Abstract
CKD progresses with fibrosis and erythropoietin (Epo)-dependent anemia, leading to increased cardiovascular complications, but the mechanisms linking Epo-dependent anemia and fibrosis remain unclear. Here, we show that the cellular phenotype of renal Epo-producing cells (REPs) alternates between a physiologic Epo-producing state and a pathologic fibrogenic state in response to microenvironmental signals. In a novel mouse model, unilateral ureteral obstruction–induced inflammatory milieu activated NFκB and Smad signaling pathways in REPs, rapidly repressed the Epo-producing potential of REPs, and led to myofibroblast transformation of these cells. Moreover, we developed a unique Cre-based cell-fate tracing method that marked current and/or previous Epo-producing cells and revealed that the majority of myofibroblasts are derived from REPs. Genetic induction of NFκB activity selectively in REPs resulted in myofibroblastic transformation, indicating that NFκB signaling elicits a phenotypic switch. Reversing the unilateral ureteral obstruction–induced inflammatory microenvironment restored the Epo-producing potential and the physiologic phenotype of REPs. This phenotypic reversion was accelerated by anti-inflammatory therapy. These findings demonstrate that REPs possess cellular plasticity, and suggest that the phenotypic transition of REPs to myofibroblasts, modulated by inflammatory molecules, underlies the connection between anemia and renal fibrosis in CKD.
Renal erythropoietin (Epo)-producing cells (REPs) are fibroblast-like cells that express neural markers and produce Epo, an indispensable erythropoietic hormone.1–3 REPs control Epo gene expression primarily through the prolyl hydroxylases/von Hippel-Lindau protein/hypoxia-inducible factor (HIF) pathway.3,4 Production of Epo at cellular level in REPs is thought to be either “on” or “off,” hence the total Epo production from kidney is regulated by the number of Epo-producing REPs, which changes markedly responding to hypoxia and/or anemia.2,5,6
Renal fibrosis is considered a final common pathway of CKD leading to ESRD.7,8 Because fibrotic kidneys have a limited Epo-producing ability at any given anemic stimuli,9,10 renal anemia develops along with the progression of fibrosis, leading to cardiovascular events.7,9,10 Interestingly, REPs in the fibrotic kidney express markers of myofibroblasts, such as desmin and α smooth muscle actin (αSMA),11,12 suggesting an etiological link between renal anemia and fibrosis by damaged REPs.
Myofibroblasts are the culprit of renal fibrosis, and their origin has been debated.8,13 Recent cell-fate mapping studies revealed that myofibroblasts are mainly derived from P0 (myelin protein 0)-Cre–labeled cells,12 or FoxD1-Cre–labeled pericytes and perivascular fibroblasts.14 However, there remains uncertainty, as these Cre-labeled cells are heterogeneous because of the shared developmental program.14 Moreover, in terms of Epo-producing ability, REPs are only <10% of P0-Cre–labeled cells, and approximately 20% of REPs are not fate-mapped by P0-Cre.6,12 Thus, precise contribution of REPs to renal fibrosis is still unclear.
There are many unanswered important questions related to REPs and renal fibrosis: (1) whether the majority of REPs contribute to total myofibroblast population or REPs rather disappear through cell death upon renal damages, (2) how REPs change their phenotype to myofibroblasts and lose their Epo-producing ability, and (3) whether this phenotypic conversion is reversible. In this regard, because CKD is characterized by sterile chronic inflammation caused by activated resident renal cells and infiltrating leukocytes,15–17 we surmised that the phenotypic changes of REPs in diseased kidneys may be attributable to inflammatory signals generated in renal microenvironments.
In this study, we utilized two novel mouse lines that manifest severe anemia (inherited super anemic mice [ISAM]) and that express Cre enzyme under the regulatory influences of Epo gene (Epo-Cre mouse) to monitor phenotypic changes of individual REPs. Utilizing unilateral ureteral obstruction (UUO), a commonly used inflammatory fibrogenic method,18,19 we analyzed how activated inflammatory signaling affects the phenotype of REPs. Through these analyses, we have discovered that REPs are the major source of myofibroblasts and myofibroblast-transformed REPs recover their normal Epo-producing potential upon removal of inflammatory stresses. These results indicate that a profound cellular plasticity resides in REPs and REPs may be ideal targets for CKD treatment.
Results
Efficient Monitoring of REPs by a Transgenic Rescue Approach
Epo-knockout mice die at around embryonic day 12.5 due to anemia.20 Because fetal secretion of Epo from liver is regulated by its 3′ enhancer,21–23 we rescued the embryonic lethality of homozygous Epo-knockout/green fluorescent protein (GFP) knock-in mice (EpoGFP/GFP) using the 3′-enhancer-driven Epo-expressing transgene (TgEpo3′). Because TgEpo3′-derived Epo was predominantly expressed in perinatal liver (S. Yamazaki et al., unpublished observations), this TgEpo3′-rescued EpoGFP/GFP mouse—designated as ISAM— showed severe Epo-deficient anemia in adulthood (Figure 1A and Supplemental Figure 1).
Figure 1.
UUO leads to rapid loss of REP potential. (A) Schematic presentation of ISAM generation. (B) Distribution patterns of G-REPs under anemic (Ht 17%) and normal (Ht 42%) conditions. In the latter case, ISAM are treated with rHuEPO. The inset shows a higher magnification of G-REPs. (C) Epo-GFP mRNA levels after administration of rHuEPO or PBS (n=4 per group). (D) UUO-induced expression changes of Epo-GFP and αSMA in ISAM kidneys. Immunofluorescence is performed for Epo-GFP (green) and αSMA (red) expressions, showing inverse expression patterns of these proteins. EM staining shows the fibrotic area as blue. (E–H) UUO-induced expression changes of Epo-GFP, Acta2, Tgfb1, and Tnfa. Real-time PCR analyses of Epo-GFP, Acta2 (encoding αSMA), Tgfb1 (encoding TGFβ1), and Tnfa (encoding TNFα) are performed using UUO-treated and control ISAM kidneys (n>4 per group). Epo-GFP mRNA level is expressed as the relative expression compared with the contralateral kidney. Data of the sham-treated group are used as the starting point (0 day), and are set as 100% (Epo-GFP mRNA) or 1 (the other mRNAs). *P<0.05; **P<0.01. Scale bars, 20 μm (inset) and 200 μm (right) in B; 100 μm in D. Cont, contralateral; EM, Elastica-Masson.
Because GFP was inserted in-frame into the endogenous Epo coding exon, the regulatory influences of the Epo gene faithfully regulated expressions of Epo-GFP. We refer to these GFP-labeled cells in ISAM kidneys as GFP-labeled renal Epo-producing cells (G-REPs). Although G-REPs did not produce Epo, we could monitor their localization and Epo-producing potential by the protein and mRNA expressions of Epo-GFP under the reproducible severe anemia. Indeed, G-REPs were widely distributed in cortex and outer medulla (Figure 1B, left panel). The distribution and Epo-GFP mRNA expression were reduced by recombinant human Epo (rHuEPO) administration (Figure 1, B right panel and C). Compared with ISAM, our previously used models had limitations: P0-Cre labeled all glomeruli and almost all interstitial fibroblasts, and a few REPs could be GFP-labeled in the juxta-medullary region of kidneys of EpoGFP/+ mice (Supplemental Figure 2A). Thus, ISAM offer significant advantages for efficient monitoring of REPs in adult mice.
Loss of Epo-Producing Potential under Fibrogenic Conditions
To test how inflammatory and fibrogenic insults affect G-REPs, we applied UUO to ISAM. UUO treatment led to progressive renal fibrosis (Figure 1, D and F–H). GFP intensities and the number of G-REPs decreased at day 7, and a scarce number of G-REPs in the juxta-medullary region was detected at day 14 (Figure 1D and Supplemental Figure 2, B–D). The Epo-GFP transcript level was rapidly reduced by 80% at day 2 despite the persistent severe anemia (Figure 1E and Table 1). Epo-GFP mRNA level is suitable for real-time monitoring of Epo-producing potential, whereas Epo-GFP protein allowed us to trace the cell-fate for a couple of days longer by virtue of its long t1/2. These findings demonstrate that Epo-producing potentials are lost in the majority of myofibroblasts even in the severe anemic conditions.
Table 1.
UUO model did not alter the levels of genetically induced severe anemia
| UUO Model | Erythrocyte Number, ×104/μl | Hemoglobin Concentration, g/dl | Hematocrit, % |
|---|---|---|---|
| Sham | 291±35 | 4.6±0.64 | 14±1.6 |
| UUO day 1 | 308±20 | 4.8±0.13 | 15±0.69 |
| UUO day 2 | 307±29 | 4.7±0.55 | 15±1.5 |
| UUO day 3 | 299±31 | 4.6±0.57 | 14±1.4 |
| UUO day 7 | 308±48 | 4.7±0.83 | 14±2.3 |
| UUO day 14 | 302±40 | 4.8±0.58 | 15±2.3 |
All indices were not statistically significant (one-way ANOVA). Data are shown as mean ± SD.
REPs as the Major Reservoir of Myofibroblasts
Closer examinations revealed that αSMA-positive myofibroblasts appeared and increased from day 2 after UUO (Figure 2A). Most G-REPs changed into myofibroblasts, as αSMA was expressed in 50% and 80% of G-REPs at day 2 and day 3 after UUO, respectively. Thirty to forty percent of αSMA-positive cells were Epo-GFP positive (Figure 2, A and B). Concomitantly, G-REPs entered into the cell cycle and started proliferation (Supplemental Figure 3). The myofibroblast-transformed G-REPs were also observed in other models of kidney disease, indicating that the myofibroblastic transformation of G-REPs is a common pathologic outcome (Supplemental Figure 4).
Figure 2.
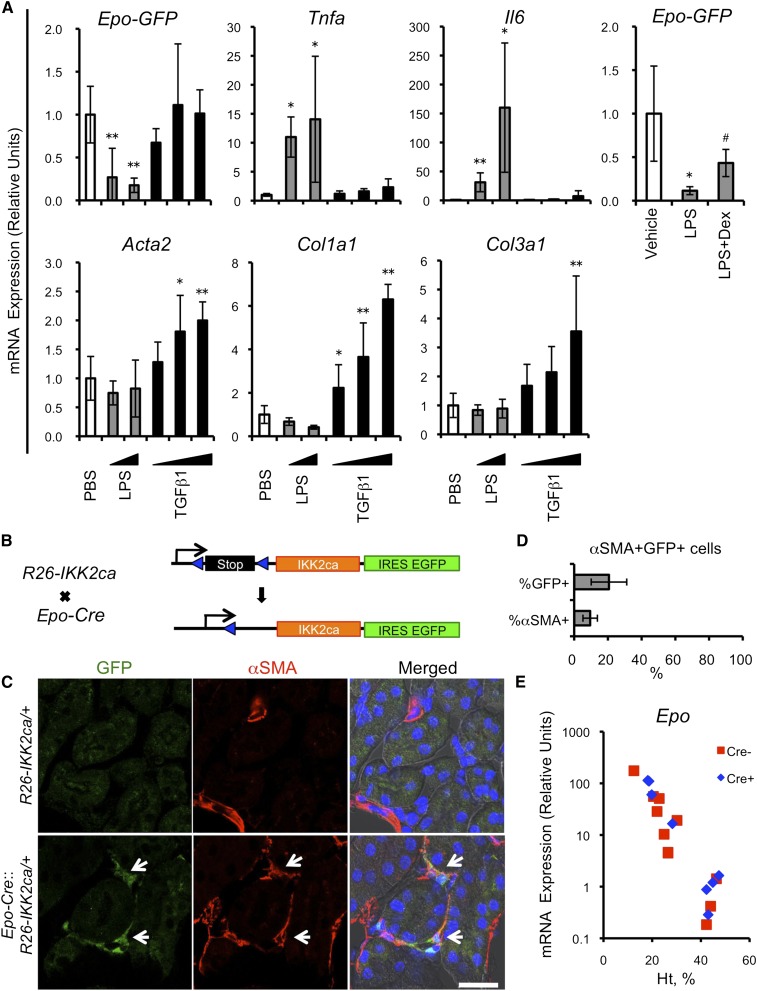
REPs are the major reservoir of myofibroblasts. (A) Expression of Epo-GFP and αSMA in early stages of UUO-treated ISAM kidneys. Immunofluorescence of Epo-GFP (green) and αSMA (red) is performed along with nuclear staining with DAPI (blue) using UUO-treated kidneys in early stages (day 1 to day 3). Note the overlapping expression of Epo-GFP and αSMA from day 2 after UUO. (B) Quantification of cells expressing both αSMA and Epo-GFP. Numbers of αSMA-positive G-REPs are compared with either those of total G-REPs (upper panel) or those of total αSMA-positive cells (lower panel) in five independent fields. **P<0.01 (n=3 per group). (C) Schematic presentation of the strategy for cell-fate analysis of REPs. A 180-kb BAC clone containing the Epo gene is used to generate an Epo-Cre mouse line. Crossing of the Epo-Cre mice with Rosa26-tdTomato (R26T) reporter mice generates Epo-Cre::R26T mice. (D) Cell-fate analysis of REPs in later stage of UUO kidneys. Native fluorescence of tdTomato (red; Epo-Cre cells) and immunofluorescence of αSMA (green) overlaps in UUO-treated kidneys at day 14, but not in sham-treated kidneys. (E) Distribution pattern of Epo-GFP and tdTomato in ISAM::Epo-Cre::R26T. Expressions of native Epo-GFP (green) and tdTomato (red) are analyzed using kidneys that underwent UUO or sham procedures (day 14). Note that tdTomato fluorescence, which represents previous and/or current expression of the Epo gene, is widely distributed in the UUO-treated kidney, but GFP fluorescence is diminished at this time point. (F) Myofibroblast transformation of Epo-Cre cells. Immunofluorescence for αSMA and tdTomato is performed using UUO kidneys at day 14. (G) Quantification of cells expressing both αSMA and tdTomato. Number of αSMA-positive Epo-Cre cells is compared with either those of total Epo-Cre cells or those of total αSMA-positive cells. (n=4). DAPI, 4',6-diamidino-2-phenylindole. Scale bars, 100 μm.
To ascertain that the G-REP–derived myofibroblasts contribute to the total population of myofibroblasts in later stages of fibrosis, we generated bacterial artificial chromosome (BAC)-transgenic mouse lines that expressed Cre recombinase under the control of a 180-kb Epo gene regulatory region (Epo-Cre mice; Figure 2C). We performed cell-fate tracing of REPs by crossing Epo-Cre mice with the tdTomato reporter line (Rosa26-tdTomato or R26T).24 We refer to Epo-Cre lineage-labeled cells as Epo-Cre cells. Epo-Cre cells recapitulated the characteristic juxta-medullary distribution of REPs, which increased with anemia, and expressed CD73, a commonly used marker of REPs (Supplemental Figure 5).1,2,25,26 To maximally label and fate-track REPs by full induction of Epo-Cre, because its induction is dependent on the level of anemia (Supplemental Figure 5G), we crossbred Epo-Cre::R26T mice with ISAM to generate ISAM::Epo-Cre::R26T. Almost all cortical and outer medullary fibroblast-like cells were labeled by tdTomato in sham-treated kidneys of ISAM::Epo-Cre::R26T. The tdTomato-positive cells were much more abundant than G-REPs (Figure 2E, sham), presumably due to labeling of cells that were previously and/or are currently producing Epo-GFP (i.e., “history of Epo production”).
Notably, the vast majority of cortical and outer medullary interstitial cells were tdTomato-positive in the UUO-treated kidney, and they lost the Epo-GFP fluorescence despite severe anemia (Figure 2E, UUO day 14). Importantly, around 80% of αSMA-positive cells (including myofibroblasts and vascular smooth muscle cells) at day 14 after UUO were tdTomato-positive (Figure 2, F and G). These results demonstrate that almost all myofibroblasts are derived from cells with a history of Epo production.
Active Contribution of REPs to Renal Inflammation and Fibrosis
To address mechanisms underlying the phenotypic transition, we focused on the inflammatory and fibrogenic signals. TNFα and TGFβ1 mRNA levels were upregulated at day 1 (Figure 1, G and H), and downstream transcriptional factors, phosphorylated p65 (p-p65) and Smad2/3 (p-Smad2/3), accumulated in the nuclei of G-REPs (Figure 3, A and B), suggesting that TGFβ1-Smad2/3 and TNFα-NFκB signaling contribute to the myofibroblastic transformation.
Figure 3.
REPs contribute to renal fibrosis and inflammation. (A and B) TGFβ and NFκB signaling in G-REPs of UUO-treated ISAM kidney. Immunofluorescence of Epo-GFP (green), p-Smad2/3 or p-p65 (red), and nucleus (blue) is performed using UUO-treated kidneys at day 1 after UUO. White arrows indicate cells that show nuclear accumulation of p-p65 or p-Smad2/3 in G-REPs. (C) Schematic presentation of the FACS protocol for isolation of G-REPs from UUO-treated (day 2) and normal ISAM kidneys. (D) Appearance of FACS-isolated G-REPs. Immunofluorescence of FACS-isolated G-REPs is performed for Epo-GFP (green) and αSMA (red), and nucleus was stained by DAPI (blue). (E) Transcriptional profiles in G-REPs upon UUO. Real-time PCR analyses are performed with FACS-isolated G-REPs to quantify Epo-GFP, Hif1a (HIF-1α), Hif2a (HIF-2α), Arnt (HIF-1β), Acta2, Col1a1 (type I collagen α1), Col3a1 (type III collagen α1), Serpine1 (plasminogen activator inhibitor 1), Rela (p65), Ccl2 (MCP1), Il6 (IL6), and Map2 (MAP2); names in parentheses indicate encoded proteins. *P<0.05; **P<0.01 (n=3, per group; one sample comprises 100,000 G-REPs collected from 5–10 mice). Scale bars, 20 μm in B.
Next, we isolated G-REPs by FACS using Epo-GFP fluorescence and analyzed transcriptional changes (Figure 3 and Supplemental Figure 3F). We found that G-REPs lost Epo-GFP expression by 90% at day 2. In contrast, mRNA expression of HIF factors (Hif1a, Hif2a, and Arnt) was maintained, suggesting that the reduction of Epo-GFP was not due to the impaired transcription of HIF factors. Expression of microtubule-associated protein 2 (Map2), a neural marker, was significantly decreased, whereas target genes of Smad2/3 (Acta2, Col1a1, Col3a1, and Serpine1) and NFκB (Il6 and Ccl2) were upregulated upon UUO (Figure 3E). Collectively, these results demonstrate that G-REP–derived myofibroblasts actively contribute to renal fibrosis and inflammation by aberrantly producing extracellular matrix proteins, an inhibitor of extracellular matrix degradation, and inflammatory cytokines and chemokines.
Repression of Epo-Producing Potential by NFκB Signaling
To explore roles that NFκB and Smad signals play in phenotypic changes of G-REPs, we administered LPS, an NFκB signal activator, or TGFβ1, and analyzed at 6 hours after the injection. LPS administration rapidly diminished the Epo-GFP expression in ISAM (Figure 4A). Coadministration of dexamethasone, which disrupts the NFκB activity, attenuated this repression, indicating the presence of an NFκB-mediated repressive regulation. However, injection of TGFβ1 did not affect the Epo-GFP expression, despite substantial inductions of its target genes (Figure 4A). Interestingly, the LPS-induced Epo-GFP repression was transient and reversible (Supplemental Figure 6). These results suggest that the rapid repression of Epo-GFP after UUO is dependent on the NFκB signaling, and imply that this repression could be reversible.
Figure 4.
Essential contribution of NFκB signaling for the dysfunction of REPs. (A) Pharmacologic activations of NFκB and Smad signals in ISAM. Real-time PCR analyses are performed using ISAM kidneys 6 hours after single intraperitoneal injection of LPS, TGFβ1, or PBS. For antagonizing NFκB signaling, dexamethasone is administered simultaneously with LPS (LPS+Dex). *P<0.05 and **P<0.01 versus PBS-injected group; #P<0.05 versus LPS-injected group (n>3 per group). (B) Schematic presentation of the strategy to selectively activate NFκB signaling in REPs. Epo-Cre mice are crossed with R26-IKK2ca mice. Expression of EGFP from the IRES cassette allows the identification of Epo-Cre cells, and indicates recombination of the R26-IKK2ca locus. (C) NFκB signals as a phenotypic switch in REPs. Immunofluorescence is performed for EGFP (green) and αSMA (red), and nucleus is stained by DAPI (blue) using kidneys from Epo-Cre::R26-IKK2ca/+ mice. White arrows indicate αSMA-positive Epo-Cre cells, indicating myofibroblastic transformation. (D) Quantification of cells expressing GFP and αSMA. The number of αSMA-positive Epo-Cre cells (GFP-positive cells) is compared with either that of total Epo-Cre cells or total αSMA-positive cells (n=4). (E) Epo mRNA expression of Epo-Cre::R26-IKK2ca/+ mice. Real-time PCR analyses are performed for quantifying Epo mRNA levels using whole kidneys. EGFP, enhanced GFP; IRES, internal ribosome entry site; Dex, dexamethasone. Scale bar, 20 μm in C.
To elucidate consequences of sustained activation of NFκB signals in REPs, we selectively activated canonical NFκB signals in REPs by crossing Epo-Cre mice with constitutively active inhibitor of IκB kinase 2 (IKK2) knock-in mice (R26-IKK2ca; Figure 4B).27 Forced expression of IKK2ca results in phosphorylation and degradation of IκB, thereby the NFκB complex activates its target genes. REPs in Epo-Cre::R26-IKK2ca/+ mice were visualized by the GFP expression from the IRES-EGFP cassette (Figure 4B). We found that approximately 20% of the GFP-positive cells expressed αSMA (Figure 4, C and D). Because Epo-Cre cells without the R26-IKK2ca allele in Epo-Cre::R26T mice were αSMA negative (Figure 2D), the observed αSMA expression could be attributable to the induced NFκB activation. Although some REPs transformed into myofibroblasts, Epo production of kidney was not attenuated, presumably due to insufficient expression of IKK2ca, and/or high reserve capacity of kidneys to produce Epo (Figure 4E). Taken together, these data suggest that NFκB signal is important not only for repressing Epo production but also for transforming REPs to myofibroblasts.
Reversal of Epo-Producing Potential by Removing Microenvironmental Cues
To elucidate whether this phenotypic transition of G-REPs is reversible, we utilized a reversible UUO model (Clip-ClipR treatment). Because most G-REPs were primed for the myofibroblastic transformation and lost their Epo-producing potential at day 2 after UUO, we reopened the obstructed ureter at this time point (Figure 5A). Notably, the Epo-GFP transcript level was restored to normal level (Figure 5B) and production of collagens are returned to the baseline at 12 days after the reopening (Figure 5C). We then analyzed the distribution and population of currently Epo-GFP producing REPs (ON-REPs or G-REPs) and total population of REPs (Epo-Cre cells) using ISAM::Epo-Cre::R26T mice that underwent Clip-ClipR treatment (Figure 5D and Supplemental Figure 7). Population of ON-REPs were comparable to those of contralateral kidneys, but the number of total REPs was increased by 2-fold (Figure 5, E–G), indicating that individual REPs restored normal Epo-producing machinery after transient proliferation through UUO-induced myofibroblastic transformation. These results thus demonstrate that G-REP–derived myofibroblasts retain functional reversibility in response to the improvement of microenvironmental cues.
Figure 5.
Elimination of UUO stress restores the normal Epo-producing potential. (A) Representative photographs of the UUO reversal model using vascular clips (Clip-ClipR treatment). We obstruct the left ureter by a vascular clip for 2 days (Clip). The vascular clip is removed 2 days after the obstruction, and then mice are euthanized 12 days after the removal (ClipR-14). (B) Recovery of Epo-producing potential of G-REPs in UUO-treated kidneys after the clip removal. Real-time PCR analysis is performed to quantify Epo-GFP mRNA levels, which are expressed as the relative expression compared with the contralateral (Cont) kidney. **P<0.01 versus sham and ClipR-14 (n=5 per group). (C) Changes of fibrogenic markers during Clip-ClipR treatment. Real-time PCR analyses are performed to quantify Acta2, Col1a1, and Col3a1 mRNA expressions using kidneys of sham, Clip, and ClipR-14 groups. ***P<0.01 versus sham and contralateral kidneys (n=5 per group). (D) Schematic presentation for the cell-fate analyses of REPs upon Clip-ClipR treatment. (E) Distribution of ON-REPs (Epo-GFP+ cells) and total REPs (tdTomato+ cells) upon Clip-ClipR treatment. Immunohistochemical analysis performed for Epo-GFP and tdTomato using kidneys of ISAM::Epo-Cre::R26T. (F and G) Changes in cell number of ON-REPs and total REPs upon Clip-ClipR treatment. FACS analyses are performed to count the number of Epo-GFP+ cells and tdTomato+ cells in ClipR-14 kidneys and contralateral (Cont) kidneys (n=5). Cont, contralateral; IHC, immunohistochemical analysis; NS, not significant. Scale bar in E, 200 μm.
Evidence for the Plasticity of Damaged REPs
We used immunoelectron microscopy to evaluate the morphologic changes of G-REPs resulting from the Clip-ClipR treatment (Figure 6A). Normal G-REPs were observed as osmium-positive angular cells that bridged capillaries and proximal tubules. At 2 days postobstruction, all G-REPs gained characteristic features of myofibroblasts, as indicated by a stellate cell shape with round nuclei and widened processes. At 12 days after the release, such changes completely disappeared. These results indicate that G-REPs possess cellular plasticity.
Figure 6.
REPs possess cellular plasticity that responds to environmental signals. (A) Morphologic changes of G-REPs upon Clip-ClipR treatment determined by immunoelectron microscopic analyses. Uninjured G-REPs appear as osmium-positive (black) cells that adhere to proximal tubular cells and capillaries (sham). In contrast, UUO alters the morphology of G-REPs to show characteristic myofibroblastic features (Clip). The morphologic features of G-REPs regain normal features 12 days after the clip release (ClipR-14). Green arrows and red arrowheads indicate G-REPs and their processes, respectively. Lower panels are higher magnification of dotted boxes in upper panels. (B) Schematic presentation for the cell-fate analyses of G-REPs. Cell-fate of G-REPs is tracked by pulse labeling using single injection of BrdU, and then mice are euthanized at day 7 (ClipR-7) and day 14 (ClipR-14). (C) BrdU incorporation into G-REPs upon Clip-ClipR treatment. Immunofluorescence of Epo-GFP (green) and BrdU (red) are performed, and nucleus is stained with DAPI (blue) using kidneys treated as depicted in B. Right panels are higher magnifications of dotted boxes in left panels. (D) Efficacy of the cell tracking using BrdU incorporation. The BrdU-incorporated G-REPs are counted in five independent fields (n≥3 per group). Approximately 7%–15% (yellow) of total G-REPs (green and yellow) are labeled with BrdU after UUO. Some tubular and GFP-negative interstitial cells also incorporate BrdU (red). PT, proximal tubular cell; C, capillary; DAPI, 4',6-diamidino-2-phenylindole. Scale bars, 10 μm (upper panels) and 0.1 μm (lower panels) in A; 20 μm in C.
To prove the plasticity of G-REPs, we performed two additional cell-fate tracking experiments. First, we tracked the cell-fate of G-REPs by detecting residual αSMA (Supplemental Figure 8, A and B). Most G-REPs retained αSMA immunoreactivity at 5 and 12 days after the release of UUO (ClipR-7 and ClipR-14, respectively; Supplemental Figure 8, C and D), indicating that the recovering G-REPs originated from myofibroblasts.
Second, we pulse labeled G-REPs with bromodeoxyuridine (BrdU), which is incorporated into the DNA of cells in S-phase and can be detected in their daughter cells,28 and determined whether BrdU-labeled myofibroblasts returned to functional G-REPs (Figure 6B). Normal G-REPs did not incorporate BrdU, indicating that the G-REPs are normally in quiescence (Figure 6C). Upon application of UUO, G-REPs incorporated BrdU, and BrdU-incorporated G-REPs were found throughout the observation period (Figure 6, C and D). These results support the notion that the UUO insults transform G-REPs into myofibroblasts, but G-REPs return to functional upon removal of UUO, and contribute to the restoration of the Epo-producing potential.
Induced DNA Methyltransferases in Myofibroblast-Transformed REPs
To further explore the molecular basis underlying the reversibility of damaged REPs, we examined expression profiles of DNA methyl transferases (DNMTs) (Dnmt1, Dnmt3a, and Dnmt3b) in UUO-treated ISAM kidneys. Expression levels of Dnmt1 and Dnmt3b were increased from early stages (from day 2), whereas Dnmt3a level was increased in later stages (from day 7; Figure 7A). Notably, expressions of Dnmt1 and Dnmt3b in UUO-treated G-REPs were increased at day 2 after UUO (Figure 7B). Clip removal reversed the expression levels of DNMTs to normal levels, suggesting that the epigenetic modifying cues were halted by this procedure (Figure 7C). These results indicate that epigenetic alteration may play a role in progression of CKD by changing the epigenetic codes of renal microenvironments as well as those of G-REPs.
Figure 7.
DNMTs are increased in myofibroblast-transformed REPs. (A) UUO-induced expression changes of DNMTs. Real-time PCR analyses of Dnmt1, Dnmt3a, and Dnmt3b are performed using UUO-treated and control ISAM kidneys (n≥3 per group). Data of the sham-treated group are used as the starting point (0 day), and set as 1. *P<0.05 and **P<0.01 versus sham. (B) Transcriptional profiles of DNMTs in G-REPs upon UUO. Real-time PCR analyses are performed with FACS-isolated G-REPs to quantify Dnmt1, Dnmt3a, and Dnmt3b. #P<0.05 and ##P<0.01 (n=3 per group; see Figure 3, C and D, for details). (C) Recovery of expression profiles of DNMTs after the clip removal. Real-time PCR analyses are performed to quantify Dnmt1, Dnmt3a, and Dnmt3b. † P<0.05 and ††P<0.01 versus sham and ClipR-14 (n=5 per group). Cont, contralateral.
Comprehensive Analyses of Microenvironmental Cues
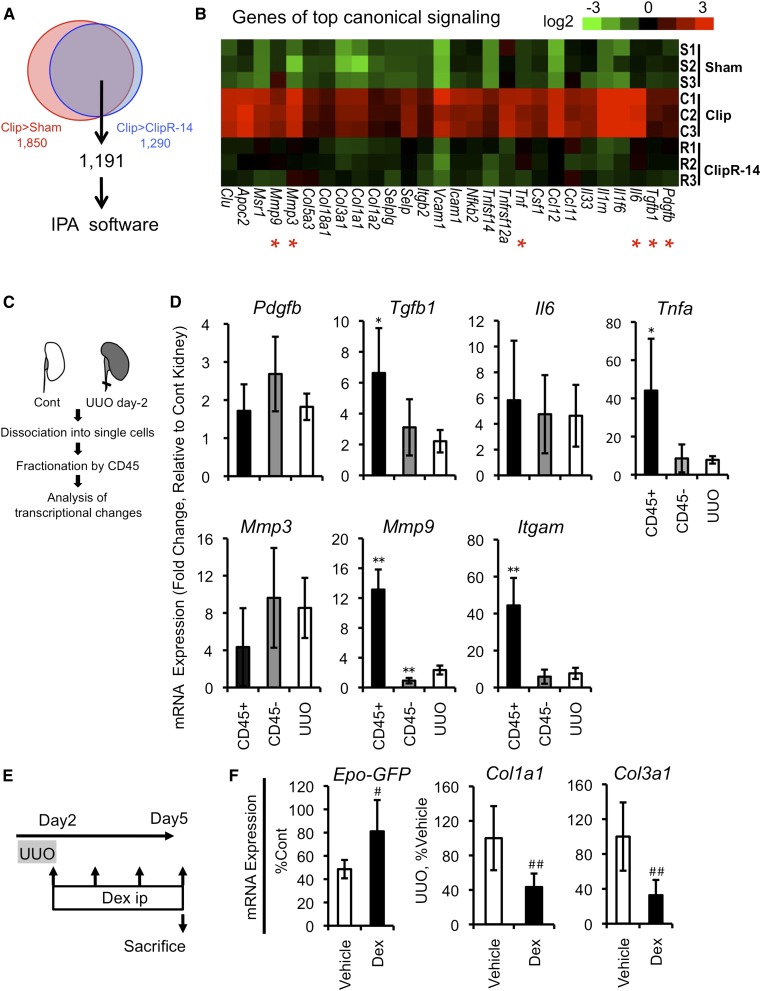
To delineate the microenvironmental cues, we performed genome-wide transcriptome analyses utilizing sham-, Clip-, and ClipR-14-treated kidneys. Pairwise comparisons of these groups revealed 1191 transcripts specifically increased in UUO-treated kidneys (Figure 8A). These were then subjected to a pathway analysis with Ingenuity Pathways Analysis (IPA), demonstrating that atherosclerosis signal and acute phase response signal were the top two key-regulated pathways (Figure 8B and Table 2). These results indicate that inflammatory cues were responsible for the UUO-induced phenotypic transition of G-REPs.
Figure 8.
Phenotypic reversions of damaged REPs are accelerated by anti-inflammatory therapy. (A) A Venn diagram summarizing the differential expression of transcripts in the kidneys with Clip-ClipR treatment. The sum of 1191 differentially regulated transcripts is identified by the microarray analyses using kidneys of sham, Clip, and ClipR-14 groups (n=3 per group, per array). (B) Top signaling pathway responsible for the myofibroblastic transformation of REPs. IPA reveals that the atherosclerosis signaling is the most significant canonical signaling pathway and a heat-map representation of genes involved in this pathway is shown. Genes marked with red asterisks are examined in D. (C) Schematic presentation of cell isolation method from UUO-treated kidneys. Infiltrating and resident leukocytes are collected from single-cell suspensions of kidneys by using anti-CD45 antibody at UUO day 2. Fractionated cells were subjected for real-time PCR analyses. (D) Infiltrated leukocytes and residual renal cells collaboratively create inflammatory microenvironments. Real-time PCR analyses are performed to quantify mRNA expression levels for Pdgfb (encoding PDGFβ), Tgfb1, Il6, Tnfa, Mmp3 (encoding matrix metalloproteinase 3), Mmp9, and Itgam (also known as CD11b, a surface marker for monocyte/macrophage). Relative mRNA levels in CD45+ fraction, CD45– fraction, and whole UUO kidneys are expressed as fold changes compared with those of contralateral kidneys (set as 1). *P<0.05 and **P<0.01 versus UUO-treated kidneys (n=5 per group). (E) Schematic presentation of dexamethasone treatment. Dex is injected daily after the clip removal for 4 days, and mice are then euthanized for the subsequent analyses. (F) Effects of dexamethasone on the recovery of G-REPs. Real-time PCR analyses are performed for quantifying mRNA levels of Epo-GFP, Col1a1, and Col3a1 using vehicle-treated and dexamethasone-treated ISAM kidneys. Epo-GFP level is expressed as the relative expression compared with the contralateral kidney. #P<0.05 and ##P<0.01 (n≥3 per group). Cont, contralateral; Dex, dexamethasone.
Table 2.
Top 10 canonical signaling pathways responsible for myofibroblastic transformation of REPs
| Ingenuity Canonical Pathway | -Log (P Value) |
|---|---|
| Atherosclerosis signaling | 1.55E01 |
| Acute phase response signaling | 1.46E01 |
| Dendritic cell maturation | 1.33E01 |
| Hepatic fibrosis/ hepatic stellate cell activation | 1.29E01 |
| LXR/RXR activation | 1.2E01 |
| Leukocyte extravasation signaling | 1.02E01 |
| Cell cycle control of chromosomal replication | 9.6E00 |
| Role of macrophages, fibroblasts, and endothelial cells in rheumatoid arthritis | 9.51E00 |
| Role of BRCA1 in DNA damage response | 9.02E00 |
| Cell cycle: G2/M DNA damage checkpoint regulation | 8.84E00 |
Accelerated Recovery of Damaged REPs by Anti-Inflammatory Therapy
To clarify cellular sources responsible for the inflammatory signals, we fractionated UUO-treated kidneys into CD45 (leukocyte common antigen)–positive leukocytes and residual renal cells (Figure 8C). Whereas Tnfa, Tgfb1, and Mmp9 were enriched in the CD45-positive fraction, others were not enriched in either fraction (Figure 8D), suggesting that the infiltrating leukocytes and other resident renal cells collaboratively created the deleterious inflammatory microenvironments.
We surmised that interventions to the inflammatory signals might exert therapeutic potentials by facilitating the recovery of myofibroblast-transformed REPs. Administration of dexamethasone after the Clip-ClipR treatment accelerated the recovery of Epo-producing potential of G-REPs (Figure 8, E and F), demonstrating the importance of inflammatory signals for the phenotypic transition and the feasibility of the therapeutic strategy (Figure 9).
Figure 9.
REPs show the dynamic phenotypic changes in health and disease. In healthy conditions, most REPs are in “OFF” state and do not produce Epo. In response to anemic and/or hypoxic stimuli, REPs turn to “ON” state and start to produce Epo. In injured kidneys, microinflammation and other stresses transform REPs into myofibroblasts, possibly due to the NFκB and Smad signals. Myofibroblast-transformed REPs lose Epo-producing potential and actively contribute to renal fibrosis and inflammation. By eliminating the inflammatory signals, myofibroblast-transformed REPs restart producing Epo, regaining their physiologic phenotypes. Persisting inflammatory signals augment expression levels of DNMTs in myofibroblast-transformed REPs and might change their epigenetic codes (“epigenetic hits”). Modulation of inflammatory signals halts the inflammatory cycles and negates the deleterious link between fibrosis and anemia. We speculate that drugs targeting epigenetic modification would be an adjunct therapeutic strategy to bring back irreversibly transformed REPs to physiologic states. In summary, REPs possess profound cellular and functional plasticity in response to microenvironmental changes. Regulating the cellular status of REPs is important for treating renal fibrosis and anemia.
Discussion
In this study, we demonstrate that REPs possess functional and cellular plasticity in response to microenvironmental changes. Damaged REPs transform into myofibroblasts, lose their Epo-producing potentials, and become the major contributor of fibrosis. However, they maintain the inherent potentials to revert to their original state when environmental cues subside.
We discovered that most myofibroblasts are derived from REPs, whereas others showed that FoxD1-labeled pericytes were a major source.14,29 These data raise a question of whether these different Cre driver lines, which are based on either cellular function or developmental origin, labeled the same or different cells. On the basis of our findings that REPs share distinctive cellular markers, such as CD73 and PDGFRβ,26 with renal pericytes and that REPs have intimate interactions with endothelium, we consider that there is a significant crossover between REPs and renal pericytes.
Inflammatory cytokines, such as TNFα, have been shown to repress hypoxia-responsive Epo-producing ability,30,31 and are assumed to be key pathogenic regulators of anemia of chronic disease32 and UUO-induced renal fibrosis.18,19,33 In turn, TGFβ/Smad signaling is the central pathway for myofibroblast activation and transformation.34,35 In progressive fibrogenesis, these two pathways amplify each other to drive inflammation and fibrosis.36–39 Here, we show that both NFκB and Smad signaling were induced in damaged REPs. By using pharmacologic activators of each signaling type, we clarified differential roles of these two signaling pathways. NFκB signaling is responsible for the rapid repression of Epo-producing potential, whereas Smad signaling is more related to the fibrogenic activity. Furthermore, we revealed that the sustained activation of NFκB signals in REPs results in a phenotypic transition. This finding is supported by the recent finding that inhibition of NFκB signaling in renal fibroblasts/myofibroblasts prevents the UUO-induced fibrosis.19 However, the fact that not all of the IKK2ca-overexpressed REPs changed into myofibroblasts implies the requirement of other coinsulting signals that might synergize with the NFκB pathway.34–39
There are growing numbers of clinical and experimental reports suggesting that structural damage of the kidney, including fibrosis, can be reversible.40–44 Therefore, elucidating whether diseased REPs in ESRD can be reversed to their physiologically normal state is an important lingering issue. Considering our findings that normal mature REPs are quiescent, but damaged REPs proliferate with a notable regenerative capacity, we assume that damaged REPs might be replenished throughout life by self-renewal under physiologic insults, such as aging, or short-term pathologic insults. Moreover, clinical observations that some patients on dialysis with ESRD become anemia-free and rHuEPO-independent during the course of treatment suggest that some REPs retain a functional reversibility, even in ESRD.45–47
TGFβ1-mediated activation of Dnmt1 is reported to be fundamental for perpetuating fibrosis.44 We found that DNMTs are increased in myofibroblast-transformed REPs. Interestingly, the Epo gene locus is known to be highly methylated in cell lines lacking Epo-producing abilities,48,49 indicating the importance of DNA methylation on epigenetic silencing of Epo gene expression. This evidence raises a possibility that sustained activation of inflammatory signaling might alter the epigenetic code of REPs and lead to difficulty in restoring myofibroblast-transformed REPs to their original state. However, considering that only a small number of REPs are required to maintain basal erythropoiesis, we surmise that a reversion of Epo-producing ability of a small number of myofibroblasts or functionally repressed REPs would be sufficient to support basal erythropoiesis of patients with ESRD.
The pathway analysis revealed the series of inflammatory signaling cascades as the most significant canonical signaling pathways responsible for the phenotypic transition. Recent reports have shown that genetic inactivation of tubule-activating signals or the depletion of macrophages improves renal microenvironments, and attenuates and even reverses fibrosis.50–52 Together with our findings that damaged REPs produce proinflammatory factors and an anti-inflammatory drug accelerates the recovery of physiologic phenotype of REPs, the pathogenic significance of a deleterious inflammatory cycle by REPs and surrounding cells is implied. We consider that the therapeutic treatments that control the proinflammatory activity of tubular epithelial cells, infiltrating leukocytes, and REPs would halt the inflammatory cycle, protect REPs, and even reverse their transformation.
In summary, our study demonstrates that REPs possess profound cellular plasticity and that the damaged REPs directly link fibrosis and anemia. Future studies are needed to elucidate the exact mechanism underlying the plasticity of REPs, which will open up new avenues for treating CKD by protecting and regenerating REPs.
Concise Methods
Mice
Mice were maintained in a specific pathogen-free facility. All experimental procedures were approved by the Animal Care Committee at Tohoku University. Mice aged 8–14 weeks were used. We used age- and sex-matched mice for each experiment. Cre-negative littermates were used as a control when phenotypic differences were examined with mice groups of different genotypes. ISAM were generated by transgenic complementary rescue of homozygous Epo GFP-knock-in mice (EpoGFP/GFP) (S. Yamazaki et al., unpublished observations). The 8-kb DNA construct containing the Epo gene was used (TgEpo3′) for this purpose. ISAM showed adult-onset Epo-deficient anemia because TgEpo3′-derived Epo in serum decreased to almost undetectable level from weaning age. Epo-Cre mice were generated using a BAC clone containing the 180-kb fragment sufficient for recapitulating endogenous Epo gene regulation.2 To prepare the Epo-Cre construct, Cre cDNA was integrated into the second exon of Epo-60K/BAC (S. Yamazaki et al., unpublished observations). Of the multiple transgenic mouse lines established, we chose one line (line #44) for further study, which showed the characteristic renal interstitial expression pattern of REPs and the anemic induction, when examined by crossing with R26T mice (from Jackson Laboratory)24 and EpoGFP/+ mice.2,23 To generate Epo-Cre::R26-IKK2ca/+ mice, we crossed Epo-Cre mice with homozygous R26-IKK2ca mice (from Jackson Laboratory).27 Genotyping was performed by PCR using primers described in Supplemental Table 1.
UUO Model
For the ligature method, left ureter was exposed by a flank incision and ligated twice by 4-0 polysorb (Syneture) at the level of the lower pole of the kidney. The ureter was cut between ligatures. For the reversible UUO model (Clip-ClipR treatment), a vascular clip (Bear Medical Corp.) was used instead of the suture. After 48 hours of the ureter clipping, the vascular clip was removed and mice were euthanized 5 and 12 days after releasing the clips (ClipR-7 and ClipR-14, respectively). Sham-operated uninjured kidneys were used as controls. For anti-inflammatory treatment, mice were intraperitoneally injected with dexamethasone (10 μg/g body weight) daily after the removal of vascular clip, and euthanized at 3 days after clip removal. The appropriate vehicle treatment group was used as a control. To evaluate the Epo-producing potential, the Epo-GFP mRNA expression of the contralateral kidney was used to normalize the anemic stimuli due to individual differences of anemic severity. Sham operation was performed in the same way except for ligating or clipping the ureter.
rHuEPO Treatment
rHuEPO (3000 U/kg, a generous gift from Chugai Pharmaceutical Co. Ltd.) was intraperitoneally administered every other day for 12 days to reverse Epo-deficient anemia. A PBS-treated group was used as a control.
LPS, hTGFβ1, and Dexamethasone Treatment
Mice were intraperitoneally injected with 0.1 and 1 μg/g body weight LPS (less than one tenth of the dose for proteinuria induction, and one fiftieth of the dose for the sepsis model; Sigma) and with 10, 20, and 40 ng/g body weight of human TGFβ1 (R&D Systems). To antagonize the NFκB signaling, dexamethasone (10 μg/g body weight; Sigma) was coadministered with LPS (0.1 μg/g body weight). At the time indicated, mice were euthanized and their kidneys were dissected out for RNA isolation. Appropriate vehicle treatment groups were used as controls.
Tissue Preparation for Histologic Analysis
To prepare frozen sections, tissues were fixed with 4% paraformaldehyde for 2 hours, washed with 20% sucrose in PBS overnight, and then embedded in optimal cutting temperature compound (Sakura FineTek). Tissues for paraffin-embedded sections were fixed with 4% paraformaldehyde for 48 hours, and then processed by conventional procedures.
Immunohistochemistry
For fluorescence labeling, 10-μm frozen or paraffin-embedded sections were used. Sections were blocked and incubated with primary antibodies. Primary antibodies against GFP (Medical & Biological Laboratories and Abcam), αSMA (Sigma), BrdU (BD Pharmingen), Ki67 (DAKO), CD73 (eBioscience), p-Smad2/3 (Santa Cruz Biotechnology), and p-p65 (Cell Signaling Technology) were used. Appropriate Alexa 488- or 555-conjugated secondary antibodies were used for fluorescent detection, and nuclei were stained with 4',6-diamidino-2-phenylindole. For paraffin-embedded sections, citrate buffer (pH 6.0) was used for antigen retrieval. For p-p65 staining, tyramide amplification was used (Invitrogen). Fluorescent images were obtained using an LSM510 confocal imaging system (Carl Zeiss).
Immunoelectron Microscopic Analyses
Frozen sections (10 μm) were used for immunoelectron microscopic analysis. Sections were incubated with an anti-GFP antibody (Medical & Biological Laboratories) to detect G-REPs. Then, 3,3-diaminobenzidine staining was performed using appropriate secondary antibody and a 3,3-diaminobenzidine staining kit (DAKO). After osmium staining, sections were embedded in resin. Embedded sections were processed to ultrathin sections for electron microscopic analysis.
Isolation of G-REPs by FACS
Kidneys were dissected after intracardiac perfusion with ice-cold PBS. Then, kidneys were decapsulated and minced. The minced whole kidney was digested with Liberase TM (Roche) with hyaluronidase (Sigma) and DNase I (Roche) for 2.5 hours at 37°C. The cell suspension was sieved through 40-μm nylon meshes. The resulting single-cell suspension was washed with 5% FBS in PBS. Dead cells were stained with 1 μg/ml propidium iodide and excluded from collection. FACS was performed using a FACSAria2 (BD Bioscience). GFP-positive cells from 5–10 kidneys were sorted and combined to collect 100,000 cells for RNA extraction.
Isolation of Leukocytes from Kidneys Using Magnetic Beads
Kidneys were digested into single-cell suspensions, and incubated with biotinylated anti-CD45 antibody (eBioscience) and with streptavidin-conjugated magnetic beads (Invitrogen). Fractionated cells were resuspended into Isogen (Nippon gene).
Real-Time PCR Analyses
Isogen was used to prepare total RNA. Total RNA (3 μg) was used for reverse transcription using random hexamers and a Superscript III polymerase kit (Invitrogen). Real-time PCR was then performed with appropriate primers (Supplemental Table 1) using an ABI prism 7300 (Applied Biosystems). Gene expression levels were calculated based on the threshold cycle (Ct) values and the efficiency of each primer set, and expressed as relative to 18S rRNA.
Microarray Analyses
Total RNA was prepared from whole kidneys using Isogen. The RNA quantity and quality were determined using an Agilent Bioanalyzer 2100 (Agilent Technologies). Gene expression data were obtained by hybridizing a total of nine ISAM kidney samples: sham, Clip, and ClipR-14 (n=3 per group) to a 4×44K whole-mouse genome array (Agilent Technologies). Data were analyzed by Gene Spring software (Agilent Technologies), normalized to the 75th percentile shift and the baseline transformed to the median of all samples to identify differentially expressed genes (P<0.01, one-way ANOVA). The data were submitted to the Gene Expression Omnibus (GSE45304). To identify factors responsible for myofibroblastic transformation of REPs, we chose >2-fold upregulated genes by comparing Clip with either sham or ClipR-14. The top canonical signaling pathways were identified using IPA software (http;//www.ingenuity.com; Ingenuity Systems). Heat maps were generated using Cluster3.0 and JAVA Treeview software.
Cell-Fate Tracking and Proliferation Analyses by BrdU
To label proliferating cells, we pulse labeled cells with BrdU (50 μg/g body weight; BD Pharmingen) 2 days after clipping the left ureter. Three groups were prepared: Clip (mice were euthanized at 3 hours after BrdU injection without releasing the clip), and ClipR-7 and ClipR-14 (mice were euthanized 5 and 12 days after the releasing the clip, respectively). Releasing of the clip was performed 3 hours after BrdU injection. For control experiments, uninjured mice were euthanized 3 hours and 12 days after BrdU injections.
Statistical Analyses
Data were expressed as means ± SD. Comparisons between groups were performed using one-way ANOVA or an unpaired t test as appropriate. For multiple comparisons, the Tukey–Kramer test was used. Data were considered statistically significant at P<0.05.
Disclosures
None.
Supplementary Material
Acknowledgments
The authors thank Professors Hozumi Motohashi and Mitchell Halperin for their invaluable advice, Ms. Maggie Patient for editing our manuscript, and Ms. Kiyomi Kisu for helping with immunoelectron microscopy. The authors also thank Biomedical Research Core and Center for Laboratory Animal Research of Tohoku University Graduate School of Medicine for technical support, and Chugai Pharmaceutical Co Ltd for comments.
This work was supported in part by JST, CREST (to M.Y.); Grants-in-Aid for Creative Scientific Research (to M.Y.), for Scientific Research (to M.Y.), and for JSPS fellows (to T.S. and S.Y.) from the JSPS; Grants-in-Aid for Scientific Research on Priority Areas (to M.Y.) from the MEXT; and a research grant from the Kidney Foundation, Japan (JKFB12-1 to T.S.).
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
This article contains supplemental material online at http://jasn.asnjournals.org/lookup/suppl/doi:10.1681/ASN.2013010030/-/DCSupplemental.
References
- 1.Maxwell PH, Osmond MK, Pugh CW, Heryet A, Nicholls LG, Tan CC, Doe BG, Ferguson DJP, Johnson MH, Ratcliffe PJ: Identification of the renal erythropoietin-producing cells using transgenic mice. Kidney Int 44: 1149–1162, 1993 [DOI] [PubMed] [Google Scholar]
- 2.Obara N, Suzuki N, Kim K, Nagasawa T, Imagawa S, Yamamoto M: Repression via the GATA box is essential for tissue-specific erythropoietin gene expression. Blood 111: 5223–5232, 2008 [DOI] [PubMed] [Google Scholar]
- 3.Wenger RH, Hoogewijs D: Regulated oxygen sensing by protein hydroxylation in renal erythropoietin-producing cells. Am J Physiol Renal Physiol 298: F1287–F1296, 2010 [DOI] [PubMed] [Google Scholar]
- 4.Haase VH: Hypoxic regulation of erythropoiesis and iron metabolism. Am J Physiol Renal Physiol 299: F1–F13, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eckardt KU, Koury ST, Tan CC, Schuster SJ, Kaissling B, Ratcliffe PJ, Kurtz A: Distribution of erythropoietin producing cells in rat kidneys during hypoxic hypoxia. Kidney Int 43: 815–823, 1993 [DOI] [PubMed] [Google Scholar]
- 6.Koury ST, Koury MJ, Bondurant MC, Caro J, Graber SE: Quantitation of erythropoietin-producing cells in kidneys of mice by in situ hybridization: Correlation with hematocrit, renal erythropoietin mRNA, and serum erythropoietin concentration. Blood 74: 645–651, 1989 [PubMed] [Google Scholar]
- 7.Risdon RA, Sloper JC, De Wardener HE: Relationship between renal function and histological changes found in renal-biopsy specimens from patients with persistent glomerular nephritis. Lancet 2: 363–366, 1968 [DOI] [PubMed] [Google Scholar]
- 8.Quaggin SE, Kapus A: Scar wars: Mapping the fate of epithelial-mesenchymal-myofibroblast transition. Kidney Int 80: 41–50, 2011 [DOI] [PubMed] [Google Scholar]
- 9.Nangaku M, Eckardt KU: Pathogenesis of renal anemia. Semin Nephrol 26: 261–268, 2006 [DOI] [PubMed] [Google Scholar]
- 10.Fishbane S: Hematological aspects of kidney disease. In: Brenner & Rector's The Kidney, 8th Ed., edited by Brenner BM, Philadelphia, Saunders, 2008, pp 1728–1743 [Google Scholar]
- 11.Maxwell PH, Ferguson DJ, Nicholls LG, Johnson MH, Ratcliffe PJ: The interstitial response to renal injury: Fibroblast-like cells show phenotypic changes and have reduced potential for erythropoietin gene expression. Kidney Int 52: 715–724, 1997 [DOI] [PubMed] [Google Scholar]
- 12.Asada N, Takase M, Nakamura J, Oguchi A, Asada M, Suzuki N, Yamamura K, Nagoshi N, Shibata S, Rao TN, Fehling HJ, Fukatsu A, Minegishi N, Kita T, Kimura T, Okano H, Yamamoto M, Yanagita M: Dysfunction of fibroblasts of extrarenal origin underlies renal fibrosis and renal anemia in mice. J Clin Invest 121: 3981–3990, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kriz W, Kaissling B, Le Hir M: Epithelial-mesenchymal transition (EMT) in kidney fibrosis: Fact or fantasy? J Clin Invest 121: 468–474, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Humphreys BD, Lin SL, Kobayashi A, Hudson TE, Nowlin BT, Bonventre JV, Valerius MT, McMahon AP, Duffield JS: Fate tracing reveals the pericyte and not epithelial origin of myofibroblasts in kidney fibrosis. Am J Pathol 176: 85–97, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mezzano SA, Barría M, Droguett MA, Burgos ME, Ardiles LG, Flores C, Egido J: Tubular NF-kappaB and AP-1 activation in human proteinuric renal disease. Kidney Int 60: 1366–1377, 2001 [DOI] [PubMed] [Google Scholar]
- 16.Schmid H, Boucherot A, Yasuda Y, Henger A, Brunner B, Eichinger F, Nitsche A, Kiss E, Bleich M, Gröne HJ, Nelson PJ, Schlöndorff D, Cohen CD, Kretzler M, European Renal cDNA Bank (ERCB) Consortium : Modular activation of nuclear factor-kappaB transcriptional programs in human diabetic nephropathy. Diabetes 55: 2993–3003, 2006 [DOI] [PubMed] [Google Scholar]
- 17.Woroniecka KI, Park AS, Mohtat D, Thomas DB, Pullman JM, Susztak K: Transcriptome analysis of human diabetic kidney disease. Diabetes 60: 2354–2369, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo GJ, Morrissey J, McCracken R, Tolley T, Klahr S: Role of TNFR1 and TNFR2 receptors in tubulointerstitial fibrosis of obstructive nephropathy. Am J Physiol 277: F766–F772, 1999 [DOI] [PubMed] [Google Scholar]
- 19.Inoue T, Takenaka T, Hayashi M, Monkawa T, Yoshino J, Shimoda K, Neilson EG, Suzuki H, Okada H: Fibroblast expression of an IκB dominant-negative transgene attenuates renal fibrosis. J Am Soc Nephrol 21: 2047–2052, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu H, Liu X, Jaenisch R, Lodish HF: Generation of committed erythroid BFU-E and CFU-E progenitors does not require erythropoietin or the erythropoietin receptor. Cell 83: 59–67, 1995 [DOI] [PubMed] [Google Scholar]
- 21.Semenza GL, Nejfelt MK, Chi SM, Antonarakis SE: Hypoxia-inducible nuclear factors bind to an enhancer element located 3′ to the human erythropoietin gene. Proc Natl Acad Sci U S A 88: 5680–5684, 1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Semenza GL, Traystman MD, Gearhart JD, Antonarakis SE: Polycythemia in transgenic mice expressing the human erythropoietin gene. Proc Natl Acad Sci U S A 86: 2301–2305, 1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Suzuki N, Obara N, Pan X, Watanabe M, Jishage K, Minegishi N, Yamamoto M: Specific contribution of the erythropoietin gene 3′ enhancer to hepatic erythropoiesis after late embryonic stages. Mol Cell Biol 31: 3896–3905, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, Lein ES, Zeng HK: A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci 13: 133–140, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaissling B, Le Hir M: The renal cortical interstitium: Morphological and functional aspects. Histochem Cell Biol 130: 247–262, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pan X, Suzuki N, Hirano I, Yamazaki S, Minegishi N, Yamamoto M: Isolation and characterization of renal erythropoietin-producing cells from genetically produced anemia mice. PLoS ONE 6: e25839, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sasaki Y, Derudder E, Hobeika E, Pelanda R, Reth M, Rajewsky K, Schmidt-Supprian M: Canonical NF-kappaB activity, dispensable for B cell development, replaces BAFF-receptor signals and promotes B cell proliferation upon activation. Immunity 24: 729–739, 2006 [DOI] [PubMed] [Google Scholar]
- 28.van Praag H, Schinder AF, Christie BR, Toni N, Palmer TD, Gage FH: Functional neurogenesis in the adult hippocampus. Nature 415: 1030–1034, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin SL, Kisseleva T, Brenner DA, Duffield JS: Pericytes and perivascular fibroblasts are the primary source of collagen-producing cells in obstructive fibrosis of the kidney. Am J Pathol 173: 1617–1627, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Frede S, Fandrey J, Pagel H, Hellwig T, Jelkmann W: Erythropoietin gene expression is suppressed after lipopolysaccharide or interleukin-1 beta injections in rats. Am J Physiol 273: R1067–R1071, 1997 [DOI] [PubMed] [Google Scholar]
- 31.Nakano Y, Imagawa S, Matsumoto K, Stockmann C, Obara N, Suzuki N, Doi T, Kodama T, Takahashi S, Nagasawa T, Yamamoto M: Oral administration of K-11706 inhibits GATA binding activity, enhances hypoxia-inducible factor 1 binding activity, and restores indicators in an in vivo mouse model of anemia of chronic disease. Blood 104: 4300–4307, 2004 [DOI] [PubMed] [Google Scholar]
- 32.Weiss G, Goodnough LT: Anemia of chronic disease. N Engl J Med 352: 1011–1023, 2005 [DOI] [PubMed] [Google Scholar]
- 33.Ernandez T, Mayadas TN: Immunoregulatory role of TNFalpha in inflammatory kidney diseases. Kidney Int 76: 262–276, 2009 [DOI] [PubMed] [Google Scholar]
- 34.Sato M, Muragaki Y, Saika S, Roberts AB, Ooshima A: Targeted disruption of TGF-beta1/Smad3 signaling protects against renal tubulointerstitial fibrosis induced by unilateral ureteral obstruction. J Clin Invest 112: 1486–1494, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koesters R, Kaissling B, Lehir M, Picard N, Theilig F, Gebhardt R, Glick AB, Hähnel B, Hosser H, Gröne HJ, Kriz W: Tubular overexpression of transforming growth factor-beta1 induces autophagy and fibrosis but not mesenchymal transition of renal epithelial cells. Am J Pathol 177: 632–643, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fukasawa H, Yamamoto T, Togawa A, Ohashi N, Fujigaki Y, Oda T, Uchida C, Kitagawa K, Hattori T, Suzuki S, Kitagawa M, Hishida A: Down-regulation of Smad7 expression by ubiquitin-dependent degradation contributes to renal fibrosis in obstructive nephropathy in mice. Proc Natl Acad Sci U S A 101: 8687–8692, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seki E, De Minicis S, Osterreicher CH, Kluwe J, Osawa Y, Brenner DA, Schwabe RF: TLR4 enhances TGF-beta signaling and hepatic fibrosis. Nat Med 13: 1324–1332, 2007 [DOI] [PubMed] [Google Scholar]
- 38.Lan HY: Diverse roles of TGF-β/Smads in renal fibrosis and inflammation. Int J Biol Sci 7: 1056–1067, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu Z, Huang XR, Chen HY, Penninger JM, Lan HY: Loss of angiotensin-converting enzyme 2 enhances TGF-β/Smad-mediated renal fibrosis and NF-κB-driven renal inflammation in a mouse model of obstructive nephropathy. Lab Invest 92: 650–661, 2012 [DOI] [PubMed] [Google Scholar]
- 40.Fioretto P, Steffes MW, Sutherland DE, Goetz FC, Mauer M: Reversal of lesions of diabetic nephropathy after pancreas transplantation. N Engl J Med 339: 69–75, 1998 [DOI] [PubMed] [Google Scholar]
- 41.Zeisberg M, Hanai J, Sugimoto H, Mammoto T, Charytan D, Strutz F, Kalluri R: BMP-7 counteracts TGF-beta1-induced epithelial-to-mesenchymal transition and reverses chronic renal injury. Nat Med 9: 964–968, 2003 [DOI] [PubMed] [Google Scholar]
- 42.Lin J, Patel SR, Cheng X, Cho EA, Levitan I, Ullenbruch M, Phan SH, Park JM, Dressler GR: Kielin/chordin-like protein, a novel enhancer of BMP signaling, attenuates renal fibrotic disease. Nat Med 11: 387–393, 2005 [DOI] [PubMed] [Google Scholar]
- 43.Fioretto P, Sutherland DE, Najafian B, Mauer M: Remodeling of renal interstitial and tubular lesions in pancreas transplant recipients. Kidney Int 69: 907–912, 2006 [DOI] [PubMed] [Google Scholar]
- 44.Bechtel W, McGoohan S, Zeisberg EM, Müller GA, Kalbacher H, Salant DJ, Müller CA, Kalluri R, Zeisberg M: Methylation determines fibroblast activation and fibrogenesis in the kidney. Nat Med 16: 544–550, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Takeda A, Toda T, Shinohara S, Mogi Y, Matsui N: Factors contributing to higher hematocrit levels in hemodialysis patients not receiving recombinant human erythropoietin. Am J Kidney Dis 40: 104–109, 2002 [DOI] [PubMed] [Google Scholar]
- 46.Kuo CC, Lee CT, Chuang CH, Su Y, Chen JB: Recombinant human erythropoietin independence in chronic hemodialysis patients: Clinical features, iron homeostasis and erythropoiesis. Clin Nephrol 63: 92–97, 2005 [DOI] [PubMed] [Google Scholar]
- 47.Schwartz DI, Pierratos A, Richardson RM, Fenton SS, Chan CT: Impact of nocturnal home hemodialysis on anemia management in patients with end-stage renal disease. Clin Nephrol 63: 202–208, 2005 [DOI] [PubMed] [Google Scholar]
- 48.Yin H, Blanchard KL: DNA methylation represses the expression of the human erythropoietin gene by two different mechanisms. Blood 95: 111–119, 2000 [PubMed] [Google Scholar]
- 49.Steinmann K, Richter AM, Dammann RH: Epigenetic silencing of erythropoietin in human cancers. Genes Cancer 2: 65–73, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lin SL, Castaño AP, Nowlin BT, Lupher ML, Jr, Duffield JS: Bone marrow Ly6Chigh monocytes are selectively recruited to injured kidney and differentiate into functionally distinct populations. J Immunol 183: 6733–6743, 2009 [DOI] [PubMed] [Google Scholar]
- 51.Bielesz B, Sirin Y, Si H, Niranjan T, Gruenwald A, Ahn S, Kato H, Pullman J, Gessler M, Haase VH, Susztak K: Epithelial Notch signaling regulates interstitial fibrosis development in the kidneys of mice and humans. J Clin Invest 120: 4040–4054, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sugimoto H, LeBleu VS, Bosukonda D, Keck P, Taduri G, Bechtel W, Okada H, Carlson W, Jr, Bey P, Rusckowski M, Tampe B, Tampe D, Kanasaki K, Zeisberg M, Kalluri R: Activin-like kinase 3 is important for kidney regeneration and reversal of fibrosis. Nat Med 18: 396–404, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.