Abstract
The purpose of this review is to give a comprehensive overview of transgenic mouse lines suitable for studying gene function and cellular lineage relationships in lung development, homeostasis, injury, and repair. Many of the mouse strains reviewed in this Perspective have been widely shared within the lung research community, and new strains are continuously being developed. There are many transgenic lines that target subsets of lung cells, but it remains a challenge for investigators to select the correct transgenic modules for their experiment. This review covers the tetracycline- and tamoxifen-inducible systems and focuses on conditional lines that target the epithelial cells. We point out the limitations of each strain so investigators can choose the system that will work best for their scientific question. Current mesenchymal and endothelial lines are limited by the fact that they are not lung specific. These lines are summarized in a brief overview. In addition, useful transgenic reporter mice for studying lineage relationships, promoter activity, and signaling pathways will complete our lung-specific conditional transgenic mouse shopping list.
Keywords: transgenic, conditional, lineage tracing, gene expression, lung, mouse
In recent years, tremendous progress has been made in understanding the cellular processes underlying lung development, homeostasis, and repair. This has been facilitated by the use of lung cell type–specific transgenic mouse lines. Initially, lung-specific promoters (1–3) were used to constitutively drive transgenes in a cell type–specific manner. This mostly resulted in embryonic lethal phenotypes and consequently limited adult studies. The development of conditional transgenic mice allowed the temporal and spatial control of gene expression, overcoming many lethal phenotypes and allowing the analysis of lung-specific gene knock-outs, identification of progenitor cells, lineage tracing, and studies of progenitor proliferation and differentiation capacity. The most widely used systems are the doxycycline (Dox) system (tTA and rtTA) and the Cre-LoxP system, but others exist, and their use is becoming more widespread. As transgenic mouse strains have been used, their strengths and limitations and the strategies required for optimal experimental design have become apparent. In this review, we discuss the mouse strains that have been shown to be the most useful for manipulating gene expression in the lung and highlight areas where new mice would be extremely beneficial for the community.
Cre Recombination
Cre-Lox technology was introduced in the 1980s (4, 5) and patented by DuPont Pharmaceuticals. It was successfully applied to mice in 1998 (6). A version of Cre recombinase, which contained the preferred mammalian codons and was more efficiently expressed in mammalian cells, was published in 2002 (7). The technology is based on the ability of the P1 bacteriophage recombinase (Cre) to direct site-specific DNA recombination between pairs of LoxP sites. Such recombination in a “Cre-lox” mouse can permanently inactivate or activate a gene of interest.
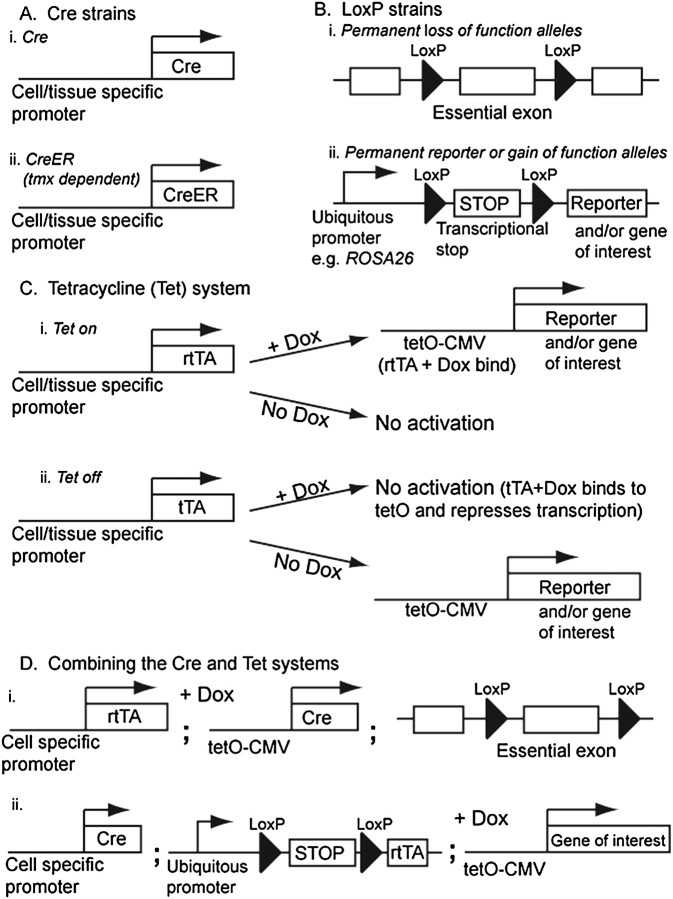
Typical Cre-Lox experiments require two transgenic animals: a Cre strain and a LoxP strain. A Cre mouse contains a Cre recombinase transgene under the control of a tissue-specific promoter (Figure 1A), whereas a LoxP mouse contains two LoxP sites that flank a genomic segment of interest, the “floxed” locus. Depending on the location and orientation of the LoxP sites in a Cre-Lox mouse, Cre recombinase can initiate deletions, inversions, and translocations of the floxed locus (8). The floxed loci can be designed to allow permanent inactivation or activation of the gene of interest (Figure 1B). Mutated LoxP sites, which allow recombination between various independent LoxP sites, have been successfully used to rapidly target genes and generate multicolor reporter mice, “brainbow” and “confetti,” which are useful reporters for clonal analysis of progenitor cells (9–11).
Figure 1.
Schematic of Cre and Tet transgenic systems. (A) Cre, or CreER, is expressed from a cell or tissue-specific promoter. (i) Cre is active in every cell in which it is expressed. Levels of activity depend on expression levels. (ii) CreER is not active (it is actively excluded from the nucleus) until tamoxifen is administered. This allows for temporal control of the system and reduces nonspecific effects of the Cre. (B) Cre catalyzes recombination between its recognition (LoxP) sites. This results in a permanent genetic rearrangement, which is inherited by daughter cells. The most common arrangement of LoxP sites are (i) flanking a crucial exon of an endogenous gene to mediate loss of function or (ii) flanking a transcriptional STOP to mediate reporter gene expression and/or overexpression. In this case, a ubiquitous promoter is usually used to drive the gene of interest (a reporter, coding sequence, or hairpin), but the transcript is disrupted by the LoxP-flanked transcriptional STOP. After Cre-mediated deletion of the stop, the gene of interest is permanently expressed. Such cassettes are frequently targeted to the ubiquitously expressed ROSA26 locus for example (18). (C) The tetracycline (Tet) system has two versions, Tet-on and Tet-off. (i) Tet-on. The reverse tetracycline transactivator (rtTA) is expressed from a cell or tissue-specific promoter. In the Tet-on system, administration of doxycycline (Dox) activates the responsive transgene. rtTA is not functional until Dox is present. Upon Dox binding, rtTA binds to its tetO recognition sites and activates transcription. The system is reversible. Once Dox has been metabolized, rtTA no longer binds to the tetO sites and the transgene turns off. Similarly, the transgene is turned off in any descendents of the cell that no longer express rtTA. (ii) Tet-off. The tetracycline transactivator (tTA) is expressed from a cell- or tissue-specific promoter. In the Tet-off system, the tetO-responsive transgene is constitutively active, and administration of Dox represses the transgene. Dox binds to tTA, which allows it to bind to the tetO elements and suppress transcription. The Tet-off system is also reversible. (D) Combining the Cre and Tet systems. (i) Dox-conditional Cre activation. rtTA is expressed from a cell- or tissue-specific promoter, and Cre is placed under the control of the tetO elements. Upon Dox administration, Cre is transcribed and enters the nucleus to recombine its LoxP target sites. In this case, deletion or activation of the LoxP-flanked target gene is permanent. Any off-target effects of the Cre can be reduced by minimizing the amount of Dox administered. However, Dox treatment itself can also affect lung development and homeostasis, and this must also be controlled for. (ii) Cre-conditional rtTA activation. Cre is expressed from a cell- or tissue-specific promoter, and rtTA is placed upstream of a LoxP-flanked STOP cassette. Cre activity results in constitutive expression of rtTA in the Cre-expressing cells and all of their descendants. However, the tetO-responsive transgene is only transcribed in the presence of Dox. In this case, activation of the gene of interest is reversible.
The site of Cre activity (cell-type specificity) is dependent on the availability of tissue-specific or cell-specific promoters. Moreover, tissue-specific Cre expression can be combined with temporal-specific activity. Time-specific Cre activation can be achieved by combination with the Dox system (Figure 1D) or by the use of Cre fusion proteins. Cell type–specific Cre strains have been widely used in the lung for lineage tracing and permanent gene activation or deletion (e.g., Refs. 12, and 13).
A very useful technique is to track the descendents of stem and progenitor cells (lineage-tracing) by crossing a Cre mouse with a reporter mouse strain that permanently expresses a reporter gene after Cre activity. The Z/AP and Z/EG reporter mice were initially used for lineage-tracing experiments (14, 15). However, studies have shown that these reporters are not expressed in all cell types and can be silenced in adult tissue. The Rosa26 reporter (Rosa26R) variants are now the most widely used reporter strains available. The ROSAβgeo26 (GtROSA26) line was initially derived from pools of embryonic stem (ES) cells infected with the retroviral gene trap vector Gen-ROSAβgeo (16). After cloning the ROSA26 locus (17), it was used to produce a variety of Cre reporter lines, starting with lacZ and expanding to a vast repertoire of cytosolic, membrane–bound, and nuclear florescent lineage tags (18). The ROSA26 locus is particularly useful for generating these strains because it is expressed robustly in most cell types and is gene targeted at high efficiency; thus, numerous other cassettes have been targeted to the ROSA26 locus.
The Tamoxifen-Inducible Cre System
To allow temporal control of Cre activity, fusion proteins have been constructed between Cre and the ligand-binding domain of steroid hormone receptors (Figure 1A). The most commonly used variant is a fusion between Cre and a mutated ligand-binding domain of the estrogen receptor (CreERT2) (19–21). Earlier CreER and CreER versions can be somewhat leaky when expressed from a strong promoter, probably due to their ability to bind endogenous estrogens. These were adapted by site-specific mutagenesis to generate CreERT2 (20, 21). ERT2 binds weakly to endogenous estrogens and strongly to 4-hydroxy tamoxifen (4OH-T), the active metabolite of the synthetic steroid tamoxifen (tmx). Administration of tmx or 4OH-T by itself can be toxic, resulting in various embryonic phenotypes if administered up to about E11.5 or abortion if administered at later stages (22). Moreover, tmx dosing can cause a transient increase in blood pressure in adult mice (23). For these reasons, it is important to titrate the tmx dose to the minimum required for each experiment. Most investigators dose their animals with tmx, which is converted to 4OH-T in the liver. 4OH-T can also be administered directly, but the kinetics of Cre activation and drug metabolism will be different.
The CreERT2 fusion protein is cytoplasmic. Upon binding to tmx, CreERT2 translocates to the nucleus where it accesses the LoxP sites. Earlier CreER and CreERT2 versions can be somewhat leaky when expressed from a strong promoter. However, such strains can be highly informative if the correct controls are performed (24). Recombination rates are very sensitive to the levels of CreERT2 expression (25) and to the length of time the protein spends in the nucleus. This is dependent on tmx dosage and frequency of dosing. Elegant studies using a cartilage-specific CreERT fusion protein demonstrated that, after a single intraperitoneal injection of tmx into a pregnant female mouse, reporter activity could be detected within 8 hours and that recombination was complete within 24 hours (26). In addition, it has been reported that administration of tmx to pregnant female mice via oral gavage, rather than via intraperitoneal injection, results in more efficient labeling and less embryonic toxicity (27). The CreER2T system has been widely used in the lung, although the extent of recombination and the most effective tmx dosing strategy must be determined empirically for each CreERT2 mouse strain (28, 29). In addition, the recombination rate between each pair of LoxP sites varies and must be determined experimentally (30). Another variant of the CreER system has the ERT fused to the amino and carboxy terminals of Cre (known as mER-Cre-mER) (31). The mER-Cre-mER has not been used widely in pulmonary research.
Multiple transgenic strains with widespread expression of tmx-inducible Cre have been generated and are potentially useful in the adult lung for deletion of genes with cell type–specific expression. Representatives of these lines are β-actin–Cre (32) and actin-CreERT (33), which have been used successfully in the developing lung mesenchyme (34) and which direct Cre expression in multiple lung cell types. Similarly, a CMV-CreERT mouse line directs expression of CreER in most cell types (20) and has been successfully used to study the role of Sox2 in the adult tracheal epithelium (35). In addition, four independent RosaCreERT2 strains exist that allow ubiquitous expression of CreERT2 from the ROSA26 locus (30, 36–39). Additional ligand-inducible Cre fusion proteins have been generated, such as Cre-Progesterone Receptor 1, which can be activated by RU486, and Cre-glucocorticoid receptor, which can be activated by dexamethosone (40, 41). The use of these constructs is less widespread than CreER2T and is not discussed in detail here.
The Split-Cre System
The Dox and Cre/CreER systems are dependent on the availability of a cell type–specific promoter to restrict gene expression to the cells of interest. However, the variety of cell type–specific promoters is limited. The split-Cre system was developed to overcome these limitations (42). In this system, the Cre protein is split into two halves that are expressed from different promoters. Individually, these two parts of the protein are inactive. When both promoters are activated in the same cell, intermolecular complementation occurs, and Cre is functional. This system has been successfully applied to the mouse brain to target populations of rare stem cells, which had previously been defined by flow cytometry only (43). An inducible split CreERT2 system has also been developed and has been shown to function in vitro (44). In vivo application of the split Cre in the lung will be beneficial to advance the field of lineage relationships and progenitor cells.
Cre Toxicity
Off-target effects of the Cre recombinase have been reported in several systems (45), including the lung (46, 47). These off-target effects are probably due to endogenous cryptic LoxP sites within the mammalian genome that cause cytotoxic chromosomal rearrangements when activated (48, 49). Not every Cre strain shows off-target effects, and this variation is likely to result from differences in levels of Cre protein expression. Cre toxicity has led to off-target phenotypes and has made interpretation of some experiments difficult. In particular, some Cre lines demonstrate Cre activity in the germline, which results in recombination of the floxed allele independent of the regulatory element driving Cre (Ref. 50 and B. R. Stripp, personal communication). Such sex-specific effects can usually be avoided by transmitting the Cre via the female or male germline. The use of Dox or tmx-dependent Cre strains limits the amount of time Cre spends in the nucleus and decreases Cre toxicity, which also highlights the importance of using the minimum dose of Dox or tmx for each experiment. In addition, it is crucial to perform the correct controls: (1) Dox and tmx treatment of single transgenic mice, (2) Dox and tmx treatment of mice with all transgenes in a nonfloxed background, and (3) untreated mice containing all transgenes in the floxed background. Cre mouse strains, often in combination with Dox or tmx, are probably the most widely used animals for manipulating gene expression in the lung.
Flipase
The FLP-FRT system is similar to the Cre-Lox system and is becoming more frequently used in mouse-based research. It involves using Flippase (FLP) recombinase, derived from the yeast Saccharomyces cerevisiae (51). FLP recognizes a pair of FLP recombinase target (FRT) sequences that flank a genomic region of interest. RosaFLPe is a mouse line with a ubiquitous FLP expression (52). A useful reporter mouse for this recombination is the Flp indicator mouse expressing alkaline phosphatase from the ROSA26 locus (53). However, despite many attempts, it has been difficult to generate lung-specific Flippase mice (A. K. Perl and J. Whitsett, unpublished data). Nevertheless, a Flp-inducible allele of K-ras was recently activated in the lung using a lentivirus-encoded FLP protein (54).
The Dox System
The tetracycline (tet) inducible system was developed independently to the Cre-LoxP system and has different advantages and limitations. The tet system in vivo consists of two transgenic mouse lines, an activator line and an operator line. The activator line expresses the tetracycline activator (TA, tet-off) or the reverse tetracycline responsive transactivator (rtTA, tet-on) in a tissue-specific manner. The operator line carries a transgene of interest under control of the (tetO)7CMV operator (tetO). In double transgenic mice, Dox causes the tTA to bind the tetO sequence, suppressing transcription (Tet-off), or causes the rtTA to bind to the tetO sequence, activating transcription of the gene of interest (Tet-on) (Figure 1C) (55, 56). The major advantage of this system is the ability to reversibly turn genes on and off and study the effects of genes at specific times. In the lung, this system was successfully used in 2000 to conditionally activate FGF7 during lung development (57). The technical aspects of using Dox to activate genes were described using a luciferase reporter mouse (58, 59), and the limitations of the system were reported in 2006 (60).
The use of the rtTA system was further expanded by the combination of the rtTA system with activation of a tetO-Cre transgene (61, 62). The cell-specific, Dox-dependent Cre expression enables permanent endogenous gene inactivation or transgene activation at any given time during development or in adult animals (Figure 1D) (61). This Dox-dependent Cre activation has been shown to be very useful for in vivo cell lineage labeling and conditional deletion of genes that otherwise would result in lethal phenotypes. It has also recently been used to conditionally deplete specific cell populations and to study epithelial regeneration (Table 4) (63).
TABLE 4.
REPORTER LINES AND OTHERS
| Mouse Strain | Function | References | Comments |
| tetO-Cre | Dox-inducible Cre activity | 61 | Makes Cre conditional, can be combined with any cell type–specific tet activator line |
| Tetoluciferase | Dox-inducible luciferase activity | 58 | Line to test new tet activator mice |
| SPC GFP | 189 | High levels of GFP in adult AT2 cells, lower levels of GFP throughout rest of adult lung epithelium. Potentially very useful for live-imaging in the embryonic or adult lung. | |
| Z/AP | Cre-activated alkaline phosphatase; transgene inactivated in adults | 14, 191 | Lineage tag for Cre recombination |
| Z/EG | Cre-activated GFP | 15 | Lineage tag for Cre recombination |
| ROSA26R-LacZ reporter | Cre-activated β-GAL | 18 | Lineage tag for Cre recombination, very reliable in embryonic and adults |
| ROSA-mT/mG | mT cassette deleted in the Cre expressing tissue(s), allowing expression of the membrane-targeted EGFP (mG) cassette | 107 | Membrane-bound lineage tag for Cre recombination |
| brainbow | GFP reporter variants placed between different sets of incompatible lox sites, recombination results in a spectrum of colors | 9 | Florescent multicolor lineage tag after Cre recombination |
| R26R-Confetti | CAG promoter driven, Brainbow 2.1 construct targeted into the Gt(ROSA)26Sor locus. These R26R-Confetti mice make it possible to label and distinguish individual and adjacent cells with nuclear localized, membrane-targeted, or cytoplasmic fluorescent proteins in Cre recombined cells in any tissue. | 10 | Florescent multicolor lineage tag after Cre recombination, allows tracing of clonal expansion of a single cell. |
| HPRT and, more recently, Cola1 | Useful loci for gene targeting of transgenic cassettes in ES cells | 74, 192 | |
| SPC-DT receptor | Diphteria toxin receptor expressed under SPC activated by injection of ligand | 193 | |
| R26-DTA | Cre activated cell death by diphtheria toxin | 63, 194 | |
| TOPGAL | Wnt reporter: epithelial | 175 | |
| BATGAL | Wnt reporter: sporadic epithelial and mesenchymal activity | 176 | |
| Axin2-lacZ | Wnt reporter for proximal lung and mesenchymal signaling | 177 | |
| TCF/Lef:H2B-GFP | wnt reporter not yet tested in the lung | 179 | |
| CP-EGFP | Notch reporter for the time of signaling | 180 | |
| N1IP::CRELOW and N1IP::CREHI are knock-ins of Cre | Notch reporter to lineage label cells that undergo notch signaling | 170, 183 |
Definition of abbreviations: EGFP = enhanced green fluorescent protein; ES = embryonic stem; GFP = green fluorescent protein; SPC = sphingosylphosphorylcholine.
Combining tissue-specific Cre lines with Rosa26rtTA transgenic mice results in Cre- mediated rtTA expression (Figure 1D and Table 4). These mice have Cre-inducible expression of rtTA and can be used to achieve spatially and temporally controlled transgene expression in a wide variety of settings simply by crossing to any existing mice carrying cell type-specific Cre recombinase and tet-O-regulatable responder genes (64, 65).
Off-target toxicity of rtTA and Dox has been reviewed previously (46, 60, 66). Briefly, these toxicities were unrelated to the effects of the transgene and varied with specific mouse strain and genetic background. Off-target effects can influence lung morphogenesis and perinatal survival and most often result in airspace enlargement. In addition, Dox is a matrix metalloproteinase inhibitor and has been shown to promote pulmonary hypertension after hypoxia and to attenuate mucin production (67–69). More recently, it has been shown that tracheal Clara cells are sensitive to Dox treatment (47). Dox is stored in tissues and is released over time. The timing and duration of treatment needed to target subsets of lung epithelial cells has been described, and we highly recommend limiting the time and dosage of Dox exposure (61, 62). With the new-generation rtTA constructs that are more sensitive to low doses of Dox (e.g., rtTA2S-M2), off-target effects of Dox will become less significant, but dosing regimens will be more important (70, 71). In all Dox experiments, it is vital to perform the correct controls. To control for phenotypes not related to the activation/inactivation of the gene of interest, single transgenic mice, and mice containing all transgenes, should be tested in the absence of Dox. Littermate control mice should be used where possible to minimize strain or age-related variability. Experiments should also be controlled for weight and gender differences.
“Knock-In” Versus Transgenic Mice
There are multiple methods for producing Cre or tTA/rtTA mouse strains. These include generating transgenic animals by pronuclear injection, bacterial artificial chromosome (BAC) transgenics, and gene targeting by homologous recombination in ES cells (“knock-ins”). The method used affects the properties of the resulting mouse strain, and it is important to be aware of the benefits and limitations of each method. (1) The “transgene” method largely depends on the availability of defined regulatory sequences to drive expression in the cell of interest. When a characterized promoter is available, transgenic mice can be generated rapidly by injection of the construct into the pronucleus of a fertilized egg. However, the transgene inserts randomly, and expression of the exogenous gene is highly susceptible to the chromosomal environment of the insertion site (heterochromatin versus euchromatin). This phenomenon is known as “position-effect variegation” and explains why founders with independent insertion sites can have different expression patterns of the transgene. Transgenes can also be silenced over an animal's lifetime or after multiple generations. In addition, the integration of the transgene can cause a mutation in a chromosomal gene (72). To overcome these problems, transgenic constructs can be targeted to a defined locus in ES cells (e.g., Hprt1; see Ref. 73), and new techniques for integrating transgenic constructs into defined loci by cassette exchange should make this a very rapid technique (11, 74, 75). (2) BAC transgenes are large and are injected into the pronucleus of a fertilized egg. BACs are an alternate way of generating transgenic constructs that overcome some of the problems of the more standard transgene method, including the lack of available defined and well characterized regulatory elements for specific cell types. For BAC transgenes, the Cre or rtTA sequences are usually placed in-frame in the initiating ATG of a known gene, which is expressed in the cells of interest. This is the preferred strategy because the Cre or rtTA sequences disrupt gene expression. This is crucial to prevent the BAC transgenic adding additional copies of the gene of interest, which may cause a phenotype. Due to their large size, the BAC constructs usually contain most, or all, of the enhancer sequences that normally regulate expression of that gene. The BAC size also makes them less susceptible to positional effects and silencing. However, their size also means that they can include extra copies of flanking genes or regulatory RNAs, which could cause an unrelated phenotype. Moreover, like a standard transgene, they can insert into the DNA at a site, which causes a mutation in an endogenous mouse chromosomal locus. (3) To generate “knock-ins,” Cre or rtTA sequences are usually placed in-frame in the initiating ATG of an endogenous chromosomal gene by homologous recombination in ES cells. This simultaneously generates a null allele, which can cause a haplo-insufficiency phenotype. An alternative technique, which maintains expression of the endogenous gene product, is to target the Cre or rtTA sequences to the 3′UTR of a locus using an internal ribosome entry site (IRES), or a 2A cleavage site, to produce two independent proteins that are cotranscribed. (Use of the 2A cleavage site is discussed in Ref. 76.) Because the 3′UTR can contain important regulatory sequences, these transgenic lines need to be tested for phenotypes and for expression pattern of the transgene. Knock-ins usually carry a single copy of Cre or rtTA, which makes them less leaky but decreases sensitivity compared with the transgenic strains that usually contain multiple copies.
The Jackson Laboratories Website
Many of the mice we mention in this review are available form the Jackson Laboratories (http://jaxmice.jax.org). The Jackson Laboratories has also developed useful databases. These include (1) Mouse Genome Informatics, which provides access to integrated data on mouse genes and genome features, from sequences and genomic maps to gene expression and disease models (http://www.informatics.jax.org/); (2) the International Mouse Strain Resource, which is a searchable online database of mouse strains and stocks available worldwide, including inbred, mutant, and genetically engineered mice (http://www.findmice.org/); and (3) the Mouse Phenome Database, which is a collaborative collection of baseline phenotypic data on inbred mouse strains. The Mouse Phenome Database includes data sets, protocols, projects and publications, and single-nucleotide polymorphisms (http://phenome.jax.org/).
Epithelial Lines
The lung buds from the foregut endoderm around embryonic day (E)9.5 in mice and continues to develop by branching morphogenesis. By contrast, the trachea and esophagus separate from the foregut just ventral to the lung buds between E10.0 and E11.5 and then increase in length and diameter during development. A number of mouse strains suitable for conditional gene activation in the developing lung have been developed using genes expressed in the foregut endoderm (Table 1). However, with the exception of Sftpc lines, these are not unique to the developing lung. The choice of line depends partly on the timing of activity required: (1) in the undivided foregut (that is, throughout the entire lung, trachea, and esophagus), or (2) more lung specific. As differentiated cell types appear during lung development, they can be targeted with cell type–specific mouse lines (Table 2).
TABLE 1.
EMBRYONIC EPITHELIAL PROGENITORS
| Mouse strain | Lung-specific subpopulation | Activity in other organs? | References |
| Shh-Cre, GFP-Cre Knock in | Ventral foregut endoderm | Yes | 77, 78 |
| Nkx2-5-Cre, knock in | Ventral foregut endoderm and mesenchyme | Yes | 88 |
| Islet1-Cre, knock in | Ventral foregut endoderm and mesenchyme | Yes | 82, 83 |
| Nkx2-1-Cre, mouse BAC | Lung and tracheal endodermal progenitors | Yes | 91 |
| Mouse Gata5-Cre transgenic | Lung endoderm | Yes | 85, 86 |
| Human SFTPC-Cre transgenic | Lung endoderm; | No published report; possible expression in testis | 96 |
| SftpC-CreERT2-rtTA knock in | Lung epithelium during development and adult alveolar type2 cells | No published report | 102 |
| Id2-CreERT2, knock in | Distal lung epithelium during development and some mesenchyme | Yes | 87 |
| Human 3.7 SFTPC-rtTA (SPCrtTA) line 1, transgenic | Development and adult lung epithelium | No | 57, 61 |
Definition of abbreviation: BAC = bacterial artificial chromosome.
TABLE 2.
MATURE EPITHELIUM
| Mouse strain | Lung-Specific Subpopulation | Activity in Other Organs? | References | Comments |
| Human 3.7kb SFTPC-rtTA (SPCrtTA) line 1, transgenic | Developing lung endoderm; alveolar type II cells | No | 57, 59 | High-expression, Dox-independent activity perinatally, lethal when homozygote |
| Human 3.7kb SFTPC-rtTA (SPCrtTA) line 2, transgenic | Alveolar type II cells | No | 95 | Lower expression than line one, can be kept homozygous |
| Sftpc-CreERT2-rtTA knock in | Developing lung endoderm; alveolar type II cells | No published report | 102 | |
| Sftpc-EGFP, | Alveolar type II cells | No published report | 189 | High levels of GFP in adult AT2 cells, lower levels of GFP throughout rest of adult lung epithelium. Potentially very useful for live-imaging in the embryonic or adult lung. |
| Aqp5-Cre-IRES-DsRed Knock in | Alveolar type 1 cells | Yes | 106 | AT2 on some genetic backgrounds |
| Rat RTIbac-BAC transgenic | Podopladin alveolar type I cells | No published report | 109 | |
| Rat 2.3 Scgb1a1-rtTA line 1, transgenic | Clara cells, subset of type II cells | No (no expression in whole uterus homogenate) | 59, 62 | Targets 70–80% of all Clara cells |
| Rat 2.3 Scgb1a1-rtTA line 2, transgenic | Clara cells, alveolar type II cells (few and dependent on background) | No published report | 46, 63 | Targets 98% of clara cells in all mice and some AT2 dependent on background |
| Scgb1a1-CreER, knock in | Clara cells, subset of type II cells | No (uterus not tested) | 24 | Some tamoxifen-independent activity |
| Rat Scgb1a1-rtTA2s-M2, transgenic | Clara cells, subset of type II cells | No published report | 71 | More sensitive Dox regulation |
| Scgb1a1 –Cre | Postnatal Clara cells only | No published report | 110 | |
| Rat Scgb1a1–Cre, transgenic | Clara cells (but effectively most of the airway epithelium as it comes on so early) | No published report | 111–113 | |
| Tgfb3-Cre, knock-in | Most Clara cells | Yes | 114, 115 | |
| Human FOXJ1-Cre | Ciliated cells | Yes | 123 | |
| Human FOXJ1-CreERT2 | Ciliated cells | Yes | 29 | |
| Human FOXJ1-GFP | Ciliated cells | Yes | 122 | |
| Human K5Cre*PR | Basal cells | Yes | 190 | This line has not been very well characterized |
| Human KRT5-CreERT2 | Basal cells | Yes | 28 | |
| Human KRT14-CreERT | Subset of basal cells at steady-state, most basal cells after injury | Yes | 132, 133 | |
| Human K14 rtTA And Human K14 rtTA/tetO-Cre | Clara cells, in combination with tetO-Cre some significant off target effects. | Yes | 134, 47 |
Definition of abbreviations: Dox = doxycycline; GFP = green fluorescent protein.
Embryonic Epithelial Progenitors
Sonic hedgehog (Shh) is expressed in a highly dynamic pattern during embryogenesis. The Shh-Cre line is a knock-in of a GFP-Cre fusion protein (77) and has been shown to activate reporter recombination in the ventral foregut endoderm by E9.5, before lung budding and tracheal/esophageal separation (78). It has successfully been used to study tracheal and lung development (12, 79, 80).
Islet1 is expressed in many tissues during embryogenesis including the limb and heart but is enriched in the lung epithelial precursors at E9.5 (81). The Islet1Cre line is a knock-in strain (82) and drives Cre expression throughout the pharyngeal endoderm, including the pulmonary epithelial precursors, and in a subset of mesenchymal cells (83). Although it is not epithelial specific, it has been successfully used to study Fgf8 function during lung development (84).
Transgenic mice driving Cre from a 4.8-kb fragment of the Gata5 promoter (Gata5Cre) mediate recombination in the developing epicardium and throughout the developing lung endoderm (85, 86). However, onset of recombination and potential activity in the developing trachea has yet to be determined.
The transcription factor Id2 is dynamically expressed in various cell types during embryogenesis and in the adult. The Id2-CreERT2 line is a knock-in of the CreERT2 fusion (87). In the developing lung, this strain displays tmx-dependent Cre-mediated recombination in the distal epithelial tips and, at a lower level, in a subset of mesenchymal cells.
Nkx2–5 is one of the earliest cardiac-specific markers in vertebrate embryos. The Nkx2–5 Cre line is a knock-in (88) and drives recombination in the developing proepicardium and subsequently throughout the myocardium and the first pharyngeal arch (89). Recombination occurs in the foregut endoderm and in the surrounding mesoderm before E9.5 (i.e., before lung budding or tracheal–esophogeal separation). Nkx2.5-Cre has successfully been used to inactivate epithelial expression of Sox2 in the developing respiratory tract (35).
Nkx2.1 (also known as Ttf1 and Titf1) is expressed in the domain of the ventral foregut, which gives rise to the lung and trachea. Nkx2.1 is also expressed in thyroid progenitors and in regions of the developing brain (90). An Nkx2.1-Cre BAC transgenic mouse (91) functions throughout the lung and tracheal epithelium and has been successfully used in a number of studies (92, 93).
Before budding, the lung epithelium transcribes Sftpc (SpC, or Surfatant associated protein C). Because the human SFTPC promoter is active once endoderm has been committed to becoming lung, it is very useful for studying early lung development. The SFTPC lines are discussed below. As lung development proceeds, more restricted progenitor cells, such as bronchiolar progenitors, exist (80, 94). Tools for manipulating gene expression specifically in such progenitor cells would be highly desirable.
Alveolar Type II Cells
The 3.7-kb fragment of the human SFTPC promoter is one of the most widely used promoters to generate constitutive and conditional transgenic mouse strains that target the respiratory epithelium (3). The advantage of the SFTPC promoter is that it is very lung specific, and expression in other organs is therefore rare (although, as discussed below, this can depend on the integration site of the transgene). In the adult lung, SFTPC promoter activity is restricted to alveolar type II cells and subsets of cuboidal bronchiolar cells. The most widely distributed strains are the SFTPC-rtTA (57, 59, 95) and the SFTPC-Cre lines (96). The SFTPC-rtTA lines are particularly useful because Dox application from E6.5 to E10.5 only targets the progenitor pool of the distal lung epithelium, the parathyroid, and the thymus. However, targeting of neuroendocrine cells with these lines was not observed (61). During organogenesis, expression levels of morphogenic genes dynamically change spatially and temporally. With the rtTA line, it was possible to activate and inactivate signaling pathways that regulate morphogenic genes in specific compartments at defined times. These studies led to a better understanding of temporal windows for FGF signaling and allowed the process of lung organogenesis to be dissected in greater detail (62, 97, 98). Most prenatal studies were done using SFTPC-rtTA line 1, which expresses rtTA at high levels but also shows Dox-independent gene activation, especially around birth and postnatally. Dox-independent expression can result in embryonic lethal phenotypes, as was the case for overexpression of FGF7 and VEGF (57, 95). To overcome these limitations, a second founder line (SFTPC-rtTA line2) was characterized (95), which expresses lower levels of rtTA and demonstrates less off-Dox effects. Although line 2 has been demonstrated to work after E14.5 and in adult mice, activation in the developing embryonic endoderm has not been tested. Both lines have been widely distributed in the scientific community.
The SFTPC-Cre transgenic strain contains the human SFTPC promoter fragment, which drives a rabbit β globin intron, followed by Cre recombinase. SFTPC-Cre directs recombination throughout the lung epithelium starting at E10.5 (96). This strain has also been widely used and has contributed significantly to our understanding of lung development (99, 100). There are reports of toxic effects on some genetic backgrounds (100, 101). More recently, it has been demonstrated that the SFTPC-Cre line directs recombination in the male germline, which may have confounded previous studies, resulting in the apparent toxicity (B.R. Stripp, personal communication). As long as this transgene is transmitted through the female germline, it is very useful to study embryonic lung development.
The Sftpc-CreERT2-rtTA is a knock-in of CreERT2 and rtTA cassettes just after the stop codon of the endogenous Sftpc gene (102). This very flexible strain drives CreERT2 and rtTA expression in mature adult type II cells. It is useful for lineage-tracing experiments and will undoubtedly be widely used for manipulating gene expression.
Other SFTPC-lines are summarized in Table 1. There are alternative promoters (e.g., ABCA3, C/EBPα) that could be potentially used to target alveolar type II cells. However, these promoters will not be lung specific.
Alveolar Type I Cells
Aquaporin 5 (Aqp5) is expressed predominantly in salivary and lacrimal glands, cornea, trachea, and distal lung (103, 104). In rat and human lungs, Aqp5 is specifically expressed in alveolar type I (AT1) cells but not in alveolar type II (AT2) cells. In mice, Aqp5 expression has also been found in AT2 cells (105). A Cre-IRES-DsRed cassette has been inserted into exon 1 of the endogenous Aqp5 locus, generating the Aqp5-Cre-IRES-DsRed, or ACID, mouse (106). Analysis with the ROSA-mT/mG reporter (107) demonstrated that recombination had occurred in a very high fraction of AT1 cells in the distal lung and not in AT2 cells. However, AT2 recombination in other genetic backgrounds cannot be ruled out. This is the first transgenic mouse engineered to express Cre in AT1 cells, and it should be very useful for studies of AT1 turnover and function.
Podopladin, or T1 α, a gene with unclear function, is expressed in mouse AT1 cells and lymphatics and also in the basal cells of pseudostratified airways (108). By contrast, the rat podopladin gene is specific for AT1 cells. A modified rat BAC containing IRES-green fluorescent protein (GFP) in the podoplanin 3′UTR has been generated (RTIbac) (109). RTIbac-transgenic mice expressed rat podoplanin in AT1 cells and in the brain, and expression in AT2 cells, airways, and vascular endothelium was not detected. Modifications of this BAC to express the rtTA or Cre recombinase could make this construct useful for targeting ATI cells.
Bronchiolar Clara Cells
Secretoglobin1a1 (Scgb1a1, also known as CCSP, CC10, and CCA) is expressed in all bronchiolar Clara cells and at lower levels in most tracheal Clara-like cells. In the rat, Scgb1a1 is also expressed in AT2 cells. A rat Scgb1a1 promoter fragment has been used for making various transgenic lines, and this directs expression to Clara cells and AT2 cells, whereas the mouse Scgb1a1 promoter fragment mainly targets Clara cells (110).
A 2.3-kb fragment of the rat Scgb1a1 promoter is sufficient to direct expression in mouse Clara cells and in a subset of AT2 cells (1). This promoter was subsequently used to generate two independent Scgb1a1-rtTA mouse lines. The first line (59) has been widely distributed within the research community and has been used successfully in multiple studies. Lineage tracing showed that this strain has efficient activity in many bronchiolar Clara cells and in a subset of AT2 cells (62). The Scgb1a1-rtTA line 2 targets most Clara cells but retains little AT2 cell activity (46, 63). Scgb1a1 is also active in the uterus. However, using luciferase reporter mice, no luciferase activity was detected in whole uterus homogenates (59).
The rat Scgb1a1 promoter has been used to generate a Scgb1a1-rtTA2S-M2, which uses the newer, more sensitive version of rtTA (71). This strain is reported to have no basal activity and increased Dox sensitivity. However, it also has some activity in AT2 cells.
An Scgb1a1-Cre transgenic strain was generated using the rat promoter fragment inserted upstream of the coding sequence for Cre (111, 112). Lineage tracing shows that this strain directs recombination in the bronchiolar cells but not in alveolar cells (113), demonstrating that the insertion site of the transgene has a strong effect on expression pattern.
An Scgb1a1-CreER “knock-in” mouse strain was generated by inserting an IRES-CreER cassette into the 3′UTR of the endogenous mouse Scgb1a1 locus (24). This line was used for detailed lineage-tracing studies, which revealed that it provides specific, tmx dose–dependent Cre activity in up to 90% of bronchiolar Clara cells and in up to 7% of AT2 cells. However, it also displays some tmx-independent activity.
A knock-in Tgfb3-Cre line (114) has recently been used to manipulate Notch signaling in the postnatal lung airways (115). Reporter analysis suggested that this strain targets the majority (~ 90%) of Clara cells.
There is evidence to suggest that not all Clara cells are functionally equivalent (116, 117). Transgenic strains that target specific subsets of Clara cells would be highly desirable for the lung research community.
Ciliated Cells
Foxj1 is a transcription factor that is expressed in all multiciliated cells, including those of the lung, oviducts, ependyma, and testes (118) and various cells with motile cilia (119–121). A 1-kb fragment of the human FOXJ1 promoter was shown to be sufficient to direct reporter gene expression specifically in all of these cell types in adult mice (122). This promoter was subsequently used to generate FOXJ1-Cre (123) and FOXJ1-CreERT2 transgenic mice (29). Both lines drive efficient recombination in ciliated cells of the respiratory tract and have been useful for gene knock-out and lineage-tracing studies. In particular, the FOXJ1-CreERT2 mice were used to determine the average half-life of ciliated cells in the mouse airways (124). The FOXJ1-CreERT2 strain also unexpectedly directs recombination very efficiently in pericytes (J.R. Rock and B.L.M. Hogan, personal communication). This may reflect a low level of endogenous pericyte Foxj1 expression or may be due to the insertion site of the transgene. Similarly, a recent paper has shown that the same human FOXJ1 promoter can drive expression in human ciliated cells but also some basal cells growing at an air–liquid interface (125). Basal cell expression was not observed in the transgenic animals. However, any future transgenic strains generated with this promoter should be screened for basal cell and pericyte activity.
Basal Cells
Published gene expression data have not identified a gene that is expressed exclusively in airway basal cells (28). A split-Cre, or viral, approach may be necessary for airway-specific basal cell genetic manipulation.
Keratin 5 (Krt5) and Keratin 14 (Krt14) promoters have been used to target basal cells in the airway epithelium. All mouse and human airway epithelial basal cells express Krt5 (126, 127). A human 6kb KRT5 promoter fragment was cloned by the Fuchs lab and successfully used to target epidermal basal cells (128, 129). Using the same promoter fragment, KRT5-CreER2T transgenic mice were generated and used for cell lineage tracing in the airways. These studies demonstrated that airway basal cells are stem cells (28). This strain has been used for studying the control of basal cell function (130) and should prove to be useful for manipulating gene expression in tracheal basal cells. However, the KRT5-CreER2T transgenic strain is limited because it directs recombination in only approximately 15% of basal cells in the adult trachea. Moreover, the high levels of transgene activity in the skin and oral epithelium make this strain extremely difficult to use for studies of oncogenes because the mice develop skin and oral tumors before the trachea is affected.
Krt14 is expressed in roughly 30% of mouse tracheal basal cells (126, 127, 131). Transgenic mice containing the human KRT14 promoter linked to CreERT were generated for use in the skin (132). In the trachea, these KRT14-CreERT transgenic mice allow tmx-induced recombination in an extremely small population of basal cells at steady-state (131). After naphthalene injury, most surviving basal cells up-regulate Krt14 and also express the transgene (133). This mouse has not been used to manipulate gene expression in airway basal cells. However, a K14-rtTA mouse (134) has been used, in combination with the tetO-Cre strain (61), to direct gene expression in the trachea (47). These data show the importance of careful control experiments because in the K14-rtTA strain tracheal Clara cells were sensitive to Dox exposure.
Neuroendocrine Cells
Pulmonary neuroendocrine (NE) cell differentiation depends on genes that are conserved in the nervous system of many organisms, for example Ascl1, NeuroD, Rb, and Gfi1 (135–138). A rat NE cell–specific promoter has been identified (139) and recently used in an adenovirus to direct gene expression specifically to NE cells (140). Although no transgenic mouse strains that specifically target NE cells have been generated, such strains could be very useful for studying NE cell function or their putative role as an airway epithelial stem cell niche (141).
Mesenchymal Lines
There is much disagreement over the numbers of different lung mesenchymal cell types and their best markers (142). The challenge of the available mesenchymal mouse strains is that they are not lung specific and that many of their expression patterns are highly variable depending on the integration of the transgene (Table 3). Pod1 (also known as Tcf21) is highly expressed in the mesenchyme of the developing lung, kidney, heart, and intestine and may be a useful promoter for more restricted mesenchymal gene manipulation (143). Tbx genes may also provide useful mesenchymal promoters. Tbx2–5 are expressed in the developing lung mesenchyme (144) and are often used as reporters of embryonic mesenchymal fate (145, 146). However, the expression patterns of Pod1 and the Tbx genes in the adult lung mesenchyme have yet to be determined.
TABLE 3.
MESENCHYMAL AND ENDOTHELIAL LINES
| Mouse Strain | Targeting Cell | References |
| Dermo1-Cre knock in | Most mesodermal progenitors, mesenchymal and mesothelial lineages in the lung | 147, 148 |
| Mesp1-Cre knock in | Anterior mesoderm from early gastrulation | 84, 151 |
| FSP1-Cre transgenic | Fibroblasts, gene is induced in epithelial–mesenchymal transition | 154 |
| AP2-Cre, AP2-CreERT2 knock in mice | Adipocytes and alveolar type II cells, aP2-cre in a subset of subset of alveolar fibroblasts | 155 |
| SM22-rtTA, transgenic | Smooth muscle: vascular and bronchiolar, mistaken for an endothelial marker | 156 |
| SM22 CreERT2 knock in | Not efficient | 157 |
| Mouse αSMA-Cre, transgenic | Expressed in peribronchiolar and perivascular smooth muscle cells in lung fibrosis after bleomycin or hyperoxia injury, expression increased in asthma. Not confirmed; in parenchymal fibroblasts during alveolarization and neoalveolarization | 164 |
| αSMA-CreERT2 BAC transgenic | See αSMA–cre above. | 165 |
| SMMHC-CreERT2 BAC transgenic | All smooth muscle, including the airway smooth muscle and lung vasculature; caution of off-target expression due to germline activity | 23 |
| Wt1-Cre YAC transgenic | Mesothelial-derived cells; caution of low-level Wt1 gene expression in the mesenchyme | 168–170 |
| Tie2-Cre transgenic | Endothelial cells; caution of off-target expression due to germline activity | 171–174 |
Definition of abbreviations: BAC = bacterial artificial chromosome; YAC = yeast artificial chromosome.
Dermo1-Cre (also known as Twist2-Cre) is a knock-in of Cre (147) that displays robust recombination in mesenchymal and mesothelial lineages in the lung (148). It has been widely used for manipulating gene expression in the developing lung mesenchyme (149, 150).
Mesp1-Cre is a knock-in of Cre replacing the Mesp1 coding region and drives expression throughout the anterior mesoderm from early gastrulation (151). It has been successfully used to manipulate gene expression in the developing lung mesoderm (84).
Fibroblast-specific protein (FSP)1, also known as S100A4, has been reported as a fibroblast specific gene but is also induced in epithelial cells during injury and tumor progression (152, 153). The FSP1 promoter has been used to generate various FSP1-Cre mice with variable success (154). The use of these mice to study mesenchymal cells in the lung remains controversial.
Adipocyte lipid-binding protein (aP)2 (also known as fatty acid binding protein 4) is expressed in alveolar type II cells and interstitial lipofibroblasts. The mouse aP2 promoter was used to generate aP2-Cre and aP2-CreERT2 mice (155). Although this aP2-Cre line does not target AT2 cells, it does target a subset of alveolar fibroblasts. Induction of recombination with the aP2-CreERT2 in adult mice was not observed (A. K. Perl, unpublished data).
The SM22-α (SM22α or transgelin) promoter was used to generate SM22-rtTA mice. This line provides a “Tet-On” tool that allows the inducible expression of genes in smooth muscle cells (156). Expression is mainly in the vascular smooth muscle, and the SM22 promoter was used to generate several transgenic and knock-in CreERT2–expressing lines with varying expression patterns, with the highest levels detected in the aorta, intestine, and uterus. However, none of these lines is particularly efficient, even in the vascular smooth muscle (157).
SMA (smooth muscle α actin, or Acta2) is expressed in all smooth muscle cells in the adult and transiently in myocardiocytes and skeletal muscle during embryonic development (158, 159). In lung parenchyma, SMA is expressed during alveolarization, realveolarization, and during the development of lung fibrosis after bleomycin or hyperoxic injury (160–163). Mice with a murine αSMA-Cre transgene express Cre in the airway smooth muscle and lung vasculature (164). This line is not inducible, which limits postnatal studies. More recently, an αSMA-CreERT2 BAC transgenic line has been generated and shown to exhibit tmx-dependent Cre activity in all adult smooth muscle, including the lung airways and vasculature (165). These mice have off-target Cre activity only in a small number of cardiomyocytes and should be very useful for future lung studies.
SMMHC (smooth muscle myosin heavy chain, or Myh11) is expressed in all smooth muscle cells. Two independent mouse strains expressing Cre from a fragment of the mouse SMMHC promoter have been generated. The expression of the transgene is somewhat variable (166, 167). This promoter leads to spurious CRE activity in some tissues due to expression in male and female germline (50). By contrast, a SMMHC-CreERT2 BAC transgenic strain shows inducible Cre activity in all smooth muscle, including the airway smooth muscle and lung vasculature (23). This strain should be useful for manipulation of gene expression in perivascular and peribronchiolar smooth muscle in the postnatal lung.
Mesothelium
To direct Cre expression to the mesothelium of internal organs including the liver, gut, and lung, a Wt1-Cre yeast artificial chromosome (YAC) transgenic strain was generated (168). This has been shown to be active throughout the lung mesothelium from early developmental stages (169, 170). However, it may be expressed at low levels in mesenchymal lineage of the embryonic lung and needs to be used with caution (B. L. M. Hogan, personal communication). An inducible version would be useful for adult studies.
Endothelial Lines
Multiple independent transgenic strains express Cre recombinase under the control of the mouse Tek promoter (Endothelial-Specific Receptor Tyrosine Kinase, also known as Tie2). Some of these have been very widely used (171–174). In these strains, reporter gene activity was detected in most endothelial cells and blood islands of the extra embryonic mesoderm by E7.5, in the dorsal aorta by E8.5, and in all blood vessels and some blood cells examined at E11.5, indicating that Cre was active in early vascular progenitors, endothelial cells, and some hematopoietic cells. This promoter leads to spurious Cre activity in some tissues due to expression in male and female germlines (50).
Reporters Of Signaling Pathway Activity
Wnt Pathway
Wnt signaling pathways play divergent roles during development, homeostasis, and repair and play a major role in stem cell proliferation and differentiation. Three transgenic reporter lines for Wnt pathway activity have been generated: (1) TOPGAL, which reports epithelial Wnt signaling (175); (2) BATGAL, which has with sporadic epithelial and mesenchymal activity (176); and (3) Axin2-lacZ, which is useful to study proximal lung and mesenchymal Wnt signaling (177). A recent study compared these lines during development and after naphthalene injury (178). A new reporter line, TCF/Lef:H2B-GFP, has not been tested in the lung (Table 4) (179).
Notch Pathway
Signals through the Notch receptors are used throughout embryonic development and in the adult to control cellular fate choices. CP-EGFP (also known as TNR) transgenic mice have a transgenic Notch reporter with an enhanced green fluorescent protein (EGFP) placed under the control of four tandem copies of a CBF1 (also known as Rbpj) responsive element (four CBF1 binding site consensus sequences and the basal SV40 promoter) (180, 181). This strain has been shown to faithfully report Notch activity in the adult trachea (182). N1IP::CRELOW and N1IP::CREHI are knock-ins of Cre, replacing the Notch1 coding region (Notch1 intramembrane proteolysis) and allow lineage studies of descendants of cells after Notch 1 activation (Table 4) (170, 183). However, this Cre line identifies each cell lineage, which has previously experienced Notch activity and does not report on current signaling events. Comparison of various Notch reporter lines will shed better light on cell fate decisions and lineage relationships and lead to a better understanding of stem cell biology and interactions of epithelium, mesenchyme, mesothelium, and endothelium during development and repair.
Virus-Mediated Transient Lung Transgenics
The lung is exposed to the external environment, and multiple groups have taken advantage of this by administering viruses to manipulate gene expression in the adult mouse lung epithelium. The most widespread system is intranasal administration of an adenovirus expressing Cre from a ubiquitous CMV promoter (AdenoCre) to activate the expression of oncogenes and model lung cancer (184). A similar adenovirus-based approach has been taken to transiently overexpress specific genes throughout the lung epithelium (e.g., Ref. 185). More recently, adenoviruses using Scgb1a1, rat SftpC, or rat CGRP promoter fragments to direct Cre expression to specific epithelial cell types have been developed (140). Lentiviral vectors containing specific promoters for manipulating gene expression in restricted adult lung epithelial cell types have also been reported (186, 187). In addition, adeno-associated virus transduction of mouse lung epithelial progenitors has been demonstrated (188). The use of viral systems is likely to become more widespread over the next few years, particularly for epithelial studies.
Conclusions
Transgenic mice have been instrumental in developing our current understanding of lung embryonic development, adult homeostasis, and repair. However, it is important to remember that all transgenic approaches have limitations, which can only be overcome by integrating findings from different lines and performing all the appropriate controls. New developments in mouse conditional genetics have the potential to further enhance our understanding of lung development and disease. Moreover, optimizing mouse strains of the existing Dox and Cre systems will increase flexibility and improve experimental design. For example, using the newer, more Dox-sensitive rtTA gene activation (rtTA2S-M2) or extremely low doses of tmx in CreER based transgenic mice will allow recombination in single cells and enable clonal cell type–specific gene manipulation. Due to the lack of lung-specific mesenchymal and endothelial gene expression, more lines need to be characterized for their usefulness in targeting specific subsets of mesenchymal and endothelial cells. On the other hand, complex targeting systems, such as the split-Cre, will be helpful to target subsets of epithelial progenitor cells or specific mesenchymal cell lineages. Recently, the applicability of the Flipase system to the lung has been demonstrated by combining Cre and FLP to independently control recombination of p53 and kRas in lung tumor progression (54). Development of tools based on flippase and viruses will further expand the combinatorial use of the existing mouse lines and help to develop newer lines, possibly overcoming the problems of off-target activation, lack of cell type specificity, and lack of adult regulation. In addition, the generation of new publicly available floxed alleles by the International Mouse Knock out Consortium (http://www.knockoutmouse.org/) and the use of transgenic mice expressing conditional RNA interference constructs should facilitate mouse conditional genetic analysis.
Acknowledgments
The authors thank Jeffrey Whitsett, Jason Rock, Barry Stripp, and Brigid Hogan for sharing unpublished data and Jeffrey Whitsett, Brigid Hogan, Jim Bridges, and members of our laboratories for critical comments on the manuscript.
Footnotes
Originally Published in Press as DOI: 10.1165/rcmb.2011-0372PS on December 22, 2011
References
- 1.Stripp BR, Sawaya PL, Luse DS, Wikenheiser KA, Wert SE, Huffman JA, Lattier DL, Singh G, Katyal SL, Whitsett JA. cis-acting elements that confer lung epithelial cell expression of the CC10 gene. J Biol Chem 1992;267:14703–14712. [PubMed] [Google Scholar]
- 2.Stripp BR, Huffman JA, Bohinski RJ. Structure and regulation of the murine Clara cell secretory protein gene. Genomics 1994;20:27–35. [DOI] [PubMed] [Google Scholar]
- 3.Wert SE, Glasser SW, Korfhagen TR, Whitsett JA. Transcriptional elements from the human SP-C gene direct expression in the primordial respiratory epithelium of transgenic mice. Dev Biol 1993;156:426–443. [DOI] [PubMed] [Google Scholar]
- 4.Sternberg N, Hamilton D. Bacteriophage P1 site-specific recombination: I. Recombination between loxP sites. J Mol Biol 1981;150:467–486. [DOI] [PubMed] [Google Scholar]
- 5.Sauer B, Henderson N. Site-specific DNA recombination in mammalian cells by the Cre recombinase of bacteriophage P1. Proc Natl Acad Sci USA 1988;85:5166–5170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sauer B. Inducible gene targeting in mice using the Cre/lox system. Methods 1998;14:381–392. [DOI] [PubMed] [Google Scholar]
- 7.Shimshek DR, Kim J, Hubner MR, Spergel DJ, Buchholz F, Casanova E, Stewart AF, Seeburg PH, Sprengel R. Codon-improved Cre recombinase (iCre) expression in the mouse. Genesis 2002;32:19–26. [DOI] [PubMed] [Google Scholar]
- 8.Nagy A. Cre recombinase: the universal reagent for genome tailoring. Genesis 2000;26:99–109. [PubMed] [Google Scholar]
- 9.Livet J, Weissman TA, Kang H, Draft RW, Lu J, Bennis RA, Sanes JR, Lichtman JW. Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature 2007;450:56–62. [DOI] [PubMed] [Google Scholar]
- 10.Snippert HJ, van der Flier LG, Sato T, van Es JH, van den Born M, Kroon-Veenboer C, Barker N, Klein AM, van Rheenen J, Simons BD, et al. Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell 2010;143:134–144. [DOI] [PubMed] [Google Scholar]
- 11.Burlison JS, Long Q, Fujitani Y, Wright CV, Magnuson MA. Pdx-1 and Ptf1a concurrently determine fate specification of pancreatic multipotent progenitor cells. Dev Biol 2008;316:74–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Domyan ET, Sun X. Patterning and plasticity in development of the respiratory lineage. Dev Dyn 2011;240:477–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu Y, Futtner C, Rock JR, Xu X, Whitworth W, Hogan BL, Onaitis MW. Evidence that SOX2 overexpression is oncogenic in the lung. PLoS ONE 2010;5:e11022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lobe CG, Koop KE, Kreppner W, Lomeli H, Gertsenstein M, Nagy A. Z/AP, a double reporter for cre-mediated recombination. Dev Biol 1999;208:281–292. [DOI] [PubMed] [Google Scholar]
- 15.Novak A, Guo C, Yang W, Nagy A, Lobe CG. Z/EG, a double reporter mouse line that expresses enhanced green fluorescent protein upon Cre-mediated excision. Genesis 2000;28:147–155. [PubMed] [Google Scholar]
- 16.Friedrich G, Soriano P. Promoter traps in embryonic stem cells: a genetic screen to identify and mutate developmental genes in mice. Genes Dev 1991;5:1513–1523. [DOI] [PubMed] [Google Scholar]
- 17.Zambrowicz BP, Imamoto A, Fiering S, Herzenberg LA, Kerr WG, Soriano P. Disruption of overlapping transcripts in the ROSA beta geo 26 gene trap strain leads to widespread expression of beta-galactosidase in mouse embryos and hematopoietic cells. Proc Natl Acad Sci USA 1997;94:3789–3794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet 1999;21:70–71. [DOI] [PubMed] [Google Scholar]
- 19.Indra AK, Warot X, Brocard J, Bornert JM, Xiao JH, Chambon P, Metzger D. Temporally-controlled site-specific mutagenesis in the basal layer of the epidermis: comparison of the recombinase activity of the tamoxifen-inducible Cre-ER(T) and Cre-ER(T2) recombinases. Nucleic Acids Res 1999;27:4324–4327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feil R, Brocard J, Mascrez B, LeMeur M, Metzger D, Chambon P. Ligand-activated site-specific recombination in mice. Proc Natl Acad Sci USA 1996;93:10887–10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feil R, Wagner J, Metzger D, Chambon P. Regulation of Cre recombinase activity by mutated estrogen receptor ligand-binding domains. Biochem Biophys Res Commun 1997;237:752–757. [DOI] [PubMed] [Google Scholar]
- 22.Hayashi S, McMahon AP. Efficient recombination in diverse tissues by a tamoxifen-inducible form of Cre: a tool for temporally regulated gene activation/inactivation in the mouse. Dev Biol 2002;244:305–318. [DOI] [PubMed] [Google Scholar]
- 23.Wirth A, Benyo Z, Lukasova M, Leutgeb B, Wettschureck N, Gorbey S, Orsy P, Horvath B, Maser-Gluth C, Greiner E, et al. G12–G13-LARG-mediated signaling in vascular smooth muscle is required for salt-induced hypertension. Nat Med 2008;14:64–68. [DOI] [PubMed] [Google Scholar]
- 24.Rawlins EL, Okubo T, Xue Y, Brass DM, Auten RL, Hasegawa H, Wang F, Hogan BL. The role of Scgb1a1+ Clara cells in the long-term maintenance and repair of lung airway, but not alveolar, epithelium. Cell Stem Cell 2009;4:525–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Buelow B, Scharenberg AM. Characterization of parameters required for effective use of tamoxifen-regulated recombination. PLoS ONE 2008;3:e3264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakamura E, Nguyen MT, Mackem S. Kinetics of tamoxifen-regulated Cre activity in mice using a cartilage-specific CreER(T) to assay temporal activity windows along the proximodistal limb skeleton. Dev Dyn 2006;235:2603–2612. [DOI] [PubMed] [Google Scholar]
- 27.Park EJ, Sun X, Nichol P, Saijoh Y, Martin JF, Moon AM. System for tamoxifen-inducible expression of cre-recombinase from the Foxa2 locus in mice. Dev Dyn 2008;237:447–453. [DOI] [PubMed] [Google Scholar]
- 28.Rock JR, Onaitis MW, Rawlins EL, Lu Y, Clark CP, Xue Y, Randell SH, Hogan BL. Basal cells as stem cells of the mouse trachea and human airway epithelium. Proc Natl Acad Sci USA 2009;106:12771–12775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rawlins EL, Ostrowski LE, Randell SH, Hogan BL. Lung development and repair: contribution of the ciliated lineage. Proc Natl Acad Sci USA 2007;104:410–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vooijs M, Jonkers J, Berns A. A highly efficient ligand-regulated Cre recombinase mouse line shows that LoxP recombination is position dependent. EMBO Rep 2001;2:292–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Y, Riesterer C, Ayrall AM, Sablitzky F, Littlewood TD, Reth M. Inducible site-directed recombination in mouse embryonic stem cells. Nucleic Acids Res 1996;24:543–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ma W, Tessarollo L, Hong SB, Baba M, Southon E, Back TC, Spence S, Lobe CG, Sharma N, Maher GW, et al. Hepatic vascular tumors, angiectasis in multiple organs, and impaired spermatogenesis in mice with conditional inactivation of the VHL gene. Cancer Res 2003;63:5320–5328. [PubMed] [Google Scholar]
- 33.Guo C, Yang W, Lobe CG. A Cre recombinase transgene with mosaic, widespread tamoxifen-inducible action. Genesis 2002;32:8–18. [DOI] [PubMed] [Google Scholar]
- 34.White AC, Lavine KJ, Ornitz DM. FGF9 and SHH regulate mesenchymal Vegfa expression and development of the pulmonary capillary network. Development 2007;134:3743–3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Que J, Luo X, Schwartz RJ, Hogan BL. Multiple roles for Sox2 in the developing and adult mouse trachea. Development 2009;136:1899–1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takeda K, Cowan A, Fong GH. Essential role for prolyl hydroxylase domain protein 2 in oxygen homeostasis of the adult vascular system. Circulation 2007;116:774–781. [DOI] [PubMed] [Google Scholar]
- 37.Cheng Y, Sudarov A, Szulc KU, Sgaier SK, Stephen D, Turnbull DH, Joyner AL. The engrailed homeobox genes determine the different foliation patterns in the vermis and hemispheres of the mammalian cerebellum. Development 2010;137:519–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.de Luca C, Kowalski TJ, Zhang Y, Elmquist JK, Lee C, Kilimann MW, Ludwig T, Liu SM, Chua SC. Complete rescue of obesity, diabetes, and infertility in db/db mice by neuron-specific LEPR-B transgenes. J Clin Invest 2005;115:3484–3493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ventura A, Kirsch DG, McLaughlin ME, Tuveson DA, Grimm J, Lintault L, Newman J, Reczek EE, Weissleder R, Jacks T. Restoration of p53 function leads to tumour regression in vivo. Nature 2007;445:661–665. [DOI] [PubMed] [Google Scholar]
- 40.Brocard J, Feil R, Chambon P, Metzger D. A chimeric Cre recombinase inducible by synthetic, but not by natural ligands of the glucocorticoid receptor. Nucleic Acids Res 1998;26:4086–4090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kellendonk C, Tronche F, Monaghan AP, Angrand PO, Stewart F, Schutz G. Regulation of Cre recombinase activity by the synthetic steroid RU 486. Nucleic Acids Res 1996;24:1404–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hirrlinger J, Requardt RP, Winkler U, Wilhelm F, Schulze C, Hirrlinger PG. Split-CreERT2: temporal control of DNA recombination mediated by split-Cre protein fragment complementation. PLoS ONE 2009;4:e8354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Beckervordersandforth R, Tripathi P, Ninkovic J, Bayam E, Lepier A, Stempfhuber B, Kirchhoff F, Hirrlinger J, Haslinger A, Lie DC, et al. In vivo fate mapping and expression analysis reveals molecular hallmarks of prospectively isolated adult neural stem cells. Cell Stem Cell 2010;7:744–758. [DOI] [PubMed] [Google Scholar]
- 44.Hirrlinger J, Scheller A, Hirrlinger PG, Kellert B, Tang W, Wehr MC, Goebbels S, Reichenbach A, Sprengel R, Rossner MJ, et al. Split-cre complementation indicates coincident activity of different genes in vivo. PLoS ONE 2009;4:e4286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Naiche LA, Papaioannou VE. Cre activity causes widespread apoptosis and lethal anemia during embryonic development. Genesis 2007;45:768–775. [DOI] [PubMed] [Google Scholar]
- 46.Perl AK, Zhang L, Whitsett JA. Conditional expression of genes in the respiratory epithelium in transgenic mice: cautionary notes and toward building a better mouse trap. Am J Respir Cell Mol Biol 2009;40:1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smith RW, Hicks DA, Reynolds SD. Roles for β-catenin and doxycycline in regulation of respiratory epithelial cell frequency and function. Am J Respir Cell Mol Biol (In press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schmidt EE, Taylor DS, Prigge JR, Barnett S, Capecchi MR. Illegitimate Cre-dependent chromosome rearrangements in transgenic mouse spermatids. Proc Natl Acad Sci USA 2000;97:13702–13707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Semprini S, Troup TJ, Kotelevtseva N, King K, Davis JR, Mullins LJ, Chapman KE, Dunbar DR, Mullins JJ. Cryptic loxP sites in mammalian genomes: genome-wide distribution and relevance for the efficiency of BAC/PAC recombineering techniques. Nucleic Acids Res 2007;35:1402–1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.de Lange WJ, Halabi CM, Beyer AM, Sigmund CD. Germ line activation of the Tie2 and SMMHC promoters causes noncell-specific deletion of floxed alleles. Physiol Genomics 2008;35:1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sadowski PD. The Flp recombinase of the 2-microns plasmid of Saccharomyces cerevisiae. Prog Nucleic Acid Res Mol Biol 1995;51:53–91. [PubMed] [Google Scholar]
- 52.Farley FW, Soriano P, Steffen LS, Dymecki SM. Widespread recombinase expression using FLPeR (flipper) mice. Genesis 2000;28:106–110. [PubMed] [Google Scholar]
- 53.Awatramani R, Soriano P, Mai JJ, Dymecki S. An Flp indicator mouse expressing alkaline phosphatase from the ROSA26 locus. Nat Genet 2001;29:257–259. [DOI] [PubMed] [Google Scholar]
- 54.Young NP, Crowley D, Jacks T. Uncoupling cancer mutations reveals critical timing of p53 loss in sarcomagenesis. Cancer Res 2011;71:4040–4047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gossen M, Bujard H. Efficacy of tetracycline-controlled gene expression is influenced by cell type. Biotechniques 1995;19:213–216. [Discussion, pp. 216–217.] [PubMed] [Google Scholar]
- 56.Kistner A, Gossen M, Zimmermann F, Jerecic J, Ullmer C, Lubbert H, Bujard H. Doxycycline-mediated quantitative and tissue-specific control of gene expression in transgenic mice. Proc Natl Acad Sci USA 1996;93:10933–10938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tichelaar JW, Lu W, Whitsett JA. Conditional expression of fibroblast growth factor-7 in the developing and mature lung. J Biol Chem 2000;275:11858–11864. [DOI] [PubMed] [Google Scholar]
- 58.Schultze N, Burki Y, Lang Y, Certa U, Bluethmann H. Efficient control of gene expression by single step integration of the tetracycline system in transgenic mice. Nat Biotechnol 1996;14:499–503. [DOI] [PubMed] [Google Scholar]
- 59.Perl AK, Tichelaar JW, Whitsett JA. Conditional gene expression in the respiratory epithelium of the mouse. Transgenic Res 2002;11:21–29. [DOI] [PubMed] [Google Scholar]
- 60.Whitsett JA, Perl AK. Conditional control of gene expression in the respiratory epithelium: a cautionary note. Am J Respir Cell Mol Biol 2006;34:519–520. [DOI] [PubMed] [Google Scholar]
- 61.Perl AK, Wert SE, Nagy A, Lobe CG, Whitsett JA. Early restriction of peripheral and proximal cell lineages during formation of the lung. Proc Natl Acad Sci USA 2002;99:10482–10487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Perl AK, Wert SE, Loudy DE, Shan Z, Blair PA, Whitsett JA. Conditional recombination reveals distinct subsets of epithelial cells in trachea, bronchi, and alveoli. Am J Respir Cell Mol Biol 2005;33:455–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Perl AK, Riethmacher D, Whitsett JA. Conditional depletion of airway progenitor cells induces peribronchiolar fibrosis. Am J Respir Crit Care Med 2011;183:511–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Belteki G, Haigh J, Kabacs N, Haigh K, Sison K, Costantini F, Whitsett J, Quaggin SE, Nagy A. Conditional and inducible transgene expression in mice through the combinatorial use of Cre-mediated recombination and tetracycline induction. Nucleic Acids Res 2005;33:e51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hochedlinger K, Yamada Y, Beard C, Jaenisch R. Ectopic expression of Oct-4 blocks progenitor-cell differentiation and causes dysplasia in epithelial tissues. Cell 2005;121:465–477. [DOI] [PubMed] [Google Scholar]
- 66.Morimoto M, Kopan R. rtTA toxicity limits the usefulness of the SP-C-rtTA transgenic mouse. Dev Biol 2009;325:171–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vieillard-Baron A, Frisdal E, Eddahibi S, Deprez I, Baker AH, Newby AC, Berger P, Levame M, Raffestin B, Adnot S, et al. Inhibition of matrix metalloproteinases by lung TIMP-1 gene transfer or doxycycline aggravates pulmonary hypertension in rats. Circ Res 2000;87:418–425. [DOI] [PubMed] [Google Scholar]
- 68.Ren S, Guo LL, Yang J, Liu DS, Wang T, Chen L, Chen YJ, Xu D, Feng YL, Wen FQ. Doxycycline attenuates acrolein-induced mucin production, in part by inhibiting MMP-9. Eur J Pharmacol 2011;650:418–423. [DOI] [PubMed] [Google Scholar]
- 69.Ohbayashi H. Matrix metalloproteinases in lung diseases. Curr Protein Pept Sci 2002;3:409–421. [DOI] [PubMed] [Google Scholar]
- 70.Urlinger S, Baron U, Thellmann M, Hasan MT, Bujard H, Hillen W. Exploring the sequence space for tetracycline-dependent transcriptional activators: novel mutations yield expanded range and sensitivity. Proc Natl Acad Sci USA 2000;97:7963–7968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Duerr J, Gruner M, Schubert SC, Haberkorn U, Bujard H, Mall MA. Use of a new generation reverse tetracycline transactivator system for quantitative control of conditional gene expression in the murine lung. Am J Respir Cell Mol Biol 2011;44:244–254. [DOI] [PubMed] [Google Scholar]
- 72.Martin DI, Whitelaw E. The vagaries of variegating transgenes. Bioessays 1996;18:919–923. [DOI] [PubMed] [Google Scholar]
- 73.Yang GS, Banks KG, Bonaguro RJ, Wilson G, Dreolini L, de Leeuw CN, Liu L, Swanson DJ, Goldowitz D, Holt RA, et al. Next generation tools for high-throughput promoter and expression analysis employing single-copy knock-ins at the Hprt1 locus. Genomics 2009;93:196–204. [DOI] [PubMed] [Google Scholar]
- 74.Premsrirut PK, Dow LE, Kim SY, Camiolo M, Malone CD, Miething C, Scuoppo C, Zuber J, Dickins RA, Kogan SC, et al. A rapid and scalable system for studying gene function in mice using conditional RNA interference. Cell 2011;145:145–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chen SX, Osipovich AB, Ustione A, Potter LA, Hipkens S, Gangula R, Yuan W, Piston DW, Magnuson MA. Quantification of factors influencing fluorescent protein expression using RMCE to generate an allelic series in the ROSA26 locus in mice. Dis Model Mech 2011;4:537–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Szymczak AL, Workman CJ, Wang Y, Vignali KM, Dilioglou S, Vanin EF, Vignali DA. Correction of multi-gene deficiency in vivo using a single ‘self-cleaving' 2A peptide-based retroviral vector. Nat Biotechnol 2004;22:589–594. [DOI] [PubMed] [Google Scholar]
- 77.Harfe BD, Scherz PJ, Nissim S, Tian H, McMahon AP, Tabin CJ. Evidence for an expansion-based temporal Shh gradient in specifying vertebrate digit identities. Cell 2004;118:517–528. [DOI] [PubMed] [Google Scholar]
- 78.Harris KS, Zhang Z, McManus MT, Harfe BD, Sun X. Dicer function is essential for lung epithelium morphogenesis. Proc Natl Acad Sci USA 2006;103:2208–2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Harris-Johnson KS, Domyan ET, Vezina CM, Sun X. beta-Catenin promotes respiratory progenitor identity in mouse foregut. Proc Natl Acad Sci USA 2009;106:16287–16292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tsao PN, Vasconcelos M, Izvolsky KI, Qian J, Lu J, Cardoso WV. Notch signaling controls the balance of ciliated and secretory cell fates in developing airways. Development 2009;136:2297–2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Millien G, Beane J, Lenburg M, Tsao PN, Lu J, Spira A, Ramirez MI. Characterization of the mid-foregut transcriptome identifies genes regulated during lung bud induction. Gene Expr Patterns 2008;8:124–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yang L, Cai CL, Lin L, Qyang Y, Chung C, Monteiro RM, Mummery CL, Fishman GI, Cogen A, Evans S. Isl1Cre reveals a common Bmp pathway in heart and limb development. Development 2006;133:1575–1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Park KS, Wells JM, Zorn AM, Wert SE, Whitsett JA. Sox17 influences the differentiation of respiratory epithelial cells. Dev Biol 2006;294:192–202. [DOI] [PubMed] [Google Scholar]
- 84.Yu S, Poe B, Schwarz M, Elliot SA, Albertine KH, Fenton S, Garg V, Moon AM. Fetal and postnatal lung defects reveal a novel and required role for Fgf8 in lung development. Dev Biol 2010;347:92–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Merki E, Zamora M, Raya A, Kawakami Y, Wang J, Zhang X, Burch J, Kubalak SW, Kaliman P, Belmonte JC, et al. Epicardial retinoid X receptor alpha is required for myocardial growth and coronary artery formation. Proc Natl Acad Sci USA 2005;102:18455–18460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Xing Y, Li C, Li A, Sridurongrit S, Tiozzo C, Bellusci S, Borok Z, Kaartinen V, Minoo P. Signaling via Alk5 controls the ontogeny of lung Clara cells. Development 2010;137:825–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rawlins EL, Clark CP, Xue Y, Hogan BL. The Id2+ distal tip lung epithelium contains individual multipotent embryonic progenitor cells. Development 2009;136:3741–3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Moses KA, DeMayo F, Braun RM, Reecy JL, Schwartz RJ. Embryonic expression of an Nkx2–5/Cre gene using ROSA26 reporter mice. Genesis 2001;31:176–180. [DOI] [PubMed] [Google Scholar]
- 89.Zhou B, Ma Q, Rajagopal S, Wu SM, Domian I, Rivera-Feliciano J, Jiang D, von Gise A, Ikeda S, Chien KR, et al. Epicardial progenitors contribute to the cardiomyocyte lineage in the developing heart. Nature 2008;454:109–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lazzaro D, Price M, de Felice M, Di Lauro R. The transcription factor TTF-1 is expressed at the onset of thyroid and lung morphogenesis and in restricted regions of the foetal brain. Development 1991;113:1093–1104. [DOI] [PubMed] [Google Scholar]
- 91.Xu Q, Tam M, Anderson SA. Fate mapping Nkx2.1-lineage cells in the mouse telencephalon. J Comp Neurol 2008;506:16–29. [DOI] [PubMed] [Google Scholar]
- 92.Li C, Li A, Li M, Xing Y, Chen H, Hu L, Tiozzo C, Anderson S, Taketo MM, Minoo P. Stabilized beta-catenin in lung epithelial cells changes cell fate and leads to tracheal and bronchial polyposis. Dev Biol 2009;334:97–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tiozzo C, De Langhe S, Yu M, Londhe VA, Carraro G, Li M, Li C, Xing Y, Anderson S, Borok Z, et al. Deletion of Pten expands lung epithelial progenitor pools and confers resistance to airway injury. Am J Respir Crit Care Med 2009;180:701–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Shannon JM. Induction of alveolar type II cell differentiation in fetal tracheal epithelium by grafted distal lung mesenchyme. Dev Biol 1994;166:600–614. [DOI] [PubMed] [Google Scholar]
- 95.Akeson AL, Greenberg JM, Cameron JE, Thompson FY, Brooks SK, Wiginton D, Whitsett JA. Temporal and spatial regulation of VEGF-A controls vascular patterning in the embryonic lung. Dev Biol 2003;264:443–455. [DOI] [PubMed] [Google Scholar]
- 96.Okubo T, Knoepfler PS, Eisenman RN, Hogan BL. Nmyc plays an essential role during lung development as a dosage-sensitive regulator of progenitor cell proliferation and differentiation. Development 2005;132:1363–1374. [DOI] [PubMed] [Google Scholar]
- 97.Perl AK, Hokuto I, Impagnatiello MA, Christofori G, Whitsett JA. Temporal effects of Sprouty on lung morphogenesis. Dev Biol 2003;258:154–168. [DOI] [PubMed] [Google Scholar]
- 98.Hokuto I, Perl AK, Whitsett JA. Prenatal, but not postnatal, inhibition of fibroblast growth factor receptor signaling causes emphysema. J Biol Chem 2003;278:415–421. [DOI] [PubMed] [Google Scholar]
- 99.Guseh JS, Bores SA, Stanger BZ, Zhou Q, Anderson WJ, Melton DA, Rajagopal J. Notch signaling promotes airway mucous metaplasia and inhibits alveolar development. Development 2009;136:1751–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Eblaghie MC, Reedy M, Oliver T, Mishina Y, Hogan BL. Evidence that autocrine signaling through Bmpr1a regulates the proliferation, survival and morphogenetic behavior of distal lung epithelial cells. Dev Biol 2006;291:67–82. [DOI] [PubMed] [Google Scholar]
- 101.Jeannotte L, Aubin J, Bourque S, Lemieux M, Montaron S, Provencher St-Pierre A. Unsuspected effects of a lung-specific cre deleter mouse line. Genesis 2011;49:152–159. [DOI] [PubMed] [Google Scholar]
- 102.Chapman HA, Li X, Alexander JP, Brumwell A, Lorizio W, Tan K, Sonnenberg A, Wei Y, Vu TH. Integrin α6β4 identifies an adult distal lung epithelial population with regenerative potential in mice. J Clin Invest 2011;121:2855–2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Funaki H, Yamamoto T, Koyama Y, Kondo D, Yaoita E, Kawasaki K, Kobayashi H, Sawaguchi S, Abe H, Kihara I. Localization and expression of AQP5 in cornea, serous salivary glands, and pulmonary epithelial cells. Am J Physiol 1998;275:C1151–C1157. [DOI] [PubMed] [Google Scholar]
- 104.Raina S, Preston GM, Guggino WB, Agre P. Molecular cloning and characterization of an aquaporin cDNA from salivary, lacrimal, and respiratory tissues. J Biol Chem 1995;270:1908–1912. [DOI] [PubMed] [Google Scholar]
- 105.Krane CM, Fortner CN, Hand AR, McGraw DW, Lorenz JN, Wert SE, Towne JE, Paul RJ, Whitsett JA, Menon AG. Aquaporin 5-deficient mouse lungs are hyperresponsive to cholinergic stimulation. Proc Natl Acad Sci USA 2001;98:14114–14119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Flodby P, Borok Z, Banfalvi A, Zhou B, Gao D, Minoo P, Ann DK, Morrisey EE, Crandall ED. Directed expression of Cre in alveolar epithelial type 1 cells. Am J Respir Cell Mol Biol 2010;43:173–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L. A global double-fluorescent Cre reporter mouse. Genesis 2007;45:593–605. [DOI] [PubMed] [Google Scholar]
- 108.Farr AG, Berry ML, Kim A, Nelson AJ, Welch MP, Aruffo A. Characterization and cloning of a novel glycoprotein expressed by stromal cells in T-dependent areas of peripheral lymphoid tissues. J Exp Med 1992;176:1477–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Vanderbilt JN, Allen L, Gonzalez RF, Tigue Z, Edmondson J, Ansaldi D, Gillespie AM, Dobbs LG. Directed expression of transgenes to alveolar type I cells in the mouse. Am J Respir Cell Mol Biol 2008;39:253–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Li H, Cho SN, Evans CM, Dickey BF, Jeong JW, DeMayo FJ. Cre-mediated recombination in mouse Clara cells. Genesis 2008;46:300–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ji H, Houghton AM, Mariani TJ, Perera S, Kim CB, Padera R, Tonon G, McNamara K, Marconcini LA, Hezel A, et al. K-ras activation generates an inflammatory response in lung tumors. Oncogene 2006;25:2105–2112. [DOI] [PubMed] [Google Scholar]
- 112.Simon DM, Arikan MC, Srisuma S, Bhattacharya S, Andalcio T, Shapiro SD, Mariani TJ. Epithelial cell PPARgamma is an endogenous regulator of normal lung maturation and maintenance. Proc Am Thorac Soc 2006;3:510–511. [DOI] [PubMed] [Google Scholar]
- 113.Reynolds SD, Zemke AC, Giangreco A, Brockway BL, Teisanu RM, Drake JA, Mariani T, Di PY, Taketo MM, Stripp BR. Conditional stabilization of beta-catenin expands the pool of lung stem cells. Stem Cells 2008;26:1337–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Yang LT, Li WY, Kaartinen V. Tissue-specific expression of Cre recombinase from the Tgfb3 locus. Genesis 2008;46:112–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Tsao PN, Wei SC, Wu MF, Huang MT, Lin HY, Lee MC, Lin KM, Wang IJ, Kaartinen V, Yang LT, et al. Notch signaling prevents mucous metaplasia in mouse conducting airways during postnatal development. Development 2011;138:3533–3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Giangreco A, Arwert EN, Rosewell IR, Snyder J, Watt FM, Stripp BR. Stem cells are dispensable for lung homeostasis but restore airways after injury. Proc Natl Acad Sci USA 2009;106:9286–9291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Hong KU, Reynolds SD, Giangreco A, Hurley CM, Stripp BR. Clara cell secretory protein-expressing cells of the airway neuroepithelial body microenvironment include a label-retaining subset and are critical for epithelial renewal after progenitor cell depletion. Am J Respir Cell Mol Biol 2001;24:671–681. [DOI] [PubMed] [Google Scholar]
- 118.Blatt EN, Yan XH, Wuerffel MK, Hamilos DL, Brody SL. Forkhead transcription factor HFH-4 expression is temporally related to ciliogenesis. Am J Respir Cell Mol Biol 1999;21:168–176. [DOI] [PubMed] [Google Scholar]
- 119.Brody BL, Williams RA, Thomas RG, Kaplan RM, Chu RM, Brown SI. Age-related macular degeneration: a randomized clinical trial of a self-management intervention. Ann Behav Med 1999;21:322–329. [DOI] [PubMed] [Google Scholar]
- 120.Cruz C, Ribes V, Kutejova E, Cayuso J, Lawson V, Norris D, Stevens J, Davey M, Blight K, Bangs F, et al. Foxj1 regulates floor plate cilia architecture and modifies the response of cells to sonic hedgehog signalling. Development 2010;137:4271–4282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Yu X, Ng CP, Habacher H, Roy S. Foxj1 transcription factors are master regulators of the motile ciliogenic program. Nat Genet 2008;40:1445–1453. [DOI] [PubMed] [Google Scholar]
- 122.Ostrowski LE, Hutchins JR, Zakel K, O'Neal WK. Targeting expression of a transgene to the airway surface epithelium using a ciliated cell-specific promoter. Mol Ther 2003;8:637–645. [DOI] [PubMed] [Google Scholar]
- 123.Zhang Y, Huang G, Shornick LP, Roswit WT, Shipley JM, Brody SL, Holtzman MJ. A transgenic FOXJ1-Cre system for gene inactivation in ciliated epithelial cells. Am J Respir Cell Mol Biol 2007;36:515–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Rawlins EL, Hogan BL. Ciliated epithelial cell lifespan in the mouse trachea and lung. Am J Physiol Lung Cell Mol Physiol 2008;295:L231–L234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Turner J, Roger J, Fitau J, Combe D, Giddings J, Heeke GV, Jones CE. Goblet cells are derived from a FOXJ1-expressing progenitor in a human airway epithelium. Am J Respir Cell Mol Biol 2011;44:276–284. [DOI] [PubMed] [Google Scholar]
- 126.Rock JR, Randell SH, Hogan BL. Airway basal stem cells: a perspective on their roles in epithelial homeostasis and remodeling. Dis Model Mech 2010;3:545–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Cole BB, Smith RW, Jenkins KM, Graham BB, Reynolds PR, Reynolds SD. Tracheal basal cells: a facultative progenitor cell pool. Am J Pathol 2010;177:362–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Byrne C, Fuchs E. Probing keratinocyte and differentiation specificity of the human K5 promoter in vitro and in transgenic mice. Mol Cell Biol 1993;13:3176–3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Bruen KJ, Campbell CA, Schooler WG, deSerres S, Cairns BA, Hultman CS, Meyer AA, Randell SH. Real-time monitoring of keratin 5 expression during burn re-epithelialization. J Surg Res 2004;120:12–20. [DOI] [PubMed] [Google Scholar]
- 130.Rock JR, Hogan BL. Epithelial progenitor cells in lung development, maintenance, repair, and disease. Annu Rev Cell Dev Biol 2011;27:493–512. [DOI] [PubMed] [Google Scholar]
- 131.Ghosh M, Brechbuhl HM, Smith RW, Li B, Hicks DA, Titchner T, Runkle CM, Reynolds SD. Context-dependent differentiation of multipotential keratin 14-expressing tracheal basal cells. Am J Respir Cell Mol Biol 2011;45:403–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Vasioukhin V, Degenstein L, Wise B, Fuchs E. The magical touch: genome targeting in epidermal stem cells induced by tamoxifen application to mouse skin. Proc Natl Acad Sci USA 1999;96:8551–8556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Hong KU, Reynolds SD, Watkins S, Fuchs E, Stripp BR. Basal cells are a multipotent progenitor capable of renewing the bronchial epithelium. Am J Pathol 2004;164:577–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Xie W, Chow LT, Paterson AJ, Chin E, Kudlow JE. Conditional expression of the ErbB2 oncogene elicits reversible hyperplasia in stratified epithelia and up-regulation of TGFalpha expression in transgenic mice. Oncogene 1999;18:3593–3607. [DOI] [PubMed] [Google Scholar]
- 135.Ito T, Udaka N, Yazawa T, Okudela K, Hayashi H, Sudo T, Guillemot F, Kageyama R, Kitamura H. Basic helix-loop-helix transcription factors regulate the neuroendocrine differentiation of fetal mouse pulmonary epithelium. Development 2000;127:3913–3921. [DOI] [PubMed] [Google Scholar]
- 136.Linnoila RI, Jensen-Taubman S, Kazanjian A, Grimes HL. Loss of GFI1 impairs pulmonary neuroendorine cell proliferation, but the neuroendocrine phenotype has limited impact on post-naphthalene airway repair. Lab Invest 2007;87:336–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Wikenheiser-Brokamp KA. Rb family proteins differentially regulate distinct cell lineages during epithelial development. Development 2004;131:4299–4310. [DOI] [PubMed] [Google Scholar]
- 138.Neptune ER, Podowski M, Calvi C, Cho JH, Garcia JG, Tuder R, Linnoila RI, Tsai MJ, Dietz HC. Targeted disruption of NeuroD, a proneural basic helix-loop-helix factor, impairs distal lung formation and neuroendocrine morphology in the neonatal lung. J Biol Chem 2008;283:21160–21169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Johnston D, Hatzis D, Sunday ME. Expression of v-Ha-ras driven by the calcitonin/calcitonin gene-related peptide promoter: a novel transgenic murine model for medullary thyroid carcinoma. Oncogene 1998;16:167–177. [DOI] [PubMed] [Google Scholar]
- 140.Sutherland KD, Proost N, Brouns I, Adriaensen D, Song JY, Berns A. Cell of origin of small cell lung cancer: inactivation of Trp53 and Rb1 in distinct cell types of adult mouse lung. Cancer Cell 2011;19:754–764. [DOI] [PubMed] [Google Scholar]
- 141.Reynolds SD, Giangreco A, Power JH, Stripp BR. Neuroepithelial bodies of pulmonary airways serve as a reservoir of progenitor cells capable of epithelial regeneration. Am J Pathol 2000;156:269–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Singh SR, Billington CK, Sayers I, Hall IP. Can lineage-specific markers be identified to characterize mesenchyme-derived cell populations in the human airways? Am J Physiol Lung Cell Mol Physiol 2010;299:L169–L183. [DOI] [PubMed] [Google Scholar]
- 143.Quaggin SE, Schwartz L, Cui S, Igarashi P, Deimling J, Post M, Rossant J. The basic-helix-loop-helix protein pod1 is critically important for kidney and lung organogenesis. Development 1999;126:5771–5783. [DOI] [PubMed] [Google Scholar]
- 144.Chapman DL, Garvey N, Hancock S, Alexiou M, Agulnik SI, Gibson-Brown JJ, Cebra-Thomas J, Bollag RJ, Silver LM, Papaioannou VE. Expression of the T-box family genes, Tbx1-Tbx5, during early mouse development. Dev Dyn 1996;206:379–390. [DOI] [PubMed] [Google Scholar]
- 145.del Moral PM, De Langhe SP, Sala FG, Veltmaat JM, Tefft D, Wang K, Warburton D, Bellusci S. Differential role of FGF9 on epithelium and mesenchyme in mouse embryonic lung. Dev Biol 2006;293:77–89. [DOI] [PubMed] [Google Scholar]
- 146.Sgantzis N, Yiakouvaki A, Remboutsika E, Kontoyiannis DL. HuR controls lung branching morphogenesis and mesenchymal FGF networks. Dev Biol 2011;354:267–279. [DOI] [PubMed] [Google Scholar]
- 147.Yu K, Xu J, Liu Z, Sosic D, Shao J, Olson EN, Towler DA, Ornitz DM. Conditional inactivation of FGF receptor 2 reveals an essential role for FGF signaling in the regulation of osteoblast function and bone growth. Development 2003;130:3063–3074. [DOI] [PubMed] [Google Scholar]
- 148.Yin Y, White AC, Huh SH, Hilton MJ, Kanazawa H, Long F, Ornitz DM. An FGF-WNT gene regulatory network controls lung mesenchyme development. Dev Biol 2008;319:426–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Abler LL, Mansour SL, Sun X. Conditional gene inactivation reveals roles for Fgf10 and Fgfr2 in establishing a normal pattern of epithelial branching in the mouse lung. Dev Dyn 2009;238:1999–2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.De Langhe SP, Carraro G, Tefft D, Li C, Xu X, Chai Y, Minoo P, Hajihosseini MK, Drouin J, Kaartinen V, et al. Formation and differentiation of multiple mesenchymal lineages during lung development is regulated by beta-catenin signaling. PLoS ONE 2008;3:e1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Saga Y, Miyagawa-Tomita S, Takagi A, Kitajima S, Miyazaki J, Inoue T. MesP1 is expressed in the heart precursor cells and required for the formation of a single heart tube. Development 1999;126:3437–3447. [DOI] [PubMed] [Google Scholar]
- 152.Strutz F, Okada H, Lo CW, Danoff T, Carone RL, Tomaszewski JE, Neilson EG. Identification and characterization of a fibroblast marker: FSP1. J Cell Biol 1995;130:393–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Iwano M, Plieth D, Danoff TM, Xue C, Okada H, Neilson EG. Evidence that fibroblasts derive from epithelium during tissue fibrosis. J Clin Invest 2002;110:341–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Trimboli AJ, Fukino K, de Bruin A, Wei G, Shen L, Tanner SM, Creasap N, Rosol TJ, Robinson ML, Eng C, et al. Direct evidence for epithelial-mesenchymal transitions in breast cancer. Cancer Res 2008;68:937–945. [DOI] [PubMed] [Google Scholar]
- 155.Imai T, Jiang M, Chambon P, Metzger D. Impaired adipogenesis and lipolysis in the mouse upon selective ablation of the retinoid X receptor alpha mediated by a tamoxifen-inducible chimeric Cre recombinase (Cre-ERT2) in adipocytes. Proc Natl Acad Sci USA 2001;98:224–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.West J, Fagan K, Steudel W, Fouty B, Lane K, Harral J, Hoedt-Miller M, Tada Y, Ozimek J, Tuder R, et al. Pulmonary hypertension in transgenic mice expressing a dominant-negative BMPRII gene in smooth muscle. Circ Res 2004;94:1109–1114. [DOI] [PubMed] [Google Scholar]
- 157.Kuhbandner S, Brummer S, Metzger D, Chambon P, Hofmann F, Feil R. Temporally controlled somatic mutagenesis in smooth muscle. Genesis 2000;28:15–22. [DOI] [PubMed] [Google Scholar]
- 158.Owens GK, Kumar MS, Wamhoff BR. Molecular regulation of vascular smooth muscle cell differentiation in development and disease. Physiol Rev 2004;84:767–801. [DOI] [PubMed] [Google Scholar]
- 159.Woodcock-Mitchell J, Mitchell JJ, Low RB, Kieny M, Sengel P, Rubbia L, Skalli O, Jackson B, Gabbiani G. Alpha-smooth muscle actin is transiently expressed in embryonic rat cardiac and skeletal muscles. Differentiation 1988;39:161–166. [DOI] [PubMed] [Google Scholar]
- 160.Mitchell JJ, Reynolds SE, Leslie KO, Low RB, Woodcock-Mitchell J. Smooth muscle cell markers in developing rat lung. Am J Respir Cell Mol Biol 1990;3:515–523. [DOI] [PubMed] [Google Scholar]
- 161.Perl AK, Gale E. FGF signaling is required for myofibroblast differentiation during alveolar regeneration. Am J Physiol Lung Cell Mol Physiol 2009;297:L299–L308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Mitchell J, Woodcock-Mitchell J, Reynolds S, Low R, Leslie K, Adler K, Gabbiani G, Skalli O. Alpha-smooth muscle actin in parenchymal cells of bleomycin-injured rat lung. Lab Invest 1989;60:643–650. [PubMed] [Google Scholar]
- 163.Thet LA, Parra SC, Shelburne JD. Sequential changes in lung morphology during the repair of acute oxygen-induced lung injury in adult rats. Exp Lung Res 1986;11:209–228. [DOI] [PubMed] [Google Scholar]
- 164.Wu Z, Yang L, Cai L, Zhang M, Cheng X, Yang X, Xu J. Detection of epithelial to mesenchymal transition in airways of a bleomycin induced pulmonary fibrosis model derived from an alpha-smooth muscle actin-Cre transgenic mouse. Respir Res 2007;8:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Wendling O, Bornert JM, Chambon P, Metzger D. Efficient temporally-controlled targeted mutagenesis in smooth muscle cells of the adult mouse. Genesis 2009;47:14–18. [DOI] [PubMed] [Google Scholar]
- 166.Regan CP, Manabe I, Owens GK. Development of a smooth muscle-targeted cre recombinase mouse reveals novel insights regarding smooth muscle myosin heavy chain promoter regulation. Circ Res 2000;87:363–369. [DOI] [PubMed] [Google Scholar]
- 167.Xin HB, Deng KY, Rishniw M, Ji G, Kotlikoff MI. Smooth muscle expression of Cre recombinase and eGFP in transgenic mice. Physiol Genomics 2002;10:211–215. [DOI] [PubMed] [Google Scholar]
- 168.Wilm B, Ipenberg A, Hastie ND, Burch JB, Bader DM. The serosal mesothelium is a major source of smooth muscle cells of the gut vasculature. Development 2005;5317–5328. [DOI] [PubMed] [Google Scholar]
- 169.Que J, Wilm B, Hasegawa H, Wang F, Bader D, Hogan BL. Mesothelium contributes to vascular smooth muscle and mesenchyme during lung development. Proc Natl Acad Sci USA 2008;105:16626–16630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Morimoto M, Liu Z, Cheng HT, Winters N, Bader D, Kopan R. Canonical Notch signaling in the developing lung is required for determination of arterial smooth muscle cells and selection of Clara versus ciliated cell fate. J Cell Sci 2010;123:213–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Braren R, Hu H, Kim YH, Beggs HE, Reichardt LF, Wang R. Endothelial FAK is essential for vascular network stability, cell survival, and lamellipodial formation. J Cell Biol 2006;172:151–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Proctor JM, Zang K, Wang D, Wang R, Reichardt LF. Vascular development of the brain requires beta8 integrin expression in the neuroepithelium. J Neurosci 2005;25:9940–9948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Koni PA, Joshi SK, Temann UA, Olson D, Burkly L, Flavell RA. Conditional vascular cell adhesion molecule 1 deletion in mice: impaired lymphocyte migration to bone marrow. J Exp Med 2001;193:741–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Kisanuki YY, Hammer RE, Miyazaki J, Williams SC, Richardson JA, Yanagisawa M. Tie2-Cre transgenic mice: a new model for endothelial cell-lineage analysis in vivo. Dev Biol 2001;230:230–242. [DOI] [PubMed] [Google Scholar]
- 175.DasGupta R, Fuchs E. Multiple roles for activated LEF/TCF transcription complexes during hair follicle development and differentiation. Development 1999;126:4557–4568. [DOI] [PubMed] [Google Scholar]
- 176.Maretto S, Cordenonsi M, Dupont S, Braghetta P, Broccoli V, Hassan AB, Volpin D, Bressan GM, Piccolo S. Mapping Wnt/beta-catenin signaling during mouse development and in colorectal tumors. Proc Natl Acad Sci USA 2003;100:3299–3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Lohi M, Tucker AS, Sharpe PT. Expression of Axin2 indicates a role for canonical Wnt signaling in development of the crown and root during pre- and postnatal tooth development. Dev Dyn 2010;239:160–167. [DOI] [PubMed] [Google Scholar]
- 178.Al Alam D, Green M, Tabatabai Irani R, Parsa S, Danopoulos S, Sala FG, Branch J, El Agha E, Tiozzo C, Voswinckel R, et al. Contrasting expression of canonical Wnt signaling reporters TOPGAL, BATGAL and Axin2 during murine lung development and repair. PLoS ONE 2011;6:e23139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Ferrer-Vaquer A, Piliszek A, Tian G, Aho RJ, Dufort D, Hadjantonakis AK. A sensitive and bright single-cell resolution live imaging reporter of Wnt/ss-catenin signaling in the mouse. BMC Dev Biol 2010;10:121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Duncan AW, Rattis FM, DiMascio LN, Congdon KL, Pazianos G, Zhao C, Yoon K, Cook JM, Willert K, Gaiano N, et al. Integration of Notch and Wnt signaling in hematopoietic stem cell maintenance. Nat Immunol 2005;6:314–322. [DOI] [PubMed] [Google Scholar]
- 181.Mizutani K, Yoon K, Dang L, Tokunaga A, Gaiano N. Differential Notch signalling distinguishes neural stem cells from intermediate progenitors. Nature 2007;449:351–355. [DOI] [PubMed] [Google Scholar]
- 182.Rock JR, Gao X, Xue Y, Randell SH, Kong YY, Hogan BL. Notch-dependent differentiation of adult airway Basal stem cells. Cell Stem Cell 2011;8:639–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Vooijs M, Ong CT, Hadland B, Huppert S, Liu Z, Korving J, van den Born M, Stappenbeck T, Wu Y, Clevers H, et al. Mapping the consequence of Notch1 proteolysis in vivo with NIP-CRE. Development 2007;134:535–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Jackson EL, Willis N, Mercer K, Bronson RT, Crowley D, Montoya R, Jacks T, Tuveson DA. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev 2001;15:3243–3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Gregory LG, Mathie SA, Walker SA, Pegorier S, Jones CP, Lloyd CM. Overexpression of Smad2 drives house dust mite-mediated airway remodeling and airway hyperresponsiveness via activin and IL-25. Am J Respir Crit Care Med 2010;182:143–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Wunderlich S, Gruh I, Winkler ME, Beier J, Radtke K, Schmiedl A, Groos S, Haverich A, Martin U. Type II pneumocyte-restricted green fluorescent protein expression after lentiviral transduction of lung epithelial cells. Hum Gene Ther 2008;19:39–52. [DOI] [PubMed] [Google Scholar]
- 187.Hendrickson B, Senadheera D, Mishra S, Bui KC, Wang X, Chan B, Petersen D, Pepper K, Lutzko C. Development of lentiviral vectors with regulated respiratory epithelial expression in vivo. Am J Respir Cell Mol Biol 2007;37:414–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Liu X, Luo M, Guo C, Yan Z, Wang Y, Lei-Butters DC, Engelhardt JF. Analysis of adeno-associated virus progenitor cell transduction in mouse lung. Mol Ther 2009;17:285–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Lo B, Hansen S, Evans K, Heath JK, Wright JR. Alveolar epithelial type II cells induce T cell tolerance to specific antigen. J Immunol 2008;180:881–888. [DOI] [PubMed] [Google Scholar]
- 190.Malkoski SP, Cleaver TG, Lu SL, Lighthall JG, Wang XJ. Keratin promoter based gene manipulation in the murine conducting airway. Int J Biol Sci 2010;6:68–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Long MA, Rossi FM. Silencing inhibits Cre-mediated recombination of the Z/AP and Z/EG reporters in adult cells. PLoS ONE 2009;4:e5435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Beard C, Hochedlinger K, Plath K, Wutz A, Jaenisch R. Efficient method to generate single-copy transgenic mice by site-specific integration in embryonic stem cells. Genesis 2006;44:23–28. [DOI] [PubMed] [Google Scholar]
- 193.Sisson TH, Mendez M, Choi K, Subbotina N, Courey A, Cunningham A, Dave A, Engelhardt JF, Liu X, White ES, et al. Targeted injury of type II alveolar epithelial cells induces pulmonary fibrosis. Am J Respir Crit Care Med 2010;181:254–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Stanger BZ, Tanaka AJ, Melton DA. Organ size is limited by the number of embryonic progenitor cells in the pancreas but not the liver. Nature 2007;445:886–891. [DOI] [PubMed] [Google Scholar]