Abstract
While several major classes of neocortical neural precursor cells have been identified, the lineal relationships and molecular profiles of these cells are still largely unknown. Furthermore, the individual contribution of each cell class to neocortical growth during normal development and in neurodevelopmental disorders has not been determined. Using a novel fate-mapping approach, we demonstrate that precursors in the embryonic ventricular (VZ) and subventricular zones (SVZ), which give rise to excitatory neurons, are divided into distinct subtypes based on lineage profile, morphology, and transcription factor expression in vivo. Using this technique, we show that short neural precursors are a unique class of VZ intermediate progenitors derived from radial glial cells and are distinct from the multipolar Tbr2(+) intermediate progenitors, which divide in the SVZ. To test whether these multiple groups of intermediate progenitors are redundant or whether they are necessary for proper neocortical growth, we measured precursor cell diversity in the Ts65Dn mouse model of Down syndrome (DS), which exhibits reduced neurogenesis and postnatal microcephaly. We report that SNP generation is markedly reduced in the Ts65Dn VZ during mid-neurogenesis, indicating that faulty specification of this progenitor pool is a central component of the neocortical abnormality in DS. Together, these findings demonstrate that neocortical neurons are produced via multiple indirect routes during embryonic development and that these parallel streams of neurogenesis collectively contribute to the proper growth and development of the neocortex.
Introduction
The stem and progenitor cells residing in the ventricular (VZ) and subventricular zones (SVZ) of the dorsal mammalian telencephalon divide to produce excitatory neurons that parcellate the neocortex into six layers. Recent work has shown that these neurons are derived simultaneously from multiple classes of proliferative cells with unique morphological and molecular properties (Breunig et al., 2011; Fietz and Huttner, 2011; Lui et al., 2011). In mammals, at least four groups of progenitors contribute to total neuronal output: radial glial cells (RGCs) and short neural precursors (SNPs) compose the VZ while intermediate progenitor cells (IPCs) and basal radial glia (bRG), which are derived from VZ cells, populate the SVZ (Noctor et al., 2001; Gal et al., 2006; Kowalczyk et al., 2009; Fietz et al., 2010; Hansen et al., 2010; Shitamukai et al., 2011; Wang et al., 2011). However, several studies suggest that these categories alone do not faithfully represent the level of heterogeneity present in the germinal zones of the developing forebrain, as additional subclasses of cells can be distinguished by antigenicity, transcriptional profiling, and lineage potential (Hartfuss et al., 2001; Mizutani et al., 2007; Kawaguchi et al., 2008; Pinto et al., 2008; Stancik et al., 2010). In addition, despite the recent increase in knowledge of VZ and SVZ cell types, details on the lineal relationships between specific neuronal progenitors and their relative contribution to the growth and function of the neocortex remain to be elucidated.
Here, we employ in utero electroporation (IUE) to perform a series of novel fate-mapping experiments designed to simultaneously identify and separate the VZ and SVZ lineages, which coexist in the developing neocortex and to examine their hierarchical relationships. Our results demonstrate that in addition to generating neurons directly, RGCs give rise to at least two classes of spatially segregated and molecularly distinct IPCs. While it is well established that RGCs divide to produce multipolar Tbr2(+) IPCs, which populate the SVZ (Noctor et al., 2004; Englund et al., 2005), we have identified an additional class of IPCs residing in the VZ. In particular, we find for the first time that SNPs are Pax6(+) IPCs lineally derived from RGCs that undergo neurogenic divisions at the ventricular surface. These data sharply demarcate the cytogenetic and molecular expression profiles of IPCs and suggest that parallel streams of indirect neurogenesis are used during the formation of the mammalian neocortex.
To test whether each of these independent intermediate progenitor groups are necessary for proper neocortical growth and whether they may be preferentially affected in developmental disorders, we measured precursor diversity in the Ts65Dn mouse model of Down syndrome (DS), which exhibits a profound neurogenesis defect during embryonic development (Chakrabarti et al., 2007). The results demonstrate that the relative distribution of VZ/SVZ cell types is altered during Ts65Dn development and is highlighted by a marked reduction in the specification of SNPs during mid-neurogenesis. To our knowledge, this is the first disturbance in a specific class of neural precursors that can be directly attributed to a neurodevelopmental disability, illustrating that precise control of precursor heterogeneity is critical for proper cerebral cortical development.
Materials and Methods
cDNA cloning and preparation.
The Cre-Stoplight plasmid (Cre-SL v2.4), a generous gift from Montana Molecular, was used to generate the pGlast-SL, pBlbp-SL, pTα1-SL, and pCAG-SL plasmids. A SpeI/SacI double digest was performed to excise the CMV promoter from the Cre-SL v2.4 plasmid. A 1.1 kb fragment of the rat Tubulin α-1 promoter or 1.7 kb fragment of the human Blbp promoter was amplified with Phusion high-fidelity polymerase (Thermo Scientific) with primers to add SpeI and SacI restriction enzyme sites to the ends of each sequence. Because the 2.1 kb fragment of the human Glast promoter contains an internal SpeI site, we instead added an XbaI site to the 5′ by PCR, which produces compatible overhangs with SpeI following digestion. PCR products were digested and then ligated with Mighty Mix DNA ligase (Takara/Clontech) into the Cre-SL v2.4 backbone. Plasmids containing each promoter fragment described previously were used as templates for PCR (Gal et al., 2006; Stancik et al., 2010). The pGlast-Cre and pTα1-Cre plasmids have been described previously (Stancik et al., 2010). The Blbp promoter (from Blbp-GFP) was subcloned into the pGlast-Cre backbone in place of the pGlast promoter. DNA for injection was amplified in DH5α chemically competent cells and purified using EndoFree DNA Maxi Prep kits (Qiagen).
IUE.
IUE was performed, as described previously (Gal et al., 2006), on timed pregnant CD-1 dams purchased from Charles River Laboratories at embryonic day 13.5 (E13.5) or E14.5 or timed pregnant Ts65Dn females generated in our colonies. Briefly, dams were anesthetized via intraperitoneal injection of a ketamine/xylazine mixture and the uterine horns were exposed by midline laparotomy. One to two microliters of plasmid DNA mixed with 0.1% fast green dye (Sigma-Aldrich) was injected intercerebrally, through the uterine wall and amniotic sac, via pulled glass micropipette. Cre and reporter plasmid vectors were mixed at a 1:1 ratio by copy number and the final concentration of each plasmid was between 2 and 3 μg/μl. The anode of a Tweezertrode (Harvard Apparatus) was placed over the dorsal telencephalon above the uterine muscle and four 35 V pulses (50 ms duration separated by a 950 ms interval) were applied with a BTX ECM830 pulse generator (Harvard Apparatus). Following electroporation, the uterine horns were returned to the abdomen and the cavity was filled with a warm 0.9% saline solution. The incisions were closed with silk sutures. The dams were then placed in a clean cage and monitored closely during recovery. These procedures were reviewed and approved by the Institutional Animal Care and Use Committee at the Boston University School of Medicine.
Ts65Dn mice.
Ts65Dn and Euploid B6EiC3 animals were generated by backcrossing Ts65Dn females to C57BL/6JEi × C3H/HeSnJ F1 hybrid (B6EiC3) males. The parental generation was obtained from The Jackson Laboratory. Quantitative PCR genotyping was performed on genomic DNA extracted from tail tips (Chakrabarti et al., 2007). Mating pairs were co-housed and checked each morning for the presence of a vaginal plug. The day a plug was detected was considered E0.5. All experiments involving animals were performed in accordance with institutional and federal guidelines.
Tissue processing, confocal imaging, and cell counting.
Embryos were killed 24 or 48 h post electroporation and heads containing electroporated fields of equivalent size and location were fixed overnight in 4% paraformaldehyde. The following day brains were dissected and embedded in 3% agar and sliced using a VT1000S vibrating blade microtome (Leica) into 50 μm sections and mounted on SuperFrost slides in Vectashield mounting medium containing DAPI. Imaging and counts were restricted to sections at the level of the future sensorimotor cortex, corresponding to coronal plates 242–256 of the mouse E15.5 reference Allen Brain Atlas.
Sections were imaged using a Zeiss LSM 710 confocal microscope. Twenty micrometer, 40× Z-stack images were acquired using the Zen software package. ZsGreen and mCherry-positive cells were manually scored and counted using the LSM image browser software. In our counting procedure, we classified cells as “ZsGreen(+)” if they expressed ZsGreen but contained no signal for mCherry. Correspondingly, cells were classified “mCherry(+)” if they contained mCherry signal, whether or not they were also ZsGreen(+) due to perdurance of the fluorescent protein or remaining copies of unrecombined reporter plasmid. Sections from a minimum of three embryos were counted and analyzed for each experiment, and surgeries for each combination of plasmids were repeated to confirm the results.
Immunohistochemistry.
The heads of positively electroporated embryos were harvested 24 h postsurgery, fixed in 4% paraformaldehyde overnight, and cryoprotected in 30% sucrose for 24–48 h. The tissue was then frozen in OCT compound in tissue molds with a dry ice/ethanol bath. Frozen tissue was cut into 18 μm sections using an HM 560 Cryostar cryostat and mounted and dried on SuperFrost slides.
Before immunostaining, antigen retrieval was performed by microwaving the cryosections in sodium citrate buffer (10 mm, pH 6) at 800 W for 1 min followed by 80 W for 10 min. Sections were then blocked in diluent (5% goat serum, 0.3% Triton X-100, 1× PBS) for 1 h at room temperature. Incubation with primary antibodies, Anti-Pax6 (1:1000; Covance) or Anti-Tbr2 (1:500; Abcam), was performed overnight at 4°C. Following three 5 min washes in PBS, sections were incubated for 2 h at room temperature in diluent containing the appropriate Alexa 633-conjugated secondary antibodies (1:250 for all). Sections were washed an additional three times for 5 min and mounted with Vectashield mounting medium containing DAPI. Then 40× Z-stack images (14 μm) were acquired using an upright Zeiss LSM 710 microscope and positive cells were identified and counted using LSM image browser software.
Slice culture/multiphoton time-lapse imaging.
Twenty-four hours post electroporation, embryos were killed and freshly dissected in complete HBSS, which consisted of 1× HBSS with 2 mm HEPES, 2 mm d-glucose, 0.2 mm CaCl2, 0.2 mm MgSO4, and 2 mm NaHCO3. Brains were screened for positive expression of ZsGreen and mCherry using an Olympus MVX10 stereomicroscope equipped with epifluorescence illumination. The brains were embedded in 3% low melting point agarose in DMEM and 300–450 μm organotypic sections were prepared using a VT1000S vibrating blade microtome. Slices were incubated at 37°C on collagen-coated transwell inserts placed in six well dishes in Neuroimaging medium (NIM) for a 1 h recovery period before imaging. NIM consisted of Neurobasal medium supplemented with B27, N2 supplement, 20% fetal bovine serum, 1% PenStrep, glutamax, 10 ng/ml bFGF, and 10 ng/ml EGF.
For imaging, the organotypic slices were placed in 35 mm dishes in NIM and positioned on a heated micro-incubation chamber (DH-40i; Warner Systems) attached to the stage of the Zeiss LSM 710 microscope coupled to a Chameleon Vision-S Ti:Sapphire laser (Coherent). ZsGreen and mCherry were simultaneously excited at 995 nm and emitted light was collected with two LSM binary GaAsP module photodetectors. Z-stack images (100–200 μm, 2 μm intervals) were collected at 6 min intervals with a 20× 1.0 NA water-dipping objective for the duration of the experiment, which typically ran overnight for >8–10 h. Individual dividing cells were analyzed further by postprocessing in both the ZEN software and Volocity (Improvision) software packages.
Statistical analysis.
Statistical analyses were performed using StatView statistical analysis software. For experiments comparing two treatment groups, an unpaired t test was performed to assess the statistical significance. For experiments in which multiple treatment groups or time points were examined, one-way ANOVA was performed to assess the statistical significance between groups. All experiments (each with a minimum n = 3 subjects) were repeated at least two times to confirm the results.
Results
Lineage-specific reporter expression segregates VZ/SVZ cells into distinct subpopulations
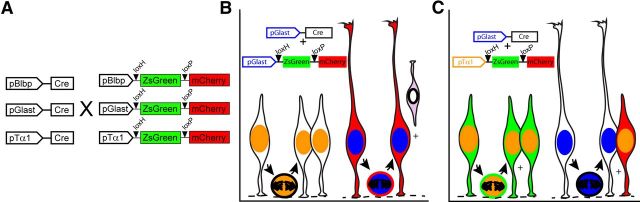
Using IUE to express fluorescent reporter molecules under the control of cell type or temporally specific DNA promoters is a powerful technique to map cell lineage and fate in the mammalian brain (Wang et al., 2007; Chen and LoTurco, 2012; Ohtaka-Maruyama et al., 2012). Previously, we demonstrated that mitotic RGCs and SNPs can be labeled and distinguished via reporter expression driven by the Blbp/Glast (pBlbp/pGlast) and Tubulin α 1 (pTα1) promoters, respectively (Gal et al., 2006). We therefore developed an intersectional approach, which employs the combinatorial activity of two promoters to determine the lineal relationships between cells independently labeled by these reporters. Specifically, we paired lineage-specific Cre driver plasmids with a series of dual fluorescent reporter plasmids that we modified from the Cre Stoplight design (Yang and Hughes, 2001). These reporter constructs, which we have named Blbp-Stoplight (pBlbp-SL), Glast-Stoplight (pGlast-SL), and Tα1-Stoplight (pTα1-SL), function as binary switches that drive the expression of ZsGreen in the absence of Cre activity or mCherry following Cre-dependent recombination (Fig. 1A).
Figure 1.
Schematic illustration of lineage-specific Cre driver and Stoplight reporter plasmids. A, In separate experiments, each Cre driver was coelectroporated in combination with each Stoplight reporter. Theoretical results of pGlast-Cre × pGlast-SL (B) and pGlast-Cre × pTα1-SL (C) coelectroporations in dorsal VZ cells. Fluorescent protein expression was restricted to subsets of cells in which the Stoplight reporter was active. In the absence of overlapping Cre expression, Stoplight+ cells were identified by the expression of ZsGreen. Cre-dependent recombination of Stoplight reporter plasmids yielded mCherry+ intracellular labeling in cells exhibiting simultaneous expression of both Cre and Stoplight plasmids, or in the Stoplight+ progeny derived from Cre-expressing parent cells.
In theory, coelectroporation of the dorsal telencephalic VZ with different combinations of Stoplight and Cre driver plasmids labels cells green or red depending on the following: (1) whether or not both reporter and Cre driver promoters are active concurrently in the same cell or (2) their lineage of origin (whether or not they are derived from parent cells expressing Cre). For example, coelectroporation of cells with pGlast-Cre × pGlast-SL highlights RGCs red due to Cre-dependent recombination and expression of mCherry, while pGlast(−) cells, such as SNPs, are excluded from the fluorescent population due to pGlast promoter inactivity (Fig. 1B). Alternatively, mixing Cre driver and Stoplight plasmids theoretically enables a hierarchical, time-dependent analysis of promoter expression in individual cell types. For instance, coelectroporation of cells with pGlast-Cre × pTα1-SL (Fig. 1C) should initially feature ZsGreen-expressing SNPs as well as unlabeled (pTα1(−)) RGCs. However, pGlast-Cre expression in RGCs nevertheless results in recombination of the silent pTα1-SL plasmid, ensuring that any pTα1(+) cells subsequently generated via RGC cell division will express mCherry (Fig. 1C). In this example, existing SNPs can be distinguished from newly generated SNPs based on fluorescent protein expression controlled by their parent cell of origin.
To test the validity of this approach (i.e., to measure recombination efficiency and to rule out differences in promoter strength), we electroporated dorsal VZ/SVZ cells with Cre and reporter plasmids containing matched promoter sequences at E14.5 in vivo. We selected 24 h as our primary short-term endpoint as it is sufficient time for electroporated cells to complete only one round of the cell cycle (Stancik et al., 2010). Importantly in these pairings, reporter plasmids approached full recombination within 24 h (Fig. 2A,B). Specifically, pGlast-Cre × pGlast-SL or pBlbp-Cre × pBlbp-SL resulted in >96% mCherry(+) cells 24 h following electroporation (Fig. 2B). Similarly, >93% of cells electroporated with pTα1-Cre × pTα1-SL were mCherry(+) in the same time frame. These results demonstrate that Cre expression and activity driven by each promoter is robust and that recombination of the target reporter plasmids occurs at the same rapid pace in all combinations.
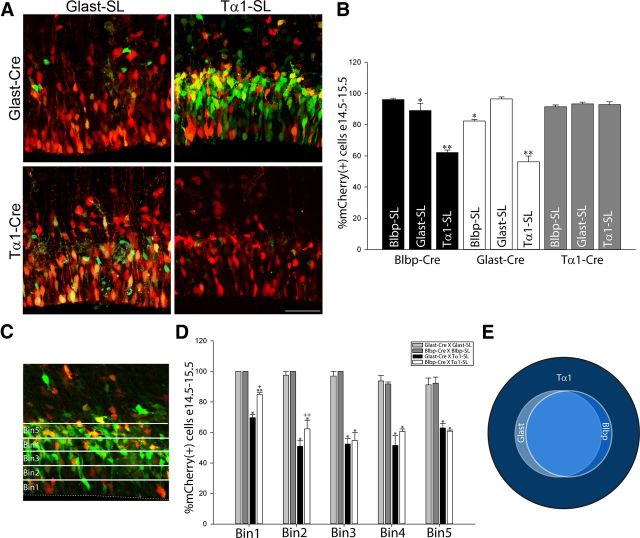
Figure 2.
Lineal fate mapping reveals heterogeneity of VZ/SVZ progenitors. A, Representative images of VZ/SVZ cells coelectroporated with pGlast-Cre or pTα1-Cre paired with pGlast-SL or pTα1-SL, 24 h after E14.5 IUE. B, Percentage of mCherry+ cells 24 h post-IUE for each plasmid combination examined. (*, 0.001 < p < 0.05 or **p < 0.0001 vs pBlbp-Cre × Blbp-SL, pGlast-Cre × pGlast-SL, and pTα1-Cre × pTα1-SL). Recombination in matched promoter pairs approached 100% in all cases. However, significant numbers of ZsGreen+ cells remained in pBlbp-Cre × pGlast-SL, pGlast-Cre × pBlbp-SL combinations. In addition, pGlast-Cre × pTα1-SL and pBlbp-Cre × pTα1-SL coelectroporations yielded comparatively fewer mCherry+ cells and a corresponding larger number of ZsGreen+ cells. Recombination for all three Stoplight reporters neared 100% when coelectroporated with pTα1-Cre, indicating that pTα1 is expressed by most electroporated precursors. C, D, Percentage of mCherry(+) cells 24 h post-IUE within each of the five 20 μm bins comprising the VZ and SVZ. (*p < 0.0001 or **, 0.0005 < p < 0.005, vs pGlast-Cre × pGlast-SL and pBlbp-Cre × pBlbp-SL; +p = 0.002 or ++p = 0.017 vs pGlast-Cre × pTα1-SL). E, Venn diagram illustrating the relationship between pBlbp, pGlast, and pTα1-labeled cells. While all pGlast+ and pBlbp+ cells coexpress pTα1, they are nested within a larger population of pTα1+ cells, which express neither pGlast nor pBlbp. Error bars, SEM.
In contrast to the results from matched promoter experiments, when VZ/SVZ cells were coelectroporated with mixed Glast and Blbp Cre and Stoplight plasmids (i.e., pGlast-Cre × Blbp-SL or pBlbp-Cre × Glast-SL) we detected a modest but significant decrease in the number of mCherry(+) cells from E14.5 to E15.5. In particular, on average 85% of the cells in these pairings expressed mCherry while 15% expressed ZsGreen, suggesting that this latter group of cells expressed pGlast or pBlbp, but not both of these promoter sequences (Fig. 2B). This finding highlights the heterogeneity among Glast- and Blbp-expressing cells, which has been previously described, and provides a new molecular labeling method with which to isolate individual cell types (Hartfuss et al., 2001; 2003).
Next, we coelectroporated pGlast-Cre × pTα1-SL. In sharp contrast to experiments with matched promoter sequences, we observed a significant decrease in the number of mCherry(+) cells (56%), mirrored by an increase in the number of ZsGreen(+) cells at E15.5 (44%) (Fig. 2A,B). Similar results were obtained following pBlbp-Cre × pTα1-SL coelectroporation (62% mCherry(+) cells, 38% ZsGreen(+) cells; Figure 2B). To determine whether these subclasses of cells were spatially segregated, we counted labeled cells based on their position relative to the ventricle by dividing the VZ/SVZ into five 20 μm bins (Fig. 2C,D). While we observed near complete recombination and an equal distribution of mCherry(+) cells following co-IUE with matched plasmid pairs (pGlast-Cre × pGlast-SL or pBlbp × Blbp-SL), coelectroporation of pGlast-Cre × pTα1-SL or pBlbp-Cre × pTα1-SL resulted in a decrease in the number of mCherry(+) cells that was apparent across all bins relative to matched promoter samples (Fig. 2D). Interestingly, a larger proportion of pTα1 cells in the apical VZ (bins 1–2) expressed mCherry due to pBlbp-Cre activity compared with pGlast-Cre, again supporting the finding that a subgroup of VZ cells exhibits nonoverlapping expression of pGlast and pBlbp. Nevertheless, pTα1-ZsGreen(+) cells that do not express Cre driven by the pGlast or pBlbp promoters are present throughout the radial axis of the VZ/SVZ and are thus likely present in all phases of the cell cycle (Takahashi et al., 1993). The pTα1-mCherry(+) cells resulting from pBlbp-Cre or pGlast-Cre recombination may reflect either overlapping expression of the pTα1, pGlast, or pBlbp promoters in the same cell or, potentially, that some pTα1(+) cells are derived from pGlast(+) or pBlbp(+) parent cells through cell division within 24 h.
Unexpectedly, when the reverse experiments were performed (pTα1-Cre × pGlast-SL or pTα1-Cre × pBlbp-SL), we observed near complete recombination 24 h post-IUE (Fig. 2B). This strongly suggests that the pTα1-Cre driver is sufficiently active to generate recombination in most pGlast(+) and pBlbp(+) RGC precursors, greatly increasing estimates of overlap in Tα1, Glast, and Blbp expression noted in our previous study (Gal et al., 2006). Nevertheless, when all of the cells labeled by these sensitive intersectional tools are considered together, the data indicate that the VZ and SVZ populations identified by the expression of pGlast or pBlbp are nested within a larger class of cells that express pTα1, almost half of which express neither pGlast nor pBlbp (Fig. 2E). In other words, while this dual reporter fate-mapping approach demonstrates that most of the VZ precursor cells exhibit pTα1 activity, our multiplex methodology can be used to subdivide this large pTα1-expressing population into pTα1(+)/pGlast(+) and pTα1(+)/pBlbp(+) populations that express mCherry, and pTα1(+)/pGlast(−) and pTα1(+)/pBlbp(−) populations that express ZsGreen. This fate-mapping approach therefore exploits the significant molecular expression differences between VZ and SVZ precursors in the mammalian telencephalon and the dual color reporter strategy enables a simultaneous cellular and molecular analysis of each defined cell type.
Blbp and Glast promoter-dependent Cre activity segregates pTα1(+) cells into morphologically distinct cohorts
To resolve whether the molecular differences we identified between precursor populations coincide with morphological distinctions, we scored fate-mapped VZ/SVZ cells as either “radial-like” or “multipolar” in 3D Z-stack reconstructions. In these counts, we confined our analysis to cells located within 100 μm of the ventricular surface and included unipolar and bipolar cells with apical processes in the radial-like group and cells with more than two processes into the multipolar group (Fig. 3A, inset). As expected, mCherry(+) radial-like VZ cells were the primary group labeled by pGlast-SL and pBlbp-SL (1.8-fold for each vs pTα1-SL; Fig. 3A), and we noted a trend toward an increase in mCherry(+) radial-like cells at the expense of mCherry(+) multipolar cells identified by Blbp-SL compared with Glast-SL. In contrast, when combined with pGlast-Cre or pBlbp-Cre, we found that the pTα1-SL reporter labeled fivefold more ZsGreen(+) VZ cells with radial-like morphology and twofold more ZsGreen(+) multipolar cells than the Blbp-SL and Glast-SL reporters (Fig. 3A). Interestingly, pGlast-Cre × pTα1-SL electroporated sections contained a 56% increase in ZsGreen radial-like cells relative to pBlbp-Cre × pTα1-SL. This finding is consistent with the observation of a greater number of mCherry(+) cells in the apical half of the VZ in pBlbp-Cre × pTα1-SL crosses (Fig. 2D). Together, these morphological counts indicate that radial-like and multipolar precursor cells were labeled to varying degrees by each Cre/Stoplight combination. However, the pTα1-SL population included sizable cohorts of ZsGreen(+) radial-like and multipolar cells that do not express pGlast and pBlbp within the 24 h time period examined, suggesting that these cells are either separate from the RGC lineage entirely or that they are a population of molecularly distinct RGCs.
Figure 3.
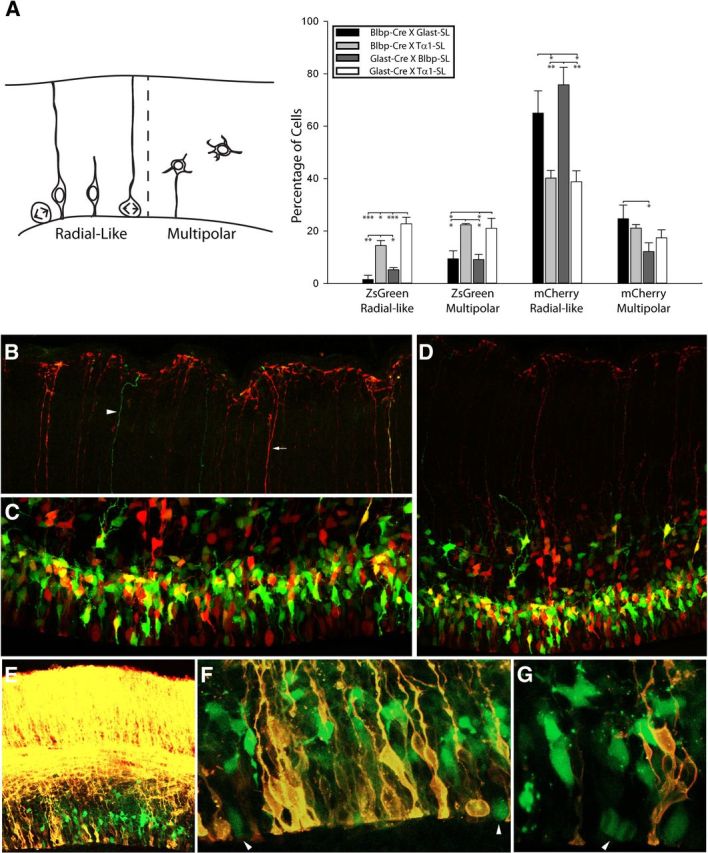
Cre-Stoplight combinations uncover morphological distinctions between VZ and SVZ cells. A, Schematic illustrating the morphology of radial-like and multipolar cells (inset) and quantification of the percentage of each morphological class 24 h following coelectroporation with Stoplight and Cre-driver plasmids at E14.5. Counts included electroporated cells within 100 μm from the ventricular surface. While all plasmid combinations yielded both ZsGreen+ and mCherry+ radial-like and multipolar cells, the pGlast-Cre × pTα1-SL and pBlbp-Cre × pTα1-SL cotransfected brains were highly enriched with ZsGreen+ radial-like and multipolar cells. In contrast, most of the cells in the pGlast-Cre × pBlbp-SL and pBlbp-Cre × pGlast-SL brains were mCherry+ radial-like cells. Significant differences within each morphological group are denoted by asterisks (*, 0.005 < p < 0.05; **, 0.0005 < p < 0.005; ***p < 0.0005). B–D, Representative images of cells electroporated with pBlbp-Cre × pTα1-SL (IUE: E14.5–E15.5). Few ZsGreen+ VZ/SVZ cells (C) extended basal processes to the pia (B). Arrow and arrowhead in B denote examples of mCherry+ and ZsGreen+ basal fibers. E–F, Many pTα1-ZsGreen+ cells fail to incorporate DiI applied to the pial surface (pGlast-Cre × pTα1-SL shown). Arrowheads in F and G denote mitotic pTα1-ZsGreen+ cells. Error bars, SEM.
To determine whether pTα1-ZsGreen(+) and pTα1-mCherry(+) radial-like cells were RGCs (i.e., possessing long basal processes contacting the pial surface), we coelectroporated either pBlbp-Cre × pTα1-SL or pGlast Cre × pTα1-SL and performed a series of 3D reconstruction confocal imaging experiments. First, we imaged pial end feet as well as cell bodies in the VZ/SVZ of the same 60-μm-thick brain section to determine whether ZsGreen(+) and mCherry(+) cells extended pial-contacting basal processes. Interestingly, despite the fact that 38% of the cells in the VZ expressed ZsGreen but not mCherry (pBlbp-Cre × Tα1-SL; Fig. 2B,C), and 14% of the ZsGreen(+) cells possessed radial-like morphology (Fig. 3A), we detected very few ZsGreen(+) basal fibers with end foot processes at the pia. Instead, the majority of pial projections were mCherry(+) (Fig. 3B,D).
As a second approach, we applied DiI to the pia of electroporated brains to trace basal fibers of RGCs and label their somas (Fig. 3E–G). For this experiment, we replaced the mCherry coding sequence in the pTα1-SL plasmid with a STOP cassette to further highlight pTα1(+) cells that express only ZsGreen but do not show pGlast or pBlbp-dependent Cre activity and expression. Consistent with the absence of ZsGreen(+) pial end feet, we observed that many pTα1-ZsGreen(+) cells failed to incorporate DiI, including cells rounded up in M-phase at the surface of the ventricle (Fig. 3F,G; pGlast-Cre × pTα1-SL shown, similar results seen with pBlbp-Cre). These results support the previous observation that many VZ cells exhibit a “short” morphology defined by the absence of a basal process that spans the cortical plate (Gal et al., 2006).
In addition to multipolar precursors in the SVZ, we noted a number of pTα1-mCherry(+) cells above the band of multipolar SVZ cells, which appeared to be bRG cells as they possessed long basal fibers but had no discernible apical processes. Interestingly, in time-lapse multiphoton imaging experiments, we were able to identify only pTα1-mCherry(+) cells dividing as bRG in the SVZ, indicating that the murine bRG population may be specifically generated from pGlast(+) RGC precursors and that our fate-mapping technique can distinguish these lineage restricted events (Movies 1, 2).
Movie 1.
Examples of intermediate progenitor cell divisions in the SVZ 24 h after Glast-Cre × Tα1-SL electroporation in an E14.5 neocortex. In this time series, an mCherry+ multipolar IPC can be seen dividing in the SVZ. Although it appears to contain a radial fiber extending both apically and basally, rotation of the 3D Z-stack demonstrates that this fiber (denoted by arrowheads) is separated from this cell and likely belongs to a neighboring RGC. In addition, an mCherry+ bRG containing a pial contacting basal fiber (arrowheads at movie start) rounds up and divides while retaining the basal process. Total elapsed imaging duration = 5 h.
Movie 2.
bRG division in the SVZ of an E15.5 neocortical wall. This movie was captured 24 h following pGlast-Cre × pTα1-SL electroporation of an E14.5 embryo. A bRG expressing both ZsGreen and mCherry (arrow) divides in the SVZ while maintaining its long basal process (arrowheads). Total elapsed imaging duration = 4.9 h.
Together, these results demonstrate that our dual fluorescent fate-mapping approach identifies all known classes of VZ and SVZ precursor cells and elucidates lineage transitions between subpopulations. For example, approximately half of the pTα1-ZsGreen(+) cells exhibit a short, radial morphology, and likely represent the SNP population, while half are multipolar. In addition, while some pTα1-mCherry(+) cells appear to be RGCs since they have basal processes contacting the pial surface, several additional non-RGC cell types express mCherry, including multipolar SVZ cells, bRG, and potentially some SNPs, which may indicate that they are the immediate progeny of Glast(+) RGCs.
pTα1 cells are heterogeneous for transcription factor expression
From this point forward, we explored the morphology, expression profiles, and division parameters of the VZ and SVZ precursors using only the pGlast-Cre × pTα1-SL vector combination as this pairing produced the clearest separation between precursor subtypes. These populations are hereafter denoted by “pTα1-ZsGreen(+)” and “pTα1-mCherry(+),” respectively. As such, pTα1-mCherry(+) cells represent cells either coexpressing pGlast or derived from a pGlast(+) parent cell; pTα1-ZsGreen(+) cells do not coexpress pGlast nor are they derived from a pGlast(+) parent during the course of the experiment. Using this approach, we asked whether the pTα1-ZsGreen(+) and pTα1-mCherry(+) populations expressed distinctive differences in transcription factor expression by staining E15.5 sections of coelectroporated brains for Pax6 and Tbr2 (Fig. 4A–C). Overall, in counts conducted within 100 μm from the ventricular surface, we found that 59% of the total electroporated population expressed Pax6 while 54% expressed Tbr2 (Fig. 4D,E). This result implies that ∼13% of the electroporated cells form an overlapping group that expresses both Pax6 and Tbr2, as described previously (Fig. 4B–E) (Englund et al., 2005).
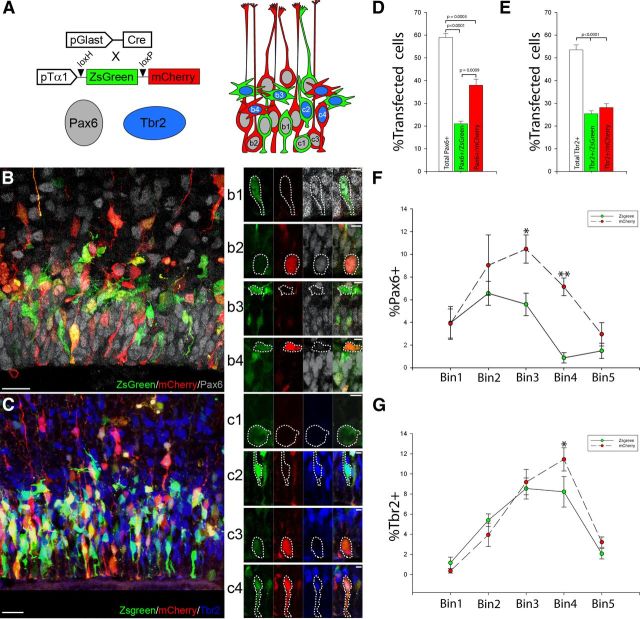
Figure 4.
Subpopulations of pTα1-ZsGreen+ and pTα1-mCherry+ cells express Pax6 or Tbr2. A, Schematic representation of the classes of cells observed 24 h post co-IUE with Glast-Cre × pTα1-SL (IUE at E14.5) immunostained for Pax6 or Tbr2. Representative images of sections stained for Pax6 (B) or Tbr2 (C) and individual cell types highlighted in b1–b4 and c1–c4. For example, Pax6+(b1)/Tbr2−(c1) ZsGreen+ cells were found in the apical region of the VZ, as were Pax6+(b2)/Tbr2−(c3) mCherry+ cells. In addition, Pax6−(b3)/Tbr2+(c2) ZsGreen+ cells were found in the basal VZ and SVZ, as were Pax6−(b4)/Tbr2+ mCherry+ cells. In addition, Tbr2+(c4) mCherry+ cells containing an apical process were noted in the VZ. The number of pTα1-ZsGreen+ and pTα1-mCherry+ cells expressing Pax6 (D) or Tbr2 expressed as a percentage of the total IUE population (E). pTα1-mCherry+ cells represented 64% of the Pax6+ population while the remaining 36% was ZsGreen+. In contrast, the Tbr2+ population was evenly split between pTα1-ZsGreen+ and pTα1-mCherry+ cells. Distribution of Pax6+ (F) or Tbr2+ (G) pTα1-ZsGreen+ and pTα1-mCherry+ cells within five 20 μm bins comprising the VZ and SVZ expressed as a percentage of the entire electroporated population (*, 0.005 < p < 0.05; **p < 0.0005). While both ZsGreen+ and mCherry+ groups exhibited similar patterns of distribution with the VZ and SVZ, the Pax6+/pTα1-ZsGreen+ population was shifted apically and found primarily within the first three VZ bins. Scale bars: B, C, 20 μm; b1–c4, 5 μm. Error bars, SEM.
Further subdividing the population by fluorescent protein expression, Pax6(+)/pTα1-ZsGreen(+) cells accounted for 21% of the total electroporated population. In comparison, the number of pTα1-mCherry(+) cells that also expressed Pax6 was significantly larger than the ZsGreen(+) cohort, representing 38% of the transfected population (Fig. 4A,D). In contrast, we did not detect a difference between the percentage of the pTα1-ZsGreen(+) and pTα1-mCherry(+) cells that express Tbr2, which constituted 25 and 28% of the electroporated cells, respectively (Fig. 4B,E). These results indicate that while pGlast-dependent recombination of the pTα1-SL plasmid separates VZ cells into RGC and non-RGC groups, the two resulting populations of pTα1-ZsGreen(+) and pTα1-mCherry(+) cells exhibit further molecular heterogeneity with respect to the expression of Pax6 and Tbr2. This finding is consistent with the observation that subsets of pTα1-ZsGreen(+) and pTα1-mCherry(+) cells exhibit either radial-like or multipolar morphologies (Fig. 3A).
To determine whether these groups were further resolved by their distribution within the germinal zone, we quantified the percentage of labeled cells by position relative to the ventricle by binning the VZ/SVZ as before (Fig. 4F,G). While the Pax6(+)/pTα1-mCherry(+) population was larger than the Pax6(+)/pTα1-ZsGreen(+) group, we noted that both groups exhibited a similar spatial profile (Fig. 4F). However, Pax6(+)/pTα1-ZsGreen(+) cells were concentrated in a narrower portion of the apical VZ. By comparison, the arrangement of Tbr2(+)/pTα1-mCherry(+) and Tbr2(+)/pTα1-ZsGreen(+) cells was nearly identical with the exception of a slight increase in the number of Tbr2(+)/pTα1-mCherry(+) in the basal VZ/SVZ (Fig. 4G).
Together, these results reveal an increased level of molecular heterogeneity among VZ/SVZ cells than previously appreciated. First, our combinatorial fate-mapping strategy shows that pBlbp and pGlast-Cre activity is able to separate pTα1 cells into defined lineage groups, which are further subdivided by morphology and transcription factor expression. In particular, we find that pTα1-ZsGreen(+) and pTα1-mCherry(+) groups are each comprised by Pax6(+) and Tbr2(+) cells with either radial-like or multipolar morphologies. As expected, the data show that the RGCs present in the VZ, which are Pax6(+)/pTα1-mCherry(+), are joined by their Tbr2(+)/pTα1-mCherry(+) IPCs progeny in the SVZ, which were likely generated from RGCs during the 24 h experimental period. This confirms the lineal origin of Tbr2(+) IPCs as suggested by other groups (Englund et al., 2005; Noctor et al., 2008). In addition, there is a cohort of multipolar Tbr2(+)/pTα1-ZsGreen(+)cells that were either generated from RGCs before the electroporation or are the progeny of Pax6(+)/pTα1-ZsGreen(+)cells. However, in addition to these cell types, which have been characterized previously, we find that a primary component of the cycling population in the VZ is a group of bipolar pTα1 cells that (1) does not exhibit active expression from the pGlast or pBlbp promoters, (2) lacks pia-contacting basal processes throughout the cell cycle, (3) expresses Pax6, and (4) appears to divide at surface of the ventricle in images of fixed tissue (Figs. 3E–G, Fig. 4C,c1). These characteristics strongly suggest that the Pax 6(+)/pTα1-ZsGreen(+) cohort of cells found in the apical half of the VZ is not equivalent to the basal Tbr2(+) IPC population, nor are they RGCs.
Radial-like pTα1-ZsGreen(+) cells divide at the ventricular surface
To conclusively demonstrate that apical pTα1-ZsGreen(+) cells are a bona fide and unique progenitor class rather than merely a transitory stage of basal Tbr2 IPCs before delamination to the SVZ, as has been previously suggested (Pontious et al., 2008; Kowalczyk et al., 2009; Miyata et al., 2001; Lui et al., 2011), we sought to describe the location and kinetics of mitotic divisions following pGlast-Cre × pTα1-SL coelectroporation. Using time-lapse multiphoton imaging in cultured slices (n = 16), we excited dividing mCherry(+) and ZsGreen(+) cells simultaneously at 995 nm and split each emission to separate detectors. As expected, we detected many pTα1-mCherry(+) RGCs dividing at the VZ surface (Fig. 5A,B; see Movie 5). In addition, we captured pTα1-ZsGreen(+) cells dividing into two separate daughter cells at the ventricular surface (Fig. 5A; Movies 3–5). These time-lapse slice experiments conclusively demonstrate that the apical pTα1-ZsGreen(+) population is bipolar and divides at the surface of the ventricle and is thus separate from the Tbr2(+) IPC population based on morphology, division dynamics, and gene expression. Thus, the fate mapping, gene expression, morphology, and time-lapse data all characterize the apical pTα1-ZsGreen(+) cells as an independent SNP population.
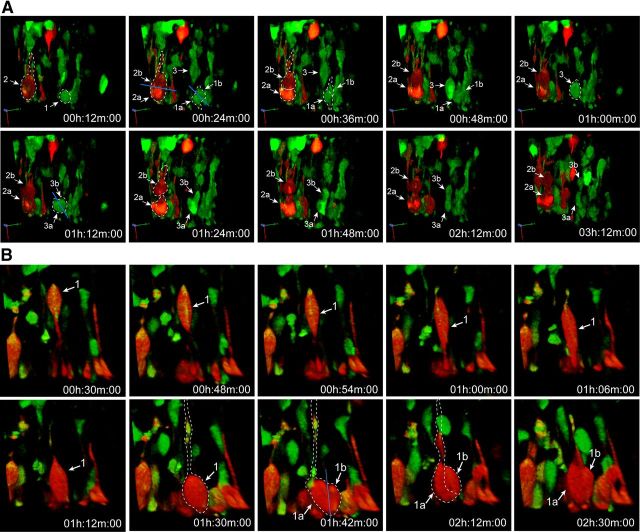
Figure 5.
Radial-like pTα1-ZsGreen+ and pTα1-mCherry+ cells divide at the surface of the ventricle. Multicolor multiphoton time-lapse imaging of organotypic brain slices (E15.5) coelectroporated with Glast-Cre × pTα1-SL (IUE at E14.5). A, In this 3 h series, three separate dividing precursors were reconstructed. Cell 1 is ZsGreen+ and lacks a basal fiber as it divides with an oblique cleavage orientation into two separate daughter cells (1a and 1b) in the 24 min frame. Cell 2 is mCherry+ and contains a basal process as it divides in a horizontal orientation to generate daughter cells 2a and 2b. Cell 3 is ZsGreen+ and descends to the VZ surface in the 24 min 1 h frames where it rounds up and divides into daughter cells 3a and 3b with an oblique cleavage plane. B, One pTα1-mCherry+ cell is depicted here dividing at the ventricle while maintaining a basal process. This cell divides with a vertical cleavage plane. (See Movies 4 and 5 for 3D time series.)
Movie 3.
Tα1-ZsGreen cell dividing at the surface of the ventricle. This movie was captured 24 h following pGlast-Cre × pTα1-SL electroporation of an E14.5 embryo. A radial-like VZ cell expressing only ZsGreen exhibits interkinetic nuclear migration and descent of the soma (arrow) to the surface of the ventricle where it divides (asterisk) into two separate daughter cells (arrows). During mitosis, this dividing cell lacks a basal process. Total elapsed imaging duration = 2.6 h.
Movie 4.
Four Tα1-ZsGreen cells dividing at the surface of the ventricle. This movie was captured 24 h following Glast-Cre × Tα1-SL electroporation of an E14.5 embryo. Four ZsGreen+ cells descend to the ventricular surface and undergo mitosis. During mitosis, these dividing cells lack basal processes. Total elapsed imaging duration = 4.2 h.
Movie 5.
mCherry+ cells dividing at the surface of the ventricle. This movie was captured 24 h following pGlast-Cre × pTα1-SL electroporation of an E14.5 embryo. A VZ cell expressing mCherry descends to the surface of the ventricle and divides. This cell maintains its basal process throughout the mitosis, indicating that it is an RGC. Total elapsed imaging duration = 5 h.
SNPs are lineally derived from RGCs
To determine the lineal origin of pTα1-ZsGreen(+) cells, and in particular whether they are derived from pGlast(+) RGCs, we performed longer term fate-mapping experiments to observe both their genesis and fate over time in vivo. Specifically, we coelectroporated pGlast-Cre × pTα1-SL into neural precursors of the dorsal telencephalic VZ at E13.5 and then imaged the distribution of mCherry(+) and ZsGreen(+) cells in the developing neocortex at E15.5. We selected E15.5 as our primary endpoint because prior experiments conducted between E14.5 and 15.5 revealed a sizable number of pTα1-ZsGreen(+) cells present in the VZ at this time point. We hypothesized two possible outcomes of this experiment: (1) if pTα1-ZsGreen(+) SNPs lineally diverge from pGlast(+) RGCs before E13.5 and self-renew as an independent population, we would detect an equal if not greater number of ZsGreen(+) SNPs in the VZ following 48 h of electroporation, relative to the number seen at 24 h post IUE (Fig. 6A); (2) if, however, SNPs are lineally derived from pGlast(+) RGCs and they do not exhibit self-renewal properties, the VZ and SVZ would be filled with pTα1-mCherry(+) cells that replaced the cohort of ZsGreen(+) SNPs as they were exhausted by terminal divisions at the ventricular surface (Fig. 6B).
Figure 6.
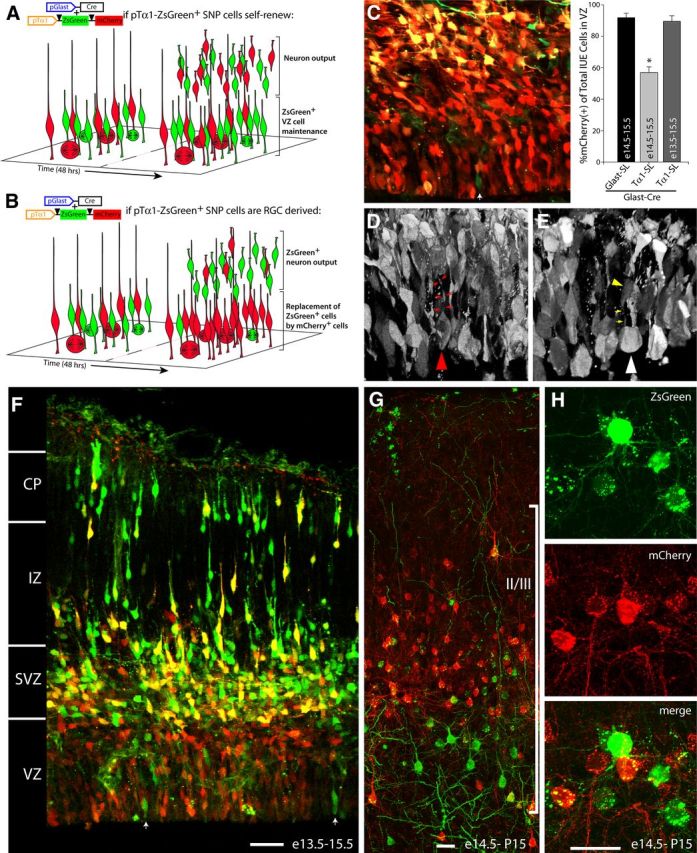
SNPs are neuronal progenitors lineally derived from RGCs. A, Theoretical model of pGlast-Cre × pTα1-SL populations if SNPs are a self-renewing precursor group. ZsGreen+ SNPs existing in the VZ at the time of (IUE) would be equal or increased in number by 48 h as they generated neuronal progeny via asymmetrical divisions. B, If SNPs do not self-renew but are generated directly from RGC parent cells, the VZ should accumulate mCherry+ SNPs by 48 h post-IUE while the neuronal progeny from the pre-existing ZsGreen+ SNPs migrate away from the VZ following symmetrical divisions. C, While 44% of pGlast-Cre × pTα1-SL transfectants were ZsGreen+ 24 h post-IUE (light gray bar), >90% of the VZ cells were mCherry+ 48 h after IUE (dark gray bar). White arrows in C and F indicate ZsGreen+ cells remaining in the VZ. The VZ cells in the pGlast-Cre × pTα1-SL transfections at 48 h post-IUE are a mix of dividing mCherry+ RGCs (D, converted to gray scale, red arrowhead) containing a basal process (red arrows) and newly generated mCherry+ SNPs dividing without a basal process (E, converted to gray scale, white arrowhead). The yellow arrowhead and arrows highlight a radial-like cell with a ventricular foot process, which is positioned behind the dividing SNP (Movie 6). F, Confocal image of neocortical wall 48 h post-IUE illustrates that the ZsGreen+ cells are a population of migrating neurons while the VZ is now composed mainly of mCherry+ precursors. Neurons born from ZsGreen+ SNPs on E14.5 migrate to cortical layer II/III and differentiate into pyramidal neurons (G, H). Scale bars, 40 μm. Error bars, SEM.
We found that virtually all of the pTα1 cells present in the VZ and SVZ expressed mCherry 48 h after pGlast-Cre × pTα1-SL electroporation (Fig. 6C). At this same time point, many pTα1-ZsGreen(+) neurons were found migrating through the intermediate zone and beginning to settle in the upper layers of the cortical plate (Fig. 6F). Importantly, two types of mCherry(+) cells were observed dividing at the ventricular surface 48 h after transfection: cells containing a basal process (Fig. 6D) and completely rounded mitotic cells lacking a basal process (Fig. 6E; Movie 6). This latter cell type is the previously determined morphology of dividing SNPs (Gal et al., 2006), which is dramatically different from the morphology of dividing RGCs that retain their basal processes (Miyata et al., 2001; Noctor et al., 2001; Noctor et al., 2002; Gal et al., 2006).
Movie 6.
SNPs are derived from RGCs. This Z-stack was captured 48 h following pGlast-Cre × pTα1-SL electroporation of an E13.5 embryo. The 3D reconstruction and rotation demonstrates that the VZ is filled with Glast+ precursor-derived mCherry+ cells that have replaced the ZsGreen+ cells exhausted by producing migrating neurons during this period (Fig. 6D). One rounded mCherry+ dividing SNP is denoted (open arrowhead). There is a radial VZ cell behind this mitotic whose soma (filled arrowhead) and foot process (open arrowhead) are clearly separated from the mitotic SNP.
While the 48 h fate map study (Fig. 6F) indicated that pTα1-ZsGreen(+) cells generate postmitotic migrating neurons, we sought to confirm that these neuronal daughter cells differentiate and integrate into postnatal neocortical circuits. We therefore performed a longer duration fate map by electroporating neocortical VZ at E14.5 and analyzing brain sections at postnatal day 15 (P15). However, since the Tα1 promoter is downregulated during postnatal neuron differentiation (Gloster et al., 1994), in this experiment we used a pCAG-SL reporter, which retains high expression in postnatal brain (Navarro-Quiroga et al., 2007). Importantly, we confirmed that both the pTα1 and pCAG reporters were interchangeable in terms of the numbers and types of cells labeled during embryonic development, which was expected since all VZ cells examined by this method express the Tα1 promoter (Fig. 2; data not shown). We found that both ZsGreen(+) and mCherry(+) neurons specified from the pGlast-Cre × pCAG-SL cotransfections on E14.5 migrated to and differentiated within layer II/III at P15 (Fig. 6G,H). Specifically, 22.5 ± 4.5% (±SEM, n = 3) of the fate mapped neurons expressed ZsGreen at P15. These findings indicate that SNPs generate neurons that differentiate and survive in the postnatal neocortex and that the progeny of the initial cohort of SNPs remains separately labeled by the multiplex fate map procedure.
Altogether, these data suggest that all pTα1(+) cells, including SNPs, are lineally derived from pGlast(+) RGCs. Furthermore, SNPs do not undergo an appreciable number of self-renewing cell divisions, as they rapidly divide to generate neurons that exit the VZ (Fig. 6; Stancik et al., 2010). Importantly, the above results also indicate that neurons generated from SNPs survive in the postnatal neocortex and are thus likely to play a significant role in brain maturation and function.
SNP production is reduced in embryonic Ts65Dn neocortex
The above results, that SNPs are a transient VZ dividing population continuously derived from RGCs, demonstrate that SNPs join Tbr2(+) IPCs and bRG as an independent class of neuronal progenitors during neocortical development. However, a question remained as to whether these three indirect streams of neuronal production are individually necessary or are perhaps redundant during neocortical development. If each precursor class is necessary for proper cortical growth, we reasoned that the relative quantities of particular neural precursors may be altered in neurodevelopmental disorders, especially those exhibiting microcephaly or dysplasia. To test this, we measured the specification of each intermediate progenitor group in a mutant animal with well characterized defects in neocortical expansion.
Previously, we determined that neocortical growth in the Ts65Dn mouse model of DS is significantly delayed from E12.5 to E18.5 due to slower cell cycle kinetics and reduced neurogenesis from VZ precursors (Chakrabarti et al., 2007). While the overall thickness of the VZ/SVZ is not remarkably different from Euploid controls over much of the neurogenic interval, differences in the cycling properties and/or proportions of VZ/SVZ cells are thought to underlie the stunted neocortical growth observed in Ts65Dn embryos. Interestingly, a compensatory expansion of Tbr2(+) abventricular divisions has been noted during late prenatal Ts65Dn development (Chakrabarti et al., 2007), but the cellular changes that underpin the differences in neurogenic output before E16.5 remain unresolved.
To determine whether specification and development of the multiple groups of VZ/SVZ neural precursors is altered during abnormal neocortical development, we measured their number and distribution in the Ts65Dn embryonic neocortex 24 h after electroporation of pGlast-Cre × pTα1-SL plasmids at E14.5. Our dual fluorescent reporter fate-mapping assay revealed a 20% increase in pTα1-mCherry(+) cells and a corresponding reduction in pTα1-ZsGreen(+) cells in Ts65Dn versus Euploid littermate embryos (Fig. 7A,B).
Figure 7.
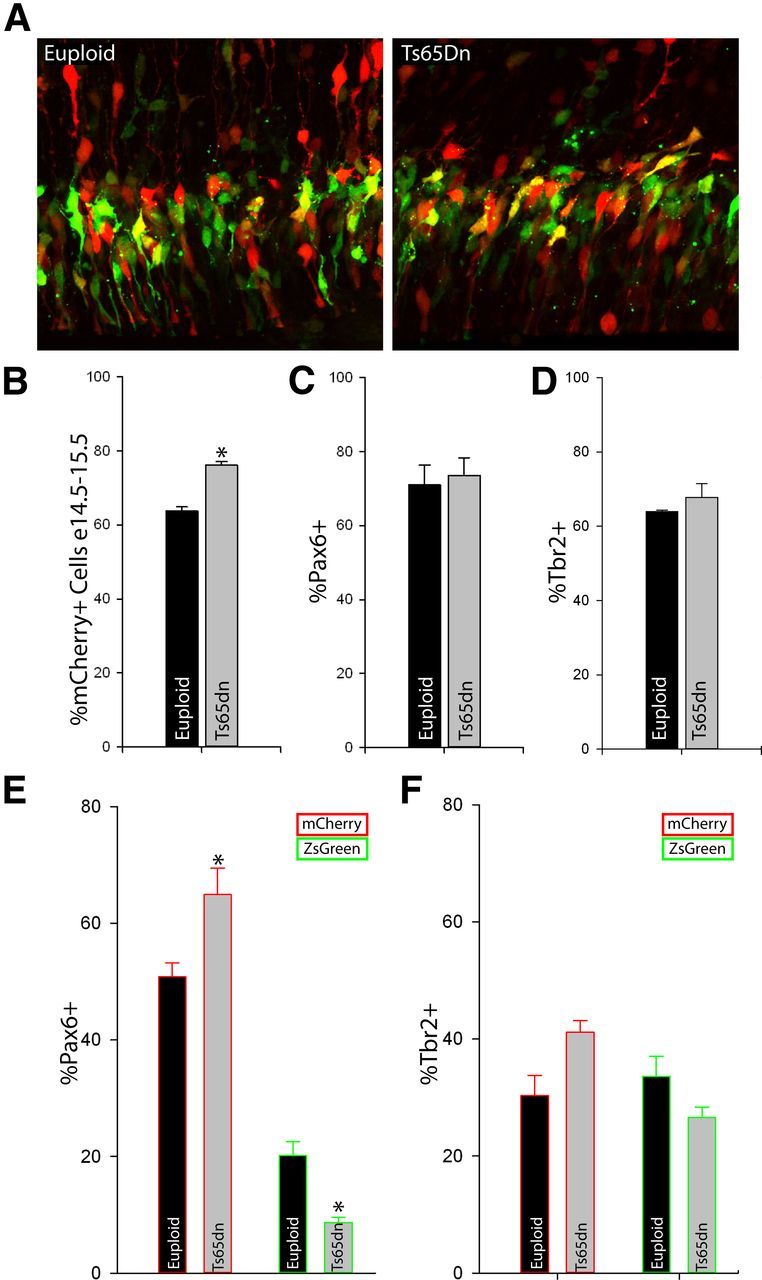
SNPs are specifically reduced during Ts65Dn neurogenesis. A, Euploid and Ts65Dn littermate embryos were cotransfected with pGlast-Cre × pTα1-SL on E14.5. Twenty-four hours later, the VZ/SVZ contained ZsGreen+ and mCherry+ cells. B, The percentage of transfected cells expressing mCherry was higher than in Euploid controls, but the overall percentage of transfected Pax6+ (C) and Tbr2+ (D) cells was not different between genotypes. However, when these transcription factor groups were further subdivided by ZsGreen and mCherry expression, the proportion of Pax6+/pTα1-mCherry+ cells was significantly higher in the Ts65Dn neocortex (E) while there was a corresponding reduction in the number of Pax6+/ZsGreen+ cells (SNPs). In contrast, there were no significant differences between Tbr2+/mCherry+ and Tbr2+/ZsGreen+ bIPCs between genotypes (F). Error bars, SEM.
Interestingly, immunohistochemistry did not reveal a difference in the total numbers of electroporated cells that expressed Pax6 or Tbr2 in the Ts65Dn neocortical wall (Fig. 7C,D). However, when the pTα1(+) cells were further subdivided based on fluorescent protein expression regulated by pGlast-Cre-dependent recombination, there was a 28% increase in the relative number of RGCs (Pax6(+)/pTα1-mCherry(+) cells) and a 57% decrease in SNPs (Pax6(+)/pTα1-ZsGreen(+) cells) in the Ts65Dn VZ (Fig. 7E). While we did not observe significant differences in the percentage of Tbr2(+)/pTα1-mCherry(+) or Tbr2(+)/ZsGreen(+) cells between Ts65Dn and Euploid samples, the groups exhibited a shift similar to that seen in the Pax6(+) populations, with more Tbr2(+)/pTα1-mCherry(+) cells at the expense of Tbr2(+)/ZsGreen(+) cells (Fig. 7F). Altogether, the data from electroporations of prenatal Ts65Dn neocortex demonstrate that subpopulations of neural precursors are specifically altered during development. In particular, Pax6(+) SNPs, and to a lesser degree Tbr2(+)/ZsGreen(+) cells, are substantially reduced. These data strongly suggest that individual streams of indirect neurogenesis are affected in the trisomic brain and that their loss contributes to improper brain development.
Discussion
Using a multiplex fate-mapping paradigm, our analysis reveals an increased level of molecular and morphological complexity among classes of neural precursors in the dorsal telencephalic VZ/SVZ (Fig. 8). During the peak of neurogenesis, the cells of the VZ/SVZ are subdivided into radial-like and multipolar morphologies. Within the radial-like group are the two apical precursor cells types, the RGCs and SNPs, which are distinguished by the presence or absence of pial-containing basal processes, respectively. While both of these cell types express Pax6 and divide at the ventricular surface, we show that they can be separated molecularly using our fate-mapping approach into a pTα1(+)/pGlast(−)/pBlbp(−) population (SNPs) and a pTα1(+)/pGlast(+)/pBlbp(+) population (RGCs). The multipolar precursor group is likewise divided into two cell types that divide in abventricular positions within the SVZ: Tbr2(+) basal intermediate progenitor cells (bIPCs) and the pia-attached bRG, which primarily express Pax6. Thus, the four types of neural precursor cells in the mammalian neocortex are sharply resolved using a combined approach of multiplex fate mapping, transcription factor immunostaining, morphological classification, and cell division properties.
Figure 8.
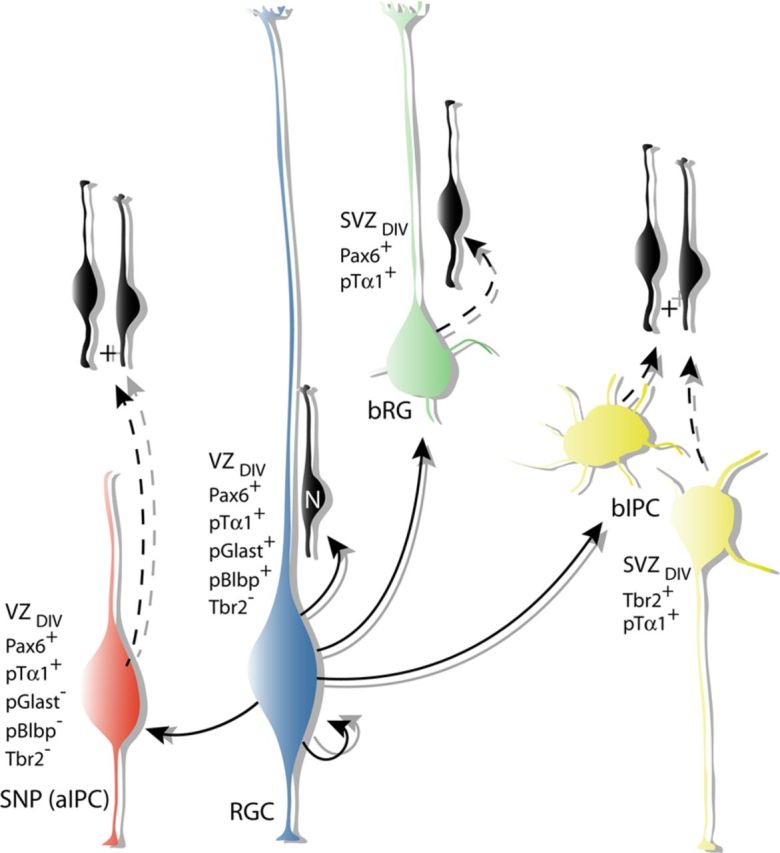
Lineal relationships, molecular profiles, and morphologies of neocortical precursor cells. In addition to generating neurons directly (black), our results indicate that RGCs generate several classes of intermediate progenitors. SNPs (red) lack basal processes and divide within the VZ at the ventricular surface after first being generated from RGC parent cells (blue). Both RGCs and SNPs express Pax6. While RGCs are thought to self-renew, SNPs divide once to generate two neuronal daughter cells which then migrate radially into the neocortical wall, indicating that they are a class of intermediate neural progenitors. Our fate mapping and time-lapse imaging study indicates that bRG (green) are derived from RGCs and express Pax6 and divide in the SVZ. While bIPCs (yellow) are also derived from RGCs, they are distinguished by their multipolar morphology, expression of Tbr2, and abventricular divisions.
Long-term intersectional fate mapping and time-lapse imaging experiments elucidate novel lineal relationships between these four cell types. In particular, the data show for the first time that SNPs are a transient group of cells derived from RGCs that undergo limited rounds of cell division to generate neurons. Importantly, the experiments also clarify that pTα1-ZsGreen(+) SNPs, which reside in the VZ and divide at the surface of the ventricle, are not equivalent to the basal Tbr2(+) IPC population, which divides in the SVZ, nor are they a transitory form of this cell type. While the possibility remains that a subset of SNPs divides to produce bIPCs, the results presented here clearly show that SNPs exist independently, albeit briefly, after arising from RGCs and before subsequent division in the VZ. By virtue of their apical process, which maintains contact with the ventricle, another defining characteristic of SNPs is their ability, unique among intermediate progenitors, to potentially respond to morphogenetic cues present in the CSF (Lehtinen et al., 2011). In light of these findings, we propose to rename SNPs “apical intermediate progenitor cells (aIPCs)” to clearly distinguish them from their Tbr2(+) bIPC counterparts. Previous transcriptional profiling of single VZ/SVZ cells predicted the existence of a VZ intermediate progenitor with an mRNA expression signature distinct from RGCs and bIPCs (Kawaguchi et al., 2008). SNPs (aIPCs) fit the description outlined in this study. Importantly, our postnatal fate-map studies also show that aIPCs produce neurons that migrate to and survive in the neocortex.
The existence of these multiple, independent groups of neural precursors indicates that each precursor subtype may be a necessary component of a system of germinal cells required for proper neocortical growth. Indeed, the data from the Ts65Dn mouse studies implicate specific changes in precursor cell specification before the development of microcephaly and the decreases in excitatory neurons and their synapses in DS and animal model forebrain (Zellweger, 1977; Sylvester, 1983; Takashima et al., 1989; Schmidt-Sidor et al., 1990; Chakrabarti et al., 2007). Since the establishment of the Ts65Dn mouse in 1995 (Reeves et al., 1995), several novel findings of altered development have emerged from basic model studies, which have then been confirmed in human DS brain. For example, the reduction in excitatory neuron number was found to be due to lower output of neurons and longer cell cycle duration in prenatal VZ/SVZ precursors (Chakrabarti et al., 2007; 2010) and this has been confirmed in fetal DS brain (Contestabile et al., 2007; Guidi et al., 2008). The results presented here demonstrate specific disturbances in VZ and SVZ neuronal progenitor subpopulations in Ts65Dn during mid-neurogenesis. While these fate-mapping studies are yet to be confirmed in human fetal brain, the results clearly indicate that neurodevelopmental disabilities presenting with malformations or dysplasia such as lissencephaly, microcephaly, autism, and epilepsy may also arise from abnormal specification or output from individual precursor populations.
The increased diversity of neural precursor cells presented here may also be important for understanding how individual rates of brain growth vary and how differences in interspecies growth may arise. Several recent studies on bIPCs and bRG indicate that their temporal and spatial distribution may be important for regulating development of neocortical areas as well as for expansion and gyrification of the forebrain during primate evolution (Fietz et al., 2010; Hansen et al., 2010; Lui et al., 2011; Reillo et al., 2011), although this is still controversial (Hevner and Haydar, 2012; Kelava et al., 2012). Since our study identifies SNPs (aIPCs) as a principal component of the neocortical germinal zone, key next steps leading to a full understanding of neocortical growth include identifying the factors that impinge on RGCs to regulate the numbers and properties of the aIPCs versus bIPCs. The new fate-mapping methods presented here, which precisely label individual RGC, aIPC, and bIPC subgroups, will allow the identification of additional molecular markers with which to quantify cell type differences across species.
A critical unresolved question in developmental neurobiology pertains to the functional significance of separate classes of neural stem and progenitors cells. Why would multiple, parallel streams of indirect neurogenesis exist? One possibility is that the diversity of precursor cells evolved as a mechanism to simply expand the proliferative zone of the developing forebrain by freeing them from the ventricular surface (Fish et al., 2008; Lui et al., 2011). As such, the collective output from these precursors would be a larger, synchronously generated population that matures into a homogeneous group of neurons within each lamina. A second intriguing possibility is that the heterogeneity present among VZ/SVZ cells is responsible for specifying neurons that exhibit as yet unrecognized differences in gene expression, connectivity, and firing properties, even within the same layer of the cortex. Regardless of the functional impact of these separate progenitor groups, results from this work and other recent studies clearly demonstrate that multiple groups of precursors contemporaneously build the neocortex, beginning from the onset of neurogenesis in the embryonic brain. As such, future work on forebrain development must incorporate measurements of this cellular diversity so that the individual roles of these cell types can be determined, especially in neurodevelopmental disabilities.
Footnotes
This work was supported by National Institutes of Health grants NS051852, NS076503, and HD057580.
The authors declare no competing financial interests.
References
- Breunig JJ, Haydar TF, Rakic P. Neural stem cells: historical perspective and future prospects. Neuron. 2011;70:614–625. doi: 10.1016/j.neuron.2011.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti L, Galdzicki Z, Haydar TF. Defects in embryonic neurogenesis and initial synapse formation in the forebrain of the Ts65Dn mouse model of Down syndrome. J Neurosci. 2007;27:11483–11495. doi: 10.1523/JNEUROSCI.3406-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti L, Best TK, Cramer NP, Carney RS, Isaac JT, Galdzicki Z, Haydar TF. Olig1 and Olig2 triplication causes developmental brain defects in Down syndrome. Nat Neurosci. 2010;13:927–934. doi: 10.1038/nn.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, LoTurco J. A method for stable transgenesis of radial glia lineage in rat neocortex by piggyBac mediated transposition. J Neurosci Methods. 2012;207:172–180. doi: 10.1016/j.jneumeth.2012.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contestabile A, Fila T, Ceccarelli C, Bonasoni P, Bonapace L, Santini D, Bartesaghi R, Ciani E. Cell cycle alteration and decreased cell proliferation in the hippocampal dentate gyrus and in the neocortical germinal matrix of fetuses with down syndrome and in Ts65Dn mice. Hippocampus. 2007;17:665–678. doi: 10.1002/hipo.20308. [DOI] [PubMed] [Google Scholar]
- Englund C, Fink A, Lau C, Pham D, Daza RA, Bulfone A, Kowalczyk T, Hevner RF. Pax6, Tbr2, and Tbr1 are expressed sequentially by radial glia, intermediate progenitor cells, and postmitotic neurons in developing neocortex. J Neurosci. 2005;25:247–251. doi: 10.1523/JNEUROSCI.2899-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fietz SA, Huttner WB. Cortical progenitor expansion, self-renewal and neurogenesis-a polarized perspective. Curr Opin Neurobiol. 2011;21:23–35. doi: 10.1016/j.conb.2010.10.002. [DOI] [PubMed] [Google Scholar]
- Fietz SA, Kelava I, Vogt J, Wilsch-Bräuninger M, Stenzel D, Fish JL, Corbeil D, Riehn A, Distler W, Nitsch R, Huttner WB. OSVZ progenitors of human and ferret neocortex are epithelial-like and expand by integrin signaling. Nat Neurosci. 2010;13:690–699. doi: 10.1038/nn.2553. [DOI] [PubMed] [Google Scholar]
- Fish JL, Dehay C, Kennedy H, Huttner WB. Making bigger brains-the evolution of neural-progenitor-cell division. J Cell Sci. 2008;121:2783–2793. doi: 10.1242/jcs.023465. [DOI] [PubMed] [Google Scholar]
- Gal JS, Morozov YM, Ayoub AE, Chatterjee M, Rakic P, Haydar TF. Molecular and morphological heterogeneity of neural precursors in the mouse neocortical proliferative zones. J Neurosci. 2006;26:1045–1056. doi: 10.1523/JNEUROSCI.4499-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gloster A, Wu W, Speelman A, Weiss S, Causing C, Pozniak C, Reynolds B, Chang E, Toma JG, Miller FD. The T alpha 1 alpha-tubulin promoter specifies gene expression as a function of neuronal growth and regeneration in transgenic mice. J Neurosci. 1994;14:7319–7330. doi: 10.1523/JNEUROSCI.14-12-07319.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guidi S, Bonasoni P, Ceccarelli C, Santini D, Gualtieri F, Ciani E, Bartesaghi R. Neurogenesis impairment and increased cell death reduce total neuron number in the hippocampal region of fetuses with Down syndrome. Brain Pathol. 2008;18:180–197. doi: 10.1111/j.1750-3639.2007.00113.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen DV, Lui JH, Parker PR, Kriegstein AR. Neurogenic radial glia in the outer subventricular zone of human neocortex. Nature. 2010;464:554–561. doi: 10.1038/nature08845. [DOI] [PubMed] [Google Scholar]
- Hartfuss E, Galli R, Heins N, Götz M. Characterization of CNS precursor subtypes and radial glia. Dev Biol. 2001;229:15–30. doi: 10.1006/dbio.2000.9962. [DOI] [PubMed] [Google Scholar]
- Hartfuss E, Förster E, Bock HH, Hack MA, Leprince P, Luque JM, Herz J, Frotscher M, Götz M. Reelin signaling directly affects radial glia morphology and biochemical maturation. Development. 2003;130:4597–4609. doi: 10.1242/dev.00654. [DOI] [PubMed] [Google Scholar]
- Hevner RF, Haydar TF. The (not necessarily) convoluted role of basal radial glia in cortical neurogenesis. Cereb Cortex. 2012;22:465–468. doi: 10.1093/cercor/bhr336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi A, Ikawa T, Kasukawa T, Ueda HR, Kurimoto K, Saitou M, Matsuzaki F. Single-cell gene profiling defines differential progenitor subclasses in mammalian neurogenesis. Development. 2008;135:3113–3124. doi: 10.1242/dev.022616. [DOI] [PubMed] [Google Scholar]
- Kelava I, Reillo I, Murayama AY, Kalinka AT, Stenzel D, Tomancak P, Matsuzaki F, Lebrand C, Sasaki E, Schwamborn JC, Okano H, Huttner WB, Borrell V. Abundant occurrence of basal radial glia in the subventricular zone of embryonic neocortex of a lissencephalic primate, the common marmoset Callithrix jacchus. Cereb Cortex. 2012;22:469–481. doi: 10.1093/cercor/bhr301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalczyk T, Pontious A, Englund C, Daza RA, Bedogni F, Hodge R, Attardo A, Bell C, Huttner WB, Hevner RF. Intermediate neuronal progenitors (basal progenitors) produce pyramidal-projection neurons for all layers of cerebral cortex. Cereb Cortex. 2009;19:2439–2450. doi: 10.1093/cercor/bhn260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehtinen MK, Zappaterra MW, Chen X, Yang YJ, Hill AD, Lun M, Maynard T, Gonzalez D, Kim S, Ye P, D'Ercole AJ, Wong ET, LaMantia AS, Walsh CA. The cerebrospinal fluid provides a proliferative niche for neural progenitor cells. Neuron. 2011;69:893–905. doi: 10.1016/j.neuron.2011.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui JH, Hansen DV, Kriegstein AR. Development and evolution of the human neocortex. Cell. 2011;146:18–36. doi: 10.1016/j.cell.2011.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata T, Kawaguchi A, Okano H, Ogawa M. Asymmetric inheritance of radial glial fibers by cortical neurons. Neuron. 2001;31:727–741. doi: 10.1016/s0896-6273(01)00420-2. [DOI] [PubMed] [Google Scholar]
- Mizutani K, Yoon K, Dang L, Tokunaga A, Gaiano N. Differential Notch signalling distinguishes neural stem cells from intermediate progenitors. Nature. 2007;449:351–355. doi: 10.1038/nature06090. [DOI] [PubMed] [Google Scholar]
- Navarro-Quiroga I, Chittajallu R, Gallo V, Haydar TF. Long-term, selective gene expression in developing and adult hippocampal pyramidal neurons using focal in utero electroporation. J Neurosci. 2007;27:5007–5011. doi: 10.1523/JNEUROSCI.0867-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noctor SC, Flint AC, Weissman TA, Dammerman RS, Kriegstein AR. Neurons derived from radial glial cells establish radial units in neocortex. Nature. 2001;409:714–720. doi: 10.1038/35055553. [DOI] [PubMed] [Google Scholar]
- Noctor SC, Flint AC, Weissman TA, Wong WS, Clinton BK, Kriegstein AR. Dividing precursor cells of the embryonic cortical ventricular zone have morphological and molecular characteristics of radial glia. J Neurosci. 2002;22:3161–3173. doi: 10.1523/JNEUROSCI.22-08-03161.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noctor SC, Martinez-Cerdeño V, Ivic L, Kriegstein AR. Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nat Neurosci. 2004;7:136–144. doi: 10.1038/nn1172. [DOI] [PubMed] [Google Scholar]
- Noctor SC, Martínez-Cerdeño V, Kriegstein AR. Distinct behaviors of neural stem and progenitor cells underlie cortical neurogenesis. J Comp Neurol. 2008;508:28–44. doi: 10.1002/cne.21669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtaka-Maruyama C, Hirai S, Miwa A, Takahashi A, Okado H. The 5′-flanking region of the RP58 coding sequence shows prominent promoter activity in multipolar cells in the subventricular zone during corticogenesis. Neuroscience. 2012;201:67–84. doi: 10.1016/j.neuroscience.2011.11.006. [DOI] [PubMed] [Google Scholar]
- Pinto L, Mader MT, Irmler M, Gentilini M, Santoni F, Drechsel D, Blum R, Stahl R, Bulfone A, Malatesta P, Beckers J, Götz M. Prospective isolation of functionally distinct radial glial subtypes–lineage and transcriptome analysis. Mol Cell Neurosci. 2008;38:15–42. doi: 10.1016/j.mcn.2008.01.012. [DOI] [PubMed] [Google Scholar]
- Pontious A, Kowalczyk T, Englund C, Hevner RF. Role of intermediate progenitor cells in cerebral cortex development. Dev Neurosci. 2008;30:24–32. doi: 10.1159/000109848. [DOI] [PubMed] [Google Scholar]
- Reeves RH, Irving NG, Moran TH, Wohn A, Kitt C, Sisodia SS, Schmidt C, Bronson RT, Davisson MT. A mouse model for Down syndrome exhibits learning and behaviour deficits. Nat Genet. 1995;11:177–184. doi: 10.1038/ng1095-177. [DOI] [PubMed] [Google Scholar]
- Reillo I, de Juan Romero C, García-Cabezas MÁ, Borrell V. A role for intermediate radial glia in the tangential expansion of the mammalian cerebral cortex. Cereb Cortex. 2011;21:1674–1694. doi: 10.1093/cercor/bhq238. [DOI] [PubMed] [Google Scholar]
- Schmidt-Sidor B, Wisniewski KE, Shepard TH, Sersen EA. Brain growth in Down syndrome subjects 15 to 22 weeks of gestational age and birth to 60 months. Clin Neuropathol. 1990;9:181–190. [PubMed] [Google Scholar]
- Shitamukai A, Konno D, Matsuzaki F. Oblique radial glial divisions in the developing mouse neocortex induce self-renewing progenitors outside the germinal zone that resemble primate outer subventricular zone progenitors. J Neurosci. 2011;31:3683–3695. doi: 10.1523/JNEUROSCI.4773-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stancik EK, Navarro-Quiroga I, Sellke R, Haydar TF. Heterogeneity in ventricular zone neural precursors contributes to neuronal fate diversity in the postnatal neocortex. J Neurosci. 2010;30:7028–7036. doi: 10.1523/JNEUROSCI.6131-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sylvester PE. The hippocampus in Down's syndrome. J Ment Defic Res. 1983;27:227–236. doi: 10.1111/j.1365-2788.1983.tb00294.x. [DOI] [PubMed] [Google Scholar]
- Takahashi T, Nowakowski RS, Caviness VS., Jr Cell cycle parameters and patterns of nuclear movement in the neocortical proliferative zone of the fetal mouse. J Neurosci. 1993;13:820–833. doi: 10.1523/JNEUROSCI.13-02-00820.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takashima S, Ieshima A, Nakamura H, Becker LE. Dendrites, dementia and the Down syndrome. Brain Dev. 1989;11:131–133. doi: 10.1016/s0387-7604(89)80082-8. [DOI] [PubMed] [Google Scholar]
- Wang X, Qiu R, Tsark W, Lu Q. Rapid promoter analysis in developing mouse brain and genetic labeling of young neurons by doublecortin-DsRed-express. J Neurosci Res. 2007;85:3567–3573. doi: 10.1002/jnr.21440. [DOI] [PubMed] [Google Scholar]
- Wang X, Tsai JW, LaMonica B, Kriegstein AR. A new subtype of progenitor cell in the mouse embryonic neocortex. Nat Neurosci. 2011;14:555–561. doi: 10.1038/nn.2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YS, Hughes TE. Cre stoplight: a red/green fluorescent reporter of Cre recombinase expression in living cells. Biotechniques. 2001;31:1036, 1038, 1040–1041. doi: 10.2144/01315st03. [DOI] [PubMed] [Google Scholar]
- Zellweger H. Handbook of clinical neurology. Amsterdam: North-Holland; 1977. Down syndrome; pp. 367–469. [Google Scholar]