Abstract
The complex genetics of human complement C4 with unusually frequent variations in the size and number of C4A and C4B, as well as their neighboring genes, in the major histocompatibility complex has been a hurdle for accurate epidemiological studies of diseases associated with C4. A comprehensive series of novel or improved techniques has been developed to determine the total gene number of C4 and the relative dosages of C4A and C4B in a diploid genome. These techniques include (1) definitive genomic restriction-fragment–length polymorphisms (RFLPs) based on the discrete duplication patterns of the RCCX (RP-C4-CYP21-TNX) modules and on the specific nucleotide changes for C4A and C4B isotypes; (2) module-specific PCR to give information on the total number of C4 genes by comparing the relative quantities of RP1- or TNXB-specific fragments with TNXA-RP2 fragments; (3) labeled-primer single-cycle DNA polymerization procedure of amplified C4d genomic DNA for diagnostic RFLP analysis of C4A and C4B; and (4) a highly reproducible long-range–mapping method that employs PmeI-digested genomic DNA for pulsed-field gel electrophoresis, to yield precise information on the number of long and short C4 genes in a haplotype. Applications of these vigorously tested techniques may clarify the roles that human C4A and C4B gene-dosage variations play in infectious and autoimmune diseases.
Introduction
Complement component C4 is a pivotal component in the activation cascades of complement pathways. It is a subunit of the C3 and C5 convertases for the classical and lectin pathways that lead to the assembly of membrane-attack complex on targets and the generation of potent anaphylatoxins C3a and C5a. The binding of activated C4 to targets also helps the solubilization of immune aggregates, opsonizes targets for phagocytosis by macrophages, and promotes the immunoadherence of immune complexes to erythrocytes and the subsequent clearance in the liver (for review, see Porter 1983; Blanchong et al. 2001; Jack et al. 2001; Kohl 2001).
Human complement C4 illustrates one of the most unusual phenomena in genetic diversity. The frequent germline variation in the number and size of C4 genes among different individuals is extraordinary. The copy number of C4 genes in a diploid human genome (i.e., the gene dosage) predominantly varies from two to six in the white population (O'Neill et al. 1978; Awdeh et al. 1979; Olaisen et al. 1980; Carroll et al. 1985; Schneider et al. 1986; Yang et al. 1999; Blanchong et al. 2000). Each of these genes encodes a C4A (MIM 120810) or C4B (MIM 120820) protein. Catalyzed by the isotypic residue His-1106, the activated C4B binds to a target rapidly but has a short half-life (<1 s) against hydrolysis. On the contrary, the reaction rate of activated C4A is 10 times slower than that of C4B, and it forms amide bonds with amino group–containing targets (Dodds et al. 1996). The longer half-life of activated C4A might have evolved to facilitate the clearance of the immune complex. In the population, the peripheral-blood plasma C4 protein levels range from 80 to 1,000 μg/ml (Welch et al. 1985; Uko et al. 1986). This large range of protein levels is determined by multiple factors, including the physiological stage of an individual, the copy number of functional C4 genes that are present in a genome (gene dosage), and, possibly, the polymorphism in cis-acting DNA elements that may affect the C4 expression levels. Plasma C4 is mainly secreted by the liver, but many extrahepatic sites—such as the thyroid gland, the adrenal glands, the kidneys and the heart—also synthesize significant levels of C4, particularly in the presence of interferon-γ (Colten and Garnier 1998; Blanchong et al. 2001).
Deficiency of C4 increases the susceptibility to or disease severity of viral and bacterial infections (Daniels et al. 1969; Bishof et al. 1990; Wessels et al. 1995; Jaatinen et al. 1999). C4 deficiency is also an important risk factor for autoimmune diseases, such as systemic lupus erythematosus (SLE [MIM 152700]) (Atkinson and Schneider 1999; Tsokos and Kammer 2000). On the contrary, excessive C4 or overactivation of C4 could aggravate an inflammatory response and render an individual more vulnerable to tissue injuries. The consumption of C4 leads to repressed C4 levels in the peripheral blood and the generation of activation product C4a and inactivation product C4d. C4a and C4d are relevant indicators of disease activity in SLE (Wild et al. 1990; Feucht et al. 1993) and complement-mediated tissue injuries in transplantation, graft rejection, ischemia-reperfusion, and myocardial infarction (Pinckard et al. 1975; Baldwin et al. 1999; Pfeifer et al. 2000).
C4 is a constituent of the four-gene module termed the “RCCX,” which duplicates as a discrete genetic unit in the class III region of the major histocompatibility complex (MHC) (Yu et al. 2000). The duplicated genomic segment is either 32.7 or 26.4 kb in size and contains the gene fragments TNXA (Gitelman et al. 1992) and RP2 (Shen et al. 1994), a complete C4 gene with or without an endogenous retrovirus HERV-K(C4) in intron 9 (Dangel et al. 1994; Schneider et al. 2001), and either a nonfunctional CYP21A or a functional CYP21B (White and Speiser 2000). The variation in the number of the RCCX modules and the size of the C4 genes leads to the presence of seven common RCCX length variants: monomodular L (long) and S (short); bimodular LL and LS; and trimodular LLL, LSS, and LLS (or LSL) (Yang et al. 1999; Blanchong et al. 2000). In addition, two rare length variants, bimodular SS and quadrimodular LLLL, have been implicated (Collier et al. 1989; Weg-Remers et al. 1997). These length variants create >20 different haplotypic combinations of RP1/RP2 (MIM 604977), C4-L/C4-S, CYP21A/CYP21B, and TNXA/TNXB genes in the white population (Blanchong et al. 2000). Definitive and efficient techniques to elucidate the number of C4A and C4B genes and polymorphism of the protein products are important to determine the roles that C4A and C4B play in disease associations, in the prognosis and therapeutic intervention of autoimmune and inflammatory diseases. Here, we present comprehensive and definitive approaches to determine both the number and length variants of the RCCX modules and the dosages of the C4A and C4B genes. We also introduce a highly reproducible method to illustrate the remarkable RCCX modular variations by PmeI-digested genomic DNA for pulsed-field gel electrophoresis (PFGE).
Material and Methods
Recruitment of Human Subjects and Preparation of Genomic DNA
Peripheral-blood samples were taken from healthy individuals and from patients with pauciarticular JRA after informed consent was obtained, after approval by the Columbus Children's Hospital institutional review board. Genomic DNA was isolated from peripheral blood by using the Puregene DNA isolation kit (Gentra Systems).
Southern Blot Analysis
Six micrograms of genomic DNA was digested to completion with the appropriate restriction enzymes. Restriction enzymes included TaqI, PshAI (New England Biolabs), and PvuII (Invitrogen). For the PshAI-PvuII RFLP, PshAI (15 U) was used for overnight digestion at 25°C, followed by an additional 6 h of 37°C incubation with PvuII (15 U). DNA fragments were resolved by electrophoresis in 0.7% agarose gels and were processed as described elsewhere (Blanchong et al. 2000; Yu et al. 2002).
The DNA probes used for hybridization (as illustrated in fig. 1) are as follows: probe A, a 600-bp NheI-EcoRI cDNA fragment corresponding to the 3′ end of the DNA sequence present both in RP1 and RP2; probe D, a 1.1-kb fragment corresponding to exons 28–31 of C4; probe 22–25, an 844-bp fragment corresponding to exons 22–25 of C4; probe E, a 757-bp fragment corresponding to exons 4–7 of CYP21; and probe F, a 500-bp fragment corresponding to exons 35–37 of TNXA.
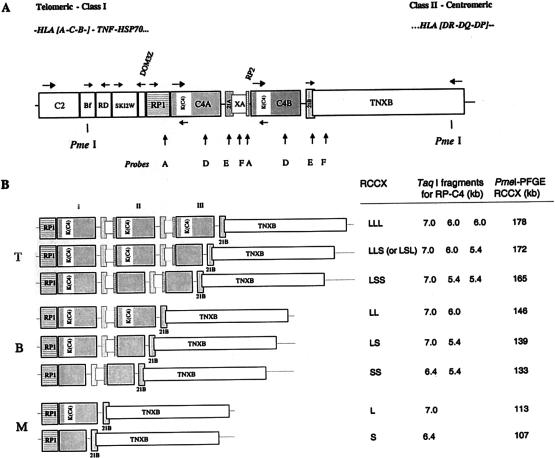
Figure 1.
Schematic diagram of the human MHC complement gene cluster (MCGC) and eight unique modular variants of the RCCX. A, The gene organization of the MCGC and the location of the two PmeI restriction sites. A horizontal arrow represents the direction of transcription of a gene. A vertical arrow represents the position at which probes A, D, E, or F hybridizes to the DNA. The position of probe 22–25 is not shown, but its location is three exons upstream of probe D. B, The alignment of eight unique RCCX length variants at the telomeric end, which are grouped into either T, B, or M haplotypes. K(C4) = endogenous retrovirus HERV-K(C4); 21A = CYP21A; 21B = CYP21B; XA = TNXA; T = trimodular; B = bimodular; M = monomodular.
Complement C4 Allotyping and Immunoblot Analysis
EDTA-blood plasma was used to test for C4 polymorphism after standard procedure (Awdeh and Alper 1980; Sim and Cross 1986). In brief, diluted plasma was treated with neuraminidase (Sigma) and carboxypeptidase B (Sigma), was resolved by high-voltage agarose electrophoresis (HVAGE) based on gross electric-charge difference of plasma proteins, was immunofixed with goat anti-human C4 serum (Incstar), was blotted and washed to remove diffusible proteins, was dried, and was stained with SimplyBlue Safestain (Invitrogen). Serological properties of C4 proteins were studied by immunoblot analysis of the C4 proteins separated by HVAGE, using anti-Ch1 and anti-Rg1 monoclonal antibodies at dilutions of 1:5,000 and 1:250, respectively. Anti-Rg1 and anti-Ch1 monoclonal antibodies were obtained from the VIIth Complement Genetic Workshop (Mauff et al. 1998). With only a few exceptions, Rg1 associates with C4A, and Ch1 usually associates with C4B (for review, see Yu et al. 2002). Immune complexes were detected by the chemiluminescence method using ECL Plus reagents (Amersham Life Science).
Module-Specific PCR
Primers a, b, c, and d were designed to hybridize to specific locations within the RCCX modules. The primer sequences are as follows: a, 5′-CAA GAG AGG AGG CCT ATC TTA CCT GG-3′; b, 5′-GCT CAA GCT GTG AGG AGA ACT-3′; c, 5′-TAT CAC AGG CTC TGG CCC CA-3′; and d, 5′-TTC GTG GTC CAG TAC AGG GA-3′. The reaction mixture was prepared as per instructions of the PLATINUM TaqPCRx DNA Polymerase (GibcoBRL). A 1 × PCRx Enhancer Solution (GibcoBRL) was used. For the RP1/TNXA-RP2 reaction, a concentration of 608 nM (200 ng) primer a, 608 nM (200 ng) primer b, and 106 nM (35 ng) primer c was used. For the TNXB/TNXA-RP2 reaction, a concentration of 304 nM (100 ng) primer b, 304 nM (100 ng) primer c, and 198 nM (65 ng) primer d was used. PCR conditions were as follows: 1 cycle at 94°C for 3 min; 32 cycles at 94°C for 45 s, 62°C for 45 s, 72°C for 1 min; and 1 cycle at 72°C for 10 min. DNA fragments were resolved by 1% agarose gels. A digital image of the gel was captured, and the intensity of each band was quantitatively analyzed (by Nucleotech-GelExpert, version 4.0, or an equivalent software).
Sequence-Specific Primer PCR (SSP-PCR)
Primers A-down and B-down specifically hybridize to the isotypic sites of C4A and C4B, respectively. Primer E27.3 hybridizes to the 3′ end of exon 27 in C4. The primer sequences are as follows: A-down, 5′-AGG ACC CCT GTC CAG TGT TAG AC-3′; B-down, 5′-AGG ACC TCT CTC CAG TGA TAC AT-3′; and E27.3, 5′-CAC TCT CTG CTT CAA TGG CT-3′. The SSP-PCR for C4A included primers A-down and E27.3, and that for C4B utilized B-down and E27.3. One microliter diluted (1:20) E26.5/E29.3 PCR product was used as the DNA template. PCR conditions were as follows: 1 cycle at 94°C for 3 min; 25 cycles at 94°C for 30 s, 60°C for 45 s, and 72°C for 1 min; and 1 cycle at 72°C for 10 min.
Labeling the 5′ End of C4 Primer E29.3
The reaction mixture included 15.2 μM primer E29.3, kinase buffer (0.5 M Tris × Cl [pH 7.6], 0.1 M MgCl2, 50 mM dithiothreitol, and 1 mM EDTA), 1 mM spermidine, 100 μCi [γ-32P] ATP, and 20 U T4 polynucleotide kinase. The reaction was incubated at 37°C for 1 h. The radiolabeled primer E29.3 was purified by size-exclusion chromatography using Sephadex G-25 columns, followed by alcohol precipitation, and was resuspended in TE (10 mM Tris and 1 mM EDTA; pH 7.4) in a final concentration of 3.8 μM.
Labeled-Primer Single-Cycle Polymerization (LSP)
Unlabeled primers E26.5 and E29.3 were designed to anneal to the 5′ end of exon 26 and the 3′ end of exon 29 in C4, respectively. The primer sequences are as follows: E26.5, 5′-GCT CAC AGC CTT TGT GTT GAA-3′, and E29.3, 5′-TTG GGT ACT GCG GAA TCC CC-3′. A 1 × Premix G with the Failsafe PCR Enzyme Mix (1.25 U) (Epicentre) was used. PCR conditions were as follows: 1 cycle at 94°C for 3 min; 34 cycles at 94°C for 30 s, 58°C for 45 s, 72°C for 1 min; and 1 cycle at 72°C for 10 min. The PCR products were purified using a Qiaquick PCR Purification Kit (Qiagen) and were resuspended in 50 μl of deionized water. A subsequent LSP reaction was performed using 0.2 μM radiolabeled primer E29.3 and the Failsafe PCR Enzyme Mix (0.625 U) with 1 × Premix G. LSP conditions were as follows: 94°C for 5 min, 58°C for 5 min, and 72°C for 10 min.
PshAI and XcmI RFLP of LSP Products
One-third of the LSP product was restriction digested by PshAI or XcmI overnight at 25°C and 37°C, respectively. DNA fragments were resolved by electrophoresis using 1% agarose gel supplemented with ethidium bromide. A digital image was captured, and the intensity of each band was quantitatively analyzed. Subsequently, the gel was processed by Southern blot technique and was subjected to autoradiography, and the radioactivity of each band was registered by phosphorimaging and was quantified by Molecular Dynamics ImageQuant Software, version 5.0.
PmeI RFLP Resolved by PFGE
DNA agarose plugs were prepared from white blood cells subjected to proteinase K digestion at 50°C. Prior to restriction-enzyme digestion, the DNA agarose plugs were washed with TE three times for 30-min intervals, followed by an additional wash overnight, and were equilibrated with restriction-enzyme buffer twice for 30 min each. Subsequently, the plugs were digested with PmeI (20 U) for 4 h at 37°C. DNA fragments were resolved by PFGE in 1% agarose in a Bio-Rad CHEF Mapper XA System under the following conditions: gradient, 6 V/cm; calibration factor, 1.4; initial switch time, 6.75 s; final switch time, 21.79 s; run time, 37 h 40 min; included angle, 120°; ramp, linear. The gel was processed by Southern blot technique, was hybridized with probe D (fig. 1), and was subjected to autoradiography.
Results
Distinguishing and Enumerating the RCCX Modular Variants using Genomic RFLPs
On the basis of differences in gene structures and nucleotide polymorphism, a series of RFLP techniques was designed to decipher the complexity of gene organizations within the RCCX modular structures (fig. 1). Five individuals with characterized HLA class I and class II alleles (fig. 2G) (Chung et al. 2002) and with total number of RCCX modules varying from two to six in each genome were selected to illustrate the RCCX patterns and the feasibility of the techniques. TaqI Southern blot bears information on the dichotomous size variation of C4 genes and the number of RCCX modules—hence, the number of C4 genes—in a diploid genome. The specific length variants and the number of RCCX modules are assigned on the basis of the presence and absence and the relative band intensities in the three groups of restriction fragments corresponding to (1) RP1-C4 (L or S) and RP2-C4 (L or S), (2) CYP21A and CYP21B (MIM 201910), and (3) TNXA and TNXB (MIM 600985). The assignment is substantiated by two additional RFLP analyses: PshAI genomic Southern blot hybridized to an RP 3′ probe, and BamHI genomic Southern blot hybridized to a TNX 3′ probe. The PshAI RFLP establishes the ratio of the number of RP1 genes to the number of TNXA-RP2 hybrids, which are represented by the 7.3-kb and 8.75-kb fragments, respectively (fig. 2B). Similarly, the band intensities in the BamHI Southern blots reflects the relative number of TNXB genes to that of TNXA-RP2 hybrids, which are represented by the 6.5-kb and 5.0-kb fragments, respectively (fig. 2C). Both the RP-PshAI and TNX-BamHI RFLPs represent gross organizational differences between RP1 and RP2, and between the TNXA and TNXB genes.
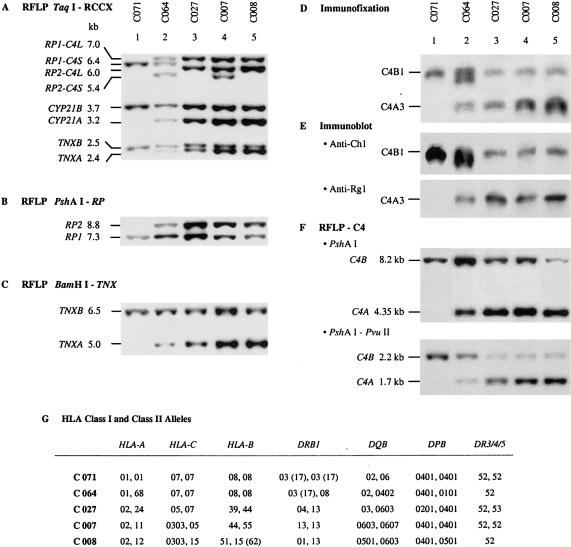
Figure 2.
Defining the RCCX modular structure using different genomic RFLP Southern blot analyses. A, Restriction patterns of TaqI RFLP for the five selected individuals on simultaneous hybridization of probes specific for 3′ RP, CYP21, and 3′ TNX. B, PshAI RFLP hybridized to a 3′ RP probe. C, BamHI RFLP hybridized to a 3′ TNX probe. D, Phenotyping of C4A and C4B proteins by immunofixation of EDTA-blood plasma resolved by HVAGE. E, Immunoblot analysis of Rg1 and Ch1 antigenic determinants in plasma C4 proteins after HVAGE. F, Genomic Southern blot analysis of C4A and C4B genes by PshAI-RFLP (using Probe D) and by PshAI-PvuII RFLP (using probe 22–25). G, HLA class I and class II alleles of the five subjects.
The results of the RFLP analyses are shown in figure 2. In brief, C071 (lane 1) has homozygous monomodular structures S/S with a total of two C4 genes in diploid. C064 (lane 2) has heterozygous bimodular LS and monomodular S structures with a total of three C4 genes. C027 (lane 3) has homozygous bimodular LL/LL with four C4 genes. C007 (lane 4) has heterozygous trimodular and bimodular structures with five C4 genes. The modular structures of C007 may be LLL/LS, LLS/LL, or LSL/LL (which are indistinguishable without parallel family segregation studies). C008 (lane 5) has homozygous trimodular structures LLL with a total of six C4 genes.
From figure 2, band intensities of the restriction fragments for RP2 relative to those of RP1 increase with the number of RCCX modules (fig. 2B, PshAI RFLP, lanes 1–5). Similarly, band intensities of the restriction fragments for TNXA relative to those of TNXB (figs. 2C, BamHI RFLP, and 2A, TaqI RFLP) and of the pseudogene CYP21A relative to the functional gene CYP21B (fig. 2A, TaqI RFLP) increase with the number of the RCCX modules that are present in an individual.
Phenotypic and Genotypic Characterization of C4A and C4B
The variations of C4A and C4B proteins and genes from the same five individuals were examined (figs. 2D–2F). The protein polymorphism of C4A and C4B was determined by the immunofixation of EDTA plasma and is resolved by HVAGE; the serological properties of the C4A and C4B allotypes are characterized by immunoblot analyses using anti-Rg1 and anti-Ch1 monoclonal antibodies. The presence and relative gene dosages of C4A and C4B were elucidated by PshAI genomic RFLP. Since the larger restriction DNA fragment for C4B (8.2 kb) can lead to relatively lower transfer efficiency to the hybridization membrane than that of C4A (4.35 kb) in a Southern blot analysis, an improved strategy employing PshAI-PvuII-digested genomic DNA was applied. By this approach, the isotype-specific fragments for C4A and C4B were 1.7 kb and 2.2 kb, respectively, which are smaller and closer in size, and the resultant relative band intensities yielded more reliable information on the C4A and C4B gene dosages.
Allotyping of C4 protein from C071 revealed the presence of C4B1 only (fig. 2D, lane 1). The PshAI and PshAI-PvuII RFLPs demonstrated the presence of single restriction fragments for C4B and the total absence of the C4A fragments (fig. 2F), which is consistent with the assignment of homozygous C4AQ0 C4B1. In C064, the observed C4A3 allotype is associated with Rg1, and the C4B1 and B2 allotypes are associated with Ch1. The PshAI (or PshAI-PvuII) RFLP reveals a 2:1 ratio of C4B:C4A. C064 has heterozygous LS/S structures totaling three C4 genes. The bimodular LS probably encodes C4A3 and C4B2, and the monomodular S probably encodes C4B1. The remaining three individuals—C027, C007, and C008—are identified as having C4A3 associated with Rg1 and C4B1 associated with Ch1 (figs. 2D and 2E, lanes 3–5). There is a gradual increase in the relative intensities of C4A versus C4B proteins beginning from C027. A parallel observation can be made for the relative dosages, of C4A:C4B genes, corresponding to C027, C007, and C008 (fig. 2F), who show C4A:C4B ratios of 3:1, 4:1, and 5:1, respectively. These results are consistent with the C4 gene dosage determined by the TaqI genomic RFLPs.
Quantifying the Number of RCCX Modules or C4 Gene Dosage using Module-Specific PCR
A semiquantitative PCR method was developed to provide a rapid and independent method to evaluate the number of RCCX modules that are present in a diploid genome (which is equivalent to the C4 gene dosage). The method is based on the unique structural features of the RCCX modular duplication: each diploid genome has two copies of the intact RP1 genes and two copies of the intact TNXB genes, regardless of the number of RCCX modules; no TNXA-RP2 hybrid is present in a monomodular RCCX structure; and a copy of the TNXA-RP2 is present in each additional duplicated RCCX module. Therefore, the relative dosages of TNXA-RP2 and RP1 or of TNXA-RP2 and TNXB serve as a reliable marker for the number of RCCX modules and, hence, the C4 gene dosage of an individual.
Two sets of PCR primers are employed to amplify the 3′ ends of RP and the 3′ ends of TNX (fig. 3A). For RP, the 5′ primer a is RP1 specific, and the 3′ primer b may anneal to both RP1 and RP2. For TNX, the 5′ primer d is TNXB specific, and the 3′ primer c may anneal to both TNXA and TNXB. This PCR technique is applied to the five selected individuals, and results are illustrated in figure 3B. Lanes 1–5 represent the amplifications of RP1 (product a-b, 1.2 kb) and TNXA-RP2 (product c-b, 1.0 kb), using primer set a–c. Similarly, lanes 6–10 show results of amplifications of TNXB (product c-d, 0.8 kb) and TNXA-RP2 (product c-b, 1.0 kb), using primer set b–d. The ratio of band intensities of the PCR products for each lane, except for lanes 1 and 6, are plotted in the graphs shown in figure 3B.
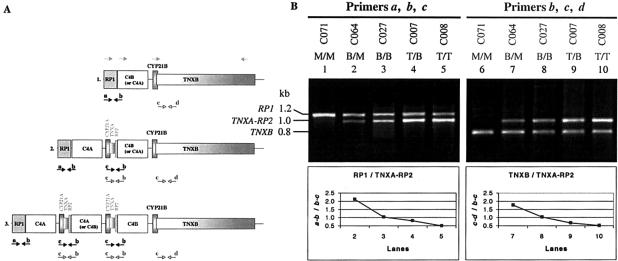
Figure 3.
Module-specific PCR analysis, to detect the modular variation of RCCX. A, Schematic diagram illustrating the approximate locations to which the primer sets a–c and b–d bind in the three modular haplotypes. B, Module-specific PCR for the RP1/TNXA-RP2 reaction, using primer set a–c (lanes 1–5, top), and for the TNXB/TNXA-RP2 reaction, using primer set b–d (lanes 6–10, top). The relative band intensities obtained from RP1 versus TNXA-RP2 or TNXB versus TNXA-RP2 were plotted in the graph and, except for lanes 1 and 6, are shown at bottom.
For C071, the presence of the solitary 1.2-kb fragment corresponding to RP1 (fig. 3B, lane 1) and the 0.8-kb fragment corresponding to TNXB (fig. 3B, lane 6) is indicative of homozygous monomodular (M/M) RCCX with a total of two C4 genes. For C064, the intensities of the 1.2-kb RP1-specific fragment (fig. 3B, lane 2) and of the 0.8-kb TNXB-specific fragment (fig. 3B, lane 7) are twice those of their corresponding 1.0-kb TNXA-RP2 fragments. In contrast, both RP1-specific (fig. 3B, lane 5) and TNXB-specific (fig. 3B, lane 10) fragments of C008 are half as intense as their corresponding TNXA-RP2 fragments. In the former scenario (C064), the ratio suggests heterozygous bimodular/monomodular (B/M) haplotypes with a total of three C4 genes, whereas the latter (C008) indicates homozygous trimodular (fig. 3B, T/T) haplotypes with a total of six C4 genes. Both RP1/TNXA-RP2 (fig. 3B, lane 3) and TNXB/TNXA-RP2 (fig. 3B, lane 8) amplifications of C027 demonstrate equal intensities between each of the two fragments present, suggesting the presence of homozygous bimodular (B/B) RCCX structures with four C4 genes. For C007, the 1.2-kb RP1-specific fragment (fig. 3B, lane 4) and the 0.8-kb TNXB-specific fragment (fig. 3B, lane 9) are slightly less intense than their corresponding 1.0-kb TNXA-RP2 fragment, by a ratio of ∼2:3, which is equivalent to heterozygous trimodular/bimodular (T/B) RCCX structures with five C4 genes.
Similar to the TaqI RFLPs, module-specific PCR yields relevant results on the number of RCCX modules or total C4 gene dosage. However, it yields information on neither the C4A and C4B isotypes nor the association between the C4 genes and Rg1 or Ch1 antigenic determinants.
Determining the Presence and Dosages of C4A and C4B Genes by SSP-PCR, PCR-RFLP, and LSP-RFLP
The C4A/C4B isotypic residues are encoded by exon 26, and the Rg1/Ch1 antigenic determinant is encoded by exon 28. For the facilitation of the quantitation of C4A and C4B genes, a general strategy was explored, to allow efficient amplification of the C4d region by PCR using different primer sets spanning exons 22–31. It was found that the primer sets corresponding to the 5′ end of exon 26 and to the 3′ end of exon 29 consistently yield robust amplification of a 1.11-kb product that is suitable for further characterization (figs. 4A and 4B).
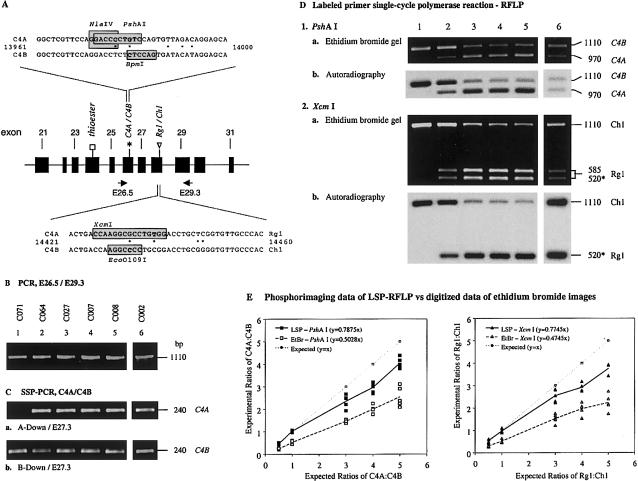
Figure 4.
PCR techniques to determine C4A/C4B gene dosage. A, Scheme depicting the intron/exon structure between exons 21 and 31 (middle) and the basis of definitive RFLP for C4A/C4B (top) and for Rg1/Ch1 (bottom). Exons are in solid boxes. Primers E26.5 and E29.3 are indicated as horizontal arrows. B, PCR products of E26.5/E29.3. C, SSP-PCR assays for the presence and absence of C4A and C4B genes. D, LSP-PshAI or XcmI RFLP, to determine the C4A/C4B and C4 genes associated with Rg1 or Ch1, respectively. An asterisk indicates the fragment for Rg1 with the labeled E29.3 primer. E, Graphic comparisons between expected results for the relative gene dosage of C4A and C4B and results obtained from LSP-RFLPs and from ethidium bromide gels. Experiments with labeled DNA products digested with PshAI that distinguishes C4A and C4B are shown at left, and results of labeled DNA products digested with XcmI that distinguishes C4 genes associated with Rg1 and Ch1 are shown at right.
SSP-PCR, to determine the presence of C4A and C4B genes.—Two different forward primers, C4A-down and C4B-down, and a common reverse primer E27.3 are used in two independent PCRs, to determine the presence of the C4A and C4B isotypic sequences, using the described 1.11-kb products from E26.5 to E29.3 as templates. The forward primers incorporated the 5-nt changes for the C4A and C4B in exon 26 (Entrez Nucleotide accession numbers M59815 and AL049547). Results of the C4A- and C4B-specific amplifications are shown in figure 4C. Among the same five samples selected, C071 demonstrates the absence of the 240-bp C4A-specific fragment and the presence of the C4B-specific fragment. All others give positive results for the presence of the C4A-specific and C4B-specific fragments, as expected. However, this SSP-PCR strategy can yield information only on the presence or absence of C4A and C4B genes but not on their polygenic variations.
“Hot-stop PCR”–PshAI RFLP, to quantify the C4A and C4B isotypes.—Theoretically, the relative dosage of the C4A and C4B genes may be determined by the PshAI or NlaIV cleavage of the E26.5/E29.3 genomic fragment amplified by PCR. However, the formation of heteroduplexes among various C4A and C4B alleles during the PCR process renders the direct PCR-RFLP results inaccurate in the quantitative analysis of C4A and C4B gene dosages. This is because the C4A/C4B heteroduplexes formed by PCR are refractory to restriction digests that define C4A/C4B isotypes or Rg1/Ch1 antigenic determinants. Therefore, the band intensities are skewed towards fragments in which the diagnostic restriction sites are absent. A recently developed hot-stop PCR technique employing a γ-32P–labeled primer in the last round of the PCR appears to be an attractive method to solve the problems incurred by heteroduplexes (Uejima et al. 2000).
For determination of the validity of the hot-stop PCR, the E29.3 primer was radiolabeled by T4 polynucleotide kinase and γ-32P-ATP. Ten nanograms of the purified, radioactive PCR primer was added to each reaction mixture, and an additional cycle of DNA synthesis was performed. The hot-stop PCR products were then subjected to PshAI-RFLP analyses and autoradiograms obtained from semidried agarose gels, or from Southern-blotted membranes. Results from the initial experiments revealed relatively weak signals that required >2 d for a visible image on an x-ray film. Optimization was achieved by purifying the PCR products with a commercial spin device and by adding fresh DNA polymerase, deoxynucleoside triphosphates, and the labeled primer, for LSP reaction.
Figure 4D shows the results of the LSP products for RFLP analyses, using the E26.5/E29.3 PCR products from the same five described samples as templates. From the PshAI restriction digests, a 970-bp fragment represents C4A, and a 1,110-bp fragment represents C4B. After electrophoresis, a digital image was obtained from ethidium bromide gel (fig. 4D, 1a and 2a), and an autoradiograph was acquired from the primer-labeled restriction fragments, after transfer to hybridization membrane by Southern blot technique. The radioactivity of each band was registered by phosphorimaging and was quantified. Five independent experiments were performed to examine the consistency of results.
Consistent in both analyses from the ethidium bromide gel and autoradiography of the digested LSP product, the PshAI-digested product of C071 indicates the presence of C4B genes only (fig. 4D, 1a and 1b, lane 1). However, each of the four remaining individuals demonstrated unequivalent results on the ratio of C4A and C4B genes that are present in each sample between the digitized data from ethidium bromide gel (apparent ratio) and from the phosphorimaging data of the LSP-PshAI products (LSP ratio).
The expected ratio of C4A:C4B for C064 is 1:2. The apparent ratio of C4A:C4B from digitized images for C064 was 1:(4.3±0.4), which deviated from the expected ratio by twofold. In contrast, the LSP ratio obtained by phosphorimaging data of the radioactive LSP-PshAI products for C064 was 1:(2.2±0.2), which is consistent with the expected result.
For C027, the expected ratio for C4A:C4B is 3:1. The apparent ratio for C4A:C4B was (1.5±0.1):1, which was again two times less than the expected ratio. The LSP ratio obtained from phosphorimaging data was (2.4±0.3):1, which was closer to the expected ratio. C007 has an expected ratio of 4:1 for C4A:C4B. The apparent ratio was (2.0±0.2):1, and the LSP ratio was (3.0±0.2):1. A C4A:C4B ratio of 5:1 is expected for C008, the apparent ratio was (2.5±0.4):1, and the LSP ratio was (4.0±0.2):1. It was noticed that, when the relative dosage of C4A:C4B is ⩾3:1, even the LSP-PshAI analysis has a tendency to yield a slight underestimation.
Since none of the five selected individuals studied above have an equal number of C4A and C4B genes, an additional analysis was performed from an individual with homozygous bimodular LL/LL, two C4A3 genes and two C4B1 genes (fig. 4, lane 6). The expected C4A:C4B ratio was 1:1. The apparent ratio of C4A:C4B was 1:(1.9±0.3), which is approximately twofold in favor of C4B. In contrast, a ratio of 1:(1.0±0.04) was obtained from the analysis of phosphorimaging data from LSP-PshAI, which accurately agrees with the expected result.
LSP-XcmI, to determine the association between C4 genes and sequences that encode Rg1/Ch1 antigenic determinants.—To determine the relative dosages of C4-Rg1 and C4-Ch1, the LSP products from E26.5/E29.3 of the five individuals were subjected to XcmI digestion. The DNA sequence encoding the Rg1 determinant is distinguishable by XcmI, and the LSP product associated with C4-Rg1 is thereby split into the 5′ fragment with 585 bp and the 3′ fragment with 520 bp. The product associated with C4-Ch1 remains as a single 1,110-bp fragment. In an autoradiograph, only the fragments that carry the radiolabeled E29.3 primers (i.e., the full-length 1,110 bp for Ch1 and the 3′ fragment of 520 bp associated with Rg1) are detectable. As in the case for LSP-PshAI, the ratios of Rg1:Ch1 for each of the five individuals were measured through five independent experiments, both from digital images of ethidium bromide gels and from the respective phosphorimaging data of the Southern blots. The results are shown in figure 4.
Similar to the scenario described for the LSP-PshAI, a skewed, higher intensity for the 1,110-bp fragment for Ch1 was observed in the ethidium bromide gels from every sample that largely reflects the effects of heteroduplexes. By contrast, the Rg1:Ch1 ratios from phosphorimaging data of LSP-XcmI for C064, C027, C007, and C008 are 1:(1.8±0.2), (2.5±0.3):1, (3.2±0.5):1, and (3.76±0.3):1, respectively. In a parallel study on an individual with two C4A and C4B genes, the apparent ratio of Rg1:Ch1 obtained from ethidium bromide gels was 1:(1.9±0.3), which is in contrast to a ratio of 1:(1.0±0.1) from the phosphorimaging data generated by an LSP-XcmI-RFLP analysis.
Figure 4E is a graphic representation comparing the expected results for the relative gene dosages of C4A and C4B with results obtained from LSP-PshAI and LSP-XcmI and with apparent results from ethidium bromide gels. The LSP-RFLP results are closer to the expected results. However, under conditions with greatly disparate proportions between C4A and C4B or between C4-Rg1 and C4-Ch1, the LSP-PshAI and LSP-XcmI methods may underevaluate the genes with the higher dosage (i.e., C4A and C4-Rg1).
A Novel Approach to Elucidate the Haplotypes of RCCX Length Variants and C4 Gene Numbers by PmeI PFGE
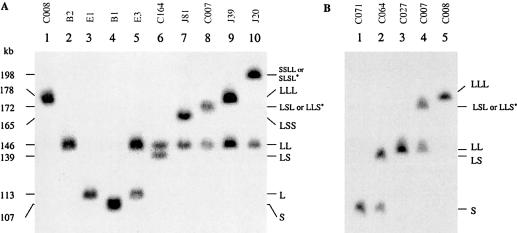
To deduce a method that allows consistent and definitive haplotypic characterization of the number and size of RCCX modules, we analyzed the MHC class III genomic DNA sequences generated in our laboratory (Yu et al. 2000) and by the Human Genome Project (MHC Sequencing Consortium 1999) for informative restriction sites. The analysis showed the presence of unique PmeI restriction sites at the 5′ region of the factor B gene and at the 5′ end of the TNXB gene but showed none in between (fig. 1). PmeI recognizes an 8-bp recognition site, GTTTAAAC, and its cleavage is not dependent on the unmethylation of CpG sequences. Therefore, the size of the PmeI fragments resolved by PFGE hybridized to a C4d probe in a Southern blot faithfully reflects the number of the RCCX modules and the size of C4 genes that are present in a haplotype. A monomodular structure with a short C4 gene is shown as a 107-kb PmeI fragment and a monomodular structure with a long C4 gene, a 113-kb fragment. The sizes of PmeI fragments for other RCCX structures are as follows: bimodular LL, 146 kb; bimodular LS, 139 kb; trimodular LLL, 178 kb; trimodular LSS, 165 kb; and trimodular LLS or LSL, 172 kb.
Figure 5A depicts the result of RCCX patterns with the PmeI PFGE from 10 individuals. Lanes 1, 2, 3, and 4 are from four individuals with homozygous structures with trimodular LLL, bimodular LL, monomodular L, and monomodular S, respectively. Lanes 5–10 are from individuals with heterozygous RCCX structures. Among these latter individuals (lanes 5–10), it is common for one of their RCCX haplotypes to be the bimodular LL; the other RCCX haplotypes among them are monomodular L (lane 5), bimodular LS (lane 6), trimodular LSS (lane 7), trimodular LLS or LSL (lane 8), trimodular LLL (lane 9), and quadrimodular SSLL/SLSL/SLLS (lane 10). The RCCX patterns of these 10 individuals were independently analyzed by TaqI RFLP, and the results (data not shown) are in good agreement with those obtained with PmeI PFGE.
Figure 5.
Elucidation of the number and size of RCCX modules present in MHC haplotypes by PmeI PFGE. A, Selected individuals, to show homozygous and heterozygous haplotypes of length variants of RCCX structures. B, PmeI RCCX patterns of the five individuals with two to six C4 genes, as characterized in figure 2.
Figure 5B further illustrates the results of a PmeI-PFGE analysis using genomic DNA from the same five individuals characterized in the previous sections. As revealed by a conventional genomic RFLP analysis, C071 has homozygous S/S structures, C064 is heterozygous for LS/S, and C027 is homozygous for LL/LL. C007 (fig. 5B, lane 4, and fig. 5A, lane 8) has heterozygous, trimodular LSL (or LLS) and bimodular LL. C008 has homozygous LLL/LLL (fig. 5B, lane 5, and fig. 5A, lane 1).
Discussion
A number of diagnostic techniques have been presented, to accurately determine the gene dosage of human complement C4A and C4B and RCCX length variants. These techniques are derived from our current knowledge of the gene organizations and the sequence data of the HLA class III region. For genomic RFLPs, we chose the restriction enzymes that yield the maximum information, and we deliberately designed length and locations of the hybridization probes so that the band intensities for each set of restriction fragments on the autoradiographs or phosphorimages faithfully reflect the relative gene dosage of the corresponding genes or gene segments. The TaqI genomic Southern blots using RP, CYP21, and TNX probes yield information on the length variants of the RCCX modules, on the total number of long and short C4 genes linked to RP1 or to RP2, on the relative dosage of CYP21B and CYP21A, and on the relative dosage of TNXB and TNXA. Results of RP-PshAI Southern blots further provided independent information, on the relative dosage of RP1 and RP2, that essentially yields the number of RCCX modules present in the diploid genome of an individual. A PshAI or PshAI-PvuII genomic Southern blot hybridized with a C4d probe gives the results on the relative dosage of the C4A and C4B genes. The latter is designed to minimize artifacts caused by uneven transfer of the C4B and C4A restriction fragments because of size difference. The limitation of genomic RFLPs in the determination C4A and C4B gene dosage and RCCX variants is their requirement of relatively large quantities of genomic DNA, which may pose constraints in multilocus genetic studies.
Two PCR-based methods to determine C4A and C4B gene dosages have been developed to facilitate genotyping when quantities of genetic materials are limited. The module-specific PCR yields information on the number of RCCX modules and, therefore, on the total number of C4 genes that are present in a diploid genome. Because two different amplicons employ one common primer and two specific primers, the PCR products are a function of the RCCX module number and also of the concentration of the available primers. A titration using standard genomic DNA samples with different C4 gene numbers is necessary to determine the appropriate quantities of the specific primers, to yield the expected results.
To further determine the relative dosages of C4A and C4B, we subjected a C4d fragment amplified by PCR to LSP-PshAI– and LSP-XcmI–RFLP analyses. The application of a labeled primer for an additional cycle of DNA synthesis circumvents the heteroduplex effect on diagnostic restriction analyses and allows for a more reliable quantitation. We refined the original hot-stop PCR protocol (Uejima et al. 2000), to increase the sensitivity of detection by purifying the PCR products and performing a single cycle of DNA synthesis in the presence of fresh reagents and a labeled DNA primer prior to the diagnostic RFLP analysis. Our experiments showed that apparent results obtained through the comparison of the band intensities of the ethidium bromide–stained images deviated significantly from the expected C4A and C4B gene dosages. The C4B-specific fragments or the C4-Ch1 fragments are usually overrepresented by approximately twofold. The results obtained by phosphorimaging analyses of LSP-RFLP images are closer to the expected C4A and C4B gene dosages. This method is most accurate when the relative gene dosages of C4A and C4B are within the range from 2:1 to 1:2. Extreme ratios of C4A:C4B gene dosages (e.g., 4:1 or 5:1), however, have tendencies of slight underestimation. A standardization curve using the range of known C4A and C4B gene dosages may be useful to correct this pitfall. Alternatively, the precise C4 gene dosage can be determined by the application of PmeI PFGE.
Unlike the PCR-RFLP analysis, the module-specific PCR described here is indifferent to artifacts of heteroduplexes. This is probably because the module-specific PCR procedure does not require a restriction-enzyme digest by which the heteroduplexes blemish the outcomes.
We attempted an alternative approach to the determination of the number of C4A and C4B genes by employing real-time PCR (Higuchi et al. 1993). The notion was to generate amplicons spanning the exon 26–intron 26 region in the presence of two fluorescent probes, one specific to C4A and the other specific to C4B, parallel to the amplification of a single-copy gene as an internal control. Thus, the results would yield the actual C4A and C4B gene numbers. Although theoretically feasible, the technique proved to be difficult and required extensive optimization because of different efficiencies and potential cross-hybridizations of the C4A and C4B oligonucleotide probes annealing to the C4 isotypic sequences.
The PmeI PFGE allows for elucidation of the number and size of the RCCX modules or C4 genes that are present in haplotypes, definitively and unambiguously. The recognition and cleavage site of PmeI has no CpG sequence that is common in most restriction enzymes used for long-range mapping by PFGE. Therefore, the PmeI-digested fragments are predictable, homogeneous, and independent of the tissue origins of the genomic DNA, and they reflect the actual size of DNA fragments with the recognition sequences. Moreover, size ranges of the restriction fragments allow a precise resolution of the RCCX length variants in a PFGE gel. The variation in the C4 gene copy number had been investigated previously by PFGE using BssHII-, SacII-, MluI-, or PvuI-digested genomic DNA. Large restriction DNA fragments were generated by cleavage of unmethylated CpG islands that may be heterogeneous in nature, leading to “partially digested” DNA fragments (Dunham et al. 1989; Zhang et al. 1990). It is expected that the application of diagnostic techniques created in the present study may advance our knowledge on the effects of C4A and C4B gene dosage and RCCX modular variations on infectious and autoimmune diseases.
Acknowledgments
We wish to sincerely thank the blood donors for their participation in this study and Dr. Maisa Lokki (Finnish Red Cross, Helsinki) for HLA typing. This work was supported by the National Institute of Arthritis and Musculoskeletal and Skin Diseases (grant R01 AR43969), the National Institute of Diabetes and Digestive and Kidney Diseases (grant P01 DK55546), an institutional grant from the Columbus Children's Research Institute (297401), and Pittsburgh Supercomputing Center (through National Institutes of Health Center for Research Resources Cooperative Agreement grant 1P41 RR06009).
Electronic-Database Information
- Entrez Nucleotide, http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=nucleotide (for C4A [accession number M59816-U07856-M58915], long C4B gene [accession number AF019413], short C4B gene [accession number AL049547 and U24578], RP1 or STK19 [accession numbers L26260 and L26261], RP2 [accession numbers L26262 and L26263], CYP21B [accession numbers M26856, M12792, M13936, and AF77974], CYP21A [accession numbers M26857, M12793, and M13935], TNXB [accession number U89337], and TNXA [accession numbers L26263 and U24488])
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for C4A [MIM 120810], C4B [MIM 120820], CYP21 and congenital adrenal hyperplasia [MIM 201910], RP1 or STK19 [MIM 604977], TNXB [MIM 600985], and SLE [MIM 152700])
References
- Atkinson JP, Schneider PM (1999) Genetic susceptibility and class III complement genes. In: Lahita RG (ed) Systemic lupus erythematosus. Academic Press, San Diego, pp 91–104 [Google Scholar]
- Awdeh ZL, Alper CA (1980) Inherited structural polymorphism of the fourth component of human complement. Proc Natl Acad Sci USA 77:3576–3580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awdeh ZL, Raum D, Alper CA (1979) Genetic polymorphism of human complement C4 and detection of heterozygotes. Nature 282:205–208 [DOI] [PubMed] [Google Scholar]
- Baldwin WM III, Samaniego-Picota M, Kasper EK, Clark AM, Czader M, Rohde C, Zachary AA, Sanfilippo F, Hruban RH (1999) Complement deposition in early cardiac transplant biopsies is associated with ischemic injury and subsequent rejection episodes. Transplantation 68:894–900 [DOI] [PubMed] [Google Scholar]
- Bishof NA, Welch TR, Beischel LS (1990) C4B deficiency: a risk factor for bacteremia with encapsulated organisms. J Infect Dis 162:248–250 [DOI] [PubMed] [Google Scholar]
- Blanchong CA, Chung EK, Rupert KL, Yang Y, Yang Z, Zhou B, Yu CY (2001) Genetic, structural and functional diversities of human complement components C4A and C4B and their mouse homologs, Slp and C4. Int Immunopharmacol 1:365–392 [DOI] [PubMed] [Google Scholar]
- Blanchong CA, Zhou B, Rupert KL, Chung EK, Jones KN, Sotos JF, Rennebohm RM, Yu CY (2000) Deficiencies of human complement component C4A and C4B and heterozygosity in length variants of RP-C4-CYP21-TNX (RCCX) modules in Caucasians: the load of RCCX genetic diversity on MHC-associated disease. J Exp Med 191:2183–2196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll MC, Palsdottir A, Belt KT, Porter RR (1985) Deletion of complement C4 and steroid 21-hydroxylase genes in the HLA class III region. EMBO J 4:2547–2552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung EK, Yang Y, Rennebohm RM, Lokki ML, Higgins GC, Jones KN, Zhou B, Blanchong CA, Yu CY (2002) Genetic sophistication of human complement components C4A and C4B and RP-C4-CYP21-TNX (RCCX) modules in the major histocompatibility complex. Am J Hum Genet 71:823–837 (in this issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier S, Sinnott PJ, Dyer PA, Price DA, Harris R, Strachan T (1989) Pulsed field gel electrophoresis identifies a high degree of variability in the number of tandem 21-hydroxylase and complement C4 gene repeats in 21-hydroxylase deficiency haplotypes. EMBO J 8:1393–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colten HR, Garnier G (1998) Regulation of complement protein gene expression. In: Volanakis JE, Frank MM (eds) The human complement system in health and disease. Marcel Dekker, New York, pp 217–240 [Google Scholar]
- Dangel AW, Mendoza AR, Baker BJ, Daniel CM, Carroll MC, Wu L-C, Yu CY (1994) The dichotomous size variation of human complement C4 gene is mediated by a novel family of endogenous retroviruses which also establishes species-specific genomic patterns among Old World primates. Immunogenetics 40:425–436 [DOI] [PubMed] [Google Scholar]
- Daniels CA, Borsos T, Rapp HJ, Snyderman R, Notkins AL (1969) Neutralization of sensitized virus by the fourth component of complement. Science 165:508–509 [DOI] [PubMed] [Google Scholar]
- Dodds AW, Ren X-D, Willis AC, Law SKA (1996) The reaction mechanism of the internal thioester in the human complement component C4. Nature 379:177–179 [DOI] [PubMed] [Google Scholar]
- Dunham I, Sargent CA, Dawkins RL, Campbell RD (1989) Direct observation of the gene organization of the complement C4 and 21-hydroxylase loci by pulsed field gel electrophoresis. J Exp Med 169:1803–1816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feucht HE, Schneeberger H, Hillebrand G, Burkhardt K, Weis M, Riethmuller G, Land W, Albert E (1993) Capillary deposition of C4d complement fragment and early renal graft loss. Kidney Int 43:1333–1338 [DOI] [PubMed] [Google Scholar]
- Gitelman SE, Bristow J, Miller WL (1992) Mechanism and consequences of the duplication of the human C4/P450c21/gene X locus. Mol Cell Biol 12:2124–2134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi R, Fockler C, Dollinger G, Watson R (1993) Kinetic PCR analysis: real-time monitoring of DNA amplification reactions. Biotechnology (NY) 11:1026–1030 [DOI] [PubMed] [Google Scholar]
- Jaatinen T, Ruuskanen O, Truedsson L, Lokki M-L (1999) Homozygous deletion of the CYP21A-TNXA-RP2-C4B gene region conferring C4B deficiency associated with recurrent respiratory infections. Hum Immunol 60:707–714 [DOI] [PubMed] [Google Scholar]
- Jack DL, Klein NJ, Turner MW (2001) Mannose-binding lectin: targeting the microbial world for complement attack and opsonophagocytosis. Immunol Rev 180:86–99 [DOI] [PubMed] [Google Scholar]
- Kohl J (2001) Anaphylatoxins and infectious and non-infectious inflammatory diseases. Mol Immunol 38:175–187 [DOI] [PubMed] [Google Scholar]
- Mauff G, Luther B, Schneider PM, Rittner C, Strandmann-Bellinghausen B, Dawkins R, Moulds JM (1998) Reference typing report for complement component C4. Exp Clin Immunogenet 15:249–260 [DOI] [PubMed] [Google Scholar]
- MHC Sequencing Consortium (1999) Complete sequence and gene map of a human major histocompatibility complex. Nature 401:921–923 [DOI] [PubMed] [Google Scholar]
- Olaisen B, Teisberg P, Jonassen R (1980) The C4 system: quantitative studies of different genotypes. Immunobiology 158:82–85 [DOI] [PubMed] [Google Scholar]
- O'Neill GJ, Yang SY, DuPont B (1978) Two HLA-linked loci controlling the fourth component of human complement. Proc Natl Acad Sci USA 75:5165–5169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer PH, Brems JJ, Brunson M, Hugli TE (2000) Plasma C3a and C4a levels in liver transplant recipients: a longitudinal study. Immunopharmacology 46:163–174 [DOI] [PubMed] [Google Scholar]
- Pinckard RN, Olson MS, Giclas PC, Terry R, Boyer JT, O'Rourke RA (1975) Consumption of classical complement components by heart subcellular membranes in vitro and in patients after acute myocardial infarction. J Clin Invest 56:740–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter RR (1983) Complement polymorphism, the major histocompatibility complex and associated diseases: a speculation. Mol Biol Med 1:161–168 [PubMed] [Google Scholar]
- Schneider PM, Carroll MC, Alper CA, Rittner C, Whitehead AS, Yunis EJ, Colten HR (1986) Polymorphism of human complement C4 and steroid 21-hydroxylase genes: restriction fragment length polymorphisms revealing structural deletions, homoduplications, and size variants. J Clin Invest 78:650–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider PM, Witzel-Schlomp K, Rittner C, Zhang L (2001) The endogenous retroviral insertion in the human complement C4 gene modulates the expression of homologous genes by antisense inhibition. Immunogenetics 53:1–9 [DOI] [PubMed] [Google Scholar]
- Shen L, Wu L-C, Sanlioglu S, Chen R, Mendoza AR, Dangel AW, Carroll MC, Zipf WB, Yu CY (1994) Structure and genetics of the partially duplicated gene RP located immediately upstream of the complement C4A and the C4B genes in the HLA class III region: molecular cloning, exon intron structure, composite retroposon, and breakpoint of gene duplication. J Biol Chem 269:8466–8476 [PubMed] [Google Scholar]
- Sim E, Cross S (1986) Phenotyping of human complement component C4, a class III HLA antigen. Biochem J 239:763–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsokos GC, Kammer GM (2000) Molecular aberrations in human systemic lupus erythematosus. Mol Med Today 6:418–424 [DOI] [PubMed] [Google Scholar]
- Uejima H, Lee MP, Cui H, Feinberg AP (2000) Hot-stop PCR: a simple and general assay for linear quantitation of allele ratios. Nat Genet 25:375–376 [DOI] [PubMed] [Google Scholar]
- Uko G, Christiansen FT, Dawkins RL, McCann VJ (1986) Reference ranges for serum C4 concentrations in subjects with and without C4 null alleles. J Clin Pathol 39:573–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weg-Remers S, Brenden M, Schwarz E, Witzel K, Schneider PM, Guerra LK, Rehfeldt IR, Lima MT, Hartmann D, Petzl-Erler ML, de Messias IJT, Mauff G (1997) Major histocompatibility complex (MHC) class III genetics in two Amerindian tribes from southern Brazil: the Kaingang and the Guarani. Hum Genet 100:548–556 [DOI] [PubMed] [Google Scholar]
- Welch TR, Beischel L, Berry A, Forristal J, West CD (1985) The effect of null C4 alleles on complement function. Clin Immunol Immunopathol 34:316–325 [DOI] [PubMed] [Google Scholar]
- Wessels MR, Butko P, Ma M, Warren HB, Lage AL, Carroll MC (1995) Studies of group B streptococcal infection in mice deficient in complement component C3 or C4 demonstrate an essential role for complement in both innate and acquired immunity. Proc Natl Acad Sci USA 92:11490–11494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White PC, Speiser PW (2000) Congenital adrenal hyperplasia due to 21-hydroxylase deficiency. Endocr Rev 21:245–291 [DOI] [PubMed] [Google Scholar]
- Wild G, Watkins J, Ward AM, Hughes P, Hume A, Rowell NR (1990) C4a anaphylatoxin levels as an indicator of disease activity in systemic lupus erythematosus. Clin Exp Immunol 80:167–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Mendoza AR, Welch TR, Zipf WB, Yu CY (1999) Modular variations of HLA class III genes for serine/threonine kinase RP, complement C4, steroid 21-hydroxylase CYP21 and tenascin TNX (RCCX): a mechanism for gene deletions and disease associations. J Biol Chem 274:12147–12156 [DOI] [PubMed] [Google Scholar]
- Yu CY, Blanchong CA, Chung EK, Rupert KL, Yang Y, Yang Z, Zhou B, Moulds JM (2002) Molecular genetic analyses of human complement components C4A and C4B. In: Rose NR, Hamilton RG, Detrick B (eds) Manuals of clinical laboratory immunology, 6th ed. ASM Press, Washington, DC, pp 117–131 [Google Scholar]
- Yu CY, Yang Z, Blanchong CA, Miller W (2000) The human and mouse MHC class III region: a parade of the centromeric segment with 21 genes. Immunol Today 21:320–328 [DOI] [PubMed] [Google Scholar]
- Zhang WJ, Degli-Esposti MA, Cobain TJ, Cameron PU, Christiansen FT, Dawkins RL (1990) Differences in gene copy number carried by different MHC ancestral haplotypes. Quantitation after physical separation of haplotypes by pulsed field gel electrophoresis. J Exp Med 171:2101–2114 [DOI] [PMC free article] [PubMed] [Google Scholar]