Introduction
Molecular machines are workshops in the cell that bring molecules and substrates together in a coordinated, processive way.1–2. The molecular complexes acting as molecular machines are often spatially localized and assembled in an ad-hoc fashion, in response to the demands of the cell’s metabolism. Products of such binding reactions can be RNA or DNA polymers, ATP, or other compounds needed at the site. Well-known examples are RNA and DNA polymerases, ATP synthase, the ribosome, and the proteasome. Visualizing molecular machines in their various states of processing poses challenges which traditional techniques of structure research find hard to meet. The complexes formed in the course of the work cycle are typically large, relatively unstable, and flexible -- all properties that disfavor X-ray crystallography as a means of visualization. In addition, molecular machines can have multiple binding partners and can go through numerous states, characterized by different conformations and binding configurations. Purification of such complexes can be done through two different routes: either from a cell extract or from an in vitro reconstituted system. In either case, it is difficult to isolate the complex by biochemical means in one of its many states with the purity required for structural research.
Cryo-electron microscopy (cryo-EM) is a fairly new technique for obtaining a 3D image of a molecule from a large set of projections, harvested from micrographs of a field of molecules captured in random orientations.3 In some cases, resolutions in the range of 4 to 5.5 Å have already been achieved for the ribosome, an RNA-containing molecular machine.4–6 Purity of the sample in terms of conformation and binding state of the molecule is evidently an absolute requirement for obtaining a meaningful 3D image from the “single particle” projection data set. In certain cases, high purity can be achieved by affinity imaging techniques.7–9 However, because of inherent difficulties of finding biochemical methods that guarantee 100% purity, it has been a goal in the development of cryo-EM to accomplish the purification of the molecules after the imaging, at the data processing step. Cryo-EM projection data have a typical signal-to-noise ratio in the order of 0.1 or even less.10 The challenge posed by classification of such noisy data is compounded by the fact that variability stemming from a change in viewing direction is intermingled with variability due to heterogeneity of composition or conformation.
Due to recent advances in cryo-EM, particularly in the methods of data analysis, it has indeed recently become possible to perform the post-imaging purification of a heterogeneous sample: by using new methods of classification, images can be divided into homogeneous classes, and thus separate reconstructions can be obtained for each of the states of a molecular machine. Since an entire spectrum of states becomes observable in a single sample, all connected by pathways along which the molecule’s trajectory unfolds, we may figuratively speak of a “Story in the Sample.”11
The following considerations were inspired by recent results in the study of the ribosome by both cryo-EM and single-molecule fluorescence resonance energy transfer techniques (smFRET),12–14 which invite a generalization to other systems, and lead to an appreciation of the potential these new techniques will have in exploring functionally relevant states of molecular machines. Cryo-EM shares with smFRET the unique property that it allows tracking of the signal coming from a single molecule and at the same time gives robust ensemble averages from thousands of molecules tracked. There is a sense that cryo-EM as a technique of structural biology is at an important juncture where application of concepts of statistical mechanics has become feasible and in all likelihood quite productive.15
Molecular Machines – The Energy Landscape, and States Observable by Cryo-EM
In the past decade, the energy landscape concept, which already proved indispensible in the analysis of protein folding,16 has been extended to describe the functional dynamics of macromolecules equilibrating among multiple states.17–18 For simplicity, a one-dimensional example is given in Fig. 1. All states represented by points on the “surface” of the landscape are in principle accessible, provided that a sufficient amount of activation energy is supplied from the thermal environment or other sources. Only local minima (or basins) in this landscape characterize conformational states having relative stability, while all other points represent unstable states which are visited only transiently.
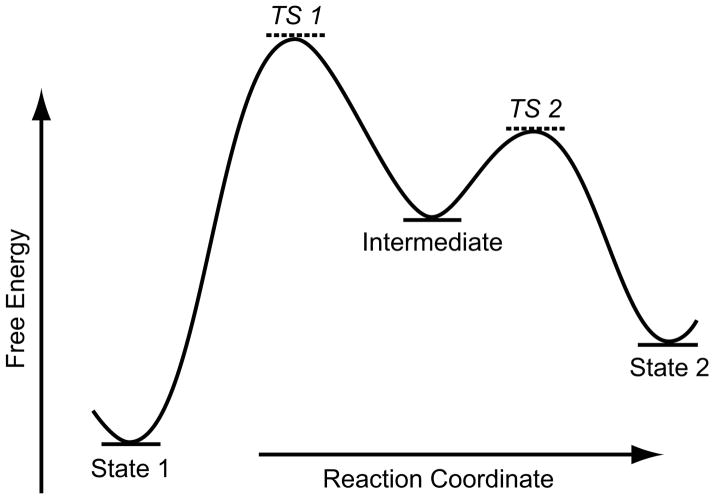
Fig. 1.
Schematic diagram of (one-dimensional) energy landscape. (It depicts either a representative slice through the conformational coordinate or an average of the conformational coordinate). Three local minima (“basins”) denoted by State 1, Intermediate, and State 2 are separated by maxima (“hills”). Sizable subpopulations of molecules will only be found in the basins (indicated by fill in each basin), signifying states of relative stability. Points TS1 and TS2 mark lowly populated labile transition states which interconvert to either of the adjacent more stable states.
We assume the existence of a population of molecules freely equilibrating in solution, which -- following the protocol of single-particle cryo-EM -- is rapidly immobilized by flash-freezing. Only those molecules which are trapped in one of the local minima of the free energy landscape will exist in numbers large enough to allow the formation of an informative, statistically significant 3D image. We may visualize this situation by having the basins being filled, to different extents, with molecules in a very similar energetic state. (The extent to which the individual basins are filled is related to their height – we come back to this below). All other molecules are in transit, with occupancies (= the number of molecules per increment of the reaction coordinate) so small that they defy attempts at visualization. This means that of the whole continuum of states on the energy landscape, only a finite number of discrete states are actually “observable,” each in the form of a three-dimensional map formed from an ensemble of molecules trapped in the same state.
This idealized “forward” model of the molecule occurring in a set of discrete states, each to be characterized by a separate 3D structure, should be juxtaposed with the experimental reality in cryo-EM which starts out with a large set of projection images showing the molecule in an unknown number of states and in lying in unknown orientations.
Formally, if the imaged portion of a given sample contains a total of N molecules, K subpopulations N1, N2, …, NK are in different, freely equilibrating states with distinct conformations. Single-particle imaging of such a population creates a set of N projections with K classes. Ideally, a classification program should divide the images into K classes on the grounds of the structural differences detectable in the images. In that case we will be able to compute a three-dimensional density map Dk for each of the classes, depicting the molecule in one of its K states. We will call the set of K density maps {Dk, k = 1 … K} a structural inventory of the sample.
Here we face the complication that the number K is unknown whereas classification programs such as ML3D19 or RELION20 require it to be known, and even to be given as input at the start, as the number of “seeds.” Moreover, the structural differences between some of the states may be so small that they escape the scrutiny of the classification program, due to the low signal-to-noise ratio of the data. Therefore the structural inventory is most often merely an approximation of the actual number of functional states. In practice, the actual number of classes K is first estimated on the basis of what is known about the system, but then some greater number K′ > K is chosen to prevent underestimation, which would have the detrimental effect of artificially merging closely related classes.
An example is provided by the case of the ribosome in the pre-translocational set of states. After peptidyl transfer, the ribosome which is bound with mRNA and two tRNAs visits several states characterized structurally by different intersubunit rotations and tRNA configurations.14, 21–27 The fact that these different intersubunit rotations and tRNA configurations can be structurally characterized can be interpreted as arising from the presence of shallow basins in the energy landscape (Fischer et al., 2010), a consequence of an architecture of the ribosome that facilitates intersubunit rotation in the thermal bath, a conformational rearrangement that is required in the first phase of the two-phase translocation process.28
Following Fischer and coworkers, we can relate the number of molecules in a given state, which is reported by the classification program as the number Nk of molecules in a class k, to the energy of that state by the Stefan-Boltzmann equation (Fischer et al., 2010; Agirrezabala et al., 2012): ΔG = −RT ln(C), where ΔG is the standard free-energy difference, R is the universal gas constant, and C is the equilibrium constant. The Boltzmann constant, b, is substituted for R (since b = R/A, where A is Avogadro’s number; i.e., b can be regarded as the gas constant for a single molecule). We rearrange the equation to obtain ΔG/bT = −ln(C). The equilibrium constant Ck for any class can be calculated as Nk/N0, where Nk is the number of particles in a particular class k, and N0 is the number of particles in the most populated class. Thus the energetic stability of each class relative to the most stable class may be obtained. In other words, the relative energies in the minima of the free-energy landscape can be calculated from a single, sufficiently large cryo-EM data set, and they are given by the proportion of molecules in each class relative to the most populated class.
The differences in the conformations observed by cryo-EM in the same sample are an indication of the molecule’s dynamic behavior. In the next section we will ask the question how to get beyond this fragmentary information, and how the detailed dynamics of the molecule can be obtained.
Dynamics
In a nutshell, what has been said in the section above implies both potential and limitations of cryo-EM. Its potential lies in the fact that multiple states, corresponding to the local energy minima, may be simultaneously recovered from the same sample – in other words, the structural inventory introduced before. The limitations relate to the transitions among the “basin” states: it must be emphasized that information about these transitions is entirely out of reach for cryo-EM.
Time-resolved cryo-EM, despite the implications that might be suggested by this term, will not give us this information either, or in a very limited form. In such techniques, two reactants are brought together to react for a defined period of time before the product is trapped by flash-freezing29–30 or photolytic uncaging of a reaction-specific quenching agent such as GTP.31–32 Thus, in contrast to the situation we have considered earlier, time-resolved cryo-EM deals with a non-equilibrium situation, in which certain states are populated only for a short time – the landscape changes such that some basins appear, exist for a short time, then disappear. For this reason, time-resolved cryo-EM techniques carry great promise for visualization of short-lived states which are otherwise decayed when cryo-EM grids are prepared in the standard way. These techniques can provide information on the “preferred pathway” of a reaction, by allowing the generation and decay of individual intermediate states (or “transiently formed basins”) to be organized in time.33 However, the fact that the molecule in states that correspond to places with high gradient in the energy landscape cannot be captured by 3D visualization remains unchanged.
Considering our inability to recover more than a finite number of 3D snapshots at pre-ordained times, we may pose the question whether, and in which way, we can obtain the additional information on how these snapshots are related to one another. This question is further complicated by the fact that the energy landscape is often more complex than illustrated by the one-dimensional diagram of Fig. 1. The consequence of a multidimensional topology, as compared to the one-dimensional one, is that there are multiple possible pathways from one basin to another (Fig. 2a). This situation can be depicted as a graph connecting the three-dimensional structures in the structural inventory with multiple reversible arrows (Fig. 2b). As in protein folding, many of these alternative pathways may actually be realized in the workings of a molecular machine.
Fig. 2.
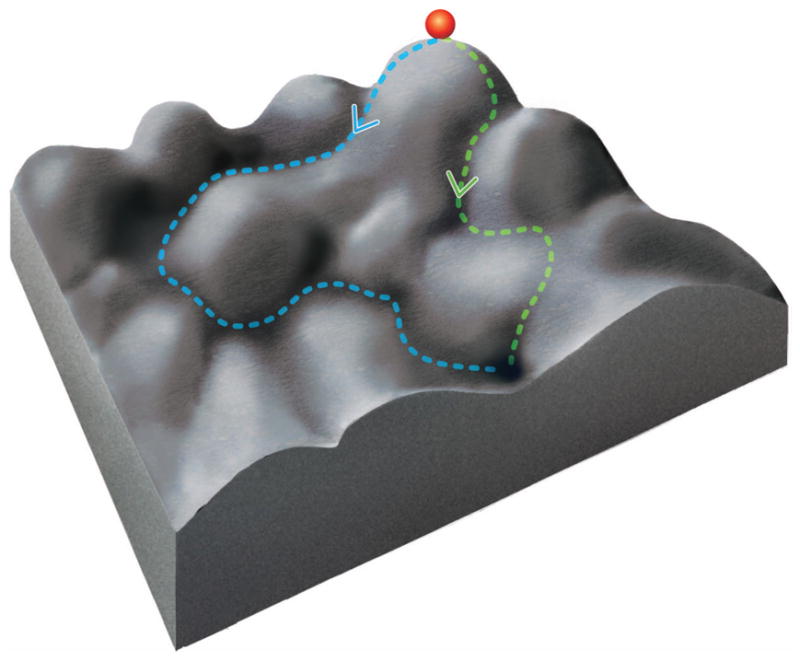
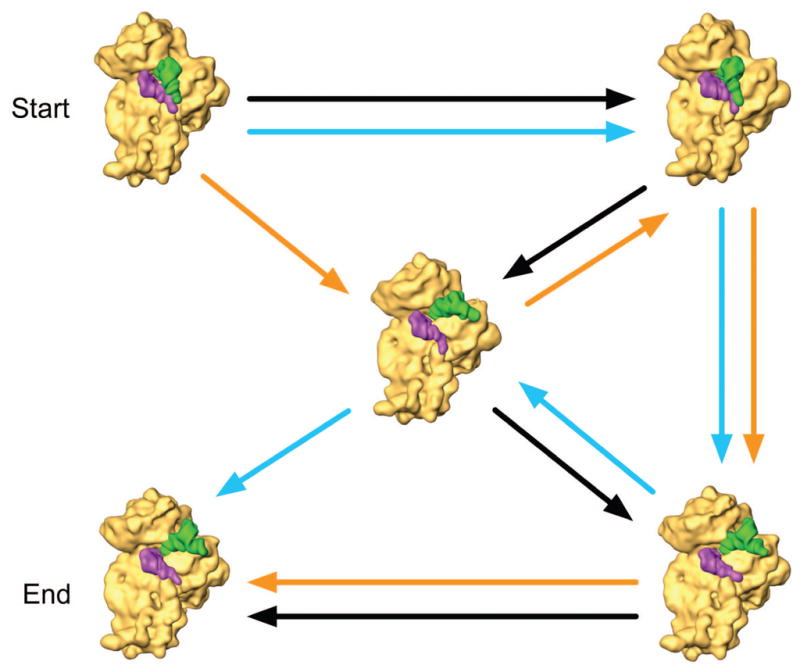
The dilemma posed by multiple pathways. (a) A two- (and higher-) dimensional energy landscape allows multiple pathways between two given states. (b) An example for ambiguities in constructing a (sequential) narrative connecting different structures. Three of many possible pathways are shown between two states designated as “beginning” and “end.” The individual density maps used to illustrate this idea are the result of classification applied to a large dataset of ribosomes in the pre-translocational state.26
The important point to make is that the comparison of two or more structures will not give us any clues on the path or sequence in which the corresponding states are visited. We can think of this problem as a puzzle calling for the arrangement of three-dimensional pieces in four dimensions, impossible to solve as the pieces are isolated and no intercalating pieces are available. A puzzle in two dimensions, by contrast, consists of a complete set of tiles, whose arrangement is unambiguously defined by the continuity of patterns across the (one-dimensional) tile boundaries (Fig. 3).
Fig. 3.

Illustration of the idea that continuity of features – here across the one-dimensional boundaries of a two-dimensional puzzle – is required to reconstruct the scene in its entirety. In contrast, no continuity exists among the reconstructions harvested from a cryo-EM sample containing a molecule in multiple states. (Photo credit Sylvia Wentzlau, http://www.webdesign-und-fotografie-leipzig.de)
The missing information about pathways connecting the different observed states evidently has to come from elsewhere, namely: (i) results of other experimental techniques, (ii) topological considerations, or (iii) computational simulations.
Among other experimental techniques, smFRET occupies a prominent position, as exemplified by studies of RNA polymerase34 and the ribosome.35 Here, guided by existing knowledge of the static structure of the molecule, derived from cryo-EM or X-ray crystallography, two fluorophores (i.e., a donor and an acceptor) are attached to different domains or ligands in the FRET-sensitive distance range (below ~70Å) so that distance changes can be monitored by recording the FRET efficiency in real time.
Topological considerations can be used in certain circumstances if the structures associated with different states allow only a single connecting pathway. For example, if two states A and B differ by rotation of a domain, an observed state with intermediate rotation of that domain is most likely on the pathway A – B. An example is the intersubunit rotation of the ribosome during translocation.14,26
Computational simulations may also be able to tell which pathways are likely feasible and which are not. An example is the molecular dynamics simulation for the accommodation of the tRNA into the A site of the ribosome following tRNA selection.36
Conclusions
Cryo-electron microscopy has come to a new juncture with the realization that in a mixture of freely equilibrating states, molecules in multiple states can be recovered and separately reconstructed. The intent of this article was to point out the opportunities this new technology will bring to the study of molecular machines, especially as the resolution of the single-particle reconstruction technique is approaching the atomic level. At the same time, it must be realized that the “story” emerging from thousands of images is fragmented and ambiguous, and that the help of other techniques must be recruited to reconstruct the dynamic pathways in their entirety.
Acknowledgments
I would like to thank Ruben Gonzalez for helpful suggestions and Melissa Thomas for assistance with the preparation of the illustrations. Supported by HHMI and R01 GM29169 and GM55440 (to J.F.).
References
- 1.Frank J, editor. Molecular Machines in Biology – Workshop of the Cell. Cambridge University Press; New York: 2011. [Google Scholar]
- 2.Roux B, editor. Molecular Machines. World Scientific; Singapore: 2011. [Google Scholar]
- 3.Frank J. Three-dimensional Electron Microscopy of Macromolocular Assemblies. Oxford University Press; New York: 2006. [Google Scholar]
- 4.Armache JP, Jarasch A, Anger AM, Villa E, Becker T, Bhushan S, Jossinet F, Habeck M, Dindar G, Franckenberg S, Marquez V, Mielke T, Thomm M, Berninghausen O, Beatrix B, Soding J, Westhof E, Wilson DN, Beckmann R. Proc Natl Acad Sci USA. 2010;107:19748–19753. doi: 10.1073/pnas.1009999107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bai X, Fernandez IS, McMullan G, Scheres SHW. eLife. 2013;2:e00461. doi: 10.7554/eLife.00461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hashem Y, des Georges A, Fu J, Buss SN, Jossinet F, Jobe A, Zhang Q, Liao HY, Grassucci RA, Bajaj C, Westhof E, Madison-Antenucci S, Frank J. Nature. 2013;494:385–389. doi: 10.1038/nature11872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kelly DF, Abeyrathne PD, Dukovski D, Walz T. J Mol Biol. 2008;382:423–433. doi: 10.1016/j.jmb.2008.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kelly DF, Dukovski D, Walz T. Meth Enzymol. 2010;481:83–107. doi: 10.1016/S0076-6879(10)81004-3. [DOI] [PubMed] [Google Scholar]
- 9.Sharma G, Pallesen J, Das S, Grassucci R, Langlois R, Hampton CM, Kelly DF, des Georges A, Frank J. J Struct Biol. 2013;181:190–194. doi: 10.1016/j.jsb.2012.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baxter WT, Grassucci RA, Haixiao Gao H, Frank J. J Struct Biol. 2009;166:126–132. doi: 10.1016/j.jsb.2009.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frank J, Gonzalez RL. Ann Rev Biochem. 2010;79:38–412. doi: 10.1146/annurev-biochem-060408-173330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Munro JB, Sanbonmatsu KY, Spahn CMT, Blanchard SC. Trends Biochem Sci. 2009;34:390–400. doi: 10.1016/j.tibs.2009.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frank J. Israel J Chem. 2010;50:95–98. doi: 10.1002/ijch.201000010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frank J. Curr Opin Struct Biol. 2012;22:778–785. doi: 10.1016/j.sbi.2012.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chowdhury D. Molecular Machines in Biology – Workshop of the Cell. Cambridge University Press; New York: 2011. Statistical mechanical treatment of molecular machines; pp. 38–58. [Google Scholar]
- 16.Dill KA, Chan HS. Nat Struct Biol. 1997;4:10–19. doi: 10.1038/nsb0197-10. [DOI] [PubMed] [Google Scholar]
- 17.Benkovic SJ, Hammes GG, Hammes-Schiffer S. Biochemistry. 2008;47:3317–3321. doi: 10.1021/bi800049z. [DOI] [PubMed] [Google Scholar]
- 18.Whitford PC, Sanbonmatsu KY, Onuchic JN. Rep Prog Phys. 2012;75 doi: 10.1088/0034-4885/75/7/076601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scheres SH, Gao H, Valle M, Herman GT, Eggermont PP, Frank J, Carazo JM. Nat Methods. 2007;4:27–29. doi: 10.1038/nmeth992. [DOI] [PubMed] [Google Scholar]
- 20.Scheres SHW. J Mol Biol. 2012;415:406–418. doi: 10.1016/j.jmb.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang W, Dunkle JA, Cate JHD. Science. 2009;325:1014–1017. doi: 10.1126/science.1175275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Agirrezabala X, Lei J, Brunelle JL, Ortiz-Meoz RF, Green R, Frank J. Mol Cell. 2008;32:190–197. doi: 10.1016/j.molcel.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Julian P, Konevega AL, Scheres SH, Lazaro M, Gil D, Wintermeyer W, Rodnina MV, Valle M. Proc Natl Acad Sci USA. 2008;105:16924–16927. doi: 10.1073/pnas.0809587105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fischer N, Konevega AL, Wintermeyer W, Rodnina MV, Stark H. Nature. 2010;466:329–333. doi: 10.1038/nature09206. [DOI] [PubMed] [Google Scholar]
- 25.Budkevich T, Giesebrecht J, Altman RB, Munro JB, Mielke T, Nierhaus KH, Blanchard SC, Spahn CMT. Mol Cell. 2011;44:214–224. doi: 10.1016/j.molcel.2011.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Agirrezabala X, Liao H, Schreiner E, Fu J, Ortiz-Meoz RF, Schulten K, Green R, Frank J. Proc Natl Acad Sci USA. 2012;109:6094–6099. doi: 10.1073/pnas.1201288109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Noeske J, Cate J. Cur Opin Struct Biol. 2012;22:743–749. doi: 10.1016/j.sbi.2012.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Frank J, Gao H, Sengupta J, Gao N, Taylor DJ. Proc Natl Acad Sci USA. 2007;104:19671–19678. doi: 10.1073/pnas.0708517104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berriman J, Unwin N. Ultramicrosc. 1994;56:241–252. doi: 10.1016/0304-3991(94)90012-4. [DOI] [PubMed] [Google Scholar]
- 30.Lu Z, Shaikh TR, Barnard D, Meng X, Mohamed H, Yassin A, Mannella CA, Agrawal RK, Lu TM, Wagenknecht T. J Struct Biol. 2009;168:388–395. doi: 10.1016/j.jsb.2009.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Subramaniam S, Gerstein M, Oesterhelt D, Henderson R. EMBO J. 1993;12:1–8. doi: 10.1002/j.1460-2075.1993.tb05625.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shaikh T, Barnard D, Meng X, Wagenknecht T. J Struct Biol. 2009;165:184–189. doi: 10.1016/j.jsb.2008.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mulder AM, Yoshioka C, Beck AH, Bunner AE, Milligan RA, Potter CS, Carragher B, Williamson JR. Science. 2010;330:673–677. doi: 10.1126/science.1193220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shi X, Ha T. Molecular Machines in Biology – Workshop of the Cell. Cambridge University Press; New York: 2011. Single-molecule FRET: Technique and applications to studies of molecular machines; pp. 4–19. [Google Scholar]
- 35.MacDougall DD, Fei J, Gonzalez RL. Molecular Machines in Biology – Workshop of the Cell. Cambridge University Press; New York: 2011. Single-molecule fluorescence resonance energy transfer investigations of ribosome-catalyzed protein synthesis; pp. 93–116. [Google Scholar]
- 36.Sanbonmatsu KY, Joseph S, Tun CS. Proc Natl Acad Sci USA. 2005;102:15854–15859. doi: 10.1073/pnas.0503456102. [DOI] [PMC free article] [PubMed] [Google Scholar]