Abstract
The aminopeptidase, ERAP1, trims peptides for MHC class I presentation, influencing the degree and specificity of CD8+ T cell responses. Single nucleotide polymorphisms (SNP) within the exons encoding ERAP1 are associated with autoimmune diseases and cervical carcinoma, but it is not known whether they act independently or as disease-associated haplotypes. We sequenced ERAP1 from 20 individuals and show that SNP occur as distinct haplotypes in the human population, and that these haplotypes encode functionally distinct ERAP1 alleles. Using a wide range of substrates, we are able to demonstrate that for any given substrate, distinct ERAP1 alleles can have “normal”, “hypo-”, or “hyper-” functional; and that each allele has a trend bias towards one of these three activities. Thus, the repertoire of peptides presented at the cell surface for recognition by CTL is likely to depend on the precise combination of both MHC class I and ERAP1 alleles expressed within an individual, and has important implications for predisposition to disease.
Introduction
Major Histocompatibility complex class I (MHC I) molecules display peptides of 8-10mer amino acids in length at the cell surface for immune surveillance by circulating cytotoxic T cells (CD8+ T cells). MHC I samples the intracellular proteome and presents peptides derived from self-proteins, including those that are aberrantly expressed in cancer, as well as proteins originating from intracellular viruses and bacteria. Cytosolic proteases, including the proteasome, generate peptides with a precise C terminus but a mixture of N-terminally extended intermediates (1-3), which are then transported into the endoplasmic reticulum (ER) by the transporter associated with antigen processing (TAP). Here, further processing in the form of N-terminal peptide trimming by ERAP1 can occur, with the net result of increasing the frequency of peptides that are of an appropriate length to bind to MHC I (4-7). Some antigenic peptides can be destroyed or “over-processed” by ERAP1 (8), indicating that ERAP1 has a role as an antigenic peptide editor, influencing the peptide repertoire displayed at the cell surface (9-11). In humans, ERAP2, a homologue of ERAP1 is also able to perform this function (4).
The ability of ERAP1 to trim N-terminal amino acids from epitope precursors has been shown to depend on the amino acids present, which are removed at vastly different rates, forming a distinct hierarchy (12). This specificity ultimately defines the abundance of presented peptide antigens which in turn can shape the immunodominance of CD8+ T cell responses to pathogens and cancer (11, 13). Recent genome wide association studies (GWAS) have identified polymorphisms encoded within ERAP1 linked to many diseases such as cervical carcinoma and the autoimmune diseases, ankylosing spondylitis (AS), multiple sclerosis and psoriasis (14-17). The link between AS and ERAP1 is interesting since a strong association between AS and HLA-B27 has been known for 40 years with 95% of AS individuals being HLA-B27 positive. Furthermore, ERAP1 is only associated with AS in HLA-B27 individuals (18). ERAP1 and HLA-B27 intersect in the antigen processing pathway supporting a role for ERAP1 peptide trimming in disease pathogenesis. Individual amino acid changes within ERAP1, corresponding to the SNPs linked with disease, resulted in a reduction in peptide trimming activity for K528R, R725Q and Q730E ERAP1 molecules, but not D575N (18-20). These studies did not examine the effect of multiple SNPs/haplotypes on the ability of ERAP1 to trim peptide precursors or their effects on amino acid specificity. Genetic association studies have identified ERAP1 SNP haplotypes (K528/D575/R725), (K528/D575/Q730E) and ERAP1/ERAP2 haplotypes (Q730/K528 ERAP1 and K392N ERAP2) as being associated with AS (21-23). These studies examined haplotypes containing only some of the five SNPs identified in the original GWAS study and did not examine their function. Therefore the extent to which these five SNPs assemble into haplotypes and whether the ERAP1 alleles encoded by the different ERAP1 haplotypes have different functions is not known. We have identified nine naturally occurring ERAP1 haplotypes from individuals, based on the five disease associated SNPs. The ERAP1 alleles encoded by these haplotypes displayed three generic activities (efficient, hypo- and hyper-functional) based on the precise substrate specificity of each allele highlighting the importance of ERAP1 alleles in the generation of the peptide repertoire.
Materials and Methods
Subjects
This study was performed, with informed consent for all samples, according to the protocol approved by National Research Ethics Services and Southampton Research Ethics committee (study reference number RHM MED 0869 and rec reference 09/H0504/88). Study title ‘ERAP1 (ERAAP) polymorphisms linked to disease susceptibility’. Samples were recruited from the Department of Rheumatology, University Hospital Southampton NHS Foundation Trust and obtained in the Southampton National Institute for Health Research Wellcome Trust Clinical Research Facility, University Hospital Southampton NHS Foundation.
ERAP1 isolation and generation of ERAP1 sequence variant E320A
RNA purified from 2 × 106 CEM (human T cell lymphoblast-like cell line) cells with RNeasy mini kit (Qiagen) or 200μl blood with ZR whole-blood RNA prep (Zymo Research) was used to generate cDNA with the Transcriptor High Fidelity cDNA synthesis kit (Roche). ERAP1 was amplified from cDNA using KOD Hot Start DNA polymerase (Merck) and the following primers: 5′ primer (EcoRI site in italics), 5′-GACGAATTCATGGTGTTTCTGCCCCTCAAATG-3′; 3′ primer (XhoI site in italics), 5′-GACCTCGAGCATACGTTCAAGCTTTTCAC-3′ (Sigma). The PCR amplification product was cloned into the vector pcDNA3.1 (Life Technologies). These cloned ERAP1 subject alleles were sequenced to identify polymorphic variants and used in functional studies. Site directed mutagenesis (SDM) was used to generate the ERAP1 E320A non-functional variant using the WT cloned ERAP1 vector construct with KOD Hot Start DNA polymerase and the following primers (mutated nucleotide in italics): E320A 5′-CTGGTGCTATGGCAAACTGGGGACTG-3′ and 5′-CAGTCCCCAGTTTGCCATAGCACCAG-3′.
DNA constructs
The ES-SHL8, ES-X5-SHL8 and ES-X6-SHL8 DNA constructs all encode the ER targeting signal sequence and have been described previously (9, 24). ES-X-SHL8 constructs were generated by the incorporation of an additional amino acid into the ES-SHL8 construct using the following primers: 5′-GCAGTCTGCAGCGCGNNSAGCATCATCAACTTCG-3′ and 5′-CGAAGTTGATGATGCTSNNCGCGCTGCAGACTGC-3′ where N = any nucleotide and S = C or G, resulting in amino acids being represented. Constructs were sequence verified and the most frequent codon for each amino acid chosen for use where possible.
Cell lines, transfection and T cell activation assays
An Erap1-deficient fibroblast cell line (a gift from Dr. Nilabh Shastri, University of California at Berkeley, Berkeley, CA) used for all transfection experiments were cultured as described previously (9). Culture conditions for B3Z T cell hybridoma and H-2Kb-L cells have been described before (24). Erap1-deficient fibroblasts were transfected with 1μg of each ERAP1 allele and ES-AIVMK-SHL8 (X5-SHL8) or ES-LEQLEK-SHL8 (X6-SHL8) minigene construct (25) (pcDNA3.1) or SCT using FuGENE 6 (Roche). Where N-terminal amino acid specificity was assessed, 0.05μg of each ERAP1 allele and 0.05μg of each X-SHL8 minigene construct were transfected together in a 96 well plate. After 48 hours, cells were harvested and incubated overnight with the LacZ inducible B3Z T cell hybridoma, specific for the recognition of SHL8/H-2Kb complexes at the cell surface. Intracellular LacZ was measured with the substrate chlorophenolred-β-D-galacto-pyrannoside (Roche) by its absorbance at 595 nm and 655 nm as reference.
Single chain trimer constructs
H-2Kb/SL8 disulfide trap single chain trimer construct (a gift from Dr. T.H. Hansen, Washington University, St. Louis, MO) was cloned into pcDNA3.1 with EcoRV and NotI. A lysine residue preceding SL8 was added by PCR using the following primers: (lysine is in italics) 5′-GACCGGTTTGTATGCTAAAAGTATCATTAATTTCG-3′ and 5′-CGAAATTAATGATACTTTTAGCATACAAACCGGTC-3′. SDM of lysine to histidine within SL8 and glycine to lysine within the linker between peptide and β2M was performed using the following primers: (mutated nucleotides in italics) K-H, 5′-CTATCATTAATTTCGAACATCTTAAATGCGGTGCTAGC-3′ and 5′-GCTAGCACCGCATTTAAGATGTTCGAAATTAATGATAG-3′; G-K, 5′-CATTAATTTCGAACATCTTAAATGCGGTGCTAGCGGTGG-3′ and 5′-CCACCGCTAGCACCGCATTTAAGATGTTCGAAATTAATG-3′.
Peptide extracts, HPLC and MS analysis
Peptides of various sequences were synthesized (GL Biochem) and their structures confirmed by mass spectrometry. Endogenous peptides were extracted from transfected Erap1-deficient fibroblasts after 48 hours. Transfected Erap1-deficient fibroblasts were lysed in 10% formic acid supplemented with 10μM irrelevant peptide, boiled for 10 min and passed through a 10KDa filter (Millipore). The filtrate was then fractionated by RP-HPLC (Shimadzu) on a 2.1mm × 250mm C18 column (Vydac) over a gradient of 15-40% acetonitrile. Flow rate was maintained at 0.25ml/min and 150μl fractions collected in 96-well plates and dried. Trypsin (50μg/ml; Sigma) was added to fractions to release SHL8 from N-terminally extended precursors and analyzed with B3Z T cell hybridoma and H-2Kb-L cells as APCs. For SCT experiments carboxypeptidase B (1U/ml; Merck) was added to fractions following RP-HPLC fractionation to remove lysine from the peptide C-terminus. For peptide mass analysis, peptide extracts or elutions were fractionated by RP-HPLC as above and detected by mass spectrometry (Shimadzu). The presence of SHL8K (m/z = 1100) and IHL7K (m/z = 1013) peptides was determined using LC solutions software (Shimadzu). Synthetic peptides and buffer only runs were analyzed in identical conditions to establish retention times and the absence of sample cross-contamination.
Immunoprecipitation and immunoblots
Expression of ERAP1 was determined by immunoblot. Erap1-deficient transfected fibroblasts were lysed in 0.5% Nonidet P-40, 150mM NaCl, 5mM EDTA and 20mM Tris pH7.4 supplemented with phenylmethylsulfonyl fluoride and iodoacetamide (Sigma). Proteins were separated by 10% SDS-PAGE and transferred to a nitrocellulose membrane (GE healthcare). Immunoblots were probed with anti-human ERAP1 (AF2334, R&D Systems) or anti-glyceraldehyde 3-phosphate dehydrogenase (Abcam) antibodies followed by HRP-conjugated secondary antibody and SuperSignal West Pico or Femto chemiluminescent substrate (Thermo Scientific). For immunoprecipitation, lysates (107 cell equivalents) were incubated with anti-H-2Kb antibody Y3 immobilized on protein G Dynabeads (10μg antibody/5mg beads; Life technologies). The beads were washed and dynabead bound SCT were incubated with trypsin (50μg/ml) for 3 hours at 37°C. Dynabeads were removed and the supernatant collected and analyzed by western blot or HPLC/MS.
Statistical Analysis
One-way ANOVA with Dunnett’s post-test was performed for analysis of differences between multiple groups and control (GraphPad prism, www.graphpad.com).
Results
ERAP1 alleles have different trimming activities
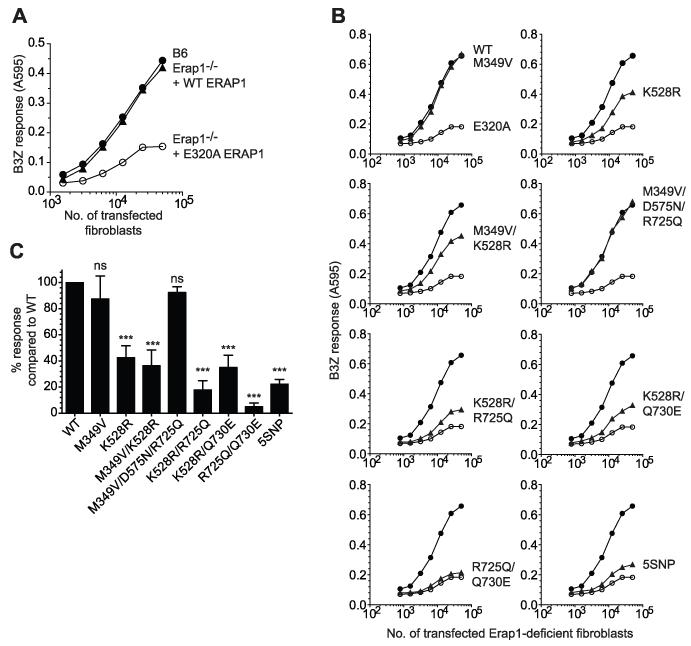
In order to determine the impact on trimming function of SNPs within ERAP1 in the context of naturally occurring haplotypes, we used molecular cloning to isolate and sequence ERAP1 genes from 20 individuals. This revealed a diverse array of ERAP1 haplotypes, mostly comprised of multiple SNP combinations based on the five SNPs with strongest disease association (Table I). The most common ERAP1 haplotype observed (cloned from CEM cells and volunteers) was identical to the previously characterized ERAP1 gene (NM 001198541.1) and termed wild-type (WT) ERAP1. To assess the trimming function of the ERAP1 alleles encoded by these haplotypes, we used the well characterized SIINFEHL (SHL8) murine model system in which an ER targeted (using an ER translocation signal) five amino acid N-terminally extended precursor AIVMK-SIINFEHL (X5-SHL8) was transfected into Erap1 deficient cells along with the ERAP1 alleles (26). The expression of trimmed SHL8 presented by H-2Kb at the cell surface was measured by coculturing transfected cells with the SHL8-specific T cell hybridoma B3Z, allowing a direct assessment of the trimming activity of ERAP1 alleles. Trimming activity in Erap1-deficient cells was <10% of that seen in WT cells following transfection with X5-SHL8 (Fig. 1A). Transfection of the WT ERAP1 sequence restored trimming activity to a level comparable to WT cells (Fig. 1A). As a negative experimental control the active site GAMEN motif, responsible for N-terminal recognition of peptide substrate, was mutated (E320A) to produce a non-functional variant (Fig. 1A and B). Some residual trimming and SHL8 presentation is observed in Erap1-deficient cells, its source is unclear but may be from aberrant signal peptidase cleavage or an ERAP1-independent pathway (26). We transfected Erap1-deficient cells with X5-SHL8 and ERAP1 alleles and confirmed expression by western blot (Supplemental Fig. 1). Figure 1B and C show that two alleles M349V and M349V/D575N/R725Q were able to trim X5-SHL8 as efficiently as WT, with other alleles showing a reduced capacity. In particular, the 5SNP, R725Q/Q730E and K528R/R725Q alleles were least able to generate the epitope, with all three showing ≤30% of WT activity (Fig. 1B and C).
Table I. Identity of ERAP1 haplotypes in the samples studied.
| SNP | ||||||
|---|---|---|---|---|---|---|
| rs2287987 (M349V) |
rs30187 (K528R) |
rs10050860 (D575N) |
rs17482078 (R725Q) |
rs27044 (Q730E) |
||
| Haplotype | WT | t/M | t/K | c/D | c/R | g/Q |
| 5SNP | c/V | c/R | t/N | t/Q | c/E | |
| K528R/Q730E | t/M | c/R | c/D | c/R | c/E | |
| K528R | t/M | c/R | c/D | c/R | g/Q | |
| M349V/D575N /R725Q |
c/V | t/K | t/N | t/Q | g/Q | |
| K528R/R725Q | t/M | c/R | c/D | t/Q | g/Q | |
| R725Q/Q730E | t/M | t/K | c/D | t/Q | c/E | |
| M349V | c/V | t/K | c/D | c/R | g/Q | |
| M349V/K528R | c/V | c/R | c/D | c/R | g/Q | |
| AS* association (P-value) |
1.4 ×10−4 |
4.9 ×10−6 |
1.2 ×10−4 |
4.0 ×10−5 |
1.6 ×10−6 |
|
| Frequency % (WT/variant) |
77/23 | 32/68 | 75/25 | 76/24 | 26/74 | |
Lower case letter denotes anti-sense strand base pair and upper case letter denotes the amino acid at this position.
Ankylosing Spondylitis
Figure 1. The trimming activity of identified ERAP1 alleles.
(A) Erap1-deficient fibroblasts were transfected with X5-SHL8 and WT or E320A ERAP1 and assayed for trimming by stimulation of the LacZ-inducible, SHL8/Kb-specific B3Z T cell hybridoma. As a control for ERAP1 trimming B6 fibroblasts were transfected with X5-SHL8 only and assayed for trimming as above. (B) Erap1-deficient fibroblasts transfected with X5-SHL8 and WT (●), E320A (○), or identified ERAP1 alleles M349V, K528R, M349V/K528R, M349V/D575N/R725Q, K528R/R725Q, K528R/Q730E, R725Q/Q730E or 5SNP (▲) and were titrated and assayed for trimming as above. Data shown in each panel is from the same experiment representative of six independent experiments. (C) Erap1-deficient fibroblasts were transfected with ERAP1 alleles and X5-SHL8, as above, and the relative maximum B3Z response compared to WT calculated. Bars show results pooled from at least six experiments ± SEM (***; P <0.001, ns; not significant).
Functional classes of ERAP1
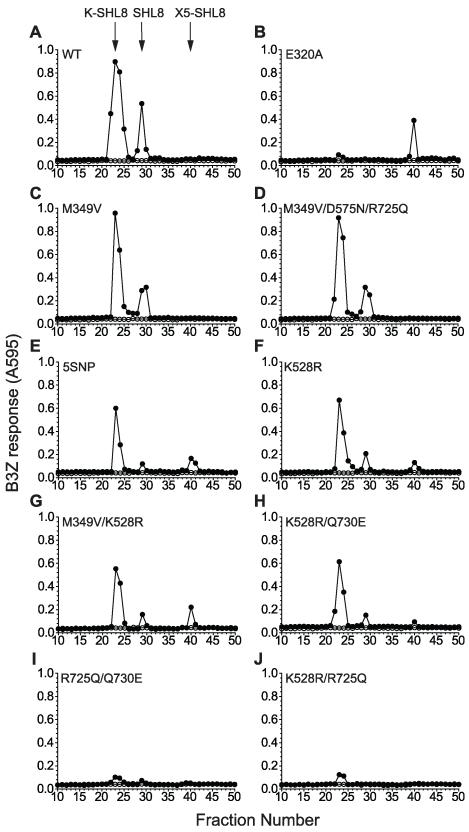
To investigate the poor trimming phenotypes and directly assess the fate of the antigenic precursors in cells, we analyzed peptide extracts by reverse-phase HPLC (Fig. 2). This method has been shown to reveal trimming intermediates at steady state, thus allowing us to identify the stage during sequential N-terminal trimming that was most affected by the polymorphisms (26). In Erap1-deficient cells transfected with X5-SHL8 and WT ERAP1, two peptide peaks were identified following RP-HPLC fractionation, corresponding to K-SHL8 (fraction 23) and SHL8 (fraction 29) (Fig. 2A). The peptide peak at fraction 23 originates from the capture of the N-terminal peptide trimming intermediate K-SHL8 by H-2Db (27). Conveniently, this allowed us to determine the relative efficiency of the cleavage of K-SHL8 to SHL8 by ERAP1 variants by assuming that more K-SHL8 would be captured by H-2Db when the K-SHL8 to SHL8 cleavage was less efficient. By contrast, when cells were transfected with E320A ERAP1 only a single peak corresponding to untrimmed precursor X5-SHL8 at fraction 40 was observed (Fig. 2B); confirming loss of function as a result of the active-site mutation. The alleles M349V and M349V/D575N/R725Q ERAP1 revealed peptide profiles consistent with trimming activity similar to WT (Figs. 2C and 2D). Analysis of 5SNP, K528R, M349V/K528R and K528R/Q730E ERAP1 revealed three peaks corresponding to untrimmed X5-SHL8, K-SHL8 and SHL8 indicating a reduced ability to trim precursor peptide (Figs. 2E-H). In all cases the ratio of K-SHL8 to SHL8 was greater than for WT (7.6, 5.4, 6.8 and 7.5 respectively compared to 4.4), consistent with their reduced ability to trim peptide precursors and indicating an inability to efficiently trim the final lysine residue. Analysis of R725Q/Q730E and K528R/R725Q ERAP1 revealed a different pattern altogether, characterized by very small K-SHL8 and SHL8 activity peaks representing <5% of the amount of trimmed product seen with WT (Figs. 2I and 2J). The absence of additional peaks corresponding to untrimmed (X5-SHL8) or partially trimmed peptides is consistent with a hyperactive function for these variants. We therefore conclude that ERAP1 alleles can be grouped into three functional classes; i) efficient, ii) hypo or iii) hyper-active trimmers.
Figure 2. RP-HPLC analysis of ERAP1 alleles reveals hypo- and hyper-active trimming phenotypes.
(A-J) Erap1-deficient fibroblasts were transfected with X5-SHL8 and ERAP1 alleles (●) identified from individuals. Peptide extracts from transfected fibroblasts were fractionated by RP-HPLC, pretreated with trypsin to allow detection of N-terminally extended SHL8 analogs and assayed with B3Z T cell hybridoma and H-2Kb-L cells as APCs. Retention times of synthetic SHL8, K-SHL8 and X5-SHL8 peptides are marked with arrows. Fractions from runs of buffer alone were assayed in parallel to exclude the possibility of sample carry over between runs (○). HPLC elution profiles are representative of individual ‘runs’ from four experiments.
Hyper-functional ERAP1
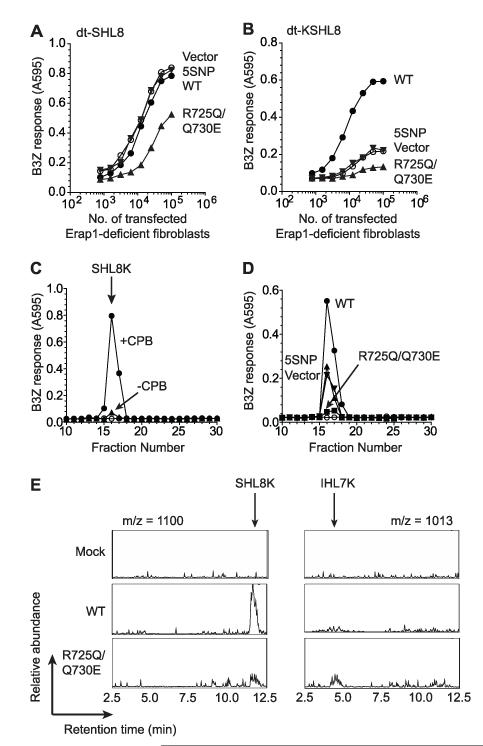
To test the hypothesis that R725Q/Q730E ERAP1 “over-trims” peptide precursors we utilized a disulfide trap single-chain MHC I construct, dt-SCT. This consists of peptide linked with β-2-microglobulin (β2M) and MHC heavy chain in which the peptide is further tethered at its C-terminus to the MHC binding groove by introducing a disulfide bond between Y84C and a second cysteine within the peptide-β2M linker (28). We transfected a construct containing SIINFEHL (SHL8) peptide, dt-SHL8 into Erap1-deficient cells, which was presented at the cell surface and stimulated B3Z T cells (Fig. 3A). When WT or 5SNP ERAP1 is co-expressed in these cells B3Z stimulation was unchanged. By contrast, R725Q/Q730E transfection resulted in a 50% decrease in B3Z stimulation suggesting SHL8 destruction by over-trimming (Fig. 3A). To further investigate this we generated a dt-SCT with an N-terminal extension (dt-KSHL8) which would require trimming in order for it to be presented to B3Z T cells. Disulfide trap-KSHL8 expressing Erap1-deficient cells stimulated B3Z poorly (Fig. 3B) consistent with the inability of B3Z to recognize N-terminally extended peptides (Supplemental Fig. 2). When WT ERAP1 was co-expressed B3Z stimulation increased (Fig. 3B), suggesting that WT ERAP1 is able to trim the N-terminal extension from the tethered KSHL8. Co-expression of 5SNP ERAP1 did not alter B3Z stimulation compared to vector, confirming its hypo-functionality. Transfection of R725Q/Q730E in dt-KSHL8 expressing cells led to a 70% reduction in B3Z stimulation compared to vector indicating barely detectable levels of optimally trimmed peptide (Fig. 3B). This is consistent with destruction of the SHL8 epitope moiety within the dt-KSHL8 construct by overtrimming.
Figure 3. Hyper-active ERAP1 alleles destroy peptides by over-trimming.
Erap1-deficient fibroblasts were transfected with dt-SHL8 (A) or dt-KSHL8 (B) and empty vector, WT, 5SNP or R725Q/Q730E ERAP1, titrated and assayed for stimulation of B3Z T cell hybridoma as in Figure 1. (C) Peptides eluted from dt-KSHL8K expressed by Erap1-deficient fibroblasts by trypsin following immunopurification by anti-H-2Kb antibody Y3 were fractionated by RP-HPLC. Fractions were untreated (▲) or pretreated with carboxypeptidase B (●) to allow detection of SHL8 and assayed with B3Z and H-2Kb-L cells as APCs. Downward arrow indicates the peak elution of SHL8K. Fractions from mock injection (○) were performed as in Figure 2. (D) Peptides eluted from Erap1-deficient fibroblasts transfected with dt-KSHL8K and WT (●), vector (▼), 5SNP (▲) or R725Q/Q730E (■) were fractionated by RP-HPLC as in C. Data are representative of four experiments. (E) Chromatogram of RP-HPLC fractionated peptides eluted from Erap1-deficient fibroblasts transfected as in (D) at mass fragments m/z = 1100; SHL8K (left panel) and m/z = 1013; IHL7K (right panel). Downward arrows indicate the retention time of SHL8K and IHL7K peptides. Data are representative of three experiments.
To gather more direct evidence for hypo and hyperfunctionality among ERAP1 variants, we introduced a trypsin cleavage site one amino acid downstream of the authentic C-terminus of SHL8 in the dt-SCT by substitution of lysine for glycine within the peptide-β2M linker. This allowed us to recover peptide from the SCT using trypsin following immunopurification from cells. Disulfide trap-KSHL8K molecules were transfected into Erap1-deficient cells, immunopurified and eluted peptides fractionated by RP-HPLC. Fractions were treated with carboxypeptidase B to remove the C terminal lysine revealing a single peak of B3Z activity corresponding to SHL8K (fraction 16; Fig. 3C). When WT ERAP1 was transfected into dt-KSHL8K expressing cells the amount of SHL8K observed was 3-fold greater than vector alone (Fig. 3D). Transfection of 5SNP ERAP1 showed an equivalent amount of SHL8K recovered compared to vector only, thus confirming the hypoactivity of 5SNP to trim peptide precursor. The use of trypsin prior to fractionation means we are unable to recover KSHL8K which, based on the results shown in Figure 2, we would expect to be elevated. When R725Q/Q730E was transfected, the amount of SHL8K observed was significantly reduced (>80% reduction; Fig. 3D) providing further evidence that R725Q/Q730E ERAP1 was over trimming the peptide precursor. We used mass spectrometry to identify peptide species eluted from the dt-KSHL8K molecules following RP-HPLC fractionation. A peak corresponding to the mass of SHL8K was the major species eluted from WT transfected cells, and was greatly reduced in the R725Q/Q730E transfectants (Fig. 3E). Further analysis of eluted peptides revealed a peak corresponding to IHL7K (IINFEHLK) as a unique product in the R725Q/Q730E transfectants (Fig. 3E) confirming the over-trimming function of this variant.
Amino acid specificity of defined ERAP1 alleles
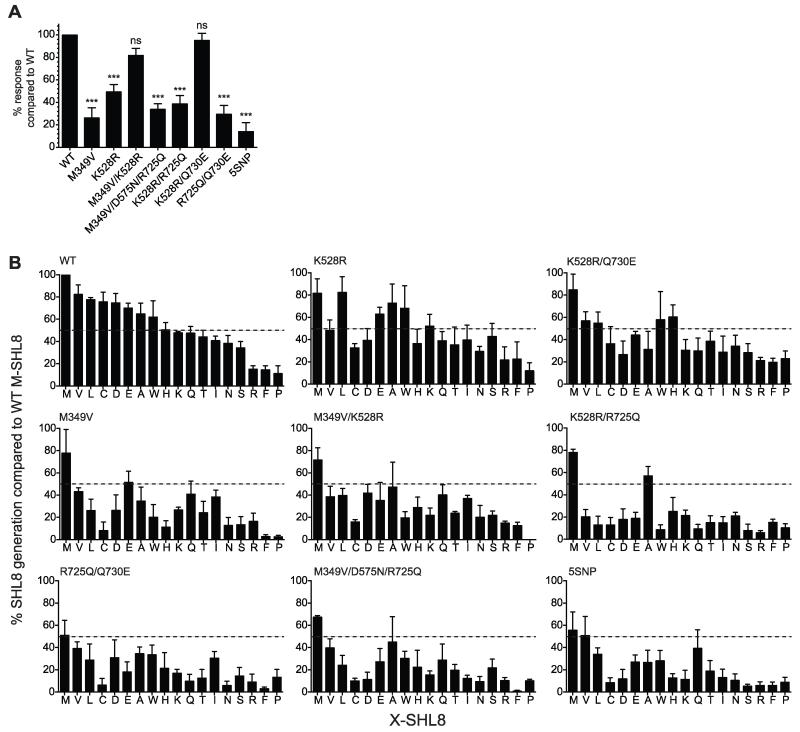
To examine whether the ability of alleles to generate SHL8 was dependent on the sequence of the N-terminal precursor, we substituted AIVMK for LEQLEK (X6-SHL8) containing one additional amino acid and consisting of mostly polar/charged amino acids compared to the mostly hydrophobic AIVMK extension. Figure 4A shows that, as for X5-SHL8, most of the alleles showed a reduced ability to generate the final SHL8 epitope from X6-SHL8 compared to WT. Interestingly, however, M349V/K528R and K528R/Q730E, which were poor trimmers of X5-SHL8 (~40% activity of WT), were able to efficiently process X6-SHL8. Conversely, M349V and M349V/D575N/R725Q, which trimmed X5-SHL8 well, showed a significant reduction in X6-SHL8 trimming (<40% of WT activity). This demonstrated that the activity of these alleles was dependent on substrate sequence, and prompted us to investigate substrate specificity in more detail.
Figure 4. Fine specificity of N-terminal amino acid trimming by ERAP1 alleles.
(A) Erap1-deficient fibroblasts transfected with X6-SHL8 and WT, E320A, or ERAP1 alleles M349V, K528R, M349V/K528R, M349V/D575N/R725Q, K528R/R725Q, K528R/Q730E, R725Q/Q730E or 5SNP were assayed for trimming and the relative maximum B3Z response compared to WT calculated. Bars show results pooled from at least six experiments ± SEM (***; P <0.001, ns; not significant). (B) Erap1-deficient fibroblasts were transfected with the indicated ERAP1 allele together with X-SHL8 minigenes representing 18 amino acids and assessed for generation of SHL8 by B3Z activation. The relative presentation of SHL8 was compared to that observed for Erap1-deficient fibroblasts transfected with WT ERAP1 and ES-M-SHL8. Dashed lines indicate 50% of ES-M-SHL8 generation. Data are pooled from three separate experiments.
To fine map amino acid trimming by alleles we utilized the ER targeted SHL8 peptide with a single amino acid extension representing 18 of the 20 amino acids (X-SHL8) transfected together with each ERAP1 allele. When we assessed the efficiency of SHL8 generation from each X-SHL8 substrate, we identified allele-specific signatures that could be broadly divided into three groups, shown in Table II and Figure 4B: i) K528R/R725Q, R725Q/Q730E, 5SNP and M349V/D575N/R725Q were unable to generate SHL8 from the majority of amino acid precursors, ii) M349V and M349V/K528R were intermediate and could generate SHL8 from some precursors well (>75% of WT activity) and others poorly (<50% of WT activity), and iii) K528R and K528R/Q730E which, like WT, generated SHL8 well from most precursors. It is important to emphasize that this assay is not able to determine whether the lack of SHL8 presentation was due to an excess or an absence of trimming. However, it is notable that the alleles R725Q/Q730E, which we have shown to over-trim K-SHL8 (Fig. 3), and K528R/R725Q, which from cell surface and RP-HPLC analysis also exhibits an over-trimming phenotype (data not shown), were found to have the lowest ability to generate SHL8 from all X-SHL8 substrates (Table II), suggesting that these alleles over-trim the majority of amino acid precursors. Interestingly, for both of these alleles, there are substrates where SHL8 is generated at similar levels to WT (Met and Ala for K528R/Q730E; Ile for R725Q/Q730E). This suggests that a given allele may have a range of activities for different N-terminal amino acids such that some are rapidly hydrolyzed and others slowly.
Table II. Relative amino acid trimming efficiency compared to WT ERAP1.
| Allele | SHL8 generation compared to WT from X-SHL8 | Total SHL8 generation+* |
||
|---|---|---|---|---|
| 75-100% | 50-75% | 0-50% | ||
| K528R/R725Q | F, M, A, P | N | E, H, K, T, I, R, V, L, C, D, W, Q, S |
345.4 |
| 5SNP | Q, P | M, V | L, E, A, W, H, T, I, N, R, F, C, D, K, S |
366.9 |
| R725Q/Q730E | P | M, A, W, I, R | V, L, D, E, H, K, T, S, C, Q, N, F |
378.7 |
| M349V/D575N /R725Q |
P | M, A, Q, S, R | V, L, E, W, H, K, T, I, C, D, N, F |
383.9 |
| M349V | R, M, Q, I | V, E, A, K, T | L, D, W, N, S, C, H, F, P |
436.9 |
| M349V/K528R | Q, I, R, F | M, L, D, E, A, H, T, N, S |
V, W, K, C, P | 533.6 |
| K528R/Q730E | H, R, F, P, M, W, T, N, S |
V, L, E, K, Q, I | C, D, A | 691.6 |
| K528R | L, A, W, K, S, R, F, P M, E, Q, T, I, N |
V, D, H | C | 862.6 |
Total SHL8 generation is the sum of SHL8 generated from all N-terminal amino acids.
WT = 950.7
Examination of the contribution of individual SNPs to X-SHL8 outcomes showed that the SNP with the strongest effect was R725Q. All alleles containing this SNP (K528R/R725Q, R725Q/Q730E, M349V/D575N/R725Q, 5SNP) showed poor trimming phenotypes when assayed across the whole range of X-SHL8 substrates, and no other SNP was uniquely associated with a poor trimming phenotype.
Discussion
Using molecular cloning, we identified nine discrete ERAP1 haplotypes based on the five disease associated SNPs. This confirms the polymorphic nature of ERAP1 and suggests that many different haplotypes may have a role in the pathology of linked diseases. Imputation and permutation haplotype studies have shown an association of ERAP1 and ERAP1/2 haplotypes with AS (21-23). Interestingly, although these studies did not examine all five SNPs examined here, the AS associated ERAP1 haplotypes (K528/D575/R725), (K528/D575/Q730E) and (Q730/K528 ERAP1 and K392N ERAP2) are represented in the haplotypes we observe, albeit being represented by more than one observed haplotype in some instances: i) K528/D575/R725 = WT or M349V; ii) K528/D575/Q730E = R725Q/Q730E; iii) Q730/K528 = WT, M349V or M349V/D575N/R725Q). Thus highlighting the importance of sequencing haplotypes to identify polymorphic variants. Examination of the trimming of model N-terminally extended substrates, X5- and X6-SHL8, revealed differences between ERAP1 alleles encoded by haplotypes and their ability to generate SHL8, in a substrate-dependent way. Mapping of N-terminal amino acid trimming by ERAP1 alleles revealed a complex picture with a range of trimming abilities found among alleles for a given substrate and within alleles for a range of substrates. WT ERAP1 was found to have the greatest capacity to generate SHL8 from N-terminally extended precursors with the hierarchy of amino acid specificity showing a similar profile to those identified in previous studies using recombinant enzyme and in living cells (4, 12, 29), with any differences most likely reflecting the particular assay of choice (living cells versus recombinant enzymes and microsomal extracts). It is worth noting also that the results of previous trimming assays using transfected HeLa cells (12) may be confounded by endogenous ERAP1 alleles (WT and K528R/Q730E; data not shown). In addition, mouse cells lack ERAP2, which is expressed in HeLa cells and has been suggested to associate with ERAP1 in human cells (4). This may therefore impact on the capacity of ERAP1 to generate peptide epitopes, and reflect differences observed with the previous study of ERAP1 activity (12). Although, no ERAP1-deficient human cells are available, any assessment of ERAP1 allele activity in human cells may be confounded by the presence of ERAP2.
Further analysis of precursor specificity shows that most variation in the generation of SHL8 between ERAP1 alleles is observed with substrates containing N-terminal Cys, His, Trp, Asn or Asp showing these amino acids to be the most sensitive to allelic variation in ERAP1. Analysis of N-terminal amino acid trimming specificity across alleles shows the amino acids Met, Val and Ala are good substrates for SHL8 generation. By contrast, Arg, Pro and Phe were poor substrates with very little SHL8 generated from these precursors. Interestingly, amino acids, Cys and Asp, were only generated well by WT ERAP1 with poor generation by all other alleles. These analyses show that the chemical property of an amino acid does not determine whether it is a good substrate or not, however in general, hydrophobic residues are hydrolyzed more efficiently.
Comparison of allele trimming profiles indicated that a range of N-terminal amino acid trimming activities may exist within individual alleles. With an array of trimming activities (some trimmed rapidly, others slowly), those alleles with activities skewed to being fast are therefore likely to over-trim whereas those skewed to being slow are likely to under-trim. This observed range in ERAP1 allele trimming activities may reflect an evolutionary process driving trimming diversity, ensuring optimal peptide epitope generation within the population to combat disease; a similar mechanism is evident for the diversity of MHC I alleles. Therefore the more extreme phenotypes we have identified, such as hypo and hyper-active trimmers, may more commonly be found with alleles that trim well in the population. Instances where aberrant trimming alleles co-exist in an individual may therefore predispose them to disease. Furthermore, our data agree with the previously identified AS associated haplotypes which also encode ERAP1 alleles with poor trimming functions (R725Q/Q730E (22), M349V or M349V/D575N/R725Q (23) and M349V (21); although the latter two may also encode the efficient WT ERAP1 allele), indicating a link between poor ERAP1 function and disease.
Recent crystal structures (20, 30) reveal an interesting link between SNPs and their effects on trimming capacity. M349V is located within the active site and although it is unlikely to directly interact with the peptide substrate, the amino acid substitution may alter the ability to form the correct catalytic conformation. Alleles which contain M349V trim amino acids poorly indicating a key role in active site maintenance. Both K528R and D575N are situated at domain junctions important for the conformation changes required for peptide trimming to occur. Similarly to M349V, alleles containing D575N have poor trimming functions, indicating its significance in allowing ERAP1 to adopt the correct conformation for trimming. By comparison, the K528R allele has an intermediate trimming phenotype suggesting a lesser role for K528R, although, like D575N, when K528R is present in multiple SNP alleles the trimming phenotype is also poor. Despite good structural data for ERAP1 very little is understood about its mechanism of action. In particular, it is not known whether the regulatory domain of ERAP1 (which contains the R725 and Q730 residues), or the MHC I peptide binding groove acts as the “molecular ruler” extracting peptides from an iterative cycle of hydrolysis when the appropriate length is reached. Relevant to this is our observation that ERAP1 trimmed the single-chain construct efficiently despite the lack of a free C-terminus, and the strong likelihood that the C-terminal amino acid of the peptide-substrate was bound tightly in the peptide-binding groove of MHC I. This does not support a model involving an interaction between ERAP1 and the free C-terminus of peptide substrate (31). Trimming of small substrates such as dipeptides (unable to engage the peptide binding pocket of ERAP1) has been shown, indicating that engagement of the peptide binding pocket is not essential for trimming to occur (4). The ability of ERAP1 to trim the tethered peptides is most likely dependent on access to the N-terminus and related to MHC I affinity. This may therefore reflect a balance between ERAP1 and MHC I for peptide binding based on affinities. For an epitope of the correct length for MHC I binding (8-10mer) the affinity is greater for MHC I than ERAP1 binding and therefore no further trimming occurs. However, an N-terminally extended peptide would have lower affinity for MHC I and allow binding to ERAP1, a mechanism similar to the model described by Kanaseki et al (26). The dt-SCT-SL8 system does not reflect the normal situation in the ER, but the identification of over-trimming in a system which should minimize the ability of ERAP1 to access peptides provides an alternative mechanism for ERAP1 trimming. The finding that R725Q/Q730E over-trims peptides tethered to MHC I suggests that SNPs may increase ERAP1 affinity for peptides allowing further trimming of cognate epitopes thus destroying them. It is worth noting that R725Q, which had the strongest negative effect on trimming and was uniquely included in all the alleles that were poor at generating SHL8 from all X-SHL8 substrates, is located within the regulatory domain of ERAP1 which has been proposed to interact with the peptide substrate (31). The observed effects of ERAP1 alleles on X-SHL8 and dt-SCT substrates may be confounded by an alteration in the competition from other endogenous peptides for ERAP1 in transfected cells. However, the data shown from peptide extracts and the trimming phenotypes for X5-SHL8 and dt-SCT experiments (where peptides are not competing for MHC) is more consistent with a direct effect of the ERAP1 alleles on peptide substrates. In addition, analysis of peptides presented in cells expressing 5SNP ERAP1 revealed longer peptides compared to cells expressing WT ERAP1; this is consistent with our observations that WT is an efficient trimmer whereas 5SNP is hypo-functional (32).
The role of disease associated SNPs on ERAP1 function has been investigated previously; single SNPs have been found to reduce trimming activity for K528R, R725Q and Q730E (18, 19), but no study has investigated their affect within naturally occurring alleles. We have found that SNPs do not act independently and that their effect on ERAP1 function when assessed individually is not an accurate predictor of their effect when in the context of a naturally occurring allele. For example we found that, when assayed on X5-SHL8, a modest reduction in function seen for R725Q was amplified when additional M349V, K528R or Q730E substitutions were introduced; and although the K528R change alone reduces activity by 50%, in combination with D575N, it generates an allele (albeit one which we have not observed in our sample of 20 genomes) with activity close to WT (Supplemental Fig. 3). Accordingly, disease association is likely to be considerably stronger when analyzed at the level of ERAP1 function than at the level of SNPs. The extent to which ERAP1 alleles exhibit the same trimming phenotypes for different peptide substrates is not known. However, examination of the K528R allele with the same amino acid extension, but different peptide backbone, shows that variation in trimming efficiency occurs when compared to WT (19). Comparison of K528R allele trimming capacity between studies shows that trimming of some amino acid substrates are similar such as Leu (equal or enhanced compared to WT; (19)), whereas others such as Typ are different (40% reduction versus equal compared to WT; (18)). These observed differences in K528R trimming may reflect the different assays employed (recombinant ERAP1 and different ERAP1 isoforms). This indicates that other factors such as the MHC binding affinity of peptides may influence the trimming phenotype of ERAP1 alleles. Further investigation using other peptide substrates is therefore required. Precisely how differential ERAP1 function might underpin disease pathology in, for example ankylosing spondylitis or psoriasis is a matter of speculation, but this study indicates how epitope presentation might be influenced by ERAP1 genotype and thus impact on CTL and NK cell function.
Supplementary Material
Acknowledgments
The authors would like to acknowledge the technical expertise of Nasia Kontouli.
This work was supported by CRUK project Grant C25722 awarded to E.J. and T.E. and CRUK Programme Grant C7056 awarded to T.E. (www.cancerresearchuk.org) and an MRC studentship awarded to E.R. (www.mrc.ac.uk). Abbreviations used in this paper: MHC I, MHC class I; ER, endoplasmic reticulum; ERAP1, endoplasmic reticulum aminopeptidase 1; SNP, single nucleotide polymorphism; SCT, single chain trimer.
References
- 1.Cascio P, Hilton C, Kisselev AF, Rock KL, Goldberg AL. 26S proteasomes and immunoproteasomes produce mainly N-extended versions of an antigenic peptide. The EMBO journal. 2001;20:2357–2366. doi: 10.1093/emboj/20.10.2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kunisawa J, Shastri N. The group II chaperonin TRiC protects proteolytic intermediates from degradation in the MHC class I antigen processing pathway. Mol Cell. 2003;12:565–576. doi: 10.1016/j.molcel.2003.08.009. [DOI] [PubMed] [Google Scholar]
- 3.Attaya M, Jameson S, Martinez CK, Hermel E, Aldrich C, Forman J, Lindahl KF, Bevan MJ, Monaco JJ. Ham-2 corrects the class I antigen-processing defect in RMA-S cells. Nature. 1992;355:647–649. doi: 10.1038/355647a0. [DOI] [PubMed] [Google Scholar]
- 4.Saveanu L, Carroll O, Lindo V, Del Val M, Lopez D, Lepelletier Y, Greer F, Schomburg L, Fruci D, Niedermann G, van Endert PM. Concerted peptide trimming by human ERAP1 and ERAP2 aminopeptidase complexes in the endoplasmic reticulum. Nature immunology. 2005;6:689–697. doi: 10.1038/ni1208. [DOI] [PubMed] [Google Scholar]
- 5.Serwold T, Gonzalez F, Kim J, Jacob R, Shastri N. ERAAP customizes peptides for MHC class I molecules in the endoplasmic reticulum. Nature. 2002;419:480–483. doi: 10.1038/nature01074. [DOI] [PubMed] [Google Scholar]
- 6.Van Kaer L, Ashton-Rickardt PG, Ploegh HL, Tonegawa S. TAP1 mutant mice are deficient in antigen presentation, surface class I molecules, and CD4-8+ T cells. Cell. 1992;71:1205–1214. doi: 10.1016/s0092-8674(05)80068-6. [DOI] [PubMed] [Google Scholar]
- 7.York IA, Chang SC, Saric T, Keys JA, Favreau JM, Goldberg AL, Rock KL. The ER aminopeptidase ERAP1 enhances or limits antigen presentation by trimming epitopes to 8-9 residues. Nature immunology. 2002;3:1177–1184. doi: 10.1038/ni860. [DOI] [PubMed] [Google Scholar]
- 8.Hammer GE, Gonzalez F, James E, Nolla H, Shastri N. In the absence of aminopeptidase ERAAP, MHC class I molecules present many unstable and highly immunogenic peptides. Nature immunology. 2007;8:101–108. doi: 10.1038/ni1409. [DOI] [PubMed] [Google Scholar]
- 9.Hammer GE, Gonzalez F, Champsaur M, Cado D, Shastri N. The aminopeptidase ERAAP shapes the peptide repertoire displayed by major histocompatibility complex class I molecules. Nature immunology. 2006;7:103–112. doi: 10.1038/ni1286. [DOI] [PubMed] [Google Scholar]
- 10.Yan J, Parekh VV, Mendez-Fernandez Y, Olivares-Villagomez D, Dragovic S, Hill T, Roopenian DC, Joyce S, Van Kaer L. In vivo role of ER-associated peptidase activity in tailoring peptides for presentation by MHC class Ia and class Ib molecules. J Exp Med. 2006;203:647–659. doi: 10.1084/jem.20052271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.York IA, Brehm MA, Zendzian S, Towne CF, Rock KL. Endoplasmic reticulum aminopeptidase 1 (ERAP1) trims MHC class I-presented peptides in vivo and plays an important role in immunodominance. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:9202–9207. doi: 10.1073/pnas.0603095103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hearn A, York IA, Rock KL. The specificity of trimming of MHC class I-presented peptides in the endoplasmic reticulum. Journal of immunology. 2009;183:5526–5536. doi: 10.4049/jimmunol.0803663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cifaldi L, Lo Monaco E, Forloni M, Giorda E, Lorenzi S, Petrini S, Tremante E, Pende D, Locatelli F, Giacomini P, Fruci D. Natural killer cells efficiently reject lymphoma silenced for the endoplasmic reticulum aminopeptidase associated with antigen processing. Cancer research. 2011;71:1597–1606. doi: 10.1158/0008-5472.CAN-10-3326. [DOI] [PubMed] [Google Scholar]
- 14.Burton PR, Clayton DG, Cardon LR, Craddock N, Deloukas P, Duncanson A, Kwiatkowski DP, McCarthy MI, Ouwehand WH, Samani NJ, Todd JA, Donnelly P, Barrett JC, Davison D, Easton D, Evans DM, Leung HT, Marchini JL, Morris AP, Spencer CC, Tobin MD, Attwood AP, Boorman JP, Cant B, Everson U, Hussey JM, Jolley JD, Knight AS, Koch K, Meech E, Nutland S, Prowse CV, Stevens HE, Taylor NC, Walters GR, Walker NM, Watkins NA, Winzer T, Jones RW, McArdle WL, Ring SM, Strachan DP, Pembrey M, Breen G, St Clair D, Caesar S, Gordon-Smith K, Jones L, Fraser C, Green EK, Grozeva D, Hamshere ML, Holmans PA, Jones IR, Kirov G, Moskivina V, Nikolov I, O’Donovan MC, Owen MJ, Collier DA, Elkin A, Farmer A, Williamson R, McGuffin P, Young AH, Ferrier IN, Ball SG, Balmforth AJ, Barrett JH, Bishop TD, Iles MM, Maqbool A, Yuldasheva N, Hall AS, Braund PS, Dixon RJ, Mangino M, Stevens S, Thompson JR, Bredin F, Tremelling M, Parkes M, Drummond H, Lees CW, Nimmo ER, Satsangi J, Fisher SA, Forbes A, Lewis CM, Onnie CM, Prescott NJ, Sanderson J, Matthew CG, Barbour J, Mohiuddin MK, Todhunter CE, Mansfield JC, Ahmad T, Cummings FR, Jewell DP, Webster J, Brown MJ, Lathrop MG, Connell J, Dominiczak A, Marcano CA, Burke B, Dobson R, Gungadoo J, Lee KL, Munroe PB, Newhouse SJ, Onipinla A, Wallace C, Xue M, Caulfield M, Farrall M, Barton A, Bruce IN, Donovan H, Eyre S, Gilbert PD, Hilder SL, Hinks AM, John SL, Potter C, Silman AJ, Symmons DP, Thomson W, Worthington J, Dunger DB, Widmer B, Frayling TM, Freathy RM, Lango H, Perry JR, Shields BM, Weedon MN, Hattersley AT, Hitman GA, Walker M, Elliott KS, Groves CJ, Lindgren CM, Rayner NW, Timpson NJ, Zeggini E, Newport M, Sirugo G, Lyons E, Vannberg F, Hill AV, Bradbury LA, Farrar C, Pointon JJ, Wordsworth P, Brown MA, Franklyn JA, Heward JM, Simmonds MJ, Gough SC, Seal S, Stratton MR, Rahman N, Ban M, Goris A, Sawcer SJ, Compston A, Conway D, Jallow M, Newport M, Sirugo G, Rockett KA, Bumpstead SJ, Chaney A, Downes K, Ghori MJ, Gwilliam R, Hunt SE, Inouye M, Keniry A, King E, McGinnis R, Potter S, Ravindrarajah R, Whittaker P, Widden C, Withers D, Cardin NJ, Davison D, Ferreira T, Pereira-Gale J, Hallgrimsdo’ttir IB, Howie BN, Su Z, Teo YY, Vukcevic D, Bentley D, Brown MA, Compston A, Farrall M, Hall AS, Hattersley AT, Hill AV, Parkes M, Pembrey M, Stratton MR, Mitchell SL, Newby PR, Brand OJ, Carr-Smith J, Pearce SH, McGinnis R, Keniry A, Deloukas P, Reveille JD, Zhou X, Sims AM, Dowling A, Taylor J, Doan T, Davis JC, Savage L, Ward MM, Learch TL, Weisman MH, Brown M. Association scan of 14,500 nonsynonymous SNPs in four diseases identifies autoimmunity variants. Nature genetics. 2007;39:1329–1337. doi: 10.1038/ng.2007.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Genetic Analysis of Psoriasis, C. C. the Wellcome Trust Case Control. Strange A, Capon F, Spencer CC, Knight J, Weale ME, Allen MH, Barton A, Band G, Bellenguez C, Bergboer JG, Blackwell JM, Bramon E, Bumpstead SJ, Casas JP, Cork MJ, Corvin A, Deloukas P, Dilthey A, Duncanson A, Edkins S, Estivill X, Fitzgerald O, Freeman C, Giardina E, Gray E, Hofer A, Huffmeier U, Hunt SE, Irvine AD, Jankowski J, Kirby B, Langford C, Lascorz J, Leman J, Leslie S, Mallbris L, Markus HS, Mathew CG, McLean WH, McManus R, Mossner R, Moutsianas L, Naluai AT, Nestle FO, Novelli G, Onoufriadis A, Palmer CN, Perricone C, Pirinen M, Plomin R, Potter SC, Pujol RM, Rautanen A, Riveira-Munoz E, Ryan AW, Salmhofer W, Samuelsson L, Sawcer SJ, Schalkwijk J, Smith CH, Stahle M, Su Z, Tazi-Ahnini R, Traupe H, Viswanathan AC, Warren RB, Weger W, Wolk K, Wood N, Worthington J, Young HS, Zeeuwen PL, Hayday A, Burden AD, Griffiths CE, Kere J, Reis A, McVean G, Evans DM, Brown MA, Barker JN, Peltonen L, Donnelly P, Trembath RC. A genome-wide association study identifies new psoriasis susceptibility loci and an interaction between HLA-C and ERAP1. Nature genetics. 2010;42:985–990. doi: 10.1038/ng.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guerini FR, Cagliani R, Forni D, Agliardi C, Caputo D, Cassinotti A, Galimberti D, Fenoglio C, Biasin M, Asselta R, Scarpini E, Comi GP, Bresolin N, Clerici M, Sironi M. A functional variant in ERAP1 predisposes to multiple sclerosis. PloS one. 2012;7:e29931. doi: 10.1371/journal.pone.0029931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mehta AM, Jordanova ES, van Wezel T, Uh HW, Corver WE, Kwappenberg KM, Verduijn W, Kenter GG, van der Burg SH, Fleuren GJ. Genetic variation of antigen processing machinery components and association with cervical carcinoma. Genes, chromosomes & cancer. 2007;46:577–586. doi: 10.1002/gcc.20441. [DOI] [PubMed] [Google Scholar]
- 18.Evans DM, Spencer CC, Pointon JJ, Su Z, Harvey D, Kochan G, Oppermann U, Dilthey A, Pirinen M, Stone MA, Appleton L, Moutsianas L, Leslie S, Wordsworth T, Kenna TJ, Karaderi T, Thomas GP, Ward MM, Weisman MH, Farrar C, Bradbury LA, Danoy P, Inman RD, Maksymowych W, Gladman D, Rahman P, Morgan A, Marzo-Ortega H, Bowness P, Gaffney K, Gaston JS, Smith M, Bruges-Armas J, Couto AR, Sorrentino R, Paladini F, Ferreira MA, Xu H, Liu Y, Jiang L, Lopez-Larrea C, Diaz-Pena R, Lopez-Vazquez A, Zayats T, Band G, Bellenguez C, Blackburn H, Blackwell JM, Bramon E, Bumpstead SJ, Casas JP, Corvin A, Craddock N, Deloukas P, Dronov S, Duncanson A, Edkins S, Freeman C, Gillman M, Gray E, Gwilliam R, Hammond N, Hunt SE, Jankowski J, Jayakumar A, Langford C, Liddle J, Markus HS, Mathew CG, McCann OT, McCarthy MI, Palmer CN, Peltonen L, Plomin R, Potter SC, Rautanen A, Ravindrarajah R, Ricketts M, Samani N, Sawcer SJ, Strange A, Trembath RC, Viswanathan AC, Waller M, Weston P, Whittaker P, Widaa S, Wood NW, McVean G, Reveille JD, Wordsworth BP, Brown MA, Donnelly P. Interaction between ERAP1 and HLA-B27 in ankylosing spondylitis implicates peptide handling in the mechanism for HLA-B27 in disease susceptibility. Nature genetics. 2011;43:761–767. doi: 10.1038/ng.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Evnouchidou I, Kamal RP, Seregin SS, Goto Y, Tsujimoto M, Hattori A, Voulgari PV, Drosos AA, Amalfitano A, York IA, Stratikos E. Cutting Edge: Coding single nucleotide polymorphisms of endoplasmic reticulum aminopeptidase 1 can affect antigenic peptide generation in vitro by influencing basic enzymatic properties of the enzyme. Journal of immunology. 2011;186:1909–1913. doi: 10.4049/jimmunol.1003337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kochan G, Krojer T, Harvey D, Fischer R, Chen L, Vollmar M, von Delft F, Kavanagh KL, Brown MA, Bowness P, Wordsworth P, Kessler BM, Oppermann U. Crystal structures of the endoplasmic reticulum aminopeptidase-1 (ERAP1) reveal the molecular basis for N-terminal peptide trimming. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:7745–7750. doi: 10.1073/pnas.1101262108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kadi A, Izac B, Said-Nahal R, Leboime A, Van Praet L, de Vlam K, Elewaut D, Chiocchia G, Breban M. Investigating the genetic association between ERAP1 and spondyloarthritis. Annals of the rheumatic diseases. 2013;72:608–613. doi: 10.1136/annrheumdis-2012-201783. [DOI] [PubMed] [Google Scholar]
- 22.Maksymowych WP, Inman RD, Gladman DD, Reeve JP, Pope A, Rahman P. Association of a specific ERAP1/ARTS1 haplotype with disease susceptibility in ankylosing spondylitis. Arthritis and rheumatism. 2009;60:1317–1323. doi: 10.1002/art.24467. [DOI] [PubMed] [Google Scholar]
- 23.Tsui FW, Haroon N, Reveille JD, Rahman P, Chiu B, Tsui HW, Inman RD. Association of an ERAP1 ERAP2 haplotype with familial ankylosing spondylitis. Annals of the rheumatic diseases. 2010;69:733–736. doi: 10.1136/ard.2008.103804. [DOI] [PubMed] [Google Scholar]
- 24.Serwold T, Gaw S, Shastri N. ER aminopeptidases generate a unique pool of peptides for MHC class I molecules. Nature immunology. 2001;2:644–651. doi: 10.1038/89800. [DOI] [PubMed] [Google Scholar]
- 25.Kanaseki T, Shastri N. Endoplasmic reticulum aminopeptidase associated with antigen processing regulates quality of processed peptides presented by MHC class I molecules. Journal of immunology. 2008;181:6275–6282. doi: 10.4049/jimmunol.181.9.6275. [DOI] [PubMed] [Google Scholar]
- 26.Kanaseki T, Blanchard N, Hammer GE, Gonzalez F, Shastri N. ERAAP synergizes with MHC class I molecules to make the final cut in the antigenic peptide precursors in the endoplasmic reticulum. Immunity. 2006;25:795–806. doi: 10.1016/j.immuni.2006.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Malarkannan S, Goth S, Buchholz DR, Shastri N. The role of MHC class I molecules in the generation of endogenous peptide/MHC complexes. Journal of immunology. 1995;154:585–598. [PubMed] [Google Scholar]
- 28.Truscott SM, Lybarger L, Martinko JM, Mitaksov VE, Kranz DM, Connolly JM, Fremont DH, Hansen TH. Disulfide bond engineering to trap peptides in the MHC class I binding groove. Journal of immunology. 2007;178:6280–6289. doi: 10.4049/jimmunol.178.10.6280. [DOI] [PubMed] [Google Scholar]
- 29.Schatz MM, Peters B, Akkad N, Ullrich N, Martinez AN, Carroll O, Bulik S, Rammensee HG, van Endert P, Holzhutter HG, Tenzer S, Schild H. Characterizing the N-terminal processing motif of MHC class I ligands. Journal of immunology. 2008;180:3210–3217. doi: 10.4049/jimmunol.180.5.3210. [DOI] [PubMed] [Google Scholar]
- 30.Nguyen TT, Chang SC, Evnouchidou I, York IA, Zikos C, Rock KL, Goldberg AL, Stratikos E, Stern LJ. Structural basis for antigenic peptide precursor processing by the endoplasmic reticulum aminopeptidase ERAP1. Nat Struct Mol Biol. 2011;18:604–613. doi: 10.1038/nsmb.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chang SC, Momburg F, Bhutani N, Goldberg AL. The ER aminopeptidase, ERAP1, trims precursors to lengths of MHC class I peptides by a “molecular ruler” mechanism. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:17107–17112. doi: 10.1073/pnas.0500721102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Garcia-Medel N, Sanz-Bravo A, Van Nguyen D, Galocha B, Gomez-Molina P, Martin-Esteban A, Alvarez-Navarro C, de Castro JA. Functional interaction of the ankylosing spondylitis-associated endoplasmic reticulum aminopeptidase 1 polymorphism and HLA-B27 in vivo. Molecular & cellular proteomics: MCP. 2012;11:1416–1429. doi: 10.1074/mcp.M112.019588. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.