Abstract
The use of iron as an enzymatic cofactor is pervasive in biological systems. Consequently most living organisms, including pathogenic bacteria, require iron to survive and replicate. To combat infection vertebrates have evolved sophisticated iron sequestration systems against which, pathogenic bacteria have concomitantly evolved equally elaborate iron acquisition mechanisms.
Keywords: Bacterial pathogenesis, Staphylococcus aureus, Isd system, nutritional immunity, Staphylococcal iron acquisition
1. Introduction
Staphylococcus aureus is a Gram-positive coccoid bacterium that innocuously colonizes the anterior nares of approximately 30% of the human population [1]. However, upon breaching this initial site of colonization, S. aureus has the capacity to infect nearly every tissue in the human body. Consequently, S. aureus can cause a spectrum of ailments ranging from superficial wound infections to more severe diseases such as septicemia, toxic shock syndrome and endocarditis [2]. S. aureus is the most frequent cause of nosocomial infections in the United States, with the percentage of infections caused by methicillin resistant Staphylococcus aureus (MRSA) growing precipitously [3]. The clinical challenge presented by MRSA is underscored by the fact that within the United States S. aureus is responsible for more deaths than HIV [4]. Furthermore, S. aureus has a remarkable capacity to resist currently available antimicrobials. Finally, in the last two decades there has been a dramatic increase in community acquired MRSA infections within populations that have not had identifiable exposure to health care institutions [2]. These facts highlight the need for the identification of new targets for the development of antimicrobials to treat this infectious threat [2].
A promising strategy to combat bacterial infections is to inhibit the procurement of nutrients that are necessary for growth. Iron is required by nearly all living organisms and S. aureus is no exception. The vertebrate host tightly regulates iron levels and sequesters this valuable nutrient intracellularly as a mechanism to prevent bacterial proliferation. Extracellular iron is rapidly removed by transferrin and lactoferrin, proteins with a high affinity for iron. Furthermore, the majority of iron within vertebrates is complexed to the porphyrin heme which is bound by hemoproteins. This process of limiting access to nutrient metal is known as nutritional immunity [5]. S. aureus responds to the iron-restricted environment of the host through the coordinated up-regulation of iron acquisition systems and other virulence factors. This review addresses the dynamic relationship between host-mediated iron sequestration and staphylococcal iron acquisition strategies.
2. Nutritional Immunity
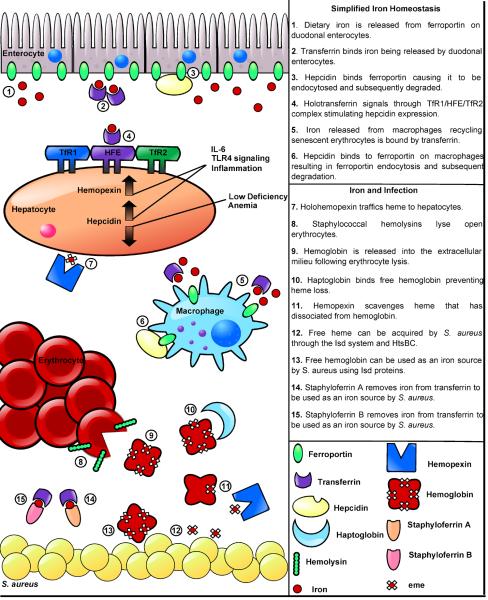
Nutritional immunity is a complex process requiring the synchronization of multiple enzymes involved in host iron regulation. Iron is insoluble at physiologic pH found within vertebrate tissues and any free iron is quickly removed by high-affinity iron binding proteins. Within the serum, free iron is bound by transferrin with an association constant of approximately 1036 [5]. In addition to binding free iron, transferrin functions as an iron transport protein, transferring iron to peripheral tissues through receptor-mediated endocytosis of transferrin receptor 1 (TfR1) upon complexation with holo-transferrin (Fig. 1) [6]. Iron levels are also limited in lymph and mucosal secretions through lactoferrin, which quickly binds all free iron. Additionally, lactoferrin is a major component within phagocytes, ensuring that engulfed pathogens have limited access to intracellular iron. In healthy individuals both lactoferrin and transferrin are only 30–40% saturated and consequently are poised to bind free iron [5].
Figure 1.
Iron homeostasis in health and disease
The overwhelming majority of iron within humans is found in the form of heme; a tetrapyrrole ring with a coordinated iron center. Heme is often bound by hemoproteins, the most abundant of which is hemoglobin. To further prevent access to iron, hemoglobin is sequestered intracellularly within erythrocytes; greater than 90% of iron within the human body is located intracellularly making it inaccessible to extracellular pathogens unless mechanisms are employed to liberate these rich sources of nutrient iron. Although hemoglobin is the most abundant hemoprotein in the body, there are additional heme-binding proteins that can serve as sources of iron for invading bacteria. Haptoglobin is a tetrachain (α2β2) glycoprotein that binds free hemoglobin following hemolysis as a means to prevent loss of iron through urinary excretion and subsequent kidney damage [7]. S. aureus binds haptoglobin-hemoglobin complexes in vitro, suggesting that this protein complex is exploited as a source of nutrient iron [8]. Myoglobin is an additional hemoprotein that is found within myocytes. S. aureus can utilize myoglobin as an iron source in vitro and this protein may represent a source of iron during colonization of muscle tissue [9]. While it is unclear if S. aureus utilizes myoglobin during infection, it is likely that the tissue damage that occurs during skin and soft tissue infections would provide S. aureus with significant access to this rich heme-iron source. In support of this contention, S. aureus readily colonizes the heart and damages surrounding cardiac myocytes [10].
Cytochromes are additional heme-binding proteins that are involved in electron transport and ATP synthesis. In eukaryotes cytochromes are localized within the mitochondria rendering them inaccessible to extracellular bacteria, and there is no precedence for the use of cytochromes as an iron source by pathogens. The ubiquitous nature of cytochromes in biological systems indicates that the ability to access cytochromes and remove the heme cofactor would be advantageous in numerous host niches. Clearly, multiple host hemoproteins have the potential to serve as iron sources to bacteria that colonize and infect vertebrates.
Nutritional immunity is a dynamic process that is capable of responding to assaults from invading bacteria. A primary coordinator of this response is the hepatic peptide hormone hepcidin, which regulates iron absorption and the distribution of iron within tissues. All iron within the body, whether it is dietary iron entering through duodenal enterocytes, or iron within macrophages that have recycled senescent erythrocytes, must leave the cells to enter circulating plasma. Although the proteins involved in intracellular trafficking are not well defined, it appears that elemental iron exits cells exclusively through the iron exporter ferroportin (Fig. 1) [11]. Upon binding to ferroportin, hepcidin causes ferroportin endocytosis and subsequent proteolytic degradation decreasing the release of iron into the plasma [12]. Hepcidin secretion by hepatocytes is itself iron-regulated and functions to maintain iron homeostasis within healthy individuals. Inflammation is one of the many stimulatory signals that affect hepcidin concentrations within the serum. Specifically, induction of IL-6 production and LPS-mediated TLR4 signaling both cause increases in circulating hepcidin (Fig. 1) [12]. Inflammation-induced increases in hepcidin represent a hypoferremic response to infection which allows the host to respond to bacterial assault by further reducing available iron and preventing bacterial replication [12]. Together hepcidin, transferrin, haptoglobin, and lactoferrin function to maintain an extracellular environment free of elemental iron.
In addition to quickly removing extracellular iron, vertebrates rapidly remove heme from plasma via hemopexin, a glycoprotein expressed mainly in the liver [13]. Hemopexin scavenges circulating heme that is generated through various processes including enucleation of mature erythroblasts, the release of heme cofactors caused by the oxidation of free hemoglobin, and dietary heme absorption [13]. Hemopexin binds free heme and delivers it to the liver where it can be taken up by hepatic parenchymal cells through receptor-mediated endocytosis (Fig. 1) [13]. When vertebrates experience a significant amount of hemolysis, the sudden increase in free hemoglobin can overwhelm available levels of haptoglobin causing the oxidation of free hemoglobin and release of heme. Under these circumstances hemopexin is aided by the serum protein albumin which also binds free heme; however it is unclear if albumin can deliver heme to the liver [13]. Although the bacterial use of albumin as a source of iron has not been clearly established, albumin exists in the highest concentration of any serum protein making it a viable target for iron acquisition systems [13]. As an acute-phase protein, hemopexin concentrations increase during inflammation and are regulated by the cytokines IL-6, IL-11 IL-1β, leukemia inhibitory factor, and tumor necrosis factor (TNF)-α [14]. The increase in hemopexin levels during infection counteracts the hemolytic activity of pathogenic bacteria. In summary, iron homeostasis within the human host requires the integration of multiple signals resulting in an environment that is iron-deplete and able to respond to bacterial assault.
3. Disruption of nutritional immunity impacts the outcome of infection
The human body allocates significant resources towards the sequestration of iron however genetic mutations exist within the human population that oppose the protective effects of nutritional immunity. Hereditary hemochromatosis (HH) is caused by mutations within the HFE gene with the two most common mutations being C282Y and H63D. HFE is a membrane protein that binds to both the TfR1 and TfR2 transferrin receptor [12]. Reduced HFE function results in decreased hepcidin responsiveness and is characterized by excessive iron in the liver and tissues and increased transferrin saturation [15]. This increase in transferrin saturation decreases the bacteriostatic nature of serum as it is easier for pathogens to attain iron from transferrin [15]. Consequently HH is associated with an increased risk of both bacterial and viral infections [15]. Moreover, infections of HH patients can occur with microbial strains that are typically avirulent [15]. HH is further complicated by the fact that iron overload compromises the efficacy of phagocytic cells by decreasing their chemotactic capabilities and increasing the ease with which engulfed bacteria can proliferate intracellularly within macrophages [15]. Together these facts underscore the importance of maintaining iron homeostasis for protection against infection.
β Thalassaemia is an inherited disorder affecting hemoglobin synthesis. Mutations within the β globin gene cause diverse degrees of defective β chain production, ineffective erythropoiesis, improper α/βchain ratios, iron overload, and a range of anemia [16]. The phenotypic manifestation of β thalassaemia is extremely diverse, ranging from the clinically silent β thalassaemia minor to the more severe β thalassaemia major, which is characterized by profound anemia. Iron overload occurs in β thalassaemia due to the increased iron demand caused by dysfunctional erythropoiesis [16]. Patients with β thalassaemia typically require medical attention within the first year of life and are subsequently required to receive regular blood transfusions and iron chelation therapy [17–18]. Infections are among the major causes of mortality in patients with thalassaemia (12–46%) along with cardiovascular disease and liver failure [18]. Common infections in thalassaemia patients are pneumonia, bilary tract infection, soft tissue infection, septicemia and liver abscesses [18]. Iron overload associated with thalassaemia patients has been implicated as a cause for the increased infections within this population [18].
4. Changes in staphylococcal metabolism in response to iron starvation
The ability to proliferate within the iron-restricted environment of the host requires a synchronized up-regulation of genes dedicated to the acquisition of iron. This orchestrated response mandates the detection of physiological changes in the environment and is critical for successful infection. Upon entering host tissues S. aureus responds to the dearth of iron by dramatically altering its gene expression profile. This change in gene expression is mediated by the canonical iron-dependent repressor Fur (ferric-uptake regulator) [9, 19]. In the presence of iron, Fur binds a consensus DNA sequence referred to as a fur box located upstream of the transcriptional start site of iron-regulated genes. The binding of Fur to the fur box typically inhibits the transcription of target genes. During conditions of iron starvation, Fur repression is alleviated and transcription is activated. The release of Fur-mediated repression during times of iron starvation results in a coordinated up-regulation of a number of genes involved in iron acquisition, glycolysis and virulence [19].
The importance of responding to iron limitation is highlighted by the diversity of genes that are controlled by Fur in an iron-dependent manner [19]. Many of the proteins within the Fur regulon are involved in central metabolic pathways suggesting that iron starvation triggers a metabolic shift in S. aureus. Specifically, either iron starvation or fur inactivation leads to a significant reduction in the abundance of numerous enzymes of the TCA cycle and a simultaneous increase in the abundance of proteins within the glycolytic pathway [19]. For example, fructose 1-P kinase (FruB), fructose biphosphate aldolase (FbaA), triosephosphate isomerase (Tpi), glyceraldehyde aldolase (FbaA), glyceraldehyde 3-phosphate dehydrogenase (Gap) and transketolase (Tkt) each increase in abundance upon iron starvation. This up-regulation facilitates a systemic activation of glycolysis resulting in increased pyruvate production. Pyruvate is subsequently used as a substrate for fermentative metabolism due to the down regulation of the TCA cycle enzymes succinate dehydrogenase (SdhA), aconitate hydratase (CitB), fumarate hydratase (CitG) and phosphoenolypyruvate carboxykinase (PckA) [19]. This metabolic redirection results in the accumulation of the fermentative end product lactate. Indeed, both Δfur mutants and wildtype S. aureus grown in iron-deplete media produce markedly more lactate than wildtype bacteria grown in iron-replete conditions, leading to a decrease in the local pH [19]. Transferrin-iron is a viable iron source for S. aureus during infection and iron dissociates from transferrin in acidic conditions. In fact, media from iron starved or Δfur staphylococcal cultures significantly increases the rate at which transferrin releases iron, providing the organism with a more readily utilized source of iron [19]. Therefore, shifting to fermentative metabolism benefits iron starved S. aureus by increasing lactate production which decreases the surrounding pH and increases the local levels of free iron. Moreover, SdhA, CitB and CitG all contain iron-sulfur clusters; therefore down-regulating the TCA cycle may decrease the dependence of S. aureus on enzymes that utilize iron.
5. Fur-mediated expression of virulence factors
Iron restriction signals entry into host tissues and triggers the secretion of a variety of virulence factors. These virulence factors include hemolysins that increase erthryocyte lysis and supply liberated hemoglobin to the iron regulated surface determinant system (Isd) for use as an iron source [9, 20–22]. Other staphylococcal exotoxins that are Fur-regulated include hydrolases such as cytolysins, nucleases, proteases, and lipases as well as proteins known to modulate the host immune response such as chemotaxis inhibiting protein (CHIP) and staphylococcal complement inhibitor (SCIN) [21]. Many virulence factors are positively regulated by Fur such that they are down-regulated in a Δfur mutant. These virulence factors include coagulase, superantigen exotoxins (Ssl 1, Ssl 2, Ssl 6, Ssl 7, Ssl 9 and Ssl 11), and two virulence factors known as Eap and Emp that are involved in biofilm formation [21]. Eap and Emp non-covalently associate with the cell wall and promote staphylococcal adhesion to host proteins fibronectin, fibrinogen, vitronectin, and collagen [23]. Interestingly, Eap was found to decrease neutrophil and T-cell recruitment by binding to endothelial ICAM-1, preventing its interaction with leukocyte LFA-1 [24–25]. The expression of Eap and Emp is not only dependent on Fur but also on the global regulators Agr, SarA and Sae underscoring the diverse signals that direct biofilm formation [26].
Fur-dependent gene regulation coordinates a staphylococcal assault on the host. In fact, S. aureus strains inactivated for fur exhibit a severe virulence defect in animal models of infection [21]. In a murine model of pneumonia, S. aureus fur mutants induce greater neutrophil recruitment despite a decreased ability to colonize and proliferate within the lung. Interestingly, S. aureus fur mutants are still able to proliferate in the lungs of neutropenic mice suggesting that Fur controls an anti-neutrophil response that protects S. aureus during infection of the vertebrate lung [21]. As exemplified from the numerous proteins that change abundance upon iron depletion, host-mediated iron sequestration is a signal that is sensed by S. aureus to alter a variety of processes that facilitate vertebrate infection.
6. The physiological role of iron in S. aureus
The conserved requirement for iron is due to the large number of proteins that utilize the versatile redox potential of this metal [27]. Like other transition metals, the redox potential of iron is greatly influenced by the surrounding protein environment allowing it to function as an oxidizing or reducing agent [27]. While a single iron atom can be used as an enzymatic cofactor, iron is often used within iron-sulfur clusters or heme groups to enable enzyme function [27].
Although the utility of iron-sulfur clusters within membrane-bound cytochromes is well established, the importance of these clusters in detecting environmental changes is only beginning to emerge. Regulatory proteins frequently utilize iron-sulfur clusters to mediate cellular responses to specific oxidants such as atmospheric oxygen, reactive oxygen species (ROS) and nitric oxide. For example, the transcription factor FNR exploits the potential of ROS to oxidize its iron-sulfur cluster, leading to FNR disassembly. Under anaerobic conditions, Escherichia coli FNR is populated with an iron sulfur cluster that promotes the dimerization and subsequent site-specific DNA-binding of FNR. Under aerobic conditions, FNR-mediated transcriptional regulation is reversed when the iron-sulfur cluster is oxidized and dissociates [28]. SoxR is a transcriptional regulator that was initially discovered for its role in sensing superoxide stress, and has since been shown to detect changes in NO [28–29]. As with FNR, SoxR is active as a homodimer and inactive as a monomer, however it is not the loss of the iron-sulfur cluster that induces the change from dimer to monomer but the redox state of the cluster [30]. The detection of NO by SoxR is the result of the nitrosylation of the iron sulfur cluster [29]. While it is unclear if S. aureus encodes a homolog of SoxR, the FNR-like transcriptional regulator ArcR has been identified within S. aureus [31]. ArcR up-regulates expression of the arginine deiminase operon under anaerobic conditions although it is unclear if the staphylococcal ArcR utilizes an iron-sulfur cluster to sense cellular oxygen levels [31].
Iron-sulfur clusters are also used as a cofactor in the bifunctional enzyme, aconitase. Holo-aconitase catalyzes the reversible isomerization of citrate to isocitrate in the TCA cycle [29]. However, the apoform of aconitase, denoted IRP (iron regulatory protein), binds and stabilizes specific mRNAs and prevents their translation [29]. The translational regulating activity of aconitase is also iron regulated as the pool of apoprotein is dependent upon intracellular iron levels as well as oxidative stress [29].
Iron-sulfur clusters are also used within the NreABC two component regulatory system to detect the environmental availability of oxygen, nitrate or nitrite all of which can be used as terminal electron acceptors in the electron transport chain of S. aureus [32]. Two component systems typically consist of a histidine sensor kinase (NreB) and a response regulator (NreC) [33–34]. Under aerobic conditions the iron-sulfur cluster in NreB is lost, resulting in a significant decrease in the autophosphorylation of NreB and subsequent phosphate transfer to NreC. When oxygen is not present the phosphotransfer from NreB to NreC results in the up-regulation of both nitrate and nitrite reductase operons as well as the nitrite extrusion protein (NarK) [33]. Furthermore, NreC derepresses the transcription of genes within the fermentative pathway such as dissimilatory lactate dehydrogenase (lctE) [33]. In this way, the two component system of NreABC utilizes an iron-sulfur cluster to detect alterations in the availability of electron acceptors and responds by up-regulating genes involved in the dissimilarity reduction of nitrate and nitrite [33]. It is apparent that the sensitivity of iron-sulfur clusters to environmental oxidants makes them ideally suited for use within bacterial two component systems.
The redox potential of heme is particularly useful for a variety of proteins that function to transfer electrons between molecules. For example cytochromes are electron transport proteins with heme prosthetic groups [34]. Cytochromes are used during aerobic and anaerobic respiration whereby ATP formation is coupled to the oxidation of reduced substrates [34]. Due to the importance of heme to staphylococcal physiology, this organism not only encodes for heme acquisition systems but can also synthesize heme de novo [35]. In fact, when the heme synthesis pathway is ablated in S. aureus through the inactivation of hemB, the result is an aberrant growth phenotype which is known as a small colony variant (SCV) [36]. SCVs have a distinct pleiotropic phenotype characterized by a slow growth rate, lack of pigmentation, reduced spectrum of carbohydrate usage, and a general lack of virulence factor production [36]. SCVs are clinically relevant as they are associated with an increase in staphylococcal persistence and obfuscate the etiology of infection which may alter the choice of antibiotic prescribed [36]. The SCV phenotype of a hemB mutant is presumably due to the uncoupling of electron transport and a resulting decrease in ATP production caused by a defect in cytochrome maturation [36]. The SCV phenotype of a hemB mutant can be reversed upon acquisition of exogenous heme [36]. In fact, even in iron replete conditions S. aureus acquires an appreciable amount of exogenous heme that can be used to populate membrane cytochromes [34, 37].
In addition to cytochromes, heme is a cofactor of nitric oxide synthetases (NOSs) and catalases. Together NOSs and catalase protect S. aureus against the antibacterial effects of reactive oxygen species (ROS) produced as part of the inflammatory response to infection [38–39]. The antibacterial activity of ROS is partly due to DNA and protein damage mediated by the Fenton reaction which creates hydroxyl radicals that react with DNA bases, sugar moieties, and amino acid side chains [40]. Catalase activity protects the cell by converting toxic hydrogen peroxide to water and oxygen. Within Bacillus subtilis, the production of nitric oxide (NO) by NOSs was found to activate a specific catalase that subsequently suppresses the deleterious effects of the Fenton reaction [38–39]. Furthermore production of NO by NOSs has been shown to provide protection from a wide range of antibiotics indicating that bacterial NOSs may be a potential target for new therapeutics.
7. Iron acquisition systems within S. aureus
While the vertebrate host has evolved sophisticated mechanisms by which it sequesters iron from invading pathogens, S. aureus has evolved equally sophisticated mechanisms to gain access to iron during infection. In addition to changes in central metabolism and virulence factor expression, Fur mediated regulation allows S. aureus to respond to iron starvation through the dramatic up-regulation of its iron acquisition systems. One such system is the iron regulated surface determinant (Isd) system which includes the cell wall anchored heme binding proteins (IsdA and IsdC), a hemoglobin receptor (IsdB), a hemoglobin-haptoglobin receptor (IsdH), a membrane transporter (IsdEF), and two cytoplasmic heme oxygenases (IsdG and IsdI) [22]. Additionally, the membrane transporter IsdEF has been predicted to act in concert with IsdD, however the function of IsdD has yet to be elucidated. The Isd system also includes sortase B, an enzyme dedicated to anchoring IsdC to the cell wall. Conversely, IsdA, IsdB and IsdH are anchored to the cell wall by sortase A [22, 41]. These proteins work in concert to enable S. aureus to utilize vertebrate hemoglobin as a source of nutrient iron.
IsdA, IsdB, IsdH and IsdC contain NEAr iron Transporter (NEAT) domains. NEAT domains are conserved stretches of amino acids that mediate heme and hemoprotein binding [42]. Heme-iron is bound within these clefts by a single axial tyrosine ligand [43]. The current model for heme acquisition via the Isd system proposes that IsdB and IsdH employ NEAT domains to initiate the process by binding hemoglobin or hemoglobin-haptoglobin, respectively. IsdB has two NEAT domains and it is the second domain that transfers heme to the single NEAT domain of either IsdA or IsdC. Heme is directly transferred from IsdA to IsdC in an affinity driven manner [44]. IsdA is unable to transfer heme to IsdE, however heme can pass directly from IsdC to IsdE [45]. Heme then traverses the cell membrane through the ABC transporter IsdEF. Upon entering the cytoplasm heme is degraded by the heme oxygenases IsdG, and IsdI [46]. The Isd system represents a paradigm of heme-iron acquisition, and illustrates a mechanism by which bacterial pathogens utilize hemoglobin as an iron source and traffic heme inside the cell.
The primary amino acid sequence of hemoglobin exhibits significant interspecies variation indicating that bacterial pathogens may bind hemoglobins from distinct species with varying affinity. S. aureus is predominantly a human pathogen and the S. aureus hemoglobin receptor IsdB binds human hemoglobin more efficiently than hemoglobin from other animal species [47]. In fact the KD of the interaction between recombinant IsdB and human hemoglobin is 5.5 × 10−8 as compared to a KD of 9.8 × 10−7 for the interaction between IsdB and mouse hemoglobin. The increased affinity of IsdB for human hemoglobin enables S. aureus to more effectively use human hemoglobin as an iron source as compared to hemoglobin from other species. Transgenic mice that express human hemoglobin are more susceptible to S. aureus infection than wildtype mice demonstrating the pathophysiological relevance of these findings. This indicates that the increased affinity of S. aureus IsdB for human hemoglobin translates to a more efficient use of hemoglobin-iron during infection [47]. Interestingly, bacterial species that primarily infect humans (Staphylococcus lugdunensis, Staphylococcus simulans, and Corynebacterium diphtheria) also display a preference for human hemoglobin over mouse hemoglobin while environmental bacterial species or less discriminate pathogens (Acinetobacter baumannii, Pseudomonas aeruginosa, Bacillus anthracis, and Bacillus cereus) do not exhibit the same hemoglobin preference [47]. Within the human population, hemoglobin is highly polymorphic and the amino acid sequence can vary at virtually any residue across individuals [48]. This raises the exciting possibility that human hemoglobin polymorphisms may impact the susceptibility of individuals to staphylococcal infections. Identifying specific hemoglobin residues that impact recognition by IsdB may allow for the identification of patients who are at an increased risk for bacterial infection. If successful, this advance could facilitate targeted prophylaxis and improved patient outcomes.
All identified heme oxygenases can be categorized into one of two families, the HO-1 family that is ubiquitous across kingdoms and the IsdG family that has only been identified in bacteria. HO-1 family heme oxygenases degrade heme to the blue-green molecule biliverdin which in vertebrates is further reduced to bilirubin by biliverdin reductase. In contrast, IsdG and IsdI degrade heme to the yellow oxo-bilirubin molecule known as staphylobilin [49]. Both bilirubin and biliverdin have potent antioxidant properties within humans however; the role of heme degradation products within bacteria has not been established. The importance of both bilirubin and biliverdin to human physiology makes it tempting to speculate that staphylobilin may serve important biological functions within bacteria. Considering the conservation of IsdG family members across numerous bacterial pathogens, identifying the function of staphylobilin may have broad implications to numerous infectious diseases.
Studies into IsdG from S. lugdunensis, a pathogenic coagulase-negative staphylococci, have revealed that staphylobilin production by IsdG family members is not restricted to S. aureus [50]. A recent survey of annotated bacterial genomes uncovered that many bacterial species which encode for a putative IsdG have no known association with any plant or animal from which they could acquire heme [50]. In fact, some of these IsdG-containing bacteria are considered extremophiles, occupying some of the earth's harshest environments. Interestingly, genomic analysis reveals that all IsdG encoding extremophiles also encode for a putative heme synthesis pathway. This raises the tempting possibility that IsdG is used to regulate endogenous heme synthesis and metabolism. Alternatively, these bacteria may synthesize heme specifically to have it catabolized to staphylobilin. The broad conservation of IsdG across multiple classes of bacteria underscores the importance of heme degradation for bacteria that acquire and synthesize heme. Despite the established utility of heme as an enzymatic cofactor and iron source, the contribution of heme homeostasis to staphylococcal pathogenesis remains poorly understood.
S. aureus encodes two seemingly redundant heme oxygenases, IsdG and IsdI. While both enzymes bind and degrade heme they are differentially regulated indicating a possible rationale for encoding two paralogous heme oxygenases [46, 51]. Both isdG and isdI transcript levels increase in iron-deplete conditions and are subjected to Fur-mediated regulation [51]. IsdG however, undergoes proteolytic degradation in the absence of the substrate heme [51]. This suggests that S. aureus differentially regulates IsdG and IsdI to precisely adjust the level of heme catabolism within the cell in response to alterations in cellular heme concentrations. Different host tissues likely have varying amounts of accessible iron and therefore S. aureus may vary in its dependency on iron from heme. Supporting this contention is the observation that S. aureus ΔisdG and ΔisdI mutants exhibit organ specific defects in virulence in a murine model of infection [51]. Interestingly, while both S. aureus ΔisdG and S. aureus ΔisdI are attenuated for virulence in murine hearts, only S. aureus ΔisdG is attenuated for virulence within murine kidneys. Taken together these data suggest that while both IsdG and IsdI are required for full virulence in S. aureus, and these enzymes are not functionally redundant during vertebrate infection [51].
IsdG and IsdI are 78% similar at the amino acid level yet they are differentially regulated through the proteolytic degradation of apo-IsdG. To avoid the accumulation of misfolded or redundant proteins, protease specificity is critical. This fact is underscored by the antimicrobial activity of acyldepsipeptides which render cytoplasmic proteases constitutively active [52]. Recently it was determined that an internal sequence located within the flexible loop region of IsdG is required for the heme-dependent degradation of this protein [53]. The process by which bacterial proteases recognize proteins that are destined for degradation remains poorly understood. Currently, the most well characterized mechanisms for substrate recognition by bacterial proteases are the N-end rule and the SsrA tag. The N-end rule relates the half-life of a protein to the N-terminal amino acid while the SsrA tag is an eleven amino acid tag that is added to the carboxy-terminus of incompletely translated proteins relegating them to degradation [54–55]. The targeted degradation of IsdG in the absence of heme does not occur through either the N-end rule or the SsrA tag as neither termini are required for proteolytic recognition [53]. Furthermore, none of the known staphylococcal proteases are responsible for the degradation of apo-IsdG, suggesting that S. aureus encodes an as-yet unidentified protease involved in protein turnover [53]. Identification of the protease responsible for the degradation of IsdG may help to elucidate the pathways involved in heme and iron homeostasis during infection.
IsdG-family heme oxygenases have been characterized in S. lugdunensis, Bacillus anthracis, Bradyrhizobium japonicum, Brucella melitensis, and Mycobacterium tuberculosis [46, 50, 56–58]. While IsdG orthologs from each of these bacteria have been shown to bind and degrade heme, differences do exist. S. lugdunensis encodes a single IsdG-family heme oxygenase which is iron regulated. Although S. lugdunensis IsdG and S. aureus IsdG are 68% identical at the amino acid level the S. lugdunensis enzyme is stable in the absence of heme. This indicates that structural differences may exist in S. lugdunensis IsdG that prevent it from being targeted for degradation. Alternatively, S. lugdunensis may not encode the protease responsible for the targeted degradation of S. aureus IsdG. Another unusual enzyme within the IsdG-family is the M. tuberculosis MhuD. Each MhuD monomer binds two heme molecules with the porphyrin rings stacked 3.45 Å apart [56]. This unusual diheme conformation does not affect the heme degrading capabilities of MhuD, however, as the product of MhuD heme degradation has not been determined; it is unclear if the diheme active site degrades heme to staphylobilin.
The use of heme and hemoglobin as an iron source was once thought to be restricted to pathogenic bacteria. The identification of heme transport systems and IsdG-family heme oxygenases within α–Proteobacteria revealed that these bacteria may acquire heme from non-vertebrate sources. Rhizobia such as B. japonicum can live as free-living soil organisms or as symbionts with leguminous plants. Rhizobia colonize the nitrogen fixing nodules of plants where they convert atmospheric nitrogen to ammonia aiding the plant in fulfilling its nitrogen requirement. Within these nodules B. japonicum acquires heme from leghemoglobin, a plant hemoglobin with structural properties similar to animal hemoglobins that can comprise up to 30% of the total soluble protein within plant nodules [59]. Together these facts illustrate that while there is great diversity in the lifestyle of bacteria encoding IsdG family heme oxygenases, the use of heme as an iron source is a ubiquitous strategy used by numerous bacteria to fulfill their nutritional requirements.
To achieve an extracellular environment virtually free of iron vertebrate hosts produce the iron-sequestering proteins transferrin and lactoferrin. To combat this sequestration S. aureus elaborates siderophores, small molecules with an impressively high affinity for iron. Siderophores target and remove iron that is bound to transferrin and lactoferrin. Siderophore production is Fur-regulated and consequently increases when the bacteria experience iron stress [19, 60]. Bacterial siderophores can be synthesized using two distinct pathways, the nonribosomal peptide synthetases (NRPS) pathway and the NRPS-independent (NIS) pathway. The NRPS pathway synthesizes siderophores using modular enzymatic platforms [61]. In contrast, the NIS pathway utilizes condensation reactions with units of dicarboxylic acids, diamines and amine alcohols to create the final siderophore structure [61]. S. aureus encodes for two siderophores, staphyloferrin A and staphyloferrin B, and both are synthesized through the NIS pathway and then secreted into the extracellular milieu. Siderophore-iron complexes are then transported back into the cytoplasm through dedicated ABC transporters [61].
Genes encoding for staphyloferrin A have been identified in all sequenced staphylococcal genomes. The sfa locus contains four genes, the three gene operon sfaABC and a divergently transcribed sfaD [60, 62]. The identification of a consensus Fur box located intergenically between sfaA and sfaD indicates that both sfA and sfaD are transcriptionally regulated by Fur. However, in some species of coagulase-negative staphylococci the sfa locus is believed to be constitutively expressed [60, 62]. Although staphyloferrin A production appears to be iron regulated the contribution of this molecule to staphylococcal iron acquisition is poorly defined. While a S. aureus strain with a chromosomal deletion of the sfa locus has no observable growth defect in vertebrate serum, the production of staphyloferrin A does appear to mitigate the growth defect of a staphyloferrin B mutant [62]. Together this suggests that staphyloferrin A contributes to staphylococcal transferrin-iron acquisition but is not the predominant means by which S. aureus procures iron from transferrin.
Staphyloferrin B is structurally distinct from staphyloferrin A but like staphyloferrin A is a hydrophilic NIS siderophore. Staphyloferrin B has been identified in the supernatants of S. aureus as well as several coagulase-negative staphylococci and Gram negative bacteria [61]. The genes encoding the enzymatic machinery required for staphyloferrin B synthesis lie within the sbn locus which has a consensus fur box within the operator region for the operon [61]. A S. aureus sbn mutant strain has impaired growth in serum, but achieves wildtype cell densities after extensive incubation, presumably due to the production of staphyloferrin A [62]. Upon binding iron, both staphyloferrin A and staphyloferrin B are transported into the cytoplasm through the ABC transporters HtsABC and the staphylococcal iron regulated transporter (SirABC), respectively [61]. Additionally, HtsBC has been found to transport heme into the cytoplasm suggesting that HtsABC may serve as a dual iron/heme transporter [37]. Siderophore production in combination with the Isd system provides S. aureus multiple routes for obtaining iron. A more thorough discussion on both siderophore synthesis and the contribution of siderophores to staphylococcal pathogenesis can be found in [61].
8. Sensing and alleviating heme toxicity
The importance of heme to staphylococcal physiology is emphasized by the numerous mechanisms used by bacteria to acquire or synthesize this molecule. Paradoxically, while heme has tremendous value as a biocatalyst is also toxic at high concentrations. Interestingly, not all bacteria are equally sensitive to heme toxicity with Gram positive bacteria generally being more sensitive than Gram negative bacteria. While the mechanism of heme toxicity remains poorly understood, it is well established that bacteria have dedicated systems in place that allow them to synthesize and acquire heme without succumbing to heme toxicity. This is evidenced by the fact that S. aureus is able to survive high concentrations of heme if previously exposed to sublethal heme concentrations [63]. This heme adaptation is mediated by a two component system known as the heme sensor system (hssRS). Like NreBC, HssRS is a paradigmatic two component system consisting of a histidine kinase (HssS) and a response regulator (HssR) [64].
Both HssR and HssS are required for heme adaptation and inactivating either hssR or hssS results in hyper-susceptibility to the lethal effects of heme. The specific ligand sensed by HssS remains unknown. While it is possible that HssS senses heme directly, there is a lack of similarity between the sensing domain of HssS and known heme binding motifs or established heme binding residues [64]. This raises the interesting possibility that HssS may sense heme indirectly, possibly sensing a metabolite produced specifically in response to heme exposure. Alternatively, HssS may bind heme through a completely novel mechanism. Heme exposure leads to the autophosphorylation of HssS which subsequently transphosphorylates the response regulator HssR. Once phosphorylated HssR binds to a direct repeat sequence within the promoter region of an ABC-type transporter system increasing its transcription. Since this ABC-type transporter is dramatically increased in response to heme it has been termed the heme regulated transporter (hrtAB) [19].
The HrtAB system is comprised of an ATPase (HrtA) and a permease (HrtB). Consistent with their assignment as the only known targets of HssRS, both HrtA and HrtB are required for protection against heme toxicity. In addition, mutants of HrtA that are unable to hydrolyze ATP render the bacteria susceptible to heme toxicity. This finding mechanistically links the ATPase activity of HrtA to the alleviation of heme toxicity. The ATPase activity of HrtA is influenced by many cellular factors including ATP concentrations, temperature, pH, and metal concentrations [65]. It remains unclear how HrtAB alleviates heme toxicity. The current model predicts that the combined action of HrtA and HrtB act as an efflux pump, removing heme or some toxic metabolite that accumulates upon heme exposure. Orthologous HrtAB systems have been identified in a number of species including L. monocytogenes, Bacillus thuringiensis,Bacillus cereus, Lactococcus lactis, Streptococcus agalactiae and S. epidermidis indicating that alleviation of heme toxicity is a common obstacle faced by many Gram positive bacteria [63, 66–67].
9. Clinical ramifications of targeting S. aureus iron acquisition systems
S. aureus is responsible for approximately 40,000 deaths per year in the United States and the incidence of multi-drug resistant strains is increasing, highlighting the need to develop new therapeutics [4]. The hemoglobin binding protein IsdB plays a pivotal role during infection and is surface exposed establishing this protein as a viable vaccine candidate [47]. Initial studies in a lethal S. aureus infection model have found that mice vaccinated with purified recombinant IsdB and adjuvant exhibited greater survival than mice that had been sham vaccinated with adjuvant alone [68]. The protective effect of the vaccine is likely due to the production of anti-IsdB antibodies as mice with higher anti-IsdB titers had a greater probability of surviving than those with low anti-IsdB titers [68]. Importantly, the vaccine was found to protect against both methicillin sensitive and resistant strains of S. aureus [68]. In a separate study affinity-purified anti-IsdB antibodies administered to mice prior to intravenous challenge with S. aureus were found to significantly reduce the bacterial load and number of abscesses formed within the kidney [69]. Furthermore, passive immunization using IsdB-specific IgG significantly improved survival outcomes of mice infected with S. aureus [69].
It remains unclear how the production of anti-IsdB antibodies provides host protection against staphylococcal challenge. Gram-positive bacteria are typically resistant to complement mediated lysis, therefore host clearance of S. aureus requires opsonophagocytic killing by immune cells [69]. Additionally, S. aureus expresses protein A which binds to the Fc region of antibodies and prevents efficient opsonization of S. aureus. Consistent with this, anti-IsdB antisera does not promote opsonization of S. aureus in mouse blood. A potential explanation for the protective effect of anti-IsdB antibodies was provided by the observation that affinity-purified rabbit antibodies directed against IsdB perturbed hemoglobin binding [69]. This raises the interesting possibility that antibodies against IsdB provide protection by inhibiting iron acquisition through the Isd system. The success of these preliminary studies has led to a phase I, double-blind study of the dose range to assess immunogenicity and safety of this novel vaccine. Within this study three dose ranges of the vaccine V710 were tested for antibody concentration and antibody persistence. Antibody titers in subjects receiving the higher doses of vaccine were similar to those achieved in the preclinical primate studies [68, 70]. Future work is needed to determine if V710 has promise as an anti-staphylococcal vaccine, however this formulation has provided a clear demonstration that targeting Isd-mediated heme uptake has significant therapeutic potential.
Concluding remarks
The ability of S. aureus to infect nearly every site of the human body combined with the increasing prevalence of antibiotic resistant strains makes S. aureus a major public health concern. Iron acquisition is a critical determinant in staphylococcal pathogenesis and one that is exploited by vertebrates through nutritional immunity. To circumvent the iron sequestration of the host, pathogenic bacteria have evolved multi-faceted iron acquisition systems to extract iron from host proteins and gain access to intracellular pools of iron. Therapeutically targeting iron acquisition systems within S. aureus may prove to be a viable strategy for combating this deadly pathogen. Further investigations elucidating the iron and heme homeostasis pathways within S. aureus may identify additional therapeutic targets.
Acknowledgements
We thank members of the Skaar laboratory for critical evaluation of this manuscript. Work in the Skaar laboratory is supported by NIH grants AI0169233, AI073843 and AI057157. Dr. Eric Skaar is a Burroughs Wellcome Fund Investigator in the Pathogenesis of Infectious Diseases. Kathryn Haley is supported by the Cellular and Molecular Microbiology Training Grant Program 5 T32 A107611-10.
References
- 1.Kuehnert MJ, et al. Prevalence of Staphylococcus aureus nasal colonization in the United States, 2001–2002. J Infect Dis. 2006;193(2):172–9. doi: 10.1086/499632. [DOI] [PubMed] [Google Scholar]
- 2.David MZ, Daum RS. Community-associated methicillin-resistant Staphylococcus aureus: epidemiology and clinical consequences of an emerging epidemic. Clin Microbiol Rev. 2010;23(3):616–87. doi: 10.1128/CMR.00081-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Todd B. Beyond MRSA: VISA and VRSA: what will ward off these pathogens in health care facilities? Am J Nurs. 2006;106(4):28–30. doi: 10.1097/00000446-200604000-00021. [DOI] [PubMed] [Google Scholar]
- 4.CDC HIV/AIDS Surveillance Report. 2007;17 [Google Scholar]
- 5.Bullen JJ, et al. Iron and infection: the heart of the matter. FEMS Immunol Med Microbiol. 2005;43(3):325–30. doi: 10.1016/j.femsim.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 6.Wessling-Resnick M. Iron homeostasis and the inflammatory response. Annu Rev Nutr. 2010;30:105–22. doi: 10.1146/annurev.nutr.012809.104804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oliviero S, Morrone G, Cortese R. The human haptoglobin gene: transcriptional regulation during development and acute phase induction. EMBO J. 1987;6(7):1905–12. doi: 10.1002/j.1460-2075.1987.tb02450.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dryla A, et al. Identification of a novel iron regulated staphylococcal surface protein with haptoglobin-haemoglobin binding activity. Mol Microbiol. 2003;49(1):37–53. doi: 10.1046/j.1365-2958.2003.03542.x. [DOI] [PubMed] [Google Scholar]
- 9.Torres VJ, et al. Staphylococcus aureus IsdB is a hemoglobin receptor required for heme iron utilization. J Bacteriol. 2006;188(24):8421–9. doi: 10.1128/JB.01335-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fowler VG, Jr., et al. Staphylococcus aureus endocarditis: a consequence of medical progress. JAMA. 2005;293(24):3012–21. doi: 10.1001/jama.293.24.3012. [DOI] [PubMed] [Google Scholar]
- 11.Donovan A, et al. The iron exporter ferroportin/Slc40a1 is essential for iron homeostasis. Cell Metab. 2005;1(3):191–200. doi: 10.1016/j.cmet.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 12.Ganz T. Hepcidin and iron regulation, 10 years later. Blood. 2011;117(17):4425–33. doi: 10.1182/blood-2011-01-258467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tolosano E, et al. Heme scavenging and the other facets of hemopexin. Antioxid Redox Signal. 2010;12(2):305–20. doi: 10.1089/ars.2009.2787. [DOI] [PubMed] [Google Scholar]
- 14.Tolosano E, Altruda F. Hemopexin: structure, function, and regulation. DNA Cell Biol. 2002;21(4):297–306. doi: 10.1089/104454902753759717. [DOI] [PubMed] [Google Scholar]
- 15.Khan FA, Fisher MA, Khakoo RA. Association of hemochromatosis with infectious diseases: expanding spectrum. Int J Infect Dis. 2007;11(6):482–7. doi: 10.1016/j.ijid.2007.04.007. [DOI] [PubMed] [Google Scholar]
- 16.Gardenghi S, et al. Hepcidin and Hfe in iron overload in beta-thalassemia. Ann N Y Acad Sci. 2010;1202:221–5. doi: 10.1111/j.1749-6632.2010.05595.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Galanello R, Origa R. Beta-thalassemia. Orphanet J Rare Dis. 2010;5:11. doi: 10.1186/1750-1172-5-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rahav G, et al. Severe infections in thalassaemic patients: prevalence and predisposing factors. Br J Haematol. 2006;133(6):667–74. doi: 10.1111/j.1365-2141.2006.06082.x. [DOI] [PubMed] [Google Scholar]
- 19.Friedman DB, et al. Staphylococcus aureus redirects central metabolism to increase iron availability. PLoS Pathog. 2006;2(8):e87. doi: 10.1371/journal.ppat.0020087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pishchany G, Dickey SE, Skaar EP. Subcellular localization of the Staphylococcus aureus heme iron transport components IsdA and IsdB. Infect Immun. 2009;77(7):2624–34. doi: 10.1128/IAI.01531-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Torres VJ, et al. Staphylococcus aureus fur regulates the expression of virulence factors that contribute to the pathogenesis of pneumonia. Infect Immun. 2010;78(4):1618–28. doi: 10.1128/IAI.01423-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grigg JC, et al. Structural biology of heme binding in the Staphylococcus aureus Isd system. J Inorg Biochem. 2010;104(3):341–8. doi: 10.1016/j.jinorgbio.2009.09.012. [DOI] [PubMed] [Google Scholar]
- 23.Hussain M, et al. Identification and characterization of a novel 38.5-kilodalton cell surface protein of Staphylococcus aureus with extended-spectrum binding activity for extracellular matrix and plasma proteins. J Bacteriol. 2001;183(23):6778–86. doi: 10.1128/JB.183.23.6778-6786.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chavakis T, et al. Staphylococcus aureus extracellular adherence protein serves as anti-inflammatory factor by inhibiting the recruitment of host leukocytes. Nat Med. 2002;8(7):687–93. doi: 10.1038/nm728. [DOI] [PubMed] [Google Scholar]
- 25.Wang H, et al. Extracellular adherence protein of Staphylococcus aureus suppresses disease by inhibiting T-cell recruitment in a mouse model of psoriasis. J Invest Dermatol. 2010;130(3):743–54. doi: 10.1038/jid.2009.310. [DOI] [PubMed] [Google Scholar]
- 26.Johnson M, Cockayne A, Morrissey JA. Iron-regulated biofilm formation in Staphylococcus aureus Newman requires ica and the secreted protein Emp. Infect Immun. 2008;76(4):1756–65. doi: 10.1128/IAI.01635-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Andreini C, et al. Metal ions in biological catalysis: from enzyme databases to general principles. J Biol Inorg Chem. 2008;13(8):1205–18. doi: 10.1007/s00775-008-0404-5. [DOI] [PubMed] [Google Scholar]
- 28.Green J, et al. Bacterial sensors of oxygen. Curr Opin Microbiol. 2009;12(2):145–51. doi: 10.1016/j.mib.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 29.Kiley PJ, Beinert H. The role of Fe-S proteins in sensing and regulation in bacteria. Curr Opin Microbiol. 2003;6(2):181–5. doi: 10.1016/s1369-5274(03)00039-0. [DOI] [PubMed] [Google Scholar]
- 30.Ding H, Demple B. In vivo kinetics of a redox-regulated transcriptional switch. Proc Natl Acad Sci U S A. 1997;94(16):8445–9. doi: 10.1073/pnas.94.16.8445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Makhlin J, et al. Staphylococcus aureus ArcR controls expression of the arginine deiminase operon. J Bacteriol. 2007;189(16):5976–86. doi: 10.1128/JB.00592-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Somerville GA, Proctor RA. At the crossroads of bacterial metabolism and virulence factor synthesis in Staphylococci. Microbiol Mol Biol Rev. 2009;73(2):233–48. doi: 10.1128/MMBR.00005-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schlag S, et al. Characterization of the oxygen-responsive NreABC regulon of Staphylococcus aureus. J Bacteriol. 2008;190(23):7847–58. doi: 10.1128/JB.00905-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kamps A, et al. Staphylococcal NreB: an O(2)-sensing histidine protein kinase with an O(2)-labile iron-sulphur cluster of the FNR type. Mol Microbiol. 2004;52(3):713–23. doi: 10.1111/j.1365-2958.2004.04024.x. [DOI] [PubMed] [Google Scholar]
- 35.Kafala B, Sasarman A. Isolation of the Staphylococcus aureus hemCDBL gene cluster coding for early steps in heme biosynthesis. Gene. 1997;199(1–2):231–9. doi: 10.1016/s0378-1119(97)00372-7. [DOI] [PubMed] [Google Scholar]
- 36.McNamara PJ, Proctor RA. Staphylococcus aureus small colony variants, electron transport and persistent infections. Int J Antimicrob Agents. 2000;14(2):117–22. doi: 10.1016/s0924-8579(99)00170-3. [DOI] [PubMed] [Google Scholar]
- 37.Skaar EP, et al. Iron-source preference of Staphylococcus aureus infections. Science. 2004;305(5690):1626–8. doi: 10.1126/science.1099930. [DOI] [PubMed] [Google Scholar]
- 38.Shatalin K, et al. Bacillus anthracis-derived nitric oxide is essential for pathogen virulence and survival in macrophages. Proc Natl Acad Sci U S A. 2008;105(3):1009–13. doi: 10.1073/pnas.0710950105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gusarov I, et al. Endogenous nitric oxide protects bacteria against a wide spectrum of antibiotics. Science. 2009;325(5946):1380–4. doi: 10.1126/science.1175439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Halliwell B. Phagocyte-derived reactive species: salvation or suicide? Trends Biochem Sci. 2006;31(9):509–15. doi: 10.1016/j.tibs.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 41.Mazmanian SK, et al. Staphylococcus aureus sortase mutants defective in the display of surface proteins and in the pathogenesis of animal infections. Proc Natl Acad Sci U S A. 2000;97(10):5510–5. doi: 10.1073/pnas.080520697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pilpa RM, et al. Solution structure of the NEAT (NEAr Transporter) domain from IsdH/HarA: the human hemoglobin receptor in Staphylococcus aureus. J Mol Biol. 2006;360(2):435–47. doi: 10.1016/j.jmb.2006.05.019. [DOI] [PubMed] [Google Scholar]
- 43.Grigg JC, et al. Haem recognition by a Staphylococcus aureus NEAT domain. Mol Microbiol. 2007;63(1):139–49. doi: 10.1111/j.1365-2958.2006.05502.x. [DOI] [PubMed] [Google Scholar]
- 44.Liu M, et al. Direct hemin transfer from IsdA to IsdC in the iron-regulated surface determinant (Isd) heme acquisition system of Staphylococcus aureus. J Biol Chem. 2008;283(11):6668–76. doi: 10.1074/jbc.M708372200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Muryoi N, et al. Demonstration of the iron-regulated surface determinant (Isd) heme transfer pathway in Staphylococcus aureus. J Biol Chem. 2008;283(42):28125–36. doi: 10.1074/jbc.M802171200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Skaar EP, Gaspar AH, Schneewind O. IsdG and IsdI, heme-degrading enzymes in the cytoplasm of Staphylococcus aureus. J Biol Chem. 2004;279(1):436–43. doi: 10.1074/jbc.M307952200. [DOI] [PubMed] [Google Scholar]
- 47.Pishchany G, et al. Specificity for human hemoglobin enhances Staphylococcus aureus infection. Cell Host Microbe. 2010;8(6):544–50. doi: 10.1016/j.chom.2010.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hardison RC, et al. HbVar: A relational database of human hemoglobin variants and thalassemia mutations at the globin gene server. Hum Mutat. 2002;19(3):225–33. doi: 10.1002/humu.10044. [DOI] [PubMed] [Google Scholar]
- 49.Reniere ML, et al. The IsdG-family of haem oxygenases degrades haem to a novel chromophore. Mol Microbiol. 2010;75(6):1529–38. doi: 10.1111/j.1365-2958.2010.07076.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Haley KP, et al. Staphylococcus lugdunensis IsdG liberates iron from host heme. J Bacteriol. 2011 doi: 10.1128/JB.00436-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reniere ML, Skaar EP. Staphylococcus aureus haem oxygenases are differentially regulated by iron and haem. Mol Microbiol. 2008;69(5):1304–15. doi: 10.1111/j.1365-2958.2008.06363.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brotz-Oesterhelt H, et al. Dysregulation of bacterial proteolytic machinery by a new class of antibiotics. Nat Med. 2005;11(10):1082–7. doi: 10.1038/nm1306. [DOI] [PubMed] [Google Scholar]
- 53.Reniere ML, Haley KP, Skaar EP. The Flexible Loop of Staphylococcus aureus IsdG Is Required for Its Degradation in the Absence of Heme. Biochemistry. 2011 doi: 10.1021/bi200999q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Varshavsky A. The N-end rule: functions, mysteries, uses. Proc Natl Acad Sci U S A. 1996;93(22):12142–9. doi: 10.1073/pnas.93.22.12142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Keiler KC, Waller PR, Sauer RT. Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science. 1996;271(5251):990–3. doi: 10.1126/science.271.5251.990. [DOI] [PubMed] [Google Scholar]
- 56.Chim N, et al. Unusual diheme conformation of the heme-degrading protein from Mycobacterium tuberculosis. J Mol Biol. 2010;395(3):595–608. doi: 10.1016/j.jmb.2009.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Puri S, O'Brian MR. The hmuQ and hmuD genes from Bradyrhizobium japonicum encode heme-degrading enzymes. J Bacteriol. 2006;188(18):6476–82. doi: 10.1128/JB.00737-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Skaar EP, Gaspar AH, Schneewind O. Bacillus anthracis IsdG, a heme-degrading monooxygenase. J Bacteriol. 2006;188(3):1071–80. doi: 10.1128/JB.188.3.1071-1080.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Noya F, Arias A, Fabiano E. Heme compounds as iron sources for nonpathogenic Rhizobium bacteria. J Bacteriol. 1997;179(9):3076–8. doi: 10.1128/jb.179.9.3076-3078.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lindsay JA, Riley TV. Staphylococcal iron requirements, siderophore production, and iron-regulated protein expression. Infect Immun. 1994;62(6):2309–14. doi: 10.1128/iai.62.6.2309-2314.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Beasley FC, Heinrichs DE. Siderophore-mediated iron acquisition in the staphylococci. J Inorg Biochem. 2010;104(3):282–8. doi: 10.1016/j.jinorgbio.2009.09.011. [DOI] [PubMed] [Google Scholar]
- 62.Beasley FC, et al. Characterization of staphyloferrin A biosynthetic and transport mutants in Staphylococcus aureus. Mol Microbiol. 2009;72(4):947–63. doi: 10.1111/j.1365-2958.2009.06698.x. [DOI] [PubMed] [Google Scholar]
- 63.Torres VJ, et al. A Staphylococcus aureus regulatory system that responds to host heme and modulates virulence. Cell Host Microbe. 2007;1(2):109–19. doi: 10.1016/j.chom.2007.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stauff DL, Torres VJ, Skaar EP. Signaling and DNA-binding activities of the Staphylococcus aureus HssR-HssS two-component system required for heme sensing. J Biol Chem. 2007;282(36):26111–21. doi: 10.1074/jbc.M703797200. [DOI] [PubMed] [Google Scholar]
- 65.Stauff DL, et al. Staphylococcus aureus HrtA is an ATPase required for protection against heme toxicity and prevention of a transcriptional heme stress response. J Bacteriol. 2008;190(10):3588–96. doi: 10.1128/JB.01921-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pedersen MB, et al. Impact of aeration and heme-activated respiration on Lactococcus lactis gene expression: identification of a heme-responsive operon. J Bacteriol. 2008;190(14):4903–11. doi: 10.1128/JB.00447-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fernandez A, et al. Two coregulated efflux transporters modulate intracellular heme and protoporphyrin IX availability in Streptococcus agalactiae. PLoS Pathog. 2010;6(4):e1000860. doi: 10.1371/journal.ppat.1000860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kuklin NA, et al. A novel Staphylococcus aureus vaccine: iron surface determinant B induces rapid antibody responses in rhesus macaques and specific increased survival in a murine S. aureus sepsis model. Infect Immun. 2006;74(4):2215–23. doi: 10.1128/IAI.74.4.2215-2223.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kim HK, et al. IsdA and IsdB antibodies protect mice against Staphylococcus aureus abscess formation and lethal challenge. Vaccine. 2010;28(38):6382–92. doi: 10.1016/j.vaccine.2010.02.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Harro C, et al. Safety and immunogenicity of a novel Staphylococcus aureus vaccine: results from the first study of the vaccine dose range in humans. Clin Vaccine Immunol. 2010;17(12):1868–74. doi: 10.1128/CVI.00356-10. [DOI] [PMC free article] [PubMed] [Google Scholar]