Abstract
The centromere is essential for the proper segregation and inheritance of genetic information. Neocentromeres are ectopic centromeres that originate occasionally from noncentromeric regions of chromosomes. Despite the complete absence of normal centromeric α-satellite DNA, human neocentromeres are able to form a primary constriction and assemble a functional kinetochore. Since the discovery and characterization of the first case of a human neocentromere in our laboratory a decade ago, 60 examples of constitutional human neocentromeres distributed widely across the genome have been described. Typically, these are located on marker chromosomes that have been detected in children with developmental delay or congenital abnormalities. Neocentromeres have also been detected in at least two types of human cancer and have been experimentally induced in Drosophila. Current evidence from human and fly studies indicates that neocentromere activity is acquired epigenetically rather than by any alteration to the DNA sequence. Since human neocentromere formation is generally detrimental to the individual, its biological value must lie beyond the individual level, such as in karyotype evolution and speciation.
Introduction
Human neocentromeres are new centromeres that appear in chromosomal locations other than that of the original centromere. The centromere is of critical importance to chromosome inheritance, with the presence of one functional centromere per chromosome being an absolute requirement. The creation of a new centromere is, therefore, an extraordinary event, with potential implications not only for the chromosome involved but also for the cell, the organism, and the species.
Centromeres are defined cytogenetically by a constriction in the chromosome, generally embedded in darkly staining heterochromatin. At the center of the constriction lies the kinetochore, a complex DNA-protein structure that attaches to microtubules and helps to direct chromosome movement along the spindle. The centromeric DNA comprises large numbers of repeat sequences in tandem arrays. In humans, centromeres typically contain 2,000–4,000 kb of a 171-bp repeat known as “α-satellite” (Choo 1997a). Although the repeat sequences themselves are poorly conserved across phylogeny, the presence of satellite DNA at centromeres is a feature of virtually all eukaryotic organisms (the one exception being the budding yeast Saccharomyces cerevisiae). Furthermore, introduction of α-satellite DNA into cultured cells can result in de novo centromere formation, indicating that the satellite sequence has a fundamental role in centromere formation (Harrington et al. 1997; Ikeno et al. 1998; Henning et al. 1999; Ebersole et al. 2000).
Against this background, the discovery a decade ago of a human neocentromere that lacked any α-satellite sequence (Voullaire et al. 1993) was startling and unexpected. This was a new type of neocentromere, quite different from the first neocentromeres described in maize half a century earlier (Rhoades and Vilkomerson 1942) (see the “Maize Knobs: a Different Type of Centromere” section). The first human neocentromere was detected on a marker chromosome during the routine karyotyping of a boy with learning difficulties. This marker, designated “mardel(10),” was derived from a de novo complex rearrangement of chromosome 10 that had resulted in loss of the original centromere. Despite the complete absence of α-satellite DNA (fig. 1A), the neocentromere was able to form a primary constriction and assemble a functional kinetochore that was stable in mitosis. In the decade following, 60 different constitutional human neocentromeres have been reported, typically located on rearranged marker chromosomes that have similarly lost their centromeres. Neocentromeres have also been detected in human cancers and have been produced experimentally in Drosophila. These cases have provided valuable insight into the structure, function, and regulation of neocentromeres, and they have provided an important model system for dissecting the function of normal centromeres. A greater understanding of the properties of neocentromeres has also opened up new fields of study, such as epigenetic mechanisms regulating centromere and neocentromere formation and propagation, the possible role of neocentromeres in karyotype and species evolution, and the use of neocentromere-based artificial chromosomes for human gene therapy.
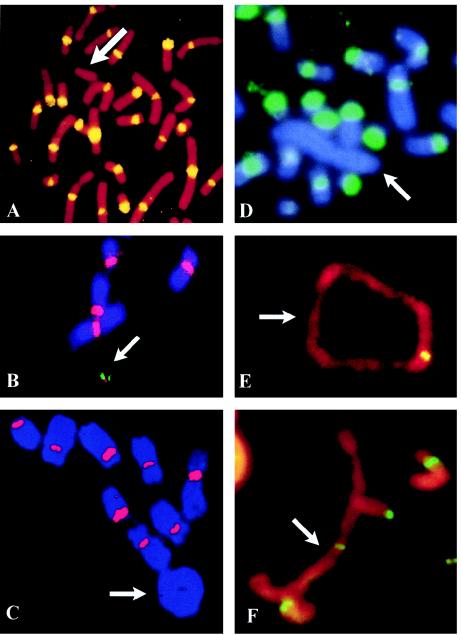
Figure 1.
FISH analysis of human neocentric chromosomes. A, Patient cells with a 10q25 neocentromere-containing mardel(10) chromosome (arrow), using pancentromeric α-satellite probe, demonstrating absence of α-satellite (yellow) on the marker chromosome. Image taken from Voullaire et al. (1993). B, A stable, <2-Mb HAC (arrow) engineered from the mardel(10) chromosome shown in A (Saffery et al. 2001). Chromosome staining is with DAPI. Image courtesy of L. Wong. C–F, Partial metaphases of well-differentiated liposarcoma cases, using FISH with a pancentromeric α-satellite probe (C and D) and immunostaining with anticentromere antibody (E and F). Arrows indicate the supernumerary rings and large rod marker chromosomes. FISH signals (red in C or green in D) with the α-satellite probe are observed on all chromosomes except the supernumerary ring (C) and large marker (D). Positive staining with the anticentromere antibody (yellow or green) is observed on all chromosomes including the supernumerary analphoid ring (E) and large marker (F). Images courtesy of F. Pedeutour and N. Sirvent.
Constitutional Human Neocentromeres
Figure 2 and table 1 summarize 60 reported cases of constitutional human neocentromeres in which chromosomal origin has been investigated. Constitutional human neocentromeres are typically analphoid (i.e., they lack detectable α-satellite), are C-band negative, contain a primary constriction, and, where tested, bind essential centromere proteins indicative of formation of a functional kinetochore. Most human neocentromeres have been ascertained either at prenatal diagnosis or by the cytogenetic analysis of individuals with congenital abnormalities, developmental delay, or intellectual disability.
Figure 2.
Sites of formation of constitutional neocentromeres within the human genome. A total of 60 cases, originating from 16 different human chromosomes, have been described. The mapped positions of each of the neocentromere cases are indicated by bars to the right of the chromosome ideograms. Longer bars indicate neocentromere sites that have not been precisely localized. Hatch marks on chromosomes 1, 9, and Y represent blocks of constitutive heterochromatin. Adapted from Choo (2001a).
Table 1.
Sixty Human Constitutional Neocentromere Cases
|
Mosaicism(% Abnormal Cells) |
|||||
| Chromosome andNeocentromere Site | Rearrangementa | Karyotype | Lymphoblast | Fibroblast | Reference(s) |
| 1: | |||||
| 1p32-p36.1 | Interstitial deletion (paracentric) | Balanced | 97 | 100 | Slater et al. 1999 |
| 1q23-q32 | Interstitial deletion | Balanced | 85 | Higgins et al. 2000 | |
| 1q43-44 | Supernumerary ring | Trisomy | 70 | 50 | Spiegel et al., in press |
| 2: | |||||
| 2p11-p21 | Interstitial deletion (paracentric) | Balanced | 100 | Petit and Fryns 1997 | |
| 3: | |||||
| 3p23 | Interstitial deletion (pericentric) | Balanced | 100 | 100 | Maraschio et al. 1996 |
| 3q26 | Interstitial deletion (pericentric) | Balanced | 100 | 100 | Wandall et al. 1998 |
| 3q27.2-qter | Supernumerary inv dup | Tetrasomy | 71 | Teshima et al. 2000 | |
| 3q26.2-qter | Supernumerary inv dup | Tetrasomy | 2 | 87 | Teshima et al. 2000 |
| 3q27.1-qter | Supernumerary inv dup | Tetrasomy | 30 | 6 | Portnoi et al. 1999 |
| 3q26.2-qter | Supernumerary inv dup | Tetrasomy | 57 | Cockwell et al. 2000 | |
| 3q21.2-qter | Supernumerary inv dup | Tetrasomy | 0 | 87 | Gimelli et al. 2000 |
| 4: | |||||
| 4q21 | Interstitial deletion (paracentric) | Balanced | 75 | Grimbacher et al. 1999 | |
| 5: | |||||
| 5p14-15 | Supernumerary inv dup | Tetrasomy | 19 | Fritz et al. 2001 | |
| 8: | |||||
| 8p23.1-pter | Supernumerary inv dup | Tetrasomy | 100 | Ohashi et al. 1994 | |
| 8q23-qter | Supernumerary inv dup | Tetrasomy | 75 | Sulcova et al. 1999 | |
| 8q23.3-qter | Supernumerary inv dup | Tetrasomy | 75 | Reddy et al. 2000 | |
| 8p23.2 | Supernumerary inv dup | Tetrasomy | 23–46 | Voullaire et al. 2001 | |
| 8p23.2 | Supernumerary inv dup | Tetrasomy | 53–60 | Voullaire et al. 2001 | |
| 8p23.1-pter | Supernumerary inv dup | Tetrasomy | 100 | Li et al. 2002 | |
| 9: | |||||
| 9p23 | Deletion + inv dup | Trisomy | 100 | Vance et al. 1997; Satinover et al. 2001b | |
| 9p23 | Supernumerary inv dup | Tetrasomy | 100 | Depinet et al. 1997; Satinover et al. 2001b | |
| 10: | |||||
| 10q25.2 | Interstitial deletion (pericentric) | Balanced | 100 | 100 | Voullaire et al. 1993 |
| 10q11-q23 | Interstitial deletion (paracentric) | Balanced | 62 | 80 | Depinet et al. 1997 |
| 10p14-pter | Supernumerary inv dup | Tetrasomy | 100 | Levy et al. 2000 | |
| 11: | |||||
| 11q22-qter | Deletion + inv dup | Trisomy | 100 | 100 | Depinet et al. 1997 |
| 12: | |||||
| 12p12.3-pter | Supernumerary inv dup | Tetrasomy | 1 | 45–57 | Dufke et al. 2001 |
| 13: | |||||
| 13q21 | Deletion + inv dup | Trisomy | 100 | Warburton et al. 2000 | |
| 13q32 | Deletion + inv dup | Trisomy | 100 | Rivera et al. 1999; Warburton et al. 2000 | |
| 13q21 | Supernumerary inv dup | Tetrasomy | 49 | Warburton et al. 2000 | |
| 13q32 | Supernumerary inv dup | Tetrasomy | 88 | Warburton et al. 1997, 2000 | |
| 13q31 | Supernumerary inv dup | Tetrasomy | 60 | Warburton et al. 2000 | |
| 13q32 | Supernumerary inv dup | Tetrasomy | 98 | 8 | Depinet et al. 1997; Warburton et al. 2000 |
| 13q32 | Supernumerary inv dup | Tetrasomy | 100 | 100 | Warburton et al. 2000 |
| 13q32 | Supernumerary inv dup | Tetrasomy | 74 | 25 | Warburton et al. 2000 |
| 13q21 | Deletion + inv dup | Trisomy | 100 | 100 | Morrissette et al. 2001 |
| 13q21 | Supernumerary inv dup | Tetrasomy | 14 | 20 | Li et al. 2002 |
| 13q32 | Supernumerary inv dup | Tetrasomy | 56 | Li et al. 2002 | |
| 13q32 | Supernumerary inv dup × 2 | Hexasomy | 12–26 | Li et al. 2002 | |
| 14: | |||||
| 14q32.1-qter | Deletion + inv dup | Trisomy | 100 | Sacchi et al. 1996 | |
| 15: | |||||
| 15q23 | Supernumerary inv dup | Tetrasomy | 70 | 11 | Blennow et al. 1994 |
| 15q24 | Supernumerary inv dup | Tetrasomy | 80 | Blennow et al. 1994 | |
| 15q25.3-qter | Supernumerary inv dup | Tetrasomy | 82 | Depinet et al. 1997 | |
| 15q25.3-qter | Supernumerary inv dup | Tetrasomy | 74 | Depinet et al. 1997 | |
| 15q26.1-qter | Supernumerary inv dup | Tetrasomy | 86 | Depinet et al. 1997 | |
| 15q25-qter | Supernumerary inv dup | Tetrasomy | 100 | Huang et al. 1998 | |
| 15q25-qter | Supernumerary inv dup | Tetrasomy | 79 | van den Enden et al. 1996 | |
| 15q26.1 | Supernumerary inv dup | Tetrasomy | 50–100 | 18 | Rowe et al. 2000 |
| 15q25.3-qter | Supernumerary inv dup | Tetrasomy | 95 | Hu et al. 2001 | |
| 15q24-qter | Supernumerary inv dup | Tetrasomy | 66 | 50 | Spiegel et al., in press |
| 17: | |||||
| 17q23 | Deletion + inv dup | Trisomy | 100 | Ravnan et al. 1999 | |
| 20: | |||||
| 20p12 | Deletion + inv dup | Trisomy | 100 | 100 | Voullaire et al. 1999 |
| 21: | |||||
| 21q21-ter | Deletion + inv dup | Trisomy/deletion 21 | 100 | Barbi et al. 2000 | |
| X: | |||||
| Xq12 | Deletion + asymmetric inv dup | Trisomy/deletion X | 82 | Kaiser-Rogers et al. 1995 | |
| Y: | |||||
| Yq12 | None | Balanced | 5 | Bukvic et al. 1996 | |
| Yq | Inversion | Recombinant Y | Mosaic | Rivera et al. 1996 | |
| Yq | None | Balanced | 94 | 100 | Tyler-Smith et al. 1999 |
| Yq11.2 | Deletion + inv dup | Disomy/deletion Y | 70 | Warburton et al. 1997 | |
| Yq11.2-qter | Deletion + inv dup | Disomy/deletion Y | 70 | Floridia et al. 2000 | |
| Distal Yp | Interstitial deletion | Deletion Y | 15 | 15 | Conde et al. 2001 |
| Yq | Deletion + inv dup | Disomy/deletion Y | 100 | Assumpcao et al., in press | |
Inv dup = inverted duplication.
The formation of a neocentromere is generally associated with a chromosomal rearrangement, such as an “inverted duplication” or an interstitial deletion, which generates a chromosome fragment lacking a conventional centromere (see fig. 3). By far the most common mechanism for the formation of a neocentric marker chromosome is the de novo inverted duplication of a distal chromosome segment (seen in 47 [78%] of the 60 reported cases; table 1). The resulting marker comprises two copies of the chromosome segment oriented as a “mirror image” around the breakpoint. Neocentromere formation occurs at an interstitial site, between the breakpoint and one of the telomeres, that is apparently unrelated to the site of the breakpoint and unaccompanied by further chromosome rearrangement. Notably, neocentromere formation occurs only once per marker chromosome, despite the fact that, in these cases, the two halves of the marker chromosome consist of apparently identical DNA sequence. In 13 of these inverted duplication cases, the marker is accompanied by a deletion chromosome complementary to the inverted duplication, resulting in partial trisomy for the duplicated region (fig. 3). In the remaining 34 inverted duplication cases, the karyotype is otherwise normal, so that the net result is partial tetrasomy for the duplicated region (fig. 3). The exact mode of origin of the partial trisomy and tetrasomy karyotype is unclear, but various mechanisms have been postulated (fig. 4) (Gardner and Sutherland 1996; Voullaire et al. 2001). In all of these cases, neocentromere formation clearly has a detrimental effect on the individual, causing the retention of a supernumerary chromosome fragment and therefore creating an unbalanced karyotype.
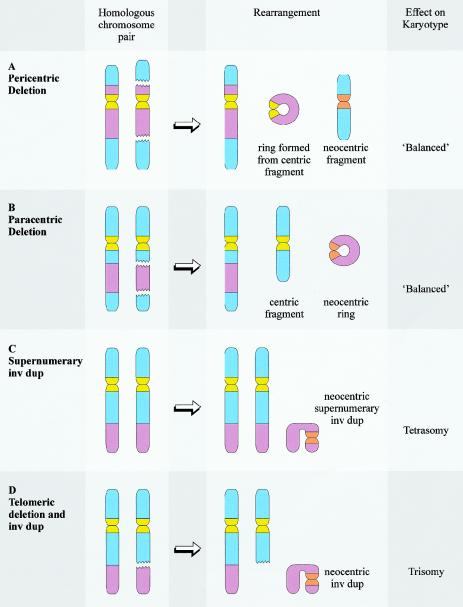
Figure 3.
Chromosome rearrangements commonly associated with neocentromere formation. Neocentromere formation is typically associated with a chromosome rearrangement resulting in the generation of a fragment that lacks a conventional centromere. The most common chromosome rearrangements are interstitial deletions (A and B) and inverted duplications (C and D). Interstitial deletions are typically associated with the formation of a ring chromosome, which may be necessary to stabilize the broken chromosome ends. The ring chromosome may be derived from the centric deletion chromosome (in the case of pericentric deletions, shown in A) or from the neocentric fragment (in the case of paracentric deletions, shown in B). When neocentromere formation results from an interstitial deletion, the resulting karyotype is usually “balanced” at a cytogenetic level. However, phenotypic effect may result from a “ring syndrome,” leading to mosaicism for one of the fragments, or from interruption of critical genes at the sites of chromosome breakage or neocentromere formation. Inverted duplications can be supernumerary to the original karyotype (C) (resulting in tetrasomy for the duplicated segment) or accompanied by a complementary deletion (D) (resulting in trisomy for the duplicated segment).
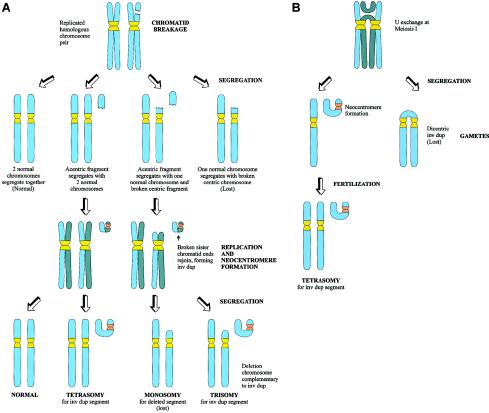
Figure 4.
Possible mechanisms of formation of neocentric inverted duplication (“inv dup”) marker chromosomes. A, Formation at mitosis. Chromatid breakage is followed by segregation of the acentric fragment and the centric fragment (or deletion chromosome). Following replication, the broken sister chromosome ends of the acentric fragment join to form a mirror image inv dup chromosome, with neocentromere formation occurring on one of the two arms of the inv dup. The centric fragment will usually be lost because of instability; however, stabilization of the broken end of the fragment will occasionally allow the fragment to survive. Segregation of the inv dup with the centric fragment will lead to trisomy for the chromosome segment involved, whereas, if the inv dup segregates with two normal chromosome homologues, tetrasomy will result. B, Formation at meiosis. Formation of an acentric inv dup fragment occurs because of anomalous crossing-over during meiosis I. After segregation, the dicentric fragment will be lost, but the acentric fragment may be “rescued” by neocentromere formation. After fertilization, zygotes containing the inv dup will be tetrasomic for the segment involved in the inv dup.
Interstitial deletions have been responsible for the generation of neocentric marker chromosomes in nine cases (table 1). Interstitial deletions are typically associated with the formation of a ring chromosome from either the centric deletion chromosome (in the case of pericentric deletions) or the acentric fragment (in the case of paracentric deletions), allowing the stabilization of the broken chromosome ends (fig. 3). Meanwhile, the generation of a neocentromere allows the recovery of the acentric fragment that would otherwise have been lost and thereby restores a “balanced” karyotype. In this circumstance, the formation of the neocentromere can be said to have “rescued” the cell or individual from chromosome imbalance, analogous to the “rescue” of trisomic embryos by the mitotic loss of one copy of a chromosome from a trisomic embryo. However, phenotypic consequences may still arise if the marker is present in mosaic form or if the activities of important genes residing at the rearrangement breakpoints and/or the site of neocentromere formation have been compromised. Nevertheless, it is possible that similar cases have not been ascertained because they do not result in an abnormal phenotype.
Four neocentromere cases cannot be classified as either inverted duplications or interstitial deletions. These comprise one neocentromere situated on a supernumerary ring chromosome (Spiegel et al., in press) and three unusual examples involving the Y chromosome (Bukvic et al. 1996; Rivera et al. 1996; Tyler-Smith et al. 1999). In the Y-chromosome cases, the original centromere is still present but has presumably been inactivated, as a result of either an intrachromosomal rearrangement, a reduction in the size of the α-satellite array, or by other mechanisms. When the neocentric Y chromosome is mitotically stable, the resulting phenotype is that of a normal male (Rivera et al. 1996; Tyler-Smith et al. 1999), but when mitotic instability leads to mosaicism for the Y chromosome, the result is gonadal dysgenesis (Bukvic et al. 1996).
Origin and Propagation of Human Neocentromeres in Mitosis and Meiosis
Understanding of the origin of human neocentromeres relies on observations in patients in whom the neocentromeres have occurred. DNA polymorphism studies performed in five cases indicate that human neocentromeres can form either during meiosis (Depinet et al. 1997; Rowe et al. 2000) or mitosis (Depinet et al. 1997). Once formed, neocentromeres can be transmitted through mitosis. Meiotic transmission has also been demonstrated, most notably by the observation, in two separate families, of a Y-derived neocentric chromosome in three generations (Rivera et al. 1996; Tyler-Smith et al. 1999). Transmission through two generations has also been described for a marker containing a neocentromere at 3q26 (Wandall et al. 1998) and for an analphoid marker of unknown origin (Winters et al. 2000).
Mitotic stability of neocentric marker chromosomes might be expected to be less than conventional centromeres, either due to suboptimal function of the neocentric kinetochore (for which there is no direct evidence at present) or selection pressure against cells containing the marker. In addition, ring chromosomes containing a neocentromere might be lost as part of a “ring syndrome,” regardless of the function of the kinetochore (e.g., because of the formation of mitotically unstable, interlocking double-ring chromosomes with two active centromeres). One expected consequence of mitotic instability is the presence of the marker in only a proportion of the patient’s cells (mosaicism), and, in fact, mosaicism has been observed in more than half of human neocentric markers (table 1). Mitotic instability is clearly responsible for the mosaicism observed in neocentric markers that have been meiotically transmitted from the previous generation (Rivera et al. 1996; Tyler-Smith et al. 1999). However, mosaicism may also result from the postzygotic formation of a marker that is mitotically 100% stable. Alternatively, mosaicism could arise for a meiotically derived marker if neocentromere function was not established at the time of meiotic rearrangement but developed subsequently after several postfertilization cell divisions, during which some of the markers may be lost (Voullaire et al. 2001). A better measure of mitotic stability of neocentric markers can be obtained by monitoring the degree of mosaicism at different points in a patient’s lifetime. Such studies reveal that the proportion of cells carrying the neocentromere can either remain stable (Dufke et al. 2001; Voullaire et al. 2001; Li et al. 2002) or decrease (Rowe et al. 2000) over time. In one study, in which five separate blood samples were collected for analysis from a patient over a period of 13 years, the level of mosaicism of a neocentric marker remained relatively unchanged, at 23%–46% (Voullaire et al. 2001). Subcloning of cultured lymphoblast cells from this patient revealed two populations, one lacking the neocentric marker and the other containing the marker chromosome that remained 100% stable through >65 cell divisions tested. This observation suggests that, in some cases, mosaicism is the result of postzygotic formation of the neocentromere rather than mitotic instability.
How Are Sites of Neocentromere Formation Determined?
Human neocentromeres typically form in euchromatic regions, with the exception of a small number of examples located in the heterochromatic region of chromosome Yq (fig. 2). However, heterochromatic proteins such as HP1 and SUV39H1 have been detected at neocentromeres formed in euchromatic regions (Aagaard et al. 2000; Saffery et al. 2000), suggesting that, regardless of their origins, neocentromeres carry certain characteristics of heterochromatin. To date, neocentromere formation has been described in 16 of the 22 autosomes and in the X and Y chromosomes, and there is no reason to believe that neocentromeres will not eventually be described on all human chromosomes. A glance at figure 2 reveals some nonrandomness in the distribution of the reported neocentromeric sites across the human genome, since those at 3q, 13q, and 15q collectively account for approximately half of all cases. In addition, distal chromosomal regions appear to be more susceptible to neocentromere formation, compared with the more proximal regions.
How can the distribution of neocentromeres be explained? One possibility is that neocentromere formation occurs randomly throughout the genome but that subsequent selection determines which changes become stabilized within a cell population. In this scenario, some putative neocentromere sites (e.g., the more proximal ones) might result in the formation of marker chromosomes that are not compatible with fetal survival, either because of their large size or their particular genomic content. An alternative hypothesis is that genomic “hotspots” that are favorable to neocentromere formation exist in certain regions of the genome. This hypothesis is supported by the repeated appearance or significant clustering of neocentromeres at a number of chromosomal sites, notably 3q, 8p, 9p, 13q, 15q, and Yq. It remains to be determined whether this clustering is due to recurrent neocentromere formation on the same genetic sequences or the utilization of different DNA sequences in the same general vicinity.
What characteristics of DNA might favor neocentromere formation? At least the neocentromeres at Yq (which consists largely of heterochromatin) may be explained by the possibility that chromosome regions with heterochromatic properties are intrinsic sites for neocentromere formation (Choo 2001a). Support for this comes from the demonstration that centromere competence appears to be an innate characteristic of DNA in heterochromatic blocks (Platero et al. 1999) and that the heterochromatic state facilitates the binding of the critical histone H3-related centromere protein CENP-A in both Drosophila and humans (Henikoff et al. 2000). The paucity or absence of neocentromere formation at the heterochromatin on the autosomes may be explained by the close juxtapositioning of this heterochromatin at the normal centromeres, which greatly reduces the probability of their separation from the normal centromeric domain.
A second and more general possibility is that neocentromere DNA shares some sequence characteristics with α-satellite DNA, which has been shown to be a preferred substrate for centromere formation (Harrington et al. 1997; Ikeno et al. 1998). Recently, the CENP-A–binding domain has been investigated in three different neocentromeres, at 10q25 (Lo et al. 2001a), 20p12 (Lo et al. 2001b), and 9p23 (Satinover et al. 2001a). The three analyzed domains have been found to range in size from 330 kb to 500 kb, and at the level of the DNA sequence the only noticeable similarity between the three domains is an increase in AT content, calculated at 65.4%, 61.1%, and 65%, respectively (compared with the genome average of 58.0% [Smit 1999] and α-satellite average of 62.6% [Lo et al. 2001b]). This suggests that increased AT content may provide a more favorable disposition for neocentromere formation. In other respects, these neocentromere-containing segments show no particular differences from bulk DNA sequences.
Protein Studies
The activity of neocentromeres is further revealed by centromere protein-binding studies using specific antibodies. Proteins that associate with mammalian centromeres can be broadly classified into two groups on the basis of their spatial positioning throughout the cell cycle (Choo 1997a). The first class comprises proteins that are constitutively associated with the centromere, such as CENP-A, CENP-B, and CENP-C, which are thought to have structural roles in kinetochore formation. The second class, known as passenger proteins, associate with the centromere transiently during the cell cycle and comprises proteins with diverse roles in cell division, such as spindle capture, metaphase-to-anaphase transition, and sister chromatin cohesion. Where analyzed, the neocentromeres have consistently demonstrated the presence of CENP-A, the centromere-specific core histone that differentiates the centromere from the rest of the chromosome at the chromatin level, as well as the proteins CENP-C and CENP-E. However, binding of CENP-B is consistently absent, a finding that is not unexpected, given that CENP-B binds specifically to α-satellite repeats (Choo 1997b). Gene-knockout experiments in mice have also shown that CENP-B is not essential for centromere function during mitosis and meiosis (Hudson et al. 1998; Kapoor et al. 1998; Perez-Castro et al. 1998).
The largest survey has involved the analysis of >20 functionally important kinetochore-associated proteins in two neocentromeres derived from 10q25 and 20p12 (Saffery et al. 2000). The pattern of protein binding at the neocentromeres was found to be indistinguishable from that of the normal centromeres in all respects other than the absence of binding of CENP-B. This suggests that neocentromeres assemble a trilaminar kinetochore and interact with other accessory proteins in a manner identical to that of normal centromeres, despite the absence of α-satellite. This conclusion is supported by evidence from electron-microscopy studies demonstrating that neocentromeres form microtubule-associated kinetochores with size and morphology identical to conventional centromeres (Wandall et al. 1998).
Replication Timing of the Neocentromere Region
Mammalian centromeric regions are known to replicate at different times during the second half of S phase (Shelby et al. 2000). Therefore, the timing of replication of neocentromere sites, both prior to and after neocentromere activation, represents an interesting area of study. A detailed analysis has been performed on the 10q25 neocentromere (Lo et al. 2001a). The results demonstrate that, prior to formation of the neocentromere, this chromosomal site replicates at mid-S phase, except for a ∼450-kb domain containing the CENP-A–binding domain that replicates during mid-to-late S phase. After neocentromere formation, replication timing in this 450-kb region is unchanged; however, timing is shifted in the areas surrounding it. This results in an overall shift of an extended domain of 1.5 Mb into the third quarter of S phase, bringing a much larger region covering the core CENP-A–binding domain into line with the replication timing of all the other centromeres. This study suggests that the assembly of a neocentromere drastically alters the overall replication timing of a genomic region. It also raises the possibility that a microgenomic site with a “centromere-correct” replication timing may be favorably predisposed to neocentromere formation (Choo 2001a).
Neocentromere Formation in Human Cancer
Chromosomal abnormalities are a common feature of a wide variety of neoplastic lesions. Primary chromosome aberrations are thought to have a causal role in tumorigenesis and are often specifically associated with particular tumor types. Secondary aberrations, on the other hand, are rarely or never found alone and may not be causally associated with the development of the cancer. To date, neocentromere formation has been observed in only two tumor categories: lipomatous tumors and acute myeloid leukemia (AML). However, this may underrepresent the true extent of neocentromere formation in cancer, because solid tumors are relatively infrequently karyotyped. Furthermore, tumors can be particularly difficult to characterize by chromosome banding techniques, owing to the complexity of rearrangements, suboptimal banding quality, and shortage of material. Specific identification of neocentromeres also requires additional techniques that may not be routinely used in a cytogenetics laboratory.
Lipomatous Tumors
Lipomatous tumors are a heterogeneous group of neoplasms arising from adipose tissue, ranging from benign lipomas to the highly malignant categories of liposarcomas (Weiss 1996). Adipocyte tumors of borderline malignancy are classified as “atypical lipomas and well-differentiated liposarcomas” (ALP-WDLPS). An apparently primary cytogenetic aberration in these tumors is the presence of supernumerary marker chromosomes, either in the form of rings or remarkable “giant rod”–shaped chromosomes (typically several times the size of chromosome 1) (Heim et al. 1987; Sreekantaiah et al. 1992; Rosai et al. 1996; Rubin and Fletcher 1997). The remainder of the karyotype is near-diploid. These rings and giant rods consistently contain amplification of the 12q14-15 region (including the MDM2 oncogene), as well as variable interspersed sequences from other chromosomes (Pedeutour et al. 1999).
The centromere properties of rings and giant rods have been analyzed in >30 cases of ALP-WDLPS (Pedeutour et al. 1994, 1999; Gisselsson et al. 1998, 1999; Sirvent et al. 2000; Forus et al. 2001). A constant and specific feature of these markers is the absence of α-satellite DNA (fig. 1C–F). However, the rings and giant rods have functional centromeres, as demonstrated by the binding of centromere proteins (such as CENP-C) and by mitotic stability. ALP-WDLPS, therefore, represents the first example of a tumor class for which the formation of analphoid neocentromeres is a predictable outcome. Neocentromere formation presumably provides a mechanism to impart mitotic stability and, thus, a selective advantage to the neoplastic cells, on what might otherwise be highly unstable acentric supernumerary marker chromosomes. Following stabilization, the neocentric ring chromosomes could be involved in further structural rearrangements and gene amplification at mitosis, through breakage-fusion-bridge cycles (Gisselsson et al. 1999). Interestingly, ring and marker chromosomes containing amplification of 12q14-15 are also found in the higher-grade forms of liposarcomas, but these markers typically contain alphoid centromeres rather than neocentromeres (Gisselsson et al. 1999; Sirvent et al. 2000). Neocentromere formation is, therefore, not a prerequisite for 12q14-15 amplification. Conversely, amplification of 12q14-15 may facilitate or precipitate the formation of the neocentromere. The nature of the DNA sequences involved in the ALP-WDLPS neocentromeres has yet to be determined, but it is possible that a site favorable for neocentromere formation is created by amplification of 12q14-15. However, it is also possible that the neocentromere forms on DNA with a different chromosomal origin, given the feasibility of neocentromere formation from a wide range of human genomic sites, as evidenced in constitutional human neocentromeres. Proper assignment of the chromosomal origin of the ALP-WDLPS neocentromeres will require colocalization of neocentromeric sites with defined subchromosomal DNA probes and/or direct molecular identification of the specific centromere protein-binding DNA sequences, through use of approaches such as chromatin immunoprecipitation and genomic array analysis (Lo et al. 2001a, 2001b).
AML
Neocentromeres have been discovered in the bone marrow cells of two patients with AML. In the first case, an analphoid inverted duplication of chromosome 10q (with constriction at 10q26) was found in the bone marrow of an 18-year-old man with AML (Abeliovich et al. 1996). The marker was present in 90% of leukemic cells and was associated with an apparently balanced t(11;17). The second neocentric marker was a mosaic ring chromosome discovered in the complex karyotype of a 71-year-old man with AML (Gisselsson et al. 1999). The ring was derived from chromosome 8 but was negative for C-bands and α-satellite, consistent with the presence of a neocentromere. Investigation of the role of neocentromere formation in the biology of AML will require the ascertainment of additional cases containing neocentric markers. However, existing evidence suggests that, in contrast to lipomatous tumors, neocentromere formation in AML is more likely to be a secondary chromosome aberration, possibly resulting from disordered cell-cycle regulation.
Neocentromere Formation in the Laboratory
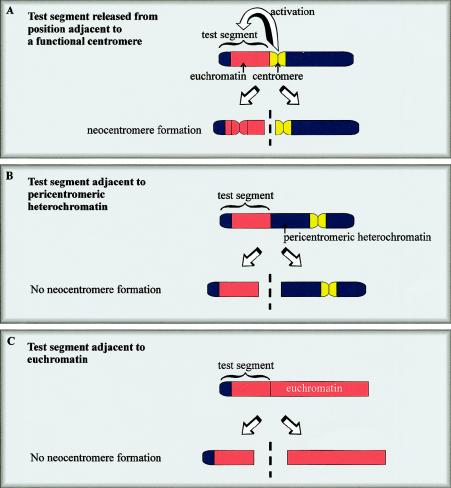
Neocentromere formation has been successfully induced in the laboratory fly, providing a useful tool for study of the underlying mechanism. Neocentromerization in Drosophila has not been described in the wild but occurred after experimentally inflicted chromosomal rearrangement in two separate studies. In the first study, a subtelomeric chromosome fragment containing a functional neocentromere was isolated after the irradiation of a 1.3-Mb minichromosome (γ238) derived from the Drosophila X chromosome (fig. 5A) (Williams et al. 1998). The γ238 minichromosome is unusual, in that 290 kb of subtelomeric DNA (on which the neocentromere forms) is translocated next to the 440-kb centromeric sequence (these entities are normally separated by 40,000 kb). The juxtaposition of these two elements is presumably critical to neocentromere formation, because similar subtelomeric fragments generated from their normal position on the wild-type X chromosome do not form neocentromeres in either mitosis or meiosis (Williams et al. 1998). In follow-up studies, an identical 290-kb segment of DNA was released from various sites within the Drosophila genome (Maggert and Karpen 2001). Neocentromere formation occurred only when the segment was released from sites immediately adjacent to the centromeric chromatin (similar to the situation shown in fig. 5A), but not from sites juxtaposed against pericentromeric heterochromatin or euchromatin (fig. 5B and 5C). These results confirm that activation of the neocentromere was dependent upon proximity to an endogenous centromere. Once established, the Drosophila neocentric minichromosome is comparable to human neocentric chromosomes in that it displays moderately efficient transmission in both meiosis and mitosis and binds a centromere-specific protein (ZW10) at a frequency and intensity indistinguishable from normal Drosophila centromeres.
Figure 5.
Generation of neocentromeres in Drosophila. A test segment comprising telomeric heterochromatin and euchromatin forms a functional neocentromere when released from a site immediately adjacent to a normal centromere (A). One model suggests that centromere activity or “centromere imprinting factor” spreads from the existing centromere to the neighboring test DNA, where it activates or imparts a stable centromeric state that can come into independent existence when this DNA is subsequently released. When the same fragment is released from sites adjacent to pericentromeric heterochromatin (B) or euchromatin (C), a neocentromere does not form.
The second study utilized another variant Drosophila chromosome. This chromosome contains a megabase-sized insertion of satellite DNA, designated “brownDominant” (bwD), near the distal tip of the right arm of chromosome 2 (Platero et al. 1999). An acentric fragment of this chromosome, containing the bwD heterochromatic element, was experimentally separated from its parent chromosome and found to display efficient centromeric behavior, as determined by formation of a kinetochore and segregation in mitosis. Centromeric activity was dependent on the presence of bwD, leading to the conclusion that bwD can act as a centromere. Furthermore, virtually all fragments containing bwD displayed centromere activity immediately upon separation, suggesting that centromeric determinants are present at all times at bwD.
Recently, neocentromere formation has also been reported in an experimental animal cell culture system. This occurred during the production of interspecies somatic cell hybrids, when a neocentromere was detected on a fusion chromosome consisting of segments of the human Y chromosome and mouse chromosomes 12 and 15 (Shen et al. 2001). The fusion chromosome lacked human α-DNA but contained a neocentromere within the human Y-derived region. The neocentromere was unstable in mouse cells but segregated faithfully after being transferred into chicken DT40 cells.
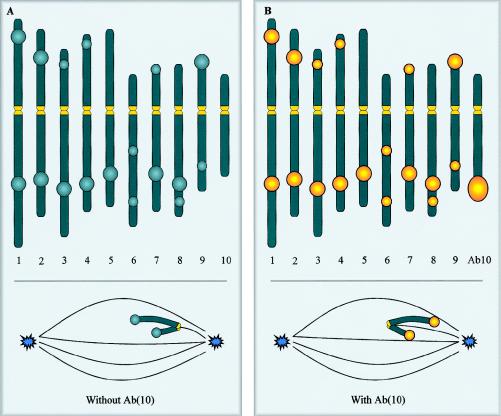
Maize Knobs: a Different Type of Neocentromere
The term “neocentromere” was first used 60 years ago, to describe the knob regions of maize chromosomes (Rhoades and Vilkomerson 1942). Plant neocentromeres have since been discovered in a number of different species (Hiatt et al. 2002), but maize knobs remain the most extensively studied. Knobs are cytologically visible heterochromatic regions that can be found at a number of distinct locations spread across all 10 maize chromosomes (fig. 6A). These regions are composed of thousands of tandemly repeating DNA sequences, such as the 180- and 350-bp repeats, which share some homology with normal maize centromeres (Peacock et al. 1981; Ananiev et al. 1998). The knobs are normally inert (i.e., they do not bind spindle microtubules or aid chromosome segregation; fig. 6A, inset), but are transformed into neocentromeres in the presence of a variant of chromosome 10, known as “Abnormal 10” (Ab10) (fig. 6B). The long arm of Ab10 has been rearranged such that it contains a large knob, as well as two neocentromere-activating cassettes that independently confer neocentromere activity at one or the other class of knob repeats (Hiatt et al. 2002). Unlike human neocentromeres, activity of maize neocentromeres is confined to meiosis. During meiosis, they are bound by the spindle (Rhoades and Vilkomerson 1942; Rhoades and Dempsey 1966), causing the knobs to move poleward in advance of the original centromere, which nonetheless remains active and attaches to spindle microtubules (fig. 6B, inset) (Yu et al. 1997). These chromosomes effectively have two or more centromeres, a circumstance that does not appear to affect the fidelity of chromosome segregation (Yu et al. 1997). When Ab10 is present, meiotic drive is observed for Ab10 and for any other chromosome carrying a knob, so that, on average, knobbed chromosomes preferentially segregate to 70% of the viable gametes instead of the expected 50% (Rhoades 1942). Given this reproductive advantage, the fact that knobs have not become fixed in most populations indicates that knobs must also have deleterious functional consequences.
Figure 6.
Maize knob neocentromeres and the maize karyotype, indicating some of the more common sites of heterochromatic knobs on the 10 chromosomes (Rhoades 1950). In the presence of a normal chromosome 10 (A), the knobs are inactive and lag behind the normal centromere at meiosis (inset). When the normal chromosome 10 is replaced by Ab(10), the knobs become neocentromeres. These neocentromeres bind the spindle microtubules in a lateral rather than an end-on manner and migrate towards the spindle pole in advance of the normal centromeres that remain active (B). Yellow indicates an active centromere or neocentromere.
The neocentromere phenomenon in maize knobs differs from that seen in humans in a number of interesting ways. Most importantly, knob neocentromeres form only at the site of large arrays of heterochromatic DNA. In this respect, knob loci are examples of facultative or accessory centromeres rather than of neocentromere function dictated by noncentromeric DNA that normally serves a totally different function, such as gene coding, as is the case for most human neocentromere sites. The activation of knob loci is triggered by a specific cue in the form of trans-acting factors encoded by Ab10 during a very specific (meiotic) stage of the organism’s developmental cycle. No such mechanistic or physiological drive is known in humans. Furthermore, knob neocentromeres occur despite the presence of a functioning normal centromere, whereas coexistence of a human neocentromere and a functional normal centromere on the same chromosome has not been described. However, genomic regions that can facultatively function as centromeres are seen in other organisms. For example, in the horse parasitic nematode Parascaris univalens, the euchromatic and heterochromatic portions of the chromosomes can differentially be used to provide centromere activity under certain physiological and developmental-cycle conditions (Goday et al. 1992). Knob neocentromeres also behave differently from both normal maize centromeres and human neocentromeres, in that (1) they do not form a typical kinetochore (as evidenced by a lack of binding of the conserved kinetochore proteins CENP-C, MAD2, and 3F3/2, [Dawe et al. 1999; Yu and Dawe 2000]), and (2) they interact with microtubules in a lateral manner (see fig. 6B, inset), instead of in the end-on manner typical of the centromeres of maize and all other organisms (see fig. 6A, inset) (Yu et al. 1997). Knob-associated neocentromeres, therefore, possess a number of unusual features that make them an interesting system for investigation to compare and contrast with conventional centromeres.
Epigenetics and Models for Neocentromere Generation
So far, we have drawn the following conclusions from the analysis of neocentromere cases in humans: (1) the formation of viable neocentric chromosomes is a rare event; (2) neocentromeres can arise either in mitosis (including in cancer) or in meiosis; (3) neocentromeres form a functional kinetochore and can be stably transmitted through meiosis and mitosis; (4) neocentromere formation can occur at many different sites across the genome that do not share a unique DNA sequence but that may have in common other features that predispose to neocentromere formation, such as increased AT content; and (5) the formation of a neocentromere is usually detrimental to the individual.
Since the generation of human neocentromeres has yet to be achieved experimentally, factors controlling the phenomenon remain largely unknown. Any explanation for how human neocentromeres form and are propagated to subsequent generations must be inferred from other sources, such as the study of the function of normal human centromeres and those in model organisms. Considerable evidence now exists that generation and propagation of centromeres is an epigenetic phenomenon (a heritable change transmitted by mechanisms other than that based entirely on DNA sequence). For example, studies in yeast have demonstrated that an inactive centromere can be converted to an active centromere without change in the content and structure of DNA and that the active centromere remains active through many generations (Steiner and Clarke 1994). An analogous human example is found in the form of pseudodicentric chromosomes, which contain only one functional centromere, despite the presence of two separate domains of centromeric DNA. This demonstrates that the mere presence of centromeric DNA does not ensure kinetochore formation and function and that an additional epigenetic factor or “mark” is required for centromere determination. This epigenetic mark would act to distinguish centromeric DNA/chromatin from normal DNA/chromatin and thereby trigger downstream effects culminating in centromere-protein acquisition and kinetochore formation. Propagation through cell division would require a strong “memory,” for the established centromeric chromatin status to allow the maintenance of the epigenetic mark. The nature of the epigenetic modification and memory is yet to be determined, but it might involve deposition of centromere-binding protein (such as CENP-A) that results in higher-order chromatin reorganization; alternatively, it might occur via chemical modification of centromeric DNA or its associated histones and nonhistone proteins, such as methylation, poly(ADP-ribosyl)ation, deacetylation, phosphorylation, and ubiquitination (Choo 2000).
Whatever epigenetic system is utilized by normal centromeres, it is likely that a closely related mechanism will apply to neocentromeres. The most convincing evidence for the epigenetic nature of neocentromerization in humans comes from extensive analysis of the 10q25 neocentromere, demonstrating that this neocentromere site is identical to that of the normal chromosome 10 (Lo et al. 2001a; N. Wong, L. Wong, and K. H. A. Choo, unpublished data). Furthermore, transfection of YAC clones containing neocentromere DNA from this region does not produce artificial chromosomes (Saffery et al. 2001), in contrast to the high frequency of artificial chromosome formation observed when transfection is performed using YACs or BACs containing α-satellite (Harrington et al. 1997; Ikeno et al. 1998; Henning et al. 1999; Ebersole et al. 2000).
If neocentromere identity is determined by an epigenetic mark, how does noncentromeric DNA acquire this mark? Two models have been proposed to explain the acquisition of neocentromere activity by noncentromeric DNA. In the first model, centromere activity spreads from existing centromeric regions to neighboring DNA, where it imparts a stable centromeric state (Murphy and Karpen 1998; Williams et al. 1998). This “activation model” is based on data from the generation of neocentromeres in the Drosophila γ238 minichromosome (fig. 5A) (Williams et al. 1998; Maggert and Karpen 2001) and predicts that sequences neighboring an active centromere will be more likely to acquire neocentromere activity than sequences elsewhere in the genome. However, observations in humans suggest that, if anything, the opposite is true, since human neocentromeres are more often than not formed in distal chromosomal regions. These regions are usually separated from existing centromeres by vast tracts of DNA that would prevent any spread of centromeric activity in cis. Similarly, in the bwD Drosophila model, the heterochromatic block that acquires neocentromeric activity lies at the opposite end of an ∼20-Mb chromosome arm from the natural centromere (Platero et al. 1999). It is, therefore, unlikely that spread of centromeric activity in cis is a common mechanism for the acquisition of the epigenetic mark. However, the epigenetic mark for centromere activity may be able to spread in trans from an existing centromere. Accordingly, neocentromeres may arise through the accidental association of normally noncentromeric DNA with an endogenous centromere, during centromere templating at meiosis or mitosis (Maggert and Karpen 2001).
The second model for neocentromere formation (i.e., the lateral inhibition model) involves the existence of many sites along a chromosome that are intrinsically capable of exhibiting centromere activity but are normally repressed in cis by the presence of a more dominant centromere. Such a mechanism might have evolved to protect against the detrimental effects of multiple centromeres forming on the same chromosome, such as dicentric bridges, chromosome breakage, and aneuploidy. On the basis of this model, neocentromeres might be expected to form whenever the resident centromeres are inactivated or deleted. This is consistent with observations in the bwD Drosophila model, where virtually all fragments containing bwD heterochromatic block display neocentromere activity (Platero et al. 1999). However, if this is also the case in humans, why are neocentromeres observed so rarely? One possible explanation is that most human neocentromeres form in euchromatic regions (other than those in Yq) that may have a significantly lower affinity for acquisition of the epigenetic mark. There appears to be a hierarchical order of DNA preference for centromere formation, with normal centromeric repeat DNA being the preferred option, followed by noncentromere heterochromatin (e.g., human Yq and Drosophila bwD), and, lastly, euchromatic DNA (Choo 2001a). Within these categories, additional variables are likely to be in play, such as a preference for DNA adjacent to the centromere or for euchromatic DNA with an increased AT content or “correct” DNA-replication time (discussed above, in the “Constitutional Human Neocentromeres” section). In this context, a large block of heterochromatin, such as bwD, may be very likely to form a neocentromere upon removal of lateral inhibition, because the epigenetic mark is strong and/or permanently present. The 290-kb neocentromeric fragment from the Drosophila γ238 minichromosome (Williams et al. 1998; Maggert and Karpen 2001), on the other hand, would require proximity to the original centromere in order to swing the balance in favor of consistent neocentromere formation. In the case of human euchromatin, the epigenetic mark may be only weakly or transiently present, so that if lateral inhibition is removed by the generation of an acentric fragment, the probability of a neocentromere forming is increased but remains low. Furthermore, in this circumstance the initial neocentromere may be less than perfect, requiring a number of cell divisions to become established. It is also possible that, in humans, lateral inhibition persists for several cell divisions, further reducing the chance that a functional neocentromere will form (Williams et al. 1998).
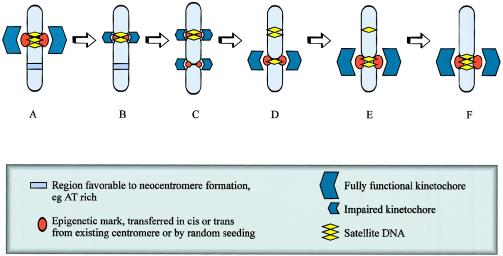
These hypotheses are based on the assumption that the epigenetic mark for neocentromere determination is acquired prior to or around the time of the chromosome rearrangement but that the neocentromere does not become functional until after the chromosome rearrangement (fig. 7A and 7B). An alternative theoretical possibility is that the occasional formation of a functional neocentromere precedes the loss or inactivation of the original centromere, such that a functional dicentric chromosome is created that is subsequently predisposed to rearrangement (see fig. 7C).
Figure 7.
Formation of human neocentromeres. Human neocentromeres can form in either meiosis or mitosis, by a mechanism that probably involves the acquisition of a centromere-specific epigenetic mark, followed by formation of a functional kinetochore. Certain chromosomal regions are predisposed to neocentromere formation, possibly because of AT content, heterochromatic qualities, or “centromere-correct” replication timing. The formation of a marker chromosome containing a neocentromere is dependent on three steps: (1) rearrangement of the chromosome, generating an acentric fragment (in this example, rearrangement is a paracentric deletion resulting in the formation of a centric deletion chromosome and a neocentric ring); (2) acquisition of the epigenetic mark required for centromere determination; and (3) formation of a functional kinetochores. The timing of these events in relation to each other is unclear. It is possible that chromosome rearrangement is the initial event (path A), followed by acquisition of the epigenetic mark and formation of the kinetochore. Alternatively, the chromosome arrangement may occur between acquisition of the epigenetic mark and formation of the kinetochore (path B), or, less likely, the chromosome rearrangement may be consequent to the formation of a functional kinetochore (path C).
Evolutionary Significance of Neocentromeres
Neocentromere formation in humans is usually detrimental to the individual (although, ironically, in at least one type of cancer, it is advantageous to the cancer cell population). Fortunately, the stability of the human genome dictates that such events are rare. In this respect, neocentromere formation is comparable to other mutations in chromosomes or DNA. If the cost of neocentromere formation is measured in terms of human disease, what biological function is served by the generation of neocentromeres? Is it possible that a certain rate of neocentromere formation, as is generally accepted for the more familiar types of mutations in chromosomes or DNA, may be beneficial or even necessary to generate sufficient genomic variation and diversity for adaptation during evolutionary time?
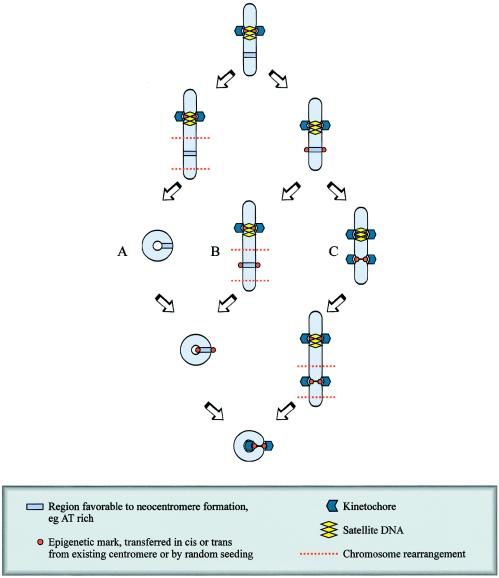
Studies in primates indicate that relocation of the centromere within a chromosome may occur via neocentromere formation (fig. 8). Ventura et al. (2001) have demonstrated that the X chromosomes of three different primate species share an identical order of genetic markers, despite the fact that these X chromosomes have different centromeric locations. The absence of rearrangement on these chromosomes suggests that centromere repositioning has resulted from the emergence of a new centromere rather than by translocation of an existing centromere into this region. More recent examples of human centromere repositioning “in progress” are suggested by the reports of two separate families in which a neocentric Y chromosome was transmitted through three generations (Rivera et al. 1996; Tyler-Smith et al. 1999). These two cases are atypical of constitutional human neocentromeres, in that centromere function is provided by an analphoid neocentromere at the expense of the inactivation and retention of the original α-satellite–containing centromere on an otherwise intact Y chromosome. Individuals with such neocentric Y chromosomes are phenotypically normal and fertile, indicating that the neocentromere is functioning in mitosis and meiosis. According to one model, an initial rearrangement or reduction in size of the α-satellite DNA array may remove kinetochore binding capacity in an amount sufficient to impair centromere function, allowing the emergence or activation of a neocentromere (Tyler-Smith et al. 1998). The initial neocentromere is imperfect, but, in subsequent generations, selection pressure improves kinetochore maturation through duplication of existing sequence or accumulation of repetitive DNA from other sources. The original α-satellite centromeric DNA would subsequently contract in the absence of selection pressure and would ultimately disappear (fig. 8). In evolutionary terms, centromere relocation would rapidly lead to the reproductive isolation of emerging species, providing a formidable mechanism for speciation (Henikoff et al. 2001).
Figure 8.
Evolutionary repositioning of centromeres by neocentromere formation. The initial event in centromere repositioning may be an impairment of function of the original centromere (B), possibly leading to a reduction of lateral inhibition. A neocentromere may then form via epigenetic mechanisms not involving alteration to the primary DNA sequence at a favorable site (C). The initial neocentromere may be imperfect, but, in subsequent generations, selection pressure improves kinetochore maturation through duplication of existing sequence or accumulation of repetitive DNA from other sources (D–F). The original satellited centromeric DNA would subsequently contract in the absence of selection pressure and ultimately disappear (D–F).
Human Artificial Chromosomes: a Therapeutic Role for the Neocentromere?
It is possible that human neocentromeres may provide new opportunity for disease treatment, in the form of neocentromere-based human artificial chromosomes (HACs) for use as vectors in gene therapy. Compared with other gene-delivery vectors, HAC-based systems are able to carry DNA segments of very large size and are unlikely to be immunogenic. HACs also avoid the unpredictable genomic events associated with random integration. All potential artificial chromosomes require an active centromere to ensure faithful mitotic segregation. Most HACs utilize α-satellite DNA centromeres, but neocentromere-based HACs, generated by telomere-associated truncation of neocentric chromosomes derived from human subjects (fig. 1B) (Saffery et al. 2001), may provide a viable alternative. Potential advantages of neocentromere-based HACs include the fact that they are fully definable in terms of DNA sequence, and they may prove to be significantly smaller and to provide a more suitable environment for expression of introduced therapeutic genes, compared with HACs containing α-satellite (Choo 2001b; Saffery and Choo 2002).
Concluding Remarks
The first decade of human neocentromere research has provided a taste for the involvement of neocentromeres in human health, disease, and evolution. It has directly unveiled the incredibly malleable nature of not only the centromeric, but also the wider genomic, chromatin. Much excitement and debate has been generated over the possible mechanisms that are responsible for the formation and propagation of the neocentromeres—mechanisms that are likely to be inseparably linked to those that underlie the typical centromeres. Further studies should now be focused on identifying these mechanisms and should capitalize on the fully known sequences of neocentromeres to study the properties of the centromeres. On the basis of the varied spectrum of information we have gained to date from studying neocentromeres in fly, plant, lower primates, and humans, the further definition of the true extent of the neocentromere phenomenon in these and other species should be quite informative. If the first decade has laid some useful foundation for this phenomenon, the next decade should see its expansion into a fuller understanding of centromere biology and, possibly, improved health outcomes.
Acknowledgments
We thank F. Pedeutour, N. Sirvent, and L. Wong, for chromosome images, and R. J. M. Gardner, for criticism of the manuscript. D.J.A. is supported by an National Health and Medical Research Council Postgraduate Medical Research Scholarship. K.H.A.C. is a Senior Principal Research Fellow of the National Health and Medical Research Council of Australia.
References
- Aagaard L, Schmid M, Warburton P, Jenuwein T (2000) Mitotic phosphorylation of SUV39H1, a novel component of active centromeres, coincides with transient accumulation at mammalian centromeres. J Cell Sci 113:817–829 [DOI] [PubMed] [Google Scholar]
- Abeliovich D, Yehuda O, Ben-Neriah S, Kapelushnik Y, Ben-Yehuda D (1996) dup(10q) lacking alpha-satellite DNA in bone marrow cells of a patient with acute myeloid leukemia. Cancer Genet Cytogenet 89:1–6 [DOI] [PubMed] [Google Scholar]
- Ananiev EV, Phillips RL, Rines HW (1998) A knob-associated tandem repeat in maize capable of forming fold-back DNA segments: are chromosome knobs megatransposons? Proc Natl Acad Sci USA 95:10785–10790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assumpção JG, Berkofsky-Fessler W, Campos NV, Macial-Guerra AT, Li S, Melaragno MI, de Mello MP, Warburton PE. Identification of a neocentromere in a rearranged Y chromosome with no detectable DY23 centromeric sequence. Am J Med Genet (in press) [DOI] [PubMed] [Google Scholar]
- Barbi G, Kennerknecht I, Wohr G, Avramopoulos D, Karadima G, Petersen MB (2000) Mirror-symmetric duplicated chromosome 21q with minor proximal deletion, and with neocentromere in a child without the classical Down syndrome phenotype. Am J Med Genet 91:116–122 [DOI] [PubMed] [Google Scholar]
- Blennow E, Telenius H, de Vos D, Larsson C, Henriksson P, Johansson O, Carter NP, Nordenskjold M (1994) Tetrasomy 15q: two marker chromosomes with no detectable alpha-satellite DNA. Am J Hum Genet 54:877–883 [PMC free article] [PubMed] [Google Scholar]
- Bukvic N, Susca F, Gentile M, Tangari E, Ianniruberto A, Guanti G (1996) An unusual dicentric Y chromosome with a functional centromere with no detectable alpha-satellite. Hum Genet 97:453–456 [DOI] [PubMed] [Google Scholar]
- Choo KHA (1997a) The centromere. Oxford University Press, New York [Google Scholar]
- ——— (1997b) Centromere DNA dynamics: latent centromeres and neocentromere formation. Am J Hum Genet 61:1225–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ——— (2000) Centromerization. Trends Cell Biol 10:182–188 [DOI] [PubMed] [Google Scholar]
- ——— (2001a) Domain organization at the centromere and neocentromere. Dev Cell 1:165–177 [DOI] [PubMed] [Google Scholar]
- ——— (2001b) Engineering human chromosomes for gene therapy studies. Trends Mol Med 7:235–237 [DOI] [PubMed] [Google Scholar]
- Cockwell AE, Gibbons B, Moore IE, Crolla JA (2000) An analphoid supernumerary marker chromosome derived from chromosome 3 ascertained in a fetus with multiple malformations. J Med Genet 37:807–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conde C, Chheng S, Wu J, Santini M, Kashork CD, Ware S, Scaglia F, Shaffer LG (2001) A novel analphoid marker of the Y chromosome. Am J Hum Genet Suppl 69:A765 [Google Scholar]
- Dawe RK, Reed LM, Yu HG, Muszynski MG, Hiatt EN (1999) A maize homolog of mammalian CENPC is a constitutive component of the inner kinetochore. Plant Cell 11:1227–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depinet TW, Zackowski JL, Earnshaw WC, Kaffe S, Sekhon GS, Stallard R, Sullivan BA, Vance GH, Van Dyke DL, Willard HF, Zinn AB, Schwartz S (1997) Characterization of neo-centromeres in marker chromosomes lacking detectable alpha-satellite DNA. Hum Mol Genet 6:1195–1204 [DOI] [PubMed] [Google Scholar]
- Dufke A, Walczak C, Liehr T, Starke H, Trifonov V, Rubtsov N, Schoning M, Enders H, Eggermann T (2001) Partial tetrasomy 12pter-12p12.3 in a girl with Pallister-Killian syndrome: extraordinary finding of an analphoid, inverted duplicated marker. Eur J Hum Genet 9:572–576 [DOI] [PubMed] [Google Scholar]
- Ebersole TA, Ross A, Clark E, McGill N, Schindelhauer D, Cooke H, Grimes B (2000) Mammalian artificial chromosome formation from circular alphoid input DNA does not require telomere repeats. Hum Mol Genet 9:1623–1631 [DOI] [PubMed] [Google Scholar]
- Floridia G, Gimelli G, Zuffardi O, Earnshaw WC, Warburton PE, Tyler-Smith C (2000) A neocentromere in the DAZ region of the human Y chromosome. Chromosoma 109:318–327 [DOI] [PubMed] [Google Scholar]
- Forus A, Bjerkehagen B, Sirvent N, Meza-Zepeda LA, Coindre JM, Berner JM, Myklebost O, Pedeutour F (2001) A well-differentiated liposarcoma with a new type of chromosome 12-derived markers. Cancer Genet Cytogenet 131:13–18 [DOI] [PubMed] [Google Scholar]
- Fritz B, Dietze I, Wandall A, Aslan M, Schmidt A, Kattner E, Schwerdtfeger R, Friedrich U (2001) A supernumerary marker chromosome with a neocentromere derived from 5p14→pter. J Med Genet 38:559–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner RJM, Sutherland GR (1996) Chromosome abnormalities and genetic counseling. Vol 2. Oxford University Press, New York [Google Scholar]
- Gimelli G, Zuffardi O, Giglio S, Zeng C, He D (2000) CENP-G in neocentromeres and inactive centromeres. Chromosoma 109:328–333 [DOI] [PubMed] [Google Scholar]
- Gisselsson D, Hoglund M, Mertens F, Johansson B, Dal Cin P, Van den Berghe H, Earnshaw WC, Mitelman F, Mandahl N (1999) The structure and dynamics of ring chromosomes in human neoplastic and non-neoplastic cells. Hum Genet 104:315–325 [DOI] [PubMed] [Google Scholar]
- Gisselsson D, Hoglund M, Mertens F, Mitelman F, Mandahl N (1998) Chromosomal organization of amplified chromosome 12 sequences in mesenchymal tumors detected by fluorescence in situ hybridization. Genes Chromosomes Cancer 23:203–212 [DOI] [PubMed] [Google Scholar]
- Goday C, Gonzalez-Garcia JM, Esteban MR, Giovinazzo G, Pimpinelli S (1992) Kinetochores and chromatin diminution in early embryos of Parascaris univalens. J Cell Biol 118:23–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimbacher B, Dutra AS, Holland SM, Fischer RE, Pao M, Gallin JI, Puck JM (1999) Analphoid marker chromosome in a patient with hyper-IgE syndrome, autism, and mild mental retardation. Genet Med 1:213–218 [DOI] [PubMed] [Google Scholar]
- Harrington JJ, Van Bokkelen G, Mays RW, Gustashaw K, Willard HF (1997) Formation of de novo centromeres and construction of first-generation human artificial microchromosomes. Nat Genet 15:345–355 [DOI] [PubMed] [Google Scholar]
- Heim S, Mandahl N, Kristoffersson U, Mitelman F, Rooser B, Rydholm A, Willen H (1987) Marker ring chromosome—a new cytogenetic abnormality characterizing lipogenic tumors? Cancer Genet Cytogenet 24:319–326 [DOI] [PubMed] [Google Scholar]
- Henikoff S, Ahmad K, Malik HS (2001) The centromere paradox: stable inheritance with rapidly evolving DNA. Science 293:1098–1102 [DOI] [PubMed] [Google Scholar]
- Henikoff S, Ahmad K, Platero JS, van Steensel B (2000) Heterochromatic deposition of centromeric histone H3-like proteins. Proc Natl Acad Sci USA 97:716–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henning KA, Novotny EA, Compton ST, Guan XY, Liu PP, Ashlock MA (1999) Human artificial chromosomes generated by modification of a yeast artificial chromosome containing both human alpha satellite and single-copy DNA sequences. Proc Natl Acad Sci USA 96:592–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiatt EN, Kentner EK, Dawe RK (2002) Independently regulated neocentromere activity of two classes of tandem repeat arrays. Plant Cell 14:407–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins RR, Wright E, Baldinger S, Tschider E, Ahmad J, Schwarz S, Curtis CA (2000) Neocentromere in a ring-shaped chromosome 1. Am J Hum Genet Suppl 67:A775 [Google Scholar]
- Hu J, McPherson E, Surti U, Hasegawa S, Gunawardena S, Gollin SM (2001) Tetrasomy 15q25.3-qter resulting from an analphoid supernumerary marker chromosome in a patient with multiple abnormalities and bilateral Wilms' tumors. Am J Hum Genet Suppl 69:A645 [DOI] [PubMed] [Google Scholar]
- Huang B, Ning Y, Lamb AN, Sandlin CJ, Jamehdor M, Ried T, Bartley J (1998) Identification of an unusual marker chromosome by spectral karyotyping. Am J Med Genet 80:368–372 [PubMed] [Google Scholar]
- Hudson DF, Fowler KJ, Earle E, Saffery R, Kalitsis P, Trowell H, Hill J, Wreford NG, deKretser DM, Cancilla MR, Howman E, Hii L, Cutts SM, Irvine DV, Choo KHA (1998) Centromere protein B null mice are mitotically and meiotically normal but have lower body and testis weights. J Cell Biol 141:309–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeno M, Grimes B, Okazaki T, Nakano M, Saitoh K, Hoshino H, McGill NI, Cooke H, Masumoto H (1998) Construction of YAC-based mammalian artificial chromosomes. Nat Biotechnol 16:431–439 [DOI] [PubMed] [Google Scholar]
- Kaiser-Rogers KA, Davenport ML, Powell CM, Rao KW (1995) A recombinant X chromosome with an atypical centromere observed in a child with Turner syndrome. Am J Hum Genet Suppl 57:A658 [Google Scholar]
- Kapoor M, MontesdeOcaLuna R, Liu G, Lozano G, Cummings C, Mancini M, Ouspenski I, Brinkley BR, May GS (1998) The cenpB gene is not essential in mice. Chromosoma 107:570–576 [DOI] [PubMed] [Google Scholar]
- Levy B, Papenhausen P, Tepperberg J, Dunn T, Fallet S, Magid M, Kardon N, Hirschhorn K, Warburton P (2000) Prenatal molecular cytogenetic diagnosis of partial tetrasomy 10p due to neocentromere formation in an inversion duplication analphoid marker chromosome. Cytogenet Cell Genet 91:165–170 [DOI] [PubMed] [Google Scholar]
- Li S, Malafiej P, Levy B, Mahmood R, Field M, Hughes M, Lockhart LH, Wu Z, Huang M, Hirschhorn K, Golpalrao VN, Velagaleti AD, Warburton PE (2002) Chromosome 13q neocentromeres: molecular cytogenetic characterization of three additional cases and clinical spectrum. Am J Med Genet 110:258–267 [DOI] [PubMed] [Google Scholar]
- Lo AW, Craig JM, Saffery R, Kalitsis P, Irvine DV, Earle E, Magliano DJ, Choo KHA (2001a) A 330 kb CENP-A binding domain and altered replication timing at a human neocentromere. EMBO J 20:2087–2096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo AW, Magliano DJ, Sibson MC, Kalitsis P, Craig JM, Choo KHA (2001b) A novel chromatin immunoprecipitation and array (CIA) analysis identifies a 460-kb CENP-A-binding neocentromere DNA. Genome Res 11:448–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maggert KA, Karpen GH (2001) The activation of a neocentromere in Drosophila requires proximity to an endogenous centromere. Genetics 158:1615–1628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maraschio P, Tupler R, Rossi E, Barbierato L, Uccellatore F, Rocchi M, Zuffardi O, Fraccaro M (1996) A novel mechanism for the origin of supernumerary marker chromosomes. Hum Genet 97:382–386 [DOI] [PubMed] [Google Scholar]
- Morrissette JD, Celle L, Owens NL, Shields CL, Zackai EH, Spinner NB (2001) Boy with bilateral retinoblastoma due to an unusual ring chromosome 13 with activation of a latent centromere. Am J Med Genet 99:21–28 [DOI] [PubMed] [Google Scholar]
- Murphy TD, Karpen GH (1998) Centromeres take flight: alpha satellite and the quest for the human centromere. Cell 93:317–320 [DOI] [PubMed] [Google Scholar]
- Ohashi H, Wakui K, Ogawa K, Okano T, Niikawa N, Fukushima Y (1994) A stable acentric marker chromosome: possible existence of an intercalary ancient centromere at distal 8p. Am J Hum Genet 55:1202–1208 [PMC free article] [PubMed] [Google Scholar]
- Peacock WJ, Dennis ES, Rhoades MM, Pryor AJ (1981) Highly repeated DNA sequence limited to knob heterochromatin in maize. Proc Natl Acad Sci USA 78:4490–4494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedeutour F, Forus A, Coindre JM, Berner JM, Nicolo G, Michiels JF, Terrier P, Ranchere-Vince D, Collin F, Myklebost O, Turc-Carel C (1999) Structure of the supernumerary ring and giant rod chromosomes in adipose tissue tumors. Genes Chromosomes Cancer 24:30–41 [PubMed] [Google Scholar]
- Pedeutour F, Suijkerbuijk RF, Forus A, Van Gaal J, Van de Klundert W, Coindre JM, Nicolo G, Collin F, Van Haelst U, Huffermann K (1994) Complex composition and co-amplification of SAS and MDM2 in ring and giant rod marker chromosomes in well-differentiated liposarcoma. Genes Chromosomes Cancer 10:85–94 [DOI] [PubMed] [Google Scholar]
- Perez-Castro AV, Shamanski FL, Meneses JJ, Lovato TL, Vogel KG, Moyzis RK, Pedersen R (1998) Centromeric protein b null mice are viable with no apparent abnormalities. Develop Biol 201:135–143 [DOI] [PubMed] [Google Scholar]
- Petit P, Fryns JP (1997) Interstitial deletion 2p accompanied by marker chromosome formation of the deleted segment resulting in a stable acentric marker chromosome. Genet Couns 8:341–343 [PubMed] [Google Scholar]
- Platero JS, Ahmad K, Henikoff S (1999) A distal heterochromatic block displays centromeric activity when detached from a natural centromere. Mol Cell 4:995–1004 [DOI] [PubMed] [Google Scholar]
- Portnoi MF, Boutchnei S, Bouscarat F, Morlier G, Nizard S, Dersarkissian H, Crickx B, Nouchy M, Taillemite JL, Belaich S (1999) Skin pigmentary anomalies and mosaicism for an acentric marker chromosome originating from 3q. J Med Genet 36:246–250 [PMC free article] [PubMed] [Google Scholar]
- Ravnan JB, Ouellette K, Fabre A, Crenshaw DC, Guillory S, Siewert R, McCoy S, Kothari J (1999) A stable acentric marker chromosome formed by interstitial deletion of 17q and subsequent inverted duplication of the deleted segment resulting in partial trisomy for 17q22 to 17q23 diagnosed in a dysmorphic newborn. Am J Hum Genet Suppl 65:A2012 [Google Scholar]
- Reddy KS, Sulcova V, Schwartz S, Noble JE, Phillips J, Brasel JA, Huff K, Lin HJ (2000) Mosaic tetrasomy 8q: inverted duplication of 8q23.3qter in an analphoid marker. Am J Med Genet 92:69–76 [PubMed] [Google Scholar]
- Rhoades MM (1942) Preferential segregation in maize. Genetics 27:395–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ——— (1950) Meiosis in maize. J Hered 41:58–67 [DOI] [PubMed] [Google Scholar]
- Rhoades MM, Dempsey E (1966) The effect of abnormal chromosome 10 on preferential segregation and crossing over in maize. Genetics 53:989–1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoades MM, Vilkomerson H (1942) On the anaphase movement of chromosomes. Proc Natl Acad Sci USA 28:433–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera H, Vasquez AI, Ayala-Madrigal ML, Ramirez-Duenas ML, Davalos MP (1996) Alphoidless centromere of a familial unstable inverted Y chromosome. Ann Genet 39:236–239 [PubMed] [Google Scholar]
- Rivera H, Vasquez AI, Garcia-Cruz D, Crolla JA (1999) Neocentromere at 13q32 in one of two stable markers derived from a 13q21 break. Am J Med Genet 85:385–388 [DOI] [PubMed] [Google Scholar]
- Rosai J, Akerman M, Dal Cin P, DeWever I, Fletcher CD, Mandahl N, Mertens F, Mitelman F, Rydholm A, Sciot R, Tallini G, Van den Berghe H, Van de Ven W, Vanni R, Willen H (1996) Combined morphologic and karyotypic study of 59 atypical lipomatous tumors. Evaluation of their relationship and differential diagnosis with other adipose tissue tumors (a report of the CHAMP Study Group). Am J Surg Path 20:1182–1189 [DOI] [PubMed] [Google Scholar]
- Rowe AG, Abrams L, Qu Y, Chen E, Cotter PD (2000) Tetrasomy 15q25→qter: cytogenetic and molecular characterization of an analphoid supernumerary marker chromosome. Am J Med Genet 93:393–398 [PubMed] [Google Scholar]
- Rubin BP, Fletcher CD (1997) The cytogenetics of lipomatous tumours. Histopathology 30:507–511 [DOI] [PubMed] [Google Scholar]
- Sacchi N, Magnani I, Fuhrman-Conti AM, Monard SP, Darfler M (1996) A stable marker chromosome with a cryptic centromere: evidence for centromeric sequences associated with an inverted duplication. Cytogenet Cell Genet 73:123–129 [DOI] [PubMed] [Google Scholar]
- Saffery R, Choo KHA (2002) Strategies for engineering human chromosomes with therapeutic potential. J Gene Med 4:5–13 [DOI] [PubMed] [Google Scholar]
- Saffery R, Irvine DV, Griffiths B, Kalitsis P, Wordeman L, Choo KHA (2000) Human centromeres and neocentromeres show identical distribution patterns of >20 functionally important kinetochore-associated proteins. Hum Mol Genet 9:175–185 [DOI] [PubMed] [Google Scholar]
- Saffery R, Wong LH, Irvine DV, Bateman MA, Griffiths B, Cutts SM, Cancilla MR, Cendron AC, Stafford AJ, Choo KHA (2001) Construction of neocentromere-based human minichromosomes by telomere-associated chromosomal truncation. Proc Natl Acad Sci USA 98:5705–5710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satinover DL, Eichler EE, Schwartz S (2001a) Identification of two distinct genomic organizations in neocentromeres: implications for neocentromere acquisition. Am J Hum Genet Suppl 69:A200 [Google Scholar]
- Satinover DL, Vance GH, Van Dyke DL, Schwartz S (2001b) Cytogenetic analysis and construction of a BAC contig across a common neocentromeric region from 9p. Chromosoma 110:275–283 [DOI] [PubMed] [Google Scholar]
- Shelby RD, Monier K, Sullivan KF (2000) Chromatin assembly at kinetochores is uncoupled from DNA replication. J Cell Biol 151:1113–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen MH, Ross A, Yang J, de las Heras JI, Cooke H (2001) Neo-centromere formation on a 2.6 Mb mini-chromosome in DT40 cells. Chromosoma 110:421–429 [DOI] [PubMed] [Google Scholar]
- Sirvent N, Forus A, Lescaut W, Burel F, Benzaken S, Chazal M, Bourgeon A, Vermeesch JR, Myklebost O, Turc-Carel C, Ayraud N, Coindre JM, Pedeutour F (2000) Characterization of centromere alterations in liposarcomas. Genes Chromosomes Cancer 29:117–129 [DOI] [PubMed] [Google Scholar]
- Slater HR, Nouri S, Earle E, Lo AW, Hale LG, Choo KHA (1999) Neocentromere formation in a stable ring 1p32-p36.1 chromosome. J Med Genet 36:914–918 [PMC free article] [PubMed] [Google Scholar]
- Smit AF (1999) Interspersed repeats and other mementos of transposable elements in mammalian genomes. Curr Opin Genet Dev 9:657–663 [DOI] [PubMed] [Google Scholar]
- Spiegel M, Hickmann G, Senger G, Kozlowski P, Bartsch O. Two new cases of analphoid marker chromosomes. Am J Med Genet (in press) [DOI] [PubMed] [Google Scholar]
- Sreekantaiah C, Karakousis CP, Leong SP, Sandberg AA (1992) Cytogenetic findings in liposarcoma correlate with histopathologic subtypes. Cancer 69:2484–2495 [DOI] [PubMed] [Google Scholar]
- Steiner NC, Clarke L (1994) A novel epigenetic effect can alter centromere function in fission yeast. Cell 79:865–874 [DOI] [PubMed] [Google Scholar]
- Sulcova V, Reddy KS, Schwartz S, Noble J, Lin H (1999) An analphoid marker chromosome shown to be an inverted duplication 8q23qter with a neocentromere. Am J Hum Genet Suppl 65:A2026 [Google Scholar]
- Teshima I, Bawle EV, Weksberg R, Shuman C, Van Dyke DL, Schwartz S (2000) Analphoid 3qter markers. Am J Med Genet 94:113–119 [DOI] [PubMed] [Google Scholar]
- Tyler-Smith C, Corish P, Burns E (1998) Neocentromeres, the Y chromosome and centromere evolution. Chromosome Res 6:65–67 [DOI] [PubMed] [Google Scholar]
- Tyler-Smith C, Gimelli G, Giglio S, Floridia G, Pandya A, Terzoli G, Warburton PE, Earnshaw WC, Zuffardi O (1999) Transmission of a fully functional human neocentromere through three generations. Am J Hum Genet 64:1440–1444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Enden A, Verschraegen-Spae MR, Van Roy N, Decaluwe W, De Praeter C, Speleman F (1996) Mosaic tetrasomy 15q25-qter in a newborn infant with multiple anomalies. Am J Med Genet 63:482–485 [DOI] [PubMed] [Google Scholar]
- Vance GH, Curtis CA, Heerema NA, Schwartz S, Palmer CG (1997) An apparently acentric marker chromosome originating from 9p with a functional centromere without detectable alpha and beta satellite sequences. Am J Med Genet 71:436–442 [DOI] [PubMed] [Google Scholar]
- Ventura M, Archidiacono N, Rocchi M (2001) Centromere emergence in evolution. Genome Res 11:595–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voullaire LE, Slater HR, Petrovic V, Choo KHA (1993) A functional marker centromere with no detectable alpha-satellite, satellite III, or CENP-B protein: activation of a latent centromere? Am J Hum Genet 52:1153–1163 [PMC free article] [PubMed] [Google Scholar]
- Voullaire L, Saffery R, Davies J, Earle E, Kalitsis P, Slater H, Irvine DV, Choo KHA (1999) Trisomy 20p resulting from inverted duplication and neocentromere formation. Am J Med Genet 85:403–408 [PubMed] [Google Scholar]
- Voullaire L, Saffery R, Earle E, Irvine DV, Slater H, Dale S, du Sart D, Fleming T, Choo KHA (2001) Mosaic inv dup(8p) marker chromosome with stable neocentromere suggests neocentromerization is a post-zygotic event. Am J Med Genet 102:86–94 [DOI] [PubMed] [Google Scholar]
- Wandall A, Tranebjaerg L, Tommerup N (1998) A neocentromere on human chromosome 3 without detectable alpha-satellite DNA forms morphologically normal kinetochores. Chromosoma 107:359–365 [DOI] [PubMed] [Google Scholar]
- Warburton PE, Cooke CA, Bourassa S, Vafa O, Sullivan BA, Stetten G, Gimelli G, Warburton D, Tyler-Smith C, Sullivan KF, Poirier GG, Earnshaw WC (1997) Immunolocalization of CENP-A suggests a distinct nucleosome structure at the inner kinetochore plate of active centromeres. Curr Biol 7:901–904 [DOI] [PubMed] [Google Scholar]
- Warburton PE, Dolled M, Mahmood R, Alonso A, Li S, Naritomi K, Tohma T, Nagai T, Hasegawa T, Ohashi H, Govaerts LC, Eussen BH, Van Hemel JO, Lozzio C, Schwartz S, Dowhanick-Morrissette JJ, Spinner NB, Rivera H, Crolla JA, Yu C, Warburton D (2000) Molecular cytogenetic analysis of eight inversion duplications of human chromosome 13q that each contain a neocentromere. Am J Hum Genet 66:1794–1806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss SW (1996) Lipomatous tumors. Monogr Pathol 38:207–239 [PubMed] [Google Scholar]
- Williams BC, Murphy TD, Goldberg ML, Karpen GH (1998) Neocentromere activity of structurally acentric mini-chromosomes in Drosophila. Nat Genet 18:30–37 [DOI] [PubMed] [Google Scholar]
- Winters J, Lipson M, Dolliver M, Owen S, Schwartz S (2000) Characterization of a familial acentric marker chromosome identified at amniocentesis. Am J Hum Genet Suppl 67:A800 [Google Scholar]
- Yu HG, Dawe RK (2000) Functional redundancy in the maize meiotic kinetochore. J Cell Biol 151:131–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu HG, Hiatt EN, Chan A, Sweeney M, Dawe RK (1997) Neocentromere-mediated chromosome movement in maize. J Cell Biol 139:831–840 [DOI] [PMC free article] [PubMed] [Google Scholar]