Abstract
Subcellular localization of messenger RNAs (mRNAs) can give precise control over where protein products are synthesized and operate. However, just 10 years ago many in the broader cell biology community would have considered this a specialized mechanism restricted to a very small fraction of transcripts. Since then, it has become clear that subcellular targeting of mRNAs is prevalent, and there is mounting evidence for central roles for this process in many cellular events. Here, we review current knowledge of the mechanisms and functions of mRNA localization in animal cells.
The asymmetric distribution of specific mRNAs in the cytoplasm was first visualized in the early 1980s, when in situ hybridization techniques were used to detect β-actin mRNA in ascidian embryos (1). The discovery of differential localization of transcripts encoding cytoskeletal proteins in cultured chicken cells soon gave further prominence to this phenomenon (2). Subsequent studies demonstrated that asymmetric mRNA localization contributes to the targeting of diverse types of protein products.
In recent years, the advent of high-throughput approaches has revealed that mRNA localization is much more common than previously assumed. Of expressed mRNA species, 70% were classified as asymmetrically distributed in a large-scale fluorescent in situ hybridization screen in early Drosophila embryos (3). In addition, large numbers of vertebrate mRNAs are specifically enriched in protrusions of migrating fibroblasts, in neuronal processes, or on spindles (table S1). Thus, mRNA localization has a prominent role in the spatial regulation of gene activity. Here, we provide an overview of the mechanisms and functions of mRNA localization in animal cells. Readers are referred elsewhere for entry points into the seminal work on mRNA localization in fungi and plants (4, 5).
Mechanisms of mRNA Localization: Illuminating a Multi-Step Process
Four mechanisms are thought to contribute to subcellular localization of specific mRNAs after their transcription: (i) vectorial export from nuclei, (ii) localized protection from degradation, (iii) polarized active transport on the cytoskeleton by using molecular motors, and (iv) localized anchorage. With the exception of vectorial nuclear export, all of these mechanisms are known to contribute to mRNA sorting in animal cells. Combinations of these mechanisms can also be used to localize a single mRNA species.
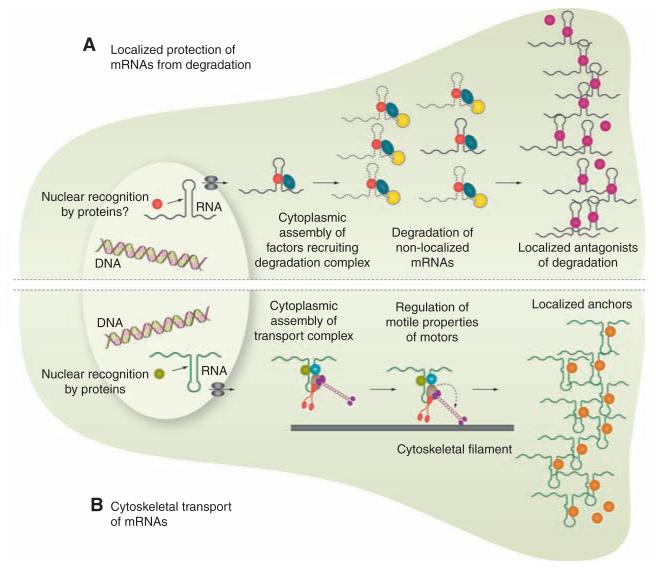
Protection of mRNAs from degradation (Fig. 1A) plays a crucial role in restricting mRNAs to the germ plasm in Drosophila and zebrafish embryos, often in conjunction with local entrapment of transcripts (6–8). There is also evidence, from the sea slug Aplysia, that mRNAs in neuronal processes can be selectively stabilized by interaction with their targets (9). However, the molecular mechanisms that locally protect specific messages remain unknown.
Fig. 1.
mRNA localization is a multi-step process. Shown is an illustration of two stylized cells, depicting mechanisms that can contribute to mRNA localization. (A) Protection of mRNAs from degradation. Red, nuclear RNA recognition factor; dark blue, cytoplasmic RNA recognition factor; yellow, ribonuclease; purple, agonist of degradation. (B) Motor-based transport. Green, nuclear RNA recognition factor; light blue and light gray, cytoplasmic RNA recognition factor; red and purple, molecular motors; orange, anchorage factor. In reality, different combinations of these mechanisms may be used to localize a single mRNA species in the same cell.
Motor-based transport (Fig. 1B) appears to be the predominant mechanism for the localization of mRNAs in animal cells probably because it provides the most rapid method for long-distance translocation of large ribonucleoprotein (RNP) particles through the crowded cellular environment. Live cell-imaging studies in recent years—involving the injection of in vitro synthesized fluorescent mRNAs or labeling transcripts by means of tethering multiple fluorescent proteins—have provided compelling evidence that mRNAs can control their own sorting by recruiting more than one kind of motor and even modulating motor properties.
For instance, in mammalian oligodendrocytes and hippocampal neurons, as well as in Drosophila embryos, mRNAs are bound to microtubule-based motor complexes that rapidly switch between bouts of motion in the minus- and plus-end directions (10–12). Specific mRNAs appear to control net sorting by increasing the relative frequency of movement in one direction through the recruitment of factors that modulate the activities of simultaneously bound opposite polarity motors (11).
In the case of delivery of oskar mRNA from the nurse cells to the posterior pole of the Drosophila oocyte, the frequency of microtubule-based movement in the minus-end and plus-end directions is also altered by specific components of messenger RNPs (mRNPs) (13). However, it appears that this comprises sequential, rather than rapidly switching, actions of motors. Localization of oskar culminates in a biased walk along a weakly polarized cytoskeleton—driven by the plus end–directed motor kinesin-1—to anchorage sites at the posterior pole (13). Vegetal localization of mRNAs in Xenopus oocytes may also be based on similar principles, although in this case the concerted action of kinesin-1 and kinesin-2 is crucial (14).
Some mRNAs, as is the case for other cellular cargoes, may simultaneously associate with actin- and microtubule-based motors, allowing transport to be fine-tuned by switching between different types of cytoskeletal tracks (15). Transcripts may also influence the choice of subsets of microtubules by motors. This mechanism has been proposed to contribute to the delivery of gurken and bicoid mRNAs to the dorso-anterior and anterior regions of the Drosophila oocyte, respectively, by the minus end–directed motor dynein and could conceivably be based on differential posttranslational modification of microtubules (16, 17).
Although our understanding of transport mechanisms is increasing, relatively little is known about the processes that contribute to mRNA anchorage. Long-distance transport of mRNPs on microtubules can be followed by transfer to the actin cytoskeleton at the cortex, with entrapment facilitated by the dense network of filaments or associated proteins (18, 19). In other cases, microtubule-based motors may act directly as anchors (20) or lead to steady-state mRNA localization through continual active transport (21).
Thus, it appears that multiple binding sites within mRNAs recruit combinations of trans-acting factors that regulate the association and activities of different molecular motors as well as mediating interplay with anchorage complexes and translational regulators (see below). Even uniformly distributed mRNAs can be transported to some extent by motors, presumably to facilitate their exploration of space (11, 22, 23). A key challenge for the future is to understand how the information within asymmetrically localized transcripts is decoded.
Recognition of Localizing mRNAs
Cis-acting RNA localization signals and interplay with translation
The cis-acting elements that mediate asymmetric localization of specific transcripts are referred to as RNA localization signals or zip codes. Depending on the nature of the transacting factor they bind, these elements can consist of single-stranded stretches or double-stranded stem loops (24). Characterizing these latter types of elements is taxing because recognition may be based on a three-dimensional structure. This is probably the case during transport of several mRNAs toward the minus ends of microtubules in Drosophila, where stem loops with relatively little in common at the primary sequence level are recognized by the same RNA-binding protein, Egalitarian (25).
Localization signals are typically found within untranslated regions of messages, where they can evolve without the constraints of retaining protein-coding sequences. In cases in which signals are found within coding sequences, their secondary structure may play a role in antagonizing the translational machinery during the mRNA localization process (26). Protein production is more commonly silenced during translocation by the recruitment of translational repressors (27). In some instances, initiation of protein synthesis at the target site is mediated by the interplay between localized translational derepressors and proteins that bind localization signals. An elegant example of this involves the phosphorylation of the β-actin zip code–binding protein ZBP1 by the localized activity of the kinase Src at the cell periphery (28). This leads to dissociation of ZBP1 from the transcript at the leading edge of migrating cells, allowing access to the translation initiation machinery.
Trans-acting factors and the assembly of mRNPs
A large number of proteins have been identified with direct roles in mRNA localization complexes. To what extent this reflects discrete pathways at work or functionally related mRNPs containing multiple proteins remains unclear. This latter scenario will be at least part of the story because there are several reports of combinations of well-characterized RNA-binding proteins, such as ZBP1, Staufen, and fragile X mental retardation protein (FMRP), being found in the same complexes. Interactions of the same RNA with multiple trans-acting factors gives scope for redundancy, which may partly explain the difficulties in identifying the molecular links between localizing transcripts and motors in animal cells. However, a complete link has recently been uncovered between mRNA localization signals and dynein in Drosophila (25), providing an opportunity to probe the molecular details of the assembly and operation of a model RNA: motor complex.
Where in the cell are mRNAs earmarked for transport? In many instances, localizing transcripts are first recognized in the nucleus. This is the case for β-actin transcripts in chicken fibroblasts, in which the cotranscriptional recruitment of the ZBP2 protein facilitates binding of ZBP1 to the mRNA and its subsequent targeting behind the leading edge (29). It has also been revealed, from elegant studies of Vg1 localization in Xenopus oocytes, that important RNA:protein interactions formed in the nucleus can be remodeled in the cytoplasm (30), and such events may regulate transitions between critical steps in localization processes. Nuclear history also plays an essential role in cytoplasmic localization of oskar mRNA. Deposition of the multicomponent exon junction complex (EJC) during splicing is essential for the translocation of this transcript to the posterior of the Drosophila oocyte (31), possibly by facilitating switching of the predominant motor activity from dynein to kinesin-1 (13). It will be fascinating to discover how the EJC regulates these motors at the molecular level, especially because components of this complex have been implicated in the localization of functionally important mRNAs within mammalian neurons (32).
Functions of mRNA Localization: Cell Polarity and Local Response to Extrinsic Cues
There are several a priori reasons why localizing an mRNA could be advantageous over targeting the protein product directly: (i) increased cost effectiveness because of the production of multiple protein copies from single localized mRNA molecules, (ii) preventing proteins from acting ectopically during translocation, (iii) facilitating the assembly of macromolecular protein complexes by producing a high local concentration of mRNA molecules in microdomains, (iv) distinct properties of newly synthesized proteins, and (v) decentralizing the control of gene expression by permitting local control of translation in response to extrinsic cues. Below, we introduce specific examples that illustrate the importance of asymmetric mRNA localization in key biological processes (see also Fig. 2).
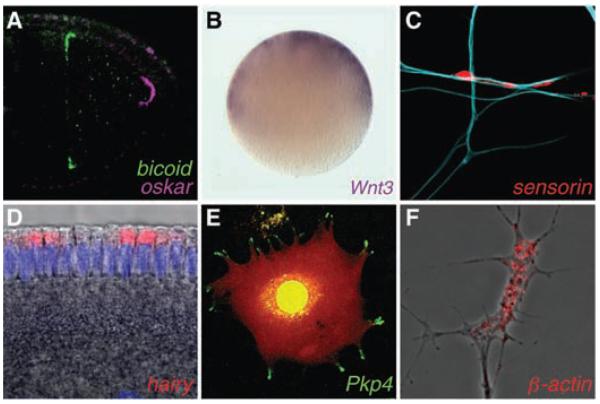
Fig. 2.
Examples of asymmetrically localized mRNAs. (A) Differential localization of mRNA determinants within the Drosophila oocyte. (B) Animal localization of a transcript encoding a signaling molecule required for axis development in the egg of a cnidarian, Clytia. (C) mRNA enrichment in synapses of an Aplysia sensory neuron in response to contact with a target motor neuron (blue). (D) Apical localization of an mRNA in the Drosophila embryo, which facilitates entry of its transcription factor product into the nuclei (purple). (E) mRNA localization in pseudopodial protrusions of a cultured mammalian fibroblast (red signal indicates the cell volume). (F) mRNA enrichment within a Xenopus axonal growth cone. mRNAs were visualized by means of in situ hybridization except in (E), in which the MS2–green fluorescent protein (GFP) system was used. Drosophila images are reproduced from (50) with permission. [Images were kindly provided by (B) T. Momose and E. Houliston, (C) D.O. Wang and K. Martin, (D) M. Dienstbier, (E) S. Mili and I. Macara, and (F) F. van Horck.]
Establishing Embryonic Organization
In Drosophila, the differential localization of maternal mRNAs plays a major role in establishing and patterning the dorsal-ventral and anterior-posterior body axes as well as in germ cell specification (table S2). During Xenopus development, localization pathways exist in early and late oogenesis that culminate in the vegetal accumulation of transcripts that are important for germline development and patterning of the mesoderm and endoderm (33). Differentially localized maternal mRNAs have also been found in ascidians and cnidarians, and many of these transcripts encode proteins with known roles in embryonic patterning (34, 35). Thus, the localization of maternal mRNAs appears to be widely used to establish embryonic organization.
In mammals, an obligatory requirement for localized mRNA determinants in the egg appears to be ruled out by the developmental lability of the early embryo. However, the recent report of apical localization of the message encoding the Cdx2 transcription factor in 8- to 16-cell embryos raises the possibility that mRNA sorting facilitates asymmetric cell fate decisions at later stages (36). A function for mRNA localization in influencing embryonic cell lineage choices is also supported by the differential inheritance of messages encoding developmental regulators in snail blastomeres, which is driven by a remarkable process of transcript enrichment at one of the two centrosomes (37).
Neurons: mRNA Localization on Demand
The critical importance of posttranscriptional regulation in neurons is illustrated by the high degree of autonomy exhibited by neuronal processes, which often extend great distances from the cell body. This autonomy permits rapid local responses to extrinsic cues and is manifest in the ability of axons and dendrites, respectively, to navigate to guidance cues and undergo certain forms of synaptic plasticity after removal of the soma. It has become increasingly evident that this “decentralization” involves the selective localization and translation of subsets of mRNAs in neuronal processes in response to external stimuli (Fig. 3).
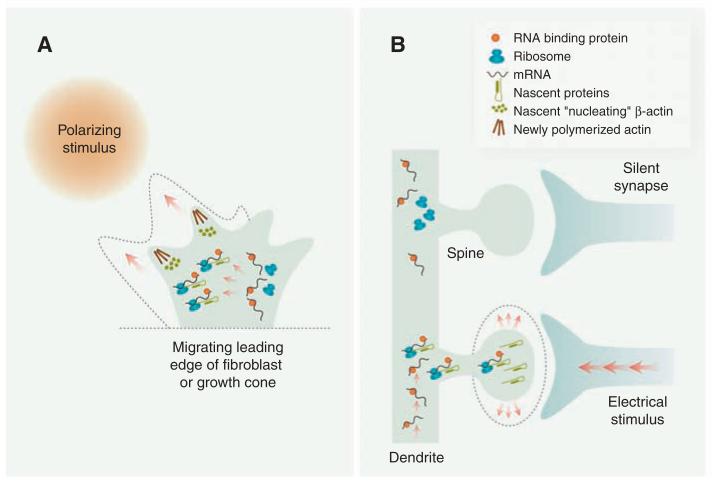
Fig. 3.
Extrinsic stimuli elicit changes in subcellular mRNA localization and translation. (A) A polarizing stimulus elicits asymmetric localization and translation of mRNAs encoding β-actin and actin regulators on the near-stimulus side of the leading edge of migrating cells, such as fibroblasts and axonal growth cones, thus contributing to polarized cell movement and directional steering. The dashed outline denotes the post-stimulus trajectory. (B) Electrical input from presynaptic contacts selectively induces localized trafficking and translation of specific mRNAs in dendrites that mediate changes in spine morphology (dashed outline) and plasticity. Several aspects of these models are speculative.
Synapse formation and plasticity
In mammalian hippocampal neurons, strong synaptic activation is accompanied by transcription of the Arc gene and rapid trafficking of its mRNA to dendrites, where it localizes selectively to active synaptic sites (38). Arc is required for the consolidation of long-term potentiation (LTP), a form of persistent synaptic change, most likely through its ability to regulate actin dynamics and to modulate dendritic spine morphogenesis (39). Direct evidence for a requirement for mRNA localization in synaptic plasticity comes from studies of CamKIIα. Disruption of dendritic targeting of this mRNA in mice, by replacing the 3′ untranslated region of the endogenous gene with one from a nonlocalizing transcript, impairs LTP and long-term memory (40).
mRNA localization is also important for the establishment of synapses. In Aplysia sensory neurons, contact with a target motor neuron triggers rapid local concentration of the neuropeptide-encoding sensorin mRNA into synapses (41). Synaptogenesis is disrupted when sensorin mRNA levels are acutely reduced, even before the total concentration of the protein is altered. This indicates not only that mRNA localization is important but that newly synthesized Sensorin could have properties distinct from those of older protein copies. Consistent with an important role for nascent Sensorin, its translation is spatially restricted to active synapses in a stimulus-specific manner (42).
Cue-induced mRNA localization in axons
Growing axons navigate in the developing brain using attractive and repulsive cues that stimulate changes in growth and directional steering. β-actin mRNA is abundant in Xenopus growth cones and is rapidly recruited to the near-stimulus side in response to an attractive gradient (43, 44). Attractive turning is abolished through the specific inhibition of local β-actin mRNA translation or disruption of the interaction of Vg1RBP (the Xenopus ZBP1 ortholog) with the zip code (43, 44). The picture that emerges of localized translation of mRNAs underlying directionally responsive cell protrusions is strikingly similar to the situation in chicken fibroblasts (see below) and suggests that common mechanisms span the two systems (Fig. 3A).
“On site” versus “distant site” for action
Proteins synthesized from spatially localized mRNAs commonly act “on site.” But this is not always the case. The mRNA encoding the transcription factor cyclic adenosine monophosphate (cAMP) response element–binding protein (CREB), which promotes cell survival in dorsal root ganglia neurons, can be translated locally in axons in response to nerve growth factor (45). The nascent CREB protein undergoes retrograde transport to the nucleus, where it activates the transcription of target genes. There is evidence that the phosphorylation status of CREB differs depending on its site of translation (45), which raises the intriguing possibility that local translation of process-targeted mRNAs controls gene expression in response to distal experience.
Polarized Functions in Other Cell Types
The functional consequences of disrupting mRNA localization have now been tested in many other cell types. These studies have revealed an important role for the localization of specific mRNAs in facilitating subcellular protein localization, helping to establish or maintain cell polarity (table S2).
A particularly intriguing example comes from primary chicken fibroblasts. Here, interference with the β-actin zip code through antisense oligonucleotides strongly reduces the persistence of cell movement (46). But given that the number of protein molecules encoded by the localizing mRNAs represents only a tiny fraction of the total β-actin protein near the leading edge, why is mRNA targeting important? It is conceivable that newly synthesized β-actin monomers polymerize more efficiently than older copies, for instance, because of differential posttranslational modifications or modulation by chaperones. An alternative explanation relates to the potential for transport along a cytoskeletal track to convey multiple β-actin mRNA molecules to a small region of the cytoplasm. This could dictate a high local concentration of the protein, aiding rapid polymerization of filaments. The finding that all seven transcripts encoding Arp2 and Arp3 components are localized behind the leading edge lends support to the notion that mRNA targeting controls actin dynamics by facilitating the local assembly of protein complexes (47).
But it is not just mRNAs encoding cytoskeletal proteins that are localized in dynamic cells. At least 50 transcript species, coding for proteins with diverse functions, are enriched in pseudopodial protrusions of mouse fibroblasts in response to migratory stimuli (48). The localization mechanism is microtubule-associated and appears to be distinct from that used to target mRNAs behind the leading edge of chicken fibroblasts, involving direct roles of the adenomatous polyposis coli (APC) tumor suppressor and FMRP. This study, together with others, opens up new perspectives for elucidating links between mRNA localization and human disease [supporting online material (SOM) text].
Perspectives
Key principles of mRNA localization mechanisms in animal cells have now been established and many players identified. An important challenge is to piece together a detailed molecular understanding of the interactions that govern the recognition and differential sorting of mRNAs as well as the interplay with translational regulators. In cases in which mRNA localization is regulated by extrinsic cues (Fig. 3), what aspects of the translocation process are being targeted and how? And what is the copy number of mRNAs within the majority of mRNPs (SOM text)? Addressing these questions will benefit from insights from genetically tractable model organisms, including flies and fungi, and from advances in the ability to visualize the composition and behavior of mRNPs in living cells. The use of unbiased genome-wide methods to identify binding sites for specific transacting factors (49) could also have profound effects on our understanding of the recognition of localizing mRNAs.
A large number of studies have now highlighted the importance of subcellular mRNA localization in diverse cellular processes. Nonetheless, several questions remain from a functional perspective. What are the relative contributions of mRNA localization and localized translation to processes such as axon guidance, synaptogenesis, and synaptic plasticity? What is the extent and importance of asymmetric targeting of microRNAs? The requirements for some localizing mRNAs are independent from their translation (table S2); could this reflect a widespread structural role for mRNA in facilitating the assembly of protein complexes?
Acknowledgments
We apologize to those whose primary work could not be cited because of space constraints. We acknowledge the continued importance of research in yeast for informing and inspiring studies on mRNA localization in animal cells. We thank C. Dix, A. Lin, and F. van Horck for comments on the manuscript, those who provided images, and many colleagues for answering queries. Work in our laboratories is supported by a Wellcome Trust Programme grant (C.H.) and the MRC and a Lister Institute Prize Fellowship (S.B.).
References and Notes
- 1.Jeffery WR, Tomlinson CR, Brodeur RD. Dev. Biol. 1983;99:408. doi: 10.1016/0012-1606(83)90290-7. [DOI] [PubMed] [Google Scholar]
- 2.Lawrence JB, Singer RH. Cell. 1986;45:407. doi: 10.1016/0092-8674(86)90326-0. [DOI] [PubMed] [Google Scholar]
- 3.Lecuyer E, et al. Cell. 2007;131:174. doi: 10.1016/j.cell.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 4.Zarnack K, Feldbrugge M. Mol. Genet. Genomics. 2007;278:347. doi: 10.1007/s00438-007-0271-8. [DOI] [PubMed] [Google Scholar]
- 5.Okita TW, Choi SB. Curr. Opin. Plant Biol. 2002;5:553. doi: 10.1016/s1369-5266(02)00304-7. [DOI] [PubMed] [Google Scholar]
- 6.Forrest KM, Gavis ER. Curr. Biol. 2003;13:1159. doi: 10.1016/s0960-9822(03)00451-2. [DOI] [PubMed] [Google Scholar]
- 7.Bashirullah A, Cooperstock RL, Lipshitz HD. Proc. Natl. Acad. Sci. U.S.A. 2001;98:7025. doi: 10.1073/pnas.111145698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wolke U, Weidinger G, Koprunner M, Raz E. Curr. Biol. 2002;12:289. doi: 10.1016/s0960-9822(02)00679-6. [DOI] [PubMed] [Google Scholar]
- 9.Hu JY, Meng X, Schacher S. J. Neurosci. 2002;22:2669. doi: 10.1523/JNEUROSCI.22-07-02669.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carson JH, Cui H, Barbarese E. Curr. Opin. Neurobiol. 2001;11:558. doi: 10.1016/s0959-4388(00)00249-x. [DOI] [PubMed] [Google Scholar]
- 11.Bullock SL, Nicol A, Gross SP, Zicha D. Curr. Biol. 2006;16:1447. doi: 10.1016/j.cub.2006.05.055. [DOI] [PubMed] [Google Scholar]
- 12.Dynes JL, Steward O. J. Comp. Neurol. 2007;500:433. doi: 10.1002/cne.21189. [DOI] [PubMed] [Google Scholar]
- 13.Zimyanin VL, et al. Cell. 2008;134:843. doi: 10.1016/j.cell.2008.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Messitt TJ, et al. Dev. Cell. 2008;15:426. doi: 10.1016/j.devcel.2008.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krauss J, Lopez de Quinto S, Nusslein-Volhard C, Ephrussi A. Curr. Biol. 2009;19:1058. doi: 10.1016/j.cub.2009.04.062. [DOI] [PubMed] [Google Scholar]
- 16.Van De Bor V, Hartswood E, Jones C, Finnegan D, Davis I. Dev. Cell. 2005;9:51. doi: 10.1016/j.devcel.2005.04.012. [DOI] [PubMed] [Google Scholar]
- 17.Vogt N, Koch I, Schwarz H, Schnorrer F, Nusslein-Volhard C. Development. 2006;133:3963. doi: 10.1242/dev.02570. [DOI] [PubMed] [Google Scholar]
- 18.Babu K, Cai Y, Bahri S, Yang X, Chia W. Genes Dev. 2004;18:138. doi: 10.1101/gad.282604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yisraeli JK, Sokol S, Melton DA. Development. 1990;108:289. doi: 10.1242/dev.108.2.289. [DOI] [PubMed] [Google Scholar]
- 20.Delanoue R, Davis I. Cell. 2005;122:97. doi: 10.1016/j.cell.2005.04.033. [DOI] [PubMed] [Google Scholar]
- 21.Weil TT, Forrest KM, Gavis ER. Dev. Cell. 2006;11:251. doi: 10.1016/j.devcel.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 22.Fusco D, et al. Curr. Biol. 2003;13:161. doi: 10.1016/s0960-9822(02)01436-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chang L, Shav-Tal Y, Trcek T, Singer RH, Goldman RD. J. Cell Biol. 2006;172:747. doi: 10.1083/jcb.200511033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jambhekar A, Derisi JL. RNA. 2007;13:625. doi: 10.1261/rna.262607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dienstbier M, Boehl F, Li X, Bullock SL. Genes Dev. 2009;23:1546. doi: 10.1101/gad.531009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chartrand P, Meng XH, Huttelmaier S, Donato D, Singer RH. Mol. Cell. 2002;10:1319. doi: 10.1016/s1097-2765(02)00694-9. [DOI] [PubMed] [Google Scholar]
- 27.Besse F, Ephrussi A. Nat. Rev. Mol. Cell Biol. 2008;9:971. doi: 10.1038/nrm2548. [DOI] [PubMed] [Google Scholar]
- 28.Huttelmaier S, et al. Nature. 2005;438:512. doi: 10.1038/nature04115. [DOI] [PubMed] [Google Scholar]
- 29.Pan F, Huttelmaier S, Singer RH, Gu W. Mol. Cell. Biol. 2007;27:8340. doi: 10.1128/MCB.00972-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lewis RA, Gagnon JA, Mowry KL. Mol. Cell. Biol. 2008;28:678. doi: 10.1128/MCB.00999-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Giorgi C, Moore MJ. Semin. Cell Dev. Biol. 2007;18:186. doi: 10.1016/j.semcdb.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 32.Giorgi C, et al. Cell. 2007;130:179. doi: 10.1016/j.cell.2007.05.028. [DOI] [PubMed] [Google Scholar]
- 33.King ML, Messitt TJ, Mowry KL. Biol. Cell. 2005;97:19. doi: 10.1042/BC20040067. [DOI] [PubMed] [Google Scholar]
- 34.Sardet C, Dru P, Prodon F. Biol. Cell. 2005;97:35. doi: 10.1042/BC20040126. [DOI] [PubMed] [Google Scholar]
- 35.Amiel A, Houliston E. Dev. Biol. 2009;327:191. doi: 10.1016/j.ydbio.2008.12.007. [DOI] [PubMed] [Google Scholar]
- 36.Jedrusik A, et al. Genes Dev. 2008;22:2692. doi: 10.1101/gad.486108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lambert JD, Nagy LM. Nature. 2002;420:682. doi: 10.1038/nature01241. [DOI] [PubMed] [Google Scholar]
- 38.Steward O, Wallace CS, Lyford GL, Worley PF. Neuron. 1998;21:741. doi: 10.1016/s0896-6273(00)80591-7. [DOI] [PubMed] [Google Scholar]
- 39.Bramham CR, Worley PF, Moore MJ, Guzowski JF. J. Neurosci. 2008;28:11760. doi: 10.1523/JNEUROSCI.3864-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miller S, et al. Neuron. 2002;36:507. doi: 10.1016/s0896-6273(02)00978-9. [DOI] [PubMed] [Google Scholar]
- 41.Lyles V, Zhao Y, Martin KC. Neuron. 2006;49:349. doi: 10.1016/j.neuron.2005.12.029. [DOI] [PubMed] [Google Scholar]
- 42.Wang DO, et al. Science. 2009;324:1536. doi: 10.1126/science.1173205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Leung KM, et al. Nat. Neurosci. 2006;9:1247. doi: 10.1038/nn1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yao J, Sasaki Y, Wen Z, Bassell GJ, Zheng JQ. Nat. Neurosci. 2006;9:1265. doi: 10.1038/nn1773. [DOI] [PubMed] [Google Scholar]
- 45.Cox LJ, Hengst U, Gurskaya NG, Lukyanov KA, Jaffrey SR. Nat. Cell Biol. 2008;10:149. doi: 10.1038/ncb1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kislauskis EH, Zhu X, Singer RH. J. Cell Biol. 1997;136:1263. doi: 10.1083/jcb.136.6.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mingle LA, et al. J. Cell Sci. 2005;118:2425. doi: 10.1242/jcs.02371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mili S, Moissoglu K, Macara IG. Nature. 2008;453:115. doi: 10.1038/nature06888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Licatalosi DD, et al. Nature. 2008;456:464. doi: 10.1038/nature07488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bullock SL. Semin. Cell Dev. Biol. 2007;18:194. doi: 10.1016/j.semcdb.2007.01.004. [DOI] [PubMed] [Google Scholar]