Abstract
Novel bioactive lipids were identified from a Guamanian cyanobacterium; the Pseudomonas aeruginosa quorum sensing inhibitor pitinoic acid A (1) and the anti-inflammatory pitinoic acids B (2) and C. The structure of 2 was confirmed by synthesis, which also allowed for biological evaluation. Since 2 is an ester of pitinoic acids A and C, it represents a prodrug strategy to liberate dual biological activity for the management of P. aeruginosa infections and their associated inflammation.
Pseudomonas aeruginosa is a Gram-negative bacterium that causes opportunistic infections in different host tissues and organs. For example, cystic fibrosis (CF) infections and microbial keratitis (MK) are two of the most serious lung and eye infections, respectively, caused by this organism.1, 2 Bacterial quorum sensing (QS) has been shown to play a major role in bacterial pathogenesis, where the infection and tissue destruction are facilitated by several QS-regulated virulence factors including elastase (LasB) and the pigment pyocyanin.2, 3 Tissue damage is also aggravated by the host’s defense response, where the release of pro-inflammatory mediators and the prolonged inflammatory response are other contributors to tissue pathology.2, 4–8 Accordingly, inhibiting QS pathways in P. aeruginosa and simultaneously controlling the associated inflammatory response is a promising strategy for the treatment of infections caused by this organism.
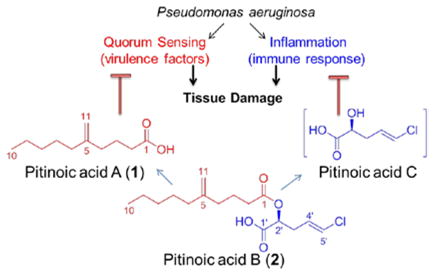
During our efforts to discover novel bioactive compounds from marine cyanobacteria, we explored a population of a marine cyanobacterium morphologically similar to Lyngbya sp. collected from a channel at the north end of Piti Bay at Guam, through NMR-guided fractionation. We isolated and characterized the novel lipids pitinoic acid A (1) (5-methylene decanoic acid) and its chlorinated ester, pitinoic acid B (2). The fatty acid 1 inhibits QS in P. aeruginosa, while the ester 2 prevents the induction of pro-inflammatory cytokine expression in LPS-induced THP-1 macrophages. In addition, we were able to identify the presence of pitinoic acid C, the alcohol moiety in 2, in one HPLC-fraction, which also maintained the anti-inflammatory effect detected for 2. Based on this, 2 appears to be a nature-designed prodrug which could deliver two essential moieties with different bioactivities upon hydrolysis; the QS-inhibitory module pitinoic acid A (1) and the anti-inflammatory module pitinoic acid C.
The EtOAc-MeOH extract was subjected to solvent-solvent partitioning followed by fractionation using silica gel chromatography. 1H NMR profiles of the generated silica fractions showed a major simple fatty acid (1) dominating most of the fractions, which was then purified by HPLC from one of the fractions that eluted with 10% i-PrOH in CH2Cl2 as colorless oil. The 1H NMR spectrum of 1 showed typical peaks for fatty acids: two methylene groups at δH ≈ 1.2 ppm, a terminal methyl group at δH 0.89 ppm, an α-methylene group at δH 2.35 ppm, and the fatty acid carbonyl carbon appeared in the 13C NMR spectrum at δC 180.2 ppm. Additionally, a methylene group at δH 4.73 ppm and δC 109.5 ppm showed an HMBC correlation to a quaternary carbon at δC 148.7 ppm, indicating the presence of an exo-double bond along the fatty acid chain. The information obtained from 1D, 2D NMR spectra and MS data (HRESIMS: m/z 183.1398 for [M-H]− corresponding to C11H19O2) led to the characterization of the simple fatty acid pitinoic acid A (1) as the major fatty acid in this sample (Table 1).
Table 1.
NMR spectroscopic data for pitinoic acid A (1) and pitinoic acid B (2) in CDCl3 (δ in ppm, J in Hz) at 600 MHz
| C/H no. | pitinoic acid A (1) | pitinoic acid B (2) | ||||
|---|---|---|---|---|---|---|
| δC | δH(J) | HMBCa | δC | δH(J) | HMBCa | |
| 1 | 180.1 | – | 2, 3 | 173.3 | – | 2, 3 |
| 2 | 33.6 | 2.35 t (7.4) | 3, 4 | 33.4 | 2.40 dt (7.8, 6.7) | 3, 4 |
| 3 | 22.5 | 1.77 m | 2, 4 | 22.7 | 1.79 m | 2, 4 |
| 4 | 35.0 | 2.06 t (7.5) | 3, 6, 11 | 35.2 | 2.06 t (7.3) | 2, 3, 6, 11 |
| 5 | 148.7 | – | 3, 4, 6, 7, 11 | 148.9 | – | 3, 4, 6, 7, 11 |
| 6 | 35.6 | 1.99 t (7.5) | 4, 7, 11 | 35.8 | 1.99 t (7.6) | 4, 7, 11 |
| 7 | 27.3 | 1.42 m | 6, 8 | 27.4 | 1.42 m | 6, 8 |
| 8 | 31.6 | 1.26 m | 7, 9, 10 | 31.7 | 1.26 m | 6, 7, 9, 10 |
| 9 | 22.4 | 1.31 m | 7, 8, 10 | 22.6 | 1.32 m | 8, 10 |
| 10 | 13.9 | 0.89 t (7.1) | 8, 9 | 14.1 | 0.89 t (7.1) | 8, 9 |
| 11 | 109.5 | 4.72 s, 4.75 s | 4, 6 | 109.8 | 4.72 s, 4.75 s | 4, 6 |
| 1′ | 173.5 | – | 2′ | |||
| 2′ | 70.7 | 5.09 t (5.6) | 3′, 4′, 5′ | |||
| 3′ | 32.5 | 2.64 m | 2′, 4′, 5′ | |||
| 4′ | 127.3 | 5.9 m | 2′, 3′, 5′ | |||
| 5′ | 121.4 | 6.12 d (13.4) | 3′, 4′ | |||
Protons showing long-range correlation to indicated carbon.
HPLC purification of another silica fraction that eluted with 40% MeOH in CH2Cl2 yielded another fatty acid compound, where the 1H and 13C NMR spectra included all the peaks corresponding to 1 as well as peaks for five additional carbons and attached protons: one carbonyl at δC 173.5 ppm, an oxygenated methine at δH 5.09 and δC 70.7 ppm, a methylene at δH 2.64 and δC 32.5 ppm and olefinic methines at δH 5.9; δC 127.3 ppm and δH 6.12 and δC 121.4 ppm (Table 1). COSY, TOCSY and HMBC data led to the assignment of the additional portion as a 2-hydroxy-pent-4-enoic acid, and a chloride attachment to the terminal olefinic methine was suggested by the remaining molecular mass and the isotopic cluster detected in the HRESIMS analysis (m/z 315.1384, 317.1348 (3:1) for [M-H]− corresponding to C16H2435ClO4 and C16H2437ClO4, respectively). Notably, the chemical shift of the carbonyl carbon in the decanoic acid part was shifted upfield compared to the free fatty acid (Table 1), and therefore the compound appeared to be a fatty acid ester, 2. The geometry of the alkene in the 2-hydroxy-5-chloro-pent-4-enoic acid moiety was determined to be E based on the large vicinal coupling constant (3JH,H 13.4 Hz) between the olefinic protons. The configuration of the stereogenic center C2′ was assigned through ozonolysis followed by oxidative workup to yield the corresponding malic acid, which was then analyzed by chiral HPLC compared to malic acid standards to reveal S configuration.
The abundance of 1 in this sample (≈ 0.3% of total dry weight) suggested that it has an important ecological role for this organism, and was sufficient for biological activity evaluation in several assays. However, the chlorinated ester 2 was only isolated in minute amounts (≈ 900 μg) and required synthetic efforts to allow for biological evaluation.
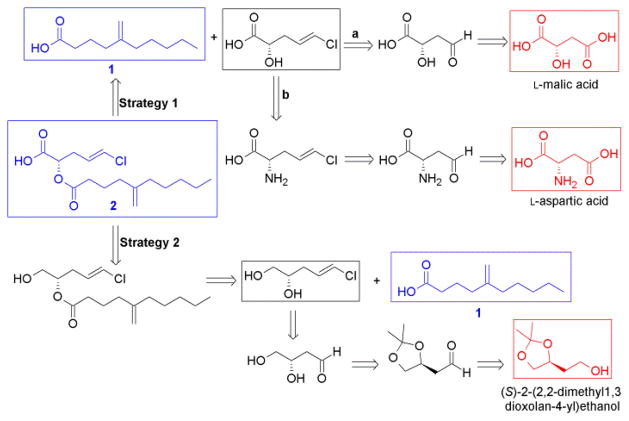
Our initial retrosynthetic strategy relied on the obvious disconnection at the ester linkage to the fatty acid 1, which could be obtained from the cyanobacterial sample, and the chlorinated α-hydroxy acid fragment pitinoic acid C. The selective introduction of the E-vinyl chloride in the alcohol moiety could be achieved through Takai-Utimoto olefination reaction9 from an α-hydroxy-4-oxobutanoic acid. The latter could be obtained by selective reduction of L-malic acid at C4 (Scheme 1, strategy 1a). Since this selective reduction of malic acid was not successful, we tried the method by Padron et al.10, to selectively reduce the di-Boc protected aspartic acid to the corresponding C4 semialdehyde (Scheme 1, strategy 1b). Indeed, the selective reduction starting from aspartic acid proceeded smoothly with a 67% yield. However, Takai-Utimoto olefination for the aspartate semialdehyde proceeded with a low yield (32%) and relatively low selectivity (E:Z ≈ 4:1). Notably, one Boc group was also lost during this reaction as determined by analyzing the 1H NMR spectrum. Trials with the acid-stable amine protecting group Fmoc did not achieve the intial selective reduction at C4 as with the di-Boc protected aspartate.
Scheme 1.
Retrosynthetic strategies to obtain pitinoic acid B (2). Natural products are shown in blue; starting points for each strategy are shown in red.
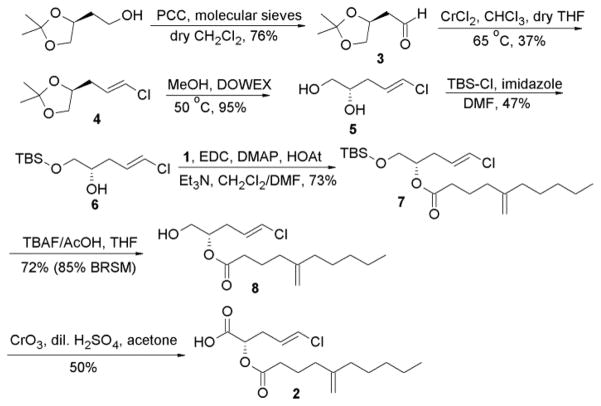
Our next retrosynthetic analysis (Scheme 1, strategy 2) aimed to primarily introduce the vinyl chloride terminus by Takai-Utimoto olefination reaction to a C4 aldehyde with a terminal vicinal diol. The secondary alcohol could be coupled first to the acid 1 before the primary alcohol at C1 could be converted to the carboxylic acid terminal. This attempt started from the commercially available acetonide (S)-2-(2,2-dimethyl-1,3-dioxolan-4-yl) ethanol (Scheme 2), where the terminal alcohol group was oxidized to the corresponding aldehyde 3 in 76% yield using PCC in CH2Cl2.11 Takai-Utimoto olefination for compound 3 appeared to proceed with good selectivity (E:Z ≈ 8:1) and reasonable yield (37%). Acetonide opening for 4 was done using DOWEX in MeOH12 to give the 1,2-diol (5) with good yield (≈ 95%). The primary alcohol in 5 was then protected as the TBDMS-ether (6, 47% yield) and the free secondary alcohol was coupled to the fatty acid 1 to furnish compound 7 (76% yield). The next step was to deprotect the primary alcohol and oxidize it to the corresponding carboxylic acid to obtain the final ester 2. First attempts to deprotect the primary alcohol in 7 using TBAF13 were faced with the challenge of acyl migration from the secondary to the primary alcohol. Minimal acyl migration was obtained by using a neutral mixture of TBAF and AcOH in THF and by limiting the reaction time to two hours giving the free primary alcohol 8 in good yield (72%; 85% BRSM). Unreacted starting material was recovered and subjected to the same conditions again.
Scheme 2.
Final scheme for the semi-synthesis of pitinoic acid B (2)
For the final oxidation step, we first pursued several mild oxidation attempts, since we were concerned about the stability of the ester linkage and the vinyl chloride functionalities under any harsh reaction conditions. The traditional PDC-mediated oxidation in DMF14 was ineffective. Additionally, two-step oxidation protocols to the aldehyde and then to the acid were also inefficient to produce the desired carboxylic acid, including Dess-Martin oxidation followed by NaClO2 or PDC treatment, and TEMPO-catalyzed oxidations with NaOCl/NaClO2 system.15 Notably, the aldehyde could be obtained in some trials, but not the desired acid. Also, the exo-methylene group in the fatty acid part appeared to be affected by the NaOCl/NaClO2 oxidizing system, as the corresponding peaks for those olefinic protons disappeared in the 1H NMR spectrum. One-pot deprotection of the silyl ether catalyzed by Bi(OTf)3 followed by TEMPO-catalyzed oxidation16 did not give desirable results either. Finally, Jones oxidation was the only method that gave the desired final product with a moderate yield (≈50%). However, in order to obtain the final pure product, HPLC purification was essential, which appeared to significantly lower the yield of the pure product (yield dropped to 20% after HPLC purification). NMR spectral data and optical rotation for the synthetic compound matched those for the natural product 2 (Supp. Fig. S28). The problems faced with the stability and the purification of this chlorinated ester could provide one explanation for the low amount of the natural product that we were able to isolate from the cyanobacterium. However, sufficient amount was successfully prepared to allow for further biological characterization.
One of the common biological roles of fatty acid compounds produced by bacteria in large quantities is the interference with quorum sensing (communication between bacteria in response to high population densities). This was shown in several previous reports, 17–20 including by our group where we reported the anti-quorum sensing activity of lyngbyoic acid.21 In Pseudomonas aeruginosa, quorum sensing (QS) system integrates two chemically distinct classes of signal molecules which act on differnet QS pathways, the N-acylhomoserine lactones which act on the LasR-LasI pathway, and the 4-quinolones which act on the Pseudomonas quinolone signaling pathway.22,23 Lyngbyoic acid was found to inhibit QS through the LasR pathway, where the cyclopropyl ring appeared to be essential for this activity.21 Based on structural similarity, 1 could also possess QS inhibitory activity similar to lyngbyoic acid, where the exo-methylene group might also be influential for this biological activity. Accordingly, we tested 1 for its ability to interfere with quorum sensing in P. aeruginosa by monitoring its effect on the transcription and the production of two virulence factors, the elastase LasB enzyme and the pigment pyocyanin. Compound 1 significantly reduced the transcript levels of lasB and the pyocyanin biosynthetic member phzG1 after 6 h at 1 mM and 100 μM as assessed by RT-qPCR (Figure 1). Additionally, after 6 h, the levels of LasB and pyocyanin in the culture supernatants were also significantly reduced by 1 at 1 mM, as evaluated by an enzymatic assay for LasB and quantitative evaluation using UV absorbance for pyocyanin (Figure 1). Measuring the bacterial cell density (OD600) showed that this fatty acid did not affect cellular growth at the tested concentrations.
Figure 1.
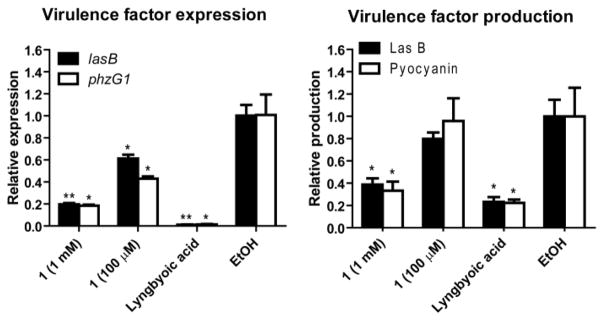
Quorum sensing inhibition by pitinoic acid A (1) in P. aeruginosa. Lyngbyoic acid (1 mM) was used as a positive control. *P < 0.05, **P < 0.01 (t-test); n = 3. Results are calculated relative to the endogenous control rpoD. Data are presented as mean ± SD.
The chlorinated ester 2 has an intriguing chemical structure. Similar structural features were also present in the recently reported anti-inflammatory marine cyanobacterial fatty acid esters honaucins, which also contain a terminal chloride.20 We initiated the biological characterization of 2 by testing its effect on LPS-induced inflammatory responses in human acute monocytic leukemia cell line THP-1 after differentiation to macrophages. Compound 2 was able to decrease the transcript levels of the pro-inflammatory cytokines TNF-α and IL-6 after 4 h of LPS stimulation at 100 μM (Figure 2).24 Additionally, the anti-inflammatory effect was also sustained after 24 h, where the mRNA levels of IL-6, IL-1β and IL-8 were significantly reduced (Figure 2). Notably, IL-6 was the most affected cytokine at all time points. Cell viability under the tested conditions was 73% based on MTT colorimetric assay.
Figure 2.
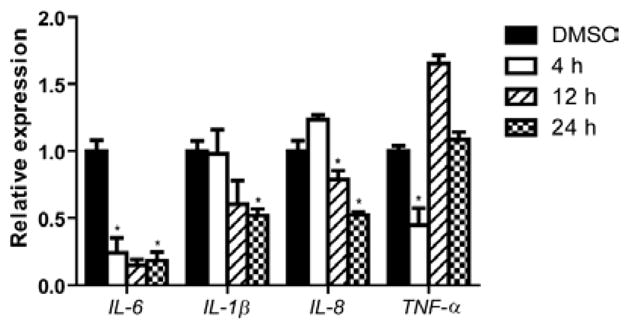
Effect of pitinoic acid B (2) (100 μM) on transcript levels of pro-inflammatory cytokines in differentiated THP-1 cells. *P-value < 0.05 (t-test); n = 3. Results are calculated relative to the endogenous control GAPDH. Data are presented as mean ± SD.
Based on its structural features, we hypothesize that the chlorinated alcohol part in the ester 2 is the major contributor to its detected anti-inflammatory effect. This notion is consistent with the structural similarity of this moiety to other previously reported anti-inflammatory compounds and to the anti-inflammatory acid part in honaucins.20 Additionally, using 1H NMR and MS analysis, we were able to confirm the presence of this α-hydroxy acid in one of the minor impure HPLC fractions obtained during the purification of 2 (Supp. Fig. 29, Supp. Fig. 30). Pitinoic acid C appeared to be the major compound in this HPLC fraction (Supp. Fig. 30), but it was unstable since purification trials led to the loss of material. Accordingly, the impure fraction containing pitinoic acid C was tested for the anti-inflammatory effects. Indeed, this fraction was also able to reduce the levels of IL-6 and TNF-α after 4 h at 15 μg/mL (Supp. Fig. 31). Notably, 1 was also able to reduce the levels of those pro-inflammatory cytokines after 4 h at 100 μM (Supp. Fig. 31). This is not surprising, since it has been shown that QS mediators could modulate the production of pro-inflammatory cytokines in mammalian cells.25, 26
In conclusion, we identified a set of interesting bioactive lipids from a marine cyanobacterial sample. This cyanobacterium appears to utilize a clever modular strategy to maintain the structure and function of those secondary metabolites. We suspect 2 to be a prodrug that utilizes the QS-inhibitor 1 as a protecting group to circumvent early reaction and deactivation of the α-hydroxy acid pitinoic acid C. One possible mechanism for the biological activity of the latter is dehydration to give a Michael acceptor fragment, which could be the reactive species. If true, then the protection of pitinoic acid C by esterification, as employed by this organism, is an optimum solution to avoid the early release of this reactive species. Additionally, if hydrolysis of 2 takes place in vivo, it will also release the anti-quorum sensing acid part 1. This could highlight 2 as a hybrid prodrug molecule with dual biological activity, which could be effective in the management of P. aeruginosa infections and associated inflammation. Further chemical and biological studies will allow for the detailed understanding of the mode of action of those molecules and the development of more potent analogues.
Supplementary Material
Acknowledgments
We thank Dr. Qi-Yin Chen (Department of Medicinal Chemistry, University of Florida) for helpful discussions regarding the synthetic procedures and Dr. Sarath Gunasekera and Vicky Pittman (Smithsonian Marine Station) for providing additional amounts of pitinoic acid A. We thank J. R. Rocca (AMRIS, McKnight Brain Institute, University of Florida) for technical assistance with NMR data acquisition. This research was supported by the National Institutes of Health, Grants P41GM086210 and R01CA172310. This is contribution 916 from the Smithsonian Marine Station.
Footnotes
Supporting Information Available. 1D and 2D NMR spectra for compounds 1 and 2, supporting figures including spectral data and characterization of synthetic intermediates, experimental procedures and additional RT-qPCR data. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Sadikot RT, Blackwell TS, Christman JW, Prince AS. Am J Respir Crit Care Med. 2005;171:1209–1223. doi: 10.1164/rccm.200408-1044SO. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Willcox MD. Optom Vis Sci. 2007;84:273–278. doi: 10.1097/OPX.0b013e3180439c3e. [DOI] [PubMed] [Google Scholar]
- 3.Willcox MD, Zhu H, Conibear TC, Hume EB, Givskov M, Kjelleberg S, Rice SA. Microbiology. 2008;154:2184–2194. doi: 10.1099/mic.0.2008/019281-0. [DOI] [PubMed] [Google Scholar]
- 4.Xue ML, Thakur A, Willcox M. Clin Experiment Ophthalmol. 2002;30:196–199. doi: 10.1046/j.1442-9071.2002.00510.x. [DOI] [PubMed] [Google Scholar]
- 5.Kernacki KA, Goebel DJ, Poosch MS, Hazlett LD. Infect Immun. 1998;66:376–379. doi: 10.1128/iai.66.1.376-379.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wölbeling F, Munder A, Kerber-Momot T, Neumann D, Hennig C, Hansen G, Tümmler B, Baumann U. Immunobiology. 2011;216:901–908. doi: 10.1016/j.imbio.2011.02.003. [DOI] [PubMed] [Google Scholar]
- 7.Rudner XL, Kernacki KA, Barrett RP, Hazlett LD. J Immunol. 2000;164:6576–6582. doi: 10.4049/jimmunol.164.12.6576. [DOI] [PubMed] [Google Scholar]
- 8.Rincon M, Irvin CG. Int J Biol Sci. 2012;8:1281–1290. doi: 10.7150/ijbs.4874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takai K, Nitta K, Utimoto K. J Am Chem Soc. 1986;108:7408–7410. doi: 10.1021/ja00279a068. [DOI] [PubMed] [Google Scholar]
- 10.Padrón JM, Kokotos G, Martín T, Markidis T, Gibbons WA, Martín VCS. Tetrahedron: Asymmetry. 1998;9:3381–3394. [Google Scholar]
- 11.Ermolenko L, Sasaki NA, Potier P. Synlett. 2001:1565–1566. [Google Scholar]
- 12.Marshall JA, Piettre A, Paige MA, Valeriote F. J Org Chem. 2003;68:1771–1779. doi: 10.1021/jo026433x. [DOI] [PubMed] [Google Scholar]
- 13.David Crouch R. Tetrahedron. 2004;60:5833–5871. [Google Scholar]
- 14.Corey EJ, Schmidt G. Tetrahedron Lett. 1979;20:399–402. [Google Scholar]
- 15.Huang L, Teumelsan N, Huang X. Chemistry. 2006;12:5246–52. doi: 10.1002/chem.200600290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barnych B, Vatèle JM. Synlett. 2011:2048–2052. [Google Scholar]
- 17.Clark BR, Engene N, Teasdale ME, Rowley DC, Matainaho T, Valeriote FA, Gerwick WH. J Nat Prod. 2008;71:1530–1537. doi: 10.1021/np800088a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kwan JC, Teplitski M, Gunasekera SP, Paul VJ, Luesch H. J Nat Prod. 2010;73:463–466. doi: 10.1021/np900614n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dobretsov S, Teplitski M, Alagely A, Gunasekera SP, Paul V. J Environ Microbiol Rep. 2010;2:739–744. doi: 10.1111/j.1758-2229.2010.00169.x. [DOI] [PubMed] [Google Scholar]
- 20.Choi H, Mascuch SJ, Villa FA, Byrum T, Teasdale ME, Smith JE, Preskitt LB, Rowley DC, Gerwick L, Gerwick WH. Chem Biol. 2012;19:589–598. doi: 10.1016/j.chembiol.2012.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kwan JC, Meickle T, Ladwa D, Teplitski M, Paul V, Luesch H. Mol Biosyst. 2011;7:1205–1216. doi: 10.1039/c0mb00180e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Suga H, Smith KM. Curr Opin Chem Biol. 2003;7:586–591. doi: 10.1016/j.cbpa.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 23.Diggle SP, Cornelis P, Williams P, Cámara M. Int J Med Microbiol. 2006;296:83–91. doi: 10.1016/j.ijmm.2006.01.038. [DOI] [PubMed] [Google Scholar]
- 24.It is not uncommon, especially for fatty acid compounds, to be tested and considered effective at this high concentration. For example, the anti-inflammatory effects of the fish oil fatty acids eicosapentaenoic acid and docosahexaenoic acid were detected at 100 μM in LPS-stimulated THP-1 macrophages (Weldon SM, Mullen AC, Loscher CE, Hurley LA, Roche HM. J Nutr Biochem. 2007;18:250–258. doi: 10.1016/j.jnutbio.2006.04.003.For other examples, see: Speranza L, Franceschelli S, Pesce M, Reale M, Menghini L, Vinciguerra I, De Lutiis MA, Felaco M, Grilli A. Phytother Res. 2010;24:1398–1404. doi: 10.1002/ptr.3173.Zhao G, Etherton TD, Martin KR, Vanden-Heuvel JP, Gillies PJ, West SG, Kris-Etherton PM. Biochem Biophys Res Commun. 2005;336:909–917. doi: 10.1016/j.bbrc.2005.08.204.Boonkaewwan C, Toskulkao C, Vongsakul M. J Agric Food Chem. 2006;54:785–789. doi: 10.1021/jf0523465.Liang Q, Wu Q, Jiang J, Duan J, Wang C, Smith MD, Lu H, Wang Q, Nagarkatti P, Fan D. J Biol Chem. 2011;286:26470–26479. doi: 10.1074/jbc.M111.227934.
- 25.Glucksam-Galnoy Y, Sananes R, Silberstein N, Krief P, Kravchenko VV, Meijler MM, Zor T. J Immunol. 2013;191:337–344. doi: 10.4049/jimmunol.1300368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kravchenko VV, Kaufmann GF, Mathison JC, Scott DA, Katz AZ, Grauer DC, Lehmann M, Meijler MM, Janda KD, Ulevitch RJ. Science. 2008;321:259–263. doi: 10.1126/science.1156499. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.