Abstract
Sex and genetic variation influence the risk of developing diabetic nephropathy and ESRD in patients with type 1 diabetes. We performed a genome-wide association study in a cohort of 3652 patients from the Finnish Diabetic Nephropathy (FinnDiane) Study with type 1 diabetes to determine whether sex-specific genetic risk factors for ESRD exist. A common variant, rs4972593 on chromosome 2q31.1, was associated with ESRD in women (P<5×10−8) but not in men (P=0.77). This association was replicated in the meta-analysis of three independent type 1 diabetes cohorts (P=0.02) and remained significant for women (P<5×10−8; odds ratio, 1.81 [95% confidence interval, 1.47 to 2.24]) upon combined meta-analysis of the discovery and replication cohorts. rs4972593 is located between the genes that code for the Sp3 transcription factor, which interacts directly with estrogen receptor α and regulates the expression of genes linked to glomerular function and the pathogenesis of nephropathy, and the CDCA7 transcription factor, which regulates cell proliferation. Further examination revealed potential transcription factor–binding sites within rs4972593 and predicted eight estrogen-responsive elements within 5 kb of this locus. Moreover, we found sex-specific differences in the glomerular expression levels of SP3 (P=0.004). Overall, these results suggest that rs4972593 is a sex-specific genetic variant associated with ESRD in patients with type 1 diabetes and may underlie the sex-specific protection against ESRD.
In the non-diabetic population, ESRD is more common in men than in women, and women seem protected from ESRD until menopause.1 Similarly, diabetic nephropathy (DN) and ESRD are more common in diabetic men than in diabetic women.2 However, the female “protection” from ESRD appears attenuated in diabetic women because women who were younger than age 15 years at onset of type 1 diabetes (T1D) do not differ from diabetic men regarding their incidence of ESRD.2 The loss of female “protection” in diabetes remains controversial.3,4 Moreover, some risk factors for DN differ between men and women,5 suggesting sex-specific mechanisms that may be related to differences in the hormonal profiles as estrogen exerts renoprotective effects in nondiabetic persons.4 Furthermore, women with T1D have lower estradiol concentrations and a hormonal profile that more closely resembles that of men.3 Nevertheless, the role of estrogen in the progression of DN still remains ambiguous.6
DN clusters in families, and the sibling risk ratio is conspicuously high for ESRD, suggesting that genetic variation influences the risk of ESRD.7 We previously identified two genetic loci that are associated with ESRD in patients with T1D, namely AFF3 and RGMA-MCTP2.8 However, no genetic variants have so far been robustly associated with ESRD in a sex-specific manner. Therefore, we studied common genetic variation affecting the risk of ESRD in men and women separately, using a genome-wide association study (GWAS) approach involving 3652 Finnish patients with T1D from the Finnish Diabetic Nephropathy (FinnDiane) Study.9
The FinnDiane discovery cohort included 258 women and 387 men with ESRD. These patients were compared with those without signs of DN despite long duration of diabetes. GWAS on ESRD was performed separately for men (Ncases=387, Ncontrols=655) and women (Ncases=258, Ncontrols=935). The main clinical characteristics of the patients are shown in Table 1.
Table 1.
Clinical characteristics of the FinnDiane patients
| Characteristic | Women | Men | ||
|---|---|---|---|---|
| ESRD (n=258) | Controls (n=935) | ESRD (n=387) | Controls (n=655) | |
| Age at T1D onset (yr) | 11.3±6.7 | 14.7±7.8 | 13.7±7.6 | 15.8±8.9 |
| Age (yr) | 44.4±8.9 | 42.9±11.1 | 47.7±8.6 | 43±11.9 |
| T1D duration (yr) | 33.1±8.8 | 28.3±9.6 | 34±8.3 | 27.2±9.2 |
| Transplantation, n (%) | 123 (47.7) | 0 (0) | 193 (49.9) | 0 (0) |
| Antihypertensive medication, n (%) | 229 (88.8) | 204 (21.9) | 361 (93.3) | 179 (27.3) |
| Lipid-lowering medication, n (%) | 99 (38.4) | 114 (12.2) | 162 (41.9) | 99 (15.1) |
| BMI (kg/m2) | 24±3.9 | 25.1±3.8 | 25.1±4.3 | 25.4±3.1 |
| Systolic BP (mmHg) | 151±24 | 133±18 | 153±25 | 136±16 |
| Diastolic BP (mmHg) | 82±13 | 78±9 | 84±12 | 79±10 |
| Hemoglobin A1c (%) | 8.8±1.8 | 8.1±1.2 | 8.7±1.7 | 8.0±1.2 |
| Total cholesterol (mmol/L) | 5.4±1.3 | 4.9±0.8 | 5.1±1.1 | 4.7±0.9 |
| Triglycerides (mmol/L) | 1.5 (1.0–1.8) | 1.0 (0.7–1.1) | 1.9 (1.1–2.3) | 1.1 (0.7–1.3) |
| HDL cholesterol (mmol/L) | 1.4±0.5 | 1.6±0.4 | 1.2±0.4 | 1.4±0.4 |
Data are mean ± SD, n (%), or mean (interquartile range).
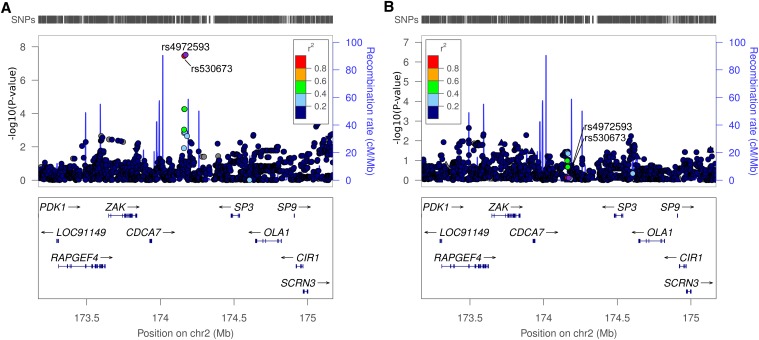
The Manhattan plots of the GWAS results for both sexes are shown in Supplemental Figure 1. GWAS on ESRD in women identified two highly correlated single-nucleotide polymorphisms (SNPs) (r2=1 in HapMap2 CEU samples) on chromosome 2q31.1 that reached genome-wide significance: rs4972593, with an odds ratio (OR) of 2.39 (95% confidence interval [CI], 1.75 to 3.25; P=3.02×10−8), and rs530673, with an OR of 2.38 (95% CI, 1.75 to 3.23; P=3.52×10−8) (Figure 1A). No association was seen for this locus in men (OR, 0.97 [95% CI, 0.78 to 1.21]; P=0.78) (Figure 1B), even though we had 99% power to detect the same association with α=0.05. Furthermore, no other loci reached genome-wide significance in men or women (Supplemental Table 1).
Figure 1.
Regional Manhattan association plot of the associated region at rs4972593 showing association in women (A) but not in men (B). The color of the SNP symbol indicates the linkage disequilibrium (r2) with the index SNP (purple) in the 1000 Genomes CEU population.
Testing for heterogeneity indicated a sex-specific difference in the association of rs4972593 with ESRD (heterogeneity P=5.8×10−5). rs4972593 was nominally associated with body mass index (BMI) in both men and women and with hemoglobin A1c in women (Supplemental Table 2). Association with ESRD was enhanced in women after adjustment for BMI (OR, 2.64 [95% CI, 1.92 to 3.63]; P=2.7×10−9) and remained even after adjustment for both BMI and hemoglobin A1c, although slightly attenuated (OR, 2.31 [95% CI, 1.65 to 3.24]; P=1.3×10−6). Thus, it is unlikely that the observed sex difference would be driven by a sex-related confounder.
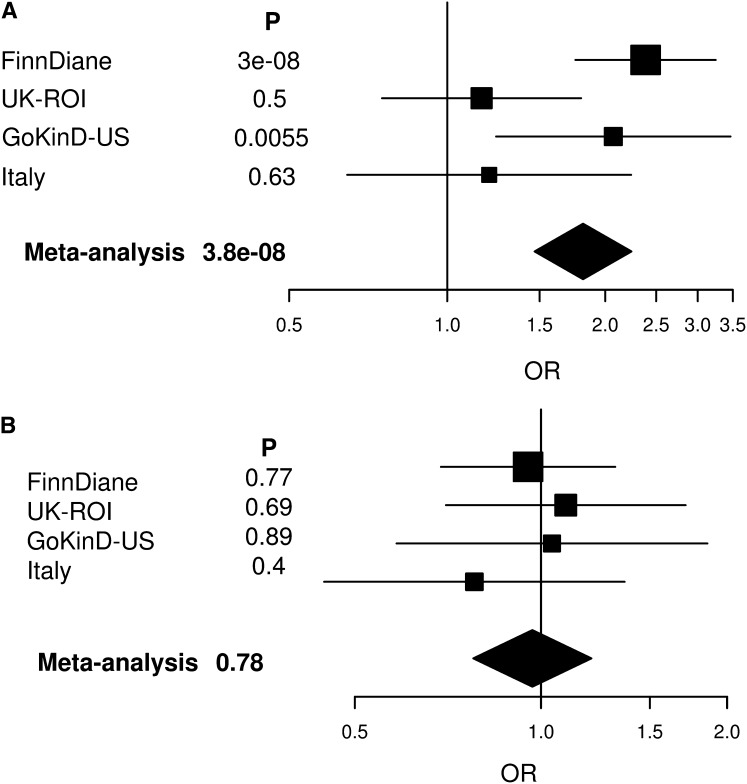
Replication of the association at rs4972593 was then sought in three additional T1D cohorts within the GENIE collaboration, with substantial numbers of women with ESRD available (Ireland-Warren 3-Genetics of Kidneys in Diabetes UK [UK-ROI], n=113; Genetics of Kidneys in Diabetes US Study [GoKinD US], n=252; Italian study, n=68).8 The association between rs4972593 and ESRD in women was replicated with a combined P value of 0.02 for the three studies (OR, 1.41 [95% CI, 1.05 to 1.90]). Even though the results did not reach statistical significance in the UK-ROI and Italian studies, the effect was in the same direction also in these studies. Moreover, all the comparisons with high power (>90%: GoKinD US and the replication studies combined) showed evidence of association. Of note, the OR of 2.07 in GoKinD US was similar to that in the FinnDiane discovery cohort. The association remained genome-wide significant after combined meta-analysis of the FinnDiane and replication cohorts (OR, 1.81 [95% CI, 1.47 to 2.24]; P=3.85×10−8) (Table 2 and Figure 2A). As seen in FinnDiane, there was no association between rs4972593 and ESRD in men in the replication cohorts (P=0.90), and the results remained nonsignificant after combined meta-analysis (OR, 0.97 [95% CI, 0.78 to 1.21]; P=0.78) (Table 2 and Figure 2B). Consequently, the meta-analysis indicated heterogeneity between men and women (P=5.7×10−5).
Table 2.
Association analysis results for rs4972593
| Variable | Women | Men | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Allele Frequency in Cases/Controls | Cases/Controls (n/n) | Power | OR (95% CI) | P Value | Allele Frequency in Cases/Controls | Cases/Controls (n/n) | Power | OR (95% CI) | P Value | |
| FinnDiane | 0.17/0.09 | 258/935 | 0.97 | 2.39 (1.75 to 3.25) | 3.02×10−8 | 0.11/0.11 | 387/655 | 0.99 | 0.95 (0.69 to 1.32) | 0.77 |
| Replication | ||||||||||
| UK-ROI | 0.18/0.16 | 113/508 | 0.86 | 1.16 (0.75 to 1.80) | 0.50 | 0.16/0.14 | 146/395 | 0.87 | 1.10 (0.70 to 1.71) | 0.68 |
| GoKinD US | 0.17/0.13 | 252/479 | 0.97 | 2.07 (1.24 to 3.46) | 0.0055 | 0.14/0.13 | 256/342 | 0.92 | 1.04 (0.58 to 1.86) | 0.89 |
| Italy | 0.18/0.15 | 65/86 | 0.48 | 1.20 (0.64 to 2.24) | 0.63 | 0.14/0.17 | 105/99 | 0.53 | 0.78 (0.44 to 1.36) | 0.40 |
| Meta replication | 0.16 | 430/1073 | 1.00 | 1.41 (1.05 to 1.90) | 0.021 | 0.14 | 507/836 | 1.00 | 0.98 (0.73 to 1.32) | 0.90 |
| Meta all | 0.14 | 688/2009 | 1.00 | 1.81 (1.47 to 2.24) | 3.85×10−8 | 0.13 | 894/1491 | 1.00 | 0.97 (0.78 to 1.21) | 0.78 |
Association analyses are calculated with the minor A allele as the effect allele. Meta replication is the meta-analysis of the three replication cohorts, and meta all is the meta-analysis of all four cohorts; for both, a weighted estimate of the minor allele frequency over all patients is given. Allele frequency refers to the frequency of the minor A allele. Power refers to the statistical power to detect association with α=0.05 and OR of 1.75 (i.e., the lower 95% CI for women in FinnDiane). Power to detect the association with α=5×10−8 was 0.08 in women and 0.12 in men in the discovery cohort and 0.93 in women and 0.92 in men in the combined meta-analysis.
Figure 2.
Forest plot of the meta-analysis of ESRD association at rs4972593 showing association in women (A) but not in men (B). Plots show the study-specific association estimates (ORs and 95% CIs) for the discovery and replication cohorts. The OR and 95% CI for the meta-analysis of the discovery and replication cohorts are denoted by the diamond.
To explore whether additional associated loci could be detected by including more patients, we performed a genome-wide meta-analysis of the FinnDiane, UK-ROI, and GoKinD US GWAS data. However, no association other than that observed between rs4972593 and ESRD in women reached genome-wide significance. Furthermore, no evidence of sex difference was found for the SNPs that have been previously reported to be associated (P<5×10−8) with ESRD in T1D (AFF3,8 RGMA-MCTP2,8 and EPO10) (Supplemental Table 3).
To ensure that the reported association with ESRD is due to diabetes, we excluded all patients with T1D known to have ESRD due to any nondiabetic cause. Supporting a diabetic background, most patients with ESRD in the FinnDiane had diabetic retinopathy, and only six patients with ESRD had no retinopathy. In both UK-ROI and GoKinD US, all patients had retinopathy as an inclusion criterion for the studies. Therefore, the genetic variation at rs4972593 appears to be associated with susceptibility to ESRD in women with T1D.
We further investigated whether the association is specific for women with T1D or is also observed in women with type 2 diabetes by exploring this association in the Family Investigation of Nephropathy and Diabetes (FIND) study.11 No association was found in African American (Ncases=570), European American (Ncases165), or Mexican American (Ncases=413) women with diabetes (Supplemental Table 4). Furthermore, we sought to investigate whether rs4972593 affects the risk for ESRD in women without diabetes by studying female patients of the Wellcome Trust Case-Control Consortium (WTCCC) study on Renal Transplant Dysfunction. However, no adequate proxies were available for rs4972593 in the WTCCC GWAS, and, thus, the role of rs4972593 in the nondiabetic women remains unclear.
The SNP rs4972593 is located in an intergenic region between the SP3 and CDCA7 genes. In silico analysis of the associated SNP for transcription factor–binding sites (TFBS) indicates that the presence of the minor A allele results in loss of several TFBSs, including binding sites for E-box and hypoxia inducible factors (Supplemental Table 5). Moreover, eight estrogen-responsive elements (EREs) were predicted within 5k base pairs (bp) up- or downstream of rs4972593 (Supplemental Table 6). rs530673, a SNP in high linkage disequilibrium with rs4972593 (D’=1; r2=1 in HapMap II CEU), indicated potential regulatory activity (GATA2 binding, SMAD4 binding motif, DNase hypersensitivity peak) in the ENCODE RegulomeDB (Supplemental Table 7).12 However, no significant expression quantitative trait loci (eQTL) association was observed between rs4972593 or any other SNP in linkage disequilibrium with rs4972593 and the expression level of the genes within a 1-Mbp region up- or downstream of rs4972593 in the human HapMap3 lymphoblastoid cell line (Supplemental Table 8).13
We then assessed DN- and sex-related differential gene expression of the flanking genes SP3 and CDCA7.14,15 Interestingly, SP3 was among the top 3\x{2030} for the sex-specific gene expression in glomeruli (fold change, −1.45 [i.e., higher expression in women]; P=0.004) Supplemental Table 9).15 No renal expression was reported for CDCA7 in studies of DN.14,15
CDCA7 encodes a transcription factor that participates in the regulation of cell proliferation, targeted by Myc, and is frequently overexpressed in human cancers.16 SP3 encodes the transcription factor Sp3, which binds to consensus GC-box and GT-box regulatory elements in target genes. Interestingly, Sp3 directly interacts with the estrogen receptor-α (ERα), and 17β-estradiol has been shown to activate GC-rich promoter constructs through the ERα/Sp3 pathway, independent of the classic EREs. Among the ERα/Sp3 pathway target genes, 17β-estradiol differentially regulates VEGFA expression in various cell models.17,18 Vascular endothelial growth factor plays a key role in the angiogenesis, and it has been suggested as one of the common pathogenic factors behind retinopathy and nephropathy.19
We recently reported an induction of the Sp1/Sp3 transcriptional network in human renal tubule epithelial cells stimulated with the profibrotic cytokine TGF-β and identified the Sp3 target gene TGFBI as the top-ranked upregulated gene.20 Other noteworthy Sp3 target genes include COL1A1,21 NOTCH1,22 and CD2AP, encoding an adapter protein essential for the glomerular filtration barrier interacting with the slit diaphragm proteins nephrin and podocin.23,24 In addition to the sex-specific glomerular expression reported for SP3, these data highlight this gene as a plausible positional and biologic candidate for future fine-mapping studies in women with T1D.
Despite the evidence that sex influences the risk of ESRD in patients with T1D, no large-scale sex-specific genetic studies have thus far been reported, and none of the earlier reported genetic susceptibility loci for ESRD in T1D showed sex-specific association. Here we were able to detect a novel locus near SP3 associated with ESRD in women. Because no association was seen in men for rs4972593, it is not surprising that this locus was not detected in our previous GWAS on ESRD, achieving a modest P value of 6.7×10−5 when men and women were analyzed together. This emphasizes the importance of analyzing well characterized, homogeneous subgroups in a GWAS setting in parallel to combining all available patients. Other sex-specific susceptibility loci for ESRD may be discovered when larger GWASs become available. In conclusion, our data point to a biologically plausible pathway and suggest that the association on 2q31.1 may be one explanation for why some women with T1D lose their protection against ESRD.
Concise Methods
Patients
Study participants have previously been described in detail.8 The discovery cohort consisted of 3652 patients with T1D from FinnDiane, a Finnish nationwide multicenter study.9 The main clinical characteristics of the patients are shown in Table 1. Replication was performed on three independent cohorts participating in the GENIE collaboration: the UK-ROI collection, with 1830 genotyped patients with T1D; theGoKinD US cohort, with 1792 patients; and an Italian cohort from the Milan region comprising of 397 patients with T1D.
We compared patients who had T1D and ESRD with T1D controls who had no evidence of diabetic kidney disease despite long duration of diabetes. ESRD was defined as the need for dialysis treatment or having received a kidney transplant, and the minimum duration of T1D was 10 years. We excluded all patients with T1D who were known to have ESRD due to any nondiabetic cause. In FinnDiane, 85% of the patients with ESRD had laser treatment (9% had missing data) and only six had no retinopathy. In UK-ROI and GoKinD US, all ESRD patients had retinopathy as an inclusion criterion. Controls were defined as patients with T1D who had stable normal urinary albumin excretion and diabetes duration of at least 15 years, as described earlier.8
Genotyping
Genotyping of the FinnDiane patients was performed using the Illumina 610Quad chip; genotype calling, quality control, and imputation procedures have been described earlier.8 In brief, genotypes with low genotyping quality or low minor allele frequency (<0.01) were discarded, as were samples with low genotyping quality or cryptic relatedness and geographic outliers based on principal component analysis. A total of 3546 samples and 549,530 SNPs remained after quality control. Imputation was based on the HapMap II CEU population and resulted in approximately 2.4 million SNPs across the autosomal genome. For the replication in UK-ROI and GoKinD US, rs4972593 was selected from the UK-ROI and GoKinD US GWAS data, where the quality control and imputation procedures were the same as described above. Quality control resulted in 1726 UK-ROI samples and 1595 GoKinD US samples. The DNA samples from Italy were genotyped using Sequenom IPLEX assays (Sequenom Inc., San Diego, CA). Imputation and genotyping quality metrics are given in Supplemental Table 10.
Patients and Genotyping in the FIND Study
The FIND cohort, consisting of 885 samples from European Americans, 1460 samples from African Americans, 889 samples from American Indians, and 1535 samples from Mexican Americans, underwent genotyping using the Affymetrix SNP 6.0 GeneChip at the Genotyping Core Facility at Affymetrix (Santa Clara, CA). Samples were plated by ethnic group, randomly assigned by case/control status; 217 pairs of duplicate samples were included for quality control. Genotype calls used the Birdsuite algorithm as implemented in GCOS software (Affymetrix). Samples with call rates >95% were subject to additional quality control procedures for sample and SNP heterozygosity, sample and SNP missingness, sex verification, expected and unexpected relatedness, and population substructure analysis via principal components analysis. After trimming, 342 cases and 404 controls of European American ancestry, 979 cases and 304 controls of African American ancestry, 538 cases and 319 controls of American Indian ancestry, and 779 cases and 594 controls of Mexican American ancestry were included in a final meta-analysis.
Statistical Analyses
The association analysis that compared ESRD cases with T1D controls with normal albumin excretion despite long duration of T1D was performed with PLINK 1.0725 using logistic regression and adjustment for age, T1D duration, and the 10 first principal components that were obtained with the EIGENSTRAT software.26 Estimated allele dosages were used rather than the most likely genotypes in order to account for the uncertainty arising from the imputation process. Women and men were analyzed separately. The quantile-quantile plots of both analyses showed good adherence to the diagonal line of expected P values, and very little excess genomic inflation was observed (λ=1.034 for women, λ=1.045 for men). The association results were adjusted for the genomic inflation. The same methods were used for the statistical analysis of UK-ROI and GoKinD US. Because of the relatively small number of women with ESRD in the Italian cohort, for those samples we used the Fisher exact test of association, which is deemed more robust for small sample numbers. Consequently, the Italian replication cohort was not adjusted for any covariates. Meta-analysis of the four cohorts was performed using a fixed-effect model based on the standard errors and P values, implemented with METAL software.27 Power calculations were performed with Genetic Power Calculator.28
TFBS and Regulatory Function
We looked for the TFBSs directly created or deleted due to rs4972593 using MatInspector (release professional 8.06; Matrix Family Library, version 8.4,) from the Genomatix software suite (Genomatix Software, GmbH, Munich, Germany). The flanking region 5 kbp up- and downstream of rs4972593 was downloaded from National Center for Biotechnology Information SNP database (http://www.ncbi.nlm.nih.gov/projects/SNP/), and MatInspector was used to detect EREs (V$EREF family) within this region. Furthermore, we sought evidence of the regulatory function of the SNPs using the RegulomeDB database, which annotates SNPs with known and predicted regulatory elements in the intergenic regions. The annotation is based on regions of DNAase hypersensitivity, TFBS, and promoter regions that have been biochemically characterized to regulate transcription.12
Renal Gene Expression
Disease- and sex-associated gene expression in published human DN microarray data sets was determined using Nephromine (www.nephromine.org). Selected data sets comprised microdissected renal biopsy specimens from patients with DN versus living donors or patients who had minimal-change disease.14,15 We performed a total of 10 various sex- and disease-specific association look-ups (Supplemental Table 9). Therefore, the P values were adjusted for multiple testing according to 10 performed tests; a P value of 0.005 was required for significance after adjustment.
eQTL Gene Expression
We studied whether rs4972593 was associated with the gene expression level of any of the genes within a 1-Mbp region up- and downstream in the HapMap3 lymphoblastoid cell lines13 using the Genevar user interface (http://www.sanger.ac.uk/resources/software/genevar/). The analysis included all the SNPs in full linkage disequilibrium (r2=1) with rs4972593 in the HapMap2 CEU samples and with data on HapMap3 eQTL in Genevar; three SNPs filled the criteria (rs530673, rs4972590, rs4972591). The P value threshold for statistical significance after multiple testing was P<0.00063 based on α=0.05 significance level, 10 studied genes, and 8 included HapMap populations. Because the selected SNPs were in strong linkage disequilibrium, we did not adjust for the number of SNPs.
Disclosures
J.C.F. has received consulting honoraria from Novartis, Lilly, and Pfizer. P.H.G. has received lecture honoraria from Abbott, Boehringer Ingelheim, Cebix, Eli Lilly, Genzyme, Novartis, Novo Nordisk, and MSD and research grants from Eli Lilly and Roche. P.H.G. is also an advisory board member of Boehringer Ingelheim and Novartis.
Supplementary Material
Acknowledgments
We acknowledge all the subjects for their participation.
FinnDiane: We acknowledge the technical assistance of Maikki Parkkonen, Anna Sandelin, Jaana Tuomikangas, Anna-Reetta Salonen and Tuula Soppela, and the physicians and nurses at each center participating in the collection of patients (Supplemental Table 11). FinnDiane was supported by grants from the Folkhälsan Research Foundation, the Wilhelm and Else Stockmann Foundation, Liv och Hälsa Foundation, Helsinki University Central Hospital Research Funds (EVO), the Sigrid Juselius Foundation, the Signe and Ane Gyllenberg Foundation, Finska Läkaresällskapet, TEKES, Academy of Finland (134379), and the European Union’s Seventh Framework Program (FP7/2007-2013) for the Innovative Medicine Initiative under grant agreement n° IMI/115006 (the SUMMIT consortium).
GENIE: The GENIE Consortium is supported by a US Ireland R&D partnership award funded by Science Foundation Ireland under grant no. SFI/08/US/B1517, The Northern Ireland Research Development office, and National Institutes of Health (NIH), National Institute of Diabetes and Digestive and Kidney Disease(NIDDK) R01 DK081923 to J.N.H., J.C.F., and P.H.G. R.M.S. was supported by a Juvenile Diabetes Research Foundation post doctoral fellowship (JDRF # 3-2011-70).
UK-ROI: The Warren 3/UK GoKinD Study Group includes the following. Belfast: A.P. Maxwell, A.J. McKnight, D.A. Savage; Edinburgh: J. Walker; London: S. Thomas, G.C. Viberti; Manchester: A.J.M. Boulton; Newcastle: S. Marshall; Plymouth: A.G. Demaine, B.A. Millward; Swansea: S.C. Bain. We are grateful to all those who participated in the collections, including clinicians, research nurses, and Jill Kilner for excellent technical support. The Warren 3/UK GoKinD Study Group was jointly funded by Diabetes UK and the Juvenile Diabetes Research Foundation. E. Swan is supported by Diabetes UK Studentship. The Golden Years cohort, established by G.V. Gill, A.H. Barnett, and S.C. Bain, was funded by Diabetes UK.
Dublin: We are grateful to our colleagues at the Renal Unit, Mater Misericordiae University Hospital, Dublin, Ireland. The ROI collection was supported by funding from the Health Research Board Ireland to Hugh R. Brady.
FIND: We thank all FIND participants and study group members. This study was supported by grants U01DK57292, U01DK57329, U01DK057300, U01DK057298, U01DK057249, U01DK57295, U01DK070657, U01DK057303, U01DK070657, U01DK57304, and DK57292-05 from the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) and, in part, by the Intramural Research Program of the NIDDK. This project has been funded in whole or in part with federal funds from the National Cancer Institute, NIH, under contract N01-CO-12400, and the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research. This work was also supported by the National Center for Research Resources for the General Clinical Research Center grants: Case Western Reserve University, M01-RR-000080; Wake Forest University, M01-RR-07122; Harbor-UCLA Medical Center, M01-RR-00425 ; College of Medicine, University of California, Irvine, M01-RR-00827–29; University of New Mexico, HSC M01-RR-00997; and Frederic C. Bartter, M01-RR-01346. Computing support provided by the Wake Forest School of Medicine Center for Public Health Genomics.
We acknowledge use of results from the WTCCC3 Renal Transplant Dysfunction study, funded by a grant the Wellcome Trust (WT090355/A/09/Z, WT090355/B/09/Z), and kindly shared by Dr. Chris Franklin on behalf of the WTCCC3.
Parts of this publication have been published as an abstract at the ASN Kidney Week 2012.
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
See related editorial, “Diabetic Nephropathy: Is ESRD Its Only Heritable Phenotype?,” on pages 1505–1507.
This article contains supplemental material online at http://jasn.asnjournals.org/lookup/suppl/doi:10.1681/ASN.2012111122/-/DCSupplemental.
References
- 1.U.S. Renal Data System: USRDS 2011 Annual Data Report: Atlas of Chronic Kidney Disease and End-Stage Renal Disease in the United States, Bethesda, MD, National Institutes of Health, National Institute of Diabetes and Digestive and 2011 [Google Scholar]
- 2.Harjutsalo V, Maric C, Forsblom C, Thorn L, Wadén J, Groop PH, FinnDiane Study Group : Sex-related differences in the long-term risk of microvascular complications by age at onset of type 1 diabetes. Diabetologia 54: 1992–1999, 2011 [DOI] [PubMed] [Google Scholar]
- 3.Maric C, Sullivan S: Estrogens and the diabetic kidney. Gend Med 5 Suppl A: S103–13, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Silbiger S, Neugarten J: Gender and human chronic renal disease. Gend Med 5 Suppl A: S3–S10, 2008 [DOI] [PubMed] [Google Scholar]
- 5.Coonrod BA, Ellis D, Becker DJ, Bunker CH, Kelsey SF, Lloyd CE, Drash AL, Kuller LH, Orchard TJ, Pittsburgh Epidemiology of Diabetes Complications Study : Predictors of microalbuminuria in individuals with IDDM. Diabetes Care 16: 1376–1383, 1993 [DOI] [PubMed] [Google Scholar]
- 6.Doublier S, Lupia E, Catanuto P, Elliot SJ: Estrogens and progression of diabetic kidney damage. Curr Diabetes Rev 7: 28–34, 2011 [DOI] [PubMed] [Google Scholar]
- 7.Harjutsalo V, Katoh S, Sarti C, Tajima N, Tuomilehto J: Population-based assessment of familial clustering of diabetic nephropathy in type 1 diabetes. Diabetes 53: 2449–2454, 2004 [DOI] [PubMed] [Google Scholar]
- 8.Sandholm N, Salem RM, McKnight AJ, Brennan EP, Forsblom C, Isakova T, McKay GJ, Williams WW, Sadlier DM, Mäkinen VP, Swan EJ, Palmer C, Boright AP, Ahlqvist E, Deshmukh HA, Keller BJ, Huang H, Ahola AJ, Fagerholm E, Gordin D, Harjutsalo V, He B, Heikkilä O, Hietala K, Kytö J, Lahermo P, Lehto M, Lithovius R, Osterholm AM, Parkkonen M, Pitkäniemi J, Rosengård-Bärlund M, Saraheimo M, Sarti C, Söderlund J, Soro-Paavonen A, Syreeni A, Thorn LM, Tikkanen H, Tolonen N, Tryggvason K, Tuomilehto J, Wadén J, Gill GV, Prior S, Guiducci C, Mirel DB, Taylor A, Hosseini SMDCCT/EDIC Research Group, Parving HH, Rossing P, Tarnow L, Ladenvall C, Alhenc-Gelas F, Lefebvre P, Rigalleau V, Roussel R, Tregouet DA, Maestroni A, Maestroni S, Falhammar H, Gu T, Möllsten A, Cimponeriu D, Ioana M, Mota M, Mota E, Serafinceanu C, Stavarachi M, Hanson RL, Nelson RG, Kretzler M, Colhoun HM, Panduru NM, Gu HF, Brismar K, Zerbini G, Hadjadj S, Marre M, Groop L, Lajer M, Bull SB, Waggott D, Paterson AD, Savage DA, Bain SC, Martin F, Hirschhorn JN, Godson C, Florez JC, Groop PH, Maxwell AP: New susceptibility loci associated with kidney disease in type 1 diabetes. PLoS Genet 8: e1002921, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thorn LM, Forsblom C, Fagerudd J, Thomas MC, Pettersson-Fernholm K, Saraheimo M, Wadén J, Rönnback M, Rosengård-Bärlund M, Björkesten CG, Taskinen MR, Groop PH, FinnDiane Study Group : Metabolic syndrome in type 1 diabetes: association with diabetic nephropathy and glycemic control (the FinnDiane study). Diabetes Care 28: 2019–2024, 2005 [DOI] [PubMed] [Google Scholar]
- 10.Tong Z, Yang Z, Patel S, Chen H, Gibbs D, Yang X, Hau VS, Kaminoh Y, Harmon J, Pearson E, Buehler J, Chen Y, Yu B, Tinkham NH, Zabriskie NA, Zeng J, Luo L, Sun JK, Prakash M, Hamam RN, Tonna S, Constantine R, Ronquillo CC, Sadda S, Avery RL, Brand JM, London N, Anduze AL, King GL, Bernstein PS, Watkins S, Jorde LB, Li DY, Aiello LP, Pollak MR, Zhang K, Genetics of Diabetes and Diabetic Complication Study Group : Promoter polymorphism of the erythropoietin gene in severe diabetic eye and kidney complications. Proc Natl Acad Sci U S A 105: 6998–7003, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Knowler WC, Coresh J, Elston RC, Freedman BI, Iyengar SK, Kimmel PL, Olson JM, Plaetke R, Sedor JR, Seldin MF, Family Investigation of Nephropathy and Diabetes Research Group : The family investigation of nephropathy and diabetes (FIND): Design and methods. J Diabetes Complications 19: 1–9, 2005 [DOI] [PubMed] [Google Scholar]
- 12.Boyle AP, Hong EL, Hariharan M, Cheng Y, Schaub MA, Kasowski M, Karczewski KJ, Park J, Hitz BC, Weng S, Cherry JM, Snyder M: Annotation of functional variation in personal genomes using RegulomeDB. Genome Res 22: 1790–1797, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stranger BE, Montgomery SB, Dimas AS, Parts L, Stegle O, Ingle CE, Sekowska M, Smith GD, Evans D, Gutierrez-Arcelus M, Price A, Raj T, Nisbett J, Nica AC, Beazley C, Durbin R, Deloukas P, Dermitzakis ET: Patterns of cis regulatory variation in diverse human populations. PLoS Genet 8: e1002639, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schmid H, Boucherot A, Yasuda Y, Henger A, Brunner B, Eichinger F, Nitsche A, Kiss E, Bleich M, Gröne HJ, Nelson PJ, Schlöndorff D, Cohen CD, Kretzler M, European Renal cDNA Bank (ERCB) Consortium : Modular activation of nuclear factor-kappaB transcriptional programs in human diabetic nephropathy. Diabetes 55: 2993–3003, 2006 [DOI] [PubMed] [Google Scholar]
- 15.Woroniecka KI, Park AS, Mohtat D, Thomas DB, Pullman JM, Susztak K: Transcriptome analysis of human diabetic kidney disease. Diabetes 60: 2354–2369, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Osthus RC, Karim B, Prescott JE, Smith BD, McDevitt M, Huso DL, Dang CV: The Myc target gene JPO1/CDCA7 is frequently overexpressed in human tumors and has limited transforming activity in vivo. Cancer Res 65: 5620–5627, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stoner M, Wang F, Wormke M, Nguyen T, Samudio I, Vyhlidal C, Marme D, Finkenzeller G, Safe S: Inhibition of vascular endothelial growth factor expression in HEC1A endometrial cancer cells through interactions of estrogen receptor alpha and Sp3 proteins. J Biol Chem 275: 22769–22779, 2000 [DOI] [PubMed] [Google Scholar]
- 18.Stoner M, Wormke M, Saville B, Samudio I, Qin C, Abdelrahim M, Safe S: Estrogen regulation of vascular endothelial growth factor gene expression in ZR-75 breast cancer cells through interaction of estrogen receptor alpha and SP proteins. Oncogene 23: 1052–1063, 2004 [DOI] [PubMed] [Google Scholar]
- 19.Tremolada G, Lattanzio R, Mazzolari G, Zerbini G: The therapeutic potential of VEGF inhibition in diabetic microvascular complications. Am J Cardiovasc Drugs 7: 393–398, 2007 [DOI] [PubMed] [Google Scholar]
- 20.Brennan EP, Morine MJ, Walsh DW, Roxburgh SA, Lindenmeyer MT, Brazil DP, Gaora PO, Roche HM, Sadlier DM, Cohen CD, Godson C, Martin F, GENIE Consortium : Next-generation sequencing identifies TGF-β1-associated gene expression profiles in renal epithelial cells reiterated in human diabetic nephropathy. Biochim Biophys Acta 1822: 589–599, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Beauchef G, Bigot N, Kypriotou M, Renard E, Porée B, Widom R, Dompmartin-Blanchere A, Oddos T, Maquart FX, Demoor M, Boumediene K, Galera P: The p65 subunit of NF-κB inhibits COL1A1 gene transcription in human dermal and scleroderma fibroblasts through its recruitment on promoter by protein interaction with transcriptional activators (c-Krox, Sp1, and Sp3). J Biol Chem 287: 3462–3478, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lambertini C, Pantano S, Dotto GP: Differential control of Notch1 gene transcription by Klf4 and Sp3 transcription factors in normal versus cancer-derived keratinocytes. PLoS ONE 5: e10369, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shih NY, Li J, Cotran R, Mundel P, Miner JH, Shaw AS: CD2AP localizes to the slit diaphragm and binds to nephrin via a novel C-terminal domain. Am J Pathol 159: 2303–2308, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schwarz K, Simons M, Reiser J, Saleem MA, Faul C, Kriz W, Shaw AS, Holzman LB, Mundel P: Podocin, a raft-associated component of the glomerular slit diaphragm, interacts with CD2AP and nephrin. J Clin Invest 108: 1621–1629, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC: PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81: 559–575, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D: Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 38: 904–909, 2006 [DOI] [PubMed] [Google Scholar]
- 27.Willer CJ, Li Y, Abecasis GR: METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26: 2190–2191, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Purcell S, Cherny SS, Sham PC: Genetic Power Calculator: Design of linkage and association genetic mapping studies of complex traits. Bioinformatics 19: 149–150, 2003 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.