Summary
Among the millions of invertebrate species with visual systems, the genetic basis of eye development and function is well understood only in Drosophila melanogaster. We describe an eye transcriptome for the planarian Schmidtea mediterranea. Planarian photoreceptors expressed orthologs of genes required for phototransduction and microvillus structure in Drosophila and vertebrates, and optic pigment cells expressed solute transporters and melanin synthesis enzymes similar to those active in the vertebrate retinal pigment epithelium. Orthologs of several planarian eye genes, such as bestrophin-1 and Usher syndrome genes, cause eye defects in mammals when perturbed and were not previously described to have roles in invertebrate eyes. Five previously undescribed planarian eye transcription factors were required for normal eye formation during head regeneration. In particular, a conserved, transcription factor-encoding ovo gene was expressed from the earliest stages of eye regeneration and was required for regeneration of all cell types of the eye.
Introduction
The vertebrate eye is a structure of striking complexity in form and function. Consequently, understanding the evolution and development of eyes has been a classic challenge. The human eye is also subject to numerous pathologies that are poorly understood, such as heritable retinopathies and age-related degeneration. Genetic studies in model invertebrates have the potential to advance the understanding of eye evolution and development, and of the functions of conserved genes associated with eye disorders.
The relevance of invertebrate models to vertebrate systems depends in part on whether homology exists between most bilaterian eyes, a difficult point to establish based on morphological studies alone. Establishing some degree of common ancestry between vertebrate and invertebrate (primarily Drosophila) visual systems has been a success of comparative molecular genetics. Two findings figure prominently in understanding the relationship between diverse animal eyes. First, two photoreceptor neuron categories, rhabdomeric (microvillar) and ciliary, exist throughout the Bilateria and rely on conserved R-opsin and C-opsin signal transduction pathways, respectively (Arendt, 2003). Second, embryonic development of multiple eye types involves Pax-6, Sine oculis, Eyes absent, and Otx gene family members. These observations have led to the suggestion that photoreceptor cell types were already present prior to the existence of the Bilateria, and that the common ancestor of the Bilateria utilized transcription factors for eye development still commonly used in extant eyes (Nilsson, 2009).
To explore many conserved features of eye biology, additional invertebrate model eyes must be established. The morphology, cell type composition, and set of gene activities present in ancestral bilaterian eyes remains largely unknown, and can only be inferred using data from multiple extant eyes belonging to diverse animal phyla. Furthermore, invertebrate models are not yet established for extensive genetic study of several common aspects of eye biology not characteristic of Drosophila eyes, such as the formation of optic cups. Finally, many genes associated with eye disease have not yet been identified and studied in invertebrate eyes, and no invertebrate system exists for studying regenerative repair of eye damage, an increasingly important therapeutic approach.
The planarian Schmidtea mediterranea is an emerging invertebrate model system that is highly amenable to gene function studies with RNAi. Planarians are ideal for the study of eye evolution because they are members of the Lophotrochozoa, the sister grouping of phyla to the Ecdysozoa (which includes Drosophila and C. elegans), and therefore exist at an important and understudied position in the animal phylogeny (Tessmar-Raible and Arendt, 2003). Planarian eyes are true cerebral eyes, as they connect via axon tracts to the brain (Agata et al., 1998), and express orthologs of Otx (Umesono et al., 1999), Sine oculis (Pineda et al., 2000), and Eyes-absent (Mannini et al., 2004). Planarians have rhabdomeric photoreceptor neurons (PRNs), pigment cells (PCs), and a pigmented optic cup structure – features that are common among cerebral eyes (Figure 1A). Photoreceptive organelles in the planarian eye face the optic cup in an inverse orientation, similar to the orientation of ciliary vertebrate photoreceptors with respect to the retinal pigment epithelium (RPE) (Figure 1A). Finally, planarians possess unique abilities to regenerate entire eyes, and replenish eye tissue throughout adulthood. These abilities require a population of adult regenerative cells (neoblasts) that includes pluripotent stem cells (Wagner et al., 2011).
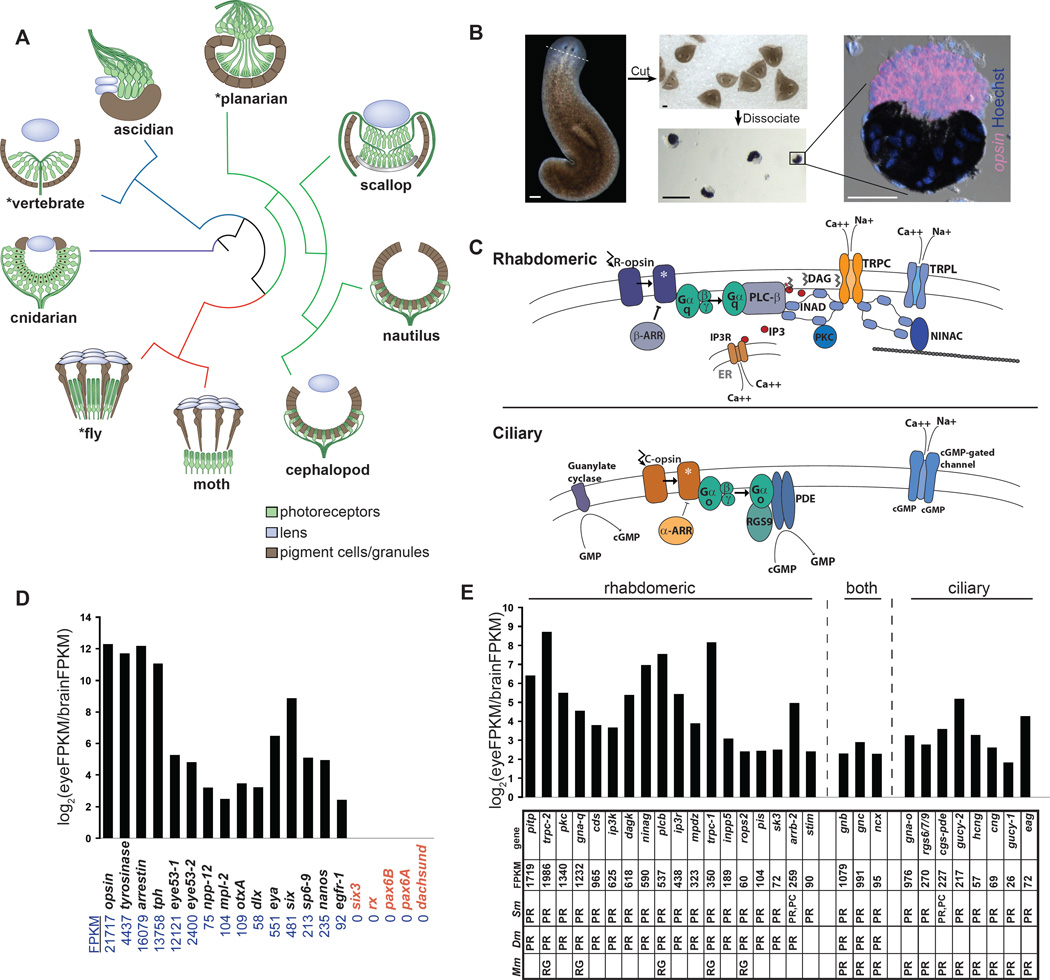
Figure 1. cDNA sequencing of purified planarian eyes.
(A) Schematic of various metazoan eye types. The red branch represents Ecdysozoa; green branch, Lophotrochozoa; blue branch, Deuterostomia; purple branch, Cnidaria. Asterisks indicate that planarians, Drosophila, and vertebrate eyes can currently be studied with a completed genome and loss of gene function tools.
(B) Planarian eye purification. opsin RNA probe and pigment (melanin) were used to assess purity of purified eyes.
(C) Schematic of signaling downstream of R-opsin and C-opsin.
(D) Plot of relative FPKM values for planarian genes previously described in the literature with expression in the eye (black), or lacking expression in the eye (red).
(E) Enriched expression was detected for genes typical of the R-opsin cascade (left set), the C-opsin cascade (right set), or either form of phototransduction (middle set). Table shows FPKM values for genes in the eye sample and summarizes expression data from mouse (Mm), Drosophila (Dm) and planarian (Sm; see also Figure 2). PR, photoreceptor neuron; PC, optic pigment cell; RG, retinal ganglion cell. Retinal ganglion cells are vertebrate retinal neurons with some photoreceptive ability, but which rely an R-opsin cascade-like phototransduction mechanism (Sexton et al., 2012).
Scale bars, 200 µm (B, left and middle); 50 µm (B, right).
Here we use eye purification and RNA-seq to identify most genes active in planarian eyes. These data demonstrate conserved features of the R-opsin signaling cascade, candidate rhabdomeric microvillus-regulating genes, and unexpected similarities between rhabdomeric and ciliary cell types. In addition, a high degree of similarity exists between the types of solute transporters expressed in planarian optic pigment cells and the vertebrate RPE, suggesting that optic pigment cells have an ancient role as an accessory eye cell type distinct from photoreceptors. We describe several new, conserved transcription factors expressed in the planarian eye and that are functionally required for eye regeneration, including a central role for the conserved gene ovo in formation of all eye progenitors.
Results
Planarian eye purification and the eye transcriptome
To obtain pure eye tissue for gene expression analysis, we developed a dissociation protocol for isolating ~200 morphologically intact planarian eyes in a 1–2 hour period (Figure 1B). Greater than 96% of cells in the purified eye preparation were indeed eye cells, as determined using an opsin RNA probe for photoreceptor neurons (PRNs) and melanin as a marker for pigmented optic cup cells (PCs) (Figure 1B). RNA from purified eyes was used to generate a cDNA library for quantitative RNA sequencing (RNA-seq). Animals were fed prior to eye harvesting to stimulate growth and incorporation of new cells in eyes. Genes previously described to be expressed in planarian eyes all displayed at least 4-fold enrichment in our eye reads, compared to reads from control tissue (ventral, anterior tissue enriched for neurons), in terms of fragments per kilobase per million fragments mapped (FPKM) values (Figure 1D) (See Experimental Procedures for details). pax6A, pax6B, rax, six3, and dachshund are orthologs of genes with roles in eye development in other organisms, but have been described to lack detectable expression and function in the planarian eye (Lapan and Reddien, 2011; Mannini et al., 2008; Pineda et al., 2002; Pineda and Salo, 2002); all of these genes displayed FPKM values of zero (Figure 1D), demonstrating specificity of the data and further suggesting that these genes have no role in planarian eye regeneration. We conclude that transcript levels in the RNA-seq dataset can be highly predictive of gene expression in vivo.
For an in situ expression screen we selected unpublished planarian genes with similarity to genes in Drosophila and vertebrates and >4-fold expression enrichment in the eye. Of these approximately 600 genes (Table S1), nearly 200 were tested by in situ hybridization (Figure 2 and Tables S2) in regenerating heads 7 days after amputation. PRNs and PCs can be identified by position and whether the eye expression domain curves toward (PRNs) or away from (PCs) the midline (Figure 2A). 93% of genes screened by in situ had detectable expression in eyes. 60% were expressed in PRNs, 21% were expressed in the pigmented optic cup cells, and 12% were detected in both cell types (Table S2).
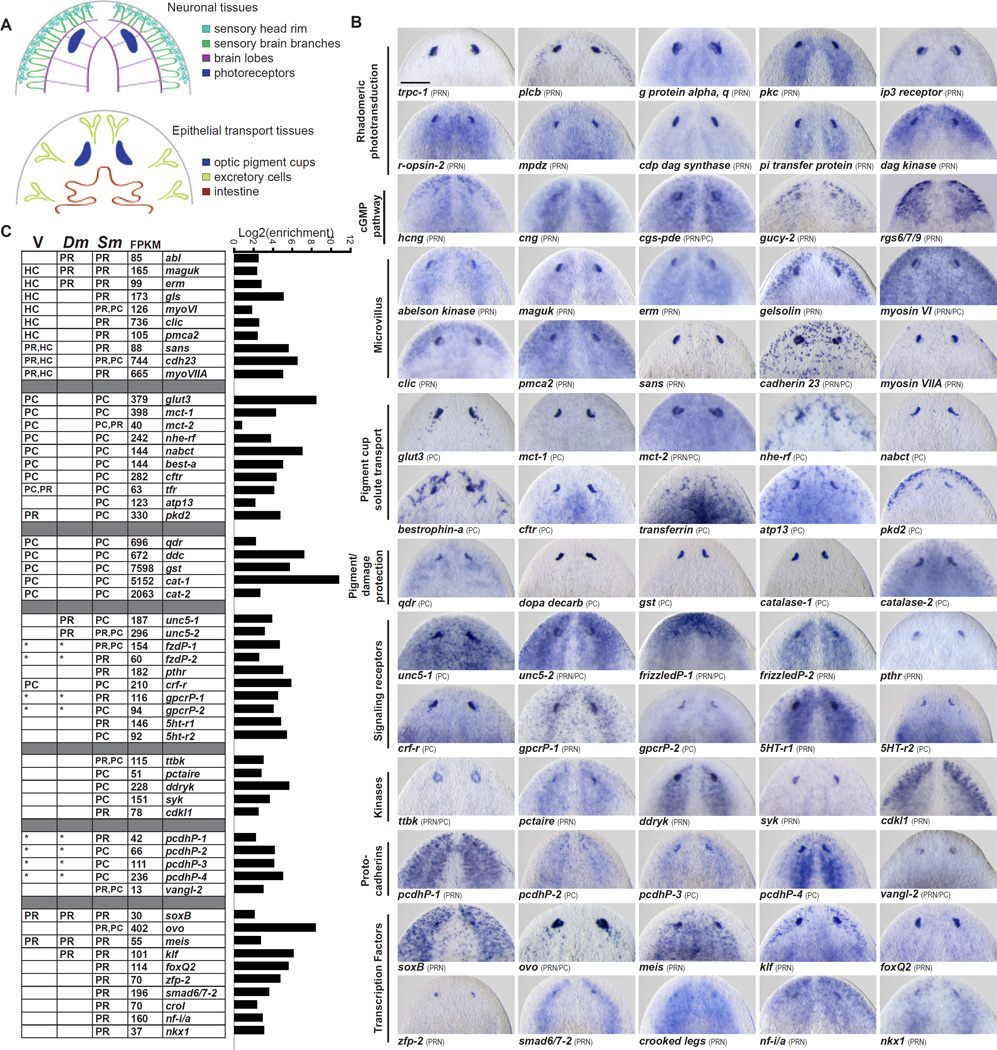
Figure 2. An in situ hybridization screen of genes with enriched expression in planarian eyes.
(A) Schematic of tissue types in the head that appear in expression patterns. PRN expression domains curve toward the midline, and PC expression domains curve away from the midline
(B) in situ hybridization of eye-enriched transcripts, grouped by predicted function. Full names and additional orthology information are given in Table S1.
(C) Plot of the log2 of fold enrichment (FPKM eye sample/FPKM control sample) for genes screened (see also Table S2). Values for rhabdomeric phototransduction and cGMP pathway genes are shown in Figure 1. Table shows FPKM values for genes in the eye sample and summarizes expression data from vertebrate (V), Drosophila (Dm), and planarian (Sm). PR, photoreceptor neuron; PC, optic pigment cell; HC, inner ear hair cell; asterisk indicates that planarian gene orthology could only be assigned to a broad gene family for which functions have been identified in the eyes of indicated species.
Scale bar, 200 µm.
Planarian phototransduction genes
Rhodopsin signaling components displayed eye-enriched expression in the RNA-seq dataset (Figure 1C, E) and in situ hybridizations revealed expression in photoreceptor neurons for two R-opsin orthologs (Sánchez Alvarado and Newmark, 1999), two β-arrestin orthologs (Agata et al., 1998), Gα-q (Smed-gna-β), PLC-β (Smed-plcb), INAD (Smed-mpdz), PKC, and two Trp channel-encoding genes (Smed-trpc-1, Smed-trpc-2) (Fain et al.) (Figures 1C,E and 2B). Planarian PRNs also had abundant expression of genes encoding enzymes of the phosphoinositide cycle (Figures 1C,E and 2B), which replenishes PIP2 after its hydrolysis by PLC (Wang and Montell, 2007). These include retinal degeneration a (Smed-dagk), retinal degeneration b (Smed-pitp), dpis (Smed-pis), and cds (Smed-cds).
Genes with predicted roles in cGMP signaling and intracellular calcium regulation, which are involved in phototransduction in multiple organisms, were also expressed in planarian eyes. Specifically, PRNs expressed orthologs of IP3-receptors (Smed-ip3r) and STIM (Smed-stim) (Figures 2B and S1), which regulate intracellular calcium levels (Smyth et al., 2010). PRNs also expressed genes encoding the cGMP pathway components guanylate cyclase (Smed-gucy-1, Smed-gucy-2), cGMP-dependent phosphodiesterase (Smed-cgs-pde), RGS7/9 (Smed-rgs6/7/9), a cGMP-gated ion channel (Smed-cng), and a hyperpolarization-activated cyclic nucleotide-gated channel (Smed-hcng) (Figures 1E, 2B and S1). Many of these components mediate signaling downstream of C-opsin in vertebrates (Arendt, 2003), but we were unable to identify an ortholog of C-opsin in planarians. GO analysis of the RNA-seq data indicated that Rhodopsin signaling, cGMP signaling, and intracellular calcium regulation were all enriched categories in the dataset of eye-expressed genes (Table S3).
Expression of microvillus-related genes in photoreceptor neurons
Unexpectedly, orthologs of genes involved in the function and development of auditory hair cells displayed enriched expression in planarian eyes and were prominently represented in GO analysis (Figure 2B, C and Table S3). Like planarian photoreceptors, hair cells are sensory neurons with abundant apical microvilli. Microvilli are actin-based structures, and we identified enriched PRN expression of genes encoding actin (Smed-actin-1), the actin-regulating proteins Ezrin/Radixin/Moesin (Smed-erm), Clic (Smed-clic), Maguk (Smed-maguk), Gelsolin (Smed-gls), and the actin-based motor Myosin VI (Smed-myoVI). These genes have all been shown to have roles in morphogenesis and function of vertebrate hair cells (Gagnon et al., 2006; Kitajiri et al., 2004; Mburu et al., 2006; Mburu et al., 2010; Self et al., 1999). Auditory hair cells of human and mouse also express Usher syndrome genes, which function in morphogenesis of microvillus bundles (Frolenkov et al., 2004). Planarian PRNs expressed orthologs of three Usher syndrome genes, Myosin VIIA/Ush1B (Smed-myoVIIA), Sans/Ush1G (Smed-sans), and Cadherin 23 (Smed-cdh23) (Figure 2B–C). These genes are also expressed in vertebrate photoreceptors, despite the fact that the latter are ciliary. The function of Usher syndrome proteins in vertebrate photoreceptors is not understood (Williams, 2008), but many localize to the periciliary region and Usher syndrome can result in aberrant photoreceptor cilia morphology (Hunter et al., 1986). Many of the hair cell-related genes described here, including Usher syndrome genes, have not been described to be expressed in Drosophila photoreceptors. Therefore, our analysis identifies unexpected similarity between a rhabdomeric photoreceptor and auditory hair cells, identifying genes that are candidates to regulate microvillus-like apical membrane specialization.
Genes of the pigmented optic cup
The vertebrate retinal pigment epithelium (RPE) supports photoreceptor function by providing organic molecules, maintaining extracellular ion concentrations, phagocytosing aging photoreceptor components, and absorbing light energy (Strauss, 2005). Many genes expressed in planarian pigment cells were orthologs of genes important for the support function of RPE cells. The metabolic rate of the vertebrate retina necessitates high levels of available glucose. Planarian PCs expressed orthologs of Glut3 (Smed-glut3) (Figure 2B–C) and MCT transporters (Smed-mct-1 and Smed-mct-2) (Figure 2B–C), which transport glucose to the retina (Ban and Rizzolo, 2000) and remove lactic acid waste (Bergersen et al., 1999), respectively. Lactic acid uptake is coupled to proton uptake (Lin et al., 1994; Lin and Miller, 1991) and two genes encoding pH-regulatory proteins, a Na+/H+ exchanger regulatory cofactor (Smed-nhe-rf) and a sodium bicarbonate cotransporter (Smed-nabct), were expressed specifically in the planarian PCs (Figure 2B–C). High metabolic activity in the retina results in excess water production, which is eliminated using chloride gradients across the RPE (Strauss, 2005). Orthologs of the RPE-expressed chloride channels Bestrophin-1 (Smed-best-a) and Cftr (Smed-cftr) were strongly expressed in the pigment cells of planarian eyes (Figure 2B–C).
Planarians are similar to vertebrates, but differ from most other invertebrates, in the use of melanin as the primary eye shading pigment (Hase et al., 2006; Strauss, 2005). Among the most abundantly expressed genes in planarian PCs were those encoding orthologs of enzymes required for melanin synthesis in vertebrates, including Tyrosinase (Lapan and Reddien, 2011) (Figure 1D), Aromatic amino acid hydroxylase (Nishimura et al., 2007) (see “tph”, Figure 1D); and Dopa decarboxylase (Smed-ddc) (Figure 2B–C). PCs also expressed orthologs of Quinoid dihydropteridine reductase (Smed-qdr), which produces an essential cofactor of tyrosine/phenylalanine hydroxylase (Schallreuter et al., 2008); and Glutathione-S-transferase (Smed-gst), which catalyzes addition of glutathione during pheomelanin polymerization (del Marmol et al., 1996) (Figures 2B–C).
Peroxide is generated by melanin-synthesizing enzymes and is required for melanin polymerization, but it can also cause cytotoxicity in melanocytic cells (Mastore et al., 2005). The GO category for “Response to hydrogen peroxide” was significantly enriched in the eye data (Table S3). Two orthologs of catalase, Smed-cat-1 and Smed-cat-2, were expressed in the planarian PCs. Similar to tyrosinase, Smed-cat-1 was one of the most strongly and specifically expressed genes in the planarian pigment cells (Figure 2B–C and Table S2). Glutathione-S-transferase might also function as an antioxidant enzyme in the eye (Giblin, 2000; Singhal et al., 1999). Therefore, planarian and vertebrate optic pigment cells express similar melanin-synthesizing enzymes and enzymes that protect from oxidative damage.
Heterogeneity of the PRN and PC populations
Recently, planarian photoreceptor neuron heterogeneity has been described, with prohormones and smad6/7-2 expressed in distinct domains of the PRN population (Collins et al., 2011; Gonzalez-Sastre et al., 2012). Smed-soxB, Smed-best-b, Smed-pcdhP-1, and Smed-pthr were expressed only in anterior photoreceptors (Figure S2A). Smed-fzdP-1 was expressed primarily in dorsal and anterior photoreceptors, as well as progenitors near the eye (Figure S2A). Smed-zfp-2, Smed-pctaire, and Smed_19866 were expressed only in posterior photoreceptors (Figure S2B). We also identified a gene, Smed-actin-2, expressed in subsets of PCs, indicating that heterogeneity also exists among pigment cup cells (Figure S2C).
Identification of candidate regulators of eye formation and an eye-specific transcription factor-encoding gene, Smed-ovo
Molecular regulators remain to be identified for key steps in planarian eye formation, including progenitor migration, homotypic cell aggregation, mesenchymal-to-epithelial transition, and cupping morphogenesis. in situ screening identified enriched expression in the planarian eye for many genes that are good candidates to encode proteins regulating these processes, including 14 kinases, 5 protocadherins, and 18 signaling receptors (Figures 2B–C and S1).
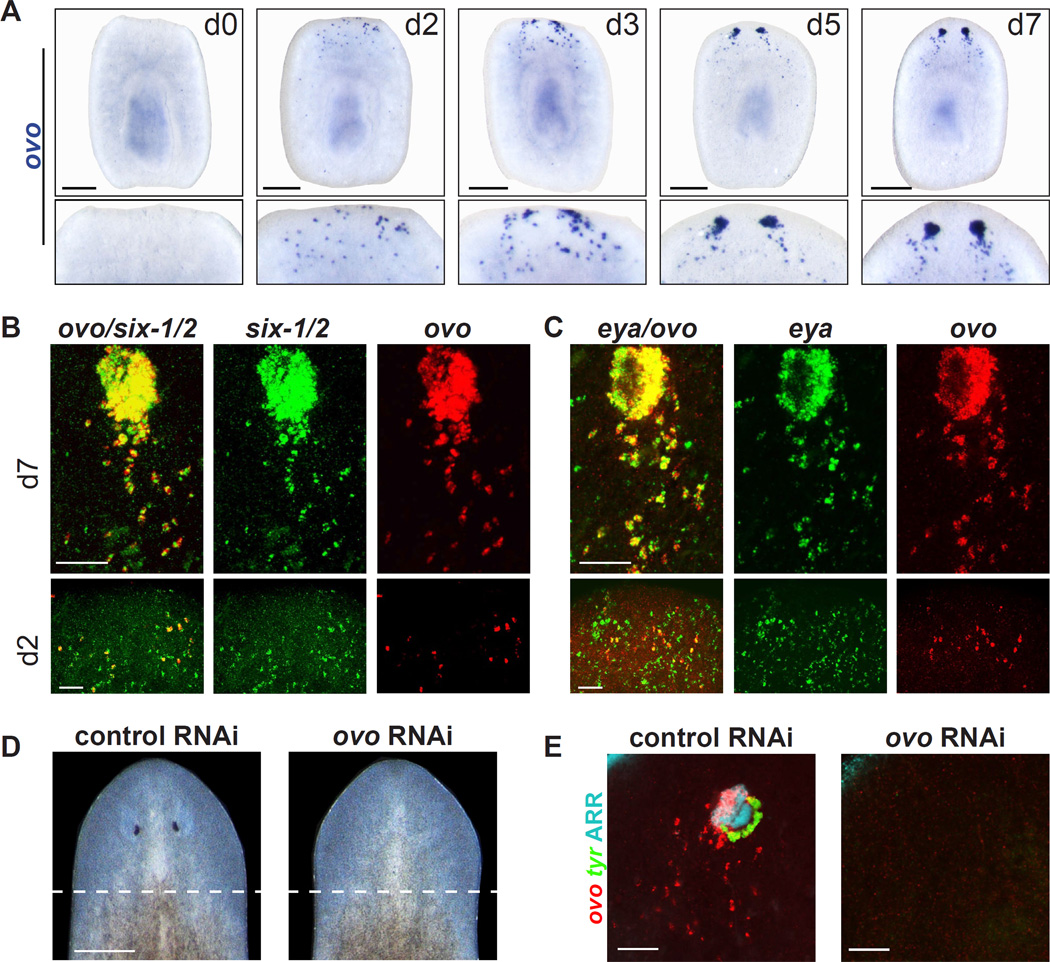
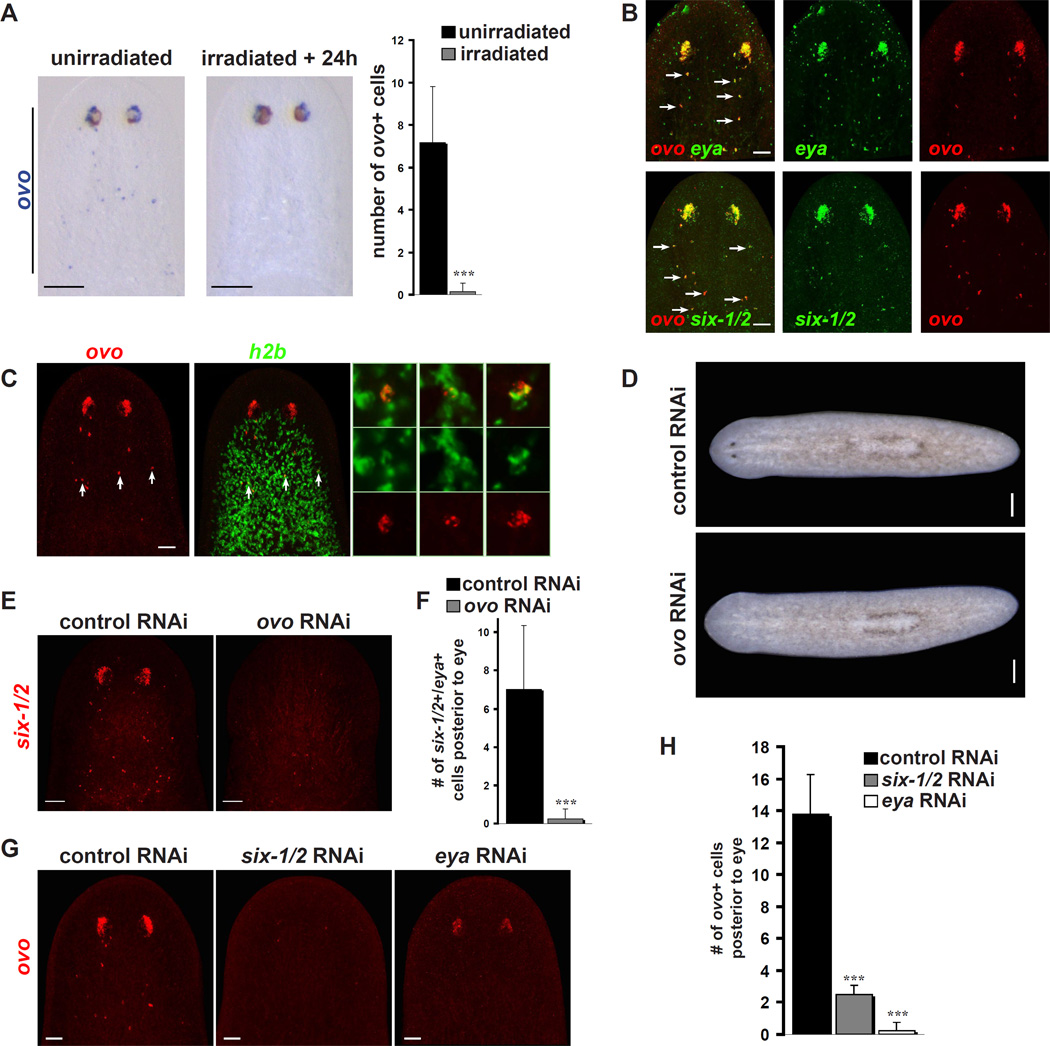
Transcription factors that control eye development in both Drosophila and vertebrates have received close attention for their capacity to elucidate the evolution of eye development. Here we identified 10 new conserved genes predicted to encode transcription factors with enriched expression in planarian eyes (Figure 2B–C). The most prominently enriched of these was a member of the Ovo family of zinc finger transcription factor-encoding genes (Figure S3), which includes the Drosophila gene ovo (shavenbaby) and mouse ovol1-3 genes. Expression of Smed-ovo (ovo) was detected in both PRNs and PCs, as well as in mesenchymal cells posterior to the eye in day 7 head blastemas (Figures 2B and 3A). After head amputation, ovo+ cells first became apparent throughout the dorsal anterior region, near the wound, by 2 days post-amputation (Figure 3A). At later regeneration time points ovo+ eye aggregates formed in the anterior blastema, while dispersed cells posterior to the eye remained visible. This pattern was similar to a recently defined population of regenerative eye progenitors in planarians (Lapan and Reddien, 2011). Consistent with this, nearly all ovo+ cells at day 2 and day 7 of regeneration also displayed expression of six-1/2 and eya, which marks eye progenitors (Figure 3B–C).
Figure 3. Smed-ovo is expressed specifically in the eye and eye progenitors, and is required for eye regeneration.
(A) Regeneration time course showing ovo expression after amputation of head and tail. Bottom panel is a magnified view of the anterior wound/blastema.
(B) ovo+ cells express the eye progenitor marker six-1/2 at early and late time points of regeneration (d2: 96% +/− 5% overlap, d7: 100% overlap, α = 0.05).
(C) ovo+ cells also express the eye progenitor marker eya (d2: 90% +/− 6% overlap, d7: 100% overlap, α =0.05).
(D) RNAi of ovo leads to failed eye regeneration (n=10/10). Dashed lines indicate approximate site of head amputation 12 days prior.
(E) ovo(RNAi) animals lack expression of ovo, tyrosinase, and Arrestin after head regeneration (n=10/10).
Scale bars, 200 µm (A), 50 µm (B,C,E), and 1 mm (D).
In regenerating or intact animals, no ovo expression was detected outside of eyes or eye progenitors (see below). Furthermore, in the control (ventral anterior tissue) sequencing data, the FPKM value for ovo was zero (Table S2 and Figure S4A). Other previously known eye-expressed transcription factors, eya, six-1/2, sp6–9, and otxA also have expression in the brain and/or elsewhere in the body (Lapan and Reddien, 2011; Pineda et al., 2000; Umesono et al., 1999). Therefore, ovo is the only known planarian transcription factor with expression exclusively in the eye and eye progenitors. It is one of only three transcription factor-encoding genes, together with six-1/2 and eya, that is expressed in all cells of the eye.
ovo expression is required for eye regeneration
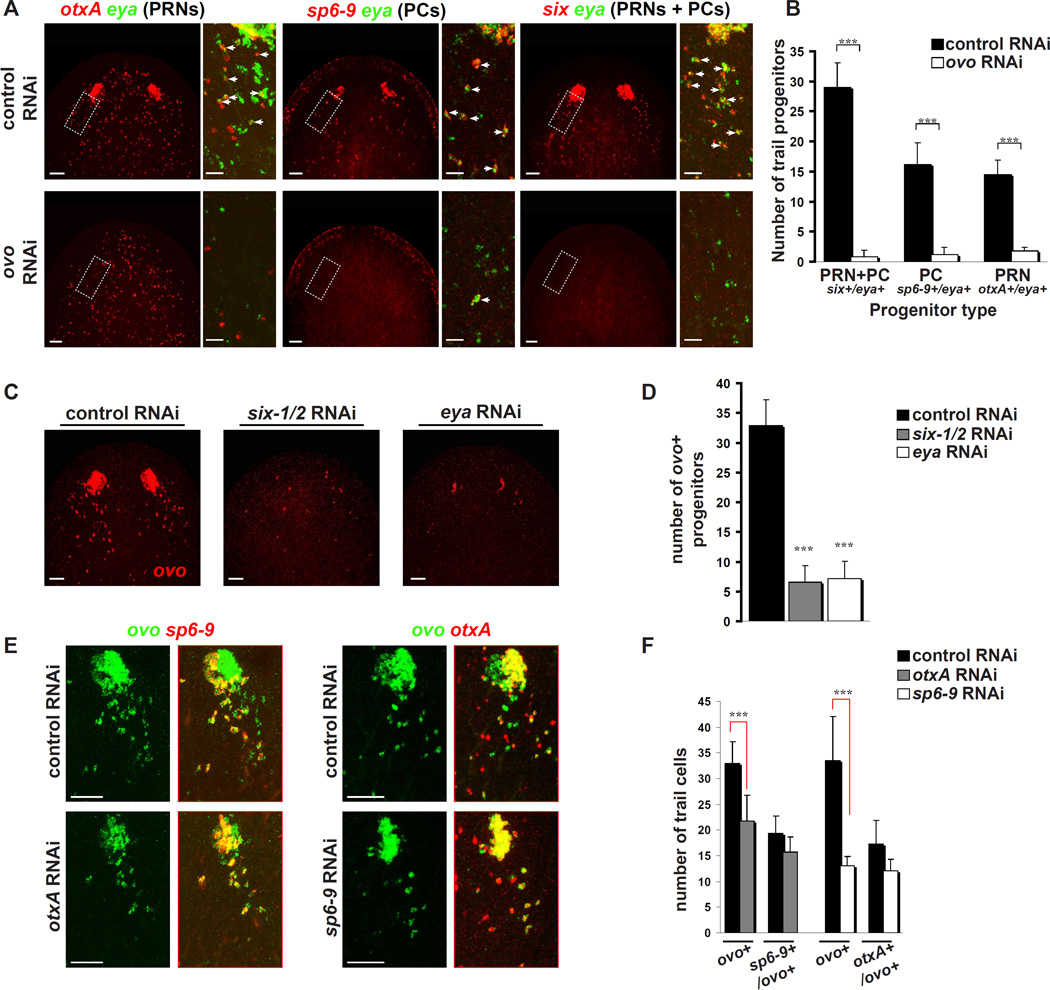
RNAi of ovo followed by decapitation resulted in animals that failed to regenerate eyes (Figure 3D). These animals also lacked detectable tyrosinase mRNA and Arrestin protein (Figure 3E), and no eye structure could be detected by differential interference contrast microscopy (DIC) (Figure S4B). We studied ovo(RNAi) animals using six-1/2/eya co-expression to label all eye progenitors, and otxA/eya and sp6–9/eya co-expression to label PRN and PC progenitors, respectively (Lapan and Reddien, 2011). ovo RNAi resulted in failure to generate any eye aggregate (primordium), and loss of most eye progenitors (Figures 4A–B). We observed no defects that were unrelated to the eyes, consistent with the highly specific expression of this gene. For example, other cell types of the head, such as sp6–9+/eya− cells of the head rim appeared unaffected (Figure 4A). Therefore, eyes and eye progenitors are strongly and specifically affected in ovo(RNAi) animals.
Figure 4. ovo is required for formation of regenerative eye progenitors, and ovo+ progenitor presence requires other eye transcription factors.
(A)–(B) ovo RNAi eliminates regenerative eye progenitors as assessed by markers for PRN (otxA+/eya+), PC (sp6-9+/eya+), and general eye progenitors (six-1/2+/eya+). Arrows indicate double-positive cells.
(C)–(D) Most regenerative ovo+ progenitor cells are eliminated by six-1/2 and eya RNAi.
(E)–(F) sp6-9 RNAi eliminates most ovo+ pigment cell progenitors (ovo+/otxA), and otxA RNAi eliminates most ovo+ photoreceptor neuron progenitors (ovo+/sp6-9).
n = 8 eyes + trails for all quantifications.
Error bars represent SD. *** p < .005. Scale bars, 50 µm (A,C,E), and 20 µm (A, insets).
Formation of ovo+ progenitors requires expression of other eye transcription factors
In gene regulatory networks involved in Drosophila and vertebrate eye development, several transcription factors that regulate eye formation are required for the expression of each other (Silver and Rebay, 2005; Zuber et al., 2003). We tested whether ovo+ progenitor cells are sensitive to expression of other planarian eye transcription factors. RNAi of six-1/2 and eya resulted in animals with greatly reduced or absent ovo signal in the eyes and eye progenitor region (Figures 4C–D). RNAi of the cell type-specific transcription factors otxA and sp6–9, required for PRN and PC regeneration respectively, also impacted the presence of ovo+ progenitors (Figures 4E–F). RNAi of otxA led to a 34% reduction in ovo+ cells; this reduction was primarily a result of loss of ovo+ PRN progenitors (Figure 4E–F), because most remaining ovo+ cells were sp6–9+. Similarly, RNAi of sp6–9 led to a 60% reduction in ovo+ cells, primarily a result of loss of ovo+ PC progenitors, because most remaining ovo+ cells were otxA+ (Figure 4E–F). These large decreases indicate that ovo-expressing cells, even in early stages of eye progenitor formation, are sensitive to the expression of six-1/2, eya, otxA, and sp6–9 transcription factors.
ovo is expressed in a population of eye progenitors during homeostasis and is required for maintenance of intact eyes
The specificity of ovo expression for eyes and eye progenitors allowed us to ask for the first time whether subsets of neoblasts in uninjured animals are specialized for particular differentiation paths. In intact animals undergoing normal adult homeostasis, ovo was expressed not only in the eye but also in sparse cells posterior to the eye and anterior to the pharynx (Figure 5A). Adult planarians undergo perpetual tissue turnover (Newmark and Sánchez Alvarado, 2000), but the identity and origin of progenitors that replenish eyes in uninjured animals is unknown. ovo+ cells posterior to the eye in intact animals expressed eya and six-1/2 (Figure 5B), and those nearer to the eye also expressed markers for differentiated photoreceptor neurons (trpc-1) or pigment cells (tyrosinase) (Figure S5A). Therefore, ovo+ cells that exist posterior to the eye in intact animals have a gene expression profile seen elsewhere only in cells of the eye.
Figure 5. ovo is expressed in a population of homeostatic eye progenitors.
(A) In starved intact animals, ovo is expressed in isolated dorsal cells between the eyes and pharynx. These cells are eliminated within 24 hours of irradiation.
(B) ovo+ cells posterior to the eyes in intact animals also express six-1/2 and eya.
(C) ovo+ cells posterior to the eyes in intact animals also express the neoblast marker histone h2b (60% +/− 15%, α =0.05).
(D) ovo(RNAi) animals that are not amputated fail to maintain eyes (n=10/10).
(E)–(F) ovo(RNAi) animals that are not amputated fail to maintain six-1/2+/eya+ cells posterior to the eyes (n=4 animals). For image clarity only six-1/2 signal is shown.
(G)–(H) six-1/2(RNAi) and eya(RNAi) animals fail to maintain ovo+ cells posterior to the eyes (n=4 animals).
Arrows indicated double-positive cells in (B) and (C). Error bars represent SD. *** p < .005. Scale bars, 100 µm (A), 50 µm (B,C,F,H), and 200 µm (D).
The localization of the ovo+ cells posterior to the eye in intact animals is consistent with the location of eye progenitors during regeneration following head amputation (Lapan and Reddien, 2011). Strikingly, these cells were eliminated within one day of irradiation (Figure 5A), a treatment known to specifically eliminate the dividing cells of the adult animal (the neoblasts) (Reddien and Sánchez Alvarado, 2004). Furthermore, many ovo+ cells in this prepharyngeal population expressed the neoblastspecific markers histone h2b (Figure 5C) and smedwi-1 (Figure S5B) (Reddien et al., 2005). It is unknown whether these ovo+ neoblasts are self-renewing or constantly produced from a more naive cell. These results suggest that cycling eye progenitors constitutively exist in the neoblast population.
ovo RNAi in intact animals resulted in gradual loss of eyes, with pigment cups undetectable following two months of RNAi (Figure 5D). These un-amputated ovo(RNAi) animals also lacked six-1/2+/eya+ cells in the region of putative homeostatic eye progenitors (Figure 5E–F). Reciprocally, homeostatic RNAi of six-1/2 and eya, in addition to causing loss of eye tissue, caused a loss of ovo+ cells in the prepharyngeal region (Figure 5G–H). Overall, these data suggest that ovo, six-1/2, and eya are expressed in a population of eye progenitors during homeostasis as well as regeneration and are required for maintenance of this population.
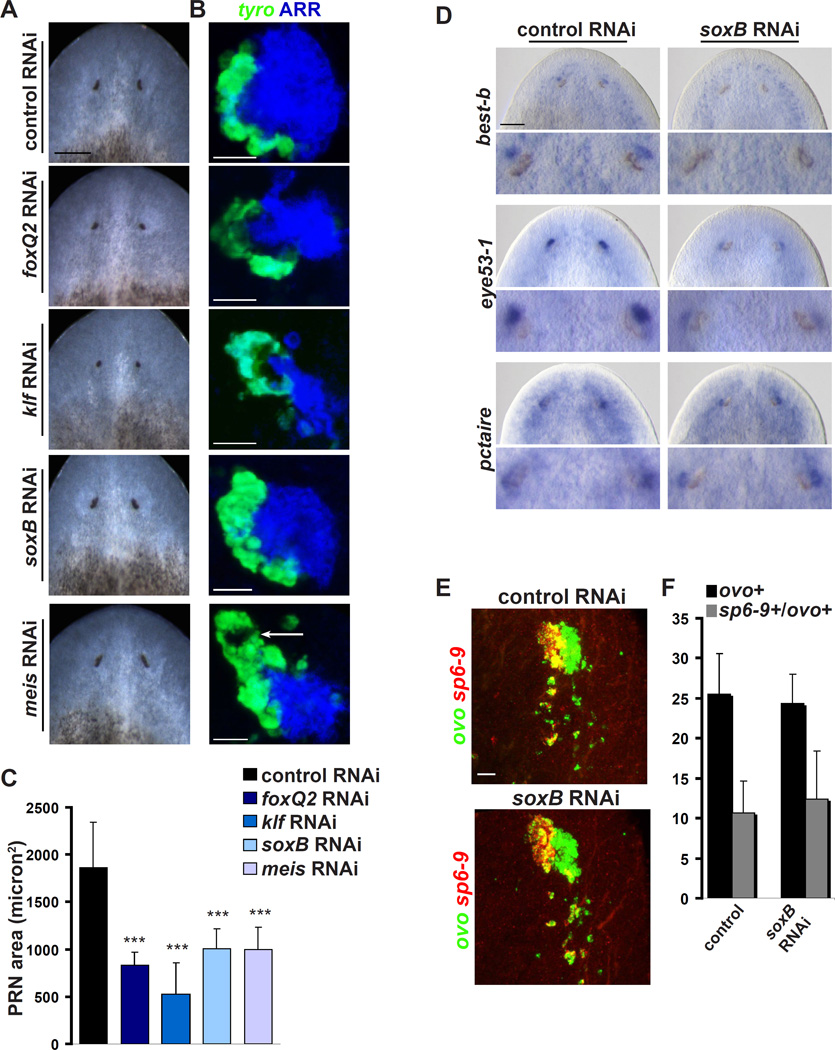
Smed-soxB, Smed-foxQ2, Smed-klf, and Smed-meis regulate eye differentiation and morphogenesis
Photoreceptor neuron differentiation
RNAi of genes encoding four additional conserved transcription factors, Smed-soxB, Smed-foxQ2, Smed-klf, and Smed-meis (Figure S3) resulted in PRN aggregates that were smaller in cross section than in the control (Figure 6A–C). All of these genes were expressed in PRNs and eye progenitors during regeneration (Figure S6A). Photoreceptor aggregates in klf(RNAi) animals were also displaced posteriorly relative to the pigment cup and descended unusually far toward the interior of the animal along the D–V axis, embedding within the brain (Figures S7A–B).
Figure 6. Smed-soxB, Smed-meis, Smed-klf, and Smed-foxq2 are required for eye regeneration.
(A) Eye regeneration defects at 7 days of regeneration in live RNAi animals.
(B) Detection of tyrosinase transcript and Arrestin protein reveals defects in optic cup formation and photoreceptor neuron regeneration. Arrow indicates duplicated optic cup in meis(RNAi) animal.
(C) Quantification of cross-section area (µm2) of photoreceptor neuron aggregate (mean + SD, n=8 eyes).
(D) soxB RNAi results in loss of anterior subtypes of photoreceptor neurons, but not posterior subtypes (n=8 eyes for each condition).
(E)–(F) soxB RNAi does not affect numbers of ovo+ progenitors, nor the ratio of PC progenitors (sp6-9+/ovo+) to PRN progenitors (sp6-9/ovo+).
Error bars represent SD. *** p < .005. Scale bars, 200 µm (A), 100 µm (D), 20 µm (B, E).
Small eye phenotypes were not accompanied by an evident decrease in the number of ovo+ progenitors in these RNAi animals (Figure S6B–C and 6F), suggesting that these genes function primarily following progenitor specification. Because soxB expression was restricted to anterior PRNs, we assessed the presence of photoreceptor neuron subsets in soxB(RNAi) animals. Anterior PRNs (eye53-1+ (Collins et al., 2011) and best-b+) were strongly reduced or eliminated in soxB(RNAi) animals (Figure 6D). By contrast, posterior PRNs (pctaire+) were not affected (Figure 6D). soxB RNAi did not significantly impact numbers of PC (sp6–9+/ovo+) or PRN (sp6–9−/ovo+) progenitors (Figure 6E–F), and soxB is not abundantly expressed in the ovo+ progenitor population (Figure S6A). However, soxB eye signal is eliminated by ovo RNAi (Figure S6D). These data indicate that whereas ovo (along with six1/2, eya, and otxA) acts in general PRN progenitor specification (Figures 4A and 4E), soxB acts subsequently to promote differentiation of an anterior subset of PRNs.
optic cup morphology
Several RNAi phenotypes affected the pigment cup shape. Loss of photoreceptor cells by RNAi of PRN-specific genes, such as otxA, results in pigment cups with diminished apertures (Lapan and Reddien, 2011). Accordingly, in klf and foxQ2 RNAi animals, optic cups were more closed than in the control, or even completely circularized (Figure 6A–B), possibly explained if rhabdomeres provide support for optic cup opening. In meis(RNAi) animals, however, optic cups were also elongated and often more than one cupped structure was visible in each eye – a defect not characteristic of photoreceptor cell loss (Figure 6B and Figure S7C). Although meis expression was not detected in the pigment cup, meis transcripts are detectable in tyrosinase+ PC progenitors (Figure S7D). Meis1 loss in mouse also disrupts optic cup morphogenesis, with optic cup duplication observed (Hisa et al., 2004).
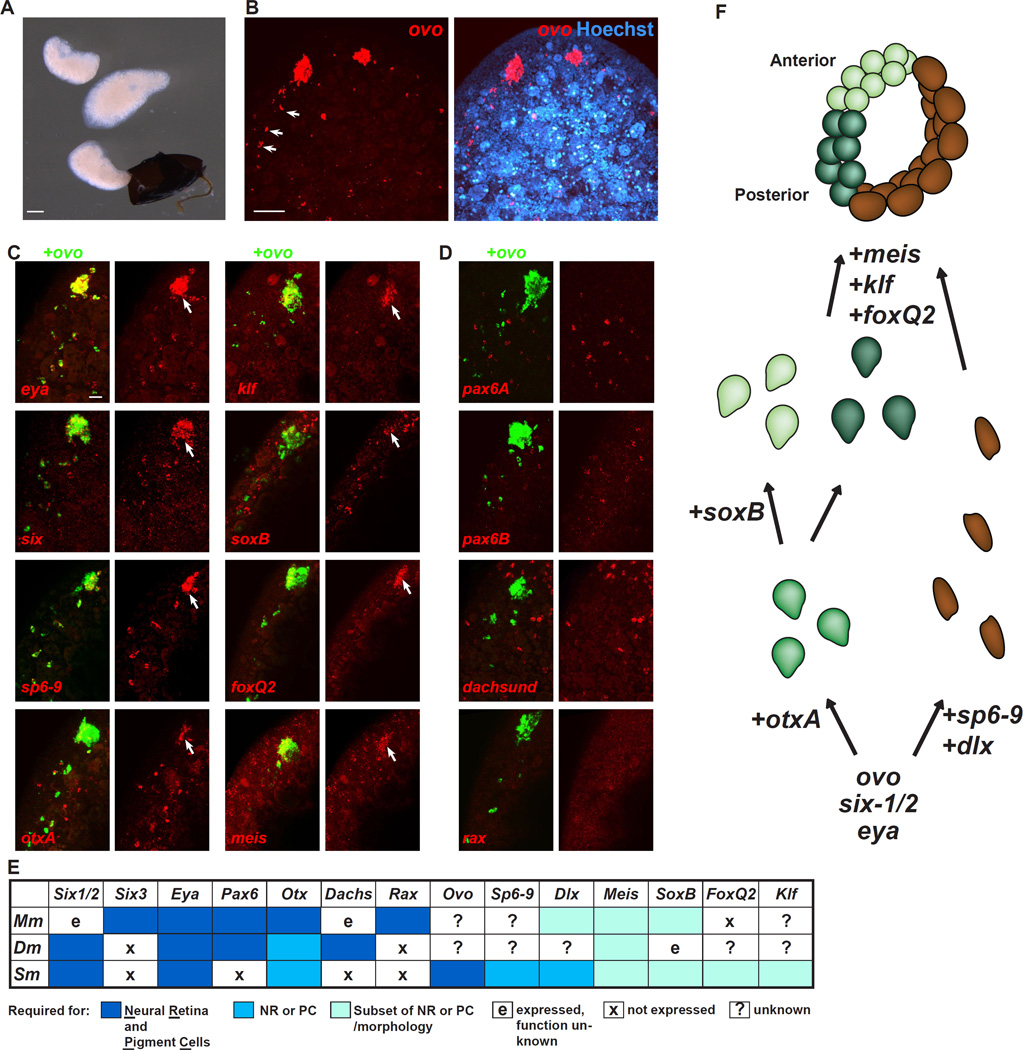
Developing eyes of planarian embryos express transcription factors involved in eye regeneration
Most eye formation studies in other organisms are performed in developing embryos. Therefore, we investigated whether the transcription factors that govern planarian eye regeneration might also be expressed during planarian embryonic development (Figure 7A). Recent work has shown that beginning at stage 6, Schmidtea polychroa embryos have expression of eya, six-1/2, otxA in the eyes, and lack expression of pax6A, rax, and dachshund in the eye domain (Martin-Duran et al., 2012), similar to the case in regeneration. We found that from early stages of S. mediterranea embryonic eye formation, ovo was expressed in the eyes and in mesenchymal cells posterior to the eye that were reminiscent of regenerative progenitor eye trails (Figure 7B). Double FISH with ovo enabled us to determine whether regeneration-related eye genes were expressed in the embryonic eye primordium and/or presumptive earlier progenitors. Embryonic eyes expressed all known transcription factors that function during eye regeneration: eya, six-1/2, otxA, sp6–9, klf, meis, foxQ2, and soxB (Figure 7C). Regionalization of expression in the embryonic eye mirrored the case in regeneration (Lapan and Reddien, 2011); for instance, otxA was expressed in the lateral eye primordium whereas sp6–9 was expressed in the medial eye primordium, and both were expressed in only a subset of ovo+ trail cells (Figure 7C). Furthermore, neither the eye primordium (Martin-Duran et al., 2012) nor the ovo+ trail cells (Figure 7D) expressed pax6A, rax, dachshund, or another Pax6 ortholog, pax6B. Therefore, findings regarding genetic regulation of eye regeneration are relevant to planarian embryonic development.
Figure 7. Expression of transcription factors in planarian embryos.
(A) Freshly decapsulated planarian embryos of the stage used for expression analyses. The dark structure is the embryo capsule.
(B) ovo is expressed during embryonic development in eyes and trails posterior to eyes (arrows)
(C) Double-fluorescent in situ hybridization with ovo is used to show presence of gene expression in eye primordium and eye progenitors during embryonic development. Arrows indicate the location of ovo+ eye primordia.
(D) Absence of gene expression for indicated genes in the eye primordium and eye progenitors during embryonic development.
(E) Table summarizing roles of transcription factors involved in eye formation in either mouse (Mm), Drosophila (Dm), or planarian (Sm). NR, neural retina; PC, optic pigment cells.
(F) Model of steps in planarian eye regeneration that are regulated by transcription factors.
Scale bars, 200 µm (A), 50 µm (B), 20 µm (C,D).
Discussion
Planarian eye formation in regeneration, homeostasis, and embryogenesis
The cellular events bridging the pluripotent state and cellular differentiation during planarian regeneration and homeostasis are poorly understood, and the eye is emerging as a system to study adult lineage specification in this animal (Lapan and Reddien, 2011). ovo is the first identified gene that is expressed in all eye progenitors and is specifically expressed in eye cells, and therefore is a marker that facilitates study of progenitor dynamics during various modes of planarian eye formation. Our investigations of ovo expression lead to three new conclusions concerning planarian eye formation. 1) Isolated eye progenitors are present early in regeneration (by two days after decapitation), prior to the appearance of an aggregated eye primordium. Therefore, specification at a distance from the aggregated eye occurs from the onset of regeneration rather than only at later stages of eye growth (Lapan and Reddien, 2011). 2) Eye progenitors exist in uninjured intact animals in a dorsal pre-pharyngeal domain, similar to regenerative progenitors but at lower density. These data indicate that some neoblasts are specialized for specific lineages during normal tissue turnover conditions of adult life. 3) During embryogenesis, ovo+ cells are present in the eye and in a trail-like formation posterior to the eye.
Further experiments are required to formally determine whether, like regenerative progenitors (Lapan and Reddien, 2011), homeostatic and embryonic eye progenitors move toward the eye primordium. However, our ovo expression analyses indicates that migration of differentiating mesenchymal cells, rather than patterning of an epithelium, might be a feature of all modes of eye formation in planarians. Our findings advance the use of the planarian eye as a system for exploring the extent of similarity between embryonic, regenerative, and homeostatic modes of organ formation within a species and between species.
Eye purification and transcriptome analysis
Canonical phototransduction cascades have been described for ciliary and rhabdomeric phototransduction based on functional investigations in vertebrates and Drosophila, respectively (Arendt, 2003). However, a diversity of phototransduction strategies likely exists among animals. For instance, horseshoe crab (Limulus) and Drosophila light sensation both begin with an R-opsin/PLC cascade, but Drosophila phototransduction culminates in Trp channel opening (Katz and Minke, 2009). By contrast, most evidence from Limulus indicates that opening of cyclic nucleotide-gated (CNG) ion channels (preceded by IP3-dependent intracellular calcium release and changes in cGMP levels) is responsible for photoreceptor depolarization (Garger et al., 2004). We observed robust expression for orthologs of nearly all central Drosophila phototransduction components in planarian photoreceptors, including two Trp channels. However, we also detected enrichment for intracellular calcium regulation and cGMP pathway-like components, including CNG channels. Based on the high levels of Trp channel gene expression, we hypothesize that these are the primary mediators of membrane depolarization following light detection. The roles of CNG channels in planarian eyes are unknown, but electrophysiological recordings combined with RNAi could be used to test possible functions, such as in the regulation of light response or light/dark adaptation.
The planarian pigmented optic cup has numerous similarities to the RPE of vertebrates. Melanin is not commonly found in protostome eyes; however, it is the primary pigment of both planarian PCs and the RPE. We expand upon this similarity by demonstrating that the enzymes used in melanin production and for protection from oxidative damage (a consequence of melanin production) are similar between these cell types. Because of the wide divergence in identity of ocular shading pigment among invertebrates (Vopalensky and Kozmik, 2009), the common synthesis of melanin in planarians and vertebrates need not indicate common ancestry of optic pigment cells. However, we identified additional similarities between planarian PCs and the vertebrate RPE with respect to expression of numerous metabolite and solute transporters, consistent with a common function of these cell types as components of a transport epithelium. This similarity supports a model in which pigmented cells already existed as an accessory cell type separate from photoreceptor neurons in the ancestral bilaterian eye.
The RPE is commonly affected in eye diseases, including age-related macular degeneration (AMD), edema, and inherited retinal degeneration syndromes. A molecular explanation for pathogenesis is lacking for most of these diseases. Planarian PCs express orthologs of two RPE chloride channels underlying eye diseases. Best-1 is mutated in Best vitelliform macular dystrophy, and Cftr mutations underlie vision defects in cystic fibrosis patients (Marmorstein et al., 2009; Xiao et al., 2010). Furthermore, RPE degeneration in AMD correlates with peroxide accumulation and declining Catalase function, which is also responsible for degeneration of other melanocytic cell types (Venza et al., 2011; Wood et al., 2009). Molecular similarities between Drosophila ocular pigment cells and the vertebrate RPE have been difficult to identify (Charlton-Perkins and Cook, 2010). The planarian eye should therefore prove useful as an invertebrate model system for investigating orthologs of mammalian RPE genes involved in pathogenesis, as well as for inferring ancestral functions of ocular pigment cells.
Transcription factor control of planarian eye formation
Comprehensive sequencing of RNA from intact eyes identified three genes that encode transcription factors and that are expressed in all cells of the eye and their progenitors – six-1/2, eya, and ovo. The requirement for six-1/2 and eya in planarian eyes and eye progenitors is previously described (Lapan and Reddien, 2011; Mannini et al., 2004; Pineda et al., 2000) and both are homologs of well-known eye development genes (Figure 7E). We propose a model for eye regeneration in which six-1/2, eya, and ovo expression specifies all eye progenitor formation, with additional expression of otxA specifying PRNs and additional expression of sp6–9 and dlx specifying PCs (Figure 7F).
How do available planarian eye data impact models about transcriptional regulators governing eye formation in the common ancester of the Bilateria? Planarian data support the inclusion of Otx and Eya as ancestral bilaterian eye genes, and of Six-1/2 as an ancestral eye gene for the protostomes (Figure 7E). Our data also support the possibility that Meis and SoxB family genes encode transcription factors with ancestral roles in eye biology. Meis genes regulate eye progenitor proliferation in Drosophila and vertebrates as well as optic cup morphology in mouse (Figure 7E) (Bessa et al., 2002; Heine et al., 2008; Hisa et al., 2004). The SoxB-family sox-neuro and fish-hook genes are expressed in the Drosophila eye disc (Mukherjee et al., 2000) and Sox2 required for neural progenitor maintenance and differentiation of retinal ganglion cells in vertebrates (Matsushima et al., 2011).
In contrast to Otx, Eya, Six-1/2, Meis, and SoxB, several transcription factors that have prominent roles in Drosophila and/or vertebrate eye formation – Pax6, Dachshund, Rax, and Six3 - have no apparent role in planarian eye formation (Figure 7E) (Lapan and Reddien, 2011; Mannini et al., 2008; Martin-Duran et al., 2012; Pineda et al., 2002), a conclusion extended by our eye transcriptome and embryonic eye expression data. Therefore, planarian data does not support the argument that these genes are ancestral regulators of eyes. Loss of a role for any of these genes in eye biology during planarian evolution is clearly a reasonable possibility, especially in cases such as Pax6 where data from multiple other phyla support an eye role for the gene. In addition to the case of the planarian eye, however, the development of Drosophila larval ocellus, branchiostome eye cups, Limulus eyes, and Platynereis adult eyes also does not depend on Pax6 (Vopalensky and Kozmik, 2009). Furthermore, orthologs of the Drosophila eye regulator Dachshund do not have major functions in mouse eye development (Davis et al., 2008), and an ortholog of the vertebrate eye regulator Rax is not found in Drosophila (Arendt et al., 2004). Six3 family transcription factors predominate in vertebrate eye development, whereas most invertebrates depend on Six-1/2 (Vopalensky and Kozmik, 2009). Analysis of additional animal eyes will therefore be an important direction for further testing evolutionary hypotheses related to these genes and eye evolution.
We describe a number of transcription factors conserved among the Bilateria that regulate planarian eye formation, but for which the functions in other animal eyes has not been extensively investigated (Figure 7E). These include ovo, klf, and foxQ2. Our prior work also indicated prominent roles for dlx and sp6–9 in optic cup formation (Lapan and Reddien, 2011). One gene encoding an Ovo-like transcription factor, shavenbaby, is known to exist in Drosophila. This gene is well-studied in epidermal morphogenesis, but roles for this gene in the eye have not been reported. The chromosomal locus responsible for the blind-sterile phenotype in mouse contains the ovol2 gene (Li et al., 2002). These mice exhibit small eye size as well as early onset cataracts; however, other genes are contained in this locus and no causal role for ovol2 in this phenotype is known. Mouse ovol2 is expressed in the surface ectoderm of the ventral forebrain at E8.75-9.5 (Mackay et al., 2006), the site of optical eminence formation. Interestingly, ovol2 knockout mice die at E10.5 without displaying any sign of optic vesicle formation typical of this stage, despite the fact that D–V expression domains examined in the neural tube are maintained and overall embryonic development is not delayed (Mackay et al., 2006; Unezaki et al., 2007).
We demonstrate an unbiased technical approach to systematically survey the molecular basis of eye formation and function. This approach identified novel and conserved genetic features of eyes in planarians, further indicating the power of planarians as a system for study of animal eyes. Few transcription factors in animals display eye-specific expression and are required for formation of all eye cells, and ovo is a new one.
Experimental procedures
Tissue isolation
Large asexual Schmidtea mediterranea were used for eye preparation. Animals were fed twice prior to eye harvest. Head tips were amputated posterior to the eyes and collected into CMFB + 1 mg/mL collagenase (Sigma) + 0.05% NAC and agitated. Freed eyes were rinsed and transferred to Trizol for RNA collection. For control (ventral anterior) tissue, heads were amputated above the pharynx and then coronal amputation was made to remove dorsal tissue, including eyes.
cDNA sequencing and analysis
~1ug of RNA was used to construct Illumina libraries for 36bp single-end sequencing on the GAII platform. Libraries were constructed using Illumina reagents and according to commercial protocol. FPKM were determined using the Cufflinks program Cuffdiff. See Supplemental Experimental Procedures for further detail.
Gene cloning
For cDNA library generation, RNA from heads was isolated using Trizol purification, and used as template for first-strand reverse transcription with Superscript III (Invitrogen). Cloning was otherwise permformed as described (Lapan and Reddien, 2011).
RNA synthesis and RNAi
For probes used in RNAi experiments, riboprobe template was produced with the same primers as were used for cloning, except that gateway adapters were not present on primers, and T7 promoter sequence was added to the 3’ primer. Probe synthesis and RNAi were otherwise performed as described (Lapan and Reddien, 2011). In homeostasis RNAi experiments, animals were fed dsRNA food every four days for six weeks.
Histology and imaging
Whole-mount fluorescent in situ hybridization (FISH) and antibody staining was performed as described (Lapan and Reddien, 2011; Pearson et al., 2009). Embryos were killed in 2.7% HCl following manual decapsulation and were otherwise treated the same as adult worms for histological analysis. Images were acquired with a Zeiss confocal microscope (LSM 700).
Supplementary Material
Highlights.
Determination of the complete transcriptome of planarian eyes
The planarian eye expresses orthologs of eye disease genes
The transcription factor Ovo is required for eye formation
Similarities in eye progenitors for regeneration, tissue turnover, and embryogenesis
Acknowledgements
Thanks to Irving E. Wang for the illustration in Figure 1A. Thanks to Jessica Witchley for generating the Illumina library. Thanks to Josien van Wolfswinkel, Mansi Srivastava and Jared Owen for bioinformatic assistance. P.W.R. is an early career scientist of the Howard Hughes Medical Institute and an associate member of the Broad Institute of Harvard and MIT. We acknowledge support from the NIH (R01GM080639) and the Keck Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agata K, Soejima Y, Kato K, Kobayashi C, Umesono Y, Watanabe K. Structure of the planarian central nervous system (CNS) revealed by neuronal cell markers. Zoological Science. 1998;15:433–440. doi: 10.2108/zsj.15.433. [DOI] [PubMed] [Google Scholar]
- Arendt D. Evolution of eyes and photoreceptor cell types. The International Journal of Developmental Biology. 2003;47:563–571. [PubMed] [Google Scholar]
- Arendt D, Tessmar-Raible K, Snyman H, Dorresteijn AW, Wittbrodt J. Ciliary photoreceptors with a vertebrate-type opsin in an invertebrate brain. Science. 2004;306:869–871. doi: 10.1126/science.1099955. [DOI] [PubMed] [Google Scholar]
- Ban Y, Rizzolo LJ. Regulation of glucose transporters during development of the retinal pigment epithelium. Brain research. 2000;121:89–95. doi: 10.1016/s0165-3806(00)00028-6. [DOI] [PubMed] [Google Scholar]
- Bergersen L, Johannsson E, Veruki ML, Nagelhus EA, Halestrap A, Sejersted OM, Ottersen OP. Cellular and subcellular expression of monocarboxylate transporters in the pigment epithelium and retina of the rat. Neuroscience. 1999;90:319–331. doi: 10.1016/s0306-4522(98)00427-8. [DOI] [PubMed] [Google Scholar]
- Bessa J, Gebelein B, Pichaud F, Casares F, Mann RS. Combinatorial control of Drosophila eye development by eyeless, homothorax, and teashirt . Genes & Development. 2002;16:2415–2427. doi: 10.1101/gad.1009002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlton-Perkins M, Cook TA. Building a fly eye: terminal differentiation events of the retina, corneal lens, and pigmented epithelia. Current topics in developmental biology. 2010;93:129–173. doi: 10.1016/B978-0-12-385044-7.00005-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins JJ, 3rd, Hou X, Romanova EV, Lambrus BG, Miller CM, Saberi A, Sweedler JV, Newmark PA. Genome-wide analyses reveal a role for peptide hormones in planarian germline development. PLoS Biology. 2011;8:e1000509. doi: 10.1371/journal.pbio.1000509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis RJ, Harding M, Moayedi Y, Mardon G. Mouse Dach1 and Dach2 are redundantly required for Mullerian duct development. Genesis. 2008;46:205–213. doi: 10.1002/dvg.20385. [DOI] [PubMed] [Google Scholar]
- del Marmol V, Ito S, Bouchard B, Libert A, Wakamatsu K, Ghanem G, Solano F. Cysteine deprivation promotes eumelanogenesis in human melanoma cells. The Journal of investigative dermatology. 1996;107:698–702. doi: 10.1111/1523-1747.ep12365591. [DOI] [PubMed] [Google Scholar]
- Fain GL, Hardie R, Laughlin SB. Phototransduction and the evolution of photoreceptors. Curr Biol. 2010;20:R114–R124. doi: 10.1016/j.cub.2009.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frolenkov GI, Belyantseva IA, Friedman TB, Griffith AJ. Genetic insights into the morphogenesis of inner ear hair cells. Nature Reviews Genetics. 2004;5:489–498. doi: 10.1038/nrg1377. [DOI] [PubMed] [Google Scholar]
- Gagnon LH, Longo-Guess CM, Berryman M, Shin JB, Saylor KW, Yu H, Gillespie PG, Johnson KR. The chloride intracellular channel protein CLIC5 is expressed at high levels in hair cell stereocilia and is essential for normal inner ear function. J Neurosci. 2006;26:10188–10198. doi: 10.1523/JNEUROSCI.2166-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garger AV, Richard EA, Lisman JE. The excitation cascade of Limulus ventral photoreceptors: guanylate cyclase as the link between InsP3-mediated Ca2+ release and the opening of cGMP-gated channels. BMC neuroscience. 2004;5:7. doi: 10.1186/1471-2202-5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giblin FJ. Glutathione: a vital lens antioxidant. J Ocul Pharmacol Ther. 2000;16:121–135. doi: 10.1089/jop.2000.16.121. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Sastre A, Molina MD, Salo E. Inhibitory Smads and bone morphogenetic protein (BMP) modulate anterior photoreceptor cell number during planarian eye regeneration. The International journal of developmental biology. 2012;56:155–163. doi: 10.1387/ijdb.123494ag. [DOI] [PubMed] [Google Scholar]
- Hase S, Wakamatsu K, Fujimoto K, Inaba A, Kobayashi K, Matsumoto M, Hoshi M, Negishi S. Characterization of the pigment produced by the planarian, Dugesia ryukyuensis . Pigment cell research / sponsored by the European Society for Pigment Cell Research and the International Pigment Cell Society. 2006;19:248–249. doi: 10.1111/j.1600-0749.2006.00306.x. [DOI] [PubMed] [Google Scholar]
- Heine P, Dohle E, Bumsted-O’Brien K, Engelkamp D, Schulte D. Evidence for an evolutionary conserved role of homothorax/Meis1/2 during vertebrate retina development. Development. 2008;135:805–811. doi: 10.1242/dev.012088. [DOI] [PubMed] [Google Scholar]
- Hisa T, Spence SE, Rachel RA, Fujita M, Nakamura T, Ward JM, Devor-Henneman DE, Saiki Y, Kutsuna H, Tessarollo L, et al. Hematopoietic, angiogenic and eye defects in Meis1 mutant animals. EMBO. 2004;23:450–459. doi: 10.1038/sj.emboj.7600038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter DG, Fishman GA, Mehta RS, Kretzer FL. Abnormal sperm and photoreceptor axonemes in Usher’s syndrome. Archives of ophthalmology. 1986;104:385–389. doi: 10.1001/archopht.1986.01050150085033. [DOI] [PubMed] [Google Scholar]
- Katz B, Minke B. Drosophila photoreceptors and signaling mechanisms. Frontiers in cellular neuroscience. 2009;3:2. doi: 10.3389/neuro.03.002.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajiri S, Fukumoto K, Hata M, Sasaki H, Katsuno T, Nakagawa T, Ito J, Tsukita S, Tsukita S. Radixin deficiency causes deafness associated with progressive degeneration of cochlear stereocilia. The Journal of Cell Biology. 2004;166:559–570. doi: 10.1083/jcb.200402007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapan SW, Reddien PW. dlx and sp6-9 Control optic cup regeneration in a prototypic eye. PLoS Genetics. 2011;7:e1002226. doi: 10.1371/journal.pgen.1002226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Dai Q, Li L, Nair M, Mackay DR, Dai X. Ovol2, a mammalian homolog of Drosophila ovo: gene structure, chromosomal mapping, and aberrant expression in blind-sterile mice. Genomics. 2002;80:319–325. doi: 10.1006/geno.2002.6831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, la Cour M, Andersen MV, Miller SS. Proton-lactate cotransport in the apical membrane of frog retinal pigment epithelium. Experimental eye research. 1994;59:679–688. doi: 10.1006/exer.1994.1153. [DOI] [PubMed] [Google Scholar]
- Lin H, Miller SS. pHi regulation in frog retinal pigment epithelium: two apical membrane mechanisms. The American Journal of Physiology. 1991;261:C132–C142. doi: 10.1152/ajpcell.1991.261.1.C132. [DOI] [PubMed] [Google Scholar]
- Mackay DR, Hu M, Li B, Rheaume C, Dai X. The mouse Ovol2 gene is required for cranial neural tube development. Developmental biology. 2006;291:38–52. doi: 10.1016/j.ydbio.2005.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannini L, Deri P, Picchi J, Batistoni R. Expression of a retinal homeobox (Rx) gene during planarian regeneration. The International journal of developmental biology. 2008;52:1113–1117. doi: 10.1387/ijdb.082616lm. [DOI] [PubMed] [Google Scholar]
- Mannini L, Rossi L, Deri P, Gremigni V, Salvetti A, Salo E, Batistoni R. Djeyes absent (Djeya) controls prototypic planarian eye regeneration by cooperating with the transcription factor Djsix-1 . Developmental biology. 2004;269:346–359. doi: 10.1016/j.ydbio.2004.01.042. [DOI] [PubMed] [Google Scholar]
- Marmorstein AD, Cross HE, Peachey NS. Functional roles of bestrophins in ocular epithelia. Progress in retinal and eye research. 2009;28:206–226. doi: 10.1016/j.preteyeres.2009.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Duran JM, Monjo F, Romero R. Morphological and molecular development of the eyes during embryogenesis of the freshwater planarian Schmidtea polychroa . Development Genes and Evolution. 2012 doi: 10.1007/s00427-012-0389-5. [DOI] [PubMed] [Google Scholar]
- Mastore M, Kohler L, Nappi AJ. Production and utilization of hydrogen peroxide associated with melanogenesis and tyrosinase-mediated oxidations of DOPA and dopamine. FEBS journal. 2005;272:2407–2415. doi: 10.1111/j.1742-4658.2005.04661.x. [DOI] [PubMed] [Google Scholar]
- Matsushima D, Heavner W, Pevny LH. Combinatorial regulation of optic cup progenitor cell fate by SOX2 and PAX6. Development. 2011;138:443–454. doi: 10.1242/dev.055178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mburu P, Kikkawa Y, Townsend S, Romero R, Yonekawa H, Brown SD. Whirlin complexes with p55 at the stereocilia tip during hair cell development. PNAS. 2006;103:10973–10978. doi: 10.1073/pnas.0600923103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mburu P, Romero MR, Hilton H, Parker A, Townsend S, Kikkawa Y, Brown SD. Gelsolin plays a role in the actin polymerization complex of hair cell stereocilia. PloS one. 2010;5:e11627. doi: 10.1371/journal.pone.0011627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee A, Shan X, Mutsuddi M, Ma Y, Nambu JR. The Drosophila sox gene, fish-hook, is required for postembryonic development. Developmental biology. 2000;217:91–106. doi: 10.1006/dbio.1999.9506. [DOI] [PubMed] [Google Scholar]
- Newmark PA, Sánchez Alvarado A. Bromodeoxyuridine specifically labels the regenerative stem cells of planarians. Developmental biology. 2000;220:142–153. doi: 10.1006/dbio.2000.9645. [DOI] [PubMed] [Google Scholar]
- Nilsson DE. The evolution of eyes and visually guided behaviour. Philosophical transactions of the Royal Society of London. 2009;364:2833–2847. doi: 10.1098/rstb.2009.0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura K, Kitamura Y, Inoue T, Umesono Y, Yoshimoto K, Takeuchi K, Taniguchi T, Agata K. Identification and distribution of tryptophan hydroxylase (TPH)-positive neurons in the planarian Dugesia japonica . Neuroscience Research. 2007;59:101–106. doi: 10.1016/j.neures.2007.05.014. [DOI] [PubMed] [Google Scholar]
- Pearson BJ, Eisenhoffer GT, Gurley KA, Rink JC, Miller DE, Sánchez Alvarado A. Formaldehyde-based whole-mount in situ hybridization method for planarians. Dev Dyn. 2009;238:443–450. doi: 10.1002/dvdy.21849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pineda D, Gonzalez J, Callaerts P, Ikeo K, Gehring WJ, Salo E. Searching for the prototypic eye genetic network: Sine oculis is essential for eye regeneration in planarians. PNAS. 2000;97:4525–4529. doi: 10.1073/pnas.97.9.4525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pineda D, Rossi L, Batistoni R, Salvetti A, Marsal M, Gremigni V, Falleni A, Gonzalez-Linares J, Deri P, Salo E. The genetic network of prototypic planarian eye regeneration is Pax6 independent. Development. 2002;129:1423–1434. doi: 10.1242/dev.129.6.1423. [DOI] [PubMed] [Google Scholar]
- Pineda D, Salo E. Planarian Gtsix3, a member of the Six/so gene family, is expressed in brain branches but not in eye cells. Mechanisms of development. 2002;1(119 Suppl):S167–S171. doi: 10.1016/s0925-4773(03)00111-4. [DOI] [PubMed] [Google Scholar]
- Reddien PW, Oviedo NJ, Jennings JR, Jenkin JC, Sánchez Alvarado A. SMEDWI-2 is a PIWI-like protein that regulates planarian stem cells. Science. 2005;310:1327–1330. doi: 10.1126/science.1116110. [DOI] [PubMed] [Google Scholar]
- Reddien PW, Sánchez Alvarado A. Fundamentals of planarian regeneration. Annual Review of Cell and Developmental Biology. 2004;20:725–757. doi: 10.1146/annurev.cellbio.20.010403.095114. [DOI] [PubMed] [Google Scholar]
- Sánchez Alvarado A, Newmark PA. Double-stranded RNA specifically disrupts gene expression during planarian regeneration. PNAS. 1999;96:5049–5054. doi: 10.1073/pnas.96.9.5049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schallreuter KU, Kothari S, Chavan B, Spencer JD. Regulation of melanogenesis--controversies and new concepts. Experimental dermatology. 2008;17:395–404. doi: 10.1111/j.1600-0625.2007.00675.x. [DOI] [PubMed] [Google Scholar]
- Self T, Sobe T, Copeland NG, Jenkins NA, Avraham KB, Steel KP. Role of myosin VI in the differentiation of cochlear hair cells. Developmental biology. 1999;214:331–341. doi: 10.1006/dbio.1999.9424. [DOI] [PubMed] [Google Scholar]
- Sexton T, Buhr E, Van Gelder RN. Melanopsin and mechanisms of non-visual ocular photoreception. The Journal of Biological Chemistry. 2012;287:1649–1656. doi: 10.1074/jbc.R111.301226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver SJ, Rebay I. Signaling circuitries in development: insights from the retinal determination gene network. Development. 2005;132:3–13. doi: 10.1242/dev.01539. [DOI] [PubMed] [Google Scholar]
- Singhal SS, Godley BF, Chandra A, Pandya U, Jin GF, Saini MK, Awasthi S, Awasthi YC. Induction of glutathione S-transferase hGST 5.8 is an early response to oxidative stress in RPE cells. Investigative ophthalmology & visual science. 1999;40:2652–2659. [PubMed] [Google Scholar]
- Smyth JT, Hwang SY, Tomita T, DeHaven WI, Mercer JC, Putney JW. Activation and regulation of store-operated calcium entry. Journal of cellular and molecular medicine. 2010;14:2337–2349. doi: 10.1111/j.1582-4934.2010.01168.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss O. The retinal pigment epithelium in visual function. Physiological reviews. 2005;85:845–881. doi: 10.1152/physrev.00021.2004. [DOI] [PubMed] [Google Scholar]
- Tessmar-Raible K, Arendt D. Emerging systems: between vertebrates and arthropods, the Lophotrochozoa. Current opinion in genetics & development. 2003;13:331–340. doi: 10.1016/s0959-437x(03)00086-8. [DOI] [PubMed] [Google Scholar]
- Umesono Y, Watanabe K, Agata K. Distinct structural domains in the planarian brain defined by the expression of evolutionarily conserved homeobox genes. Development Genes and Evolution. 1999;209:31–39. doi: 10.1007/s004270050224. [DOI] [PubMed] [Google Scholar]
- Unezaki S, Horai R, Sudo K, Iwakura Y, Ito S. Ovol2/Movo, a homologue of Drosophila ovo, is required for angiogenesis, heart formation and placental development in mice. Genes Cells. 2007;12:773–785. doi: 10.1111/j.1365-2443.2007.01084.x. [DOI] [PubMed] [Google Scholar]
- Venza I, Visalli M, Cucinotta M, Teti D, Venza M. Association between oxidative stress and macromolecular damage in elderly patients with age-related macular degeneration. Aging clinical and experimental research. 2011;24:21–27. doi: 10.3275/7659. [DOI] [PubMed] [Google Scholar]
- Vopalensky P, Kozmik Z. Eye evolution: common use and independent recruitment of genetic components. Philosophical transactions of the Royal Society of London. 2009;364:2819–2832. doi: 10.1098/rstb.2009.0079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner DE, Wang IE, Reddien PW. Clonogenic neoblasts are pluripotent adult stem cells that underlie planarian regeneration. Science. 2011;332:811–816. doi: 10.1126/science.1203983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Montell C. Phototransduction and retinal degeneration in Drosophila . Pflugers Arch. 2007;454:821–847. doi: 10.1007/s00424-007-0251-1. [DOI] [PubMed] [Google Scholar]
- Williams DS. Usher syndrome: animal models, retinal function of Usher proteins, and prospects for gene therapy. Vision research. 2008;48:433–441. doi: 10.1016/j.visres.2007.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood JM, Decker H, Hartmann H, Chavan B, Rokos H, Spencer JD, Hasse S, Thornton MJ, Shalbaf M, Paus R, et al. Senile hair graying: H2O2-mediated oxidative stress affects human hair color by blunting methionine sulfoxide repair. Faseb J. 2009;23:2065–2075. doi: 10.1096/fj.08-125435. [DOI] [PubMed] [Google Scholar]
- Xiao Q, Hartzell HC, Yu K. Bestrophins and retinopathies. Pflugers Arch. 2010;460:559–569. doi: 10.1007/s00424-010-0821-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber ME, Gestri G, Viczian AS, Barsacchi G, Harris WA. Specification of the vertebrate eye by a network of eye field transcription factors. Development. 2003;130:5155–5167. doi: 10.1242/dev.00723. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.