Abstract
Human populations are endowed with a sophisticated genetic diversity of complement C4 and its flanking genes RP, CYP21, and TNX in the RCCX modules of the major histocompatibility complex class III region. We applied definitive techniques to elucidate (a) the complement C4 polymorphisms in gene sizes, gene numbers, and protein isotypes and (b) their gene orders. Several intriguing features are unraveled, including (1) a trimodular RCCX haplotype with three long C4 genes expressing C4A protein only, (2) two trimodular haplotypes with two long (L) and one short (S) C4 genes organized in LSL configurations, (3) a quadrimodular haplotype with four C4 genes organized in a SLSL configuration, and (4) another quadrimodular structure, with four long C4 genes (LLLL), that has the human leukocyte antigen haplotype that is identical to ancestral haplotype 7.2 in the Japanese population. Long-range PCR and PshAI-RFLP analyses conclusively revealed that the short genes from the LSL and SLSL haplotypes are C4A. In four informative families, an astonishingly complex pattern of genetic diversity for RCCX haplotypes with one, two, three and four C4 genes is demonstrated; each C4 gene may be long or short, encoding a C4A or C4B protein. Such diversity may be related to different intrinsic strengths among humans to defend against infections and susceptibilities to autoimmune diseases.
Introduction
Human populations are endowed with a distinct genetic diversity of complement component C4 that may confer varying intrinsic strengths on different individuals in the defense against infections, but also may confer vulnerabilities for autoimmunity (Porter 1983; Blanchong et al. 2001; Martinez et al. 2001). Since 1969, when Rosenfeld et al. (1969), using the antibody-antigen crossed electrophoresis, illustrated, for the first time, complement C4 polymorphisms in the hemolytic activities and electrophoretic mobilities, the progress toward understanding the sophisticated genetics of human C4 has taken a somewhat meandering course. It appears that there are multiple allotypes of C4 proteins (Mauff et al. 1998). These protein allotypes may be grouped into two classes: the acidic C4A (MIM 120810), with lower hemolytic activities, and the basic C4B (MIM 120820) with higher hemolytic activities (Awdeh and Alper 1980). There is a wide spectrum of C4A and C4B protein levels in the population—from complete absence of C4A or C4B, more C4A than C4B, and more C4B than C4A to equal quantities of C4A and C4B (Moulds et al. 1990; Rebmann et al. 1992). A single locus with codominant expressions of the fast- and slow-migrating forms of C4 was first proposed (Teisberg et al. 1976) but was soon replaced by a two-locus model with one C4A gene and one C4B gene (O'Neill et al. 1978). The differential levels of C4A and C4B proteins in the plasma were explained by the presence of null or silent alleles in either the C4A locus or the C4B locus. The model was supported by work from independent laboratories (Awdeh et al. 1979; Roos et al. 1982). However, there are abundant exceptions to a rigid C4A-C4B, two-locus model of C4 genes in the major histocompatibility complex (MHC) class III region on chromosome 6 (Teisberg et al. 1976; Carroll et al. 1984). Family segregation studies showed considerable frequencies of single-locus haplotypes encoding C4A or C4B proteins and of three-locus haplotypes encoding one C4A and two different C4B allotypes (Olaisen et al. 1980; Teisberg et al. 1988). There is also mounting evidence for the homoexpression of C4A proteins from two consecutive C4 loci (e.g., C4A3 A2) (Raum et al. 1984; Rittner et al. 1984; Schendel et al. 1985). The phenomenon was observed because, in those cases, each C4 gene encodes a polymorphic variant distinguishable in an allotyping gel (Awdeh and Alper 1980; Sim and Cross 1986).
Molecular characterization of various C4A and C4B genes from individuals with defined phenotypes allowed identification of amino acid residues contributed to the differential chemical reactivities of the activated C4A and C4B, as well as their associated Rg and Ch blood-group antigens (Carroll et al. 1984, 1990; Yu et al. 1986, 1988; Dodds et al. 1996). It enabled the creation of molecular biologic techniques to determine the presence and number of C4A and C4B genes in the MHC (Chung et al. 2002; Yu et al. 2002) and led to the elucidation of dichotomous features of C4 and other constituent genes of the RP-C4-CYP21-TNX (RCCX) modules (fig. 1). First, there are long and short forms of the C4 gene, 20.6 and 14.2 kb in size, respectively. The long C4 gene has an endogenous retrovirus of 6.36 kb integrated into intron 9 (Dangel et al. 1994; Schneider et al. 2001). The size of intron 9 in the short C4 gene is only 416 bp. Second, a C4 gene may encode a C4A or C4B protein; the basis for this variation is five nucleotide substitutions in exon 26, leading to four isotype-specific amino acid residues (i.e., PCPVLD 1101-6 in C4A and LSPVIH 1101-6 in C4B) (Belt et al. 1985; Yu et al. 1986). In haplotypes with two or more C4 genes, duplication is discretely modular of 32.7 kb (long [L]) or 26.3 kb (short [S]) including an intact C4A or C4B gene, a gene for the steroid CYP21 (MIM 201910) that is frequently the pseudogene CYP21A, and partially duplicated gene fragments TNXA (MIM 600985) and RP2 (MIM 604977).
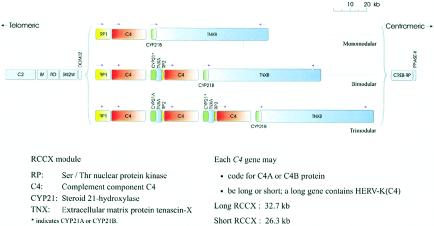
Figure 1.
The 1-2-3–locus concept of human C4 and RCCX modular variations. This map of the human MHC class III region shows the physical length variants among the monomodular, bimodular, and trimodular RCCX structures. The MHC class I genes are located at the telomeric end; the MHC class II genes are located at the centromeric end.
Initial structural characterization for a limited number of clones revealed that the C4 gene that is linked to factor B at the 5′ region (i.e., located at the telomeric end) is a long gene encoding C4A; a short C4B gene is present at the centromeric region linked to the functional CYP21B gene (Carroll et al. 1984, 1985). However, population studies using an informative or definitive RFLP analysis coupled with phenotypic characterization of C4A and C4B proteins revealed a great repertoire of length variants for the RCCX modules with different combinations of genotypic features for the RCCX constituents (Blanchong et al. 2000). A dynamic 1-2-3–locus model for C4 genes is proposed as close to one-third of the chromosomes 6 in the normal white population that has either monomodular RCCX haplotypes with a single copy of C4 gene or trimodular haplotypes with three copies of C4 genes in one chromosome. Only 55% of the white MHC haplotypes have the two-locus structures with C4A-C4B configurations in the MHC. The remainder have different combinations (or unequal numbers) of C4A and C4B genes. It is speculated that the diversities in C4A and C4B protein levels and in functional properties were one of the intrinsic immune responses to defend against a vast variety of bacterial, viral, and other microbial infections. The high frequency of heterozygosity in RCCX length variants is probably a mechanism to achieve or maintain diversities of C4A and C4B proteins in the population, since it would allow the exchange of genetic information among homologs of the RCCX constituents. However, heterozygous length variants of RCCX modules could also promote unequal crossovers during meiosis, leading to gene deletions, gene duplications, and disease associations (Rupert et al. 1999; Z. Yang et al. 1999; Blanchong et al. 2000, 2001; Jaatinen et al. 2002).
To define the dosage of C4A and C4B genes together with the other RCCX constituents, we have applied novel, more accurate techniques to directly address the number of C4 genes present in a diploid genome and in an MHC haplotype. Thus, we further demonstrate the sophistications of C4A and C4B genetics. New varieties of RCCX modular haplotypes are shown. These include two trimodular structures with LSL configurations, a trimodular LLL haplotype with homoexpression of C4A, and two quadrimodular haplotypes, each with four C4 genes, from pediatric patients with juvenile rheumatoid arthritis (JRA) or with systemic lupus erythematosus (SLE [MIM 152700]).
Material and Methods
Human Subjects, Genomic DNA, and Southern Blot Analysis
Peripheral-blood samples were taken from healthy individuals, patients with SLE, and patients with pauciarticular JRA and relatives after informed consent was obtained, after approval by the Columbus Children's Hospital institutional review board. The isolation of genomic DNA and Southern blot analysis were performed as described elsewhere (Blanchong et al. 2000; Chung et al. 2002).
Labeled-Primer Single-Cycle Polymerization (LSP)–RFLP Analysis of C4A and C4B
A 1.1-kb genomic fragment spanning complement C4 exons 26–29 was amplified by PCR and was subjected to PshAI-LSP RFLP and XcmI-LSP RFLP, as described elsewhere (Chung et al. 2002).
Human Leukocyte Antigen (HLA) Typing
HLA class I genes A, B, C, and class II genes DRB1, DRB3/4/5, DPB1, and DQB1 were typed by sequence-specific primer PCR of genomic DNA, using commercial kits from One Lambda (for HLA A, B, and DR), Pel-Freez (for HLA C), and INNO-LiPA (Innogenetics; for HLA DQB1 and DPB1) and standardized protocols.
Complement C4 and C3
Polymorphisms of complement C4A and C4B proteins were determined by immunofixation of EDTA-plasma after published protocols (Yu et al. 2002). Complement C3 typing was performed by immunofixation using a method similar to that of C4, except that the quantity of plasma was reduced by fivefold. Goat antiserum for human C3 was purchased from Diasorin.
The concentrations of human C4 in EDTA-plasma were determined by a sandwiched-ELISA method. All dilutions were performed using PBS (pH 7.4). In brief, diluted goat anti-human C4 sera (dilution, 1:300) in PBS was coated on ELISA plate overnight at room temperature. Washing with PBS and treatment with blocking buffer (Pierce) were performed four times after coating, reactions with diluted plasma, primary antibody, and secondary antibody. Plasma samples at 1:4,000 dilution was added to each well and incubated at room temperature for 1.5 h. The primary antibody used was a rabbit antiserum against human C4 (immunoglobulin G fraction; Sigma; 1:50 dilution). The secondary antibody used was a mouse monoclonal against rabbit immunoglobulin G conjugated with alkaline phosphatase (Sigma; 1:40,000 dilution). After addition of the substrate PNPP (obtained from Pierce) for 1 h, the reaction was terminated by addition of 0.5 M NaOH, and optical density at 405 nm was determined with an ELISA plate reader. A standardization curve of C4 protein levels was determined using purified C4 protein (Calbiochem), with quantities varying from 1.56 ng to 100 ng and subjected to the same experimental procedures in the same microtiter plate. C4 protein level for each plasma sample was assayed twice and each time in triplicate. The validity of the assay was verified with a radial immunodiffusion method using a kit purchased from the Binding Site.
Pulsed-Field Gel Electrophoresis (PFGE) of PmeI- and PacI-Digested Genomic DNA
Genomic DNA in agarose plugs were digested with PmeI or PacI restriction enzymes and processed as described elsewhere (Chung et al. 2002; Yu et al. 2002). The following conditions were optimized for PacI PFGE: gradient, 6 V/cm; calibration factor, 1.4; initial switch time, 1.79 s; final switch time, 12.91 s; run time, 37 h 40 min; included angle, 120°; ramp, linear. The PmeI- and PacI-PFGE gels were processed by Southern blot; were hybridized with probes specific to C4d and to the 3′ region of TNX, respectively; and were subjected to autoradiography.
Long-Range PCR of the Short C4 Genes and PshAI-RFLP Analysis
Primers E9.5 and E31.3 were designed to anneal to the 5′ end of exon 9 and to the 3′ end of exon 31 in C4, respectively. The sequence for primer E9.5 is 5′-CCC TGG AGA AGC TGA ATA TGG-3′ and E31.3 is 5′-CTT CAG GGT TCC TTT GCT GT-3′. A 1 × Premix D with the Failsafe PCR Enzyme Mix (1.25 U) (Epicentre) was used. PCR conditions were as follows: 1 cycle at 94°C for 3 min; 8 cycles at 94°C for 45 s, 66°C for 1 min (decrease 0.5°C/cycle), 72°C for 9 min; 30 cycles at 94°C for 45 s, 60°C for 1 min, 72°C for 9 min; and 1 cycle at 72°C for 15 min. For the RFLP analysis, one-fifth of the PCR product was digested with PshAI (15 U) overnight at 25°C and was resolved by electrophoresis using 1% agarose gel.
Results
Homoexpression of C4A3 from a Trimodular RCCX Haplotype with Three Long C4 Genes
Previous molecular genetic analysis revealed that white female C008 has a total of six C4 genes in a diploid genome. PmeI-PFGE and TaqI-RFLP analyses showed that C008 has the LLL/LLL structures for the RCCX modules. In genomic PshAI or PshAI-PvuII Southern blots hybridized to a C4d-specific probe, C008 exhibited an unusual C4A:C4B ratio of 5:1 (Chung et al. 2002). Together with C4-allotyping data, heterozygous C4A3 A3 A3/C4A3 A3 B1 haplotypes are predicted. The presence of three C4A genes on a chromosome (or haplotype) has never been reported. To confirm this observation, we recruited family members (two parents and two children) of C008 for segregation analysis (fig. 2A).
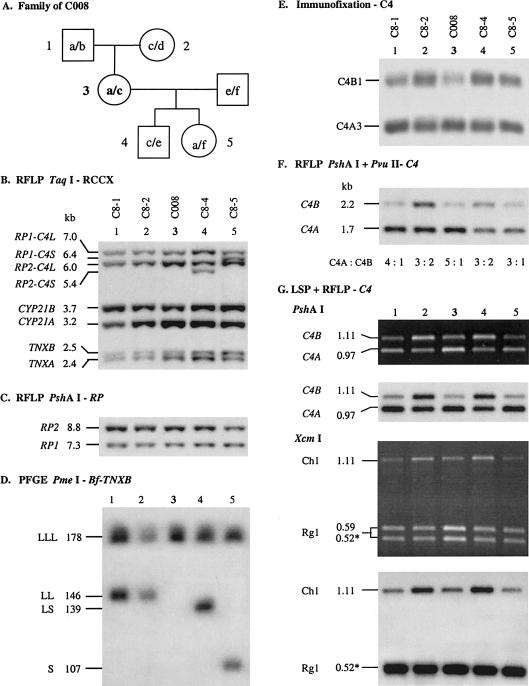
Figure 2.
Homoexpression of C4A in a trimodular LLL haplotype from a white family. A, Pedigree of the family C008. B, Genomic TaqI RFLP, to show the RCCX modular variations. C, Genomic PshAI-RFLP, to show the number of RCCX modules by determining the relative dosages of RP2 and RP1. D, PmeI-PFGE analysis, to show the length variants of RCCX haplotypes. E, Immunofixation, to show the relative quantities of C4A and C4B allotypes. F, PshAI-PvuII RFLP, to show the relative dosage of C4A and C4B genes in each individual. G, LSP-RFLP analysis, to determine the relative dosage of C4A and C4B. The dark-background panels are ethidium bromide–stained pictures that contain artifact results because of heteroduplexes. The light-background pictures are results of autoradiography corresponding to LSP-RFLP images free of heteroduplex effects.
Consistent results from the TaqI and PshAI RFLPs and from the PmeI PFGE (figs. 2B–2D) indicate that the parents, C8-1 and C8-2, each carry the heterozygous LLL/LL RCCX structures and a total of five C4 genes. The two children, C8-4 and C8-5, have the heterozygous LLL/LS and LLL/S structures and a total of five and four C4 genes, respectively. Interestingly, the ratio of C4A:C4B as measured in the genomic PshAI-PvuII RFLP (fig. 2F), LSP-PshAI RFLP, and LSP-XcmI RFLP (fig. 2G) for C8-1 was 4:1, whereas that for C8-2 was 3:2. Therefore, it was deduced that C8-1 has haplotypes of C4A3 A3 A3/C4A3 B1, and that C8-2 has haplotypes of C4A3 A3 B1/C4A3 B1. This suggests that the C4A3 A3 A3 haplotype of C008 originates from C8-1. The dosage of C4A and C4B were faithfully reflected in the corresponding C4 allotypes (fig. 2E). By contrast, the ratio of C4A:C4B as determined by the same techniques for C8-4 and C8-5 were 3:2 and 3:1, respectively. Therefore, it was deduced that C8-4 has haplotypes of C4A3 A3 B1/C4A3 B1 and that C8-5 has haplotypes of C4A3 A3 A3/C4AQ0 C4B1. In essence, the family segregation study provides informative support for the presence of homoexpression of C4A3 from a trimodular LLL structure that is present in all three generations: C8-1, C008, and C8-5.
TaqI RFLP revealed that C8-1 has three CYP21B and two CYP21A genes. On examination of the segregation of CYP21B and CYP21A (fig. 2B, lane 1), it becomes clear that the nontransmitted haplotype from C8-1 (i.e., bimodular LL) has the unusual configuration with 21B-21B. It is of interest that, among the six RCCX haplotypes present in the family of C008, there are six different combinations of C4A, C4B, CYP21A, and CYP21B genes.
Trimodular RCCX with Two Long and One Short C4 Genes
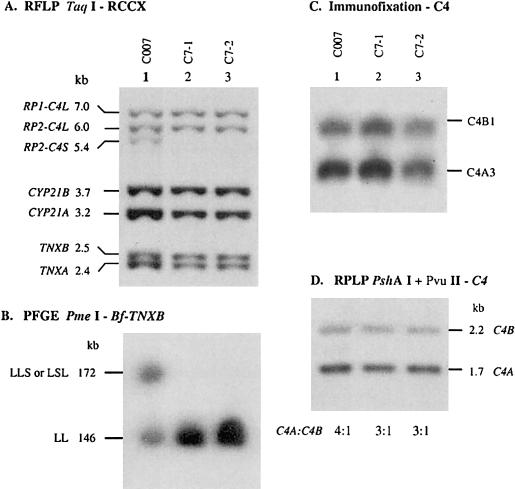
An RFLP analysis established that individual C007 has heterozygous bimodular and trimodular RCCX structures with a total of four long and one short C4 genes in her genome. These C4 genes could be organized as LLL/LS or LLS/LL or LSL/LL. PmeI-PFGE analysis of C007 revealed a 146-kb fragment corresponding to the bimodular LL haplotype that is also shared by her two siblings, C7-1 and C7-2 (figs. 3A and 3B, lanes 2 and 3). C007 also has a 172-kb PmeI fragment corresponding to a trimodular structure with two long and one short C4 genes, implying the presence of a novel trimodular RCCX structure that may be either LLS or LSL.
Figure 3.
A family with a trimodular RCCX haplotype consisting of two long and one short C4 genes. A, Genomic TaqI-RFLP analysis of RCCX modules. B, PmeI-PFGE analysis, to show haplotypes of RCCX length variants. C, Immunofixation of EDTA-plasma, to show the qualitative and quantitative variations of C4A and C4B proteins. D, Genomic PshAI-PvuII RFLP, to determine the relative dosage of C4A and C4B.
Quadrimodular RCCX with Two Short and Two Long C4 Genes in a Family with JRA
Case report.—J20 is a white female who developed definite pauciarticular-onset JRA at 21 mo of age, when she presented with left-knee synovitis. She has now been followed for 10 years since the onset of her disease. Her JRA has followed a polycyclic pauciarticular course. The only joint that has ever been affected has been her left knee. She has experienced three remissions and three exacerbations of left-knee synovitis since her initial episode. She has been ANA positive but has never been noted to have uveitis. Family history reveals a maternal aunt with adult-onset rheumatoid arthritis.
Molecular genetic analysis.—The TaqI-RFLP analysis of J20 shows the presence of the 7.0-, 6.4-, 6.0-, and 5.4-kb restriction fragments with respective relative band intensities of 1:1:3:1, indicating the presence of six C4 genes in total but a puzzled combination of long and short C4 genes. PshAI-RFLP analysis for the relative dosage of RP2 and RP1 revealed a 2:1 ratio with six RCCX modules on the two chromosomes (fig. 4B). PmeI PFGE clearly shows that one of the chromosomes has the bimodular LL and that the other chromosome has a quadrimodular RCCX structure with two short C4 genes and two long C4 genes (fig. 4E). The configuration of these four C4 genes is undetermined. Since the first RCCX module (i.e., module I) on chromosome 6 always starts with the RP1 that may be linked to a long C4 gene (characterized by a 7.0-kb TaqI fragment) or to a short C4 gene (characterized by a 6.4-kb TaqI fragment), the quadrimodular RCCX chromosome in J20 starts with RP1 linked to a short C4 gene because of the presence of the 6.4-kb TaqI fragment. Therefore, the configuration of C4 genes in the quadrimodular structure has one of the following three combinations: SSLL, SLSL, or SLLS.
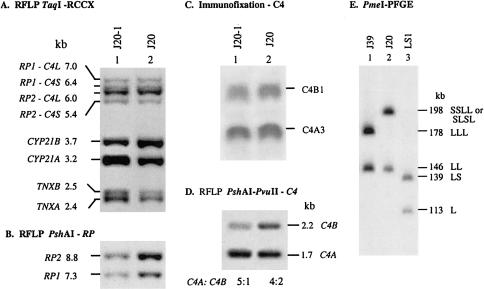
Figure 4.
A patient with JRA who had the quadrimodular RCCX haplotype consisting of two short C4 and two long C4 genes. A, Genomic TaqI-RFLP analysis of RCCX modules. B, PshAI RFLP, to show the relative dosage of RP2 and RP1. C, Immunofixation, to determine the C4A and C4B allotypes. D, Genomic PshAI-PvuII RFLP, to determine the relative gene dosage of C4A and C4B. E, PmeI PFGE, to show the quadrimodular RCCX in J20 (lane 2). Lanes 1 and 3 are references for LLL/LL and LS/L RCCX structures, respectively.
One of the parents of J20, J20-1, was recruited for analysis. PmeI PFGE revealed that J20-1 and J20 have identical patterns with the quadrimodular and bimodular haplotypes (data not shown). In TaqI RFLP, J20-1 has the same 1:1:3:1 RP-C4 gene dosages (fig. 4A). However, there are two notable differences. First, instead of an equal ratio of CYP21A:CYP21B, as seen in J20, a 2:1 ratio was observed in J20-1, suggesting that the CYP21 haplotypes for J20 are 21A-21A-21A-21B/21B-21B and that those for J20-1 are 21A-21A-21A-21B/21A-21B. A PshAI-PvuII Southern blot analysis showed that the ratio of C4A:C4B genes in J20 was 2:1 (or 4:2) but showed an astounding 5:1 ratio in J20-1 (fig. 4D, lane 1). Phenotyping experiments revealed the expression of more C4A3 allotype than the C4B1 allotype in the blood plasma of both individuals (fig. 4C). It is therefore predicted that J20 has haplotypes C4A3 A3 A3 B1/A3 B1, whereas J20-1 has haplotypes C4A3 A3 A3 B1/C4A3 A3.
Quadrimodular RCCX with Four Long C4 Genes in a Family with Pediatric Lupus
Case report.—CS-P4 is an Asian American female who developed definite SLE at age 12 years. Six of the 11 American College of Rheumatology 1997 revised criteria for classification of SLE were fulfilled (Tan et al. 1982; Hochberg 1997). The first manifestation of her lupus was the development of multiple small-vessel vasculitic skin lesions on her fingers and toes (at distal digital tufts, along the sides of her fingers, and within palmar flexor creases on the fingers). Shortly thereafter, she developed a malar rash, polyarthritis, fatigue, a mouth sore on her hard palate, and mild lymphadenopathy.
Initial laboratory testing revealed positive results for the following: antinuclear antibody (1:1,280, speckled pattern), anti-dsDNA antibody (1:40, Crithidia assay), anti-Sm antibody (1:32), anti-RNP antibody (1:64), anti-SS-A antibody (1:128), rheumatoid factor (1:320), anti-Ribosomal P (130, with normal being <20), Coombs, and lupus anticoagulant. Other abnormalities included the following: low C3 (20, with normal being >85), low C4 (<10, with normal being >15), thrombocytopenia (107,000), anemia (10.8), low white-blood-cell counts (3,000), and elevated erythrocyte-sedimentation rate (50, Wintrobe). Her urinalysis results and renal function were normal.
The patient has now been followed for 14 mo since onset of symptoms. Her disease promptly came under control and has remained under good control while she was being treated, cumulatively with corticosteroid, hydroxychloroquine, and methotrexate. No additional lupus features have developed, and she has remained free of renal disease. At her last visit, her C3 and C4 were normal (95 and 23 mg/dl, respectively). The patient’s father is Chinese American. He is aware of no family history of lupus or any other autoimmune disease. The patient’s mother is Japanese. She knows little of her family history.
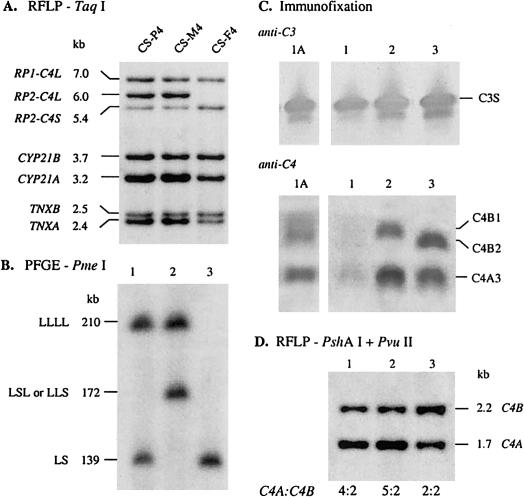
Molecular genetic analysis.—A TaqI-RFLP analysis of the RCCX gene organizations in CS-P4 revealed a 2-0-3-1 pattern for the restriction fragments corresponding to RP1-C4L, RP1-C4S, RP2-C4L, and RP2-C4S, respectively. The band intensity for CYP21B is half that of CYP21A, as is the band intensity of TNXB to TNXA (fig. 5A, lane 1). These data suggested the presence of six C4 genes or RCCX modules in this patient, among them four C4A and two C4B genes, as their corresponding PshAI-PvuII restriction fragments have a ratio of 4:2 (fig. 5D). The blood sample was taken from the patient during the lupus disease flare stage, when her C4A and C4B proteins dropped to an almost undetectable level (fig. 5C, lane 1), despite the presence of a considerable amount of complement C3 (fig. 5C, anti-C3). A fresh blood sample was taken 6 mo after her disease flare. The C4 allotyping experiment showed the presence of C4A3, B1, and B2 (fig. 5C, anti-C4, lane 1A). A sandwich ELISA using goat anti-human C4 as capture antibodies and rabbit anti-human C4 as primary antibodies confirmed a very low C4 concentration of 7.4 mg/dl during the relapse, and a partial recovery to 19.7 mg/dl during remission.
Figure 5.
Quadrimodular and trimodular RCCX structures in an Asian family with a pediatric patient with SLE. A, Genomic TaqI-RFLP analysis of RCCX modules. B, PmeI PFGE of the length variants in RCCX haplotypes. C, Immunofixation of human complement C3 (anti-C3) and C4 (anti-C4). The sample from lane 1A was taken from the pediatric patient with SLE (CS-P4) while her disease was in remission; the sample from lane 1 was taken from the patient in disease flare. Lanes 2 and 3 were samples from CS-M4 and CS-F4, respectively. D, Genomic PshAI-PvuII RFLP, to determine the relative dosage of C4A and C4B.
Analyses of the RCCX modular structures of CS-P4 revealed a quadrimodular structure with four long C4 genes (LLLL) in one chromosome and a bimodular structure (LS) with two C4 genes in the other chromosome, as suggested by the presence of the 210-kb and the 139-kb PmeI fragments (fig. 5B, lane 1), respectively. A PshAI-PvuII–RFLP analysis further suggested that CS-P4 has four C4A and two C4B genes (fig. 5D, lane 1).
The quadrimodular LLLL structure is inherited from the mother (CS-M4), who also has the 210-kb PmeI fragment (fig. 5B, lane 2). Unexpectedly, CS-M4 has a 172-kb PmeI fragment corresponding to a trimodular LLS or LSL, as in C007 (see the “Trimodular RCCX with Two Long and One Short C4 Genes” subsection, above). In other words, CS-M4 has a total of seven C4 genes or RCCX modules. Among them are probably five C4A genes (encoding C4A3) and two C4B genes (encoding C4B1) (figs. 5C and 5D, lanes 2). The combined data from TaqI-RFLP, PmeI-PFGE, PshAI-PvuII–RFLP, and alloytping experiments showed that the patient's father, CS-F4, has the homozygous bimodular LS/LS structures that encode C4A3 and C4B2.
Configurations of Long and Short C4 Genes in Trimodular and Quadrimodular RCCX Haplotypes
It has been established that C007 and CS-M4 each have a trimodular RCCX structure that is either LLS or LSL, and J20 has a quadrimodular structure that may be SSLL, SLSL, or SLLS. We further seek to resolve the configuration of the RCCX structures by determining whether a long or short C4 gene is located immediately upstream from TNXB. Deliberate analyses of rare restriction sites in the centromeric end of the HLA class III region (for accession numbers, see the Entrez Nucleotide Web site) revealed the presence of one PacI restriction site in TNXB, one site in the endogenous retrovirus HERV-K(C4) in each long C4 gene, and none in CYP21A/CYP21B and RP1/RP2. The next PacI restriction site telomeric to RCCX is located at the intergenic region between complement C2 and novel gene G10 (MHC Sequencing Consortium 1999; Milner and Campbell 2001).
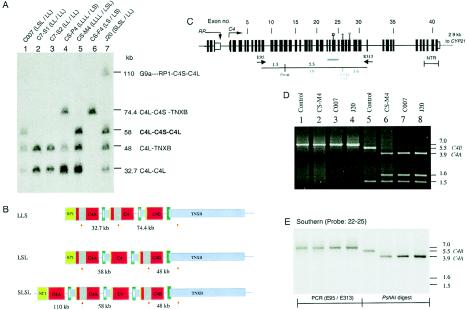
Figure 6 shows results of a PacI-PFGE analysis, performed using a TNX probe for hybridization, of C007 and siblings, CS-P4 and parents, and J20. Lanes 1, 5, and 7 respectively show that C007, CS-M4, and J20 each has a 58-kb restriction fragment representing the presence of three linked C4 genes with a configuration of LSL, a 48-kb restriction fragment representing a long C4 gene linked to TNXB (C4L- TNXB), and a 32.7-kb fragment corresponding to a long C4 gene linked to another long C4 gene. In addition, J20 has a 110-kb fragment corresponding to a PacI fragment spanning from the intergenic region of G10 and C2 to RP1-C4S-CYP21A-TNXA-RP2-C4L. From these observations, it was deduced that C007 has a trimodular LSL and a bimodular LL, that CS-M4 has the quadrimodular LLLL and trimodular LSL (lane 5), and that J20 has the quadrimodular SLSL and the bimodular LL (lane 7). The PacI restriction patterns in C7-1 (lane 2), C7-2 (lane 3), CS-P4 (lane 4), and CS-F4 (lane 4) are consistent with previous assignments of RCCX structures.
Figure 6.
Determining the configurations of long and short C4A and C4B genes in multi-modular RCCX structures. A, PacI-PFGE analysis of long or short C4 genes linked to TNXA and TNXB. B, A scheme illustrating the locations (upward arrows) and size of the PacI restriction fragments containing the C4 and TNX in three different length variants. C, Exon-intron structure of a short C4 gene and PCR strategy to amplify the genomic region between exons 9 and 31. E9.5 and E31.3 are primers used for amplification of the 7.0-kb genomic fragment for the short C4 gene. The PshAI restriction maps for the C4A and C4B genes are shown in red and blue, respectively. D, Restriction analysis of the PCR fragments from four different individuals with short C4. Lanes 1–4 are undigested PCR products of E9.5-E31.3. Lanes 5–8 are digested with PshAI. Lanes 1 and 5 are from an individual with homozygous, monomodular short C4B genes. E, Southern blot analysis of the genomic fragments in panel D, using a C4d exons 22–25 probe.
The Short Genes in LSL and SLSL Haplotypes Are C4A Genes
Although C007 (LSL/LL) and CS-M4 (LLLL/LSL) have C4 gene dosages of five and seven, respectively, there is only one short C4 gene in each of their genomes. The quadrimodular RCCX haplotype SLSL in J20 consists of two short C4 genes, and the homologous chromosome has bimodular LL. For determination of whether the short C4 genes in the unusual RCCX structures are encoding C4A or C4B proteins, specific genomic DNA fragments containing the short intron 9—that is, without the endogenous retrovirus HERV-K(C4)—and the C4A/C4B isotypic sequences at exon 26 were amplified by PCR, using the primer set E9.5 and E31.3 (fig. 6C). A 7.0-kb genomic DNA fragment spanning exons 9–31 of the short C4 gene was produced from each of these three individuals (fig. 6D, lanes 2–4). A positive control from an individual with homozygous S/S that encodes C4B1 allotype was included (fig. 6D, lane 1). Aliquots of these PCR fragments were digested with PshAI (fig. 6D, lanes 5–8) and were subjected to Southern blot analysis using a C4d probe that spans exons 22–25. In the amplified 7-kb DNA fragment, there is a PshAI site, in exon 14, that is common to the C4A and C4B genes, and there is an additional site, in exon 26, that is specific to C4A (fig. 6C). The probe hybridized to the 7-kb undigested PCR fragments (fig. 6E, lanes 1–4), the 5.5-kb fragment characteristic of C4B (fig. 6E, lane 5), and the 3.9-kb fragment specific to C4A in C007, CS-M4, and J20 (fig. 6E, lanes 6–8). Therefore, the short genes in LSL haplotypes of C007 and CS-M4 and in SLSL haplotypes of J20 are C4A genes. Results of C4-allotyping experiments indicated that these C4A genes encode C4A3.
The HLA Haplotypes with the Unusual RCCX Structures
The class I and class II alleles of the MHC (i.e., HLA) in the four described families were determined and segregated into haplotypes. The results are shown in table 1table 1 and are categorized as follows:
Table 1.
HLA Haplotypes in Families with Trimodular and Quadrimodular RCCX Structures
|
Loci |
|||||
| Family, Haplotype, and MHC Class | 1st | 2nd | 3rd | 4th | Remarks |
| Family C008:a | |||||
| a: | |||||
| I—HLA | A*2 | C*0303 | B*15(62) | ||
| III—C4 | A3 | A3 | A3 | LLL | |
| III—CYP | 21A | 21A | 21B | ||
| II—HLA | DRB1*13 | DQB1*0603 | DPB1*0401 | DR52 group | |
| b: | |||||
| I—HLA | A*32 | C*05 | B*15(62) | ||
| III—C4 | A3 | B1 | LL | ||
| III—CYP | 21B | 21B | |||
| II—HLA | DRB1*04 | DQB1*0301 | DPB1*0401 | DR53 group | |
| c: | |||||
| I—HLA | A*11 | C*15 | B*51 | ||
| III—C4 | A3 | A3 | B1 | LLL | |
| III—CYP | 21A | 21A | 21B | ||
| II—HLA | DRB1*01 | DQB1*0501 | DPB1*0501 | DR1 group | |
| d: | |||||
| I—HLA | A*03 | C*07 | B*07 | ||
| III—C4 | A3 | B1 | LL | ||
| III—CYP | 21A | 21B | |||
| II—HLA | DRB1*15 | DQB1*0602 | DPB1*0401 | DR51 group | |
| e: | |||||
| I—HLA | A*31 | C*08 | B*65 | ||
| III—C4 | A3 | B1 | LS | ||
| III—CYP | 21A | 21B | |||
| II—HLA | DRB1*07 | DQB1*02 | DPB1*0301 | DR53 group | |
| f: | |||||
| I—HLA | A*01 | C*07 | B*08 | ||
| III—C4 | B1 | S | |||
| III—CYP | 21B | ||||
| II—HLA | DRB1*03 | DQB1*02 | DPB1*0201 | DR52 group | |
| Family C007:b | |||||
| a: | |||||
| I—HLA | A*2 | C*05 | B*44 | ||
| III—C4 | A3 | A3 | LL | ||
| III—CYP | 21A | 21B | |||
| II—HLA | DRB1*13 | DQB1*0603 | DPB1*0401 | DR52 group | |
| bc | |||||
| c: | |||||
| I—HLA | A*11 | C*0303 | B*55 | ||
| III—C4 | A3 | A3 | B1 | LSL | |
| III—CYP | 21A | 21A | 21B | ||
| II—HLA | DRB1*13 | DQB1*0607 | DPB1*0401 | DR52 group | |
| d: | |||||
| I—HLA | A*68 | C*15 | B*51 | ||
| III—C4 | A3 | B1 | LL | ||
| III—CYP | 21A | 21B | |||
| II—HLA | DRB1*04 | DQB1*0302 | DPB1*0301 | DR53 group | |
| a: | |||||
| I—HLA | A*68 | C*04 | B*35 | ||
| III—C4 | A3 | B1 | LL | ||
| III—CYP | 21B | 21B | |||
| II—HLA | DRB1*13 | DQB1*0603 or DQB1*0607e | DPB1*1901 | DR52 group | |
| bc | |||||
| c: | |||||
| I—HLA | A*31 | C*0303 | B*15(62) | ||
| III—C4 | A3 | A3 | A3 | B1 | SLSL |
| III—CYP | 21A | 21A | 21A | 21B | |
| II—HLA | DRB1*13 | DQB1*0603 or DQB1*0607e | DPB1*0401 | DR52 group | |
| d: | |||||
| I—HLA | A*02 | C*03 | B*15(62) | ||
| III—C4 | A3 | A3 | LL | ||
| III—CYP | 21A | 21B | |||
| II—HLA | DRB1*13 | DQB1*0603 or DQB1*0607e | DPB1*0401 | DR52 group | |
| Family CS4:f | |||||
| a: | |||||
| I—HLA | A*11 | C*01 | B*46 | ||
| III—C4 | A3 | B2 | LS | ||
| III—CYP | 21A | 21B | |||
| II—HLA | DRB1*09 | DQB1*0303 | DPB1*0501 | DR53 group | |
| b: | |||||
| I—HLA | A*11 | C*0304 | B*13 | ||
| III—C4 | A3 | B2 | LS | ||
| III—CYP | 21A | 21B | |||
| II—HLA | DRB1*12 | DQB1*0301 | DPB1*1301 | DR52 group | |
| c: | |||||
| I—HLA | A*24 | C*07 | B*07 | ||
| III—C4 | A3 | A3 | A3 | B1 | LLLL |
| III—CYP | 21A | 21A | 21A | 21B | |
| II—HLA | DRB1*01 | DQB1*0501 | DPB1*0402 | DR1 group | |
| d: | |||||
| I—HLA | A*02 | C*03 | B*61 | ||
| III—C4 | A3 | A3 | B1 | LSL | |
| III—CYP | 21A | 21A | 21B | ||
| II—HLA | DRB1*11 | DQB1*0301 | DPB1*0201 | DR52 group | |
C8-1: a/b; C8-2: c/d; C008: a/c; C8-4: c/e; C8-5: a/f.
C007: a/c; C7-1: a/d; C7-2: a/d.
Not available.
J20: a/c; J20-1: c/d.
Not determined.
CS-F4: a/b; CS-M4: c/d; CS-P4: a/c.
Table 1.
HLA Haplotypes in Families with Trimodular and Quadrimodular RCCX Structures
|
Loci |
|||||
| Family, Haplotype, and MHC Class | 1st | 2nd | 3rd | 4th | Remarks |
| Family C008:a | |||||
| a: | |||||
| I—HLA | A*2 | C*0303 | B*15(62) | ||
| III—C4 | A3 | A3 | A3 | LLL | |
| III—CYP | 21A | 21A | 21B | ||
| II—HLA | DRB1*13 | DQB1*0603 | DPB1*0401 | DR52 group | |
| b: | |||||
| I—HLA | A*32 | C*05 | B*15(62) | ||
| III—C4 | A3 | B1 | LL | ||
| III—CYP | 21B | 21B | |||
| II—HLA | DRB1*04 | DQB1*0301 | DPB1*0401 | DR53 group | |
| c: | |||||
| I—HLA | A*11 | C*15 | B*51 | ||
| III—C4 | A3 | A3 | B1 | LLL | |
| III—CYP | 21A | 21A | 21B | ||
| II—HLA | DRB1*01 | DQB1*0501 | DPB1*0501 | DR1 group | |
| d: | |||||
| I—HLA | A*03 | C*07 | B*07 | ||
| III—C4 | A3 | B1 | LL | ||
| III—CYP | 21A | 21B | |||
| II—HLA | DRB1*15 | DQB1*0602 | DPB1*0401 | DR51 group | |
| e: | |||||
| I—HLA | A*31 | C*08 | B*65 | ||
| III—C4 | A3 | B1 | LS | ||
| III—CYP | 21A | 21B | |||
| II—HLA | DRB1*07 | DQB1*02 | DPB1*0301 | DR53 group | |
| f: | |||||
| I—HLA | A*01 | C*07 | B*08 | ||
| III—C4 | B1 | S | |||
| III—CYP | 21B | ||||
| II—HLA | DRB1*03 | DQB1*02 | DPB1*0201 | DR52 group | |
| Family C007:b | |||||
| a: | |||||
| I—HLA | A*2 | C*05 | B*44 | ||
| III—C4 | A3 | A3 | LL | ||
| III—CYP | 21A | 21B | |||
| II—HLA | DRB1*13 | DQB1*0603 | DPB1*0401 | DR52 group | |
| bc | |||||
| c: | |||||
| I—HLA | A*11 | C*0303 | B*55 | ||
| III—C4 | A3 | A3 | B1 | LSL | |
| III—CYP | 21A | 21A | 21B | ||
| II—HLA | DRB1*13 | DQB1*0607 | DPB1*0401 | DR52 group | |
| d: | |||||
| I—HLA | A*68 | C*15 | B*51 | ||
| III—C4 | A3 | B1 | LL | ||
| III—CYP | 21A | 21B | |||
| II—HLA | DRB1*04 | DQB1*0302 | DPB1*0301 | DR53 group | |
| (continued) | |||||
Table 1.
HLA Haplotypes in Families with Trimodular and Quadrimodular RCCX Structures
|
Loci |
|||||
| Family, Haplotype, and MHC Class | 1st | 2nd | 3rd | 4th | Remarks |
| Family JRA 20:d | |||||
| a: | |||||
| I—HLA | A*68 | C*04 | B*35 | ||
| III—C4 | A3 | B1 | LL | ||
| III—CYP | 21B | 21B | |||
| II—HLA | DRB1*13 | DQB1*0603 or DQB1*0607e | DPB1*1901 | DR52 group | |
| bc | |||||
| c: | |||||
| I—HLA | A*31 | C*0303 | B*15(62) | ||
| III—C4 | A3 | A3 | A3 | B1 | SLSL |
| III—CYP | 21A | 21A | 21A | 21B | |
| II—HLA | DRB1*13 | DQB1*0603 or DQB1*0607e | DPB1*0401 | DR52 group | |
| d: | |||||
| I—HLA | A*02 | C*03 | B*15(62) | ||
| III—C4 | A3 | A3 | LL | ||
| III—CYP | 21A | 21B | |||
| II—HLA | DRB1*13 | DQB1*0603 or DQB1*0607e | DPB1*0401 | DR52 group | |
| Family CS4:f | |||||
| a: | |||||
| I—HLA | A*11 | C*01 | B*46 | ||
| III—C4 | A3 | B2 | LS | ||
| III—CYP | 21A | 21B | |||
| II—HLA | DRB1*09 | DQB1*0303 | DPB1*0501 | DR53 group | |
| b: | |||||
| I—HLA | A*11 | C*0304 | B*13 | ||
| III—C4 | A3 | B2 | LS | ||
| III—CYP | 21A | 21B | |||
| II—HLA | DRB1*12 | DQB1*0301 | DPB1*1301 | DR52 group | |
| c: | |||||
| I—HLA | A*24 | C*07 | B*07 | ||
| III—C4 | A3 | A3 | A3 | B1 | LLLL |
| III—CYP | 21A | 21A | 21A | 21B | |
| II—HLA | DRB1*01 | DQB1*0501 | DPB1*0402 | DR1 group | |
| d: | |||||
| I—HLA | A*02 | C*03 | B*61 | ||
| III—C4 | A3 | A3 | B1 | LSL | |
| III—CYP | 21A | 21A | 21B | ||
| II—HLA | DRB1*11 | DQB1*0301 | DPB1*0201 | DR52 group | |
C8-1: a/b; C8-2: c/d; C008: a/c; C8-4: c/e; C8-5: a/f.
C007: a/c; C7-1: a/d; C7-2: a/d.
Not available.
J20: a/c; J20-1: c/d.
Not determined.
CS-F4: a/b; CS-M4: c/d; CS-P4: a/c.
Monomodular S RCCX.—The monomodular S structure with a single short C4B gene present in C8-5 is typed as having HLA A*01 C*07 B*08, in the class I region, and DRB1*03 DQB1*02 DPB1*0201, in the class II region.
Homoexpression of CYP21B.—Two bimodular LL structures are shown to have the infrequent CYP21B-CYP21B configuration. The first is present in C8-1, with HLA A*32 C*05 B*15(62) DRB1*04 DQB1*0301 DPB1*0401. The second is present in J20, with HLA haplotype A*68 C*04 B*35 DRB1*13 DQB1*0603 DPB1*1901.
Homoexpression of C4A.—Bimodular LL structures with homoexpression of C4A3 (i.e., C4A3 A3) is detected in two haplotypes. The first is in C007 who has HLA A*2 C*05 B*44 DRB1*13 DQB1*0603 DPB1*0401. The second is present in J20-1 who has HLA haplotype A*02 C*03 B*15(62) DRB1*13 DQB1*(0603 or 0607; not determined) DPB1*0401. The trimodular LLL structure with homoexpression of C4A3 in C008 is present between HLA A*2 C*0303 B*15(62) and DRB1*13 DQB1*0603 DPB1*0401.
Trimodular structures expressing C4A3 A3 B1.—Three haplotypes are shown that express two C4As and one C4B. The first is present in a trimodular LLL of C008 and is located between HLA A*11 C*15 B*51 and DRB1*01 DQB1*0501 DPB1*0501. The second is present in C007, who has an LSL structure in HLA A*11 C*0303 B*55 and DRB1*13 DQB1*0607 DPB1*0401. The third is in CS-M4, who also has an LSL structure, but her HLA is A*02 C*0303 B*61 and DRB1*11 DQB1*0301 DPB1*0201.
Quadrimodular RCCX haplotypes.—The quadrimodular LLLL in the pediatric patient with SLE CS-P4, encoding C4A3 A3 A3 B1, is present between HLA A*24 C*07 B*07 and DRB1*01 DQB1*0501 DPB1*0402. The quadrimodular SLSL structure in J20, encoding C4A3 A3 A3 B1, has HLA haplotype A*31 C*0303 B*15(62) DRB1*13 DQB1*0607 DPB1*0401.
Discussion
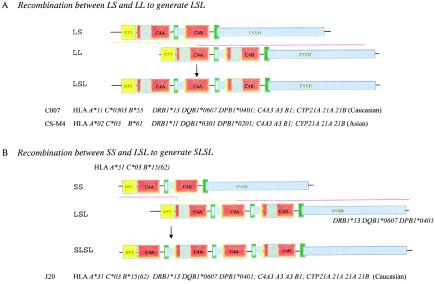
Studies on the molecular genetics of complement C4A and complement C4B gene dosages and RCCX modular variations continue to yield surprises. Here, we elucidated novel haplotypes with three and four RCCX modules that contain long and short C4 genes. In an unaffected white individual (C007) with HLA haplotype A*11 C*0303 B*55 DRB1*13 DQB1*0607 DPB1*0401 and in an unaffected Asian individual (CS-M4) with HLA haplotype A*02 C*03 B*61 DRB1*11 DQB1*0301 DPB1*0201, trimodular RCCX haplotypes with two long and one short C4 genes were demonstrated by TaqI RFLP and PmeI PFGE. PacI PFGE further shows that the configuration of the C4 genes is LSL, with respect to RP1 at the telomeric end and TNXB at the centromeric end. Long-range PCR amplification of the single short C4 genes in C007 and in CS-M4 and the subsequent PshAI-RFLP analysis definitively showed that the short gene (in the middle) encodes C4A. Thus, the order of the three C4 genes in each case is probably C4A3, A3, and B1.
By similar approaches, a peculiar quadrimodular RCCX structure with two short and two long C4 genes with a configuration of SLSL is elucidated in a white pediatric patient with pauciarticulate JRA (J20). PshAI-PvuII RFLP showed that three of these C4 genes encode C4A and that the remaining gene encodes C4B. This SLSL structure is located between HLA A*31 C*03 B*15(62) and DRB1*13 DQB1*0607 DPB1*0401. It is noteworthy that the haplotype of the centromeric HLA class II alleles in this quadrimodular SLSL structure is identical to that of the trimodular LSL structure in C007. A model to illustrate the generation of LSL and SLSL is presented in figure 7. We suggest that the trimodular LSL haplotype could have derived from an unequal crossover between a bimodular LS and a bimodular LL (fig. 7A). Another unequal crossover between the trimodular LSL and a bimodular SS would give rise to a quadrimodular SLSL and a monomodular S (fig. 7B).
Figure 7.
Mechanistic models to generate LSL (A) and SLSL (B) modules of RCCX by unequal crossovers
A different quadrimodular RCCX haplotype with LLLL structure containing three C4A3 genes and one C4B1 gene was detected in an Asian family with a pediatric patient with SLE. The RCCX modules contain an intact RP1 gene at one end and the intact CYP21B- TNXB genes at the other end. HLA typing and a segregation analysis showed that the LLLL structure is present in the HLA haplotype A*24 C*07 B*07 DRB1*01 DQB1*0501 DPB1*0402. This is a relatively common haplotype in the Japanese population, having a frequency of 4.2% (Tokunaga et al. 1985). Tokunaga et al. (1991) described it as ancestral haplotype (AH) 7.2. Their earlier phenotypic and genotypic characterizations were in close agreement with our observation: plasma proteins from individuals with AH 7.2 had a substantially larger quantity of C4A than of C4B; a TaqI-RFLP analysis of the C4 genes yielded much higher intensities of the 6.0-kb fragment corresponding to the RP2-C4L compared to the 7.0-kb fragment corresponding to the RP1-C4L; and PFGE of MluI-digested genomic DNA revealed a genomic fragment ∼50 kb larger than that from individuals with two C4 genes. Therefore, it was suggested—conservatively, in retrospect—that three long C4 genes were present in AH 7.2 (Tokunaga et al. 1991). The present study definitively shows that AH 7.2 from the Asian patient with SLE has four long C4 genes. In keeping with the ethnicity of AH 7.2, the LLLL RCCX haplotype was inherited from the patient's mother, who immigrated to the United States from Japan. Another HLA haplotype with four repeating units of long C4 genes and CYP21A pseudogenes was discovered in a patient with congenital adrenal hyperplasia (CAH [MIM 201910]) who had HLA haplotype A31 B51 DR4 (Collier et al. 1989).
Each of the two quadrimodular LLLL structures described above could have been generated by an unequal crossover between a trimodular LLL chromosome and a bimodular LL chromosome. A crossover between LLL and a regular bimodular LL partner with one CYP21A gene and one CYP21B gene would give rise to the LLLL(CYP21B) haplotype; a recombination between LLL and a bimodular LL with two CYP21A genes (which is a CAH-carrier haplotype) would generate the LLLL(CYP21A) haplotype.
A third unexpected observation from the present study is the homoexpression of C4A3 protein from a trimodular RCCX structure with three long C4 genes in a white female. This was unraveled through a combination of precise genotyping experiments to determine the total number and the relative dosage of C4A and C4B genes, phenotyping experiments to detect the qualitative and quantitative variations of plasma C4A and C4B levels, and family segregation studies. This may not be the only case for the homoexpression of C4A proteins from a trimodular LLL haplotype, since we noticed a similar pattern in a published study of duplicated C4A genes in the French family St (Giles et al. 1987). On the basis of the genotyping data from TaqI-RFLP analysis, phenotypic analysis of C4A and C4B allotypes and α-chains of C4A and C4B proteins, and hemolytic activities of the C4A and C4B proteins, it is clear to us that the haplotype with HLA A24 B18 DR1 in the St family consists of an LLL structure that encodes C4A3, A3, and A2, although the specific order of these three C4 genes could not be deduced.
The phenomena described above reveal the plasticity of the RCCX length variants, which leads to frequent C4A and C4B gene-dosage variation in human populations. Just from the 13 members in four families, we have demonstrated the magnitude of genetic diversities in C4 and in RCCX. We observed the homoexpression of C4A from bimodular LL (haplotype a, C007's family; haplotype d, J20's family) and trimodular LLL (haplotype a, C008's family), the presence of LSL in a white individual (haplotype c, C007's family) and in Asian American individuals (haplotype d, CS-P4's family), and the presence of CYP21B-CYP21B in bimodular LL from two different HLA haplotypes (haplotype a, J20's family; haplotype b, C008's family). Although it is well established that deficiency of CYP21B is the main cause of CAH (White and Speiser 2000), it is not clear whether high dosage of CYP21B has any influence on the production of corticols and sex steroids in an individual. In other words, we have elucidated haplotypes consisting of one, two, three, and four C4 genes, with different combinations of long and short C4 genes encoding C4A or C4B, and we have observed the variation in the dosage of pseudogene CYP21A and functional gene CYP21B. The C4A and C4B gene-dosage variation is a main contributing factor for a wide range of C4A and C4B protein levels in the circulation among different individuals. A study of 125 white individuals has shown a significant correlation between C4 gene dosages and serum C4 levels and hemolytic activities (Y. Yang et al. 2001).
The sophistication in human complement C4 genetic diversity demands special attention to data interpretation when studying disease associations with C4A or C4B deficiencies and protein polymorphisms. Of particular interest is the presence of six C4 genes in the pediatric patient with lupus (CS-P4) and in the patient with JRA (J20). It has been well documented that complete deficiency of C4A and C4B or deficiency of C4A is an important risk factor of SLE (Rupert et al. 2002). It is not clear, however, what role high C4 gene dosage plays in the disease pathogenesis. One possibility is that high C4 gene dosage could lead to the production of larger quantities of C4 proteins at local tissues during an inflammatory process that could exacerbate complement-mediated tissue injuries.
Acknowledgments
We are indebted to the blood donors and patients who participated in this study. This work was supported by the National Institutes of Health (National Institute of Arthritis and Musculoskeletal and Skin Diseases grant R01 AR43969 and National Institute of Diabetes and Digestive and Kidney Diseases grant P01 DK55546), a pilot grant from the Columbus Children’s Research Institute (297401), and Pittsburgh Supercomputing Center (through National Institutes of Health Center for Research Resources Cooperative Agreement grant 1P41 RR06009).
Electronic-Database Information
Accession numbers and URLs for data presented herein are as follows:
- Entrez Nucleotide, http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=nucleotide (for C4A [accession number M59816-U07856-M58915], long C4B gene [accession number AF019413], short C4B gene [accession numbers AL049547 and U24578], RP1 or STK19 [accession numbers L26260 and L26261], RP2 [accession numbers L26262 and L26263], CYP21B [accession numbers M26856, M12792, M13936, and AF77974], CYP21A [accession numbers M26857, M12793, and M13935], TNXB [accession number U89337], and TNXA [accession numbers L26263 and U24488])
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for C4A [MIM 120810], C4B [MIM 120820], CYP21 and CAH [MIM 201910], TNX [MIM 600985], RP1 or STK19 [MIM 604977], and SLE [MIM 152700])
References
- Awdeh ZL, Alper CA (1980) Inherited structural polymorphism of the fourth component of human complement. Proc Natl Acad Sci USA 77:3576–3580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awdeh ZL, Raum D, Alper CA (1979) Genetic polymorphism of human complement C4 and detection of heterozygotes. Nature 282:205–208 [DOI] [PubMed] [Google Scholar]
- Belt KT, Yu CY, Carroll MC, Porter RR (1985) Polymorphism of human complement component C4. Immunogenetics 21:173–180 [DOI] [PubMed] [Google Scholar]
- Blanchong CA, Chung EK, Rupert KL, Yang Y, Yang Z, Zhou B, Yu CY (2001) Genetic, structural and functional diversities of human complement components C4A and C4B and their mouse homologs, Slp and C4. Int Immunopharmacol 1:365–392 [DOI] [PubMed] [Google Scholar]
- Blanchong CA, Zhou B, Rupert KL, Chung EK, Jones KN, Sotos JF, Rennebohm RM, Yu CY (2000) Deficiencies of human complement component C4A and C4B and heterozygosity in length variants of RP-C4-CYP21-TNX (RCCX) modules in Caucasians: the load of RCCX genetic diversity on MHC-associated disease. J Exp Med 191:2183–2196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll MC, Belt KT, Palsdottir A, Yu Y (1985) Molecular genetics of the fourth component of human complement and the steroid 21-hydroxylase. Immunol Rev 87:39–60 [DOI] [PubMed] [Google Scholar]
- Carroll MC, Campbell RD, Bentley DR, Porter RR (1984) A molecular map of the human major histocompatibility complex class III region linking complement genes C4, C2 and factor B. Nature 307:237–241 [DOI] [PubMed] [Google Scholar]
- Carroll MC, Fathallah DM, Bergamaschini L, Alicot EM, Isenman DE (1990) Substitution of a single amino acid (aspartic acid for histidine) converts the functional activity of human complement C4B to C4A. Proc Natl Acad Sci USA 87:6868–6872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung EK, Yang Y, Rupert KL, Rennebohm RM, Blanchong CA, Yu CY (2002) Determining the one, two, three, or four long and short loci of human complement C4 in a major histocompatibility complex haplotype encoding C4A or C4B proteins. Am J Hum Genet 71:810–822 (in this issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier S, Sinnott PJ, Dyer PA, Price DA, Harris R, Strachan T (1989) Pulsed field gel electrophoresis identifies a high degree of variability in the number of tandem 21-hydroxylase and complement C4 gene repeats in 21-hydroxylase deficiency haplotypes. EMBO J 8:1393–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangel AW, Mendoza AR, Baker BJ, Daniel CM, Carroll MC, Wu L-C, Yu CY (1994) The dichotomous size variation of human complement C4 gene is mediated by a novel family of endogenous retroviruses which also establishes species-specific genomic patterns among Old World primates. Immunogenetics 40:425–436 [DOI] [PubMed] [Google Scholar]
- Dodds AW, Ren X-D, Willis AC, Law SKA (1996) The reaction mechanism of the internal thioester in the human complement component C4. Nature 379:177–179 [DOI] [PubMed] [Google Scholar]
- Giles CM, Uring-Lambert B, Boksch W, Braun M, Goetz J, Neumann R, Mauff G, Hauptmann G (1987) The study of a French family with two duplicated C4A haplotypes. Hum Genet 77:359–365 [DOI] [PubMed] [Google Scholar]
- Hochberg MC (1997) Updating the American College of Rheumatology revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum 40:1725 [DOI] [PubMed] [Google Scholar]
- Jaatinen T, Eholouto M, Laitinen T, Lokki M-L (2002) Characterization of a de novo conversion in human complement C4 gene producing a C4B5-like protein. J Immunol 168:5652–5658 [DOI] [PubMed] [Google Scholar]
- Martinez OP, Lomgman-Jacobsen N, Davies R, Chung EK, Gaudieri S, Dawkins RL, Yu CY (2001) Genetics of human complement C4 and evolution of the central MHC. Front Biosci 6:d904–d913 [DOI] [PubMed] [Google Scholar]
- Mauff G, Luther B, Schneider PM, Rittner C, Strandmann-Bellinghausen B, Dawkins R, Moulds JM (1998) Reference typing report for complement component C4. Exp Clin Immunogenet 15:249–260 [DOI] [PubMed] [Google Scholar]
- MHC Sequencing Consortium (1999) Complete sequence and gene map of a human major histocompatibility complex. Nature 401:921–923 [DOI] [PubMed] [Google Scholar]
- Milner CM, Campbell RD (2001) Genetic organization of the human MHC class III region. Front Biosci 6:d914–d926 [DOI] [PubMed] [Google Scholar]
- Moulds JM, Arnett FC, Giles CM, Hamilton RG (1990) A novel immunoassay for the quantitation of human C4 gene products. Complement Inflamm 7:95–101 [DOI] [PubMed] [Google Scholar]
- Olaisen B, Teisberg P, Jonassen R (1980) The C4 system: quantitative studies of different genotypes. Immunobiology 158:82–85 [DOI] [PubMed] [Google Scholar]
- O'Neill GJ, Yang SY, DuPont B (1978) Two HLA-linked loci controlling the fourth component of human complement. Proc Natl Acad Sci USA 75:5165–5169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter RR (1983) Complement polymorphism, the major histocompatibility complex and associated diseases: a speculation. Mol Biol Med 1:161–168 [PubMed] [Google Scholar]
- Raum D, Awdeh Z, Anderson J, Strong L, Granados J, Teran L, Giblett E, Yunis EJ, Alper CA (1984) Human C4 haplotypes with duplicated C4A or C4B. Am J Hum Genet 36:72–79 [PMC free article] [PubMed] [Google Scholar]
- Rebmann V, Doxiadis I, Kubens BS, Grosse-Wilde H (1992) Quantitation of the human component C4: definition of C4 Q0 alleles and C4A duplications. Vox Sang 62:117–123 [DOI] [PubMed] [Google Scholar]
- Rittner C, Giles CM, Roos CM, Demant P, Mollenhauer E (1984) Genetics of human C4 polymorphism: detection and segregation of rare and duplicated haplotypes. Immunogenetics 19:321–333 [DOI] [PubMed] [Google Scholar]
- Roos MH, Mollenhauer E, Demant P, Rittner C (1982) A molecular basis for the two locus model of human complement component C4. Nature 298:854–856 [DOI] [PubMed] [Google Scholar]
- Rosenfeld SI, Ruddy S, Austin KF (1969) Structural polymorphism of the fourth component of human complement. J Clin Invest 48:2283–2292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupert KL, Moulds JM, Yang Y, Arnett FC, Warren RW, Reveille JD, Myones BL, Blanchong CA, Yu CY (2002) The molecular basis of complete C4A and C4B deficiencies in a systemic lupus erythematosus (SLE) patient with homozygous C4A and C4B mutant genes. J Immunol 169:1570–1578 [DOI] [PubMed] [Google Scholar]
- Rupert KL, Rennebohm RM, Yu CY (1999) An unequal crossover between the RCCX modules of the human MHC leading to the presence of a CYP21B gene and a tenascin TNXB/TNXA-RP2 recombinant between C4A and C4B genes in a patient with juvenile rheumatoid arthritis. Exp Clin Immunogenet 16:81–97 [DOI] [PubMed] [Google Scholar]
- Schendel DJ, Wank R, O'Neill GJ (1985) C4 phenotypic variation suggests an unusual class III gene organization. Vox Sang 48:110–115 [DOI] [PubMed] [Google Scholar]
- Schneider PM, Witzel-Schlomp K, Rittner C, Zhang L (2001) The endogenous retroviral insertion in the human complement C4 gene modulates the expression of homologous genes by antisense inhibition. Immunogenetics 53:1–9 [DOI] [PubMed] [Google Scholar]
- Sim E, Cross S (1986) Phenotyping of human complement component C4, a class III HLA antigen. Biochem J 239:763–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan EM, Cohen AS, Fries JF, Masi AT, McShane DJ, Rothfield NF, Schaller JG, Talal N, Winchester RJ (1982) The 1982 revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum 25:1271–1277 [DOI] [PubMed] [Google Scholar]
- Teisberg P, Akesson B, Olaisen T, Gedde-Dahl T Jr, Thorsby E (1976) Genetic polymorphism of C4 in man and localisation of a structural C4 locus to the HLA gene complex of chromosome 6. Nature 264:253–254 [DOI] [PubMed] [Google Scholar]
- Teisberg P, Jonassen R, Mevag B, Gedde-Dahl T Jr, Olaisen B (1988) Restriction fragment length polymorphisms of the complement component C4 loci on chromosome 6: studies with emphasis on the determination of gene number. Ann Hum Genet 52:77–84 [DOI] [PubMed] [Google Scholar]
- Tokunaga K, Omoto K, Akaza T, Akiyama N, Amemiya H, Naito S, Sasazuki T, Satoh H, Juji T (1985) Haplotype study on C4 polymorphism in Japanese. Associations with MHC alleles, complotypes, and HLA-complement haplotypes. Immunogenetics 22:359–365 [DOI] [PubMed] [Google Scholar]
- Tokunaga K, Zhang WJ, Christiansen FT, Dawkins RL (1991) The genomic structure to two ancestral haplotypes carrying C4A duplications. Immunogenetics 34:247–251 [DOI] [PubMed] [Google Scholar]
- White PC, Speiser PW (2000) Congenital adrenal hyperplasia due to 21-hydroxylase deficiency. Endocr Rev 21:245–291 [DOI] [PubMed] [Google Scholar]
- Yang Y, Chung EK, Zhou B, Blanchong CA, Yu CY, Kovacs M, Karadi I, Fust G, Varga L (2001) Genetic diversity of complement components C4A and C4B and correlations of C4 gene dosages with C4 haemolytic activities and protein levels in a central European population. Mol Immunol 38:129 [Google Scholar]
- Yang Z, Mendoza AR, Welch TR, Zipf WB, Yu CY (1999) Modular variations of HLA class III genes for serine/threonine kinase RP, complement C4, steroid 21-hydroxylase CYP21 and tenascin TNX (RCCX): a mechanism for gene deletions and disease associations. J Biol Chem 274:12147–12156 [DOI] [PubMed] [Google Scholar]
- Yu CY, Belt KT, Giles CM, Campbell RD, Porter RR (1986) Structural basis of the polymorphism of human complement component C4A and C4B: gene size, reactivity and antigenicity. EMBO J 5:2873–2881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu CY, Blanchong CA, Chung EK, Rupert KL, Yang Y, Yang Z, Zhou B, Moulds JM (2002) Molecular genetic analyses of human complement components C4A and C4B. In: Rose NR, Hamilton RG, Detrick B (eds) Manuals of clinical laboratory immunology, 6th ed. ASM Press, Washington, DC, pp 117–131 [Google Scholar]
- Yu CY, Campbell RD, Porter RR (1988) A structural model for the location of the Rodgers and the Chido antigenic determinants and their correlation with the human complement C4A/C4B isotypes. Immunogenetics 27:399–405 [DOI] [PubMed] [Google Scholar]