Abstract
DNA polymerase delta, whose catalytic subunit is encoded by POLD1, is responsible for lagging strand DNA synthesis during DNA replication1. It achieves this with high fidelity due to its intrinsic 3′ to 5′ exonuclease activity, which confers proofreading ability. Missense mutations in the exonuclease domain of POLD1 have recently been shown to predispose to colorectal and endometrial cancer2. Here we report a recurring heterozygous single amino acid deletion at the polymerase active site of POLD1 that abolishes DNA polymerase activity but only mildly impairs 3′ to 5′ exonuclease activity. This mutation causes a distinct multisystem disorder that includes subcutaneous lipodystrophy, deafness, mandibular hypoplasia and hypogonadism in males. This suggests that perturbation of function of the ubiquitously expressed POLD1 polymerase has surprisingly tissue-specific effects in man, and argues for an important role for POLD1 function in adipose tissue homeostasis.
Progressive loss of subcutaneous adipose tissue, or lipodystrophy, with concomitant severe insulin resistance is a major feature of the recently described mandibular hypoplasia, deafness, and progeroid features (MDP) syndrome3. Defining the genetic basis of this novel form of progressive partial lipodystrophy would give new insights into the regulation of adipose tissue. Several genetic causes of partial lipodystrophy have previously been described4. Some of these affect genes with firmly established roles in adipocyte differentiation or function, including PPARG, PLIN1 and CIDEC. However for other commonly implicated genes, including LMNA and ZMPSTE24, the link between the cellular defect and the adipose phenotype is poorly understood4. The mechanism underlying selective loss of only some adipose depots across all these conditions is also currently unclear.
As all reported MDP patients had unrelated parents and no other affected family members we hypothesised that the cause of the syndrome was a heterozygous de novo mutation in a single gene. We therefore performed exome sequencing on two MDP probands (Figure 1 and Supplementary Table 1) and their unaffected parents to look for de novo disease-causing mutations. Exonic sequences were enriched from genomic DNA using Agilent’s SureSelect Human All Exon kit (version 4) and then sequenced on an Illumina HiSeq2000 sequencer using 100bp paired end reads. We used BWA (v0.6.2)5 to align sequence reads to the hg19 reference genome and GATK (v2.2-10)6 to call SNVs and indels. Eighty-eight percent of targeted bases were covered at >20X.
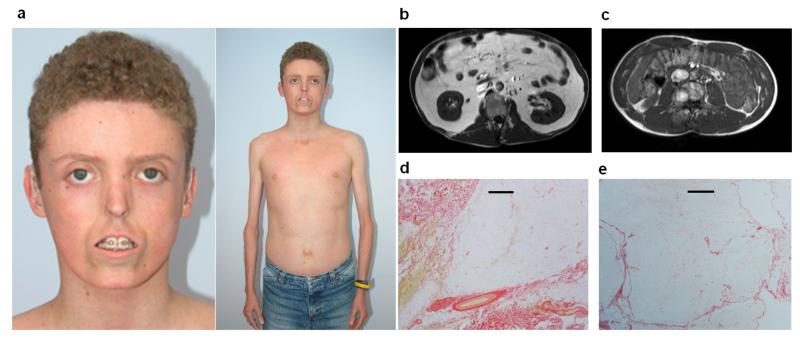
Figure 1. Clinical characteristics of patients (full details in Supplementary Table 1).
(a) Patient 1 (aged 15yrs), demonstrating prominent lipodystrophy, small nose, “pseudo”-proptosis, secondary to lack of subcutaneous periorbital fat, tight skin, mandibular hypoplasia, bilateral hearing aids and reduced limb muscles. Informed consent was obtained to use Patient 1’s photo (b and c) Abnormal fat distribution shown by abdominal MRI at level L3 from patient 1 (b) and an age, gender and BMI matched control (c) showing the striking reduction of subcutaneous fat and marked increase in intra-abdominal fat as demonstrated by an increase in high intensity signal. (d and e) Picrosirius red stain of an abdominal subcutaneous adipose tissue image of (d) patient 1 with S605del POLD1 mutation and (e) a representative comparative image of a BMI matched subject. The black scale bars = 500μm. Adipose tissue of the subject with POLD1 mutation shows a considerable amount of fibrosis (in red).
An in-frame deletion (c.1812_1814del, p.S605del) of a single serine residue in the polymerase active site of POLD1 was identified in both probands but in none of their four parents. These results were confirmed by Sanger sequencing (Supplementary Methods). This variant was not present in dbSNP137, the 1000 genomes project7 nor the exome variant server8. We then tested two additional patients with MDP syndrome by Sanger sequencing and identified the identical in-frame deletion in both patients. This provides overwhelming genetic evidence that S605del is the disease causing mutation in these four unrelated patients.
The most prominent phenotype of all four patients is lack of subcutaneous adipose tissue which was first noted in early childhood, although each proband was born with normal birth weight and appearance (Supplementary Table 1). The minimal subcutaneous adipose tissue (SAT) at all sites contrasted with the marked increase in visceral adipose tissue (VAT; Figure 1 and Supplementary Table 1). The VAT/SAT ratio was greatly elevated at 15.21 in patient 1 in contrast to values of a median of 0.84 (interquartile range 0.64–1.10) in 1,680 middle aged men9. All patients had clinical and biochemical evidence of insulin resistance despite having body mass index (BMI) values < 20kg/m2. Other common clinical features included skin scleroderma and telangiectasia, ligament contractures, reduced limb muscle mass, mandibular hypoplasia, hypogonadism and undescended testes in males, and sensorineural deafness (Supplementary Table 1). None of the patients in our relatively young series, nor in the previous description3, has been diagnosed with cancer.
DNA polymerase delta (Pol delta) has been shown to cooperate with WRN, a DNA helicase, to help maintain genome stability10. Loss of WRN function in humans leads to Werner syndrome, which presents with the premature onset of features associated with normal aging11. Werner syndrome includes progressive loss of limb adipose tissue and fat with severe insulin resistance, prominent joint contractures, scleroderma, hypogonadism and pinched nose, similar to MDP syndrome. However, MDP syndrome patients lack several key signs of Werner syndrome including short stature, premature balding/greying and cataracts (Supplementary Table 1 and Werner Syndrome GeneReview11). Similarly there are clear differences from the progeroid syndrome mandibular acral dysplasia (MAD) caused by mutations in LMNA12 or ZMPSTE2413 as MDP patients do not have short stature, premature hair loss, clavicle hypoplasia or acroosteolysis and MAD patients do not have hearing loss.
DNA replication is fundamental to successful division of all nucleated cells, and so POLD1, which encodes the catalytic subunit of the key, processive lagging strand polymerase, serves a ubiquitous and critical cellular function1. Correspondingly, knockout of the mouse orthologue does not produce viable animals14. The tissue-specific effect of the heterozygous deletion mutation we have found is thus a surprise. POLD1 mRNA is well expressed across a panel of human tissues, including muscle and fat, but differential expression does not account for the phenotype (Supplementary Figure 1). Moreover, there is no evidence in the commonly used murine 3T3-L1 model of adipogenesis that POLD1 expression changes during differentiation of pre-adipocytes to fat cells (Supplementary Figure 2).
Subcutaneous abdominal adipose tissue of patient 1 (BMI 19.1kg/m2) shows abundant fibrosis dividing the remaining fat tissue, but no obvious inflammatory cell infiltrates (Figure 1 and Supplementary Figure 3). Consistent with this, expression of key extracellular matrix genes was markedly different from that of five lean controls (BMI 20-21.8 kg/m2), with increased expression of Transforming Growth Factor beta (TGFB), a hallmark of tissue fibrosis and a strikingly higher expression of fibronectin (Supplementary Figure 4). Increased fibrosis has been correlated with adipose tissue dysfunction and insulin resistance in a variety of settings, including patients with lipodystrophy due to PLIN1 mutations15 and common obesity16.
The S605del mutation occurs in motif A, a highly conserved region of the Pol delta catalytic subunit. This motif comprises 11 amino acids that are involved in the alignment of the incoming complementary deoxynucleotide (dNTP) with the primer terminus, as well as coordination and catalysis of phosphodiester bond formation17. Alterations in this domain have been shown to affect base selection, and, consequently, polymerase fidelity, in nearly every family of DNA polymerase from viruses to mammals18,19. In silico modelling predicted that the S605 deletion profoundly affects the structure of the polymerase active site, in particular by disrupting the hydrogen bonding between the substrate trinucleotide and the catalytic aspartate residues. The proofreading domain is not predicted to be affected (in contrast to the previously reported cancer predisposing mutations2; Figure 2 and Supplementary Methods). We therefore hypothesised that the S605del mutant polymerase would be able to bind DNA, but not catalyse polymerisation.
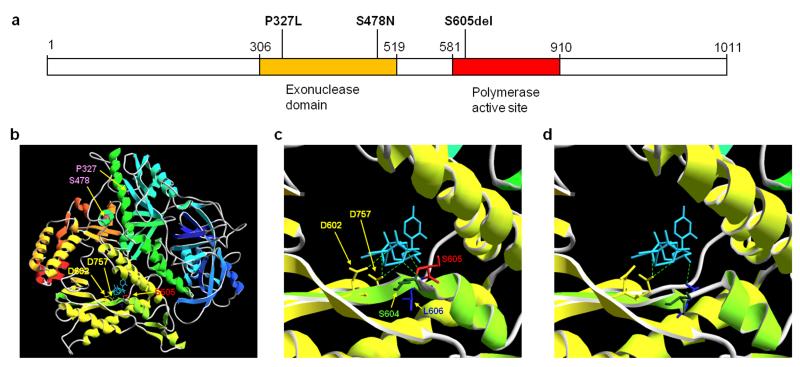
Figure 2. Gene Schematic and mutational modelling of POLD1 mutations.
(a) Gene Schematic of POLD1. Mutations predisposing to cancer (P327L and S478N) affect residues within the exonuclease (proofreading) domain whereas the S605del (found in all our 4 patients) is within the polymerase active site. (b) Global structure of POLD1 and sites of relevant mutations. The structure of POLD1 (residues 77-983, modelled on template 3iayA) is shown in ribbon form, coloured by secondary structure succession (blue, N-terminal, to red, C-terminal). Backbone and sidechains are displayed in stick format for residues P327 and S478 (pink), D602 and D757 (yellow), S604 (dark green), S605 (red) and L606 (blue), as indicated. The trinucleotide substrate is shown in stick format (light blue), with hydrogen bonds to the polymerase catalytic site indicated by green broken lines. Template DNA has been omitted for clarity. (c and d) Detail of the polymerase catalytic site, shown from the same viewpoint as in (b); (c) wild type POLD1, and (d) mutant S605del. Note the predicted deformation of the catalytic domain and loss of hydrogen bonding to substrate in S605del compared to normal POLD1; in contrast, the predicted structure of the catalytic site, and bonding to the trinucleotide substrate, was identical to normal POLD1 for both P327L and S478N mutants (not shown).
To test this hypothesis we generated the S605 deletion mutant by PCR mutagenesis, and purified the four-subunit human DNA Pol delta (S605del) holoenzyme from E. coli (Supplementary Methods). Polymerase activity of the purified enzyme was quantified by measuring primer elongation on a 23/46 primer/template duplex in the presence of all four dNTPs (Figure 3a). Under conditions in which wild type Pol delta (150 fmol) extended 81% of the primer, DNA extension with the same concentration of the mutant enzyme was not detected.
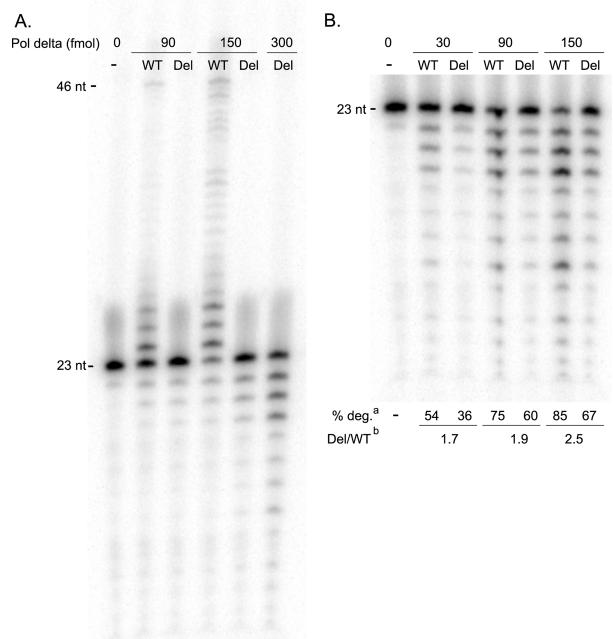
Figure 3. Pol delta S605del has no detectable polymerase activity, but robust exonuclease activity.
(a) Polymerase reactions using increasing concentrations of either wild type (WT) or S605del (Del) Pol delta reveal that the mutant is incapable of extending the primer band (23 nt) in a 2 minute reaction with 25 μM dNTPs. (b) Exonuclease reactions on the same 23/46 primed duplex DNA, in the absence of dNTPs, show that S605del exhibits a 2- to 3-fold decrease in exonuclease activity when compared to wild type. aValues for percent of primer band degraded (% deg.) reflect three independent experiments, and are derived from a comparison of the intensity of the primer band (P) and the exonuclease product bands (E), given by the following equation: % deg = 1-(P/(P+E)). bThe % degradation values of Pol delta-S605del at 90, 150, and 300 fmol were plotted against the line obtained from measurements with the wild type polymerase (y=0.0029+0.445; R2=0.96). The values derived indicate that ~1.7 to 2.5-fold more mutant enzyme is required to observe the same amount of primer degradation as the wild type enzyme under these experimental conditions.
To assess the exonuclease activity of wild type and S605del Pol delta, the extension assay was adapted by excluding dNTPs. The exonuclease activity of the wild type enzyme was linear over three concentrations (30, 90, and 150 fmol), and a comparison of the amount of primer degradation at different concentrations (90, 150, and 300 fmol) of mutant enzyme against this linear range revealed that Pol delta-S605del exhibits 1.7- to 2.5-fold lower exonuclease activity than wild type (Figure 3b). Despite this difference, the exonuclease activity of the mutant Pol delta enzyme is considerable. Collectively these findings demonstrate decoupling of the enzymatic activities of the mutant enzyme. Its preserved exonuclease activity indicates that it can bind DNA, and suggests that the inability to extend DNA primers is due to an inability to interact with and incorporate dNTPs efficiently.
These results provide a possible molecular explanation for the strikingly different phenotype caused by the S605 deletion compared to the previously described cancer predisposition mutations2. Whereas the cancer mutations occur in the proofreading domain and lead to increased base substitution error rates2, our results show that Pol delta-S605del can bind to DNA but not catalyze polymerization. This is likely to result in an increase in stalled replication forks, which are associated with double stranded DNA breaks, genomic instability, cell cycle checkpoint response and cell senescence and death20. Preserved DNA binding by the mutant also raises the possibility that it may be able to compete with co-expressed wild type polymerase, and previous work in yeast has shown that a reduction in the levels of DNA polymerase delta leads to an increase in deletions and genomic instability21,22. However, further work is required to understand how this mutation leads to the specific phenotype observed in these patients.
There is some support for distinct phenotypes caused by mutations in different POLD1 domains in mouse models. A homozygous D400A mutation in the proofreading domain of Pold1 led to a 10-fold increase in base-substitution error rate and 94% of mice developed cancer by the age of 18 months23. In contrast, a heterozygous L604K substitution in mice (equivalent to human L606K and neighbouring the S605 residue deleted in MDP syndrome patients) reduced life span by 18%, led to a small increase in tumorigenesis and a clear increase in chromosome aberrations19. Subsequent work suggested that this phenotype is due to an increase in stalled replication forks, particularly at sites of DNA damage24. Lipodystrophy was not reported in L604K mice; however, it has proved very challenging to replicate other forms of partial lipodystrophy in mice, which may require extreme caloric loads to manifest metabolic disease25.
In conclusion, whereas missense mutations in the proofreading domain of POLD1 predispose to cancer, our data show that a single amino acid deletion in the polymerase active site of POLD1 causes a markedly different phenotype including lipodystrophy. We show that the mutation results in complete loss of polymerase activity, but very little loss of exonuclease capacity. In addition to aiding in understanding the basis of a complex syndrome, this protein could serve as an important tool to explore more fundamental aspects of DNA polymerase biology.
Online Methods
Sanger sequencing confirmation
We amplified POLD1 exon 15 using primers shown in Supplementary Table 2. PCR products were sequenced on an ABI3730 capillary machine (Applied Biosystems, Warrington, UK) and analyzed using Mutation Surveyor v3.98 (SoftGenetics, Pennsylvania, USA).
Protein Modelling
The sequence of full-length POLD1 was submitted for automated modelling and template selection using the SWISS-MODEL web interface (http://swissmodel.expasy.org/). This returned a structure for POLD1 residues 77-983 based on template 3iay, chain A (S. cerevisiae DNA pol delta, catalytic subunit; 49% sequence identity), which is the same template used for the model of human POLD1 in the SWISS-MODEL Repository (entry P28340). Subsequently, all modelling of mutant sequences was carried out using template 3iayA, and structures visualized in Swiss-PdbViewer.
Human Tissue Expression Profiling
A human tissue whole RNA panel was purchased from AMS biotechnology Abingdon, and was provided by Dr. Loraine Tung, University of Cambridge, UK. cDNA synthesis was undertaken using the ImProm-IITM Reverse Transcription System (Promega) according to the manufacturer’s recommendation. Human POLD1 mRNA and 18S rRNA levels were determined using TaqMan® Gene Expression assays (Hs01100821_m1 for POLD1 and custom designed for 18S (sequences available on request); both Applied Biosystems). Serially diluted human cDNA was used to generate a standard curve for each assay, and all RNA determinations were undertaken in duplicate using TaqMan® PCR Master Mix on an ABI 7900 detection system (both Applied Biosystems). POLD1 mRNA expression levels were finally normalised to levels of 18S rRNA for comparison among tissues.
3T3-L1 Differentiation Timecourse
3T3-L1 cells (American Tissue Culture Collection) were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% neonatal calf serum, 100U/l penicillin, 100μg/ml streptomycin, 4mM L-glutamine and 4 mg/ml puromycin (Sigma, St Louis, MO, USA) in a humidified 95% air and 5% CO2 at 37°C incubator. Preadipocyte differentiation was undertaken by growing cells to 2 days post confluence followed by exposure to medium in which foetal bovine serum replaced neonatal calf serum, and which was supplemented for the first three days with 500μM IBMX 1 μM dexamethasone (all Sigma-Aldrich) and 100nmol/l soluble human insulin (Novo Nordisk, Bagsvard), and for the next three days with 100nM insulin only, with no supplements thereafter. Total RNA was extracted using an RNeasy mini kit (Qiagen) at time points indicated. mRNA levels of Pold1 were determined using an inventoried TaqMan® Gene Expression assays (Hs01100821_m1; Applied Biosystems), while levels of Adipoq, and cyclophilin A were determined using custom designed TaqMan® Gene Expression assays (sequences available on request). qRT-PCR assays were undertaken as described above, and levels of genes of interest were normalised to those of cyclophilin A for comparison. Three time courses were undertaken in duplicate. Results show mean levels and standard errors of the mean from these biological replicates.
Expression and fibrosis studies from fat
Abdominal subcutaneous adipose tissue biopsies were obtained from patient 1 (BMI 19.1kg/m2) from below and lateral to the umbilicus and five healthy, lean Caucasian controls (BMI range 20-21.8 kg/m2) obtained during elective surgery and with consent and ethical approval from the NIHR Exeter Tissue Bank. Biopsies were immediately snap frozen in liquid nitrogen and stored at −80°C until processing. Total RNA was extracted using the phenol-chloroform-guanidinium-thiocyanate method26. Briefly ~100mg tissue was homogenised in Tri reagent (Life Technologies, Paisley, UK) using a Retsch Mixer Mill MM400 in accordance with manufacturer’s instructions before quantification was determined spectrophotometrically using Nanodrop technology. RNA was DNase treated to remove residual genomic contamination before 500ng was reverse transcribed in a random primed single strand synthesis reaction using high capacity reverse transcriptase (Life Technologies, Paisley, UK).
cDNA was diluted 1:20 in 0.01M Tris HCl before amplification was performed using Universal PCR mastermix (Life Technologies, Paisley, UK) and extracellular matrix gene specific probes (probe sequences available on request). All samples were amplified in triplicate for each assay and were run alongside a standard curve to allow assay efficiency determination. Samples amplified in the absence of the reverse transcriptase enzyme and samples without template were used as negative controls. Amplification was performed using an ABI7900 thermal cycler with the following cycling parameters: 50°C for 2minutes, 95°C-10 minutes, 40 cycles(95°C for 15 seconds,60°C for 1 minute). Gene expression levels were calculated using the 2^ΔΔCt analysis method modified by Pfaffl27, normalising to the adipose specific housekeeping genes PPIA and UBC28.
For the fibrosis studies, the fat tissue biopsy from patient 1, a lean individual (BMI=20kg/m2) and an obese individual (BMI=43.6kg/m2) obtained during elective surgery were formalin fixed, paraffin-embedded and stained with picrosirius red (Sigma Aldrich, UK) for fibrosis as previously described16.
MRI imaging
Abdominal images were collected via a MRI T1 (spin-lattice relaxation time) weighted breath-hold sequence in order to obtain images with high fat contrast. Subcutaneous and visceral fat volumes were calculated via semi-automated processing based upon signal intensity thresholding between the L5/S1 disk and the bottom of the lung.
Site Directed Mutagenesis
The 3-nucleotide deletion was introduced into the pET303-hpold1 plasmid29 by using 5′ phosphorylated primers flanking the deletion site (Supplementary Table 3). Products of the PCR were gel purified and ligated prior to transformation into XL 10 Gold Ultracompetent Cells (Stratagene). A region between the Age1 and Cla1 sites of POLD1 was amplified and sequence-verified, and then subcloned back into Age1/Cla1-digested pET303-hpold1 vector that had not been PCR amplified.
Purification of Pol delta S605del
The mutant Pol delta holoenzyme was purified as described30. Briefly, the pET303-hpold1 plasmid and pCOLA-2,3,4, harboring the p66, p55, and p12 subunits of the Pol delta holoenzyme, were co-transformed into BL21-tRARE cells by electroporation. Protein expression was induced in mid-log cells by the addition of β-D-1-thiogalactopyranoside (IPTG). Following sonication, the crude extract was subject to nickel column (Novagen) chromatography by binding via the polyhistidine tag on the P12 subunit. Bound enzyme was eluted with 300mM imidazole, and was further purified by ion exchange chromatography on a HiTrap SP HP column (GE) using a linear (0.2-1.0 M) NaCl gradient.
Polymerase and Exonuclease Assays
The polymerase and exonuclease activities of wild type and S605del Pol delta were measured using the same reaction mixtures and enzyme dilutions. Reactions included indicated concentrations of enzyme, 40 mM Tris-HCl (pH 7.4), 4 mM MgCl2, 0.1 mg/ml bovine serum albumin, 5 mM dithiothreitol, 25 μM of each dNTP (polymerase reactions only), and 10 nM 5′-end labeled primer-template DNA duplex (Supplementary Table 4). The reactions were incubated for 2 minutes at 37°C, and stopped by the addition of 95% formamide/20mM EDTA. Samples were resolved on a 14% polyacrylamide TBE-urea gel; reaction products were visualized following imaging on a PhosphorImager (Molecular Dynamics), and analyzed using ImageJ64 software (http://imagej.nih.gov/ij).
Supplementary Material
Acknowledgements
This work was supported by NIHR Exeter Clinical Research Facility through funding for SE and ATH and general infrastructure. The authors thank Michael Day, Annet Damhuis and Richard Gilbert for technical assistance. We thank Karen Knapp for providing the data for the DEXA calculations. SE, ATH, SO are supported by Wellcome Trust Senior Investigator awards. DS and RKS (098498/Z/12/Z) are supported by Wellcome Trust Senior Research Fellowships in Clinical Science. MNW is supported by the Wellcome Trust as part of the WT Biomedical Informatics Hub funding. RO is supported by Diabetes UK. DS, RKS and SO are supported by the UK National Institute for Health Research (NIHR) Cambridge Biomedical Research Centre. KJG is supported by the Agency for Science, Technology and Research, Singapore (A*STAR). LAL and MJP are supported by grants NCI-61-6845 and 62-4860. The opinions in this paper are those of the authors and do not necessarily represent the views of the funders or the department of health.
Footnotes
Accession Codes NCBI reference sequence: NM_002691.2
Protein Data Bank: 3iay
Competing Financial Interests The authors declare no competing financial interests
References
- 1.Prindle MJ, Loeb LA. DNA polymerase delta in DNA replication and genome maintenance. Environmental and molecular mutagenesis. 2012;53:666–82. doi: 10.1002/em.21745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Palles C, et al. Germline mutations affecting the proofreading domains of POLE and POLD1 predispose to colorectal adenomas and carcinomas. Nature genetics. 2012;45:136–144. doi: 10.1038/ng.2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shastry S, et al. A novel syndrome of mandibular hypoplasia, deafness, and progeroid features associated with lipodystrophy, undescended testes, and male hypogonadism. The Journal of clinical endocrinology and metabolism. 2010;95:E192–7. doi: 10.1210/jc.2010-0419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Semple RK, Savage DB, Cochran EK, Gorden P, O’Rahilly S. Genetic syndromes of severe insulin resistance. Endocrine reviews. 2011;32:498–514. doi: 10.1210/er.2010-0020. [DOI] [PubMed] [Google Scholar]
- 5.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–60. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McKenna A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome research. 2010;20:1297–303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abecasis GR, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fu W, et al. Analysis of 6,515 exomes reveals the recent origin of most human proteincoding variants. Nature. 2013;493:216–20. doi: 10.1038/nature11690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaess BM, et al. The ratio of visceral to subcutaneous fat, a metric of body fat distribution, is a unique correlate of cardiometabolic risk. Diabetologia. 2012;55:2622–30. doi: 10.1007/s00125-012-2639-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kamath-Loeb AS, Shen JC, Schmitt MW, Loeb LA. The Werner syndrome exonuclease facilitates DNA degradation and high fidelity DNA polymerization by human DNA polymerase delta. The Journal of biological chemistry. 2012;287:12480–90. doi: 10.1074/jbc.M111.332577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oshima J, Martin GM, Hisama FM. Werner Syndrome. In: Pagon RA, Bird TD, Dolan CR, et al., editors. GeneReviews™ [Internet] University of Washington; Seattle (WA): [Updated 2012 Dec 13]. Dec 2, 2002. Seattle; 1993-. Available from: http://www.ncbi.nlm.nih.gov/books/NBK1514/. [Google Scholar]
- 12.Novelli G, et al. Mandibuloacral dysplasia is caused by a mutation in LMNA-encoding lamin A/C. American journal of human genetics. 2002;71:426–31. doi: 10.1086/341908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Agarwal AK, Fryns JP, Auchus RJ, Garg A. Zinc metalloproteinase, ZMPSTE24, is mutated in mandibuloacral dysplasia. Human molecular genetics. 2003;12:1995–2001. doi: 10.1093/hmg/ddg213. [DOI] [PubMed] [Google Scholar]
- 14.Uchimura A, Hidaka Y, Hirabayashi T, Hirabayashi M, Yagi T. DNA polymerase delta is required for early mammalian embryogenesis. PloS one. 2009;4:e4184. doi: 10.1371/journal.pone.0004184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gandotra S, et al. Perilipin deficiency and autosomal dominant partial lipodystrophy. The New England journal of medicine. 2011;364:740–8. doi: 10.1056/NEJMoa1007487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Henegar C, et al. Adipose tissue transcriptomic signature highlights the pathological relevance of extracellular matrix in human obesity. Genome biology. 2008;9:R14. doi: 10.1186/gb-2008-9-1-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Swan MK, Johnson RE, Prakash L, Prakash S, Aggarwal AK. Structural basis of high-fidelity DNA synthesis by yeast DNA polymerase delta. Nature structural & molecular biology. 2009;16:979–86. doi: 10.1038/nsmb.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reha-Krantz LJ, Nonay RL. Motif A of bacteriophage T4 DNA polymerase: role in primer extension and DNA replication fidelity. Isolation of new antimutator and mutator DNA polymerases. The Journal of biological chemistry. 1994;269:5635–43. [PubMed] [Google Scholar]
- 19.Venkatesan RN, et al. Mutation at the polymerase active site of mouse DNA polymerase delta increases genomic instability and accelerates tumorigenesis. Molecular and cellular biology. 2007;27:7669–82. doi: 10.1128/MCB.00002-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Branzei D, Foiani M. Maintaining genome stability at the replication fork. Nature reviews. Molecular cell biology. 2010;11:208–19. doi: 10.1038/nrm2852. [DOI] [PubMed] [Google Scholar]
- 21.Kokoska RJ, Stefanovic L, DeMai J, Petes TD. Increased rates of genomic deletions generated by mutations in the yeast gene encoding DNA polymerase delta or by decreases in the cellular levels of DNA polymerase delta. Molecular and cellular biology. 2000;20:7490–504. doi: 10.1128/mcb.20.20.7490-7504.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lemoine FJ, Degtyareva NP, Kokoska RJ, Petes TD. Reduced levels of DNA polymerase delta induce chromosome fragile site instability in yeast. Molecular and cellular biology. 2008;28:5359–68. doi: 10.1128/MCB.02084-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goldsby RE, et al. High incidence of epithelial cancers in mice deficient for DNA polymerase delta proofreading. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:15560–5. doi: 10.1073/pnas.232340999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schmitt MW, et al. Active site mutations in mammalian DNA polymerase delta alter accuracy and replication fork progression. The Journal of biological chemistry. 2010;285:32264–72. doi: 10.1074/jbc.M110.147017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Savage DB. Mouse models of inherited lipodystrophy. Disease models & mechanisms. 2009;2:554–62. doi: 10.1242/dmm.002907. [DOI] [PubMed] [Google Scholar]
- 26.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Analytical biochemistry. 1987;162:156–9. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 27.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic acids research. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Neville MJ, Collins JM, Gloyn AL, McCarthy MI, Karpe F. Comprehensive human adipose tissue mRNA and microRNA endogenous control selection for quantitative real-time-PCR normalization. Obesity. 2011;19:888–92. doi: 10.1038/oby.2010.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fazlieva R, et al. Proofreading exonuclease activity of human DNA polymerase delta and its effects on lesion-bypass DNA synthesis. Nucleic acids research. 2009;37:2854–66. doi: 10.1093/nar/gkp155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schmitt MW, Matsumoto Y, Loeb LA. High fidelity and lesion bypass capability of human DNA polymerase delta. Biochimie. 2009;91:1163–72. doi: 10.1016/j.biochi.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.