Abstract
Background
Expression of multiple graft-protective proteins targeted to different locations (i.e., intracellular, cell surface, and secreted) has become an increasingly important goal in xenotransplantation. The 2A “ribosome skip” signal is used as a linker to enable transgene co-expression, but some studies have shown that post-translational modification and trafficking of 2A-linked proteins may be adversely affected depending on their position relative to 2A. We tested whether several relevant proteins, subject to a range of processing and localization mechanisms, could be efficiently co-expressed using the 2A system.
Methods
Six expression cassettes were constructed, each containing up to four 2A-linked open reading frames, encoding combinations of human CD55, thrombomodulin (TBM), CD39, CTLA4-Ig and hygromycin resistance. Each linker incorporated a furin cleavage site to remove the carboxy-terminal extension that remains on upstream proteins after 2A processing. The cassettes were used to produce vectors for transfection, adenoviral transduction and transgenesis. Expression was detected by flow cytometry and/or Western blotting.
Results
All proteins were expressed in the appropriate location following transient transfection of COS-7 cells, irrespective of the number of linked genes. The percentage of stable transfectants expressing a linked gene was increased 10-fold (from 4–5% to 58–67%) by incorporating the hygromycin resistance gene into the cassette. Stable transfection of transgenic GalT KO pig fibroblasts with a hygromycin- TBM-CD39 construct resulted in surface expression of both TBM and CD39 by the majority of hygromycin-resistant cells. Expression was maintained after flow cytometric sorting and expansion. Adenoviral transduction of NIT-1 mouse insulinoma cells with a TBM-CD39 construct resulted in strong expression of both genes on the cell surface. Mice transgenic for 3-gene (CD55- TBM-CD39) or 4-gene (CD55- TBM-CTLA4Ig-CD39) constructs expressed all genes except CD55.
Conclusions
These results confirm the versatility of the 2A system, and demonstrate that careful construct design can minimize potential problems with post-translational modification and trafficking. In addition, incorporation of a selection marker into the 2A-linked chain can dramatically increase the proportion of stable transfectants expressing proteins of interest. This provides a powerful method for the rapid modification of existing genetically modified pigs.
Keywords: 2A, genetic modification, transgene co-expression, xenotransplantation
Introduction
Combining multiple independent genetic modifications in pigs by breeding is time-consuming and expensive. To circumvent this problem, new transgenes can be rapidly added by stably transfecting fibroblasts from existing pigs and subsequently cloning by somatic cell nuclear transfer [1]. The 2A “ribosome skip” system lends itself to this approach because multiple genes can be efficiently expressed using a single construct. The 2A element has been used for the co-expression of several linked open reading frames (ORFs) for a range of applications [2–4]. According to the current model, ribosomes pause near the end of the 2A coding sequence and release the nascent upstream protein before continuing on to translate the downstream protein [5]. This is particularly useful for expressing proteins that benefit from co-ordinated equimolar synthesis, such as immunoglobulin (Ig) heavy and light chains [6,7]. The most widely used 2A sequence is derived from the foot-and-mouthdisease virus and is often referred to as F2A [8]. Other 2A peptides used successfully include T2A from Thosea asigna virus and E2A from equine rhinitis A virus [2,9].
The 18 to 24 amino acid 2A signal concludes with the motif D-V/I-E-X-N-P-G-P, where X is any amino acid. Ribosome skipping occurs at the glycine-proline junction, leaving residual 2A sequence on both the C-terminus of the upstream protein and the N-terminus of the downstream protein [5]. While the N-terminal addition of a single proline to the latter does not appear to be problematic [10], the longer C-terminal extension on the upstream protein might have unpredictable side effects, potentially interfering with post-translational modification, trafficking or function, or inducing an immune response that could limit protein half-life in vivo. In the case of F2A, many proteins have been successfully expressed from the upstream position, and no overt immunogenicity or toxicity of F2A peptides has been observed, at least in mice [11,12]. In addition, correct targeting of F2A-linked proteins to different subcellular compartments has been demonstrated in a range of cells from different species [13,14]. However, position-dependent effects on the separation, stability and localization of F2A-linked proteins have also been reported [15–17]. For example, secretion of functional TGFβ was abolished when its coding region was moved from downstream to upstream of the linker [17]. A potential solution to this problem, at least for secreted and membrane-associated proteins, is to incorporate a furin cleavage site immediately upstream of 2A. Furin is a ubiquitously expressed enzyme that cleaves proproteins after the recognition site R-X-K/R-R, with carboxypeptidases subsequently removing basic amino acid residues from the C-terminus [18]. Furin is predominantly located in the trans- Golgi network, which is responsible for directing secretory pathway proteins to their final destinations. Addition of an RAKR furin motif to 2A-linked Ig chains caused the removal of residual 2A C-terminal extensions, leaving only a dipeptide (RA) tail, and resulted in a marked increase in Ig expression levels [11].
Another approach is to order genes within a 2A-linked cassette such that their endogenous processing/trafficking signals eliminate residual extensions. For example, the proprotein form of membrane proteins tethered by a glycophosphatidyl inositol (GPI) moiety contain a C-terminal region that is post-translationally replaced with a GPI anchor in the endoplasmic reticulum (ER), prior to translocation to the cell surface [19]. Locating the ORF for a GPI-linked protein upstream of a 2A or furin-2A (fu2A) signal should therefore, result in complete removal of any C-terminal extensions.
We and others have used transgenic mouse and pig models to identify several candidate xenograft-protective human proteins, including inhibitors of complement activation such as CD55 and CD59, regulators of thrombosis and inflammation such as thrombomodulin (TBM), endothelial protein C receptor (EPCR) and CD39, and immunosuppressive molecules such as CTLA4-Ig (reviewed in [20]). CTLA4-Ig is secreted, whereas the others are attached to the cell surface either by a GPI anchor (CD55, CD59) or by one (TBM, EPCR) or two (CD39) integral transmembrane domains. Generation of transgenic pigs by cloning also requires the use of an antibiotic selection marker, which is normally intracellular. In this study, we examined whether the fu2A system would allow correct processing and trafficking of up to four of this disparate group of proteins in mammalian cells. We also tested whether including the antibiotic resistance gene in the cassette would increase the efficiency of obtaining stable transfectants expressing the genes of interest.
Methods
Construction of multi-gene cassettes
Oligonucleotides and primers used in the construction of multi-gene cassettes were purchased from Sigma-Aldrich (Sydney, Australia) and are listed in Table 1. Molecular biology enzymes were from New England Biolabs and were purchased from Genesearch Pty Ltd (Arundel, Australia). All cassettes were fully sequenced before use.
Table 1.
Oligonucleotides and primers used in the construction of multi-gene cassettes
| Name | Sequence |
|---|---|
| fuF2A-1 | CTAGAGCCAAACGCGCTCCCGTGAAGCAGACCCTGAACTTTGACCTTCTGAAACTTGCCGGCGACGTCGAGTCCAACCCTGGCC |
| fuF2A-2 | CCGGGGCCAGGGTTGGACTCGACGTCGCCGGCAAGTTTCAGAAGGTCAAAGTTCAGGGTCTGCTTCACGGGAGCGCGTTTGGCT |
| 55Xba-F | CCCTTCTAGAATTCGCTGCGACTCGGCGGAGT |
| 55Xba-R | CCCTTCTAGAAGTCAGCAAGCCCATGGTTACTA |
| 39Xma-F | CCCCCGGGATGGAAGATACAAAGGAGT |
| 39Xho-R | CCCCCTCGAGAATTCATACCATATCTTTCCAGAAAT |
| TBM-F | ATGCTTGGGGTCCTGGTCCTTGGCGCGCTGGCCCTGGCCGGCCTGGGGTTCCCCGCACCCGCAGAGCCGCAGCCGGGTGGCAGCCAGTGCGTCGAGCACGACTGCTTCGCGCTCTATCCGGGCCCCGCG |
| TBM-R | GGGTCCGGGATTCTCCTCCACGTCGCCGCAGGTCAGCAGGCTGCCGCGGCCCTCGCGCTTGGCTCTGAGTCTCTGCGGCGTCCGCTCGGT |
| LEA-F1 | ATGGCTTGCCTTGGATTTCAGCGGCA |
| LEA-F2 | TCTCCAGGCAAATACACTGAGGTCCGGGT |
| LEA-F3 | ACCGCCATACTACGAGGGCATAGGCA |
| LEA-F4 | TTTGGTACCATGGCTTGCCTTGGATTTCAGCGGCA |
| LEA-R1 | ACCCGGACCTCAGTGTATTTGCCTGGAGA |
| LEA-R2 | TGCCTATGCCCTCGTAGTATGGCGGT |
| LEA-R3 | AGGTCCGGGGTTGCTTTCCACATCGCCAGCGAGTTTCAACAAAGCGTAGTTAGTACACTGCCGCTTAGCCCTTTTACCCGGAGACAGGGAGAGGCT |
| LEA-R4 | AGGTCCGGGGTTGCTTTCCACATCGCCA |
| LEA-R5 | GGGATCCGGGTCAGAATCTGGGCACGGTTCTGGATCAATTACATAAATCTGGGTTCCGTTGCCTATGCCCTCGTAGT |
TBM, thrombomodulin.
Cassettes #1–3
The fuF2A coding sequence (RAKRAPVKQTLNFDLLKLAGDVESNPGP) was cloned by annealing oligos fuF2A-1 and fuF2A-2 and ligating them with pBluescript II SK+ (Stratagene, La Jolla, CA, USA) cut with XbaI/XmaI. The human CD55 ORF, minus the stop codon, was PCR-amplified using primers 55Xba-F/55Xba-R and cloned into the XbaI site upstream of fuF2A, and the human CD39 ORF (with stop codon) was PCR-amplified using primers 39Xma-F/39Xho-R and cloned into XmaI/XhoI downstream of fuF2A, generating cassette #1 (CD55-CD39; Fig. 1).
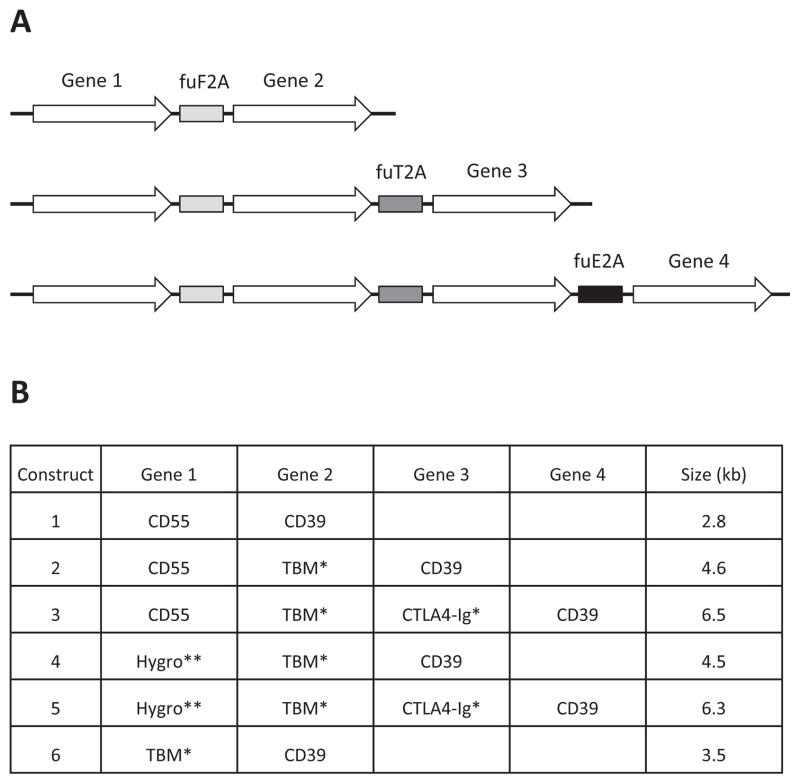
Fig. 1.
(A) Layout of fu2A-linked multi-cistronic cassettes containing 2, 3 or 4 coding regions (not to scale). (B) Details of the six constructs. Hygro, hygromycin resistance. Single and double asterisks indicate short and longer predicted C-terminal extensions after processing (see text for details).
A human TBM ORF (i) modified with a silent mutation to remove a SmaI site and (ii) fused with a downstream sequence for fuT2A (RAKREGRGSLLTCGDVEENPGP) was PCR-amplified using primers TBM-F and TBM-R. This fragment was cloned into the SmaI site between fuF2A and CD39 in cassette #1 to generate cassette #2 (CD55-TBM-CD39).
A splice/overlap PCR method was used to mutate human CTLA4-Ig to the high-affinity variant LEA29Y [21]. Primer sets LEA-F1/LEA-R1 and LEA-F2/LEA-R2 were used to PCR-amplify overlapping mutated fragments of CTLA4, which were mixed and re-amplified using primers LEAF1/LEA-R2. Further amplification with primers LEA-F4/LEA-R5 introduced KpnI and BamHI sites necessary for subsequent steps. A PCR product consisting of the human IgG1 portion of CTLA4-Ig fused to fuE2A (RAKRQCTNYALLKLAGDVESNPGP) was amplified using primers LEA-F3/R3. and ligated with the mutated CTLA4. The composite fragment (LEA29YfuE2A) was then amplified using primers LEAF1/LEA-R4 and cloned into the SmaI site between fuT2A and CD39 in cassette #2 to generate cassette #3 (CD55-TBM-CTLA4Ig-CD39).
Cassettes #4 & 5
A synthetic hygromycin-resistance ORF flanked by XbaI sites was purchased from GenScript Corporation (Piscataway, NJ, USA) and cloned into the XbaI sites of cassettes #2 and #3 to replace the CD55 ORF, producing cassettes #4 (Hygro-TBMCD39) and #5 (Hygro-TBM-CTLA4Ig-CD39), respectively.
Cassette #6
A human TBM cDNA containing 101 base pairs of 5′ untranslated region and the ORF (minus stop codon) was cloned as an XbaI fragment into the XbaI site of cassette #1 to replace the CD55 ORF, producing cassette #6 (TBM-CD39).
Cell culture
All cells were grown in 5% CO2 at 37 °C. COS-7 and NIT-1 cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM; Life Technologies, Melbourne, Australia) supplemented with 10% heat-inactivated fetal calf serum (FCS; Thermo Fisher Scientific Australia Pty Ltd, Scoresby, Australia). Fibroblasts were isolated from an ear punch biopsy taken from a GalT KO pig transgenic for human CD55, CD59 and α1,2- fucosyltransferase. The pig was generated by crossbreeding the triple transgenic line [22] onto an outbred GalT KO background [23]. The fibroblasts were isolated and cultured based on the method of Brunetti et al. [24]. They were grown on 0.01% gelatin coated flasks in 50% DMEM/50% M199c (Life Technologies), 10% heat-inactivated FCS, 1× penicillin/streptomycin and 5 ng/ml bFGF (Life Technologies).
Antibodies
The following fluorescein isothiocyanate- (FITC-) conjugated antibodies were used: mouse antihuman CD55 clone IH4 [25], mouse anti-human TBM clone IA4 (kind gift of Dr Phillip Bird, Monash University, Melbourne, Australia), mouse anti-human CD39 clone BU-61 (Ancell, Bayport, MN, USA), and rabbit polyclonal anti-human IgG (code F0202, Dako, Vital Diagnostics, Noble Park, Australia). Horseradish peroxidase-conjugated secondary antibodies used were rabbit anti-human IgG (code P0214, Dako) and rabbit anti-mouse immunoglobulins (code P0260, Dako). For adenovirus- transduced cells, biotinylated IA4 followed by streptavidin-phycoerythrin (BD Biosciences, San Jose, CA, USA) was used to detect human TBM expression.
Transfection
Cassettes were cloned as EcoRI fragments into the mammalian expression vector pCI-Neo (Promega, Madison, WI, USA). COS-7 cells and pig primary fibroblasts were transfected with 10 μg of plasmid DNAby electroporation essentially as described [26] using a BioRad Gene Pulser (BioRad, Gladesville, Australia). Stable transfectants were selected by adding 300 μg/ml hygromycin or 800 μg/ml neomycin (Life Technologies) 48 h after electroporation.
Adenoviral transduction
Replication-defective recombinant adenovirus (Adv) vectors (human serotype 5 containing E1/E3 deletions) were constructed using the AdMax kit (Microbix Biosystems Inc., Toronto, Canada) following the manufacturer’s instructions, and were grown, purified and titrated as previously described [27]. The control adenovirus expressed an irrelevant protein (CD5 leader sequence fused to the Fc of mouse IgG2c) [28]. NIT-1 cells were transduced at a multiplicity of infection of 20.
Transgenic mice
Selected cassettes were cloned under the control of the murine H-2Kb promoter and used to generate transgenic mice on the C57BL/6 background as described [29]. Tail genomicDNAwas prepared and screened by PCR using primers for human TBM as described [29]. Mice were maintained in individually ventilated cages and bred on the C57BL/6 background. All work with mice was approved by the Animal Ethics Committee, St Vincent’s Hospital Melbourne, in accordance with the Australian Code of Practice for the Care and Use of Animals for Scientific Purposes, 7th Edition, 2004.
Flow cytometry
For detection of cell surface proteins, transfected cells or mouse peripheral blood leukocytes were incubated with FITC-labelled antibodies and analyzed on a FACS Calibur flow cytometer (Becton Dickinson, North Ryde, Australia), with exclusion of dead cells using 1 μg/ml propidium iodide. Secreted CTLA4-Ig was detected by incubating culture supernatant or mouse serum with COS-7 cells transiently transfected with an expression vector for baboon CD86, followed by FITClabelled rabbit polyclonal anti-human IgG and flow cytometry as above. Stable transfectants were sorted on the basis of TBM expression using a FACS Aria flow cytometer (Becton Dickinson). Adenovirus-transduced cells were stained with the appropriate antibodies before being fixed with Cytofix (BD Biosciences) and analyzed using a FACS Scan flow cytometer (Becton Dickinson).
Western blotting
Transfectants were lyzed in lysis buffer (20 mM Tris pH 7.6, 150 mM NaCl, 1% Triton X-100, 2 μg/ml aprotinin, 2 μg/ml leupeptin, 1 mM Benzomidine, 1 mM phenylmethylsulfonyl fluoride). Protein concentration was estimated using the Pierce Coomassie Plus (Bradford) Assay Kit (Thermo Fisher Scientific Australia Pty Ltd, Scoresby, Australia). 100 μg total protein was mixed with an equal volume of 2x non-reducing buffer, heated to 100 °C for 5 min, and electrophoresed on a 7.5% polyacrylamide gel. The gel was blotted at 250 mAmp for 1 h at 4 °C onto a Hybond-P membrane (GE Healthcare, Rydalmere, Australia). Non-specific binding was blocked for 2 h in Tris-buffered saline containing 5% Blotto and 0.05% Tween-20. Membranes were incubated overnight at 4 °C with primary antibody (anti-CD55, anti-TBM, or anti- CD39), diluted 1/1000, followed after washing by incubation for 30 min with horseradish peroxidaseconjugated anti-mouse Ig, diluted 1/5000. The blot was developed using the Pierce SuperSignal West Pico Chemiluminescent System (Thermo Fisher Scientific).
Results
Design and construction of fu2A-linked expression cassettes
Six fu2A-linked multi-cistronic cassettes containing 2, 3 or 4 coding regions (Fig. 1) were constructed by a series of PCR and cloning steps (see Methods). The cassettes ranged in size from 2.8 to 6.5 kb. To minimize the potential for instability due to fu2A sequence repetition, three different 2A signals (F2A, T2A and E2A) were used and alternative codons were utilized when amino acid stretches were repeated. The genes were placed in an order designed to maximize the likelihood that residual fu2A-derived sequences would be removed by endogenous processing signals, although this was not always possible. TBM and CTLA4-Ig were predicted to retain a C-terminal dipeptide (RA) tail after furin cleavage. Because the hygromycin resistance protein does not enter the trans-Golgi network, it is unlikely to encounter furin, and was therefore, predicted to contain a longer C-terminal tail consisting of the fuF2A signal after 2A processing only (RAKRAPVKQTLNFDLLKLAGDVESNPG).
Transient transfection of COS-7 cells
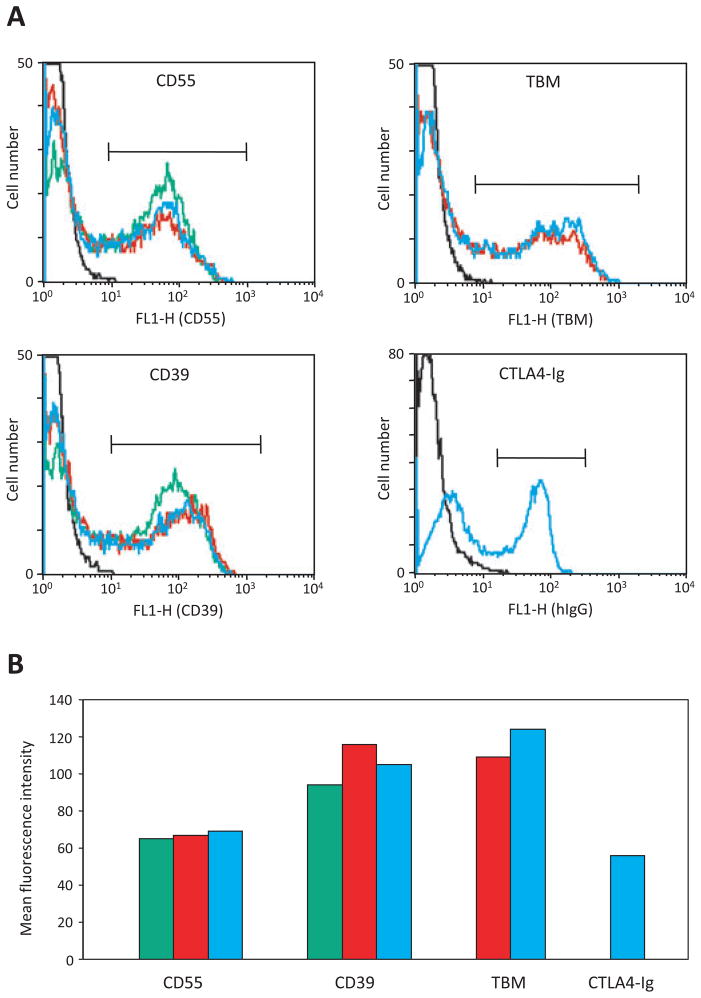
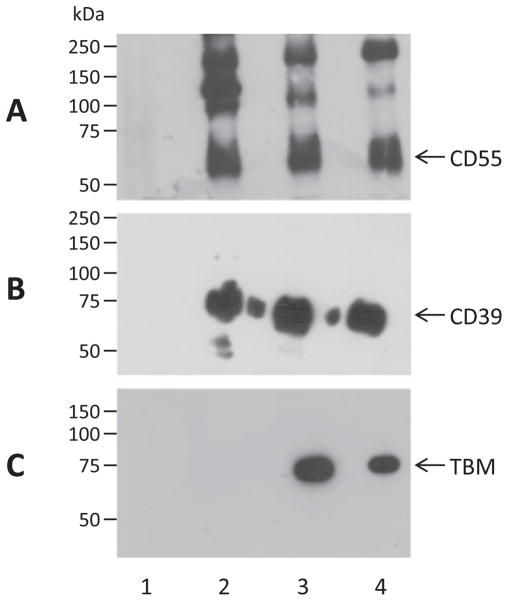
Six cassettes were cloned under the control of the CMV promoter in the expression vector pCI-Neo and electroporated into COS-7 cells. Expression of cell surface proteins was assessed directly by flow cytometry 48 h after transfection. Secretion of CTLA4-Ig was detected by incubation of culture supernatants with COS-7 cells transiently transfected with baboon CD86 (a ligand for CTLA4- Ig), followed by flow cytometric detection using a labelled anti-human IgG antibody. All proteins were expressed at significant levels that were unaffected by the number of genes in the construct. A representative example comparing the related cassettes #1–3 (CD55-CD39, CD55-TBM-CD39 and CD55-TBM-CTLA4Ig-CD39) is shown in Fig. 2. Consistent with the flow cytometric analysis, Western blot of cell lysates prepared from these transfectants demonstrated strong and similar expression of the common genes CD55 (Fig. 3A) and CD39 (Fig. 3B), and TBM was strongly expressed in cells transfected with constructs #2 and #3 (Fig. 3C). The CD55 blot showed several bands of higher than expected molecular weight, but these were not observed in either the CD39 or TBM blots, suggesting that they probably represent multimeric forms of CD55 rather than uncleaved intermediates of fu2A processing. Additional cassettes that were successfully validated by transient and/or stable transfection included CD55-CD59, CD55-CTLA4Ig, Hygro-CD39 and Hygro-TBM-EPCR (data not shown).
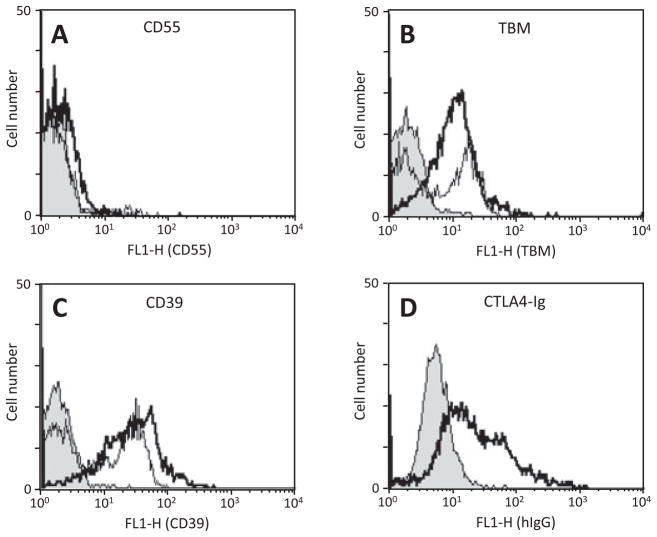
Fig. 2.
(A) Expression of human CD55, thrombomodulin (TBM), CD39 and CTLA4-Ig by COS-7 cells transiently transfected with vector (black line), 2-gene construct #1 (CMV/CD55-CD39, green line), 3-gene construct #2 (CMV/CD55- TBM-CD39, red line), or 4-gene construct #3 (CMV/CD55-TBM-CTLA4Ig-CD39, blue line). (B) Mean fluorescence intensity of transfectants within the regions indicated by the markers on each histogram.
Fig. 3.
Western blot detection of human CD55 (A), CD39 (B) and thrombomodulin (TBM) (C) in lysates of COS-7 cells transiently transfected with vector (lane 1), 2-gene construct #1 (CMV/CD55-CD39; lane 2), 3-gene construct #2 (CMV/CD55-TBM-CD39; lane 3) or 4-gene construct #3 (CMV/CD55-TBM-CTLA4Ig-CD39; lane 4).
Stable transfection of COS-7 cells and porcine fibroblasts
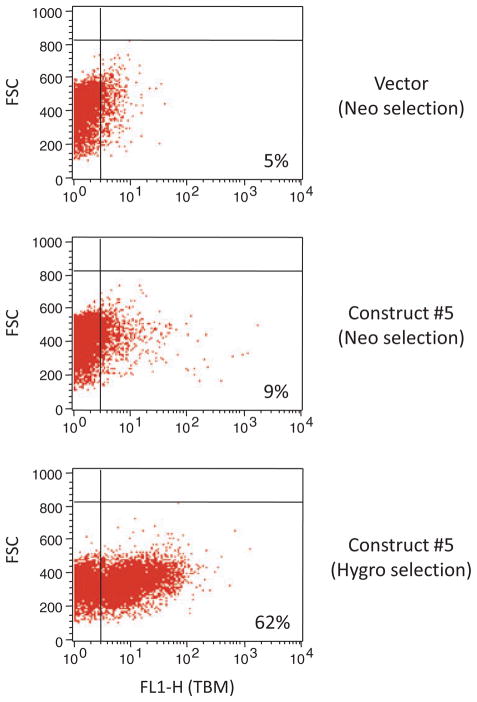
We next tested whether incorporation of a selection marker within a fu2A cassette would increase the frequency of stable transfectants expressing linked genes of interest. COS-7 cells were transfected with pCI-Neo containing cassette #5 (Hygro-TBM-CTLA4Ig-CD39; Fig. 1). In addition to the hygromycin resistance gene within the cassette, this plasmid contains neomycin resistance as a separately transcribed gene. Selection was applied with either neomycin or hygromycin, and pooled stable transfectants were screened by flow cytometry for expression of the second gene in the cassette (TBM). After correcting for background (vector-transfected cells), the proportion of TBMexpressing transfectants was much higher with hygromycin selection (58 to 67%) than with neomycin selection (4 to 5%) (Fig. 4).
Fig. 4.
Expression of human thrombomodulin (TBM) by COS-7 cells stably transfected with 4-gene construct #5 (CMV/Hygro-TBM-CTLA4Ig-CD39) after selection with neomycin (middle panel) or hygromycin (bottom panel). Control vectortransfected cells selected with neomycin are shown in the top panel. A representative example of two separate experiments is shown.
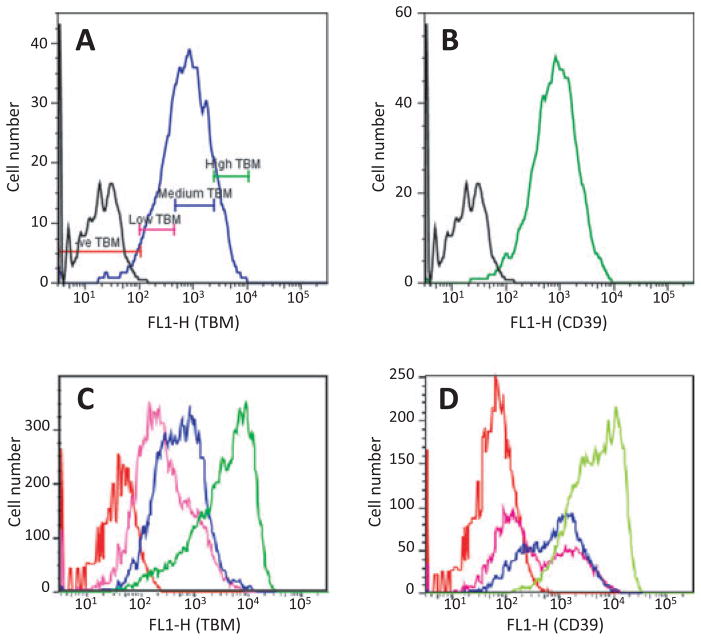
To determine whether the 2A system was applicable in pig cells that can be used for cloning, a construct containing cassette #4 (Hygro- TBM-CD39; Fig. 1) driven by the mouse H-2Kb promoter was stably transfected into fibroblasts isolated from a GalT KO pig transgenic for human CD55, CD59 and α1,2-fucosyltransferase (H-transferase or HTF). Following selection with hygromycin, the majority of pooled transfectants expressed both TBM(Fig. 5A) and CD39 (Fig. 5B). Expression of the transfected genes did not affect the level of expression of the existing transgenes (data not shown). The pooled transfectants were FACS-sorted into four different populations on the basis of TBM expression, using the gates indicated in Fig. 5A. After expansion, each population maintained TBM expression consistent with the original sort profile (Fig. 5C) and expressed CD39 at roughly corresponding levels (Fig. 5D).
Fig. 5.
Expression of human thrombomodulin (TBM) and CD39 by GalT KO/CD55/CD59/HTF pig fibroblasts stably transfected with 3-gene construct #4 (H-2Kb/Hygro-TBM-CD39). (A) pooled transfectants stained for TBM, showing the gates used to sort populations for expansion. (B) pooled transfectants stained for CD39. Black line, untransfected cells (C) and (D), expanded sorted populations stained for expression of TBM and CD39, respectively. High, medium, low and negative TBM-expressing pools are indicated by green, blue, purple and red lines.
Adenoviral transduction of NIT-1 cells
Culturing of pig islets prior to transplantation provides the opportunity for viral delivery of protective transgenes as an alternative to transgenesis. We therefore examined the utility of the 2A system for adenoviral transduction of the immortalized pancreatic beta cell line NIT-1, which retains several features characteristic of native islet beta cells [30]. We used cassette #6 (TBM-CD39) as a test case because of the likely value of these proteins in protecting against the instant blood mediated inflammatory reaction (IBMIR). NIT-1 cells were transduced with the TBM-CD39 adenovirus at a multiplicity of infection of 20, and analyzed by flow cytometry after 48 h. Almost 100% of cells transduced with the 2-gene construct strongly expressed both TBM (Fig. 6A) and CD39 (Fig. 6B) on the cell surface.
Fig. 6.
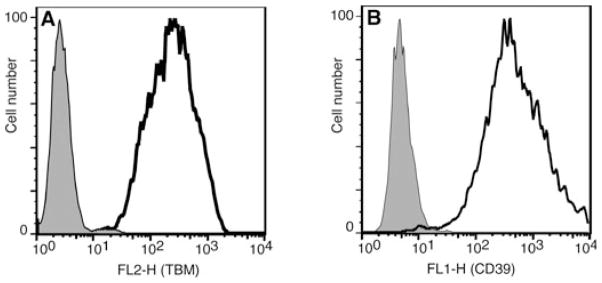
Expression of human thrombomodulin (TBM) (A) and CD39 (B) by NIT-1 insulinoma cells 2 days after transduction with an adenovirus carrying 2-gene construct #6 (CMV/TBM-CD39, unfilled line) or a control adenovirus (filled line).
Generation of transgenic mice
Constructs containing cassette #2 (CD55-TBMCD39) or cassette #3 (CD55-TBM-CTLA4Ig- CD39) driven by the mouse H-2Kb promoter were used to generate transgenic mice. Flow cytometric analysis of peripheral blood lymphocytes revealed expression of TBM and CD39 in both lines of transgenic mice, but expression of CD55 was not detected in either line (Fig. 7). Line #3 transgenic mice were shown to express human CTLA4-Ig by incubating their sera with transiently transfected COS-7 cells expressing the CTLA4-Ig ligand CD86, and detecting bound CTLA4-Ig (Fig. 7). Thus correct in vivo processing and localization of up to three proteins, two transmembrane-anchored and one secreted, was demonstrated. Surprisingly, however, co-expression of the GPI-anchored protein CD55 was not observed.
Fig. 7.
(A–C), flow cytometric analysis of peripheral blood lymphocytes from transgenic mice for expression of human CD55, thrombomodulin (TBM) and CD39. Thin line, mouse transgenic for 3-gene construct #2 (H-2Kb/CD55-TBMCD39); thick line, mouse transgenic for 4-gene construct #3 (H-2Kb/CD55- TBM-CTLA4Ig-CD39); filled line, non-transgenic littermates. (D) detection of CTLA4-Ig in serum by incubation with COS-7 cells transiently transfected with baboon CD86. Key as above.
Discussion
Compared with other means of co-expression such as internal ribosome entry sites, 2A has a number of practical advantages including its compactness, suitability for expression of more than two genes, and capacity for production of proteins in roughly equal proportions [10]. Although part of the 2A signal remains on the C-terminus of upstream proteins after processing, it can be removed (in the case of cell surface and secreted proteins) by incorporating a furin cleavage site adjacent to 2A (fu2A). In this study, we used the fu2A system to co-express up to four genes relevant to xenotransplantation. Careful attention was given to the order of the genes. CD55 was placed first so that GPI anchor addition in the ER would remove the 2A C-terminal extension. The hygromycin resistance gene was kept in this position when substituted for CD55, but for a different reason: to eliminate the possibility of “slipstreaming”, in which an intracellular protein encoded downstream of a protein containing a signal sequence can be translocated into the ER [10]. A consequence of this positioning was that the hygromycin resistance protein was predicted to retain residual 2A sequence uncleaved by furin; however, this had no apparent effect on function, as hygromycin-resistant stable transfectants were readily obtained. TBM and CTLA4-Ig were predicted to retain a short C-terminal extension (RA) after furin cleavage, but little or no functional impact was anticipated in either case. The 36-residue C-terminal cytoplasmic domain of TBM is already rich in basic amino acids, and the RA extension has been demonstrated not to affect the biological activity of an antibody expressed using a fu2A-based construct [6], suggesting that it was unlikely to affect CTLA4-Ig. Interestingly, a recent study has shown that even the RA tail can be eliminated by modifying the furin cleavage site in fuF2A to contain all basic residues (e.g. RKRR) [11].
All gene combinations were strongly expressed in transiently transfected cells. Expression levels were unaffected by the number of genes in the construct, and there was no evidence of incomplete separation of individual proteins. To our knowledge, the CD55-TBM-CTLA4Ig-CD39 construct encodes the longest (1939 amino acids) and most complex polyprotein successfully translated and processed using a 2A-based construct. Furthermore, we demonstrated that integrating an antibiotic resistance gene into the chain was a powerful method to enrich for stable transfectants expressing other linked genes. Although not tested here, it is conceivable that the expression level in such transfectants can be increased by using a higher antibiotic concentration for selection. We also showed that the fu2A system could be adapted for efficient adenoviral delivery of two anti-IBMIR proteins (TBM and CD39) to an islet beta cell line.
The 2A system has been used successfully in transgenic mice [12]. We generated transgenic mouse lines using fu2A-linked 3-gene (CD55- TBM-CD39) and 4-gene (CD55-TBM-CTLA4Ig- CD39) constructs under the control of the H-2Kb promoter. However, the first gene in the chain (CD55) was not expressed in either line. Expression of the downstream proteins implies that CD55 must also have been translated; suggesting a failure in folding and/or localization in vivo, although why CD55 could be correctly expressed from the same constructs in vitro is puzzling. It should also be noted that while the 2A system is appropriate for delivering co-expression of particular sets of transgenes, it is not suited to situations where individual transgenes require a different tissue distribution.
Importantly, the H-2Kb promoter was sufficiently strong to drive selectable levels of antibiotic resistance in transfected primary pig fibroblasts, enabling us to use a fu2A-linked Hygro construct to generate GalT KO cells expressing TBM and CD39 in addition to three existing transgenes. These cells were sorted on the basis of TBM expression and are currently being used for somatic cell nuclear transfer to clone new pigs. While the discrepancy between in vitro and in vivo outcomes using the CD55-containing constructs sounds a note of caution, this is balanced by the recent demonstration of functional 2A-mediated transgene co-expression in cloned pigs [31].
Acknowledgments
This work was supported by the Australian National Health and Medical Research Council, the National Institutes of Health USA (to SCR and AJFD; U01 AI066331 and P01AI045897), and the Juvenile Diabetes Research Foundation. We thank Helen Barlow and the staff of the Bioresources Centre, St Vincent’s Hospital Melbourne, for the generation and care of transgenic mice.
Abbreviations
- DMEM
Dulbecco’s Modified Eagle’s Medium
- ER
endoplasmic reticulum
- EPCR
endothelial protein C inhibitor
- FCS
fetal calf serum
- FITC
fluorescein isothiocyanate
- fu2A
furin-2A
- GPI
glycophosphatidyl inositol
- Hygro
hygromycin resistance
- IBMIR
instant blood mediated inflammatory reaction
- Ig
immunoglobulin
- ORF
open reading frame
- PE
phycoerythrin
- TBM
thrombomodulin
References
- 1.Galli C, Perota A, Brunetti D, Lagutina I, Lazzari G, Lucchini F. Genetic engineering including superseding microinjection: new ways to make GM pigs. Xenotransplantation. 2010;17:397–410. doi: 10.1111/j.1399-3089.2010.00590.x. [DOI] [PubMed] [Google Scholar]
- 2.Szymczak AL, Workman CJ, Wang Y, et al. Correction of multi-gene deficiency in vivo using a single ‘self-cleaving’ 2A peptide-based retroviral vector. Nat Biotechnol. 2004;22:589–594. doi: 10.1038/nbt957. [DOI] [PubMed] [Google Scholar]
- 3.Casales E, Aranda A, Quetglas JI, et al. A novel system for the production of high levels of functional human therapeutic proteins in stable cells with a Semliki Forest virus noncytopathic vector. Nat Biotechnol. 2010;27:138–148. doi: 10.1016/j.nbt.2010.02.005. [DOI] [PubMed] [Google Scholar]
- 4.Carey BW, Markoulaki S, Hanna J, et al. Reprogramming of murine and human somatic cells using a single polycistronic vector. Proc Natl Acad Sci U S A. 2009;106:157–162. doi: 10.1073/pnas.0811426106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Doronina VA, Wu C, de Felipe P, Sachs MS, Ryan MD, Brown JD. Site-specific release of nascent chains from ribosomes at a sense codon. Mol Cell Biol. 2008;28:4227–4239. doi: 10.1128/MCB.00421-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fang J, Qian JJ, Yi S, et al. Stable antibody expression at therapeutic levels using the 2A peptide. Nat Biotechnol. 2005;23:584–590. doi: 10.1038/nbt1087. [DOI] [PubMed] [Google Scholar]
- 7.Ho DT, Wykoff-Clary S, Gross CS, et al. Growth inhibition of an established A431 xenograft tumor by a full-length anti-EGFR antibody following gene delivery by AAV. Cancer Gene Ther. 2009;16:184–194. doi: 10.1038/cgt.2008.68. [DOI] [PubMed] [Google Scholar]
- 8.de Felipe P. Skipping the co-expression problem: the new 2A “CHYSEL” technology. Genet Vaccines Ther. 2004;2:13. doi: 10.1186/1479-0556-2-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu T, Fu Q, Chen P, Zhang K, Guo D. Generation of a stable mammalian cell line for simultaneous expression of multiple genes by using 2A peptide-based lentiviral vector. Biotechnol Lett. 2009;31:353–359. doi: 10.1007/s10529-008-9882-3. [DOI] [PubMed] [Google Scholar]
- 10.de Felipe P, Luke GA, Hughes LE, Gani D, Halpin C, Ryan MD. E unum pluribus: multiple proteins from a self-processing polyprotein. Trends Biotechnol. 2006;24:68–75. doi: 10.1016/j.tibtech.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 11.Fang J, Yi S, Simmons A, et al. Anantibody delivery system for regulated expression of therapeutic levels of monoclonal antibodies in vivo. Mol Ther. 2007;15:1153–1159. doi: 10.1038/sj.mt.6300142. [DOI] [PubMed] [Google Scholar]
- 12.Trichas G, Begbie J, Srinivas S. Use of the viral 2A peptide for bicistronic expression in transgenic mice. BMC Biol. 2008;6:40. doi: 10.1186/1741-7007-6-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lorens JB, Pearsall DM, Swift SE, et al. Stable, stoichiometric delivery of diverse protein functions. J Biochem Biophys Methods. 2004;58:101–110. doi: 10.1016/S0165-022X(03)00147-7. [DOI] [PubMed] [Google Scholar]
- 14.Provost E, Rhee J, Leach SD. Viral 2A peptides allow expression of multiple proteins from a single ORF in transgenic zebrafish embryos. Genesis. 2007;45:625– 629. doi: 10.1002/dvg.20338. [DOI] [PubMed] [Google Scholar]
- 15.Lengler J, Holzmuller H, Salmons B, Gunzburg WH, Renner M. FMDV-2A sequence and protein arrangement contribute to functionality of CYP2B1- reporter fusion protein. Anal Biochem. 2005;343:116–124. doi: 10.1016/j.ab.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 16.Funston GM, Kallioinen SE, de Felipe P, Ryan MD, Iggo RD. Expression of heterologous genes in oncolytic adenoviruses using picornaviral 2A sequences that trigger ribosome skipping. J Gen Virol. 2008;89:389–396. doi: 10.1099/vir.0.83444-0. [DOI] [PubMed] [Google Scholar]
- 17.Rothwell DG, Crossley R, Bridgeman JS, et al. Functional expression of secreted proteins from a bicistronic retroviral cassette based on foot-and-mouth disease virus 2A can be position dependent. Hum Gene Ther. 2010;21:1631–1637. doi: 10.1089/hum.2009.197. [DOI] [PubMed] [Google Scholar]
- 18.Thomas G. Furin at the cutting edge: from protein traffic to embryogenesis and disease. Nat Rev Mol Cell Biol. 2002;3:753–766. doi: 10.1038/nrm934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eisenhaber B, Maurer-Stroh S, Novatchkova M, Schneider G, Eisenhaber F. Enzymes and auxiliary factors for GPI lipid anchor biosynthesis and posttranslational transfer to proteins. Bioessays. 2003;25:367– 385. doi: 10.1002/bies.10254. [DOI] [PubMed] [Google Scholar]
- 20.Cowan PJ, Nottle MB, d’Apice AJ. Molecular regulation of xenoreactivity. Curr Opin Organ Transplant. 2007;12:30–36. doi: 10.1097/MOT.0b013e328012b0bb. [DOI] [PubMed] [Google Scholar]
- 21.Larsen CP, Pearson TC, Adams AB, et al. Rational development of LEA29Y (belatacept), a high-affinity variant of CTLA4-Ig with potent immunosuppressive properties. Am J Transplant. 2005;5:443–453. doi: 10.1111/j.1600-6143.2005.00749.x. [DOI] [PubMed] [Google Scholar]
- 22.Cowan PJ, Aminian A, Barlow H, et al. Renal xenografts from triple-transgenic pigs are not hyperacutely rejected but cause coagulopathy in non-immunosuppressed baboons. Transplantation. 2000;69:2504–2515. doi: 10.1097/00007890-200006270-00008. [DOI] [PubMed] [Google Scholar]
- 23.Nottle MB, Beebe LF, Harrison SJ, et al. Production of homozygous alpha-1,3-galactosyltransferase knockout pigs by breeding and somatic cell nuclear transfer. Xenotransplantation. 2007;14:339–344. doi: 10.1111/j.1399-3089.2007.00417.x. [DOI] [PubMed] [Google Scholar]
- 24.Brunetti D, Perota A, Lagutina I, et al. Transgene expression of green fluorescent protein and germ line transmission in cloned pigs derived from in vitro transfected adult fibroblasts. Cloning Stem Cells. 2008;10:409– 419. doi: 10.1089/clo.2008.0036. [DOI] [PubMed] [Google Scholar]
- 25.Coyne KE, Hall SE, Thompson S, et al. Mapping of epitopes, glycosylation sites, and complement regulatory domains in human decay accelerating factor. J Immunol. 1992;149:2906–2913. [PubMed] [Google Scholar]
- 26.Roussel JC, Moran CJ, Salvaris EJ, Nandurkar HH, d’Apice AJ, Cowan PJ. Pig thrombomodulin binds human thrombin but is a poor cofactor for activation of human protein C and TAFI. Am J Transplant. 2008;8:1101–1112. doi: 10.1111/j.1600-6143.2008.02210.x. [DOI] [PubMed] [Google Scholar]
- 27.Londrigan SL, Sutherland RM, Brady JL, et al. Prolonged local expression of anti-CD4 antibody by adenovirally transduced allografts can promote long-term graft survival. J Gene Med. 2006;8:42–52. doi: 10.1002/jgm.818. [DOI] [PubMed] [Google Scholar]
- 28.Londrigan SL, Sutherland RM, Brady JL, et al. In situ protection against islet allograft rejection by CTLA4Ig transduction. Transplantation. 2010;90:951–957. doi: 10.1097/TP.0b013e3181f54728. [DOI] [PubMed] [Google Scholar]
- 29.Crikis S, Zhang XM, Dezfouli S, et al. Anti-inflammatory and anticoagulant effects of transgenic expression of human thrombomodulin in mice. Am J Transplant. 2010;10:242–250. doi: 10.1111/j.1600-6143.2009.02939.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hamaguchi K, Gaskins HR, Leiter EH. NIT-1, a pancreatic beta-cell line established from a transgenic NOD/Lt mouse. Diabetes. 1991;40:842–849. doi: 10.2337/diab.40.7.842. [DOI] [PubMed] [Google Scholar]
- 31.Yang D, Wang CE, Zhao B, et al. Expression of Huntington’s disease protein results in apoptotic neurons in the brains of cloned transgenic pigs. Hum Mol Genet. 2010;19:3983–3994. doi: 10.1093/hmg/ddq313. [DOI] [PMC free article] [PubMed] [Google Scholar]