrated that M-CPT I expression is controlled at the level of gene transcription by long-chain fatty acids (9, 10, 26). Basal and oleate-induced M-CPT I mRNA levels were determined by RNA blot analysis. This was performed with total RNA isolated from myocytes exposed to vehicle, oleate, PE, or oleate plus PE. M-CPT I mRNA levels were 55–65% lower in myocytes exposed to PE than in vehicle-treated control cells (P < 0.05; Figure 2). As expected, exposure to oleate induced M-CPT I gene expression approximately sevenfold compared with vehicle-treated controls (Figure 2). The oleate-stimulated induction of M-CPT I gene expression was blunted by more than 50% after exposure to PE (P < 0.05; Figure 2). Thus, basal and fatty acid–induced M-CPT I gene expression is significantly repressed in the hypertrophied myocyte.
Figure 2.
Basal and fatty acid–activated M-CPT I gene expression is repressed during cardiac myocyte hypertrophy. (a) Representative autoradiograph of Northern blot analyses performed with total RNA isolated from rat neonatal cardiac myocytes in culture. Each lane contained 10 μg of total RNA isolated from cardiac myocytes incubated in the presence of 100 μM PE, 250 μM oleate complexed to BSA, or vehicle control (water, BSA, or both). The blot was sequentially hybridized with radiolabeled cDNA probes encoding M-CPT I or β-actin. (b) Bars represent mean (± SEM) steady-state M-CPT I mRNA levels as determined by phosphorimage analysis of bands on Northern blots of RNA obtained from at least four separate experiments. Values shown are arbitrary units corrected to actin signal intensity and normalized (= 1.0) to the value obtained with vehicle alone. ASignificantly different (P < 0.05; ANOVA coupled to Scheffe test) from the values obtained from samples prepared from cells exposed to vehicle alone. BSignificantly different from values obtained with cells treated with oleate alone.
PPARα-mediated activation of M-CPT I gene expression is repressed in the hypertrophied cardiac myocyte.
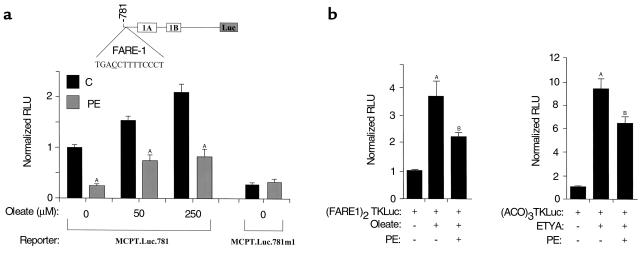
Activation of M-CPT I gene expression by oleate occurs at the transcriptional level via the action of the lipid-activated nuclear receptor PPARα (9). To determine whether α1-adrenergic agonist–mediated repression of M-CPT I gene expression occurs because of interference with the PPARα regulatory pathway, we performed transfection experiments with a fragment of the human M-CPT I gene promoter that contains the oleate/PPARα-responsive element FARE-1 (9), fused to a luciferase reporter (MCPT.Luc.781; Figure 3a). Cardiac myocytes were incubated for 24 hours in serum-free medium containing PE or vehicle before transient transfection with MCPT.Luc.781. Twelve hours after transfection, the cells were incubated with media containing oleate (complexed to BSA) at several concentrations in the presence or absence of PE. As expected, MCPT.Luc.781 activity was induced in cells treated with oleate (at 50 μM and 250 μM) relative to the activity in cells treated with BSA alone (Figure 3a). PE repressed both basal and oleate-stimulated MCPTI.Luc.781 activity by 50–75% at each of the oleate concentrations tested, and ablated the incremental increase in reporter activity that occurred in the presence of 250 μM oleate compared with 50 μM oleate (Figure 3a). In contrast, a construct containing a substitution mutation in the FARE-1 element known to abolish PPAR binding and oleate-mediated activation (MCPT.Luc.781.M1) (9) was not repressed by PE (Figure 3a).
Figure 3.
PPARα-mediated transcriptional control of MCPT.Luc.781 is blocked in the hypertrophied cardiac myocyte. (a) Shown is the homologous promoter–reporter plasmid, MCPT.Luc.781, containing the PPARα response element, FARE-1, located upstream of two untranslated exons, 1A and 1B (9, 26) (top). Either MCPT.Luc.781 or a construct containing FARE-1 mutated at the position underlined in the FARE-1 DNA sequence (MCPT.Luc.781.m1) was transfected into rat neonatal cardiac myocytes in serum-free media, followed by a 60-hour exposure to either vehicle (water) control or PE. Exposure to oleate (50 μM or 250 μM) or vehicle (0) began 12 hours after transfection and was continued for 48 hours. Bars represent mean (± SEM) luciferase activity (in relative luciferase units, or RLU) in cardiac myocytes exposed to the indicated concentrations of oleate, and incubated in the absence (C) or presence of PE. Values shown were corrected for transfection efficiency using the activity of cotransfected pSV40–β-Gal plasmid and normalized (= 1.0) to the values obtained with cells exposed to vehicle alone. ASignificantly different from control cardiac myocytes. (b) Activity of the heterologous promoter–luciferase gene reporter plasmid (FARE1)2TKLuc (left) or (ACO)3TKLuc (right) in the presence of pCDM.PPAR. The values shown are RLU corrected for the activity of cotransfected pSV40–β-Gal and are normalized (= 1.0) to the activity of the reporter construct in identically treated cells cotransfected with vector backbone [pCDM(-)] and exposed to vehicle. The data shown represent the mean (± SEM) of three independent experiments. ASignificantly different (P < 0.05) from the control value. BSignificantly different from value obtained in the presence of oleate or ETYA without PE added.
To determine whether FARE-1 was sufficient to confer the hypertrophy-mediated repression of the M-CPT I gene reporter construct, and to further investigate the direct involvement of the PPARα regulatory pathway in this response, myocyte cotransfection experiments were repeated using a luciferase reporter construct containing two copies of FARE-1 upstream of a heterologous promoter driving the luciferase gene [(FARE1)2TKLuc]. For these experiments, cardiac myocytes were cotransfected with pCDM.PPAR (or the expression vector backbone alone), as the amount of endogenous PPARα in neonatal cardiac myocytes in culture is limiting with regard to activation of this PPARα target reporter plasmid (9). Activity of (FARE1)2TKLuc was induced approximately fourfold in the presence of overexpressed PPARα and its exogenously added activator, oleate. Exposure to PE reduced the level of PPARα/oleate-mediated activation of (FARE1)2TKLuc by approximately 40% (P < 0.05; Figure 3b).
To determine whether the antagonistic effect of PE-induced myocyte hypertrophy on PPARα-mediated activation of the M-CPT I gene could be generalized to other PPARα response elements, the transfection experiments were repeated using (ACO)3TKLuc, a luciferase reporter construct containing three copies of a PPARα response element derived from the peroxisomal ACO gene. For these latter experiments, the PPARα ligand, ETYA, was used to determine whether the hypertrophy effect generalized to PPARα ligands in addition to oleate. As expected, (ACO)3TKLuc activity was activated by PPARα + ETYA (increasing nearly tenfold). As with the FARE-1–containing reporter, the PPARα-mediated activation of (ACO)3TKLuc was significantly blunted by exposure to PE (Figure 3b). Collectively, these data indicate that during hypertrophic growth of the cardiac myocyte, the activity of PPARα is diminished.
A role for ERK-MAPK in the deactivation of the PPARα regulatory pathway during cardiac myocyte hypertrophic growth.
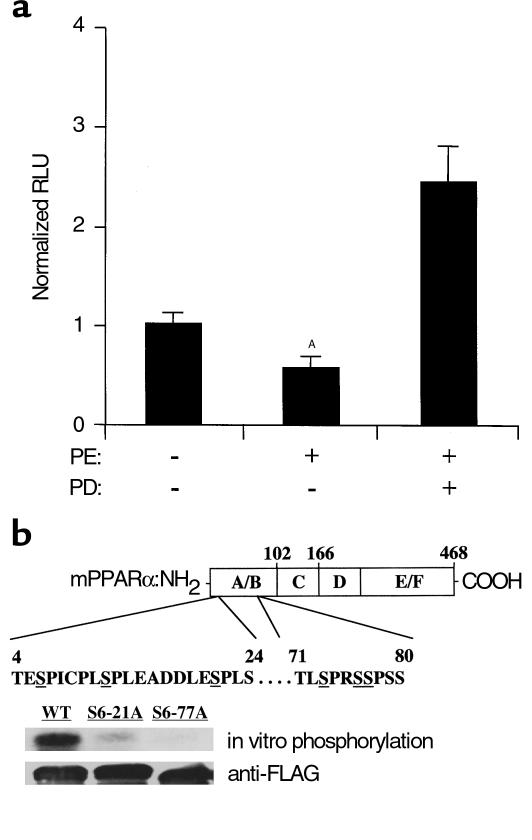
We sought to identify the upstream signaling pathway involved in the downregulation of M-CPT I gene transcription during α1-adrenergic agonist–induced myocyte hypertrophy. MAPK pathways are known to be activated during myocyte hypertrophic growth (27). To explore the possibility that one of the MAPK pathways serves as a link between G protein–coupled receptor activation and reduced activity of PPARα during PE-induced myocyte hypertrophy, transfection experiments were repeated with MCPT.Luc.781 in the presence or absence of MAPK pathway–selective inhibitors. An inhibitor of the p38-MAPK pathway (SB202190) had no effect on the repressive effects of PE on MCPT.Luc.781 (data not shown). In contrast, PD98059, a known inhibitor of the ERK-MAPK pathway, prevented the PE-mediated repression of MCPT.Luc.781 and resulted in an approximately 2.5-fold activation of the reporter above basal levels (Figure 4a).
Figure 4.
Inhibition of the ERK-MAPK pathway blocks the repressive effects of PE on PPARα-mediated control of MCPT.Luc.781. (a) Rat neonatal cardiac myocytes in serum-free media were transfected with MCPT.Luc.781, followed 12 hours later by exposure to PE, PD98059 (PD; 50 μM), or vehicle (DMSO and water). Bars represent mean luciferase activity (RLU) corrected for pSV40–β-Gal activity. ASignificantly different from control. (b) Top: Schematic diagram of mouse PPARα showing the location of putative MAPK recognition sites (potential target serines are underlined). Center: Gel autoradiograph containing samples from the in vitro phosphorylation studies performed with activated ERK2 and bacterially expressed, FLAG epitope–tagged, wild-type (WT) PPARα or mutant PPARα proteins containing serine-to-alanine mutations, either at amino acids 6, 12, and 21 (S6-21A), or at amino acids 6, 12, 21, 73, 76, and 77 (S6-77A). Bottom: Western blot analysis of the phosphoprotein samples using anti-FLAG antisera (to control for loading).
To determine whether PPARα is phosphorylated by ERK, in vitro kinase experiments were performed with PPARα protein produced in a bacterial expression system. Activated ERK2 phosphorylated PPARα, as demonstrated by SDS-PAGE autoradiography (Figure 4b). Examination of the mouse PPARα amino acid sequence revealed a series of potential MAPK phosphorylation sites (consensus PxS/TP) located in the NH2-terminal AB domain between amino acids 4 and 80 (Figure 4b). To confirm that these sequences serve as MAPK phosphorylation sites, the kinase experiments were repeated with mutant PPARα proteins containing a serine-to-alanine substitution, either in three (S6-21A) or in all six (S6-77A) of the putative sites. ERK-mediated phosphorylation was significantly reduced with the S6-21A mutant and was abolished with the S6-77A mutant, compared with the wild-type protein (Figure 4b). These results confirm that PPARα contains ERK phosphorylation recognition sites.
PPARα gene expression is downregulated in vivo in response to ventricular pressure overload.
The results shown above indicate that the activity of PPARα is altered at the posttranslational level during myocyte hypertrophy. However, we have shown previously that the nuclear levels of PPARα fall in the hypertrophied heart (20), suggesting that its expression is also reduced in the hypertrophied myocyte. To explore this possibility and to establish an in vivo correlate of the myocyte hypertrophy experiments described above, PPARα gene expression was evaluated in an established in vivo murine pressure-overload model. For these experiments, TAC was achieved by surgical placement of a ligature (20, 24). Northern blot analysis was performed with total RNA isolated from the left ventricles of mice 7 days after TAC or sham operation. We have shown previously that the ratio of left ventricle to body weight increases significantly within 7 days of the TAC procedure (24). Expression of the gene encoding atrial natriuretic factor, a known marker of ventricular hypertrophy, was higher in the pressure-overloaded ventricles than in control ventricles (Figure 5). As expected, the expression of several PPARα target genes encoding the mitochondrial FAO enzymes M-CPT I and MCAD was downregulated in the pressure-overloaded ventricles (Figure 5). The left ventricle expression of the PPARα target gene encoding ACO, a peroxisomal FAO enzyme, was also modestly reduced in the TAC group compared with the sham-operated group (Figure 5). Importantly, mean PPARα mRNA levels were 39% lower in the pressure-overloaded left ventricle of the banded mice than in sham-operated control mice (P < 0.05; representative autoradiograph shown in Figure 5). These results demonstrate that in addition to posttranslational deactivation of the PPARα regulatory pathway, the expression of the PPARα gene is downregulated during pressure overload–induced hypertrophy.
Altered lipid homeostasis in the hypertrophied cardiac myocyte.
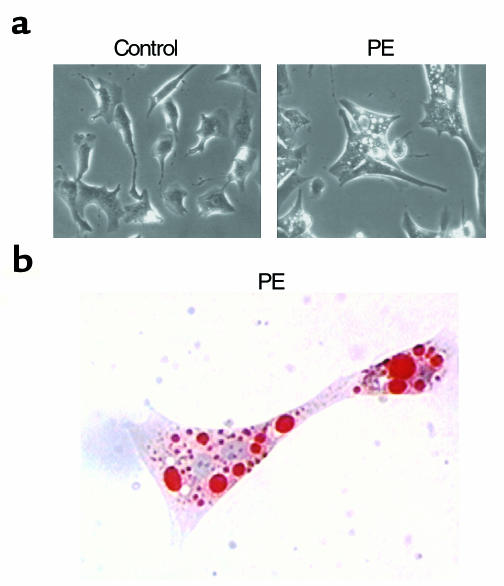
Previous studies have shown that PPARα-null mice have a reduced capacity for myocardial and hepatic lipid homeostasis in conditions where cellular lipid import exceeds utilization capacity. Fasting (15) or pharmacologic inhibition of mitochondrial FAO flux (14) results in a dramatic phenotype in PPARα–/– mice, characterized by accumulation of triglyceride-containing lipid droplets in cardiac myocytes and hepatocytes. To determine whether a similar defect in cellular lipid homeostatic capacity occurs in hypertrophied myocytes in culture due to reduced activity of PPARα, fatty acid loading experiments were performed. Cardiac myocytes were exposed to a medium containing 500 μM oleate in the presence or absence of PE. As demonstrated by phase-contrast microscopy (Figure 6), the hypertrophied, oleate-treated myocytes contained numerous cytoplasmic droplets compared with control, oleate-treated cells that exhibited minimal lipid droplet accumulation in the absence of PE-induced hypertrophy (Figure 6, top). Oil red O staining of the oleate-loaded, PE-treated cardiac myocytes confirmed that the intracellular droplets contained neutral lipid (Figure 6, bottom). These results provide a histologic correlate of the [1-14C]palmitate oxidation studies shown in Figure 1, and demonstrate that the hypertrophied cardiac myocyte exhibits reduced capacity to catabolize long-chain fatty acids, consistent with a defect in the PPARα regulatory pathway.
Figure 6.
Neutral lipid accumulates in hypertrophied cardiac myocytes. (a) Phase-contrast photomicrographs (×320) of cardiac myocytes incubated for 90 hours in serum-free medium containing 500 μM oleate and either PE (100 μM) or vehicle control. (b) Myocytes incubated in 500 μM oleate and stained with oil red O after exposure to PE (×400).
Discussion
The expression of genes encoding mitochondrial FAO cycle enzymes is downregulated in parallel with the reprogramming of cardiac energy metabolism during the development of cardiac hypertrophy (19, 20). In this study, we sought to define the regulatory pathway involved in this response, and to characterize the lipid metabolic alterations that occur in the hypertrophied cardiac myocyte. Our results demonstrate that the nuclear receptor PPARα is deactivated at pre- and posttranslational levels during cardiac hypertrophic growth, leading to a reduction in the capacity for FAO and cellular lipid homeostasis.
Our data indicate that the PPARα-mediated control of M-CPT I gene expression is altered at several levels during hypertrophic growth of the cardiac myocyte. Northern blotting studies revealed that PPARα gene expression is reduced within 7 days of left ventricular pressure overload. In addition, α1-adrenergic agonist–induced myocyte hypertrophy was associated with a rapid reduction in PPARα activity in a system in which recombinant PPARα was overexpressed, implicating posttranslational regulatory mechanisms. Examination of the putative upstream signaling events involved in the posttranslational reduction in PPARα activity during myocyte hypertrophy identified a role for the ERK-MAPK pathway. Our results are consistent with a previous study demonstrating phosphorylation of PPARα by ERK in hepatoma cells in culture (28). The ERK-MAPK pathway has also been shown to phosphorylate and inhibit PPARγ activity in noncardiac cell lines (29). Possible mechanisms for the decrease in PPARα activity in response to phosphorylation by ERK include effects on ligand binding, nuclear localization, or interaction with transcriptional coactivators or corepressors. It is possible that during the early stages of hypertrophic growth, PPARα activity is reduced through posttranslational mechanisms, including ERK-mediated phosphorylation of PPARα, followed by long-term effects on PPARα gene expression.
The results of previous studies have demonstrated that during cardiac hypertrophic growth, a number of protein kinase cascades are activated, including the ERK-, JNK-, and p38-MAPKs (27). The role of ERK-MAPK activation in the cardiac hypertrophy program is controversial. Overexpression of constitutively active Ras, an upstream activator of the ERK-MAPK pathway, induces changes in gene expression and cellular morphology typical of the hypertrophy phenotype in cultured myocytes (30). Ras activity was shown to be required for α1-adrenergic agonist–induced hypertrophy, as demonstrated by dominant-negative Ras experiments (31). Constitutively active MEK, the kinase that activates ERK, has also been shown to lead to typical hypertrophic changes in gene expression (32). However, other signal transduction circuits have also been implicated as components of the cardiac hypertrophy regulatory program, including p38-MAPK (33), JNK-MAPK (34), and the calcineurin-NFAT (35) pathway. Accordingly, the collective results of previous studies indicate that multiple signaling pathways are activated during cardiac hypertrophic growth. An unanswered question is whether specific signal transduction pathways activate distinct target genes. Our results suggest that the ERK limb of the MAPK pathway regulates metabolic gene targets during cardiac hypertrophic growth. Specifically, our results from experiments with the MEK inhibitor PD98059 indicate that during α1-adrenergic agonist–induced myocyte growth, activated ERK phosphorylates PPARα, leading to downregulated expression of FAO enzyme genes, an effect not observed with inhibition of the p38-MAPK pathway (data not shown).
Pressure overload–induced cardiac hypertrophy is associated with reduced myocardial FAO rates. This metabolic response is probably initially adaptive from an oxygen consumption standpoint, given that ATP production via FAO requires greater oxygen utilization per mole of substrate than does glucose oxidation. However, given the pivotal role played by PPARα in the maintenance of cellular lipid and energy balance, the metabolic regulatory response described here would be predicted to become maladaptive in vivo for several reasons. First, energy reserves may become limited given that the amount of ATP produced per mole of energy substrate is greater for fatty acids than for glucose. Second, as PPARα activity falls, leading to reduced expression of key enzymes involved in cellular mitochondrial and peroxisomal FAO, the capacity for maintenance of cellular lipid homeostasis becomes limited. This prediction is supported by the results shown here demonstrating marked intracellular lipid accumulation in response to oleate loading in hypertrophied cardiac myocytes. The pivotal role played by PPARα in the maintenance of cellular lipid homeostasis is further underscored by the observation that PPARα-null mice develop massive myocyte lipid accumulation in response to stressors known to increase myocardial intracellular lipid (14, 15). Third, reduced mitochondrial FAO enzyme expression could lead to the accumulation of long-chain fatty acid intermediates, such as acylcarnitines, which have been implicated in the genesis of ventricular rhythm disorders during myocardial ischemia (36) and in patients with cardiomyopathy due to inborn errors of FAO (37). Taken together, these results suggest the intriguing possibility that reduced capacity to maintain lipid and energy homeostasis in the hypertrophied myocyte leads to contractile dysfunction or cardiac rhythm disturbances, two signatures of pathologic remodeling of the hypertrophied heart.
Acknowledgments
Special thanks to Kelly Hall for assistance with preparation of the manuscript. This work was supported by the National Institutes of Health (NIH) (grants RO1 HL-58493 and P50 HL-61006). D.P. Kelly is an Established Investigator of the American Heart Association. J.M. Brandt was supported by an individual National Research Service Award grant (NHLBI F32-HL09799) during this work. P.M. Barger is supported by an NIH K08 award (K08 HL03808).
Footnotes
Philip M. Barger and Jon M. Brandt contributed equally to this work.
References
- 1.Bing RJ. The metabolism of the heart. Harvey Lect. 1955;50:27–70. [PubMed] [Google Scholar]
- 2.Bremer, J., and Osmundson, H. 1984. Fatty acid oxidation and its regulation. In Fatty acid metabolism and its regulation. S. Numa, editor. Elsevier Science. Amsterdam, The Netherlands. 113–154.
- 3.Wittels B, Bressler R. Lipid metabolism in the newborn heart. J Clin Invest. 1965;44:1639–1646. doi: 10.1172/JCI105270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kelly DP, Strauss AW. Inherited cardiomyopathies. N Engl J Med. 1994;330:913–919. doi: 10.1056/NEJM199403313301308. [DOI] [PubMed] [Google Scholar]
- 5.Gulick T, Cresci S, Caira T, Moore DD, Kelly DP. The peroxisome proliferator activated receptor regulates mitochondrial fatty acid oxidative enzyme gene expression. Proc Natl Acad Sci USA. 1994;91:11012–11016. doi: 10.1073/pnas.91.23.11012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carter ME, Gulick T, Moore DD, Kelly DP. A pleiotropic element in the medium-chain acyl coenzyme A dehydrogenase gene promoter mediates transcriptional regulation by multiple nuclear receptor transcription factors and defines novel receptor-DNA binding motifs. Mol Cell Biol. 1994;14:4360–4372. doi: 10.1128/mcb.14.7.4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Disch DL, et al. Transcriptional control of a nuclear gene encoding a mitochondrial fatty acid oxidation enzyme in transgenic mice: role for nuclear receptors in cardiac and brown adipose expression. Mol Cell Biol. 1996;16:4043–4051. doi: 10.1128/mcb.16.8.4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Issemann I, Green S. Activation of a member of the steroid hormone receptor superfamily by peroxisome proliferators. Nature. 1990;347:645–650. doi: 10.1038/347645a0. [DOI] [PubMed] [Google Scholar]
- 9.Brandt J, Djouadi F, Kelly DP. Fatty acids activate transcription of the muscle carnitine palmitoyltransferase I gene in cardiac myocytes via the peroxisome proliferator-activated receptor α. J Biol Chem. 1998;273:23786–23793. doi: 10.1074/jbc.273.37.23786. [DOI] [PubMed] [Google Scholar]
- 10.Mascaró C, et al. Control of human muscle-type carnitine palmitoyltransferase I gene transcription by peroxisome proliferator-activated receptor. J Biol Chem. 1998;273:8560–8563. doi: 10.1074/jbc.273.15.8560. [DOI] [PubMed] [Google Scholar]
- 11.Forman BM, Chen J, Evans RM. Hypolipidemic drugs, polyunsaturated fatty acids, and eicosanoids are ligands for peroxisome proliferator-activated receptors α and δ. Proc Natl Acad Sci USA. 1997;94:4312–4317. doi: 10.1073/pnas.94.9.4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee SST, et al. Targeted disruption of the α isoform of the peroxisome proliferator-activated receptor gene in mice results in abolishment of the pleiotropic effects of peroxisome proliferators. Mol Cell Biol. 1995;15:3012–3022. doi: 10.1128/mcb.15.6.3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aoyama T, et al. Altered constitutive expression of fatty acid-metabolizing enzymes in mice lacking the peroxisome proliferator-activated receptor α (PPARα) J Biol Chem. 1998;273:5678–5684. doi: 10.1074/jbc.273.10.5678. [DOI] [PubMed] [Google Scholar]
- 14.Djouadi F, et al. A gender-related defect in lipid metabolism and glucose homeostasis in peroxisome proliferator-activated receptor α-deficient mice. J Clin Invest. 1998;102:1083–1091. doi: 10.1172/JCI3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leone TC, Weinheimer CJ, Kelly DP. A critical role for the peroxisome proliferator-activated receptor alpha (PPARα) in the cellular fasting response: the PPARα-null mouse as a model of fatty acid oxidation disorders. Proc Natl Acad Sci USA. 1999;96:7473–7478. doi: 10.1073/pnas.96.13.7473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bishop SP, Altschuld RA. Increased glycolytic metabolism in cardiac hypertrophy and congestive failure. Am J Physiol. 1970;218:153–159. doi: 10.1152/ajplegacy.1970.218.1.153. [DOI] [PubMed] [Google Scholar]
- 17.Taegtmeyer H, Overturf ML. Effects of moderate hypertension on cardiac function and metabolism in the rabbit. Hypertension. 1988;11:416–426. doi: 10.1161/01.hyp.11.5.416. [DOI] [PubMed] [Google Scholar]
- 18.Christe MD, Rodgers RL. Altered glucose and fatty acid oxidation in hearts of the spontaneously hypertensive rat. J Mol Cell Cardiol. 1994;26:1371–1375. doi: 10.1006/jmcc.1994.1155. [DOI] [PubMed] [Google Scholar]
- 19.Sack MN, et al. Fatty acid oxidation enzyme is downregulated in the failing heart. Circulation. 1996;94:2837–2842. doi: 10.1161/01.cir.94.11.2837. [DOI] [PubMed] [Google Scholar]
- 20.Sack MN, Disch DL, Rockman HA, Kelly DP. A role for Sp and nuclear receptor transcription factors in a cardiac hypertrophic growth program. Proc Natl Acad Sci USA. 1997;94:6438–6443. doi: 10.1073/pnas.94.12.6438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marcus SL, et al. Diverse peroxisome proliferator-activated receptors bind to the peroxisome proliferator-responsive elements of the rat hydratase/dehydrogenase and fatty acyl-CoA oxidase genes but differentially induce expression. Proc Natl Acad Sci USA. 1993;90:5723–5727. doi: 10.1073/pnas.90.12.5723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vega R, Huss JM, Kelly DP. The coactivator PGC-1 cooperates with PPARα in transcriptional control of nuclear genes encoding mitochondrial fatty acid oxidation enzymes. Mol Cell Biol. 2000;20:1868–1876. doi: 10.1128/mcb.20.5.1868-1876.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Strauss AW, et al. Molecular basis of human mitochondrial very-long-chain acyl-CoA dehydrogenase deficiency causing cardiomyopathy and sudden death in childhood. Proc Natl Acad Sci USA. 1995;92:10496–10500. doi: 10.1073/pnas.92.23.10496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rogers JH, et al. RGS4 causes increased mortality and reduced cardiac hypertrophy in response to pressure overload. J Clin Invest. 1999;104:567–576. doi: 10.1172/JCI6713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McGarry JD, Woeltje KF, Kuwajima M, Foster DW. Regulation of ketogenesis and the renaissance of carnitine palmitoyltransferase. Diabetes Metab Rev. 1989;5:271–284. doi: 10.1002/dmr.5610050305. [DOI] [PubMed] [Google Scholar]
- 26.Yu GS, Lu YC, Gulick T. Co-regulation of tissue-specific alternative human carnitine palmitoyltransferase Iβ gene promoters by fatty acid enzyme substrate. J Biol Chem. 1998;273:32901–32909. doi: 10.1074/jbc.273.49.32901. [DOI] [PubMed] [Google Scholar]
- 27.Sugden PH. Signaling in myocardial hypertrophy. Life after calcineurin? Circ Res. 1999;84:633–646. doi: 10.1161/01.res.84.6.633. [DOI] [PubMed] [Google Scholar]
- 28.Juge-Aubry CE, et al. Regulation of the transcriptional activity of the peroxisome proliferator-activated receptor α by phosphorylation of a ligand-independent trans-activating domain. J Biol Chem. 1999;274:10505–10510. doi: 10.1074/jbc.274.15.10505. [DOI] [PubMed] [Google Scholar]
- 29.Hu E, Kim JB, Sarraf P, Spiegelman BM. Inhibition of adipogenesis through MAP kinase-mediated phosphorylation of PPARγ. Science. 1996;274:2100–2103. doi: 10.1126/science.274.5295.2100. [DOI] [PubMed] [Google Scholar]
- 30.Thorburn A, et al. HRas-dependent pathways can activate morphological and genetic markers of cardiac muscle cell hypertrophy. J Biol Chem. 1993;268:2244–2249. [PubMed] [Google Scholar]
- 31.Thorburn A. Ras activity is required for phenylephrine-induced activation of mitogen-activated protein kinase in cardiac muscle cells. Biochem Biophys Res Commun. 1994;205:1417–1422. doi: 10.1006/bbrc.1994.2823. [DOI] [PubMed] [Google Scholar]
- 32.Gillespie-Brown J, Fuller SJ, Bogoyevitch MA, Cowley S, Sugden PH. The mitogen-activated protein kinase kinase MEK1 stimulates a pattern of gene expression typical of the hypertrophic phenotype in rat ventricular cardiomyocytes. J Biol Chem. 1995;270:28092–28096. doi: 10.1074/jbc.270.47.28092. [DOI] [PubMed] [Google Scholar]
- 33.Zechner D, Thuerauf DJ, Hanford DS, McDonough PM, Glembotski CC. A role for the p38 mitogen-activated protein kinase pathway in myocardial cell growth, sarcomeric organization, and cardiac-specific gene expression. J Cell Biol. 1997;139:115–127. doi: 10.1083/jcb.139.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Choukroun G, et al. Regulation of cardiac hypertrophy in vivo by the stress-activated protein kinases/c-Jun NH2-terminal kinases. J Clin Invest. 1999;104:391–398. doi: 10.1172/JCI6350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Molkentin JD, et al. A calcineurin-dependent transcriptional pathway for cardiac hypertrophy. Cell. 1998;93:215–228. doi: 10.1016/s0092-8674(00)81573-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Corr PB, Creer MH, Yamada KA, Saffitz JE, Sobel BE. Prophylaxis of early ventricular fibrillation by inhibition of acylcarnitine accumulation. J Clin Invest. 1989;83:927–936. doi: 10.1172/JCI113978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stanley CA, et al. A deficiency of carnitine-acylcarnitine translocase in the inner mitochondrial membrane. N Engl J Med. 1992;327:19–23. doi: 10.1056/NEJM199207023270104. [DOI] [PubMed] [Google Scholar]