Abstract
Cleidocranial dysplasia (CCD) is an autosomal dominant heritable skeletal disease caused by heterozygous mutations in the osteoblast-specific transcription factor RUNX2. We have performed mutational analysis of RUNX2 on 24 unrelated patients with CCD. In 17 patients, 16 distinct mutations were detected in the coding region of RUNX2: 4 frameshift, 3 nonsense, 6 missense, and 2 splicing mutations, in addition to 1 polymorphism. The missense mutations were all clustered within the Runt domain, and their protein products were severely impaired in DNA binding and transactivation. In contrast, two RUNX2 mutants had the Runt domain intact and remained partially competent for transactivation. One criterion of CCD, short stature, was much milder in the patients with the intact Runt domain than in those without. Furthermore, a significant correlation was found between short stature and the number of supernumerary teeth. On the one hand, these genotype-phenotype correlations highlight a general, quantitative dependency, by skeleto-dental developments, on the gene dosage of RUNX2, which has hitherto been obscured by extreme clinical diversities of CCD; this gene-dosage effect is presumed to manifest on small reductions in the total RUNX2 activity, by approximately one-fourth of the normal level at minimum. On the other hand, the classic CCD phenotype, hypoplastic clavicles or open fontanelles, was invariably observed in all patients, including those with normal height. Thus, the cleidocranial bone formation, as mediated by intramembranous ossification, may require a higher level of RUNX2 than does skeletogenesis (mediated by endochondral ossification), as well as odontogenesis (involving still different complex processes). Overall, these results suggest that CCD could result from much smaller losses in the RUNX2 function than has been envisioned on the basis of the conventional haploinsufficiency model.
Introduction
Cleidocranial dysplasia (CCD [MIM 119600]) is a dominantly inherited autosomal bone disease that is characterized by persistently open sutures or delayed closure of sutures, hypoplastic or aplastic clavicles, short stature, delayed eruption of permanent dentition, supernumerary teeth, and other skeletal anomalies (Jarvis and Keats 1974). Considerable phenotypic variation has been reported, even within families (Chitayat et al. 1992). The phenotypic spectrum ranges from mildly affected individuals with mere dental abnormalities to severely affected patients with generalized osteoporosis, although tooth anomalies and some degrees of clavicular hypoplasia seem to be consistent features of the disease (Mundlos 1999; Quack et al. 1999).
Recently, it has been established that CCD results from heterozygous mutations or deletion of an osteoblast-specific transcription factor, core-binding factor A1 (CBFA1), leading to the proposal that haploinsufficiency in CBFA1 is responsible for this disease (Lee et al. 1997; Mundlos et al. 1997). Furthermore, a radiation-induced mutant mouse that carries similarities to CCD (Sillence et al. 1987) has also been demonstrated to contain a deletion in one Cbfa1 allele (Otto et al. 1997). CBFA1 (also variously called “PEBP2αA,” AML3,” or “OSF2”) is one of the three mammalian genes that encode the α subunit of the heterodimeric transcription factor PEBP2/CBF (Ogawa et al. 1993b; Levanon et al. 1994; Ducy et al. 1997). PEBP2/CBF is composed of two structurally unrelated subunits, α and β (Kamachi et al. 1990). The α subunit is characterized by a highly conserved 128-amino-acid region termed the “Runt domain” (Kagoshima et al. 1993), which shares a high degree of homology with the products of the Drosophila genes runt (Kania et al. 1990) and lozenge (Daga et al. 1996). Herein, we refer to this gene as “RUNX2” (GenBank accession number AF001450), according to the recently introduced standard naming for the mammalian runt-related genes. The other two α-subunit–encoding genes are RUNX1/AML1/PEBP2αB/CBFA2 (Miyoshi et al. 1991; Bae et al. 1993) and RUNX3/PEBP2αC/AML2/CBFA3 (Levanon et al. 1994; Bae et al. 1995; Wijmenga et al. 1995). Mouse knockout models have demonstrated that these three genes have indispensable roles in the master regulation of distinct developmental pathways: Runx1, for definitive hematopoiesis (Okuda et al. 1996; Q. Wang et al. 1996a); Runx2, for osteogenesis (Komori et al. 1997; Otto et al. 1997); and Runx3, for gastrointestinal organogenesis and function (Li et al. 2002). In contrast, only one gene is known to encode the mammalian β subunit, termed “PEBP2β/CBFβ” (designated “PEBP2β” hereafter) (Ogawa et al. 1993a; S. Wang et al. 1993). Although PEBP2β can heterodimerize with all three RUNX paralogs, until very recently, its biological significance has been shown only for RUNX1. Homozygous disruption of PEBP2β in mice completely blocked definitive hematopoiesis in a manner virtually identical to that observed with RUNX1 knockout mice, but their premature death in utero made it difficult to investigate potential alterations in other late-developing processes (Sasaki et al. 1996; Q. Wang et al. 1996b). However, PEBP2β has also proved to be essential at least for osteogenesis by the recent convergent demonstrations, from three different laboratories, that its conditional or partial deficiency in mice causes severe bone defects that are similar to Ccd (Yoshida et al., in press).
The Runt domain is responsible for DNA binding and heterodimerization with the β subunit (Kagoshima et al. 1993). The Runt domain also contains a nuclear-localization signal (NLS) on its C-terminal border (T. Kanno et al. 1998). The C-terminus of RUNX2 is a region rich in proline, serine, and threonine, which are necessary for RUNX2-mediated transcriptional regulation and are involved in functional interactions with various other transcription factors, coactivators, and corepressors.
The β subunit does not bind to DNA but allosterically enhances DNA binding by the α subunit (Ogawa et al. 1993b; Kagoshima et al. 1996; Tahirov et al. 2001). Recently, PEBP2β has been found to have another important role in the protection of the α subunit from intracellular proteolytic degradation mediated by the ubiquitin-proteasome system (Huang et al. 2001). Within cells, PEBP2β itself is localized in the cytoplasm and can enter the nucleus only through association with Runx proteins. Notably, however, the nuclear colocalization of PEBP2β with Runx proteins is normally regulated to proceed only partly, supposedly because the Runt domain is largely masked by its intramolecular interactions with regions N- or C-terminal of the Runt domain (Lu et al. 1995; T. Kanno et al. 1998; Y. Kanno et al. 1998; Kim et al. 1999). Nevertheless, this intramolecular masking can be relieved when the N- or C-terminal region flanking the Runt domain is truncated artificially or is replaced with foreign sequences, as seen in leukemia-associated chimeric products of RUNX1 such as AML1-ETO and AML1-Evi1, or when PEBP2β is altered to gain an increased ability to heterodimerize with Runx1 again, as seen in its leukemia-associated chimeric product, CBFβ-SMMHC (Lu et al. 1995; Adya et al. 1998; T. Kanno et al. 1998; Y. Kanno et al. 1998; Tanaka et al. 1998). Thus, mutation-mediated perturbations in the intracellular interaction between PEBP2β and RUNX2 have been regarded to have important pathogenic implications.
The haploinsufficiency model for CCD implies that bone formation is highly sensitive to the gene dosage of RUNX2. As a matter of fact, the regulatory importance of gene dosage is a common theme across most Runt-family proteins. The earliest known precedent is the Drosophila gene runt, which contributes to sex determination in a dosage-dependent manner (Duffy and Gergen 1991). Another more recent example is the association between RUNX1 and a human blood disease called “familial platelet disorder with predisposition to acute myelogenous leukemia” (FPD/AML1). In a close analogy to CCD, FPD/AML, as well as some sporadic cases of leukemias, has been shown to be due to haploinsufficiency in RUNX1—caused by its heterozygous mutation (Osato et al. 1999; Song et al. 1999).
Recently, additional cases of heterozygous mutation in RUNX2 have been identified in nearly 60 families with CCD, including both familial and sporadic cases (Quack et al. 1999; Zhou et al. 1999; Golan et al. 2000; Yokozeki et al. 2000; Zhang et al. 2000; Yamachika et al. 2001; for review, see Otto et al. 2002). Despite these accumulating mutational data, it has largely remained obscure exactly what range and extent of functional loss could be conferred by the various mutations identified and also how such putative diversities in mutational effects would be correlated with the phenotypic variability of CCD. To address these questions, we have performed screening and detailed functional analyses of RUNX2 mutations in 24 Japanese patients with CCD. We describe herein the results of these analyses and some novel genotype-phenotype correlations revealed therefrom. Although preliminary information from this study has frequently been quoted in several previous articles (Nagata et al. 1999; Werner et al. 1999; Warren et al. 2000; Bravo et al. 2001; Nagata and Werner 2001; Tahirov et al. 2001; Otto et al. 2002), the present report provides the first full account of our results, with corrections to earlier quotations in a few important details.
Patients, Material, and Methods
Patients
Twenty-four unrelated families with the clinical diagnosis of CCD (for review of diagnostic criteria, see Mundlos 1999) were investigated in the present study. Informed consent was obtained from all individuals. Genomic DNA and RNA were prepared from blood lymphocytes by standard procedures.
Mutation Detection
Exons 1–7 of the RUNX2 gene were amplified by PCR under standard conditions. The primers used for genomic PCR amplification and sequencing have been described elsewhere (Zhang et al. 2000). The amplification products were checked by agarose gel electrophoresis. For SSCP, PCR products were added to 5 μl of sample buffer, were denatured at 95°C for 5 min, and were applied to the gel (GeneGel Excel 12.5/24Kit; Pharmacia Biotech). After electrophoresis, PCR products were visualized by silver staining, and those showing altered mobilities were selected and sequenced using Big Dye Terminator on an ABI310 automated sequencer (PE Biosystems). Every mutation was confirmed by sequencing of the products from several independent PCRs. To determine exon 3 skipping in patient 17, RT-PCR was performed as described elsewhere (Zhang et al. 2000). In patient 15, RT-PCR was done with primers 5′-TCAGATTACAGACCCCAGGC-3′ (sense) and 5′-TCTCAGTGAGGGATGAAATG-3′ (antisense). The product was cloned into the pCRII TA cloning vector (Invitrogen) and was sequenced.
Plasmids and Mutagenesis
For expression of the Runt domain–containing fragment as a fusion N-terminally tagged with hexahistidines, the nucleotide sequences for amino acids 74–273 of human RUNX2 were cloned in frame into the BamHI and PstI sites of the pQE9 vector (Qiagen). RUNX2 mutations were introduced into this construct by site-directed mutagenesis through use of PCR. The integrity of all the PCR constructs was confirmed by sequencing.
Electrophoretic Mobility Shift Assay (EMSA)
The hexahistidine-tagged Runt domain was expressed in Escherichia coli, was purified in a nickel nitrilotriacetic resin (Ni-NTA) (Qiagen), and was subjected to EMSA with a probe carrying a polyomavirus enhancer-derived PEBP2 site, essentially as described elsewhere (Kagoshima et al. 1996). Expression and purification of the PEBP2β protein has been described elsewhere (Ogawa et al. 1993a).
Affinity Assay of RUNX2-PEBP2β Association
The hexahistidine-tagged RUNX2 fragment (10 μg) was incubated with tagless PEBP2β (5 μg) and a Ni-NTA resin. The resin was successively washed with buffers containing 16 mmol/liter and was eluted with 250 mmol/liter imidazole. Proteins in each fraction were analyzed by SDS-PAGE followed by staining with Coomassie brilliant blue.
Transcriptional Assay
For in vivo functional studies of mutant RUNX2 proteins, the NotI-HindIII fragments from pQE9-RUNX2 (eight mutants in the Runt domain) were substituted into the compatible sites of pEF-RUNX2 (sites 1–507) (Zhang et al. 2000). For Q266X, the NotI-Tth111I fragments from pQE9-RUNX2 were substituted into the compatible sites of pEF-RUNX2 (sites 1–507). The remaining mutants were constructed by site-directed mutagenesis. NIH3T3 cells were maintained in Dulbecco modified Eagle medium supplemented with 10% (v/v) fetal bovine serum. For the luciferase assay, NIH3T3 cells seeded into six-well plates were transfected by using the nonliposomal transfection reagent FuGENE 6 (Boehringer Mannheim). The luciferase-reporter plasmid wild-type 1050.rOC-luc (Towler et al. 1994; Javed et al. 1999) (0.5 μg), the effector plasmid pEF-RUNX2 (1-507) (0.5 μg) containing mutation, and pEF-PEBP2β2 (0.2 μg) were transfected into NIH3T3 cells by using FuGENE 6. As an internal control, 1 ng of pEF-RL luciferase reporter also was included. On the one hand, to avoid an excessive production of RUNX2, we adjusted the amount of pEF-RUNX2 to slightly less (80%) than minimally saturating level (0.6 μg), as determined by prior titration experiments. On the other hand, PEBP2β2 was used at an oversaturating dose. Cells were harvested 24 h after transfection, and luciferase activities were determined by the Dual-Luciferase Reporter Assay System (Promega) in a luminometer. Firefly luciferase activities were normalized by Renilla luciferase activities. All transfection experiments were done at least three times.
Subcellular Localization
The pEF-RUNX2 plasmids bearing wild type and mutant were transfected into NIH3T3 cells by using the FuGENE 6 reagent. Immunofluorescence labeling of RUNX2 and PEBP2β were performed as described elsewhere (Lu et al. 1995), by using mouse anti-PEBP2αA, fluorescein isothiocyanate–conjugated goat anti-mouse immunoglobulin G (IgG), rabbit anti-PEBP2β, and Texas Red–conjugated AffiniPure Donkey anti-Rabbit IgG. The cells were visualized and photographed with a fluorescent microscope.
Statistical Analysis
Statistical analysis of present height SD scores was performed on 31 patients. The height was evaluated by the longitudinal growth standards for the Japanese. The height SD score between the Runt domain–impaired group (15 families; 27 patients) and the Runt domain–intact group (2 families; 4 patients) were compared using the Mann-Whitney test. The difference between the two groups was accepted as significant at P<.01. The Kruskal-Wallis H test was performed, to compare three groups of clavicle phenotypes. Correlation between the number of supernumerary teeth and height was evaluated with the Spearman correlation in nine patients.
Results
Clinical Data
Twenty-four unrelated Japanese families with clinical diagnosis of CCD were included in the present study (table 1). The patient group contained both male and female patients, in approximately equal proportions, ranging in age from 0 to 30 years, with a median of 2 years. Of the 24 families, 9 (37.5%) represented sporadic cases. A CCD phenotype was defined by the presence of hypoplastic clavicles and delayed closure of the anterior fontanelle in addition to the observation of classic craniofacial features. Although dental anomalies were not included as a fixed criterion because of their age-related penetrance, this feature was consistently observed in older patients. Stature was found to be significantly reduced in both male and female patients with CCD, in agreement with previous observation (Jensen 1990).
Table 1.
Clinical Features of Patients with CCD Who Were Tested for RUNX2 Gene Mutations
|
Mutation |
||||||||
| Family (No.of IndividualsAffected) | DelayedClosureof Suturesa | HypoplasticClaviclesb | SupernumeraryTeethc | Height SD Scoreof IndividualsAffectedd | Nucleotide | Codon | Type | Reference(s) |
| 1 (5) | + | + | NA | −4.03 | 92ins(4) | 31 | Insertion/frameshift | Present study |
| 2 (2) | + | + | 3 | −1.44 | 136delC | 46 | Deletion/frameshift | Present study |
| 3 (3) | NA | + | 3 | −1.73 | 154C→T | Q52X | Nonsense | Present study |
| 4 (1) | + | ++ | + | −4.04 | 453delT | 151 | Deletion/frameshift | Present study |
| 5 (1) | + | ++ | 0 | −.60 | 453delT | 151 | Deletion/frameshift | Present study |
| 6 (1) | + | ++ | 11 | −4.60 | 526C→T | R176W | Missense | Present study, Zhou et al. 1999 |
| 7 (4) | + | + | NA | −1.78 | 535C→T | R179X | Nonsense | Present study, Quack et al. 1999 |
| 8 (3) | + | + | NA | −1.50 | 548T→C | F183S | Missense | Present study, Zhang et al. 2000 |
| 9 (4) | + | + | NA | −2.21 | 612A→T | K204N | Missense | Present study |
| 10 (3) | + | ++ | NA | −2.01 | 617C→T | T206I | Missense | Present study |
| 11 (1) | + | + | 2 | −1.48 | 631C→T | R211W | Missense | Present study, Quack et al. 1999 |
| 12 (1) | + | + | 6 | −3.41 | 631C→T | R211W | Missense | Present study, Quack et al. 1999 |
| 13 (1) | + | + | 4 | −1.98 | 632G→A | R211Q | Missense | Present study, Quack et al. 1999 |
| 14 (4) | + | + | NA | −.74 | 793C→T, 510T→C | Q266X, 170 | Nonsense, polymorphism | Present study (for both) |
| 15 (1) | NA | NA | 0 | 0 | 1043C→T | 348 | Splicing/frameshift | Present study |
| 16 (2) | NA | + | NA | −3.02 | IVS3+2T→C | Exon skip/in frame | Present study | |
| 17 (2) | + | + | NA | −2.57 | IVS3+3delGTAA | Exon skip/in frame | Present study | |
+ = Present; NA = not available.
+ = Hypoplastic clavicles; ++ = aplastic clavicles; NA = not available.
+ = Present; NA = not available. Arabic numbers indicate the total number of supernumerary teeth in a representative patient.
When multiple patients were examined within one family, their average score is presented.
Identification of Heterozygous Mutations in RUNX2
Among the 24 families examined, we were able to detect a total of 15 heterozygous mutations and 1 additional polymorphism in 17 patients (table 1 and fig. 1). Of these mutations, 11 are novel, and the other 5 have been reported previously (Quack et al. 1999; Zhou et al. 1999; Zhang et al. 2000). In the remaining seven patients, no mutation was detected by the SSCP method combined with direct sequencing. In four of these patients, we also searched for larger chromosomal deletions in the RUNX2 gene, by Southern blotting of genomic DNA with human RUNX2 cDNA as a probe; however, no such deletion was detected (data not shown). The other three patients were not analyzed, because the DNA material was inaccessible. The mutations thus identified can be classified into five groups, according to their effects on the final protein coding frame, as follows:
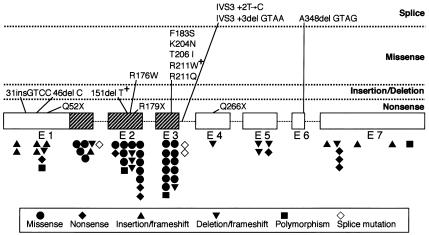
Figure 1.
Schematic representation of RUNX2, showing the localization of mutations in patients with CCD. The seven exons of RUNX2 are represented by boxes, and introns are represented by lines; hatched boxes indicate the Runt domain. The mutations identified in the present study and previous studies are indicated above and below the diagrammatic structure of RUNX2 (the 507-amino-acid isoform starting with MRIPVD).
Missense mutations.—Six missense mutations were detected in seven patients. Of these mutations, two (612A→T [K204N] and 617C→T [T206I]) were novel, and the other four (526C→T [R176W], 548T→C [F183S], 631C→T [R211W], and 632G→A [R211Q]) have been described previously. Notably, R211W was shared by two unrelated de novo patients in the present study. All missense mutations were located within the Runt domain, reconfirming the general trend observed previously (Quack et al. 1999; Zhou et al. 1999).
Nonsense mutations.—Nonsense mutations were detected in three patients. Of these mutations, two (154C→T [Q52X] and 793C→T [Q266X]) were novel, and the third (535C→T [R179X]) has been reported previously (Quack et al. 1999).
Frameshift mutations.—Four frameshift mutations were identified in five patients. Of these mutations, three were simply due to single-base deletions (136delC [46fs] and 453delT [151fs]) and a four-base insertion (92insCGGT [31fs]), respectively; notably, 453delT recurred in two unrelated de novo patients. The fourth mutation was caused by a single-base substitution, 1043C→T, which was initially supposed to cause a missense mutation, A348V, and was described as such in the recent review by Otto et al. (2002). However, this mutation simultaneously generated a new alternative splice-donor site within exon 6 at the position 4 bases upstream from the normal donor site (fig. 2a). RT-PCR analysis revealed that this new donor site was predominantly used in place of the normal one, resulting in a frameshift after Q347.
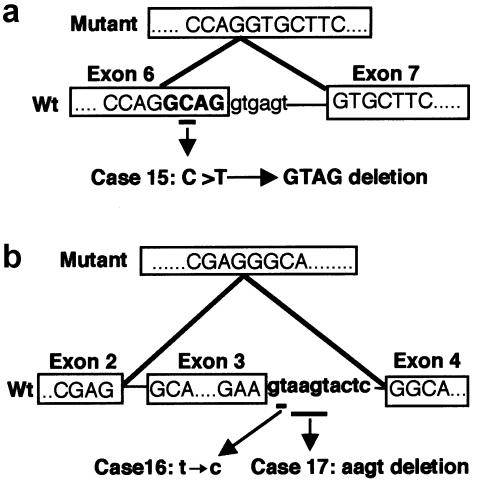
Figure 2.
Sequence analysis of splicing mutations. A, Point mutation with eventual 4-base skipping in patient 15. The boundary sequences of exons 6 and 7 in the wild-type cDNA are shown in the lower boxes. The mutated nucleotide is underlined and annotated as indicated by arrows. The resultant cDNA sequence causing frameshift is shown in the upper box. B, Exon-skipping mutations found in patients 16 and 17. The boundary sequences of exons 2–4 in the wild-type cDNA are shown in the lower boxes. The mutated nucleotide(s) at the upstream boundary of intron 3 are underlined and annotated as indicated by arrows. The resultant cDNA sequence lacking exon 3 is shown in the upper box. RT-PCR was performed as described elsewhere (Zhang et al. 2000). Wt = wild type.
Exon-skipping mutations.—Two mutations were detected on the boundary between exon 3 and intron 3 (fig. 2b). In one of them, a 4-nt deletion (IVS3+3delAAGT) had taken place within the splice-donor signal at the boundary between exon 3 and intron 3 (AAGTAAGT), resulting in a less optimal donor site in which the canonical GT motif was followed by an unfavorable pyrimidine-rich segment (AAGTACTC). RT-PCR analysis on RNA from the patient revealed that this mutation ultimately led to an in-frame skipping of exon 3 as a whole (amino acids 190–214), rather than to a simple inactivation of that donor site alone. The other mutation, IVS3+2T→C, directly hit the invariant GT signal at the same splice donor site. Although RT-PCR analysis could not be performed in this case, the mutation is also supposed to result in skipping of exon 3. In fact, an analogous mutation of GT→TT in RUNX2 has been reported to elicit the skipping of exon 3 for two independent cases of CCD (Zhou et al. 1999; Zhang et al. 2000).
Polymorphism.—One novel polymorphism, 510T→C (D170, synonymous), was found within exon 2 on one allele in one patient, whose healthy mother carried the same mutation on both alleles. Incidentally, this patient also possessed one of the aforementioned nonsense mutations, Q266X, which was supposed to be responsible for CCD.
Mutational Effects on Runt Domain Function
To examine mutational effects on Runt domain function, we overproduced partial RUNX2 proteins (amino acids 74–273 for the wild type and 8 mutants) in E. coli and subjected them to EMSA (fig. 3a). In this assay, the DNA-binding and heterodimerization activities can readily be detected by shifting and supershifting of the DNA band in the absence and presence, respectively, of PEBP2β, as typically seen with the wild type (fig. 3a, Wt − and +, respectively). Of the eight mutants tested (151fs, R179X, R176W, K204N, T206I, R211W, and R211Q), all but F183S showed no DNA binding, regardless of the presence or absence of PEBP2β. F183S gave a barely visible supershifted band in a PEBP2β-dependent manner, suggesting that it retained the heterodimerization activity together with a trace potential for DNA binding. The six missense mutants were further tested for the heterodimerization ability by an affinity assay (fig. 3b). They were all capable of binding to PEBP2β as proficiently as the wild type. Thus, the previous supposition that F183S would induce a global change in protein conformation (Bravo et al. 2001; Nagata and Werner 2001; Tahirov et al. 2001; Otto et al. 2002) has to be amended.
Figure 3.
Mutational alterations in DNA-binding and heterodimerization activities of the Runt domain. β = PEBP2β; Wt = wild type. a, Partial RUNX2 proteins (indicated at top) produced in E. coli and subjected to EMSA. The “+” and “−” symbols signify the presence and absence, respectively, of PEBP2β. b, Partial RUNX2 proteins subjected to affinity assay. A = input RUNX2 protein; W = unbound proteins in the third wash; E = bound proteins eluted at 250 mmol/liter imidazole.
Transcription Activation Abilities of the RUNX2 Mutants
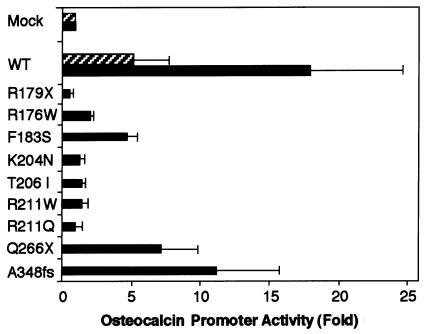
We measured the transactivation potential of the mutant RUNX2 proteins, using a reporter construct based on the rat osteocalcin promoter, which has been well characterized as an osteoblast-specific target of RUNX2 (Geoffroy et al. 1995; Merriman et al. 1995; Zhang et al. 2000). With NIH3T3 cells as host, the transfection of the wild-type RUNX2 at a near-optimal dose together with a saturating amount of PEBP2β effected a strong transactivation up to ∼20-fold over the mock-transfected control. Note here that a much weaker transcriptional stimulation was observed without coexpression of PEBP2β (fig. 4, cf. shaded bar vs. solid bar marked “WT”). This indicates that endogenous PEBP2β in NIH3T3 is limiting for the RUNX2-dependent transcription and that the coexpressed RUNX2 and PEBP2β can intracellularly interact with each other in a productive manner. Of the mutants tested, those impaired in the Runt domain displayed either no transactivation activity or markedly reduced transactivation activities, showing good parallelism with their impaired DNA binding. Notable in particular is F183S, which retained a low but still substantial activity, as though reflecting its cryptic DNA-binding potential noted above. In contrast, Q266X and 348fs, the two mutants that have the intact Runt domain and part of the C-terminal region, showed even higher residual transactivation abilities that amount to nearly half the normal level (fig. 4).
Figure 4.
Transactivation of the osteocalcin promoter by exogenously expressed RUNX2 proteins in NIH3T3 cells. Cells were transfected with a reporter plasmid (0.5 μg), indicated RUNX2 expression constructs (0.5 μg), and PEBP2β (0.2 μg). Luciferase activities were measured and presented as the fold increase relative to the control mock-transfected with the backbone expression vector. The mean values from three separate measurements are given with SDs (thin vertical bars). Results obtained with and without coexpression of PEBP2β are denoted by solid and shaded bars, respectively. WT = wild type.
Subcellular Localization of the RUNX2 Mutants
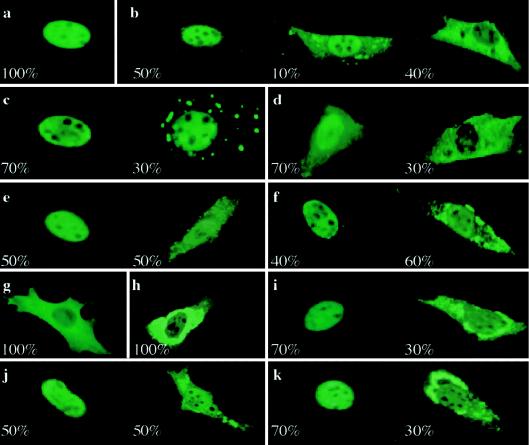
RUNX proteins, in general, have an NLS at the C-terminal border of the Runt domain (amino acids 204–220 in RUNX2 [KVTVDGPREPRREHRQKL]) (T. Kanno et al. 1998). They also promote nuclear cotranslocation of PEBP2β, which has no NLS element and tends to show cytoplasmic localization on its own (Lu et al. 1995). For the examination of whether the mutant RUNX2 proteins were affected in these nuclear-localization functions, they were overexpressed in NIH3T3 cells and were visualized by immunostaining. So far, 10 mutants have been tested, with the exception of those that either terminate before the Runt domain or involve exon skipping, which were obviously supposed to lack the nuclear-localization potential. The wild-type RUNX2 showed a clear nuclear-localization pattern with exclusion from the nucleoli in virtually every cell stained significantly (fig. 5a). In contrast, the RUNX2 mutants all exhibited altered subcellular localization with varying degrees and patterns regardless of the presence or absence of the intact NLS. Most mutants (R176W, F183S, T206I, R211Q, R211W, 151fs, Q266X, and 348fs) showed nonuniform distributions: nuclear localization in 30%–50% of the observed cells and dual localization to both the cytoplasm and the nucleus in the remaining 50%–70% (figs. 5b–5f and 5i–5k). Among the mutants impaired in the NLS, only K204N and R179X were completely localized in the cytoplasm (figs. 5g and 5h). Overall, these results are consistent with the previous proposal that the nuclear accumulation of the RUNX2 protein depends on the interplay between multiple nuclear-localization elements spread over the Runt domain and the C-terminal region (Lu et al. 1995; for more details, see the “Discussion” section). Furthermore, the mutants that show cytoplasmic localization produced speckled staining patterns in the cytoplasm in virtually all cases, except that the two nonsense mutants (R179X and Q266X) were stained diffusely in the cytoplasm (figs. 5d and 5g).
Figure 5.
Subcellular localization of RUNX2. The indicated RUNX2 proteins were overexpressed in NIH3T3 cells and were made visible by immunofluorescence staining with an anti-PEBP2αA antibody: wild type (a), R176W (b), F183S (c), Q266X (d), 348fs (e), 151fs (f), R179X (g), K204N (h), T206I (i), R211Q (j), and R211W (k). For each mutant, at least 100 well-stained cells were counted and were classified into a few subpopulations according to their pattern of staining. A typical image of each distinct subpopulation is presented together with its fractional percentage.
For the further examination of mutational effects on the subcellular localization of PEBP2β, RUNX2 and PEBP2β were coexpressed in NIH3T3 cells and were subjected to double immunofluorescent staining with antibodies specific to the respective proteins. In the control experiment, the wild-type RUNX2 and PEBP2β showed quite different distribution patterns. Whereas RUNX2 was almost completely localized to the nucleus, PEBP2β was distributed in both the cytoplasm and the nucleus, although it was visibly more abundant in the former than in the latter (figs. 6a and 6c). This reconfirmed the earlier observations that the wild-type RUNX2 and also RUNX1 displayed only a limited capacity to colocalize PEBP2β, at least under the coexpression regimen employed (Lu et al. 1995; Y. Kanno et al. 1998; Tanaka et al. 1998). In contrast, T206I and PEBP2β were colocalized in both the cytoplasm and the nucleus, showing almost identical patterns as characterized by bright cytoplasmic speckles and nucleoli-like dark spots that punctuated low diffuse backgrounds (figs. 6b and 6d). This speckled pattern is very similar to that observed when T206I was expressed alone. Thus, the mutant RUNX2 protein accumulated in the aberrant cytoplasmic speckles must retain a high heterodimerization activity and hence could avidly sequester PEBP2β therein.
Figure 6.
Colocalization of RUNX2 and PEBP2β. PEBP2β was coexpressed with either the wild-type RUNX2 (a and c) or T206I RUNX2 (b and d) in NIH3T3 cells, and the two subunits were visualized by double staining with anti-RUNX and anti-β antibodies, respectively. a, Wild-type RUNX2. b, T206I RUNX2. c, PEBP2β coexpressed with wild-type RUNX2. d, PEBP2β coexpressed with T206I RUNX2.
Genotype-Phenotype Correlation
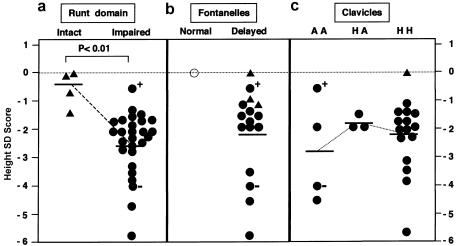
Taking into account the result of transactivation analysis, we compared the various phenotypes of CCD between two groups of patients that were clustered according to whether the Runt domain was or was not mutationally impaired. A significant difference was found for the height SD score, whose mean value was much lower in the impaired group of 27 patients (−2.56) than in the intact group of 4 patients (−0.55) (fig. 7a). As an apparent deviation from this correlation, one patient (151fs with 47,XXX [●+ in fig. 7]) in the Runt domain–impaired group showed a nearly normal physical growth (−0.5), whereas the other female patient with the same mutation having a normal karyotype (46,XX [●− in fig. 7]) had a very severe short stature (−4.04). However, it is known that the standard height of female patients with the 47,XXX karyotype is higher than that of the unaffected control individual, owing to the presence of SHOX (short stature homeobox-containing gene) on the X chromosome (Ogata et al. 2001). Thus, it is possible that an overdosage of SHOX masked the growth-retarding effect due to mutation 151fs in this particular patient (fig. 7a).
Figure 7.
Comparison of height SD scores between groups of patients with different genotypes or phenotypic variations. ○ = Void; ● = impaired in the Runt domain; ▴ = intact in the Runt domain; ●+ = 151fs with 47,XXX; ●− = 151fs with 46,XX. a, Mutational effects on the Runt domain, impaired (n=27) and intact (n=4) (P<.01). b, Fontanelles, normal (none) and delayed (n=16). c, Clavicles. AA = aplastic on both sides (n=4); AH = aplastic on one side and hyopoplastic on the other side (n=3); HH = hypoplastic on both sides (n=16).
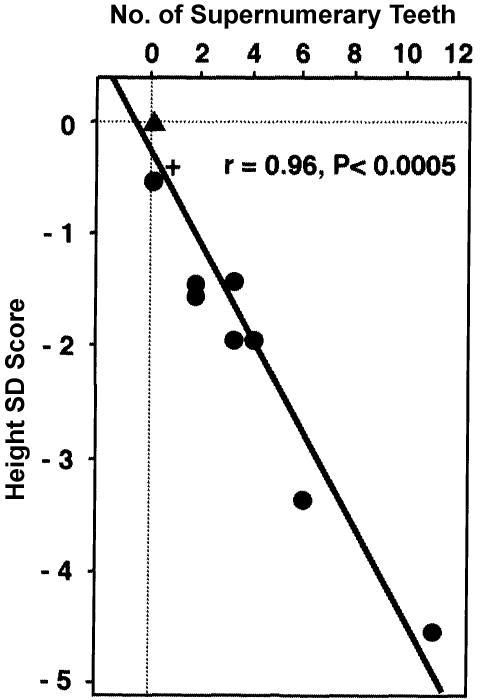
Prompted by the preceding result, we further compared the height SD score with other functional and phenotypic parameters. No recognizable correlation was observed with the classic CCD phenotype, cranial and clavicular anomalies (figs. 7b and 7c). However, a remarkable correlation was found between the height SD score and the transactivation potential of RUNX2 mutants (fig. 8). The regression curve suggests that the skeletal growth begins to deteriorate when the RUNX2 activity from one allele goes down just by one-half and undergoes accelerated worsening with its further loss. In addition, another intriguing correlation was demonstrated between the height SD score and the number of supernumerary teeth (fig. 9). Note that all the data points fit a linear regression curve with surprisingly small deviations. This implies an inherent mechanistic commonality between the skeletal growth and the dental development in their dependencies on the RUNX2 activity, despite the tremendous apparent dissimilarities of these two organogenic processes.
Figure 8.
Correlation between height SD score and transactivation ability of each RUNX2 mutant (% wild-type control), as determined in figure 4 (n=16; r=0.71; P<.01). Symbols are as defined in figure 7.
Figure 9.
Correlation between number of supernumerary teeth and height SD score (n=9; r=0.96; P<.0005). Symbols are as defined in figure 7.
Discussion
In the present study, we have identified 15 nonpolymorphic mutations in 17 unrelated families with CCD in Japan. Of these mutations, 10 are novel, and 5 have previously been reported for multiple cases. Detailed functional analyses of these mutations have provided novel insights into various critical issues, ranging from the structure-function relationships of RUNX2 to the genotype-phenotype correlation of CCD.
Among the mutations identified, missense mutations were exclusively clustered within the Runt domain, reconfirming previous reports (Quack et al. 1999; Zhou et al. 1999; Zhang et al. 2000). In contrast, other types of mutations that cause premature terminations or internal deletions/insertions were widely scattered over the entire protein sequence. This extreme bias in the distribution of missense mutations is taken to indicate that the function of the Runt domain with its highly conserved sequence is very susceptible to single-amino-acid changes, whereas the less-well-conserved N- and C-terminal regions are functionally more robust and require larger structural changes for their pathogenically significant alterations. Indeed, the Runt domain that carries each missense mutation was severely impaired in DNA binding and hence also in transactivation. The observed mutational effects on Runt domain function are consistent with and further serve to complement the information provided by recent nuclear-magnetic-resonance and x-ray crystallographic analyses of the Runt domain (Bravo et al. 2001; Nagata and Werner 2001; Tahirov et al. 2001). In contrast, the two nonsense or frameshift mutations located within the C-terminal region retained substantial transactivation potentials.
When the mutations identified in both the present study and previous studies are compared, several positions emerge as mutational hotspots (fig. 1) (Otto et al. 2002). Of these, most conspicuous are the four arginine residues: R211 (eight times), R176 and R179 (five times each), and R377 (three times). Coincidentally, these arginine residues are all encoded by a CGA or CGG codon, which should be particularly prone to mutagenic events through a CpG-directed methylation of C followed by deamination (Quack et al. 1999). Consistent with this supposition, no mutation has ever been identified for an AGA-encoded arginine, R214, even though this residue is directly involved in DNA binding, as is R211. Moreover, a converse situation occurs in RUNX1, in which recurrent mutations have been reported for the R214-equivalent residue encoded by CGA, whereas no mutation has been detected at the R179-equivalent residue encoded by AGA (Osato et al. 1999; Song et al. 1999; Preudhomme et al. 2000). Taken together, these observations suggest that the mutation rate of arginine is definitely facilitated, plausibly by one order of magnitude (or more), when its codon contains CpG. A lesson from these statistical considerations is that mutational screening on RUNX2 is still far from saturation, except for the aforementioned hotspots, even though a total of nearly 80 cases have so far been identified. This warrants further systematic mutational screening, by which more novel mutations will be identified to enrich our insights into the molecular basis for pathogenesis of CCD.
The RUNX2 mutants identified in the present study also affected additional important functions—in particular, the abilities to mediate the nuclear localization of RUNX2 itself, as well as of the partner subunit, PEBP2β. This finding may merit keen attention, since it makes an interesting parallel to the recurrent theme of perturbed intracellular interplay between RUNX1 and PEBP2β as incurred by leukemia-associated chromosomal rearrangements involving the genes that encode the respective proteins (Lu et al. 1995; Adya et al. 1998; T. Kanno et al. 1998; Y. Kanno et al. 1998; Tanaka et al. 1998). Enigmatically, the mutants showed surprisingly complex responses regardless of the presence or absence of the intact NLS, causing variable localizations in either the nucleus or the cytoplasm, from cell to cell in most cases—with the exceptions of R179X and K204N, which displayed cytoplasmic localization in all cells. An additional strange feature found here is the occurrence of cytoplasmic speckles in substantial fractions of cells transfected with most RUNX2 mutants except the two nonsense mutants, R179X and Q266X. One may wonder whether these anomalies simply represent artifactual cellular responses to overproduction of mutant proteins. However, it should be emphasized that the wild-type RUNX2 never showed such aberrant distribution patterns, not even when their expression vector was transfected at oversaturating doses during our preliminary titration experiments (data not shown). Conversely, moreover, mutant RUNX2 invariably showed altered subcellular distributions in nearly constant fractions of visibly stained cells when they were expressed at much lower dosages than that used in the standard assay. Thus, we may reason that some intrinsic molecular or functional properties unique to mutant proteins should be responsible for their aberrant behaviors. Notable in this regard is the previous deletion analysis (Lu et al. 1995), which has suggested that Runx2 contains additional elements that promote nuclear localization as coarsely mapped to the N-proximal half of the Runt domain (positions 93–158) and the C-terminal region spanning amino acids 272–502, in addition to the best-characterized NLS on the C-terminal border of the Runt domain (T. Kanno et al. 1998). A mutational change in one or more of these elements would cause reduced nuclear-localization potential to some extent, but total inhibition would seldom result unless all of them should be physically destroyed or conformationally perturbed. An additional strange feature found in the above analysis is the occurrence of cytoplasmic speckles in substantial fractions of cells transfected with most RUNX2 mutants except the two nonsense mutants, R179X and Q266X. Furthermore, the double-staining analysis revealed that PEBP2β was colocalized with the T206I RUNX2—and probably other mutants as well—in those cytoplasmic speckles. Of interest, a very similar observation has also been made with missense mutants of RUNX1 that are associated with leukemias (Michaud et al. 2002). Thus, the mutation-induced cytoplasmic colocalization of RUNX proteins and PEBP2β could have a general pathogenic significance. One attractive possibility may be sequestration of PEBP2β away from the normal RUNX protein encoded by the single residual wild-type allele (Michaud et al. 2002). Since PEBP2β acts in the protection of RUNX proteins from ubiquitin-proteasome–mediated proteolytic degradation (Huang et al. 2001), its sequestration may lead to the more-enhanced reduction of the residual RUNX2 activity. Accordingly, RUNX mutants would have more-severe deleterious effects when they retain the heterodimerization activity than they otherwise would. This may be the reason why missense RUNX mutations predictably affecting heterodimerization have scarcely been detected thus far—with only three found in RUNX2 (Quack et al. 1999; Zhou et al. 1999; Otto et al. 2002) and none found in RUNX1. In fact, one such mutant, T176A (T200A in the original notation), was reported to have only mild effects on the transactivation potential and the CCD phenotype (Zhou et al. 1999).
The most crucial question remaining is how the various functional defects conferred by RUNX2 mutations would correlate with the CCD phenotype. On the one hand, the classic CCD phenotype, hypoplastic clavicles and open sutures, was observed in virtually all patients examined. On the other hand, skeletal and dental features, as represented by short stature and supernumerary teeth, respectively, showed significant differences, depending on whether the Runt domain was impaired (figs. 7a and 9) and, in more quantitative terms, on how much residual transactivation potential remained in mutant RUNX2 proteins (fig. 8). By a simple extrapolation of this correlation, the RUNX2 activity in heterozygously affected cells relative to the normal level would be reduced by approximately one-fourth in the Runt domain–intact group and by approximately one-half in the Runt domain–impaired group. This means that the skeletal growth and the dental development could readily and proportionally be affected by such fractional changes in the RUNX2 activity. By this line of reasoning, we suppose that the omnipresent cleidocranial phenotype must be even more sensitive to the RUNX2 deficiency. In other words, the cleidocranial bone formation, which is mediated by intramembranous ossification, may require a higher level of RUNX2 than does the skeletal bone formation as mediated by intrachondral ossification. Unlike the skeleto-dental phenotype, however, the cleidocranial phenotype was extremely variable in pattern and severity either between or within individual families, thereby thwarting attempts to find any further genotype-phenotype correlation. This discrepancy may reflect a possible difference between these two distinct osteogenic processes in their sensitivities to genetic backgrounds other than RUNX, or in fluctuations of developmental and physiological conditions among individuals. The importance of genetic backgrounds was exemplified for the case of skeletal growth by the observation that the short stature phenotype due to mutation 151fs was much milder in one female patient with a 47,XXX karyotype than in the corresponding patient with a normal karyotype.
To make an apparent contrast to human patients with CCD, mice carrying one Runx2 null allele have been reported to develop to normal size and to have no supernumerary teeth, although they recapitulate all other aspects of CCD (Sillence et al. 1987; Otto et al. 1997). On the one hand, since supernumerary teeth are specific to permanent teeth in humans, it is no surprise that mice, which possess solely primary teeth, lack this symptom. On the other hand, the lack of short stature has thus far been regarded to suggest that the skeletal growth in mice could generally be more resistant to Runx2 deficiency than that in humans. Analogous to the taller female patient with mutation 151fs, however, an alternative culprit may be lurking somewhere in the genetic background of the Ccd model mice—C57- or C3H-related lineages have only been employed thus far (Sillence et al. 1987; Otto et al. 1997). Indeed, Choi et al. (2001) have recently described a suggestive observation that male mice (C57BL/6) heterozygous for Runx2 with a C-terminal deletion exhibit a significantly lower weight and weight gain than the wild type, whereas the corresponding female heterozygotes show normal growth. Most recently, transgenic mice made conditionally or partially deficient for PEBP2β/Cbfb have emerged as a new Ccd model that is severely defective in bone formation for both cleidocranial and skeletal features (Yoshida et al., in press).
The genotype-phenotype correlation found in the present study appears to be somewhat different from what has been observed in two previous large-scale mutational studies (Quack et al. 1999; Zhou et al. 1999). Zhou et al. (1999) examined 26 sporadic and familial cases of CCD recruited from the United States, Canada, and Europe, and found that most mutations resulting in premature termination in the Runt domain produced a classic CCD phenotype, whereas three mutations (R377X, T176A, and 48insC in our notation) with considerable residual transactivation showed a nonuniform clinical spectrum including classic and mild CCD, as well as an isolated dental phenotype characterized by delayed eruption of permanent teeth. On the other hand, Quack et al. (1999) screened 42 unrelated families with CCD of different ethnic backgrounds recruited from Europe and other continents but were unable to find a significant genotype-phenotype correlation. In these families, clavicular involvement was almost invariably present, but the number and presence of supernumerary teeth was highly variable and did not correlate with either the rest of the phenotype or the type of mutation. In both of these studies, functional analysis of the mutations was done on only several cases in limited aspects, and no data were presented for height and the number of supernumerary teeth. Thus, it is rather difficult to compare these previous results with ours, but the apparent discrepancies between the results from the three groups could be due to differences in either the exact diagnostic criteria used or the genetic backgrounds among various ethnic groups studied.
The consistent quantitative correlation found between the number of supernumerary teeth and short stature may have important implications in prognosis and treatment of CCD, although its generality remains to be established by further studies. It has been proposed that supernumerary teeth in patients with CCD should be diagnosed and removed as early as possible, because the supernumerary teeth will always impede normal eruption of the permanent teeth (Jensen and Kreiborg 1992). If the type of mutation in RUNX2 is known in advance, we could predict the supernumerary teeth for an individual, to allow the early initiation of the necessary treatment. Furthermore, we could predict the final height of patients from the number of supernumerary teeth, to judge the necessity of prospective intervention, such as a recombinant human-growth-hormone (hGH) therapy, for improvement of their height. However, there has been controversy over whether the hGH therapy really improves growth outcomes in short non–GH-deficient children, because of its prolonged and expensive treatments (Kawai et al. 1997; Kaplowitz 2001). One major difficulty in the resolution of this issue may be the lack of knowledge on underlying genetic factors in most cases. Under such circumstances, it would be tempting and justifiable to perform prospective studies on whether the hGH therapy is beneficial for patients with CCD, particularly when they have many supernumerary teeth.
In the present study, no mutation was detected in seven families. Now that CCD has been shown potentially to result from any subtle deficiency in RUNX2, it would be interesting and important in such cases to extend mutational screening to nonexon and regulatory regions of the RUNX2 gene. In addition, the possibility also remains that certain other factors known to cooperate with RUNX2 during bone formation could be potential targets of mutation in these patients with CCD. In light of the aforementioned gene-targeting studies in mice, PEBP2β in particular should be worth looking at. Further explorations are awaited to address all these possibilities.
Acknowledgments
We thank the many patients and their families for their cooperation. We are grateful to Drs. N. Okamoto (Osaka Medical Center and Research Institute for Maternal and Child Health), T. Momoi (Japanese Red Cross Society Wakayama Medical Center), H. Ohashi and H. Motizuki (Saitama Children's Medical Center), Y. Ito and Y. Makita (Asahikawa medical college), T. Nagai (Dokkyo Medical Koshigaya School), K. Yano (Nayoro City Hospital), Y. Tanaka (Tokyo Dental University Ichikawa Hospital), S. Nishimaki (Yokohama City University School of Medicine), H. Yamakawa (Yokohama Municipal Citizen's Hospital), S. Teramoto (Fuji City Central Hospital), S. Tamai (Yamato City Hospital), Y. Nishi (Hiroshima Red Cross and Atomic-Bomb Survivors Hospital), N. Yasui (Osaka University Medical School), N. Natsume (Aichi-Gakuin University), K. Takahashi (Kyoto University), K. Kouzai (Hiroshima University), T. Fujiwaki (Matsue Red Cross Hospital), K. Sueishi (Suidobashi Hospital), and Y. Adachi (Toyama Medical and Pharmaceutical University) for providing clinical data and blood samples from patients with CCD. We are deeply indebted to Dr. Yue Wang (Toyama Medical and Pharmaceutical University) for technical assistance and Drs. T. Momoi and N. Okamoto for critical discussion. We also thank Drs. T. Komori (Osaka University), P. P. Liu (National Human Genome Institute), and N. A. Speck (Dartmouth University) for sharing their valuable observations on Cbfb transgenic mice before publication. This work was supported, in part, by Grants-in-Aid 12213064 and 13214052 for Priority Areas in Cancer Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan (to K.S.).
Electronic-Database Information
Accession numbers and URLs for data presented herein are as follows:
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for RUNX2 [accession number AF001450])
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for CCD [MIM 119600])
References
- Adya N, Stacy T, Speck NA, Liu PP (1998) The leukemic protein core binding factor β (CBFβ)-smooth-muscle myosin heavy chain sequesters CBFα2 into cytoskeletal filaments and aggregates. Mol Cell Biol 18:7432–7443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae SC, Takahashi E, Zhang YW, Ogawa E, Shigesada K, Namba Y, Satake M, Ito Y (1995) Cloning, mapping and expression of PEBP2αC, a third gene encoding the mammalian Runt domain. Gene 159:245–248 [DOI] [PubMed] [Google Scholar]
- Bae SC, Yamaguchi-Iwai Y, Ogawa E, Maruyama M, Inuzuka M, Kagoshima H, Shigesada K, Satake M, Ito Y (1993) Isolation of PEBP2αB cDNA representing the mouse homolog of human acute myeloid leukemia gene, AML1. Oncogene 8:809–814 [PubMed] [Google Scholar]
- Bravo J, Li Z, Speck NA, Warren AJ (2001) The leukemia-associated AML1 (Runx1)–CBFβ complex functions as a DNA-induced molecular clamp. Nat Struct Biol 8:371–378 [DOI] [PubMed] [Google Scholar]
- Chitayat D, Hodgkinson KA, Azouz EM (1992) Intrafamilial variability in cleidocranial dysplasia: a three generation family. Am J Med Genet 42:298–303 [DOI] [PubMed] [Google Scholar]
- Choi JY, Pratap J, Javed A, Zaidi SK, Xing L, Balint E, Dalamangas S, Boyce B, van Wijnen AJ, Lian JB, Stein JL, Jones SN, Stein G (2001) Subnuclear targeting of Runx/Cbfα/AML factors is essential for tissue-specific differentiation during embryonic development. Proc Natl Acad Sci USA 98:8650–8655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daga A, Karlovich CA, Dumstrei K, Banerjee U (1996) Patterning of cells in the Drosophila eye by Lozenge, which shares homologous domains with AML1. Genes Dev 10:1194–1205 [DOI] [PubMed] [Google Scholar]
- Ducy P, Zhang R, Geoffroy V, Ridall AL, Karsenty G (1997) Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation. Cell 89:747–754 [DOI] [PubMed] [Google Scholar]
- Duffy JB, Gergen JP (1991) The Drosophila segmentation gene runt acts as a position-specific numerator element necessary for the uniform expression of the sex-determining gene Sex-lethal. Genes Dev 5:2176–2187 [DOI] [PubMed] [Google Scholar]
- Geoffroy V, Ducy P, Karsenty G (1995) A PEBP2α/AML-1-related factor increases osteocalcin promoter activity through its binding to an osteoblast-specific cis-acting element. J Biol Chem 270:30973–30979 [DOI] [PubMed] [Google Scholar]
- Golan I, Preising M, Wagener H, Baumert U, Niederdellmann H, Lorenz B, Mussig D (2000) A novel missense mutation of the CBFA1 gene in a family with cleidocranial dysplasia (CCD) and variable expressivity. J Craniofac Genet Dev Biol 20:113–120 [PubMed] [Google Scholar]
- Huang G, Shigesada K, Ito K, Wee HJ, Yokomizo T, Ito Y (2001) Dimerization with PEBP2β protects RUNX1/AML1 from ubiquitin-proteasome-mediated degradation. EMBO J 20:723–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis JL, Keats TE (1974) Cleidocranial dysostosis: a review of 40 new cases. Am J Roentgenol Radium Ther Nucl Med 121:5–16 [DOI] [PubMed] [Google Scholar]
- Javed A, Gutierrez S, Montecino M, van Wijnen AJ, Stein JL, Stein GS, Lian JB (1999) Multiple Cbfa/AML sites in the rat osteocalcin promoter are required for basal and vitamin D-responsive transcription and contribute to chromatin organization. Mol Cell Biol 19:7491–7500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen BL (1990) Somatic development in cleidocranial dysplasia. Am J Med Genet 35:69–74 [DOI] [PubMed] [Google Scholar]
- Jensen BL, Kreiborg S (1992) Dental treatment strategies in cleidocranial dysplasia. Br Dent J 172:243–247 [DOI] [PubMed] [Google Scholar]
- Kagoshima H, Akamatsu Y, Ito Y, Shigesada K (1996) Functional dissection of the α and β subunits of transcription factor PEBP2 and the redox susceptibility of its DNA binding activity. J Biol Chem 271:33074–33082 [DOI] [PubMed] [Google Scholar]
- Kagoshima H, Shigesada K, Satake M, Ito Y, Miyoshi H, Ohki M, Pepling M, Gergen P (1993) The Runt domain identifies a new family of heteromeric transcriptional regulators. Trends Genet 9:338–341 [DOI] [PubMed] [Google Scholar]
- Kamachi Y, Ogawa E, Asano M, Ishida S, Murakami Y, Satake M, Ito Y, Shigesada K (1990) Purification of a mouse nuclear factor that binds to both the A and B cores of the polyomavirus enhancer. J Virol 64:4808–4819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kania MA, Bonner AS, Duffy JB, Gergen JP (1990) The Drosophila segmentation gene runt encodes a novel nuclear regulatory protein that is also expressed in the developing nervous system. Genes Dev 4:1701–1713 [DOI] [PubMed] [Google Scholar]
- Kanno T, Kanno Y, Chen LF, Ogawa E, Kim WY, Ito Y (1998) Intrinsic transcriptional activation-inhibition domains of the polyomavirus enhancer binding protein 2/core binding factor α subunit revealed in the presence of the β subunit. Mol Cell Biol 18:2444–2454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno Y, Kanno T, Sakakura C, Bae SC, Ito Y (1998) Cytoplasmic sequestration of the polyomavirus enhancer binding protein 2 (PEBP2)/core binding factor α (CBFα) subunit by the leukemia-related PEBP2/CBFβ-SMMHC fusion protein inhibits PEBP2/CBF-mediated transactivation. Mol Cell Biol 18:4252–4261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplowitz PB (2001) If gonadotropin-releasing hormone plus growth hormone (GH) really improves growth outcomes in short non-GH-deficient children, then what? J Clin Endocrinol Metab 86:2965–2963 [DOI] [PubMed] [Google Scholar]
- Kawai M, Momoi T, Yorifuji T, Yamanaka C, Sasaki H, Furusho K (1997) Unfavorable effects of growth hormone therapy on the final height of boys with short stature not caused by growth hormone deficiency. J Pediatr 130:205–209 [DOI] [PubMed] [Google Scholar]
- Kim WY, Sieweke M, Ogawa E, Wee HJ, Englmeier U, Graf T, Ito Y (1999) Mutual activation of Ets-1 and AML1 DNA binding by direct interaction of their autoinhibitory domains. EMBO J 18:1609–1620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komori T, Yagi H, Nomura S, Yamaguchi A, Sasaki K, Deguchi K, Shimizu Y, Bronson RT, Gao YH, Inada M, Sato M, Okamoto R, Kitamura Y, Yoshiki S, Kishimoto T (1997) Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell 89:755–764 [DOI] [PubMed] [Google Scholar]
- Lee B, Thirunavukkarasu K, Zhou L, Pastore L, Baldini A, Hecht J, Geoffroy V, Ducy P, Karsenty G (1997) Missense mutations abolishing DNA binding of the osteoblast-specific transcription factor OSF2/CBFA1 in cleidocranial dysplasia. Nat Genet 16:307–310 [DOI] [PubMed] [Google Scholar]
- Levanon D, Negreanu V, Bernstein Y, Bar-Am I, Avivi L, Groner Y (1994) AML1, AML2, and AML3, the human members of the Runt domain gene-family: cDNA structure, expression, and chromosomal localization. Genomics 23:425–432 [DOI] [PubMed] [Google Scholar]
- Li QL, Ito K, Sakakura C, Fukamachi H, Inoue K, Chi XZ, Lee KY, et al (2002) Causal relationship between the loss of RUNX3 expression and gastric cancer. Cell 109:113–124 [DOI] [PubMed] [Google Scholar]
- Lu J, Maruyama M, Satake M, Bae SC, Ogawa E, Kagoshima H, Shigesada K, Ito Y (1995) Subcellular localization of the α and β subunits of the acute myeloid leukemia-linked transcription factor PEBP2/CBF. Mol Cell Biol 15:1651–1661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merriman HL, van Wijnen AJ, Hiebert S, Bidwell JP, Fey E, Lian J, Stein J, Stein GS (1995) The tissue-specific nuclear matrix protein, NMP-2, is a member of the AML/CBF/PEBP2/Runt domain transcription factor family: interactions with the osteocalcin gene promoter. Biochemistry 34:13125–13132 [DOI] [PubMed] [Google Scholar]
- Michaud J, Wu F, Osato M, Cottles GM, Yanagida M, Asou N, Shigesada K, Ito Y, Benson KF, Raskind WH, Rossier C, Antonarakis SE, Israels S, McNicol A, Weiss H, Horwitz M, Scott HS (2002) In vitro analyses of known and novel RUNX1/AML1 mutations in dominant familial platelet disorder with predisposition to acute myelogenous leukemia: implications for mechanisms of pathogenesis. Blood 99:1364–1372 [DOI] [PubMed] [Google Scholar]
- Miyoshi H, Shimizu K, Kozu T, Maseki N, Kaneko Y, Ohki M (1991) t(8;21) breakpoints on chromosome 21 in acute myeloid leukemia are clustered within a limited region of a single gene, AML1. Proc Natl Acad Sci USA 88:10431–10434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mundlos S (1999) Cleidocranial dysplasia: clinical and molecular genetics. J Med Genet 36:177–182 [PMC free article] [PubMed] [Google Scholar]
- Mundlos S, Otto F, Mundlos C, Mulliken JB, Aylsworth AS, Albright S, Lindhout D, Cole WG, Henn W, Knoll JH, Owen MJ, Mertelsmann R, Zabel BU, Olsen BR (1997) Mutations involving the transcription factor CBFA1 cause cleidocranial dysplasia. Cell 89:773–779 [DOI] [PubMed] [Google Scholar]
- Nagata T, Gupta V, Sorce D, Kim WY, Sali A, Chait BT, Shigesada K, Ito Y, Werner MH (1999) Immunoglobulin motif DNA recognition and heterodimerization of the PEBP2/CBF Runt domain. Nat Struct Biol 6:615–619 [DOI] [PubMed] [Google Scholar]
- Nagata T, Werner MH (2001) Functional mutagenesis of AML1/RUNX1 and PEBP2β/CBFβ define distinct, non-overlapping sites for DNA recognition and heterodimerization by the Runt domain. J Mol Biol 308:191–203 [DOI] [PubMed] [Google Scholar]
- Ogata T, Matsuo N, Nishimura G (2001) SHOX haploinsufficiency and overdosage: impact of gonadal function status. J Med Genet 38:1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa E, Inuzuka M, Maruyama M, Satake M, Naito-Fujimoto M, Ito Y, Shigesada K (1993a) Molecular cloning and characterization of PEBP2β, the heterodimeric partner of a novel Drosophila runt-related DNA binding protein PEBP2α. Virology 194:314–331 [DOI] [PubMed] [Google Scholar]
- Ogawa E, Maruyama M, Kagoshima H, Inuzuka M, Lu J, Satake M, Shigesada K, Ito Y (1993b) PEBP2/PEA2 represents a family of transcription factors homologous to the products of the Drosophila runt gene and the human AML1 gene. Proc Natl Acad Sci USA 90:6859–6863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuda T, van Deursen J, Hiebert SW, Grosveld G, Dowing JR (1996) AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell 84:321–330 [DOI] [PubMed] [Google Scholar]
- Osato M, Asou N, Abdalla E, Hoshino K, Yamasaki H, Okubo T, Suzushima H, Takatsuki K, Kanno T, Shigesada K, Ito Y (1999) Biallelic and heterozygous point mutations in the Runt domain of the AML1/PEBP2αB gene associated with myeloblastic leukemias. Blood 93:1817–1824 [PubMed] [Google Scholar]
- Otto F, Thornell AP, Crompton T, Denzel A, Gilmour KC, Rosewell IR, Stamp GW, Beddington RS, Mundlos S, Olsen BR, Selby PB, Owen MJ (1997) Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell 89:765–771 [DOI] [PubMed] [Google Scholar]
- Otto F, Kanegane H, Mundlos S (2002) Mutations in the RUNX2 gene in patients with cleidocranial dysplasia. Hum Mutat 19:209–216 [DOI] [PubMed] [Google Scholar]
- Preudhomme C, Warot-Loze D, Roumier C, Grardel-Duflos N, Garand R, Lai JL, Dastugue N, Macintyre E, Denis C, Bauters F, Kerckaert JP, Cosson A, Fenaux P (2000) High incidence of biallelic point mutations in the Runt domain of the AML1/PEBP2αB gene in Mo acute myeloid leukemia and in myeloid malignancies with acquired trisomy 21. Blood 96:2862–2869 [PubMed] [Google Scholar]
- Quack I, Vonderstrass B, Stock M, Aylsworth AS, Becker A, Brueton L, Lee PJ, Majewski F, Mulliken JB, Suri M, Zenker M, Mundlos S, Otto F (1999) Mutation analysis of core binding factor A1 in patients with cleidocranial dysplasia. Am J Hum Genet 65:1268–1278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki K, Yagi H, Bronson RT, Tominaga K, Matsunashi T, Deguchi K, Tani Y, Kishimoto T, Komori T (1996) Absence of fetal liver hematopoiesis in mice deficient in transcriptional coactivator core binding factor β. Proc Natl Acad Sci USA 93:12359–12363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sillence DO, Ritchie HE, Selby PB (1987) Animal model: skeletal anomalies in mice with cleidocranial dysplasia. Am J Med Genet 27:75–85 [DOI] [PubMed] [Google Scholar]
- Song WJ, Sullivan MG, Legare RD, Hutchings S, Tan X, Kufrin D, Ratajczak J, Resende IC, Haworth C, Hock R, Loh M, Felix C, Roy DC, Busque L, Kurnit D, Willman C, Gewirtz AM, Speck NA, Bushweller JH, Li FP, Gardiner K, Poncz M, Maris JM, Gilliland DG (1999) Haploinsufficiency of CBFA2 causes familial thrombocytopenia with propensity to develop acute myelogenous leukaemia. Nat Genet 23:166–175 [DOI] [PubMed] [Google Scholar]
- Tahirov TH, Inoue-Bungo T, Morii H, Fujikawa A, Sasaki M, Kimura K, Shiina M, Sato K, Kumasaka T, Yamamoto M, Ishii S, Ogata K (2001) Structural analyses of DNA recognition by the AML1/Runx-1 Runt domain and its allosteric control by CBFβ. Cell 104:755–767 [DOI] [PubMed] [Google Scholar]
- Tanaka K, Tanaka T, Kurokawa M, Imai Y, Ogawa S, Mitani K, Yazaki Y, Hirai H (1998) The AML1/ETO(MTG8) and AML1/Evi-1 leukemia-associated chimeric oncoproteins accumulate PEBP2β(CBFβ) in the nucleus more efficiently than wild-type AML1. Blood 91:1688–1699 [PubMed] [Google Scholar]
- Towler DA, Bennett CD, Rodan GA (1994) Activity of the rat osteocalcin basal promoter in osteoblastic cells is dependent upon homeodomain and CP1 binding motifs. Mol Endocrinol 8:614–624 [DOI] [PubMed] [Google Scholar]
- Wang Q, Stacy T, Binder M, Marin-Padilla M, Sharpe AH, Speck NA (1996a) Disruption of the cbfa2 gene causes necrosis and hemorrhaging in the central nervous system and blocks definitive hematopoiesis. Proc Natl Acad Sci USA 93:3444–3449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Stacy T, Miller D, Lewis AF, Gu TL, Huang X, Bushweller JH, Marin-Padilla M, Sharpe AH, Speck NA (1996b) The CBFβ subunit is essential for CBFα2 (AML1) function in vivo. Cell 87:697–708 [DOI] [PubMed] [Google Scholar]
- Wang S, Wang Q, Crute BE, Melnikova IN, Keller SR, Speck NA (1993) Cloning and characterization of subunits of the T-cell receptor and murine leukemia virus enhancer core-binding factor. Mol Cell Biol 13:3324–3339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren AJ, Bravo J, Williams RL, Rabbitts TH (2000) Structural basis for the heterodimeric interaction between the acute leukaemia-associated transcription factors AML1 and CBFβ. EMBO J 19:3004–3015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner MH, Shigesada K, Ito Y (1999) Runt domains take the lead in hematopoiesis and osteogenesis. Nat Med 5:1356–1357 [DOI] [PubMed] [Google Scholar]
- Wijmenga C, Speck NA, Dracopoli NC, Hofker MH, Liu P, Collins FS (1995) Identification of a new murine Runt domain-containing gene, Cbfa3, and localization of the human homolog, CBFA3, to chromosome 1p35-pter. Genomics 26:611–614 [DOI] [PubMed] [Google Scholar]
- Yamachika E, Tsujigiwa H, Ishiwari Y, Mizukawa N, Nagai N, Sugahara T (2001) Identification of a stop codon mutation in the CBFA1 Runt domain from a patient with cleidocranial dysplasia and cleft lip. J Oral Pathol Med 30:381–383 [DOI] [PubMed] [Google Scholar]
- Yokozeki M, Ohyama K, Tsuji M, Goseki-Sone M, Oida S, Orimo H, Moriyama K, Kuroda T (2000) A case of Japanese cleidocranial dysplasia with a CBFA1 frameshift mutation. J Craniofac Genet Dev Biol 20:121–126 [PubMed] [Google Scholar]
- Yoshida CA, Furuichi T, Fujita T, Fukuyama R, Kanatani N, Kobayashi S, Satake M, Takada K, Komori T. Core-binding factor β interacts with Runx2 and is required for skeletal development. Nat Genet (in press) [DOI] [PubMed] [Google Scholar]
- Zhang YW, Yasui N, Kakazu N, Abe T, Takada K, Imai S, Sato M, Nomura S, Ochi T, Okuzumi S, Nogami H, Nagai T, Ohashi H, Ito Y (2000) PEBP2αA/CBFA1 mutations in Japanese cleidocranial dysplasia patients. Gene 244:21–28 [DOI] [PubMed] [Google Scholar]
- Zhou G, Chen Y, Zhou L, Thirunavukkarasu K, Hecht J, Chitayat D, Gelb BD, Pirinen S, Berry SA, Greenberg CR, Karsenty G, Lee B (1999) CBFA1 mutation analysis and functional correlation with phenotypic variability in cleidocranial dysplasia. Hum Mol Genet 8:2311–2316 [DOI] [PubMed] [Google Scholar]