SUMMARY
The evolution of human anatomical features likely involved changes in gene regulation during development. However, the nature and extent of human-specific developmental regulatory functions remain unknown. We obtained a genome-wide view of cis-regulatory evolution in human embryonic tissues by comparing the histone modification H3K27ac, which provides a quantitative readout of promoter and enhancer activity, during human, rhesus, and mouse limb development. Based on increased H3K27ac, we find that 13% of promoters and 11% of enhancers have gained activity on the human lineage since the human-rhesus divergence. These gains largely arose by modification of ancestral regulatory activities in the limb or potential co-option from other tissues and are likely to have heterogeneous genetic causes. Most enhancers that exhibit gain of activity in humans originated in mammals. Gains at promoters and enhancers in the human limb are associated with increased gene expression, suggesting they include molecular drivers of human morphological evolution.
INTRODUCTION
Morphological change is a central feature of human evolution. Humans exhibit characteristic physical differences, such as increased brain size and specialization of the limbs, relative to other primate species. Many of these differences emerge early in human development; for example, the distinct morphologies of the human hand and foot are evident by gestational day 54 (Schoenwolf et al., 2009). The genetic drivers of most human-specific traits remain unknown. It has long been thought that changes in gene regulation contributed to human-specific morphological adaptations by altering the level and distribution of gene expression during development (King and Wilson, 1975; Wray, 2007). Changes in developmental enhancer function have been shown to underlie evolutionary differences in model organisms and also contribute to human morphological defects (Lettice et al., 2003; Wittkopp and Kalay, 2012). However, changes in the activity of developmental regulatory elements during human evolution have yet to be characterized on a genome-wide scale, in large part because comprehensive maps of promoters and enhancers active in human embryonic tissues are lacking.
Previous efforts to identify developmental regulatory elements with human-specific functions have largely relied on indirect computational approaches. Comparative analyses of conserved noncoding sequences have identified putative enhancers showing accelerated rates of sequence change on the human lineage or that have been lost from the human genome (McLean et al., 2011; Pollard et al., 2006; Prabhakar et al., 2006). A handful of these sequences have associated experimental evidence supporting human-specific gain or loss of enhancer function during embryonic development (McLean et al., 2011; Prabhakar et al., 2008). These studies are restricted to regulatory elements that are highly conserved across species and cannot detect human-specific regulatory functions arising from a small number of sequence changes with large effects. They also do not provide a means to predict tissue-specific changes in regulatory activity.
Global analyses of transcription factor binding and histone modifications have been employed to compare cis-regulatory activity in a variety of human and nonhuman cell lines and adult tissues (Kunarso et al., 2010; Mikkelsen et al., 2010; Schmidt et al., 2010). Although these studies illustrate the potential advantages of a comparative functional genomics strategy for identifying regulatory changes on the human lineage, they provide limited insight into development. The experimental systems used do not recapitulate embryonic tissues, and in comparisons of cultured cell systems, the cell types utilized may not represent an unambiguously homologous biological state across species (Kunarso et al., 2010; Mikkelsen et al., 2010). Understanding the nature and rate of human-specific developmental regulatory change ultimately requires direct functional comparisons of human and nonhuman embryonic structures. However, such studies have not been feasible due to the extremely limited availability of relevant primate specimens.
We address this issue by using comparative analysis of the histone modification H3K27ac in human, rhesus macaque, and mouse embryonic limb at multiple stages of development. The limb is well suited for this purpose: its homology is unambiguous across all three species, its development has been extensively studied in model systems (Zeller et al., 2009), and it exhibits human-specific morphology that likely originates during the stages we examined. The scarcity of human and rhesus embryonic material required us to target a single, highly sensitive marker of promoter and enhancer activity. Previous studies have established that H3K27ac chromatin immunoprecipitation (ChIP)-seq profiling identifies active promoters and enhancers in embryonic and adult tissues and can be efficiently carried out with small amounts of input material (Cotney et al., 2012; Rada-Iglesias et al., 2012). Moreover, the level of H3K27ac marking at promoters and enhancers correlates with the level of nearby gene expression, so lineage-specific increases in H3K27ac may identify lineage-specific increases in regulatory element activity (Cotney et al., 2012). In this study, we developed and applied a statistical framework for quantitative phylogenetic comparisons of histone modification signatures. We find that increases in H3K27ac are associated with increases in gene expression and enhancer activity in the human limb and reveal thousands of promoters and enhancers with potential lineage-specific functions.
RESULTS
Mapping Active Promoters and Enhancers in the Human, Rhesus, and Mouse Embryonic Limb
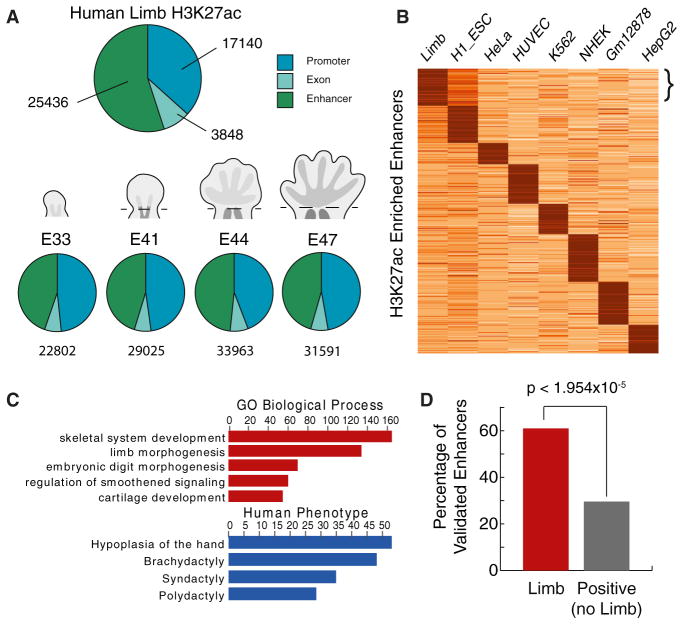
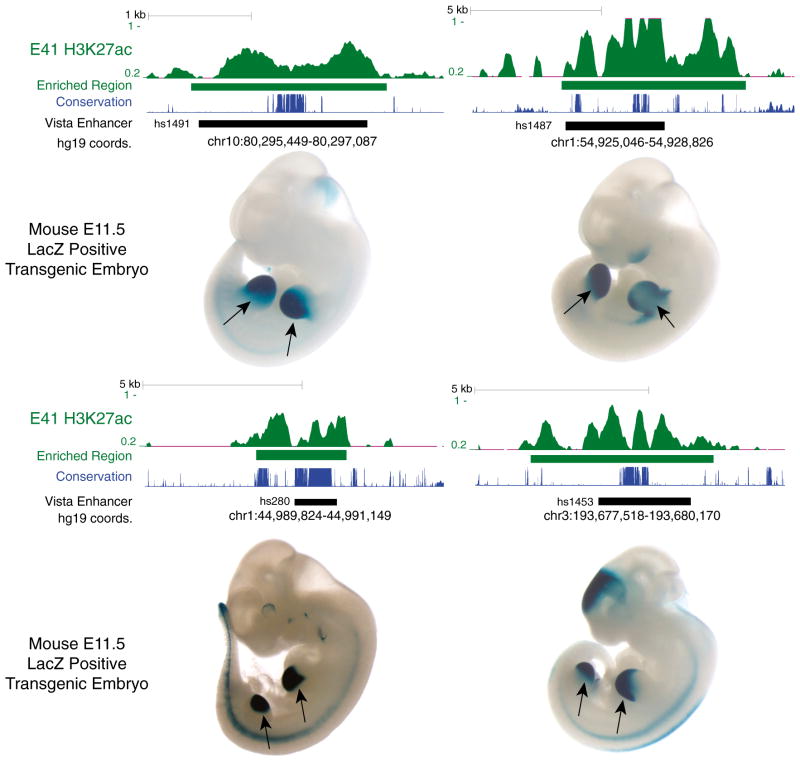
To identify promoters and enhancers with a potential gain of activity on the human lineage since the human-rhesus divergence, hereafter referred to as “human lineage gains,” we performed ChIP-seq of H3K27ac in human, rhesus, and mouse embryonic limb. In each species, we examined individual time points from the bud stage through the onset of digit separation: embryonic day 33 (E33) through E47 in human, E31–E36 in rhesus and E10.5–E13.5 in mouse (Figure S1A available online). The morphology of these human, rhesus, and mouse specimens indicates they encompass comparable developmental time frames across all three species. At early stages (E33 and E41 in human), we mapped H3K27ac in the entire limb bud. At later stages (E44 and E47), we focused on the handplate and footplate. We combined forelimb and hindlimb tissues in all experiments and included two biological replicates at each time point for human and mouse (Extended Experimental Procedures). We identified thousands of H3K27ac-enriched regions at all stages in all three species (Figures 1A and S1B; Table S1). H3K27ac regions within 1 kb upstream of a transcription start site were annotated as promoters, whereas intergenic or intronic regions were annotated as putative enhancers. We identified a subset of putative enhancers that are strongly marked in limb compared to other human cell types and are overrepresented near genes with strong limb-specific expression or known functions in limb development (Figures 1B, 1C, and S1C). Similar enrichments were obtained in mouse (Figure S1D) (Shen et al., 2012). Moreover, 60% of human sequences known to act as limb enhancers in mouse transgenic assays are marked by H3K27ac in the human limb, demonstrating that H3K27ac enriches for tissue-specific enhancer activity (Figures 1D and 2) (Visel et al., 2007). Comparisons among individual human time points revealed temporal patterns of enhancer activation (Figure S1E). Enhancers active at early stages of human limb development are associated with genes involved in pattern formation along the anterior-posterior axis of the limb, whereas enhancers activated later are enriched near genes contributing to the function and growth of differentiated tissues such as muscle (Figure S1E) (Taher et al., 2011).
Figure 1. Genome-wide Identification of Promoters and Enhancers Active in Human Embryonic Limb.
(A) Annotating reproducible H3K27ac-enriched regions in limb. Top: union of all H3K27ac regions identified at one or more time points. Bottom: annotation at each time point. A schematic of limb morphology is shown for each stage (not to scale). See also Figures S1A and S1B and Table S1.
(B) K-means clustering of H3K27ac signals across 104,228 putative enhancer regions identified in human limb or ENCODE cell lines (k = 8). The bracket indicates a cluster of enhancers strongly marked in limb compared to the other data sets. See also Figure S1D.
(C) Gene ontology and human phenotype enrichments calculated by GREAT (McLean et al., 2010) (y axis = −log10 binomial p value) for the strong limb-specific enhancers in (B). See also Figure S1D.
(D) Percentage of human enhancers annotated in the VISTA Enhancer Browser as active in limb or other tissues that are marked by H3K27ac in human limb (Visel et al., 2007). The p value was calculated using Fisher’s exact test.
Figure 2. H3K27ac Signal Profiles in Human Embryonic Limb at Experimentally Validated Enhancers.
The normalized H3K27ac signal (Extended Experimental Procedures) and corresponding enriched region (green), in human E41 limb bud at four sequences demonstrated in a mouse transgenic enhancer assay to drive reproducible LacZ expression in E11.5 mouse limb (indicated by arrows). Images and enhancer locations were obtained from the VISTA Enhancer Browser (http://enhancer.lbl.gov) (Visel et al., 2007). The location of each sequence tested in the enhancer assay is shown as a black bar, accompanied by the corresponding genomic coordinates (UCSC hg19 assembly) and VISTA identifier. The level of sequence conservation (phastCons scores) at each site in placental mammals is shown in blue.
To evaluate at a molecular level whether the human, rhesus, and mouse limb specimens we selected represent comparable developmental time frames, we considered temporal changes in the distribution of H3K27ac at the HOXD cluster in all three species. As mouse limb development proceeds, genes located at the 5′ end of the cluster (e.g., HoxD13) are preferentially activated, whereas genes located at the 3′ end become progressively less transcribed (Spitz et al., 2003; Tarchini and Duboule, 2006). The deep evolutionary conservation of HOXD cluster genes suggests their spatiotemporal activation patterns are likely to be conserved across mammals. The spatial and temporal distribution of H3K27ac signal at the HOXD locus is highly consistent in our human, mouse, and rhesus limb specimens: H3K27ac marking shifts toward HOXD13 as limb development progresses in each species (Figure S2). This provides further evidence that we are comparing similar developmental windows in our analysis.
The Extent of Human Lineage Gains in H3K27ac at Promoters and Enhancers in the Limb
We first considered potential increases in promoter and enhancer activity on the human lineage. To detect these events, we focused on sites in the human genome that showed reproducible enrichment of H3K27ac in two biological replicates at each time point and exhibited a significant increase in H3K27ac signal compared to the orthologous locations in rhesus and mouse (Figures 3 and S3A). Comparing signal at orthologous regions mitigates false positives due to sequence copy number changes among species, which include lineage-specific paralogous elements and repeats that are difficult to study using short read alignments (Alkan et al., 2011). In all comparisons, we used the number of reads mapped to each orthologous site in each native genome, normalized for sequencing depth and species differences in orthologous element length. We did not require that an H3K27ac-enriched region be called at the orthologous site in rhesus or mouse at any stage; there may or may not be an enriched region present in these species.
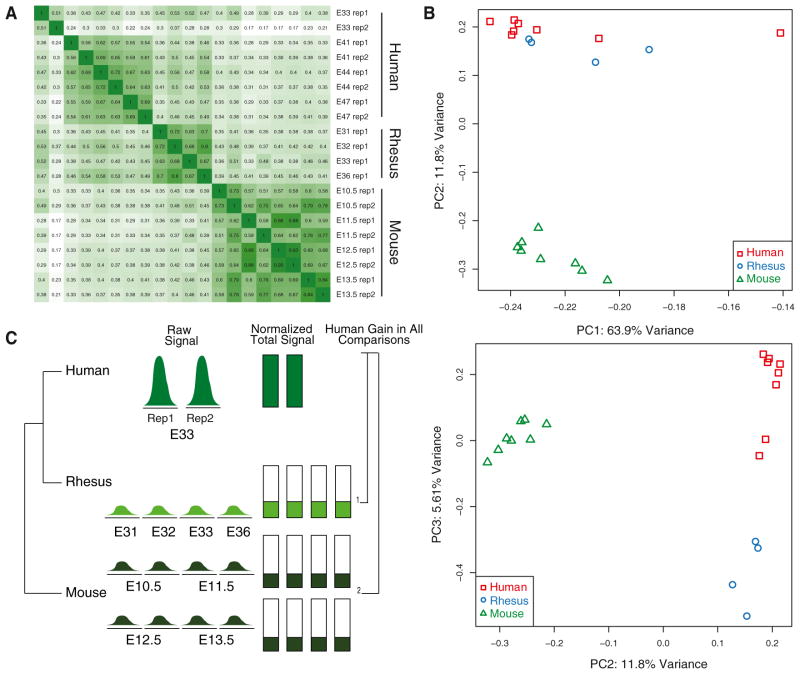
Figure 3. Cross-Species Comparisons of H3K27ac Marking in the Embryonic Limb.
(A) Spearman correlation matrix of H3K27ac signal at orthologous sites in all three species generated by this study.
(B) Principal component analysis of H3K27ac signal at orthologous sites in all data sets. The proportion of variance explained by the first three components is shown. See also Figure S3B.
(C) A schematic illustrating the cross-species H3K27ac signal comparison strategy used in this study. An idealized example of human lineage gain at E33 is shown.
See also Figures S2 and S3A and Table S2.
H3K27ac signals from orthologous regions were well correlated between replicates in each species (Figure 3A). In addition, essential regulators of limb development, including PITX1 and TBX5 (Duboc and Logan, 2011), exhibit similar H3K27ac profiles at promoters and putative enhancers across all three species, consistent with their highly conserved functions and further reinforcing the overall quality of our data (Figure S4A). To gain an initial global view of cross-species similarities and differences in H3K27ac profiles we performed principal component analysis (PCA). In the PCA, H3K27ac data sets are separated according to evolutionary distance: human and rhesus, although clearly distinct, are more similar to each other than either is to mouse (Figure 3B). PCA of human and mouse H3K27ac limb data with published human and mouse embryonic stem cell and adipocyte H3K27ac data separated each data set by tissue or cell type (Figure S3B) (Mikkelsen et al., 2010; Shen et al., 2012).
To identify potential human lineage gains, we required that reproducibly marked H3K27ac sites at each time point in human show a significant increase in H3K27ac signal compared to all 12 mouse and rhesus limb data sets in our study (Figure 3C; Extended Experimental Procedures). We independently identified gains at each human time point in light of the temporal changes in H3K27ac activation we identified in human limb development (Figure S1E). Moreover, given the uncertainty in precisely matching developmental stages across species, we chose to identify H3K27ac gains at each human time point relative to a broad range of developmental stages in rhesus and mouse rather than attempting to compare specific stages. In total, we identified 2,175 promoters and 2,915 enhancers with a significant increase in H3K27ac on the human lineage at one or more time points (Figure 4A; Tables S2 and S3). We considered each time point-specific set of promoters and enhancers with human gains independently in subsequent analyses. The distribution of orthologous sequence length changes in mouse and rhesus were similar for all human H3K27ac regions and regions with human lineage gain of marking, indicating that cross-species changes in sequence length are not producing overestimates of human lineage changes in H3K27ac (Figure S4B).
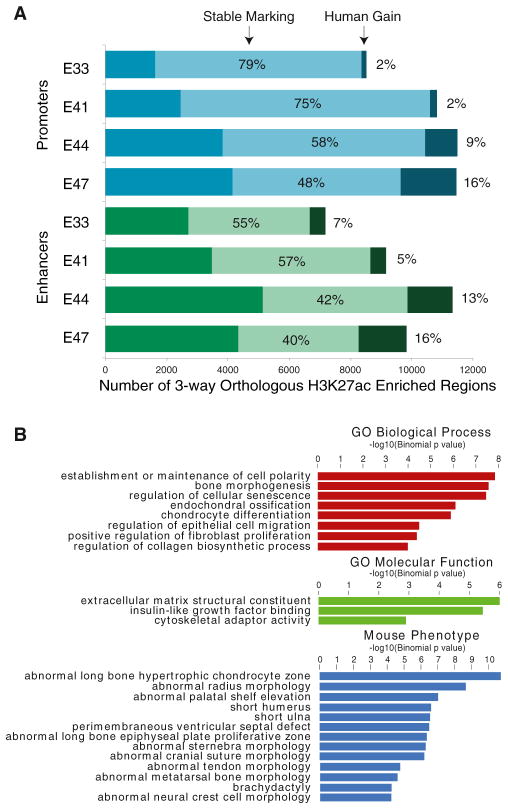
Figure 4. Identification of Promoters and Enhancers with Human Lineage Gain of H3K27ac.
(A) Number of orthologous promoters and enhancers marked by H3K27ac at each time point. The percentage exhibiting stable marking or human lineage gain of marking is indicated in each bar.
(B) Gene ontology and mouse phenotype enrichments for human gain enhancers calculated using GREAT (McLean et al., 2010) (y axis = −log10 binomial p value).
Human lineage gains at promoters (“human gain promoters”) represent 2% (at E33) to 16% (at E47) of all orthologous promoters in our analysis. Human lineage gains at putative enhancers (“human gain enhancers”) occurred at 7% (at E33) to 16% (at E47) of orthologous enhancers. These gains reflect regulatory changes arising after divergence of the human and rhesus lineages and likely include changes on the human lineage that postdate the human-chimpanzee split. Genes associated with promoters and enhancers exhibiting human gains of H3K27ac were enriched for functions related to proliferation and growth, bone morphogenesis, and connective tissue (Figures 4B and S4C). Human gain enhancers are also enriched near genes with mouse limb phenotypes, suggesting some human gain events influence genes with conserved limb functions. We confirmed increases of H3K27ac at 21 of 28 regions (75%) by ChIP-qPCR in human E44 limb versus both E11.5 and E13.5 in mouse (Figure S4D).
We also considered potential losses of promoter and enhancer activity on the human lineage. We only identified 74 orthologous regions exhibiting loss of H3K27ac in human compared to rhesus and mouse (Table S4). The asymmetry between gains and losses we observed is consistent with a previous comparative study of DNase I hypersensitivity sites in human, chimpanzee, and rhesus cultured cells and is likely due to our study design (Shibata et al., 2012). Human gain of H3K27ac may in principle occur at any site whether or not that site exhibits H3K27ac marking in any other species or has a conserved regulatory function. However, our study design only allows us to identify human losses that entail reduction in H3K27ac compared to rhesus and mouse. These sites are more likely to include cis-regulatory functions conserved in both species and thus under deep evolutionary constraint in mammals.
Evolutionary Origins of Human Lineage Increases in H3K27ac at Promoters and Enhancers
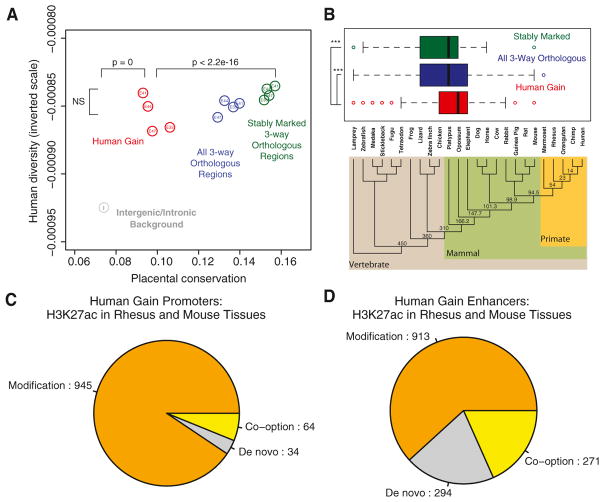
We next sought to determine the evolutionary origin of human lineage gains in promoter and enhancer marking. We first compared sequence conservation levels of three subsets of enhancers active in human limb: all enhancers with orthologous sequences in rhesus and mouse, enhancers with similar levels of H3K27ac across all three species (“stably marked” enhancers), and human gain enhancers at each time point. Many known developmental enhancers are deeply conserved in vertebrates, and previous efforts to identify human-specific enhancer functions have consequently targeted highly conserved noncoding sequences that exhibit increased rates of human-specific sequence change (McLean et al., 2011; Pollard et al., 2006; Prabhakar et al., 2006). However, the majority of human lineage increases in H3K27ac we detected do not involve extremely conserved elements. Although human gain enhancers are significantly more conserved than background sequence, they are less conserved than stably marked enhancers (Figure 5A). Human gain enhancers and stably marked enhancers do not show a significant difference in overall genetic diversity, based on single-nucleotide variation data obtained in the 1000 Genomes Project (1000 Genomes Project Consortium, 2012) (Figure 5A; Extended Experimental Procedures). This suggests that human gain enhancers are potentially under similar functional constraint in modern human populations as stably marked enhancers. However, we note that recently identified ascertainment biases and false positives in 1000 Genomes variant data may confound this analysis (Ward and Kellis, 2012, 2013; Green and Ewing, 2013).
Figure 5. Conservation and Putative Ancestral Activities of Promoters and Enhancers with Human Lineage Increases in H3K27ac.
(A) Mean per-element conservation and hetero-zygosity values (1000 Genomes Project Consortium et al., 2012) for all orthologous human limb enhancers (blue), stably marked enhancers (green), human gain enhancers (red), and background intergenic or intronic regions (gray). p values are shown for comparisons discussed in the text.
(B) Distribution of age estimates for all orthologous, stably marked, and human gain enhancers at human E44, mapped onto the known phylogeny of vertebrate genomes used in the analysis. We estimated the age of each enhancer by identifying the most distant vertebrate lineage with an orthologous sequence (Extended Experimental Procedures). Estimated ages (in millions of years) for internal nodes in the phylogeny are shown. In the box plots, the right, middle and left bars of the boxes represent the 25th, 50th, and 75th percentiles of the data, respectively. Whiskers extend from the box to the most extreme data point that is <1.5 times the interquartile range. Human gain enhancers show a significantly more recent origin compared to all orthologous and stably marked regions (***, Wilcoxon rank-sum p < 0.0001). See also Figure S5A.
(C and D) Inferred human gain mechanisms based on human E44 H3K27ac marking at the orthologous rhesus and mouse position in a total of 19 tissues and cell lines, for promoters (C) and enhancers (D).
See also Figures S5B and S6 and Tables S3 and S6.
The median age of E44 human gain enhancers dates to the common ancestor of opossum and placental mammals, although a subset of enhancers is more ancient (Figure 5B). In contrast, both stably marked and all orthologous enhancer sequences in E44 human limb are significantly older: a greater fraction of elements in each class are present in nonmammal vertebrate genomes. Enhancers at other time points showed similar trends, validating the robustness of our observations (Figure S5A). Human gain enhancers are also significantly less conserved than stably marked enhancers both in primate-rodent comparisons and across placental mammals, suggesting they have a more recent evolutionary history and are less conserved overall (Figure S5A). The median age of human gain promoter sequences identified at each time point predates the origin of mammals (Figure S5A). Although human gain promoters at E44 and E47 show a significantly more recent evolutionary origin than stably marked promoters, human gain promoters at E33 and E41 are not significantly younger. Despite their age, human gain promoters at each time point are significantly less conserved across placental mammals than stably marked promoters (Figure S5A).
To evaluate the potential ancestral regulatory activity of regions with human lineage gains, we examined H3K27ac enrichment at orthologs of human gain promoters and enhancers in rhesus and mouse limb, as well as in 18 nonlimb mouse tissues (Shen et al., 2012). Although the quantitative approach we used to identify human gains does not require that the orthologous site in rhesus or mouse show significant H3K27ac enrichment, there may be a significant peak present, indicating an ancestral regulatory function. Human lineage gains may thus have arisen by modification of an existing regulatory element in the limb, co-option of a regulatory element active in another tissue, or de novo. At E44, we find that 91% of human gain promoters show significant H3K27ac enrichment in the rhesus or mouse limb, indicating human gain through modification (Figure 5C). A smaller fraction, 6%, are not marked in rhesus or mouse limb, but are marked in at least one other mouse tissue, suggesting they may have been co-opted into human limb development. Finally, 3% of human gain promoters show no marking in any mouse or rhesus tissue we examined, suggesting they may involve de novo gain of activity. Similar trends were observed for the remaining human limb time points (Figure S5B). A greater fraction of human gain enhancers showed evidence of potential co-option or de novo gain compared to promoters. Modifications of ancestral limb regulatory activity may have occurred at 62% of E44 human gain enhancers, 18% are potential co-option events, and 20% are potential de novo gains (Figure 5D; Table S3). Similar trends were observed for other human limb time points, although at earlier stages a larger proportion of enhancers may arise from de novo gains (Figure S5B).
The genetic basis of the human lineage gains we identified remains to be determined. Sixteen conserved noncoding sequences exhibiting accelerated rates of sequence change in human evolution identified in previous studies show gain of H3K27ac on the human lineage (Lindblad-Toh et al., 2011; Prabhakar et al., 2006) (Table S5). However, our analysis suggests that, in general, promoters or enhancers showing human lineage gains do not exhibit a human-specific increased substitution rate relative to other primate species. Rates of human-specific, chimpanzee-specific, ape-specific, and rhesus-specific change were all elevated in these sequences, potentially due to their lower level of sequence conservation (Figure S6; Extended Experimental Procedures). Human lineage gain sites also did not show human-lineage enrichment for particular transcription factor motifs or repetitive elements (Table S6). These results suggest human lineage H3K27ac gains may have heterogeneous genetic origins and highlight the difficulty of identifying regulatory gains using purely sequence-based comparative metrics.
Lineage-specific sequence gains and changes in copy number could also contribute to human lineage gains of regulatory function. Approximately 8% and 22% of H3K27ac-enriched sites identified at each time point in human limb do not have a clearly identifiable ortholog in rhesus or mouse, respectively (Table S1). We have excluded these sequences from our analysis. Based on multiple sequence alignments, we could only identify 14 (0.06%) and 123 (~0.5%) putative limb enhancers with sequences specific to human or ape respectively (Table S3). However, our analysis may underestimate the contribution of lineage-specific sequence change, as many human lineage gains of sequence are driven by recent duplication and accurately mapping 75 bp reads to such sequences is challenging (Alkan et al., 2011).
Human Lineage Increases in H3K27ac Reveal Human Lineage Promoter and Enhancer Activities
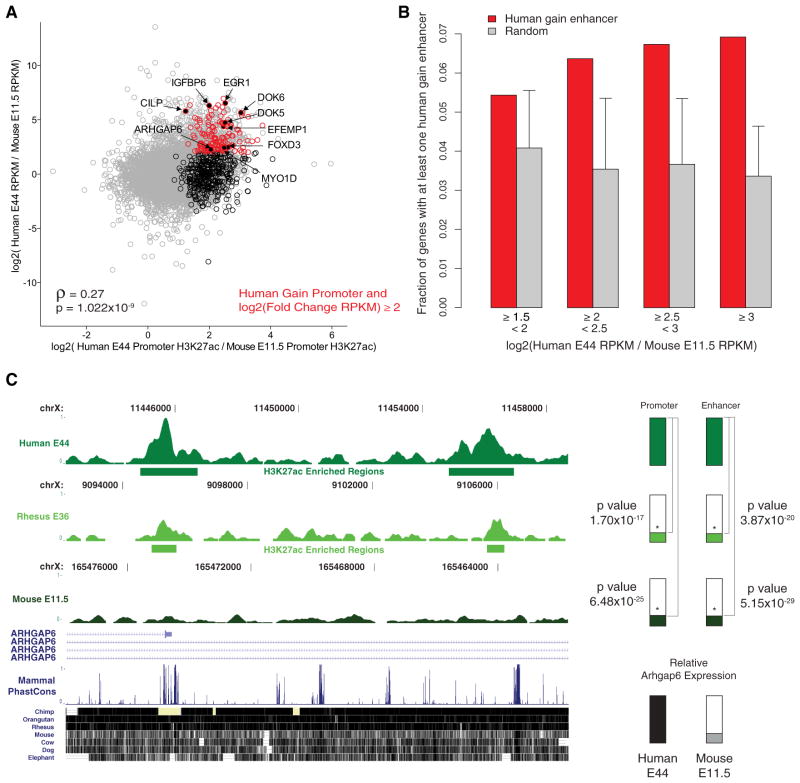
Our results suggest human gain of H3K27ac reveals regulatory activities arising on the human lineage that may impact gene expression during development. Human lineage increases at promoters in E44 limb were significantly correlated with increased gene expression in human relative to the orthologous genes in mouse (Figure 6A; Table S7). Nearly 30% (142 of 502) of the orthologous genes associated with a human gain promoter exhibit a 4-fold or greater increase in gene expression in the human limb compared to mouse. This represents a significant enrichment compared to the fraction of 7,484 orthologous genes that show a 4-fold or greater expression increase in human (943 of 7,484, Fisher’s exact test p value = 1.23 ×10−22). Human lineage gain enhancers were also significantly enriched near genes with ≥4-fold increases in expression level in human limb compared to mouse (Figures 6B and S7A).
Figure 6. Human Lineage Increases in H3K27ac Correlate with Human Lineage Regulatory Activities.
(A) Comparison of differential promoter H3K27ac marking and differential gene expression in human E44 and mouse E11.5 limb for 7,484 orthologous genes. Spearman rank correlation coefficient and p value, ρ and p respectively, indicate a significant positive correlation between H3K27ac level and gene expression. Grey dots represent 7,484 one-to-one orthologous genes between human and mouse. Black circles indicate 502 human genes associated with human gain promoters. Red circles indicate 142 human genes associated with gain promoters that also show a ≥4-fold change of gene expression in human compared to mouse. Genes with potential roles in human limb phenotypes are named and labeled in black. See also Table S7.
(B) Fraction of human gain or the same number of randomly sampled stably marked regions assigned to a gene with the indicated level of increased expression in human E44 limb compared to mouse E11.5 limb (see also Figure S7A). Error bars represent 99.9% quantile values from 1,000 sampling iterations (Extended Experimental Procedures).
(C) Limb H3K27ac signal from the indicated human, rhesus, and mouse time points at the promoter (left) and a potential enhancer (right) of the ARGHAP6 gene. Horizontal bars indicate regions of H3K27ac enrichment. Coordinates for human, rhesus, and mouse are for hg19, rheMac2, and mm9 genomes respectively. Vertical green bars indicate total signal in human region and relative signal in orthologous regions from rhesus and mouse; p values indicating significant human lineage increased marking are shown. Vertical black and gray bars indicate relative ARGHAP6 expression determined by human E44 and mouse E11.5 RNA-seq.
See also Figure S7B and Table S7.
Genes associated with human gains of H3K27ac at promoters and enhancers and that exhibit increased expression in human limb, are candidates for producing human-specific limb phenotypes. One such gene, ARHGAP6, shows increased marking in human at both of its known promoters as well as a nearby enhancer and is highly expressed in human limb compared to mouse (Figures 6C and S7B). The mouse ortholog resides in the Xpl locus, which includes an unknown dominant regulatory mutation that gives rise to polydactyly and elongation and thickening of digit one in hindlimb (Cormier et al., 2001; Masuya et al., 1997) (Figure S7C).
An enhancer with human-specific activity in the limb, HACNS1, also shows significantly increased H3K27ac marking on the human lineage (Figure 7A) (Prabhakar et al., 2008). Human-specific sequence changes in this element confer strong limb expression in a mouse transgenic enhancer assay compared to the orthologous chimpanzee and rhesus elements (Figure 7B). H3K27ac marking at HACNS1 is strongest in E33 human limb, consistent with its robust limb enhancer activity at mouse E11.5. These results suggest that human lineage increases in H3K27ac marking may help identify enhancers with potential human-specific developmental functions and provide a means to prioritize candidates for downstream experimental analyses.
Figure 7. Human Lineage Increases in H3K27ac Identify an Enhancer with Human-Specific Regulatory Function In Vivo.
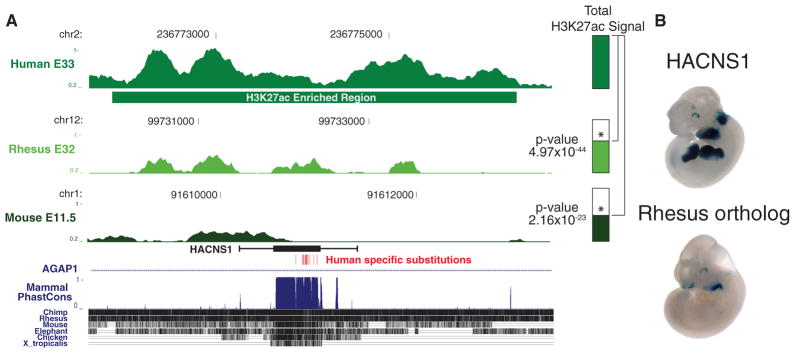
(A) Normalized H3K27ac signal in human E33 limb at a conserved noncoding sequence exhibiting human-specific accelerated evolution (HACNS1) and at the orthologous positions in rhesus and mouse. The horizontal green bar indicates the H3K27ac-enriched region called in human. The H3K27ac signals at the orthologous positions in rhesus and mouse are also shown for the indicated time points. The location of HACNS1 is indicated in black; whiskers indicate the sequence shown to have human-specific enhancer activity in a mouse transgenic assay. p values indicate significantly increased marking in human compared to rhesus and mouse. See also Tables S2 and S5.
(B) Activity of HACNS1 and its rhesus ortholog, indicated by LacZ reporter activity in mouse E11.5 transgenic embryos.
DISCUSSION
The systematic identification of genetic and molecular changes that distinguish humans from other species is essential for understanding the evolution of uniquely human traits. In this study, we globally mapped promoters and enhancers active during critical stages of limb development in human, rhesus, and mouse, from patterning of the limb bud through emergence and separation of the digits. Our results provide a direct, genome-wide view of both conserved and human lineage-specific cis-regulatory activities in the embryonic limb. Using quantitative phylogenetic analysis of H3K27ac marking, we identified several thousand promoters and enhancers that show increased activity on the human lineage. Not all of the human lineage gains are likely to affect gene expression, or be relevant to human limb phenotypes. However, our results provide several starting points to identify candidates for downstream studies and enable functional studies of regulatory change in human limb evolution.
First, we identified 302 genes that show 4-fold or greater expression in human limb relative to mouse and that are associated with human promoter or enhancer gains (Figures 6A and 6B; Table S7). These results support that H3K27ac gains at promoters and enhancers in human do contribute to increased gene expression. Enhancers showing lineage-specific gain of activity in human and that are associated with genes showing increased expression, are candidates for initial in vivo analysis in the mouse using comparative transgenic enhancer assays (Figure 7). Characterizing morphological changes in the limb or other structures arising due to lineage-specific enhancer functions identified by this approach will then require humanized mouse models, in which enhancers showing human-specific activity are introduced by homologous recombination into the mouse genome. These studies would also provide insight into potential systemic effects of human lineage regulatory changes in limb development, including increases or decreases of downstream gene expression due to direct (e.g., enhancers acting to up-regulate promoters) or indirect mechanisms (e.g., increased expression of a repressive factor that downregulates multiple genes). We provide a list of all 2,304 known human and mouse orthologous genes associated with human gain promoters or enhancers in Table S7.
Second, our results suggest that modification of existing limb regulatory programs may contribute to unique human limb morphology. The human hand is distinguished from that of other primates by the increased relative length of the first digit and reduced curvature of the digits, which have served to increase its dexterity (Marzke and Marzke, 2000). The human foot has evolved a rigid structure that allows it to act as a lever during walking, and the first digit is no longer opposable to the other digits (Aiello and Dean, 2002). At a genome-wide level, we show that human lineage gains in H3K27ac marking are associated with genes important for limb development (Figures 4B and 6). These analyses also revealed specific genes that show human lineage regulatory changes in limb and have known limb phenotypes in mouse. We identified human gains of promoter activation and increased gene expression of EGR1 and CILP, which are involved in tendon (Lejard et al., 2011) and cartilage formation (Lorenzo et al., 1998), respectively. We also detected multiple human regulatory gains and increased gene expression at ARHGAP6. In the mouse, this gene resides in a locus that includes a putative cis-regulatory mutation resulting in a hindlimb phenotype that closely mirrors evolutionary changes in the human foot, such as thickening of the first digit (Figure S7C) (Masuya et al., 1997, Bramble and Lieberman, 2004). We also identified an enhancer gain associated with a human lineage increase in expression for EFEMP1. This gene has been implicated in regulating chondrocyte differentiation (Wakabayashi et al., 2010) and has been associated with variation in human height (Weedon et al., 2008; Zhao et al., 2010). These genes and their associated regulatory changes are high-priority candidates for experimental characterization.
Third, our results enable direct interpretation of human accelerated regions (HARs or HACNSs) identified by previous studies and allow their prioritization for downstream analysis (Pollard et al., 2006; Prabhakar et al., 2006). These elements were defined by purely computational means and have generally resisted large-scale efforts to identify their functions due to a lack of associated, relevant experimental data. We show that human lineage increases in H3K27ac may be used to identify human accelerated sequences with human-specific regulatory activity (Figure 7). We have identified 16 human accelerated regions that show significant increases in H3K27ac on the human lineage. The results shown in Figure 7 provide a proof of principle for using mouse transgenic assays to characterize potential human-specific regulatory functions in this larger set of elements. As discussed above, characterizing the phenotypic effects of regulatory changes in human accelerated regions will require humanized mouse models.
In addition to their utility for identifying candidates for experimental study, our results provide general insights into the origin and abundance of human lineage gains of developmental enhancer activity. We estimate that 2%–16% of enhancers show evidence of human lineage gain of H3K27ac marking relative to rhesus and mouse, depending on the time point examined. It is not feasible to obtain embryonic tissues from chimpanzee, so we could not directly determine the rate of human enhancer gains since the human-chimpanzee split. However, because these events are a subset of the human-rhesus differences we identified, the rate of human lineage gain of activity at enhancers relative to chimpanzee is likely to be less than the estimates we obtained from human-rhesus comparisons.
We note that our estimates are based on a small number of replicates in each species, due to the difficulty in obtaining human and rhesus embryonic tissues. A subset of the human gains we identified are likely to be false positives due to technical issues arising from our limited sample size, or may be polymorphic in human populations. Moreover, we may lack power to reliably detect small increases in H3K27ac marking due to our sample size, so we may be underestimating weak human lineage gains. Additional biological replicates in human and rhesus would refine the estimates we provide here and better distinguish between gains that are fixed on the human lineage and epigenetic variation within humans. We also note that our human gain estimates are specific to promoters and enhancers that are marked by H3K27ac. Comparative analysis of other histone modifications, or of other factors that mark regulatory sequences, may reveal a greater or lesser number of human gains.
Many of the human lineage gains we identified appear to involve modification of ancestral enhancer activities, based on patterns of H3K27ac marking at their rhesus and mouse orthologs. Our analysis also revealed a subset of human gain enhancers that may involve de novo genesis of regulatory function. However, we have not exhaustively characterized H3K27ac in all rhesus and mouse tissues, so we may be overestimating the frequency of de novo gains. In addition, human gain enhancers are less conserved and have a more recent evolutionary origin than the average enhancer active in the human limb, suggesting that regulatory changes contributing to human limb morphology generally involve modification of mammal-specific enhancers, rather than enhancers deeply conserved in vertebrates. Combined, these results point to a conservative process of regulatory change in the evolution of the human limb.
The human lineage gains in enhancer activity we identified are likely to have diverse genetic causes. We found no evidence of human-specific enrichment for any known transcription factor motif in these elements, and they exhibited no significant human-specific or ape-specific sequence acceleration compared to chimpanzee-specific or rhesus-specific substitution rates. This suggests many human lineage gains may be due to a small number of substitutions with large effects in each element and introduce or remove binding sites for a broad set of transcription factors overall. Novel regulatory sequences may also be generated by exaptation of transposable elements (Feschotte, 2008). However, repeat exaptation at orthologous sites does not appear to substantially contribute to the generation of human lineage-specific enhancer activities in the limb and neither do lineage-specific sequences (Tables S3 and S6). Nevertheless, we may be underestimating the abundance of human lineage regulatory changes at lineage-specific and paralogous sequences due to the inherent computational challenges in mapping short reads to recently duplicated sequences. We were unable to clearly identify a single orthologous sequence in rhesus for 8% of enriched H3K27ac sites in human limb. These sequences may encode human-specific regulatory functions relevant to human limb evolution, but this cannot be determined using the strategy we employ here.
We observed a higher degree of regulatory conservation among human, rhesus, and mouse limb compared to several previous studies of transcription factor binding in human and mouse embryonic stem cells and adult liver and histone modifications in human and mouse adipocytes (Figure 4A; Table S1) (Kunarso et al., 2010; Mikkelsen et al., 2010; Schmidt et al., 2010). This may be due to the conservative approach we used to identify human lineage quantitative changes in H3K27ac. It may also reflect the increased specificity of H3K27ac compared to binding of single transcription factors for detecting active regulatory elements. Deposition of H3K27ac is likely a terminal step in enhancer activation (Creyghton et al., 2010; Rada-Iglesias et al., 2011), whereas many transcription factor binding events appear to be incidental to regulatory function and thus may be poorly constrained (Göke et al., 2011; Junion et al., 2012). Tissue-specific variation in the frequency of species-specific regulatory change may also be a factor, arising due to different constraints on regulatory functions among developing and adult tissues (Brawand et al., 2011). Ensuring homology is another major consideration: the limb is unambiguously homologous (Figure S1A), whereas cell culture models may exhibit very different biological characteristics that confound cross-species analysis, even if they are ostensibly derived from homologous tissues. Analysis of regulatory divergence in multiple homologous embryonic tissues, such as the brain and the heart, using the strategy we have implemented here would yield insight into these questions and enable comprehensive experimental studies of uniquely human biology.
EXPERIMENTAL PROCEDURES
Tissue Collection, ChIP-Seq, and RNA-Seq
Human embryonic limb tissue was collected, staged, and provided by the Joint MRC/Wellcome Trust Human Developmental Biology Resource. Tissues were flash frozen upon collection and stored at −80°C. Human limbs were staged using the Carnegie staging system (O’Rahilly et al., 1987) and only tissues matching these criteria were used in our experiments. The use of human embryonic tissue in this study was reviewed and approved by the Yale Human Investigation Committee. For each time point replicate, a single forelimb and single hindlimb autopod from the same embryo were combined, homogenized, and crosslinked as described for mouse tissue (Cotney et al., 2012). Fetal rhesus limb tissue from gestational days 31 to 36 was harvested according to approved Yale IACUC protocols (Dominguez et al., 2012). Single forelimb and single hindlimb rhesus autopods were combined as for human experiments. Mouse embryos were harvested in accordance with approved Yale IACUC protocols. Only autopod tissue from forelimb and hindlimb were used for E12.5 and E13.5 experiments. Chromatin extraction, shearing, immunoprecipitation, and sequencing were performed as previously described (Cotney et al., 2012). For RNA-Seq experiments, digit one was separated from remaining digits of E44 forelimb or hindlimb autopod tissue and placed directly in Qiazol. RNA was extracted and sequenced as previously described (Cotney et al., 2012). Data from these experiments was combined computationally to generate composite limb expression values (Extended Experimental Procedures).
Validation of Human-Specific H3K27ac Regions
Human gain H3K27ac regions at E44 were chosen for testing that had an average reads per kilobase per million (RPKM) ≥1 and when lifted to the mouse genome changed <10% in overall length to enable meaningful comparison by QPCR. Human DNA sequence (300 bp) surrounding the peak of signal within each enriched region were selected and QPCR amplicons were designed using BatchPrimer3 (You et al., 2008). These 300 bp regions were lifted to the mouse genome and QPCR amplicons were designed as above (Table S3). Two independent human E44 limb samples were subjected to ChIP as above. Resulting enriched genomic DNA was used as template for QPCR. Human gain was determined by normalizing human E44 H3K27ac ChIP Ct values versus input Ct values in a fashion similar to ΔΔCt values typically utilized in detecting gene expression differences via RT-QPCR. p values were calculated by two-way ANOVA with Prism v5 (GraphPad).
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health grants GM094780 (to J.P.N.), DA023999 (to P.R.), and NS014841 (to P.R.); a career award from the Edward J. Mallinckrodt Jr. Foundation (to J.P.N.); a Rudolph Andersen Fellowship (to J.C.); a Howard Hughes Medical Research Scholars Fellowship (to L.E.D.); and an NSF Graduate Research Fellowship (to S.K.R.). The human embryonic and fetal material was provided by the Joint MRC (G0700089)/Wellcome Trust (GR082557) Human Developmental Biology Resource. We thank Marianne Horn and Steve R. Wilson for technical assistance and veterinary expertise. We also thank Shrikant Mane, John Overton, Sheila Umlauf, and Alex Lopez at the Yale Center for Genome Analysis for generating sequencing data and Nicholas Carriero and Robert Bjornson at the Yale University Biomedical Performance Computing Center for computer resource support.
Footnotes
ACCESSION NUMBERS
The Gene Expression Omnibus accession numbers for the raw sequencing reads and alignment data reported in this paper are GSE42413 and GSE42237.
Supplemental Information includes Extended Experimental Procedures, seven figures, and seven tables and can be found with this article online at http://dx.doi.org/10.1016/j.cell.2013.05.056.
References
- 1000 Genomes Project Consortium. Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, Kang HM, Marth GT, McVean GA. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aiello L, Dean C. An Introduction to Human Evolutionary Anatomy. San Diego: Academic Press; 2002. [Google Scholar]
- Alkan C, Coe BP, Eichler EE. Genome structural variation discovery and genotyping. Nat Rev Genet. 2011;12:363–376. doi: 10.1038/nrg2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bramble DM, Lieberman DE. Endurance running and the evolution of Homo. Nature. 2004;432:345–352. doi: 10.1038/nature03052. [DOI] [PubMed] [Google Scholar]
- Brawand D, Soumillon M, Necsulea A, Julien P, Csárdi G, Harrigan P, Weier M, Liechti A, Aximu-Petri A, Kircher M, et al. The evolution of gene expression levels in mammalian organs. Nature. 2011;478:343–348. doi: 10.1038/nature10532. [DOI] [PubMed] [Google Scholar]
- Cormier TA, Prakash SK, Magner DB, Zoghbi HY, Van den Veyver IB. Analysis of Mid1, Hccs, Arhgap6, and Msl3l1 in X-linked polydactyly (Xpl) and Patchy-fur (Paf) mutant mice. Mamm Genome. 2001;12:796–798. doi: 10.1007/s00335-001-1006-5. [DOI] [PubMed] [Google Scholar]
- Cotney J, Leng J, Oh S, DeMare LE, Reilly SK, Gerstein MB, Noonan JP. Chromatin state signatures associated with tissue-specific gene expression and enhancer activity in the embryonic limb. Genome Res. 2012;22:1069–1080. doi: 10.1101/gr.129817.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci USA. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez MH, Ayoub AE, Rakic P. POU-III transcription factors (Brn1, Brn2, and Oct6) influence neurogenesis, molecular identity, and migratory destination of upper-layer cells of the cerebral cortex. Cereb Cortex Published online August. 2012;14:2012. doi: 10.1093/cercor/bhs252. http://dx.doi.org/10.1093/cercor/bhs252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duboc V, Logan MPO. Regulation of limb bud initiation and limb-type morphology. Dev Dyn. 2011;240:1017–1027. doi: 10.1002/dvdy.22582. [DOI] [PubMed] [Google Scholar]
- Feschotte C. Transposable elements and the evolution of regulatory networks. Nat Rev Genet. 2008;9:397–405. doi: 10.1038/nrg2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göke J, Jung M, Behrens S, Chavez L, O’Keeffe S, Timmermann B, Lehrach H, Adjaye J, Vingron M. Combinatorial binding in human and mouse embryonic stem cells identifies conserved enhancers active in early embryonic development. PLoS Comput Biol. 2011;7:e1002304. doi: 10.1371/journal.pcbi.1002304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green P, Ewing B. Comment on “Evidence of abundant purifying selection in humans for recently acquired regulatory functions”. Science. 2013;340:682. doi: 10.1126/science.1233195. discussion 682. [DOI] [PubMed] [Google Scholar]
- Junion G, Spivakov M, Girardot C, Braun M, Gustafson EH, Birney E, Furlong EEM. A transcription factor collective defines cardiac cell fate and reflects lineage history. Cell. 2012;148:473–486. doi: 10.1016/j.cell.2012.01.030. [DOI] [PubMed] [Google Scholar]
- King MC, Wilson AC. Evolution at two levels in humans and chimpanzees. Science. 1975;188:107–116. doi: 10.1126/science.1090005. [DOI] [PubMed] [Google Scholar]
- Kunarso G, Chia NY, Jeyakani J, Hwang C, Lu X, Chan YS, Ng HH, Bourque G. Transposable elements have rewired the core regulatory network of human embryonic stem cells. Nat Genet. 2010;42:631–634. doi: 10.1038/ng.600. [DOI] [PubMed] [Google Scholar]
- Lejard V, Blais F, Guerquin MJ, Bonnet A, Bonnin MA, Havis E, Malbouyres M, Bidaud CB, Maro G, Gilardi-Hebenstreit P, et al. EGR1 and EGR2 involvement in vertebrate tendon differentiation. J Biol Chem. 2011;286:5855–5867. doi: 10.1074/jbc.M110.153106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lettice LA, Heaney SJH, Purdie LA, Li L, de Beer P, Oostra BA, Goode D, Elgar G, Hill RE, de Graaff E. A long-range Shh enhancer regulates expression in the developing limb and fin and is associated with preaxial polydactyly. Hum Mol Genet. 2003;12:1725–1735. doi: 10.1093/hmg/ddg180. [DOI] [PubMed] [Google Scholar]
- Lindblad-Toh K, Garber M, Zuk O, Lin MF, Parker BJ, Washietl S, Kheradpour P, Ernst J, Jordan G, Mauceli E, et al. Whole Genome Assembly Team; Baylor College of Medicine Human Genome Sequencing Center Sequencing Team; Genome Institute at Washington University. A high-resolution map of human evolutionary constraint using 29 mammals. Nature. 2011;478:476–482. doi: 10.1038/nature10530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo P, Bayliss MT, Heinegård D. A novel cartilage protein (CILP) present in the mid-zone of human articular cartilage increases with age. J Biol Chem. 1998;273:23463–23468. doi: 10.1074/jbc.273.36.23463. [DOI] [PubMed] [Google Scholar]
- Marzke MW, Marzke RF. Evolution of the human hand: approaches to acquiring, analysing and interpreting the anatomical evidence. J Anat. 2000;197:121–140. doi: 10.1046/j.1469-7580.2000.19710121.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuya H, Sagai T, Moriwaki K, Shiroishi T. Multigenic control of the localization of the zone of polarizing activity in limb morphogenesis in the mouse. Dev Biol. 1997;182:42–51. doi: 10.1006/dbio.1996.8457. [DOI] [PubMed] [Google Scholar]
- McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, Wenger AM, Bejerano G. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol. 2010;28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean CY, Reno PL, Pollen AA, Bassan AI, Capellini TD, Guenther C, Indjeian VB, Lim X, Menke DB, Schaar BT, et al. Human-specific loss of regulatory DNA and the evolution of human-specific traits. Nature. 2011;471:216–219. doi: 10.1038/nature09774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Xu Z, Zhang X, Wang L, Gimble JM, Lander ES, Rosen ED. Comparative epigenomic analysis of murine and human adipogenesis. Cell. 2010;143:156–169. doi: 10.1016/j.cell.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Rahilly R, Müller F, Streeter GL. Developmental Stages in Human Embryos. Washington, DC: Carnegie Institution of Washington; 1987. [Google Scholar]
- Pollard KS, Salama SR, Lambert N, Lambot MA, Coppens S, Pedersen JS, Katzman S, King B, Onodera C, Siepel A, et al. An RNA gene expressed during cortical development evolved rapidly in humans. Nature. 2006;443:167–172. doi: 10.1038/nature05113. [DOI] [PubMed] [Google Scholar]
- Prabhakar S, Noonan JP, Pääbo S, Rubin EM. Accelerated evolution of conserved noncoding sequences in humans. Science. 2006;314:786. doi: 10.1126/science.1130738. [DOI] [PubMed] [Google Scholar]
- Prabhakar S, Visel A, Akiyama JA, Shoukry M, Lewis KD, Holt A, Plajzer-Frick I, Morrison H, Fitzpatrick DR, Afzal V, et al. Human-specific gain of function in a developmental enhancer. Science. 2008;321:1346–1350. doi: 10.1126/science.1159974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada-Iglesias A, Bajpai R, Prescott S, Brugmann SA, Swigut T, Wysocka J. Epigenomic annotation of enhancers predicts transcriptional regulators of human neural crest. Cell Stem Cell. 2012;11:633–648. doi: 10.1016/j.stem.2012.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt D, Wilson MD, Ballester B, Schwalie PC, Brown GD, Marshall A, Kutter C, Watt S, Martinez-Jimenez CP, Mackay S, et al. Five-vertebrate ChIP-seq reveals the evolutionary dynamics of transcription factor binding. Science. 2010;328:1036–1040. doi: 10.1126/science.1186176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenwolf GC, Bleyl SB, Brauer PR, Francis-West PH. Larsen’s Human Embryology. Ann Arbor, MI: Churchill Livingstone/Elsevier; 2009. [Google Scholar]
- Shen Y, Yue F, McCleary DF, Ye Z, Edsall L, Kuan S, Wagner U, Dixon J, Lee L, Lobanenkov VV, Ren B. A map of the cis-regulatory sequences in the mouse genome. Nature. 2012;488:116–120. doi: 10.1038/nature11243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata Y, Sheffield NC, Fedrigo O, Babbitt CC, Wortham M, Tewari AK, London D, Song L, Lee BK, Iyer VR, et al. Extensive evolutionary changes in regulatory element activity during human origins are associated with altered gene expression and positive selection. PLoS Genet. 2012;8:e1002789. doi: 10.1371/journal.pgen.1002789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitz F, Gonzalez F, Duboule D. A global control region defines a chromosomal regulatory landscape containing the HoxD cluster. Cell. 2003;113:405–417. doi: 10.1016/s0092-8674(03)00310-6. [DOI] [PubMed] [Google Scholar]
- Taher L, Collette NM, Murugesh D, Maxwell E, Ovcharenko I, Loots GG. Global gene expression analysis of murine limb development. PLoS ONE. 2011;6:e28358. doi: 10.1371/journal.pone.0028358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarchini B, Duboule D. Control of Hoxd genes’ collinearity during early limb development. Dev Cell. 2006;10:93–103. doi: 10.1016/j.devcel.2005.11.014. [DOI] [PubMed] [Google Scholar]
- Visel A, Minovitsky S, Dubchak I, Pennacchio LA. VISTA Enhancer Browser—a database of tissue-specific human enhancers. Nucleic Acids Res. 2007;35(Database issue):D88–D92. doi: 10.1093/nar/gkl822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakabayashi T, Matsumine A, Nakazora S, Hasegawa M, Iino T, Ota H, Sonoda H, Sudo A, Uchida A. Fibulin-3 negatively regulates chondrocyte differentiation. Biochem Biophys Res Commun. 2010;391:1116–1121. doi: 10.1016/j.bbrc.2009.12.034. [DOI] [PubMed] [Google Scholar]
- Ward LD, Kellis M. Evidence of abundant purifying selection in humans for recently acquired regulatory functions. Science. 2012;337:1675–1678. doi: 10.1126/science.1225057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward LD, Kellis M. Response to comment on “Evidence of Abundant Purifying Selection in Humans for Recently Acquired Regulatory Functions. Science. 2013;340:682b. doi: 10.1126/science.1233366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weedon MN, Lango H, Lindgren CM, Wallace C, Evans DM, Mangino M, Freathy RM, Perry JRB, Stevens S, Hall AS, et al. Genome-wide association analysis identifies 20 loci that influence adult height. Nat Genet. 2008;40:575–583. doi: 10.1038/ng.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittkopp PJ, Kalay G. Cis-regulatory elements: molecular mechanisms and evolutionary processes underlying divergence. Nat Rev Genet. 2012;13:59–69. doi: 10.1038/nrg3095. [DOI] [PubMed] [Google Scholar]
- Wray GA. The evolutionary significance of cis-regulatory mutations. Nat Rev Genet. 2007;8:206–216. doi: 10.1038/nrg2063. [DOI] [PubMed] [Google Scholar]
- You FM, Huo N, Gu YQ, Luo MC, Ma Y, Hane D, Lazo GR, Dvorak J, Anderson OD. BatchPrimer3: a high throughput web application for PCR and sequencing primer design. BMC Bioinformatics. 2008;9:253. doi: 10.1186/1471-2105-9-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeller R, López-Ríos J, Zuniga A. Vertebrate limb bud development: moving towards integrative analysis of organogenesis. Nat Rev Genet. 2009;10:845–858. doi: 10.1038/nrg2681. [DOI] [PubMed] [Google Scholar]
- Zhao J, Li M, Bradfield JP, Zhang H, Mentch FD, Wang K, Sleiman PM, Kim CE, Glessner JT, Hou C, et al. The role of height-associated loci identified in genome wide association studies in the determination of pediatric stature. BMC Med Genet. 2010;11:96. doi: 10.1186/1471-2350-11-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.